Abstract
The systematic position of 16 yeast strains isolated from Thailand, Hungary, The Netherlands, and the Republic of Poland were evaluated using morphological, physiological, and phylogenetic analyses. Based on the similarity of the D1/D2 domain of the LSU rRNA gene, the strains were assigned to two distinct species, Trichosporiella flavificans and representatives of a new yeast species. Phylogenetic analyses revealed that Candida ghanaensis CBS 8798T showed a strong relationship with the aforementioned two species. The more fascinating issue is that Candida and Trichosporiella genera have been placed in different subphyla, Saccharomycotina and Pezizomycotina, respectively. The close relationship between Trichosporiella flavificans, Candida ghanaensis and the undescribed species was unexpected and needed to be clarified. As for morphological and physiological characteristics, the three yeast species shared a hairy colony appearance and an ability to assimilate 18 carbon sources. Based on phylogenetic analyses carried out in the present study, Crinitomyces gen. nov. was proposed to accommodate the new yeast species, Crinitomyces reliqui sp. nov. (Holotype: TBRC 15054, Isotypes: DMKU-FW23-23 and PYCC 9001). In addition, the two species Trichosporiella flavificans and Candida ghanaensis were reassigned to the genus Crinitomyces as, Crinitomyces flavificans (Type: CBS 760.79) comb. nov. and Crinitomyces ghanaensis (Type: CBS 8798) comb. nov., respectively.
Keywords: ascomycetous yeast, Crinitomyces flavificans comb. nov., Crinitomyces ghanaensis comb. nov., four new taxa, Crinitomyces reliqui gen. nov., sp. nov.
1. Introduction
Several species concepts have been applied for yeast identification. A phenotypic species concept and growth profiles were initially used, while a biological species concept including data from mating experiments was later employed [1]. However, the phenotypic concept is limited due to the simplicity of fungal features, such as spore characters, which may lead to phenotypically cryptic taxa [2], and the lack of phenotypic divergence may also occur from the failure of accurate diagnosis [3,4,5]. With the introduction of sequencing technology, the sequence-based species concepts, including the phylogenetic analysis, became broadly applied and extensively employed in fungal taxonomy [1]. As a result, a more accurate classification of yeasts has been obtained.
A number of yeasts have been identified by phenotypic approach, often leading to misidentification or incorrect taxonomic assignment. One of these species-rich and polyphyletic yeast genera within Saccharomycetales order is the genus Candida. In the past, asexual yeasts with multilateral budding but showing nondistinctive cellular morphology were temporarily assigned to the genus Candida [6]. Currently, the placement of many Candida species is still unclear although sequencing technology has been frequently employed in yeast identification and classification. Phylogenetic analysis led to the recognition that various Candida species are distributed throughout the subphylum Saccharomycotina [7]. This situation is aggravated by the observation that even now some novel species are described in the Candida genus as a temporary placement based on short sequences and/or a small number of genes in phylogenetic analyses and, in addition, the lack of taxonomic characters needed to classify them elsewhere [8]. Fortunately, due to the availability of sufficient DNA sequence datasets, various Candida species have been transferred to new or already existing genera such as Scheffersomyces [9], Danielozyma, Deakozyma, Middelhovenomyces [10], Diutina [11], Saturnispora [12], Groenewaldozyma [13], Teunomyces [7] and Limtongozyma [14]. However, many Candida species are awaiting more analysis and accurate classification.
During our investigation of yeast communities in food waste, the strain DMKU-FW23-23 was found. The initial search of the GenBank using BLASTn search of the D1/D2 domain of the large subunit (LSU) of ribosomal RNA (rRNA) gene revealed that this yeast strain was distinct from the described yeast species in the database, but related with Trichosporiella flavificans CBS 760.79T and Candida ghanaensis CBS 8798T. It is surprising that Trichosporiella and Candida are placed in different subphyla i.e., Pezizomycotina and Saccharomycotina, therefore it is unlikely that they are closely related to each other. Obtaining the correct placement and description of the new yeast species represented by strain DMKU-FW23-23 based on an integrative (polyphasic) taxonomic approach and reassignment of Trichosporiella flavificans and Candida ghanaensis were accomplished in this study.
2. Materials and Methods
2.1. Yeast Isolation
Table 1 presents the list of strains considered in this study. The strains from Thailand were isolated by the direct isolation method as described by Sakpuntoon et al. [15]. Yeast extract peptone dextrose (YPD) agar supplemented with 0.025% (w/v) sodium propionate and 0.02% (w/v) chloramphenicol was used as yeast isolation medium. The inoculated agar plates were incubated at 30 ± 2 °C until colonies appeared. Yeast colonies were selected based on different colony morphologies and then purified by cross streaking on YPD agar without antibiotics. The strains CBS 15,014 and CBS 142,641 were isolated from soil and sediment from wastewater treatment facility in The Netherlands, respectively, while the strain CBS 161.94 was isolated from sewage sludge in Katowice, the Republic of Poland. All Hungarian strains were isolated from Danube water. The water samples were taken from the surface of the river from the riverbank by sterile wide mouth screw capped bottles. The samples were kept in a refrigerator until they were processed within 24 h. Hundred milliliter aliquots of the samples were enriched in 500 mL yeast nitrogen base (YNB) medium supplemented with 0.5% (v/v) carbon-source (methanol or hexadecane) and incubated on a horizontal shaker for seven days (25 °C, 100 rpm), then 0.1 mL of each culture was transferred to a 16 mm culture tube containing 5 mL of liquid medium with the same composition. Following an additional week of incubation on a rotary shaker (25 °C, 30 rpm) the enriched cultures were serially diluted and surface plated on Rose-Bengal Chloramphenicol (RBC) agar. Representative strains were isolated on glucose (2%)-peptone-yeast extract (GPY) agar after 7 days of incubation at 25 °C in darkness and purified by repeated streaking.
Table 1.
Yeast strains and isolation sources investigated in this study.
| Yeast | Source of Isolation | Geographical Origin |
|---|---|---|
| Crinitomycesreliqui sp. nov. | ||
| DMKU-FW23-23T | Domestic food waste trap | Thailand |
| CBS 15014 | Soil taken from 2 cm deep in Utrecht | The Netherlands |
| CBS 161.94 | Sewage sludge in Katowice | Republic of Poland |
| CBS 142641 | Sediment from wastewater treatment facility in Zeewolde | The Netherlands |
| CBS 15,240 (=NCAIM Y.01958) |
Water of Danube Budapest (Location 1, 47.484163; 19.054271) |
Hungary |
| CBS 15,241 (=NCAIM Y.02184) |
Water of Danube Budapest (Location 2, 47.594721; 19.070331) |
Hungary |
| CBS 15242 | Water of Danube Budapest (Location 2, 47.594721; 19.070331) |
Hungary |
| CBS 15,243 (=NCAIM Y.02185) |
Water of Danube Budapest (Location 3, 47.592204; 19.069164) |
Hungary |
| Trichosporiella flavificans | ||
| CBS 760.79T | Washings of ion-exchange resin in a guanosine monophosphate manufacturing plant | Japan |
| DMKU-GTSC2-8 | Food waste trap of Faculty of Science, KU canteen | Thailand |
| DMKU-GTSC2-2 | Food waste trap of Faculty of Science, KU canteen | Thailand |
| DMKU-GTCC5-6 | Food waste trap of KU central canteen | Thailand |
| DMKU-GTCC5-12 | Food waste trap of KU central canteen | Thailand |
| DMKU-GTCC5-19 | Food waste trap of KU central canteen | Thailand |
| CBS 15,244 (=NCAIM Y.02186) |
Water of Danube Budapest (Location 3, 47.592204; 19.069164) |
Hungary |
| CBS 15245 | Water of Danube Budapest (Location 3, 47.592204; 19.069164) |
Hungary |
| Candida ghanaensis | ||
| CBS 8798T | Soil in Ghana | Ghana |
KU: Kasetsart University, Bangkok.
