Abstract
Background
Both changes in circulating lipids represented by a validated poor prognostic 3-lipid signature (3LS) and somatic tumour genetic aberrations are individually associated with worse clinical outcomes in men with metastatic castration-resistant prostate cancer (mCRPC). A key question is how the lipid environment and the cancer genome are interrelated in order to exploit this therapeutically. We assessed the association between the poor prognostic 3-lipid signature (3LS), somatic genetic aberrations and clinical outcomes in mCRPC.
Methods
We performed plasma lipidomic analysis and cell-free DNA (cfDNA) sequencing on 106 men with mCRPC commencing docetaxel, cabazitaxel, abiraterone or enzalutamide (discovery cohort) and 94 men with mCRPC commencing docetaxel (validation cohort). Differences in lipid levels between men ± somatic genetic aberrations were assessed with t-tests. Associations between the 3LS and genetic aberrations with overall survival (OS) were examined using Kaplan-Meier methods and Cox proportional hazard models.
Results
The 3LS was associated with shorter OS in the discovery (hazard ratio [HR] 2.15, 95% confidence interval [CI] 1.4-3.3, p < 0.001) and validation cohorts (HR 2.32, 95% CI 1.59–3.38, p < 0.001). Elevated plasma sphingolipids were associated with AR, TP53, RB1 and PI3K aberrations (p < 0.05). Men with both the 3LS and aberrations in AR, TP53, RB1 or PI3K had shorter OS than men with neither in both cohorts (p ≤ 0.001). The presence of 3LS and/or genetic aberration was independently associated with shorter OS for men with AR, TP53, RB1 and PI3K aberrations (p < 0.02). Furthermore, aggressive-variant prostate cancer (AVPC), defined as 2 or more aberrations in TP53, RB1 and/or PTEN, was associated with elevated sphingolipids. The combination of AVPC and 3LS predicted for a median survival of ~12 months. The relatively small sample size of the cohorts limits clinical applicability and warrants future studies.
Conclusions
Elevated circulating sphingolipids were associated with AR, TP53, RB1, PI3K and AVPC aberrations in mCRPC, and the combination of lipid and genetic abnormalities conferred a worse prognosis. These findings suggest that certain genotypes in mCRPC may benefit from metabolic therapies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-022-02298-0.
Keywords: Androgen receptor, Biomarker, Castration-resistant prostate cancer, Genomics, Lipid, PI3K, RB1, TP53
Background
Prostate cancer (PC) is the second most frequent cancer and fifth leading cause of cancer death in men worldwide [1]. Therapies such as taxanes, androgen receptor signalling inhibitors (ARSI), poly-ADP ribose polymerase inhibitors (PARPi) and targeted radioisotopes have significantly increased survival in metastatic castration-resistant prostate cancer (mCRPC); however, treatments driven by molecular subtyping are currently limited to PARPi in the setting of DNA repair gene aberrations and pembrolizumab in men with microsatellite instability [2]. Moreover, long-term control of mCRPC requires a combined approach targeting multiple hallmarks of cancer, encompassing the cancer genome, the immune system and metabolic factors including lipid metabolism, all of which contribute to cancer progression and treatment resistance [3].
The genomic landscape of mCRPC is well characterised [4], with numerous somatic genetic aberrations linked to poor clinical outcomes. Androgen receptor (AR) aberrations (50–70%), TP53 aberrations (50%), PTEN deletion (30%) and RB1 deletion (20%) are all associated with shorter time on ARSI [4–7]. RB1 deletion is also correlated with shorter overall survival (OS) in mCRPC [6]. Aggressive-variant prostate cancer (AVPC), a subset of mCRPC which is rapidly progressive with a poor prognosis, is morphologically heterogenous, but has been shown to correlate with genetic aberrations in two or more of TP53, RB1 and/or PTEN [8].
Altered lipid metabolism and its impact on PC is increasingly recognised through epidemiological [9–11] and molecular [12, 13] studies. Lipidomic profiling of plasma from men with PC has demonstrated that the plasma lipid profiles including elevated sphingolipids are associated with a poorer prognosis [14–17]. Ceramide, a key sphingolipid, can be metabolised into sphingosine-1-phosphate (S1P) or sphingomyelin to promote cancer cell growth, tumour metastasis and treatment resistance [18–20]. Elevated circulating sphingolipids, in particular ceramides, are associated with poorer clinical outcomes across the natural history of PC, including metastatic relapse in localised PC, earlier androgen deprivation failure in metastatic hormone-sensitive PC, and shorter OS and radiographic progression-free survival (rPFS) in mCRPC [14–17]. We have derived and validated a poor prognostic 3-lipid signature (3LS) consisting of ceramide(d18:1/24:1), sphingomyelin(d18:2/16:0) and phosphatidylcholine(16:0/16:0) to represent the poor prognostic lipidomic profile [14]. The 3LS is independently associated with shorter OS in men with mCRPC commencing docetaxel in internal and external validation cohorts [14, 15].
A key question arises from lipidomic profiling studies—how do circulating lipid aberrations relate to the genomic landscape of prostate cancer? Genetic alterations can lead to metabolic reprogramming in cancer, and conversely, metabolic dysregulation can be an initiator of malignant cellular transformation [21]. The current study is uniquely placed to address this question and provide insights into new potential therapeutic strategies due to the multidisciplinary approach of parallel metabolic and genomic profiling. Thus, we aimed to assess the relationship between the poor prognostic 3-lipid signature, somatic genetic aberrations and clinical outcomes in mCRPC.
Methods
Study population and sample collection
The discovery cohort consisted of 149 men with mCRPC commencing taxanes (docetaxel or cabazitaxel) or ARSI (abiraterone or enzalutamide) at seven sites in New South Wales and Victoria, Australia, who were prospectively enrolled between June 2016 and February 2020. All participants provided written informed consent, with ethics approval obtained from Monash Health Institutional Review Board (15571X) and Royal Prince Alfred Hospital Human Research Ethics Committee (X19-0320). Plasma samples were collected prior to starting treatment according to a standardised blood collection protocol [22].
The validation cohort comprised of 142 men with mCRPC commencing docetaxel at a single US tertiary cancer centre (Mayo Clinic), who were prospectively enrolled between September 2009 to August 2013. Details of ethics approval and sample collection for this cohort have been published previously [23].
Plasma lipidomic analysis
Lipidomic profiling of plasma samples was performed by liquid chromatography-mass spectrometry as previously described [24]. Data was normalised using the Probabilistic Quotient Normalisation method as outlined previously [15], with final lipid levels transformed to logarithm-2 of pmol/mL for statistical analysis. More detail is provided in Additional File 1: Section S1 [14, 15, 24–26].
Targeted cfDNA sequencing
Plasma cfDNA extraction, next-generation sequencing and bioinformatics analysis was performed as previously reported [22, 27]. Briefly, extracted cfDNA underwent library preparation, panel-based hybridisation (Predicine, Inc.) and enrichment, followed by paired-end sequencing on the Illumina HiSeq XTen. Somatic point mutations, insertions/deletions and copy number alterations were identified using Predicine’s proprietary GeneRADAR technology and DeepSea machine learning bioinformatics algorithm. Circulating tumour DNA (ctDNA) fraction was calculated as previously described [22]. More detail is included in Additional File 1: Section S2 [7, 22, 23, 27].
Statistical analysis
Time to rPFS and OS were calculated from the date of treatment commencement to event and censored at date of last follow-up if the event had not occurred. Data regarding rPFS was not available for the validation cohort.
