Key Points
-
•
Structured illumination microscopy (SIM) imaging reveals kidney tubule cell mitochondria form continuous networks in healthy mouse and human tissue, which break down with injury.
-
•
SIM imaging can be applied to various organelles and structures, including autophagosomes, endoplasmic reticulum, brush border, and cell morphology.
-
•
Promoting mitochondrial fusion maintains mitochondrial interconnectivity and ameliorates cisplatin-induced kidney injury.
Keywords: acute kidney injury and ICU nephrology, acute renal failure, basic science, cell adhesion, cell biology and structure, electron microscopy, endoplasmic reticulum, ischemia-reperfusion, mitochondria, pathology, renal morphology, renal proximal tubule cell
Visual Abstract
Abstract
Background
The root of many kidney diseases in humans can be traced to alterations or damage to subcellular organelles. Mitochondrial fragmentation, endoplasmic reticulum (ER) stress, and lysosomal inhibition, among others, ultimately contribute to kidney injury and are the target of therapeutics in development. Although recent technological advancements allow for the understanding of disease states at the cellular level, investigating changes in subcellular organelles from kidney tissue remains challenging.
Methods
Using structured illumination microscopy, we imaged mitochondria and other organelles from paraffin sections of mouse tissue and human kidney biopsy specimens. The resulting images were 3D rendered to quantify mitochondrial size, content, and morphology. Results were compared with those from transmission electron microscopy and segmentation.
Results
Super-resolution imaging reveals kidney tubular epithelial cell mitochondria in rodent and human kidney tissue form large, interconnected networks under basal conditions, which are fragmented with injury. This approach can be expanded to other organelles and cellular structures including autophagosomes, ER, brush border, and cell morphology. We find that, during unilateral ischemia, mitochondrial fragmentation occurs in most tubule cells and they remain fragmented for >96 hours. Promoting mitochondrial fusion with the fusion promotor M1 preserves mitochondrial morphology and interconnectivity and protects against cisplatin-induced kidney injury.
Conclusions
We provide, for the first time, a nonbiased, semiautomated approach for quantification of the 3D morphology of mitochondria in kidney tissue. Maintaining mitochondrial interconnectivity and morphology protects against kidney injury. Super-resolution imaging has the potential to both drive discovery of novel pathobiologic mechanisms in kidney tissue and broaden the diagnoses that can be made on human biopsy specimens.
Introduction
CKD is estimated to affect >10% of the world’s population (1). Studies have shown that AKI and CKD progression often start with or involve stress to subcellular organelles, such as endoplasmic reticulum (ER) and mitochondria (2–8). Subcellular organelle health and function are closely associated with their morphology (8–13). Although once thought to be the direct result of injury, studies have demonstrated changes in mitochondrial morphology occur due to regulation of conserved mitochondrial fission (involving dynamin related protein 1 [Drp-1], fission 1 [Fis-1]) and fusion (involving mitofusin [Mfn] 1, Mfn2, and optic atrophy 1 [Opa1]) pathways that balance to control morphology, divide mitochondria during cell division, distribute mitochondria to areas of high metabolic demands, and eliminate dysfunctional mitochondria (2,14–17). Under basal conditions, mitochondrial fusion predominates, resulting in long, interconnected mitochondria (2,14,15). This interconnectivity allows for the transfer of mitochondrial DNA and other components, helping to maintain mitochondrial health (2,18–20). Under pathologic conditions, however, hyperactivation of mitochondrial fission and suppression of fusion lead to fragmentation of the mitochondria, sensitizing cells to injury and death (2,14,15).
Proximal tubule cells (PTCs) of the kidney have one of the highest concentrations of mitochondria among mammalian cells. It is estimated that mitochondria occupy approximately 25% of the volume of a PTC, compared with 9% for the nucleus (21). PTC mitochondria are tightly packed perpendicularly to the basolateral plasma membrane (21). The gold standard for analyzing mitochondria in kidney epithelial cells is electron microscopy (EM) because mitochondria are too closely packed to resolve by traditional light microscopy. Using EM, the number and size of mitochondrial cross-sections can be quantified, but not the network size or 3D shape (22). Thus, although it is known that mitochondria form continuous, filamentous networks in cultured kidney cells, the mitochondrial morphology of cells in kidney tissue is not well understood (6,7,15,16,22).
Recent advances in light microscopy techniques have made super-resolution imaging more accessible. These techniques include stochastic optical reconstruction microscopy, stimulated emission depletion microscopy, super-resolution radial fluctuations (SRRF), and structured illumination microscopy (SIM) (23). Of these techniques, SIM is appealing because it can be performed on samples stained by standard immunofluorescence techniques (23,24). SIM illuminates the sample with patterned illumination, usually stripes. The sample is imaged multiple times with the stripes changing orientation. This process creates Moiré patterns with high-frequency spatial information that can be processed by specialized algorithms to generate higher resolution images, effectively doubling the resolving power compared with standard light microscopy (25). SIM imaging has been used to study glomerular injuries, such as podocyte effacement, in animal models and human tissue (26–28).
In this study, we demonstrate that super-resolution imaging can be used to image and create 3D models of mitochondria from cells in kidney tissue, allowing for the quantification of mitochondrial network size and morphology. We also demonstrate super-resolution imaging can quantify mitochondrial morphology in human biopsy specimens. Combining organelle markers with injury- or cell type–specific markers can further enhance the analysis. Our data suggest that promoting mitochondrial fusion maintains mitochondrial morphology and reduces kidney injury.
Materials and Methods
Animals
C57BL6J mice obtained from Jackson Laboratory were used to establish a breeding colony at Vanderbilt University Medical Center. Mice were provided with rodent diet and water ad libitum. All mice were maintained in the same room on a 12‐hour light/dark photoperiod at an ambient temperature. Male mice, aged 8–12 weeks old, were used for all experiments.
Mouse Models
In all of the following models, kidneys and hearts were fixed in 4% paraformaldehyde for 48 hours, embedded in paraffin, and sections were cut.
All procedures involving mice were performed in accordance with National Institutes of Health (NIH) guidelines for the use and care of live animals and were approved by the Vanderbilt University Institutional Animal Care and Use Committee.
Aristolochic Acid Nephropathy Model
Aristolochic acid (AA) nephropathy was induced by intraperitoneal injection of three doses of AA (5 mg/kg body wt, catalog number A5512; Sigma) in PBS every other day for 5 days. The control mice were injected with the same amount of PBS. Kidneys were harvested at 42 days after the last dose of AA.
Unilateral Ureteral Obstruction Model
Mice were anesthetized with isoflurane using a low-flow anesthesia system (SomnoSuite; Kent Scientific). Mice were kept on a heating pad and body temperature was monitored by a rectal probe with a feedback system to the heating pad to maintain body temperature (PhysioSuite; Kent Scientific). The left kidney was externalized through a flank incision and the ureter was ligated by 3-0 suture.
Ischemic-Reperfusion Injury Model
Mice were anesthetized and body temperature monitored as described above. Kidneys were externalized through dorsal incisions, and the renal pedicle was clamped using a nontraumatic microclamp for 26 minutes for bilateral ischemic-reperfusion injury (IRI). For unilateral IRI (UIRI), only the left kidney was clamped for 32 minutes. Kidneys were isolated at 0, 24, 48, and 96 hours after 32 minutes of ischemia (n=3).
Cisplatin-Induced Injury Model
Cisplatin-induced AKI was induced by an intraperitoneal injection of cisplatin (20 mg/kg body wt, catalog number P4394; Sigma) in PBS. Sham mice were injected with the same amount of PBS. Kidneys were harvested 96 hours after cisplatin administration. M1 (catalog number 475859; EMD Millipore), a mitochondrial fusion promoter, was dissolved in 1 vol of DMSO and 9 vol of corn oil and injected intraperitoneally at 10 mg/kg body wt every 24 hours until day 3 after cisplatin injection. Vehicle (comparable amount of DMSO and corn oil) was injected in the same manner as M1.
Diabetic Injury Model (db/db)
db/db mice, a widely used mouse model of type 2 diabetes mellitus, and age-matched nondiabetic db/m littermates were fed with regular chow and heart and kidneys were isolated at 18 weeks old.
Kim1-GFPCreERt2/R26tdTomato mice were kindly provided by Dr. Benjamin Humphreys. Mice were injured via IRI as described above and injected with 5 mg/kg tamoxifen after injury to induce Cre mediated recombination.
Antibodies
Antibodies directed against cytochrome c oxidase IV (COX IV; catalog number ab14744, RRID:AB_301443; Abcam) (catalog number 4850, RRID:AB_2085424; Cell Signaling Technology), VDAC (catalog number 4661, RRID:AB_10557420; Cell Signaling Technology), Na+K+ATPase (catalog number a5, RRID:AB_2166869; Developmental Studies Hybridoma Bank), GRP94 (catalog number sc-393402; Santa Cruz Biotechnology), GRP78 (catalog number sc-166490, RRID:AB_2264290; Santa Cruz Biotechnology), laminin (catalog number AB19012, RRID:AB_2133893; Millipore), collagen type IV (catalog number AB769, RRID:AB_92262; Millipore), and kidney injury molecule-1 (KIM-1; catalog number AF1817, RRID:AB_2116446; R&D Systems) were used as primary antibodies. Alexa Fluor 488 donkey anti-rabbit antibody (catalog number 711-545-152, RRID:AB_2313584; Jackson ImmunoResearch Labs), Cy3-Donkey anti-rabbit IgG antibody (catalog number 711-165-152, RRID:AB_2307443; Jackson ImmunoResearch Labs), Alexa Fluor 488 donkey anti-mouse antibody (catalog number 715-546-151, RRID:AB_2340850; Jackson ImmunoResearch Labs), Cy3-donkey anti-mouse antibody (catalog number 705-165-147, RRID:AB_2307351; Jackson ImmunoResearch Labs), and Cy5-donkey anti-mouse IgG antibody (catalog number 715-175-150, RRID:AB_2340819; Jackson ImmunoResearch Labs) were used as secondary antibodies.