For preservation of the isolated strains, a single yeast colony was cultured in a yeast extract malt extract (YM) broth for 18–24 h. Cell pellets were then collected by centrifugation, washed twice with sterile distilled water, and resuspended in fresh YM medium. The active yeast was then preserved in a metabolically inactive state by storing at −80 °C in YM broth supplemented with 30% (v/v) glycerol for long-term preservation.
2.2. DNA Sequencing and Phylogenetic Analysis
Yeasts were grown in YM broth for 18–24 h. The cell pellets were then collected and used for DNA extraction by enzymatic method [16]. The small subunit (SSU) rRNA gene, internal transcribed spacer (ITS) region and the D1/D2 domain of the large subunit (LSU) rRNA gene were amplified with the primer pairs SSU1f/SSU4r [17], SSU3f/SSU2r [17], NL5A/NS7A [18] and NL1/NL4 [19] respectively. The PCR products were purified with a FavorPrepTM Gel/PCR Purification Mini Kit (Favorgen, Austria) and were then sent for DNA sequencing to First BASE Laboratories located at Seri Kembangan in Selangor state, Malaysia. Sequence assembly and alignment were conducted by the BioEdit version 7.0.5.3 program [20]. Aligned sequences were compared with the sequences in the GenBank database (http://www.ncbi.nlm.nih.gov/, accessed on 20 January 2022) using a BLASTn search. Phylogenetic trees were constructed based on the neighbor-joining method with the MEGA version 7.0.26 program [21]. Bootstrap analysis for the estimation of confidence levels of the clades was performed on 1000 bootstrap replications [22], and only values greater than 50% were shown. Table 2 shows the accession numbers of reference sequences retrieved from the GenBank database.
Table 2.
The accession numbers of studied yeasts and reference sequences retrieved from the GenBank database.
| Yeasts | Strain | SSU | ITS | D1/D2 |
|---|---|---|---|---|
| Botryozyma nematodophila | CBS 7426T | NG061133 | NR111167 | NG042439 |
| Candida caryicola | CBS 8847T | - | NR077194 | NG055176 |
| Candida galis | CBS 8842T | - | NR151797 | NG058980 |
| Candida ghanaensis | CBS 8798T | NG065532 | KY102101 | NG055180 |
| Candida haemulonii | CBS 5149T | NG063413 | NR130669 | JX459759 |
| Candida maltosa | CBS 5611T | - | NR138346 | KY106554 |
| Candida tropicalis | CBS 94T | EU348785 | NR111250 | NG054834 |
| Clavispora lusitaniae | CBS 6936T | NG065595 | NR130677 | NG055408 |
| Crinitomyces reliqui | DMKU-FW23-23T | OK275053 | MW720560 | OK298472 |
| Crinitomyces reliqui | CBS 15014 | OK275054 | GU195664 | OK298471 |
| Crinitomyces reliqui | CBS 161.94 | OK275055 | MG250346 | OK298463 |
| Crinitomyces reliqui | CBS 142641 | OK275056 | MG250347 | OK298464 |
| Crinitomyces reliqui | CBS 15,240 (NCAIM Y.01958) | OK275057 | MZ331539 | OK298466 |
| Crinitomyces reliqui | CBS 15,241 (NCAIM Y.02184) | OK275058 | MZ312239 | OK298468 |
| Crinitomyces reliqui | CBS 15242 | OK275059 | MZ312240 | OK298469 |
| Crinitomyces reliqui | CBS 15,243 (NCAIM Y.02185) | OK275060 | MZ312241 | OK298470 |
| Davidhawksworthia ilicicola | CBS 734.94T | - | NR154008 | NG067307 |
| Deakozyma indianensis | NRRL YB-1937T | NG061171 | KJ476205 | NG064315 |
| Debaryomyces hansenii | NRRL Y-7426T | NG063420 | NR120016 | NG042634 |
| Dermea chinensis | CFCC 53008T | - | NR171069 | NG073667 |
| Diddensiella santjacobensis | CBS 8183T | NG063433 | NR151808 | NG058985 |
| Dipodascus albidus | CBS 766.85T | MK834548 | AY788342 | NG066154 |
| Dipodascus carabidarus | CBS 9891T | - | NR155144 | NG058292 |
| Dipodascus cucujoidarus | NRRL Y-27731T | - | NR111352 | NG055370 |
| Dipodascus geniculatus | CBS 184.80T | NG064797 | AY788301 | NG066466 |
| Dipodascus histeridarus | CBS 9892T | - | NR111351 | NG042466 |
| Dipodascus tetrasporeus | CBS 10071T | AB300502 | AB300502 | AB300502 |
| Diutina catenulata | CBS 565T | - | NR077200 | NG059158 |
| Diutina siamensis | DMKU-RE43T | - | KT336715 | KT336715 |
| Geotrichum candidum | CBS 772.71T | JQ698930 | HE663404 | JQ689071 |
| Hanseniaspora valbyensis | CBS 479T | NG063247 | NR111113 | NG042630 |
| Kluyveromyces marxianus | CBS 712T | - | NR111251 | NG042627 |
| Kockiozyma suomiensis | CBS 7251T | NG062713 | NR155335 | NG055355 |
| Kregervanrija fluxuum | CBS 2287T | NG063291 | NR111196 | NG042445 |
| Lachancea thermotolerans | NRRL Y-8284T | NG061071 | NR111334 | NG042626 |
| Limtongia smithiae | CBS 7407T | NG062712 | NR138235 | NG055354 |
| Lipomyces anomalus | CBS 6740T | NG062697 | KT923624 | NG055345 |
| Lipomyces starkeyi | CBS 1807T | - | NG055350 | NR077109 |
| Magnusiomyces magnusii | NRRL Y-17563T | MK834553 | AY788307 | MK834532 |
| Metschnikowia bicuspidata | CBS 5575T | NG065596 | KY104192 | KY108455 |
| Meyerozyma guilliermondii | CBS 2030T | NG063363 | NR111247 | NG042640 |
| Middelhovenomyces petrohuensis | CBS 8173T | NG063431 | NR156314 | NG055211 |
| Middelhovenomyces tepae | CBS 5115T | NG063435 | NR154200 | NG055181 |
| Mollisia caesia | CBS 220.56T | - | MH857591 | MT026503 |
| Mollisia dextrinospora | CBS 401.78T | - | NR119489 | MH872917 |
| Mollisia rosae | CBS 230.71T | - | MH860088 | MH871865 |
| Phlyctema phoenicis | CPC 29372T | - | NR155690 | NG067319 |
| Phlyctema vincetoxici | CBS 123727T | - | NR145310 | NG067282 |
| Pichia membranifaciens | CBS 107T | NG064813 | NR111195 | NG042444 |
| Pseudofabraea citricarpa | CBS 130297T | - | NR154319 | NG069282 |
| Saccharomyces cerevisiae | CBS 1171T | NG063315 | NR111007 | NG042623 |
| Saccharomycodes ludwigii | CBS 821T | NG063254 | NR165986 | NG042629 |
| Saprochaete chiloensis | CBS 8187T | NG070306 | AY788349 | MK834538 |
| Saprochaete saccharophila | CBS 252.91T | NG070310 | AY788309 | MK834545 |
