Abstract
We applied double gradient-denaturing gradient gel electrophoresis (DG-DGGE) for the rapid detection of rifampin (RMP) resistance from rpoB PCR products of Mycobacterium tuberculosis isolates and clinical samples. The results of this method were fully concordant with those of DNA sequencing and susceptibility testing analyses. DG-DGGE is a valid alternative to the other methods of detecting mutations for predicting RMP resistance.
Rifampin (RMP) resistance in mycobacteria is most frequently due to point mutations as well as small insertions and deletions located in an 81-nucleotide region of the rpoB gene encoding the β-subunit of RNA polymerase (7, 10, 12, 19, 24). The genotype assessment of RMP resistance offers a number of advantages over conventional culture susceptibility testing. (i) The turnaround time is days, rather than weeks or months. (ii) It can be automated. (iii) It leads to fewer laboratory biohazards.
This study made use of a modified version of the original denaturing gradient gel electrophoresis (DGGE) methodology described by Fisher and Lerman (5) and Myers et al. (14). In order to obtain maximum DGGE sensitivity, heteroduplexes of wild-type and mutant sequences were generated, and to improve the analysis of these molecules (which often produce curtains and smears instead of sharp zones), we used a second (porous) gradient over the denaturing gradient (3, 6). This double gradient-DGGE (DG-DGGE) was used to detect mutations in the rpoB region of the PCR products obtained from mycobacterial isolates and clinical samples.
(Part of this work was presented at the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., 24 to 27 September 1998 [18]).
We processed 117 Mycobacterium tuberculosis (81 RMP-resistant and 36 RMP-susceptible) clinical strains; RMP-susceptible H37Rv M. tuberculosis ATCC 27294 was used as a control. RMP susceptibility was determined by the BACTEC radiometric method (8). The critical RMP concentration in BACTEC 12B vials was 2 μg/ml. The clinical isolates were classified on the basis of IS6110 restriction fragment length polymorphism (RFLP) pattern subtypes (22). Two groups of clinical specimens were also processed by means of PCR–DG-DGGE: (i) 84 IS6110 PCR-positive clinical specimens from a variety of anatomical sources obtained from 62 patients with microbiologically proved tuberculosis, and (ii) 48 cerebrospinal fluid (CSF) samples collected from 28 patients with confirmed or highly probable tuberculous meningitis.
Bacterial suspensions containing approximately 105 acid-fast bacilli in 250 μl of sterile deionized H2O and 250 to 500 μl of each clinical specimen underwent DNA extraction by the chaotrope-silica method (2). Aliquots of DNA were used for time release PCR (1) of the rpoB region (GenBank accession no. L27989) by means of primers TR9 (5′-TCGCCGCGATCAAGGAGT) and rpoB-2643GC (5′-CGTCGCTAACCACGCCGT). The latter oligonucleotide carries an additional 30 bp of a GC-rich sequence at its 5′ end. The annealing temperatures were 63°C for the first 20 cycles and 61°C for the subsequent 30 cycles. The presence of a 339-bp band on a 1.8% agarose gel indicated successful amplification. The heteroduplex molecules were obtained as previously described (3).
In order to obtain some indications for the best choice of the DNA sequence containing the rpoB core region susceptible to DGGE analysis, we used a computer algorithm (MacMelt 1.0; Bio-Rad Laboratories, Hercules, Calif.) derived from the original computer program developed by Lerman and Silverstein (11).
The denaturing gradient gels were electrophoresed in an apparatus purchased from Bio-Rad Laboratories (D GENE Denaturing Gel Electrophoresis System); a two-chamber gradient maker (Pharmacia, Uppsala, Sweden) was used to create the linear-gradient gels. The PCR products were electrophoresed in a colinearly increasing double gradient of 50 to 75% denaturant and 6.5 to 12% polyacrylamide gels in 1× TAE buffer (20× TAE buffer is 0.8 M Tris base–0.4 M sodium acetate–0.02 M EDTA, pH 7.4) at a constant temperature of 60°C at 50 V overnight. The gels were stained with Sybr green II (FMC Bio Products, Rockland, Maine) and photographed with a UV transilluminator.