Statistical analyses were performed with R version 4.0.2 and SPSS version 27. Principal components analysis of baseline lipidomic profiles was used to assess whether there were any underlying baseline metabolic differences that could confound our subsequent survival analyses. To determine the presence of the circulating 3LS of poor prognosis, lipidomic datasets were aligned to the original cohort in Lin et al [14] from which the 3LS was derived using the ComBat algorithm (R package sva, v3.34.0) and then calculated as previously described (Additional File 1: Section S1.5) [14, 15]. Somatic gene aberrations were defined as copy number variations or mutations.
Differences in lipid levels between men with or without genetic aberrations were assessed with t-tests (R package rstatix, v0.7.0). Survival analyses were performed using Kaplan-Meier methods (R package survival, v3.2-10). Cox regression was used to determine associations between the combined presence of 3LS and genetic aberrations, established clinicopathologic factors (Tables 2 and 3) and OS. Men with mCRPC were grouped according to the biomarker combination of 3LS and genetic aberration in the Cox regression analyses as follows: Group 0 = absence of both 3LS and the genetic aberration, Group 1 = presence of one abnormality (either 3LS or the genetic aberration), and Group 2 = presence of both 3LS and the genetic aberration. P-values < 0.05 were considered statistically significant.
Table 2.
Cox proportional hazards analysis of overall survival based on biomarker combination in the discovery cohort
| Variable | Univariable Cox regression | Multivariable Cox regression using AR aberration and 3-lipid signature | Multivariable Cox regression using TP53 aberration and 3-lipid signature | Multivariable Cox regression using RB1 deletion and 3-lipid signature | Multivariable Cox regression using PI3K aberration and 3-lipid signature | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Biomarker combinations | AR aberration and/or 3-lipid signature (Groups 1 and 2 vs 0) | 4.55 (2.27–9.12) | < 0.001 | 2.89 (1.39–6.00) | 0.005 | – | – | – | – | – | – |
| TP53 aberration and/or 3-lipid signature (Groups 1 and 2 vs 0) | 5.22 (2.62–10.41) | < 0.001 | – | – | 3.26 (1.57–6.77) | 0.001 | – | – | – | – | |
| RB1 deletion and/or 3-lipid signature (Groups 1 and 2 vs 0) | 3.05 (1.79–5.20) | < 0.001 | – | – | – | – | 1.98 (1.12–3.50) | 0.019 | – | – | |
| PI3K aberration and 3-lipid signature (Group 2 vs 0 and 1) | 3.02 (1.75–5.21) | < 0.001 | – | – | – | – | – | – | 2.15 (1.21–3.82) | 0.009 | |
| Clinicopatholgic factors | Albumin, g/La | 0.86 (0.82–0.91) | < 0.001 | 0.89 (0.84–0.95) | < 0.001 | 0.89 (0.84–0.95) | < 0.001 | 0.89 (0.84–0.95) | < 0.001 | 0.88 (0.83–0.94) | < 0.001 |
| ECOG performance status (≥ 2 vs 0–1) | 3.94 (1.75–8.87) | < 0.001 | 1.74 (0.73–4.13) | 0.212 | 1.64 (0.69–3.91) | 0.263 | 1.72 (0.72–4.11) | 0.222 | 2.36 (0.98–5.67) | 0.054 | |
| Pain at baseline (present vs absent) | 1.89 (1.12–3.19) | 0.018 | 1.73 (0.99–3.04) | 0.056 | 1.50 (0.86–2.63) | 0.152 | 1.77 (1.01–3.09) | 0.046 | 1.64 (0.93–2.90) | 0.089 | |
| Haemoglobin (<90 g/L vs ≥ 90 g/L) | 2.70 (0.84–8.73) | 0.096 | – | – | – | – | – | – | – | – | |
| PSA, ng/mLa | 1.00 (1.00–1.00) | 0.081 | – | – | – | – | – | – | – | – | |
| ALP, IU/La | 1.00 (1.00–1.00) | 0.197 | – | – | – | – | – | – | – | – | |
| Treatment type (taxane vs ARSI) | 1.05 (0.62–1.79) | 0.844 | – | – | – | – | – | – | – | – | |
| Treatment line (second line vs first line) | 0.87 (0.51–1.48) | 0.597 | – | – | – | – | – | – | – | – | |
| Visceral metastases (present vs absent) | 1.59 (0.75–3.37) | 0.225 | – | – | – | – | – | – | – | – | |
| ctDNA Fraction | ctDNA fraction > 2% | 1.97 (1.06–3.66) | 0.032 | 1.74 (0.91–3.35) | 0.097 | 1.76 (0.93–3.33) | 0.083 | 2.22 (1.17–4.18) | 0.014 | 2.24 (1.18–4.24) | 0.014 |
Univariable and multivariable Cox regression based on biomarker combination, clinicopathologic factors and ctDNA fraction. Only variables with p < 0.05 in univariable analysis were included in multivariable analysis
Group 0 = absence of both 3LS and the genetic aberration; Group 1 = presence of one abnormality (either 3LS or the genetic aberration); Group 2 = presence of both 3LS and the genetic aberration
ALP alkaline phosphatase, ARSI androgen receptor signalling inhibitor, CI confidence interval, ctDNA circulating tumour DNA, ECOG Eastern Cooperative Oncology Group, HR hazard ratio, PI3K phosphatidylinositol-3-kinase, PSA prostate-specific antigen
All p-values < 0.05 are highlighted in bold
aContinuous variable
Table 3.
Cox proportional hazards analysis of overall survival based on biomarker combination in the validation cohort
| Variable | Univariable Cox regression | Multivariable Cox regression using AR aberration and 3-lipid signature | Multivariable Cox regression using TP53 aberration and 3-lipid signature | Multivariable Cox regression using RB1 deletion and 3-lipid signature | Multivariable Cox regression using PI3K aberration and 3-lipid signature | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Biomarker combinations | AR aberration and/or 3-lipid signature (Groups 1 and 2 vs 0) | 2.92 (1.79–4.75) | < 0.001 | 2.45 (1.42–4.20) | 0.001 | – | – | – | – | – | – |
| TP53 aberration and/or 3-lipid signature (Groups 1 and 2 vs 0) | 2.77 (1.67–4.61) | < 0.001 | – | – | 2.82 (1.64–4.83) | < 0.001 | – | – | – | – | |
| RB1 deletion and/or 3-lipid signature (Groups 1 and 2 vs 0) | 2.53 (1.58–4.07) | < 0.001 | – | – | – | – | 1.95 (0.98–3.86) | < 0.001 | – | – | |
| PI3K aberration and 3-lipid signature (Group 2 vs 0 and 1) | 1.78 (1.03–3.10) | 0.04 | – | – | – | – | – | – | 1.32 (0.70–2.50) | 0.396 | |
| Clinicopatholgic factors | PSA, ng/mLa | 1.00 (1.00–1.00) | < 0.001 | 1.00 (1.00–1.00) | < 0.001 | 1.00 (1.00–1.00) | < 0.001 | 1.00 (1.00–1.00) | < 0.001 | 1.00 (1.00–1.00) | < 0.001 |
| ALP, IU/La | 1.00 (1.00–1.00) | 0.002 | 1.00 (1.00–1.00) | 0.024 | 1.00 (1.00–1.00) | 0.03 | 1.00 (1.00–1.00) | 0.069 | 1.00 (1.00–1.00) | 0.034 | |
| ctDNA Fraction | ctDNA fraction > 2% | 1.91 (1.18–3.08) | 0.008 | 1.29 (0.76–2.17) | 0.347 | 1.37 (0.81–2.30) | 0.238 | 1.74 (1.05–2.87) | 0.062 | 1.67 (1.01–2.78) | 0.048 |
Univariable and multivariable Cox regression based on biomarker combination, clinicopathologic factors and ctDNA fraction. Only variables with p < 0.05 in univariable analysis were included in multivariable analysis
Group 0 = absence of both 3LS and the genetic aberration; Group 1 = presence of one abnormality (either 3LS or the genetic aberration); Group 2 = presence of both 3LS and the genetic aberration
ALP alkaline phosphatase, CI confidence interval, ctDNA circulating tumour DNA, HR hazard ratio, PI3K phosphatidylinositol-3-kinase, PSA prostate-specific antigen
All p-values < 0.05 are highlighted in bold
aContinuous variable
Results
Study cohorts
In the discovery cohort, 149 men with mCRPC commencing taxanes (27%) or ARSI (73%) underwent plasma lipidomic analysis. Their baseline plasma lipidomic profiles did not show major variations between treatment type (taxane vs ARSI) or line of treatment (first vs second) according to principal components analysis (Additional File 1: Fig S3.1-S3.2).
