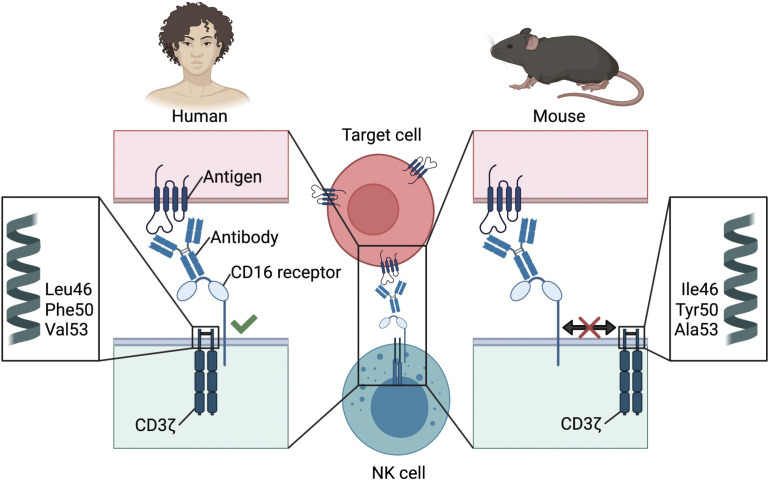
CD16 is a potent activating receptor in human NK cells but is weakly responsive in mouse NK cells. Aguilar et al. demonstrate that this is due to differences in amino acids in the transmembrane of CD3ζ that facilitate receptor assembly and function.
Abstract
Natural killer (NK) cells can detect antibody-coated cells through recognition by the CD16 Fc receptor. The importance of CD16 in human NK cell biology has long been appreciated, but how CD16 functions in mouse NK cells remains poorly understood. Here, we report drastic differences between human and mouse CD16 functions in NK cells. We demonstrate that one of the adaptor molecules that CD16 associates with and signals through, CD3ζ, plays a critical role in these functional differences. Using a systematic approach, we demonstrate that residues in the transmembrane domain of the mouse CD3ζ molecule prevent efficient complex formation with mouse CD16, thereby dampening receptor function. Mutating these residues in mouse CD3ζ to those encoded by human CD3ζ resulted in rescue of CD16 receptor function. We reveal that the mouse CD3ζ transmembrane domain adopts a tightly packed confirmation, preventing association with CD16, whereas human CD3ζ adopts a versatile configuration that accommodates receptor assembly.
Graphical Abstract
Introduction
Natural killer (NK) cells are innate lymphoid cells that can eliminate several pathologic cells, including cancerous, virally infected, and antibody-coated cells. As members of the innate immune system, they have the capability to respond to stimuli in the absence of prior sensitization (Chiossone et al., 2018; Lanier, 2005; Lanier, 2008; Mujal et al., 2021). NK cells rely on cell surface receptors to distinguish between healthy (self) and abnormal (nonself) cells (Raulet and Vance, 2006). Although NK cells largely rely on germline-encoded receptors for target cell recognition, they are also equipped with activating Fc receptors that can harness the antigen-specific humoral response to detect pathogenic cells.
In human NK cells, the Fc receptor CD16 (FcγRIIIA/CD16A/FCGR3A) is one of the most potent activating receptors (Bryceson et al., 2006). CD16 is expressed in 90% of blood NK cells and marks the mature NK subset (CD56dimCD16+; Lanier et al., 1983). CD16 receptor–mediated antibody-dependent cellular cytotoxicity (ADCC) through NK cells has been implicated in therapies involving targeting antibodies to pathogenic cells (Germain et al., 2010; Miller and Lanier, 2019; Pahl et al., 2018). Engagement of CD16 on human NK cells with the Fc portion of IgG on an antibody-coated cell yields powerful stimulatory signals resulting in release of cytotoxic granules and cytokine secretion (Lanier, 2005). Like other activating receptors expressed on NK cells, CD16 has a short cytoplasmic tail incapable of delivering intracellular signals and therefore relies on adaptor molecules for surface expression and signaling. In human NK cells, CD16 associates with homo- or heterodimers of FcεR1γ and CD3ζ adaptor molecules which contain immunoreceptor tyrosine-based activating motifs (ITAM) in their intracellular domains, responsible for mediating downstream signals (Kurosaki et al., 1991; Lanier, 2005; Lanier et al., 1988; Lanier et al., 1989; Lanier et al., 1991). Cross-linking of CD16 yields phosphorylation of the ITAMs by Src kinase family members, which generates docking sites for Syk and ZAP70, thereby propagating the signaling cascade that ultimately results in the release of Ca2+, paving the way for effector functions (Lanier, 2005; Nimmerjahn and Ravetch, 2008).
Recent studies have discovered that some individuals seropositive for human CMV acquire specialized NK cells that exhibit adaptive-like features (Lee et al., 2015; Schlums et al., 2015). These specialized NK cells are analogous to their counterparts identified during mouse CMV infection that respond to the viral antigen m157 through the stimulatory Ly49H receptor (Sun et al., 2009). These adaptive (or memory) NK cells are a subset of mature NK cells (CD56dimCD16+CD57+) that express the activating NKG2C receptor and characteristically lose expression of FcεR1γ (Lee et al., 2015; Lopez-Verges et al., 2011; Schlums et al., 2015). Expansion of the NKG2C+ NK cell subsets has been attributed to detection of a human CMV–encoded UL40 peptide presented on HLA-E, a mechanism evolved to evade NK recognition by engaging the inhibitory NKG2A receptor but circumvented by the activating NKG2C receptor on NK cells (Guma et al., 2004; Hammer et al., 2018; Heatley et al., 2013; Wang et al., 2002). Of note, these FcεR1γ− NK cells display epigenetic signatures unique from conventional NK cells and exhibit enhanced CD16 functional responses that may be due to the dependence on signaling through CD3ζ, which possesses three ITAMs rather than the one ITAM in FcεR1γ (Lee et al., 2015).
Although CD16 receptor function has been extensively investigated on human NK cells, studies of CD16 on mouse NK cells have been lacking, partly owing to lack of reagents, including mouse CD16-specific antibodies. Interestingly, on mouse NK cells, CD16 does not associate with CD3ζ and instead associates only with the FcεR1γ chain (Kurosaki and Ravetch, 1989; Lanier et al., 1989). Therefore, we investigated the differences between CD16 responses in mouse and human NK cells. Specifically, we focused on the molecular mechanisms underpinning the functional differences between the Fcγ receptor complexes in these species. We demonstrate that the transmembrane domain of the signaling adaptor CD3ζ plays a critical role in properly associating with the CD16 receptor, and we report that key amino acid residues present in human but not in mouse CD3ζ are essential for assembly and function.
Results
Ex vivo mouse NK cells fail to elicit potent CD16 responses
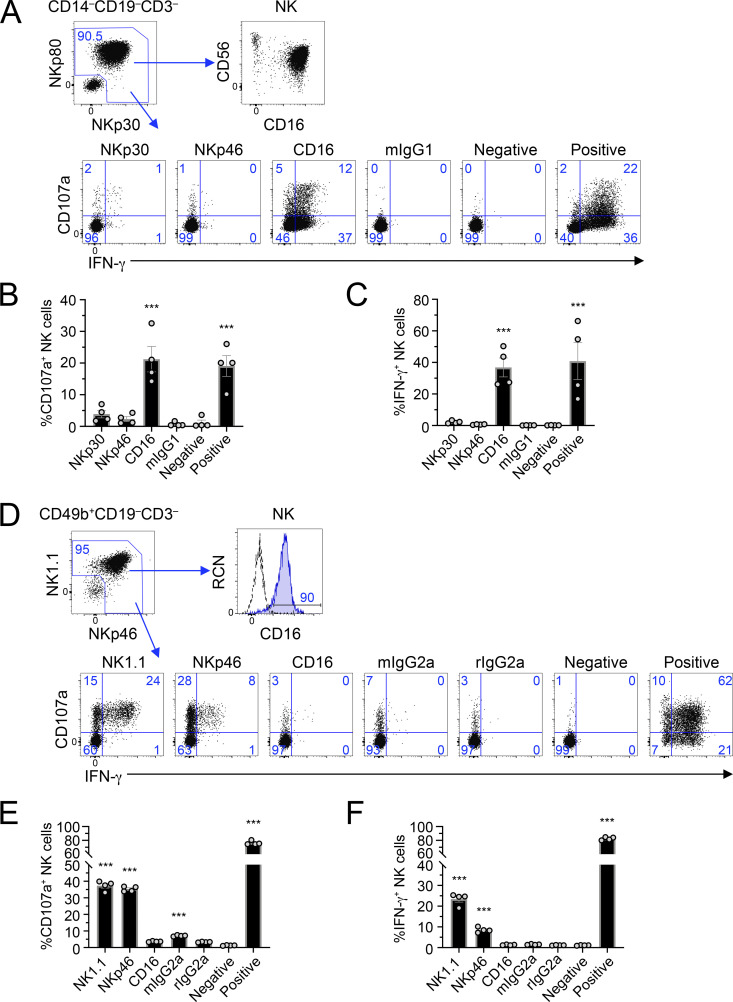
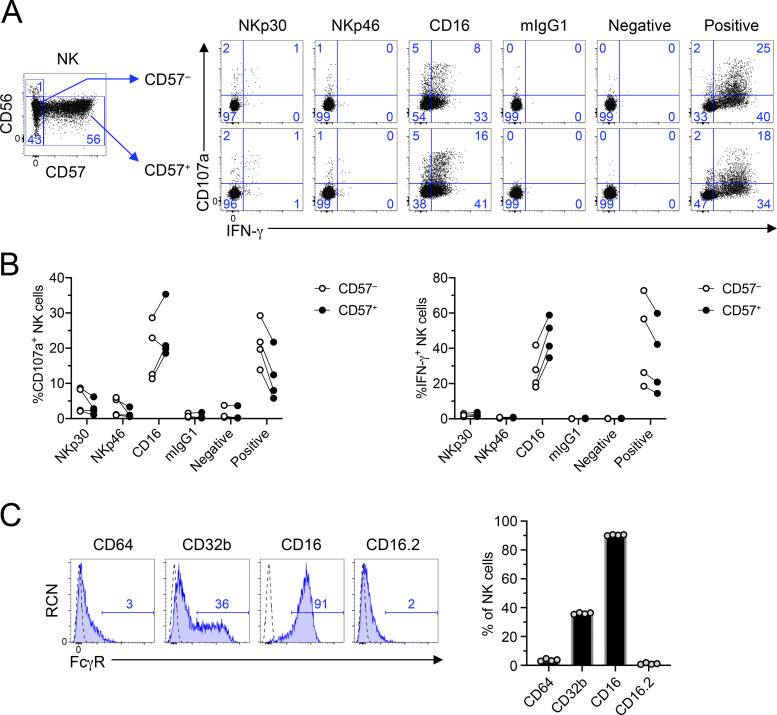
NK cells have long been known for their ability to be activated in the absence of prior sensitization. This is especially the case in human NK cells stimulated through CD16. We first wanted to determine if this was also observed in mice and compare NK cells between the two species. Therefore, we cross-linked activating receptors on human NK cells using plate-bound agonist antibody stimulations, stained the cells, and analyzed them by flow cytometry. Because CD16 is typically used as a marker to identify NK cells, and because we stimulated the CD16 receptor, we used NKp80 and NKp30 to identify NK cells in the lineage− (CD3−CD19−CD14−) gate (Fig. 1 A). As shown in Fig. 1 A, this gate captures the CD56+CD16+/− NK cell subsets. Freshly isolated human NK cells were potently activated by stimulation of the CD16 receptor, as determined by their ability to degranulate cytotoxic granules (CD107a+; Fig. 1 B) and release cytokines (IFN-γ+ production; Fig. 1 C), but were not stimulated with isotype-matched control antibodies (Fig. 1, A–C). Cross-linking of NKp30 or NKp46 only weakly activated ex vivo NK cells (Fig. 1, A–C). Consistent with prior reports, the CD56dimCD16+CD57+ subset of NK cells showed greater activation than the CD57− subset (Fig. S1; Lopez-Verges et al., 2010).
Figure 1.
