Abstract
The effects of itraconazole on ergosterol biosynthesis were investigated in a series of 16 matched clinical Candida albicans isolates which had been previously analyzed for mechanisms of resistance to azoles (D. Sanglard, K. Kuchler, F. Ischer, J. L. Pagani, M. Monod, and J. Bille, Antimicrob. Agents Chemother., 39:2378–2386, 1995). Under control conditions, all isolates contained ergosterol as the predominant sterol, except two strains (C48 and C56). In isolates C48 and C56, both less susceptible to azoles than their parent, C43, substantial concentrations (20 to 30%) of 14α-methyl-ergosta-8,24(28)-diene-3β,6α-diol (3,6-diol) were found. Itraconazole treatment of C43 resulted in a dose-dependent inhibition of ergosterol biosynthesis (50% inhibitory concentration, 2 nM) and accumulation of 3,6-diol (up to 60% of the total sterols) together with eburicol, lanosterol, obtusifoliol, 14α-methyl-ergosta-5,7,22,24(28)-tetraene-3βol, and 14α-methyl-fecosterol. In strains C48 and C56, no further increase of 3,6-diol was observed after exposure to itraconazole. Ergosterol synthesis was less sensitive to itraconazole inhibition, as was expected for these azole-resistant isolates which overexpress ATP-binding cassette transporter genes CDR1 and CDR2. In addition to 3,6-diol, substantial amounts of obtusifolione were found after exposure to itraconazole. This toxic 3-ketosteroid was demonstrated previously to accumulate after itraconazole treatment in Cryptococcus neoformans and Histoplasma capsulatum but has not been reported in Candida isolates. Accumulation of obtusifolione correlated with nearly complete growth inhibition in these azole-resistant strains compared to that found in the susceptible parent strain, although the onset of growth inhibition only occurred at higher concentrations of itraconazole. ERG25 and ERG26 are the only genes assigned to the 4-demethylation process, of which the 3-ketoreductase is part. To verify whether mutations in these ERG25 genes contributed to obtusifolione accumulation, their nucleotide sequences were determined in all three related isolates. No mutations in ERG25 alleles of isolates C48 and C56 were found, suggesting that this gene is not involved in obtusifolione accumulation. The molecular basis for the accumulation of this sterol in these two strains remains to be established.
The incidence of fungal infections has increased during the last decade. Only a few classes of antifungal compounds are available to treat these infections. One important class consists of inhibitors of ergosterol biosynthesis. These include the allylamines, inhibitors of squalene epoxidase; the morpholines, which inhibit both the Δ14-reductase and the Δ8,7-isomerase; and the imidazoles and triazoles (azoles), which interfere with the cytochrome P-450s catalyzing the lanosterol 14α-demethylase and sterol Δ22-desaturation. In patients with impaired immune responses, e.g., AIDS patients, neutropenic patients, and patients receiving bone marrow or organ transplants, a higher incidence of nonresponse to treatment is found (40). Treatment failures may result from host-related factors, abnormal drug pharmacokinetics, or resistance of the infecting fungus to the agent used. Over the last 5 years, many studies have been published to elucidate the underlying causes of clinical resistance to azoles. These studies have been extensively reviewed (2–4, 6, 9, 11, 13, 15, 21, 25, 26, 38–40, 44). Several mechanisms have been identified that contribute to fungal resistance to azoles. Probably the most common mechanism is to effect a diminution of the active-compound concentration at the target site. In the majority of cases studied recently, this was the result of overexpression of efflux pumps (8, 20, 27), but in earlier studies, permeability changes in the plasma membrane were also found (12). Two types of efflux transporters have been reported. The ATP-binding cassette type (ABC transporters, e.g., CDR1 and CDR2) can export a wide variety of azoles and unrelated chemicals, including antifungal sterol synthesis inhibitors such as amorolfine and terbinafine, which do not inhibit the action of cytochrome P-450s (18, 28, 29). This group of proteins uses the energy freed by the hydrolysis of ATP to effect the efflux of compounds out of the cell. The so-called major facilitators (e.g., CaMDR1, previously described as Benr) identified so far in fungi have a much narrower substrate spectrum, so that only hydrophilic azole compounds such as fluconazole are exported whereas lipophilic azoles such as itraconazole are not affected (27). The energy for this second class of pump is provided by the proton gradient across the membrane.
