Abstract
Secretion and quality control of large extracellular matrix proteins remain poorly understood and debated, particularly transport intermediates delivering folded proteins from the ER to Golgi and misfolded ones to lysosomes. Discrepancies between different studies are related to utilization of exogenous cargo, off-target effects of experimental conditions and cell manipulation, and identification of transport intermediates without tracing their origin and destination. To address these issues, here we imaged secretory and degradative trafficking of type I procollagen in live MC3T3 osteoblasts by replacing a region encoding N-propeptide in endogenous Col1a2 gDNA with GFP cDNA. We selected clones that produced the resulting fluorescent procollagen yet had normal expression of key osteoblast and ER/cell stress genes, normal procollagen folding, and normal deposition and mineralization of extracellular matrix. Live-cell imaging of these clones revealed ARF1-dependent transport intermediates, which had no COPII coat and delivered procollagen from ER exit sites (ERESs) to Golgi without stopping at ER–Golgi intermediate compartment (ERGIC). It also confirmed ERES microautophagy, i.e., lysosomes engulfing ERESs containing misfolded procollagen. Beyond validating these trafficking models for endogenous procollagen, we uncovered a probable cause of noncanonical cell stress response to procollagen misfolding. Recognized and retained only at ERESs, misfolded procollagen does not directly activate the canonical UPR, yet it disrupts the ER lumen by blocking normal secretory export from the ER.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00018-021-04017-z.
Keywords: Collagen, Trafficking, Autophagy, Quality control, Live-cell imaging
Introduction
Type I collagen is by far the most abundant and structurally important protein in all vertebrates, fibers of which provide an organic scaffold for bone, form matrix of skin and many other tissues, and even function on their own as tendons and ligaments [1]. Its procollagen precursor is a heterotrimer of two proα1(I) and one proα2(I) chains. It contains a ~ 300-nm-long (> 3000 amino acids) central triple helical domain, in which the three chains are wound together. The helix is flanked by large N- (> 300 AA) and C-terminal (> 700 AA) propeptides. Procollagen is folded within the endoplasmic reticulum (ER), transported through Golgi, and secreted [2–5]. Mature α1(I)2α2(I) triple helices of type I collagen are produced by cleavage of the propeptides, which occurs primarily after secretion, but might begin inside the cell after procollagen passes through Golgi [2]. Because of unique features of procollagen folding, ~ 10–15% of the molecules are misfolded and must be delivered from the ER to lysosomes for degradation even under normal conditions [6, 7]. Mutations causing additional misfolding or affecting trafficking of type I procollagen through the cell result in severe skeletal dysplasias and bone fragility [8–10]. Moreover, deficient folding, quality control, or trafficking of procollagen might contribute to malfunction of bone-producing cells (osteoblasts) in common age-related osteoporosis [11].
Procollagens have been used as prototypical large cargoes, yet neither their secretory nor degradative trafficking under physiological conditions are well understood. One question is how molecules much larger than 100 nm in size could be delivered from the ER to Golgi or to an intermediate ER–Golgi compartment (ERGIC), given that 60–90 nm COPII vesicles are widely considered to be early transport intermediates for such trafficking [12, 13]. Several models for early ER–Golgi transport intermediates were proposed: (a) enlarged (~ 1 µm or larger) vesicles with monoubiquitinated COPII coat [14–16]; (b) vesicles formed upon fusion of ERGIC membranes with ERES [17]; (c) ERGIC-like structures maturing into cis-Golgi without moving away from ERES or even forming direct ERES–ERGIC–Golgi connections [18]; and (d) ~ 0.5 µm vesicles distinguished by rapid (~ 1 µm/s), directional movement along microtubules [19]. The latter model was established in our previous study of osteoblasts transiently transfected with GFP-proα1(I) or GFP-proα2(I) chains. We observed that the moving vesicles had no COPII coat, formed at ERES after GDP to GTP exchange at ARF1 GTPase, contained ERGIC53, and delivered GFP-proα1(I) or GFP-proα2(I) chains from ERES to Golgi without stopping at a distinct ERGIC compartment. These observations suggested formation of COPI rather than COPII procollagen carriers at ERES, but it remained unclear whether the cells might utilize different carriers for endogenous procollagen molecules.
Another unanswered question is how misfolded procollagen is recognized and removed from the ER for degradation. Only unassociated proα chains with defective C-propeptides are degraded by proteasomes via the canonical ERAD pathway [20–23]. Misfolded procollagen heterotrimers with intact chains are degraded by lysosomes [24], but the quality control mechanism and autophagy pathway delivering misfolded procollagen to lysosomes remain unclear. One model proposed recognition of misfolded procollagen aggregates by calnexin complexes with an ER-phagy receptor FAM134B, resulting in isolation of the corresponding ER regions inside autophagosomes and subsequent autophagosome–lysosome fusion (ER-phagy pathway) [25–27]. However, our visualization of autophagic structures containing transiently transfected GFP-proα1(I) or GFP-proα2(I) chains suggests that osteoblasts recycle misfolded procollagen primarily from ERESs, which are engulfed by lysosomes without autophagosome formation (ERES microautophagy pathway) [28]. ERES microautophagy might dominate in cells that are not overwhelmed by misfolded procollagen accumulation in the ER [28], yet it could also be responsible specifically for degradation of transiently transfected GFP-proα1(I) or GFP-proα2(I) rather than endogenous procollagen [29].
A major obstacle to resolving these questions is visualization of bona fide transport and autophagy intermediates without disrupting cellular function. Traditional analysis of transport and autophagy intermediates in fixed or frozen cells suffers from assuming rather than identifying their origin and destination as well as from altering cellular function and ultrastructure when testing these assumptions. Tracing their origin, dynamics, and destination in live cells helps prevent misidentification of transport intermediates, but it requires fluorescent cargo [30]. Typically, such cargo is an exogenous protein, which might cause formation of nonphysiological intermediates. Compounding the uncertainty, most live-cell imaging studies provide little or no information on how the fluorescent cargo expression affects cellular differentiation and function.
Here we describe the first live-cell imaging study of transport and autophagy intermediates based on fluorescent labeling of endogenous type I procollagen and validating normal function and collagen matrix deposition by the resulting cells. We created Col1a2GFP mouse osteoblasts by replacing gDNA encoding most of the proα2(I) N-propeptide with GFP cDNA in one of the two alleles of Col1a2 in the MC3T3 mouse osteoblast cell line. We selected the clones with unaltered expression of key genes and extracellular matrix (ECM) deposition and mineralization. We measured procollagen synthesis, folding, secretion, and clearance from the cells, validating that the resulting GFP-proα2(I) chain was incorporated into procollagen and trafficked through the cell as efficiently as unmodified proα2(I). While the GFP tag caused excessive procollagen accumulation in the ER of some cells, imaging of other, healthy cells revealed transport and autophagy intermediates consistent with earlier studies of MC3T3 cells transiently transfected with GFP-proα2(I) or GFP-proα1(I). Besides clarifying intermediates delivering endogenous procollagen from the ER to Golgi or lysosomes in MC3T3 osteoblasts, the present study reveals how even subtle perturbation of procollagen export from the ER might affect cellular function.
Results
MC3T3 osteoblasts expressing GFP-tagged endogenous proα2(I) chain of type I collagen
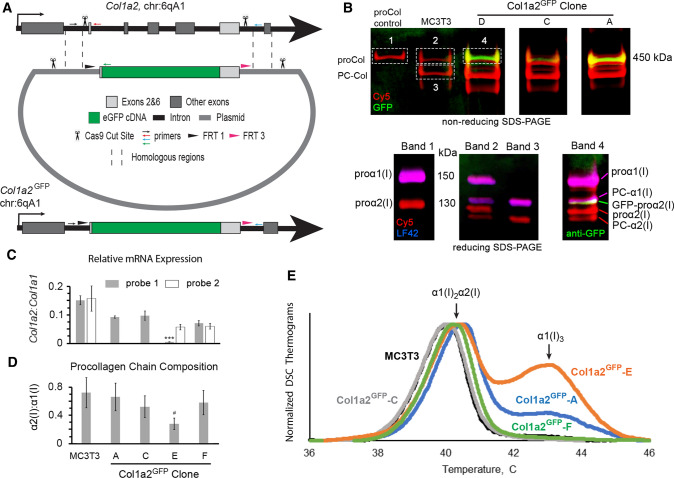
To examine intracellular trafficking of GFP-tagged endogenous procollagen molecules, we edited endogenous Col1a2 gDNA in MC3T3-E1 (subclone 4) cells, creating GFP-tagged procollagen molecules identical to those previously studied by transient transfection [19, 28]. Specifically, we replaced exons 2–6 and flanking regions of introns 1 and 6 with an insert, in which exon 2 of Col1a2 was fused to the 5’ end and exon 6 to the 3’ end of the eGFP cDNA as shown in Fig. 1a. CAS9 enzyme was targeted to genomic Col1a2 by guide RNAs (gRNAs) to intron 1 and intron 6 (Supp. Table S1). In addition to the fused coding sequence, the insert contained flanking regions of introns 1 and 6 with the endogenous splice sites. It also contained FRT1 and FRT3 sites for subsequent insert swapping with Flp recombinase, e.g., GFP replacement with different color fluorescent proteins or restoration of endogenous N-propeptide coding sequence. The corresponding template for homology directed repair contained 1 kb homologous arms upstream and downstream of the insert and CAS9 cleavage sites outside of the homologous arms (to facilitate template linearization and increase repair efficiency [31]).
Fig. 1.
