Abstract
Beauveria bassiana, as a well-studied entomopathogenic fungus, has a great potential for the biological control of insect pests. Lipid metabolism has been linked to the life cycle of B. bassiana; however, the underlying mechanisms remain unknown. In this study, a homolog of yeast steryl acetyl hydrolase 1 (Say1) was functionally characterized. The loss of B. bassianaSAY1 (BbSAY1) impaired the lipid homeostasis in conidia, with a significant reduction in oleic acid content. The ΔBbsay1 mutant strain displayed anelevated accumulation of lipid bodies and aweakened membrane permeability. As for phenotypic aspects, gene loss resulted in significant defects in germination, conidiation, and virulence. Our findings highlight that Say1, involved in lipid homeostasis, contributes to the cytomembrane integrity, development, and virulence in B. bassiana.
Keywords: steryl acetyl hydrolase 1, lipid homeostasis, entomopathogenic fungus, development, virulence
1. Introduction
Beauveria bassiana is one of the most prevalent insect pathogens in the eco-system and has great potential for the biological control of pests [1,2]. Conidia, the major form of infectious cells, germinate and ingress the host body through the trans-cuticular route. In the host hemocoel, fungal cells undergo replication and overcome the host defense [3,4]. After the host dies, hyphae grow outside the host body and produce a lot of conidia on the cadaver. The newly-born conidia will initiate the subsequent infection cycle when encountering the sensitive hosts [5].
In B. bassiana, conidia form on the ‘zig-zag’-shaped conidiophores and accumulate a plethora of fatty acids (FAs) and lipids [6,7]. The homeostasis of lipid metabolism is finely tuned by a series of processes of synthesis and degradation. FAs act as the synthetic precursors of storage lipids and membranes [8] and can be recycled from the degradation of lipids and membranes [9]. Oleic acid (OA) is an important unsaturated FA in fungal cells and is synthesized through the OLE pathway, in which the Δ9-fatty acid desaturase gene (Ole1) catalyzes the desaturation of palmitic acid (PA), critical for the synthesis of unsaturated fatty acids [10]. HapX represents a family of basic leucine zipper (bZIP) transcription factors, which are indispensable for iron acquisition in fungi [7,11]. In B. bassiana, the OLE pathway also contributes to conidial storage of OA and is transcriptionally regulated by HapX (BbHapX). In addition, BbHapX has a greater influence on lipid homeostasis than BbOle1 [7]. However, the mechanisms underlying the lipid homeostasis remains enigmatic. In budding yeast, steryl acetyl hydrolase 1 (Say1) is required for lipid homeostasis [12]. We supposed that the ortholog of yeast Say1 might be required for lipid homeostasis in B. bassiana.
In this study, we functionally characterized Say1 in B. bassiana by constructing the gene disruption and complementation mutant strains. Our data revealed that BbSAY1 contributes to conidial lipid reserves, formation, and virulence.
2. Materials and Methods
2.1. Microbial Strains and Cultivation
The wild-type strain of B. bassiana ARSEF2860 was cultured on Sabouraud dextrose agar (SDAY: 4% glucose, 1% peptone, 1.5% agar, and 1% yeast extract) for routine maintenance. Escherichia coli DH5α (Invitrogen, Carlsbad, CA, USA) for plasmid proliferation was cultured in Luria-Bertani (LB) medium with appropriate antibiotics. A yeast extract broth (w/v: 0.5% sucrose, 1% protein, 0.1% yeast extract, 0.05% MgSO4, and 1.5% agar) was used to culture Agrobacterium tumefaciens AGL-1, which was used in fungal transformation. Czapek-Dox agar (CZA) (3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4, and 0.001% FeSO4 plus 1.5% agar) was used as a chemical-defined medium.
2.2. Bioinformatic and Transcriptional Analyses of BbSay1 in B. bassiana
The protein sequence of S. cerevisiae Say1 (GenBank No.: DAA08353) was used as a query to search the potential ortholog in the B. bassiana genome [13]. After mapping the cDNA sequence of BbSay1, a complete open reading frame with upstream and downstream flanking sequences was obtained. Domain annotation was conducted through an online portal SMART (http://smart.embl-heidelberg.de/) (last accessed on: 9 September 2020). The ortholog of yeast Say1 in B. bassiana was determined with the method of reciprocal best alignment between two species [14]. After searching all the Say1-domain proteins in B. bassiana, their homologs in other fungi were retrieved via BLAST analyses and proofed via domain analysis. Tested fungal species included Aspergillus nidulans FGSC A4, A. niger CBS 513.88, A. fumigatus Af293, Candida albicans SC5314, Cordyceps militaris CM01, Magnaporthe grisea 70-15, Metarhizium acridumCQMa 102, M. robertsii ARSEF 23, S. cerevisiae S288C, and Yarrowia lipolytica CLIB122.
The transcriptional analyses of the Say1-domain protein genes were performed as the methods documented [15]. Conidia of the wild-type were cultured on a SDAY plate at 25 °C, and mycelia were sampled at 2, 3, 4, and 5 d. Total RNA was extracted from the mycelial samples with the RNAisoTM Plus Reagent (TaKaRa, Dalian, China), and cDNA was reverse transcribed using the PrimeScript® RT reagent Kit (TaKaRa). The resultant cDNA was used as templates to perform the qRT-PCR reaction on a Mastercycler® EP Realplex (Eppendorf, Hamburg, Germany) cycler. All primers are listed in Table S1. The relative transcript level of each gene was calculated as the relative expression of different time points over 2 d using the fungal 18S rRNA as an internal reference and the 2−ΔΔCt method [16].