A subset of the discovery cohort (n = 106) had available cfDNA for genomic sequencing (Additional File 1: Fig S4). In the validation cohort, 142 men with mCRPC commencing docetaxel underwent plasma lipidomic analysis. cfDNA for genomic sequencing was available from 94 participants (Additional File 1: Fig S4). The clinical characteristics were consistent between the full cohorts and the subsets with lipidomic and genomic analyses (Additional File 1: Table S5).
Clinical outcomes by 3-lipid signature
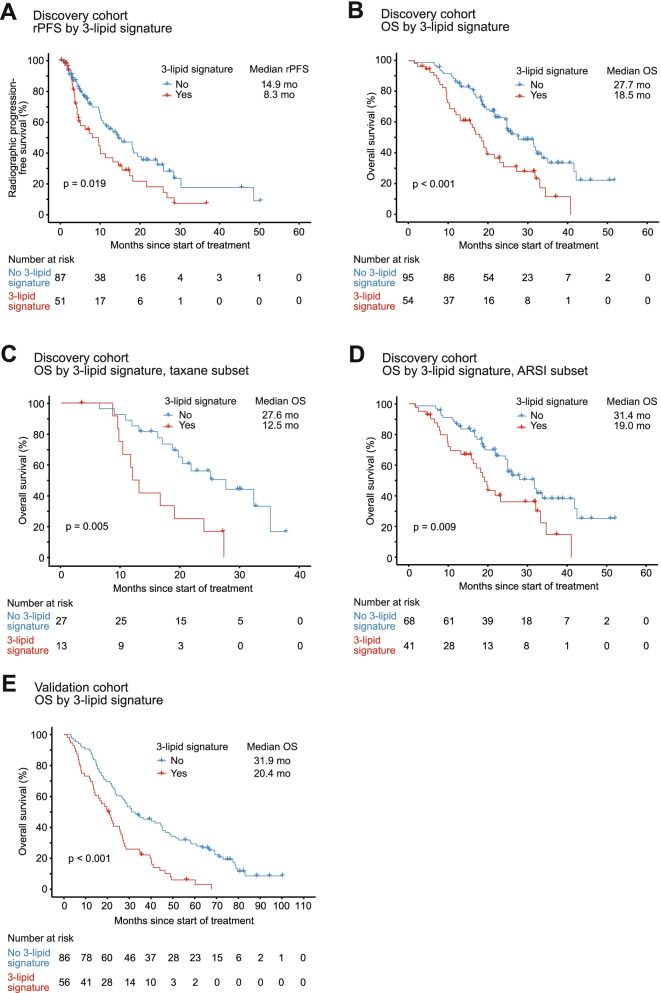
The 3LS was present in 41% of men in both cohorts. Consistent with our previous studies [14, 15], the 3LS was associated with shorter rPFS (hazard ratio [HR] 1.71, 95% confidence interval [CI] 1.09–2.66, p = 0.019) and OS (HR 2.15, 95% CI 1.4–3.3, p < 0.001) in the discovery cohort (Fig. 1A, B). The association with shorter OS remained significant in both the taxane subset (HR 3.29, 95% CI 1.44–7.54, p = 0.005; Fig. 1C) and ARSI subset (HR 1.99, 95% CI 1.19–3.34, p = 0.009; Fig. 1D). Furthermore, the 3LS was associated with shorter OS (HR 2.32, 95% CI 1.59–3.38, p < 0.001) in the validation cohort (Fig. 1E).
Fig. 1.
Kaplan-Meier analysis according to 3-lipid signature status in the discovery and validation cohorts. Using A radiographic progression-free survival (rPFS) in the discovery cohort, B overall survival (OS) in the discovery cohort, C OS in the taxane subset of the discovery cohort, D OS in the androgen receptor signalling inhibitor (ARSI) subset of the discovery cohort and E OS in the validation cohort
Clinical outcomes by genetic aberrations
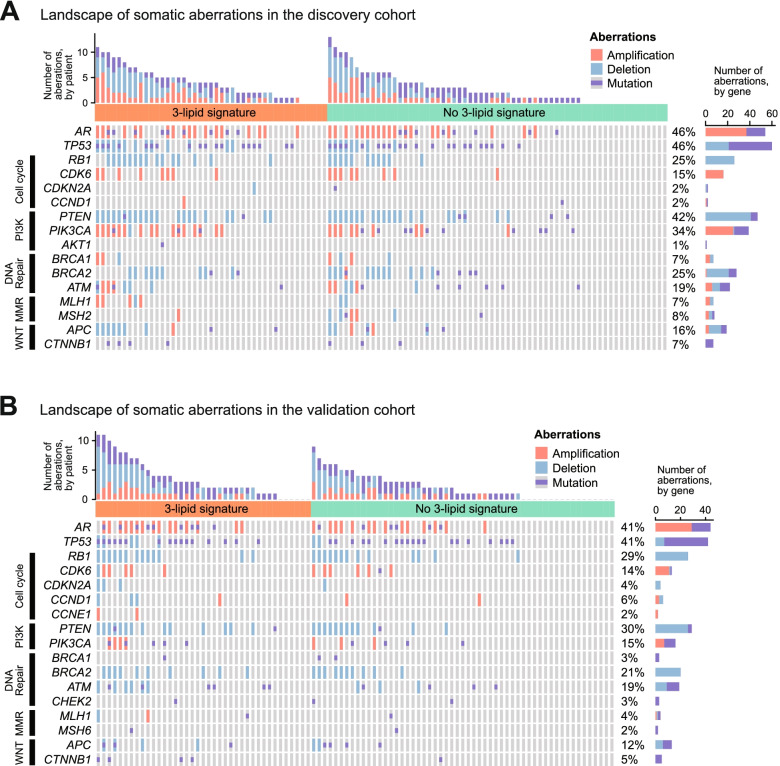
The four most common genetic aberrations seen in both cohorts of men with mCRPC were AR aberrations (discovery 46%; validation 41%), TP53 aberrations (discovery 46%, validation 41%), RB1 deletion (discovery 25%, validation 29%) and phosphatidylinositol-3-kinase (PI3K) pathway aberrations (discovery 55%, validation 35%) (Fig. 2). All four of these aberrations were associated with shorter OS in the discovery and validation cohorts and with shorter rPFS in the discovery cohort (Table 1).
Fig. 2.
Landscape of somatic aberrations in the A discovery cohort and B validation cohort
Table 1.
