Abstract
Shrimp is one of the most valuable aquaculture species globally, and the most internationally traded seafood product. Consequently, shrimp aquaculture practices have received increasing attention due to their high value and levels of demand, and this has contributed to economic growth in many developing countries. The global production of shrimp reached approximately 6.5 million t in 2019 and the shrimp aquaculture industry has consequently become a large-scale operation. However, the expansion of shrimp aquaculture has also been accompanied by various disease outbreaks, leading to large losses in shrimp production. Among the diseases, there are various viral diseases which can cause serious damage when compared to bacterial and fungi-based illness. In addition, new viral diseases occur rapidly, and existing diseases can evolve into new types. To address this, the review presented here will provide information on the DNA and RNA of shrimp viral diseases that have been designated by the World Organization for Animal Health and identify the latest shrimp disease trends.
Keywords: shrimp disease, OIE, viral disease, DNA and RNA virus
1. Introduction
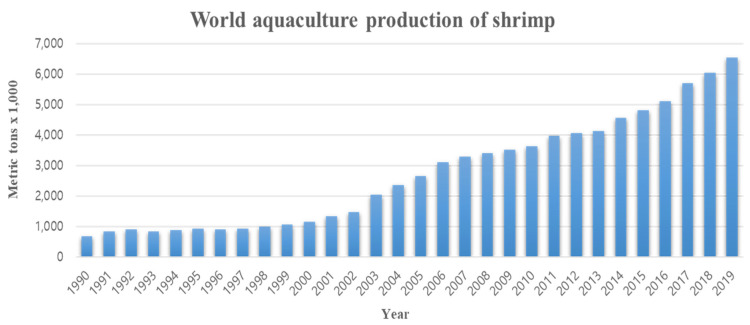
The shrimp aquaculture industry has grown rapidly in previous decades due to increasing consumer demand, and it has consequently contributed significantly to the socio-economic development of coastal communities in many developing countries [1]. Production by the shrimp farming industry has steadily increased to approximately 3.6 million t in 2008, accounting for more than 50% of the global shrimp market, with the main production areas being in Southeast Asia, such as China, Thailand, Vietnam, Indonesia, and India, while in the Americas, the major producers are Ecuador, Mexico, and Brazil [2]. Shrimp production has steadily grown from 0.673 million t in 1990 to 6.004 million t in 2019, which is a nearly tenfold increase (Figure 1). Until recently, shrimp aquaculture production was most widespread in Latin America and East and Southeast Asian countries, but consumption is concentrated in various developed countries. Consequently, this industry is helping to reduce the economic gaps between countries by generating high levels of income in developing countries [3]. Indeed, in Southeast Asia, penaeid shrimp have contributed significantly to the economies of Indonesia, the Philippines, Vietnam, and Thailand [4].
Figure 1.
World aquaculture production of shrimp from 1990 to 2019 (Source: FAO yearbook of Fishery and Aquaculture Statistics).
The shrimp aquaculture industry is growing in many regions of the world, including Asia and Latin America, and it accounts for 17% of the total value of aquatic products [5]. Globally, 67% of shrimp production is from aquaculture and 33% is caught naturally, and the most common species used in shrimp aquaculture are the whiteleg shrimp, Penaeus vannamei, and Giant tiger prawn, the marine shrimp Penaeus monodon, and the freshwater prawns Macrobrachium rosenbergii and Macrobrachium. nipponense [6]. Crustacean production totaled 8.4 million t in 2017, representing an average annual increase of 9.92% since 2000, and more than 30 crustacean species were valued at 61.06 billion USD in 2017 [7]. However, with the increase in global shrimp aquaculture production, mass mortality caused by frequent disease outbreaks has become a major obstacle for the industry. Worldwide losses from disease in shrimp aquaculture in the last 15 years to 2005 were estimated to be approximately 15 billion USD, 80% of which occurred in Asia [8].
Until the 1980s, marine viruses were considered ecologically insignificant, because their concentrations were underestimated, but subsequent studies have confirmed that the ocean contains an abundance of organisms, including millions of virus particles per milliliter of seawater [9]. Most shrimp diseases are caused by viral infection, and they have an approximately four times more negative impact than bacterial diseases. In most cases, diseases caused by bacterial pathogens and parasites can be prevented through the proper management of shrimp farms (biosecurity, water quality control, stocking density, aeration, fresh feed, shrimp seed quality, and proper breeding environment), which is in contrast to viral diseases [8,10].
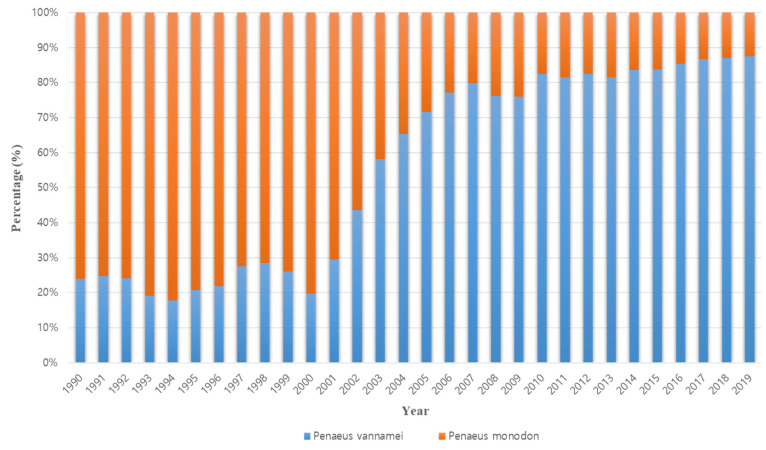
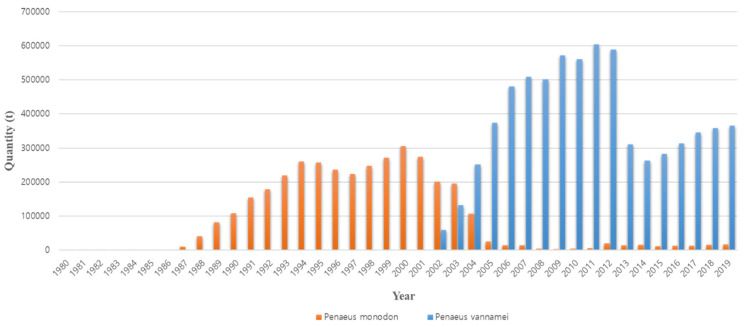
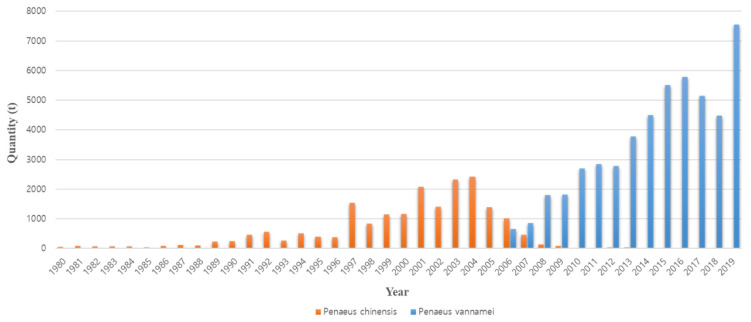
The occurrence of disease is the reason that existing farmed shrimp species are replaced with other species. The cause of the conversion from P. monodon in the 1990s to P. vannamei in the 2000s is also closely related to disease occurrence (Figure 2). Thailand’s P. monodon production increased rapidly from 1987 to the early 1990s, but thereafter, until the early 2000s, there was a large loss in production due to YHV (yellow head virus), WSSV (white spot syndrome virus), and then MSGS (monodon slow growth syndrome) [11]. Prior to 2000, P. monodon was the predominant aquaculture shrimp species in Asia, but the disease-free SPF (specific pathogen free) species P. vannamei began to increase as a replacement species (Figure 3). In Korea, the reason for the rapid replacement of P. vannamei from P. chinensis, which had been cultured since 2006, is also due to the damage caused by the frequent occurrence of WSSV (Figure 4). Ultimately, P. vannamei has now become the dominant shrimp aquaculture species worldwide as it is less susceptible to WSD (white spot disease) outbreaks, which had a major impact on many other shrimp species [12]. The replacement of shrimp species with P. vannamei in Asia has led to an increase in shrimp production from approximately 900,000 t in 2004 to 2.9 million t in 2009.
Figure 2.
Proportion of the major shrimp species Penaeus monodon and Penaeus vannamei in aquaculture production from 1990 to 2019 (Source: FAO yearbook of Fishery and Aquaculture Statistics).
Figure 3.
Total shrimp aquaculture production for Penaeus monodon and Penaeus vannamei in Thailand from 1980 to 2019 (Source: FAO Global Aquaculture Production Statistics from FishstatJ Software for Fishery and Aquaculture Statistical Time Series).
Figure 4.
Total shrimp aquaculture production for Penaeus chinensis and Penaeus vannamei in the Republic of Korea from 1980 to 2019 (Source: FAO Global Aquaculture Production Statistics from FishstatJ Software for Fishery and Aquaculture Statistical Time Series).
Managing the health of farmed shrimp species and developing new methods for disease prevention and treatment, preventing the illegal transboundary movement of live shrimp species, and controlling disease outbreaks through the supply of fresh food worldwide, requires an immense amount of effort. To address these issues, Flegel (2012) [8] suggested the following: (1) the development of pathogen-free SPF shrimp seeds; (2) widespread use and standardization of diagnostic tests; (3) development of biosecurity-applied breeding techniques; (4) control efforts to reduce the risk of disease transmission through cross-border movement; (5) investigations into the efficacy of immune-stimulants and vaccines; (6) a complete understanding of the specificity of shrimp species by pathogen; (7) rich epidemiologic studies of shrimp diseases; (8) molecular ecology studies to control pathogenic microorganisms in shrimp hatcheries and breeding grounds; (9) conducting virus tests through strict cross-border quarantine procedures; and (10) restricting indiscriminate imports of exotic crustaceans. This review aims to analyze the viral OIE shrimp diseases that occur frequently around the world, by examining the disease occurrence trends and diagnostic methods and providing basic data for future alternatives to shrimp diseases using the latest trend analyses and treatment plans.
2. DNA Viral Diseases
2.1. White Spot Syndrome Disease (WSSD)
Aquaculture practices are responsible for approximately 75% of the world’s shrimp production, and the predominant species used are black tiger shrimp, P. monodon and white Pacific shrimp, P. vannamei [13,14]. In the past 20 years, shrimp diseases have caused critical economic losses that seriously threaten farming practices, of which white spot syndrome (WSS) is the deadliest viral disease caused by white spot syndrome virus (WSSV) [15]. WSSV causes up to 80–100% mortality of infected shrimp within 5–10 days, thus leading to great economic loss [16]; the total economic loss from this disease is estimated to be approximately 8–15 billion USD, and this continues to increase by 1 billion USD (10% of global shrimp production) annually [17,18].
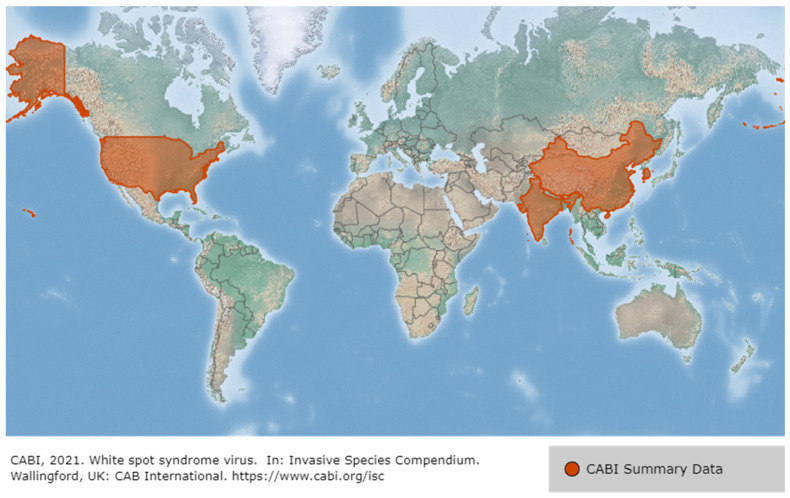
The first reports of WSD in penaeid shrimp occurred in China and Taiwan in 1992, and then spread to Korea (1993), Japan (1993), Vietnam, Thailand (1994), Malaysia (1995) and Indonesia. WSSV also occurred in America (Latin America, such as Ecuador, Mexico, and Brazil in 1999 and North America in 1995), the Middle East in 2001, and Africa (such as Mozambique and Madagascar in 2011), and most recently at an Australian shrimp farm in 2016 [19] (Figure 5). WSSV presumably reached America through the importation of P. monodon from Asia and became rapidly endemic in American native species such as P. vannamei. In Asia, during the early 2000s, the SPF species P. vannamei was imported from the Americas to avoid disease problems such as WSSV, resulting in the conversion of the predominant farmed species from P. monodon to P. vannamei. However, the translocation of broodstock that are unscreened or inadequately tested for WSSV has led to the spread of WSSV back to Asia from the Americas [12,19]. White spot syndrome disease (WSSD) has been listed by the World Organization for Animal Health since 1997 [20]. WSSV is considered the most serious of approximately 20 viral pathogens in shrimp, and in 2018, 46.3% of farmed crayfish in 13 provinces in China were WSSV-positive. Of note, however, is that the WSSV mortality rate in farmed crayfish is less sensitive than for shrimp, at approximately 5–90%, and it does not always lead to mortality [21].
Figure 5.
Distribution map showing the geographical occurrence of white spot syndrome disease (WSSD) (Reprinted from CABI, 2019, White spot syndrome virus. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
WSSV is the only member of the genus Whispovirus in the family Nimaviridae (initially included in the family Baculoviridae) and has a double-stranded DNA genome with a virion size of 80–120 × 250–380 nm, which is rod-shaped to elliptical, and surrounded by a trilaminar envelop with a tail-like appendage [16] (Table 1). The naked viral nucleocapsid is about 80 × 350 nm and has 15 spiral and cylindrical helices of 14 spherical capsomers along its long axis, with a ‘ring’ structure at one end [22]. On the outer surface of the viral envelope, there are many tadpole-shaped spikes (5–6 nm long, 4–5 nm head diameter) to which host cells can easily attach [20]. WSSV has been reported to be approximately 300–305 kbp in length according to the isolates with 180 open reading frames (ORFs) and nine repeated sequence regions in tandem, and minisatellites (ORF 94, ORF75 and ORF125) are used for WSSV genomic and epidemiological studies [20] (Table 2). As a result, of sequencing the genes isolated from China and Taiwan, significant variations were confirmed in WSSV isolates from Vietnam and Thailand, due to the insertion of major ORF14/15 and ORF23/24 variable regions [23].
Table 1.
Summary information for DNA and RNA viral diseases infections.
| Virus Type | Pathogen | Taxonomy | Morphology | Reference | ||
|---|---|---|---|---|---|---|
| DNA virus | ds DNA | WSSV | Family | Nimaviridae |
|
[9,13,16,18,20,22,52] |
| Genus | Whispovirus | |||||
| ss DNA | IHHNV (Decapod penstylhamaparvovirus 1) |
Family | Parvoviridae |
|
[57,59,60,65,66,85,161,162] | |
| subfamily | Hamaparvovirinae | |||||
| Genus | Penstylhamaparvovirus | |||||
| RNA virus | ds RNA | IMNV (PsIMNV) |
Family | Totiviridae |
|
[73,77,79,83,85] |
| Genus | Similar Giardiavirus |
|||||
| ss RNA | YHV | Order | Nidovirales |
|
[68,95,104] | |
| Family | Roniviridae | |||||
| Genus | Okavirus | |||||
| TSV | Order | Picornavirales |
|
[2,62,95,116,117] | ||
| Family | Dicistroviridae | |||||
| Genus | Aparavirus | |||||
| WTD (MrNV) |
Famliy | Nodaviridae |
|
[56,140,141,142,144,145,146,149,150,157,163] | ||
| Genus | Gammanodavirus | |||||
| WTD (XSV) |
Unassigned |
|
||||
Table 2.
Summary of the DNA and RNA viral diseases ORF characteristics.
| Virus Type | Pathogen | ORF | Characteristics | Reference | |
|---|---|---|---|---|---|
| DNA virus | ds DNA | WSSV | ORF75 |
|
[9,13,20,24,164,165,166,167,168,169,170,171,172] |
| ORF94 |
|
||||
| ORF125 |
|
||||
| ORF14/15 |
|
||||
| ORF23/24 |
|
||||
| ORF109 |
|
||||
| ORF182 |
|
||||
| ORF153 |
|
||||
| ORF-wsv002 |
|
||||
| ORF-wsv421 |
|
||||
| ORF-wsv308 |
|
||||
| ss DNA | IHHNV (Decapod penstylhamaparvovirus 1) |
ORF1 |
|
[57,58,59,85,173,174] | |
| ORF2 |
|
||||
| ORF3 |
|
||||
| RNA virus | ds RNA | IMNV (PsIMNV) |
ORF1 (59 ORF) |
|
[62,73,74,76,83,86] |
| ORF2 (39 ORF) |
|
||||
| ss RNA | YHV | ORF1a |
|
[1,92,100,104,107,111,114,163,175,176] | |
| ORF1a/ORF1b |
|
||||
| ORF1b |
|
||||
| ORF2 |
|
||||
| ORF3 |
|
||||
| ORF4 |
|
||||
| GAV | ORF1a |
|
[1,92,100,107,114,177] | ||
| ORF1b |
|
||||
| ORF1a/ORF1b |
|
||||
| ORF2 |
|
||||
| ORF3 |
|
||||
| ORF4 |
|
||||
| TSV | ORF1 |
|
[95,128,130,178] | ||
| ORF2 |
|
||||
| WTD (MrNV) |
ORF1 (RNA-1) |
|
[56,140,150,153,156,157] | ||
| ORF2 (RNA-2) |
|
||||
| WTD (XSV) |
XSV genome |
|
|||
Structural proteins play important roles in cell targeting, viral entry, assembly, and budding, which is highly related to WSSV infection. Envelope protein function has a particularly critical role in viral entry to the host cell [24]. Interactions between structural proteins are common in enveloped viruses such as WSSV, but this kind of interaction involves nine WSSV virion proteins (VP19, VP24, VP26, VP28, VP37 or VP281, VP38A or VP38, VP51C or VP51, VP51A and WSV010), some of which (VP19, VP24 and VP51A) prefer self-interaction [22]. Of the envelope proteins, VP19, VP24, VP26 and VP28 are the main proteins, and VP28 and VP26 account for approximately 60% (VP28, VP26, VP24 and VP19 account for about 90%) of the envelope as the most abundant proteins [20,25]. VP28 has a critical role in the early stages of viral infection by binding WSSV to shrimp cellular receptors, and the structural protein VP24 is a key protein that directly binds to VP26, VP28, VP38A, VP51A, and WSV010 to form a membrane-associated protein complex [22]. WSSV VP28 is an adhesion protein that helps the virus to bind to shrimp cells and enter the cytoplasm during infection, and VP26 may bind to actin or actin-related proteins and help WSSV translocate to the nucleus [9]. In addition to VP28, VP37 is a viral envelope protein known to promote WSSV infection through binding to shrimp cells, resulting in virus binding to the hemocytes [26]. Furthermore, structural proteins of the virion envelope such as VP26, VP31, VP37, VP90, and VP136 interact with integrin receptors to stimulate the binding of viruses to the extracellular matrix (or intercellular adhesion) [13,27].
WSSV isolates from several regions with different genotypes [Thailand (GenBank no. AF369029), Taiwan (GenBank no. AF440570), China (GenBank no. AF332093), and South Korea (GenBank no. JX515788)] have been sequenced, but they are all classified as a single species of the genus Whispovirus (family Nimaviridae) [24,28]. The complete genome sequence of WSSV isolates was reported in 2001 (WSSV-TH, GenBank no. AF369029; WSSV-CN, GenBank no. AF332093), 2002 (WSSV-TW, GenBank no. AF440570), 2013 (WSSV-KR, GenBank no. JX515788), 2016 (WSSV-MX08, GenBank no. KU216744), 2017 (WSSV-CN02, CN01 and CN03, GenBank no. KT995470-995472; WSSV-CN04, GenBank no., KY827813; WSSV-CN-Pc, GenBank no. KX686117) and 2018 (WSSV-AU, GenBank no. MF768985; IN_AP4RU, GenBank no. MG702567; WSSV-EC-15098, GenBank no. MH090824; WSSV-chimera, GenBank no. MG264599) and 2020 (CN_95_DFPE, GenBank no. MN840357) [29,30,31,32,33,34,35,36,37,38,39,40,41] (Table 3). The major deletion region at ORF23/24, variable region at ORF14/15, and variable number tandem repeats (VNTRs) located in ORF75, ORF94, and ORF125 are used as genetic marker to differentiate WSSV genotypes [23,36,42]. Mx-F, Mx-H, Mx-C, and Mx-G strains (GenBank no. HQ257380-257383) have 99–100% identity to each other in the ORF14/15 region and all four contain a 314 bp region present only in isolated WSSV-In-07-I (GenBank no. EF468499). The low-virulence strain Mx-G has additional repeat units (RUs) in ORF94 when compared to the highly virulent strain Mx-H, and both have 100% identity in the variable number of tandem repeats (VNTR) in ORF75 and ORF125 [28]. During the spread of WSSV in Asia, significant changes were observed in the ORF14/15 and ORF23/24 regions, and consequently, WSSV strains increased host mortality, shortened host survival, and developed increased competencies in host competition [43].
Table 3.
Summary isolation and GenBank accession number information for the DNA and RNA viral disease infections.