Localization of BbSay1 was performed as described previously [17]. Primers sequences were listed in Table S2. The coding sequence of BbSay1 was amplified with primers PL1 and PL2, and then, it was fused to the green fluorescent protein gene (GFP) by ligating into the NcoI/EcoRI sites of pBMGB. The resulting vector was transformed into the wild-type strain. The transformant was screened on CZA plates with 200 µg/mL glufosinate and cultured on an SDAY plate at 25 °C. Fungal cells were sampled, and the fluorescent signals were observed under a laser scanning confocal microscope (LSM 710, Carl Zeiss Microscopy GmbH, Jena, Germany).
2.3. Gene Disruption and Complementation
The methods for gene disruption and complementation were the same as those described previously [18]. All primers arelisted in Supporting Information Table S2. Upstream (0.81 kb) and downstream (1.14 kb) flanking sequences of the BbSAY1 open reading frame (ORF) were amplified by PCR with the paired primers P1/P2 and P3/P4, respectively, using genomic DNA as the template. The amplified PCR fragments were digested with EcoRⅠ/BamHⅠ and XbaⅠ/HpaⅠ, respectively, and then successively cloned into the EcoRI/BamHI and XbaⅠ/HpaⅠ sites of p0380-bar [19]. The resulting plasmid was named p0380-bar-Bbsay1 and used for gene disruption. The candidate disruptants were screened by PCR with the primers P5 and P6. To complement the gene loss, the intact BbSAY1 was ectopically integrated in the gene disruption mutant strain. The BbSAY1 ORF plus 1.85 kb of upstream and 0.34 kb of downstream sequences was amplified with primers P7/P8, and the resultant fragment was recombined into the vector p0380-sur-gateway with a sulfonylurea resistance cassette [19], generating the plasmid p0380-sur-Bbsay1. The plasmids were transformed into the B. bassiana strain using an Agrobacterium-mediated transformation method. Putative disruption mutants were screened on CZA supplemented with phosphinothricin (200 μg/mL), and the complemented strains were screened on CZA plates with 10 mg/mL chlorimuron ethyl.
Southern blotting analyses were used to further confirm the transformants. Fungal genomic DNA (30 μg) digested by SacⅠ/NcoⅠ were resolved on a 0.7% agarose gel and then electronically transferred to a Biodyne B nylon membrane (Gelman Laboratory, Shelton, WA, USA). Target fragments were hybridized with the probes prepared from the templates amplified with the primer pair P9/P10 and visualized with the DIG-High Prime DNA Labeling and Detection Starter Kit II (Roche, Penzberg, Germany).
2.4. Biochemical and Phenotypic Assays
The indicated strain was cultured on SDAY plates at 25 °C for 7 days, and the resultant conidia were suspended in a 0.02% Tween-80 solution. All phenotypes were compared among the wild-type, ΔBbsay1, and complementation mutant strains with three parallel experiments [7,20].
2.4.1. Quantification of Conidial FAs
Conidia (100 mg) were suspended in 10 mLCMW mixture (chloroform:methanol:water = 2:1:1, v/v/v) and placed at −20 °C for 2 h. Then, 3 mLCM mixture (chloroform:methanol = 2:1, v/v) was added and the conidial suspension stood for 1 h. The bottom layer was carefully collected and dried. FAs were methylated and analyzed on a Focus series gas chromatograph (Thermo Scientific, Waltham, MA, USA), using a standard mixture of fatty acid methyl esters (47885U) (Sigma, St. Louis, MO, USA) as the internal references.
2.4.2. Vegetative Growth
Fungal radial growth was determined on CZA and SDAY plates. Conidial suspension (1 µL, 106 conidia/mL) was inoculated on the plates. After 7 d of incubation at 25 °C, the colony diameter was examined. A feeding assay was conducted by adding oleic acid (0.3%) in medium. Oxidative stress was caused by including manedione (0.02 mM) into CZA plates. Conidial suspension was point inoculated on the plates and cultured at 25 °C.The colony diameter was determined at 7 d. Relative growth inhibition was calculated using the diameter on the CZA plate as a control.
2.4.3. Conidial Production
An aliquot of conidial suspension (100 μL, 107 conidia/mL) was uniformly smeared on SDAY plates and incubated at 25 °C for 9 d. From the sixth day after incubation, a mycelial disc (5 mm in diameter) was sampled, and conidia on the disc were washed into a 0.02% Tween-80 solution by vigorous vortexing. The conidial concentration of the suspension was quantified and used to calculate the conidial yield as the number of conidia per square centimeter. In the feeding assay, 0.3% oleic acid was added to the SDAY plates, and conidial production was also quantified as described above. Additionally, a transformant for the sub-cellular localization of BbSay1 was used to evaluate the effect of the over-expression of BbSAY1 on conidiation.
