Abstract
Burkholderia pseudomallei is a gram-negative bacterium that causes the disease known as melioidosis. This pathogen is endemic to Southeast Asia and northern Australia and is particularly problematic in northeastern Thailand. It has been previously reported that B. pseudomallei is resistant to the killing action of cationic antimicrobial peptides, including human neutrophil peptide, protamine sulfate, poly-l-lysine, magainins, and polymyxins. Recently, we have also found that the virulent clinical isolate B. pseudomallei 1026b is capable of replicating in media containing polymyxin B at concentrations of >100 mg/ml. In order to identify genetic loci that are associated with this particular resistance phenotype, we employed a Tn5-OT182 mutagenesis system in coordination with a replica plating screen to isolate polymyxin B-susceptible mutants. Of the 17,000 Tn5-OT182 mutants screened via this approach, five polymyxin B-susceptible mutants were obtained. Three of these mutants harbored Tn5-OT182 insertions within a genetic locus demonstrating strong homology to the lytB gene present in other gram-negative bacteria. Of the remaining two mutants, one contained a transposon insertion in a locus involved in lipopolysaccharide core biosynthesis (waaF), while the other contained an insertion in an open reading frame homologous to UDP-glucose dehydrogenase genes. Isogenic mutants were also constructed via allelic exchange and used in complementation analysis studies to further characterize the relative importance of each of the various genetic loci with respect to the polymyxin B resistance phenotype exhibited by B. pseudomallei 1026b.
Burkholderia pseudomallei is the causative agent of melioidosis. This bacterial pathogen is endemic to Southeast Asia, northern Australia, and temperate areas that border the equator (23). B. pseudomallei is found as a natural inhabitant of moist soils, stagnant waters, and rice paddies that predominate in regions of endemicity such as northeastern Thailand (8, 35).
The clinical manifestations of melioidosis may be observed as inapparent infection, asymptomatic pulmonary infiltration, acute localized supprative infection, acute pulmonary infection, acute septicemic infection, or chronic supprative infection (9, 39). B. pseudomallei is a common cause of opportunistic infections in areas of endemicity, and individuals particularly susceptible include diabetics and those with renal disease (8). In addition, it has been shown that in some areas this pathogen is a major cause of community-acquired sepsis, resulting in up to 70% mortality even with treatment (8).
B. pseudomallei strains are intrinsically resistant to a broad spectrum of antibiotics, a feature that can often complicate the treatment of melioidosis (14). This organism is resistant to a variety of antibiotics, including penicillin, ampicillin (AMP), narrow-spectrum cephalosporins, streptomycin (STR), tobramycin, and gentamicin (GEN) (14, 20, 23). Recently, Moore et al. (28) have demonstrated the presence of an efflux pump involved in aminoglycoside resistance. In addition, B. pseudomallei demonstrates high levels of resistance to the action of cationic antimicrobial peptides such as polylysine, protamine sulfate, human neutrophil peptides (HNP-1), and polymyxins (14, 21).
In the present studies we have chosen polymyxin B (PMB) as a model in an attempt to elucidate the mechanisms by which B. pseudomallei resists the killing action imparted by cationic antimicrobial peptides.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Cultures were grown at 37°C on Luria-Bertani (LB) base agar plates or in LB broth. For Escherichia coli, antibiotics were used, when appropriate, at the following concentrations: AMP, 100 μg/ml; kanamycin (KAN), 25 μg/ml; chloramphenicol (CHL), 25 μg/ml; STR, 100 μg/ml; tetracycline (TET), 15 μg/ml; trimethoprim (TMP), 1.5 mg/ml; and Zeocin (ZEO), 25 μg/ml. For B. pseudomallei, antibiotic concentrations were as follows: KAN, 50 μg/ml; TET, 50 μg/ml; TMP, 100 μg/ml; and ZEO, 100 μg/ml. Antibiotics were purchased from Sigma Chemical Co. and Invitrogen.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| SM10 | Mobilizing strain, transfer genes of RP4 integrated in chromosome; Kanr Strs | 36 |
| SM10 λ pir | SM10 with a λ prophage carrying the pir gene | 25 |
| SURE | e14− (mcrA) Δ(mcrCB-hsdSMR-mrr)171 endA1 supE44 thi-1 gyrA96 relA1 lac recB recJ sbcC umuC::Tn5 uvrC [F′ proAB lacIqZΔM15 Tn10]; Kanr Tetr | Stratagene |
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1 | Bethesda Research Laboratories |
| B. thailandensis E264 | Environmental isolate; Kanr Genr Pmbr Strr Tetr | 5 |
| B. pseudomallei | ||
| 1026b | Clinical isolate; Kanr Genr Pmbr Strr Tets Tmps Zeos | 12 |
| DD503 | 1026b derivative; allelic exchange strain; Δ(amrR-oprA) (Kans Gens Strs); rpsL (Strr) | 28 |
| SRM 117 | 1026b derivative; wbiI::Tn5-OT182; Tetr | 13 |
| PMB-7 | 1026b derivative; waaF::Tn5-OT182; Tetr | This study |
| PMB-20 | 1026b derivative; udg::Tn5-OT182; Tetr | This study |
| PMB-4 | 1026b derivative; lytB::Tn5-OT182; Tetr | This study |
| MB100 | DD503 derivative; waaF::dhfrIIb-p15AoriV; Zeor | This study |
| MB203 | DD503 derivative; lytB::dhfrIIb-p15AoriV; Tmpr | This study |
| MB300 | DD503 derivative; udg::dhfrIIb-p15AoriV; Tmpr | This study |
| MB301 | DD503 derivative; waaE::dhfrIIb-p15AoriV; Zeor | This study |
| MB100C | MB100(pUCP28T::waaF); Strs Zeor Tmpr | This study |
| MB203C | MB203(pRK415::lytB); Strs Tmpr Tetr | This study |
| MB300C | MB300(pRK415::udg); Strs Tmpr Tetr | This study |
| Plasmids | ||
| pOT182 | pSUP102(Gm)::Tn5-OT182; Chlr Genr Ampr Tetr | 24 |
| p34E-oriTP | Vector containing self-cloning Tmpr cassette; dhfrIIb-p15AoriV | 6 |
| p34E-oriZeo | Vector containing self-cloning Zeor cassette; dhfrIIb-p15oriV | 6 |
| pPMB-7B | 4.0-kb BamHI fragment from PMB-7 obtained by self-cloning; contains partial waaF gene; Ampr Tetr | This study |
| pPMB-4Ss | 4.5-kb SstI fragment from PMB-4 obtained by self-cloning; contains partial lytB gene; Ampr Tetr | This study |
| pPMB-20H | 8.0-kb HindIII fragment from PMB-20 obtained by self-cloning; contains partial udg gene; Ampr Tetr | This study |
| pPMB-20Ss | 5.2-kb SstI fragment from PMB-20 obtained by self-cloning; contains waaE gene; Ampr Tetr | This study |
| pUCP28T | Broad-host-range vector; IncP OriT; pRO1600ori; Tmpr | 34 |
| pRK415 | Broad-host-range vector; Tetr | 22 |
Plasmids were purified by using Wizard minipreps for plasmid DNA (Promega) or QIAprep spin plasmid minipreps (Qiagen). Conjugations were performed essentially as previously described (12).