| Saturnispora dispora | CBS 794T | EF550358 | NR155832 | NG055103 |
| Savitreea pentosicarens | DMKU-GTCP10-8T | NG073529 | NR172171 | NG073813 |
| Scheffersomyces stipitis | NRRL Y-7124T | NG063362 | NR165947 | NG042637 |
| Sporopachydermia lactativora | CBS 6192T | - | NR111310 | KY109772 |
| Starmerella bombicola | NRRL Y-17069T | JQ698924 | NR121483 | NG042648 |
| Starmerella geochares | CBS 6870T | NG065473 | KJ630497 | NG060806 |
| Tortispora caseinolytica | CBS 7781T | NG065577 | NR154482 | NG055343 |
| Torulaspora delbrueckii | CBS 1146T | NG061300 | NR111257 | NG058413 |
| Tremella mesenterica | CBS 6973T | - | NR155937 | NG069419 |
| Trichomonascus petasosporus | CBS 9602T | NG062797 | NR155940 | NG055332 |
| Trichosporiella cerebriformis | CBS 135.68T | - | NR155940 | MH859089 |
| Trichosporiella flavificans | CBS 760.79T | OK275050 | MH873011 | OK298462 |
| Trichosporiella flavificans | DMKU-GTSC2-8 | OK275046 | MN460331 | OK283398 |
| Trichosporiella flavificans | DMKU-GTSC2-2 | OK275045 | MN460330 | OK283396 |
| Trichosporiella flavificans | DMKU-GTCC5-6 | OK275047 | MN460342 | OK283393 |
| Trichosporiella flavificans | DMKU-GTCC5-12 | OK275048 | MN460340 | OK283395 |
| Trichosporiella flavificans | DMKU-GTCC5-19 | OK275049 | MN460339 | OK283397 |
| Trichosporiella flavificans | CBS 15,244 (NCAIM Y.02186) | OK275052 | MG250348 | OK298465 |
| Trichosporiella flavificans | CBS 15245 | OK275051 | MZ331540 | OK298467 |
| Trigonopsis variabilis | CBS 1040T | NG061132 | NR154506 | NG055341 |
| Wickerhamiella domercqiae | CBS 4351T | NG061104 | DQ911462 | NG055328 |
| Wickerhamiella infanticola | CBS 7922T | - | NR155985 | NG058278 |
| Wickerhamiella osmotolerans | DMKU VGT1-14T | MN192121 | MN194615 | MH141490 |
| Wickerhamiella sorbophila | CBS 6739T | - | NR155987 | NG055325 |
| Zygoascus hellenicus | CBS 5839T | NG063434 | NR111258 | AY447007 |
T: Type strain of species.
2.3. Phenotypic Characterization
The investigated yeasts were morphologically and physiologically characterized by standard methods described by Kurtzman et al. [23]. Yeasts were grown for three days in YM broth and YM agar at 25 °C for morphological study. Pseudo-hyphae and true hyphae formation were investigated on corn meal agar slide cultures at 25 °C for three days. Growth at different temperatures (15, 25, 30, 35, 37, 40, 42 and 45 °C) was determined in YM broth. The strains were examined individually or mixed in pairs for ascospore formation using different media including PDA, YM agar, YPD agar, corn meal agar, 5% malt extract agar, Gorodkowa agar, V8 agar, Fowell’s acetate agar [24] and yeast carbon base ammonium sulfate (YCBAS) agar [25] at 25 °C for up to twelve weeks with periodic microscopic inspection. Carbon and nitrogen source assimilation, carbohydrate fermentation, starch-like compounds production, and cycloheximide resistance tests were conducted in liquid media. A urea hydrolysis test was performed on a urea slant medium. Acid production and Diazonium Blue B (DBB) tests were conducted on solid medium in Petri dishes. All experiments were carried out with three replicates.
3. Results
3.1. Species Delineation and Molecular Phylogeny
BLASTn search analysis of the D1/D2 domain of the LSU rRNA gene against the GenBank database was performed to identify the yeast strain DMKU-FW23-23 found during a study of yeast community in food waste. The result showed that the top two results from a BLASTn search hit with the currently recognized species Trichosporiella flavificans CBS 760.79T and Candida ghanaensis CBS 8798T, respectively. Surprisingly, these two species are described in different subphyla i.e., T. flavificans was placed in the subphylum Pezizomycotina and C. ghanaensis in the subphylum Saccharomycotina. The relationship between these two species was unexpected and needed to be clarified. Thus, a placement of the strains DMKU-FW23-23, T. flavificans CBS 760.79T and C. ghanaensis CBS 8798T was thoroughly investigated in this study. Seven additional strains, CBS 15240, CBS 15241, CBS 15242, CBS 15243, CBS 15014, CBS 161.94, and CBS 142641, that were similar to the strain DMKU-FW23-23, were found from BLASTn search analysis. Pairwise alignment revealed that the strain DMKU-FW23-23 and its companions differed from each other at 0–2 nucleotide substitutions without gaps in the D1/D2 domain of the LSU rRNA gene, while their ITS region showed no nucleotide substitutions and 0–1 gap (Table 3.).
Table 3.
Pairwise DNA sequence comparisons between the strain DMKU-FW23-23 and its related strains.
| Yeasts | Nucleotide Substitution (bp)/Gap (bp)/Percentage of Sequence Similarity (%) | ||
|---|---|---|---|
| SSU | ITS | D1/D2 | |
| CBS 15,240 (NCAIM Y.01958) | 2/0/99.9 | 0/0/100 | 0/0/100 |
| CBS 15,241 (NCAIM Y.02184) | 0/0/100 | 0/0/100 | 0/0/100 |
| CBS 15,243 (NCAIM Y.02185) | 0/0/100 | 0/0/100 | 0/0/100 |
| CBS 15242 | 0/0/100 | 0/1/99.9 | 0/0/100 |
| CBS 15014 | 0/0/100 | 0/1/99.9 | 2/0/99.6 |
| CBS 161.94 | 0/0/100 | 0/0/100 | 0/0/100 |
| CBS 142641 | 0/0/100 | 0/1/99.9 | 0/0/100 |
All available strains with similar sequences to that of T. flavificans CBS 760.79T in the GenBank database, DMKU-GTSC2-8, DMKU-GTSC2-2, DMKU-GTCC5-6, DMKU-GTCC5-12, DMKU-GTCC5-19, CBS 15244, and CBS 15245, were subjected to physiological and molecular analyses. The results of the pairwise alignment of T. flavificans CBS 760.79T and its related strains are shown in Table 4. Identical sequences (0 nucleotide substitution with 0–1 gap) were found in the D1/D2 domain of the LSU rRNA gene and 0–3 nucleotide substitutions with 0–6 gaps were found in the ITS region among T. flavificans CBS 760.79T and its related strains.