Design of Col1a2GFP MC3T3 murine osteoblasts and procollagen synthesis by different clones. a CRISPR/CAS9 editing scheme at exons 1–7 (gray boxes). Homologous repair of CAS9 cleavage sites in introns 1 and 6 (scissors) based on a donor plasmid with two large homologous arms (dashed lines) replaces exons 2–6 with the following cDNA sequence (5’ → 3’): a Flp recombinase FRT1 site → endogenous intron 1 splice site with full-length exon 2 → eGFP without start and stop codons → full-length exon 6 with endogenous intron 6 splice site → a second, unique Flp recombinase site FRT3. Homologous repair was facilitated by introducing the same CRISPR/CAS9 cleavage sites into the donor plasmid (scissors). The editing was validated by PCR with a common forward primer (small black arrow) and reverse primers for uncut sequence (red arrow), eGFP insertion (green arrow), and additional insertions/deletions (blue arrow). b Top gel shows GFP fluorescence (green) in secreted procollagen molecules labeled with Cy5 (red), which migrate close to procollagen control (proCol) on nonreducing SDS-PAGE. Bottom panel is reducing SDS-PAGE of the main bands cut out from the top gel followed by Western blotting. It shows proα2(I) (Cy5, red) and GFP-proα2(I) chains (red Cy5 and green anti-GFP) integrated into procollagen heterotrimers with proα1(I) (red Cy5 and blue LF42 anti-proα1(I)). PC-Col, PC-α1(I), and PC-α2(I) denote procollagen molecules and chains after cleavage of N-propeptides, respectively. For the first gel, procollagen was denatured at 65 °C to preserve GFP fluorescence. c Relative Col1a2:Col1a1 mRNA ratio (2−ΔCT) in different Col1a2GFP clones measured by qPCR in N = 3 cultures of each clone assuming 100% PCR efficiency. For Col1a2, probe 1 binding at exon 1–2 boundary junction (gray, all cells) and probe 2 binding at exon 34–35 boundary junction (white, MC3T3 and clones E,F) were used. In clone E, probe 1 detected only the Col1a2GFP+ allele, expression of which was ~ 10 times lower than one Col1a2 allele in MC3T3 (***, p < 0.001). d Fluorescence intensity ratio of Cy5-labeled α2(I):α1(I) SDS-PAGE gel bands in pepsin-resistant triple helices purified from cell culture media (N = 6–12), indicating secretion of proα1(I)3 homotrimers by clone E. e Differential scanning calorimetry (DSC) thermograms of pepsin-purified triple helices secreted by wild-type MC3T3 cells and Col1a2GFP clones (0.125 °C/min heating rate in 0.2 M Na-phosphate, 0.5 M glycerol, pH 7.5). The main DSC peak at 40–41 °C represents denaturation of α1(I)2α2(I) triple helices. The second peak at ~ 43 °C is denaturation of α1(I)3 triple helices from proα1(I)3 homotrimers, synthesis of which is caused by deficient expression of Col1a2 [33, 61, 62]. In c, error bars show standard error of the mean (SEM). In d, error bars show estimated assay accuracy (approximately ± 30%), since it was larger than the observed standard deviation. The estimate is based on α2(I):α1(I) ratio variation in control experiments with different cells that produce only α1(I)2α2(I) heterotrimers; # indicates estimated significance. Variation analysis and significance (p value) evaluation are described in Statistical analysis section of Materials and Methods
Edited MC3T3 cells were isolated by single cell flow sorting based on GFP fluorescence and used for generation of Col1a2GFP clones expressing the desired GFP-proα2(I) chains. 16 heterozygous Col1a2GFP clones with correct GFP insertion in one Col1a2 allele (Col1a2GFP+) were generated. No homozygous clones were obtained, likely because of much higher probability of nonhomologous end joining (NHEJ) compared to homology directed repair after CAS9 cleavage. Homozygous cells could also have low viability given that even some heterozygous clones failed to thrive. Several Col1a2GFP clones had excision of exons 2–6 in the second Col1a2 allele (Col1a2GFP−, Supp. Fig. S1b). All but one of the latter clones were discarded. Six clones (A–F) were selected for detailed characterization (Table 1).
Table 1.
Basic properties of Col1a2GFP clones selected for this study
| Clone | Col1a2 genotype | Homotrimer secretion | Mineralization | Hspa5/Cryab/Ddit3 upregulationa | SP7/Bglap expression |
|---|---|---|---|---|---|
| MC3T3 | + / + | − | + | − | + |
| Col1a2GFP-A | GFP/ + | + | + | ± | + |
| Col1a2GFP-B | GFP/ + | + | − | ± | + |
| Col1a2GFP-C | GFP/ + | − | − | ± | + |
| Col1a2GFP-D | GFP/ + | − | − | NDb | NDb |
| Col1a2GFP-E | GFP/- | + | + | ± | + |
| Col1a2GFP-F | GFP/ + | − | + | − | + |
aThe ± sign indicates a statistically significant yet relatively small (< two-fold) increase in just one of these three markers of cell stress in osteoblasts (Hspa5 in clone E, Cryab in clones B,C, and Ddit3 in clone A)
bND not determined. The cells had significantly altered morphology and were discarded after the mineralization experiment
We validated GFP-proα2(I) chain synthesis and incorporation into procollagen by SDS polyacrylamide gel electrophoresis (SDS-PAGE) as illustrated in Fig. 1b. These chains were detected within proα1(I)2proα2(I) heterotrimers secreted into cell culture media. Their GFP fluorescence was detectable on a nonreducing gel after denaturation of collagen triple helices at 65 °C, and their electrophoretic migration was consistent with the construct design. However, SDS-PAGE pointed to possible synthesis of homotrimeric proα1(I)3 molecules in addition to the normal proα1(I)2proα2(I) in some of the clones. One cause of the homotrimer synthesis could be altered expression of the Col1a2GFP+ allele due to potential effects of the genomic DNA editing on mRNA transcription and splicing. Another cause could be altered expression of the Col1a2GFP− allele due to indel mutations at NHEJ sites after CAS9 cuts.
Therefore, we examined relative expression of Col1a2 and Col1a1 mRNA transcripts by qPCR (Fig. 1c). We measured Col1a2/Col1a1 mRNA ratio with two different Col1a2 qPCR probes. Probe 1 binding at the exon 1–2 junction was designed to detect only Col1a2GFP+ in clone E (due to exon 2–6 excision in Col1a2GFP−) while detecting both alleles in all other clones. Probe 2 binding at exon 34–35 junction was designed to detect both alleles in all clones. It was used in unedited MC3T3 cells (hereafter referred to as MC3T3 cells), clone E, and clone F. Analysis of clone E with probe 1 revealed ~ 10 times lower transcription of the Col1a2GFP+ mRNA (p < 0.001) than unaltered Col1a2 in MC3T3 cells (½ of total Col1a2 mRNA), likely because of the significant change in gDNA of this allele between exons 2 and 6. Since homology directed repair should produce the same gDNA, we assumed the same transcription of the Col1a2GFP+ allele in all clones and estimated Col1a2GFP− mRNA as Col1a2GFP− = Col1a2—Col1a2GFP+ ≈ Col1a2 (where Col1a2 is total mRNA for both alleles). In clones C–F, Col1a2GFP− mRNA was close to 50% of total Col1a2 mRNA in MC3T3 cells, i.e., unaltered single allele expression. Apparently, exon 2–6 excision in clone E did not affect transcription of the shortened Col1a2GFP− allele (Col1a2ΔE2−6) or the resulting transcript stability. In clone A, Col1a2GFP− transcription was slightly reduced, but the change was not statistically significant. It is noted that NHEJ repair in Col1a2GFP− could introduce different indel mutations and have variable effects on the transcription in different clones.
We further examined the chain composition and stability of procollagen secreted by different clones into cell culture media by SDS-PAGE and differential scanning calorimetry (DSC) after purification of pepsin-resistant triple helical domains of these molecules. The α2(I):α1(I) chain ratio measured by SDS-PAGE was significantly reduced only in clone E (≥ two-fold, see Statistical analysis), suggesting synthesis of α1(I)3 homotrimers (Fig. 1d). Homotrimer secretion was confirmed by DSC (Fig. 1e). A DSC thermogram shows the heat associated with triple helix denaturation, the peak of the thermogram representing apparent denaturation temperature, and the area under the thermogram being proportional to collagen concentration. The thermograms of molecules produced by clones C and F were indistinguishable from unedited cells, indicating that partial replacement of the N-propeptide with GFP in proα2(I) had no effect on the thermal stability and therefore integrity of the triple helix [32]. The thermograms of molecules produced by clones A and E had a second peak at higher temperature consistent with denaturation of triple helices in α1(I)3 homotrimers [33]. The fraction of the homotrimers was larger in clone E, consistent with the significantly reduced SDS-PAGE α2(I):α1(I) ratio.
Taken together, gel electrophoresis, qPCR, and DSC experiments demonstrate successful generation of heterozygous Col1a2GFP cells producing fluorescent GFP-proα2(I) chains. Expression of these chains by the Col1a2GFP+ allele is reduced compared to expression of proα2(I) by unaltered Col1a2, but they seem to be efficiently incorporated into procollagen heterotrimers without affecting triple helix integrity and subsequently secreted into cell culture media (Supp. Fig. S2). Synthesis of proα1(I)3 homotrimers by some of the clones is caused by disruptions in the Col1a2GFP− allele rather than reduced expression of GFP-proα2(I).
The clones were confirmed to be derived from MC3T3-E1 (subclone 4) cells by short tandem repeat (STR) profiling at 18 loci [34, 35]. The STR profiling performed by ATCC (US patent #9,556,482) showed 100% identity of clones A and E to MC3T3-E1 (subclone 4). Clone F had an extra satellite signal at locus 3–2 STR allele (Supp. Fig. S3), resulting in 97% identity.
Cellular and extracellular localization of GFP-proα2(I)
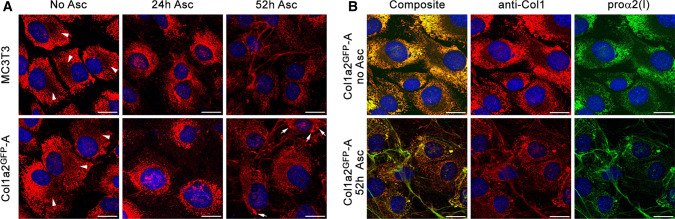
Next, we compared localization of GFP-proα2(I) chains, GFP-procollagen heterotrimers containing these chains, and unedited procollagen inside and outside the cells by imaging fixed-cell cultures for GFP and type I collagen triple helix (Fig. 2). We observed similar patterns of the triple helix antibody (anti-Col1) fluorescence in Col1a2GFP and MC3T3 cells (Fig. 2a). Without ascorbic acid 2-phosphate (Asc2P), GFP and anti-Col1 fluorescence were fully colocalized inside the ER and absent in Golgi or outside the cells (Fig. 2b, arrowheads). 24 h after Asc2P addition, more procollagen appeared to be localized in Golgi in Col1a2GFP and MC3T3 cells. 52 h after Asc2P addition, significant accumulation of extracellular collagen fibers was observed, and procollagen was relatively evenly distributed between the ER and Golgi inside the cells. GFP and anti-Col1 fluorescence signals were fully colocalized both inside and outside the cells, although fewer anti-Col1 antibodies were colocalized with GFP at extracellular collagen fibers. The latter effect was likely related to deficient cleavage of GFP-containing N-propeptides and less efficient binding of anti-Col1 to collagen fibers compared to individual procollagen molecules. The only notable difference between Col1a2GFP and MC3T3 cells was higher incidence of procollagen pools (Fig. 2a, arrows) that appeared to be consistent with dilated ER areas previously reported after transient transfection of MC3T3 cells with GFP-proα2(I) [19, 28].
Fig. 2.
