Abstract
Most cases of invasive aspergillosis are caused by Aspergillus fumigatus, whose conidia are ubiquitous in the environment. Additionally, in indoor environments, such as houses or hospitals, conidia are frequently detected too. Hospital-acquired aspergillosis is usually associated with airborne fungal contamination of the hospital air, especially after building construction events. A. fumigatus strain typing can fulfill many needs both in clinical settings and otherwise. The high incidence of aspergillosis in COVID patients from our hospital, made us wonder if they were hospital-acquired aspergillosis. The purpose of this study was to evaluate whether the hospital environment was the source of aspergillosis infection in CAPA patients, admitted to the Hospital Universitario Central de Asturias, during the first and second wave of the COVID-19 pandemic, or whether it was community-acquired aspergillosis before admission. During 2020, sixty-nine A. fumigatus strains were collected for this study: 59 were clinical isolates from 28 COVID-19 patients, and 10 strains were environmentally isolated from seven hospital rooms and intensive care units. A diagnosis of pulmonary aspergillosis was based on the ECCM/ISHAM criteria. Strains were genotyped by PCR amplification and sequencing of a panel of four hypervariable tandem repeats within exons of surface protein coding genes (TRESPERG). A total of seven genotypes among the 10 environmental strains and 28 genotypes among the 59 clinical strains were identified. Genotyping revealed that only one environmental A. fumigatus from UCI 5 (box 54) isolated in October (30 October 2020) and one A. fumigatus isolated from a COVID-19 patient admitted in Pneumology (Room 532-B) in November (24 November 2020) had the same genotype, but there was a significant difference in time and location. There was also no relationship in time and location between similar A. fumigatus genotypes of patients. The global A. fumigatus, environmental and clinical isolates, showed a wide diversity of genotypes. To our knowledge, this is the first study monitoring and genotyping A. fumigatus isolates obtained from hospital air and COVID-19 patients, admitted with aspergillosis, during one year. Our work shows that patients do not acquire A. fumigatus in the hospital. This proves that COVID-associated aspergillosis in our hospital is not a nosocomial infection, but supports the hypothesis of “community aspergillosis” acquisition outside the hospital, having the home environment (pandemic period at home) as the main suspected focus of infection.
Keywords: COVID-19, aspergillosis, CAPA community-acquired, CAPA hospital-acquired
1. Introduction
Exposure to some fungal species, such as Aspergillus fumigatus, has been associated with opportunistic invasive fungal infections, a significant cause of mortality and morbidity in patients with severe neutropenia or immunosuppression. In these immunosuppressed patients, invasive aspergillosis (IA) is easy to suspect because given the typical and well-described risk factors and forms of presentation [1]. In recent years, several case series have been published about patients with severe influenza virus disease and without other risk factors who develop IA. These are generally patients with severe acute respiratory distress syndrome, in which incidence of IA depends on geographical differences, vaccination rates, and diagnostic tests used, varying from 11 to 28% [2,3,4]. More recently, in relation to the COVID-19 pandemic, an increasing number of patients with SARS-CoV-2 pneumonia and respiratory distress have been diagnosed with IA [2]. According to the different studies published, CAPA prevalence is highly variable, ranging between 3 and 33% [5,6,7,8,9]. These differences can be attributed to different thresholds for clinical evaluation, diagnostic methods, and differences in criteria for CAPA definition [6,10]. Prattes et al. [5], in a multicenter study, described a median prevalence of 10.7%. Our group has published a recent study with a prevalence of 11.7% during the first and second waves of pandemic COVID-19 [11]. This entity has been reported to significantly increase the severity of the COVID-19 with worse outcome and it has an important impact on the prognosis of the SARS-CoV-2 infection [2,6,7,8,9], so an early diagnosis can influence survival.
The growing number of cases has led to the belief in the possibility of nosocomial transmission of IA. Nosocomial acquisition of invasive aspergillosis has been proved in epidemic situations such as construction and renovation works, and also in non-epidemic cases [1,4,12,13,14], with studies showing how environment concentrations of Aspergillus and other fungi can be correlated with the presence of IA [1,14]. For this reason, during hospitalization, severely immunocompromised patients are believed to be at high risk of IA and, therefore, should receive care in units with rooms equipped with high-efficiency filters, laminar flow, and positive pressure [15] while maintaining good air quality in hospitals also contributes significantly to reducing the incidence of these Aspergillus infections [16]. However, the presence of filamentous fungi in the external environment has also been reported to have a significant impact on the disease [17,18], and therefore the influence of external isolates, not related to hospital nosocomial infections, as a source of IA has raised interest.
Our hospital saw a rapid increase of IA in patients with COVID-19 (CAPA) during the first and second waves of the pandemic, which led to an active search for cases and an interest to discover whether there was an element of nosocomial transmission to these infections [11]. Different publications have reported different incidences of CAPA among hospitalized patients [5,6,7,8,9], and none of them have been able to differentiate nosocomial and community infections, which is important for case investigation and infection control since hospital-acquired infections require adequate control measures to prevent subsequent cases. However, it is not easy to find an association between a hospital stay and the appearance of IA since it depends on the timing and the biological possibility. Therefore, epidemiological criteria must be assessed, as well as the time between admission and the onset of symptoms and microbiological criteria.
It is also important to bear in mind that while it is already difficult to establish a nosocomial origin in the case of common bacterial pathogens, it is even more complicated in the case of Aspergillus, in which patients have underlying conditions that lead to severe immunosuppression that puts them at higher risk of both community and hospital-acquired infections [19]. It should also be noted that IA incubation is influenced by individual differences and environmental determinants, including the severity of immunosuppression and air quality. Knowing better the early events related to IA will help prevent this disease, for which prognosis continues to be poor.
The purpose of our study was to evaluate the source of infection in CAPA patients admitted to Hospital Universitario Central de Asturias during the first and second wave of the COVID-19 pandemic, analyzing the distribution of Aspergillus spp. species among the samples of patients diagnosed with CAPA and hospital environmental isolates and carrying out the genotyping of the A. fumigatus isolates, to assess whether the aspergillosis was hospital or community-acquired.