Mutagenesis and screening.
B. pseudomallei 1026b was mutagenized with Tn5-OT182 as previously described (12). PMB-susceptible mutants were isolated by using a replica plating method in which Tn5-OT182 mutants were screened on LB agar plates with or without 200 μg of PMB per ml. Those mutants that failed to grow on the medium containing PMB were retested and retained for further analyses.
MIC determination.
MICs were determined by using Mueller-Hinton (MH) agar-based plates or MH broth (Becton Dickinson Microbiology Systems). Standard MIC tests were used for a variety of antimicrobials and included both agar and broth dilution assays (29). For MIC testing in excess of 10,000 μg/ml, PMB sulfate was solubilized directly in MH broth, filter sterilized, and then inoculated with mid-log-phase cultures of the B. pseudomallei strain. In addition, when appropriate, E-tests (AB Biodisk, Solna, Sweden) were used as per the manufacturer’s instructions.
DPX binding assay.
The interaction of dansyl polymyxin (DPX) with B. pseudomallei was examined under standard assay conditions as previously described (26, 27). The DPX used in this study was generously provided by R. E. W. Hancock, University of British Columbia, Vancouver, Canada. A 1.5 mM stock solution of DPX was stored at −20°C and diluted appropriately for assays. Fluorescence was measured with an F-2000 fluorescence spectrophotometer (Hitachi).
DNA manipulation and electroporation.
Restriction endonucleases and T4 DNA ligase were purchased from Gibco BRL, Boehringer Mannheim, and New England BioLabs and were used according to the manufacturer’s instructions. A Gene Clean II kit (Bio 101) was used for purification of DNA fragments that were excised from agarose gels and used in cloning procedures. Isolation of chromosomal DNA and cloning of DNA immediately flanking Tn5-OT182 insertions were performed as previously described (12, 43).
ELISA.
B. pseudomallei strains were assayed for the presence of type II O-polysaccharide (O-PS) moieties via enzyme-linked immunosorbent assay (ELISA) (13) with a type II O-PS-specific monoclonal antibody (MAb) (19).
LPS purification and immunoblot analysis.
Lipopolysaccharide (LPS) was purified as previously described (4). Immunoblot analyses were performed with the type II O-PS-specific MAb (7, 19). In addition, polyclonal rabbit sera recognizing type I and II O-PSs as well as flagellin proteins were used for immunoblot analysis as previously described (4).
LPS silver stain analysis.
LPS from whole cells of B. pseudomallei was silver stained by using a previously described method and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (18, 42).
Outer membrane protein isolation and analysis.
Outer membrane proteins were prepared as previously described (15). The protein samples were subjected to SDS-polyacrylamide gel electrophoresis analysis (40) with a 5% stacking gel and a 12% separating gel. Protein was visualized with Coomassie blue staining.
Construction of allelic exchange knockouts.
Allelic exchange was performed in this study as previously described for B. pseudomallei (13), with the rpsL-based vector pKAS46 (38). B. pseudomallei DD503 was used for gene replacement experiments (13, 28). A typical allelic exchange procedure consisted of first transforming SM10 λ pir with a pKAS46 derivative harboring an insertionally inactivated allele. Each gene was disrupted with an antibiotic cassette next to an origin of replication (6). This was followed by conjugation of this SM10 λ pir strain to DD503 as previously described (13). Transconjugates were selected for on LB plates containing PMB (50 μg/ml) and ZEO (100 μg/ml) or TMP (100 μg/ml). The Pmbr and Zeor or Tmpr transconjugates were then plated on STR (100 μg/ml) and either ZEO (100 μg/ml) or TMP (100 μg/ml) to select for loss of the vector. These mutants were then tested on plates containing KAN (50 μg/ml) to confirm loss of the vector; this was indicated by absence of growth. Mutations were confirmed by either Southern blot analysis or self-cloning and sequencing of the DNA flanking the oriZeo or oriTp cassette.
PCR amplification and cloning of PCR products.