Ex vivo mouse NK cells fail to elicit potent CD16 responses, unlike ex vivo human NK cells. (A–C) Human PBMCs were stimulated by receptor-specific antibodies in plate-bound stimulation assays; 5 h later, they were analyzed by flow cytometry. (A) Gating strategy revealing identification of NK cells, CD16 expression, and representative flow plots of NK cell activation (CD107a and IFN-γ) by different stimuli. Blue numbers in flow plots represent the cell percentage within gates. (B and C) Graphical representation of data in A as measured by percentage CD107a+ NK cells (B) or percentage IFN-γ+ NK cells (C). (D–F) Splenocytes were isolated from C57BL/6 mice and used in plate-bound stimulation assays; 5 h later, they were analyzed by flow cytometry. (D) Gating strategy revealing identification of NK cells, CD16 expression, and representative flow plots of NK activation (CD107a and IFN-γ) by different stimuli. (E and F) Graphical representation of data in D as measured by percentage CD107a+ NK cells (E) or percentage IFN-γ+ NK cells (F). Graphs show mean ± SEM; ***, P < 0.001 represents statistical significance from negative control treatments as determined by one-way ANOVA. All data in A–F are representative of at least two independent experiments with cohorts of four biological samples.
Figure S1.
Functional characterization of CD57+ human NK cells upon stimulation by CD16 and analysis of Fcγ receptor expression on mouse NK cells. Human PBMCs were stimulated by antibodies in plate-bound antibody stimulation assays; 5 h later, they were analyzed by flow cytometry. (A) Gating strategy revealing identification of NK cell subsets and representative flow plots of NK activation (CD107a and IFN-γ) by different stimuli. Blue numbers in flow plots represent the cell percentage within the gates. (B) Graphical representation of data in A as measured by percentage of CD107a+ (top) or percentage of IFN-γ+ (bottom) in CD57− (left) or CD57+ (right) human NK cells. Graphs show mean ± SEM. All data include n = 4 biological samples and are representative of at least two independent experiments. (C) Splenocytes were isolated from B6 mice and stained with fluorophore-conjugated antibodies to assess the expression of CD64, CD32b, CD16, and CD16.2 Fcγ receptors on NK cells (NK1.1+NKp46+CD3ε−CD19−) using flow cytometry. Dashed lines represent isotype-matched control antibody, and blue numbers in histograms represent the percentage of FcγR+ cells. Graphs show mean ± SEM. All data in A–D are representative of at least two independent experiments with cohorts of four biological samples.
We first assessed the expression of Fcγ receptors on mouse NK cells and confirmed that they express CD16 (Fcgr3) and CD32b (Fcgr2b), but not CD16.2 (Fcgr4) or CD64 (Fcgr1; Fig. S1 C). In plate-bound stimulation assays, we observed robust activation upon cross-linking of NK1.1 or NKp46 as measured by CD107a+ (Fig. 1, D and E) and IFN-γ+ (Fig. 1, D and F) production. However, stimulation through CD16 or using isotype-matched control antibodies failed to produce any significant signal (Fig. 1, D–F). In fact, even IL-2–activated splenic NK cells fail to produce strong ADCC responses in cytotoxicity assays (Aguilar et al., 2020). Therefore, these data demonstrate that while ex vivo human NK cells are efficient at being activated through the CD16 Fc receptor without prior priming, mouse NK cells fail to exhibit significant activation ex vivo and in vitro.
Human NK cells transduced with mouse CD16 can functionally respond to stimuli
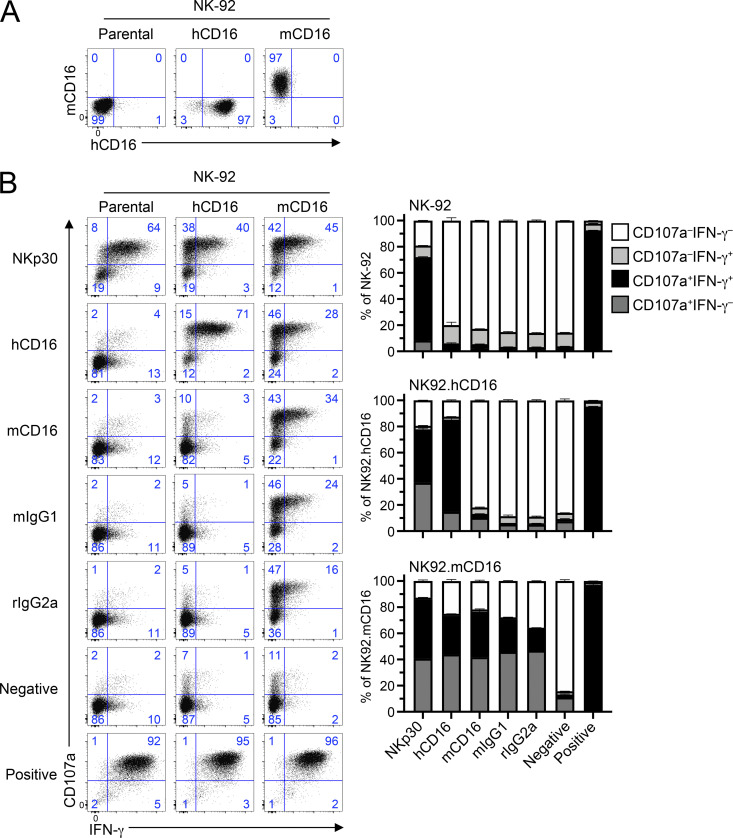
Given that both human and mouse CD16 can signal through FcεR1γ, but only human CD16 can use CD3ζ, we hypothesized that the CD3ζ adaptor molecule plays a role in these different phenotypic outcomes. We tested if human CD3ζ could assemble with mouse CD16. Here, we used the human NK cell line NK-92, which expresses human CD3ζ, and importantly is CD16− and FcεR1γ− (data not shown). We cloned mouse CD16 (mCD16) and human CD16A (hCD16) into the pMXs-Puro retroviral vector, transduced NK-92 with these constructs, and sorted them for CD16+ expression. As shown in Fig. 2 A, NK-92 cells transduced with hCD16 or mCD16 expressed these Fc receptors at the cell surface, showing that human CD3ζ associates with these Fc receptors.
Figure 2.
Human NK cells transduced with mouse CD16 functionally respond to stimuli. (A) Human NK-92 cells were transduced with a retroviral vector expressing human or mouse CD16 and selected for surface expression of CD16. Data are representative of three independent experiments. (B) Parental NK-92 or cells transduced with human CD16 (hCD16) or mouse CD16 (mCD16) were stimulated in plate-bound stimulation assays using several antibodies against NK receptors. Blue numbers in flow plots represent the cell percentage within gate. Graphs on the right represent graphical summary data and show mean ± SD. All data are representative of two independent experiments performed in duplicate.
To determine whether mCD16 was functional on these NK-92 cells, we performed plate-bound stimulations using antibodies specific for hCD16, mCD16, NKp30 (positive control for NK92 activation), or isotype-matched antibody controls. Here, we used NK92-expressing hCD16 as a positive control for Fc receptor activation and for comparison with its mouse counterpart. Consistent with findings in Fig. 1 A, NK92.hCD16 cells were efficiently activated by agonist anti-human CD16 antibodies, but not isotype-matched controls (Fig. 2 B). In comparison, cells expressing mCD16 were activated by all antibody stimulations tested, as mouse CD16 recognizes these antibody isotypes (Fig. 2 B and data not shown). NK92 cells lacking CD16 were activated only by anti-NKp30 antibodies or PMA + ionomycin treatment.
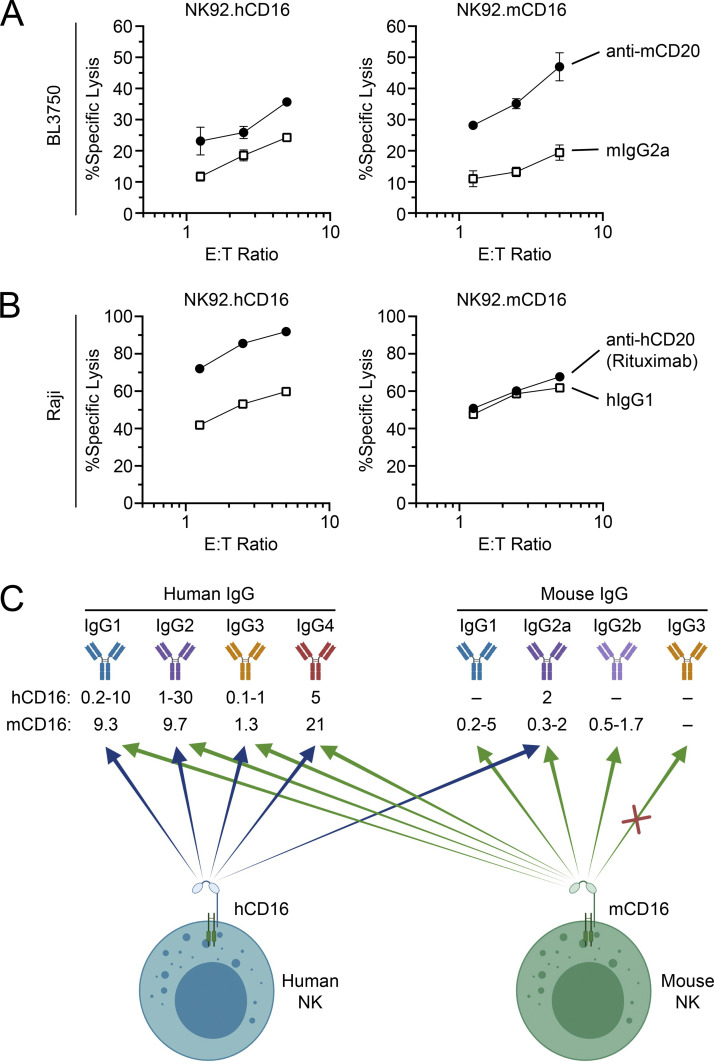
To confirm that the mouse CD16 receptor expressed on our NK92 cell lines was capable of mediating ADCC, we performed cytotoxicity assays using B cell lymphoma cell lines. We used the mouse CD20+ BL3750 B cell lymphoma line as targets and NK92 cells expressing mouse or human CD16 as effectors and performed ADCC assays in the presence of anti-mouse CD20 antibody or isotype-matched control antibody (Fig. 3 A). NK92.mCD16 were capable of more efficiently killing antibody-coated BL750 cells in comparison to the isotype-matched control antibody. NK92.hCD16 cells also showed a slight improvement in ADCC over the isotype-matched control antibody, as human CD16 can recognize mouse IgG2a isotype antibodies (Fig. 3 C; Temming et al., 2020). We also conducted reciprocal experiments using the human CD20+ B cell lymphoma Raji cell line. Here, NK92.hCD16 cells potently killed rituximab-coated Raji cells in comparison to isotype-matched control antibody (Fig. 3 B). Although mouse CD16 can detect human IgG1 isotypes (Fig. 3 C; Dekkers et al., 2017), NK92.mCD16 cells did not show enhanced ADCC against rituximab-coated Raji cells. We also observed that both mouse and human CD16 receptors on NK92 cells responded in redirected lysis assays using FcR+ P815 as target cells (data not shown). Therefore, these data show that indeed mouse CD16 is capable of associating with and transmitting signals through human CD3ζ.
Figure 3.
Mouse or human CD16 Fcγ receptor on NK92 cells induces strong ADCC. (A) The mouse CD20+ B cell lymphoma line BL3750 was used as target in ADCC assays using NK92.mCD16 or NK92.hCD16 at different effector-to-target (E:T) ratios in the presence of anti-mCD20 antibody (clone 5D2) or isotype-matched control antibody (mIgG2a). (B) Human CD20+ B cell lymphoma cells, Raji, were used as targets in ADCC assays using NK92.mCD16 or NK92.hCD16 effectors at different E:T ratios in the presence of anti-hCD20 (Rituximab) or isotype-matched control antibody (hIgG1). Graphs on the right represent graphical summary data and show mean ± SD. All data are representative of two independent experiments performed in triplicate. (C) Diagram summarizing affinities (Kd in µM) of human and mouse CD16 for human and mouse IgG isotypes, as previously reported (Baudino et al., 2008; Bruhns et al., 2009; Dekkers et al., 2017; Guilliams et al., 2014; Nimmerjahn et al., 2005; Takai, 2002; Temming et al., 2020). These affinities are ranges that can be influenced by glycosylation and allelic differences, among other factors.
