Toho-2 is a non-TEM, non-SHV-type extended-spectrum β-lactamase, produced by Escherichia coli TUM 1083 isolated in 1995 in Japan, a cefotaxime-resistant clinical isolate (8). Toho-2, like Toho-1 (7), belongs to a β-lactamase cluster which includes plasmid-mediated β-lactamases such as MEN-1 (3, 6), also referred as CTX-M-1 (4, 5), and chromosomally mediated β-lactamases such as those produced by Proteus vulgaris (9), Klebsiella oxytoca (11), and Serratia fonticola (10).
The cefotaxime resistance gene for Toho-2 was sequenced by Ma et al. (8). The deduced protein sequence consisted of a precursor of 327 residues. As reported by the authors (8), the Toho-2 β-lactamase protein sequence is close to that of Toho-1, but the sequence located at Ala-185 to Ala-219 had “almost no homology with the other class A β-lactamases. The sequence may constitute a loop structure at the substrate-binding site and has a deletion of 2 amino acid residues…”
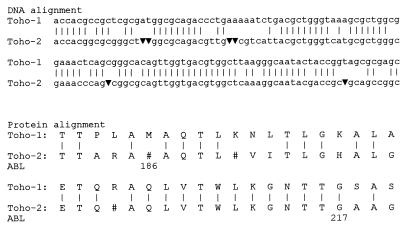
In order to better understand the possible evolution of Toho-2 from an unknown precursor, I tried to introduce a few putative bases in the blatoho gene, as six were possibly lost (Fig. 1). I started at base 676, using the numbering of Ma et al. This corresponds to amino acid residue 186, following the numbering scheme of Ambler et al. (2). The last introduced base corresponds to amino acid residue 217. Thus, for the 32 residues of the novel deduced protein sequence, and in comparison with the Toho-1 β-lactamase sequence, 25 residues are conserved, 3 cannot be determined because of lack of information, and 4 are substituted. This shows that the corrected sequence is very close to that of Toho-1. Next, a Blast search of the National Center for Biotechnology Information database (8a) was performed by using a 90-base sequence starting at base 676 of the blatoho2 gene (1). High alignment scores were obtained only for β-lactamases of the previously defined cluster. Two 17-base identities were obtained, respectively, for the Mus cookii DNA sequence, GenBank accession number M97512 (3′-tggtgacgtggctcaaa-5′, starting at base 735 of the blatoho2 gene following the numbering of Ma et al. [8]), and Streptomyces coelicolor cosmid 2E1, GenBank accession number AL023797 (3′-gggtcatgcgctgggcg-5′, starting at base 701). Nevertheless, these two 17-base identities are meaningless.
FIG. 1.
Local alignment of the blatoho1 and blatoho2 genes. Toho-1 starts at base 640 and Toho-2 starts at base 661, following the numbering system of Ma et al. (8). Six presumed bases, starting at position 676 of blatoho2, were introduced. The protein alignment is deduced from the blatoho1 gene and the blatoho2 gene after introduction of the six bases. “ABL” represents Ambler’s numbering scheme for amino acids (2). ▾, putative bases. Note that the four codons “gc▾” code for glycine (Gly-217).
These results suggest that the blatoho2 gene sequence should be revised.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P, Coulson A F W, Frère J M, Ghuysen J M, Joris B, Forsman M, Levesque R C, Tirabi G, Waley S G. A standard numbering scheme for the class A β-lactamases. Biochem J. 1991;276:269–272. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barthélémy M, Péduzzi J, Bernard H, Tancrède C, Labia R. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim Biophys Acta. 1992;1122:15–22. doi: 10.1016/0167-4838(92)90121-s. [DOI] [PubMed] [Google Scholar]
- 4.Bauernfeind A, Cassellas J M, Goldberg M, Holley R, Jungwirth R, Mangold P, Röhnisch T, Schweighart S, Wilhelm R. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection. 1992;20:158–163. doi: 10.1007/BF01704610. [DOI] [PubMed] [Google Scholar]
- 5.Bauernfeind A, Stemplinger I, Jungwirth R, Ernst S, Casellas J M. Sequences of β-lactamase genes encoding CTX-M-1 (MEN-1) and CTX-M-2 and relationship of their amino acid sequences with those of other β-lactamases. Antimicrob Agents Chemother. 1996;40:509–513. doi: 10.1128/aac.40.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard H, Tancrède C, Livrelli V, Morand A, Barthélémy M, Labia R. A novel plasmid-mediated extended-spectrum β-lactamase not derived from TEM- or SHV-type enzymes. J Antimicrob Chemother. 1992;29:590–592. doi: 10.1093/jac/29.5.590. [DOI] [PubMed] [Google Scholar]
- 7.Ishii Y, Ohno A, Taguchi H, Imajo S, Ishiguro M, Matsuzawa H. Cloning and sequence of the gene encoding a cefotaxime-hydrolyzing class A β-lactamase isolated from Escherichia coli. Antimicrob Agents Chemother. 1995;39:2269–2275. doi: 10.1128/aac.39.10.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, Ishii Y, Ishiguro M, Matsuzawa H, Yamaguchi K. Cloning and sequencing of the gene encoding Toho-2, a class A β-lactamase preferentially inhibited by tazobactam. Antimicrob Agents Chemother. 1998;42:1181–1186. doi: 10.1128/aac.42.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.National Center for Biotechnology Information Website. 5 January 1999, posting date. BLASTN 2.0.8. [Online.] www.ncbi.nlmnih.gov/BLAST. [23 April 1999, last date accessed.]
- 9.Pèduzzi J, Reynaud A, Baron P, Barthélémy M, Labia R. Chromosomally encoded cephalosporinase of Proteus vulgarisRo104 belongs to Ambler’s class A. Biochim Biophys Acta. 1994;1207:31–39. doi: 10.1016/0167-4838(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 10.Péduzzi J, Farzaneh S, Reynaud A, Barthélémy M, Labia R. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A β-lactamase from Serratia fonticolaCUV. Biochim Biophys Acta. 1997;1341:58–70. doi: 10.1016/s0167-4838(97)00020-4. [DOI] [PubMed] [Google Scholar]
- 11.Reynaud A, Péduzzi J, Barthélémy M, Labia R. Cefotaxime-hydrolysing activity of the β-lactamase of Klebsiella oxytocaD488 could be related to a threonine residue at position 140. FEMS Microbiol Lett. 1991;81:185–192. doi: 10.1016/0378-1097(91)90301-p. [DOI] [PubMed] [Google Scholar]