Sectioning and Immunofluorescence Staining
Coverslips were cleaned and activated using a plasma cleaner and treated with 0.1 N sodium hydroxide and (3-aminopropyl)trimethoxysilane. Kidney sections from fixed tissue were cut at 4–6 μm and mounted onto the treated coverslips. After deparaffinization, the tissue sections were incubated in TE buffer (pH 9.0) at high temperature and pressure using a pressure cooker for permeabilization and antigen retrieval. Mitochondria were labeled with COX IV or VDAC antibodies, followed by appropriate secondary antibody conjugated with fluorophore. PHEM buffer (60 mM piperazine-N,N′-bis[2-ethanesulfonic acid], 25 mM HEPES, 10 mM EGTA, 2 mM magnesium chloride, pH 6.9) was used for washing and dilution of antibodies.
Widefield Fluorescent Imaging
Stained kidneys were sequentially imaged using a Nikon Eclipse Ti2-E automated microscope at ×20, and the images were stitched using NIS-Elements to create a transverse large image of the whole kidney section (Nikon, New York, NY). For quantification, staining was analyzed in a blinded and automated manner. Large images were batch analyzed using NIS-Elements. Within an experimental set, the staining was performed on all samples simultaneously, and microscope and analysis settings were the same for all samples.
SIM
Tissues were imaged on an N-SIM microscope (Nikon structured illumination microscope) using a ×100 TIRF super-resolution objective (with minimal chromatic aberration) and taken as a z-stack at 0.12 μm per z step. To automate image capture, multiple tubules were chosen at random and imaged using NIS Elements multi-dimensional image acquisition. The resulting z-stack was processed and reconstructed in Nikon NIS software to generate the super-resolution SIM image. Images were reconstructed using the slice reconstruction with illumination modulation contrast of 1.22, high resolution noise suppression of 1.56, and out of focus blur suppression of 0.14. A total of 5–9 tubules per sample were imaged. SIM imaging and image processing was performed, in part, through the Vanderbilt Cell Imaging Shared Resource and Nikon Center of Excellence (supported by NIH grants CA68485, DK20593, DK58404, DK59637, and EY08126).
SoRa
Kidney tissues were imaged using the Nikon Ti2 microscope partnered with the Yokogawa CSU-W1 SoRa system using Apo TIRF ×100 oil DICN2 as an objective and taken as a z-stack at 0.1 μm per z step. The images were reconstructed and processed using the same procedure as for the SIM images.
SRRF
Kidney tissues were captured using an Andor IXon Back-Illuminated sCMOS Camera (Andor Technology, Belfast, UK) equipped with the Nikon TiE microscope using a CFI Plan Apochromat VC ×100 Oil objective (Nikon). SRRF images were obtained from 100 frames per second diffraction-limited images using NanoJ-SRRF plugged in to FIJI.
Mitochondrial Volume, Surface Area, and Morphology
Mitochondrial networks were quantified by analyzing the network volume using Imaris software (Bitplane, Concord, MA). SIM images of tubule cross-sections were taken as a z-stack and 3D reconstructed in Imaris. The resulting images were converted to surface renderings. Imaris identifies individual mitochondria and interconnected networks as surfaces. To automate image analysis, parameters for surface rendering were set at the begining of the analysis for each data set and the remaining samples were rendered using batch processing. Once the surfaces are rendered, the volume, area, intensity, and other parameters can be generated for quantification. Mitochondria were analyzed using the following criteria: maximum mitochondrial size (μm3; the largest mitochondrial network in a given field), total volume per PTC (μm3; total mitochondrial volume in a field/the number of PTC nuclei), and number of mitochondria per PTC (n; number of mitochondria/PTC nuclei). To investigate the proportion of fragmented mitochondria after injury, PTCs’ mitochondria were divided into three groups by their sphericity as follows: fragmented, sphericity >0.451; intermediate, sphericity between 0.450 and 0.311; and filamentous, sphericity <0.310. Proportion (%) of each group was defined by each group’s mitochondrial volume divided by the total mitochondrial volume. For each sample, 5–9 tubules were analyzed and averaged. To investigate the morphologic changes of mitochondria in the kidneys of patients with AKI, the same analysis was performed in biopsy tissue scored by a pathologist as having minimal tubule injury (control) or AKI with tubule injury (n=3; Supplemental Table 1).
Tissue Culture
LLC-PK1 cells were obtained from ATCC (ATCC-CL101) and cultured in MEM containing 3% FBS in a humidified incubator at 37°C and 5% carbon dioxide.
Human Tissues
Formalin-fixed, paraffin-embedded tissue sections, scored by a pathologist as having minimal tubule injury (control) or AKI with tubule injury (Supplemental Table 1), were obtained from deidentified biopsy specimens through the Department of Pathology, Microbiology and Immunology at Vanderbilt University Medical Center.
Statistical Analysis
All data are presented as means±SD. One-way ANOVA with Tukey post hoc test was used to evaluate differences when more than three groups were compared. When two groups were analyzed, the unpaired two-tailed t test was performed. All statistical analysis was carried out with GraphPad (San Diego, CA) Prism 7 software (RRID:SCR_002798). For all comparisons, P<0.05 was considered to indicate statistical significance.
Transmission EM
Fixed kidney cortex was trimmed into approximately 1-mm3 samples, postfixed with osmium tetroxide, dehydrated serially in ethanol, and embedded in epoxy resin. Semithin sections were cut at 1 μm and stained with toluidine blue for light microscopic examination. Ultrathin sections were cut at 80 nm, mounted on 200‐mesh copper grids, and stained with uranyl acetate and lead citrate. Sections were examined with a JEOL JEM‐1010 transmission electron microscope.
EM Segmentation and Volume Rendering
After euthanasia, mice were cardiac perfused with 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M cacodylate (pH 7.4). After the initial perfusion, the kidneys were dissected and fixed for an additional 24 hours. Samples were postfixed with tannic acid, followed by osmium tetroxide, and then they were en bloc stained with uranyl acetate. Dehydration was performed using a graded ethanol series followed by infiltration with Epon-812 using propylene oxide as the transition solvent. The Epon-812 was polymerized at 60°C for 48 hours. Samples were serial sectioned by applying a thin coating of 1% rubber cement dissolved in chloroform over the block face to facilitate section ribbons. Sections were taken at a nominal thickness of 100 nm on a Leica UC6 ultramicrotome and collected onto slot grids. The sections were stained with uranyl acetate and lead citrate.
Imaging was performed on a Tecnai T12 equipped with a LaB6 filament operating at 100 kV using an AMT nanosprint5 CMOS. Imaging was performed using SerialEM for acquisition and etomo/IMOD to align the images in z-stacks. Segmentation was performed in Dragonfly (Object Research Systems) by manually annotating the mitochondria and nucleus in each image in the z-stack, then creating a 3D mesh from the 2D regions of interest. Dragonfly was also used to create the 3D renders. Data from the segmentation were analyzed in Imaris for surface rendering and analysis.
Results
Tissue Super-Resolution Imaging Resolves Kidney Tubular Cell Mitochondria
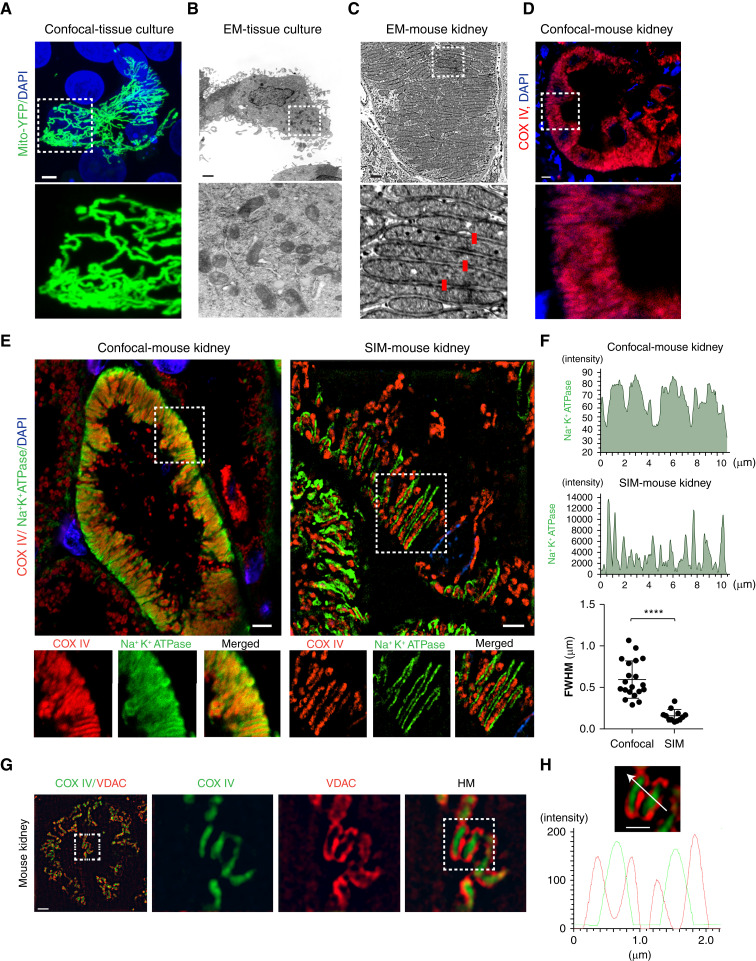
Whereas studies have shown some organs, such as muscle, have interconnected mitochondria, other organs, such as the brain, often have mitochondria that do not form a network (29,30). To determine the morphology of kidney tubular cell mitochondria, we first analyzed tubular cells in vitro. Similar to our earlier findings (16,22), we found cultured PTCs (LLC-PK1) have large interconnected mitochondrial networks (Figure 1A). Whereas these mitochondrial networks are readily imaged by confocal microscopy, the 3D architecture is hard to appreciate from EM micrographs (Figure 1B). Unlike cultured cells, tubular cells in vivo have densely packed mitochondria that take up a large portion of the cell body (Figure 1C). Because we previously demonstrated that mitochondria around the nucleus have an altered arrangement (22), we imaged an area of the tubule without nuclei. The tight arrangement of mitochondria prevents resolving individual mitochondria by conventional light microscopy, because they are separated by less than the diffraction limit of light microscopy, approximately 200 nm (Figure 1, C and D).