2.4.4. Conidia Germination
To simulate conidial germination under an oligotrophic condition, the conidial suspension was inoculated on a water agar plate (WA) (1.2% agarose). After an incubation of 24 h at 25 °C, the germination percentage was calculated. For each sample, at least 100 conidia were counted. A chemical feeding test was performed by adding oleic acid (0.3%) into the WA plates. To simulate conidial germination on the host cuticle, a conidial suspension (107 cells/mL) was sprayed on the locust hindwings. After an incubation of 24 h at 25 °C, the conidial morphology was observed under a microscope.
2.4.5. Insect Bioassay
To determine fungal virulence, the larvae of Galleria mellonella were used as the insect hosts. Last-instar larvae (~300 mg in weight) were used in this study [21]. In the trans-cuticular assay, larvae (30−35 individuals) were dipped in a conidial suspension (40 mL, 107 conidia/mL) for 15 s. All infected insects were reared at 25 °C, and the mortality was recorded daily. The median lethal time (LT50) was calculated by a Probit analysis for three replicates of each bioassay. In the feeding test, oleic acid (0.3%) was added into the conidial suspension.
2.5. Lipidomic Analyses
The total lipid sample was prepared with the methods used in FA extraction as described previously [7]. Lipids in the sample were resolved and analyzed on UPLC-MS/MS (QE Plus™) (Thermo Scientific). A non-targeted lipidomics analysis platform combined with the Lipid Search™ software was applied for lipid identification and data processing. Data for the molecule intensity were plotted with the CV% method, which indicatedthe difference in the relative content in the total lipids between the WT and ΔBbsay1 mutant strains.
To view the variation of total lipids in mycelia, a conidial suspension was grown on the SDAY for 3 d at 25 °C. The resulting mycelia were stained with observed hyphae with the lipid-specific dye BODIPY493/503 (Thermo Fisher Scientific, Waltham, MA, USA).
2.6. Assay for Membrane Integrity
Nucleus staining with SYTOX Green was applied as previously described [7]. The conidia suspension was inoculated onto the SDAY plates and cultured for 12 h at 25 °C. The resultant germlings were collected and stained with SYTOX Green for 10 min away from light. The green fluorescence was detected under a laser scanning confocal microscope (LSM 710, Carl Zeiss Microscopy GmbH, Jena, Germany), and the percentage of stained cells (PSC) was calculated by counting at least 100 cells.
2.7. Data Analyses
All measurements from biochemical and phenotypic assays were subjected to a one-way ANOVA. The significant difference was determined by a Tukey’s honest significance test (Tukey’s-HSD).
3. Results
3.1. Bioinformatic Analysis and Molecular Manipulation of BbSay1
Based on the results of the BLAST search, the gene of BBA_02920 was significantly homologous to steryl acetyl hydrolase 1 in S. cerevisiae (GenBank No.: DAA08353) (E-value: 1 × 1010) and was designated as BbSay1. The open rearing frame of BbSAY1 was 1453 bp in length with 3 introns, encoding a 386-amino acid protein. The domain analysis indicated that BbSay1 contained a domain of Say1_Mug180 (PF10340.9), and the domain annotation revealed that there were 10 Say1-domain-containing proteins in the B. bassiana genome. Their orthologs in other fungi were revealed via sequence alignment and domain annotation analyses (Table S3). Phylogenic analyses indicated that BbSay1 has a closer relationship to the orthologs from filamentous fungi than those of yeast species (Figure S1A). qRT-PCR analyses indicated that the Say1-domain protein genes displayed different expression patterns during growth in the nutrient medium. (Figure S1B). Under a fluorescent field, the green signals of the BbSay-Gfp protein distributed evenly in cytoplam (Figure S2). This indicated that BbSay1 dominated in the cytoplasm.
To uncover the physiological functions of BbSay1, its disruption mutant was constructed with a strategy of homologous replacement. To complement the gene loss, the entire ORF of BbSAY1 and its promoter region was ectopically integrated in the genome of the ΔBbsay1 mutant strain. All candidate strains were screened, verified with PCR, and successfully confirmed by Southern blotting analyses (Figure S3).
3.2. Effects of Gene Loss on Fungal Phenotypes
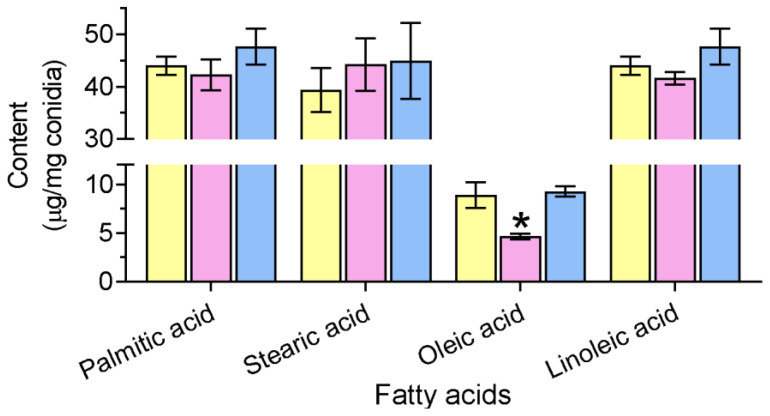
The B. bassiana conidia mainly accumulated four FAs, including stearic, palmitic, oleic, and linoleic acids. The ablation of BbSAY1 resulted in a significant reduction in the content of oleic acid, and the reduction was 50.2%. However, the contents of the other three FAs were not significantly influenced (Figure 1).