A second general type of azole resistance mechanism involves changes at the level of the antifungal target. The primary target for azole antifungals is the cytochrome P-450-catalyzed 14α-demethylation of ergosterol precursors, encoded by ERG11 (also called ERG16 or CYP51). Overexpression of this enzyme, induced either by enhanced transcription or by gene or chromosomal amplification, results in decreased susceptibility to azole antifungals (5, 22). Point mutations in ERG11, such as Y132H (tyrosine 132 is replaced with histidine), T315A (threonine 315 is replaced with alanine), or R476K (arginine 476 is replaced with lysine), have been shown to decrease the affinity of the target for azoles (19, 30, 43). Numerous publications have listed other ERG11 mutations but unfortunately do not include data on the effect of the mutation on azole sensitivity.
The third way in which fungi achieve effective resistance to azoles is to circumvent or compensate for the toxic consequences of the azole-induced depletion of ergosterol and concomitant accumulation of 14-methylated precursors. For example, it was shown in Saccharomyces cerevisiae that a strain deficient in Erg11p activity only survived in the presence of a defect in Δ5,6-desaturase, encoded by the ERG3 gene (16). ERG3-deficient strains of Candida albicans were found to be azole resistant (17). It was hypothesized that resistance to fluconazole was due to the combination of the presence of substantial quantities of 14α-methyl-fecosterol and the absence of 14α-methyl-ergosta-8,24(28)-diene-3β,6α-diol (3,6-diol).
Despite the numerous recent molecular genetic studies on azole resistance, little attention has been paid to the details of inhibition of the ergosterol biosynthetic pathway in clinical azole-resistant C. albicans strains. In this study, we have investigated the effects of itraconazole on ergosterol biosynthesis in a series of matched clinical isolates that were biochemically characterized previously in the pivotal study of Sanglard et al. (27).
MATERIALS AND METHODS
Strains.
The C. albicans strains used in this study originated at the Institute of Microbiology (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) and were isolated from AIDS patients with oropharyngeal Candida infections. Identification of the yeast isolates and their clonal relatedness were described by Sanglard et al. (27). Yeasts were maintained as glycerol stocks at −80°C. The inocula for each individual experiment were prepared from these glycerol stocks to minimize the possible influences of genotypic or phenotypic instability. Table 1 summarizes the biochemical characterization of the isolates.
TABLE 1.
Strains tested in this study and their previously publisheda resistance mechanisms
| Patient | Isolate | Efflux pumpb expression level | ERG11 point mutation(s) |
|---|---|---|---|
| I | C23 | S405F heterozygote | |
| I | C32 | No increase relative to C23 | No data |
| I | C39 | Increased CDR1 and CDR2 signals relative to C23 | S405F homozygote |
| II | C45 | ||
| II | C18 | No increase relative to C45 | |
| II | C46 | No increase relative to C45 | |
| III | C33 | ||
| III | C34 | No increase relative to C33 | S405F homozygote |
| III | C26 | Increased CDR1 and CDR2 signals relative to C33 | S405F and Y132H homozygote |
| III | C82 | Less pronounced increase in CDR1 and CDR2 signals relative to C33 | S405F homozygote |
| IV | C27 | ||
| IV | C37 | No increase relative to C27 | G464S and R467K homozygote |
| IV | C40 | Increased CaMDR1 signal relative to C27 | G464S and R467K homozygote, Y132H heterozygote |
| V | C43 | G129A heterozygote | |
| V | C48 | Increased CDR1 and CDR2 signals relative to C43 | No data |
| V | C56 | Increased CDR1 and CDR2 signals relative to C43 | G129A and G464S homozygote |
MIC determination.
To verify that the strains showed azole susceptibility phenotypes similar to the original published characteristics, MICs were determined spectrophotometrically by a broth microdilution method (24) based on the National Committee for Clinical Laboratory Standards (NCCLS) M27A protocol (23). The MIC was the lowest concentration that inhibited growth by more than 50%; this endpoint showed the best reproducibility and correlation with results from the NCCLS broth macrodilution method. Quality control yeasts Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were tested in parallel, and for these organisms the MICs of the antifungals tested were in the correct ranges (23).
Sterol synthesis experiments.