Procollagen, collagen, and GFP-proα2(I) localization in Col1a2GFP cells and extracellular matrix. a Immunofluorescence of procollagen and collagen molecules in fixed MC3T3 and Col1a2GFP-A cells imaged with collagen triple helix antibody (anti-Col1) before and after treatment with ascorbic acid 2-phopsphate (Asc2P). Bright ER labeling combined with distinct perinuclear shadows (arrowheads) indicates procollagen retention in the ER and empty Golgi before Asc2P addition. Most procollagen is translocated from the ER to Golgi 24 h after Asc2P addition. Normal steady-state procollagen trafficking and collagen fiber formation is observed 52 h after initial addition of Asc2P. Arrows show procollagen pools likely within dilated ER regions (see Fig. 7). b Colocalization of anti-Col1 with GFP-proα2(I) inside Col1a2GFP-A cells and in extracellular fibers. Complete colocalization indicates efficient incorporation of GFP-proα2(I) chains followed by folding and secretion of the resulting procollagen molecules. Less efficient staining of extracellular fibers by anti-Col1 relative to GFP-proα2(I) is likely related to reduced binding of this antibody to collagen triple helices at fiber surfaces. Scale bars = 20 µm
Osteoblastic differentiation of Col1a2GFP clones
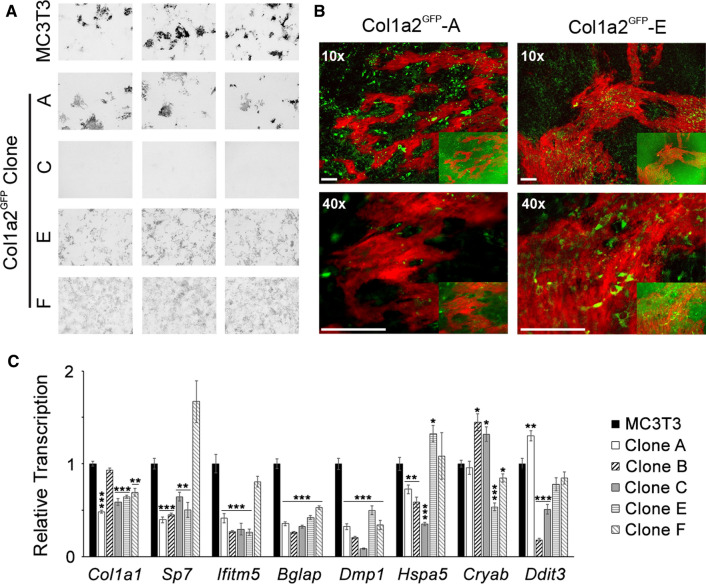
Having established synthesis, secretion, and extracellular fiber incorporation of GFP-procollagen, we examined osteoblastic differentiation of Col1a2GFP clones A–F after stimulation with 100 µM Asc2P and 2.5 mM β-glycerophosphate (βGP). After 8 weeks in culture, formation of mineralized ECM was observed in MC3T3 cell culture and Col1a2GFP-A, E, and F but not in clones B–D (Table 1, Fig. 3a). After removing fluorescent background associated with GFP-proα2(I) incorporation into ECM by a Gaussian blur filter, we visualized localization and relative GFP-proα2(I) expression by Col1a2GFP cells in partially mineralized areas of the ECM (Fig. 3b). There did not seem to be any correlation between GFP-proα2(I) expression and mineralization. All Col1a2GFP clones expressed key osteoblast genes (Sp7, Ifitm5, Bglap, and Dmp1; Fig. 3c). Clone D had altered cell morphology and was not examined further. Clones B and C had strongly reduced (> four-fold) transcription of Dmp1 (expressed by osteoblasts transitioning to osteocytes and osteocytes). Somewhat altered expression of cell stress genes (Hspa5, Cryab, or Ddit3) was observed in clones A–C and E but not clone F (Fig. 3c). Clones C and E had similar expression of Col1a1 and osteoblast maturation markers, Sp7, Ifitm5, and Bglap. Nonetheless, clone E cells with damaged Col1a2GFP− DNA produced mineralized matrix, yet clone C cells with seemingly normal Col1a2GFP− had strongly reduced Dmp1 expression and deposited no mineral.
Fig. 3.
Osteoblastic differentiation of Col1a2GFP cells. a Low-resolution (10 µm/pixel) images of extracellular matrix (ECM) in cell culture plates (8 week culture with 100 µM Asc2P and 2.5 mM βGP) after mineral labeling with Alizarin Complexone (N = 3 wells for each clone). Given identical appearance of no mineral (no Alizarin Complexone staining) in clones B–D, only clones A, C, E, and F are shown. Multiple mineralization nodules (dark areas) are observed in unedited MC3T3 cells and Col1a2GFP clones A, E, and F. b Higher-resolution images of mineralization nodules in clones A and E, showing localization of primarily intracellular GFP-proα2(I) (green) relative to mineralized ECM (red). Insets and main panels are the same images before and after removal of background ECM fluorescence by a Gaussian blur (σ = 70) filtering of the GFP channel. Scale bars = 100 µm. c Relative transcription of Col1a1, osteogenic (Sp7, Ifitm5, Bglap, and Dmp1) and cell stress (Hspa5, Cryab, and Ddit3) genes after 8 weeks with 100 µM Asc2P and 2.5 mM βGP in the ECM mineralization experiment (2−ΔΔCT with Hprt1 and B2m used as endogenous controls). Error bars show SEM (N = 3 wells); * p < 0.05, ** p < 0.01, *** p < 0.001
All these observations pointed to deficient mineralization in clones B–D being caused either by CAS9 effects outside of the targeted Col1a2 sequences or by effects of the single cell clone selection process (known to produce distinct subclones). Clones A, E, and F appeared to be sufficiently close to unedited MC3T3 cells for procollagen trafficking studies (formed mineralized matrix and had ≤ two-fold changes in expression of osteoblast and cell stress markers). Subtle difference in the appearance of mineralization pattern in clones E and F from MC3T3 and clone A at low resolution (Fig. 3a) did not correlate with microscopic appearance of mineralized structures (Fig. 3b) or expression of osteoblastic marker genes (Fig. 3c). Hence, it did not appear to result from altered osteoblast function but likely from a variation in the local cell density.
Trafficking of GFP-procollagen through the cell
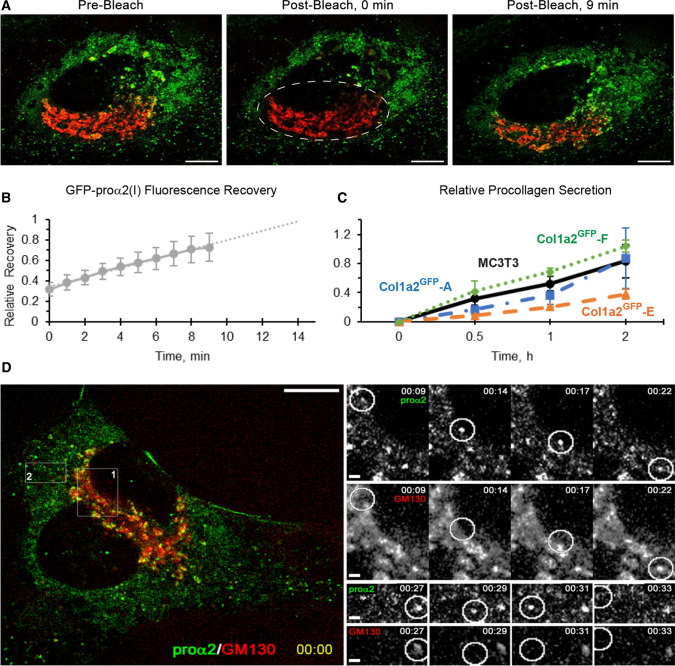
After selecting the Col1a2GFP clones, we examined GFP-procollagen trafficking through the cell (Fig. 4). The observed Golgi refill rate after photobleaching of GFP-procollagen in Col1a2GFP cells (Fig. 4a, b and Movie 1) was ~ 10–15 min. It was similar to the refill kinetics in MC3T3 cells transiently transfected with GFP-proα2(I) [19] and consistent with 20–30 min needed for folded procollagen to travel from the ER to extracellular space in mouse osteoblasts [36]. The overall kinetics of procollagen secretion into the cell culture media by Col1a2GFP-A and -F was similar to MC3T3 cells (Fig. 4c). Reduced procollagen secretion by Col1a2GFP-E cells was apparently caused by inefficient Col1a2ΔE2−6 translation and/or the truncated proα2(I) affecting procollagen folding. The size, directional movement, and dynamics of secretory procollagen transport intermediates as well as lysosomes in Col1a2GFP-A (Fig. 4d, Movie 2) and Col1a2GFP-F (Fig. 5, Movie 3) cells were consistent with the previous transient transfection study. Thus, the GFP tag did not appear to affect either secretory or degradative procollagen trafficking.
Fig. 4.
Procollagen trafficking through the cell. a and Movie 1 Golgi (red) refill by fluorescent GFP-proα2(I) (green) in a Col1a2GFP-A cell after photobleaching of GFP-proα2(I) in the Golgi area (dashed oval in the middle panel). Scale bars = 10 µm. b Kinetics of GFP-proα2(I) fluorescence recovery after photobleaching averaged over 5 different cells (1 = full recovery). c Relative kinetics of procollagen secretion per cell (1 = maximum average secretion). Each experiment was done in triplicate with one or two Col1a2GFP clones and MC3T3 control at the same time. Since all experiments produced consistent results, the values were averaged over all cultures: Col1a2GFP- A (N = 9), E (N = 6), F (N = 3), and MC3T3 (N = 12). d and Movie 2 Time-lapse images of rapidly moving transport vesicles delivering GFP-proα2(I) from the ER to Golgi (box 1) and from Golgi to the plasma membrane (box 2) of a Col1a2GFP-A cell. In a and d, cis-Golgi cisternae (red) were marked by transfecting the cells with cis-Golgi marker mCherry-GM130 in the presence of Asc2P. Scale bars = 10 µm (whole cell) and = 1 µm (zoomed regions). In b and c, error bars show SEM
Fig. 5.