Figure 1.
Super-resolution imaging resolves kidney tubular cell mitochondria. (A) LLC-PK1 cells transfected with a mitochondrial-targeting yellow fluorescent protein (YFP) construct (green) and imaged by confocal microscopy. For clarity, a field containing one transfected cell was chosen. Scale bar, 2 μm. (B) Electron microscopy (EM) micrograph of an LLC-PK1 cell. Scale bar, 5 μm. (C) EM micrograph of kidney tubular cell mitochondria in mouse kidneys. An area away from the nucleus was chosen. Red bars are 200 nm in length. Scale bar, 1 μm. (D) Confocal imaging of mouse kidney tubular cells stained with cytochrome c oxidase IV (COX IV; red) to label mitochondria. Taken with Zeiss 780 confocal and ×63 objective; 1024×1024 pixels. Scale bar, 5 μm. (E) Uninjured mouse kidneys were stained with COX IV (red), to label mitochondria, and Na+K+ATPase (green), to label the basolateral plasma membrane, and imaged by confocal microscopy at high resolution (Zeiss 780, ×63, 4096×4096 pixels, average of four images; left panel) or structured illumination microscopy (SIM; Nikon N-SIM, 1024×1024, ×100; right panel). Scale bar, 3 μm. (F) Line scan profile of Na+K+ATPase (green) and the quantification of full width at half maximum (FWHM) of confocal images or SIM. (G) Kidney cell mitochondria were stained with voltage dependent anion channel (VDAC) (red) or COX IV (green) and imaged by SIM. Scale bar, 3 μm. (H) Line scan profile of Figure 1G. Scale bar, 1 μm. Data are presented as means±SD. ****P<0.0001. Dashed insets delineate the regions shown at higher magnification. DAPI, 4′,6-diamidino-2-phenylindole; mito, mitochondria; HM, high magnification.
To test if super-resolution microscopy could be used to image PTC mitochondria in tissue, we imaged mouse kidneys stained with COX IV and Na+K+ATPase using SIM. The major limitation of SIM is the Moiré patterns become diffuse as a function of distance, limiting the working distance (25). To circumvent this issue, we mounted the tissue directly to the coverslip, instead of the slide (Supplemental Figure 1A). Compared with high-resolution confocal images (4096×4096), SIM images could resolve the tightly packed side-by-side mitochondria (Figure 1E). When combined with staining for the basolateral plasma membrane, Na+K+ATPase, SIM imaging identified the involutions of the plasma membrane that are found in between mitochondria (Figure 1E). To further test the resolution of SIM, we measured the full width half maximum (FWHM) resolution. Compared with confocal microscopy, the full width half maximum is considerably smaller with SIM imaging (Figure 1F). Next, we stained for the inner and outer mitochondrial membranes using COX IV and VDAC antibodies. The inner mitochondrial membrane was clearly distinguishable from the outer membrane (Figure 1, G and H).
As mentioned above, there are multiple imaging techniques that can overcome the diffraction limit of light microscopy. To test if other super-resolution techniques yielded similar results, we imaged samples by SRRF imaging with a widefield microscope and a Nikon SoRa super-resolution spinning disk microscope, both of which achieve resolutions similar to SIM. Both SRRF and SoRa could resolve kidney tubular cell mitochondria (Supplemental Figure 1, B–D). These data demonstrate that SIM and other tissue super-resolution imaging techniques have the resolving power to image tubule cell mitochondria in kidney tissue.
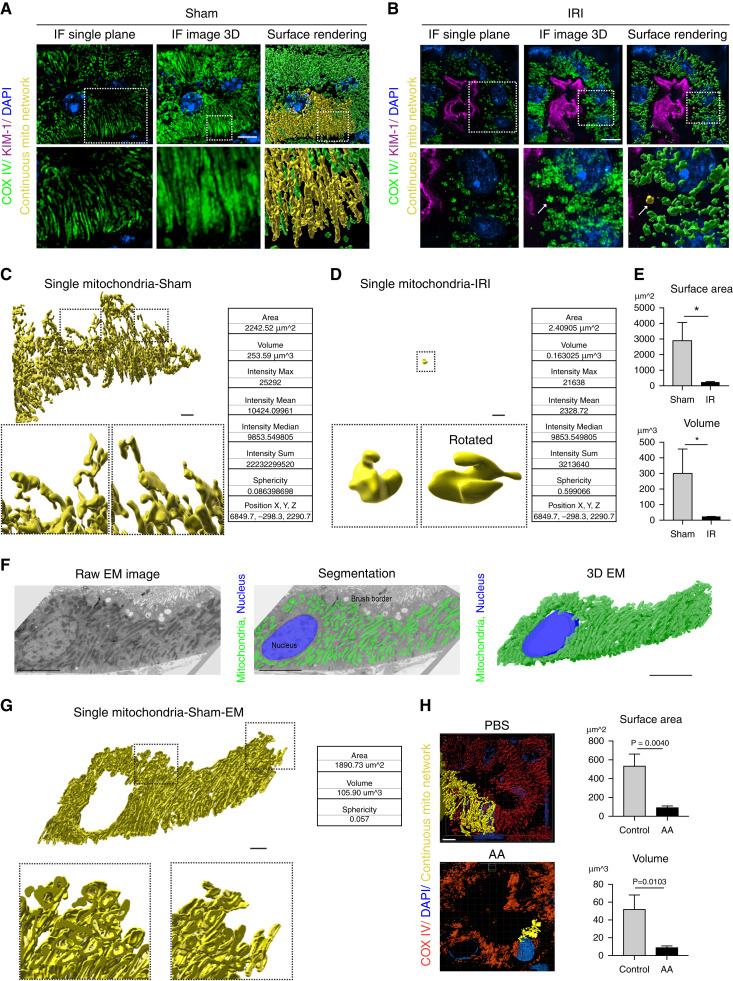
3D Reconstruction Reveals Kidney Tubular Cell Mitochondria Form a Large, Interconnected Network
We have previously demonstrated that kidney tubule cell mitochondria are longer than shown in thin sections imaged by segmentation of transmission EM serial sections (22). The full extent of the mitochondrial network, however, has not been determined. To examine the mitochondrial network morphology, we imaged kidney tubule cells from sham- or IRI-treated mouse kidneys stained for COX IV as a z-stack with 0.12-μm steps using SIM (Figure 2, A and B, left panels). The resulting images were combined into a maximum intensity projection for visualization (Figure 2, A and B, middle panels). To quantify changes to the mitochondrial networks, we used Imaris software to create surface renderings of the mitochondria (Figure 2, A and B, right panel). This identifies each individual mitochondrion or mitochondrial network as a separate rendered surface. As shown in Figure 2A (right panel), highlighting individual mitochondrial networks (yellow) in healthy tubule cells reveals one large mitochondrial network comprising most of the mitochondrial mass within a given tubular cell (Supplemental Video 1). This interconnected network breaks down after injury (Figure 2B, right panel). Figure 2C shows a single, interconnected mitochondrion from surface rendering images in Figure 2A and higher magnification of the surface. Figure 2D highlights a single mitochondrion after IRI injury. In addition to visualizing the interconnectivity of mitochondria, surface rendering provides quantitative information about each object, including the surface area, volume, intensity, sphericity, and position of each structure (among other values). Comparing a single mitochondrial network from sham kidneys with kidneys exposed to IRI demonstrates the large difference in the size of the networks (Figure 2, C, D, and E); of note, surface area (2242 μm2 versus 2 μm2), volume (253.59 μm3 versus 0.16 μm3), and sphericity (0.08 versus 0.59) are dramatically altered with injury. Similar results were achieved using the SoRa imaging system (Supplemental Figure 1D).
Figure 2.