Analysis of the transmembrane domains of mouse and human FcγR complexes
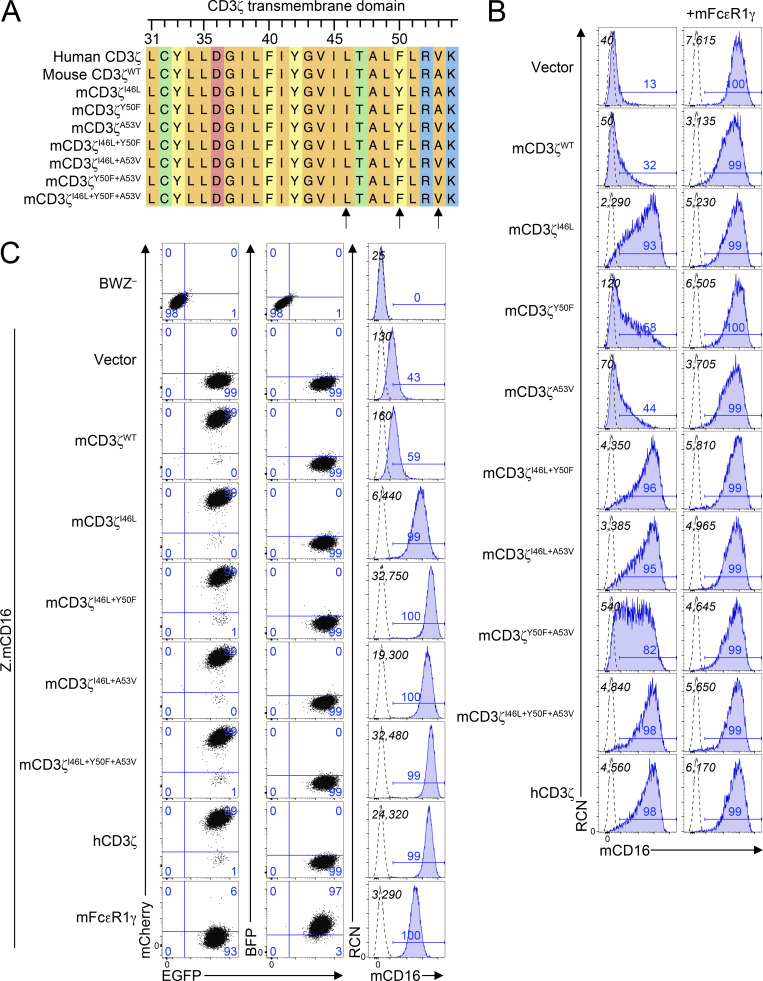
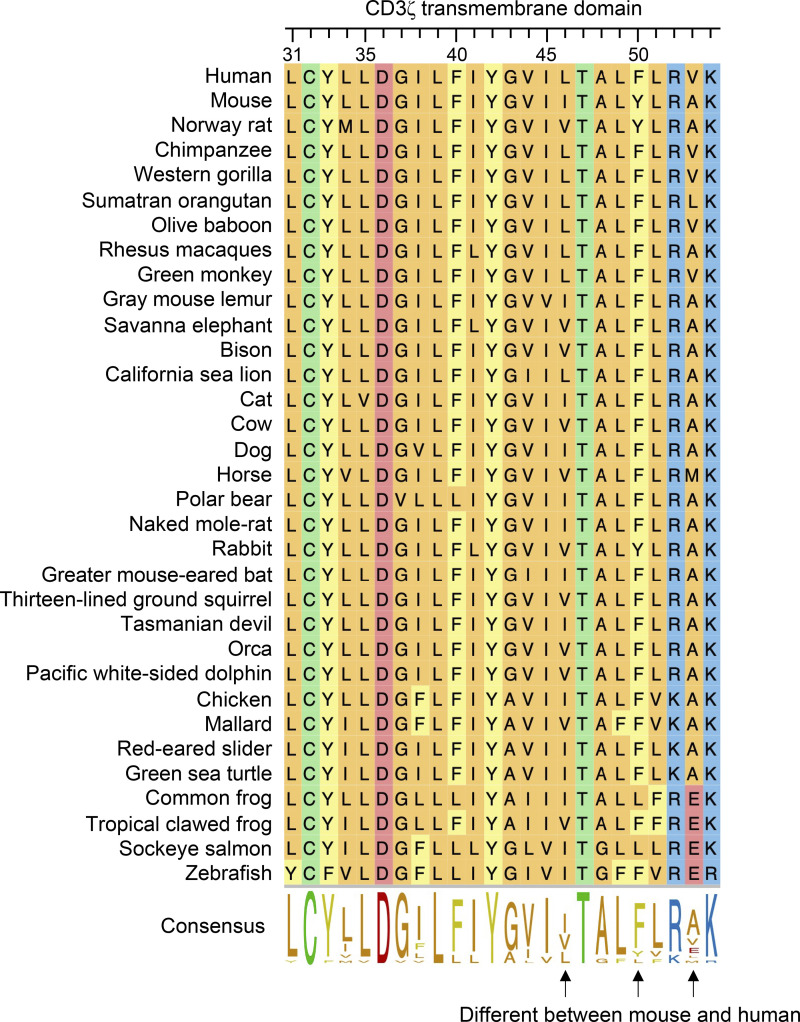
Because human CD3ζ can form functional complexes with mouse CD16, we performed in silico analysis of human and mouse CD3ζ in order to underpin the molecular explanation for this difference in function. As seen in Fig. 4 A, human and mouse CD3ζ are very well conserved, with 31 amino acid differences out of 164 (81.1% identity), 11 of which map to the signal peptide. Upon closer inspection of the transmembrane domains, we observed that there are three amino acid substitutions at positions 46, 50, and 53. One of these, isoleucine 46 (I46) in mouse, leucine (L46) in humans, has previously been described to play a role in the interaction with CD16 (Arase et al., 2001; Kurosaki et al., 1991). Interestingly, this 46L polymorphism present in human has only been observed in primates (Fig. S2). In addition, there are two additional polymorphisms that yield Tyr50Phe (Y50F) and Ala53Val (A5V) amino acid substitutions in human CD3ζ that differ between mice and humans (Fig. 4 A). Leucine 46 and phenylalanine 50 are the only residues conserved in primate species analyzed, while position 53 showed variability between valine, alanine, and leucine (Fig. S2). FcεR1γ transmembrane on the other hand has only one amino acid different between mouse and human (V31I; not shown).
Figure 4.
Analysis of residues in the transmembrane domain of CD3ζ highlights key residues involved in surface expression of CD16. (A) Analysis of the transmembrane domain of human and mouse CD3ζ, along with mutant mCD3ζ constructs generated. Colors represent amino acid properties as follows: green, polar (neutral); orange, nonpolar; blue, basic; red, acidic; yellow, aromatic. (B) HEK293T cells were cotransfected with mCD16 in combination with variants of CD3ζ in the absence (left) or presence (right) of mFcεR1γ. Italicized numbers in black represent the mean fluorescence intensity (MFI) of CD16; those in blue represent the percentage of CD16+ cells. (C) Expression of mouse CD16 on BWZ transductants. BWZ− were transduced with retroviral expression vectors expressing mouse CD16 along with IRES-EGFP as reporter (Z.mCD16) and with vectors expressing either CD3ζ mutants (mCherry+) or mFcεR1γ (BFP+). 2D plots on the left depict the EGFP and mCherry expression (left) or EGFP and BFP expression (center), and the histograms on the right represent the expression of mCD16 on BWZ transductants. Numbers in quadrants correspond to percentage of cells in the gate. Dotted lines represent unstained cells, and shaded blue histograms show cells stained with anti-mCD16 antibody. Black numbers in the top left corner show MFI values of mCD16, and numbers in blue above the gate represent the percentage of mCD16+. Data in B and C are representative of two independent experiments.
Figure S2.
Analysis of transmembrane domains of CD3ζ across vertebrate species highlights conserved residues in primates. Amino acid sequences of CD3ζ from various vertebrate species were aligned and analyzed using ClustalW algorithm. Numbers on top represent amino acid coordinates of mouse CD3ζ. Colors represent amino acid properties as follows: green, polar (neutral); orange, nonpolar; blue, basic; red, acidic; yellow, aromatic.
Expression of mouse CD3ζ variants improves mouse CD16 cell surface expression
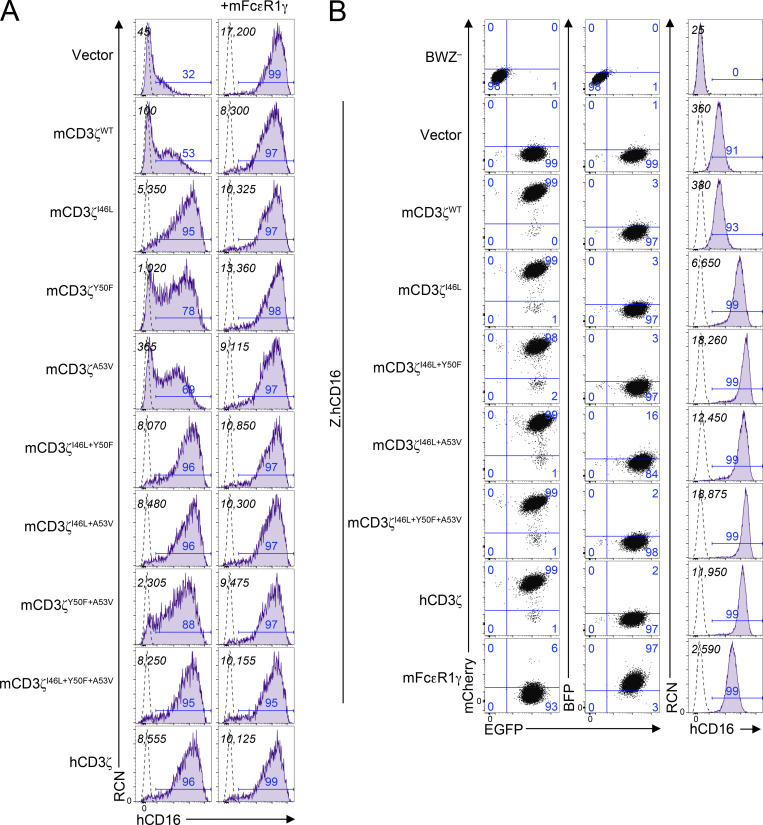
To determine whether the additional polymorphisms in mCD3ζ play a role in receptor complex assembly, we used site-directed mutagenesis to generate mammalian expression vectors (pIRES2) that contain mouse or human CD16, CD3ζ, or FcεR1γ along with EGFP, mCherry, or BFP fluorescent reporters, respectively. We then transfected HEK293T with combinations of CD16 with CD3ζ, FcεR1γ, or both. Using this approach, we validated that mouse CD16 can reach the cell surface only when coexpressed with FcεR1γ, but not mCD3ζWT; indeed, coexpression of CD3ζ with FcεR1γ resulted in lower CD16 expression (Fig. 4 B). The introduction of I46L improved cell surface expression, but this was not as high as in transfection of mCD16 with hCD3ζ (positive control). However, co-expression of mCD16 with CD3ζI46L+Y50F or CD3ζI46L+Y50F+A53V yielded cell surface levels similar to hCD3ζ. Interestingly, mouse versus human FcεR1γ did not appear to have any drastic differences (data not shown). Similar results were observed with cotransfections of the hCD16 receptor with CD3ζ or FcεR1γ adapter molecules (Fig. S3).
Figure S3.