The waaF, udg, and lytB genes were amplified from B. pseudomallei 1026b chromosomal DNA by PCR. The oligodeoxyribonucleotide primers used to amplify the waaF gene were rfaF-5′ (5′GGGGTACCGAGCGTCGCGTTTATTACG3′) and rfaF-3′ (5′CGGGATCCTGAATCGGGTGCGGGTGCGC3′). The waaF gene was PCR amplified in a 100-μl reaction mixture containing 500 ng of genomic DNA, 1× PCR buffer (Gibco BRL), a 200 μM concentration of each deoxynucleoside triphosphate, a 0.5 μM concentration of each primer, 1.5 mM MgCl2 (Gibco BRL), and 5 U of Taq DNA polymerase (Gibco BRL) per μl. This mixture was placed in a GeneAmp PCR system 9600 (Perkin-Elmer Cetus) thermal cycler and subjected to a 5-min denaturation step at 97°C followed by 30 cycles at 97°C for 45 s, 53°C for 30 s, and 72°C for 90 s. The reaction mixture was then held at 72°C for 10 min. The oligodeoxyribonucleotide primers used to amplify the lytB gene were lytB-5′ (5′GGGGTACCATCCAAGTTGGGGCGATCGG3′) and lytB-3′ (5′GCTCTAGAGCGGATAGCGTTTTGTTGCC3′). The PCR conditions were the same as those described above except that the annealing step was performed at 55°C rather than 53°C. The primer sequences used for the amplification of the udg gene were udg2-5′ (5′GGGTACCAGCCGGGCGGACGCCGTTCG3′) and udg2-3′ (5′GCTCTAGAGACTTCGCGATCTGCTCGCG3′). The PCR conditions were the same as those used for amplification of the waaF gene. The PCR products were cloned into pCR2.1-TOPO (Invitrogen) by using the TOPO TA Cloning Kit (Invitrogen). The cloned PCR products were sequenced to confirm that the desired gene was obtained. Each gene was then cloned into a broad-host-range vector, either pUCP28T (waaF) or pRK415 (lytB and udg).
Complementation of allelic exchange mutants.
Broad-host-range vectors (either pUCP28T or pRK415) containing wild-type copies of the waaF, lytB, and udg genes were used for complementation analyses. E. coli SM10 λ pir strains containing the appropriate vectors were conjugated to B. pseudomallei MB100, MB203, or MB300, followed by selection on appropriate antibiotics. MICs and LPS profiles were then determined for the complemented strains.
DNA sequencing and sequence analysis.
DNA sequencing of both strands was performed by University Core DNA Services (University of Calgary). Fragments that were sequenced were further analyzed by performing database searches to establish homology to known gene sequences with the gapped BLASTX and BLASTP programs (2). DNA and protein sequences were analyzed by using DNASIS version 2.5 (Hitachi).
Nucleotide sequence accession numbers.
The waaF gene sequence was submitted to GenBank under accession no. AF097748. The lytB gene sequence was submitted under accession no. AF098521. The udg, waaE, and gmhD gene sequences were submitted to GenBank under accession no. AF159428.
RESULTS
Isolation of PMB-susceptible mutants.
In order to identify specific genes necessary for resistance of B. pseudomallei to PMB, the virulent clinical isolate 1026b was mutagenized with the transposon Tn5-OT182. Transposon mutants were replica plated onto LB agar with TET (50 μg/ml) and onto LB agar with 200 μg of PMB per ml. A total of 17,000 colonies from five separate mutagenesis experiments were screened, and five PMB-susceptible mutants were obtained. These mutants were designated PMB-4, -7, -14, -17, and -20. The MICs for the PMB-susceptible mutants were determined and proved to be 500- to 4,000-fold lower than those for the parent strain 1026b (Table 2). Additionally, for all of the PMB-susceptible mutants, the colistin (polymyxin E) and PMB MICs were similar (Table 2). The MIC for B. pseudomallei SRM 117, carrying a Tn5-OT182 mutation in the wbiI gene and lacking the O-antigen moiety of the type II LPS, was determined to be similar to that for 1026b in the agar plate dilution assay. The MIC for B. thailandensis E264, an organism that possesses only type II LPS, was the same as that for B. pseudomallei 1026b.
TABLE 2.
MICs for B. pseudomallei and B. thailandensis strains in this study
| Strain | MIC
|
||||||
|---|---|---|---|---|---|---|---|
| PMB (μg/ml) | Colistin (μg/ml) | GEN (μg/ml) | KAN (μg/ml) | Tobramycin (μg/ml) | Benalkonium chloride (μg/ml) | SDS (%) | |
| 1026b | >128,000 | >128,000 | >256 | 32 | 48 | 64 | 0.12 |
| E264 | >128,000 | >128,000 | >256 | 48 | 48 | 64 | 0.12 |
| DD503 | >10,000 | >10,000 | 1.5 | 0.75 | 1.5 | 64 | 0.12 |
| SRM 117 | >10,000 | >10,000 | NDa | ND | ND | 64 | 0.12 |
| PMB-7 | 256 | 512 | 64 | 3 | 8 | 32 | 0.06 |
| PMB-17 | 128 | 128 | <1 | 32 | 8 | 32 | 0.12 |
| PMB-14 | 128 | 256 | 32 | 32 | 12 | 32 | 0.06 |
| PMB-4 | 32 | 128 | 64 | 32 | 16 | 32 | 0.12 |
| PMB-20 | 64 | 128 | 2 | 6 | <1 | 32 | 0.06 |
| MB100 | 256 | 256 | —b | — | — | ND | ND |
| MB203 | 128 | 256 | — | — | — | ND | ND |
| MB300 | 64 | 128 | — | — | — | ND | ND |
| MB301 | 64 | 32 | — | — | — | ND | ND |
| MB100C | 5,000 | 10,000 | — | — | — | ND | ND |
| MB203C | 512 | 1024 | — | — | — | ND | ND |
| MB300C | 2,500 | 5,000 | — | — | — | ND | ND |
ND, not determined.
—, not determined because these strains were constructed in DD503, which is susceptible to the aminoglycosides due to a deletion in the amrRAB-oprA operon.
The MICs of a number of antimicrobial substances, including GEN, tobramycin, KAN, benzalkonium chloride, and SDS, were also decreased for the PMB-susceptible mutants (Table 2). These mutants did not show increased susceptibility to the action of cationic peptides such as protamine sulfate and HNP-1 (data not shown).
DPX interacts with lipid A of PMB-susceptible mutants.