2. Materials and Methods
2.1. Study Design
A prospective study was conducted at a tertiary university hospital in Asturias (Spain) during the COVID-19 pandemic between 1 January and 31 December 2020. Our institution is a large teaching hospital attending a population of 1,000,000 inhabitants. All the 3 intensive care units have positive pressure and HEPA filters. Patients in the ICUs are followed up by a team of specialized intensivists, anesthetists, pneumologists, and cardiovascular surgeons. Microbiologists, and infectious disease and preventive specialists perform daily rounds to collaborate in diagnosing and treating infectious complications and implementing preventive measures (HUCAPA Group).
The study was conducted among 300 patients tested for COVID-19 infection who were admitted to the hospital at ICU during the first and second waves (March–May 2020 and October–December 2020).
A diagnosis of COVID-19-associated pulmonary aspergillosis (CAPA) was defined based on the 2020 European Confederation of Medical Mycology/International Society for Human and Animal Mycology (ECCM/ISHAM) [20] consensus criteria, and according to these criteria, patients were classified as proven CAPA, probable CAPA, possible CAPA or no evidence of CAPA. CAPA diagnosis was defined as the earliest date when a diagnostic feature was identified.
The study was in accordance with the Helsinki declaration and national ethical standards. The hospital research ethics committee approved the study protocol.
2.2. Microbiology Data Collection
Starting in January 2020, institutional recommendations were to screen patients in ICUs for fungal infections by means of:
1. Fungal cultures from respiratory samples on Sabouraud dextrose agar plates (BioMerieux, Mercy, L’Etoile, France). Fungus identification was performed by a matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry instrument (Bruker, Madrid, Spain), following the manufacturer’s instructions.
2. The two commercially manufactured lateral flow devices (LFDs) (AspLFD, OLM Diagnostics, Newcastle upon Tyne, UK) and Lateral Flow Assays (LFAs) (IMMY sona Aspergillus Galactomannan Lateral Flow Assay, IMMY, Norman, OK, USA) tests, with a visual reader that provides a semiquantitative reading and removes subjectivity when interpreting results.
3. Quantitative real-time PCR for Aspergillus genus as follows: DNA extraction for PCR analysis was performed on an ELITe InGenius automated platform as well as RT-PCR using the Aspergillus spp. ELITe MGB kit (Elitegroup, Palex, Barcelona, Spain). The DNA was extracted from a 1-mL volume of BAL fluid and was eluted in a 200-µL saline solution before DNA amplification in the same platform. RT-PCR for the Aspergillus genus was performed by an Aspergillus spp. ELITe MGB kit, which was CE-in vitro diagnostic (CE-IVD) validated on a diverse range of sample types. The target region was the ribosomal DNA18S (rDNA18S), and the human B-globin gene was used as an internal standard. The fungal DNA copy number was expressed as copies/mL in relation to a rDNA18s standard curve.
To elucidate whether CAPA patients acquired aspergillosis in the community or the hospital, we looked for the presence of Aspergillus in the first respiratory sample obtained from the COVID patient, even if they were not yet in the ICU. In several available patients, we even perform Aspergillus PCR on the same sample used for the diagnosis of COVID.
4. GM testing was performed using Platelia™ Aspergillus (Bio-Rad Laboratories, Madrid, Spain) with a cut-off value of ≥0.5 in serum and ≥1.0 in BAL or ≥4 in tracheal and bronchial aspirates.
5. Detection of 1,3-β-d-glucan (βDG) in serum was performed with the Wako β-glucan test (Fujifilm Wako Pure Chemical Corporation, Vircell Microbiologists, Granada, Spain). A cut-off value of 7 pg/mL was used.
2.3. Antifungal Drugs Susceptibility Testing
Antifungal susceptibility testing (AFST) was performed following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution reference method 9.3.1 [21]. Antifungals used were amphotericin B (Sigma-Aldrich Química, Madrid, Spain) and the azoles itraconazole (Janssen Pharmaceutica, Madrid, Spain), voriconazole (Pfizer SA, Madrid, Spain), posaconazole (Schering-Plough Research Institute, Kenilworth, NJ, USA), and isavuconazole (Basilea Pharmaceutica, Basel, Switzerland). The final concentrations tested ranged from 0.03 to 16 mg/L for amphotericin B and 0.015 to 8 mg/L for the four azoles. A. flavus ATCC 204304 and A. fumigatus ATCC 204305 were used as quality control strains in all tests performed. Minimal inhibitory concentrations (MICs) were visually read after 24 and 48 h of incubation at 37 °C in a humid atmosphere. MICs were performed at least twice for each isolate. Clinical breakpoints for interpreting AFST results established by EUCAST [22] were used for classifying the A. fumigatus strains as susceptible or resistant.
2.4. Environmental Surveillance
Our local surveillance program consists of monthly environmental air sampling in operating rooms, ICUs, and high risks units, including the hematology (adult and pediatric) units and the transplantation units (heart, kidney, and liver) for quantitative and qualitative identification of filamentous fungi. Additional samples were also obtained when a suspicious case of Aspergillus infection was detected.
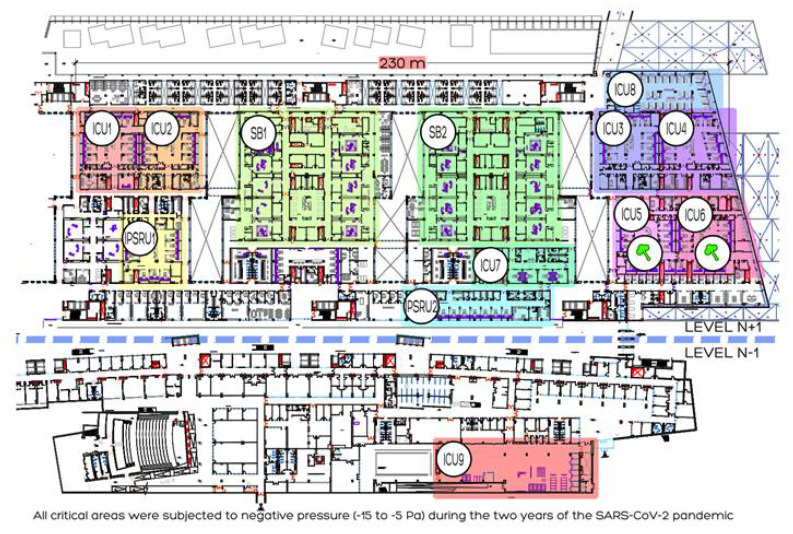
The distance between the west walls of UCI 1 and the east walls of UCI 4 is 230 m. All ICUs are located at level +1, with the exception of ICU 9, which was located at level −1.