| Type | Pathogen | Origin | Host Species | Isolation | ORF Region | GenBank No. | Year | Reference | |
|---|---|---|---|---|---|---|---|---|---|
| DNA virus | ds DNA | WSSV | Mexico | Penaeus vannamei | Mx-F | Hypothetical protein (ORF13 and ORF16) gene; Nonfunctional hypothetical protein gene |
HQ257380 | 2001 | [179] |
| Mx-H | HQ257381 | 2004 | |||||||
| Mx-C | Nonfunctional hypothetical protein genes | HQ257382 | 2005 | ||||||
| Mx-G | HQ257383 | 2004 | |||||||
| Mx-L1 | HQ257384 | 2001 | |||||||
| WSSV-MX08 | Complete genome | KU216744 | 2008 | [33] | |||||
| Penaeus vannamei | LG | Partial genome | MG432482 | 2012 | [180] | ||||
| JP | MG432479 | 2011 | |||||||
| AC1 | MG432474 | 2011 | |||||||
| DV1 | MG432477 | 2011 | |||||||
| LC1 | MG432481 | 2011 | |||||||
| LC10 | MG432480 | 2011 | |||||||
| ACF2 | MG432475 | 2012 | |||||||
| ACF4 | MG432476 | 2012 | |||||||
| GVE05 | MG432478 | 2005 | |||||||
| India | Penaeus monodon | ANI | wsv285 gene | KX980155 | 2016 | [181] | |||
| WSSV-IN-07-I | Unknown gene | EF468499 | 2007 | [182] | |||||
| WSSV-IN-06-I | EF468498 | 2006 | |||||||
| WSSV-IN-05-I | EU327499 | 2005 | |||||||
| WSSV-IN-05-II | ORF23/ORF24 region genomic sequence | EU327500 | 2005 | ||||||
| Penaeus vannamei | IN_AP4RU | Complete genome | MG702567 | 2013 | [38] | ||||
| Iran | Penaeus vannamei | IRWSSVKH2 | Hypothetical protein 75 gene | KF157839 | 2012 | [183] | |||
| IRWSSVKH4 | ORF75 gene | KC906268 | 2011 | ||||||
| IRWSSVKH5 | KF157833 | 2012 | |||||||
| IRWSSVKH3 | KF157832 | 2012 | |||||||
| IRWSSVSIS3 | KP455493 | 2014 | |||||||
| IRWSSVSIS2 | KF956791 | 2013 | |||||||
|
Penaeus indicus;
Penaeus vannamei |
IWV-MS21 | ORF75 gene | KX694234 | 2013 | |||||
| IWV-MS24 | KX694236 | 2014 | |||||||
| IWV-MS25 | KX694237 | 2014 | |||||||
| IWV-MS26 | KX694238 | 2014 | |||||||
| IWV-MS19 | KX694242 | 2013 | |||||||
| IWV-MS18 | KX584741 | 2013 | |||||||
| China | Penaeus japonicus | WSSV-CN | Complete genome | AF332093 | 1996 | [30] | |||
| WSSV-CN01 | KT995472 | 1994 | [34] | ||||||
| Procambarus clarkii | WSSV-CN02 | Complete genome | KT995470 | 2010 | [34] | ||||
| WSSV-CN-Pc | KX686117 | 2015 | [36] | ||||||
| Penaeus vannamei | WSSV-CN03 | Complete genome | KT995471 | 2010 | [34] | ||||
| Marsupenaeus japonicus | WSSV-CN04 | Complete genome | KY827813 | 2012 | [35] | ||||
| Thailand | Penaeus monodon | WSSV-TH | Complete genome | AF369029 | 1996 | [29] | |||
| TH-96-II | Nonfunctional ORF14 gene; ORFI, ORFII, ORFIII, ORFIV, and ORFV genes; ORF15 and ORF16 gene |
AY753327 | 2005 | [184] | |||||
| Taiwan | Penaeus monodon | WSSV-TW | Complete genome | AF440570 | 1994 | [31] | |||
| South Korea | Penaeus vannamei | WSSV-KR | Complete genome | JX515788 | 2011 | [32,34] | |||
| Australia | Penaeus monodon | WSSV-AU | Complete genome | MF768985 | 2016 | [37] | |||
| USA | Penaeus vannamei | CN_95_DFPE | Complete genome | MN840357 | 2017 | [41] | |||
| Ecuador | Penaeus vannamei | WSSV-EC-15098 | Complete genome | MH090824 | 2015 | [39] | |||
| Brazil | Penaeus vannamei | WSSV-chimera | Complete genome | MG264599 | 2015 | [40] | |||
| FSL39 | Partial genome | MF784752 | |||||||
| ss DNA | IHHNV (Type I) |
Australia | Penaeus monodon | Australian | Non-structural protein gene Non-structural protein 1 gene Capsid protein genes |
GQ475529 | 2008 | [60] | |
| IHHNV (Type II) |
Thailand | Penaeus monodon | - | Non-structural protein 2 gene Non-structural protein 1 gene Capsid protein genes |
AY362547 | 2003 | [173] | ||
| IHHNV_TH | AY102034 | 2000 | [185] | ||||||
| Taiwan | Penaeus monodon | Taiwan B | Non-structural protein 2 gene Non-structural protein 1 gene; Capsid protein genes |
AY355307 | 2003 | [186] | |||
| Vietnam | Penaeus monodon | IHHNV-VN | Non-structural protein 2 gene Non-structural protein 1 gene Capsid protein genes |
JN616415 | 2009 | [60] | |||
| ST | KC513422 | 2011 | |||||||
|
Penaeus monodon;
Penaeus vannamei |
KK-Lv-VIET1 | Non-structural protein 1 gene | MN481525 | 2019 | [187] | ||||
| Penaeus stylirostris | VN2007 | Complete genome | KF031144 | 2007 | [57] | ||||
| India | Penaeus monodon | IN-07 | Complete genome | GQ411199 | 2007 | [60] | |||
| IHHNV | Capsid protein gene | FJ169961 | 2007 | [173] | |||||
| IHHNV (Type III) |
Vietnam | Penaeus monodon | KG | Complete genome | JX840067 | 2012 | [57] | ||
| Taiwan | Penaeus monodon | Taiwan A | Non-structural protein 2 gene; Non-structural protein 1 gene; Capsid protein genes |
AY355306 | 2003 | [186] | |||
| Taiwan C | AY355308 | 2003 | |||||||
| Ecuador | Penaeus vannamei | IHHNV | Non-structural protein 2 gene; Non-structural protein 1 gene; Capsid protein genes |
AY362548 | 2003 | [186] | |||
| Brazil | Penaeus vannamei | IHHNV_BR | Partial genome | KJ862253 | 2013 | [60] | |||
| China | Penaeus penicillatus | IHHNV | Complete genome | KJ830753 | - | [60] | |||
| Penaeus monodon | Fujian | EF633688 | 2007 | [188] | |||||
| Ganyu | JX258653 | 2009 | [57] | ||||||
| Penaeus vannamei | CSH-1 | KF907320 | 2012 | ||||||
| Penaeus vannamei | Sheyang | KF214742 | 2011 | ||||||
| Hawaii | Penaeus stylirostris | Hawaii A | Complete genome | NC_002190 | 1990 | [60] | |||
| Hawaii B | AF218266 | 1990 | |||||||
| Malaysia | Macrobrachium rosenbergii | IHHNV | Non-structural protein genome | HM536212 | 2009 | [189] | |||
| Taiwan | Macrobrachium rosenbergii | AC-04-367 | Non-structural protein 1 gene | DQ057982 | - | ||||
| AC-05-005 | DQ057983 | - | |||||||
| Mexico | Penaeus stylirostris | IHHNV | Non-structural protein 2 gene; Non-structural protein 1 gene; Capsid protein genes |
AF273215 | 2000 | [190] | |||
| South Korea | Penaeus vannamei | K1 | Structural protein gene | HQ699073 | 2010 | [191] | |||
| K2 | HQ699074 | 2010 | |||||||
| KLV-2010-01 | Complete genome | JN377975 | 2010 | [58] | |||||
| IHHNV(Type A) | Madagascar | Penaeus monodon | IHHNV | Non-structural protein 1 gene; Structural protein genes; Unnamed retrotransposon reverse transcriptase gene |
DQ228358 | - | [191] | ||
| Australia | Penaeus monodon | Au2005 | Non-structural protein 2 gene; Non-structural protein 1-like gene; Viral capsid protein gene |
EU675312 | - | [188] | |||
| IHHNV (Type B) |
Tanzania Mozambique |
Penaeus monodon | East Africa | Non-structural protein 1 gene; Structural protein genes |
AY124937 | 2000 | [185] | ||
| RNA virus | ds RNA | IMNV (PsIMNV) |
Indonesia | Penaeus vannamei | ID-EJ-12-1 | ORF1/ORF2 and ORF1 polyprotein genes | KJ636783 | 2012 | [40,77] |
| ID-EJ-12-1 | ORF1 polyprotein | AIC34743 | 2012 | ||||||
| ID-EJ-12-2 | ORF1/ORF2 | AIC34746 | 2012 | ||||||
| ID-EJ-12-3 | ORF1 polyprotein | AIC34749 | 2012 | ||||||
| ID-LP-12-2 | AIC34750 | 2012 | |||||||
| ID-BB-12 | AIC34752 | 2012 | |||||||
| ID-EJ-06 | Structural protein | ABN05324 | - | ||||||
| ID-LP-11 | Complete genome | KJ636782 | 2011 | ||||||
| ID-LP-11 | ORF1 polyprotein | AIC34741 | 2011 | ||||||
| ID-LP-12-1 | ORF1/ORF2 | AIC34748 | 2012 | ||||||
| IMNV | Complete genome | EF061744 | - | [74] | |||||
| Indonesia | KF836757 | 2013 | [192] | ||||||
| Brazil | Penaeus vannamei | BZ-03 | Structural protein | AAT67230 | - | [77] | |||
| ZS2011001 | Capsid protein | AGF33812 | 2004 | ||||||
| Brazil 01 | Structural protein | ADG37656 | 2007 | ||||||
| Brazil 02 | ADN43996 | 2007 | |||||||
| IMNV-BZ-11-UAZ219 | ORF1 polyprotein | AIC34754 | 2011 | ||||||
| IMNV | Complete genome | AY570982 | - | [74] | |||||
| ss RNA | YHV (genotype 1) |
Thailand | Penaeus monodon | YHV1992 | Complete genome | FJ848673 | 1992 | [98,101] | |
| YHV1995 | Complete genome | FJ848674 | 1995 | ||||||
| Chachoengsao 1998 | Complete genome | EU487200 | 1988 | [98,108] | |||||
| YHA-98-Ref | pp1ab gene | EU785033 | 1998 | [98,114] | |||||
| Thailand: Cholburi | Envelope structural glycoprotein gene | EF156405 | 1999 | [108] | |||||
| YHV1999 | Complete genome | FJ848675 | [98,101] | ||||||
| YHV-PmA | 3C-like protease gene | EU977577 | - | [108] | |||||
| Replicase polyprotein 1ab gene | EU977578 | ||||||||
| RNA polymerase gene | EU977579 | ||||||||
| Helicase gene | EU977580 | ||||||||
| Nucleocapsid gene | EU977581 | ||||||||
| Glycoprotein 116 gene | EU977582 | ||||||||
| Glycoprotein 64 gene | EU977583 | ||||||||
| Genomic sequence | EU977584 | ||||||||
| THA-00-DRH | pp1ab gene | EU785032 | 2000 | [98,114] | |||||
| THA-01-D4 | EU785004 | 2001 | |||||||
| THA-01-D8 | EU785034 | 2001 | |||||||
| THA-01-D9 | EU785019 | 2001 | |||||||
| THA-01-D10 | EU784984 | 2001 | |||||||
| THA-02-D34 | EU785001 | 2002 | |||||||
| THA-03-D1 | EU784982 | 2003 | |||||||
| THA-03-D2 | EU784991 | 2003 | |||||||
| THA-03-D3 | EU784998 | 2003 | |||||||
| THA-03-DB1 | EU785023 | 2003 | |||||||
| THA-03-D29 | EU785035 | 2003 | |||||||
| THA-03-D30 | EU784999 | 2003 | |||||||
| THA-03-D33 | EU785000 | 2003 | |||||||
| Penaeus vannamei | YHV | ORF1b genes | FJ627274 | 2007 | [106] | ||||
| Mexico | Penaeus vannamei | YHV | 3C-like protease gene | DQ978355 | 2000 | [108] | |||
| ORF1a and ORF1b polyprotein gene | DQ978356 | ||||||||
| Nonfunctional ORF1b polyprotein gene | DQ978357 | ||||||||
| ORF1b polyprotein gene | DQ978358 | ||||||||
| Helicase gene | DQ978359 | ||||||||
| Nucleocapsid gene | DQ978360 | ||||||||
| Glycoprotein 116 gene | DQ978361 | ||||||||
| Glycoprotein 64 gene | DQ978362 | ||||||||
| ORF4-like gene | DQ978363 | ||||||||
| China | Fenneropenaeus chinensis | Hb2012 | Replicase polyprotein 1b mRNA | KF278563 | 2012 | [98] | |||
| GAV (genotype 2) |
Australia | Penaeus monodon | GAV | Complete genome | AF227196 | - | [98,101,108] | ||
| NC_010306 | - | [101] | |||||||
| AUS-97-MCMS1 | pp1ab gene | EU784980 | 1997 | [98,114] | |||||
| AUS-97-MCMS2 | EU784989 | 1997 | |||||||
| AUS-97-MCMS3 | EU785038 | 1997 | |||||||
| AUS-00-H2 | EU785029 | 2000 | |||||||
| AUS-00-HL4 | EU785030 | 2000 | |||||||
| AUS-00-HL5 | EU785031 | 2000 | |||||||
| AUS-00-HL11 | EU785028 | 2000 | |||||||
| AUS-96-Ref | EU785026 | 1996 | |||||||
| Vietnam | Penaeus monodon | VNT-01-H65 | pp1ab gene | EU785039 | 2001 | [114] | |||
| VNT-01-H77 | EU785013 | 2001 | |||||||
| VNM-02-H6 | EU785009 | 2002 | |||||||
| VNM-02-H64 | EU785008 | 2002 | |||||||
| Thailand | Penaeus monodon | THA-03-HB3 | pp1ab gene | EU785024 | 2003 | [114] | |||
| THA-03-HG | EU785025 | 2003 | |||||||
| THA-03-HA | EU785021 | 2003 | |||||||
| THA-03-HN | EU785022 | 2003 | |||||||
| THA-04-H20 | EU784992 | 2004 | |||||||
| THA-04-HK | EU785027 | 2004 | |||||||
| YHV (genotype 3) |
Vietnam | Penaeus monodon | VNM-02-H5 | pp1ab gene | EU785006 | 2002 | [98,114] | ||
| VNM-02-H258 | EU784994 | 2002 | |||||||
| VNM-02-H81 | EU785016 | 2002 | |||||||
| VNM-02-H70 | EU785012 | 2002 | |||||||
| VNM-01-H41 | EU785040 | 2001 | |||||||
| VNM-01-H42 | EU785041 | 2001 | |||||||
| VNM-02-H278 | EU784996 | 2002 | |||||||
| VNM-02-H264 | EU784995 | 2002 | |||||||
| VNM-02-H93 | EU785020 | 2002 | |||||||
| VNM-02-H93 | p20 gene; pp3 gene |
EU785042 | 2002 | [114] | |||||
| Indonesia | Penaeus monodon | IDN-04-H7 | pp1ab gene | EU785011 | 2004 | [114] | |||
| IDN-04-H11 | EU784985 | 2004 | |||||||
| IDN-04-H10 | EU784983 | 2004 | |||||||
| IDN-04-H4 | EU785002 | 2004 | [98,114] | ||||||
| Malaysia | Penaeus monodon | MYS-03-H1 | pp1ab gene | EU784981 | 2003 | [114] | |||
| MYS-03-H2 | EU784990 | 2003 | |||||||
| MYS-03-H3 | EU784997 | 2003 | |||||||
| Mozambique | Penaeus monodon | MOZ-04-H1 | pp1ab gene | EU784986 | 2004 | ||||
| YHV (genotype 4) |
Thailand | Penaeus monodon | YHV type 4 | ORF1b polyprotein gene | EU170438 | - | [98,193] | ||
| gp116 gene | EU123854 | ||||||||
| Indonesia | Penaeus monodon | IND-02-H9 | pp1ab gene | EU785017 | 2002 | [98,114] | |||
| IND-02-H5 | EU785005 | 2002 | |||||||
| IND-02-H7 | EU785010 | 2002 | |||||||
| India | Penaeus monodon | IND-02-H9 | p20 gene; pp3 gene |
EU785043 | 2002 | [114] | |||
| YHV (genotype 5) |
Thailand | Penaeus monodon | THA-03-SG21 | pp1ab gene | EU784993 | 2003 | |||
| YHV | ORF1b polyprotein gene | EU853170 | 2005 | [193] | |||||
| Malaysia | Penaeus monodon | MYS-03-H4 | pp1ab gene | EU785003 | 2003 | [114] | |||
| Philippines | Penaeus monodon | PHL-03-H8 | EU785015 | 2003 | |||||
| YHV (genotype 6) |
Mozambique | Penaeus monodon | MOZ-04-H6 | pp1ab gene | EU785007 | 2004 | |||
| MOZ-04-H8 | EU785014 | 2004 | |||||||
| MOZ-04-H9 | EU785018 | 2004 | |||||||
| MOZ-04-H11 | EU785036 | 2004 | |||||||
| MOZ-04-H12 | EU785037 | 2004 | |||||||
| YHV (genotype 7) |
Australia | Penaeus monodon | YHV7 (13-00169-01) PCR1 | ORF1b polyprotein gene | KP738160 | 2012 | [98,105] | ||
| YHV7 (13-00169-01) PCR2 | KP738161 | ||||||||
| YHV7 (13-00169-02) PCR2 | KP738162 | ||||||||
| YHV7 (13-00169-03) PCR2 | KP738163 | ||||||||
| YHV7 (13-00169-02) PCR3 | KP738164 | ||||||||
| YHV (genotype 8) |
China | Fenneropenaeus chinensis | 20120706 | Complete genome | KX947267 | 2012 | [101] | ||
| TSV | Ecuador | Penaeid shrimp | EC1993a | Capsid protein 2 gene | FJ876460 | 1993 | [127] | ||
| EC1993b | FJ876461 | ||||||||
| EC1994 | FJ876466 | 1994 | |||||||
| EC2006a | FJ876512 | 2006 | |||||||
| EC2006b | FJ876513 | ||||||||
| Columbia | Penaeid shrimp | CO1994a | FJ876462 | 1994 | |||||
| CO1994b | FJ876463 | ||||||||
| CO1994c | FJ876464 | ||||||||
| CO1994d | FJ876465 | ||||||||
| CO1998 | FJ876477 | 1998 | |||||||
| Penaeus vannamei | CO-06A | JN194141 | 2006 | [138] | |||||
| CO-06B | JN194142 | ||||||||
| CO-06C | JN194143 | ||||||||
| CO-07A | JN194144 | 2007 | |||||||
| CO-07B | JN194145 | ||||||||
| CO-10 | JN194146 | 2010 | |||||||
| CO10 | Complete genome | JF966384 | 2010 | ||||||
| USA | Penaeus vannamei | 94USHI | Complete genome | AF277675 | 1994 | [62,132,194,195] | |||
| HI94TSV | Viral coat protein 2 gene | AY826054 | 1994 | [117] | |||||
| Viral coat protein 3 gene | AY826055 | ||||||||
| US-TX04 | Complete genome | GQ502201 | 2004 | [132] | |||||
| 2005-334 | MT877007 | 2019 | [119] | ||||||
| Penaeid shrimp | US1994 | Capsid protein 2 gene | FJ876468 | 1994 | [127] | ||||
| US1995 | FJ876469 | 1995 | |||||||
| US1996 | FJ876474 | 1996 | |||||||
| US1998 | FJ876476 | 1998 | |||||||
| US2004 | FJ876492 | 2004 | |||||||
| US2007 | FJ876517 | 2007 | |||||||
| Honduras | Penaeid shrimp | HO1994 | Capsid protein 2 gene | FJ876467 | 1994 | ||||
| HO1998 | FJ876475 | 1998 | |||||||
| HO2003 | FJ876483 | 2003 | |||||||
| Mexico | Penaeid shrimp | MX1995a | Capsid protein 2 gene | FJ876470 | 1995 | [127] | |||
| MX1995b | FJ876471 | ||||||||
| MX1995c | FJ876472 | ||||||||
| MX1996 | FJ876473 | 1996 | |||||||
| MX1998 | FJ876478 | 1998 | |||||||
| MX1999a | FJ876479 | 1999 | |||||||
| MX2000 | FJ876480 | 2000 | |||||||
| MX2004 | FJ876493 | 2004 | |||||||
| MX2005a | FJ876504 | 2005 | |||||||
| MX2005b | FJ876505 | ||||||||
| MX2005c | FJ876506 | ||||||||
| MX2006 | FJ876514 | 2006 | |||||||
| MX2007 | FJ876521 | 2007 | |||||||
| Penaeus vannamei | SIN98TSV | Viral coat protein 1 gene | AF510515 | 1998 | [125,195] | ||||
| MX99 | Coat protein gene | AF277378 | 1999 | [126,127] | |||||
| Mexico 10 | Capsid protein 2 gene | JN194147 | 2010 | [138] | |||||
| Penaeus stylirostris | MX99TSV | Viral coat protein 1 gene | AF510516 | 1999 | [125,195] | ||||
| SON2KTSV | AF510517 | 2000 | [131,195] | ||||||
| Penaeus stylirostris | HI94TSV | Viral coat protein 1 gene | AF510518 | 2000 | [117,125] | ||||
| Taiwan | Penaeus vannamei | TW99 | Coat protein gene | AF406789 | 1999 | [62,126,195] | |||
| Penaeus monodon | Tw2KPmTSV | Capsid protein precursor | AY355309 | 2000 | [126] | ||||
| Metapenaeus ensis | Tw2KMeTSV | Capsid protein precursor | AY355310 | 2000 | [196] | ||||
| Penaeus vannamei | Tw02PvTSV | Capsid protein precursor | AY355311 | 2002 | [127] | ||||
| Penaeid shrimp | TW2007 | Capsid protein 2 gene | FJ876520 | 2007 | |||||
| Thailand | Penaeus vannamei | Th03-1TSV | Capsid protein 2 gene | DQ000304 | 2003 | [196] | |||
| Th03-2TSV | DQ000305 | ||||||||
| ThOct03LvTSV | VP1 gene | AY912503 | 2003 | [126] | |||||
| ThMar04LvTSV | AY912504 | 2004 | |||||||
| ThJul04LvTSV | AY912508 | ||||||||
| Penaeus monodon | ThMar04Pm1TSV | VP1 gene | AY912505 | 2004 | |||||
| ThMar04Pm2TSV | AY912506 | ||||||||
|
Penaeus monodon (post-larvae) |
ThMay04PmPLTSV | VP1 gene | AY912507 | 2004 | |||||
| Penaeus vannamei | TH03-1 | Capsid protein 1 gene | AY755587 | 2003 | [125,196] | ||||
| TH03-2 | AY755588 | ||||||||
| TH03-3 | AY755589 | ||||||||
| TH03-4 | AY755590 | ||||||||
| TH03-5 | AY755591 | ||||||||
| TH03-7 | AY755593 | ||||||||
| TH03-9 | AY755595 | ||||||||
| TH04Lv | Complete genome | AY997025 | 2005 | [132,197] | |||||
| Macrobrachium rosenbergii | TH03-6 | Capsid protein 1 gene | AY755592 | 2003 | [125] | ||||
| Penaeus monodon | TH04Pm | Capsid protein 2 gene | DQ000306 | 2004 | [196] | ||||
| TH03-8 | Capsid protein 1 gene | AY755594 | 2003 | [125] | |||||
| Penaeid shrimp | TH2003a | Capsid protein 2 gene | FJ876484 | 2003 | [127] | ||||
| TH2003b | FJ876485 | ||||||||
| TH2004a | FJ876496 | 2004 | |||||||
| TH2004b | FJ876497 | ||||||||
| TH2006 | FJ876515 | 2006 | |||||||
| Myanmar | Penaeus monodon | Mm03Pm | Capsid protein 1 gene | AY755596 | 2003 | [125,196] | |||
| Vietnam | Penaeus vannamei | VN-TSV | Capsid protein gene | AY694136 | - | [198] | |||
| Belize | Penaeus vannamei | BZ01 | Non-structural polyprotein gene; Capsid protein precursor gene |
AY590471 | 2001 | [62,124,132] | |||
| Penaeus vannamei | 2005-175 | Complete gene | MT877008 | 2019 | [119] | ||||
| BLZ02TSV | Viral coat protein 1 gene | AY826051 | 2002 | [117] | |||||
| Viral coat protein 2 gene | AY826052 | ||||||||
| Viral coat protein 3 gene | AY826053 | ||||||||
| Penaeid shrimp | BH2001 | Capsid protein 2 gene | FJ876481 | 2001 | [127] | ||||
| BH2002 | FJ876482 | 2002 | |||||||
| BH2004a | FJ876490 | 2004 | |||||||
| BH2004b | FJ876491 | ||||||||
| BH2005a | FJ876498 | 2005 | |||||||
| BH2005b | FJ876499 | ||||||||
| BH2005c | FJ876500 | ||||||||
| BH2008 | FJ876522 | 2008 | |||||||
| Indonesia | Penaeus vannamei | Id03TSV | Capsid protein 2 gene | DQ000303 | 2003 | [196] | |||
| Penaeus vannamei | Indonesia 10 | JN194148 | 2010 | [138] | |||||
| Penaeid shrimp | ID2003a | FJ876486 | 2003 | [127] | |||||
| ID2003b | FJ876487 | ||||||||
| ID2003c | FJ876488 | ||||||||
| ID2005 | FJ876501 | 2005 | |||||||
| ID2006 | FJ876510 | 2006 | |||||||
| China | Penaeus vannamei | ZHZC3TSV | Complete genome | DQ104696 | 2005 | [132,199] | |||
| Cn03TSV | Capsid protein 2 gene | DQ000301 | 2003 | [196] | |||||
| Ch-1 | Capsid protein 1 gene | AY755597 | [125,200] | ||||||
| Ch-2 | AY755598 | ||||||||
| Ch-3 | AY755599 | ||||||||
| Ch-4 | AY755600 | ||||||||
| Ch-6 | AY755602 | ||||||||
| Penaeus japonicus | Ch-5 | Capsid protein 1 gene | AY755601 | 2003 | [125] | ||||
| Penaeid shrimp | CH2003a | Capsid protein 2 gene | FJ876489 | 2003 | [127] | ||||
| CH2004 | FJ876494 | 2004 | |||||||
| CH2005a | FJ876509 | 2005 | |||||||
| CH2007 | FJ876518 | 2007 | |||||||
| Korea | Penaeus vannamei | KOR-CsPv04TSV | Capsid protein 1 mRNA | DQ099912 | 2004 | [131] | |||
| KOR-ImPv05TSV | DQ099913 | ||||||||
| Eritrea | Penaeus monodon | Er04PmTSV | Capsid protein 2 gene | DQ000302 | 2004 | [196] | |||
| Penaeid shrimp | ER2004 | FJ876495 | 2004 | [127] | |||||
| Venezuela | Penaeus vannamei | VE05 | Complete genome | DQ212790 | 2005 | [124] | |||
| 2005-194 | MT877006 | 2019 | [119] | ||||||
| Penaeid shrimp | VE2005a | Capsid protein 2 gene | FJ876502 | 2005 | [127] | ||||
| VE2005b | FJ876503 | ||||||||
| Saudi Arabia | Penaeid shrimp | SA2007 | Capsid protein 2 gene | FJ876519 | 2007 | ||||
| Penaeus indicus | SAPi | Complete genome | JX094350 | 2010 | [118] | ||||
| SA2010a | Capsid protein 2 gene | JQ356858 | |||||||
| SA2010b | JQ356859 | ||||||||
| SA2010c | JQ356860 | ||||||||
| SA2011a | JQ356861 | 2011 | |||||||
| SA2011b | JQ356862 | ||||||||
| SA2011c | JQ356863 | ||||||||
| SA2011d | JQ356864 | ||||||||
| SA2011e | JQ356865 | ||||||||
| Aruba | Penaeid shrimp | AW2005 | Capsid protein 2 gene | FJ876508 | 2005 | [127] | |||
| AW2006 | FJ876511 | 2006 | |||||||
| Nicaragua | Penaeid shrimp | NI2005 | Capsid protein 2 gene | FJ876507 | 2005 | ||||
| NI2006 | FJ876516 | 2006 | |||||||
| WTD (MrNV) |
French West Indies | Macrobrachium rosenbergii | MrNV | Segment RNA-1 | AY222839 | 2003 | [143,154] | ||
| Segment RNA-2 | AY222840 | ||||||||
| RNA-1 | NC_005094 | 2009 | [201] | ||||||
| RNA-2 | NC_005095 | - | |||||||
| MrNV-Ant | Putative RNA-dependent RNA-polymerase gene | AY313773 | 2005 | [141] | |||||
| China | MrNV | RNA-directed RNA polymerase gene | AAQ54758 | - | [202] | ||||
| Chinese 1 | AY231436 | 2006 | [143,202] | ||||||
| Chinese 2 | Segment RNA-2 | FJ751225 | - | [143] | |||||
| MrNV | Segment RNA-1 RNA-dependent RNA polymerase gene; B2 protein gene |
FJ751226 | 2006 | [201] | |||||
| Capsid protein gene | AY231437 | - | [143] | ||||||
| India | Nellore | Capsid protein gene | GU300102 | - | [203] | ||||
| B2 protein gene | GU300103 | 2011 | |||||||
| MrNV | Capsid protein-like gene | HM565741 | 2010 | [143] | |||||
| RNA-1 RNA-dependent RNA polymerase gene; B2 protein gene |
JQ418295 | - | [153] | ||||||
| RNA-2 capsid protein gene | JQ418298 | - | [149,200] | ||||||
| Capsid protein | AM114036 | - | |||||||
| RNA-dependent RNA polymerase gene | AAO60068 | - | [152] | ||||||
| RNA-directed RNA polymerase gene | DQ146969 | - | [201] | ||||||
| Kakinada 1MrNV | Isolate Kakinada 1MrNV capsid protein gene | HQ637179 | 2008 | [149] | |||||
| Taiwan | AC06-016 | RNA-directed RNA polymerase gene | DQ459203 | - | [143] | ||||
| AC06-017 | DQ459204 | ||||||||
| AC06-024 | DQ459205 | ||||||||
| AC06-86 | DQ459206 | ||||||||
| AC06-088 | DQ459207 | ||||||||
| AC06-89 | DQ459208 | ||||||||
| MrNV | Segment RNA-1 nonfunctional polymerase gene | DQ521574 | - | [201] | |||||
| Segment RNA-2 capsid protein gene | DQ521575 | - | |||||||
| Malaysia | MrNV | Dependent RNA polymerase gene | JN187416 | 2009 | [143] | ||||
| Australia | 07-265.1 | Capsid protein gene | FJ379530 | 2007 | [204] | ||||
| 07-265.2 | A protein gene | FJ379531 | |||||||
| Australian | Segment RNA 1 | JN619369 | 2004 | [143] | |||||
| Segment RNA 2 | JN619370 | ||||||||
| Thailand | M298 | Capsid protein gene | EU150126 | - | [143] | ||||
| M299 | EU150127 | ||||||||
| M308 | EU150128 | ||||||||
| M12 | EU150129 | ||||||||
| MrNV | Capsid protein mRNA | DQ189990 | - | [201] | |||||
| WTD (XSV) |
Taiwan | Macrobrachium rosenbergii | XSV | Nucleocapsid protein CP17 gene | DQ521573 | - | [205] | ||
| Thailand | M23 | Capsid protein gene | EU150133 | - | [204] | ||||
| M309 | EU150132 | - | |||||||
| 07-265.3 | FJ379532 | 2007 | |||||||
| India | Kakinada 1XSV | Isolate Kakinada 1XSV capsid protein gene | HQ637180 | 2008 | [149] | ||||
| XSV | Capsid protein gene | JQ418299 | - | ||||||
| Capsid protein, genomic RNA | AM114037 | - | |||||||
| Capsid protein gene | NC_043494 | - | |||||||
| Capsid protein gene | AY247793 | - | [198] | ||||||
| China | XSV | Nucleocapsid protein CP17 and CP16 genes | DQ174318 | - | [206] | ||||
WSSV is known to be highly pathogenic to crabs, copepods, and other arthropods, including penaeid shrimp (P. monodon, P. indicus, P. japonicus, P. chinensis, P. penicillatus, P. semisulcatus, P. aztecus, P. vannamei, P. merguiensis, P. duorarum, P. stylirostris, Trachypenaeus curvirostris, and Metapenaeus ensis), caridean shrimp (Exopalaemon orientalis and M. rosenbergii) and crayfish, Procambarus clarkii [44] (Table 4). Of the more than 100 potential host species for WSSV, it is particularly lethal to all marine aquaculture shrimp which are more vulnerable to WSSV than freshwater shrimp and other species, even though the susceptibility of a potential host to WSSV may vary from species to species [20,45]. During all stages of development, from egg to adult, species are vulnerable to WSSV [46].
Table 4.
Summary of host species following DNA and RNA viral disease infections.
| Type | Pathogen | Host Species | Characteristics | Reference | |
|---|---|---|---|---|---|
| DNA virus | ds DNA | WSSV | Penaeus monodon |
|
[9,13,18,20,44,49,51,53,207,208,209,210] |
| Penaeus indicus | |||||
| Penaeus japonicas | |||||
| Penaeus chinensis | |||||
| Penaeus penicillatus | |||||
| Penaeus semisulcatus | |||||
| Penaeus aztecus | |||||
| Penaeus vannamei | |||||
| Penaeus merguiensis | |||||
| Penaeus duorarum | |||||
| Penaeus stylirostris | |||||
| Trachypenaeus curvirostris | |||||
| Metapenaeus ensis | |||||
| Exopalaemon orientalis | |||||
| Macrobrachium rosenbergii | |||||
| Marsupenaeus japonicus | |||||
| Metapenaeus dobsoni | |||||
| Parapenaeopsis stylifera | |||||
| Solenocera indica | |||||
| Squilla mantis | |||||
| Procambarus clarkii |
|
[48,50,207,211] | |||
| Pacifastacus leniusculus | |||||
| Orconectes punctimanus | |||||
| Austropotamobius pallipes | |||||
| Panulirus versicolor |
|
[212,213] | |||
| Panulirus penicillatus | |||||
| Panulirus homarus | |||||
| Panulirus ornatus | |||||
| Charybdis feriatus |
|
[48,50,207,210,212,214] | |||
| Charybdis cruciata | |||||
| Portunus pelagicus | |||||
| Portunus sanguinolentus | |||||
| Charybdis granulata | |||||
| Scylla serrata | |||||
| Helice tridens | |||||
| Carcinus maenas | |||||
| Calappa lophos | |||||
| Paratelphusa hydrodomous | |||||
| Paratelphusa pulvinata | |||||
| Matuta planipes | |||||
| ss DNA | IHHNV | Penaeus vannamei |
|
[57,58,59,66,85] | |
| Penaeus stylirostris | |||||
| Penaeus occidentalis | |||||
| Penaeus monodon | |||||
| Penaeus semisulcatus | |||||
| Penaeus californiensis | |||||
| Penaeus schmitti | |||||
| Penaeus japonicus | |||||
| Penaeus latisulcatus | |||||
| Penaeus chinensis | |||||
| Penaeus setiferus | |||||
| Penaeus aztecus | |||||
| Penaeus duorarum | |||||
| Penaeus subtilis | |||||
| Artemesia longinaris | |||||
| Macrobrachium rosenbergii | |||||
| Palaemon macrodactylus | |||||
| Procambarus clarkii | |||||
| Hemigrapsus penicillatus | |||||
| Neohelice granulate | |||||
| Corydoras arcuatus | |||||
| Mytilus edulis | |||||
| Mactra chinensis | |||||
| Tegillarca granosa | |||||
| Ruditapes philippinarum | |||||
| Sinonovacula constricta | |||||
| Meretrix meretrix | |||||
| Mactra veneriformis | |||||
| RNA virus | ds RNA | IMNV | Penaeus vannamei |
|
[62,73,76,83,84,85,86,195] |
| Penaeus stylirostris | |||||
| Penaeus monodon | |||||
| Farfantepenaeus subtiltis | |||||
| ss RNA | YHD | Penaeus stylirostris |
|
[68,91,100,101,106,113,215] | |
| Penaeus aztecus | |||||
| Penaeus duorarum | |||||
| Penaeus setiferus | |||||
| Penaeus vannamei | |||||
| Penaeus esculentus | |||||
| Penaeus stylirostris | |||||
| Penaeus monodon | |||||
| Fenneropenaeus merguiensis | |||||
| Farfantepenaeus aztecus | |||||
| Farfantepenaeus duorarum | |||||
| Metapenaeus ensis | |||||
| Metapenaeus affinis | |||||
| Marsupenaeus japonicus | |||||
| TSV | Penaeus stylirostris |
|
[52,95,129,130,136,216] | ||
| Penaeus schmitti | |||||
| Penaeus setiferus | |||||
| Penaeus duorarum | |||||
| Penaeus aztecus | |||||
| Penaeus monodon | |||||
| Penaeus japonicus | |||||
| Penaeus chinensis | |||||
| WTD | Macrobrachium rosenbergii |
|
[56,140,144,160,163] | ||
| Penaeus indicus | |||||
| Penaeus japonicus | |||||
| Penaeus monodon | |||||
| Penaeus vannamei | |||||
| Cherax quadricarinatus | |||||
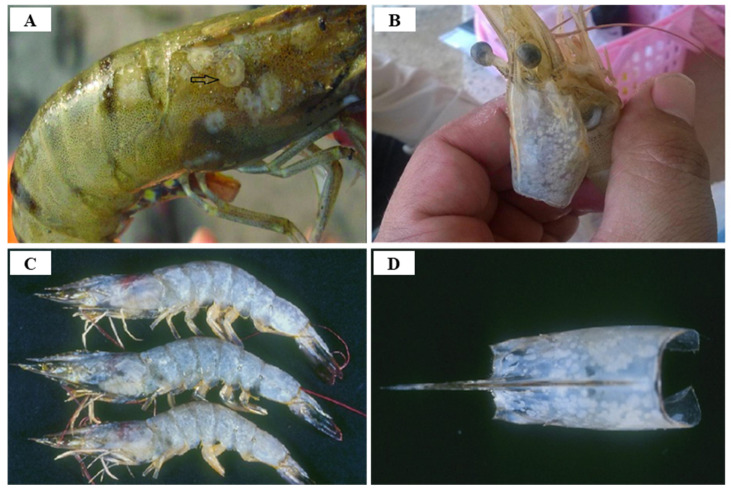
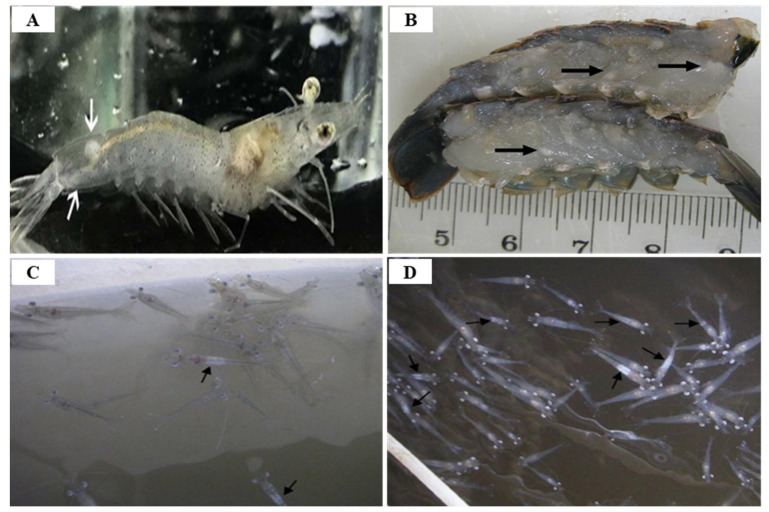
Shrimp infected with WSSV are characterized by anorexia, lethargy, abnormal behavior (decreased swimming ability, disorientation and swimming on one side), red discoloration of the body surface (uropods, telson, pereiopods, and pleopods), swelling of branchiostegites, a loosening of the cuticle, enlargement and yellowish discoloration of the hepatopancreas, thinning and delayed clotting of hemolymph, and characteristic white spots with a diameter of 1–2 mm (or 0.5–3.0 mm) on the carapace, appendages, and internal surfaces during disease progression [47] (Figure 6). WSSV infection in shrimp is easily recognized by the characteristic white spots on the carapace, but WSSV infection does not always show symptoms of white spots and cannot be considered as a reliable indication for the diagnosis of disease, as some bacterial infections, high alkalinity, and stress can also produce similar spots [48]. Although the exact mechanism of white spot formation by WSSV infection is not known, WSSV infection can cause integumentary dysfunction, resulting in accumulation of calcium salts in the cuticle, resulting in white spot formation [49]. WSSV proliferates in the nucleus of the target cell in the subcuticular epithelium, gills, lymphoid organs, antennal glands, hematopoietic tissue, connective tissue, ovaries, and ventral nerve cord. In the later stages of infection, the infected cell is degraded and the tissue destroyed [50].