Figure 1.
Fatty acid content in the B. bassiana conidia. The indicated strain was cultured on SDAY plates for 8 d, and the levels of fatty acids were determined in the conidia. The B. bassiana conidia accumulated four free fatty acids (stearic, palmitic, oleic, and linoleic acid) in the conidia. The gene disruption of BbSAY1 led to a significant decrease in the level of oleic acid. The asterisk on the column indicates the significant difference in the fatty acid content between the ΔBbsay1 and the wild-type or complemented strains (Tukey’s HSD: p < 0.05). Error bars: standard deviation.
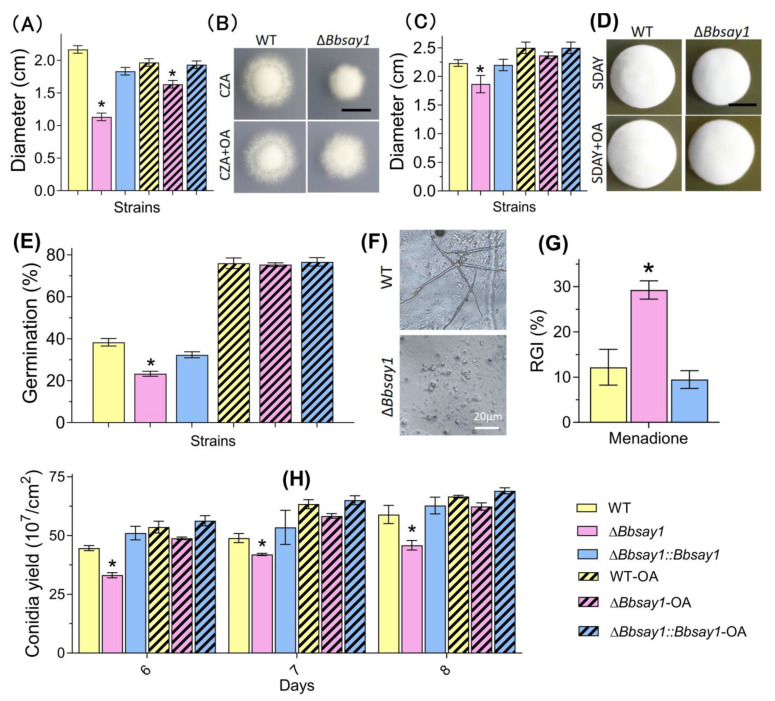
As shown in Figure 2A,C, the ΔBbsay1 mutant strain exhibit a significant decrease in colony size, with approximatelya 48.6% and 9.1% reduction on the CZA and SDAY plates, respectively, when compared with those of the wild-type (2.17 ± 0.05 cm (CZA) and 2.23 ± 0.05 cm (SDAY)) (mean ± standard deviation). Feeding oleic acid could increase the colony sizes on both the CZA and SDAY plates. This indicated that exogenous oleic acid could restore vegetative growth under an aerial condition. The colony morphologies on the CZA and SDAY plates areshown in Figure 2B and Figure 2D, respectively.
Figure 2.
Fungal vegetative growth, germination, and conidiation. Fungal strains were inoculated on CZA (A) and SDAY (C) plates. After an incubation of 7 d at 25 °C, the colony diameter was examined. Photographs were taken for the colonies on the CZA (B) and SDAY (D) plates. Bars: 1 cm. (E) Conidial germination under an oligotrophic condition. The conidia were inoculated on a water agar plate (only with agarose) and cultured for 24 h at 25 °C, followed by determining the germination percentage. (F) Microscopic view of germlings on the hindwing of a locust. Bar: 20 μm. (G) Growth under oxidative stress. The conidial suspension was inoculated on a CZA plate plus menadione and cultured at 25 °C. Seven days later, the colony diameter was measured and used to calculate the relative growth inhibition (RGI). A CZA plate without manedione was used as a control. (H) Conidial production. Fugal strains were inoculated on SDAY plates and cultured at 25 °C. The conidial yield was examined at 6, 7, and 8 d post incubation. A chemical feeding assay was conducted by adding oleic acid (OA) to the indicated medium. The asterisks on the bars indicate the significant difference in the indicated phenotype between the ΔBbsay1 and the wild-type (WT) or complemented strains (Tukey’s HSD: p < 0.05). Error bars: standard deviation.
The conidial germination on WA plates was significantly decreased by gene disruption (Figure 2E). The germination percentage for the ΔBbsay1 mutant strain was 23.3 ± 2.1%; however, the wide-type and complementation strains displayed a germination percentage of approximately 35%. After adding oleic acid, there was no significant difference in the germination percentage among the wild-type, ΔBbsay1, and complementation strains. On the hindwing of a locust, the conidia of the wild-type germinated well and developed into a germling; however, the ΔBbsay1 mutant displayed aretarded germination (F). Under oxidative stress by menadione, the gene disruption mutant displayed a significant defect in vegetative growth (G). The conidial yield was evaluated on SDAY plates (Figure 2H). Gene disruption caused a slight reduction in the conidial yield at three sampling time points. At 8 d post incubation, the conidial yield for the ΔBbsay1 mutant strain was 4.58 ± 0.35 × 108 conidia/cm2, and that of the wild-type was 5.89 ± 0.67 × 108 conidia/cm2. Exogenous oleic acid could reduce and eliminate this defect in the ΔBbsay1 mutant strain. A transformant with over-expression of BbSAY1 generated 5.76 ± 4.48 × 108 conidia/cm2, which did not significantly differ fromthe wild-type.