3D reconstruction reveals kidney tubular cell mitochondria form a large interconnected network. (A, B) Representative images of kidney tubular cell mitochondria stained for COX IV (green) with kidney injury molecule-1 (KIM-1; magenta), imaged by SIM and displayed as a single z-plane (left column), 3D maximum intensity projection (middle), and surface-rendering images (right column) after (A) sham or (B) ischemic-reperfusion injury (IRI) surgery. A continuous mitochondrion is labeled yellow. Arrow indicates fragmented mitochondria. Scale bar, 3 μm. (C, D) Individual mitochondria from the surface-rendering images of (A, B) and sample statistics from the rendered structures. Scale bar, 2 μm. (E) Quantification of total mitochondrial surface area and volume on the basis of surface-rendered images in sham and IRI kidneys. Five tubules per animal were analyzed (n=3). (F) Representative EM image of kidney tubular cells in sham kidney (left). Mitochondria are labeled green in EM image segmentation (middle). 3D structure of mitochondria created by EM serial sections (right). Scale bar, 5 μm. (G) Representative surface rendering of a continuous mitochondrion from the EM segmentation in (F). Scale bar, 2 μm. (H) Representative surface renderings of COX IV–stained (red) mitochondria in kidney tubular cells 6 weeks after PBS or aristolochic acid (AA) injection, and quantification of the total mitochondrial surface area and volume on the basis of surface-rendered images. Five tubules per animal were analyzed (n=3). Scale bar, 4 μm. Data are presented as means±SD. *P<0.05. Dashed insets deliniate the regions shown at higher magnification. (A low-magnification overview of the tubule where the cell was chosen is in Supplemental Figure 1E.) DAPI, 4′,6-diamidino-2-phenylindole; IF, immunofluorescence.
To confirm that kidney tubule cell mitochondria are interconnected, we performed segmentation of mitochondria in EM serial sections. Compared with our earlier study, which focused on a few mitochondria (22), in this study we analyzed all of the mitochondria in a tubule cell (see low-magnification overview of the tubule in Supplemental Figure 1E). Twenty 100-nm serial sections were imaged and segmented, for a total depth of 2 μm (Figure 2F). To determine the area, volume, and interconnectivity of the mitochondrial network, we used Imaris to analyze the segmentation data. The tubule cell mitochondria imaged by EM form a highly interconnected network consisting of most of the mitochondria within a cell (Figure 2G, Supplemental Video 2). The area, volume, and sphericity of the single interconnected network was similar to the results seen with SIM, given the EM data is 2 μm in depth and the SIM data are 4 μm (Figure 2, C and G). (Note that mitochondria imaged by EM were segmented by tracing the outer mitochondrial membrane and filling the interior, whereas SIM imaging was performed on samples stained for COX IV on the inner mitochondrial membrane. Therefore, the mitochondria reconstructed by these two techniques have slightly different structures.)
We confirmed a similar pattern of mitochondrial breakdown in the chronic phase of AA-induced nephropathy (Figure 2H). Quantification of mitochondria network size revealed that the networks in AA-treated mice were approximately 1/5 the size of control mice, which is larger than mitochondria after ischemic injury (Figure 2F versus Figure 2H). These data demonstrate that the mitochondria in an uninjured tubule cell form one large network consisting of most of the mitochondria in the cell, which breaks down in acute or chronically injured tubule cells.
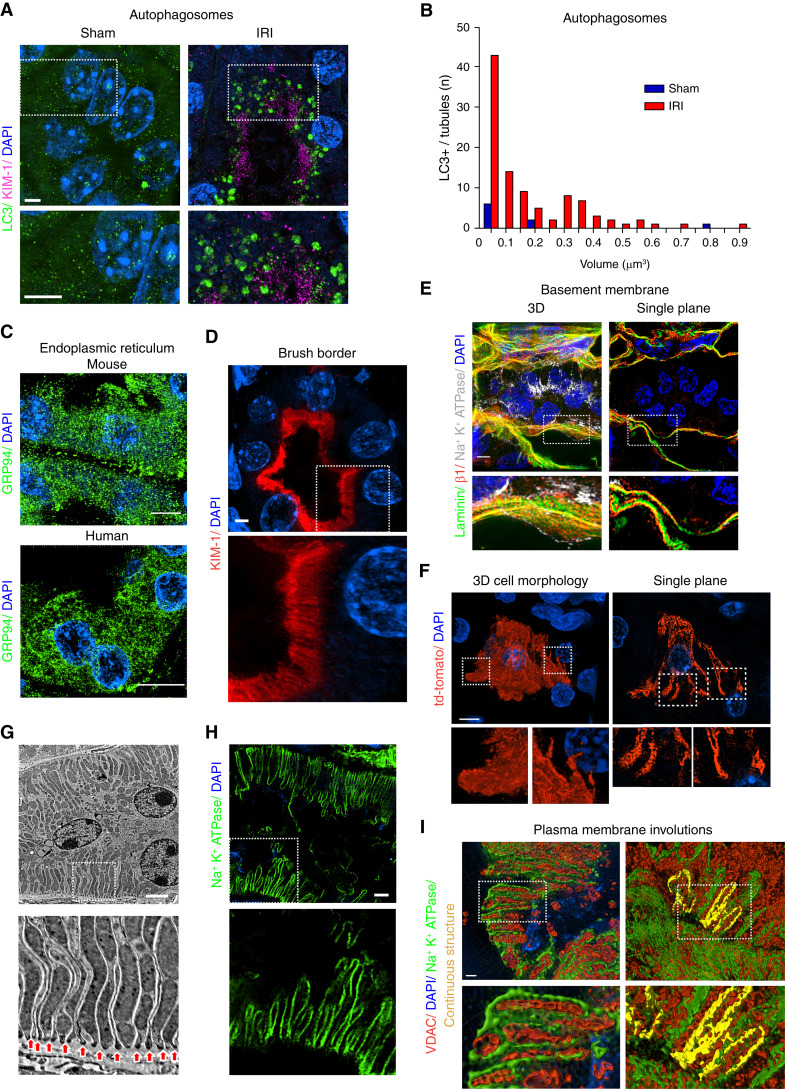
SIM Can Image Multiple Organelles and Cellular Structures in Tissue
Next, we sought to test whether super-resolution imaging could be used to image other subcellular organelles and structures. First, we examined staining of autophagosomes. Staining for LC3 revealed an increase in large LC3-positive structures after IRI (Figure 3A). Quantification revealed autophagosome number and size were increased in injured mice compared with control mice (Figure 3B). Next, we tested if the ER could be imaged by SIM. The staining for GRP94 revealed a large ER network in both mouse and human kidney samples (Figure 3C). Similarly, staining for KIM-1 (known to localize to the brush boarder [31]) revealed the structure of the brush border (Figure 3D). Also, we stained for components of the tubular basement membrane. Staining for laminin and integrin β1 revealed a complex structure of the extracellular matrix (Figure 3E).
Figure 3.
SIM imaging can resolve multiple organelles and cellular structures in tissue. (A) Representative SIM images of kidney tubules from mice subjected to sham or bilateral IRI surgery stained for LC3 (green). Scale bar, 3 μm. (B) Histogram comparing autophagosome number and size between sham and IRI kidneys. (C) SIM imaging of mouse or human kidney tissue stained for endoplasmic reticulum (ER) using GRP94 (green). (D) Staining of unilateral ureteral obstruction (UUO)–injured mouse kidney tissue for KIM-1 (red), highlighting the brush border. (E) Staining of basement membrane–associated proteins. Uninjured kidneys were stained for laminin (green), integrin β1 (red), and Na+K+ATPase (gray) and imaged by SIM. Maximum intensity projection of a kidney tubule and surrounding basement membrane is shown in left panel. A single z-plane from the same image is shown in right panel. Cutouts show the spatial separation of the proteins. (F) Left panel shows maximum intensity projection of a tdTomato (cytosolic)–positive proximal tubule cell (PTC). Right panels show a single z-plane from a tdTomato-positive tubular cell. (G) EM micrograph of kidney tubular cells highlighting interdigitations of the basolateral plasma membrane (arrows). (H) SIM imaging of tubular cell basolateral membranes. Basolateral plasma membrane is identified by staining for Na+K+ATPase (green). Scale bar, 3 μm. (I) Representative surface renderings of VDAC- (red) and Na+K+ATPase-stained (green) kidneys. Left panel shows high magnification of the basolateral side of a tubule with close association between the Na+K+ATPase and mitochondria. Right panel represents highlighted Na+K+ATPase positive volume showing continuous plasma membrane interdigitation. DAPI, 4′,6-diamidino-2-phenylindole. Dashed insets delineate the regions shown at higher magnification.
Next, we tested if we could image plasma membrane involutions. Although PTCs are often described as “simple cuboidal cells,” careful studies using EM to reconstruct the 3D architecture of PTCs confirmed a complex structure with many interdigitations between neighboring PTCs (32). By EM, the interdigitations are observed as numerous involutions of the basolateral plasma membrane (Figure 3G). Using sporadically labeled cells expressing a cytosolic tdTomato fluorescent protein, we imaged individual PTCs, revealing a complex morphology with many cellular processes (Figure 3F). Staining for Na+K+ATPase identifies the basolateral plasma membrane (33,34), resulting in a pattern very similar to the membrane involutions seen by EM (Figure 3, G and H). Surface renderings of the Na+K+ATPase- and VDAC-positive structures revealed an extremely close relationship between the Na+K+ATPase-positive membrane and the mitochondrial outer membrane (Figure 3I, left panel). Further, we highlighted the Na+K+ATPase membranes in surface renderings to determine if they were continuous. The Na+K+ATPase-positive membrane structures form a continuous loop that completely envelops some of the mitochondria, suggesting these mitochondria are in a cellular process from a neighboring cell (Figure 3I, right panel). Thus, super-resolution microscopy can resolve many different organelles and structures in kidney tissue, allowing for more detailed studies of kidney tissue at the subcellular level.