Table 4.
Pairwise DNA sequence comparisons between Trichosporiella flavificans CBS 760.79T and its related strains.
| Yeasts | Nucleotide Substitutions (bp)/Gaps (bp)/Percentage of Sequence Similarity (%) | ||
|---|---|---|---|
| SSU | ITS | D1/D2 | |
| T. flavificans DMKU-GTSC2-8 | 0/0/100 | 0/0/100 | 0/0/100 |
| T. flavificans DMKU-GTSC2-2 | 1/0/99.9 | 1/0/99.8 | 0/0/100 |
| T. flavificans DMKU-GTCC5-6 | 1/0/99.9 | 1/1/99.8 | 0/1/99.8 |
| T. flavificans DMKU-GTCC5-12 | 0/0/100 | 3/6/98.6 | 0/0/100 |
| T. flavificans DMKU-GTCC5-19 | 0/0/100 | 1/2/99.8 | 0/1/99.8 |
| T. flavificans CBS 15,244 (NCAIM Y.02186) | 0/0/100 | 0/0/100 | 0/0/100 |
| T. flavificans CBS 15245 | 0/0/100 | 1/1/99.9 | 0/0/100 |
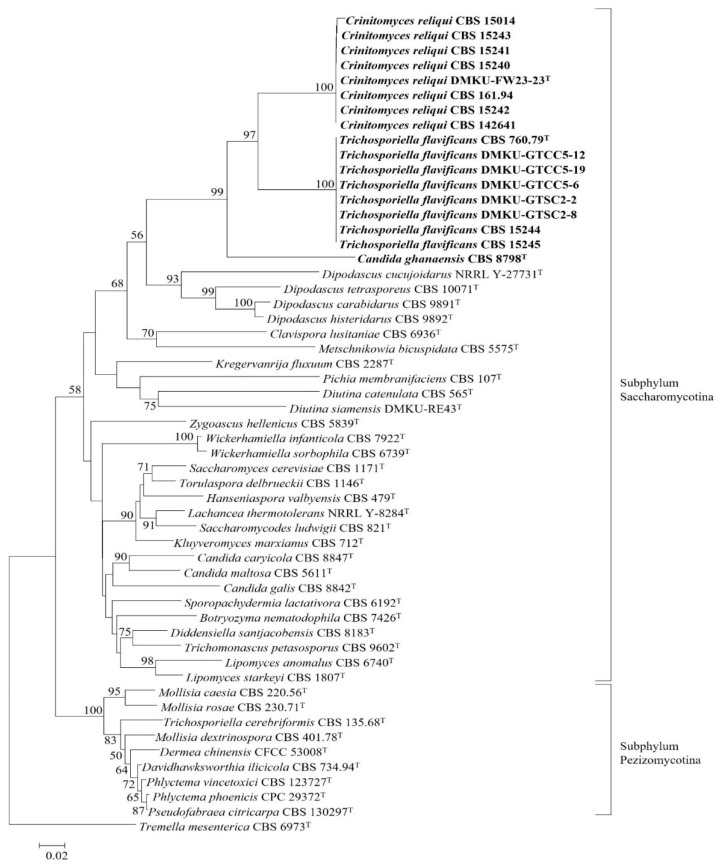
To find the accurate taxonomic placement of the strain DMKU-FW23-23 and its companions, T. flavificans CBS 760.79T and C. ghanaensis CBS 8798T, a phylogenetic tree based on the D1/D2 domain of the LSU rRNA gene was constructed. In addition to the above-noted strains, related species were included in the analysis from the subphyla Saccharomycotina and Pezizomycotina. The results revealed that the strain DMKU-FW23-23 clustered with T. flavificans CBS 760.79T and C. ghanaensis CBS 8798T and their placements were found within the subphylum Saccharomycotina (Figure 1).
Figure 1.
Phylogenetic tree based on the D1/D2 domain of the LSU rRNA gene showing an overview placement of Trichosporiella flavificans, T. cerebriformis, Candida ghanaensis and the novel species Crinitomyces reliqui. The phylogenetic tree was constructed using the neighbor-joining (NJ) method. The numbers provided on branches are frequencies with which a given branch appeared in 1000 bootstrap replications. Bootstrap values of less than 50% are not shown. Tremella mesenterica CBS 6973T served as an outgroup species. Bar, 0.02 substitutions per site.
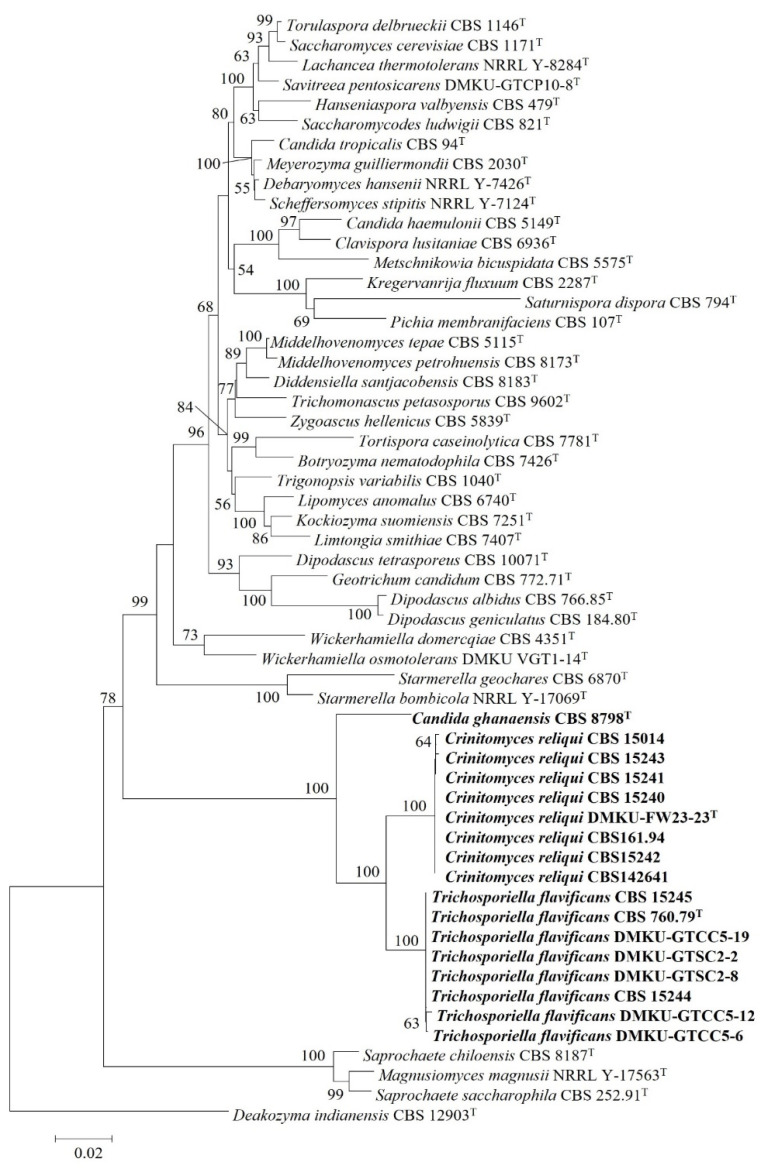
This result suggested that the assignment of T. flavificans to the genus Trichosporiella, which is nested in the subphylum Pezizomycotina, must be revised, and it should be transferred to the subphylum Saccharomycotina. In order to find its accurate placement within the subphylum Saccharomycotina, a phylogenetic tree based on a concatenated sequence of three genes including the small subunit (SSU) rRNA gene, the ITS region and the D1/D2 domain of the LSU rRNA gene was constructed (Figure 2 and Figure S1). The result demonstrated that the strain DMKU-FW23-23 and its companions formed a single lineage and were placed next to T. flavificans CBS 760.79T and also grouped together with C. ghanaensis CBS 8798T. These three species formed a distinct monophyletic clade that is clearly separated from other described yeast species. Hence, a novel yeast genus namely Crinitomyces is proposed to accommodate T. flavificans and C. ghanaensis which are reassigned as Crinitomyces flavificans and Crinitomyces ghanaensis, respectively. Moreover, the strain DMKU-FW23-23 and its companion strains are also proposed as a novel yeast species within this novel genus and the name Crinitomyces reliqui sp. nov. is proposed.