All the Units in the Critical Areas (2 IPSRU and 9 ICUs) and all the Operating Rooms have independent and double air ducts. All of these double-ducted facilities have been under negative pressure (−15 to −5 Pa) since the start of the SARS-CoV2 pandemic (Figure 1).
Figure 1.
Map of Intensive Care Units (ICU), Intensive Post-Surgical Resuscitation Units (PSRU) and Operating Rooms (SB). Level of A. fumigatus conidia load obtained in ICUs 5 and 6 with 1 CFU/m3 each one during the study period.
During the study period, the attending physicians worked independently in each intensive care unit.
Volumetric air samples from environmental rooms and intensive care units were obtained using a volumetric sampler (Merck Air Sampler MAS100) as previously described [23].
Sealed Sabouraud-dextrose irradiated plates were incubated at 30 °C for 5 days. The plates were examined daily to check for fungal growth. Colonies of A. fumigatus growing on the plates were isolated, identified and stored.
2.5. Aspergillus fumigatus Strains
In this study, a total of 43 A. fumigatus strains were analyzed, 35 clinical and 8 environmental isolates. Strains identification was confirmed by amplification and sequencing of the ITS1-5.8S-ITS2 rDNA regions and a portion of the β-tubulin gene [24].
2.5.1. Cyp51A Amplification, PCR Conditions and Sequencing
For DNA extraction, conidia from each strain were cultured in glucose-yeast extractpeptone (GYEP) liquid medium (0.3% yeast extract, 1% peptone; Difco, Soria Melguizo, Madrid, Spain) with 2% glucose (Sigma-Aldrich Química, Madrid, Spain) for 24 h at 37 °C. After mechanical disruption of the mycelium by vortex-mixing with glass beads, genomic DNA of isolates was extracted using the phenol-chloroform method [25,26]. The full coding sequence of cyp51A including its promoter was amplified and sequenced. To exclude the possibility that any change identified in the sequences was due to PCR-induced errors, each isolate was independently analyzed twice. PCR reaction mixtures contained 0.5 µM of each primer, 0.2 µM of deoxynucleoside triphosphate (Roche, Madrid, Spain), 5 µL of PCR 10× buffer, 2 mM of MgCl2, DMSO 5.2%, 2.5 U of Taq DNA polymerase (Applied Biosystems, Foster City, CA, USA), and 100–200 ng of DNA in a final volume of 50 µL. A DNA 1-kb molecular ladder (Promega, Madrid, Spain) was used for all electrophoresis analyses. Samples were amplified in a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA). The parameters used were 1 cycle of 5 min at 94 °C and then 35 cycles of 30 s at 94 °C, 45 s at 56 °C for cyp51A promoter and 58 °C for cyp51A gene, and 2 min at 72 °C, followed by a 1 final cycle of 5 min at 72 °C. The amplified products were purified using IllustraExoProStar 1–step (GE Healthcare Life Science, Buckinghamshire, UK), and both strands were sequenced with the Big-Dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. All gene sequences were edited and assembled using the Lasergene software package (DNAStar Inc., Madison, WI, USA). Primers used to amplify and sequence cyp51A and its promoter have been previously described [27].
2.5.2. Strains Genotyping
All of the strains included in this study were genotyped following the previously described typing method TRESPERG [28,29]. Four markers were used: (i) Afu2g05150 encoding an MP-2 antigenic galactomannan protein (MP2); (ii) Afu6g14090 encoding a hypothetical protein with a CFEM domain (CFEM); (iii) Afu3g08990 encoding a cell surface protein A (CSP) and (iv) Afu1g07140 (ERG), which encodes a putative C-24 (28) sterol reductase. The combination of the genotypes obtained with each marker has a discriminatory value (D) of 0.9972 using the Simpson index [30].
3. Results
3.1. Clinical A. fumigatus Genotypes
The different genotypes isolated from 28 patients during the study period are shown in Table 1. Among the 56 clinical A. fumigatus strains isolated from patients, 28 different genotypes were detected. Strains from the same patient hosting the same genotype are summarized only in one representative isolate and similar isolates obtained from the same sample are excluded in the table.
Table 1.