Analysis of Mitochondrial Content, Network Volume, and Morphology from Super-Resolution Images
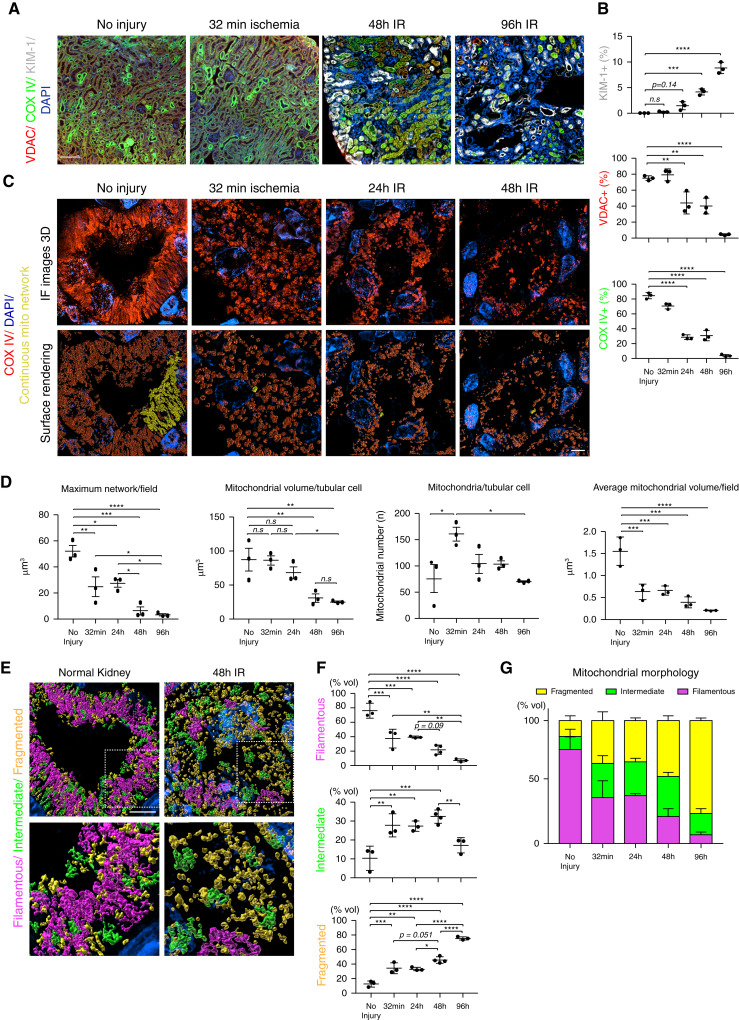
We next sought to determine if image analysis of tissue super-resolution images could identify changes in mitochondrial dynamics at greater detail than traditional techniques. To test this approach, we performed UIRI on wild-type C57BL/6 mice and collected kidneys immediately after injury (before reperfusion), and at 24, 48, and 96 hours post injury, and imaged the mitochondria as described above. Unlike bilateral IRI, kidneys subjected to UIRI have diminished recovery and atrophy over time. At low magnification, it is apparent that positive staining for mitochondrial markers VDAC and COX IV diminished over the course of the experiment, whereas expression of KIM-1, a marker of tubular injury, increased (Figure 4, A and B). Quantifying the VDAC- and COX IV–positive area revealed that, by 96 hours, <10% of the positive area remained, suggesting a loss of mitochondrial mass (Figure 4B). SIM imaging and analysis confirmed a dramatic decrease in mitochondrial volume with injury, which further decreased with time (Figure 4, C and D). Moreover, SIM imaging allowed for the analysis of individual mitochondrial network size and mitochondrial content on a per-cell basis (Figure 4D). To test if this technique is applicable to other organs, we imaged mitochondria in cardiomyocytes. Cardiomyocyte mitochondria also form large networks that are broken down with stress (Supplemental Figure 2, A and B).
Figure 4.
Analysis of SIM imaging can quantify mitochondrial content, network volume, and morphology. (A) Mouse kidney sections were stained for VDAC (red), COX IV (green), and KIM-1 (gray) and imaged at ×20 magnification. The resulting images were stitched together to reconstruct the whole kidney section. Scale bar, 100 μm. (B) Quantification of the images in (A) (n=3). (C) Representative maximum intensity projections (top panels) and surface renderings (bottom panels) of uninjured kidneys, 32 minutes and 24, 48, and 96 hours after unilateral IRI, stained for COX IV. Scale bar, 3 μm. (D) Quantification of mitochondrial volume and number from surface renderings in (C) (n=3). (E) Representative surface renderings color coded for network morphology on the basis of sphericity: fragmented, yellow; intermediate, green; and filamentous, magenta. Scale bar, 4 μm. (F) Quantification of mitochondrial morphology on the basis of sphericity. n=3–4. (G) Distribution of mitochondrial morphology over the time course of unilateral IRI. Data are presented as means±SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. DAPI, 4′,6-diamidino-2-phenylindole.
Whereas quantification of network size and content suggests alterations to mitochondrial morphology, it does not directly measure filamentous versus fragmented mitochondria. We hypothesized analyzing mitochondrial networks as a function of sphericity, i.e., greater sphericity indicates more fragmented mitochondria, would allow for a more direct quantification of mitochondrial morphology. As shown in Figure 4E, PTC mitochondria were divided into three groups by their sphericity and color labeled as follows: fragmented, sphericity >0.451 (yellow); intermediate, sphericity between 0.450 and 0.311 (green); and filamentous, sphericity <0.310 (magenta). We found that 32 minutes of ischemia induced significant mitochondrial fragmentation (Figure 4, E, F, and G). The percentage of fragmented mitochondria increased over time, with a corresponding decrease in filamentous mitochondria (Figure 4, F and G). A number of mitochondria were identified as having an intermediate morphology. These intermediate mitochondria tended to be filamentous in structure, but were not part of a larger network (Figure 4, F and G). These data demonstrate that super-resolution imaging and analysis can provide more detailed quantification of mitochondrial content and morphology.
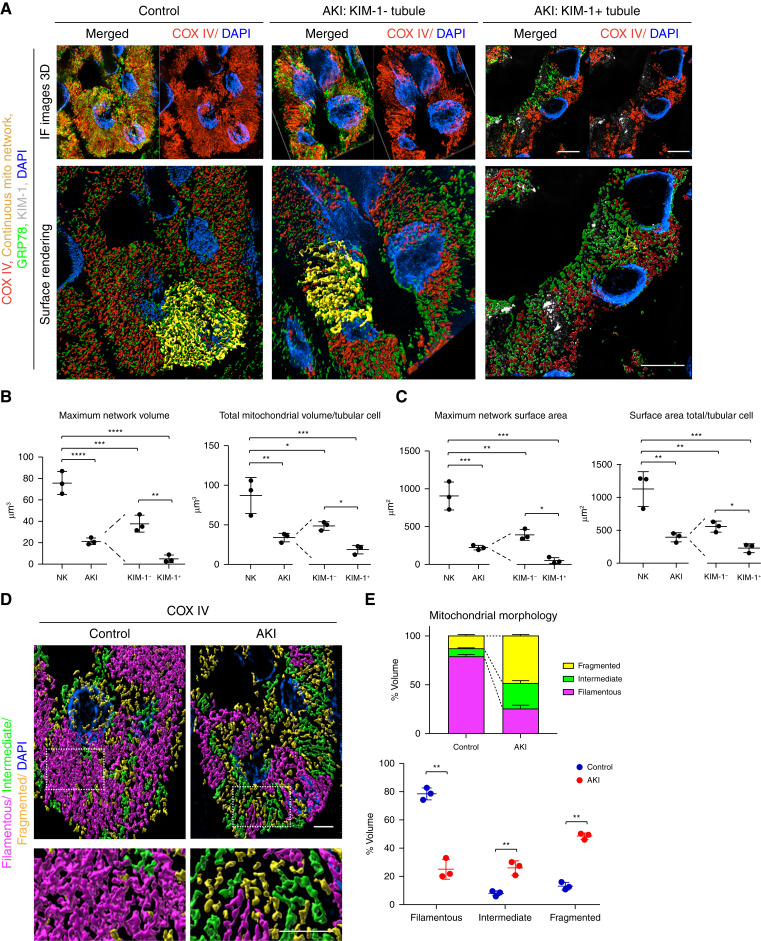
Quantification of Mitochondrial Morphology in Human Biopsy Specimens with Super-Resolution Microscopy
Mitochondrial fragmentation has been proposed as a driver of kidney disease in humans; however, quantifying mitochondrial morphology in human kidney tissue is challenging. To test if tissue super-resolution microscopy could be used to quantify mitochondrial changes in human samples, we stained biopsy samples with minimal acute tubular injury (control) or AKI with significant tubular injury (Supplemental Table 1) with COX IV. SIM resolved mitochondria in biopsy specimens similarly to that in mouse models (Figure 5A). Mitochondria from patients with AKI have reduced mitochondrial network volume and mitochondrial surface area compared with controls (Figure 5, B and C). Although there was an overall reduction in mitochondrial volume and surface area compared with control, it was unclear if this was due to a subset of injured tubules or if all tubules had reduced mitochondrial size. To test if mitochondria in injured tubules are more likely to be fragmented, we costained for KIM-1 to label injured PTCs. We found both KIM-1+ and KIM-1− tubule cells in the AKI samples had reduced mitochondrial volume and surface area, compared to controls (Figure 5, B and C). KIM-1+ tubules, however, had smaller mitochondria than tubules negative for KIM-1 (Figure 5, B and C). Next, we examined if the mitochondrial networks in the biopsy specimens could be categorized and quantified on the basis of sphericity. In the kidneys with minimal tubular damage, the mitochondrial networks were mostly filamentous and formed large networks (Figure 5, D and E). Biopsy specimens from patients with acute tubular injury had mostly fragmented or intermediate mitochondria (Figure 5, D and E). These data demonstrate that mitochondria in uninjured human kidney tubular cells form large networks that break down with kidney injury, and super-resolution imaging can be used to analyze organellar changes in human biopsy specimens.