To study sterol synthesis in C. albicans, yeasts were grown in 100 ml of CYG medium (casein hydrolysate [Merck, Darmstadt, Germany], yeast extract [Difco, Detroit, Mich.], and glucose, each at a concentration of 5 g liter−1) in 500-ml Erlenmeyer flasks in a reciprocating shaker set at 100 strokes/min at 37°C as described before (33). Radiolabeled [2-14C]acetate (5 μCi; specific activity, 58 μCi mmol−1) was added immediately prior to inoculation. Itraconazole was dissolved in dimethyl sulfoxide (DMSO) (10 mM stock solution; 7.05 mg ml−1), further diluted in 100% DMSO, and added to the incubation mixtures at a final solvent concentration of 0.1%. In control experiments, a similar amount of DMSO was added. After 24 h of growth, cells were collected by centrifugation at 1,500 × g and the cell pellet was washed with saline (0.15 M NaCl) and homogenized with acid-washed glass beads (diameter, 0.40 to 0.45 mm) in a 20-ml scintillation vial in a Retsch laboratory mixer mill 2000 set at maximum speed for 5 min. The homogenate was quantitatively separated from the glass beads by sequential washing. The homogenates were supplemented with 1 volume of 15% KOH dissolved in 90% ethanol and saponified at 84°C. Nonsaponifiable lipids were extracted with 1 volume of n-heptane. The heptane extracts were scanned spectrophotometrically from 200 to 330 nm to quantify sterols with a Δ5,7-conjugated double-bond system. At a wavelength of 281 nm, Δ5,7-desaturated sterols could be specifically detected and quantified by using an ɛ281 of 11,200. The linearity of this method was checked against a standard curve obtained with authentic ergosterol. Heptane extracts were then dried with a stream of N2 and sterols were separated by both thin-layer chromatography and high-performance liquid chromatography analysis as described previously (36). Sterol fractions were identified by reference to authentic standards and by gas chromatography-mass spectrometry. To quantify the effect of itraconazole on growth under these conditions, cell concentrations were determined with a Coulter counter as described earlier (34). Cell clustering and size were examined with a Becton Dickinson FACScalibur flow cytometer and by phase-contrast microscopy.
PCR amplification and sequence analysis of C. albicans ERG25 and ERG3 genes.
To amplify the ERG25 gene encoding at least part of the 4-demethylase, we used primers adjacent to the open reading frame using the sequence deposited by Johnson et al. (GenBank accession no. AF051914). All subsequent numbering is according to this sequence. Vector NTi was used to detect optimal primer pairs to amplify the entire open reading frame. As a sense primer, 5′ATTGTTATATTTCAACATATACATATTCC3′ was used (nucleotides 7 to 35 of AF051914), whereas the antisense primer was 5′AAACATTGAGAAGTTGTACACATATACT3′ (nucleotides 1065 to 1038 of AF051914). Heat-activatable AmpliTaq Gold (Perkin-Elmer, Foster City, Calif.; 0.5 U) was used with 2.5 mM MgCl2. DNA from C. albicans strains was prepared by the Qiagen DNA extraction method according to the procedures of the manufacturer with Zymolyase (60 U; 5,000 U g−1; Arthrobacter lutens; Seikagaku Kogyo, Tokyo, Japan) used as the cell wall-degrading enzyme. The PCR parameters were 10 min at 94°C to activate the polymerase and then 30 cycles of 1 min of annealing at 48°C, 2 min of elongation at 72°C, and 1 min of denaturation at 92°C. After the reaction, the 1,059-bp PCR product was cleaned up with a Qiagen PCR cleanup kit and a sample was separated on a 1% agarose gel with Boehringer molecular weight standard VI. The 1,059-bp amplification products from the different isolates were sequenced on both strands by using the following PCR primers and internal primers every 300 bp (name, sequence, nucleotide position, and direction): Ca ERG25-01, TTCCATCCATTATGTC, 458, sense; Ca ERG25-02, CCGATTGTTTGGTGTC, 716, sense; Ca ERG25-03, GTTACCAGTGATAAGAC, 745, antisense; Ca ERG25-04, CATGGTAAACATCTACC, 318, antisense; and Ca ERG25-05, GTCTTCCATTAGTAATG, 103, sense. Primers were designed by visual inspection of the sequence for stretches of 16 to 18 