Figure 6.
External white spot symptoms indicating white spot syndrome virus (WSSV) infection. (A) Penaeus monodon and (B–D) Penaeus vannamei infected with WSSV. (A) Reprinted from Letter in Applied Microbiology, Vol. 60 (2), Hossain, A., Nandi, S.P., Siddique, M.A., Sanyal, S.K., Sultana, M., Hossain, M.A., Prevalence and distribution of White Spot Syndrome Virus in cultured shrimp, p. 7, Copyright (2014), with permission from John Wiley and Sons; (B) Reprinted from Elsevier Books, Dashtiannasab, A., Emerging and Reemerging Viral Pathogens, p. 12, Copyright (2020), with permission from Elsevier; (C,D) Reprinted from Journal of Fish Diseases, Vol. 36 (12), Cheng, L., Lin, W.H., Wang, P.C., Tsai, M.A., Hsu, J.P., Chen, S.C., White spot syndrome virus epizootic in cultured Pacific white shrimp Litopenaeus vannamei (Boone) in Taiwan, p. 9, Copyright (2013), with permission from John Wiley and Sons).
WSSV replicates rapidly in the host’s cells after infecting the host, and usually causes host death within one week [51]. WSSV frequency can be influenced by a variety of environmental stressors, such as temperature changes, salinity reductions, and pH fluctuations [27]. The transmission of WSSV disease can occur through the feeding of infected individuals, and horizontal transmission through the water-borne route has also been demonstrated. Individuals surviving WSSV infection can carry the virus for life and transmit it to their offspring through vertical transmission via oocytes [52]. Aquatic and benthic organisms such as polychaete worms, microalgae, and rotifer eggs are known vectors of WSSV, and 43 arthropods have been reported as hosts and vectors of WSSV in culture facilities, aquatic systems, and experiment [18]. Shrimp infected with WSSV usually congregate near the edge of the pond and show clinical signs one to two days before death occurs [20]. WSSV disease susceptibility in crabs, crayfish, freshwater prawns, spiny lobster, and clawed lobsters is highly variable, but in penaeid shrimp, the cumulative mortality rate is typically 90–100%, 3–10 days post-infection and WSSV is fatal to penaeid shrimp [18]. WSSV usually shows clinical signs in farmed penaeid shrimp at 14–40 days and shows a high mortality rate with up to 100% mortality in sensitive hosts.
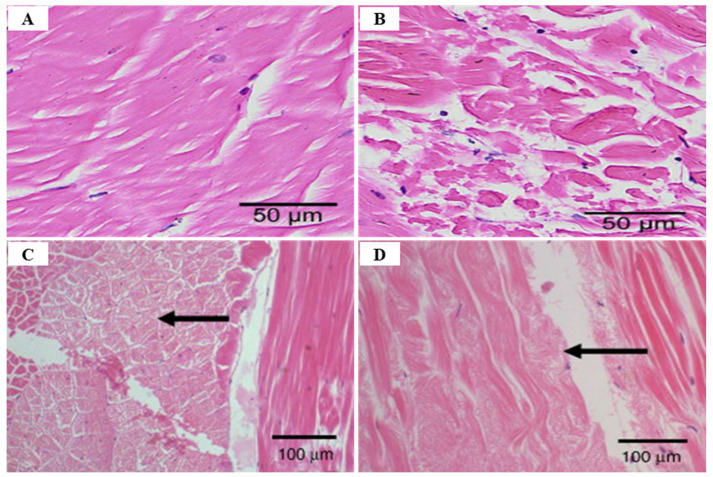
WSSV diagnostic technology is evolving from the previous, morphology-based identification to more highly sensitive immunological and molecular technologies that can detect viruses, even in asymptomatic carriers, using electron microscopy (EM) [53]. Among various diagnostic methods, PCR is used as the most sensitive method by which to detect WSSV infection, by targeting the VP28 gene [27] (Table 5). There are several PCR methods available for the diagnosis of WSSV, such as one-step PCR, nested-PCR, and real-time PCR [54]. One-step PCR can be used to detect the presence of WSSV in shrimp with high levels of infection, and nested-PCR can increase the sensitivity level when compared to one-step [55] to detect low levels of infection in the broodstock, nauplii, post-larvae, and juvenile stages [54]. Therefore, the pathogen can be easily detected using one-step PCR when clinical signs such as lethargy, reduced feeding and white spots on the exoskeleton appear, but can only be detected by nested-PCR when asymptomatic [55]. In addition, real-time PCR is a reliable technique by which to monitor the entire analysis in actual time through the detection and quantification of WSSV virion copy number [27]. Hematoxylin and eosin (H & E) histology is an important diagnosis method that is used to verify WSSV infection in shrimp [56]. Histological diagnosis following WSSV infection occurs in all tissues of mesodermal and ectodermal origin such as gills, lymphoid organ, cuticular epithelium, and sub-cuticular connective tissues, and infected nuclei are enlarged with alienated chromatin and contain inclusion bodies with strongly stained eosinophils in early infection and basophils in more advanced infections [18] (Figure 7). Biosecurity measures (specific pathogen-free (SPF) broodstock, complete dry-out of culture tanks after harvest, low water exchange systems such as RAS), restricting access to vectors and pathogens (through crab fence, bird blocking, and foot baths in shrimp farm entrance), and improving disease resistance (immunostimulants, neutralization, environmental management and vaccines) in shrimp are effective management methods, as there is currently no way to treat WSSV infection [20].
Table 5.
Summary of the DNA and RNA viral diseases PCR analyses.
| Type | Pathogen | PCR | Host | Tissue | Primer | Sequence 5′-3′ | Annealing Temperature (°C) | Amplicons (bp) |
Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA virus | ds DNA | WSSV | Conventional PCR | Cherax quadricarinatus; Procambarus clarkii | Hepatopancreas, gills, cuticle, muscle | WSI3 | GTA ACT CCT TCC ATC TCC A | 62 | 941 | [217] | |
| WSI4 | TAC GGC AGC TGC TGC ACC TTG T | ||||||||||
| Penaeus monodon | Muscle | WSSV-VP28 F | TGT GAC CAA GAC CAT CGA AAC | 52 | 516 | [27] | |||||
| WSSV-VP28 R | TCG GTC TCA GTG CCA GAG TA | ||||||||||
| Real-time qPCR (EVA green) |
Penaeus vannamei | Gills | VP24 F1 | AGG ACC CGA TCG CTT ACT TTG | - | 240 | [218] | ||||
| VP24 R1 | CTC CCT CCC TTG CGA ACT T | ||||||||||
| β-Actin F1 | GAA GTA GCC GCC CTG GTT G | 416 | |||||||||
| β-Actin R1 | CGG TTA GCC TTG GGG TTG AG | ||||||||||
| Real-time PCR (BRYT Green) |
Penaeus monodon | Muscle | WSSV-qVP28 F | TGT GAC CAA GAC CAT CGA AA | 53 | 148 | [27] | ||||
| WSSV-qVP28 R | CTT GAT TTT GCC CAA GGT GT | ||||||||||
| Real-time PCR (TaqMan) |
Cherax quadricarinatus; Procambarus clarkii | Hepatopancreas, gills, cuticle, muscle | WSS1011F | TGG TCC CGT CCT CAT CTC AG | 60 | 69 | [217] | ||||
| WSS1079R | GCT GCC TTG CCG GAA ATT A | ||||||||||
| Nested PCR | Fenneropenaeus indicus | Pleopod | 146F1 | First | ACT ACT AAC TTC AGC CTA TCT AG | 55 | 1447 | [150] | |||
| Second | GTA ACT GCC CCT TCC ATC TCC A | 941 | |||||||||
| ss DNA | IHHNV | Conventional PCR | Penaeus monodon | Tissues of infected samples | 77012F | ATC GGT GCA CTA CTC GGA | 53 | 356 | [58] | ||
| 77353R | TCG TAC TGG CTG TTC ATC | ||||||||||
| Penaeus vannamei | IHHNV389F | CGG AAC ACA ACC CGA CTT TA | 55 | 389 | |||||||
| IHHNV389R | GGC CAA GAC CAA AAT ACG AA | ||||||||||
| IHHNV392F | GGG CGA ACC AGA ATC ACT TA | 392 | |||||||||
| IHHNV392R | ATC CGG AGG AAT CTG ATG TG | ||||||||||
| Penaeus stylirostris; Penaeus vannamei | IHHNV721F | TCT ACT GCC TCT GCA ACG AG | 2000 | ||||||||
| IHHNV2860R | GTG GGT CTG GTC CAC TTG AT | ||||||||||
| Penaeus monodon | IHHNV3065F | GAC GAC GAA GAA TGG ACA GA | 3000 | ||||||||
| IHHNV3065R | TGC CTG GGT AGC TGG TAT GTA TA | ||||||||||
| IHHNV309F | TCC AAC ACT TAG TCA AAA CCA A | 309 | |||||||||
| IHHNV309R | TGT CTG CTA CGA TGA TTA TCC A | ||||||||||
| Penaeus vannamei | Hepatopancreas | IHHNV REPF | CGA TGT GCA ATA TAT ACC CGA TT | 52 | 442 | [57] | |||||
| IHHNV REPR | CTT CGC AGA AAC CGT TAA CTT | ||||||||||
| IHHNV472F | ACG AAC GAC CAC CCA TGG CA | 57 | 472 | ||||||||
| IHHNV472R | TCT GGT TCG CCC TGA CGT GT | ||||||||||
| IHHNV447F | CGA AGC GCG AGT ATC CAT CA | 55 | 447 | ||||||||
| IHHNV447R | TGA GTG ATG GAC GAA AGC GG | ||||||||||
| IHHNV-F | TCA TGA AGC GCG AGT ATC CAT CAT | 54 | 228 | ||||||||
| IHHNV-R1 | TGG GTG GTC GTT CGT ATC TT | ||||||||||
| Real-time PCR (TaqMan) |
Penaeus monodon | Gills | IHHNV-q309F1 | CCT AAA GAA AAC AGT GCA GAA TAT GAC | 60.7 | 98 | [219] | ||||
| IHHNV-q309R1 | TCA TCG TCA AGT TTA TTG ACA AGT TC | 60.8 | |||||||||
| IHHNV-qEVEF1 | CCC ACA AAA AGC AAA TAT ATC TCA CTA T | 61.1 | 106 | ||||||||
| IHHNV-qEVER1 | GTC ATT ATG AGA TTA TTG TCC CAC CTT | 61.7 | |||||||||
| Pmon-EF1qF1 | GGC CGT GTG GAG ACT GGT AT | 62.3 | 110 | ||||||||
| Pmon-EF1qR1 | CGT GGT GCA TCT CCA CAG A | 62.0 | |||||||||
| Real-time PCR (SYBR Green) |
Penaeus vannamei | Gillsm muscle, hepatopancreas, hemolymph | IHHNV 195F | GGG AGT TAC CTT TGC TGC | 56 | 195 | [220] | ||||
| IHHNV 195R | GGT CCG TCT ACT GCG TCT | ||||||||||
| RNA virus | ds RNA | IMNV | Reverse transcriptase PCR | Penaeus vannamei | Muscle | 389F | CGG AAC ACA ACC CGA CTT TA | 55 | 284 | [62] | |
| 389R | GGC CAA GAC CAA AAT ACG AA | ||||||||||
| Penaeus vannamei | Muscle | IMNV 105-297-F |
CAT ATG GGG CAA TTA CGG TTA CAG GG | 60 | 600 | [74] | |||||
| IMNV 105-297-R |
CGG GAT CCG TAT ACA TAC CAA ATG GCC | ||||||||||
| IMNV 300-527-F |
CTC GAG ACT AAA CAA ACA ACA GAC AAT GC | 55 | 700 | [87] | |||||||
| IMNV 300-527-R |
GGA TCC GGA GTC CCA TCA TAT AAC TGG | ||||||||||
| IMNVF22 | C CAT ATG ATT GTT TCA ATG GAA AAT C | 57 | 811 | [84] | |||||||
| IMNVR819 | G GAA TTC TTG TAG TGC AGT TGC TGG | ||||||||||
| IMNVF820 | CGG GA TCC GCT GCA AAA GAG GGT GCT CG |
924 | |||||||||
| IMNVR1728 | G GAA TTC TTG CAT TGA ACTCCACGAAAA C | ||||||||||
| IMNVF1729 | CG GGA TCC GGT AGT ATT GCA CCA GCA ATG | 1041 | |||||||||
| IMNVR | GGA ATT CTT ATA CTG TTG CTG T CG CTT G | ||||||||||
| IMNV 99372G09- F |
CGA CGC TGC TAA CCA TAC A A | 62 | 372 | [221] | |||||||
| IMNV 99372 G10-R | ACT CGC CTG TTC GAT CAA GT | ||||||||||
| IMNV-NF | GGC ACA TGC TCA GAG ACA | 60 | 139 | [89] | |||||||
| IMNV-NR | AGC GCT GAG TCC AGT CTT G | ||||||||||
| ss RNA | YHD | RT-PCR | Penaeus monodon | Gills, hemolymph | YHV5f | CGT ATT GCA TCG AAC GTC ACT G | 60 | 885 | [222] | ||
| YHV5r | CAA GAT CAC TAA TAA CGC CTG ATG C | ||||||||||
| Nested PCR | YHV2s | CGG GGT TAC CCG CTT ATA TT | 400 | ||||||||
| YHV2as | GCC TGA GGT GAA GTC CAT GT | ||||||||||
| RT-PCR | Penaeus monodon | Gills, epidermis | YCF1a | ATC GTC GTC AGC TAC CGC AAT ACT GC | 60 | 359 | [98] | ||||
| YCF1b | ATC GTC GTC AGY TAY CGT AAC ACC GC | ||||||||||
| YCR1a | TCT TCR CGT GTG AAC ACY TTC TTR GC | ||||||||||
| YCR1b | TCT GCG TGG GTG AAC ACC TTC TTG GC | ||||||||||
| Nested PCR | YCF2a | CGC TTC CAA TGT ATC TGY ATG CAC CA | 66 | 147 | |||||||
| YCF2b | CGC TTY CAR TGT ATC TGC ATG CAC CA | ||||||||||
| YCR2a | RTC DGT GTA CAT GTT TGA GAG TTT GTT | ||||||||||
| YCR2b | GTC AGT GTA CAT ATT GGA GAG TTT RTT | ||||||||||
| Real time RT-qPCR (TaqMan) |
Penaeus monodon | Pleopod | GAVQPF1 | GGG ATC CTA ACA TCG TCA ACG T | 60 | - | [223] | ||||
| GAVQPR1 | AGT AGT ATG GAT TAC CCT GGT GCA T | ||||||||||
| 6FAM-TAMRA probe | 6FAM-TCA GCC GCT TCC GCT TCC AAT G | ||||||||||
| RT-LAMP PCR | Penaeus vannamei | Pleopods | YHV-F3 | ACC CTG TAA TTG GCG ATG TT | 65 | 186 | [113] | ||||
| YHV-B3 | TGC AGT TAA GAT GGT CAC AG | ||||||||||
| YHV-FIP | AGA GCA CTG TAG ACT GGT GGG TTT TTG TGG AAC CTG AAG AAT GC | ||||||||||
| YHV-BIP-Biotin | Biotin-TCA GCA CCT GGG CTC GTC TCT TTT CGA CAG TGA TTG AAG ACT CG | ||||||||||
| YHV-LF | AAC TGT TGC AGA TCG GAT T | ||||||||||
| YHV-LB | ATG TGT CAT GAT ATT CTC | ||||||||||
| YHV FITC probe | CTC CAT CCA GAA A | ||||||||||
| YHV7-qPCR (TaqMan) |
Penaeus monodon | Pleopods, gills | qYHV-F1 | CAT CCA ACC TAT CGC CTA CA | - | 79 | [91] | ||||
| qYHV-F2 | ACC TAT CGC CTA CAC AGC TA | 73 | |||||||||
| qYHV-R1 | TGT GAA GTC CAT GTG AAC GA | - | |||||||||
| qYHV7-Pr1 | 6FAM- CAA CGA CAG ACA CCT CAT CCG TGA-BHQ1 | - | |||||||||
| YH7-PCR | YHV7-F1a | CCT ACA CGC ATG CTC TCT CTA TG | - | 788 | |||||||
| YHV7-R1b | GGT GTC TGT CGT TGT GTA TAG CT | ||||||||||
| YHV7-nPCR | YHV7-F2a | CAA ACA CCA ACC GAC ATT CAG T | 58 | 412 | |||||||
| YHV7-R2a | GCG ACA GTG CTT GAA GAC TTT AG | ||||||||||
| TSV | ConventionalPCR | Penaeus monodon | Gills, tail, body cuticles, swimming feet |
9992F | AAG TAG ACA GCC GCG CTT | 60 | 231 | [129] | |||
| Real-time RT-PCR (TaqMan) |
Davidson’s-fixed paraffin-embedded (DFPE) shrimp tissue | TSV1004F | TTG GGC ACC AAA CGA CAT T | 60 | 417 | [119] | |||||
| TSV1075R | GGG AGC TTA AAC TGG ACA CAC TGT | ||||||||||
| TSV-P1 | FAM-CAG CAC TGA CGC ACA ATA TTC GAG CAT C-TAMARA | ||||||||||
| TSV1004F | TTG GGC ACC AAA CGA CAT T | 122 | [120] | ||||||||
| TSV1075R | GGG AGC TTA AAC TGG ACA CAC TGT | ||||||||||
| TSV-probe | FAM-CAG CAC TGA CGC ACA ATA TTC GAG CAT C-TAMARA | ||||||||||
| Penaeus vannamei | Pleopods | TSV-55P1 | GGC GTA GTG AGT AAT GTA GC | 60 | 955 | [116] | |||||
| TSV-55P2 | CTT CAG TGA CCA CGG TAT AG | ||||||||||
| Real-time RT-PCR (SYBR green) |
Penaeus vannamei | Cephalothorax | TSV-306F | CGT AAA TAG ACG GCC CAC AAA | 60 | 79 | [138] | ||||
| TSV384R | TGC ATC TAT ATA TCC AGG GAC TTA TCC | ||||||||||
| TSV-285F | TTC TAT AGG TCT GGT TTA AAA CGT AAA | 232 | |||||||||
| TSV-516R | CGG TTT TCT CCA TCA TCG TT | ||||||||||
| WTD | Reverse transcriptase PCR | Macrobrachium rosenbergii | Infected sample | Mr-RdRp-F | GCA TTT GTG AAG AAT GAA CCG | 50 | 729 | [56] | |||
| Mr-RdRp-R | CAT GTT CAACTTTCTCCACGT | ||||||||||
| qMrNV-F | AGG ATC CAC TAA GAA CGT GG | 211 | |||||||||
| qMrNV-R | CACGGTCACAATCCTTGCG | ||||||||||
| MrNv2F | GAT ACA GAT CCA CTA GAT GAC C | 55 | 681 | ||||||||
| MrNv2R | GAC GAT AGC TCT GAT AAT CC | ||||||||||
| Muscle | 1A775 | CCA CGT TCT TAG TGG ATC CT | 55 | 850 | [147] | ||||||
| 1B690 | CGT CCG CCT GGT AGT TCC | ||||||||||
| MrNV DBHF | ATG GCT AGA GGT AAA CAA AAT TC | 50 | 564 | [149] | |||||||
| MrNV DBHR | TCA TTG ATC ATC ACG CCT GAC A | ||||||||||
| MrNV PEF | GGG CCG GAT CCA TGG CTA GAG GTA AAC AAA ATT C | ||||||||||
| MrNV PER | GGC CAA GCT TTC ATT GAT CAT CAC GCC TGA CA | ||||||||||
| Infected sample | FL-XSV-F | CCA CGT CTA GCT GCT GAC GTT | 50 | 796 | [56] | ||||||
| FL-XSV-R | AAG GTC TTT ATT TAT CGA CGC | ||||||||||
| XSV-F | GGA GAA CCA TGA GAT CAC G | 55 | 507 | ||||||||
| XSV-R | CTG CTC ATT ACT GTT CGG AGT C | ||||||||||
| qXSV-F | AGC CAC ACT CTC GCA TCT GA | 50 | 68 | ||||||||
| qXSV-R | CTC CAG CAA AGT GCG ATA CG | ||||||||||
| Muscle | XSV DBHF | ATG AAT AAG CGC ATT AAT AAT | 50 | 525 | [149] | ||||||
| XSV DBHR | TTA CTG TTC GGA GTC CCA ATA | ||||||||||
| XSV PEF | GGG CCG GAT CCA TGA ATA AGC GCA TTA ATA AT | ||||||||||
| XSV PER | GGC CAA GCT TTT ACT GTT CGG AGT CCC AAT A | ||||||||||
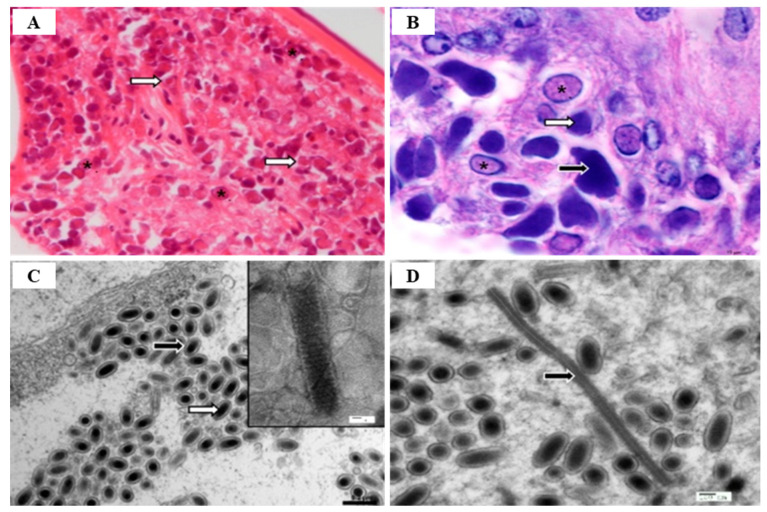
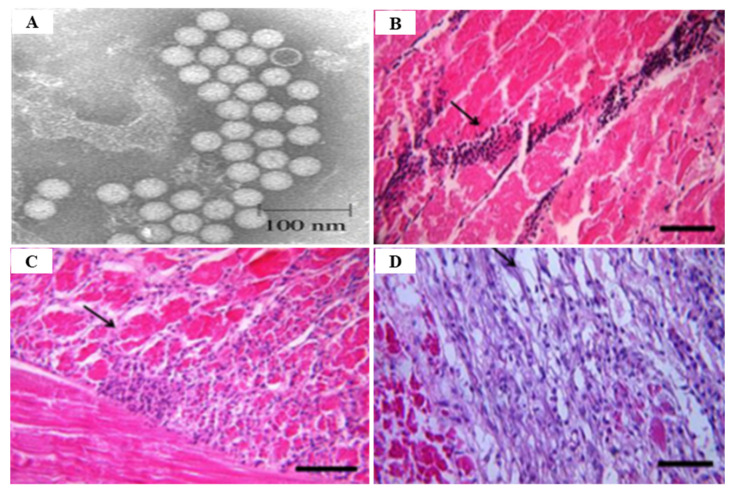
Figure 7.
Penaeusvannamei infected with white spot syndrome virus (WSSV). The infection progresses through different stages that can be seen in the nucleus via histology. (A) Early-stage infected cells display enlarged nuclei with marginalized chromatin and a homogenous eosinophilic central region. These then develop an intranuclear eosinophilic Cowdry A-type inclusion (*); this can be surrounded by a clear halo beneath the nuclear membrane (white arrow). Scale bar = 25 µm; (B) The eosinophilic inclusion usually expands to fill the nucleus (*). This inclusion becomes basophilic when staining and denser in color as the infection progresses (white arrow). Nuclei then disintegrate so that the content fuses with the cytoplasm (black arrow). Scale bar = 10 µm. H & E stain; (C) WSSV virions appear ovoid in shape and contain an electron-dense nucleocapsid (white arrow) within a trilaminar envelope (black arrow). Scale bar = 0.2 µm. Inset. Negatively stained WSSV nucleocapsid, showing the presence of cross-hatched or striated material that is structured as a series of stacked rings of subunits and is a key diagnostic feature of WSSV. Scale bar = 20 nm; (D) Presumptive nucleocapsid material within the nucleus prior to envelopment. This material is cross-hatched or striated in appearance and linear prior to its incorporation in the formation of mature WSSV particles. This linear nucleocapsid material is observed sporadically in the manufacture of the WSSV particles. Scale bar = 100 nm. Transmission electron microscopy images (Source: Verbruggen et al., 2016, https://doi.org/10.3390/v8010023 accessed on 11 May 2018).
2.2. Infectious Hypodermal and Hematopoietic Necrosis Virus (IHHNV)
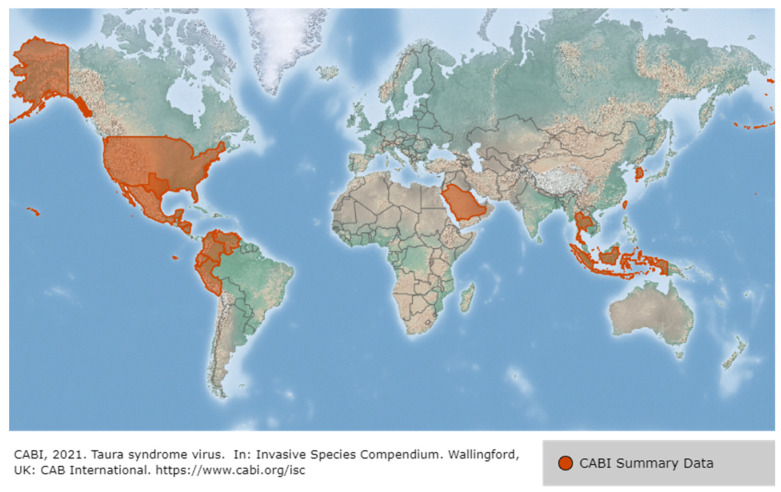
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) is a critical viral pathogen of penaeid shrimp, causing serious economic loss to the shrimp aquaculture industry (up to 50% of the overall economic loss in shrimp aquaculture), and has been listed as a reportable crustacean disease pathogen by the World Organisation for Animal Health (OIE) since the year of 1995 [57]. IHHNV was first detected in blue shrimp, P. stylirostris post-larvae and juvenile imported from Costa Rica and Ecuador at a shrimp farm of Hawaii in 1981, causing up to 90% mortality, and it was discovered in the quarantine process of imported white leg shrimp, P. vannamei at a shrimp farming facility in Taiwan in 1986, and in giant tiger prawn, P. monodon aquaculture of Australia in 2008 [58].