A. fumigatus TRESPERg genotypes of isolates obtained from patient samples and their location at the hospital.
| SAMPLE | PATIENT | DATE | LOCATION | GENOTYPE |
|---|---|---|---|---|
| H-1880 | 1 | 23 March 2020 | BOX 45 | t18bm6.3g09.e09 |
| H-1882 | 2 | 30 March 2020 | BOX 10 | t02m1.1g09.e05 |
| H-1883 | 3 | 31 March 2020 | BOX 44 | t01m1.1g09.e07 |
| H-1885 | 4 | 4 April 2020 | 915 B | t01m5.1g09.e07 |
| H-1891 | 5 | 4 April 2020 | BOX 29 | t09m1.1g08A.e07 |
| H-1918 | 6 | 27 April 2020 | BOX 54 | t28m1.1g09.e20 |
| H-1935 | 7 | 16 May 2020 | 513 A | t03m1.3g08A.e07 |
| H-2096 | 8 | 8 October 2020 | SE 2° B | t03m1.1g05A.e09 |
| H-2097 | 9 | 8 October 2020 | BOX 39 | t02m1.1g09.e16 |
| H-2104 | 10 | 14 October 2020 | BOX 54 | t03m3.3g05A.e07 |
| H-2129 | 11 | 23 October 2020 | BOX 32 | t04Am1.1g05A.e07 |
| H-2135 | 12 | 25 October 2020 | BOX 14 | t11m1.2g09.e13 |
| H-2136 | 12 | 25 October 2020 | BOX 14 | t03m1.1g10.e06 |
| H-2159 | 12 | 3 November 2020 | BOX 14 | t02m1.8g09.e05 |
| H-2152 | 13 | 30 October 2020 | BOX 49 | t06Bm6.1g08A.e09 |
| H-2158 | 14 | 4 November 2020 | BOX 47 | t06Bm3.4g08A.e11 |
| H-2169 | 14 | 7 November 2020 | BOX 47 | t02m1.1g09.e16 |
| H-2167 | 15 | 5 November 2020 | BOX 33 | t03m1.1g04.e07 |
| H-2175 | 16 | 9 November 2020 | BOX 24 | t04Bm1.2g12.e15 |
| H-2184 | 17 | 13 November 2020 | BOX 89 | t04Am3.4g08A.e11 |
| H-2185 | 17 | 13 November 2020 | BOX 89 | t01m5.1g09.e06 |
| H-2186 | 18 | 13 November 2020 | BOX 91 | t02m1.1g09.e05 |
| H-2194 | 19 | 20 November 2020 | BOX 36 | t03m1.1g05A.e09 |
| H-2203 | 20 | 19 November 2020 | 925 A | t03m1.1g10.e09 |
| H-2211 | 21 | 31 November 2020 | 631 B | t02m14.1g09.e05 |
| H-2217 | 22 | 24 November 2020 | 532 B | t01m1.1g08A.e07 |
| H-2224 | 22 | 27 November 2020 | 306 B | t03m1.1g09.e07 |
| H-2238 | 23 | 3 December 2020 | BOX 67 | t02m1.2g09.e05 |
| H-2239 | 24 | 4 December 2020 | BOX 26 | t04Am3.3g17.eND |
| H-2267 | 24 | 16 December 2020 | BOX 31 | t04Am3.3g24.eND |
| H-2244 | 25 | 8 December 2020 | BOX 84 | t01m3.4g05A.e07 |
| H-2245 | 26 | 9 December 2020 | BOX 40S | t11m13.1g08A.e16 |
| H-2257 | 26 | 13 December 2020 | BOX 40S | t01m5.1g09.e13 |
| H-2246 | 27 | 8 December 2020 | BOX 57 | t02m1.1g09.e05 |
| H-2279 | 28 | 1 January 2021 | BOX 18 | t02m1.1g09.e05 |
Of the 28 patients analyzed, 22 (78.6%) had only one genotype of A. fumigatus. However; the remaining patients harbored more than one genotype of A. fumigatus; specifically, five (17.8%) of the patients (14, 17, 22, 24, and 26) harbor in their lungs two different genotypes; and in patient 12 (3.6%) coexisted three different genotypes of A. fumigatus. (Table 1).
3.2. Hospital Environmental A. fumigatus Genotypes
The chronological distribution of the environmental A. fumigatus isolates and level of A. fumigatus conidia obtained in the different rooms and ICUs are summarized in Table 2. Out of a total of 336 environmental samples taken during 2020 at the hospital, only seven showed positive growth of A. fumigatus (2%) with a minimum count of 1 CFU/m3 each one (Figure 1).
Table 2.
A. fumigatus TRESPERg genotypes of isolates from environmental samples.
| SAMPLE | DATE | LOCATION | CFU/m3 | GENOTYPE |
|---|---|---|---|---|
| HUCA-1800 | 23 January 2020 | HB 902 | 1 | 04Am1.3g08A.e07 |
| HUCA-1801 | 23 January 2020 | HB 902 | 1 | t04Am1.1g04.e07 |
| HUCA-1903 | 14 April 2020 | UCI 6 | 1 | t03m1.1g09.e09 |
| HUCA-2011 | 29 July 2020 | HB 902 | 1 | t02m11.1g09.e16 |
| HUCA-2061 | 10 September 2020 | Cytogenetics | 1 | t03m1.3g08A.e09 |
| HUCA-2130 | 19 October 2020 | Hb 903 | 1 | t02m1.1g09.e13 |
| HUCA-2151 | 30 October 2020 | UCI 5 | 1 | t01m1.1g08A.e07 |
During all the study period, only 1 CFU/m3 of A. fumigatus was isolated in ICU 5 and ICU 6. The remaining ICUs did not show any growth of Aspergillus.
Among the seven environmental A. fumigatus isolates, seven different genotypes were identified (Table 2).
3.3. Correlation between Clinical and Environmental A. fumigatus Genotypes
There was only one coincidence of genotypes between patient and environment during the study period; although there was no correlation in time or location. Specifically, patient 22 (diagnosed on 20 November 2020, on the fifth floor, room 532 B) in whom two different genotypes of A. fumigatus coexisted, one of them (t01m1.1g08A.e07) matched with the one isolated in the hospital environment (low level, box 54 on 30 October 2020), being the time difference of 25 days and a location difference of five floors (Table 3).
Table 3.
A. fumigatus isolates with the same TRESPERg genotype.
| SAMPLE | ORIGIN | DATE | LOCATION | GENOTYPE |
|---|---|---|---|---|
| HUCA-2151 | Air Sample | 30 October 2020 | UCI-5 BOX 54 | t01m1.1g08A.e07 |
| H-2217 | Patient 22 | 24 November 2020 | 532 B | t01m1.1g08A.e07 |
| H-2096 | Patient 8 | 8 October 2020 | SE 2° B | t03m1.1g05A.e09 |
| H-2194 | Patient 19 | 20 November 2020 | BOX 36 | t03m1.1g05A.e09 |
| H-2097 | Patient 9 | 8 October 2020 | BOX 39 | t02m1.1g09.e16 |
| H-2169 | Patient 14 | 7 November 2020 | BOX 47 | t02m1.1g09.e16 |
| H-1882 | Patient 2 | 30 March 2020 | BOX 10 | t02m1.1g09.e05 |
| H-2186 | Patient 18 | 13 November 2020 | BOX 91 | t02m1.1g09.e05 |
| H-2246 | Patient 27 | 8 December 2020 | BOX 57 | t02m1.1g09.e05 |
| H-2279 | Patient 28 | 1 January 2021 | BOX 18 | t02m1.1g09.e05 |
Regarding matches between patients were as follows:
Patient 8 admitted to Pneumology SE 2° B on 8 October 2020, exhibited the same A. fumigatus genotype (t03m1.1g05A.e09) as patient 19 admitted to box 36 on 20 November 2020; however, there was a significant difference over time (44 days) and in unit location (2 floors).