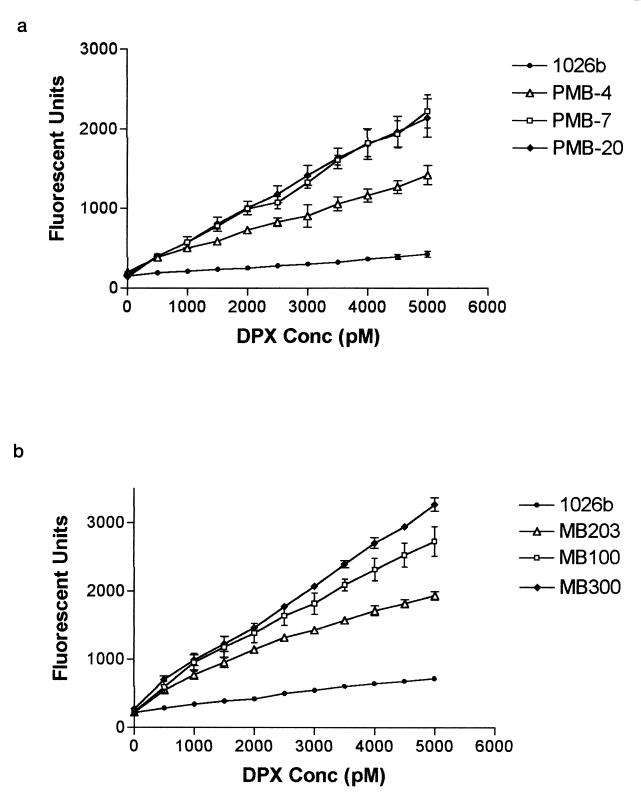
In order to explore the interaction of polymyxin with lipid A moieties, DPX binding assays were conducted with B. pseudomallei 1026b, PMB-7, PMB-20, and PMB-4 (a representative LytB mutant). The PMB-susceptible mutants bound significantly more DPX, as indicated by an increase in fluorescence compared to that for the wild type (Fig. 1a). Allelic exchange mutants MB100, MB203, and MB300 also showed increased binding of DPX (Fig. 1b). These results indicate that the DPX is able to permeabilize the outer membrane and interact with the lipid A moieties and phospholipids of the Tn5-OT182 and allelic exchange mutants.
FIG. 1.
DPX binding assay results. (a) DPX binding of 1026b and Tn5-OT182 mutants. (b) DPX binding of 1026b and allelic exchange mutants. Conc, concentration. Error bars indicate standard deviations.
Analysis of DNA flanking Tn5-OT182 integrations in PMB-susceptible mutants.
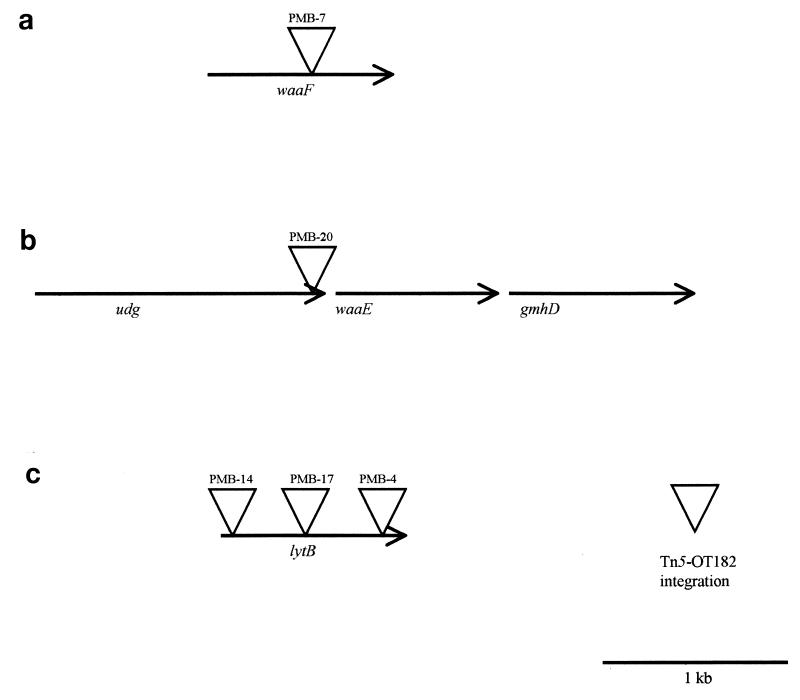
The DNA sequences immediately flanking Tn5-OT182 integrations in all five mutants were obtained by self-cloning (12, 24). The resultant plasmids were sequenced, and database searches for homology to known gene sequences were performed. The Tn5-OT182 integrations in PMB-7, PMB-20, and PMB-4, -14, and -17 map to three physically distinct loci (Table 3; Fig. 2). Each gene was fully sequenced through primer walking or subcloning, allowing for the identification of putative start codons and ribosome binding sites for each open reading frame.
TABLE 3.
Homology search results for DNA sequences flanking Tn5-OT182 integrations in this study
| Gene (product) | Gene size (bp) | Similar gene products (species) | Putative function | GenBank accession no. |
|---|---|---|---|---|
| waaF (WaaF) | 1,040 | Heptosyl transferase II (P. aeruginosa) | Heptosyl II transferase | U70983 |
| ADP-heptose:heptosyl transferase homolog (P. aeruginosa) | U63816 | |||
| ADP-heptose:heptosyl transferase II (Neisseria gonorrhoeae) | Z37141 | |||
| RfaF (R. solanacaerum) | X95498 | |||
| ADP-heptose–LPS heptosyltransferase II (E. coli) | AE000440 | |||
| udg (Udg) | 1,401 | UDP-glucose dehydrogenase (Xanthomonas campestris) | UDP-glucose dehydrogenase | X79772 |
| UDP-glucose dehydrogenase (Sinorhizobium meliloti) | AJ222661 | |||
| Nucleotide sugar dehydrogenase (Aquifex aeolicus) | AE000669 | |||
| UDP-glucose dehydrogenase (B. subtilis) | AF015609 | |||
| Product similar to P. aeruginosa GDP-mannose-6-dehydrogenase protein | Z92952 | |||
| waaE (WaaE) | 924 | Autotrophic growth protein homolog (Haemophilus influenzae) | ADP-heptose synthase | U32828 |
| ADP-heptose synthase (H. influenzae) | U17642 | |||
| Putative kinase (E. coli) | AE000387 | |||
| Probable LPS core synthesis (V. cholerae) | AB012957 | |||
| ADP-heptose synthase (A. aeolicus) | AE000696 | |||
| gmhD (GmhD) | 993 | ADP-l-glycero-d-mannoheptose (N. gonorrhoeae) | ADP-glyceromannoheptose epimerase | L07845 |
| ADP-l-glycero-d-mannoheptose-6-epimerase (E. coli) | X54492 | |||
| ADP-l-glycero-d-mannoheptose-6-epimerase (S. typhimurium) | U06472 | |||
| ADP-l-glycero-d-mannoheptose-6-epimerase (H. influenzae) | F64183 | |||
| ADP-l-glycero-d-mannoheptose-6-epimerase (Vibrio cholerae) | X90547 | |||
| lytB (LytB) | 941 | LytB (E. coli) | Regulator of the relA gene product | AE000113 |
| Hypothetical protein HI1007 (H. influenzae) | A64164 | |||
| LytB (H. influenzae) | U32781 | |||
| LytB (Mycobacterium tuberculosis) | AL009198 | |||
| LytB′ (M. tuberculosis) | AL021897 |
FIG. 2.