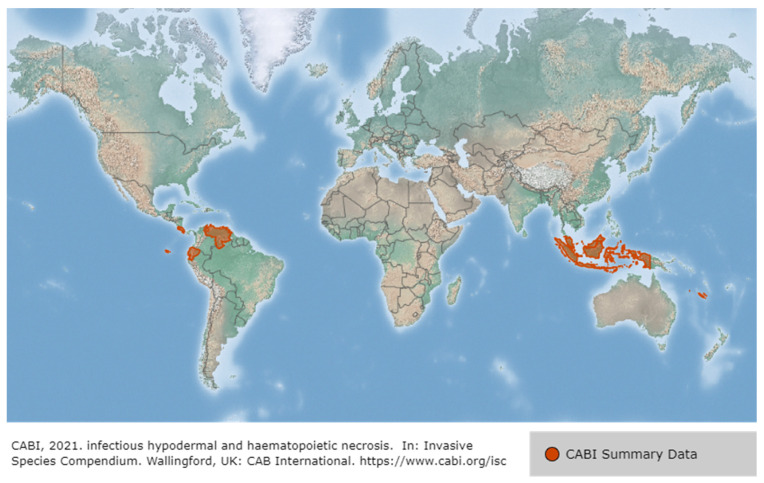
Since IHHNV was first reported in blue shrimp, P. stylirostris, IHHNV disease outbreaks had been reported in more than 20 countries in Asia, America, Africa and Oceania, such as Korea, Philippines, Singapore, Malaysia, Thailand, Indonesia, USA, Brazil, Mexico, Argentina, India, Venezuela, Mozambique, Madagascar, Tanzania and Australia [59] (Figure 8). IHHNV infects the major aquaculture shrimp species, P. stylirostris and P. vannamei, in North America, which is causing economic losses [60]. IHHNV is lethal in juvenile P. stylirostris with 90% mortality (acute disease), whereas it causes runt deformity syndrome (RDS; asymptomatic carrier of the virus) in P. monodon and P. vannamei, reducing the market value by 10–50% [61]. IHHNV causes the RDS in juvenile P. vannamei and P. monodon, which causes stunting in growth, and accounts for 50% of the economic loss in the shrimp industry [59,62] (Figure 9). IHHNV causes economic damage by reducing the marketability of shrimp due to poor growth, irregular growth, and epidermal malformation during harvest by RDS (cuticular deformities of the rostrum, antennae, thoracic and abdominal areas) [63,64] (Figure 10).
Figure 8.
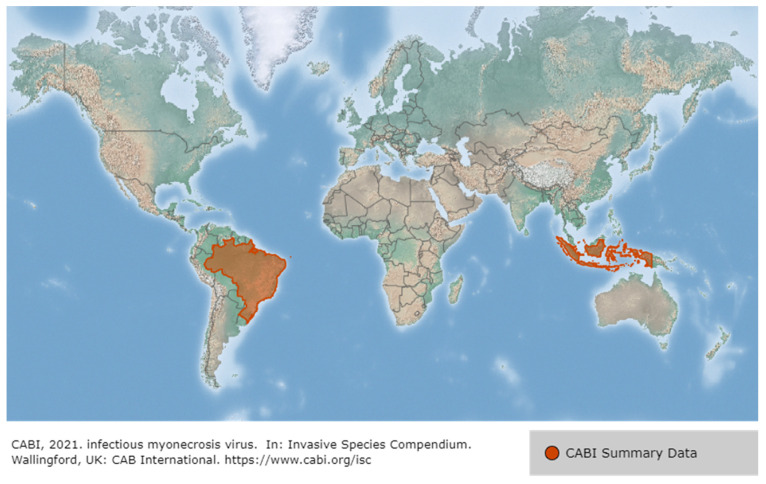
Distribution maps showing the geographical occurrence of infectious hypodermal and hematopoietic necrosis virus (Reprinted from CABI, 2019, Infectious hypodermal and hematopoietic necrosis. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
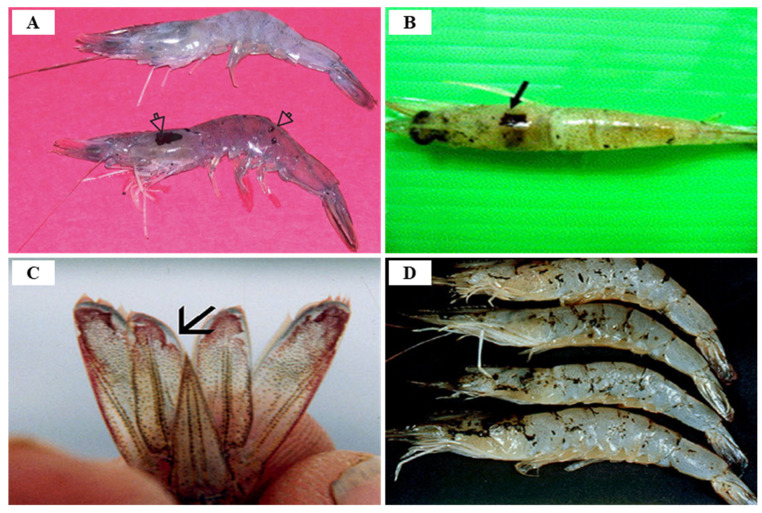
Figure 9.
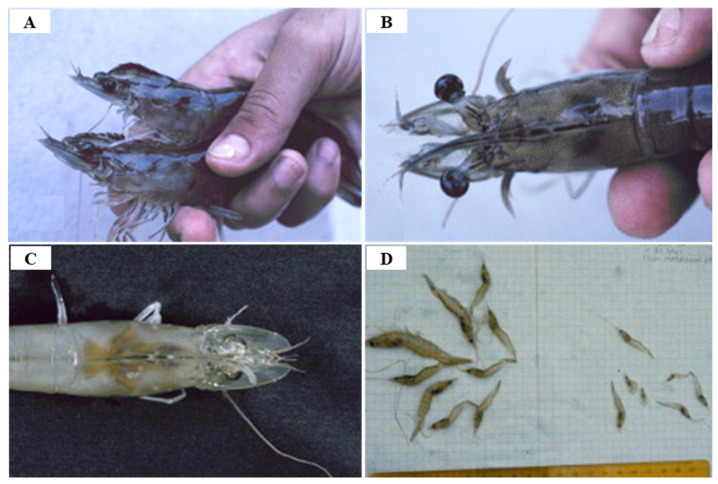
External symptoms of infectious hypodermal and hematopoietic necrosis virus (IHHNV) on shrimp. (A,B) subadult Penaeus vannamei with bent (to the left) rostrums, a classic sign of ‘runt deformity syndrome’ (RDS); (C) a juvenile P. vannamei with RDS. In this specimen the rostrum is bent to the right and the antennal flagella are wrinkled, brittle and mostly broken-off; (D) juvenile P. vannamei with RDS from a nursery population at approximately 60 days post stocking (Reprinted from Journal of Invertebrate Pathology, Vol. 106 (1), Lightner D.V., Virus diseases of farmed shrimp in the Western Hemisphere (the Americas) A rieview, p. 21, Copyright (2011), with permission from Elsevier).
Figure 10.
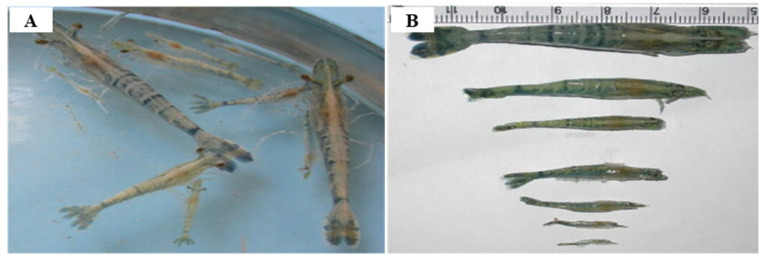
Size variations observed in 50-day-old Penaeus monodon with infectious hypodermal and hematopoietic necrosis virus (IHHNV) (A,B) (Reprinted from Aquaculture, Vol. 289 (3–4), Rai, P., Pradeep, B., Karunasagar, I., Karunasagar, I., Detection of viruses in Penaeus monodon from India showing signs of slow growth syndrome, p. 5, Copyright (2009), with permission from Elsevier).
IHHNV is a linear single stranded DNA virus of 3.9 kb in length and the smallest penaeid shrimp virus that is non-enveloped and icosahedral linear virion with an average diameter of 22–23 nm [60]. IHHNV was taxonomically a Penaeus stylirostris densovirus (PstDNV) from the Parvoviridae family, Densovirinae subfamily, but in July 2019, ICTV (International Committee on Taxonomy of Viruses) reconstituted the Parvoviriae family as the Parvoviridae family, Hamaparvovirinae subfamily, and Penstylhamaparvovirus [59]. IHHNV has a capsid made up of four polypeptides with molecular weights of 74 k, 47 k, 39 k and 37.5 k [65]. IHHNV may exhibit different virulence due to differences in genotype of IHHNV, host susceptibility and developmental stage of infected shrimp; (i) Acute infection: IHHNV-infected post-larvae and juveniles P. stylirostris sink to the bottom without swimming and can cause up to 90% of shrimp mortality in a short period of time; (ii) Chronic infection: Mass mortality does not usually occur in IHHNV-infected juvenile P. vannamei and P. monodon, and sub-adults M. rosenbergii, which can cause RDS such as growth and rostrum retardation, abdominal and tail fan deformation, cuticular roughness, and wrinkled antennal flagella, resulting in 30–90% growth retardation; (iii) Asymptomatic carriers: Mytilus edulis and adult M. rosenbergii can carry the infectious IHHNV type, but do not show major clinical and pathological symptoms and serve only as carriers; (iv) non-infectious IHHNV insertion into shrimp host genome: Exposure to IHHNV was not infectious in P. monodon and P. vannamei individuals injected with crude extracts of P. monodon carrying the IHHNV sequence through feeding and injection [59].
Genetic characterization of multiple IHHNV strains isolated from multiple regions can determine whether the virus has evolved or not and the existence of other strains in the region with exogenous sources [58] (Table 3). The IHHNV genome consists of three ORFs (open reading frames): two encoding nonstructural proteins (NS1; 2001 bp and NS2; 1092 pb) and one encoding viral capsid proteins (CP; 990 bp) [57,59] (Table 2). Of five genotypes classified in IHHNV, type I, type II, and type III are infectious types, and type A and type B are non-infectious. Type I was found in P. monodon of Australia (GenBank no. CQ475529.1); type II was mainly found in the United States and Southeast Asia (GenBank no. AY102034.1, JN616415.1, AY362547.1, etc.), and type III was mainly distributed in East Asia (GenBank no. AY355308.1, EF633688.1, KF214742.1, and JX258653.1, etc.) [59] (Table 3). Two IHHNV virus sequences were found in P. monodon in Africa (Type A was found in Madagascar and Australia, and type B was found in Tanzania). Type A and type B sequences have three ORFs with high similarity, which has the identical replication initiator motif and NTP-binding and helicase domains with IHHNV virus, but both type A and type B IHHNV-related sequences are non-infectious genotypes [66].
IHHNV was found in P. monodon in Southeast Asia (Thailand, Taiwan, and the Philippines), and only about 30 animal species are known to be IHHNV-susceptible or carriers of IHHNV [59]. IHHNV mainly affects Penaeid shrimp, but Artemesia longinaris, Palaemon macrodactylus and post-larvae and subadults of M. rosenbergii as well as P. clarkii are also known to be naturally infected with IHHNV. Bivalve shellfish and adults of M. rosenbergii act as carriers in IHHNV without infection-related symptoms [57]. For example, in the IHHNV PCR test on the coast of China, the positive rate of IHHNV in the gills, muscles and gonads of Mytilus edulis was more than 80%, but the pathogenicity of IHHNV infection was not shown. In addition, the pathogenicity of IHHNV infection was closely related to the age and size of the host, and in general, young shrimp are more susceptible to IHHNV infection [59]. Larval and juvenile P. stylirostris at 0.05–2 g is more susceptible to IHHNV, especially P. stylirostris at 0.08 g is most susceptible to IHHNV, whereas P. stylirostris at 2 g or more significantly weakens IHHNV pathogenicity. Adults of M. rogenbergii do not show obvious symptoms of IHHNV infection, but IHHNV infection in subadults can cause slow growth and cause RDS also in juvenile of P. vannamei and P. monodon, whereas adult P. vannamei showed no obvious pathological symptoms [62]. IHHNV shows a marked difference in pathogenicity according to the infecting shrimp species; While P. sylirostris is highly pathogenic, P. vannamei causes RDS, a chronic disease [67].
Because IHHNV does not encode a DNA polymerase and is dependent on the host cell for DNA replication and proliferation, it requires the host’s rapidly proliferating cells for replication; the main target organs for IHHNV infection contains tissues of ectodermal (cuticular epidermis, nerve cord and ganglia, hypodermal epithelium of the fore and hind gut) and mesodermal (antennal gland, lymphoid organ, hematopoietic organs, striated muscles, tubule epithelium and connective tissue) origin, but IHHNV does not affect tissues of endodermal origin such as hepatopancreas, anterior mid-gut caecum, midgut epithelium or posterior midgut caecum [58] (Table 4). It is the post-larvae and juvenile shrimp that are susceptible to IHHNV owing to the reason that they have actively dividing cells. The P. stylirostris presents acute symptoms of IHHNV such as white or buff-colored spots at the junction of the tergal plates in the abdomen, whereas IHHNV in the P. vannamei appears as a chronic disease, RDS, showing symptoms such as wrinkled antennal flagella, ‘bubble-heads’, deformed rostrum, cuticular roughness and deformation in 6th abdominal segment and tail fan [59].
Shellfish, as an important carrier of IHHNV disease, have a very high risk of transmission, but the mechanisms of infection and pathogenicity are still unclear in many respects [59]. In the case of horizontal transfer of infection, the P. sytlirostris surviving IHHNV infection can become life-long carriers of the virus and cause spread through vertical and horizontal propagation. In the natural environment, IHHNV transmission can occur horizontally through shrimp feeding and water, and vertical transmission can occur from mother to offspring [58]. IHHNV was detected in the ovaries of IHHNV-infected females, whereas the IHHNV did not appear in the sperm of infected males, so vertical transmission of IHHNV from infected females was clearly established [67]. Post-larvae M. rosenbergii with IHHNV infection showed a high mortality rate of up to 80–100, and juvenile and subadult P. stylirostris showed a mortality rate of up to 90% (however, P. stylirostris also has increased resistance to IHHNV infection, and no significant mortality has recently been reported.); on the other hand, in P. vannamei and P. monodon, IHHNV was less virulent with no death, just including RDS such as stunting and cuticular deformities [58,66].
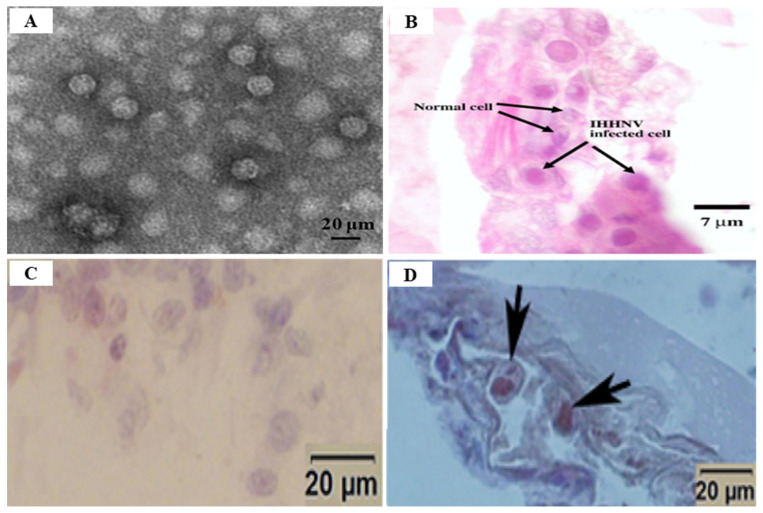
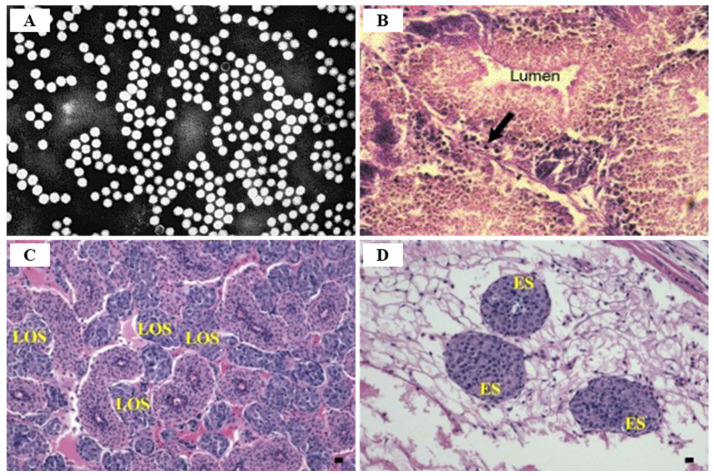
In an epidemiological survey, the IHHNV prevalence of shrimp in aquaculture areas was 51.5% and 8.3% for shrimp and crab in China, 9.4~81% for shrimp in northeastern Brazil, 14.1%for P. monodon in Brunei Barussalam and 30% for Artemesia longinaris in Argentina, 1.1~3.3% for P. vannamei in Venezuela, 20% for M. rosenbergii in Malaysia [58]. Currently, the most reliable techniques used for IHHNV detection are conventional PCR and real-time PCR. However, since the existing PCR cannot quantify the virus in the infected sample, the real-time PCR technique (probe-based and dye-based methods) is more useful [68] (Table 5). TaqMan probe-based real-time PCR is also a sensitive technique for IHHNV detection (Table 5). Encinas-García et al. (2015) [69] developed SYBR Green-based real-time PCR for the detection and quantification of IHHNV in P. sylirostris, which is much cheaper and simpler than TaqMan probe real-time PCR (Table 5). Histologically, the diagnosis of IHHNV infection is made through the identification of prominent Cowdry type A, eosinophilic, intra-nuclear inclusion bodies enclosed by marginated chromatin in hypertrophied nuclei of cells in tissues of ectodermal and mesodermal origin [58]. In electron microscopy of negatively stained IHHNV VLPs in P. vannamei, IHHNV-VLPs were uniformly spherical and 23 ± 3 nm in diameter, similar to native IHHNV particles [70] (Figure 11A). H&E staining of P. monodon infected with IHHNV showed intra-nuclear Cowdry type A eosinophilic inclusion bodies [64] (Figure 11B). Several hypertrophied nuclei were observed in the gill tissues of IHHNV-infected P. clarkii [71] (Figure 11D). An effective vaccination strategy for IHHNV has not been developed, and there are no confirmed reports of effective chemotherapy and immune-stimulation treatment [72]. As there is currently no effective treatment for IHHNV, the best management strategy is to screen SPF shrimp for IHHNV, but when IHHNV cannot be completely controlled, IHHNV-resistant shrimp populations may be used.
Figure 11.
Electron microscopy and histological analysis of the changes in shrimp with infectious hypodermal and hematopoietic necrosis virus (IHHNV). (A) Electron microscopy of negatively stained IHHNV VLPs under self-assembly and disassembly conditions in Penaeus vannamei; (B) Cowdry type A eosinophilic inclusion of IHHNV in a nucleus of subcuticular epithelial cells of the pleopod of P. monodon (H & E, 1000×); (C) Histological detection of Procambarus clarkii gills negative to IHHNV detected by PCR. The gill cells were normal, no hypertrophied nucleus was observed; (D) Histological detection of P. clarkii gills positive to IHHNV detected by PCR. Several hypertrophied nuclei (arrow) were observed. ((A) Reprinted from Journal of Invertebrate Pathology, Vol. 166, Zhu, Y.P., Li, C., Wan, X.Y., Yang, Q., Xie, G.S., Huang, J., Delivery of plasmid DNA to shrimp hemocytes by infectious hypodermal and hematopoietic necrosis virus (IHHNV) nanoparticles expressed from a baculovirus insect cell system, p. 1, Copyright (2019), with permission from Elsevier; (B) Reprinted from Aquaculture, Vol. 289 (3–4), Rai, P., Pradeep, B., Karunasagar, I., Karunasagar, I., Detection of viruses in Penaeus monodon from India showing signs of slow growth syndrome, p. 5, Copyright (2009), with permission from Elsevier; (C,D) Reprinted from Aquaculture, Vol. 477, Chen, B.K., Dong, Z., Liu, D.P., Yan, Y.B., Pang, N.Y., Nian, Y.Y., Yan, D.C., Infectious hypodermal and hematopoietic necrosis virus (IHHNV) infection in freshwater crayfish Procambarus clarkii, p. 4, Copyright (2017), with permission from Elsevier).
3. RNA Viral Diseases
3.1. Infectious Myonecrosis Virus (IMNV)
Infectious myonecrosis (IMN), also known as Penaeid shrimp myonecrosis virus (PsIMNV), is a major disease caused by the infectious myonecrosis virus (IMNV), which adversely affects the shrimp aquaculture industry [73,74]. IMN was first identified in Piaui state, Brazil in August 2002, and then rapidly spread through the coastal areas of northeastern Brazil, which significantly reduced the productivity of the Brazilian shrimp aquaculture industry in 2004 and 2005 [75]. In the Asia-Pacific region, P. vannamei is steadily increasing in importance as a major aquaculture species. Furthermore, IMNV was added to the World Organization for Animal Health in 2005 and NACA (Network of Aquaculture Centres in Asia-Pacific)/FAO (Food and Agriculture Organisation) in January 2006 due to large-scale transboundary movements of the disease and its impacts on aquaculture species [62,76]. In Brazil this pathogen caused an economic loss of approximately 20 million USD with 40–60% mortality in 2003. By the end of 2005 the economic losses as a result of the IMNV outbreak had reached 430 million USD, and by the end of 2011, Brazil and Indonesia had suffered a combined economic loss of approximately 1 billion USD in Brazil and Indonesia [76,77].
IMNV was first reported in 2003 in P. vannamei cultured in northeastern Brazil, then in Indonesia (2006), and most recently in India (2016), Malaysia (2018) and Indonesia (2018) [78,79] (Figure 12). Until the IMNV virus was reported in India in 2016, it had only occurred in Brazil and Indonesia [80]. IMNV occurs in P. vannamei, its infectious host, and causes infective myonecrosis. The occurrence of this disease is thought to be related to certain types of environmental and physical stress (extreme temperature and salinity, collection by cast-net) and the use of low-quality shrimp feed [62]. Although IMNV can induce an increase in mortality due to an acute infection in P. vannamei, the infection is usually detected by observing chronic symptoms in the host rather than a rapid mortality. The symptoms displayed by P. vannamei infected with IMNV include focal to extensive white necrotic areas in the striated muscle, especially the distal abdominal segments and tail fan [79], as well as a slow mortality that persists during the culture period (cumulative mortality reaching up to 70%) [81].
Figure 12.
Distribution map showing the geographical occurrence of infectious myonecrosis virus (IMNV) (Reprinted from CABI, 2019, Infectious myonecrosis virus. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
IMNV is a single molecule of double-stranded RNA forming a monopartite genome that is 7561–8230 bp in length with two open reading frames (ORFs). It is a non-enveloped icosahedral virus with a diameter of 40 nm and fiber-like protrusions on the surface [74,82] (Table 1). IMNV is taxonomically a totivirus belonging to Totiviridae family that is similar to Protozoa and Fungal viruses. In a phylogenetic analysis based on RdRp, IMNV was identified as a member of the Totiviridae family in 2008 [74,83]. The Totiviridae family consists of five genera (Giardiavirus, Leishmaniavirus and Trichomonasvirus, which infect protozoa; and Totivirus, and Victorivirus, which infect fungi) recognized by the ICTV (International Committee on Taxonomy of Viruses), but many researchers have recently suggested that the Arthropod Totiviruses should be classified separately as an Artivirus genus within the Totiviridae family [76].
Whole-genome sequencing of IMNV revealed two ORFs such as ORF1, encoding RNA binding and capsid proteins and ORF2, encoding putative RNA-dependent RNA polymerase (RdRp) [83] (Table 2). The coding region of the RNA-binding protein is situated in the first half of ORF 1 (including a dsRNA-binding motif). The second half of ORF1 encodes a capsid protein with a molecular mass of 106 kDa [77]. The function of the dsRBM (dsRNA binding motif) is critical for modulation and viral replication in the immune system of the shrimp host. However, the functions of small proteins are still unclear, but hypotheses have been suggested in which they may be connected to assembly, cell entry, and extracellular transmission of the virus [76]. ORF2 demonstrates high similarity to the RdRp of the Totiviridae family, and ORF2 coding strategies of IMNV are similar to the strategies of GLV (Giardia lamblia virus) and other members of the Totiviridae family, which indicates that RdRp is a conserved domain [76].
IMNV strains identified in Brazil (six strains) and Indonesia (ten strains) showed high similarity with the alignment of a 372 bp fragment encoding the major capsid protein (MCP) of IMNV strains isolated from the two regions. This suggests that the MCP could be used as a target gene to track the movement of IMNV [77] (Table 3). Through subsequent analysis, it was confirmed that the IMNV in Brazil and Indonesian reported by GenBank had nucleic acid sequence identity of 99.6% [82]. The capsid protein has a major role in virus adhesion, virulence, and cell entry, and the MCP gene (nt 2248~4953) of IMNV also contains a variable region with 72 polymorphic sites, so that the MCP gene sequence can be used to trace the origin of a new strain [82].
IMNV not only infects P. vannamei, which are naturally susceptible to it, but also P. stylirostris and P. monodon, which have been found to be experimentally susceptible. Furthermore, the wild Southern brown shrimp, Penaeus (Farfantepenaeus) subtilis is also susceptible to IMNV infection [76,84]. IMNV is known to only infect Penaeid shrimp (4 shrimp species: P. vannamei, P. sylirostris, P. monodon, P. subtiltis), but can do so at all life stages including post larvae, juvenile, and adult, but mortality was observed only in the juveniles and adults showing symptoms of a cooked appearance [76,79] (Figure 13). In IMNV-infected shrimp, extensive white necrosis of the striated muscle, especially the distal part of the abdomen and tail fan, may progress, and dissection of moribund shrimp may show enlarged lymphoid organs more than twice the normal size [62] (Figure 13C,D). Clinical signs of IMNV are prominent in the acute phase of infection, and although the main target organ is the skeletal muscle, gills and lymphatic organs may also be affected. IMNV infection in the chronic stage can be identified by necrotic muscle liquefaction exhibiting coagulative muscle necrosis [76]. Typical symptoms of IMNV infection include transparency loss, abdominal and cephalothorax necrosis, tail coloration, hepatopancreas volume loss, and progressive tail fan necrosis [85]. Shrimp infected with IMNV are characterized by whitish or reddish discolorations in the tail muscle and opaque, whitish discolorations in the abdominal muscle due to white necrosis in the striated muscle [86]. Coelho et al. (2009) [75] suggest that shrimp infected with IMNV lose transparency, and this symptom starts at around the second or third segment and then extends towards the telson.
Figure 13.
External symptoms of infectious myonecrosis virus (IMNV) on shrimp. (A) IMNV-infected Penaeus vannamei with reddish opaque muscles at the distal abdominal segments; (B) P. vannamei injected with IMNV propagated in a C6/36 cell line with reddish opaque muscle at the distal abdominal segments as observed in the natural infection; (C,D) P. vannamei infected with IMNV and displaying focal to extensive white necrotic areas in the striated muscle, especially of the distal abdominal segments and tail fan, and exposure of the paired lymphoid organs (LO) by simple dissection will show that the paired LO are hypertrophic to twice or more their normal size. ((A) Reprinted from Journal of Fish Diseases, Vol. 40 (12), Sahul Hameed, A.S., Abdul Majeed, S., Vimal, S., Madan, N., Rajkumar, T., Santhoshkumar, S., Sivakumar, S., Studies on the occurrence of infectious myonecrosis virus in pond-reared Litopenaeus vannamei (Boone, 1931) in India, p. 8, Copyright (2017), with permission from John Wiley and Sons; (B) Reprinted from Journal of Fish Diseases, Vol. 44 (7), Santhosh Kumar, S., Sivakumar, S., Abdul Majeed, S., Vimal, S., Taju, G., Sahul Hameed, A.S., In vitro propagation of infectious myonecrosis virus in C6/36 mosquito cell line, p. 6, Copyright (2021), with permission from John Wiley and Sons; (C,D) Reprinted from Journal of Invertebrate Pathology, Vol. 106(1), Lightner, D.V., Virus diseases of farmed shrimp in the Western Hemisphere (the Americas) a review, p. 21, Copyright (2011), with permission from Elsevier).
The first report of IMNV occurred in a shrimp farm in northeastern Brazil in 2002 and it then spread to a shrimp farm in Indonesia in 2006. The cause of IMNV transmission is believed to be the uncontrolled movement of brood stocks and post larvae shrimp across borders [87]. Since it was first reported from Brazil, the origin of IMNV is thought to be South America, and the geographical distribution of the disease is limited. Although the exact mechanism for IMNV transmission is unknown, there is also the possibility of horizontal transmission through cannibalistic behavior or the water via infected shrimp, and vertical transmission from broodstock to progeny [76]. The source of vertical transmission is assumed to be maternal based on the low sperm cell survival rate of naturally infected males and the 100% positive occurrence in the ovaries of female shrimp infected with IMNV [82]. Although specific data on the vector of IMNV are lacking, it has a non-envelope particle structure like TSV (non-enveloped virus particles have high survival rates in the gastrointestinal tracts of animals), and thus has the potential to maintain infectivity in the intestines and feces of seabirds that feed on IMNV-infected dead or dying shrimp [88].
IMNV infection progresses slowly throughout the growing season with low mortality, but cumulative shrimp mortality in ponds during harvest can reach up to 70% [86]. In general, the mortality rate due to IMNV infection is between 20–50%, and the mortality rate gradually increases, resulting in 40–70% mortality during the growing season [83]. Given that the major target tissues of IMNV are the striated skeletal muscles which are not considered vital tissue, the virulence following IMNV infection is less lethal, when compared to other viruses such as WSSV, YHV, and TSV. In addition, the damage at the early steps of IMNV infection can be repaired in the muscle tissues [76]. Although IMNV is not fatal when compared to WSSV and YHV, this virus is a stress-dependent virus, which is lethal to P. vannamei when there are rapid changes in water quality parameters such as pH, temperature, plankton, and dissolved oxygen [82]. Due to its slow disease progression, IMNV can cause significant economic losses due to high feed conversion efficiency as the infected individuals consume feed continuously [76].