Univariable Cox proportional hazards analysis of survival based on genetic aberration in the two cohorts
| Genetic aberration | Overall survival in discovery cohort | Overall survival in validation cohort | Radiographic progression-free survival in discovery cohort | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| AR aberration | 3.87 (2.21–6.77) | < 0.001 | 2.26 (1.43–3.58) | <0.001 | 1.86 (1.07–3.24) | 0.028 |
| TP53 aberration | 3.55 (2.07–6.08) | < 0.001 | 2.18 (1.37–3.47) | 0.001 | 1.80 (1.01–3.13) | 0.039 |
| RB1 deletion | 4.10 (2.33–7.22) | < 0.001 | 1.79 (1.09–2.94) | 0.021 | 3.08 (1.64–5.79) | < 0.001 |
| PI3K aberration | 2.66 (1.55–4.58) | < 0.001 | 2.11 (1.33–3.34) | 0.002 | 2.53 (1.42–4.49) | 0.001 |
All p-values < 0.05 are highlighted in bold
Data regarding radiographic progression-free survival was not available for the validation cohort
CI confidence interval, HR hazard ratio, PI3K phosphatidylinositol-3-kinase
The 3-lipid signature and genetic aberrations
The overall frequency of somatic aberrations within the AR, TP53, cell cycle, PI3K, DNA repair, mismatch repair (MMR) and WNT pathways was increased in men with the 3LS (Fig. 2). In the discovery cohort, 88% of men with the 3LS had ≥ 1 genetic aberration, compared to 75% of men without the 3LS. In total 40% of men with the 3LS had ≥ 5 aberrations, compared to 21% of men without the 3LS. In the validation cohort, 85% of men with the 3LS had ≥ 1 genetic aberration, compared to 69% of men without the 3LS. In total, 26% of men with the 3LS had ≥ 5 aberrations, compared to 15% of men without the 3LS. In both cohorts, increased genomic heterogeneity was associated with the presence of the 3LS.
Plasma sphingolipids and genetic aberrations
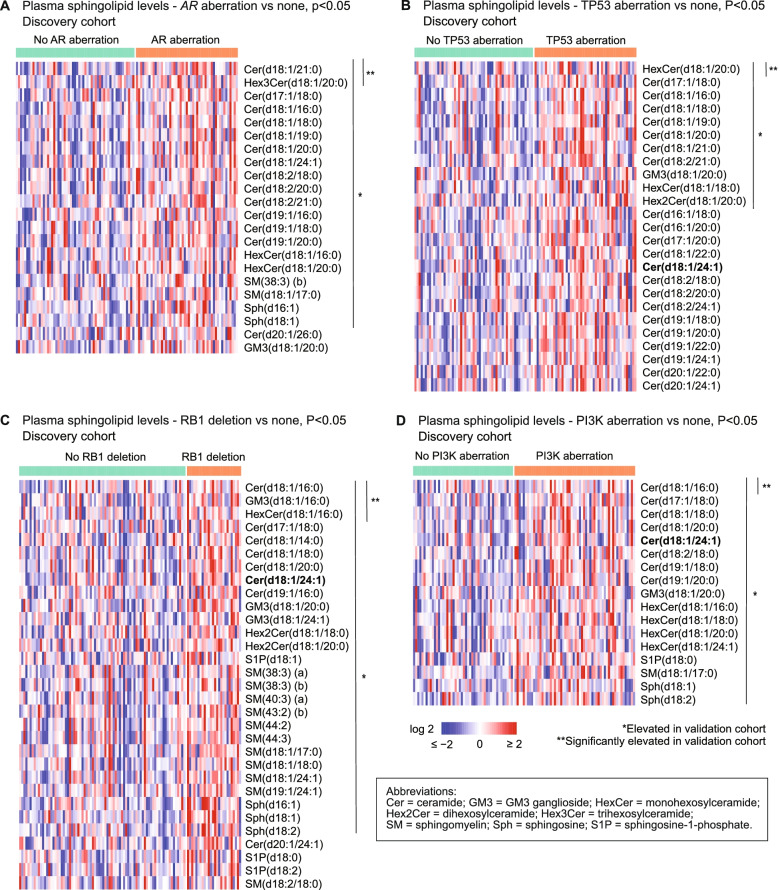
Elevated circulating sphingolipids were associated with AR aberrations, TP53 aberrations, RB1 deletion and PI3K pathway aberrations in both cohorts. In the discovery cohort, 22 sphingolipids were significantly elevated in men with any AR aberration compared to men without (p < 0.05), including ceramide(d18:1/24:1), a key component of the 3LS (Fig. 3A, Additional File 1: Table S6.1). Twenty of these sphingolipids were also elevated in the validation cohort (Fig. 3A, Additional File 1: Table S6.1). Similarly, 17–31 sphingolipids were significantly elevated in men with TP53 aberrations, RB1 deletion or PI3K aberrations (p < 0.05), with a proportion of these also elevated in the validation cohort (Fig. 3B–D, Additional File 1: Tables S6.2-6.4).
Fig. 3.
Significantly elevated sphingolipids in men with genetic aberrations in the discovery cohort. Heatmaps show elevated sphingolipids in men with A AR aberration, B TP53 aberration, C RB1 deletion, and D PI3K aberration; compared to men without the aberration. Sphingolipids which are also elevated in the validation cohort are asterisked. Cer(d18:1/24:1), which is a component of the 3-lipid signature, is bolded
Aberrations in the DNA repair pathway (BRCA1/2, ATM, CHEK2), MMR genes (MLH1, MSH2, MSH6) or WNT pathway (APC, CTNNB1) were not significantly associated with elevated circulating sphingolipids in either cohort (Additional File 1: Tables S6.5-6.7). This demonstrates that not all genotypes are associated with the poor prognostic metabolic profile.
3-lipid signature and genetic aberrations as a biomarker combination
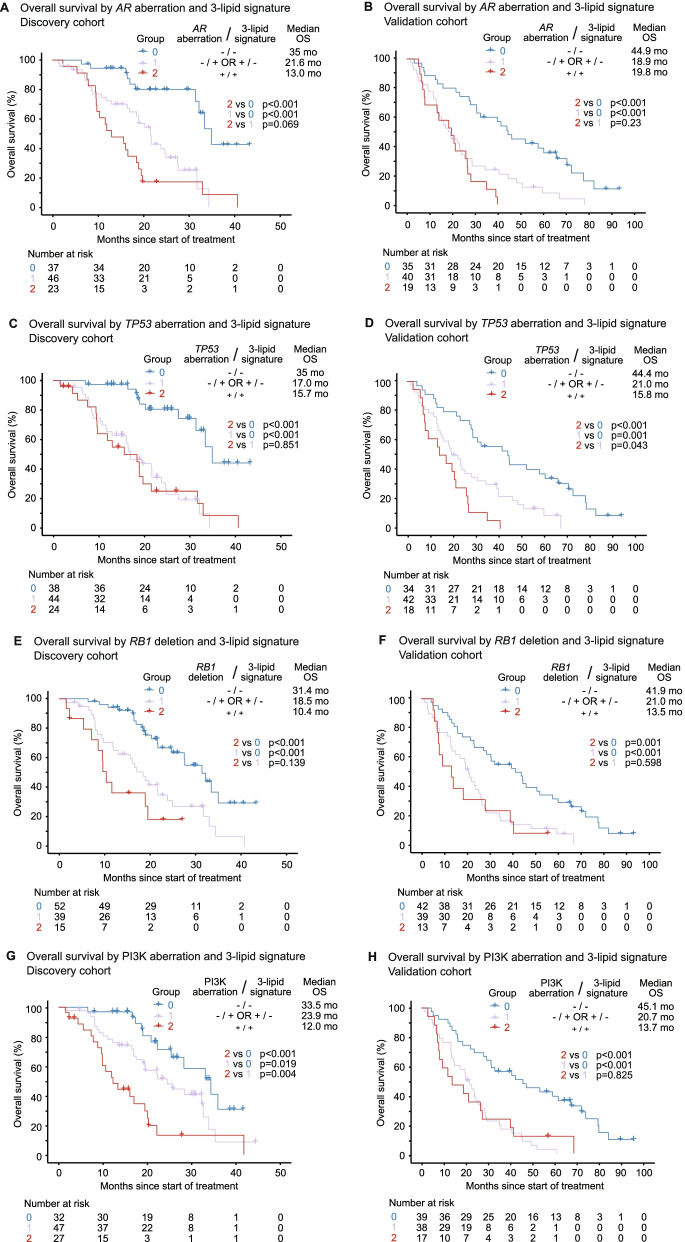
The combined presence of the 3LS with aberrations in AR, TP53, RB1 or PI3K in men with mCRPC was associated with poorer prognosis. Men with the 3LS and a genetic aberration (AR, TP53, RB1 or PI3K) (Group 2) had worse OS than men with neither characteristic (Group 0) in both cohorts (p ≤ 0.001; Fig. 4).