Map of genetic loci involved in PMB resistance. Locations of Tn5-OT182 integrations in the mutants PMB-7, PMB-20, PMB-4, PMB-14, and PMB-17 are shown. (a) waaF gene, with approximate position of the Tn5-OT182 integration. (b) udg, waaE, and gmhD genes, with approximate position of the Tn5-OT182 integration in the udg gene. The direction of transcription of these genes relative to one another is shown by arrows. (c) lytB gene, with approximate positions of the three different Tn5-OT182 integrations.
PMB-7 has a Tn5-OT182 integration in a gene whose product shows homology to heptosyl transferases of a number of gram-negative bacteria, specifically the waaF (rfaF) gene of Pseudomonas aeruginosa. Previous studies have shown that a mutation in the waaF homolog of B. pseudomallei results in the loss of type II O-PS and serum susceptibility (13). Interestingly, it has been previously shown that the rfaF gene of Ralstonia solanacaerum plays an important role in resistance to the action of antimicrobial peptides produced by a variety of plants (41).
PMB-20 has a Tn5-OT182 integration in an open reading frame whose product showed homology to UDP-glucose dehydrogenases of other species. The pmrE locus of Salmonella typhimurium (formerly pagA or udg) has been predicted to encode a UDP-glucose dehydrogenase and has recently been implicated in the resistance of S. typhimurium to PMB (16). This locus is involved in complex carbohydrate and capsule synthesis in a variety of bacterial species and is thought to be involved in LPS modifications that lead to PMB resistance in normally susceptible bacteria. In addition, the wlbA gene homolog of Bordetella bronchiseptica has been shown to be involved in cationic peptide resistance, and it is thought to encode a dehydrogenase involved in LPS biosynthesis (3). It is therefore not surprising that the udg gene homolog of wild-type B. pseudomallei may be in part responsible for conferring a PMB-resistant phenotype.
Analysis of the sequence immediately downstream of the udg gene revealed open reading frames whose products demonstrate homology to those of the waaE (rfaE) and gmhD (rfaD) genes from a variety of gram-negative bacteria. These genes are involved in LPS core biosynthesis, and it is possible that these genes may contribute to PMB resistance in this organism. It will be interesting to see whether these genes are part of an LPS core biosynthetic operon. It will also be useful to determine the arrangement of the waaE and gmhD genes with respect to the waaF gene.
PMB-4, -14, and -17 had Tn5-OT182 integrations in an open reading frame whose product shows homology to that of the lytB locus found in a variety of gram-negative bacteria. The product (LytB) has previously been shown to be involved in the stringent response (17) and has recently been identified as a regulator of the relA gene product (31). In addition, it has been shown to affect both peptidoglycan and phospholipid biosynthesis in E. coli (32, 33), likely leading to alterations in cell permeability.
Genetic loci required for a PMB-resistant phenotype.
In order to confirm the role of each genetic locus identified in the PMB-susceptible mutants, isogenic mutants were constructed by allelic exchange. We found that insertional inactivation of the waaF, lytB, udg, and waaE genes leads to a PMB-susceptible phenotype. MICs for each of the mutants were similar to those for the corresponding Tn5-OT182 mutants (Table 2).
In strain MB100, the waaF gene was insertionally inactivated by using an oriZeo cassette. This gene encodes a protein similar to a heptosyl transferase involved in LPS inner core biosynthesis (11, 37), resulting in a strain with an altered LPS profile. The PMB MIC is the same as that for PMB-7. In MB203, the lytB gene was inactivated through the insertion of an oriTp cassette. This resulted in a PMB MIC that is significantly reduced compared to that for the parent strain and was determined to be similar to that for the Tn5-OT182 mutants. This confirmed that disruption of the lytB gene was responsible for the observed phenotype. Inactivation of the udg locus resulted in strain MB300. The MIC for this strain was also decreased compared to that for the wild type.
Insertional inactivation of the waaE locus resulted in strain MB301. Studies confirmed an MIC similar to that for MB300, suggesting that these genes may be coordinately regulated and expressed. Additional sequence analysis in this region of the B. pseudomallei genome along with specific inactivation of each gene identified is necessary to determine if, in fact, core oligosaccharide genes form an operon. In any case, it is clear that genes affecting LPS core biosynthesis in this organism are involved in maintenance of a PMB-resistant phenotype.
Complementation.
Complementation analyses further support the fact that the waaF, lytB, and udg genes play a critical role in maintenance of a PMB-resistant phenotype. Complementation in trans with wild-type copies of the waaF gene, the lytB gene, or the udg gene partially restored the phenotypes of the allelic exchange mutants MB100, MB203, and MB300, respectively, to various degrees; the complemented mutants were designated MB100C, MB203C, and MB300C (Table 2).
Mutations in the waaF, udg, and waaE genes result in alterations of LPS moieties.