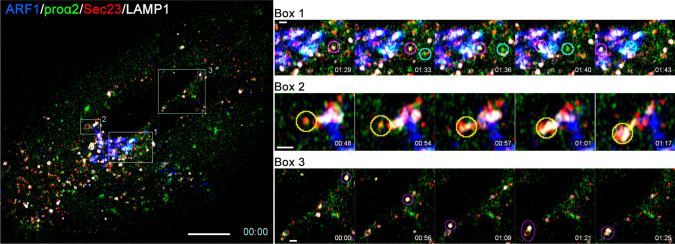
Secretory and degradative procollagen export from ERES. Left panel, first frame of 1 s/frame time-lapse image series (Movie 3) of a Col1a2GFP-F cell transfected with mTagBFP2-ARF1, mCherry-Sec23, and LAMP1-Halo. Right panels, selected time-lapse frames of areas marked by boxes 1–3 in the left panel. Box 1 frames show a transport vesicle delivering procollagen from ERES to Golgi (encircled in cyan) and a lysosome (encircled in magenta). Box 2 shows ERES being engulfed by a lysosome (encircled in yellow). Box 3 shows lysosome (encircled in magenta) movement and fusion with a quasi-stationary vesicular structure (likely a lysosome as well). Fast mode (1 s/frame), 4-color, super-resolution Airyscan imaging was necessary for visualizing and distinguishing rapidly moving procollagen ER–Golgi intermediates and lysosomes. Some crosstalk between fluorescence channels in this imaging mode was unavoidable, causing ambiguity in interpretation of weaker fluorescence signals (e.g., the vesicular structure in Box 3). Scale bars = 10 µm (whole cell) and = 1 µm (zoomed regions)
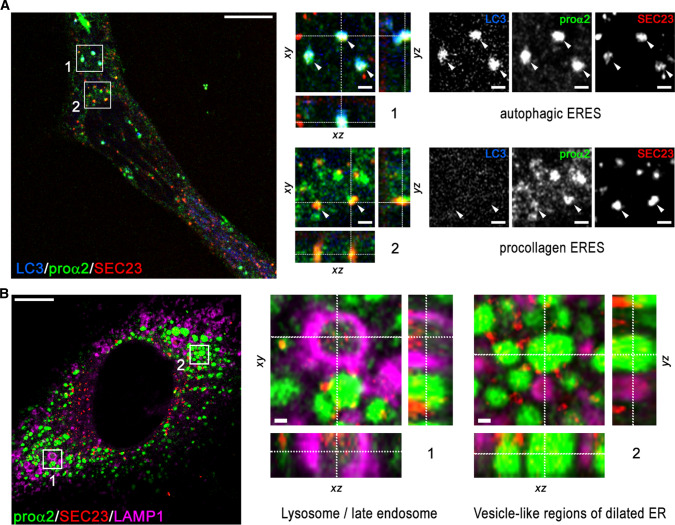
Like in transient transfection experiments [19, 28], ER exit sites (ERESs) appeared to be the sorting stations from which GFP-procollagen was routed to the secretory pathway or rerouted to autophagy and lysosomal degradation (Figs. 5, 6). More detailed super-resolution 3D imaging of Col1a2GFP cells exhibiting normal trafficking confirmed GFP-procollagen at multiple ERESs marked with mCherry-Sec23 (Fig. 6a). Some of the ERESs also contained mTagBFP2-LC3, which marks autophagic structures. After blocking the secretory pathway with brefeldin A and inhibiting lysosomal hydrolases with leupeptin, we observed GFP-procollagen colocalized with mTagBFP2-Sec23 inside lysosomal (late endosomal) membranes marked with LAMP1-Halo (Fig. 6b). Similar appearance of these structures to the ones observed after transient transfection with GFP-proα2(I) [28] suggests that the ERES microautophagy pathway of misfolded procollagen degradation delineated in the latter study is not specific to exogenous, overexpressed GFP-proα2(I). Figure 5 (Box 2) and Movie 3 (Box2) visualize the microautophagy-like engulfment of a procollagen-filled ERES by a lysosome or late endosome marked with LAMP1.
Fig. 6.
Procollagen sorting at ERES. a Super-resolution (Airyscan) images of procollagen ERESs marked by GFP-proα2(I) and COPII coat protein Sec23 (box 2, arrowheads) and autophagic ERES structures colocalized with autophagy marker LC3 (box 1, arrowheads) in a Col1a2GFP-A cell transfected with mCherry-Sec23 and mTagBFP2-LC3 in the presence of Asc2P. b Airyscan images of an autophagic procollagen ERES surrounded by lysosomal membrane marked with LAMP1 (box 1) and multiple vesicle-like procollagen pools in dilated ER studded with Sec23 (region 2) in a Col1a2GFP-A cell transfected with mTagBFP2-Sec23 and LAMP1-Halo. The cells were treated for 8 h with leupeptin and 3 h with brefeldin A before being fixed, to prevent procollagen export from the ER and rapid degradation of Sec23 in lysosomes [19]. It is noted that the Sec23 signal inside the lysosome is just as strong as at ERESs (outside the lysosome), while the procollagen signal is fainter due to reduced GFP fluorescence at acidic pH [63]. Scale bars in a and b = 10 µm (whole cell) and = 1 µm (zoomed regions)
Also similar to transient transfection experiments [19], the treatment with brefeldin A induced formation of multiple vesicle-like structures filled with GFP-procollagen and studded with mTagBFP2-Sec23 (Fig. 6b). Based on their appearance and dynamics being similar to transiently transfected cells, we identified these structures as dilated ER regions, confirming that inhibition of GDP → GTP exchange at ARF1 by brefeldin A blocks procollagen export from ERES into the secretory pathway [19]. While we cannot completely exclude the possibility of GFP cleaved from GFP-proα2(I) accumulating in the ER, such cleavage is unlikely because of very limited proteolytic activity within the ER lumen containing numerous unfolded polypeptides. Brefeldin A could cause an increase in this proteolytic activity. However, in cells transiently expressing fluorescent protein (FP)-tagged proα2(I), including the same GFP-proα2(I) as here, the fluorescence of the procollagen tag within the ER recovered after photobleaching much slower than the fluorescence of a free FP with C-terminal KDEL or FP-tagged HSP47 [19]. These experiments (performed with and without brefeldin A treatment) indicated that it was the FP-proα2(I) rather than a cleaved FP tag that was accumulating within dilated ER.
Overall, we observed no difference between clone A and F cells in procollagen localization within subcellular compartments or in structure and dynamics of procollagen carriers, as long as the cells had no large regions of dilated ER. In the cells that appeared normal, the GFP tag did not affect either secretory of degradative procollagen trafficking.
GFP-procollagen accumulation in dilated regions of ER lumen
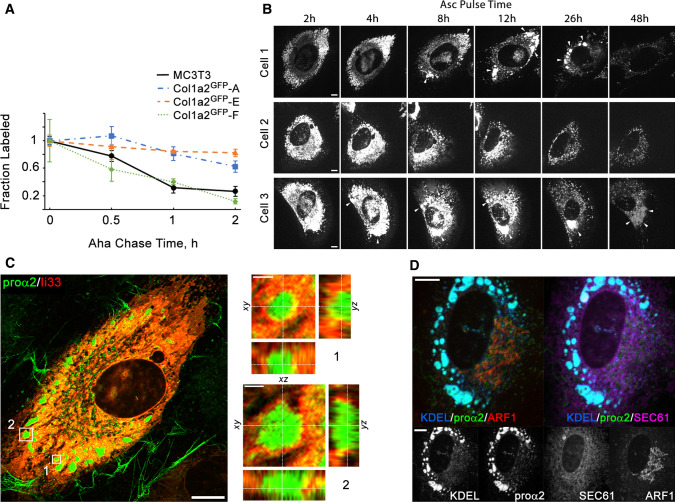
While many Col1a2GFP cells appeared to exhibit normal procollagen trafficking, slower clearance of molecules pulse labeled with azidohomoalanine (Aha) from the cells in all clones except F suggested procollagen retention in a fraction of the cells (Fig. 7a). Within 60 min after the Aha labeling pulse, 70 ± 9% of labeled procollagen was cleared from MC3T3 and Col1a2GFP-F cells, but only 30 ± 7.5% was cleared from Col1a2GFP clones A and E.
Fig. 7.
Procollagen trafficking and accumulation in the ER. a Kinetics of procollagen clearance from cells measured by pulse chase labeling with Aha that replaces Met in proteins synthesized during Aha pulse in Met-free media. Plotted is the fraction of Aha-labeled chains relative to the start of Aha chase in Aha-free, Met-containing media. The measurements of procollagen clearance kinetics were performed in the same experiments as measurements of secretion kinetics described in Fig. 4c. Error bars show SEM based on the same number of replicates as in Fig. 4c. b and Movie 4 Time-lapse images of procollagen clearance, formation of procollagen pools, and dynamics of these pools in the ER of Col1a2GFP-A cells after addition of Asc2P. Cells 1–3 illustrate different observed behaviors, including formation and slow clearance of large procollagen pools in the ER (cell 1), no formation of such pools (cell 2), and accumulation and enlargement of dilated ER containing procollagen (cell 3). c Airyscan imaging of ER membrane marker ii33 [64] surrounding large procollagen pools in a Col1a2GFP-A cell transfected with ii33-RFP. Zoomed orthogonal cross-sections of regions 1 and 2 show ER membrane localization in 3D. d Colocalization of procollagen pools with ER lumen in a Col1a2GFP-F cell co-transfected with ss-RFP-KDEL (ER lumen marker), Halo-Sec61 (ER membrane marker), and mTagBFP2-ARF1 (COPI vesicles, ERGIC, and Golgi marker). Scale bars in b–d = 10 µm (whole cells) and = 1 µm (zoomed regions in c)
Time-lapse imaging of Col1a2GFP cells after Asc2P addition (Fig. 7b) and colocalization with ER membrane (Fig. 7c,d) and lumen (Fig. 7d) by super-resolution (Fig. 7c) and regular confocal microscopy (Fig. 7d) revealed GFP-procollagen accumulation in vesicle-like dilated regions of the ER lumen in some Col1a2GFP cells from each of the clones. Dilation of the ER by accumulating GFP-procollagen was more pronounced in clones A (Fig. 7c) and E consistent with the pulse-chase labeling experiments. It still occurred in some Col1a2GFP-F cells (Fig. 7d), yet the fraction of such cells in this clone was smaller. Asc2P is essential for proline hydroxylation by collagen prolyl hydroxylases in the ER [37]. Without Asc2P, just as much (or even more) procollagen is present in the ER of MC3T3 cells, but procollagen translation, triple helix folding, and export from the ER are strongly inhibited. Prior to Asc2P addition, the fluorescence patterns of GFP-proα2(I) in Col1a2GFP cells were consistent with the reticular structure of the ER and similar to anti-Col1 (Fig. 2). Within several hours of Asc2P addition, GFP-procollagen was released from the ER to Golgi (Fig. 7b, Movie 4). At the same time, many (cells 1 and 3) but not all (cell 2) cells formed vesicle-like pools of GFP-proα2(I), which were similar to vesicle-like dilated ER regions observed after brefeldin A treatment and were ~ 1 µm in size or larger. Time-lapse imaging (Fig. 7b, Movie 4) showed slow dissipation of these pools in some cells (cell 1) as well as slow growth and fusion of these pools in other cells (cell 3). These pools were surrounded by the ER membrane (Fig. 7c, d), colocalized with an ER lumen marker (Fig. 7d), and colocalized with anti-Col1 antibody (Fig. 2b). Apparently, once procollagen synthesis was stimulated with Asc2P, a fraction of GFP-procollagen slowly accumulated in the ER lumen and caused ER dilation in some Col1a2GFP cells, similar to GFP-proα2(I) accumulation in MC3T3 cells more than 24 h after transient transfection (which is why the transient transfection studies were limited to ~ 18 h post transfection). Reduced synthesis (clone A) or truncation (clone E) of the unlabeled proα2(I) appeared to increase this accumulation by increasing the fraction and interaction between GFP-proα2(I) chains or by affecting folding of procollagen with truncated proα2(I).