Analysis of cell surface expression of human CD16 in HEK293T transfectants and BWZ transductants. (A) HEK293T cells were cotransfected with mammalian expression vectors expressing human CD16 in combination with variants of CD3ζ in the absence (left) or presence (right) of mouse FcεR1γ. Italicized numbers in black represent the MFI of CD16; those in blue represent the percentage of CD16+ cells. (B) Expression of human CD16 on BWZ transductants. BWZ− were transduced with retroviral expression vectors expressing hCD16 along with IRES-EGFP as reporter (Z.hCD16), then with vectors expressing either CD3ζ mutants (mCherry+) or mFcεR1γ (BFP+). 2D plots on the left depict the EGFP and mCherry expression (left) or EGFP and BFP expression (center), and the histograms on the right represent the expression of hCD16 on BWZ transductants. Numbers in quadrants correspond to percentage of cells in the gate. Dotted lines represent unstained cells, and shaded purple histograms show cells stained with anti-hCD16 antibody. Black numbers in the top left corner show MFI values of CD16, and numbers in blue above gate represent the percentage of CD16+. Data in A and B are representative of two independent experiments.
We addressed if these results were consistent upon stable transduction using constructs that showed greater CD16 expression. Here, we used the T cell lymphoma line, BWZ. These cells are negative for FcεR1γ and CD3ζ; however, to ensure that CD3ζ would not get induced during cell manipulation, we ablated endogenous mCD3ζ expression by CRISPR-Cas9 ribonucleoprotein targeting exon 2 of Cd247 (BWZ.∆ζ−; data not shown). We then subcloned the pIRES2 constructs along with fluorescent markers into pMSCV retroviral vectors. These constructs were used to transduce BWZ.∆ζ− cells with retroviruses expressing CD16-IRES-EGFP and sorted for EGFP+ expression. Once EGFP+ cells were established, these cells were transduced with retroviruses expressing CD3ζ-IRES-mCherry or FcεR1γ-IRES-BFP and sorted for the appropriate fluorescent marker. As shown in Fig. 4 C, BWZ.∆ζ transduced with mCD16 alone fail to express mCD16 at the cell surface. This was also the case with cells cotransduced with mCD3ζWT. However, cells expressing the mCD3ζI46L mutant had improved cell surface expression that was further enhanced when Y50F, A53V, or both polymorphisms were introduced. This was also observed in cells expressing WT hCD3ζ. Human CD16 coexpressed with mCD3ζ mutants or hCD3ζ revealed similar trends (Fig. S3). Transductants of mFcεR1γWT allowed cell surface expression of CD16, albeit lower than in some of the mCD3ζ mutants and hCD3ζ. These data indicate that Y50F and A53V, in addition to I46L, residues on CD3ζ are important for the stability of the FcγR complex involving CD16 and CD3ζ.
Mouse CD3ζ mutants improve CD16 function in reporter cells
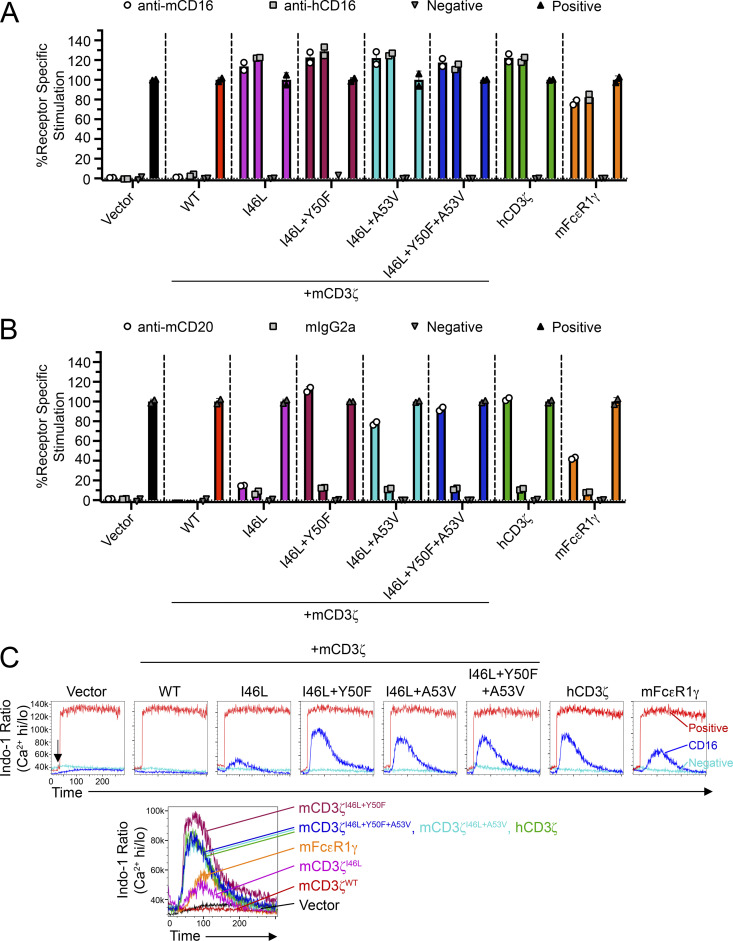
To determine if these residues on CD3ζ played a role in formation of functional FcγRIII complex, we used the endogenous reporter function of the BWZ cells. BWZ− cells express LacZ under a tandem NFAT promoter, thereby efficiently allowing the readout of receptor–ligand interactions using a colorimetric assay involving the β-gal substrate, CPRG (Aguilar et al., 2017). To determine whether the CD16 receptor complexes were functional in BWZ.∆ζ transductants, we tested these cells in plate-bound stimulation assays. Here, all mCD3ζ mutants showed strong stimulation (Fig. 5 A). However, to mimic CD16 function in a cellular context, we performed coculture assays using BL3750 lymphoma cells as stimulators to BWZ.∆ζ transductants in the presence of anti-CD20 mAb (Fig. 5 B). Here, the mCD3ζI46L+Y50F and mCD3ζI46L+Y50F+A53V revealed the greatest signals; however, mCD3ζI46L+A53V was also capable of detecting antibody on stimulator cells. Similar results were observed using antibodies targeting CD19 or IgM, antigens expressed by BL3750 cells (data not shown).
Figure 5.
CD3ζ mutants improve CD16 function in reporter cells. (A) Z.mCD16 cells transduced with CD3ζ mutants or mFcεR1γ were stimulated using plate-bound antibody stimulation assays. (B) Z.mCD16 transductants in A were cocultured with BL3750 lymphoma cells (CD20+) in the presence of anti-CD20 mAb or isotype-matched control antibody. Media alone or PMA + ionomycin were used as negative or positive controls, respectively. Data in A and B are representative of two independent experiments performed in duplicate. (C) Calcium influx assay using Z.mCD16 transductants. Cells were labeled with Indo-1 and stained with biotinylated anti-mCD16 antibody, and CD16 was cross-linked by adding streptavidin while running flow cytometry (arrow). Addition of streptavidin or PMA + ionomycin served as negative and positive controls, respectively. Histogram overlays on top demonstrate representative treatments for each cell line; histograms on the bottom demonstrate the overlays of mCD16 stimulations. Data in C are representative of two independent experiments.
Given that the reporter assays measured stimulation after an overnight culture, we investigated whether similar observations were seen in early signaling events. Here, we conducted calcium (Ca2+) influx assays using the BWZ.∆ζ transductants and flow cytometry. Cells were labeled with Indo-1, stained with biotinylated anti-CD16 antibody, and then receptor was cross-linked using streptavidin while being analyzed on a flow cytometer. Consistent with the coculture reporter assays, BWZ cells coexpressing mCD16 with mCD3ζI46L+Y50F showed the greatest intracellular Ca2+ influx, followed by cells expressing CD3ζI46L+Y50F+A53V or human CD3ζ (Fig. 5 C). No signal was detected with cells expressing only mCD16 or mCD16 with mCD3ζWT. Therefore, in summary, these data revealed that the I46L and Y50F residue substitutions on mouse CD3ζ are critical for efficient and stable associations with mouse CD16 and eliciting activating downstream signals.
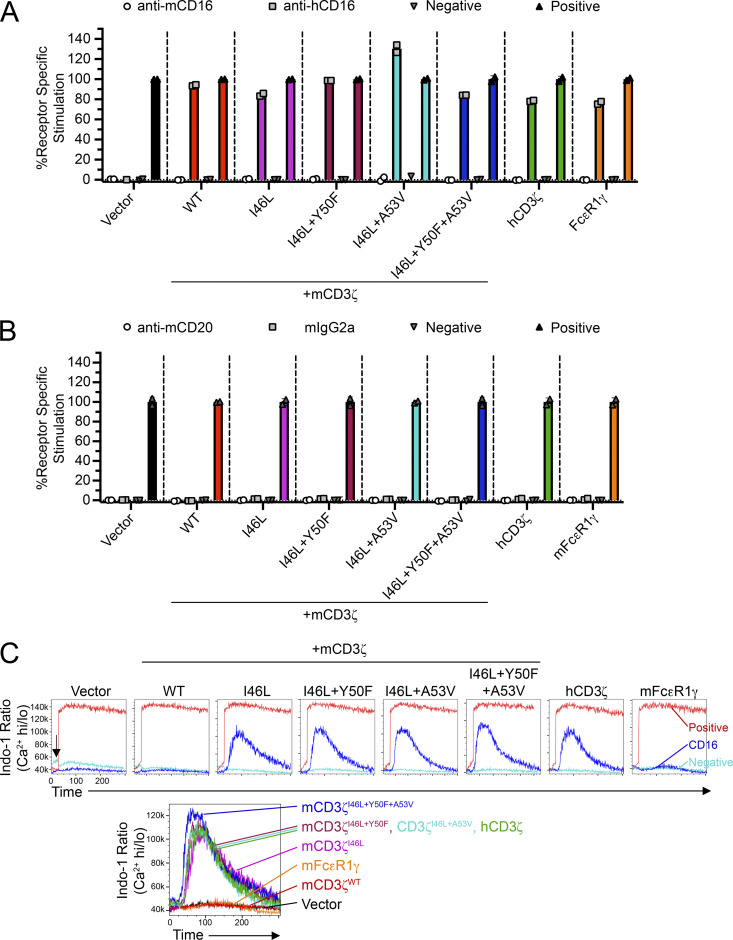
Interestingly, the hCD16 receptor is more resistant to modifications in the transmembrane domain of CD3ζ. In plate-bound antibody stimulations, hCD16 was capable of being stimulated by all CD3ζ variants (Fig. S4 A). Although human CD16A has been reported to bind mouse IgG2a antibodies, we did not detect binding or receptor stimulation in co-cultures with mouse BL3750 coated with the mouse IgG2a anti-CD20 mAb (Fig. S4 B). In addition, apart from mCD3ζWT and mFcεR1γ, we detected similar levels of Ca2+ influx in cells expressing mCD3ζ mutants that were comparable to hCD3ζ (Fig. S4 C). Thus, it is likely that residues on the transmembrane domain of CD16 also play a role in the association and signaling through CD3ζ.
Figure S4.
CD3ζ mutants improve CD16 function in reporter cells. (A) Z.hCD16 cells transduced with CD3ζ mutants or mFcεR1γ were stimulated using plate-bound antibody stimulation assays. (B) Z.hCD16 transductants in A were co-cultured with BL3750 lymphoma cells (CD20+) in the presence of anti-CD20 mAb or isotype-matched control antibody. Media alone or PMA + ionomycin were used as negative or positive controls, respectively. Data in A and B are representative of two independent experiments performed in duplicate. (C) Calcium influx assay using Z.hCD16 transductants. Cells were labeled with Indo-1 and stained with biotinylated anti-hCD16 antibody, and CD16 was cross-linked by adding streptavidin while running flow cytometry (arrow). Addition of streptavidin or PMA + ionomycin served as negative and positive controls, respectively. Histogram overlays on top demonstrate representative treatments for each cell line; histogram on bottom demonstrates the overlays of hCD16 stimulations. Data representative of two independent experiments.