Figure 5.
Quantification of mitochondrial morphology in human biopsy specimens with super-resolution microscopy. (A) Human biopsy specimens from patients with minimal tubular injury (control) and patients with AKI with pathologically confirmed tubule injury were stained for COX IV (red), GRP78 (green), and KIM-1 (gray) and imaged via SIM (maximum intensity projections, top panels). The resulting images were surface rendered (bottom panels). Scale bar, 5 μm (B) Quantification of mitochondrial volume on the basis of surface renderings. Maximum network volume=largest network/field. Total mitochondrial volume/tubular cell=total volume mitochondrial volume of a tubule/the number of tubular cell nuclei. Results were further subcharacterized into KIM-1+ and KIM-1− tubules (n=3). (C) Quantification of mitochondrial surface area on the basis of surface renderings. Maximum network surface area=surface area of largest network in a field. Surface area total/tubular cell=sum of the surface area of networks in a tubule/the number of tubular cell nuclei. Results were further subcharacterized into KIM-1+ and KIM-1− tubules (n=3). (D) Representative surface renderings color coded for mitochondrial morphology on the basis of the sphericity of the network: fragmented, yellow; intermediate, green; and filamentous, magenta. Dashed insets deliniate the regions shown at higher magnification. Scale bar, 4 μm. (E) Quantification of mitochondrial morphology on the basis of sphericity (n=3). Data are presented as means±SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. DAPI, 4′,6-diamidino-2-phenylindole; NK, normal kidney.
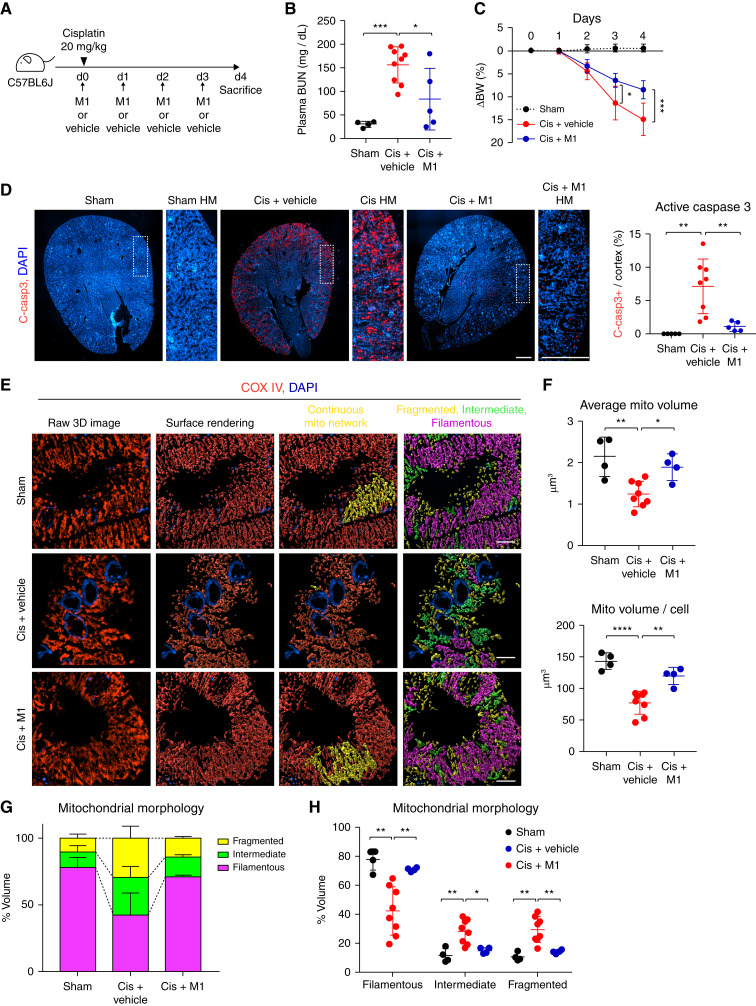
Promoting Mitochondrial Interconnectivity Protects against Cisplatin-Induced Kidney Injury
Our results thus far indicate that mitochondria in healthy kidney tubule cells form highly interconnected networks, which are lost in kidney injury. We hypothesized that preventing loss of mitochondrial interconnectivity would be protective in the setting of AKI. To test this hypothesis, we treated animals with cisplatin and added the mitochondrial fusion promotor M1, a compound that activates the mitochondrial fusion pathway (35). M1 was given at the same time as 20 mg/kg cisplatin, every 24 hours for 3 days (Figure 6A). M1 injection significantly reduced kidney injury, as measured by BUN andretained body weight, compared with the cisplatin plus vehicle group (Figure 6, B and C). M1 also significantly reduced apoptosis, as measured by active-caspase 3 positivity, in cisplatin-treated kidneys (Figure 6D). Quantification of mitochondrial volume revealed that M1 treatment preserved the average mitochondrial volume and mitochondrial volume per cell at or near baseline levels, despite the cisplatin exposure (Figure 6E). Mitochondrial morphology analysis revealed M1 treatment maintained mitochondrial network morphology and interconnectivity at similar levels as uninjured cells (Figure 6, E, F, G, and H). These data suggest that promoting mitochondrial fusion can maintain mitochondrial network morphology and interconnectivity and protect against kidney injury.
Figure 6.
Promoting mitochondrial interconnectivity protects against cisplatin-induced kidney injury. (A) A schematic diagram of cisplatin-induced kidney injury in the presence of M1 or vehicle. (B) Plasma BUN level on day 4 after administration with cisplatin (20 mg/kg) or vehicle: sham (vehicle; n=4), cisplatin plus vehicle (cis+vehicle; n=9), and cisplatin plus M1 (cis+M1; n=5). (C) Body weight (BW) (compared with day 0 [d0]) after injection of cisplatin in sham (n=4), cis+vehicle (n=8), and cis+M1 (n=5). (D) Representative images of cleaved caspase 3 (c-casp3)–stained (red) kidney and the corresponding data of c-casp3+ area/kidney cortex in sham (n=4), cis+vehicle (n=8), and cis+M1 (n=5). Dashed insets delineate the regions shown at higher magnification. Scale bar, 500 μm. (E) Representative maximum intensity projections (left panels) and surface renderings (middle and right panels) of kidney tubular mitochondria stained for COX IV (red) in control, cis+vehicle, and cis+M1 group. Scale bar, 3 μm. (F) Quantification of mitochondrial volume from images as in (E). Sham (n=4), cis+vehicle (n=8), and cis+M1 (n=4). (G) Distribution of mitochondrial morphology on the basis of sphericity, presented as percent volume of filamentous, fragmented, or intermediate mitochondria. (H) Quantification of mitochondrial morphology on the basis of sphericity. Sham (n=4), cis+vehicle (n=8), and cis+M1 (n=4). Data are presented as means±SD. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. DAPI, 4′,6-diamidino-2-phenylindole; mito, mitochondrial.
Discussion
The morphology of mitochondria and other organelles is known to change early during kidney injury (6,8,11,22,36). Although organellar morphologic dynamics are known to be both a marker of and contributor to kidney disease, they remain difficult to evaluate in kidney tissue. Recent advances have led to the development of numerous super-resolution imaging techniques that use optical and/or computational approaches to break the resolution limit of light microcopy. Here, we use tissue SIM–based super-resolution imaging to demonstrate that mitochondria form large, interconnected networks in kidney tubule cells, indicating that, under basal conditions, kidney tubule cell mitochondria represent a single, continuous membrane structure. Although this finding seems inconsistent with the classic textbook description of mitochondria, it is in line with findings in other organs and cultured mammalian cells (2,7,10,14,22,30). Our analysis used surface renderings of organelles, allowing for the digitization of each organellar unit and providing values/statistics of various features of the organelle. Using these data, we were not only able to demonstrate a loss of mitochondrial volume with injury, but also quantify the level of fragmentation of the mitochondria network in mouse and human kidney tissue. Costaining with KIM-1 revealed that, although injured PTCs have the largest reduction in mitochondrial volume and most mitochondrial fragmentation, KIM-1− (and presumably healthier) PTCs have a significant reduction in mitochondrial volume. Promoting mitochondrial fusion not only maintained mitochondrial morphology, volume, and interconnectivity, it also ameliorated cisplatin-induced kidney injury. Thus, the imaging and analysis techniques described here can be used to quantify organellar dynamics of multiple organelles in kidney tissue and other organs, such as the heart.