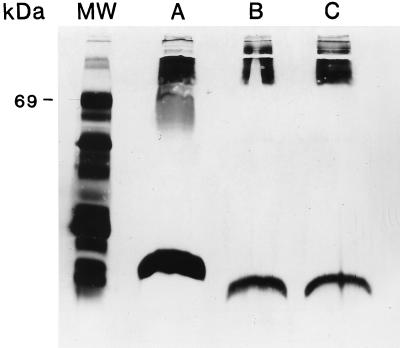
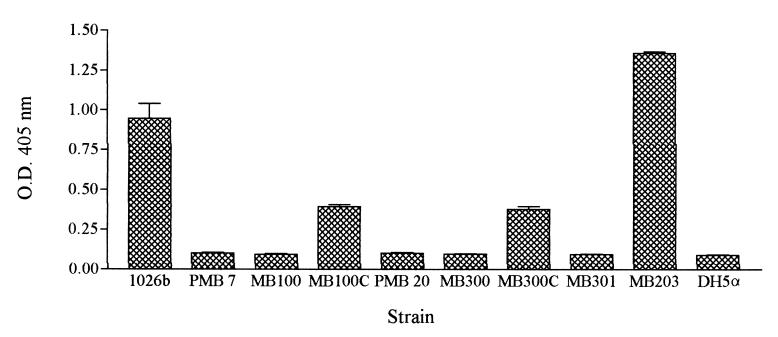
Silver stain analysis of PMB-7 and MB100 revealed alteration of the LPS type II moiety, specifically, truncation of the core oligosaccharide-lipid A region (Fig. 3). ELISAs and Western blot analyses of both purified LPS and whole cells of PMB-7 demonstrated a loss of LPS type II O-PS (Fig. 4 and 5). Figure 4 shows the lack of LPS type II O antigen in PMB-7 as indicated by decreased optical density. In addition, 13C nuclear magnetic resonance analysis demonstrated that PMB-7 had only LPS type I O antigen (data not shown). These results indicate that a disruption in the waaF gene leads to production of an incomplete LPS type II molecule.
FIG. 3.
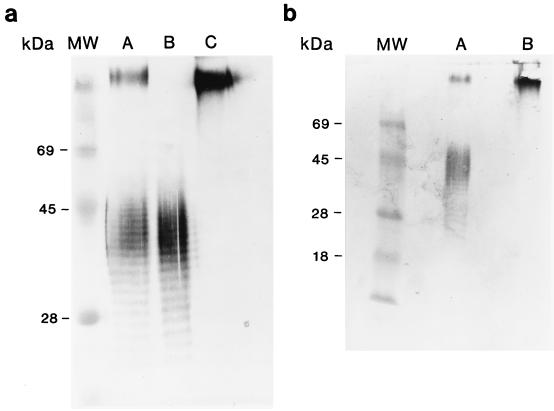
Silver stain analysis of 1026b, PMB-7, and MB100. Approximately 50 μl from overnight cultures of 1026b (lane A), PMB-7 (lane B), and MB100 (lane C) was proteinase K treated, boiled, and silver stained. The power supply was shut off with the dye front approximately 2 cm from the bottom of the gel. Lane MW, protein molecular mass standards (Gibco BRL) (the apparent molecular mass of bovine serum albumin is marked).
FIG. 4.
ELISA with type II O-PS-specific MAb. The values are the means and standard deviations from a single experiment performed in triplicate. O.D. 405 nm, optical density at 405 nm.
FIG. 5.
(a) Immunoblot analysis of LPSs from 1026b, E264, and PMB-7. Approximately 3 μg of purified LPS from 1026b (lane A), 3 μg of purified LPS from E264 (lane B), and 30 μg of purified LPS from PMB-7 (lane C) were reacted with a 1:250 dilution of polyclonal rabbit sera containing antibodies specific for type I and II O-PS. (b) Immunoblot analysis of LPSs from 1026b and PMB-20. Approximately 50 μl from overnight cultures of 1026b (lane A) and PMB-20 (lane B) was proteinase K treated, boiled, and reacted with a 1:250 dilution of polyclonal rabbit sera containing antibodies specific for type I and II O-PS. Lanes MW, prestained protein molecular mass standards (Gibco BRL).
Analysis of whole cells of PMB-20 clearly demonstrates the loss of LPS type II O-PS in both ELISA and Western blot analyses (Fig. 4 and 5). Several attempts were made to purify PMB-20 LPS by the previously described methods, but without success; it is suspected that the LPS molecules of this mutant are deep rough moieties and are therefore too small to purify by the method used for PMB-7. MB300 and MB301 also demonstrate a lack of LPS type II O antigen (Fig. 4).
It is clear from the analysis of the LPS profiles of PMB-7 and MB100 that mutations causing LPS to become truncated in the core region lead to PMB susceptibility. This is clearly exhibited by SRM 117, a mutant lacking only the LPS II O-PS and maintaining an intact core oligosaccharide region (13); SRM 117 is not susceptible to the action of PMB. Therefore, alterations in LPS moieties such that both the outer core and O antigen are lost are necessary to increase susceptibility to the action of PMB.
The complemented mutants MB100C and MB300C demonstrated increased optical density in the ELISA, indicating that LPS type II O antigen was expressed in these mutants, although not to the level in the parent strain (Fig. 4). In addition, these strains clearly showed an LPS type II O-antigen banding pattern in Western blot analysis (data not shown).
PMB-susceptible mutants have altered outer membrane protein profiles.
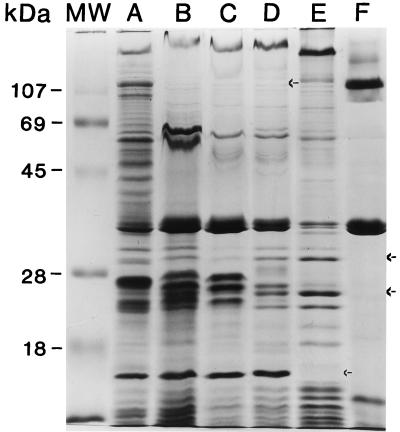
All of the PMB-susceptible mutants isolated showed alterations in their outer membrane protein profile compared to wild-type B. pseudomallei 1026b (Fig. 6). PMB-4, -14, and -17 are all missing a protein band with a molecular mass of 110 kDa. PMB-7 clearly lacks a well-conserved band at approximately 15 kDa, and PMB-20 is missing a number of bands throughout its outer membrane protein profile. These changes potentially affect the permeability of the bacterial cell by compromising its effectiveness as a barrier. The outer membranes of the allelic exchange mutants were not analyzed.