Figure 2.
Phylogenetic tree based on concatenated sequences of the SSU rRNA gene, the ITS region and the D1/D2 domain of the LSU rRNA gene showing the placement of Trichosporiella flavificans, Candida ghanaensis and Crinitomyces reliqui within the subphylum Saccharomycotina. The phylogenetic tree was constructed using the neighbor-joining (NJ) method. The numbers provided on branches are frequencies with which a given branch appeared in 1000 bootstrap replications. Bootstrap values of less than 50% are not shown. Deakozyma indianensis CBS 12903T served as an outgroup species. Bar, 0.02 substitutions per site.
Phenotypic characters of the three yeast species: Crinitomyces reliqui sp. nov., Crinitomyces flavificans comb. nov. and Crinitomyces ghanaensis comb. nov. were compared and are summarized in Table 5.
Table 5.
Physiological characteristics of Crinitomyces reliqui in comparison to its closely related species.
| Physiological Characteristics | 1 | 2 | 3 |
|---|---|---|---|
| Fermentation | |||
| Glucose | v | + | - |
| Galactose | v | v | - |
| Lactose | - | - | - |
| Maltose | v | - | - |
| Melibiose | - | - | - |
| Sucrose | - | - | - |
| Trehalose | - | v | - |
| Raffinose | - | - | - |
| Carbon assimilation | |||
| D-glucose | + | + | + |
| Galactose | + | + | + |
| Sorbose | + | v | v |
| Cellobiose | + | + | + |
| Lactose | w | + | - |
| Maltose | w | + | + |
| Melibiose | w | - | - |
| Sucrose | w | v | + |
| Trehalose | + | + | + |
| Melezitose | w | v | + |
| Raffinose | w | v | - |
| Inulin | - | - | - |
| Starch | v | v | - |
| D-arabinose | + | v | - |
| L-arabinose | + | + | + |
| D-ribose | + | + | - |
| L-rhamnose | + | - | - |
| D-xylose | + | + | + |
| Galactitol | w | - | - |
| Erythritol | + | + | + |
| D-glucitol | s/w | v | + |
| Inositol | s/w | - | - |
| D-mannitol | s/w | v | + |
| Glycerol | + | + | + |
| Ribitol | + | v | - |
| Ethanol | + | s/w | + |
| Methanol | s/w | s/w | - |
| Citric acid | - | - | - |
| Lactic acid | + | + | - |
| Succinic acid | + | + | + |
| D-gluconic acid | + | v | - |
| D-glucuronic acid | - | - | nd |
| D-galacturonic acid | - | - | nd |
| α-Met-D-glucoside | + | - | + |
| Salicin | w | + | + |
| N-acetyl-D-glucosamine | + | + | + |
| D-Glucono-5-lactone | w | + | nd |
| 2-Keto-D-gluconate | w | v | - |
| 5-Keto-D-gluconate | w | - | - |
| Nitrogen assimilation | |||
| (NH4)2SO4 | + | + | nd |
| KNO3 | w | v | - |
| NaNO2 | w | w | nd |
| Ethylamine-HCl | + | + | nd |
| L-lysine | + | + | nd |
| Cadaverine | + | + | + |
| Others | |||
| Diazonium Blue B | - | - | nd |
| Starch-like compounds | - | - | - |
| Growth at vitamin free medium | - | - | - |
| Urea hydrolysis | - | - | nd |
| Growth at 15 °C | + | + | nd |
| Growth at 25 °C | + | + | + |
| Growth at 30 °C | + | + | + |
| Growth at 35 °C | v | + | + |
| Growth at 37 °C | v | + | + |
| Growth at 40 °C | - | + | - |
| Growth at 42 °C | - | - | - |
| Growth at 45 °C | - | - | - |
| 0.1% Cycloheximide resistance | v | - | + |
| 0.01% Cycloheximide resistance | v | - | + |
| Growth in medium supplemented with 50% glucose | + | v | nd |
| Growth in medium supplemented with 60% glucose | + | + | nd |
| Growth in medium supplemented with 5% glucose +10%NaCl | + | + | + |
| Growth in medium supplemented with 5% glucose +16%NaCl | - | - | nd |
| Acid production | v | v | nd |
Species: 1, Crinitomyces reliqui sp. nov. (DMKU-FW23-23T and seven additional strains); 2, Crinitomyces flavificans comb. nov. (CBS 760.79T and seven additional strains) and 3, Crinitomyces ghanaensis comb. nov. CBS 8798T [26] and data obtained from CBS. Abbreviation: +, positive; -, negative; s, slow positive; w, weak positive; v, variable (some strains are positive, others negative); nd, no data.