The rpoB PCR products were sequenced with the same primers in an automated DNA sequencer (model 377; Applied Biosystems, Foster City, Calif.). Both strands were sequenced.
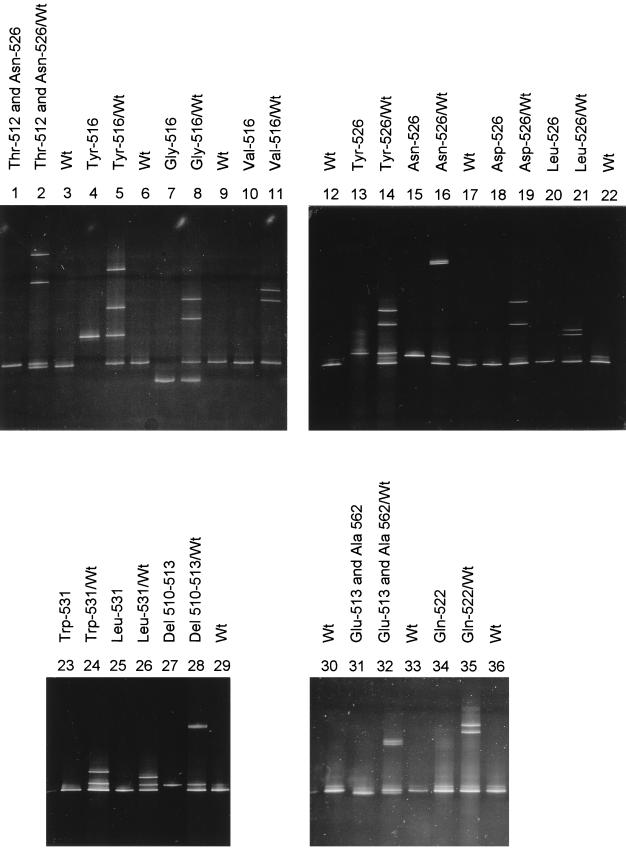
RFLP analysis of the 117 clinical isolates revealed 93 different patterns corresponding to single isolates, multiple samples from the same patient, or epidemic strains (Table 1). DG-DGGE analysis revealed altered homoduplex mobility and/or heteroduplex molecule formation in the PCR products of all of the RMP-resistant strains. Fourteen different patterns were observed in these samples, consisting of sets of three or four bands when heteroduplexes were produced (Fig. 1). The PCR products of the RMP-susceptible strains did not show any heteroduplex formation or abnormal homoduplex mobility. DNA sequencing results confirmed the DG-DGGE results. Fourteen alleles were detected in the RMP-resistant strains (Table 1). Eleven of the mutations found in this study have already been reported in the literature. Four new mutations were found in three strains: one in codon 522 in one strain, two in codons 513 and 562 in another strain, and a deletion of codons 511 to 513 in the third strain (codon 510 changed to CAA still codes for Gln) (Table 1).
TABLE 1.