Patient 9 admitted to box 39 on 8 October 2020, exhibited the same A. fumigatus genotype (t02m1.1g09.e16) as patient 14 was admitted to box 47 on 7 November 2020; however, there was a significant difference over time (31 days) and in unit location (different ICUs).
Four patients (2, 18, 27 and 28) share the same genotype (t02m1.1g09.e05). Analyzing the patients individually, we observed that there is no correlation neither in time nor in their location in the hospital. The first patient (2) diagnosed on 30 March 2020, in box 10 presents a significant difference of 228 days with the second patient (18) admitted in box 91. The difference with the third patient (27) admitted in box 57 was 254 days, and the difference in time with the fourth patient (28) admitted in box 18 was 279 days. The four patients were admitted to different ICUs.
In summary, although several patients harbored the same A. fumigatus genotype, none of them were at the same time and/or in the same unit.
3.4. Aspergillus PCR in the Primary Respiratory Sample
Aspergillus PCR could be performed on the primary respiratory sample (sputum, tracheal aspirate, and/or bronchoalveolar lavage) obtained after admission, either on the ward and/or in the ICU.
Of the primary samples available from 20 patients, in 18 patients the Aspergillus PCR was positive with a CT < 36; and in the other two patients in whom the Aspergillus PCR was negative with a cycle greater than 36, the presence of Aspergillus was detected with an average of 110 copies/mL, an amount that increased in the following respiratory sample whose Aspergillus PCR was already positive.
This data supports the presence and/or co-infection of SARS-CoV-2 and Aspergillus since the first day of hospital admission.
3.5. Antifungal Susceptibility and Cyp51A Amplification
All isolates were susceptible to all antifungals tested and none of them showed any mutation responsible for azole resistance.
4. Discussion
Our study supports the idea that, in our population, COVID-associated aspergillosis is not a nosocomial infection, but a community acquired one, with home environment being the main suspected source of infection.
There are several findings that directly and indirectly back this claim. Firstly, the lack of epidemiological relationship between A. fumigatus isolates from hospital environmental air and clinical genotypes reinforce the role of community environmental air in the acquisition of CAPA. In our study, there was only one identical air-patient genotype that, however, did not coincide in time nor in location.
Secondly, during the study period, our hospital environment monitoring has shown excellent air quality, with an Aspergillus load of <0.005 CFU/m3. Only two ICUs had 1 CFU/m3 Aspergillus conidia, and they were not related to any of the cases.
It is important to highlight that all the isolates were prospectively collected, which is one of the strengths of our study. We have been able to genotype environmental and clinical isolates, and we have also used a high-discriminatory molecular typing tool (TRESPERG), without finding any matches between environmental hospital air and clinical genotypes. The wide diversity of A. fumigatus genotypes among patients and hospital environmental air shows an absence of specific clonal populations in the clinical setting, and thus also supports the hypothesis of a community acquired infection.
There is, as yet, no agreement about how to define a nosocomial case of IA, and different authors have adopted different standards. Patterson et al. [31] defined a nosocomial case of the disease as one that occurred > 1 week after admission to the hospital or <2 weeks after discharge. Although nosocomial outbreaks of IA have contributed to the current perception that most cases of IA are hospital acquired, the short time interval between admission and infection suggests that the patients are colonized with Aspergillus before they enter to the hospital, and a significant number of Aspergillus infections are acquired on an outpatient basis.
Indeed, the potential impact of Aspergillus colonization before hospitalization remains an issue that is still broadly discussed [17,32]; and the role of previous colonization with Aspergillus needs to be taken into account while also looking at risk factors related to the host (i.e., immunodepression, underlying diseases, etc.) as well as environmental factors (i.e., airway ventilation, home environment, etc.).
One of the complexities of this diagnosis is that the incubation period for CAPA patients is usually unknown and probably varies among different patients, making it hard to standardize a definition. One of the variables to consider is the fungal load of the previously colonized patient. Logically, with a higher fungal load of Aspergillus colonization and a greater viral load of SARS-CoV-2, the progression from colonization to clinical infection could be faster; and, if the patient also had immunosuppression (older, chronic lung disease, etc.) the time elapsed until clinical presentation of invasive aspergillosis could be shorter. Another risk factor to be considered is previous chronic lung damage: in the study by Prattes et al. [5], lung damage was described as a risk factor for the appearance of CAPA, and in the study published by García-Clemente et al. [11], 23% of patients with CAPA had some prior chronic respiratory disease, with these diseases remaining an independent risk factor for CAPA diagnosis in multivariate analysis. It is possible that previous colonization by Aspergillus in the lungs of patients with chronic pulmonary diseases could lead to invasive aspergillosis by adding the immune dysregulation and immunosuppression derived from severe SARS-CoV-2 disease. Based on these previous hypothesis, our study shows frequent co-infection of SARS-CoV-2 and Aspergillus in the first respiratory sample obtained from a patient on his first hospital admission day, which proves that there are patients previously colonized and/or infected by Aspegillus, in their own home and/or outside the hospital, whose symptoms appeared when, due to the severity of the viral infection, they became immunosuppressed.