IMNV infection is diagnosed primarily through clinical symptoms, histopathological examination, and molecular techniques [74]. Since there are no effective drugs or vaccines available for IMNV, a sensitive and reliable diagnosis is required for appropriate control measures. The TaqMan real-time RT-PCR assay provides a rapid and sensitive method for clinical diagnosis of IMNV [89] (Table 5). Histological lesions due to IMNV infection are characterized by coagulative myonecrosis, with hemocytic infiltration, fibrosis, and fluid accumulation in muscle fiber (edema) [90] (Figure 14). Among shrimp challenged with IMNV, 10% showed a light coagulation and hemocyte infiltration [75]. During the acute phase of IMNV, the main target organs are the striated muscles, hemocytes, connective tissues, and lymphoid organ tubule parenchyma cells, whereas the major tissues targeted during the chronic phase are the lymphoid organs [76]. During the acute or chronic phase of IMNV, considerable hypertrophy of the lymphoid organs, induced by the accumulation of lymphoid organ spheroids (LOS), results in the development of consistent lesions [62].
Figure 14.
Electron microscopy and histological changes in shrimp with infectious myonecrosis virus (IMNV). (A) TEM of a purified preparation of IMNV from naturally infected Penaeus vannamei from Brazil. Photomicrographs of tissue sections from P. vannamei examined for IMNV lesions (B–D) (Scale bar = 50 μm); (B) Focal hemocytic infiltration in muscle tissue; (C) Muscle coagulation necrosis accompanied by infiltration of hemocytes; (D) Muscle liquefactive necrosis and fibrosis. ((A) Reprinted from Journal of Invertebrate Pathology, Vol. 106 (1), Lightner, D.V., Virus diseases of farmed shrimp in the Western Hemisphere (the Americas) a review, p. 21, Copyright (2011), with permission from Elsevier; (B–D) Reprinted from Aquaculture, Vol. 380, Feijó, R.G., Kamimura, M.T., Oliveira-Neto, J.M., Vila-Nova, C.M., Gomes, A.C., Maria das Graças, L.C., Maggioni, R., Infectious myonecrosis virus and white spot syndrome virus co-infection in Pacific white shrimp (Litopenaeus vannamei) farmed in Brazil, p. 5, Copyright (2013), with permission from Elsevier).
As there is currently no effective method by which to control the spread of or treat IMNV, prevention, management, and prompt diagnosis are the most effective tools [87]. Experimental infections showed that here was 20% mortality in P. vannamei, but 0% mortality in P. stylirostris and P. monodon. Therefore, restocking with IMNV-resistant individuals such as P. monodon and P. stylirostris could be a useful method to reduce mortality losses [76]. To prevent the vertical transmission of IMNV, eggs and larvae must be disinfected, and biological security measures, appropriate quarantine, and SPF (specific pathogen free) bloodstocks procedures implemented, in addition to stocking density decreases, stress reduction in the culture environment, and immune-stimulant administration [82].
3.2. Yellow Head Virus Genotype 1 (YHV Genotype 1)
Yellow head virus (YHV-1) and gill-associated virus (YHV-2; GAV) first emerged in the early to mid-1990s and are serious pathogens of the giant tiger shrimp, P. monodon farmed in Thailand and Australia, respectively [91]. Although YHV-1 and YHV-2 (GAV) share the same susceptible host, P. monodon, they have geographically distant natural distributions and show significant differences in virulence and pathogenicity [92]. Of the eight identified genotypes, typical symptoms of YHV infection in shrimp are known only for the YHV genotype 1 [93], and losses due to YHV were estimated to be between 30 to 40 million USD in Thailand in 1995, before the outbreak of WSSV [94].
The YHV genotype 1 is the most virulent, was first identified in P. monodon cultured in Thailand in 1990 [95] (Figure 15), and it caused mass mortality of the species and significant economic losses to the shrimp industry. It was designated as a notifiable disease by the World Organisation for Animal Health (OIE) in 1995 [68]. It was first observed in cultured black tiger shrimp, P. monodon in central Thailand in 1990, and by 1992 had spread to shrimp farming areas on the eastern and western coasts of the Gulf of Thailand. In 1993, a virus morphologically identical to YHV genotype 1 was detected in the lymphoid organs of healthy wild and farmed P. monodon in Queensland, Australia, and was thereafter named the lymphoid organ virus (LOV). YHV was then detected at high levels in gills with YHD (yellow head disease)-like histopathology in the gills of moribund aquaculture P. monodon between 1995 and 1996 and was named GAV (gill-associated virus) [95].
Figure 15.
Distribution map showing the geographical occurrence of yellow head virus genotype 1 (YHV genotype 1) (Reprinted from CABI, 2019, Yellow head virus. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
There have been reports of YHD infection in farmed P. vannamei and P. stylirostris in Mexico, but it has not been confirmed, and there are no official reports of YHV infection in the Americas [96]. YHD has also been reported in P. monodon in Asian countries such as Vietnam, Philippines, Sri Lanka, Indonesia, Malaysia, India, and China, but has rarely been confirmed by laboratory analysis [97]. GAV, a YHV strain in Australia (YHV genotype 2), is related to a disease called mid-crop mortality syndrome (MCMS) in P. monodon in Australia, which was also detected in black tiger shrimp, P. monodon farmed in Vietnam and Thailand [98]. GAV is a chronic infection in Australia, causing significant economic losses to the Australian shrimp aquaculture industry since 1996, and GAV infections have been reported in farmed and wild P. monodon along the eastern coast of Australia [99]. YHV genotype 3 was detected in Taiwan, Vietnam, Indonesia, Malaysia, Thailand, and Mozambique, and YHV genotype 4 was found in India, which is the most frequently detected genotype. YHV genotype 5 was detected in the Philippines, Malaysia, and Thailand, and YHV genotype 6 was detected in Mozambique [100]. YHV genotype 7 was detected in P. monodon infected with the disease in Australia in 2012 [101]. In China, YHV genotype 1 was first detected in P. monodon imported from Thailand by the Shanghai Entry-Exit Inspection and Quarantine Bureau in 2005, and a new genotype YHV 8 was discovered in Hebei, China in July of 2012 [68].
YHV genotype 1 is a positive sense, rod-shaped, enveloped single-stranded RNA genome with virions of 40–60 nm × 150–200 nm and internal helical nucleocapsids of 15 nm in diameter 80–450 nm in length [94,100]. YHV is taxonomically classified in the Okavirus genus belonging to the Roniviridae family within the Nidovirales order [102] (Table 1). The virions of YHV include a polyadenylated 26.6 kDa genome and three structural proteins with transmembrane glycoproteins gp64 and gp116, the components on the virion surface [100]. YHV virions include three structural proteins, such as two transmembrane glycoproteins (gp116 and gp64) and a nucleoprotein (p20), and the envelope glycoprotein (gp116) has been shown to be the main virulence factor of YHV genotype 1 [103]. The genotypes that have evolved from P. monodon individuals are geographically separated from YHV and have evolved into YHV (YHV genotype 1) and GAV (YHV genotype 2) forms, which are indistinguishable [91].
The genome includes five canonical long ORFs (ORF1a, ORF1b, ORF2, ORF3, and ORF4), in order from the 5′-end: encoding replicase enzymes (ORF1a, overlapping ORF1b); encoding the nucleoprotein, p20 (ORF2); encoding the precursor polyprotein, pp3 that is processed to produce envelope glycoproteins such as gp116 and gp64 (ORF3) [104] (Table 2). YHV (YHV genotype 1) and GAV (YHV genotype 2) share a similar genome as the level of nucleotide sequence identity between them is approximately 79% overall (approximately 74% for ORF3 and 82% for ORF1b); the level of amino acid sequence identity between the genomes is 73% for gp116 and 84% for pp1ab [92]. The YHV genome (26,662 nt) is larger than the GAV genome (26,235 nt) owing to the sequence insertions occurring in several large blocks, whereas the GAV genome has few sequence insertions [92]. After YHV was first reported in Thailand in 1990, eight geographic types of genotypes have been reported, with genotypes differing by up to 20% in virulence and whole genome sequence [105] (Table 3). The mutant YHV genotype was also detected in healthy P. monodon broodstock in Thailand and was reported in P. monodon and P. japonicus which were cultured in Taiwan [97]. YHV genotype 1, the only virulence genotype of YHV was first reported in 1990 with typical signs of yellow head disease, which caused the mass mortality of P. monodon in Thailand [68]. YHV genotype 2 (GAV) is the only disease-associated YHV gene line other than YHV genotype 1 and is associated with a less severe form of the disease in Australian farmed shrimp [98]. Senapin et al. (2010) [106] suggests that GAV induces MCMS, which have lower virulence levels than those for YHV genotype 1 which is 106 times more virulent.
Most aquacultured species of penaeid shrimp, including P. stylirostris, P. aztecus, P. duorarum, P. setiferus, and P. vannamei, are susceptible to YHV-1 infection, while P. esculentus, P. merguiensis, and P. japonicus are susceptible to GAV [107] (Table 4). YHV infection also caused high mortality in Marsupenaeus japonicus, P. vannamei, P. stylirostris, P. esculentus, P. merguiensis, P. setiferus, P. aztecus, P. duorarum, M. ensis, and M. affinis [100], but P. monodon was the most affected overall [108]. It was observed that juvenile and sub-adult shrimp are susceptible to YHD and mortality within a few hours after showing clinical symptoms [95]. The GAV and YHV genotypes (YHV 3~8) have also been reported in healthy P. monodon from Indonesia, Malaysia, the Philippines, Vietnam, Thailand, Taiwan, Brunei, India, Mozambique, and Fiji [100].
YHV genotype 1 infection presents typical disease symptoms with yellow coloration of the cephalothorax and gills, but YHV-1 infection can exist for long periods without any signs of disease, such as with the WSSV outbreaks [102]. Samocha (2019) [109] also reported yellow discoloration of the cephalothorax and gills of P. monodon infected with YHV-1 (Figure 16). YHV-1 infection faded the overall body color of the shrimp, and mortality progressed after about 45–60 days of culture, resulting in a cumulative mortality rate of 60–70% [106]. Prapavorarat et al. (2010) [110] reported that after the initial clinical signs of YHV-1 disease (the development of yellow discoloration of the cephalothorax and gills), 100% mortality occurred within 3–9 days, resulting in rapid damage to shrimp production. As a result, of dissecting moribund shrimp due to YHV-1 infection, hepatopancreatic atrophy was reported [68]. YHV-1 affects tissues of ectodermal and mesodermal origin, and leads to critical lymphoid organ and gills necrosis [1]. In acute GAV infection, yellow cephalothorax lesions were not clearly seen, and general redness of the body and gills was observed, which was reproduced in artificial GAV challenge infection experiments in the laboratory [95]. GAV is very prevalent in penaeid shrimp and does not cause disease in healthy shrimp, other than a chronic infection [99]. Acute infection with YHV-1 and GAV can affect all mesodermal and ectodermal tissues containing lymphoid organs, circulating hemocytes, neural ganglia, nerve fibers, neurosecretory, glial cells, gonads, stomach subcuticulum, heart, and antennal gland [111].
Figure 16.
External symptoms on yellow head virus genotype 1 (YHV genotype 1)-infected shrimp. (A) P. monodon showing signs of yellow head disease (YHD) Yellow (light gray in print version) to yellow-brown (dark gray in print version) discoloration of the cephalothorax. Three shrimp with (left) and without (right) YHD; (B) discoloration of the gill region. ((A,B) Reprinted from Elsevier Books, Samocha, Sustainable biofloc systems for marine shrimp, p. 23, Copyright (2019), with permission from Elsevier).
YHV-1 can cause lethal infections in farmed penaeid shrimp species, but some wild shrimp and crab species can be YHV-1 carriers and transmit the disease without showing serious symptoms themselves [102]. YHV-1 can be horizontally transferred when the YHV-1 virus is released into the water, or through a formula of the infected shrimp individual [95]. It has been reported that YHV-1 can remain infectious for at least 72 h in seawater, and that approximately 30 ppm of calcium hypochlorite is an effective disinfectant [103]. YHV-1 is combined with a specific receptor, YRP65 on the cell membrane of lymphocyte cells as its primary target organ [92]. Although there is no direct report that YHV-1 propagates vertically, it has been experimentally verified for GAV [1]. GAV was detected in infected mature ovarian and spermatophores in broodstock, fertilized eggs and nauplii from shrimp infected with GAV, which demonstrated efficient vertical propagation from both males and females [100].
Mortality in shrimp infected with YHV-1 occurs a few days after the onset of symptoms. Generally, individuals die within 1–2 days, and mass death (70–100%) occurs within 2–3 days [102,112]. YHV-1 infection can occur from the late post-larvae stage of development, but mass mortality usually occurs in the early to late juvenile stages [100]. In contrast, GAV causes death after 7–14 days in experimentally infected P. monodon, and mainly occurs as a chronic farm disease [95]. It was reported that there was 100% prevalence of GAV infection in healthy P. monodon in eastern Australia and common prevalence in healthy P. monodon in Vietnam and Thailand [108]. GAV-infections are much less lethal for shrimp than YHV-1, and mortality progresses more slowly, with100% mortality being rare. GAV-infected moribund shrimp do not show the pale discoloration typical of yellow head disease and are reddish [1]. Walker and Mohan (2009) [1] reported that YHV-1 was 106 times more virulent than GAV at lethal concentrations of 50% in an artificial YHV-1 and GAV challenge experiment.
There are various techniques for YHV detection, including reverse transcriptase-polymerase chain reaction (RT-PCR), nested RT-PCR (IQ2000™ YHV Detection and Prevention System), loop mediated isothermal amplification (RT-LAMP), in situ hybridization, and real time RT-LAMP, all of which are currently being used [113] (Table 5). PCR-based methods for detecting YHV-1 and GAV have high efficiency in terms of speed, sensitivity and specificity, and quantitative real-time RT-PCR using a TaqMan probe or SYBR Green chemistry are effective detection methods [114] (Table 5). The OIE manual recommends detection using the YHV ORF1b gene region to diagnose YHV [91]. YHV infection is histologically accompanied by the observation of pyknotic and karyorrhectic nuclei and dense basophilic cytoplasmic inclusions in the lymphoid organs and gills, as well as the target tissues such as hepatopancreas, hematopoietic tissue, heart, midgut, nerve cord, eyestalks, abdominal muscle, and soft head tissues [102,110] (Figure 17).
Figure 17.
Electron microscopy and histological changes in shrimp infected with yellow head virus genotype 1 (YHV). (A) TEM of negative-strained YHV virions (Scale bars = 100 nm); (B) LO tissue of moribund shrimp from YHV immersion challenged P. vannamei at day 5 showing numerous pyknotic nuclei (arrows), karyorrhectic nucleic and cytoplasmic inclusion (arrow heads); (C) Hemolymph from normal and YHV infected shrimp identified by staining hemolymph smears; (D) Gills of YHV infected shrimp stained with H&E in rapidly fixed and stained (3 h) whole mounts. ((A) Reprinted from Advances in virus research, Vol. 63, Dhar, A.K., Cowley, J.A., Hasson, K.W., Walker, P.J., Genomic organization, biology, and diagnosis of Taura syndrome virus and yellow head virus of penaeid shrimp, p. 69, Copyright (2004), with permission from Elsevier; (B) Reprinted from Developmental & Comparative Immunology, Vol. 32 (6), Anantasomboon, G., Poonkhum, R., Sittidilokratna, N., Flegel, T.W., Withyachumnarnkul, B., Low viral loads and lymphoid organ spheroids are associated with yellow head virus (YHV) tolerance in whiteleg shrimp Penaeus vannamei, p. 14, Copyright (2008), with permission from Elsevier; (C,D) Reprinted from Aquaculture, Vol. 258 (1–4), Flegel, T.W., Detection of major penaeid shrimp viruses in Asia, a historical perspective with emphasis on Thailand, p. 33, Copyright (2006), with permission from Elsevier).
Prevention of YHV gene expression is considered a major method to control YHV infection; the method by RNA interference (RNAi)-based anti-YHV efficiency through dsRNA injection was reported to specifically inhibit YHV infection by inducing the sequence-specific degradation of mRNA [112]. Sanitt et al. (2014) [115] confirmed that three types of orally delivered dsRNA (dsRab7, dsYHV, combined dsRab7 + dsYHV) were effective in reducing mortality by YHV infection up to 70% compared to control (dsRab7: 70%, dsYHV: 40%, combined dsRab7 + dsYHV: 56%). YHV disease control should mainly be done through the selection of YHV-1 SPF individuals through PCR screening of broodstock and seeds, strengthening of biological security and sanitation measures in the farm, and management of the water environment [100].
3.3. Taura Syndrome Virus (TSV)
TSV (Taura syndrome virus) is known as one of the three most critical shrimp viruses alongside WSSV and YHV, as it has seriously damaged the shrimp aquaculture industry worldwide over the past two decades [95,116]. The name, TSV disease, comes from the Taura River in Ecuador, where it was first reported [52] in the P. vannamei of Ecuador in June 1992 (viral etiology confirmation in 1995). It has since spread to the Americas (Ecuador, Columbia, Honduras, USA, and Mexico), Asia (Thailand, Indonesia, China, Taiwan, and Myanmar), Africa, and the Middle East (Saudi Arabia), with new TSV strains continuing to appear as the virus adapts to new penaeid species and environments [117]. It is estimated that TSV in the Americas has resulted in 1.2 to 2 billion USD in economic losses from 1992–1996 [118].
TSV causes severe mortality in P. vannamei raised in the Americas. It is transmitted through regional and international migration of live host-larvae and broodstock [119]. TSV was originally limited to the Americas, but after P. vannamei was introduced to Asia, it was reported across Asia, in countries such as Thailand, Taiwan, and China and was spread via infected P. vannamei from Latin America [52]. TSV was first reported in juvenile P. vannamei in Ecuador in 1992 and then spread to Colombia in 1993, Honduras and Hawaii in 1994, Mexico and Guatemalan borders in 1995, Taiwan in 1998–1999, Thailand 2003, Korea and Texas coastal countries in 2004, Venezuela in 2005, Saudi Arabia in 2010–2011 and Venezuela in 2016 [1,120,121,122,123,124,125,126,127]. Since the first case of TSV infection in Asia was reported in P. vannamei imported for aquaculture from Taiwan in 1998, it has been reported in all Asian countries that import P. vannamei [62]. TSV was listed as an OIE-designated disease in 2000 and is widespread especially in the Americas and Asia [128] (Figure 18). TSV occurs in all regions except Australia, Africa and some specific regions according to the guidelines of the OIE Aquatic Animal Health Code, and it is the second most damaging disease in the shrimp aquaculture industry after WSSV, in terms of economic loss [2]. However, recently, through enhanced biological security measures, the introduction of TSV-SPF (specific pathogen free) species, and the production of TSV-resistant P. vannamei, the occurrence and damage caused by TSV infection has greatly been reduced [118].
Figure 18.
Distribution map showing the geographical occurrence of Taura syndrome virus (TSV) (Reprinted from CABI, 2019, Taura syndrome virus. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
TSV is a positive-sense, icosahedral-shaped, non-enveloped single-stranded RNA genome of 10.2 kb with a diameter of 32 nm [129] (Table 1). TSV is taxonomically classified in the Aparavirus genus belonging to the Dicistroviridae family [117]. TSV infects tissues of ectodermal and mesodermal origin, particularly hematopoietic tissue, epidermal epithelium, antennal glands, subcuticular connective tissue, lymphoid organs, and striated muscle [1]. The TSV viral capsid consists of three major polypeptides, VP1 (55 kDa), VP2 (40 kDa), and VP3 (24 kDa), and a minor polypeptide, VP0 (58 kDa) [130]. The TSV genome includes ORF 1 [the sequence motifs for non-structural proteins containing protease, helicase, and RNA-dependent RNA polymerase (RdRp); 6324 nt long, encoding a 2107 amino acid polyprotein with a 324 kDa molecular mass] and ORF 2 [the sequences for TSV structural proteins such as three major capsid proteins [VP1 (55 kDa), VP2 (40 kDa), and VP3 (24 kDa)]; 3036 nt long, encoding a 1011 amino acid polypeptide with a 112 kDa molecular mass [2] (Table 2). As the VP2 (40 kDa) gene among the capsid protein genes exhibits the highest genetic variation, it is widely used to determine the genetic relationship between TSV geographical isolates [117].
Phylogenetic analysis of TSV isolates has identified seven lineages, corresponding to geographic origins: (1) America such as Ecuador, Columbia, Honduras, USA, and Mexico from 1993–1998; (2) Southeast Asia (Thailand, Indonesia, China, Taiwan, snd Myanmar); (3) Mexico; (4) Belize; (5) Venezuela, (6) Colombia, and (7) Saudi Arabia [116] (Table 3). Based on the sequence of the VP1 (55 kDa) structural protein, three genotypic variants were identified: the American group, the Southeast Asian group, and the Belize group [52]. When the TSV isolate from Belize (GenBank no. AY826051-826053) in 2002 was compared with the reference isolate from Hawaiian (GenBank no. AY826054-826055), it was confirmed that the Belize isolate was a unique variant of TSV [117]. A new TSV genotype was observed in Saudi Arabia (GenBank no. JX094350), which was a distinct TSV isolate when compared to those from Southeast Asia and Latin America, and it shared 90% sequence identity with a reference isolate in Hawaii (GenBank no. AF277675) [122]. Phylogenetic analysis of Korean TSV strains based on the partial nucleotide sequence of VP1 (55 kDa) determined that Korean isolates (GenBank no. DQ099912-DQ099913) are closely associated with Thailand TSV types (GenBank no. AY912503-9125038) [131]. Sequence identity of TSV isolates for the Texas isolate (GQ502201) were very high in the Chinese and Thai isolates (GenBank no. DQ104696 and AY997025, respectively) and the Hawaii and Belize isolates (GenBank no. AF277675 and AY590471, respectively) (sequence identities for the Texas isolate ORF 1: 98% for the China and Thailand isolates, 97% for Hawaii and Belize isolates, sequence identities for the Texas isolate, an intergenic region (IGR) sequence: 98% for the Hawaii, China, Belize and Thailand isolates, sequence identities for the Texas isolate ORF 2: 97% for the Hawaii, China, and Thailand isolates, 96% for the Belize isolate) [132].
Other species susceptible to TSV infection include the Gulf white shrimp, P. setiferus and Pacific blue shrimp, P. stylirostris, which has been shown to be affected by TSV disease in the juvenile and adults, as well as in the nursery or post larval stages [52]. Although P. vannamei is known to be the main infective host for TSV, several other penaeid species (P. stylirostris, P. setiferus, P. aztecus, P. duorarum, P. chinensis, and P. monodon) have also been identified as susceptibility through experimental challenge infections. In addition, natural infections of TSV were found in various species including P. stylirostris, P. monodon, P. japonicus, M. ensis and the freshwater shrimp, M. rosenbergii [1]. Dhar and Allnutt (2008) [130] reported that the susceptibility of penaeid shrimp species to TSV differs from species to species, and P. vannamei and P. schmitti cultured in the Americas are highly susceptible, whereas other penaeid shrimp species in the Americas such as P. stylirostris, P. setiferus, P. duorarum, and P. aztecus reported less sensitivity to TSV infection. TSV usually causes serious disease as it infects P. vannamei in the late post larval to early juvenile stages, between 15–40 days, but it can also induce serious diseases in both sub-adult and adult P. vannamei [95].
TSV infection in P. vannamei is divided into three stages: acute (7 days after infection with an asymptomatic phase of 2–5 days), transition (lasting 5 days after the acute stage), and chronic (survivors after molting) stages, with a mortality rate of 60–90% [86,133]. Clinical symptoms of acute TSV infection in farmed P. vannamei are characterized by a reddish body color (especially on the tail; uropods, and appendages induced by chromatophore expansion) and irregular black (melanization) spots under the cuticle layer, in addition to anorexia, an erratic swimming behavior, lethargy, soft cuticles, anorexia, flaccid bodies and opaque musculature [95,129] (Figure 19). Shrimp acutely infected with TSV persist for 1–10 days after infection, and exhibit TSV-specific histological lesions, and mortality occurs during or immediately after molting [134,135]. According to Dhar and Allnutt (2008) [130], TSV infection begins within 24 h and death peaks between 7–10 days, and naturally or experimentally surviving individuals with acute infections develop grossly visible, multifocal, melanized lesions on the cephalothorax, tail, and appendages [95]. The main target organs following TSV infection are the cuticular epithelium of the gills, appendages, hindgut, foregut, and general body cuticle, and the lesion can spread to the underlying subcuticular connective tissue and striated muscle, and even the hematopoietic tissue, antennal gland, testes, and ovaries can become infected.
Figure 19.
External symptoms of Taura syndrome virus (TSV) on infected shrimp. (A,B) Penaeus vannamei showing typical signs of TSV at the end of the acute phase: Multifocal and melanized lesions on the thorax and tail (indicated by arrow); (C,D) P. vannamei showing signs of TSV: red tail fan with rough edges on the cuticular epithelium of uropods (indicated by arrow) and multiple melanized cuticular lesions. ((A) Reprinted from Elsevier Books, Dhar, A.K., Allnutt, F.T., Taura Syndrome Virus. In Encyclopedia of virology, p. 8, Copyright (2008), with permission from Elsevier; (B) Reprinted from Aquaculture, Vol. 260 (1–4), Phalitakul, S., Wongtawatchai, J., Sarikaputi, M., Viseshakul, N., The molecular detection of Taura syndrome virus emerging with White spot syndrome virus in penaeid shrimps of Thailand, p. 9, Copyright (2006), with permission from Elsevier; (C,D) Reprinted from Elsevier Books, Samocha, Sustainable biofloc system for marine shrimp, p. 23, Copyright (2019), with permission from Elsevier).
The transition stage of TSV infection is characterized by melanized multifocal lesions of the cephalothorax and tail with reduced mortality, lethargy, and anorexia [95]. Histological features of TSV infected shrimp at the transition stage show the initiation of spheroid developments within the lymphoid organ (LO), normal-appearing LO arterioles (tubules) that demonstrate a diffuse TSV probe positive signal by in situ hybridization (ISH), and infrequent scattered acute phase epithelial lesions [95] (Figure 20). The stage from transition infection to chronic infection begins with the shedding of the melanized exoskeleton and resumption of the molt cycle [136].
Figure 20.
Electron microscopy and histological changes in shrimp infected with Taura syndrome virus (TSV). (A) TEM of CsCl gradient-purified and negative-strained (with 2% PTA) TSV particle isolated from Penaeus vannamei in Ecuador; (B) the section of intestine with 400 × magnification has cytoplasmic inclusion bodies in the lymphoid organ of Penaeus monodon (arrow); (C,D) spheroids (LOS) in the lymphoid organ tissue and ectopic spheroids in the connective tissue of P. vannamei from Venezuela, when stained with H&E, respectively (Scale bar = 25 μm). ((A) Reprinted from Advances in virus research, Vol. 63, Dhar, A.K., Cowley, J.A., Hasson, K.W., Walker, P.J., Genomic organization, biology, and diagnosis of Taura syndrome virus and yellowhead virus of penaeid shrimp, p. 69, Copyright (2004), with permission from Elsevier; (B) Reprinted from Aquaculture, Vol. 260 (1–4), Phalitakul, S., Wongtawatchai, J., Sarikaputi, M., Viseshakul, N., The molecular detection of Taura syndrome virus emerging with White spot syndrome virus in penaeid shrimps of Thailand, p. 9, Copyright (2006), with permission from Elsevier; (C,D) Reprinted from Aquaculture, Vol. 480, Tang, K.F., Aranguren, L.F., Piamsomboon, P., Han, J.E., Maskaykina, I.Y., Schmidt, M.M., Detection of the microsporidian Enterocytozoon hepatopenaei (EHP) and Taura syndrome virus in Penaeus vannamei cultured in Venezuela, p. 5, Copyright (2017), with permission from Elsevier).
The TSV chronic infection stage (or ‘recovery stage’) appears from 6 days after TSV infection and lasts for a period of 8–12 months in experimentally infected P. vannamei with no disease symptoms, normal swimming behavior, and feeding, and no mortality [95]. During chronic TSV infection, there can be complete removal of TSV through apoptosis or there can be continued infection in a chronic state due to continuous virus replication, which is determined by the host’s immunity, nutritional status, and overall health condition [129]. In the chronic stage of TSV infection, shrimp are asymptomatic, and the only histologically identifiable lesions are numerous lymphoid organ spheroids (LOS) [133]. Surviving individuals after TSV infection can act as life-long carriers of TSV infection, and the prevalence of TSV infection in farms can vary from 0–100% [134].
TSV can maintain pathogenicity in dead shrimp for up to 3 weeks, and transmission of TSV can occur when healthy shrimp ingest infected moribund or dead P. vannamei through formula. The water-borne transmission of TSV has been experimentally shown to occur for up to 48 h after the period of maximum mortality, and it is known that TSV infection can be transmitted to other farms through the excrement of birds including seagulls, Larus atricilla that eat TSV-infected shrimp, as well as a flying aquatic insects such as water boatmen, Trichocorixa reticulata [52,130]. Transboundary transport of TSV occurs primarily through the sale and export of live post-larvae or adult shrimp infected with acute or chronic TSV, while frozen shrimp can also be potential carriers due to the ability of TSV to remain infective during prolonged freezing [95]. Although studies on the survival and resistance of TSV under environmental conditions are insufficient, it has commonly been shown to be very resistant, especially in seawater [52]. Although it is hypothesized that vertical transmission from TSV-infected broodstock to offspring is possible, it has not been experimentally verified [137].