EM remains the gold standard for analyzing organellar morphology in kidney tissue and is vital to measure mitochondrial swelling, cristae alterations, autophagy, lysosomal disruptions, and other features. Understanding the 3D architecture of mitochondria by EM, however, remains challenging. The three most common approaches to using EM to quantify mitochondrial fragmentation in solid organs involve imaging thousands of cells to quantify rare long cross-sections of mitochondria, serial sections followed by segmentation and 3D reconstructions, or focused-ion-beam scanning EM with artificial intelligence–based segmentation (22,37,38). All of these processes are labor intensive and have low throughput, and focused-ion-beam scanning EM is only available at a small number of institutions. In kidney tubular cells, it is somewhat easier to visualize mitochondrial fragmentation by EM because the mitochondria tend to align perpendicularly to the basement membrane. In this study, we demonstrated that super-resolution imaging can fill a gap between traditional light microscopy techniques, such as confocal microscopy, and EM by providing enough resolution to quantify changes in organelle morphology while maintaining the high-throughput image analysis associated with light microscopy. Using a microscope with motorized x, y, and z controls, the image acquisition and analysis can be largely automated, removing potential bias from the experiment. The result is a semiautomated method to quantify changes in organelles and other structures at a subcellular level in kidney tissue. The super-resolution techniques used in this study, however, cannot provide detailed information with regard to cristae structure and other fine ultrastructural details. Thus, super-resolution imaging can be used in parallel with or to augment EM studies.
Perhaps the biggest advantage of this fluorescent imaging–based method is that it allows for the quantification of organellar morphology plus additional markers. For instance, one could combine the mitochondrial morphology analysis with cell type–specific markers or markers of injury. Another application would be to use genetic tracers to label cells that have been genetically modified. Combined with staining for up to four different targets, this technique can image multiple organelles at the same time in kidney tissue from mice or human biopsy specimens, or, when combined with genetic labeling approaches, can analyze organelles in genetically manipulated cells versus normal cells in the same tissue. For image analysis and quantification, we use Imaris software from Bitplane. This is a leading 3D analysis software for subcellular imaging. Using Imaris, we generated digital renderings of all mitochondria and/or other organelles (ER, brush border, etc.) within the kidney cells, allowing for the quantification of size, shape, intensity, colocalization, and position of each organelle in an unbiased manner.
Although we used SIM as the primary imaging technique in this study, it is important to note that many super-resolution imaging techniques can achieve the same level of resolution at a lower cost. The SIM instrument we used, Nikon N-SIM, has approximately double the resolution (115 nm) of traditional confocal or other light microscopy techniques. We found that a Nikon SoRa spinning disk confocal microscope, which achieves a similar resolution, can resolve tubular cell mitochondria and be quantified using the same analysis. Similarly, we were able to generate images of tubular cell mitochondria of sufficient resolution with a Nikon TiE widefield fluorescence microscope using SRRF imaging, which uses a software-based approach to generate super-resolution images at approximately double the resolution of widefield images. In addition, much of the analysis performed in Imaris can be performed in ImageJ, including surface rendering. Thus, with the combination of SRRF imaging and ImageJ analysis, a laboratory could perform the same level of imaging and analysis as described in this study using a standard widefield fluorescent microscope.
One key finding of this study is that kidney tubule cell mitochondria form large, nearly continuous networks within the cell. The overall 3D morphology of these networks was previously unknown; however, it is known that cells in culture form large networks, and interconnected mitochondria are healthier and have increased energy production (39–41). Thus, it makes sense for highly metabolic cells, like PTCs, to maintain interconnected mitochondrial networks. To demonstrate the importance of these interconnected networks, we enhanced network formation by promoting mitochondrial fusion. Promoting mitochondrial fusion both prevented the breakdown of mitochondrial networks and ameliorated development of cisplatin-induced kidney injury. Promoting mitochondrial fusion is an attractive alternative to methods that inhibit mitochondrial fission used in previous studies, because blocking mitochondrial fragmentation can have a negative effect on cell division. Taken together, these data indicate promoting mitochondrial fusion helps maintain mitochondrial interconnectivity and can be a potential therapeutic approach to treating AKI.
Here, we demonstrate the ability of super-resolution microscopy to capture the dynamics of kidney biology and pathobiology at the subcellular level. Applying super-resolution imaging to kidney tissue opens fields of study in animal models and human kidney tissue that have previously been relegated to tissue culture models. Now the structure, size, position, morphology, and interactions of multiple organelles can be analyzed at the same time. Applying tissue super-resolution imaging to human kidney tissue can provide unparalleled insight into the organellar alterations that occur in kidney disease, allowing researchers to confirm hypotheses in human kidney tissue and, potentially, aid in clinical diagnoses. Thus, tissue super-resolution imaging has the potential to revolutionize the way researchers and clinicians interrogate kidney disease.
Disclosures
C.R. Brooks reports having ownership interest in DermYoung LLC. A.B. Fogo reports serving as speaker at various national nephrology meetings, Associate Editor Kidney International, and as president elect of the International Society of Nephrology (president as of April 19, 2021); serving on the editorial board for American Journal of Physiology-Renal, Human Pathology, and JASN, on an advisory committee for Bayer, as guest editor for the yearly pathology focus of Current Opinion in Nephrology and Hypertension, as associate editor of Kidney International and Laboratory Investigation, as subject editor of Nephrology Dialysis Transplantation, and as a scientific advisor for/member of The American Journal of Pathology; receiving honoraria from Amgen, GlaxoSmithKline, and Novartis; receiving research funding from Bayer, Gilead, and Novartis; and having consultancy agreements with Novartis. All remaining authors have nothing to disclose.
Funding
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grants DK114809 and DK121101 (to C.R. Brooks) and American Heart Association 20POST35200221 (to K. Taguchi).
Acknowledgments
We thank Dr. Benjamin Humphreys for the use of the KIM-1 Cre mice and Vanderbilt Cell Imaging Shared Resource and Nikon Center of Excellence (supported by NIH grants DK020593, CA68485, DK20593, DK58404, DK59637, and EY08126).
Footnotes
This article contains a podcast at https://www.asn-online.org/media/podcast/K360/2021_12_30_KID0001602021.mp3.
Author Contributions
C.R. Brooks was responsible for funding acquisition and project administration; C.R. Brooks, B.C. Elias, S. Qian, S. Sant, and K. Taguchi were responsible for formal analysis and investigation; C.R. Brooks, A.B. Fogo, E. Krystofiak, K. Taguchi, and H. Yang were responsible for methodology; C.R. Brooks, A.B. Fogo, and K. Taguchi conceptualized the study; C.R. Brooks, A.B. Fogo, K. Taguchi, B.C. Elias and H. Yang reviewed and edited the manuscript; C.R. Brooks, A.B. Fogo, and H. Yang were responsible for resources; C.R. Brooks and K. Taguchi wrote the original draft and were responsible for data curation, validation, and visualization; and E. Krystofiak provided supervision.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0001602021/-/DCSupplemental.