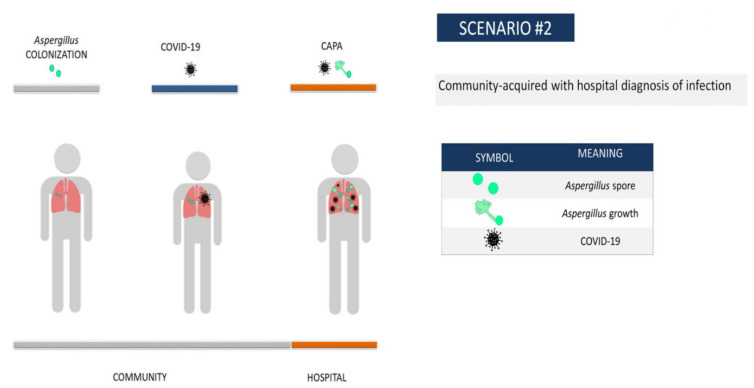
If we considered how the progression of invasive aspergillosis can be divided into four stages [33,34,35], the majority of CAPA patients in our hospital would be in scenario 2 (Figure 2), namely, community Aspergillus colonization, community COVID disease, and hospital diagnosis of community acquired-CAPA. In some patients, the diagnosis of CAPA was even diagnosed post-mortem and there was not time to implement antifungal treatment. This is the main reason why we must advocate for performing a fungal screening to look for an early mycological diagnosis in the primary respiratory sample in all patients with risk factors (older, with chronic lung disease, …) [11] in order to improve the survival of CAPA patients.
Figure 2.
Scenario 2—Community Aspergillus colonization, community COVID disease, and hospital diagnosis of community acquired-CAPA.
Finally, the last indirect evidence that supports our hypothesis that CAPA patients acquire aspergillosis in the community is that, in the first wave of COVID-19 in Spain, all people were housebound and therefore, the main focus of exposure and subsequent colonization would be the patients’ own homes, outside of the hospital setting.
Our study emphasizes the need for preventive measures outside of the hospital and, since the incidence of community Aspergillus infections will probably continue rising as high risk patients spend more time outside the hospital setting, we believe that taking care of the home environment of these patients would be essential.
The main limitation of our study is that we have not taken environmental samples at patients’ homes, but we have obtained direct evidence that the IA of our patients was not hospital-acquired since there were no matches between hospital-patient genotypes. Specific host and environmental risk factors still need to be studied in greater detail for CAPA patients.
5. Conclusions
To the best of our knowledge, this is the first study monitoring and genotyping A. fumigatus isolates from hospital air and COVID-19 patients admitted with aspergillosis, obtained during one year. Our study reveals a wide diversity of A. fumigatus genotypes among patients and the absence of specific clonal populations in the clinical setting, and highlights that COVID-associated aspergillosis is not a nosocomial infection; on the other hand, our data support the hypothesis of community acquisition, having home environment (pandemic period at home) as the main suspected focus of infection.
In the future, the role of prior colonization by Aspergillus needs prospective studies, since when IA appears during hospitalization, it is difficult to decide whether it is an acquired infection or really an infection that manifests itself during hospitalization starting from an underlying state of previous colonization, that progresses due to the situation of immunosuppression of the admitted patient, and studies must be carried out that take into account the host (immunosuppression, underlying disease, etc.) and environmental situation (ventilation, exposure to water supplies, etc.) characteristics that may be related to the risk of IA.
Acknowledgments
Members of the HUCAPA Group: Enrique García Carús, Tomás Suárez-Zarracina Secades, Mercedes Rodríguez Pérez, Marta Elena Álvarez-Argüelles, Susana Rojo, Jose Antonio Boga Riveiro, Marta Mateos Mazón, Eva Fernández Bretón, Lorena Forcelledo-Espina, Belén Rivaya, Ana Fernández-Verdugo, Blanca Leoz-Gordillo, Rodrigo Albillos-Almaraz, Cristina Iglesias, Marta Sandoval, Cristian Castelló-Abietar, Isabel Díaz Costales, Edurne Rodrigo Arrazola, Pablo González Moreno, Guillermo Albaiceta, Lisardo Iglesias, Diego Parra, Cecilia del Busto, Amparo Templado Barroso, María Magdalena Ramallal García, María Cristina Riestra Martínez, María Esther Díaz Díaz, María Isabel Díaz Zurrón, Francisco Carreño Alonso, Sandra Martínez Fernández, Martina Sánchez Araujo, Davinia Bravo Álvarez, Gonzalo Solís García, Jose Iraola García, Jorge González Díaz, Josu Jiménez Idoeta.
Abbreviations
| ARDS | Acute respiratory distress syndrome |
| BAL | Bronchoalveolar lavage. |
| B-D-G | Beta-D glucan. |
| CAPA | COVID-19 Associated Pulmonary Aspergillosis) |
| COVID | Coronavirus disease. |
| ECMM/ISHAM | European Confederation of Medical Mycology/International Society for Human and Animal Mycology |
| Study | Group Education and Research Consortium. |
| GM | Galactomannan. |
| ICU | Intensive care unit. |
| LFA | Lateral flow assays |
| LFD | Lateral flow device. |
| MALDI-TOF | Matrix-assisted laser desorption ionization-time of -light |
| NBL | Non-bronchoscopic lavage |
| OR | Odds ratio |
| PCR | Polymerase chain reaction. |
| TA | Tracheal aspirate. |
Author Contributions
Conceptualization, T.P.-G.d.l.R. and E.M.; methodology, I.G.-J., A.R., J.L.C.-A. and A.F.-A.; software, I.G.-J., A.R. and E.M.; validation, T.P.-G.d.l.R., E.M., M.T.-A. and M.G.-C.; formal analysis, E.M. and S.M.-G.; investigation, T.P.-G.d.l.R., E.M., M.G.-C., E.G.-P., L.C.-M., I.F.-S. and S.M.-G.; resources, T.P.-G.d.l.R. and M.G.-C.; writing—original draft preparation, T.P.-G.d.l.R., M.G.-C., A.F.-A. and E.M.; writing—review and editing, T.P.-G.d.l.R., M.G.-C., A.F.-A. and E.M.; visualization, T.P.-G.d.l.R., M.G.-C., A.F.-A., E.M. and J.L.C.-A.; supervision, T.P.-G.d.l.R., F.V.-V., M.M.-S. and M.L.S.-N.; project administration, T.P.-G.d.l.R. and M.G.-C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Principado of Asturias (protocol code 2020.472 and date of approval: 26 March 2020).