Sequence analysis of the rpoB gene of M. tuberculosis isolates from a region in Northern Italy
| RMP phenotype | Affected codon(s) (mutation)a | Amino acid change(s) | No. of isolates | No. of individual strains (frequency [%]) |
|---|---|---|---|---|
| Resistantb | 533 (CCG) | Leu→Pro | 1 | 1 |
| Resistant | 531 (TTG) | Ser→Leu | 43 | 35 (57.4) |
| Resistant | 531 (TGG) | Ser→Trp | 1 | 1 |
| Resistant | 526 (GAC) | His→Asp | 14 | 4 (6.6) |
| Resistant | 526 (TAC) | His→Tyr | 7 | 6 (9.8) |
| Resistant | 526 (CTC) | His→Leu | 1 | 1 |
| Resistant | 526 (AAC) | His→Asn | 1 | 1 |
| Resistantc | 522 (CAG) | Ser→Gln | 1 | 1 |
| Resistant | 516 (TAC) | Asp→Tyr | 6 | 5 (8.2) |
| Resistant | 516 (GGC) | Asp→Gly | 1 | 1 |
| Resistant | 516 (GTC) | Asp→Val | 2 | 2 |
| Resistantd | 512 (ACC), 526 (AAC) | Ser→Thr, His→Asn | 1 | 1 |
| Resistante | 513 (GAA), 562 (GCA) | Gln→Glu, Glu→Ala | 1 | 1 |
| Resistant | 510–513 deletion (GCTGAGCCA) | Leu-Ser-Gln deletion | 1 | 1 |
| Susceptible | None | None | 36 | 32 |
| Total | 117 | 93 |
Numbers correspond to E. coli numbering system for the RNA polymerase β-subunit. Nucleotide substitutions are underlined.
Resistant to RMP at 2 μg/ml by the BACTEC method.
Two mutations in the same codon.
Two mutations in separate codons.
Two mutations in separate codons, one of which is located outside the rpoB core region.
FIG. 1.
DG-DGGE analysis. Photograph of DG-DGGE patterns showing the electrophoretic mobility of homoduplex and heteroduplex PCR fragments from RMP-susceptible H37Rv M. tuberculosis ATCC 27294 (lanes 3, 6, 9, 12, 17, 22, 29, 30, and 36) and RMP-resistant strains (all other lanes). For each resistant strain, both the homoduplex and heteroduplex patterns are shown. Thirteen of the 14 alleles listed in Table 1 are shown (mutation affecting codon 533 is not shown). Wt, wild type; Del, deletion.
Amplified rpoB products were generated from 54 of 84 IS6110 PCR-positive specimens (64.3%) collected from 42 (67.7%) of the 62 patients with non-central nervous system (non-CNS) tuberculosis (Table 2). From all 30 of the specimens with the wild-type DG-DGGE pattern, a RMP-susceptible strain was cultured. From the 24 specimens with an abnormal DG-DGGE pattern, 1 RMP-susceptible and 23 RMP-resistant strains were cultured. In the case of discordance, DG-DGGE analysis showed the presence of at least two clones, one of which carried a mutation in the rpoB gene; as a matter of fact, that heteroduplex formation was observed without mixing with wild-type DNA. Direct DNA sequencing showed a single point mutation located in codon 526 reading GAC instead of CAC (His→Asp) or in codon 531 reading TTG instead of TCG (Ser→Leu), in all of the strains with an abnormal DG-DGGE pattern and a resistant phenotype.
TABLE 2.
Direct detection of M. tuberculosis rpoB in 84 clinical specimens collected from 62 patients suffering from tuberculosis
| Specimen source (no. of specimens)a | Resultb
|
|||
|---|---|---|---|---|
| Smear | Culture | IS6110 PCRc | rpoB PCR | |
| Respiratory (39) | + | + | + | + |
| Respiratory (5) | − | + | + | + |
| Respiratory (1) | − | − | + | + |
| Respiratory (15) | + | + | + | − |
| Respiratory (7) | − | + | + | − |
| Respiratory (5) | − | − | + | − |
| Nonrespiratory GW and L (6) | + | + | + | + |
| Nonrespiratory GW, PE, and U (3) | − | + | + | + |
| Nonrespiratory GW and L (2) | + | + | + | − |
| Nonrespiratory PE (1) | − | − | + | − |
Respiratory specimens include sputum and bronchial wash or lavage specimens. Abbreviations: GW, gastric wash; L, lymph node; PE, pleural effusion; U, urine.
+, positive; −, negative.
A previously described homemade nested PCR protocol was used to obtain these results (16).