P. vannamei infected with TSV exhibits a cumulative mortality rate of 60–95% (cumulative loss 80–95%, survival rate of ≥60%) within one week of TSV disease onset [52,95]. In the years following the first outbreak of TSV in Colombia, the mortality rate from TSV reached 100% [138]. According to Wertheim et al. (2009) [127], it was reported that mortality rates ranged from 40% to 100% when TSV infection occurred in P. vannamei farms. TSV infection occurs most frequently in P. vannamei in the nursery- the grow-out-stage post-larvae or in juveniles weighing <0.05–5 g within 14–40 days [62]. Efforts of several research and commercial breeding programs through TSV-SPR (specific pathogen resistance) selective breeding to control TSV disease since the mid-1990s have significantly reduced TSV incidence (Sookruksawong et al. 2013). Indeed, from 1999 to 2004, there were no TSV outbreaks in the shrimp farms of Colombia, indicating the success of a TSV-resistant breeding program in which 100% of the animals raised were TSV-SPR [138].
Diagnosis of pathogens following TSV disease infection is important to control, predict, and prevent potential outbreaks and significant economic losses [120]. TSV infection at acute, transition, and early chronic stages can be accurately diagnosed using histological or molecular methods, but it is difficult to detect low virus levels during the chronic stage, when the symptoms and most histological lesions disappear [86]. TSV virus testing is carried out using PCR assays, such as a commercial nested RT-PCR kits and reverse transcriptase PCR (RT-PCR) using TSV virus target organs such as uropods, gills, body cuticles, and swimming feet; the OIE recommends using a one-step PCR method for TSV testing [129,139] (Table 5).
In the acute stage of TSV, the cuticular epithelium of the appendages, gills, hindgut, foregut, and general body cuticle are infected as major target tissues, and infected cells appear to have highly basophilic pyknotic, karyorrhectic nuclei, and vivid cytoplasmic eosinophilia, with staining and sized cytoplasmic inclusion bodies in a variable manner [130]. The TSV at the transition stage histologically represents the onset of lymphoid organ (LO) arterioles (tubules) and spheroid development within the LO, and the marked histological characteristic during the chronic stage of infection is the LO spheroid appearance; spheroids include phagocytic semigranular and granular hemocytes undergoing apoptosis [130]. TSV control methods would be effective using farm-level biological security and TSV-specific pathogen free (SPF) and TSV-specific pathogen resistance (SPR) shrimp, a clean environment, and strict seed selection in addition to the immune system improvements for shrimp, could help to reduce the rate of TSV infection [123].
3.4. White Tail Disease (WTD)
WTD (white tail disease) is caused by Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV), and it induces critical economic losses, especially at the hatchery and nursery stages [140]. WTD was first reported in Guadeloupe (French West Indies) in 1995 or 1997 (named white tail disease from Pointe Noire, Guadeloupe in 1997) and later in Martinique (French West Indies) (1999), China (2003), India (2004), Thailand (2006), Taiwan (2006), Australia (2008), Malaysia (2012) [141,142,143,144] (Figure 21). White-tailed disease occurs in the freshwater shrimp M. rosenbergii, which is cultivated in many countries, and has an extremely high mortality rate (often reaching 100%) and causes enormous economic loss [145].
Figure 21.
Distribution map of the geographical occurrence of White tail disease (WTD). (Reprinted from CABI, 2019, Macrobrachium rosenbergii nodavirus. In: Invasive Species Compendium. Wallingford, UK: CAB International, with permission from CABI).
Natural infection of WTD was also observed in P. monodon and P. indicus hatcheries, which are geographically close to the freshwater shrimp M. rosenbergii hatcheries with reported WTD infections; the transmission of MrNV and XSV from M. rosenbergii to P. monodon and P. indicus [144]. Mass mortality due to WTD occurs frequently in M. rosenbergii hatcheries in India, and the cumulative losses are estimated to be worth of millions of dollars [146]. WTD causes high mortality (up to 100%) in M. rosenbergii post-larvae within 2–3 days after infection. In India, WTD caused more than 50 freshwater shrimp hatcheries to have losses of 50%, which resulted in economic losses of approximately 15 million USD per year [147]. WTD (MrNV) causes large amounts of damage in all countries with aquaculture practices for M. rosenbergii, including the world’s largest producer, China [148]. This disease has the potential to disrupt the M. rosenbergii aquaculture industry in the future, and it was listed as the OIE-designated disease of 2009 [149].
WTD is caused by MrNV (Macrobrachium rosenbergii nodavirus) which is accompanied by another virus, XSV (extra small virus) [142] (Table 1). MrNV is a small icosahedral with non-enveloped two single-stranded RNA virus (RNA1: size 2.9 kb, RNA2: size 1.26 kb) approximately 26–27 nm in diameter and was observed in the cytoplasm of connective cells classified into the family Nodaviridae, which consists of two genera, Alphanodavirus and Betanodavirus, Nodaviruses have T = 3 capsids of a single polypeptide that is 43 kDa [54,144]. The phylogenetic tree obtained from RdRp demonstrates that MrNV is more related to alphanodaviruses, whereas in the capsid-based phylogenetic tree, MrNV and PvNV (a second prawn nodavirus; Penaeus vannamei nodavirus) are more closely related to betanodaviruses (MrNV and PvNV: 69% homology in the capsid protein genes) [150,151]. Since it is difficult to classify MrNV as an Alphanodavirus as it mainly infects insects and Betanodavirus which mainly infects fish, it has been proposed that it be classifies as a Gammanodavirus genus belonging to the Nodaviridae family [146,150]. Shrimp infected with MrNV target hemocytes and myonuclei in the lower abdomen, they then spread to the rest of the abdomen, and subsequently, throughout the body via the hemolymph circulatory system, thereby observing the almost tissues of infected shrimp except for hepatopancreas and eyestalks [142]. MrNV, a viral particle with an initial diameter of 27 nm, was observed in WTD-infected shrimp, and shortly thereafter, a second type of virus particle with an abnormally small diameter of 15 nm was observed in the WTD-infected shrimp tissue, which was named XSV [152]. Although there is evidence that MrNV has a critical role in the pathogenesis of WTD, the role of XSV is also important in its pathogenesis [149]. XSV is an icosahedral and linear single stranded positive-sense RNA genome of 0.9 kb (approximately 700–1200 nucleotides) coding for a capsid protein, cp-17 with a 15 nm diameter that was identified in the cytoplasm of connective tissue cells [140]. MrNV and XSV are found to be related in WTD-infected M. rosenbergii, but the interactions between the two pathogens and their effects on pathogenicity are currently unknown [149,150].
MrNV genomic nucleotide sequencing suggested that RNA-1 contained 3202 nucleotides (GenBank no. AY222839) and RNA-2 consisted of 1175 nucleotides (GenBank no. AY222840) [153] (Table 2). RNA-1 included two nonstructural proteins such as A protein [RNA-dependent RNA polymerase (RdRp) containing approximately 1000 amino acids (ca. 100 kDa)] and B protein [13 kDa encoding 30 region of RNA-1 (2725–3126 nucleotides)], whereas RNA-2 included a single polypeptide in the capsid protein [54]. XSV genomic nucleotide sequencing indicated that it consisted of 796 nucleotides such as the coding sequence of the capsid protein CP-17 (17 kDa) and CP-16 (16 kDa) [137]. The MrNV structural protein consisted of a single protein of approximately CP-43 (43 kDa), whereas two polypeptides of approximately CP-17 (17 kDa) and CP-16 (16 kDa) were observed in the XSV particles [150].
Phylogenetic analysis of the WTD isolates was divided into groups for the French West Indies, China, India, Taiwan, Malaysia, Australia, Thailand, and France. The complete genome sequence of MrNV RNA-1 and RNA-2 was reported in 2003 (French West Indies, Gen bank no. AY222839 and AY222840, respectively) in 2004 (Australia, GenBank no. JN619369 and JN619370) [143,154]. Analysis of the nucleotide sequence was used to determine identity with other MrNV. The nucleotide sequence of MrNV (RNA-1) isolated India (GenBank no. AAO60068) has 98% identity with MrNV isolated from French West Indies (GenBank no. AY222839). Similar to MrNV, the nucleotide sequence of XSV isolated from Taiwan (GenBank no. DQ521573) has 97% and 98% identity with the XSV isolated from India (GenBank no. AY247793) and China (GenBank no DQ147318), respectively [151]. In addition, that isolated from Australia (Australian, GenBank no. JN619369) has 94%, 95%, 95%, and 97% identity with MrNV isolated French West Indies (GenBank no. AY222839), China (Chinese 1, GenBank no. AY231436; Chinese 2, GenBank no. FJ751226) and Malaysia (GenBank no. JN187416), respectively. The nucleotide sequence of MrNV (RNA-2) isolated from Australia (GenBank no. JN619370) has 92% identity with French West Indies (GenBank no. AY222840), Chinese 2 (GenBank no. FJ751225), China (GenBank no. AY231437), and Thailand (GenBank no. EU150126-150129) [143].
M. rosenbergii is more susceptible to WTD than other shrimp species, and especially in the larvae, post-larvae, and juvenile stages of development, it has a high mortality. In post-larvae infected M. rosenbergii, the striated muscles of the cephalothorax, abdomen and tail are the most targeted tissues, and adults of M. rosenbergii infected with WTD are resistant to WTD and function only as carriers [140]. Although M. rosenbergii was initially reported as the only host species for the onset of WTD induced by MrNV and XSV, subsequent reports confirmed that marine shrimp species such as P. indicus, P. japonicus, P. monodon, and P. vannamei at the post-larval (PL) stage are also susceptible and capable of high mortality [150] (Table 4). However, according to Bonami and Widada (2011) [150], in the WTD challenge test by the oral route and injection, marine shrimp such as P. indicus, P. japonicus, and P. monodon did not show high susceptibility to the WTD and had no clinical signs or mortality.
Clinical signs of WTD-infected shrimp include lethargy, degeneration of the telson and uropods, opaqueness of the abdominal muscle, reaching up to 100% within 4 days of onset [150,155] (Figure 22). WTD-infected shrimp at post-larvae stage develop symptoms in the second or third abdominal region, gradually extending from the center of the muscle to the anterior and posterior parts of the muscle, showing lethargy and opaqueness of the abdominal muscle [156]. WTD infection begins in some areas of the tail, extends to the tail muscles (abdomen), and causes whitish pigmentation in all muscles in the final stage, including the head (cephalothorax) muscles; in severe cases, degeneration of telsons and uropods is observed [147,150]. WTD symptoms mainly appeared when MrNV values were high, suggesting that MrNV plays an important role in WTD [140].
Figure 22.
External symptoms of shrimps with White tail disease (WTD). (A) MrNV-infected Penaeus vannamei showing signs of whitish muscle in the tail (arrows); (B) Cherax quadricarinatus showing signs of WTD with necrosis and myositis (arrows); (C,D) Clinical signs, whitish abdominal muscles (arrows), in the infected post-larvae of Penaeus indicus ((A) Reprinted from Aquaculture, Vol. 483, Jariyapong, P., Pudgerd, A., Weerachatyanukul, W., Hirono, I., Senapin, S., Dhar, A.K., Chotwiwatthanakun, C., Construction of an infectious Macrobrachium rosenbergii nodavirus from cDNA clones in Sf9 cells and improved recovery of viral RNA with AZT treatment, p. 9, Copyright (2018), with permission from Elsevier; (B) Reprinted from Aquaculture, Vol. 319 (1–2), Hayakijkosol, O., La Fauce, K., Owens, L., Experimental infection of redclaw crayfish (Cherax quadricarinatus) with Macrobrachium rosenbergii nodavirus, the aetiological agent of white tail disease, p. 5, Copyright (2011), with permission from Elsevier; (C,D) Reprinted from Aquaculture, Vol. 292(1–2), Ravi, M., Basha, A.N., Sarathi, M., Idalia, H.R., Widada, J.S., Bonami, J.R., Hameed, A.S., Studies on the occurrence of white tail disease (WTD) caused by MrNV and XSV in hatchery-reared post-larvae of Penaeus indicus and P. monodon, p. 4, Copyright (2009), with permission from Elsevier).
MrNV and XSV can be transmitted horizontally in the form of dead tissue, live carriers, and free virions through formulas of M. rosenbergii infected with WTD, and natural hosts of adjacent ecosystems and culture systems [142,146]. In the WTD horizontal transmission experiment, artemia was exposed to MrNV and XSV by immersion and oral routes, confirming that it could act as a reservoir or carrier for the MrNV and XSV [140]. A high prevalence of WTD induced by MrNV and XSV has been reported in hatchery larvae and post-larvae of M. rosenbergii, suggesting that vertical transmission may occur from infected brooders to offspring during spawning [157]. Murwantoko et al. (2016) [147] also reported the vertical transmission of MrNV and XSV in M. rosenbergii, suggesting that this is the main disease transmission mechanism of WTD. Vectors of WTD include penaeid shrimp (P. japonicus, P. indicus, and P. monodon), aquatic insects (Cybister sp., Aesohna sp., Belostoma sp., and Notonecta sp.), and artemia [158]. A WTD challenge experiment using both oral and intramuscular routes in M. malcolmsonii and M. rude did not cause clinical symptoms or mortality but indicated that it could serve as a reservoir as the toxicity of MrNV and XSV were maintained [147].
Mortality due to WTD infection reaches its maximum 5–6 days after the first severe symptoms appear, and infected post-larvae die within 15 days, and surviving post-larvae can grow to market size just like normal individuals [140]. MrNV infection of M. rosenbergii at the post-larvae stage results in a high mortality rate of almost 100% but it is not fatal for adults [139]. Bonami and Widada (2011) [150] reported that mortality started to occur 1–3 days after the first clinical signs of post-larvae M. rosenbergii infection with WTD, and the cumulative mortality rate reached 100%, 8–14 days post-infection.
To confirm WTD infection, real-time RT-PCR is one of the most sensitive diagnostic methods and has been used to detect the presence of both MrNV and XSV [149] (Table 5). Of the many samples infected with WTD, the majority of MrNV-positive samples were also positive for XSV, but some samples did not have XSV, and in some cases XSV was detected without MrNV [150]. Histological features of WTD-infected shrimp include large oval or irregular basophilic cytoplasmic inclusions with a diameter of 1–4 µm in the infected muscles of the abdomen, cephalothorax, and intratubular connective tissue of the hepatopancreas [140]. Murwantoko et al. (2016) [147] also found lesions in the muscle and connective tissues upon histological examination of the shrimp infected with WTD, and these lesions corresponded to the dense basophilic inclusions that had a diameter of 0.5–3.0 µm, and were located in the cytoplasm. Jariyapong et al. (2018) [159] confirmed coagulation necrosis of skeletal muscle in P. vannamei infected with MrNV (Figure 23B). Hayakijkosol et al. (2011) [160] reported that muscle degeneration, tissue necrosis, and myolysis with hemocytic infiltration were found in MrNV-infected redclaw crayfish, Cherax quadricarinatus (Figure 23C,D).
Figure 23.
Histological changes in shrimp tissues when infected with White tail disease (WTD) and stained with H&E. (A) Uninfected shrimp; (B) Histological detection included the aggregation of cells into clumps of various sizes and coagulative necrosis in P. vannamei skeletal muscle (72 h post-infection); (C,D) Muscle degeneration and necrotic muscle tissues in MrNV-infected C. quadricarinatus (arrow). ((A,B) Reprinted from Aquaculture, Vol. 483, Jariyapong, P., Pudgerd, A., Weerachatyanukul, W., Hirono, I., Senapin, S., Dhar, A.K., Chotwiwatthanakun, C., Construction of an infectious Macrobrachium rosenbergii nodavirus from cDNA clones in Sf9 cells and improved recovery of viral RNA with AZT treatment, p. 9, Copyright (2018), with permission from Elsevier; (C,D) Reprinted from Aquaculture, Vol. 319 (1–2), Hayakijkosol, O., La Fauce, K., Owens, L., Experimental infection of redclaw crayfish (Cherax quadricarinatus) with Macrobrachium rosenbergii nodavirus, the aetiological agent of white tail disease, p. 5, Copyright (2011), with permission from Elsevier).
To control the spread of WTD, it is essential to develop highly sensitive and rapid diagnostic methods that can detect pathogens early, because effective methods such as vaccines or treatment for controlling and preventing WTD have not been presented [142]. Screening using sensitive diagnostic methods such as reverse-transcription polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) to select specific pathogen free (SPF) brood stock and post-larvae can be an effective method [153]. Since virus-borne infections such as WTD are difficult to control, only preventive measures, including daily monitoring of shrimp health and early diagnosis, are critical and can help manage the WTD occurrence [149].
4. Conclusions
In this review, we have looked at the DNA and RNA viral diseases affecting shrimp, which are listed by the World Organization for Animal Health. We have provided an overview of the basic characteristics of the viral disease pathogens that can be fatal to farmed shrimp, as well as the disease distribution range, information on the specific hosts, apparent clinical symptoms, disease transmission methods and vectors, mortality rates, diagnostic techniques, as well as strategies for control and prevention. The legal or illegal cross-border movement of living aquatic species for aquaculture has accelerated the spread of diseases and the demand for vaccines and therapeutics for their prevention. However, to find a fundamental solution, various studies on the etiology of these diseases are needed, and breeding organism-friendly aquaculture methods will be required, which consider animal welfare, such as maintaining an appropriate breeding density and a clean breeding environment, using SPF (specific pathogen free) or SPR (specific pathogen resistance), and nature-friendly breeding and nurturing for a disease-free and sustainable shrimp farming industry. The material in this review will help researchers and those working in the industry to better understand the major viral diseases of shrimp, and can be used as a basic data document to help prepare policy measures to prevent and control shrimp viral diseases in the future.
Author Contributions
Conceptualization, Y.-B.Y. and J.-H.C.; Visualization, A.-H.J. and S.-M.H.; Supervision, J.-H.K. and J.-C.K.; Investigation, A.-H.J. and S.-M.H.; Writing-original draft, D.L., Y.-B.Y. and J.-H.C.; Writing-review & editing, J.-H.K. and J.-C.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the project ‘Development and industrialization of genetically advanced aquatic organism using genetic breeding technology’ (grant number, R2022001) of the National Institute of Fisheries Science.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Walker P.J., Mohan C.V. Viral disease emergence in shrimp aquaculture: Origins, impact and the effectiveness of health management strategies. Rev. Aquac. 2009;1:125–154. doi: 10.1111/j.1753-5131.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lightner D.V., Redman R.M., Pantoja C.R., Tang K.F.J., Noble B.L., Schofield P., Navarro S.A. Historic emergence, impact and current status of shrimp pathogens in the Americas. J. Invertebr. Pathol. 2012;110:174–183. doi: 10.1016/j.jip.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Roy S., Bossier P., Norouzitallab P., Vanrompay D. Trained immunity and perspectives for shrimp aquaculture. Rev. Aquac. 2020;12:2351–2370. doi: 10.1111/raq.12438. [DOI] [Google Scholar]
- 4.Manan H., Ikhwanuddin M. Triploid induction in penaeid shrimps aquaculture: A review. Rev. Aquac. 2021;13:619–631. doi: 10.1111/raq.12489. [DOI] [Google Scholar]
- 5.Morshed M., Islam M.S., Lohano H.D., Shyamsundar P. Production externalities of shrimp aquaculture on paddy farming in coastal Bangladesh. Agric. Water. Manag. 2020;238:106213. doi: 10.1016/j.agwat.2020.106213. [DOI] [Google Scholar]
- 6.Thornber K., Verner-Jeffreys D., Hinchliffe S., Rahman M.M., Bass D., Tyler C.R. Evaluating antimicrobial resistance in the global shrimp industry. Rev Aquac. 2020;12:966–986. doi: 10.1111/raq.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tacon A.G. Trends in global aquaculture and aquafeed production: 2000–2017. Rev. Fish. Sci. Aquac. 2020;28:43–56. doi: 10.1080/23308249.2019.1649634. [DOI] [Google Scholar]
- 8.Flegel T.W. Historic emergence, impact and current status of shrimp pathogens in Asia. J. Invertebr. Pathol. 2012;110:166–173. doi: 10.1016/j.jip.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Sánchez-Paz A. White spot syndrome virus: An overview on an emergent concern. Vet. Res. 2010;41:43. doi: 10.1051/vetres/2010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiong J. Progress in the gut microbiota in exploring shrimp disease pathogenesis and incidence. Appl. Microbiol. Biotechnol. 2018;102:7343–7350. doi: 10.1007/s00253-018-9199-7. [DOI] [PubMed] [Google Scholar]
- 11.Flegel T.W. Current status of viral diseases in Asian shrimp aquaculture. Isr. J. Aquac. Bamidgeh. 2009;60:229–239. doi: 10.46989/001c.20556. [DOI] [Google Scholar]
- 12.Thitamadee S., Prachumwat A., Srisala J., Jaroenlak P., Salachan P.V., Sritunyalucksana K., Itsathiphaisarn O. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture. 2016;452:69–87. doi: 10.1016/j.aquaculture.2015.10.028. [DOI] [Google Scholar]
- 13.Boonyakida J., Xu J., Satoh J., Nakanishi T., Mekata T., Kato T., Park E.Y. Antigenic properties of VP15 from white spot syndrome virus in kuruma shrimp Marsupenaeus japonicus. Fish Shellfish. Immunol. 2020;101:152–158. doi: 10.1016/j.fsi.2020.03.061. [DOI] [PubMed] [Google Scholar]
- 14.Pradeep B., Rai P., Mohan S.A., Shekhar M.S., Karunasagar I. Biology, host range, pathogenesis and diagnosis of white spot syndrome virus. Indian. J. Virol. 2012;23:161–174. doi: 10.1007/s13337-012-0079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L., Zhang S., Hou C., Liang X., Dehwah M.A.S., Tan B., Shi L. The T cell factor, pangolin, from Litopenaeus vannamei play a positive role in the immune responses against white spot syndrome virus infection. Dev. Comp. Immunol. 2021;119:104041. doi: 10.1016/j.dci.2021.104041. [DOI] [PubMed] [Google Scholar]
- 16.Qiu W., Geng R., Zuo H., Weng S., He J., Xu X. Toll receptor 2 (Toll2) positively regulates antibacterial immunity but promotes white spot syndrome virus (WSSV) infection in shrimp. Dev. Comp. Immunol. 2021;115:103878. doi: 10.1016/j.dci.2020.103878. [DOI] [PubMed] [Google Scholar]
- 17.Panchal V., Kumar S., Hossain S.N., Vasudevan D. Structure analysis of thymidylate synthase from white spot syndrome virus reveals WSSV-specific structural elements. Int. J. Biol. Macromol. 2021;167:1168–1175. doi: 10.1016/j.ijbiomac.2020.11.071. [DOI] [PubMed] [Google Scholar]
- 18.Verbruggen B., Bickley L.K., Van Aerle R., Bateman K.S., Stentiford G.D., Santos E.M., Tyler C.R. Molecular mechanisms of white spot syndrome virus infection and perspectives on treatments. Viruses. 2016;8:23. doi: 10.3390/v8010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oakey J., Smith C., Underwood D., Afsharnasab M., Alday-Sanz V., Dhar A., Crook A. Global distribution of white spot syndrome virus genotypes determined using a novel genotyping assay. Arch. Virol. 2019;164:2061–2082. doi: 10.1007/s00705-019-04265-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dey B.K., Dugassa G.H., Hinzano S.M., Bossier P. Causative agent, diagnosis and management of white spot disease in shrimp: A review. Rev. Aquac. 2020;12:822–865. doi: 10.1111/raq.12352. [DOI] [Google Scholar]
- 21.Chen H., Wang Y., Zhang J., Bao J. Intestinal microbiota in white spot syndrome virus infected red swamp crayfish (Procambarus clarkii) at different health statuses. Aquaculture. 2021;542:736826. doi: 10.1016/j.aquaculture.2021.736826. [DOI] [Google Scholar]
- 22.Lai Y., Zhu F., Xu Y. WSSV proteins and DNA genome released by ultrasonic rupture can infect cray fish as effectively as intact virions. J. Virol. Methods. 2020;283:113917. doi: 10.1016/j.jviromet.2020.113917. [DOI] [PubMed] [Google Scholar]
- 23.Pereira J.M.P., de Souza E.N.V., Candido J.R., Dantas M.D., Nunes A.R., Ribeiro K., Lanza D.C. Alternative PCR primers for genotyping of Brazilian WSSV isolates. J. Invertebr. Pathol. 2019;162:55–63. doi: 10.1016/j.jip.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Sathyabhama A.B., Puthumana J., Kombiyil S., Philip R., Singh I.S.B. ‘PmLyO-Sf9-WSSV complex’could be a platform for elucidating the mechanism of viral entry, cellular apoptosis and replication impediments. Virology. 2021;553:102–110. doi: 10.1016/j.virol.2020.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Weerachatyanukul W., Chotwiwatthanakun C., Jariyapong P. Dual VP28 and VP37 dsRNA encapsulation in IHHNV virus-like particles enhances shrimp protection against white spot syndrome virus. Fish Shellfish. Immunol. 2021;113:89–95. doi: 10.1016/j.fsi.2021.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Li J., Xu L., Li F., Yang F. Low-abundance envelope protein VP12 of white spot syndrome virus interacts with envelope protein VP150 and capsid protein VP51. Virus Res. 2013;178:206–210. doi: 10.1016/j.virusres.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Talukder A.S., Punom N.J., Eshik M.M.E., Begum M.K., Islam H.R., Hossain Z., Rahman M.S. Molecular identification of white spot syndrome virus (WSSV) and associated risk factors for white spot disease (WSD) prevalence in shrimp (Penaeus monodon) aquaculture in Bangladesh. J. Invertebr. Pathol. 2021;179:107535. doi: 10.1016/j.jip.2021.107535. [DOI] [PubMed] [Google Scholar]
- 28.Ramos-Paredes J., Grijalva-Chon J.M., Ibarra-Gámez J.C. Virulence and genotypes of white spot syndrome virus infecting Pacific white shrimp Litopenaeus vannamei in north-western Mexico. J. Fish Dis. 2017;40:425–435. doi: 10.1111/jfd.12598. [DOI] [PubMed] [Google Scholar]
- 29.van Hulten M.C., Witteveldt J., Peters S., Kloosterboer N., Tarchini R., Fiers M., Vlak J.M. The white spot syndrome virus DNA genome sequence. Virology. 2001;286:7–22. doi: 10.1006/viro.2001.1002. [DOI] [PubMed] [Google Scholar]
- 30.Yang F., He J., Lin X., Li Q., Pan D., Zhang X., Xu X. Complete genome sequence of the shrimp white spot bacilliform virus. J. Virol. 2001;75:11811–11820. doi: 10.1128/JVI.75.23.11811-11820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L.L., Wang H.C., Huang C.J., Peng S.E., Chen Y.G., Lin S.J., Chen W.Y., Dai C.F., Yu H.T., Wang C.H., et al. Transcriptional analysis of the DNA polymerase gene of shrimp white spot syndrome virus. Virology. 2002;301:136–147. doi: 10.1006/viro.2002.1536. [DOI] [PubMed] [Google Scholar]