Discussion
Imaging of proteins of interest (POIs) in live cells is revolutionizing cell biology [30, 38–41], yet it necessitates accounting for side effects of the required fluorescent protein (FP) tags and overexpression of exogenous FP-POI, which is particularly important for procollagen trafficking. Exogenous FP-proα chains might overwhelm the cell and alter the trafficking because procollagen naturally tends to misfold (causing degradation up to ~ 10–15% of all proα chains even under normal conditions [6, 7]). Procollagen assembly from 3 chains, slow C-to-N terminal folding, low stability, and high proα chain concentration inside the ER (particularly in osteoblasts) enables (Gly-X–Y)n stretches of any 3 chains to form a triple helix together, even when the chains are misaligned or belong to different molecules. Such nonspecific assembly might produce insoluble gelatin-like aggregates containing few to many molecules [21, 36], triggering excessive cell stress and potentially activating alternative trafficking pathways upon overexpression of exogenous FP-proα chains. The introduction of a swappable GFP tag into one of the two endogenous Col1a2 alleles in the present study is a first step toward resolving this problem, eliminating the uncontrolled overexpression and enabling future analysis of other side effects by swapping the tag.
Col1a2GFP clones of MC3T3 osteoblasts
Using CRISPR/CAS9 editing of Col1a2 in MC3T3 cells, we generated three Col1a2GFP clones (A, E, and F) that produced mineralized matrix and therefore retained osteoblastic nature. Other clones were discarded based on genotyping or cell appearance, were not viable, or did not produce mineralized matrix (likely because of off-target CAS9 cuts or mutations acquired during the clonal selection process). Each of the three osteoblastic clones had one Col1a2 allele with the desired GFP insertion (Col1a2GFP+), and the second allele without the insertion (Col1a2GFP−). In clone E, the Col1a2GFP− allele (Col1a2ΔE2−6) had a 4.6 kb excision of exons 2–6, yet the resulting mRNA was stable. The Col1a2GFP+ allele results from homologous recombination and therefore should have similar gDNA, transcription, and GFP-proα2(I) synthesis in all clones. However, Col1a2GFP+ expression might differ from Col1a2 in unaltered MC3T3 cells due to the gDNA changes (e.g., loss of intronic regulatory elements, altered binding of enhancer and promoter complexes, and altered transcript structure and splicing). The Col1a2GFP− allele is either uncut Col1a2 or a NHEJ product after CAS9 cuts. Even though the cuts occur within introns, the error-prone NHEJ might cause some variation in Col1a2GFP− expression. The loss of exons 2–6 might further affect the expression of this allele in clone E.
The measured transcription of Col1a2GFP+ was ~ 10 times lower than unaltered Col1a2 (Fig. 1c), consistent with SDS-PAGE and immunoprecipitation of secreted procollagen that indicated only ~ 10% of the molecules containing the GFP tag (Supp. Fig. S2). Col1a2GFP− transcription in clones C, E, and F was close to unaltered Co1a2 despite the exon 2–6 excision in clone E (Fig. 1c). It was lower (although not significantly) in clone A. The overall reduction in untagged and GFP-tagged proα2(I) synthesis caused formation of a small fraction of proα1(I)3 homotrimers in clone A but not in clones C and F (Fig. 1c–e). A much larger (~ 50%, Fig. 1e) fraction of the homotrimers in clone E indicates inefficient translation of Col1a2ΔE2−6 and/or inefficient integration of proα2(I)ΔE2−6 into procollagen. Although procollagen chain assembly begins at the C-terminal end, Col1a2ΔE2−6 mRNA could be directed to wrong ribosomes, affecting the translation and translocation of proα2(I)ΔE2−6 into the ER. It is noted that low GFP-proα2(I) synthesis is beneficial for reducing GFP side effects on procollagen trafficking and cellular function, despite the homotrimer formation in clones A and E.
Procollagen folding and trafficking
As expected, the GFP tag increased procollagen misfolding, resulting in a noticeable accumulation of the molecules in the ER lumen and their delayed clearance from the cells in clones A and E (Fig. 7a). Only in clone F, in which cells appeared to produce the lowest fraction of proα2(I) that was tagged with GFP (< 10%, Supp. Fig. S2), was the average clearance rate indistinguishable from unedited MC3T3. Still, dilated ER regions filled with GFP-procollagen were observed in some cells even in clone F within 24 h after stimulation of procollagen synthesis with Asc2P (Fig. 7d). Such ER dilation shortly after Asc2P addition appears to be caused by Asc2P-initiated triple helix folding in the ER lumen overfilled with unfolded proα chains (Fig. 7b–d), since Asc2P is required for Pro hydroxylation that precedes the folding [37]. More pronounced ER dilation in clones A and E vs. clone F indicates that it is enhanced either by the larger fraction of GFP-tagged chains or by proα1(I)3 homotrimer synthesis in the former two clones.
Most Col1a2GFP cells, even in clones A and E, clear the dilated ER regions within 12–24 h and appear to function normally afterward, with no differences in either secretory or degradative trafficking between clones A and F that we examined in greater detail. Nonetheless, malfunction of some of the cells (Fig. 7a, b) underscores the significance of even small assay-related disruptions for procollagen trafficking. For instance, cells appear to function normally only up to ~ 24 h after even low-to-moderate transfection with FP-proα, limiting transient transfection studies to a short window until FP-proα accumulation, ER dilation, and resulting cellular malfunction become prevalent [19, 28]. Since cells might adapt by altering procollagen biosynthesis and trafficking pathways, even more caution should be exercised in interpreting observations based on stable overexpression of GFP-tagged procollagen [18]. The same applies to utilization of exogenously and endogenously expressed procollagen with various tags that significantly elongate or induce nonphysiological interactions/function of procollagen chains [42–45]. Synchronized ER export assays based on deliberate procollagen retention in the ER [18, 46, 47] and assays based on inhibition of secretory or degradative trafficking [25] might also be expected to exacerbate ER disruption and cellular malfunction. It is important to keep in mind that unintentional enhancement of ER disruption in any of these assays might activate cell stress response pathways that would be mostly dormant otherwise. Therefore, it is important to validate any conclusions by using several different assays and approaches. With this in mind, we reexamined previously reported procollagen trafficking pathways by performing experiments in the newly created Col1a2GFP clones, using the cells that had no significant ER dilation more than 16 h after Asc2P addition.
These experiments revealed procollagen being delivered from ERES to Golgi in rapidly and directionally moving transport intermediates that did not appear to stop at any intermediate compartment (Figs. 4d, 5 and Movies 2, 3). This observation was consistent with findings in MC3T3 cells transiently transfected with GFP-proα2(I) or GFP-proα1(I) [19, 28] but different from the study of synchronized GFP-proα1(I) export from the ER in human fibroblasts after stable transfection [18]. The latter study proposed short-loop ER–Golgi trafficking because it did not detect such intermediates by 15–30 s/frame live-cell imaging. We cannot estimate the importance of such a short-loop pathway in MC3T3 cells. Even our ~ 1 s/frame imaging is not fast enough for capturing all or even most transport vesicles [19]. Nonetheless, similar observations with exogenous and endogenous GFP-procollagen support the importance of procollagen transport by rapidly moving vesicular intermediates that have no COPII coat and require GDP → GTP exchange at ARF1 for their formation [19]. They are also consistent with transport intermediates recently observed by FIB-SEM (focused ion beam scanning electron microscopy) in cells that do not produce significant amounts of type I collagen [48]. We still do not know the structure of these intermediates in osteoblasts, their cargo specificity, and the mechanism of their formation at ERES. These questions, however, will have to be answered by future studies.
We also found that a fraction of procollagen was rerouted from ERES to lysosomal degradation through direct ERES engulfment by lysosomes (Figs. 5, 6). These observations were consistent with the ERES microautophagy pathway observed in transiently transfected MC3T3 cells [28] and different from the FAM134-mediated ER-phagy pathway described in [25–27]. We do not know whether the latter discrepancy is due to an inherent difference between MC3T3 osteoblasts and cells used in the latter studies, effects of prolonged bafilomycin A treatment on cellular function in Refs. [25, 27], potential effects of FAM134 knockdown on overall ER function and secretory pathway (not evaluated in Refs. [25–27]), or some other factors. However, we can now exclude the hypothesis of procollagen ERES microautophagy being activated only for degradation of transiently transfected exogenous GFP-proα2(I) [29]. One source of nonphysiological degradation of the transfected chains could be their poor incorporation into procollagen molecules due to the general excess of proα2(I) in the ER lumen. In the present study, the proα2(I) chains are in deficit in all three clones, and the endogenous GFP-proa2(I) chains are incorporated into procollagen heterotrimers with the same efficiency as proα2(I). Another source of nonphysiological degradation of transiently transfected GFP-proα2(I) could be abnormal targeting of exogenous Col1a2GFP mRNA lacking physiological 3’ and 5’ UTRs to ER ribosomes [49]. In the present study, the UTRs of the endogenous Col1a2GFP mRNA are the same as in MC3T3 cells (see the editing scheme in Fig. 1). ERES microautophagy could still be specifically triggered by the GFP tag, yet GFP could not induce an entirely novel degradation pathway in cells that are not familiar with this molecule. The presence of the GFP tag in just a small fraction of procollagen (Supp. Fig. S2) taken together with the observation of ERES COPII coat in most lysosomes degrading procollagen (Fig. 6b) supports the physiological importance of ERES microautophagy.