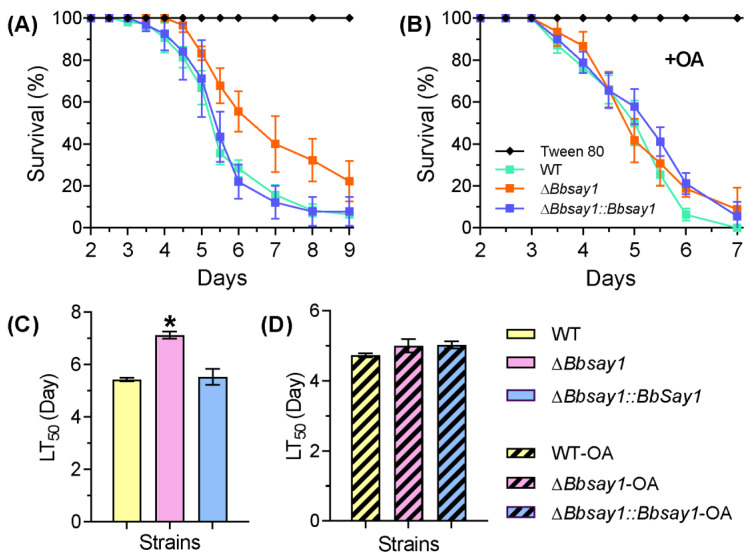
In a cuticle penetration bioassay, the survival trend for the ΔBbsay1 mutant showed a significant delay when compared to the wild-type (Figure 3A). The LT50 for ΔBbsay1 mutant was calculated as 7.13 ± 0.23 d (mean ± standard deviation), with a slight delay, when compared with that of the wild-type (5.43 ± 0.11 d) (Figure 3C). When adding oleic acid, there was no significant difference in the survival trend and LT50 between the wild-type and ΔBbsay1 mutant strains (Figure 3B,D).
Figure 3.
Fungal virulence. Conidial virulence was assayed for the wild-type (WT), ΔBbsay1, and ΔBbsay1::Bbsay1 strains. The conidial suspension was flooded onto the host cuticle, and then, the inoculated insects were cultured at 25 °C. The survival trend was recorded daily (A), and the accumulative mortality was used to calculate the median lethal time (LT50) with a Probit analysis (C). In the feeding test, oleic acid (OA) was included in the conidial suspension, and accumulative mortality was used to calculate the survival trend (B) and LT50 (D). A Tween-80 solution was used as blank control. The asterisk on the column indicates the significant difference in the LT50 value between the ΔBbsay1 and the wild-type or complemented strains (Tukey’s HSD: p < 0.05). Error bars: standard deviation.
3.3. Gene Loss Affects Cellular Lipidomics and Membrane Integrity
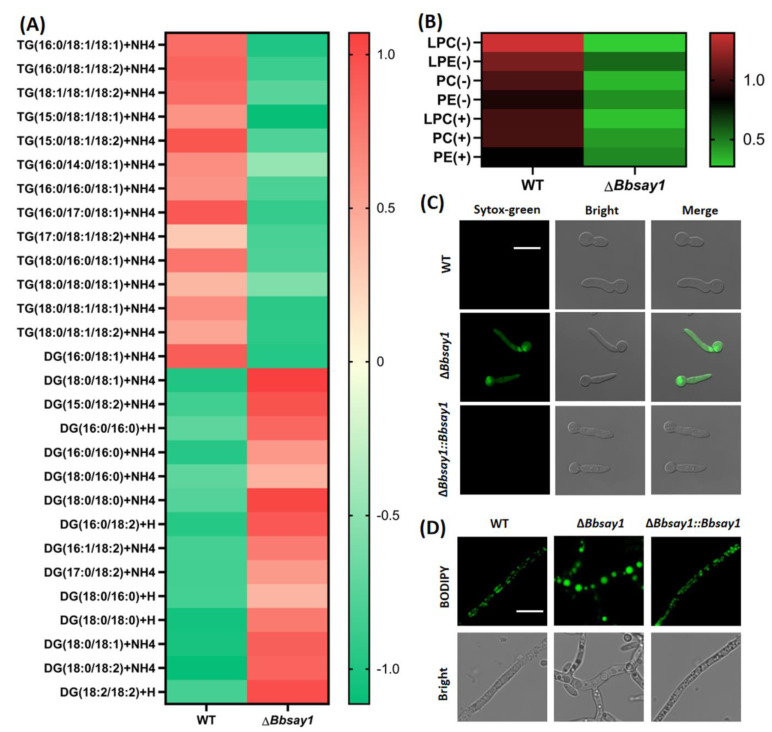
As shown in Figure 4A, the ablation of BbSAY1 resulted in an enhanced accumulation of diacylglycerols (DG) and a decrease in the content of triacylglycerols (TG). Four kinds of phospholipids were detected in the B. bassiana conidia, including lysophosphatidylcholine (LPC), lysophosphatidylethanolamine (LPE), phosphatidylcholine (PC), and phosphatidylethanolamine (PE), in their cationic or anionic forms. Gene disruption resulted in a significant decrease in the content of phospholipids (e.g., LPC) (Figure 4B). The loss of Bbsay1 led to poor membrane integrity in germlings. Without oleic acid, nearly all germlings were stained by SYTOX, while only 1% of the WT cells were stained (Figure 4C). Interestingly, exogenous oleic acids significantly reduced the percentage of the stained cells (PSC) in the ΔBbsay1 mutant strain. The microscopic view indicated that the size of lipid bodies was significantly increased when compared with that in the wild-type and complemented strains (Figure 4D).