- 32.Chai C.Y., Yoon J., Lee Y.S., Kim Y.B., Choi T.J. Analysis of the complete nucleotide sequence of a white spot syndrome virus isolated from pacific white shrimp. J. Microbiol. 2013;51:695–699. doi: 10.1007/s12275-013-3171-0. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Anaya L.Z., Gonzalez-Galaviz J.R., Casillas-Hernandez R. Draft genome sequence of white spot syndrome virus isolated from cultured Litopenaeus vannamei in Mexico. Genome Announc. 2016;4:e01674-15. doi: 10.1128/genomeA.01674-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li F., Gao M., Xu L., Yang F. Comparative genomic analysis of three white spot syndrome virus isolates of different virulence. Virus Genes. 2017;53:249–258. doi: 10.1007/s11262-016-1421-z. [DOI] [PubMed] [Google Scholar]
- 35.Han Y., Li F., Xu L., Yang F.A. VP24-truncated isolate of white spot syndrome virus is inefficient in per os infection. Vet. Res. 2017;48:1–11. doi: 10.1186/s13567-017-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang L., Xiao J., Liu L., Pan Y., Yan S., Wang Y. Characterization and prevalence of a novel white spot syndrome viral genotype in naturally infected wild crayfish, Procambarus clarkii, in Shanghai, China. Virusdisease. 2017;28:250–261. doi: 10.1007/s13337-017-0394-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oakey H.J., Smith C.S. Complete genome sequence of a white spot syndrome virus associated with a disease incursion in Australia. Aquaculture. 2018;484:152–159. doi: 10.1016/j.aquaculture.2017.11.009. [DOI] [Google Scholar]
- 38.Vinaya Kumar K., Shekhar M.S., Otta S.K., Karthic K., Ashok Kumar J., Gopikrishna G., Vijayan K.K. First Report of a Complete Genome Sequence of White spot syndrome virus from India. Genome Announc. 2018;6:e00055-18. doi: 10.1128/genomeA.00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Restrepo L., Reyes A., Bajaña L., Betancourt I., Bayot B. Draft genome sequence of a white spot syndrome virus isolate obtained in Ecuador. Genome Announc. 2018;6:e00605-18. doi: 10.1128/genomeA.00605-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dantas M.D.A., Teixeira D.G., Silva-Portela R.C.B., Soares P.E.T., Lima J.P.M.S., Agnez-Lima L.F., Lanza D.C.F. Direct sequencing of the white spot syndrome virus from Brazil: Genome assembly and new insights on phylogeny. Virus Res. 2018;245:52–61. doi: 10.1016/j.virusres.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Cruz-Flores R., Mai H.N., Kanrar S., Caro L.F.A., Dhar A.K. Genome reconstruction of white spot syndrome virus (WSSV) from archival Davidson’s-fixed paraffin embedded shrimp (Penaeus vannamei) tissue. Sci. Rep. 2020;10:13425. doi: 10.1038/s41598-020-70435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dashtiannasab A. White Spot Syndrome Virus. In: Ennaji M.M., editor. Emerging and Reemerging Viral Pathogens. Academic Press; Cambridge, MA, USA: 2020. pp. 717–728. Volume 1: Fundamental and Basic Viology Aspects of Human, Animal and Plant Pathogens. [DOI] [Google Scholar]
- 43.Zwart M.P., Dieu B.T.M., Hemerik L., Vlak J.M. Evolutionary trajectory of white spot syndrome virus (WSSV) genome shrinkage during spread in Asia. PLoS ONE. 2010;5:e13400. doi: 10.1371/journal.pone.0013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoganandhan K., Thirupathi S., Hameed A.S. Biochemical, physiological and hematological changes in white spot syndrome virus-infected shrimp, Penaeus indicus. Aquaculture. 2003;221:1–11. doi: 10.1016/S0044-8486(02)00220-X. [DOI] [Google Scholar]
- 45.Tuyen N.X., Verreth J., Vlak J.M., de Jong M.C.M. Horizontal transmission dynamics of White spot syndrome virus by cohabitation trials in juvenile Penaeus monodon and P. vannamei. Prev. Vet. Med. 2014;117:286–294. doi: 10.1016/j.prevetmed.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Arulmoorthy M.P., Anandajothi E., Vasudevan S., Suresh E. Major viral diseases in culturable penaeid shrimps: A review. Aquac. Int. 2020;28:1939–1967. doi: 10.1007/s10499-020-00568-3. [DOI] [Google Scholar]
- 47.Patil P.K., Geetha R., Ravisankar T., Avunje S., Solanki H.G., Abraham T.J., Vijayan K.K. Economic loss due to diseases in Indian shrimp farming with special reference to Enterocytozoon hepatopenaei (EHP) and white spot syndrome virus (WSSV) Aquaculture. 2021;533:736231. doi: 10.1016/j.aquaculture.2020.736231. [DOI] [Google Scholar]
- 48.Hameed A.S., Yoganandhan K., Sathish S., Rasheed M., Murugan V., Jayaraman K. White spot syndrome virus (WSSV) in two species of freshwater crabs (Paratelphusa hydrodomous and P. pulvinate) Aquaculture. 2001;201:179–186. doi: 10.1016/S0044-8486(01)00525-7. [DOI] [Google Scholar]
- 49.Escobedo-Bonilla C.M., Alday-Sanz V., Wille M., Sorgeloos P., Pensaert M.B., Nauwynck H.J. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J. Fish Dis. 2008;31:1–18. doi: 10.1111/j.1365-2761.2007.00877.x. [DOI] [PubMed] [Google Scholar]
- 50.Qian X., Zhu F. Use of glycerol monolaurate as a treatment against white spot syndrome virus in crayfish (Procambarus clarkii) Aquaculture. 2021;541:736853. doi: 10.1016/j.aquaculture.2021.736853. [DOI] [Google Scholar]
- 51.Xiao C., Zhang Y., Zhu F. Effect of dietary sodium butyrate on the innate immune response of Procambarus clarkii and disease resistance against white spot syndrome virus. Aquaculture. 2021;541:736784. doi: 10.1016/j.aquaculture.2021.736784. [DOI] [Google Scholar]
- 52.Stentiford G.D., Bonami J.R., Alday-Sanz V. A critical review of susceptibility of crustaceans to Taura syndrome, Yellowhead disease and White Spot Disease and implications of inclusion of these diseases in European legislation. Aquaculture. 2009;291:1–17. doi: 10.1016/j.aquaculture.2009.02.042. [DOI] [Google Scholar]
- 53.Hossain A., Nandi S.P., Siddique M.A., Sanyal S.K., Sultana M., Hossain M.A. Prevalence and distribution of White Spot Syndrome Virus in cultured shrimp. Lett. Appl. Microbiol. 2015;60:128–134. doi: 10.1111/lam.12353. [DOI] [PubMed] [Google Scholar]
- 54.Gholamhosseini A., Mohammadi A., Akbari S., Banaee M. Molecular, histopathologic and electron microscopic analysis of white spot syndrome virus in wild shrimp (Fenneropenaeus indicus) in the coastal waters of Iran. Arch. Virol. 2020;165:1433–1440. doi: 10.1007/s00705-020-04625-3. [DOI] [PubMed] [Google Scholar]
- 55.Sritunyalucksana K., Srisala J., McColl K., Nielsen L., Flegel T.W. Comparison of PCR testing methods for white spot syndrome virus (WSSV) infections in penaeid shrimp. Aquaculture. 2006;255:95–104. doi: 10.1016/j.aquaculture.2005.12.002. [DOI] [Google Scholar]
- 56.Gangnonngiw W., Bunnontae M., Phiwsaiya K., Senapin S., Dhar A.K. In experimental challenge with infectious clones of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV), MrNV alone can cause mortality in freshwater prawn (Macrobrachium rosenbergii) Virology. 2020;540:30–37. doi: 10.1016/j.virol.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Chai C., Liu Y., Xia X., Wang H., Pan Y., Yan S., Wang Y. Prevalence and genomic analysis of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei shrimp farmed in Shanghai, China. Arch. Virol. 2016;161:3189–3201. doi: 10.1007/s00705-016-3022-5. [DOI] [PubMed] [Google Scholar]
- 58.Rai P., Safeena M.P., Krabsetsve K., La Fauce K., Owens L., Karunasagar I. Genomics, molecular epidemiology and diagnostics of infectious hypodermal and hematopoietic necrosis virus. Indian J. Virol. 2012;23:203–214. doi: 10.1007/s13337-012-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J.Y., Yang N., Hou Z.H. Research progress on hosts and carriers, prevalence, virulence of infectious hypodermal and hematopoietic necrosis virus (IHHNV) J. Invertebr. Pathol. 2021;183:107556. doi: 10.1016/j.jip.2021.107556. [DOI] [PubMed] [Google Scholar]
- 60.Shen H., Zhang W., Shao S. Phylogenetic and recombination analysis of genomic sequences of IHHNV. J. Basic Microbiol. 2015;55:1048–1052. doi: 10.1002/jobm.201400900. [DOI] [PubMed] [Google Scholar]
- 61.Nita M.K.H., Kua B.C., Bhassu S., Othman R.Y. Detection and genetic profiling of infectious hypodermal and haematopoietic necrosis virus (IHHNV) infections in wild berried freshwater prawn, Macrobrachium rosenbergii collected for hatchery production. Mol. Biol. Rep. 2012;39:3785–3790. doi: 10.1007/s11033-011-1155-x. [DOI] [PubMed] [Google Scholar]
- 62.Lightner D.V. Virus diseases of farmed shrimp in the Western Hemisphere (the Americas): A review. J. Invertebr. Pathol. 2011;106:110–130. doi: 10.1016/j.jip.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leyva-Madrigal K.Y., Luna-González A., Escobedo-Bonilla C.M., Fierro-Coronado J.A., Maldonado-Mendoza I.E. Screening for potential probiotic bacteria to reduce prevalence of WSSV and IHHNV in whiteleg shrimp (Litopenaeus vannamei) under experimental conditions. Aquaculture. 2011;322:16–22. doi: 10.1016/j.aquaculture.2011.09.033. [DOI] [Google Scholar]
- 64.Rai P., Pradeep B., Karunasagar I., Karunasagar I. Detection of viruses in Penaeus monodon from India showing signs of slow growth syndrome. Aquaculture. 2009;289:231–235. doi: 10.1016/j.aquaculture.2008.12.035. [DOI] [Google Scholar]
- 65.Montgomery-Brock D., Tacon A.G.J., Poulos B., Lightner D. Reduced replication of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei held in warm water. Aquaculture. 2007;265:41–48. doi: 10.1016/j.aquaculture.2007.01.025. [DOI] [Google Scholar]
- 66.Tang K.F., Lightner D.V. Infectious hypodermal and hematopoietic necrosis virus (IHHNV)-related sequences in the genome of the black tiger prawn Penaeus monodon from Africa and Australia. Virus Res. 2006;118:185–191. doi: 10.1016/j.virusres.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Motte E., Yugcha E., Luzardo J., Castro F., Leclercq G., Rodríguez J., Boulo V. Prevention of IHHNV vertical transmission in the white shrimp Litopenaeus vannamei. Aquaculture. 2003;219:57–70. doi: 10.1016/S0044-8486(02)00631-2. [DOI] [Google Scholar]
- 68.Chen J., Wang W., Wang X., Zhang Q., Ren Y., Song J., Wang X., Dong X., Huang J. First detection of yellow head virus genotype 3 (YHV-3) in cultured Penaeus monodon, mainland China. J. Fish Dis. 2018;41:1449–1451. doi: 10.1111/jfd.12826. [DOI] [PubMed] [Google Scholar]
- 69.Encinas-García T., Mendoza-Cano F., Enríquez-Espinoza T., Luken-Vega L., Vichido-Chávez R., Sánchez-Paz A. An improved validated SYBR green-based real-time quantitative PCR assay for the detection of the Penaeus stylirostris densovirus in penaeid shrimp. J. Virol. Methods. 2015;212:53–58. doi: 10.1016/j.jviromet.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Y.P., Li C., Wan X.Y., Yang Q., Xie G.S., Huang J. Delivery of plasmid DNA to shrimp hemocytes by Infectious hypodermal and hematopoietic necrosis virus (IHHNV) nanoparticles expressed from a baculovirus insect cell system. J. Invertebr. Pathol. 2019;166:107231. doi: 10.1016/j.jip.2019.107231. [DOI] [PubMed] [Google Scholar]
- 71.Chen B.K., Dong Z., Liu D.P., Yan Y.B., Pang N.Y., Nian Y.Y., Yan D.C. Infectious hypodermal and haematopoietic necrosis virus (IHHNV) infection in freshwater crayfish Procambarus clarkii. Aquaculture. 2017;477:76–79. doi: 10.1016/j.aquaculture.2017.05.002. [DOI] [Google Scholar]
- 72.OIE Manual of Diagnostic Tests for Aquatic Animals. Chapter 2.2.4, Infection with Infectious Hypodermal and Haematopoietic Necrosis Virus. 2019. [(accessed on 11 May 2018)]. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/aquatic-manual-online-access/
- 73.Andrade T.P.D., Redman R.M., Lightner D.V. Evaluation of the preservation of shrimp samples with Davidson’s AFA fixative for infectious myonecrosis virus (IMNV) in situ hybridization. Aquaculture. 2008;278:179–183. doi: 10.1016/j.aquaculture.2008.03.024. [DOI] [Google Scholar]
- 74.Borsa M., Seibert C.H., Rosa R.D., Stoco P.H., Cargnin-Ferreira E., Pereira A.M.L., Grisar E.C., Zanetti C.R., Pinto A.R. Detection of infectious myonecrosis virus in penaeid shrimps using immunoassays: Usefulness of monoclonal antibodies directed to the viral major capsid protein. Arch. Virol. 2011;156:9–16. doi: 10.1007/s00705-010-0810-1. [DOI] [PubMed] [Google Scholar]
- 75.Coelho M.G.L., Silva A.C.G., Nova C.M.V.V., Neto J.M.O., Lima A.C.N., Feijó R.G., Apolinário D.F., Maggioni R., Gesteira T.C.V. Susceptibility of the wild southern brown shrimp (Farfantepenaeus subtilis) to infectious hypodermal and hematopoietic necrosis (IHHN) and infectious myonecrosis (IMN) Aquaculture. 2009;294:1–4. doi: 10.1016/j.aquaculture.2009.05.023. [DOI] [Google Scholar]
- 76.Prasad K.P., Shyam K.U., Banu H., Jeena K., Krishnan R. Infectious Myonecrosis Virus (IMNV)–An alarming viral pathogen to Penaeid shrimps. Aquaculture. 2017;477:99–105. doi: 10.1016/j.aquaculture.2016.12.021. [DOI] [Google Scholar]
- 77.Mai H.N., Hanggono B., Caro L.F.A., Komaruddin U., Nur’aini Y.L., Dhar A.K. Novel infectious myonecrosis virus (IMNV) genotypes associated with disease outbreaks on Penaeus vannamei shrimp farms in Indonesia. Arch. Virol. 2019;164:3051–3057. doi: 10.1007/s00705-019-04408-5. [DOI] [PubMed] [Google Scholar]
- 78.Jithendran K.P., Krishnan A.N., Jagadeesan V., Anandaraja R., Ezhil Praveena P., Anushya S., Bhuvaneswari T. Co-infection of infectious myonecrosis virus and Enterocytozoon hepatopenaei in Penaeus vannamei farms in the east coast of India. Aquac. Res. 2021;52:4701–4710. doi: 10.1111/are.15304. [DOI] [Google Scholar]
- 79.Santhosh Kumar S., Sivakumar S., Abdul Majeed S., Vimal S., Taju G., Sahul Hameed A.S. In vitro propagation of infectious myonecrosis virus in C6/36 mosquito cell line. J. Fish Dis. 2021;44:987–992. doi: 10.1111/jfd.13359. [DOI] [PubMed] [Google Scholar]
- 80.Kokkattunivarthil S., Krishnan R., Kezhedath J., Prasad K.P., Naik T.V. New set of PCR primers for SYBR green-based qPCR detection of IMNV in India. Aquaculture. 2018;495:726–730. doi: 10.1016/j.aquaculture.2018.06.061. [DOI] [Google Scholar]
- 81.Senapin S., Phiwsaiya K., Gangnonngiw W., Flegel T.W. False rumours of disease outbreaks caused by infectious myonecrosis virus (IMNV) in the whiteleg shrimp in Asia. J. Negat. Results. Biomed. 2011;10:10. doi: 10.1186/1477-5751-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jha R.K., Babikian H., Kristina, Srisombat S. Managing infectious myonecrosis virus (IMNV) in Vannamei shrimp culture: Learning by doing. Int. J. Fish. Aquat. Stud. 2021;9:385–391. doi: 10.22271/fish.2021.v9.i1e.2424. [DOI] [Google Scholar]
- 83.Sahul Hameed A.S., Abdul Majeed S., Vimal S., Madan N., Rajkumar T., Santhoshkumar S., Sivakumar S. Studies on the occurrence of infectious myonecrosis virus in pond-reared Litopenaeus vannamei (Boone, 1931) in India. J. Fish Dis. 2017;40:1823–1830. doi: 10.1111/jfd.12655. [DOI] [PubMed] [Google Scholar]
- 84.Kunanopparat A., Chaivisuthangkura P., Senapin S., Longyant S., Rukpratanporn S., Flegel T.W., Sithigorngul P. Detection of infectious myonecrosis virus using monoclonal antibody specific to N and C fragments of the capsid protein expressed heterologously. J. Virol. Methods. 2011;171:141–148. doi: 10.1016/j.jviromet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 85.Teixeira-Lopes M.A., Vieira-Girão P.R.N., da Cruze Freire J.E., Rocha I.R.C.B., Costa F.H.F., Rádis-Baptista G. Natural co-infection with infectious hypodermal and hematopoietic necrosis virus (IHHNV) and infectious myonecrosis virus (IMNV) in Litopenaeus vannamei in Brazil. Aquaculture. 2011;312:212–216. doi: 10.1016/j.aquaculture.2011.01.005. [DOI] [Google Scholar]
- 86.Poulos B.T., Tang K.F.J., Pantoja C.R., Bonami J.R., Lightner D.V. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J. Gen. Virol. 2006;87:987–996. doi: 10.1099/vir.0.81127-0. [DOI] [PubMed] [Google Scholar]
- 87.Seibert C.H., Borsa M., Rosa R.D., Cargnin-Ferreira E., Pereira A.M.L., Grisard E.C., Zanetti C.R., Pinto A.R. Detection of major capsid protein of infectious myonecrosis virus in shrimps using monoclonal antibodies. J. Virol. Methods. 2010;169:169–175. doi: 10.1016/j.jviromet.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 88.Vanpatten K.A., Nunan L.M., Lightner D.V. Seabirds as potential vectors of penaeid shrimp viruses and the development of a surrogate laboratory model utilizing domestic chickens. Aquaculture. 2004;241:31–46. doi: 10.1016/j.aquaculture.2004.08.012. [DOI] [Google Scholar]
- 89.Srisala J., Sanguanrut P., Laiphrom S., Siriwattano J., Khudet J., Thaiue D., Sritunyalucksana K. Infectious myonecrosis virus (IMNV) and decapod iridescent virus 1 (DIV1) detected in Penaeus monodon from the Indian Ocean. Aquaculture. 2021;545:1–26. doi: 10.1016/j.aquaculture.2021.737262. [DOI] [Google Scholar]
- 90.Feijó R.G., Kamimura M.T., Oliveira-Neto J.M., Vila-Nova C.M.V.M., Gomes A.C.S., Coelho M.G.L., Vasconcelos R.F., Gesteira T.C.V., Marins L.F., Maggioni R. Infectious myonecrosis virus and white spot syndrome virus co-infection in Pacific white shrimp (Litopenaeus vannamei) farmed in Brazil. Aquaculture. 2013;380:1–5. doi: 10.1016/j.aquaculture.2012.11.026. [DOI] [Google Scholar]
- 91.Cowley J.A., Rao M., Mohr P., Moody N.J., Sellars M.J., Crane M.S.J. TaqMan real-time and conventional nested PCR tests specific to yellow head virus genotype 7 (YHV7) identified in giant tiger shrimp in Australia. J. Virol. Methods. 2019;273:113689. doi: 10.1016/j.jviromet.2019.113689. [DOI] [PubMed] [Google Scholar]
- 92.Sittidilokratna N., Dangtip S., Cowley J.A., Walker P.J. RNA transcription analysis and completion of the genome sequence of yellow head nidovirus. Virus Res. 2008;136:157–165. doi: 10.1016/j.virusres.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li C., Ren Y., Dong X., Wang C., Huang J. Extraction of assembling complexes of viral capsomers from shrimp tissue infected with yellow head virus genotype 8 (YHV-8) J. Fish Dis. 2019;42:613–616. doi: 10.1111/jfd.12929. [DOI] [PubMed] [Google Scholar]
- 94.Soowannayan C., Nguyen G.T., Pham L.N., Phanthura M., Nakthong N. Australian red claw crayfish (Cherax quadricarinatus) is susceptible to yellow head virus (YHV) infection and can transmit it to the black tiger shrimp (Penaeus monodon) Aquaculture. 2015;445:63–69. doi: 10.1016/j.aquaculture.2015.04.015. [DOI] [Google Scholar]
- 95.Dhar A.K., Cowley J.A., Hasson K.W., Walker P.J. Genomic organization, biology, and diagnosis of Taura syndrome virus and yellowhead virus of penaeid shrimp. Adv. Virus Res. 2004;63:353. doi: 10.1016/S0065-3527(04)63006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bateman K.S., Stentiford G.D. A taxonomic review of viruses infecting crustaceans with an emphasis on wild hosts. J. Invertebr. Pathol. 2017;147:86–110. doi: 10.1016/j.jip.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 97.Wijegoonawardane P.K., Cowley J.A., Phan T., Hodgson R.A.J., Nielsen L., Kiatpathomchai W., Walker P.J. Genetic diversity in the yellow head nidovirus complex. Virology. 2008;380:213–225. doi: 10.1016/j.virol.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mohr P.G., Moody N.J.G., Hoad J., Williams L.M., Bowater R.O., Cummins D.M., Crane M.S.J. New yellow head virus genotype (YHV7) in giant tiger shrimp Penaeus monodon indigenous to northern Australia. Dis. Aquat. Org. 2015;115:263–268. doi: 10.3354/dao02894. [DOI] [PubMed] [Google Scholar]
- 99.Elliott L., Owens L. CART analysis of environmental factors, biomarkers and gill-associated virus to predict production outcomes for farmed Penaeus monodon. Aquaculture. 2015;448:298–305. doi: 10.1016/j.aquaculture.2015.05.006. [DOI] [Google Scholar]
- 100.Walker P.J., Sittidilokratna N. Yellow Head Virus. In: Mahy B.W.J., Van Regenmortel M.H.V., editors. Encyclopedia of Virology. 3rd ed. Academic Press; Cambridge, MA, USA: 2008. pp. 476–483. [DOI] [Google Scholar]
- 101.Dong X., Liu S., Zhu L., Wan X., Liu Q., Qiu L., Zou P., Zhang Q., Huang J. Complete genome sequence of an isolate of a novel genotype of yellow head virus from Fenneropenaeus chinensis indigenous in China. Arch. Virol. 2017;162:1149–1152. doi: 10.1007/s00705-016-3203-2. [DOI] [PubMed] [Google Scholar]
- 102.Anantasomboon G., Poonkhum R., Sittidilokratna N., Flegel T.W., Withyachumnarnkul B. Low viral loads and lymphoid organ spheroids are associated with yellow head virus (YHV) tolerance in whiteleg shrimp Penaeus vannamei. Dev. Comp. Immunol. 2008;32:613–626. doi: 10.1016/j.dci.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Havanapan P.O., Taengchaiyaphum S., Paemanee A., Phungthanom N., Roytrakul S., Sritunyalucksana K., Krittanai C. Caspase-3, a shrimp phosphorylated hemocytic protein is necessary to control YHV infection. Fish Shellfish Immunol. 2021;114:36–48. doi: 10.1016/j.fsi.2021.04.007. [DOI] [PubMed] [Google Scholar]
- 104.Walker P.J., Cowley J.A., Dong X., Huang J., Moody N., Ziebuhr J. ICTV Virus Taxonomy Profile: Roniviridae. J. Gen. Virol. 2021;102:001514. doi: 10.1099/jgv.0.001514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang H.L., Qiu L., Liu Q., Wan X.Y., Liu S., Zhu L.L., Huang J. A novel method of real-time reverse-transcription loop-mediated isothermal amplification developed for rapid and quantitative detection of a new genotype (YHV-8) of yellow head virus. Lett. Appl. Microbiol. 2016;63:103–110. doi: 10.1111/lam.12591. [DOI] [PubMed] [Google Scholar]
- 106.Senapin S., Thaowbut Y., Gangnonngiw W., Chuchird N., Sriurairatana S., Flegel T.W. Impact of yellow head virus outbreaks in the whiteleg shrimp, Penaeus vannamei (Boone), in Thailand. J. Fish Dis. 2010;33:421–430. doi: 10.1111/j.1365-2761.2009.01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cowley J.A., Cadogan L.C., Spann K.M., Sittidilokratna N., Walker P.J. The Gene Encoding the Nucleocapsid Protein of Gill-Associated Nidovirus of Penaeus monodon Prawns Is Located Upstream of the Glycoprotein Gene. J. Virol. 2004;78:8935–8941. doi: 10.1128/JVI.78.16.8935-8941.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cedano-Thomas Y., de la Rosa-Vélez J., Bonami J.R., Vargas-Albores F. Gene expression kinetics of the yellow head virus in experimentally infected Litopenaeus vannamei. Aquac. Res. 2010;41:1432–1443. doi: 10.1111/j.1365-2109.2009.02434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Samocha T.M. Sustainable Biofloc Systems for Marine Shrimp. Academic Press; Cambridge, MA, USA: 2019. [Google Scholar]
- 110.Prapavorarat A., Pongsomboon S., Tassanakajon A. Identification of genes expressed in response to yellow head virus infection in the black tiger shrimp, Penaeus monodon, by suppression subtractive hybridization. Dev. Comp. Immunol. 2010;34:611–617. doi: 10.1016/j.dci.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 111.Cowley J.A. Nidoviruses of Fish and Crustaceans. In: Kibenge F.S.B., Godoy M.G., editors. Aquaculture Virology. Academic Press; Cambridge, MA, USA: 2016. pp. 443–472. [DOI] [Google Scholar]
- 112.Thedcharoen P., Pewkliang Y., Kiem H.K.T., Nuntakarn L., Taengchaiyaphum S., Sritunyalucksana K., Borwornpinyo S. Effective suppression of yellow head virus replication in Penaeus monodon hemocytes using constitutive expression vector for long-hairpin RNA (lhRNA) J. Invertebr. Pathol. 2020;175:107442. doi: 10.1016/j.jip.2020.107442. [DOI] [PubMed] [Google Scholar]
- 113.Khunthong S., Jaroenram W., Arunrut N., Suebsing R. Rapid and sensitive detection of shrimp yellow head virus by loop-mediated isothermal amplification combined with a lateral flow dipstick. J. Virol. Methods. 2013;188:51–56. doi: 10.1016/j.jviromet.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 114.Wijegoonawardane P.K.M., Cowley J.A., Walker P.J. Consensus RT-nested PCR detection of yellow head complex genotypes in penaeid shrimp. J. Virol. Methods. 2008;153:168–175. doi: 10.1016/j.jviromet.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 115.Sanitt P., Attasart P., Panyim S. Protection of yellow head virus infection in shrimp by feeding of bacteria expressing dsRNAs. J. Biotechnol. 2014;179:26–31. doi: 10.1016/j.jbiotec.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 116.Tang K.F., Aranguren L.F., Piamsomboon P., Han J.E., Maskaykina I.Y., Schmidt M.M. Detection of the microsporidian Enterocytozoon hepatopenaei (EHP) and Taura syndrome virus in Penaeus vannamei cultured in Venezuela. Aquaculture. 2017;480:17–21. doi: 10.1016/j.aquaculture.2017.07.043. [DOI] [Google Scholar]
- 117.Cruz-Flores R., Mai H.N., Dhar A.K. Complete genome reconstruction and genetic analysis of Taura syndrome virus of shrimp from archival Davidson’s-fixed paraffin embedded tissue. Virology. 2021;553:117–121. doi: 10.1016/j.virol.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 118.Ochoa L.M., Cruz-Flores R., Dhar A.K. Detection and Phylogenetic Analyses of Taura Syndrome Virus from Archived Davidson’s-Fixed Paraffin-Embedded Shrimp Tissue. Viruses. 2020;12:1030. doi: 10.3390/v12091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kiatpathomchai W., Jareonram W., Jitrapakdee S., Flegel T.W. Rapid and sensitive detection of Taura syndrome virus by reverse transcription loop-mediated isothermal amplification. J. Virol. Methods. 2007;146:125–128. doi: 10.1016/j.jviromet.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 120.Boube I., Lotz J.M., Pozhitkov A.E., Li S., Griffitt R.J. Identification of genes involved in Taura Syndrome Virus resistance in Litopenaeus vannamei. J. Aquat. Anim. Health. 2014;26:137–143. doi: 10.1080/08997659.2013.860058. [DOI] [PubMed] [Google Scholar]
- 121.Cheng L.T., Lin W.H., Wang P.C., Tsai M.A., Ho P.Y., Hsu J.P., Chern R.S., Chen S.C. Epidemiology and phylogenetic analysis of Taura syndrome virus in cultured Pacific white shrimp Litopenaeus vannamei B. in Taiwan. Dis. Aquat. Org. 2011;97:17–23. doi: 10.3354/dao02407. [DOI] [PubMed] [Google Scholar]
- 122.Tang K.F., Navarro S.A., Pantoja C.R., Aranguren F.L., Lightner D.V. New genotypes of white spot syndrome virus (WSSV) and Taura syndrome virus (TSV) from the Kingdom of Saudi Arabia. Dis. Aquat. Org. 2012;99:179–185. doi: 10.3354/dao02470. [DOI] [PubMed] [Google Scholar]
- 123.Moss D.R., Moss S.M., Lotz J.M. Estimation of genetic parameters for survival to multiple isolates of Taura syndrome virus in a selected population of Pacific white shrimp Penaeus (Litopenaeus) vannamei. Aquaculture. 2013;416:78–84. doi: 10.1016/j.aquaculture.2013.07.037. [DOI] [Google Scholar]
- 124.Nielsen L., Sang-Oum W., Cheevadhanarak S., Flegel T.W. Taura syndrome virus (TSV) in Thailand and its relationship to TSV in China and the Americas. Dis. Aquat. Org. 2005;63:101–106. doi: 10.3354/dao063101. [DOI] [PubMed] [Google Scholar]
- 125.Phalitakul S., Wongtawatchai J., Sarikaputi M., Viseshakul N. The molecular detection of Taura syndrome virus emerging with White spot syndrome virus in penaeid shrimps of Thailand. Aquaculture. 2006;260:77–85. doi: 10.1016/j.aquaculture.2006.05.040. [DOI] [Google Scholar]
- 126.Erickson H.S., Poulos B.T., Tang K.F.J., Bradley-dunlop D., Lightner D.V. Taura syndrome virus from Belize represents a unique variant. Dis. Aquat. Org. 2005;64:91–98. doi: 10.3354/dao064091. [DOI] [PubMed] [Google Scholar]
- 127.Wertheim J.O., Tang K.F., Navarro S.A., Lightner D.V. A quick fuse and the emergence of Taura syndrome virus. Virology. 2009;390:324–329. doi: 10.1016/j.virol.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 128.George S.K., Kaizer K.N., Betz Y.M., Dhar A.K. Multiplication of Taura syndrome virus in primary hemocyte culture of shrimp (Penaeus vannamei) J. Virol. Methods. 2011;172:54–59. doi: 10.1016/j.jviromet.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 129.Fadilah A.N., Fasya A.H. Examination of Taura Syndrome Virus (TSV) in white shrimp (Litopenaeus vannamei) and tiger prawn (Penaeus monodon) with Polymerase Chain Reaction (PCR) method. IOP Conf. Ser. Earth Environ. Sci. 2021;679:0212069. doi: 10.1088/1755-1315/679/1/012069. [DOI] [Google Scholar]