FIG. 6.
Outer membrane protein preparations. Approximately 20 μg of protein was solubilized in sample buffer with β-mercaptoethanol and boiled for 5 min. Lane A, 1026b; lane B, PMB-4; lane C, PMB-14; lane D, PMB-17; lane E, PMB-7; lane F, PMB-20; lane MW, prestained protein molecular mass standards (Gibco BRL). Arrows indicate the positions of apparent missing protein bands.
DISCUSSION
B. pseudomallei is intrinsically resistant to a variety of antibiotics, including most β-lactams, aminoglycosides, and polymyxins (20). Despite aggressive therapy with antimicrobials effective against this pathogen, the mortality rate due to acute septicemic B. pseudomallei infection remains unacceptably high (10). The ability of B. pseudomallei to survive intracellularly and to resist the killing action of many antibiotics, including cationic antimicrobial peptides, is clearly important in the pathogenesis of this infection. In this study, we have isolated PMB-susceptible mutants of B. pseudomallei and identified genetic loci involved in the resistance phenotype of wild-type B. pseudomallei.
We have found that B. pseudomallei 1026b survived and multiplied at all achievable concentrations of PMB. The data shown here indicate that PMB is unable to permeabilize the outer membrane of wild-type B. pseudomallei 1026b. The mutants obtained in this study possess alterations such that PMB had an increased ability to permeabilize their outer membranes, as shown by the DPX binding assay results. In addition, for these mutants the MICs of a number of other antimicrobial compounds were decreased, suggesting increased permeability of the bacterial outer membranes.
In these studies two distinct groups of mutants were identified: (i) those with disruptions in genes predicted to be involved in LPS core oligosaccharide biosynthesis, resulting in incomplete LPS molecules, and (ii) those with disruptions in the lytB gene. Both groups of mutants exhibited differences in their outer membrane protein profiles compared to wild-type B. pseudomallei, suggesting that the integrity of this permeability barrier may be compromised. In addition, it is well established that bacterial LPS is a permeability barrier that confers resistance to a variety of antimicrobial agents (30). Alteration of this barrier often leads to increased sensitivity to hydrophobic and cationic compounds (30).
The data collected in the present study indicates that it is the architecture of the B. pseudomallei cell which prevents PMB molecules from interacting with target groups on LPS molecules and phospholipids. It seems that the O-antigen and outer core components of B. pseudomallei LPS act as a protective barrier in order to prevent PMB molecules from interacting with potential binding sites found in the inner core and lipid A regions of the moiety. This is clearly seen in mutants of B. pseudomallei that possess incomplete LPS molecules, specifically those with disruptions in genes involved in LPS core biosynthesis. Thus, in order to achieve a PMB-susceptible phenotype in this organism, LPS alterations must be such that both outer core and O-antigen moieties of LPS type II are absent.
It appears that there may be another mechanism of PMB resistance that is LPS independent. This is seen in the mutants that contain mutations in their lytB genes. These mutants seem to possess a wild-type LPS profile, as no obvious changes were identified, yet the PMB MICs are significantly reduced. This may be explained in part by the fact that the lytB gene product affects phospholipid and peptidoglycan biosynthesis (32, 33), leading to changes in membrane permeability. Possible factors affecting membrane integrity include changes in phospholipid arrangement and stability as well as the destabilization of cross bridges between neighboring LPS molecules. In any case, there are complex changes that occur due to disruption of the lytB gene, leading to PMB susceptibility. These changes are not yet fully understood and are being further investigated.
Following screening for PMB-susceptible mutants of B. pseudomallei, we have confirmed that LPS core biosynthesis genes are implicated in cationic peptide resistance, and this is consistent with the findings of a number of previous studies (1, 3, 41). Further investigation is required in order to determine the exact lengths of the truncated LPS molecules, the presence or absence of potential PMB binding sites, and the charges on the LPS molecules. With this information it can be assessed whether PMB and possibly other antibiotics can efficiently attack the membrane of B. pseudomallei cells. In addition, we have identified a possible LPS-independent mechanism of PMB resistance, but the exact nature of this susceptibility requires further attention.
Topics for future studies include investigation of the arrangement of the LPS core oligosaccharide biosynthesis genes and their functions. Also, the role of the lytB gene in B. pseudomallei pathogenesis has yet to be determined. In addition, the mechanism(s) responsible for the resistance to other cationic peptides, such as protamine sulfate, poly-l-lysine, and HNP-1, in B. pseudomallei is currently being studied. It is hoped that these studies will lead to the elucidation of a novel mechanism(s) of cationic peptide resistance that will lead to the identification of new targets of therapy against this organism.
ACKNOWLEDGMENTS
This work was funded by the Canadian Bacterial Diseases Network Centers for Excellence Program. M.N.B. is the recipient of an Alberta Heritage Foundation for Medical Research Studentship Award.
We are grateful to Malcolm B. Perry for 13C nuclear magnetic resonance results.
REFERENCES
- 1.Allen C A, Adams L G, Ficht T A. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect Immun. 1998;66:1008–1016. doi: 10.1128/iai.66.3.1008-1016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altshul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banemann A, Deppeisch H, Gross R. The lipopolysaccharide of Bordetella brochiseptica acts as protective shield against antimicrobial peptides. Infect Immun. 1998;66:5607–5612. doi: 10.1128/iai.66.12.5607-5612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett P J, Woods D E. Structural and immunological characterization of Burkholderia pseudomallei O-polysaccharide–flagellin protein conjugates. Infect Immun. 1996;64:2824–2828. doi: 10.1128/iai.64.7.2824-2828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett P J, DeShazer D, Woods D E. Burkholderia thailandensis sp. nov., description of a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 6.Brett, P. J., D. DeShazer, and D. E. Woods. Self-cloning cassette vectors. Unpublished.