Table 3 summarizes the results obtained by processing the CSF samples. In brief, the results of PCR–DG-DGGE analysis of the CSF specimens correlated in all cases with the RMP phenotypes of isolates cultured from the same patients. No differences in the DG-DGGE patterns of the amplified products obtained from suspensions of M. tuberculosis isolates and from CSF specimens collected from the same patients were observed (data not shown). No mutations in the rpoB segment sequenced from the RMP-susceptible strains were revealed, whereas three different kinds of nucleotide substitution were found in all of the RMP-resistant strains. The mutations were located in codon 531, coding TTG instead of TCG (Ser→Leu), and in codon 526, coding GAC or TAC instead of CAC (His→Asp or His→Tyr).
TABLE 3.
DG-DGGE analysis of CSF samples, patient data, the results obtained by two PCR protocols, PCR–DG-DGGE patterns, and susceptibilities of clinical isolates to RMP
| Infection status and patient no. | Age (yr) | Sexa | No. of CSF samples
|
CSF PCR–DG-DGGE patternb | RMP phenotype | ||
|---|---|---|---|---|---|---|---|
| Total | Positive by IS6110 PCR | Positive by rpoB PCR (evaluable by DG-DGGE) | |||||
| Confirmed CNS TBf by CSF-positive culture | |||||||
| 1 | 32 | Md | 2 | 2 | 1 | Wild type | Susceptible |
| 2 | 72 | M | 2 | 2 | 2 | Wild type | Susceptible |
| 3 | 42 | Fd | 5 | 4 | 4 | Wild type | Susceptible |
| 4 | 31 | F | 2 | 1 | 1 | Wild type | Susceptible |
| 5 | 63 | M | 4 | 3 | 1 | Wild type | Susceptible |
| 6 | 30 | M | 1 | 1 | 1 | Wild type | Susceptible |
| 7 | 30 | Md | 1 | 1 | 1 | Wild type | Not available |
| 8 | 35 | Md | 1 | 1 | 1 | Abnormal (A) | Resistant |
| 9 | 30 | Md | 1 | 1 | 1 | Abnormal (B) | Resistant |
| 10 | 59 | Md | 1 | 1 | 1 | Abnormal (A) | Resistant |
| 11 | 56 | Md | 1 | 1 | 1 | Abnormal (A) | Resistant |
| 12 | 27 | Fd | 2 | 2 | 0 | Susceptible | |
| 13 | 40 | F | 1 | 1 | 0 | Susceptible | |
| 14 | 34 | Md | 1 | 1 | 0 | Resistant | |
| 15 | 30 | Md | 2 | 2 | 0 | Resistant | |
| 16 | 83 | Md | 1 | 1 | 0 | Not available | |
| 17 | 80 | F | 2 | 2 | 0 | Not available | |
| Confirmed CNS TB at autopsy | |||||||
| 18 | 30 | Md | 1 | 1 | 1 | Abnormal (B) | Resistante |
| 19 | 33 | Md | 3 | 3 | 2 | Abnormal (B) | Resistante |
| 20 | 36 | Md | 2 | 2 | 1 | Abnormal (B) | Resistante |
| Highly probablec | |||||||
| 21 | 27 | Fd | 2 | 2 | 2 | Wild type | Not available |
| 22 | 30 | Fd | 2 | 2 | 1 | Abnormal (C) | Resistante |
| 23 | 51 | F | 2 | 1 | 1 | Abnormal (C) | Not available |
| 24 | 75 | M | 1 | 1 | 1 | Wild type | Not available |
| 25 | 30 | Md | 1 | 1 | 1 | Wild type | Not available |
| 26 | 29 | M | 1 | 1 | 1 | Wild type | Not available |
| 27 | 29 | Md | 1 | 0 | 0 | Not available | |
| 28 | 41 | F | 2 | 0 | 0 | Not available | |
| Total no. of samples (no. of patients) | 48 (28) | 41 (26) | 26 (20) | ||||
| % of positive samples (% of patients) | 85.4 (92.9) | 54.2 (71.4) | |||||
M, male; F, female.