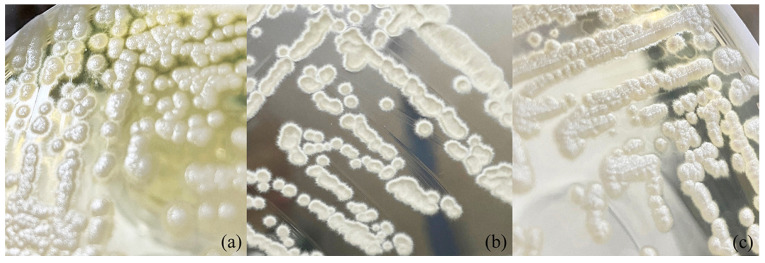
A broad range of carbon sources was found to be assimilated, namely glucose, galactose, sorbose, cellobiose, maltose, sucrose, trehalose, melezitose, L-arabinose, D-xylose, erythritol, glucitol, mannitol, glycerol, ethanol, succinic acid, salicin and N-acetyl-glucosamine and only two carbon sources, inulin, and citric acid, were not assimilated by any of these yeasts. However, Crinitomyces ghanaensis CBS 8798T did not show fermentation ability. It should be noted that, by all strains of Crinitomyces flavificans, a yellow pigment was exuded onto the agar medium during cultivation. For morphological characteristics, yeast colonies of the three species are white to cream, convex and butyrous, with a dull surface. All of the three yeast species showed a hairy colony morphology (Figure 3) which is the origin of the genus name, Crinitomyces.
Figure 3.
A hairy colony appearance of three yeast species on YM agar. (a) Crinitomyces flavificans CBS 760.79T; (b) Crinitomyces ghanaensis CBS 8798T and (c) Crinitomyces reliqui DMKU-FW23-23T sp. nov.
3.2. Ecology
The type strain of Crinitomyces reliqui DMKU-FW23-23T was isolated from domestic food waste in Thailand while the two related strains, CBS 161.94 and CBS 142641, were isolated from similar types of habitats, sewage sludge and wastewater in Republic of Poland and The Netherlands, respectively. However, the strain CBS 15,014 was isolated from soil and the four related strains were isolated from water surface of a river in The Netherlands and Hungary, respectively. These habitats showed a contrast in terms of amount and type of nutrients available. The occurrence of this species in food waste, sewage sludge and rivers raised the possibility that the water of the rivers might have been polluted at the time of sampling. Crinitomyces reliqui is suggested to be a cosmopolitan species because all eight strains of this species were found from four different countries.
The yeast Crinitomyces flavificans CBS 760.79T was isolated from washings of ion-exchange resin in a guanosine monophosphate manufacturing plant in Japan whereas its seven companion strains were isolated from food waste and water in different countries. Therefore, C. flavificans should also be claimed as a cosmopolitan species.
The isolation sources and geographical origin of all investigated strains are summarized in Table 1.
3.3. Taxonomy
Description of Crinitomyces V. Sakpuntoon, G. Péter, M. Groenew., D. Dlauchy, S. Limtong & N. Srisuk, gen. nov.
MycoBank number: 842461.
Crinitomyces (Cri.ni.to.my’ces. N.L. fem. n. Crinitomyces refers to the hairy colony appearance of yeast within the genus).
Cells are spherical or ellipsoidal and asexual reproduction proceeds by multilateral budding. Septate hyphae are produced. Ascospore formation is not observed. DBB reaction is negative, starch-like compounds are not produced, and urea hydrolysis is negative.
Phylogenetic placement: Saccharomycetales, Saccharomycotina, Ascomycota.
Type species: Crinitomyces flavificans (Nakase) V. Sakpuntoon, G. Péter, M. Groenew., D. Dlauchy, S. Limtong & N. Srisuk comb. nov.
Description of Crinitomyces reliqui V. Sakpuntoon, G. Péter, M. Groenew, D. Dlauchy, S. Limtong & N. Srisuk, sp. nov.
Crinitomyces reliqui (re.li.qu’i. L. fam. adj. reliquum of the residue; reliquum indicating that the type strain was isolated from residue of food or food waste).
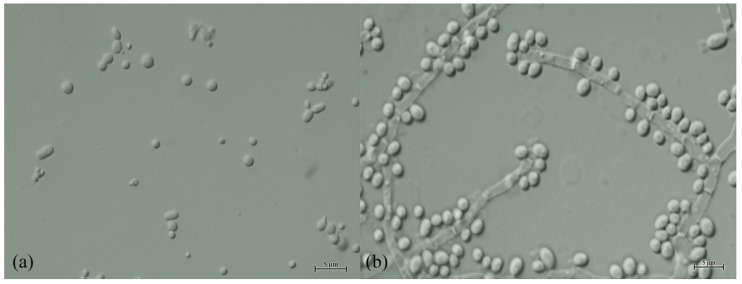
After 3 days growth in YM broth at 25 °C, cells are spherical (1.5–3 μm) or ellipsoidal (1.5–2.0 × 2–3 μm). Colonies are white to cream, convex and butyrous, with a dull surface and filamentous margins (Figure 3c). True hyphae and branching lateral hyphae are observed on corn meal agar at 25 °C after 3 days (Figure 4b). Blasto-conidia are formed randomly from both hyphal types, globose to sub-globose, 1.8–3.0 μm. Ascospores are not found in individual cultures or in mixed cultures on PDA, YM agar, YPD agar, corn meal agar, 5% malt extract agar, Gorodkowa agar, Fowell’s acetate agar, V8 agar and YCBAS medium after 12 weeks at 25 °C. Glucose (viable), galactose (viable) and maltose (viable) are fermented. but lactose, sucrose, trehalose, melibiose and raffinose are not. Glucose, galactose, sorbose, cellobiose, lactose (weak), maltose (weak), melibiose (weak), sucrose (weak), trehalose, melezitose (weak), raffinose (weak), starch (variable), D-arabinose, L-arabinose, D-ribose, L-rhamnose, D-xylose, galactitol (weak), erythritol, D-glucitol (slow and weak), inositol (slow and weak), D-mannitol (slow and weak), glycerol, ribitol, ethanol, methanol (slow and weak), lactic acid, succinic acid, D-gluconic acid, α-Met-D-Glucoside, salicin (weak), N-Acetyl-D-Glucosamine, D-Glucono-5-lactone (weak), 2-Keto-D-gluconate (weak) and 5-Keto-D-gluconate (weak) are assimilated as the sole carbon sources, while inulin, citric acid, D-glucuronic acid and D-galacturonic acid are not assimilated. Ammonium sulfate, potassium nitrate (weak), sodium nitrite (weak), ethylamine hydrochloride, L-lysine and cadaverine dihydrochloride are utilized as sole nitrogen sources. Growth occurs in media containing 10% (w/v) sodium chloride/5% (w/v) glucose but not in 16% (w/v) sodium chloride/5% (w/v) glucose. Growth at 37 °C is positive for all strains except the strain CBS 15,014 of which growth occurs at 15 –30 °C. Growth is not observed in vitamin-free medium but variable results were found in medium supplemented with 0.01% (w/v) and 0.1% (w/v) cycloheximide. Acid production is variable. Urea hydrolysis, starch-like compounds production and diazonium blue B reaction are negative.
Figure 4.
Morphology of Crinitomyces reliqui DMKU-FW23-23T (a) Cells in YM broth after 3 days at 25 °C (bar, 5 μm) and (b) True hypha formation on corn meal agar after incubated at 25 °C for 3 days (bar, 5 μm).