Molecular dynamics (MD) simulations of the CD3ζ transmembrane domain reveal conformational differences between human and mouse proteins
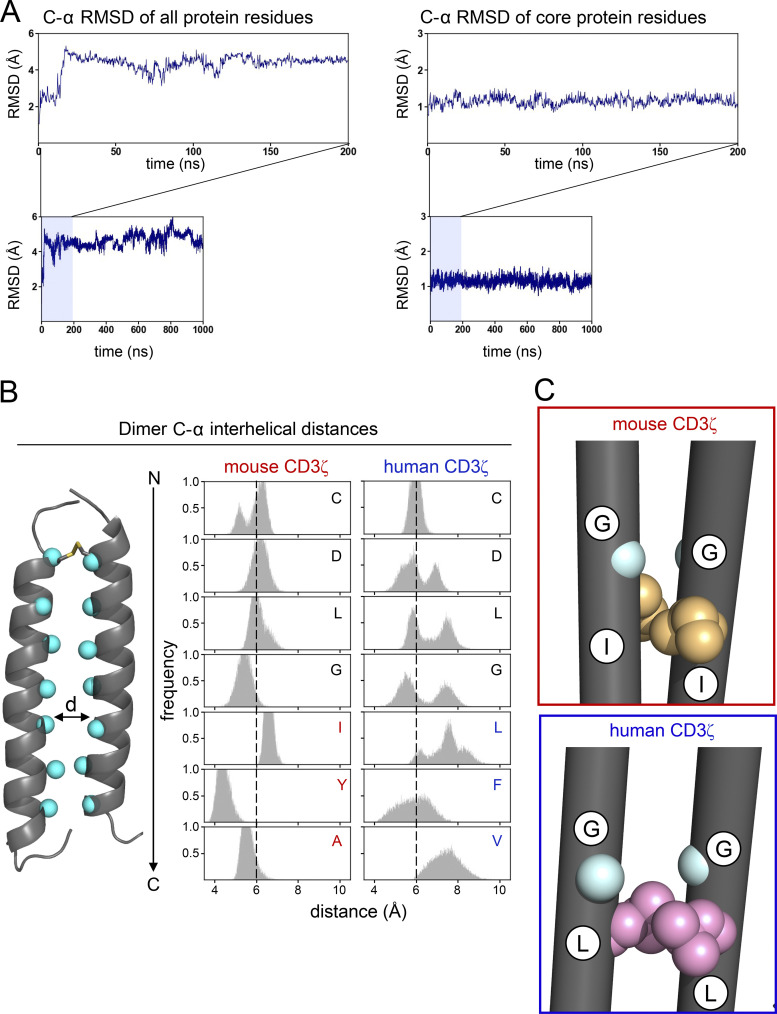
To understand how differences in the three key amino acid residues changed the dimer interface of CD3ζ and infer the effect on assembly with CD16, we performed atomistic MD simulations on the human CD3ζ dimer, starting from the solution nuclear magnetic resonance (NMR) structure of the transmembrane domain (Call et al., 2006). For comparison, we also simulated a triple mutant with the mouse sequence (L46I, F50Y, and V53A). Three independent 1.05-μs-long simulations were performed for each dimer. To ensure structural stability in simulation, the C-α root-mean-square deviation (RMSD) from the starting NMR structure as a function of time was calculated for each dimer. RMSD values plateaued within the first 50 ns of simulation and then remained relatively stable throughout the remainder of simulation time, indicating equilibration in this time regime (Fig. S5 A). The first 50 ns of each simulation were excluded for subsequent analysis.
Figure S5.
Conformational stability of CD3ζ dimer and comparison between mouse and human sequences. (A) C-α RMSD of the CD3ζ human homodimer (as an example) from the starting NMR structure to measure conformational stability after equilibration. RMSD is calculated for all protein residues (left) as well as the 24 core residues in each of the helices to demonstrate that most of the conformational variation after equilibration comes from the N- and C-terminal helical residues. (B) Distribution of minimum distance between the C-α of a residue on one CD3ζ helix and the same residue on the second helix. Distances are calculated for all residues at the dimer interface for the mouse (left) and human (right) sequences. (C) Comparison of the dominant conformers explored by CD3ζ with the mouse (top) and human (bottom) sequences. Residues G43 and I/L46 are highlighted to show the role of amino acid identity at position 46 on the packing of glycine at position 43.
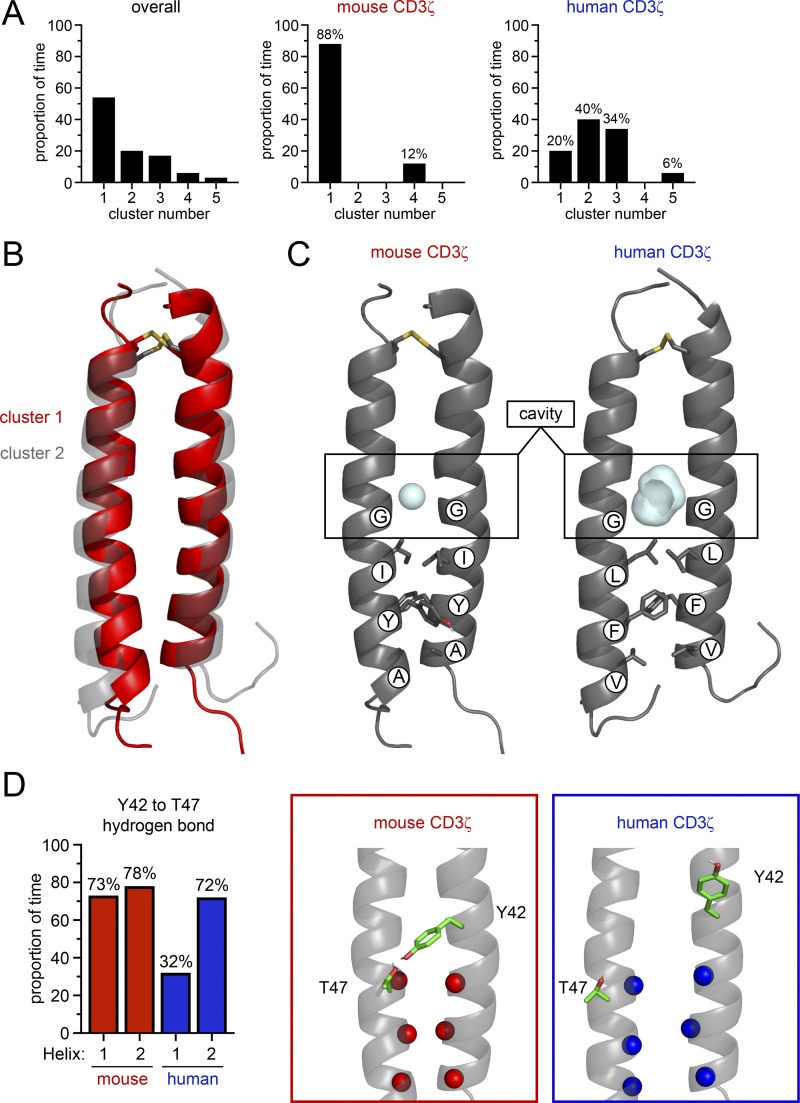
We first calculated the C-α interhelical distances of each dimer to get a sense for the dynamics of the dimer interface. This analysis revealed that human CD3ζ homodimer has a dynamic arrangement, as evidenced by the broad distribution of C-α interhelical distances, particularly in the C-terminus near the three key hydrophobic residues (Fig. S5 B). In contrast, the mouse CD3ζ sequence exhibited a tighter distribution of C-α interhelical distances, indicating a less dynamic interface. We then performed a combined cluster analysis at the helical level to understand the conformational variation of the ζ-ζ dimers (Fig. 6 A). Similar structures were grouped based on the C-α RMSD of the 24 core residues of each helix with a 1.2 Å RMSD cutoff. The overall clustering analysis revealed multiple clusters, which indicate that multiple conformations of the dimer are explored in simulation. Clustering was further refined to reveal conformational variation as function of mutation state. Interestingly, simulations of mouse CD3ζ were dominated by the conformer in cluster 1 (88% of time) while human CD3ζ populated multiple conformational states during the simulation, consistent with the broad distribution of C-α interhelical distances. Although human CD3ζ explores many conformations, including that of cluster 1, the dimer spent most of the time (40%) in the conformer of cluster 2. A superposition of the centroids of clusters 1 and 2 shows that the predominant conformation explored by the human dimer is less compact than the mouse dimer conformation (Fig. 6 B). This less compact interface is caused by a small translation and outward rotation of one of the helices such that the three key residues (L46, F50, V53) project onto the other helix in order to maximize favorable knobs-into-holes packing interactions. The asymmetry created by this new packing interface in the human dimer offsets a pair of glycine residues (G43) that lie N-terminal to the three key residues and generates a large cavity. This cavity is smaller in the more symmetric and compact mouse dimer conformation due to closer stacking of the glycine residues (Fig. 6 C and Fig. S5 C).
Figure 6.
Key residues change the conformations explored by mouse and human CD3ζ. (A) Combined clustering analysis at the helical level of conformations explored by human and mouse CD3ζ during simulation. Quantification of structural clustering overall (left) and further broken down as a function of mutation state for the mouse (middle) and human (right) sequences. (B) Comparison of the structural centroids of clusters one and two that correspond to the dominant conformers explored by CD3ζ with the mouse and human sequences, respectively. (C) Comparison of the cavities formed at the CD3ζ interface with either the mouse (left, red) or human (right, blue) sequence. (D) Quantification of hydrogen bond persistence given as the proportion of time hydrogen spent bonded in a total of three simulations for the hydrogen bonds formed between Y42 and T47 (left). H1 is the hydrogen bond formed between Y42 on helix 1 and T47 on helix 2, and H2 is the inverse. Structural depiction of the Y42-T47 hydrogen bonding architecture (data for mouse and human sequences are given in red and blue, respectively).
We further explored how mutations of these three key residues influenced other residues on the CD3ζ dimer that have been proposed to be essential for association with CD16. Previous studies have shown that the CD3ζ dimer is stabilized by two interchain hydrogen bonds between a tyrosine residue (Y42) on one helix and a threonine residue (T47) on the other helix (Call et al., 2006). Each Y42-T47 pair makes an interchain hydrogen bond that is essential for dimer assembly. Importantly, the breaking of one of these Y42-T47 hydrogen bonds enables association of CD16 with CD3ζ in a model of receptor assembly (Blazquez-Moreno et al., 2017). We therefore reasoned that the packing of the three key residues would play a role on the stability of the Y42-T47 hydrogen bonds. Hydrogen bonding analysis of the Y42-T47 pair revealed that the mouse CD3ζ Y42-T47 hydrogen bonds persist for 78% and 73% of simulation time ensuring the dimer is clamped shut for the majority of simulation time (Fig. 6 D). In contrast, for human CD3ζ, this hydrogen bonding network is disrupted asymmetrically such that on one side the hydrogen bond persists for 72% of simulation time but is only formed 32% of the time on the other side. These combined data show that conservative differences between the mouse and human CD3ζ sequences change the conformational landscape of the dimer interface. The mouse CD3ζ dimer adopts a symmetric and compact structure where the side chains of the key residues (I46, Y50, and A53) do not pack into the dimer interface but instead rotate outward to allow the helix backbones to come together. This compact architecture is reinforced by a pair of interhelical hydrogen bonds between Y42 on one helix and T47 on the other. Conversely, the human CD3ζ dimer rearranges to maximize packing interactions of the key residues (L46, F50, and V53) at the dimer interface. This packing rearrangement pushes the helical backbones apart such that the dimer adopts a more open conformation where one of the interhelical hydrogen bonds that clamps the mouse dimer together is broken and a cavity forms at the dimer interface above the key residues. In conclusion, our analysis reveals that the three amino acids found on human CD3ζ facilitate a more dynamic conformational state that can readily accommodate assembly with CD16, whereas mouse CD3ζ adopts a more restrictive compact structure that inhibits association with CD16.
Discussion
The importance of CD16 function on human NK cells has historically been appreciated and serves as an attractive receptor to target therapeutically (Bournazos et al., 2020; Hogarth and Pietersz, 2012; Miller and Lanier, 2019; Pahl et al., 2018). Yet the role that CD16 plays in mouse NK cells has received less scrutiny. In this study, we investigated the functional differences between human and mouse NK cells and conclude that resting mouse NK cells, unlike human NK cells, fail to be strongly activated by CD16 stimulation. This characteristic of mouse NK cells is also observed in IL-2–activated mouse NK cells (Aguilar et al., 2020). We further investigated the molecular mechanisms underpinning these differences and identified that three amino acids in the transmembrane domain of the CD3ζ adaptor molecule are instrumental for the association and function of CD16. Thus, this demonstrates a divergence of Fc receptor function between human and mouse NK cells.