Three different abnormal patterns were found and classified as A, B, or C.
CNS TB was considered highly probable because the suggestive clinical picture was accompanied by at least three of the following supporting tests: (i) compatibly abnormal CSF findings; (ii) computed tomography findings suggesting CNS TB; (iii) evidence of tuberculosis outside the CNS; (iv) clinical response to antituberculosis therapy.
Patients coinfected with human immunodeficiency virus.
Susceptibility testing determined with isolates cultured from respiratory specimens.
TB, tuberculosis.
The detection of RMP resistance at the genotype level by molecular biology techniques designed to search for rpoB gene mutations has been the subject of various publications since 1993 (4, 7, 10, 15, 17, 19–21, 23, 24). Once these methods have been optimized and partially automated, it is possible that they will be adopted at least by reference laboratories (21).
The main aim of this study was to verify the applicability of DG-DGGE to investigating the sequence variations arising in the core region of rpoB. In comparison with other methods, DG-DGGE has a number of practical advantages. It is certainly less laborious than the manual single-strand conformation polymorphism (SSCP) method used by us in a previous study (17) because it does not require the casting of ultrathin gels or the use of radioactive isotopes. Furthermore, DG-DGGE is performed by means of a relatively inexpensive machine that allows controlled experimental conditions and optimum reproducibility.
By using this procedure, the amplified products obtained from all of the RMP-resistant strains unequivocally demonstrated homoduplex mobility alterations and/or the formation of heteroduplex molecules expressing the presence of mutations. This result was confirmed by automated sequence analysis of the entire sequence under investigation.
The mutations in the rpoB core region found in this study affect 7 of the 11 amino acids involved and constitute about one-third of the mutations found in that region previously reported in the literature; nevertheless, in terms of frequency, the mutations detected by us represent about 84% of the alterations reported in a recent review (13). Moreover, the involved codons found in this study are uniformly distributed throughout the rpoB core region, thus suggesting that DG-DGGE analysis may be capable of detecting mutations located anywhere in this sequence.
A RMP-resistant strain of our collection was shown have a double mutation in the rpoB gene, one of which (Glu562Ala) was located outside the core region. The significance of this mutation in the genesis of RMP resistance in M. tuberculosis is still unclear. However, in Escherichia coli, a cluster of mutations in rpoB gene located between codons 563 and 572 has been shown to cause RMP resistance in vitro (9).
The results were never difficult to interpret: the most frequently detected mutation (Leu-531), which is not always easily detectable by means of SSCP analysis (17), leads to the formation of an unmistakable three-band pattern (Fig. 1).
DG-DGGE analysis of the PCR products obtained from pathological clinical samples was also performed. The rpoB PCR protocol used in this study seems to be less sensitive than a similar protocol we optimized in order to produce a shorter sequence for SSCP analysis (17) and also less sensitive than the single-tube heminested protocol used by Whelen et al. (23). The possibility of fragmentation of the DNA target contained in a clinical sample stored at −20°C for a long time could partially justify the less sensitive nature of a method designed to amplify a considerably longer sequence.
In conclusion, the use of DG-DGGE analysis can reasonably be considered a valid alternative to other indirect methods of detecting sequence variations in the rpoB core region. Its excellent reproducibility, together with the fact that it is easy to perform, means that it can be used in clinical microbacteriology laboratories.
Acknowledgments
We are grateful to Cecilia Gelfi of ITBA (CNR, Milan, Italy) for excellent technical assistance.
This study was supported in part by a grant from the Istituto Superiore di Sanità (Progetto Nazionale Tubercolosi, grant 96/D/T57).