Quality control and cell stress response
The spatial separation of the site where misfolded procollagen is recognized (directed to degradation) from the site where it accumulates (ERES vs. ER lumen) is unlike other protein folding quality control (QC), pointing to a probable noncanonical cell stress response. Misfolded procollagen that does not bind BIP [50] escapes luminal QC and enters ERESs for sorting and rerouting to degradation [19, 28], causing congestion at these ER exit gates and resulting in delayed export and luminal accumulation of all secretory proteins. The accumulation of proteins that do not sequester luminal QC chaperones might disrupt the ER without activating the canonical unfolded protein response (UPR) until the altered ER lumen environment causes general protein misfolding and secondary UPR. Secondary vs. primary UPR would explain ER disruption without UPR in osteoblasts from a mouse model of osteogenesis imperfecta (OI) [36], UPR in hypertrophic chondrocytes from the same mice [51], and ER disruption with and without UPR in cells from different OI patients [52]. Understanding this noncanonical ER disruption by procollagen molecules with misfolded triple helices that do not bind BIP [50] and the resulting cell stress response is likely to be important for developing better treatments not only for OI but also for common bone pathologies [36, 52].
Our Col1a2GFP cells enhance stress response studies by adding the option of visualizing accumulation, trafficking, and degradation of endogenous procollagen in live osteoblasts. The design of these cells with FRT sites for swapping the GFP insert for any other cDNA by Flp-recombinase enables one to perform many additional experiments, some of which are ongoing in our laboratory. To facilitate such experiments (including independent testing of our findings), all three osteoblastic Col1a2GFP clones (A, E, and F) are available for general nonprofit research use upon request. In addition to the flexibility of the insert swapping in the Col1a2GFP+ allele, different expression of the second, Col1a2GFP− allele among these clones and the expression of proα1(I)3 homotrimers in two of the three clones create further opportunities for various studies.
Materials and methods
Design of HDR/gRNA
Targeting sites for CRISPR/Cas9 were identified using the Broad Institute online web application (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design). The top three scoring gRNAs were used for intron 1 and intron 6. Target sites were located > 100 bp from exon/intron boundaries to avoid disturbing endogenous splice sites. Sequences encoding gRNAs (Supp. Table S1) were purchased as 5’ phosphorylated single stranded oligos (Sigma), annealed to form double-stranded inserts, and ligated into a CAS9 expressing plasmid (pSpCas9(BB)-2A-GFP, Addgene #48,138) downstream of a U6 promoter and upstream of the gRNA scaffolding sequence as described in Ref. [53]. Six plasmids were generated, each encoding one of the six gRNAs. All plasmids were sequenced using a U6 promoter primer (GCTTACCGTAACTTGAAAGT) to ensure correct insertion of the gRNA (ACGT, Inc.).
The homology directed repair (HDR) template was designed in silico (SnapGene) and made de novo by GenScript Biotech (Fig. 1a). To prevent cleavage of homologous arms within the HDR template by CAS9, the corresponding NGG sequences were mutated [54]. This construct was ligated into the EcoRV site of Puc57 per GenScript protocol.
Cell culture and clone authentication
The murine osteoblast cell line MC3T3-E1 subclone 4 was obtained from ATCC (ATCC CRL-2593). Cells were cultured at 37 °C, 5% CO2 in growth medium containing αMEM + Glutamax (32571-036; Gibco) supplemented with 1% Pen-Strep (Corning) and 10% FBS tested for supporting osteoblastic differentiation (Gemini, GemCell™, Lot #A83F821). Media were replaced every 48–72 h. Cells were maintained in a proliferative stage (< 80% confluency). Collagen synthesis was stimulated using 100 µM ascorbic acid 2-phosphate (Asc2P, Sigma-Aldrich), which was refreshed every 48 h. Final Col1a2GFP clones were authenticated by STR profiling using ATCC mouse cell line authentication service.
CRISPR/CAS9 editing
MC3T3 cells were transfected in a 96-well plate with a mix of 0.3 µl Fugene 6 (Promega), 50 ng of HDR, 25 ng intron 1 CRISPR/CAS9 plasmid, and 25 ng of intron 6 CRISPR/CAS9 plasmid in 160 µl of cell culture media without Asc2P. The media were replaced with the standard culture media 24–36 h after the transfection. The cells were dissociated with 0.05% Trypsin–EDTA (Gibco), pooled, and seeded in a T25 flask 96 h after transfection. Once 80% confluent, GFP-positive cells were sorted into one well of a 6-well plate using a custom-built flow sorter (NHLBI Flow Cytometry Core) to increase the efficiency of subsequent single cell sorting. Once the well reached 80% confluency, GFP-positive cells were sorted into a 96-well plate, one cell per well. Each colony was expanded and treated as an individual clone. Once cells reached 80% confluency, they were transferred into 12-well plates and subsequently T25 flasks. During the latter passage, ~ 10,000 cells were retained for genotyping. The genotyping was performed with a Phire Direct PCR kit (Thermo Fischer) following the manufacturer’s protocol with the final concentrations of 500 nM common forward primer, 250 nM reverse Col1a2 primer, and 250 nM reverse Col1a2GFP primer (Supp. Table S2). After 100-fold dilution of DNA with water, genotyping fragments were amplified using 30 cycles of 5 s 98 °C, 5 s 68 °C, and 1 min 72 °C, which were preceded by 5 min 98 °C DNA denaturation and followed by 1 min final extension at 72 °C. The fragments were separated by electrophoresis on precast 2% agarose ethidium bromide gels (e-Gel, Invitrogen).
Procollagen analysis
Confluent cells were pretreated with 100 µM Asc2P for at least 24 h. Secreted procollagen was collected by incubating cells for 8 h with FBS-free media (to reduce contamination with bovine collagen) containing fresh Asc2P. Multiple cycles of 8 h FBS-free media followed by overnight 10% FBS media were used to collect enough protein. Procollagen was precipitated with 176 mg/mL of (NH4)2SO4 and resuspended in a Cy5 labeling buffer (0.1 M Na2CO3, 0.5 M NaCl, pH 9.0). After removing (NH4)2SO4 with Amersham G50 buffer exchange columns (Cytvia), procollagen was conjugated with Cy5 NHS ester (Cytvia), denatured at 65 °C in a nonreducing LDS loading buffer (Invitrogen), and separated on a precast 3–8% Tris–Acetate SDS polyacrylamide mini-gel (Invitrogen) as previously described [55]. Procollagen and PC-collagen bands cut out from the gel were re-denatured in a reducing loading LDS buffer (50 mM DTT) at 95 °C, separated on a precast 10% Tris–Glycine mini-gel (Invitrogen), and transferred to nitrocellulose for Western blotting with LF-42 [1:1000 dilution, ENH017-FP, Kerafast, recognizes C-propeptide of proα1(I)] and anti-GFP (1:1000 dilution, GTX628528, GeneTex) antibodies [36, 56].
To evaluate relative transcription of Col1a1 and Col1a2, confluent cells were treated with 100 µM Asc2P for 7 days with 3 changes of Asc2P-supplemented media. RNA was isolated and analyzed by quantitative, real-time PCR (qPCR) as described in the qPCR section below. To estimate the chain composition of secreted molecules, procollagen precipitated with (NH4)2SO4 was resuspended in 0.5 M acetic acid, digested with 0.1 mg/ml pepsin (Millipore) overnight, precipitated with 0.7 M NaCl (final concentration), resuspended in the Cy5 labeling buffer, conjugated with Cy5, and separated by SDS-PAGE on precast 3–8% Tris–Acetate gels. The fluorescence intensity ratio of α2(I):α1(I) bands was measured by imaging the gels on FLA9500 fluorescence scanner (Cytvia).
Differential scanning calorimetry
NaCl precipitates of pepsin-treated procollagen described above were resuspended in 0.2 M sodium phosphate, 0.5 M glycerol, and pH 7.4 and dialyzed against the latter buffer. Differential scanning calorimetry was performed in a Nano III calorimeter (TA Instruments) from 20 to 60 ℃ at 0.125 ℃/min heating rate as previously described [57]. The resulting thermograms were corrected by 1.7 °C to represent physiological conditions [57].
qPCR
RNA was purified with a Direct-zol kit (Zymo Research) and reverse transcribed with a random hexamer primer mix using SuperScript III kit (ThermoFisher). Relative mRNA transcription was measured on a 7500 Fast Real Time PCR system (Applied Biosystems) with TaqMan gene expression assays (Applied Biosystems) for Sp7 (Mm00504574_m1), Bglap (Mm03413826_mH), Col1a1 (Mm00801666_g1), Col1a2 (Mm00483888_m1, probe 1 and Mm00483921_m1, probe 2), Cryab (Mm00515567_m1), Ddit3 (Mm01135937_g1), Hprt1 (Mm01545399_m1), B2m (Mm00437762_m1), and Gapdh (Mm99999915_g1). Hprt1 and B2m were used as endogenous controls for calculating the values of ΔΔCT.
Immunofluorescence
Cells were fixed for 15–20 min in methanol-free formaldehyde (Pierce), which was freshly diluted to 2% final concentration with PBS, pH 7.4. Fixed cells were washed three times with PBS, permeabilized and blocked with 0.3% Triton-X and 3% BSA in PBS for 1 h at room temperature, incubated overnight at 4 °C with primary antibodies diluted in the same buffer, washed twice with PBS, incubated with secondary antibodies in 3% BSA in PBS for 30–60 min at room temperature, and mounted with Prolong Diamond Antifade with DAPI (ThermoFisher). We used the following antibodies: Rabbit anti-Col1 (AB765P, 1:400; Millipore), Mouse anti-GFP (GTX628528, 1:400; GeneTex), Goat anti-Rabbit IgG (DyLight 550, SA5-10,033, 1:10,000; ThermoFisher), and Goat anti-Mouse IgG (AlexaFluor 647, A-21236, 1:10,000; ThermoFisher).
Differentiation and mineralization
Cells were seeded in 24-well plates (6 wells per clone) in growth medium. At confluency, the medium was supplemented with 100 µM Asc2P. Four days after initial stimulation with Asc2P, the medium was additionally supplemented with 2.5 mM β-glycerol phosphate (βGP). The Asc2P- and βGP-supplemented medium was changed every two days for 8 weeks. RNA was extracted from 3 wells per clone for qPCR analysis. The remaining 3 wells per clone were fixed with freshly prepared 2% formaldehyde in PBS, stained with Alizarin Complexone (Sigma), and imaged at low resolution on FLA9500 scanner (Cytvia) as previously described [36].
Pulse chase experiments
Confluent cell cultures were pretreated with 100 µM Asc2P for 24–48 h, incubated in Met, Cys, and Gln-free DMEM (21013-024, Gibco) supplemented with Glutamax (Gibco) for 30 min, pulse labeled with 0.5 mM azidohomoalanine (Aha) in the same medium for 2 h, and chased with 10 mM Met in the same medium. Collagen triple helices were separately isolated from the chase medium and cell layer after adding internal standard (long fragments of MMP-1 cleaved rat tail tendon collagen labeled with Alexa Fluor 488, 0.7 µg /mL final concentration) [58]. The fraction of triple helices labeled with Aha was determined by conjugation of Aha with Alexa Fluor 555 DIBO alkyne and conjugation of Lys/Hyl residues with Cy5 followed by SDS-PAGE analysis [58]. The total amount of secreted collagen was calculated based on the Cy5 labeling intensity ratios for the α1(I) chains in secreted collagen and internal standard [58]. Cell nuclei remaining after the lysis were labeled with Hoechst 33,342 stain (ThermoFisher) and counted to determine collagen secretion per cell.