Figure 4.
Assay for lipid homeostasis in B. bassiana. The lipidomic assay indicated that the ablation of BbSAY1 resulted in the impaired homeostasis of a neutral lipid (A) and phospholipid (B). (C) Sytox-green nucleus staining. The ΔBbsay1 mutant strain displayed anincreased cytomembrane permeability, and its nuclei were easily stained by fluorescent dye. (D) Staining lipid bodies. The fluorescent dye BODIPY was used to stain the lipid bodies in fungal cells. The ΔBbsay1 mutant strain accumulated the enlarged lipid bodies, which indicatedthe unbalanced lipid metabolism after the ablating of BbSAY1.
4. Discussion
FA/lipid metabolism plays an important role in the interaction between entomopathogenic fungi and insect hosts [22]. This study has revealed that the ortholog of yeast steryl acetyl hydrolase 1 (Say1) contributes to lipid homeostasis, which is required for development and virulence in B. bassiana.
Lipid homeostasis is maintained by a series of biochemical reactions of esterification and transesterification, as well as the interaction among different catalytic pathways [9]. In S. cerevisiae, Say1 catalyzes the reaction of sterol deacetylation, which is required for sterol homeostasis [12,23]. In B. bassiana, BbSay1 contributes to the homeostasis of lipid metabolism. Its loss leads to a comprehensive fluctuation in the cellular lipidome, although more investigations are needed to reveal the direct effects of BbSay1 in B. bassiana. This finding suggests the conservative role of Say1 in lipid metabolism. It is well known that triacylglycerol is synthesized by the conjugation of acyl coenzyme A (acyl-CoA) to diacylglycerol, which is catalyzed by acyl-CoA:diacylglycerol acyltransferase [24]. After disrupting BbSAY1, B. bassiana accumulated a high level of diacylglycerol and a low level of triacylglycerol. Additionally, BbSay1 contributes to maintaining the morphologies of lipid bodies and the conidial reserve of oleic acid. Lipid droplets are organelles that store neutral lipids (i.e., triacylglycerols and steryl esters) and maintain the homeostasis of stored lipids and free fatty acids [25]. These results suggest that BbSay1 is further involved in lipid metabolism via modulating the biogenesis of lipid bodies. Considering the Say1 role in sterol homeostasis [23], our findings further suggest that there might exist an interaction between triacylglycerol and sterol homeostasis.
The conidial capacity to germinate and penetrate through the host cuticle is determinant for successful infection by a fungal entomopathogen [3]. Particularly, conidial germination on the nutrient-limited cuticle is critical for infection initiation [26]. Additionally, the cytomembrane functionality of germlings is critical for fungal virulence [7]. BbSay1 contributes to fungal virulence, which is due to its roles in conidial germination under the oligotrophic condition and the cytomembrane integrity. Several physiological processes have been linked to the cytomembrane integrity of the germling in B. bassiana, including the biosynthesis of unsaturated FA (e.g., BbOle1) [7]), lipid trafficking (e.g., BbScp2) [27], and thioesterification of FA (e.g., BbFaa1) [28]. In addition, a lectin-like protein functions at the interface between the cell membrane and wall and contributes to the cytomembrane functionality [29]. These findings reinforce that the homeostasis of lipid metabolism and the cell wall is essential for membrane integrity.
Conidia, generated through asexual development, are essential for entomopathogenic fungi to survive and disperse in the environment [14,30].BbSay1 contributes to conidiation in anoleic acid-dependent manner, and it maintains the morphology of lipid bodies during vegetative growth. In A. nidulans, two Δ9-stearic acid desaturases catalyze the biosynthesis of oleic acid, which is required for fungal development [31]. In filamentous fungi (e.g., Blumeria graminis), the oxidation of unsaturated fatty acids is involved in the conidiation process [32]. In Magnaporthe oryzea, the elevated accumulation of lipid bodies leads to severe conidiation defects [33]. Considering the roles of oleic acid in energy supply and the membrane integrity, our findings suggest that BbSay1 might affect multiple aspects in B. bassiana conidiation. In addition, BbSNF1, an AMPK protein kinase, mediates the amino-acid homeostasis during conidiation in B. bassiana [30]. These findings suggest that primary metabolism is essential for conidiation in filamentous fungi.
5. Conclusions
In conclusion, B. bassiana contains 10 Say1-domain-containing proteins. BbSay1, the ortholog of yeast Say1, maintains lipid homeostasis, which contributes to the cytomembrane integrity, development, and virulence in B. bassiana. In future, more investigations are needed to functionally identify other Say1 proteins, which should provide novel insights into the lipid metabolisms involved in the differentiation and virulence of entomopathogenic fungi.