REFERENCES
- 1.Birch D E, Kolmodin L, Laird W J, McKinney N, Wong J, Young K K J, Zangenberg G A, Zoccoli M A. Simplified hot start PCR. Nature. 1996;381:445–446. doi: 10.1038/381445a0. [DOI] [PubMed] [Google Scholar]
- 2.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cremonesi L, Firpo S, Ferrari M, Righetti P G, Gelfi C. Double-gradient DGGE for optimization detection of DNA point mutations. BioTechniques. 1997;22:326–330. doi: 10.2144/97222rr01. [DOI] [PubMed] [Google Scholar]
- 4.Felmlee T A, Liu Q, Whelen A C, Williams D, Sommer S S, Persing D H. Genotypic detection of Mycobacterium tuberculosis rifampin resistance: comparison of single-strand conformation polymorphism and dideoxy fingerprinting. J Clin Microbiol. 1995;33:1617–1623. doi: 10.1128/jcm.33.6.1617-1623.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fischer S G, Lerman L. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci USA. 1983;80:1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gelfi C, Righetti S C, Zunino F, Della Torre G, Pierotti M A, Righetti P G. Detection of p53 point mutations by double-gradient, denaturing gradient gel electrophoresis. Electrophoresis. 1997;18:2921–2927. doi: 10.1002/elps.1150181533. [DOI] [PubMed] [Google Scholar]
- 7.Hunt J M, Roberts G D, Stockman L, Felmlee T A, Persing D H. Detection of a genetic locus encoding resistance to rifampin in mycobacterial cultures and in clinical specimens. Diagn Microbiol Infect Dis. 1994;18:219–227. doi: 10.1016/0732-8893(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 8.Inderlied C B, Salfinger M. Antimicrobial agents and susceptibility tests: mycobacteria. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. Washington, D.C: American Society for Microbiology; 1995. pp. 1385–1404. [Google Scholar]
- 9.Jin D J, Gross C A. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 10.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreinswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lerman L S, Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- 12.Morris S, Bai G H, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 13.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 15.Rossau R, Traore H, De Beenhouwer H, Mijs W, Jannes G, De Rijk P, Portaels F. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob Agents Chemother. 1997;41:2093–2098. doi: 10.1128/aac.41.10.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarpellini P, Racca S, Cinque P, Delfanti F, Gianotti N, Terreni M R, Vago L, Lazzarin A. Nested polymerase chain reaction for diagnosis and monitoring treatment response in AIDS patients with tuberculous meningitis. AIDS. 1995;9:895–900. doi: 10.1097/00002030-199508000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Scarpellini P, Braglia S, Brambilla A M, Dalessandro M, Cichero P, Gori A, Lazzarin A. Detection of rifampin resistance by single-strand conformation polymorphism analysis of cerebrospinal fluid of patients with tuberculosis of the central nervous system. J Clin Microbiol. 1997;35:2802–2806. doi: 10.1128/jcm.35.11.2802-2806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scarpellini P, Braglia S, Carrera P, Cedri M, Cichero P, Crucianelli R, Ferrari M, Mapelli B, Lazzarin A. Program and Abstracts of the 38th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Detection of rifampin (RMP) resistance in Mycobacterium tuberculosis by double gradient-denaturing gradient gel electrophoresis (DG-DGGE), abstr. D-94; p. 155. [Google Scholar]
- 19.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 20.Telenti A, Imboden P, Marchesi F, Schmidheini T, Bodmer T. Direct, automated detection of rifampin-resistant Mycobacterium tuberculosis by polymerase chain reaction and single-strand conformation polymorphism analysis. Antimicrob Agents Chemother. 1993;37:2054–2058. doi: 10.1128/aac.37.10.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telenti A, Honoré N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whelen A C, Felmlee T A, Hunt J M, Williams D L, Roberts G D, Stockman L, Persing D H. Direct genotypic detection of Mycobacterium tuberculosis rifampin resistance in clinical specimens by using single-tube heminested PCR assay. J Clin Microbiol. 1995;33:556–561. doi: 10.1128/jcm.33.3.556-561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]