Imaging and image processing
High-resolution fixed-cell, live-cell, and fluorescence recovery after photobleaching (FRAP) imaging was performed at NICHD Microscopy and Imaging Core (MIC) on LSM 780 (standard confocal resolution,) or LSM 880 with Airyscan (super-resolution) microscopes (Zeiss) with 63 × oil-immersion objectives as previously described [19]. Cell transfection with fluorescent (FP-conjugated) markers of ER lumen (ss-FP-KDEL), ER membrane (ii33-FP or FP-Sec61), ERES (FP-Sec23), COPI coat (FP-ARF1, mostly Golgi but also ERGIC and transport vesicles), cis-Golgi (FP-GM130), and lysosome/late-endosome membrane (LAMP1-FP) was performed as previously described [19, 28]. Live cells were imaged using a temperature-controlled incubation stage set to maintain 37 °C and 5% CO2. Time-lapse imaging of procollagen trafficking in live cells was performed on LSM 880 in fast linescan (Airyscan) imaging mode at ~ 1 s/frame resolution to capture rapidly moving (~ 1 µm/s) transport vesicles and lysosomes. Only cells with sufficiently high expression of fluorescent markers that could be unambiguously identified with high spatial resolution in the fast mode (which precludes switching of emission filters) were analyzed. Long-term time-lapse imaging after Asc2P addition was performed on a Nikon spinning disk confocal microscope with a 60 × oil immersion objective at NICHD MIC. Long time-lapse experiments were corrected for photobleaching when needed using a Bleach Correction plugin with histogram matching within the FIJI image processing package [59]. Selected areas of fixed, mineralized cell cultures stained with Alizarin Complexone were imaged on Evos FL Auto fluorescence microscope (ThermoFisher) with 10 × and 40 × air objectives. Background fluorescence of thick GFP-labeled ECM in the latter cultures was removed by a Gaussian blur filter (σ = 70). The original images are shown as insets in Fig. 3b.
Statistical analysis
All measurements were performed with ≥ 3 biological replicates (different cell cultures in biochemical experiments or different cells in imaging experiments). The N values in figure legends are biological replicate numbers. Standard errors of the mean (SEM) and statistical significance (p values) were calculated from analysis of variations in independent experiments [except for α2(I):α1(I) chain ratio measurements by SDS-PAGE]. Experiments were treated as independent for different biological samples. Different analyses of the same sample were also treated as independent whenever variations between these analyses were comparable or larger than between different samples. Within the same measurement, replicates from the same sample (e.g., common triplicate qPCR) were treated as technical. Their average value was used as a single input for SEM and p-value calculation. Statistical significance was determined from the two-tailed heteroscedastic Student’s t test.
Analysis of α2(I):α1(I) chain ratios measured by SDS-PAGE (Fig. 1d) was based on estimated assay accuracy of approximately ± 30% rather than measured reproducibility of independent experiments (< 5–20% standard deviation). The estimated accuracy was based on our experience of the SDS-PAGE α2(I):α1(I) ratio varying from ~ 1:3 to ~ 2:3 in different cultures of cells that produce only type I procollagen heterotrimers [1:2 chain ratio confirmed by more accurate DSC (Fig. 1e) or MMP1 assays [60]]. Based on this estimated accuracy, only more than two-fold variation in the SDS-PAGE α2(I):α1(I) ratio was considered significant (clone E vs MC3T3 cells). Consistently, DSC (Fig. 1e) confirmed purely heterotrimeric procollagen in MC3T3 cells [α2(I):α1(I) = 0.71 ± 0.04], clone C [α2(I):α1(I) = 0.51 ± 0.01], and clone F [α2(I):α1(I) = 0.57 ± 0.04]. It confirmed a significant α1(I)3 homotrimer fraction in clone E [α2(I):α1(I) = 0.27 ± 0.05] and revealed a small homotrimer fraction in clone A [α2(I):α1(I) = 0.65 ± 0.12].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Intramural Research Program of NICHD, NIH. The authors thank Juan Bonifacino (NICHD) and Jennifer Lippincott-Schwartz (Howard Hughes Medical Institute, Janelia Research Campus) for access to super-resolution microscopes in their laboratories. They also thank Vincent Schram for assistance in imaging at NICHD Microscopy and Imaging Core.
Author contribution
LG, SO, EM, and SL contributed to Study design; LG, SO, and EM performed experiments; LG, SO, EM, and SL performed data analysis; LG, SO, EM, and SL were involved in writing and revising the paper.
Funding
This work was funded by the Intramural Research Program of NICHD, NIH.
Data availability
All cell lines, plasmids, and data generated in this study will be deposited to public repositories or freely provided upon request after acceptance of the paper for publication.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Prockop DJ, Kivirikko KI. Collagens: molecular biology, diseases, and potentials for therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 2.Canty EG, Kadler KE. Procollagen trafficking, processing and fibrillogenesis. J Cell Sci. 2005;118:1341–1353. doi: 10.1242/jcs.01731. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa Y, Bachinger HP. A molecular ensemble in the rER for procollagen maturation. Biochim Biophys Acta. 2013;1833:2479–2491. doi: 10.1016/j.bbamcr.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 4.Koide T, Nagata K. Collagen biosynthesis. In: Brinckmann J, Notbohm H, Muller PK, editors. Collagen: primer in structure, processing and assembly. Topics in current chemistry. Heidelberg: Springer; 2005. pp. 85–114. [Google Scholar]
- 5.Makareeva E, Aviles NA, Leikin S. Chaperoning osteogenesis: new protein-folding disease paradigms. Trends Cell Biol. 2011;21:168–176. doi: 10.1016/j.tcb.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barile FA, Guzowski DE, Ripley C, Siddiqi ZA, Bienkowski RS. Ammonium chloride inhibits basal degradation of newly synthesized collagen in human fetal lung fibroblasts. Arch Biochem Biophys. 1990;276:125–131. doi: 10.1016/0003-9861(90)90018-t. [DOI] [PubMed] [Google Scholar]
- 7.Berg RA, Schwartz ML, Crystal RG. Regulation of the production of secretory proteins: intracellular degradation of newly synthesized "defective" collagen. Proc Natl Acad Sci USA. 1980;77:4746–4750. doi: 10.1073/pnas.77.8.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateman JF, Boot-Handford RP, Lamande SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat Rev Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 9.Forlino A, Marini JC. Osteogenesis imperfecta. Lancet. 2016;387:1657–1671. doi: 10.1016/S0140-6736(15)00728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omari S, Makareeva E, Leikin S. Procollagen trafficking and its implications in OI. In: Ruggiero F, editor. Collagen superfamily and collagenopathies. Heidelberg: Springer; 2021. [Google Scholar]
- 11.Yin X, Zhou C, Li J, Liu R, Shi B, Yuan Q, Zou S. Autophagy in bone homeostasis and the onset of osteoporosis. Bone Res. 2019;7:28. doi: 10.1038/s41413-019-0058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venditti R, Wilson C, De Matteis MA. Exiting the ER: what we know and what we don't. Trends Cell Biol. 2014;24:9–18. doi: 10.1016/j.tcb.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 13.Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat Cell Biol. 2011;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- 14.Gorur A, Yuan L, Kenny SJ, Baba S, Xu K, Schekman R. COPII-coated membranes function as transport carriers of intracellular procollagen I. J Cell Biol. 2017;216:1745–1759. doi: 10.1083/jcb.201702135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgartel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan L, Kenny SJ, Hemmati J, Xu K, Schekman R. TANGO1 and SEC12 are copackaged with procollagen I to facilitate the generation of large COPII carriers. Proc Natl Acad Sci USA. 2018;115:E12255–E12264. doi: 10.1073/pnas.1814810115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogueira C, Erlmann P, Villeneuve J, Santos AJ, Martinez-Alonso E, Martinez-Menarguez JA, Malhotra V. SLY1 and syntaxin 18 specify a distinct pathway for procollagen VII export from the endoplasmic reticulum. Elife. 2014;3:e02784. doi: 10.7554/eLife.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCaughey J, Stevenson NL, Cross S, Stephens DJ. ER-to-Golgi trafficking of procollagen in the absence of large carriers. J Cell Biol. 2019;218:929–948. doi: 10.1083/jcb.201806035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omari S, Makareeva E, Gorrell L, Jarnik M, Lippincott-Schwartz J, Leikin S. Mechanisms of procollagen and HSP47 sorting during ER-to-Golgi trafficking. Matrix Biol. 2020;93:79–94. doi: 10.1016/j.matbio.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitzgerald J, Lamande SR, Bateman JF. Proteasomal degradation of unassembled mutant type I collagen pro-alpha1(I) chains. J Biol Chem. 1999;274:27392–27398. doi: 10.1074/jbc.274.39.27392. [DOI] [PubMed] [Google Scholar]
- 21.Ishida Y, Yamamoto A, Kitamura A, Lamande SR, Yoshimori T, Bateman JF, Kubota H, Nagata K. Autophagic elimination of misfolded procollagen aggregates in the endoplasmic reticulum as a means of cell protection. Mol Biol Cell. 2009;20:2744–2754. doi: 10.1091/mbc.E08-11-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamande SR, Chessler SD, Golub SB, Byers PH, Chan D, Cole WG, Sillence DO, Bateman JF. Endoplasmic reticulum-mediated quality control of type I collagen production by cells from osteogenesis imperfecta patients with mutations in the pro alpha 1 (I) chain carboxyl-terminal propeptide which impair subunit assembly. J Biol Chem. 1995;270:8642–8649. doi: 10.1074/jbc.270.15.8642. [DOI] [PubMed] [Google Scholar]
- 23.Doan ND, Hosseini AS, Bikovtseva AA, Huang MS, DiChiara AS, Papa LJ, 3rd, Koller A, Shoulders MD. Elucidation of proteostasis defects caused by osteogenesis imperfecta mutations in the collagen-alpha2(I) C-propeptide domain. J Biol Chem. 2020;295:9959–9973. doi: 10.1074/jbc.RA120.014071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishida Y, Nagata K. Autophagy eliminates a specific species of misfolded procollagen and plays a protective role in cell survival against ER stress. Autophagy. 2009;5:1217–1219. doi: 10.4161/auto.5.8.10168. [DOI] [PubMed] [Google Scholar]