nucleotides of normal composition (40 to 60% GC, no palindromes, no homopolymeric stretches). Primers were ordered from Eurogentec (Seraing, Belgium) and synthesized by the β-cyanoethylphosphoramidite method. Sequencing reactions were performed with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit used according to the instructions of the manufacturer (Perkin-Elmer), except that half of the volume of terminator mix was replaced with HalfTerm (GenPak Ltd., Brighton, United Kingdom). Sequencing reactions were run on an Applied Biosystems 377 XL DNA sequencer (Perkin-Elmer). Sequences were assembled from the individual runs into single contig sequences with the aid of the Sequencher software (Gene Codes Corporation, Ann Arbor, Mich.). Ambiguity positions were scored by setting the threshold as low as 30% (i.e., secondary peaks at 30% of the primary peak result in an ambiguity call) and by inspecting all of the ambiguity calls on all available readings. To amplify the ERG3 gene from the three related strains, similar techniques were used. Numbering of nucleotides is according to the sequence deposited by Miyazaki et al. (GenBank accession no. AF069752). As a sense primer, 5′ACAGTTTVVVATTTTCCTTCCAA3′ was used (nucleotides 208 to 230 of AF069752), whereas the antisense primer was 5′CATCTTTGTTTTGGACCATTGACTAGAGCTCHHH3′ (SacI restriction site plus nucleotides 1578 to 1554 of AF069752). PCR was performed as described above. The 1,380-bp amplification products were sequenced on both strands using the following PCR primers and internal primers every 300 bp (name, sequence, nucleotide position, and direction): Ca ERG3-01, CCCAGCTACTGATTTC, 683, sense; Ca ERG3-02; TGAAATCAGTAGCTGG, 699, antisense; Ca ERG3-03, TTACACTGGCCATCTG, 1071, sense; and Ca ERG3-04, GCATGAGAAGCAAATGG, 1150, antisense.
RESULTS
MIC determination.
In previous papers, Sanglard et al. described different resistance mechanisms in series of sequential isogenic C. albicans isolates from five AIDS patients with oropharyngeal candidiasis (27, 29, 31). The results from these studies are summarized in Table 1. To verify that no changes in susceptibility were induced during shipment or with the initial subcultivation to prepare the glycerol stock, MICs of fluconazole, ketoconazole, and itraconazole for all 16 isolates were redetermined. In addition, the amphotericin B sensitivity was also measured. The results obtained are summarized in Table 2. Taking the well-documented variation of azole susceptibility testing into account (23, 41), an excellent correlation between the two determinations with a correlation coefficient of 0.87 was found, indicating that the sensitivities of the strains were not significantly altered. Only minor differences in amphotericin B sensitivity were found among the 16 isolates, demonstrating that they were not cross-resistant to this polyene antifungal.
TABLE 2.
In vitro susceptibilities of C. albicans isolates
| Patient | Isolate | MIC (μg/ml) of:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Fluconazolea | Ketoconazolea | Itraconazolea | Fluconazole | Ketoconazole | Itraconazole | Amphotericin B | ||
| I | C23 | 1 | 0.015 | 0.0625 | 1.6 | 0.05 | 0.05 | 0.63 |
| I | C32 | 8 | 0.0312 | 0.125 | 6.3 | 0.1 | 0.2 | 0.63 |
| I | C39 | 32 | 0.125 | 0.125 | 25 | 0.2 | 0.4 | 0.63 |
| II | C45 | 1 | 0.015 | 0.0312 | 0.8 | 0.025 | 0.025 | 0.63 |
| II | C18 | 2 | 0.015 | 0.0625 | 0.4 | ≤0.025 | ≤0.025 | 0.63 |
| II | C46 | 2 | 0.015 | 0.0625 | 0.8 | ≤0.025 | 0.025 | 0.63 |
| III | C33 | 0.25 | 0.015 | 0.0312 | 0.2 | ≤0.025 | ≤0.025 | 0.32 |
| III | C34 | 2 | 0.015 | 0.0625 | 1.6 | 0.05 | 0.05 | 0.32 |
| III | C26 | >128 | 4 | >2 | >100 | 6.3 | >25 | 0.16 |
| III | C82 | 32 | 0.5 | 1 | 25 | 0.4 | 0.4 | 0.63 |
| IV | C27 | 1 | 0.015 | 0.0312 | 0.4 | ≤0.025 | 0.05 | 0.63 |
| IV | C37 | 8 | 0.0325 | 0.0625 | 6.3 | 0.1 | 0.05 | 0.63 |
| IV | C40 | 128 | 2 | 1 | 50 | 1.6 | 0.8 | 0.63 |
| V | C43 | 0.25 | 0.015 | 0.0312 | 0.2 | ≤0.025 | ≤0.025 | 0.63 |
| V | C48 | 64 | 2 | 1 | 100 | 0.4 | 0.4 | 0.63 |
| V | C56 | 128 | 4 | >2 | 100 | 1.6 | 0.8 | 0.63 |
MICs reported by Sanglard et al. (27).