Informed Consent Statement
Patient consent was waived because it was the usual clinical practice in patients admitted to ICU, and most patients were intubated.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Brenier-Pinchart M.P., Lebeau B., Borel J.L., Quesada J.L., Mallaret M.R., Garban F., Brion J.P., Molina L., Bosson J.L., Thiebaut-Bertrand A., et al. Community-acquired invasive aspergillosis and outdoor filamentous fungal spore load: A relationship? Clin. Microbiol. Infect. 2011;17:1387–1390. doi: 10.1111/j.1469-0691.2011.03523.x. [DOI] [PubMed] [Google Scholar]
- 2.García Clemente M., Madrid Carbajal C., Iscar Urrutia M. Influenza, SARS-CoV-2 y aspergilosis pulmonar invasiva [Influenza, SARS-CoV-2 and Invasive Pulmonary Aspergillosis] Arch. Bronconeumol. 2021;57:11–12. doi: 10.1016/j.arbres.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schauwvlieghe A.F.A.D., Rijnders B.J.A., Philips N., Verwijs R., Vanderbeke L., Van Tienen C., Lagrou K., Verweij P.E., Van de Veerdonk F.L., Gommers D., et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: A retrospective cohort study. Lancet Respir. Med. 2018;6:782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 4.Wauters J., Baar I., Meersseman P., Meersseman W., Dams K., De Paep R., Lagrou K., Wilmer A., Jorens P., Hermans G. Invasive pulmonary aspergillosis is a frequent complication of critically ill H1N1 patients: A retrospective study. Intensiv. Care Med. 2012;38:1761–1768. doi: 10.1007/s00134-012-2673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prattes J., Wauters J., Giacobbe D.R., Salmanton-García J., Maertens J., Bourgeois M., Reynders M., Rutsaert L., Van Regenmortel N., Lormans P., et al. Risk factors and outcome of pulmonary aspergillosis in critically ill coronavirus disease 2019 patients-a multinational observational study by the European Confederation of Medical Mycology. Clin. Microbiol. Infect. 2021;26:S1198-743X(21)00474-2. doi: 10.1016/j.cmi.2021.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verweij P.E., Rijnders B.J.A., Brüggemann R.J.M., Azoulay E., Bassetti M., Blot S., Calandra T., Clancy C.J., Cornely O.A., Chiller T., et al. Review of influenza-associated pulmonary aspergillosis in ICU patients and proposal for a case definition: An expert opinion. Intensiv. Care Med. 2020;46:1524–1535. doi: 10.1007/s00134-020-06091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alanio A., Dellière S., Fodil S., Bretagne S., Mégarbane B. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respir. Med. 2020;8:e48–e49. doi: 10.1016/S2213-2600(20)30237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segrelles-Calvo G., Araújo G.R.S., Llopis-Pastor E., Carrillo J., Hernández-Hernández M., Rey L., Rodríguez Melean N., Escribano I., Antón E., Zamarro C., et al. Prevalence of opportunistic invasive aspergillosis in COVID-19 patients with severe pneumonia. Mycoses. 2021;64:144–151. doi: 10.1111/myc.13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machado M., Valerio M., Álvarez-Uría A., Olmedo M., Veintimilla C., Padilla B., De la Villa S., Guinea J., Escribano P., Ruiz-Serrano M.J., et al. Invasive pulmonary aspergillosis in the COVID-19 era: An expected new entity. Mycoses. 2021;64:132–143. doi: 10.1111/myc.13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jenks J.D., Nam H.H., Hoenigl M. Invasive aspergillosis in critically ill patients: Review of definitions and diagnostic approaches. Mycoses. 2021;64:1002–1014. doi: 10.1111/myc.13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marta G.-C., Lorena F.-E., Laura M.-V., Angela L.-M., Blanca L.-G., Rodrigo A.-A., Marta S.-G., Santiago M.-G., Liliana P.-M., Luisa S.-N.M., et al. COVID-19 Associated Pulmonary Aspergillosis in a tertiary hospital. J. Fungi. 2022;82:97. doi: 10.3390/jof8020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waldeck F., Boroli F., Suh N., Wendel Garcia P.D., Flury D., Notter J., Iten A., Kaiser L., Schrenzel J., Boggian K., et al. Influenza-associated aspergillosis in a critically-ill patients-a retrospective bicentric cohort study. Eur. J. Clin. Microbiol. Infect. Dis. 2020;39:1915–1923. doi: 10.1007/s10096-020-03923-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahieu L.M., De Dooy J.J., Van Laer F.A., Jansens H., Ieven M.M. A prospective study on factors influencing aspergillus spore load in the air during renovation works in a neonatal intensive care unit. J. Hosp. Infect. 2000;45:191–197. doi: 10.1053/jhin.2000.0773. [DOI] [PubMed] [Google Scholar]
- 14.Alberti C., Bouakline A., Ribaud P., Lacroix C., Rousselot P., Leblanc T., Derouin F. Relationship between environmental fungal contamination and the incidence of invasive aspergillosis in hematology patients. J. Hosp. Infect. 2001;48:198–206. doi: 10.1053/jhin.2001.0998. [DOI] [PubMed] [Google Scholar]
- 15.Gangneux J.P., Bretagne S., Cordonnier C., Datry A., Derouin F., Grillot R., Kauffmann-Lacroix C., Lebeau B., Morin O., Nicolle M.C., et al. Prevention of nosocomial fungal infection: The French approach. Clin. Infect. Dis. 2002;35:343–346. doi: 10.1086/341318. [DOI] [PubMed] [Google Scholar]
- 16.Peláez T., Muñoz P., Guinea J., Valerio M., Giannella M., Klaasen H.W., Bouza E. Outbreak of Invasive Aspergillosis after Major Heart Surgery Caused by Spores in the Air of the Intensive Care Unit. Clin. Infect. Dis. 2012;54:e24–e31. doi: 10.1093/cid/cir771. [DOI] [PubMed] [Google Scholar]