- 25.Forrester A, De Leonibus C, Grumati P, Fasana E, Piemontese M, Staiano L, et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 2019 doi: 10.15252/embj.201899847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fregno I, Fasana E, Solda T, Galli C, Molinari M. N-glycan processing selects ERAD-resistant misfolded proteins for ER-to-lysosome-associated degradation. EMBO J. 2021 doi: 10.15252/embj.2020107240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reggio A, Buonomo V, Berkane R, Bhaskara RM, Tellechea M, Peluso I, et al. Role of FAM134 paralogues in endoplasmic reticulum remodeling, ER-phagy, and Collagen quality control. EMBO Rep. 2021 doi: 10.15252/embr.202052289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Omari S, Makareeva E, Roberts-Pilgrim A, Mirigian L, Jarnik M, Ott C, Lippincott-Schwartz J, Leikin S. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc Natl Acad Sci USA. 2018;115:E10099–E10108. doi: 10.1073/pnas.1814552115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fregno I, Molinari M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit Rev Biochem Mol Biol. 2019;54:153–163. doi: 10.1080/10409238.2019.1610351. [DOI] [PubMed] [Google Scholar]
- 30.Lippincott-Schwartz J, Roberts TH, Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song F, Stieger K. Optimizing the DNA donor template for homology-directed repair of double-strand breaks. Mol Ther Nucl Acids. 2017;7:53–60. doi: 10.1016/j.omtn.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makareeva E, Mertz EL, Kuznetsova NV, Sutter MB, DeRidder AM, Cabral WA, Barnes AM, McBride DJ, Marini JC, Leikin S. Structural heterogeneity of type I collagen triple helix and its role in osteogenesis imperfecta. J Biol Chem. 2008;283:4787–4798. doi: 10.1074/jbc.M705773200. [DOI] [PubMed] [Google Scholar]
- 33.Daley E, Streeten EA, Sorkin JD, Kuznetsova N, Shapses SA, Carleton SM, Shuldiner AR, Marini JC, Phillips CL, Goldstein SA, Leikin S, McBride DJ., Jr Variable bone fragility associated with an Amish COL1A2 variant and a knock-in mouse model. J Bone Miner Res. 2010;25:247–261. doi: 10.1359/jbmr.090720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.American Type Culture Collection Standards Development Organization Workgroup ASN Cell line misidentification: the beginning of the end. Nat Rev Cancer. 2010;10:441–8. doi: 10.1038/nrc2852. [DOI] [PubMed] [Google Scholar]
- 35.Tanabe H, Takada Y, Minegishi D, Kurematsu M, Masui T, Mizusawa H. Cell line individualization by STR multiplex system in the cell bank found cross-contamination between ECV304 and EJ-1/T24. Tiss Cult Res Commun. 1999;18:329–338. doi: 10.11418/jtca1981.18.4_329. [DOI] [Google Scholar]
- 36.Mirigian LS, Makareeva E, Mertz EL, Omari S, Roberts-Pilgrim AM, Oestreich AK, Phillips CL, Leikin S. Osteoblast malfunction caused by cell stress response to procollagen misfolding in alpha2(I)-G610C mouse model of osteogenesis imperfecta. J Bone Miner Res. 2016;31:1608–1616. doi: 10.1002/jbmr.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterkofsky B. Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am J Clin Nutr. 1991;54:1135s–s1140. doi: 10.1093/ajcn/54.6.1135s. [DOI] [PubMed] [Google Scholar]
- 38.Stephens DJ, Allan VJ. Light microscopy techniques for live cell imaging. Science. 2003;300:82–86. doi: 10.1126/science.1082160. [DOI] [PubMed] [Google Scholar]
- 39.Lippincott-Schwartz J. Emerging in vivo analyses of cell function using fluorescence imaging (*) Annu Rev Biochem. 2011;80:327–332. doi: 10.1146/annurev-biochem-121010-125553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lemon WC, McDole K. Live-cell imaging in the era of too many microscopes. Curr Opin Cell Biol. 2020;66:34–42. doi: 10.1016/j.ceb.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Specht EA, Braselmann E, Palmer AE. A critical and comparative review of fluorescent tools for live-cell imaging. Annu Rev Physiol. 2017;79:93–117. doi: 10.1146/annurev-physiol-022516-034055. [DOI] [PubMed] [Google Scholar]
- 42.Calverley BC, Kadler KE, Pickard A. Dynamic high-sensitivity quantitation of procollagen-I by endogenous CRISPR-cas9 nanoluciferase tagging. Cells. 2020;9:2070. doi: 10.3390/cells9092070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickard A, Adamson A, Lu Y, Chang J, Garva R, Hodson N, Kadler KE. Collagen assembly and turnover imaged with a CRISPR-Cas9 engineered dendra2 tag. bioRxiv. 2018 doi: 10.1101/331496. [DOI] [Google Scholar]
- 44.Omachi K, Kamura M, Teramoto K, Kojima H, Yokota T, Kaseda S, Kuwazuru J, Fukuda R, Koyama K, Matsuyama S, Motomura K, Shuto T, Suico MA, Kai H. A split-luciferase-based trimer formation assay as a high-throughput screening platform for therapeutics in alport syndrome. Cell Chem Biol. 2018;25(634–43):e4. doi: 10.1016/j.chembiol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Wong MY, Doan ND, DiChiara AS, Papa LJ, 3rd, Cheah JH, Soule CK, Watson N, Hulleman JD, Shoulders MD. A high-throughput assay for collagen secretion suggests an unanticipated role for Hsp90 in collagen production. Biochemistry. 2018;57:2814–2827. doi: 10.1021/acs.biochem.8b00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonfanti L, Mironov AA, Jr, Martinez-Menarguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 47.Mironov AA, Mironov AA, Jr, Beznoussenko GV, Trucco A, Lupetti P, Smith JD, Geerts WJ, Koster AJ, Burger KN, Martone ME, Deerinck TJ, Ellisman MH, Luini A. ER-to-Golgi carriers arise through direct en bloc protrusion and multistage maturation of specialized ER exit domains. Dev Cell. 2003;5:583–594. doi: 10.1016/s1534-5807(03)00294-6. [DOI] [PubMed] [Google Scholar]
- 48.Weigel AV, Chang CL, Shtengel G, Xu CS, Hoffman DP, Freeman M, Iyer N, Aaron J, Khuon S, Bogovic J, Qiu W, Hess HF, Lippincott-Schwartz J. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell. 2021;184(2412–29):e16. doi: 10.1016/j.cell.2021.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Stefanovic B. LARP6 meets collagen mRNA: specific regulation of type I collagen expression. Int J Mol Sci. 2016;17:419. doi: 10.3390/ijms17030419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chessler SD, Byers PH. BiP binds type I procollagen pro alpha chains with mutations in the carboxyl-terminal propeptide synthesized by cells from patients with osteogenesis imperfecta. J Biol Chem. 1993;268:18226–18233. doi: 10.1016/S0021-9258(17)46834-7. [DOI] [PubMed] [Google Scholar]
- 51.Scheiber AL, Guess AJ, Kaito T, Abzug JM, Enomoto-Iwamoto M, Leikin S, Iwamoto M, Otsuru S. Endoplasmic reticulum stress is induced in growth plate hypertrophic chondrocytes in G610C mouse model of osteogenesis imperfecta. Biochem Biophys Res Commun. 2019;509:235–240. doi: 10.1016/j.bbrc.2018.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Besio R, Iula G, Garibaldi N, Cipolla L, Sabbioneda S, Biggiogera M, Marini JC, Rossi A, Forlino A. 4-PBA ameliorates cellular homeostasis in fibroblasts from osteogenesis imperfecta patients by enhancing autophagy and stimulating protein secretion. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1642–1652. doi: 10.1016/j.bbadis.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18:35. doi: 10.1186/s13059-017-1164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Makareeva E, Cabral WA, Marini JC, Leikin S. Molecular mechanism of alpha 1(I)-osteogenesis imperfecta/ehlers-danlos syndrome: unfolding of an N-anchor domain at the N-terminal end of the type I collagen triple helix. J Biol Chem. 2006;281:6463–6470. doi: 10.1074/jbc.M511830200. [DOI] [PubMed] [Google Scholar]
- 56.Makareeva E, Sun G, Mirigian LS, Mertz EL, Vera JC, Espinoza NA, Yang K, Chen D, Klein TE, Byers PH, Leikin S. Substitutions for arginine at position 780 in triple helical domain of the alpha1(I) chain alter folding of the type I procollagen molecule and cause osteogenesis imperfecta. PLoS One. 2018;13:e0200264. doi: 10.1371/journal.pone.0200264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leikina E, Mertts MV, Kuznetsova N, Leikin S. Type I collagen is thermally unstable at body temperature. Proc Natl Acad Sci USA. 2002;99:1314–1318. doi: 10.1073/pnas.032307099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mirigian LS, Makareeva E, Leikin S. Pulse-chase analysis of procollagen biosynthesis by azidohomoalanine labeling. Connect Tissue Res. 2014;55:403–410. doi: 10.3109/03008207.2014.959120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miura K. Bleach correction ImageJ plugin for compensating the photobleaching of time-lapse sequences. F100Research. 2020 doi: 10.12688/f1000research.27171.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Makareeva E, Han S, Vera JC, Sackett DL, Holmbeck K, Phillips CL, Visse R, Nagase H, Leikin S. Carcinomas contain a matrix metalloproteinase-resistant isoform of type I collagen exerting selective support to invasion. Cancer Res. 2010;70:4366–4374. doi: 10.1158/0008-5472.CAN-09-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miles CA, Sims TJ, Camacho NP, Bailey AJ. The role of the α2 chain in the stabilization of the collagen type I heterotrimer: a study of the type I homotrimer in oim mouse tissues. J Mol Biol. 2002;321:797–805. doi: 10.1016/S0022-2836(02)00703-9. [DOI] [PubMed] [Google Scholar]
- 62.Kuznetsova NV, McBride DJ, Leikin S. Changes in thermal stability and microunfolding pattern of collagen helix resulting from the loss of alpha2(I) chain in osteogenesis imperfecta murine. J Mol Biol. 2003;331:191–200. doi: 10.1016/s0022-2836(03)00715-0. [DOI] [PubMed] [Google Scholar]
- 63.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sengupta P, Satpute-Krishnan P, Seo AY, Burnette DT, Patterson GH, Lippincott-Schwartz J. ER trapping reveals Golgi enzymes continually revisit the ER through a recycling pathway that controls Golgi organization. Proc Natl Acad Sci USA. 2015;112:E6752–E6761. doi: 10.1073/pnas.1520957112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All cell lines, plasmids, and data generated in this study will be deposited to public repositories or freely provided upon request after acceptance of the paper for publication.