Acknowledgments
We greatly appreciate Hai-Chun Gao (College of Life Sciences, Zhejiang University) for the constructive advice on the fatty acid analysis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof8030292/s1, Figure S1: Bioinformatic and transcriptional analyses. Figure S2: Sub-cellular localization of Say1 in B. bassiana. Figure S3: Molecular manipulation for gene function analysis of B. bassiana. Table S1: Primers for the qRT-PCR analyses. Table S2: Primers used for the gene disruption and complementation of BbSAY1 in B. bassiana. Table S3: Homologs of B. bassiana Say1-domain-containing proteins in other fungal species.
Author Contributions
Y.-J.P.: Investigation, Formal analysis, Validation, Visualization, and Writing—original draft. H.Z.: Formal analysis and Investigation. M.-G.F.: Writing—review and editing. S.-H.Y.: Conceptualization, Funding acquisition, Project administration, Supervision, and Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (32170027) and the Zhejiang Provincial Natural Science Foundation of China (LZ19C140001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De la Cruz Q.R., Roussos S., Hernandez D., Rodriguez R., Castillo F., Aguilar C.N. Challenges and opportunities of the bio-pesticides production by solid-state fermentation: Filamentous Fungi as a Model. Crit. Rev. Biotechnol. 2015;35:326–333. doi: 10.3109/07388551.2013.857292. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz-Urquiza A., Luo Z., Keyhani N.O. Improving mycoinsecticides for insect biological control. Appl. Microbiol. Biotechnol. 2015;99:1057–1068. doi: 10.1007/s00253-014-6270-x. [DOI] [PubMed] [Google Scholar]
- 3.Ortiz-Urquiza A., Keyhani N.O. Action on the surface: Entomopathogenic Fungi Versus the Insect Cuticle. Insects. 2013;4:357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wanchoo A., Lewis M.W., Keyhani N.O. Lectin mapping reveals stage-specific display of surface carbohydrates in in vitro and haemolymph-derived cells of the entomopathogenic fungus Beauveria Bassiana. Microbiology. 2009;155:3121–3133. doi: 10.1099/mic.0.029157-0. [DOI] [PubMed] [Google Scholar]
- 5.Shah P.A., Pell J.K. Entomopathogenic fungi as biological control agents. Appl. Microbiol. Biotechnol. 2003;61:413–423. doi: 10.1007/s00253-003-1240-8. [DOI] [PubMed] [Google Scholar]
- 6.Palma-Guerrero J., Lopez-Jimenez J.A., Perez-Berna A.J., Huang I.-C., Jansson H., Salinas B.J., Villalain J., Read N.D., Lopez-Llorca L.V. Membrane fluidity determines sensitivity of filamentous fungi to chitosan. Mol. Microbiol. 2010;75:1021–1032. doi: 10.1111/j.1365-2958.2009.07039.x. [DOI] [PubMed] [Google Scholar]
- 7.Peng Y.-J., Wang J.-J., Lin H.-Y., Ding J.-L., Feng M.-G., Ying S.-H. HapX, an indispensable bZIP transcription factor for iron acquisition, regulates infection initiation by orchestrating conidial oleic acid homeostasis and cytomembrane functionality in mycopathogen Beauveria bassiana. Msystems. 2020;5:e00695. doi: 10.1128/mSystems.00695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo A.M., Gardner H.W., Keller N.P. Genetic connection between fatty acid metabolism and sporulation in Aspergillus nidulans. J. Biol. Chem. 2001;276:25766–25774. doi: 10.1074/jbc.M100732200. [DOI] [PubMed] [Google Scholar]
- 9.Borrelli G.M., Trono D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015;16:20774–20840. doi: 10.3390/ijms160920774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ballweg S., Ernst R. Control of membrane fluidity: The OLE Pathway in Focus. Biol. Chem. 2017;398:215–228. doi: 10.1515/hsz-2016-0277. [DOI] [PubMed] [Google Scholar]
- 11.Gsaller F., Hortschansky P., Beattie S.R., Klammer V., Tuppatsch K., Lechner B.E., Rietzschel N., Werner E.R., Vogan A.A., Chung D., et al. The Janus transcription factor HapX controls fungal adaptation to both iron starvation and iron excess. EMBO J. 2014;33:2261–2276. doi: 10.15252/embj.201489468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari R., Koffel R., Schneiter R. An acetylation/deacetylation cycle controls the export of sterols and steroids from S. cerevisiae. EMBO J. 2007;26:5109–5119. doi: 10.1038/sj.emboj.7601924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao G., Ying S.-H., Zheng P., Wang Z.-L., Zhang S., Xie X.-Q., Shang Y., St Leger R.J., Zhao G.-P., Wang C. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012;2:483. doi: 10.1038/srep00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lechner M., Findeis S., Steiner L., Marz M., Stadler P.F., Prohaska S.J. Proteinortho: Detection of (Co-)orthologs in Large-Scale Analysis. BMC Bioinform. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X.-X., He P.-H., Feng M.-G., Ying S.-H. BbSNF1 contributes to cell differentiation, extracellular acidification, and virulence in Beauveria bassiana, a filamentous entomopathogenic fungus. Appl. Microbiol. Biotechnol. 2014;98:8657–8673. doi: 10.1007/s00253-014-5907-0. [DOI] [PubMed] [Google Scholar]