The holotype was isolated from domestic food waste in Bangkok, Thailand. The food waste sample was randomly collected via aseptic technique, and it was used for isolation process as previous described in materials and methods within 24 h. The obtained yeast colony was purified by cross-steaking on YM medium. After an initial BLASTn search analysis, additional representatives of the novel species were found. The strain was preserved at −80 °C in YM broth supplemented with 30% (v/v) glycerol. The holotype has been deposited and permanently preserved in a metabolically-inactive state in the Thailand Bioresource Research Centre (TBRC), Thailand, as TBRC 15054. An isotype has been permanently preserved in a metabolically-inactive state at the Department of Microbiology, Faculty of Science, Kasetsart University, Bangkok, Thailand as strain DMKU-FW23-23 and in the collection of the Portuguese Yeast Culture Collection (PYCC), Caparica, Portugal, as strain PYCC 9001. MycoBank number 842462.
New combinations.
Crinitomyces flavificans (Nakase) V. Sakpuntoon, G. Péter, M. Groenew., D. Dlauchy, S. Limtong & N. Srisuk, comb. nov.
MycoBank number: 842463.
Basionym: Candida flavificans, T. Nakase (1975). Antonie van Leeuwenhoek 41:202
Holotype: CBS 760.79, from washings of ion-exchange resin in a guanosine monophosphate manufacturing plant.
Note: Crinitomyces flavificans was first described as Candida flavificans based on an analysis of physiological and biochemical characteristics in 1975 [27]. Later, based on morphological characters it was reclassified as Trichosporiella flavificans [27]. However, the type species of Trichosporiella, T. cerebriformis, is nested in the subphylum Pezizomycotina. Based on the phylogenetic analyses carried out in this study, T. flavificans is reclassified here in Saccharomycotina as Crinitomyces flavificans. All phenotypic characters of the type strain, CBS 760.79, were re-examined in this study and results were found consistent with those of the first report. Nevertheless, some characters were differed among the companion strains and were then reported as “variable” as shown in the Table 5.
Crinitomyces ghanaensis (Kurtzman) V. Sakpuntoon, G. Péter, M. Groenew., D. Dlauchy, S. Limtong & N. Srisuk, comb. nov.
MycoBank number: 842464.
Basionym: Candida ghanaensis, C.P. Kurtzman (2001). Antonie van Leeuwenhoek 79:355.
Holotype: CBS 8798, from soil in Ghana.
4. Conclusions and Discussion
Candida flavificans CBS 760.79T was isolated from washing of the ion-exchange resin in a guanosine monophosphate manufacturing plant and classified based on DNA base composition, proton-magnetic-resonance spectrum of polysaccharide, and serological characteristics [27]. However, in 1985, it was reclassified as Trichosporiella flavificans CBS 760.79T due to a stronger coherence between hyphal cells and an absence of arthroconidia that split Trichosporiella from Candida [28]. Sequence analysis of this yeast genus has not been accomplished since then. In the era in which molecular study and sequencing technology play an important role in taxonomic study, yeast classification by the aforementioned analyses may not be sufficient and may also cause misidentifications.
Even if molecular methods and phylogenetic analyses have been used to identify yeast, unstable placement in the evolution line may occur, since they may be characterized based on short sequences and/or a small number of genes in phylogenetic analysis. Candida ghanaensis CBS 8798T isolated from soil in Ghana, was first described with a weakly supported phylogenetic placement based on the D1/D2 domain of the LSU of rRNA gene by Kurtzman [26]. Subsequent analysis of the SSU of the rRNA gene sequence revealed that C. ghanaensis had a weak and probably insignificant affinity with C. incommunis, but the highest matches in the GenBank database were found with members of the Dipodascus/Geotrichum clade [8]. However, a well-supported placement of this yeast species has not yet established and a phylogenetic reconstruction from additional data was required.
Accurate classification of yeasts may not be achieved by a single method. Although molecular methods are irreplaceable, yeast identification requires combined application of several approaches (polyphasic taxonomy). Similarly, multiple conspecific strains are more reliable to propose a new yeast species. Nevertheless, it will also be more reliable if conspecific strains are isolated from different samples and/or geographical regions.
In this study we described a new yeast species, Crinitomyces reliqui, which is closely related to T. flavificans and C. ghanaensis as they formed a well-supported clade in the phylogenetic trees and they also shared morphological and some physiological characteristics. We proposed here a novel yeast genus, Crinitomyces, to accommodate the novel species Crinitomyces reliqui as well as T. flavificans and C. ghanaensis, which were reassigned as Crinitomyces flavificans and Crinitomyces ghanaensis, respectively.
Acknowledgments
We would like to thank Kasetsart University Research and Development Institute (KURDI) for the grant no. FF(KU)18.64, the Royal Golden Jubilee PhD programme, the Hungarian Ministry for Innovation and Technology and by the European Union and co-financed by the European Social Fund (grant agreement no. EFOP-3.6.3-VEKOP-16-2017-00005), UGSAS-GU via the “Microbiology Laboratory Station for IC-GU12” at Kasetsart University and International SciKU Branding (ISB), Faculty of Science, Kasetsart University.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jof8030224/s1, Figure S1: Phylogenetic tree based on concatenated sequences of the SSU rRNA gene, the ITS region and the D1/D2 domain of the LSU rRNA gene showing the placement of Trichosporiella flavificans, Candida ghanaensis and Crinitomyces reliqui within the subphylum Saccharomycotina. The phylogenetic tree was constructed using the maximum likelihood (ML) method with optimized models for 3 partitions (SSU, ITS region and LSU) in IQ-TREE. The numbers provided on branches are frequencies with which a given branch appeared in 1000 bootstrap replications. Bootstrap values of less than 50% are not shown. Deakozyma indianensis CBS 12903T was served as an outgroup species. Bar, 0.1 substitutions per site.