- 17.Fourneret-Vivier A., Lebeau B., Mallaret M.R., Brenier-Pinchart M.P., Brion J.P., Pinel C., Garban F., Pison C., Hamidfar R., Plantaz D., et al. Hospital-wide prospective mandatory surveillance of invasive aspergillosis in a French teaching hospital (2000–2002) J. Hosp. Infect. 2006;62:22–28. doi: 10.1016/j.jhin.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 18.Hajjeh R.A., Warnock D.W. Counterpoint: Invasive aspergillosis and the environment—rethinking our approach to prevention. Clin. Infect. Dis. 2001;33:1549–1552. doi: 10.1086/322970. [DOI] [PubMed] [Google Scholar]
- 19.Panackal A.A., Li H., Kontoyiannis D.P., Mori M., Perego C.A., Boeckh M., Marr K.A. Geoclimatic influences on invasive aspergillosis after hematopoietic stem cell transplantation. Clin. Infect. Dis. 2010;50:1588–1597. doi: 10.1086/652761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verweij P.E., Gangneux J.P., Bassetti M., Brüggemann R.J.M., Cornely O.A., Koehler P., Lass-Flörl C., van de Veerdonk F.L., Chakrabarti A., Hoenigl M. Diagnosing COVID-19-associated pulmonary aspergillosis. Lancet Microbe. 2020;1:e53–e55. doi: 10.1016/S2666-5247(20)30027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arendrup M.C., Cuenca-Estrella M., Lass-Flörl C., Hope W.W. European Committee on Antimicrobial Susceptibility Testing Subcommittee on Antifungal Susceptibility Testing (EUCAST-AFST). EUCAST technical note on Aspergillusand amphotericin B, itraconazole, and posaconazole. Clin. Microbiol. Infect. 2012;18:E248–E250. doi: 10.1111/j.1469-0691.2012.03890.x. [DOI] [PubMed] [Google Scholar]
- 22.European Committee on Antimicrobial Susceptibility Testing Antifungal Agents. Breakpoint Tables for Interpretation of MICs. Version 9.0. [(accessed on 7 December 2020)]. Available online: https://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals/
- 23.Vandecasteele S.J., Boelaert J.R., Verrelst P., Graulus E., Gordts B.Z. Diagnosis and treatment of Aspergillus flavus sternal wound infections after cardiac surgery. Clin. Infect. Dis. 2002;35:887–890. doi: 10.1086/342699. [DOI] [PubMed] [Google Scholar]
- 24.Alcazar-Fuoli L., Mellado E., Alastruey-Izquierdo A., Cuenca-Estrella M., Rodriguez-Tudela J.L. Aspergillus section Fumigati: Antifungal susceptibility patterns and sequence-based identification. Antimicrob. Agents Chemother. 2008;52:1244–1251. doi: 10.1128/AAC.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco J.L., Hontecillas R., Bouza E., Blanco I., Pelaez T., Muñoz P., Molina J.P., Garcia M.E. Correlation between the Elastase Activity Index and Invasiveness of Clinical Isolates of Aspergillus fumigatus. J. Clin. Microbiol. 2002;40:1811–1813. doi: 10.1128/JCM.40.5.1811-1813.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang C.M., Cohen J., Holden D.W. An Aspergillusfumigatus alkaline protease mutant constructed by gene disruption is deficient in extracellular elastase activity. Mol. Microbiol. 1992;6:1663–1671. doi: 10.1111/j.1365-2958.1992.tb00891.x. [DOI] [PubMed] [Google Scholar]
- 27.Mellado E., Diaz-Guerra T.M., Cuenca-Estrella M., Rodriguez-Tudela J.L. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillusfumigatus and other Aspergillus species. J. Clin. Microbiol. 2001;39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Rubio R., Gil H., Monteiro M.C., Peláez T., Mellado E. A new Aspergillus fumigatus typing method based on hypervariable tandem repeats located within exons of surface protein-coding genes (TRESP) PLoS ONE. 2016;11:e0163869. doi: 10.1371/journal.pone.0163869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Rubio R., Escribano P., Gomez A., Guinea J., Mellado E. Comparison of Two Highly Discriminatory Typing Methods to Analyze Aspergillus fumigatus Azole Resistance. Front. Microbiol. 2018;9:1626. doi: 10.3389/fmicb.2018.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson J.E., Zidouh A., Miniter P., Andriole V.T., Patterson T.F. Hospital epidemiological surveillancefor invasive aspergillosis: Patient demographics and the utility of antigen detection. Infect. Control. Hosp. Epidemiol. 1997;18:104–108. doi: 10.2307/30142398. [DOI] [PubMed] [Google Scholar]
- 32.Vandewoude K.H., I Blot S., Depuydt P., Benoit D., Temmerman W., Colardyn F., Vogelaers D. Clinical relevance of Aspergillus isolation from respiratory tract samples in critically ill patients. Crit. Care. 2006;10:R31. doi: 10.1186/cc4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolle M.C., Benet T., Vanhems P. Aspergillosis: Nosocomial or community-acquired? Med. Micol. 2011;49:S24–S29. doi: 10.3109/13693786.2010.509335. [DOI] [PubMed] [Google Scholar]
- 34.Sulahian A., Boutboul F., Ribaud P., Leblanc T., Lacroix C., Derouin F. Value of antigen detection using an enzyme immunoassay in diagnosing and predicting invasive aspergillosis in two adult and pediatric hematology units during a 4-year prospective study. Cancer. 2001;91:311–318. doi: 10.1002/1097-0142(20010115)91:2<311::AID-CNCR1003>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 35.Hope W.W., Walsh T.J., Denning D.W. Laboratory diagnosis of invasive aspergillosis. Lancet Infect. Dis. 2005;50:609–622. doi: 10.1016/S1473-3099(05)70238-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.