- 16.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Wang J.-J., Peng Y.-J., Feng M.-G., Ying S.-H. Functional analysis of the mitochondrial gene mitofilin in the filamentous entomopathogenic fungus Beauveria bassiana. Fungal Genet. Biol. 2019;132:103250. doi: 10.1016/j.fgb.2019.103250. [DOI] [PubMed] [Google Scholar]
- 18.He P.-H., Dong W.-X., Chu X.-L., Feng M.-G., Ying S.-H. The cellular proteome is affected by a gelsolin (BbGEL1) during morphological transitions in aerobic surface versus liquid growth in the entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2016;18:4153–4169. doi: 10.1111/1462-2920.13500. [DOI] [PubMed] [Google Scholar]
- 19.Xie X.-Q., Li F., Ying S.-H., Feng M.-G. Additive contributions of two manganese-cored superoxide dismutases (MnSODs) to antioxidation, UV tolerance and virulence of Beauveria bassiana. PLoS ONE. 2012;7:e30298. doi: 10.1371/journal.pone.0030298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J.-L., Hou J., Li X.-H., Feng M.-G., Ying S.-H. Transcription activator Swi6 interacts with Mbp1 in MluI cell cycle box-binding complex and regulates hyphal differentiation and virulence in Beauveria bassiana. J. Fungi. 2021;7:411. doi: 10.3390/jof7060411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding J.-L., Hou J., Feng M.-G., Ying S.-H. Transcriptomic analyses reveal comprehensive responses of insect hemocytes to mycopathogen Beauveria bassiana, and fungal virulence-related cell wall protein assists pathogen to evade host cellular defense. Virulence. 2020;11:1352–1365. doi: 10.1080/21505594.2020.1827886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keyhani N.O. Lipid biology in fungal stress and virulence: Entomopathogenic Fungi. Fungal Biol. 2018;122:420–429. doi: 10.1016/j.funbio.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T.C., Farese R.J. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 2014;55:1465–1477. doi: 10.1194/jlr.M050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Siloto R.M., Lehner R., Stone S.J., Weselake R.J. Acyl-CoA:diacylglycerol acyltransferase: Molecular Biology, Biochemistry and Biotechnology. Prog. Lipid Res. 2012;51:350–377. doi: 10.1016/j.plipres.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Walther T.C., Farese R.J. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 2012;81:687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying S.-H., Liu J., Chu X.-L., Xie X.-Q., Feng M.-G. The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana. Sci. Rep. 2016;6:e26376. doi: 10.1038/srep26376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin H.-Y., Pang M.-Y., Feng M.-G., Ying S.-H. A peroxisomal sterol carrier protein 2 (Scp2) contributes to lipid trafficking in differentiation and virulence of the insect pathogenic fungus Beauveria bassiana. Fungal Genet. Biol. 2022;158:e103651. doi: 10.1016/j.fgb.2021.103651. [DOI] [PubMed] [Google Scholar]
- 28.Li X.-H., Peng Y.-J., Ding J.-L., Feng M.-G., Ying S.-H. A homologue of yeast acyl-CoA synthetase Faa1 contributes to cytomembrane functionality involved in development and virulence in the insect pathogenic fungus Beauveria bassiana. Microb. Pathog. 2022;164:e105419. doi: 10.1016/j.micpath.2022.105419. [DOI] [PubMed] [Google Scholar]
- 29.Peng Y.-J., Ding J.-L., Lin H.-Y., Feng M.-G., Ying S.-H. A virulence-related lectin traffics into eisosome and contributes to functionality of cytomembrane and cell-wall in the insect-pathogenic fungus Beauveria bassiana. Fungal Biol. 2021;125:914–922. doi: 10.1016/j.funbio.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 30.He P.-H., Wang X.-X., Chu X.-L., Feng M.-G., Ying S.-H. RNA sequencing analysis identifies the metabolic and developmental genes regulated by BbSNF1 during conidiation of the entomopathogenic fungus Beauveria bassiana. Curr. Genet. 2015;61:143–152. doi: 10.1007/s00294-014-0462-x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson R.A., Chang P.K., Dobrzyn A., Ntambi J.M., Zarnowski R., Keller N.P. Two Δ9-stearic acid desaturases are required for Aspergillus nidulans growth and development. Fungal Genet. Biol. 2004;41:501–509. doi: 10.1016/j.fgb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 32.Zeng F.-S., Menardo F., Xue M.-F., Zhang X.-J., Gong S.-J., Yang L.-J., Shi W.-Q., Yu D.-Z. Transcriptome analyses shed new insights into primary metabolism and regulation of Blumeria graminis f. sp. tritici during conidiation. Front. Plant Sci. 2017;8:e1146. doi: 10.3389/fpls.2017.01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y., Zhu J., Hu J., Meng X., Zhang Q., Zhu K., Chen X., Chen X., Li G., Wang Z., et al. Functional characterization of electron-transferring flavoprotein and its dehydrogenase required for fungal development and plant infection by the rice blast fungus. Sci. Rep. 2016;6:e24911. doi: 10.1038/srep24911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.