Author Contributions
Data curation, formal analysis, investigation, and writing—original draft preparation, V.S.; Supervision and validation, G.P. and M.G.; Methodology, D.D.; Supervision, S.L.; Conceptualization, funding acquisition, resources, supervision, writing—review and editing, N.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Kasetsart University Research and Development Institute (KURDI) grant no. FF(KU)18.64 to N.S., the Royal Golden Jubilee PhD programme grant no. PHD/0070/2560 to V.S. This work was also supported by the Hungarian Ministry for Innovation and Technology and by the European Union and co-financed by the European Social Fund (grant agreement no. EFOP-3.6.3-VEKOP-16-2017-00005 to G.P. and D.D.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Conflicts of Interest
The authors declare that there is no conflict of interest in terms of the publication.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Boekhout T., Aime M.C., Begerow D., Gabaldon T., Heitman J., Kemler M., Khayhan K., Lachance M.A., Louis E.J., Sun S., et al. The evolving species concepts used for yeasts: From phenotypes and genomes to speciation networks. Fungal Divers. 2021;109:27–55. doi: 10.1007/s13225-021-00475-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lucking R., Aime M.C., Robbertse B., Miller A.N., Ariyawansa H.A., Aoki T., Cardinali G., Crous P.W., Druzhinina I.S., Geiser D.M., et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus. 2020;11:14. doi: 10.1186/s43008-020-00033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moncada B., Lücking R., Suárez A. Molecular phylogeny of the genus Sticta (lichenized Ascomycota: Lobariaceae) in Colombia. Fungal Divers. 2014;64:205–231. doi: 10.1007/s13225-013-0230-0. [DOI] [PubMed] [Google Scholar]
- 4.Lücking R., Forno M.D., Moncada B., Coca L.F., Vargas-Mendoza L.Y., Aptroot A., Arias L.J., Besal B., Bungartz F., Cabrera-Amaya D.M., et al. Turbo-taxonomy to assemble a megadiverse lichen genus: Seventy new species of Cora (Basidiomycota: Agaricales: Hygrophoraceae), honouring David Leslie Hawksworth’s seventieth birthday. Fungal Divers. 2017;84:139–207. doi: 10.1007/s13225-016-0374-9. [DOI] [Google Scholar]
- 5.Merényi Z., Varga T., Hubai A.G., Pitlik P., Erős Á., Trappe J.M., Bratek Z. Challenges in the delimitation of morphologically similar species: A case study of Tuber brumale agg. (Ascomycota, Pezizales) Mycol. Prog. 2017;16:613–624. doi: 10.1007/s11557-017-1296-y. [DOI] [Google Scholar]
- 6.Daniel H.M., Lachance M.A., Kurtzman C.P. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek. 2014;106:67–84. doi: 10.1007/s10482-014-0170-z. [DOI] [PubMed] [Google Scholar]
- 7.Kurtzman C.P., Robnett C.J., Blackwell M. Description of Teunomyces gen. nov. for the Candida kruisii clade, Suhomyces gen. nov. for the Candida tanzawaensis clade and Suhomyces kilbournensis sp. nov. FEMS Yeast Res. 2016;16:fow041. doi: 10.1093/femsyr/fow041. [DOI] [PubMed] [Google Scholar]
- 8.Lachance M., Boekhout T., Scorzetti G., Fell J.W., Kurtzman C.P. Candida Berkhout The Yeasts, a Taxonomic Study. Elsevier; Amsterdam, The Netherlands: 1923. pp. 987–1278. [Google Scholar]
- 9.Urbina H., Blackwell M. Multilocus Phylogenetic Study of the Scheffersomyces Yeast Clade and Characterization of the N-Terminal Region of Xylose Reductase Gene. PLoS ONE. 2012;7:e39128. doi: 10.1371/journal.pone.0039128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurtzman C.P., Robnett C.J. Three new anascosporic genera of the Saccharomycotina: Danielozyma gen. nov., Deakozyma gen. nov. and Middelhovenomyces gen. nov. Antonie Van Leeuwenhoek. 2014;105:933–942. doi: 10.1007/s10482-014-0149-9. [DOI] [PubMed] [Google Scholar]
- 11.Khunnamwong P., Lertwattanasakul N., Jindamorakot S., Limtong S., Lachance M.A. Description of Diutina gen. nov., Diutina siamensis, f.a. sp. nov., and reassignment of Candida catenulata, Candida mesorugosa, Candida neorugosa, Candida pseudorugosa, Candida ranongensis, Candida rugosa and Candida scorzettiae to the genus Diutina. Int. J. Syst. Evol. Microbiol. 2015;65:4701–4709. doi: 10.1099/ijsem.0.000634. [DOI] [PubMed] [Google Scholar]
- 12.Kurtzman C.P. Description of Martiniozyma gen. nov. and transfer of seven Candida species to Saturnispora as new combinations. Antonie Van Leeuwenhoek. 2015;108:803–809. doi: 10.1007/s10482-015-0536-x. [DOI] [PubMed] [Google Scholar]
- 13.Kurtzman C.P. Description of Groenewaldozyma gen. nov. for placement of Candida auringiensis, Candida salmanticensis and Candida tartarivorans. Antonie Van Leeuwenhoek. 2016;109:1041–1045. doi: 10.1007/s10482-016-0703-8. [DOI] [PubMed] [Google Scholar]
- 14.Boontham W., Angchuan J., Boonmak C., Srisuk N. Limtongozyma siamensis gen. nov., sp. nov., a yeast species in the Saccharomycetales and reassignment of Candida cylindracea to the genus Limtongozyma. Int. J. Syst. Evol. Microbiol. 2020;70:199–203. doi: 10.1099/ijsem.0.003735. [DOI] [PubMed] [Google Scholar]
- 15.Sakpuntoon V., Angchuan J., Boontham W., Khunnamwong P., Boonmak C., Srisuk N. Grease Waste as a Reservoir of Lipase-Producing Yeast and Description of Limtongella siamensis gen. nov., sp. nov. Microorganisms. 2019;8:27. doi: 10.3390/microorganisms8010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles D.J., Busser K., Stalder C., Higgins D.R. Isolation of Nucleic Acids. In: Higgins D.R., Cregg J.M., editors. Pichia Protocols. Volume 103. Humana Press; Totowa, NJ, USA: 1998. pp. 73–80. [DOI] [PubMed] [Google Scholar]
- 17.Rosa C.A., Lachance M.A., Teixeira L.C., Pimenta R.S., Morais P.B. Metschnikowia cerradonensis sp. nov., a yeast species isolated from ephemeral flowers and their nitidulid beetles in Brazil. Int. J. Syst. Evol. Microbiol. 2007;57:161–165. doi: 10.1099/ijs.0.64624-0. [DOI] [PubMed] [Google Scholar]
- 18.Kurtzman C., Robnett C. Phylogenetic relationships among yeasts of the complex determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzman C.P., Robnett C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/A:1001761008817. [DOI] [PubMed] [Google Scholar]
- 20.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 21.Kumar S., Stecher G., Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Kurtzman C.P., Fell J.W., Boekhout T., Robert V. Methods for Isolation, Phenotypic Characterization and Maintenance of Yeasts. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The Yeasts, a Taxonomic Study. 5th ed. Elsevier; Amsterdam, The Netherlands: 2011. pp. 87–110. [Google Scholar]
- 24.Fowell R.R. Sodium acetate agar as a sporulation medium for yeasts. Nature. 1952;170:578. doi: 10.1038/170578a0. [DOI] [PubMed] [Google Scholar]
- 25.Kurtzman C.P., Fell J.W., Boekhout T. The Yeasts, a Taxonomic Study. 5th ed. Volume 1 Elsevier; Amsterdam, The Netherlands: 2011. [Google Scholar]
- 26.Kurtzman C.P. Four new Candida species from geographically diverse locations. Antonie Van Leeuwenhoek. 2001;79:353–361. doi: 10.1023/A:1012097023978. [DOI] [PubMed] [Google Scholar]
- 27.Nakase T. Three new asporogenous yeasts found in industrial waste water. Antonie Van Leeuwenhoek. 1975;41:201–210. doi: 10.1007/BF02565052. [DOI] [PubMed] [Google Scholar]
- 28.de Hoog G.S., Rantio-Lehtimaki A.H., Smith M.T. Blastobotrys, Sporothrix and Trichosporiella: Generic delimitation, new species, and a Stephanoascus teleomorph. Antonie Van Leeuwenhoek. 1985;51:79–109. doi: 10.1007/BF00444231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.