Fig. 4.
Kaplan-Meier analysis of overall survival (OS) by genetic aberration and 3-lipid signature. OS by AR aberration and 3-lipid signature in the A discovery and B validation cohorts. OS by TP53 aberration and 3-lipid signature in the C discovery and D validation cohorts. OS by RB1 deletion and 3-lipid signature in the E discovery and F validation cohorts. OS by PI3K aberration and 3-lipid signature in the G discovery and H validation cohorts
In multivariable analysis with clinicopathologic factors, presence of an AR aberration and/or the 3LS (Groups 1 and 2) was independently associated with worse OS compared to men with neither characteristic (Group 0) in the discovery cohort (p = 0.005; Table 2) and validation cohort (p = 0.001; Table 3). The association with shorter OS was also seen with the TP53 aberration and/or 3LS combination (discovery p = 0.001; validation p < 0.001)), the RB1 deletion and/or the 3LS combination (discovery p = 0.019; validation p < 0.001) and the PI3K and 3LS combination (discovery p = 0.009; validation p = 0.396) (Tables 2 and 3).
3-lipid signature and aggressive-variant prostate cancer
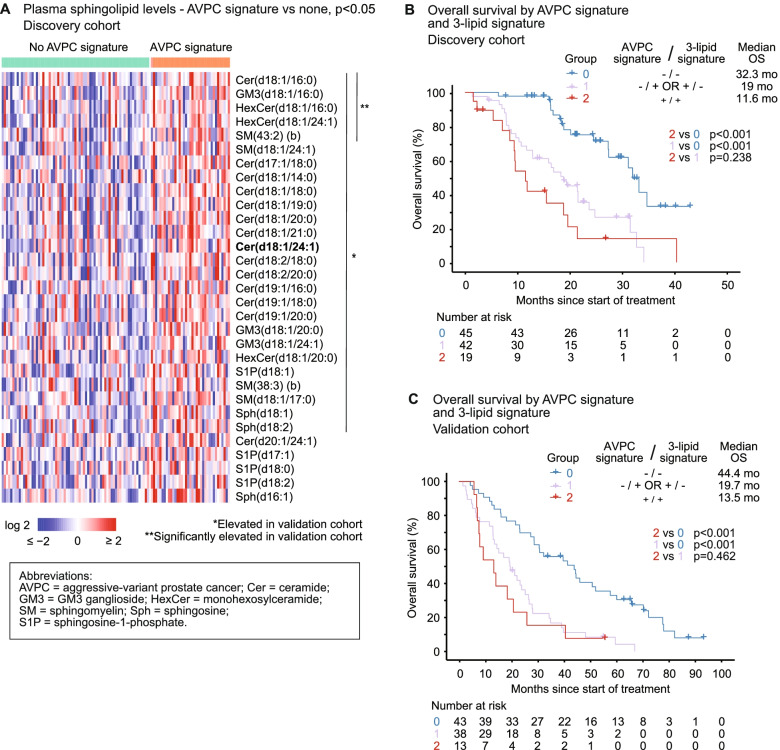
AVPC, as defined by the presence of genetic aberrations in two or more of TP53, RB1 and/ or PTEN [8], was present in 35% of men in the discovery cohort and 27% of men in the validation cohort. As expected, men with the molecular AVPC signature had shorter OS (discovery: HR 3.57, 95% CI 2.12–6.00, p < 0.001; validation: HR 2.40, 95% CI 1.44–3.98, p < 0.001). Elevated circulating sphingolipids were associated with AVPC in both cohorts (Fig. 5A).
Fig. 5.
Association between elevated sphingolipids and aggressive-variant prostate cancer (AVPC), and their combined impact on clinical outcomes. A Heatmap of sphingolipids with significantly elevated levels in men with the AVPC signature, compared to men without. Sphingolipids which are also elevated in the validation cohort are asterisked. Cer(d18:1/24:1), which is a component of the 3-lipid signature, is bolded. Kaplan-Meier analysis of overall survival by molecular AVPC signature and 3-lipid signature in B the discovery cohort and C the validation cohort
Men with the combination of 3LS and AVPC had significantly shorter OS in both cohorts, with median survival of ~12 months compared to > 2 years for men with neither signature (discovery 11.6 months vs 32.3 months, p < 0.001; validation 13.5 months vs 44.4 months, p < 0.001) (Fig. 5B, C). The genetic aberrations of AVPC are the main drivers of poor prognosis in these men, as men with AVPC (regardless of the presence of 3LS) have shorter OS than non-AVPC men with 3LS (Additional File 1: Fig S7.5). However, in both cohorts, a higher proportion of men with AVPC have the 3LS compared to men without AVPC (52% versus 35–39%). The molecular AVPC signature and/or 3LS was independently associated with shorter OS in multivariable analysis with clinicopathological features (Additional File 1: Tables S8.1-S8.2).
Discussion
This study provides new insights into the correlation between circulating lipids and tumour genotypes in mCRPC. Elevated circulating sphingolipids were correlated with AR, RB1 and PI3K aberrations in two independent cohorts and associated with TP53 aberrations in the discovery cohort, but not with DNA repair, MMR or WNT pathway aberrations. The presence of a somatic aberration (AR, TP53, RB1, PI3K) and/or the 3LS was independently associated with shorter OS in multivariable analysis in both cohorts. In addition, AVPC was associated with elevated sphingolipids and the combination of AVPC and 3LS predicted for a shorter median survival of ~12 months. Taken together, these findings provide the first understanding into the correlation between circulating lipid and somatic gene alterations in mCRPC and suggest that men with AR, TP53, RB1, PI3K and AVPC aberrations are more likely to benefit from therapies targeting sphingolipid metabolism than men with DNA repair, MMR or WNT pathway anomalies.
A key question arising from this data is how the genomic and lipidomic changes are related biologically. Not all genotypes are associated with the sphingolipid profiles, suggesting that certain genetic aberrations (AR, TP53, RB1, PI3K) are more likely to be dysregulating lipid metabolism. Bivariate analysis of the genotypes with the 3LS was inconclusive regarding whether the genetics was driving the lipid metabolism or vice versa (Additional File 1: Tables S9.1-9.4).