SRRF and SoRa imaging can resolve tubule cell mitochondria and be used for quantitative analysis of mitochondrial volume. Download Supplemental Figure 1, PDF file, 6.4 MB (6.3MB, pdf)
Tissue SIM can resolve cardiomyocyte mitochondria and quantify mitochondrial surface area alterations with injury. Download Supplemental Figure 2, PDF file, 6.4 MB (6.3MB, pdf)
Characteristics and diagnosis of patients with or without tubular injury. Download Supplemental Table 1, PDF file, 6.4 MB (6.3MB, pdf)
Video showing a full 3D rotation of the single mitochondrion imaged by SIM in Figure 2C. Download Supplemental Video 1, MP4 file, 7.5 MB (7.4MB, mp4)
Video showing a full 3D rotation of the single mitochondrion imaged in Figure 2G. Download Supplemental Video 2, MP4 file, 5.4 MB (5.3MB, mp4)
References
- 1.World Kidney Day : Chronic Kidney Disease. Available at: http://www.worldkidneyday.org/faqs/chronic-kidney-disease/. Accessed July 29, 2021
- 2.Tang C, Cai J, Yin XM, Weinberg JM, Venkatachalam MA, Dong Z: Mitochondrial quality control in kidney injury and repair. Nat Rev Nephrol 17: 299–318, 2020. 10.1038/s41581-020-00369-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison SJ: Uromodulin in ER stress and apoptosis. Nat Rev Nephrol 13: 722, 2017. 10.1038/nrneph.2017.152 [DOI] [PubMed] [Google Scholar]
- 4.Maekawa H, Inagi R: Stress signal network between hypoxia and ER stress in chronic kidney disease. Front Physiol 8: 74, 2017. 10.3389/fphys.2017.00074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, do Carmo JM, da Silva AA, Fu Y, Hall JE: Mechanisms of synergistic interactions of diabetes and hypertension in chronic kidney disease: Role of mitochondrial dysfunction and ER stress. Curr Hypertens Rep 22: 15, 2020. 10.1007/s11906-020-1016-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks C, Dong Z: Regulation of mitochondrial morphological dynamics during apoptosis by Bcl-2 family proteins: A key in Bak? Cell Cycle 6: 3043–3047, 2007. 10.4161/cc.6.24.5115 [DOI] [PubMed] [Google Scholar]
- 7.Brooks C, Wei Q, Feng L, Dong G, Tao Y, Mei L, Xie ZJ, Dong Z: Bak regulates mitochondrial morphology and pathology during apoptosis by interacting with mitofusins. Proc Natl Acad Sci U S A 104: 11649–11654, 2007. 10.1073/pnas.0703976104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inoue T, Maekawa H, Inagi R: Organelle crosstalk in the kidney. Kidney Int 95: 1318–1325, 2019. 10.1016/j.kint.2018.11.035 [DOI] [PubMed] [Google Scholar]
- 9.Ward JM, Stoyas CA, Switonski PM, Ichou F, Fan W, Collins B, Wall CE, Adanyeguh I, Niu C, Sopher BL, Kinoshita C, Morrison RS, Durr A, Muotri AR, Evans RM, Mochel F, La Spada AR: Metabolic and organelle morphology defects in mice and human patients define spinocerebellar ataxia type 7 as a mitochondrial disease. Cell Rep 26: 1189–1202.e6, 2019. 10.1016/j.celrep.2019.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wienke DC, Knetsch ML, Neuhaus EM, Reedy MC, Manstein DJ: Disruption of a dynamin homologue affects endocytosis, organelle morphology, and cytokinesis in Dictyostelium discoideum. Mol Biol Cell 10: 225–243, 1999. 10.1091/mbc.10.1.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen S, Valm AM, Lippincott-Schwartz J: Interacting organelles. Curr Opin Cell Biol 53: 84–91, 2018. 10.1016/j.ceb.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vernier RL, Papermaster BW, Good RA: Aminonucleoside nephrosis. I. Electron microscopic study of the renal lesion in rats. J Exp Med 109: 115–126, 1959. 10.1084/jem.109.1.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandra S: The reversal of mitochondrial membrane. J Cell Biol 12: 503–513, 1962. 10.1083/jcb.12.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamano K, Youle RJ: Coupling mitochondrial and cell division. Nat Cell Biol 13: 1026–1027, 2011. 10.1038/ncb2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhan M, Brooks C, Liu F, Sun L, Dong Z: Mitochondrial dynamics: Regulatory mechanisms and emerging role in renal pathophysiology. Kidney Int 83: 568–581, 2013. 10.1038/ki.2012.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks C, Cho SG, Wang CY, Yang T, Dong Z: Fragmented mitochondria are sensitized to Bax insertion and activation during apoptosis. Am J Physiol Cell Physiol 300: C447–C455, 2011. 10.1152/ajpcell.00402.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abrisch RG, Gumbin SC, Wisniewski BT, Lackner LL, Voeltz GK: Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J Cell Biol 219: e201911122, 2020. 10.1083/jcb.201911122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, Duncan SR, Rojas M, Shiva S, Chu CT, Mora AL: PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. J Clin Invest 125: 521–538, 2015. 10.1172/JCI74942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Brito OM, Scorrano L: Mitofusin 2 tethers endoplasmic reticulum to mitochondria [published correction appears in Nature 516: 266, 2014]. Nature 456: 605–610, 2008. 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- 20.Kawakami T, Gomez IG, Ren S, Hudkins K, Roach A, Alpers CE, Shankland SJ, D’Agati VD, Duffield JS: Deficient autophagy results in mitochondrial dysfunction and FSGS. J Am Soc Nephrol 26: 1040–1052, 2015. 10.1681/ASN.2013111202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson L: The ultrastructure of the developing proximal tubule in the rat kidney. J Ultrastruct Res 51: 119–139, 1975. 10.1016/S0022-5320(75)80013-X [DOI] [PubMed] [Google Scholar]
- 22.Brooks C, Wei Q, Cho SG, Dong Z: Regulation of mitochondrial dynamics in acute kidney injury in cell culture and rodent models. J Clin Invest 119: 1275–1285, 2009. 10.1172/JCI37829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranjit S, Lanzano L, Libby AE, Gratton E, Levi M: Advances in fluorescence microscopy techniques to study kidney function. Nat Rev Nephrol 17: 128–144, 2020. 10.1038/s41581-020-00337-8 [DOI] [PubMed] [Google Scholar]
- 24.Pullman JM, Nylk J, Campbell EC, Gunn-Moore FJ, Prystowsky MB, Dholakia K: Visualization of podocyte substructure with structured illumination microscopy (SIM): A new approach to nephrotic disease. Biomed Opt Express 7: 302–311, 2016. 10.1364/BOE.7.000302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Y, Shroff H: Faster, sharper, and deeper: Structured illumination microscopy for biological imaging [published correction appears in Nat Methods 16: 205, 2019]. Nat Methods 15: 1011–1019, 2018. 10.1038/s41592-018-0211-z [DOI] [PubMed] [Google Scholar]
- 26.Siegerist F, Ribback S, Dombrowski F, Amann K, Zimmermann U, Endlich K, Endlich N: Structured illumination microscopy and automatized image processing as a rapid diagnostic tool for podocyte effacement. Sci Rep 7: 11473, 2017. 10.1038/s41598-017-11553-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angelotti ML, Antonelli G, Conte C, Romagnani P: Imaging the kidney: From light to super-resolution microscopy. Nephrol Dial Transplant 36: 19–28, 2019. 10.1093/ndt/gfz136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wunderlich LCS, Ströhl F, Ströhl S, Vanderpoorten O, Mascheroni L, Kaminski CF: Superresolving the kidney-A practical comparison of fluorescence nanoscopy of the glomerular filtration barrier. Anal Bioanal Chem 413: 1203–1214, 2020. 10.1007/s00216-020-03084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheung EC, McBride HM, Slack RS: Mitochondrial dynamics in the regulation of neuronal cell death. Apoptosis 12: 979–992, 2007. 10.1007/s10495-007-0745-5 [DOI] [PubMed] [Google Scholar]
- 30.Mishra P, Varuzhanyan G, Pham AH, Chan DC: Mitochondrial dynamics is a distinguishing feature of skeletal muscle fiber types and regulates organellar compartmentalization. Cell Metab 22: 1033–1044, 2015. 10.1016/j.cmet.2015.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks CR, Yeung MY, Brooks YS, Chen H, Ichimura T, Henderson JM, Bonventre JV: KIM-1-/TIM-1-mediated phagocytosis links ATG5-/ULK1-dependent clearance of apoptotic cells to antigen presentation. EMBO J 34: 2441–2464, 2015. 10.15252/embj.201489838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welling LW, Welling DJ: Shape of epithelial cells and intercellular channels in the rabbit proximal nephron. Kidney Int 9: 385–394, 1976. 10.1038/ki.1976.48 [DOI] [PubMed] [Google Scholar]
- 33.Jørgensen PL: Structure, function and regulation of Na,K-ATPase in the kidney. Kidney Int 29: 10–20, 1986. 10.1038/ki.1986.3 [DOI] [PubMed] [Google Scholar]
- 34.Xie JX, Li X, Xie Z: Regulation of renal function and structure by the signaling Na/K-ATPase. IUBMB Life 65: 991–998, 2013. 10.1002/iub.1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D, Wang J, Bonamy GM, Meeusen S, Brusch RG, Turk C, Yang P, Schultz PG: A small molecule promotes mitochondrial fusion in mammalian cells. Angew Chem Int Ed Engl 51: 9302–9305, 2012. 10.1002/anie.201204589 [DOI] [PubMed] [Google Scholar]
- 36.Ricciardi CA, Gnudi L: The endoplasmic reticulum stress and the unfolded protein response in kidney disease: Implications for vascular growth factors. J Cell Mol Med 24: 12910–12919, 2020. 10.1111/jcmm.15999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Gräber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E: Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J 25: 3900–3911, 2006. 10.1038/sj.emboj.7601253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C, Chen X, Li W, Li L, Wang L, Xie Q, Han H: Automatic mitochondria segmentation for EM data using a 3D supervised convolutional network. Front Neuroanat 12: 92, 2018. 10.3389/fnana.2018.00092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattie S, Krols M, McBride HM: The enigma of an interconnected mitochondrial reticulum: new insights into mitochondrial fusion. Curr Opin Cell Biol 59: 159–166, 2019. 10.1016/j.ceb.2019.05.004 [DOI] [PubMed] [Google Scholar]
- 40.Rafelski SM: Mitochondrial network morphology: Building an integrative, geometrical view. BMC Biol 11: 71, 2013. 10.1186/1741-7007-11-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westermann B: Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta 1817: 1833–1838, 2012. 10.1016/j.bbabio.2012.02.033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SRRF and SoRa imaging can resolve tubule cell mitochondria and be used for quantitative analysis of mitochondrial volume. Download Supplemental Figure 1, PDF file, 6.4 MB (6.3MB, pdf)
Tissue SIM can resolve cardiomyocyte mitochondria and quantify mitochondrial surface area alterations with injury. Download Supplemental Figure 2, PDF file, 6.4 MB (6.3MB, pdf)
Characteristics and diagnosis of patients with or without tubular injury. Download Supplemental Table 1, PDF file, 6.4 MB (6.3MB, pdf)
Video showing a full 3D rotation of the single mitochondrion imaged by SIM in Figure 2C. Download Supplemental Video 1, MP4 file, 7.5 MB (7.4MB, mp4)
Video showing a full 3D rotation of the single mitochondrion imaged in Figure 2G. Download Supplemental Video 2, MP4 file, 5.4 MB (5.3MB, mp4)