- 7.Bryan L E, Wong S, Woods D E, Dance D A B, Chaowagul W. Passive protection of diabetic rats with antisera specific for the polysaccharide portion of the lipopolysaccharide from Pseudomonas pseudomallei. Can J Infect Dis. 1994;5:170–178. doi: 10.1155/1994/856850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaowagul W, White N J, Dance D A B, Wattanagoon Y, Naigowit P, Davis T M E, Looareesuwan S, Pitawatchara N. Melioidosis: a major cause of community acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 9.Dance D A B. Melioidosis: tip of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dance D A B. Melioidosis. In: Cook G C, editor. Manson’s tropical diseases. 20th ed. London, England: W. B. Saunders Co. Ltd.; 1996. pp. 925–930. [Google Scholar]
- 11.De Kievet T R, Lam J S. Isolation and characterization of two genes, waaC (rfaC) and waaF (rfaF), involved in Pseudomonas aeruginosa serotype O5 inner-core biosynthesis. J Bacteriol. 1997;179:3451–3457. doi: 10.1128/jb.179.11.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeShazer D, Brett P J, Carlyon R, Woods D E. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J Bacteriol. 1997;179:2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeShazer D, Brett P J, Woods D E. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 14.Eickhoff T C, Bennett J V, Hayes P S, Feeley J. Pseudomonas pseudomallei: susceptibility to chemotherapeutic agents. J Infect Dis. 1970;121:95–102. doi: 10.1093/infdis/121.2.95. [DOI] [PubMed] [Google Scholar]
- 15.Gotoh N, White N J, Choawagul W, Woods D E. Isolation and characterization of the outer-membrane proteins of Burkholderia (Pseudomonas) pseudomallei. Microbiology. 1994;140:797–805. doi: 10.1099/00221287-140-4-797. [DOI] [PubMed] [Google Scholar]
- 16.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin B resistance. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 17.Gustafson C E, Kaul S, Ishiguro E E. Identification of the Escherichia coli lytB gene, which is involved in penicillin tolerance and control of the stringent response. J Bacteriol. 1993;175:1203–1205. doi: 10.1128/jb.175.4.1203-1205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho M, Schollaardt T, Smith M D, Perry M B, Brett P J, Chaowagul W, Bryan L E. Specificity and functional activity of anti-Burkholderia pseudomallei polysaccharide antibodies. Infect Immun. 1997;65:3648–3653. doi: 10.1128/iai.65.9.3648-3653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howe C, Sampath A, Spotnitz M. The pseudomallei group: a review. J Infect Dis. 1971;124:598–606. doi: 10.1093/infdis/124.6.598. [DOI] [PubMed] [Google Scholar]
- 21.Jones A L, Beveridge T J, Woods D E. Intracellular survival of Burkholderia pseudomallei. Infect Immun. 1996;64:782–790. doi: 10.1128/iai.64.3.782-790.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 23.Leelarasamee A, Bovornkitti S. Melioidosis: review and update. Rev Infect Dis. 1989;11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 24.Merriman T R, Lamont I L. Construction and use of a self cloning promoter probe vector for Gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 25.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore R A, Hancock R E W. Involvement of outer membrane of Pseudomonas cepacia in aminoglycoside and polymyxin resistance. Antimicrob Agents Chemother. 1986;30:923–926. doi: 10.1128/aac.30.6.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore R A, Bates N C, Hancock R E W. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied using dansyl-polymyxin. Antimicrob Agents Chemother. 1986;29:496–500. doi: 10.1128/aac.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 2nd ed. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 30.Nikaido H. Outer membrane barrier as a mechanism of antimicrobial resistance. Antimicrob Agents Chemother. 1989;33:1831–1836. doi: 10.1128/aac.33.11.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potter S, Yang X, Boulanger M J, Ishiguro E E. Occurrence of homologs of the Escherichia coli lytB gene in gram-negative bacterial species. J Bacteriol. 1998;180:1959–1965. doi: 10.1128/jb.180.7.1959-1961.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodinov D G, Pisabarro A G, De Pedro M A, Kusser W, Ishiguro E E. β-Lactam-induced bacteriolysis of amino acid-deprived Escherichia coli is dependent on phospholipid synthesis. J Bacteriol. 1995;177:992–997. doi: 10.1128/jb.177.4.992-997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodinov D G, Ishiguro E E. Dependence of peptidoglycan metabolism on phospholipid synthesis during growth of Escherichia coli. Microbiology. 1996;142:2871–2877. doi: 10.1099/13500872-142-10-2871. [DOI] [PubMed] [Google Scholar]
- 34.Schweizer H P, Klassen T, Hoang T. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: American Society for Microbiology; 1996. pp. 229–237. [Google Scholar]
- 35.Sexton M M, Goebel L A, Godfrey A J, Chaowagul W, White N J, Woods D E. Ribotype analysis of Pseudomonas pseudomallei isolates. J Clin Microbiol. 1993;31:238–243. doi: 10.1128/jcm.31.2.238-243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 37.Sirisena D M, MacLachlan P R, Liu S L, Hessel A, Sanderson K E. Molecular analysis of the rfaD gene, for heptose synthesis, and the rfaF gene, for heptose transfer, in lipopolysaccharide synthesis in Salmonella typhimurium. J Bacteriol. 1994;176:2379–2385. doi: 10.1128/jb.176.8.2379-2385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 39.Smith C J, Allen J C, Embi M N, Othman O, Ratzak N, Ismail G. Human melioidosis: an emerging medical problem. MIRCEN. 1987;3:343–366. [Google Scholar]
- 40.Smith J A. Electrophoretic separation of proteins. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 10.2.1–10.2.7. [Google Scholar]
- 41.Titarenko E, Lopez-Solanilla E, Garcia-Olmedo F, Rodriguez-Palenzuela P. Mutants of Ralstonia (Pseudomonas) solanacearum sensitive to antimicrobial peptides are altered in their lipopolysaccharide structure and are avirulent in tobacco. J Bacteriol. 1997;179:6699–6704. doi: 10.1128/jb.179.21.6699-6704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai C, Frasch C E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 43.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. pp. 2.4.1–2.4.5. [DOI] [PubMed] [Google Scholar]