In addition to the functional differences, there are genetic differences between human and mouse genes that encode the Fc receptor complexes. For example, the genes that encode the CD16 receptor (Fcgr3 in mouse and FCGR3A in human) are not orthologous, as mCD16 shares greater homology to human CD32A (FCGR2A) and CD32C (FCGR3C), while hCD16 more closely resembles mouse CD16.2 (Fcgr4; Akula et al., 2014; Nimmerjahn and Ravetch, 2006; Nimmerjahn and Ravetch, 2008). Despite these differences in homology, the expression patterns and function of these receptors are shared between mouse and human, with both species expressing CD16 and CD32b on NK cells, although mouse NK cells do not express CD16.2 (Fcgr4). Thus, of all the activating Fc receptors, mouse NK cells only express CD16, whereas human NK cells express CD32A and CD32C in addition to CD16, albeit at much lower frequencies (Dutertre et al., 2008).
Given that the CD16 genes are not orthologous, they also have different residues in the transmembrane domains. A recent study by Blazquez-Moreno et al. (2017) highlighted important residues in the transmembrane of the hCD16 receptor. The authors reported that Q208, F211, F219, D222, T223, Y226, and F227 all contribute to the assembly with FcεR1γ and CD3ζ. Importantly, apart from Q208, all of these residues are conserved in the transmembrane of the mCD16 receptor (H214 in mouse CD16). Interestingly, mutagenesis involving H214Q in mCD16 did not improve mCD16 expression, and in fact appeared to suppress surface levels compared to the WT allele (data not shown).
NK cells in human and mice also differ in adaptor molecule usage for assembly with CD16. Since the discovery of these receptor complexes, it was observed that human CD16 associates with CD3ζ and FcεR1γ, whereas mouse CD16 only associates with FcεR1γ (Blazquez-Moreno et al., 2017; Kurosaki et al., 1991; Lanier et al., 1989; Wirthmueller et al., 1992). In fact, mouse CD3ζ negatively influences FcεRγ-dependent CD16 functions in mouse NK cells, as mouse NK cells deficient in CD3ζ displayed enhanced CD16 function in vitro (Arase et al., 2001). We reveal the molecular interactions underpinning these differences and highlight that phenylalanine 50 and valine 53, in addition to leucine 46, are critical for efficient CD16 receptor complex assembly. Notably, L46 and F50 are the only residues that are shared in all primate species, while position 53 showed some variability, with alanine, valine, and leucine being encoded among primates analyzed. These findings were striking given that these amino acids are all hydrophobic, yet functionally are not interchangeable. Yet, MD simulations revealed that conservative mutations at positions 46, 50, and 53 make the mouse CD3ζ dimer interface more compact and less dynamic than the human dimer. Conformational restriction of the mouse dimer is primarily due to the mutation at position 46 of leucine to its β-branched isomer, isoleucine. β-Branching places steric bulk near the protein backbone that restricts the conformations isoleucine can adopt. In simulation we observed that isoleucine does not pack at the dimer interface but rotates outward, likely to avoid steric clash and prevent energetically costly distortions to the helix backbone locally. This outward rotation is also observed for Y50 and A53 in the mouse dimer. Leucine, in contrast, can adopt several rotamers that allow it to maximize favorable van der Waals packing interactions between the helix monomers. All three key hydrophobic residues (L46, F50, and V53) in human CD3ζ optimize knobs-into-holes packing interactions at the dimer interface and induce asymmetry. This new asymmetric packing generates a cavity N-terminal to the key residues and breaks a Y42-T47 hydrogen bond. The energetic cost of breaking this hydrogen bond is likely offset by maximized packing of the key residues. In sum, these features open the human dimer for association with a third helix.
Assembly of CD16 with CD3ζ has been shown to rely on a polar and aromatic face on the CD16 transmembrane domain focused on three residues (F219, D222, and T223; Blazquez-Moreno et al., 2017). The predominant dimer conformation explored by human CD3ζ likely assembles with CD16 with little disruption. The cavity that forms can accommodate F219 from CD16 and a break in the Y42-T47 hydrogen bond ensures that Y42 and T47 on CD3ζ are available to hydrogen bond with T223 and D222 on CD16, as has been previously proposed (Blazquez-Moreno et al., 2017). CD16-CD3ζ complex formation can, therefore, conceivably occur without the energetic penalty of unpacking the CD3ζ dimer to create a cavity or breaking of the hydrogen bond. Conversely, persistence of the mouse CD3ζ dimer hydrogen bonding network and the absence of a large cavity at the dimer interface create an energetic barrier for association with CD16, which would have to disrupt dimer packing to assemble. Exactly how these key residues on CD3ζ impact spatial packing or influence van der Waals interactions with the transmembrane domain of CD16 within the plasma membrane will require structural analysis in combination with additional modeling. It would be interesting to visualize the interaction between CD16 and CD3ζ transmembrane domains as this would also address how CD16 is capable of associating with CD3ζ while lacking a basic residue in its transmembrane. Importantly, because disruption of the Y42-T47 hydrogen bonding network by mutation to alanine has been shown to abrogate TCR-CD3ζ assembly (Call et al., 2006), our simulation suggests that the residues at positions 46, 50, and 53 also have an impact on the association with the TCR and merits investigation.
In conclusion, this study demonstrates that mouse NK cells fail to generate robust activation upon triggering of their CD16 receptor and that residues in the transmembrane of the CD3ζ adaptor molecule play a pivotal role. Further investigating the interactions that influence Fcγ receptor function will provide an insight to enhancing antibody-directed therapies in NK cells.
Materials and methods
Human samples
Blood samples were collected from healthy adult volunteers (with informed consent), and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation using Ficoll-Plaque (GE Healthcare). All volunteers gave informed consent to participate in this study. Experiments performed in this study were approved by the University of California Committee on Human Research IRB #10-00265.
Mice
C57BL/6J (B6) mice were purchased from The Jackson Laboratory. Mice were bred and housed, and experiments were conducted, according to approved University of California San Francisco (UCSF) Institutional Animal Care and Use Committee and guidelines set by the National Institutes of Health (NIH).
Cells
BWZ.36 (BWZ) cells were generously provided by Dr. N. Shastri (University of California, Berkeley, Berkeley, CA; Sanderson and Shastri, 1994). HEK293T were obtained from American Type Culture Collection. BL3750 B cell lymphoma line was generously provided by Dr. Ronald Levy (Stanford University, Stanford, CA; Minard-Colin et al., 2008). Raji cells were obtained from the UCSF Cell and Genome Engineering Core. Cells were cultured in complete DMEM-HG (DMEM) or RPMI-1640 (RPMI) supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 50 µg/ml gentamicin, 110 µg/ml sodium pyruvate, 50 µM 2-ME, 10 mM Hepes, and 10% FBS. Human PBMCs and mouse splenocytes were cultured in complete RPMI.
NK-92 cells were obtained from the American Type Culture Collection and were cultured with α-MEM supplemented with 2 mM glutamine, 100 U/ml penicillin, 100 µg/ml streptomycin, 200 µM myo-inositol, 20 µM folic acid, 100 µM 2-ME, 10 mM Hepes, 12.5% FBS, and 12.5% horse serum, and 200 U/ml recombinant human IL-2 (teceleukin; generously provided by NCI Biological Resources Branch).
PCR and cloning
The mammalian expression vector pIRES2-EGFP was used to generate pIRES2-mCherry and pIRES2-BFP by replacing EGFP. Mouse and human alleles of CD16 (mouse, Fcgr3; human, FCGR3A), CD3ζ (Cd247; CD247) and FcεR1γ (Fcer1g; FCER1G) were cloned into pIRES2-EGFP, pIRES2-mCherry, and pIRES2-BFP, respectively, using Q5 high fidelity DNA polymerase (New England Biolabs). Mutant alleles of mouse CD3ζ were generated by using site-directed mutagenesis through gene splicing by overlap extension or by using gBlocks (IDT). These constructs were subcloned into pMSCV-IRES-EGFP plasmid to generate retroviral variants in the aforementioned vectors. Human CD16 and mouse CD16 were also cloned into pMXs-Puro retroviral vector. All vectors were sequenced for validation of desired inserts.
Transfections and transductions
For transfections, HEK293T cells were plated 1 d prior to transfection in 6-well plates (6 × 105/well). Transfections were performed using Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Scientific). Cells were analyzed 48 h after transfection using flow cytometry. All mammalian expression vectors used have been described previously (Aguilar et al., 2017).
Retroviruses were generated by transfecting pMSCV-IRES-EGFP/mCherry/BFP or pMXs-Puro vectors into HEK293T along with packaging plasmids, and viral supernatant was then used to transduce BWZ or NK-92 cells. Cells were then sorted for expression of EGFP, mCherry, BFP (BWZ), or selected using media containing puromycin prior to flow cytometric sorting for CD16 expression (NK-92).
BWZ reporter assays
BWZ cells were first CRISPR edited to disrupt the endogenous Cd247 locus using purified Cas9-RNP and using crRNA guides flanking exon 2 (upstream: 5′-TCTGTCACAGAATTGACCTG-3′, downstream: 5′-GTATAGGTTCCTCCACTGGG-3′; IDT) as previously described (Roth et al., 2018). A clone of CD247-deficient BWZ cells (BWZ.∆CD247) was identified using limiting dilution method and validated using PCR and flow cytometry. These CD3ζ−/− cells were transduced with MSCV retrovirus expressing mouse or human CD16, and then sorted for EGFP+ expression (BWZ.mCD16 or BWZ.hCD16). These EGFP+ BWZ cells were then transduced with MSCV retroviruses expressing mouse CD3ζ variants, human CD3ζ, or mouse FcεR1γWT and then sorted for either mCherry+ (CD3ζ) or BFP+ (FcεR1γ).
For plate-bound antibody stimulations, high-binding enzyme immunoassay/radio immunoassay plates (Corning) were precoated with purified antibodies (20–30 µg/ml in PBS) overnight. The next day, wells were thoroughly washed with PBS, BWZ reporters (5 × 104) were added to the wells, and plates were incubated overnight. Stimulations using 30.2 nM PMA and 0.5 µM ionomycin served as positive controls, while media alone served as negative controls. The following day, cells were pelleted by centrifugation, washed with PBS, resuspended in 150 μl of CPRG buffer (90 mg/l chlorophenol-red-β-D-galactopyranoside [Roche], 9 mM MgCl2, 0.1% NP-40 in PBS), and assays were developed at room temperature. Readings were measured using a microplate reader (Tecan Life Sciences) at OD 595–655 nm. Data were normalized to controls values using the following formula: % receptor-specific stimulation = (treatment – negative control)/(positive control – negative control) × 100%. For cocultures with stimulators, BL3750 cells (5 × 104) were added into 96 flat-well plates with BWZ cells (5 × 104) in the presence of antibodies at a concentration of 10 µg/ml and incubated overnight at 37°C. The following day, cells were washed, resuspended in CPRG buffer, and analyzed as described above.
Antibodies and flow cytometry
Cells were stained with primary mAb in FACS buffer (2.5% FBS, 1 mM EDTA, and 0.03% NaN3, in PBS) on ice for 25 mins, washed, incubated with secondary antibodies for 25 min, and analyzed using a LSRII (BD Biosciences). Cells were gated by forward and side light scatter properties, and then for cell viability using propidium iodide, DAPI, Zombie Red, or Zombie NIR (BioLegend) exclusion. Data were analyzed using FlowJo software (FlowJo). The anti-mouse CD20 antibody (mouse IgG2a, clone 5D2) was generously provided by Dr. Jeffrey Venstrom (Genentech, Inc.; Marino et al., 2016; Montalvao et al., 2013). Anti-human CD20 (human IgG1, Rituximab) was a kind gift from Dr. James Rubenstein (UCSF). Anti-mouse CD16 (S17014E or AT154-2), mouse CD16/32 (93 or 2.4G2), mouse NK1.1/NKR-P1CB6 (PK136), mouse NKp46 (29A1.4), mouse CD3ε (145-2C11), mouse CD19 (clone 6D5), mouse CD107a (1D4B), mouse IFN-γ (XMG1.2), human CD16 (3G8 or B73.1), human NKp30 (P30-15), human NKp46 (9E2), human NKp80 (5D12), human CD56 (HCD56), human CD57 (HNK-1), human CD3 (UCHT1), human CD19 (HIB19), human CD14 (63D3), human CD107a (H4A3), human IFN-γ (4S.B3), mouse IgG1 isotype control (MOPC-21), mouse IgG2a isotype control (MOPC-173), rat IgG2a isotype control (RTK2758), and human IgG1 mAbs were purchased from BioLegend, BD Biosciences, Thermo Fisher Scientific, BioXCell, or Bio-Rad. All secondary PE or APC-conjugated streptavidin reagents were purchased from BioLegend.