There is emerging evidence of a biological link between lipid metabolism and genetic aberrations including AR, RB1 and PI3K on PC growth. Enhanced ceramide-S1P signalling may be mediating ARSI resistance induced by AR gain, as men with mCRPC had significantly shorter ARSI treatment duration if their tumours had AR gain in combination with increased expression of sphingolipid genes (involved in ceramide-S1P signalling) [16]. Furthermore, de novo resistance to enzalutamide in androgen-independent cells can be reversed with sphingosine kinase inhibitors in vitro [16]. A recent study demonstrated that a novel fatty acid synthase inhibitor antagonised CRPC growth through metabolic reprogramming and inhibited expression of AR and its variants, including AR-V7 [28]. Another study found that 3-hydroxy-3-methyl-glutaryl–CoA reductase (HMGCR), a key enzyme in the cholesterol synthesis pathway, was elevated in enzalutamide-resistant PC cell lines and that simvastatin, a HMGCR inhibitor, blocked AR synthesis and inhibited growth in vitro and in vivo [29]. There is also recent evidence suggesting a role for RB1 in regulating sphingolipid metabolism in advanced PC. In an in vitro model of mCRPC with RB1 deletion utilising the androgen-independent PC cell line C4-2 after RB knockdown, 27% of upregulated genes were involved in metabolic pathways, including sphingolipid metabolism [30]. In addition, ceramide and its metabolite S1P play key roles in the PI3K pathway, as a negative regulator and activator respectively [31]. Overall, these studies suggest that lipid metabolism may play a biological and possibly clinically relevant role in some of these molecular pathways.
These findings raise the question of whether manipulation of the lipidome might be an effective therapeutic strategy in addition to existing drugs, particularly for patients with unfavourable lipidomic and somatic genetic aberrations. For example in the IPATential150 study, men with mCRPC with PTEN-loss tumours had improved rPFS on treatment with abiraterone and an AKT inhibitor versus abiraterone alone (18.5 months vs 16.5 months, p = 0.034) [32]. Our data supports the hypothesis that the addition of a metabolic modifier in men with both a PI3K aberration and 3LS may increase the efficacy of the AKT inhibitor, and this warrants further investigation.
Metabolic therapies are currently not the standard of care for PC, but are used in the treatment of other diseases such as cardiovascular disease and diabetes. Non-pharmacological interventions such as exercise and diet can reduce ceramides [33, 34]. The cholesterol lowering drugs statins and PCSK9 inhibitors reduce levels of circulating ceramides and other sphingolipids in patients with dyslipidaemia [35, 36]. Several studies have already indicated that statin therapy is associated with improved OS in PC [37–40]; however, blood cholesterol levels were not associated with prognosis in prostate cancer [41–43]. Therefore, the beneficial effects of statin therapy may be related to other lipids such as ceramides given that statin can lower the circulating levels of these lipids.
Drugs specifically targeting sphingolipid metabolism are mostly in pre-clinical development in cancer [18], but a specific inhibitor of sphingosine kinase 2, Opaganib (ABC294640), has been tested in patients with solid tumours [44]. Recently, we showed that Opaganib can overcome de novo enzalutamide resistance in androgen-independent PC cells in vitro [16]. Opaganib is currently undergoing a Phase 2 study in combination with AR inhibitors in patients with mCRPC (NCT04207255). Clinical trials are required to determine if repurposing statins or other sphingolipid-targeting therapies for prostate cancer in combination with standard of care can improve clinical outcomes.
Furthermore, plasma lipidomic signatures predicting for high-risk patients in cardiovascular disease already exist [45] and have been translated into a rapid-turnover plasma ceramide test by the Mayo Clinic Laboratories [46]. Overall, the availability of metabolic therapies that can target sphingolipid metabolism in combination with genotyping and lipidomic analysis for patient selection, demonstrates the feasibility of personalised metabolic therapy in a clinical setting for men with mCRPC.
A strength of this study is the analysis of two independent cohorts of prospectively enrolled men with mCRPC using the previously validated 3LS [14, 15], with similar observations in both the discovery and validation cohorts. This study was limited by exclusion of men without available cfDNA for sequencing (29% of discovery cohort, 34% of validation cohort). High levels of ctDNA are associated with poorer clinical outcomes and likely to represent a higher tumour burden [7]. Therefore, the exclusion of men with undetectable/low cfDNA from our cohorts may have resulted in the cohorts having worse clinical outcomes compared to the average population of men with mCRPC. Inadvertent skewing of the cohort characteristics might account for differences between the discovery and validation cohorts (e.g. less significant differences in sphingolipid levels between men with and without genetic aberrations in the validation cohort). The cohorts also had differences in baseline PSA levels and Gleason grade—the median PSA levels were two times higher in the discovery cohort (30 vs 14 ng/mL), and a higher proportion of the discovery cohort had Gleason Grade ≥9 (49% vs 32%). Overall, the relatively small sample size of the cohorts limits clinical applicability and will need to be addressed in the future with studies of larger cohorts. Another potential confounder was the presence of co-occurring genetic aberrations.
Conclusion
Elevated circulating sphingolipids, especially ceramides, were associated with AR, TP53, RB1, PI3K and AVPC aberrations in mCRPC, and the combination of lipid and genetic abnormalities conferred a worse prognosis. This suggests that approaches targeting the aberrant lipid metabolism defined by the 3LS should be considered in men with these mCRPC genotypes in prospective clinical trials.
Supplementary Information
Additional file 1. Supplementary information for ‘Combined impact of lipidomic and genetic aberrations on clinical outcomes in metastatic castration-resistant prostate cancer’. Supplementary information, figures and tables for ‘Combined impact of lipidomic and genetic aberrations on clinical outcomes in metastatic castration-resistant prostate cancer’.
Acknowledgements
We gratefully acknowledge Lisa-Jane Graham (Chris O’Brien Lifehouse), Anne-Maree Haynes (Garvan Institute of Medical Research), Daniela Barreto (Garvan Institute of Medical Research) and research nurses for collection of blood/tissue specimens and clinical data; and Tony Maxwell as our consumer representative.
Abbreviations
- AR
Androgen receptor
- ARSI
Androgen receptor signalling inhibitors
- AVPC
Aggressive-variant prostate cancer
- cfDNA
Cell-free DNA
- ctDNA
Circulating tumour DNA
- CI
Confidence interval
- HMGCR
3-Hydroxy-3-methyl-glutaryl–CoA reductase
- HR
Hazard ratio
- mCRPC
Metastatic castration-resistant prostate cancer
- MMR
Mismatch repair
- OS
Overall survival
- PARPi
Poly-ADP ribose polymerase inhibitors
- PC
Prostate cancer
- PI3K
Phosphatidylinositol-3-kinase
- rPFS
Radiographic progression-free survival
- S1P
Sphingosine-1-phosphate
- 3LS
3-lipid signature
Authors’ contributions
Study concept and design: LGH. Acquisition of data: EMK, HF, BT, IDD, KM, MRS, KB, GM, AZ, MC, WT, KH, TGM, NAM, PD, JY, SJ, AMJ, MK, AAA, LGH. Analysis and interpretation of data: BM, HML, LGH, AAA, PJM. Drafting of the manuscript: BM, HML, LGH. Critical revision of the manuscript for important intellectual content: BM, HML, BT, AMJ, IDD, MRS, AJH, LMB, MK, DJW, PJK, AAA, LGH. Statistical analysis: BM, HML. Obtaining funding: LGH, AAA. Supervision: LGH. All authors read and approved the final manuscript
Funding
National Health and Medical Research Council of Australia (GNT1196225 to LGH, GNT1098647 to AAA); Cancer Institute New South Wales (10/TPG/1-04, 2018/TPG001); Australian Prostate Cancer Research Centre-New South Wales; Australian Department of Health and Aging; the Movember Foundation and the Prostate Cancer Foundation of Australia (Revolutionary Team Award MRTA3); Cancer Council New South Wales (PG 10-01); Cancer Council South Australia (Beat Cancer Project Principal Cancer Research Fellowship, PRF1117 to LMB); The Victorian Government’s Operational Infrastructure Support Program; National Institutes of Health grant award to MK (RO1-CA212097); Australian Government Research Training Program (RTP) Scholarship and University of Sydney Merit Award to BM; NHMRC Postgraduate Scholarship and Monash University Postgraduate Publications Award to EMK; Australian Government RTP Scholarship and Monash University Postgraduate Scholarship to HF; Victorian Cancer Agency Clinical Research Fellowship (CRF14009) and Astellas Investigator-Initiated Grant to AAA; ANZUP Noel Castan Fellowship to HML; Twin Towns Services Community Foundation to LGH.