- 130.Dhar A.K., Allnutt F.T. Encyclopedia of Virology. Academic Press; Cambridge, MA, USA: 2008. Taura Syndrome Virus; pp. 1–8. [DOI] [Google Scholar]
- 131.Do J.W., Cha S.J., Lee N.S., Kim Y.C., Kim J.D., Park J.W. Taura syndrome virus from Penaeus vannamei shrimp cultured in Korea. Dis. Aquat. Org. 2006;70:171–174. doi: 10.3354/dao070171. [DOI] [PubMed] [Google Scholar]
- 132.Dhar A.K., Lakshman D.K., Amundsen K., Robles-Sikisaka R., Kaizer K.N., Roy S., Hasson K.W., Allnutt F.C.T. Characterization of a Taura syndrome virus isolate originating from the 2004 Texas epizootic in cultured shrimp. Arch. Virol. 2010;155:315–327. doi: 10.1007/s00705-009-0584-5. [DOI] [PubMed] [Google Scholar]
- 133.Tang K.F., Wang J., Lightner D.V. Quantitation of Taura syndrome virus by real-time RT-PCR with a TaqMan assay. J. Virol. Methods. 2004;115:109–114. doi: 10.1016/j.jviromet.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 134.Cao Z., Wang S.Y., Breeland V., Moore A.M., Lotz J.M. Taura syndrome virus loads in Litopenaeus vannamei hemolymph following infection are related to differential mortality. Dis. Aquat. Org. 2010;91:97–103. doi: 10.3354/dao02258. [DOI] [PubMed] [Google Scholar]
- 135.Tumburu L., Shepard E.F., Strand A.E., Browdy C.L. Effects of endosulfan exposure and Taura Syndrome Virus infection on the survival and molting of the marine penaeid shrimp, Litopenaeus vannamei. Chemosphere. 2012;86:912–918. doi: 10.1016/j.chemosphere.2011.10.057. [DOI] [PubMed] [Google Scholar]
- 136.Côté I., Navarro S., Tang K.F.J., Noble B., Lightner D.V. Taura syndrome virus from Venezuela is a new genetic variant. Aquaculture. 2008;284:62–67. doi: 10.1016/j.aquaculture.2008.07.059. [DOI] [Google Scholar]
- 137.Vergel J.C.V., Cabawatan L.D.P., Madrona V.A.C., Rosario A.F.T., Tare M.V.R., Maningas M.B.B. Detection of Taura Syndrome Virus (TSV) in Litopenaeus vannamei in the Philippines. Philipp. J. Fish. 2019;26:8–14. doi: 10.31398/tpjf/25.2.2018-0003. [DOI] [Google Scholar]
- 138.Aranguren L.F., Salazar M., Tang K., Caraballo X., Lightner D. Characterization of a new strain of Taura syndrome virus (TSV) from Colombian shrimp farms and the implication in the selection of TSV resistant lines. J. Invertebr. Pathol. 2013;112:68–73. doi: 10.1016/j.jip.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 139.Phromjai J., Mathuros T., Phokharatkul D., Prombun P., Suebsing R., Tuantranont A., Kiatpathomchai W. RT-LAMP detection of shrimp Taura syndrome virus (TSV) by combination with a nanogold-oligo probe. Aquac. Res. 2015;46:1902–1913. doi: 10.1111/are.12345. [DOI] [Google Scholar]
- 140.Hameed A.S., Bonami J.R. White tail disease of freshwater prawn, macrobrachium rosenbergii. Indian. J. Virol. 2012;23:134–140. doi: 10.1007/s13337-012-0087-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bonami J.R., Shi Z., Qian D., Widada J.S. White tail disease of the giant freshwater prawn, Macrobrachium rosenbergii: Separation of the associated virions and characterization of MrNV as a new type of nodavirus. J. Fish Dis. 2005;28:23–31. doi: 10.1111/j.1365-2761.2004.00595.x. [DOI] [PubMed] [Google Scholar]
- 142.Chen K.F., Tan W.S., Ong L.K., Abidin S.A.Z., Othman I., Tey B.T., Lee R.F.S. The Macrobrachium rosenbergii nodavirus: A detailed review of structure, infectivity, host immunity, diagnosis and prevention. Rev Aquac. 2021;13:2117–2141. doi: 10.1111/raq.12562. [DOI] [Google Scholar]
- 143.Hayakijkosol O., Burgess G., La Fauce K., Owens L. The complete sequence of the Australia recognizate of Macrobrachium rosenbergii nodavirus which causes white tail disease. Aquaculture. 2012;366:98–104. doi: 10.1016/j.aquaculture.2012.08.049. [DOI] [Google Scholar]
- 144.Ravi M., Nazeer Basha A., Sarathi M., Idalia H.R., Widada J.S., Bonami J.R., Hameed A.S. Studies on the occurrence of white tail disease (WTD) caused by MrNV and XSV in hatchery-reared post-larvae of Penaeus indicus and P. monodon. Aquaculture. 2009;292:117–120. doi: 10.1016/j.aquaculture.2009.03.051. [DOI] [Google Scholar]
- 145.Pillai D., Bonami J.R., Sri Widada J. Rapid detection of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV), the pathogenic agents of white tail disease of Macrobrachium rosenbergii (De Man), by loop-mediated isothermal amplification. J. Fish Dis. 2006;29:275–283. doi: 10.1111/j.1365-2761.2006.00718.x. [DOI] [PubMed] [Google Scholar]
- 146.Sudhakaran R., Haribabu P., Kumar S.R., Sarathi M., Ahmed V.I., Babu V.S., Hameed A.S. Natural aquatic insect carriers of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV) Dis. Aquat. Org. 2008;79:141–145. doi: 10.3354/dao01886. [DOI] [PubMed] [Google Scholar]
- 147.Murwantoko M., Bimantara A., Roosmanto R., Kawaichi M. Macrobrachium rosenbergii nodavirus infection in a giant freshwater prawn hatchery in Indonesia. Springerplus. 2016;5:2–9. doi: 10.1186/s40064-016-3127-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lin F., Liu L., Hao G.J., Sheng P.C., Cao Z., Zhou Y., Chen K. The development and application of a duplex reverse transcription loop-mediated isothermal amplification assay combined with a lateral flow dipstick method for Macrobrachium rosenbergii nodavirus and extra small virus isolated in China. Mol. Cell. Probes. 2018;40:1–7. doi: 10.1016/j.mcp.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 149.Kumar S.N., Rai P., Karunasagar I., Karunasagar I. Genomic and antibody-based assays for the detection of Indian strains of Macrobrachium rosenbergii nodavirus and extra small virus associated with white tail disease of Macrobrachium rosenbergii. VirusDisease. 2020;31:459–469. doi: 10.1007/s13337-020-00641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bonami J.R., Widada J.S. Viral diseases of the giant fresh water prawn Macrobrachium rosenbergii: A review. J. Invertebr. Pathol. 2011;106:131–142. doi: 10.1016/j.jip.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 151.Pillai D., Bonami J.R. A review on the diseases of freshwater prawns with special focus on white tail disease of Macrobrachium rosenbergii. Aquac. Res. 2012;43:1029–1037. doi: 10.1111/j.1365-2109.2011.03061.x. [DOI] [Google Scholar]
- 152.NaveenKumar S., Shekar M., Karunasagar I., Karunasagar I. Genetic analysis of RNA1 and RNA2 of Macrobrachium rosenbergii nodavirus (MrNV) isolated from India. Virus Res. 2013;173:377–385. doi: 10.1016/j.virusres.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 153.Pasookhush P., Hindmarch C., Sithigorngul P., Longyant S., Bendena W.G., Chaivisuthangkura P. Transcriptomic analysis of Macrobrachium rosenbergii (giant fresh water prawn) post-larvae in response to M. rosenbergii nodavirus (MrNV) infection: De novo assembly and functional annotation. BMC Genom. 2019;20:762. doi: 10.1186/s12864-019-6102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Widada J.S., Durand S., Cambournac I., Qian D., Shi Z., Dejonghe E., Bonami J.R. Genome-based detection methods of Macrobrachium rosenbergii nodavirus, a pathogen of the giant freshwater prawn, Macrobrachium rosenbergii: Dot-blot, in situ hybridization and RT-PCR. J. Fish Dis. 2003;26:583–590. doi: 10.1046/j.1365-2761.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 155.Kumar B.T.N., Murthy H.S., Patil P., Doddamani P.L., Patil R. Enhanced immune response and resistance to white tail disease in chitin-diet fed freshwater prawn, Macrobrachium rosenbergii. Aquac. Rep. 2015;2:34–38. doi: 10.1016/j.aqrep.2015.06.001. [DOI] [Google Scholar]
- 156.Saedi T.A., Moeini H., Tan W.S., Yusoff K., Daud H.M., Chu K.B., Bhassu S. Detection and phylogenetic profiling of nodavirus associated with white tail disease in Malaysian Macrobrachium rosenbergii de Man. Mol. Biol. Rep. 2012;39:5785–5790. doi: 10.1007/s11033-011-1389-7. [DOI] [PubMed] [Google Scholar]
- 157.Sudhakaran R., Ishaq Ahmed V.P., Haribabu P., Mukherjee S.C., Sri Widada J., Bonami J.R., Sahul Hameed A.S. Experimental vertical transmission of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV) from brooders to progeny in Macrobrachium rosenbergii and Artemia. J. Fish Dis. 2007;30:27–35. doi: 10.1111/j.1365-2761.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- 158.OIE Manual of Diagnostic Tests for Aquatic Animals. Chapter 2.2.6, White Tail Disease. 2018. [(accessed on 11 May 2017)]. Available online: https://www.oie.int/en/what-we-do/standards/codes-and-manuals/aquatic-manual-online-access/
- 159.Jariyapong P., Pudgerd A., Weerachatyanukul W., Hirono I., Senapin S., Dhar A.K., Chotwiwatthanakun C. Construction of an infectious Macrobrachium rosenbergii nodavirus from cDNA clones in Sf9 cells and improved recovery of viral RNA with AZT treatment. Aquaculture. 2018;483:111–119. doi: 10.1016/j.aquaculture.2017.10.008. [DOI] [Google Scholar]
- 160.Hayakijkosol O., La Fauce K., Owens L. Experimental infection of redclaw crayfish (Cherax quadricarinatus) with Macrobrachium rosenbergii nodavirus, the aetiological agent of white tail disease. Aquaculture. 2011;319:25–29. doi: 10.1016/j.aquaculture.2011.06.023. [DOI] [Google Scholar]
- 161.Palmer P.J., Rao M., Cowley J.A. Reduced transmission of IHHNV to Penaeus monodon from shrimp pond wastewater filtered through a polychaete-assisted sand filter (PASF) system. Aquaculture. 2021;535:736359. doi: 10.1016/j.aquaculture.2021.736359. [DOI] [Google Scholar]
- 162.Sun Z.F., Hu C.Q., Ren C.H., Shen Q. Sensitive and rapid detection of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in shrimps by loop-mediated isothermal amplification. J. Virol. Methods. 2006;131:41–46. doi: 10.1016/j.jviromet.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 163.Sudhakaran R., Syed Musthaq S., Haribabu P., Mukherjee S.C., Gopal C., Sahul Hameed A.S. Experimental transmission of Macrobrachium rosenbergii nodavirus (MrNV) and extra small virus (XSV) in three species of marine shrimp (Penaeus indicus, Penaeus japonicus and Penaeus monodon) Aquaculture. 2006;257:136–141. doi: 10.1016/j.aquaculture.2006.02.053. [DOI] [Google Scholar]
- 164.Chaivisuthangkura P., Phattanapaijitkul P., Thammapalerd N., Rukpratanporn S., Longyant S., Sithigorngul W., Sithigorngul P. Development of a polyclonal antibody specific to VP19 envelope protein of white spot syndrome virus (WSSV) using a recombinant protein preparation. J. Virol. Methods. 2006;133:180–184. doi: 10.1016/j.jviromet.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 165.de Jesús Durán-Avelar M., Pérez-Enríquez R., Zambrano-Zaragoza J.F., Montoya-Rodríguez L., Vázquez-Juárez R., Vibanco-Pérez N. Genotyping WSSV isolates from northwestern Mexican shrimp farms affected by white spot disease outbreaks in 2010-2012. Dis. Aquat. Org. 2015;114:11–20. doi: 10.3354/dao02844. [DOI] [PubMed] [Google Scholar]
- 166.Li Z., Li F., Han Y., Xu L., Yang F. VP24 is a chitin-binding protein involved in white spot syndrome virus infection. J. Virol. 2016;90:842–850. doi: 10.1128/JVI.02357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu Q.H., Ma F.F., Guan G.K., Wang X.F., Li C., Huang J. White spot syndrome virus VP51 interact with ribosomal protein L7 of Litopenaeus vannamei. Fish Shellfish Immunol. 2015;44:382–388. doi: 10.1016/j.fsi.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 168.Mendoza-cano F., Sánchez-paz A. Development and validation of a quantitative real-time polymerase chain assay for universal detection of the White Spot Syndrome Virus in marine crustaceans. Virol. J. 2013;10:1–11. doi: 10.1186/1743-422X-10-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Muller I.C., Andrade T.P.D., Tang-Nelson K.F.J., Marques M.R.F., Lightner D.V. Genotyping of White spot syndrome virus (WSSV) geographical isolates from Brazil and comparison to other isolates from the Americas. Dis. Aquat. Org. 2010;88:91–98. doi: 10.3354/dao02142. [DOI] [PubMed] [Google Scholar]
- 170.Sindhupriya M., Saravanan P., Otta S.wK., Amarnath C.B., Arulraj R., Bhuvaneswari T., Ponniah A.G. White spot syndrome virus (WSSV) genome stability maintained over six passages through three different penaeid shrimp species. Dis. Aquat. Org. 2014;111:23–29. doi: 10.3354/dao02786. [DOI] [PubMed] [Google Scholar]
- 171.Tang K.F., Groumellec L.M., Lightner D.V. Novel, closely related, white spot syndrome virus (WSSV) genotypes from Madagascar, Mozambique and the Kingdom of Saudi Arabia. Dis. Aquat. Org. 2013;106:1–6. doi: 10.3354/dao02645. [DOI] [PubMed] [Google Scholar]
- 172.Wan Q., Xu L., Yang F. VP26 of white spot syndrome virus functions as a linker protein between the envelope and nucleocapsid of virions by binding with VP51. J. Virol. 2008;82:12598–12601. doi: 10.1128/JVI.01732-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kim J.H., Kim H.K., Nguyen V.G., Park B.K., Choresca C.H., Shin S.P., Park S.C. Genomic sequence of infectious hypodermal and hematopoietic necrosis virus (IHHNV) KLV-2010-01 originating from the first Korean outbreak in cultured Litopenaeus vannamei. Arch. Virol. 2012;157:369–373. doi: 10.1007/s00705-011-1155-0. [DOI] [PubMed] [Google Scholar]
- 174.Rai P., Safeena M.P., Karunasagar I., Karunasagar I. Complete nucleic acid sequence of Penaeus stylirostris densovirus (PstDNV) from India. Virus Res. 2011;158:37–45. doi: 10.1016/j.virusres.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 175.Sittidilokratna N., Chotwiwatthanakun C., Wijegoonawardane P.K., Unajak S., Boonnad A., Wangnai W., Walker P.J. A virulent isolate of yellow head nidovirus contains a deformed envelope glycoprotein gp116. Virology. 2009;384:192–200. doi: 10.1016/j.virol.2008.10.042. [DOI] [PubMed] [Google Scholar]
- 176.Sriphaijit T., Flegel T.W., Senapin S. Characterization of a shrimp serine protease homolog, a binding protein of yellow head virus. Dev. Comp. Immunol. 2007;31:1145–1158. doi: 10.1016/j.dci.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 177.Cowley J.A., Walker P.J. The complete genome sequence of gill-associated virus of Penaeus monodon prawns indicates a gene organisation unique among nidoviruses. Arch. Virol. 2002;147:1977–1987. doi: 10.1007/s00705-002-0847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Aranguren L.F., Tang K.F.J., Lightner D.V. Protection from yellow head virus (YHV) infection in Penaeus vannamei pre-infected with Taura syndrome virus (TSV) Dis. Aquat. Org. 2012;98:185–192. doi: 10.3354/dao02448. [DOI] [PubMed] [Google Scholar]
- 179.Ramos-Paredes J., Grijalva-Chon J.M., Rosa-Vélez J.D.L., Enríquez-Paredes L.M. New genetic recombination in hypervariable regions of the white spot syndrome virus isolated from Litopenaeus vannamei (Boone) in northwest Mexico. Aquac. Res. 2012;43:339–348. doi: 10.1111/j.1365-2109.2011.02836.x. [DOI] [Google Scholar]
- 180.Parrilla-Taylor D.P., Vibanco-Porez N., Durán-Avelar M.D.J., Gomez-Gil B., Llera-Herrera R., Vázquez-Juárez R. Molecular variability and genetic structure of white spot syndrome virus strains from northwest Mexico based on the analysis of genomes. FEMS Microbiol. Lett. 2018;365:fny216. doi: 10.1093/femsle/fny216. [DOI] [PubMed] [Google Scholar]
- 181.Saravanan K., Kumar P.P., Praveenraj J., Baruah A., Sivaramakrishnan T., Kumar T.S., Roy S.D. Investigation and confirmation of white spot syndrome virus (WSSV) infection in wild caught penaeid shrimps of Andaman and Nicobar Islands, India. VirusDisease. 2017;28:368–372. doi: 10.1007/s13337-017-0406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pradeep B., Shekar M., Karunasagar I., Karunasagar I. Characterization of variable genomic regions of Indian white spot syndrome virus. Virology. 2008;376:24–30. doi: 10.1016/j.virol.2008.02.037. [DOI] [PubMed] [Google Scholar]
- 183.Zeng Y. Molecular epidemiology of white spot syndrome virus in the world. Aquaculture. 2021;537:736509. doi: 10.1016/j.aquaculture.2021.736509. [DOI] [Google Scholar]
- 184.Marks H., van Duijse J.J.A., Zuidema D., van Hulten M.C.W., Vlak J.M. Fitness and virulence of an ancestral White Spot Syndrome Virus isolate from shrimp. Virus Res. 2005;110:9–20. doi: 10.1016/j.virusres.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 185.Tang K.F., Poulos B.T., Wang J., Redman R.M., Shih H.H., Lightner D.V. Geographic variations among infectious hypodermal and hematopoietic necrosis virus (IHHNV) isolates and characteristics of their infection. Dis. Aquat. Org. 2003;53:91–99. doi: 10.3354/dao053091. [DOI] [PubMed] [Google Scholar]
- 186.Hsia H.L., Chen L.L., Peng S.E., Yu H.T., Lo C.F., Kou G.H. Comparison of genomic sequence of infectious hypodermal and hematopoietic necrosis virus (IHHNV) between taiwan and other geographical isolates. Fish Pathol. 2003;38:177–179. doi: 10.3147/jsfp.38.177. [DOI] [Google Scholar]
- 187.Park S.C., Choi S.K., Han S.H., Park S., Jeon H.J., Lee S.C., Han J.E. Detection of infectious hypodermal and hematopoietic necrosis virus and white spot syndrome virus in whiteleg shrimp (Penaeus vannamei) imported from Vietnam to South Korea. J. Vet. Sci. 2020;21:e31. doi: 10.4142/jvs.2020.21.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Saksmerprome V., Puiprom O., Noonin C., Flegel T.W. Detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in farmed Australian Penaeus monodon by PCR analysis and DNA sequencing. Aquaculture. 2010;298:190–193. doi: 10.1016/j.aquaculture.2009.11.012. [DOI] [Google Scholar]
- 189.Wei Y.W., Fan D.D., Chen J. The mussel Mytilus edulis L. as an important reservoir of infectious hypodermal and hematopoietic necrosis virus (IHHNV) Aquac. Res. 2017;48:1346–1350. doi: 10.1111/are.12918. [DOI] [Google Scholar]
- 190.Hou L., Wu H., Xu L., Yang F. Expression and self-assembly of virus-like particles of infectious hypodermal and hematopoietic necrosis virus in Escherichia coli. Arch. Virol. 2009;154:547–553. doi: 10.1007/s00705-009-0336-6. [DOI] [PubMed] [Google Scholar]
- 191.Kim J.H., Choresca Jr C.H., Shin S.P., Han J.E., Jun J.W., Han S.Y., Park S.C. Detection of infectious hypodermal and hematopoietic necrosis virus (IHHNV) in Litopenaeus vannamei shrimp cultured in South Korea. Aquaculture. 2011;313:161–164. doi: 10.1016/j.aquaculture.2011.01.019. [DOI] [Google Scholar]
- 192.Dantas M.D.A., Chavante S.F., Teixeira D.I.A., Lima J.P.M., Lanza D.C. Analysis of new isolates reveals new genome organization and a hypervariable region in infectious myonecrosis virus (IMNV) Virus Res. 2015;203:66–71. doi: 10.1016/j.virusres.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 193.Gangnonngiw W., Anantasomboon G., Sang-oum W., Sriurairatana S., Sritunyalucksana K., Flegel T.W. Non-virulence of a recombinant shrimp nidovirus is associated with its non structural gene sequence and not a large structural gene deletion. Virology. 2009;385:161–168. doi: 10.1016/j.virol.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 194.Mari J., Poulos B.T., Lightner D.V., Bonami J. Shrimp Taura syndrome virus: Genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 2002;83:915–926. doi: 10.1099/0022-1317-83-4-915. [DOI] [PubMed] [Google Scholar]
- 195.Tang K.F., Lightner D.V. Phylogenetic analysis of Taura syndrome virus isolates collected between 1993 and 2004 and virulence comparison between two isolates representing different genetic variants. Virus Res. 2005;112:69–76. doi: 10.1016/j.virusres.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 196.Srisuvan T., Tang K.F., Lightner D.V. Experimental infection of Penaeus monodon with Taura syndrome virus (TSV) Dis. Aquat. Org. 2005;67:1–8. doi: 10.3354/dao067001. [DOI] [PubMed] [Google Scholar]
- 197.Ongvarrasopone C., Saejia P., Chanasakulniyom M., Panyim S. Inhibition of Taura syndrome virus replication in Litopenaeus vannamei through silencing the LvRab7 gene using double-stranded RNA. Arch. Virol. 2011;156:1117–1123. doi: 10.1007/s00705-011-0952-9. [DOI] [PubMed] [Google Scholar]
- 198.Widada J.S., Bonami J.R. Characteristics of the monocistronic genome of extra small virus, a virus-like particle associated with Macrobrachium rosenbergii nodavirus: Possible candidate for a new species of satellite virus. J. Gen. Virol. 2004;85:643–646. doi: 10.1099/vir.0.79777-0. [DOI] [PubMed] [Google Scholar]
- 199.Ueda R., Krabsetsve K., Owens L. Polymerase chain reaction detection of Taura Syndrome Virus and infectious hypodermal and haematopoietic necrosis virus in frozen commodity tails of Penaeus vannamei Boone. Aquac. Res. 2008;39:1606–1611. doi: 10.1111/j.1365-2109.2008.02033.x. [DOI] [Google Scholar]
- 200.Tripathy S., Sahoo P.K., Kumari J., Mishra B.K., Sarangi N., Ayyappan S. Multiplex RT-PCR detection and sequence comparison of viruses MrNV and XSV associated with white tail disease in Macrobrachium rosenbergii. Aquaculture. 2006;258:134–139. doi: 10.1016/j.aquaculture.2006.04.016. [DOI] [Google Scholar]
- 201.Senapin S., Jaengsanong C., Phiwsaiya K., Prasertsri S., Laisutisan K., Chuchird N., Flegel T.W. Infections of MrNV (Macrobrachium rosenbergii nodavirus) in cultivated whiteleg shrimp Penaeus vannamei in Asia. Aquaculture. 2012;338:41–46. doi: 10.1016/j.aquaculture.2012.01.019. [DOI] [Google Scholar]
- 202.Shekhar M.S., Azad I.S., Jithendran K.P. RT-PCR and sequence analysis of Macrobrachium rosenbergii nodavirus: Indian isolate. Aquaculture. 2006;252:128–132. doi: 10.1016/j.aquaculture.2005.07.026. [DOI] [Google Scholar]
- 203.Shekhar M.S., Sahoo P.K., Dillikumar M., Das A. Cloning, expression and sequence analysis of Macrobrachium rosenbergii nodavirus genes: Indian isolate. Aquac. Res. 2011;42:1778–1788. doi: 10.1111/j.1365-2109.2010.02775.x. [DOI] [Google Scholar]
- 204.Owens L., Fauce L.K., Juntunen K., Hayakijkosol O., Zeng C. Macrobrachium rosenbergii nodavirus disease (white tail disease) in Australia. Dis. Aquat. Org. 2009;85:175–180. doi: 10.3354/dao02086. [DOI] [PubMed] [Google Scholar]
- 205.Wang C.S., Chang J.S., Shih H.H., Chen S.N. RT-PCR amplification and sequence analysis of extra small virus associated with white tail disease of Macrobrachium rosenbergii (de Man) cultured in Taiwan. J. Fish Dis. 2007;30:127–132. doi: 10.1111/j.1365-2761.2007.00793.x. [DOI] [PubMed] [Google Scholar]
- 206.Wang J.M., Zhang H.J., Shi Z.L. Expression and assembly mechanism of the capsid proteins of a satellite virus (XSV) associated with Macrobrachium rosenbergii nodavirus. Virol. Sin. 2008;23:73–77. doi: 10.1007/s12250-008-2926-y. [DOI] [Google Scholar]
- 207.Bateman K.S., Tew I., French C., Hicks R.J., Martin P., Munro J., Setentiford G.D. Susceptibility to infection and pathogenicity of White Spot Disease (WSD) in non-model crustacean host taxa from temperate regions. J. Invertebr. Pathol. 2012;110:340–351. doi: 10.1016/j.jip.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 208.Hameed A.S., Charles M.X., Anilkumar M. Tolerance of Macrobrachium rosenbergii to white spot syndrome virus. Aquaculture. 2000;183:207–213. doi: 10.1016/S0044-8486(99)00305-1. [DOI] [Google Scholar]
- 209.Jin W., Lai Y., Zhu F. Effect of dietary fucoidan on innate immune response of Procambarus clarkii and disease resistance against white spot syndrome virus. Aquaculture. 2021;534:736233. doi: 10.1016/j.aquaculture.2020.736233. [DOI] [Google Scholar]
- 210.Kono T., Savan R., Sakai M., Itami T. Detection of white spot syndrome virus in shrimp by loop-mediated isothermal amplification. J. Virol. Methods. 2004;115:59–65. doi: 10.1016/j.jviromet.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 211.Yuan G., Zhu L., Jiang X., Zhang J., Pei C., Zhao X., Kong X. Diagnosis of co-infection with white spot syndrome virus and Aeromonas veronii in red swamp crayfish Procambarus clarkii. Aquaculture. 2021;532:736010. doi: 10.1016/j.aquaculture.2020.736010. [DOI] [Google Scholar]
- 212.Chang P.S., Chen L.J., Wang Y.C. The effect of ultraviolet irradiation, heat, pH, ozone, salinity and chemical disinfectants on the infectivity of white spot syndrome baculovirus. Aquaculture. 1998;166:1–17. doi: 10.1016/S0044-8486(97)00238-X. [DOI] [Google Scholar]
- 213.Musthaq S.S., Sudhakaran R., Ahmed V.I., Balasubramanian G., Hameed A.S. Variability in the tandem repetitive DNA sequences of white spot syndrome virus (WSSV) genome and suitability of VP28 gene to detect different isolates of WSSV from India. Aquaculture. 2006;256:34–41. doi: 10.1016/j.aquaculture.2006.01.036. [DOI] [Google Scholar]
- 214.Mathew S., Kumar K.A., Anandan R., Nair P.G.V., Devadasan K. Changes in tissue defence system in white spot syndrome virus (WSSV) infected Penaeus monodon. Comp. Biochem. Phys. C. 2007;145:315–320. doi: 10.1016/j.cbpc.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 215.Jatuyosporn T., Supungul P., Tassanakajon A., Krusong K. The essential role of clathrin-mediated endocytosis in yellow head virus propagation in the black tiger shrimp Penaeus monodon. Dev. Comp. Immunol. 2014;44:100–110. doi: 10.1016/j.dci.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 216.Senapin S., Phongdara A. Binding of shrimp cellular proteins to Taura syndrome viral capsid proteins VP1, VP2 and VP3. Virus Res. 2006;122:69–77. doi: 10.1016/j.virusres.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 217.Lee C., Kim J.H., Choi S.K., Jeon H.J., Lee S.H., Kim B.K., Kim Y.K., Lee K.J., Han J.E. Detection of infectious white spot syndrome virus in red claw crayfish (Cherax quadricarinatus) and red swamp crayfish (Procambarus clarkii) imported into Korea. Aquaculture. 2021;544:737117. doi: 10.1016/j.aquaculture.2021.737117. [DOI] [Google Scholar]
- 218.Ochoa-Meza A.R., Álvarez-Sánchez A.R., Romo-Quiñonez C.R., Barraza A., Magallón-Barajas F.J., Chávez-Sánchez A., Mejía-Ruiz C.H. Silver nanoparticles enhance survival of white spot syndrome virus infected Penaeus vannamei shrimps by activation of its immunological system. Fish Shellfish Immunol. 2019;84:1083–1089. doi: 10.1016/j.fsi.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 219.Cowley J.A., Rao M., Coman G.J. Real-time PCR tests to specifically detect Infectious hypodermal and haemopoietic necrosis virus (IHHNV) lineages and an IHHNV endogenous viral element (EVE) integrated in the genome of Black Tiger shrimp (Penaeus monodon) Dis. Aquat. Org. 2018;129:145–158. doi: 10.3354/dao03243. [DOI] [PubMed] [Google Scholar]
- 220.Chen B.K., Dong Z., Pang N.Y., Nian Y.Y., Yan D.C. A novel real-time PCR approach for detection of infectious hypodermal and haematopoietic necrosis virus (IHHNV) in the freshwater crayfish Procambarus clarkii. J. Invertebr. Pathol. 2018;157:100–103. doi: 10.1016/j.jip.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 221.Coelho-Melo M.V., Florindo-Guedes M.I., Rodriguez-Málaga S., de Almeida L.M., de Freitas Moreira M., de Oliveira T.R. Molecular characterization of Infectious Myonecrosis Virus (IMNV) isolated from the shrimp Litopenaeus vannamei farmed in Ceará state, Brazil. Lat. Am. J. Aquat. Res. 2014;42:649–652. doi: 10.3856/vol42-issue3-fulltext-22. [DOI] [Google Scholar]
- 222.Hamano K., Miyoshi T., Aue-umneoy D., Srisapoome P., Maeno Y., Tsutsui I. Waterborne and cannibalism-mediated transmission of the Yellow head virus in Penaeus monodon. Aquaculture. 2015;437:161–166. doi: 10.1016/j.aquaculture.2014.11.038. [DOI] [Google Scholar]
- 223.Noble T.H., Coman G.J., Cowley J.A., Wade N., Sellars M.J., Jerry D.R. Comparison of methods for uniformly challenging Black Tiger shrimp (Penaeus monodon) with gill-associated virus. Aquaculture. 2017;473:191–196. doi: 10.1016/j.aquaculture.2017.02.017. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.