NK cell stimulation assays
Mouse splenocytes, human PBMCs, or NK-92 cells were cultured in 96-well high binding enzyme immunoassay/radio immunoassay plates (Corning) pre-coated with purified antibodies (20–30 µg/ml in PBS) for 5 h in the presence of anti-CD107a antibody and monensin (BioLegend). Cells were then stained for surface markers, fixed, permeabilized, stained for intracellular IFN-γ, and then analyzed using flow cytometry. Stimulations using 30.2 nM PMA and 0.5 µM ionomycin served as positive controls, while media alone served as negative controls.
Cytotoxicity assays
Target cells (BL3750 and Raji) were labeled with CellTrace Violet (Thermo Fisher Scientific) and used as targets in cytotoxicity assays with NK92 effectors in the presence of antibodies (10 µg/ml). These cells were cocultured for 4 h and then washed, resuspended in FACS buffer containing propidium iodide, and transferred to tubes for analysis by flow cytometry. Cell counts were obtained by adding CountBright Absolute Counting Beads (Thermo Fisher Scientific) to samples according to the manufacturer’s protocol. Percentage of specific lysis was calculated by determining the number of live cells in experimental samples relative to control samples as follows:
Calcium influx assays
BWZ cells were loaded with 7 µg/ml of Indo-1 AM (Thermo Fisher Scientific) for 45 min at room temperature, washed thoroughly, and then stained with biotinylated anti-CD16 antibody. Cells were then analyzed on a flow cytometer (using appropriate UV fluorescence channels) by first assessing baseline Ca2+ and then measuring influx upon addition of streptavidin (500 µg/ml, 100×) while maintaining 37°C temperature using a warm water pump.
In silico sequence alignments
Nucleotide and protein sequences were obtained from the NCBI (https://www.ncbi.nlm.nih.gov/). These sequences were then analyzed, and alignments were conducted using ClustalW tool on MegAlign Pro (DNASTAR Lasergene 17).
Statistical analysis
Data were analyzed using Prism 9 (GraphPad) with one-way ANOVA analysis. Graphs show mean ± SEM or mean ± SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001. All data are representative of at least two independent experiments or three to four biological replicates. See figure legends for details.
MD simulations: System setup
The coordinates for the human CD3ζ transmembrane homodimer (ζ-ζ) were obtained from the NMR structure (PDB ID: 2HAC; Call et al., 2006). The pair of membrane-embedded Asp residues necessary for assembly (D36 on each monomer) was monoprotonated with an overall charge of −1 based on previous findings (Fong et al., 2021). All other residues were assigned their standard protonation states at pH 7. A disulfide bond was formed between C32 in one monomer with C32 in the other. For both helices, the N-termini were acetylated and the C-termini were amidated. Using the CHARMM-GUI webserver (Jo et al., 2007), the ζ-ζ dimer was embedded in a palmitoyl-oleoyl phosphatidylcholine bilayer and hydrated (minimum water height of 20 Å). Cl− ions were added to neutralize and the NaCl concentration was brought to 150 mM. The system was converted to the Amber format with a combination of in-house scripts and the Amber LEaP program in AmberTools 16 (Case et al., 2016). Mutations to the mouse sequence were also made in LEaP. Details of the simulations performed are listed in Table 1.
Table 1.
Description of atomistic simulations performed of ζ-ζ dimer
| System | Protonation/mutation state | Total atoms | Simulation time |
|---|---|---|---|
| Mouse S1, S2, S3 | AspH-Asp, I46, Y50, A53 | 31,633/31,669/31,711 | 3× 1 μs |
| Human S1, S2, S3 | AspH-Asp, L46, F50, V53 | 31,632/31,733/31,724 | 3× 1 μs |
MD simulations: Atomistic simulation procedure
Atomistic MD simulations were performed with the Amber ff14SB protein force field and the lipid17 lipid force field parameter sets (Dickson et al., 2014; Maier et al., 2015). Water molecules were described using the TIP3P model. Simulations were carried out on GPUs with the PMEMD MD engine (Salomon-Ferrer et al., 2013). To remove high-energy contacts, minimization consisted of 5,000 steps, 2,500 steps of steepest descent algorithm with a switch to 2,500 steps of the conjugated gradient algorithm. For NVT equilibration, the systems were gradually heated from 0 to 310 K over 300 ps. Temperature was maintained with the Langevin thermostat with a friction coefficient of γ = 1 ps−1. The proteins and lipids were restrained with a harmonic potential and force constant of 5 kcal/(mol ⋅ Å2). Once a temperature of 310 K was reached, the system was switched to NPT, and pressure was maintained at 1 atm using a semi-isotropic pressure tensor and the Monte Carlo barostat. The 5 kcal/(mol ⋅ Å2) restraint on the proteins and lipids was gradually removed over 4 ns. During the production period, the system was simulated for a total of 1.05 μs. Simulations were carried out under periodic boundary conditions. The SHAKE algorithm was used to constrain bonds involving hydrogen atoms, enabling a 2-fs timestep (Miyamoto and Kollman, 1992). The particle-mesh Ewald method was used to compute long-range electrostatics, and non-bonded interactions were cut off at 12 Å with force-based switching at 10 Å. Three independent simulations were performed per mutation state (human and mouse) starting from the same initial position of the dimer but different starting velocities. Atomic coordinates were saved every 200 ps for subsequent trajectory analysis.
MD simulations: Clustering analysis
Clustering analysis of MD simulations is used to group similar conformations of a biomolecule together in order to understand molecule sample conformational space during simulation time (Shao et al., 2007). Because sampling of conformational space should occur in accordance with a Boltzmann distribution such that higher energy states are sampled less than lower energy states, clustering can be used to discern stability of a particular conformation.
All structural clustering analysis was performed using a hierarchical agglomerative (i.e., bottom-up) average-linkage approach. In this approach, each structure (simulation frame) begins as its own cluster. At each step, the two closest clusters are merged based on a distance metric, coordinate RMSD in this case. With average-linkage, the distance from one cluster to another is defined by determining the RMSD between every structure in cluster 1 and every structure in cluster 2 and averaging the distances. Structures that are more similar have a smaller distance between clusters. Clustering occurs until the distance between all clusters exceeds a cutoff value.
For clustering analysis of helical conformations, structures were taken every 200 ps for a total of 15,000 structures per dimer. Structures for both the human and mouse dimer sequences were combined prior to clustering for a total of 30,000 structures. The C-α RMSD of 48 residues (24 residues per helix in the dimer) was used with a 1.2-Å cutoff.
MD simulations: Hydrogen bonding analysis
A hydrogen bond was considered if the distance between the acceptor and donor heavy atoms (example: O-O distance in the case of a Tyr-Thr hydrogen bond) was <3.0 Å and the angle between the acceptor heavy atom, the donor proton, and the donor heavy atom was >135°. For time series data, hydrogen bonds were given a binary classification: either a hydrogen bond was present or it was not. All analysis of MD simulations was performed using the CPPTRAJ analysis package in AmberTools 16 (Roe and Cheatham, 2013).
Online supplemental material
Fig. S1 demonstrates the enhanced CD16 activation of CD57+ human NK cells and characterizes the expression pattern of Fcγ receptors on mouse NK cells. Fig. S2 shows a phylogenetic analysis of the transmembrane of CD3ζ across vertebrate species. Fig. S3 demonstrates the surface expression of human CD16 when coexpressed with different mouse CD3ζ mutants. Fig. S4 shows the functional responses of BWZ cells transduced with human CD16 and mouse CD3ζ mutants in reporter assays and in calcium influx assays. Fig. S5 demonstrates the C-α RMSD of the human CD3ζ homodimer depicting conformational stability after equilibration, the distance between Cα residues on the CD3ζ helices, and a comparison of the dominant conformers explored by mouse and human CD3ζ, highlighting the packing of glycine at position 43.
Acknowledgments
We thank the Lanier lab members for critical discussion of this work. We thank Dr. Andrei Goga for use of the Tecan spectrophotometer and Drs. Wan-Lin Lo and Arthur Weiss for advice on calcium influx assays.
We thank the UCSF Parnassus Flow Core (RRID:SCR_018206), which is supported by Diabetes Research Center NIH grant P30 DK063720, NIH grant P30 DK063720, and NIH instrumentation grant S10 1S10OD021822-01, for help and advice. O.A. Aguilar holds a Postdoctoral Enrichment Program Award from the Burroughs Wellcome Fund and is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. L.-K. Fong is supported by NIH grant K12GM081266 and the UC President’s Postdoctoral Fellowship. O.A. Aguilar, K. Ishiyama, and L.L. Lanier are supported by the Parker Institute for Cancer Immunotherapy and NIH grants AI068129 and AI146581.
Author contributions: O.A. Aguilar designed, performed, and analyzed experiments and contributed to the manuscript writing. L.-K. Fong designed, performed, and analyzed molecular dynamics simulations and contributed to the manuscript writing. K. Ishiyama helped design and perform experiments. O.A. Aguilar, L.-K. Fong, W.F. DeGrado, and L.L. Lanier contributed to the direction of the study. This work was done in the labs of W.F. DeGrado and L.L. Lanier. L.L. Lanier contributed to the design and interpretation of experiments, project management, and manuscript writing.