The funders had no roles in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
All participants provided written informed consent, with ethics approval obtained from Monash Health Institutional Review Board (15571X), Royal Prince Alfred Hospital Human Research Ethics Committee (X19-0320), and Mayo Clinic Institutional Review Board (09-1889).
Consent for publication
Not applicable.
Competing interests
EMK: Honoraria—Janssen, Ipsen, Astellas Pharma, Research Review; Consulting or Advisory Role—Astellas Pharma, Janssen, Ipsen; Research Funding—Astellas Pharma, AstraZeneca; Travel & accommodation—Astellas Pharma, Pfizer, Ipsen, Roche.
BT: Grants and personal fees—Amgen, AstraZeneca, Astellas, BMS, Janssen, Pfizer, MSD, Ipsen, Bayer; Personal fees—IQVIA, Sanofi, Tolmar, Novartis, Roche.
IDD: Institutional funding—Pfizer, ANZUP Cancer Trials Group, Bayer, Astellas, Janssen, Movember Foundation, Merck Sharp & Dohme; AstraZeneca, Bayer, Bristol Myers Squibb, Eisai, Exelixis, Ipsen, Roche/Genentech, Seagen; Unremunerated chair of ANZUP Cancer Trials Group.
AZ: Advisory Role—Astellas; Honoraria—Astellas; Grants and personal fees—Astra Zeneca, Pfizer; Personal fees—Merck Sharp & Dome, Bayer, Mundipharma, Janssen.
AMJ.: Institutional funding—Pfizer, Astellas.
AAA: Consultant—Astellas, Janssen, Novartis; Speakers Bureau—Astellas, Janssen, Novartis, Amgen, Ipsen, Bristol Myers Squibb; Merck Serono, Bayer; Honoraria—Astellas, Novartis, Sanofi, AstraZeneca, Tolmar, Telix, Merck Serono; Janssen, Bristol Myers Squibb, Ipsen, Bayer, Pfizer, Amgen, Noxopharm, Merck Sharpe Dome; Scientific Advisory Board—Astellas, Novartis, Sanofi, AstraZeneca, Tolmar, Pfizer, Telix, Merck Serono; Janssen, Bristol Myers Squibb, Ipsen, Bayer, Merck Sharpe Dome , Amgen, Noxopharm; Travel & accommodation—Astellas, Merck Serono, Amgen, Novartis, Janssen, Tolmar, Pfizer; Investigator research funding—Astellas, Merck Serono, Astra Zeneca; Institutional research funding—Bristol Myers Squibb, Astra Zeneca, Aptevo Therapeutics, Glaxo Smith Kline, Pfizer, MedImmune, Astellas, SYNthorx, Bionomics, Sanofi Aventis, Novartis, Ipsen.
LGH: Research funding—Astellas; Travel sponsorship—Janssen, Pfizer; Honoraria—Imagion Biosystems.
All other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Sartor O, de Bono JS. Metastatic Prostate Cancer. N Engl J Med. 2018;378(7):645–657. doi: 10.1056/NEJMra1701695. [DOI] [PubMed] [Google Scholar]
- 3.Iyengar NM, Gucalp A, Dannenberg AJ, Hudis CA. Obesity and cancer mechanisms: tumor microenvironment and inflammation. J Clin Oncol. 2016;34(35):4270–4276. doi: 10.1200/JCO.2016.67.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ku SY, Gleave ME, Beltran H. Towards precision oncology in advanced prostate cancer. Nat Rev Urol. 2019;16(11):645–654. doi: 10.1038/s41585-019-0237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A. 2019;116(23):11428–11436. doi: 10.1073/pnas.1902651116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Annala M, Vandekerkhove G, Khalaf D, Taavitsainen S, Beja K, Warner EW, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018;8(4):444–457. doi: 10.1158/2159-8290.CD-17-0937. [DOI] [PubMed] [Google Scholar]
- 8.Spetsieris N, Boukovala M, Patsakis G, Alafis I, Efstathiou E. Neuroendocrine and aggressive-variant prostate cancer. Cancers (Basel). 2020;12(12):3792. [DOI] [PMC free article] [PubMed]
- 9.Allott EH, Masko EM, Freedland SJ. Obesity and prostate cancer: weighing the evidence. Eur Urol. 2013;63(5):800–809. doi: 10.1016/j.eururo.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Y, Ma J. Body mass index, prostate cancer-specific mortality, and biochemical recurrence: a systematic review and meta-analysis. Cancer Prev Res (Phila). 2011;4(4):486–501. doi: 10.1158/1940-6207.CAPR-10-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flanagan J, Gray PK, Hahn N, Hayes J, Myers LJ, Carney-Doebbeling C, et al. Presence of the metabolic syndrome is associated with shorter time to castration-resistant prostate cancer. Ann Oncol. 2011;22(4):801–807. doi: 10.1093/annonc/mdq443. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Daniels G, Lee P, Monaco ME. Lipid metabolism in prostate cancer. Am J Clin Exp Urol. 2014;2(2):111–120. [PMC free article] [PubMed] [Google Scholar]
- 13.Nagarajan SR, Butler LM, Hoy AJ. The diversity and breadth of cancer cell fatty acid metabolism. Cancer Metab. 2021;9(1):2. doi: 10.1186/s40170-020-00237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin HM, Mahon KL, Weir JM, Mundra PA, Spielman C, Briscoe K, et al. A distinct plasma lipid signature associated with poor prognosis in castration-resistant prostate cancer. Int J Cancer. 2017;141(10):2112–2120. doi: 10.1002/ijc.30903. [DOI] [PubMed] [Google Scholar]
- 15.Lin HM, Huynh K, Kohli M, Tan W, Azad AA, Yeung N, et al. Aberrations in circulating ceramide levels are associated with poor clinical outcomes across localised and metastatic prostate cancer. Prostate Cancer Prostatic Dis. 2021;24(3):860–870. doi: 10.1038/s41391-021-00338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin HM, Mak B, Yeung N, Huynh K, Meikle TG, Mellett NA, et al. Overcoming enzalutamide resistance in metastatic prostate cancer by targeting sphingosine kinase. EBioMedicine. 2021;72:103625. doi: 10.1016/j.ebiom.2021.103625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin HM, Yeung N, Hastings JF, Croucher DR, Huynh K, Meikle TG, et al. Relationship between circulating lipids and cytokines in metastatic castration-resistant prostate cancer. Cancers (Basel). 2021;13(19):4964. [DOI] [PMC free article] [PubMed]
- 18.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nat Rev Cancer. 2010;10(7):489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- 20.Vykoukal J, Fahrmann JF, Gregg JR, Tang Z, Basourakos S, Irajizad E, et al. Caveolin-1-mediated sphingolipid oncometabolism underlies a metabolic vulnerability of prostate cancer. Nat Commun. 2020;11(1):4279. doi: 10.1038/s41467-020-17645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth Nanda C, Venkateswaran SV, Patani N, Yuneva M. Defining a metabolic landscape of tumours: genome meets metabolism. Br J Cancer. 2020;122(2):136–149. doi: 10.1038/s41416-019-0663-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fettke H, Kwan EM, Docanto MM, Bukczynska P, Ng N, Graham LK, et al. Combined cell-free DNA and RNA profiling of the androgen receptor: clinical utility of a novel multianalyte liquid biopsy assay for metastatic prostate cancer. Eur Urol. 2020;78(2):173–180. doi: 10.1016/j.eururo.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohli M, Tan W, Zheng T, Wang A, Montesinos C, Wong C, et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine. 2020;54:102728. doi: 10.1016/j.ebiom.2020.102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh K, Barlow CK, Jayawardana KS, Weir JM, Mellett NA, Cinel M, et al. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem Biol. 2019;26(1):71–84.e4. doi: 10.1016/j.chembiol.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Alshehry ZH, Barlow CK, Weir JM, Zhou Y, McConville MJ, Meikle PJ. An efficient single phase method for the extraction of plasma lipids. Metabolites. 2015;5(2):389–403. doi: 10.3390/metabo5020389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal Chem. 2006;78(13):4281–4290. doi: 10.1021/ac051632c. [DOI] [PubMed] [Google Scholar]