References
- Aguilar, O.A., Berry R., Rahim M.M.A., Reichel J.J., Popovic B., Tanaka M., Fu Z., Balaji G.R., Lau T.N.H., Tu M.M., et al. 2017. A viral immunoevasin controls innate immunity by targeting the prototypical natural killer cell receptor family. Cell. 169:58–71.e14. 10.1016/j.cell.2017.03.002 [DOI] [PubMed] [Google Scholar]
- Aguilar, O.A., Tanaka M., Balaji G.R., Berry R., Rossjohn J., Lanier L.L., and Carlyle J.R.. 2020. Tetramer immunization and selection followed by CELLISA screening to generate monoclonal antibodies against the mouse cytomegalovirus m12 immunoevasin. J. Immunol. 205:1709–1717. 10.4049/jimmunol.2000687 [DOI] [PubMed] [Google Scholar]
- Akula, S., Mohammadamin S., and Hellman L.. 2014. Fc receptors for immunoglobulins and their appearance during vertebrate evolution. PLoS One. 9:e96903. 10.1371/journal.pone.0096903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arase, H., Suenaga T., Arase N., Kimura Y., Ito K., Shiina R., Ohno H., and Saito T.. 2001. Negative regulation of expression and function of Fc gamma RIII by CD3 zeta in murine NK cells. J. Immunol. 166:21–25. 10.4049/jimmunol.166.1.21 [DOI] [PubMed] [Google Scholar]
- Baudino, L., Nimmerjahn F., Shinohara Y., Furukawa J.-I., Petry F., Sjef Verbeek J., Nishimura S.-I., Ravetch J.V., and Izui S.. 2008. Impact of a three amino acid deletion in the CH2 domain of murine IgG1 on Fc-associated effector functions. J. Immunol. 181:4107–4112. 10.4049/jimmunol.181.6.4107 [DOI] [PubMed] [Google Scholar]
- Blazquez-Moreno, A., Park S., Im W., Call M.J., Call M.E., and Reyburn H.T.. 2017. Transmembrane features governing Fc receptor CD16A assembly with CD16A signaling adaptor molecules. Proc. Natl. Acad. Sci. USA. 114:E5645–E5654. 10.1073/pnas.1706483114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos, S., Gupta A., and Ravetch J.V.. 2020. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 20:633–643. 10.1038/s41577-020s4100410-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhns, P., Iannascoli B., England P., Mancardi D.A., Fernandez N., Jorieux S., and Daëron M.. 2009. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 113:3716–3725. 10.1182/blood-2008b09-179754 [DOI] [PubMed] [Google Scholar]
- Bryceson, Y.T., March M.E., Ljunggren H.G., and Long E.O.. 2006. Synergy among receptors on resting NK cells for the activation of natural cytotoxicity and cytokine secretion. Blood. 107:159–166. 10.1182/blood-2005blo04-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call, M.E., Schnell J.R., Xu C., Lutz R.A., Chou J.J., and Wucherpfennig K.W.. 2006. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 127:355–368. 10.1016/j.cell.2006.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiossone, L., Dumas P.Y., Vienne M., and Vivier E.. 2018. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18:671–688. 10.1038/s41577-018-0061-z [DOI] [PubMed] [Google Scholar]
- Case, D.A., Cerutti D.S., Cheatham I.T.E., Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., Homeyer N., Izadi S., et al. 2016. AMBER 2016. University of California, San Francisco, CA. [Google Scholar]
- Dekkers, G., Bentlage A.E.H., Stegmann T.C., Howie H.L., Lissenberg-Thunnissen S., Zimring J., Rispens T., and Vidarsson G.. 2017. Affinity of human IgG subclasses to mouse Fc gamma receptors. MAbs. 9:767–773. 10.1080/19420862.2017.1323159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson, C.J., Madej B.D., Skjevik A.A., Betz R.M., Teigen K., Gould I.R., and Walker R.C.. 2014. Lipid14: The amber lipid force field. J. Chem. Theory Comput. 10:865–879. 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre, C.A., Bonnin-Gelize E., Pulford K., Bourel D., Fridman W.H., and Teillaud J.L.. 2008. A novel subset of NK cells expressing high levels of inhibitory FcgammaRIIB modulating antibody-dependent function. J. Leukoc. Biol. 84:1511–1520. 10.1189/jlb.0608343 [DOI] [PubMed] [Google Scholar]
- Fong, L.K., Chalkley M.J., Tan S.K., Grabe M., and DeGrado W.F.. 2021. Elucidation of the molecular interactions that enable stable assembly and structural diversity in multicomponent immune receptors. Proc. Natl. Acad. Sci. USA. 118:e2026318118. 10.1073/pnas.2026318118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain, C., Bihl F., Zahn S., Poupon G., Dumaurier M.J., Rampanarivo H.H., Padkjær S.B., Spee P., and Braud V.M.. 2010. Characterization of alternatively spliced transcript variants of CLEC2D gene. J. Biol. Chem. 285:36207–36215. 10.1074/jbc.M110.179622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams, M., Bruhns P., Saeys Y., Hammad H., and Lambrecht B.N.. 2014. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 14:94–108. 10.1038/nri3582 [DOI] [PubMed] [Google Scholar]
- Guma, M., Angulo A., Vilches C., Gomez-Lozano N., Malats N., and Lopez-Botet M.. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 104:3664–3671. 10.1182/blood-2004blo05-2058 [DOI] [PubMed] [Google Scholar]
- Hammer, Q., Ruckert T., Borst E.M., Dunst J., Haubner A., Durek P., Heinrich F., Gasparoni G., Babic M., Tomic A., et al. 2018. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 19:453–463. 10.1038/s41590-018s4150082-6 [DOI] [PubMed] [Google Scholar]
- Heatley, S.L., Pietra G., Lin J., Widjaja J.M.L., Harpur C.M., Lester S., Rossjohn J., Szer J., Schwarer A., Bradstock K., et al. 2013. Polymorphism in human cytomegalovirus UL40 impacts on recognition of human leukocyte antigen-E (HLA-E) by natural killer cells. J. Biol. Chem. 288:8679–8690. 10.1074/jbc.m112.409672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth, P.M., and Pietersz G.A.. 2012. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov. 11:311–331. 10.1038/nrd2909 [DOI] [PubMed] [Google Scholar]
- Jo, S., Kim T., and Im W.. 2007. Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS One. 2:e880. 10.1371/journal.pone.0000880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, T., Gander I., and Ravetch J.V.. 1991. A subunit common to an IgG Fc receptor and the T-cell receptor mediates assembly through different interactions. Proc. Natl. Acad. Sci. USA. 88:3837–3841. 10.1073/pnas.88.9.3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, T., and Ravetch J.V.. 1989. A single amino acid in the glycosyl phosphatidylinositol attachment domain determines the membrane topology of Fc gamma RIII. Nature. 342:805–807. 10.1038/342805a0 [DOI] [PubMed] [Google Scholar]
- Lanier, L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- Lanier, L.L. 2008. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 8:259–268. 10.1038/nri2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, L.L., Le A.M., Phillips J.H., Warner N.L., and Babcock G.F.. 1983. Subpopulations of human natural killer cells defined by expression of the Leu-7 (HNK-1) and Leu-11 (NK-15) antigens. J. Immunol. 131:1789–1796. [PubMed] [Google Scholar]
- Lanier, L.L., Ruitenberg J.J., and Phillips J.H.. 1988. Functional and biochemical analysis of CD16 antigen on natural killer cells and granulocytes. J. Immunol. 141:3478–3485. [PubMed] [Google Scholar]
- Lanier, L.L., Yu G., and Phillips J.H.. 1989. Co-association of CD3 zeta with a receptor (CD16) for IgG Fc on human natural killer cells. Nature. 342:803–805. 10.1038/342803a0 [DOI] [PubMed] [Google Scholar]
- Lanier, L.L., Yu G., and Phillips J.H.. 1991. Analysis of Fc gamma RIII (CD16) membrane expression and association with CD3 zeta and Fc epsilon RI-gamma by site-directed mutation. J. Immunol. 146:1571–1576. [PubMed] [Google Scholar]
- Lee, J., Zhang T., Hwang I., Kim A., Nitschke L., Kim M., Scott J.M., Kamimura Y., Lanier L.L., and Kim S.. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity. 42:431–442. 10.1016/j.immuni.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Verges, S., Milush J.M., Pandey S., York V.A., Arakawa-Hoyt J., Pircher H., Norris P.J., Nixon D.F., and Lanier L.L.. 2010. CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK-cell subset. Blood. 116:3865–3874. 10.1182/blood-2010b04-282301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Verges, S., Milush J.M., Schwartz B.S., Pando M.J., Jarjoura J., York V.A., Houchins J.P., Miller S., Kang S.M., Norris P.J., et al. 2011. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 108:14725–14732. 10.1073/pnas.1110900108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., and Simmerling C.. 2015. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11:3696–3713. 10.1021/acs.jctc.5b00255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, J., Paster J.T., Trowell A., Maxwell L., Briggs K.H., Crosby Bertorini P., and Benichou G.. 2016. B cell depletion with an anti-CD20 antibody enhances alloreactive memory T cell responses after Transplantation. Am. J. Transpl. 16:672–678. 10.1111/ajt.13483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.S., and Lanier L.L.. 2019. Natural killer cells in cancer immunotherapy. Annu. Rev. Cancer Biol. 3:77–103. 10.1146/annurev-cancerbio-030518-055653 [DOI] [Google Scholar]
- Minard-Colin, V., Xiu Y., Poe J.C., Horikawa M., Magro C.M., Hamaguchi Y., Haas K.M., and Tedder T.F.. 2008. Lymphoma depletion during CD20 immunotherapy in mice is mediated by macrophage FcgammaRI, FcgammaRIII, and FcgammaRIV. Blood. 112:1205–1213. 10.1182/blood-2008b01-135160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, S., and Kollman P.A.. 1992. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13:952–962. 10.1002/jcc.540130805 [DOI] [Google Scholar]
- Montalvao, F., Garcia Z., Celli S., Breart B., Deguine J., Van Rooijen N., and Bousso P.. 2013. The mechanism of anti-CD20-mediated B cell depletion revealed by intravital imaging. J. Clin. Invest. 123:5098–5103. 10.1172/JCI70972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujal, A.M., Delconte R.B., and Sun J.C.. 2021. Natural killer cells: From innate to adaptive features. Annu. Rev. Immunol. 39:417–447. 10.1146/annurev-immunolan101819-074948 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn, F., Bruhns P., Horiuchi K., and Ravetch J.V.. 2005. FcgammaRIV: A novel FcR with distinct IgG subclass specificity. Immunity. 23:41–51. 10.1016/j.immuni.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn, F., and Ravetch J.V.. 2006. Fcgamma receptors: Old friends and new family members. Immunity. 24:19–28. 10.1016/j.immuni.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn, F., and Ravetch J.V.. 2008. Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 8:34–47. 10.1038/nri2206 [DOI] [PubMed] [Google Scholar]
- Pahl, J.H.W., Koch J., Gotz J.J., Arnold A., Reusch U., Gantke T., Rajkovic E., Treder M., and Cerwenka A.. 2018. CD16A activation of NK cells promotes NK cell proliferation and memory-like cytotoxicity against cancer cells. Cancer Immunol. Res. 6:517–527. 10.1158/2326-6066.CIR2326-617-0550 [DOI] [PubMed] [Google Scholar]
- Raulet, D.H., and Vance R.E.. 2006. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 6:520–531. 10.1038/nri1863 [DOI] [PubMed] [Google Scholar]
- Roe, D.R., and Cheatham T.E. III. 2013. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9:3084–3095. 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Roth, T.L., Puig-Saus C., Yu R., Shifrut E., Carnevale J., Li P.J., Hiatt J., Saco J., Krystofinski P., Li H., et al. 2018. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 559:405–409. 10.1038/s41586-018s4150326-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Ferrer, R., Gotz A.W., Poole D., Le Grand S., and Walker R.C.. 2013. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J. Chem. Theory Comput. 9:3878–3888. 10.1021/ct400314y [DOI] [PubMed] [Google Scholar]
- Sanderson, S., and Shastri N.. 1994. LacZ inducible, antigen/MHC-specific T cell hybrids. Int. Immunol. 6:369–376. 10.1093/intimm/6.3.369 [DOI] [PubMed] [Google Scholar]
- Schlums, H., Cichocki F., Tesi B., Theorell J., Beziat V., Holmes T.D., Han H., Chiang S.C.C., Foley B., Mattsson K., et al. 2015. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity. 42:443–456. 10.1016/j.immuni.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, J., Tanner S.W., Thompson N., and Cheatham T.E.. 2007. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theory Comput. 3:2312–2334. 10.1021/ct700119m [DOI] [PubMed] [Google Scholar]
- Sun, J.C., Beilke J.N., and Lanier L.L.. 2009. Adaptive immune features of natural killer cells. Nature. 457:557–561. 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai, T. 2002. Roles of Fc receptors in autoimmunity. Nat. Rev. Immunol. 2:580–592. 10.1038/nri856 [DOI] [PubMed] [Google Scholar]
- Temming, A.R., Bentlage A.E.H., de Taeye S.W., Bosman G.P., Lissenberg-Thunnissen S.N., Derksen N.I.L., Brasser G., Mok J.Y., van Esch W.J.E., Howie H.L., et al. 2020. Cross-reactivity of mouse IgG subclasses to human Fc gamma receptors: Antibody deglycosylation only eliminates IgG2b binding. Mol. Immunol. 127:79–86. 10.1016/j.molimm.2020.08.015 [DOI] [PubMed] [Google Scholar]
- Wang, E.C.Y., McSharry B., Retiere C., Tomasec P., Williams S., Borysiewicz L.K., Braud V.M., and Wilkinson G.W.G.. 2002. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc. Natl. Acad. Sci. USA. 99:7570–7575. 10.1073/pnas.112680099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller, U., Kurosaki T., Murakami M.S., and Ravetch J.V.. 1992. Signal transduction by Fc gamma RIII (CD16) is mediated through the gamma chain. J. Exp. Med. 175:1381–1390. 10.1084/jem.175.5.1381 [DOI] [PMC free article] [PubMed] [Google Scholar]