- 27.Kwan EM, Dai C, Fettke H, Hauser C, Docanto MM, Bukczynska P, et al. Plasma cell-free DNA profiling of PTEN-PI3K-AKT pathway aberrations in metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2021;5:622–37. [DOI] [PMC free article] [PubMed]
- 28.Zadra G, Ribeiro CF, Chetta P, Ho Y, Cacciatore S, Gao X, et al. Inhibition of de novo lipogenesis targets androgen receptor signaling in castration-resistant prostate cancer. Proc Natl Acad Sci U S A. 2019;116(2):631–640. doi: 10.1073/pnas.1808834116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong Y, Cheng L, Mao F, Zhang Z, Zhang Y, Farah E, et al. Inhibition of cholesterol biosynthesis overcomes enzalutamide resistance in castration-resistant prostate cancer (CRPC) J Biol Chem. 2018;293(37):14328–14341. doi: 10.1074/jbc.RA118.004442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandigo AC, Yuan W, Xu K, Gallagher P, Pang A, Guan YF, et al. RB/E2F1 as a master regulator of cancer cell metabolism in advanced disease. Cancer Discov. 2021;11(9):2334–53. [DOI] [PMC free article] [PubMed]
- 31.Oskouian B, Saba JD. Cancer treatment strategies targeting sphingolipid metabolism. Adv Exp Med Biol. 2010;688:185–205. doi: 10.1007/978-1-4419-6741-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sweeney C, Bracarda S, Sternberg CN, Chi KN, Olmos D, Sandhu S, et al. Ipatasertib plus abiraterone and prednisolone in metastatic castration-resistant prostate cancer (IPATential150): a multicentre, randomised, double-blind, phase 3 trial. Lancet. 2021;398(10295):131–142. doi: 10.1016/S0140-6736(21)00580-8. [DOI] [PubMed] [Google Scholar]
- 33.Kasumov T, Solomon TP, Hwang C, Huang H, Haus JM, Zhang R, et al. Improved insulin sensitivity after exercise training is linked to reduced plasma C14:0 ceramide in obesity and type 2 diabetes. Obesity (Silver Spring). 2015;23(7):1414–1421. doi: 10.1002/oby.21117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah C, Yang G, Lee I, Bielawski J, Hannun YA, Samad F. Protection from high fat diet-induced increase in ceramide in mice lacking plasminogen activator inhibitor 1. J Biol Chem. 2008;283(20):13538–13548. doi: 10.1074/jbc.M709950200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meikle PJ, Wong G, Tan R, Giral P, Robillard P, Orsoni A, et al. Statin action favors normalization of the plasma lipidome in the atherogenic mixed dyslipidemia of MetS: potential relevance to statin-associated dysglycemia. J Lipid Res. 2015;56(12):2381–2392. doi: 10.1194/jlr.P061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilvo M, Simolin H, Metso J, Ruuth M, Öörni K, Jauhiainen M, et al. PCSK9 inhibition alters the lipidome of plasma and lipoprotein fractions. Atherosclerosis. 2018;269:159–165. doi: 10.1016/j.atherosclerosis.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Raval AD, Thakker D, Negi H, Vyas A, Salkini MW. Association between statins and clinical outcomes among men with prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2016;19(2):151–162. doi: 10.1038/pcan.2015.58. [DOI] [PubMed] [Google Scholar]
- 38.Van Rompay MI, Solomon KR, Nickel JC, Ranganathan G, Kantoff PW, McKinlay JB. Prostate cancer incidence and mortality among men using statins and non-statin lipid-lowering medications. European Journal of Cancer. 2019;112:118–126. doi: 10.1016/j.ejca.2018.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carretero-González A, Lora D, Manneh R, Lorente D, Castellano D, de Velasco G. Combination of statin/vitamin D and metastatic castration-resistant prostate cancer (CRPC): a post hoc analysis of two randomized clinical trials. Clin Transl Oncol. 2020;22(11):2126–2129. doi: 10.1007/s12094-020-02334-6. [DOI] [PubMed] [Google Scholar]
- 40.Wu S-Y, Fang S-C, Shih H-J, Wen Y-C, Shao Y-HJ. Mortality associated with statins in men with advanced prostate cancer treated with androgen deprivation therapy. Eur J Cancer. 2019;112:109–117. doi: 10.1016/j.ejca.2018.11.032. [DOI] [PubMed] [Google Scholar]
- 41.Arthur R, Møller H, Garmo H, Häggström C, Holmberg L, Stattin P, et al. Serum glucose, triglycerides, and cholesterol in relation to prostate cancer death in the Swedish AMORIS study. Cancer Causes Control. 2019;30(2):195–206. doi: 10.1007/s10552-018-1093-1. [DOI] [PubMed] [Google Scholar]
- 42.Allott EH, Howard LE, Cooperberg MR, Kane CJ, Aronson WJ, Terris MK, et al. Serum lipid profile and risk of prostate cancer recurrence: Results from the SEARCH database. Cancer Epidemiol Biomarkers Prev. 2014;23(11):2349–2356. doi: 10.1158/1055-9965.EPI-14-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng S, Zheng Q, Ding G, Li G. Influence of serum total cholesterol, LDL, HDL, and triglyceride on prostate cancer recurrence after radical prostatectomy. Cancer Manag Res. 2019;11:6651–6661. doi: 10.2147/CMAR.S204947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Britten CD, Garrett-Mayer E, Chin SH, Shirai K, Ogretmen B, Bentz TA, et al. A phase I study of ABC294640, a first-in-class sphingosine kinase-2 inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2017;23(16):4642–4650. doi: 10.1158/1078-0432.CCR-16-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hilvo M, Meikle PJ, Pedersen ER, Tell GS, Dhar I, Brenner H, et al. Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J. 2020;41(3):371–380. doi: 10.1093/eurheartj/ehz387. [DOI] [PubMed] [Google Scholar]
- 46.Mayo Clinic Laboratories Test ID: CERAM. https://www.mayocliniclabs.com/test-catalog/Overview/606777. Accessed 15 Jul 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary information for ‘Combined impact of lipidomic and genetic aberrations on clinical outcomes in metastatic castration-resistant prostate cancer’. Supplementary information, figures and tables for ‘Combined impact of lipidomic and genetic aberrations on clinical outcomes in metastatic castration-resistant prostate cancer’.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.