Sterol synthesis experiments.
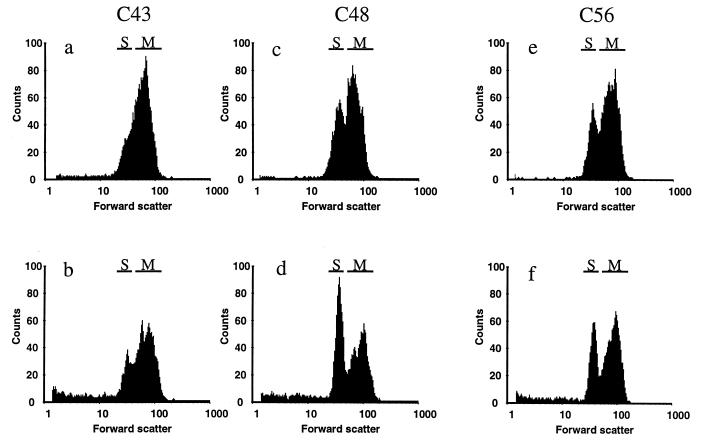
In a pilot study, the sterol compositions of all 16 isolates grown under control conditions were analyzed. The results are presented in Table 3. Under control conditions, most isolates, except C48, C56, and to a lesser extent C26, contained ergosterol as the only predominant sterol. In isolates C48 and C56, both related to C43, substantial accumulation of 3,6-diol at a level as high as 20 to 30% of the total sterols was found. In the related azole-sensitive isolate C43, this sterol comprised only 2.5% of the sterols isolated. This 14-methylated dihydroxysterol is normally found only after treatment with an azole as described initially by Ebert et al. in Ustilago maydis treated with etaconazole (7) and by Vanden Bossche et al. in C. albicans (34). The three isolates from patient V were selected for further sterol analysis, and the impact of itraconazole treatment on these isolates was investigated. In Table 4, the growth yields and the effects of itraconazole on the cell counts and ergosterol contents of the three isolates are summarized. The inhibition profiles for both of the parameters measured are shown in Fig. 1. Under control conditions, C43 reached a higher yield of cells as measured by Coulter counter. At stationary phase, C43 samples (100-ml culture) contained (201 ± 24) × 108 cells compared to (127 ± 10) × 108 cells for C48 and (111 ± 6) × 108 cells for C56. The Wilcoxon rank-sum statistical analysis suggests that isolates C48 and C56 behaved significantly differently from isolate C43. It could be argued that this decrease in yield was the result of a more pronounced clustering because the Coulter counter does not discriminate between single cells and multicellular clusters. To eliminate this possibility, we checked the size distribution of the cultures with flow cytometry and looked at the morphology in a microscope. In Fig. 2, the forward-scatter histograms are shown for all three isolates grown for 24 h in CYG medium. Under control conditions, both isolates C48 and C56 contained more single cells than did the C43 strain, represented on the graph by the S region. The positions of the multicellular particle region (M) were similar for all three isolates, indicating similar degrees of clustering. This was confirmed by the microscopical examination. The contents of Δ5,7diene-containing sterols (under control conditions, ergosterol for all three strains) extracted from the cultures were found to be similar in all three isolates (Table 4). This implies a higher cellular ergosterol content in C48 (82 fg · cell−1) and C56 (83 fg · cell−1) than in C43 (53 fg · cell−1). This latter value matches that published by Hitchcock et al. (12). These authors found total lipid and total sterol contents of 670 and 52 fg · cell−1, respectively, in C. albicans isolate A.
TABLE 3.
Sterols found in cultures of the C. albicans isolatesa
| Patient | Isolate | % of sterols
|
|||
|---|---|---|---|---|---|
| 3,6-Diol | Ergosterol | Obtusifoliol | Trimethylsterolsb | ||
| I | C23 | 2.4 | 90.7 | 2.5 | 4.4 |
| I | C32 | 2.5 | 91.7 | 1.6 | 4.2 |
| I | C39 | 1.6 | 91.9 | 1.0 | 5.5 |
| II | C45 | 1.5 | 89.9 | 4.3 | 4.3 |
| II | C18 | 3.4 | 88.6 | 3.8 | 4.2 |
| II | C46 | 4.9 | 89.6 | 2.6 | 2.9 |
| III | C33 | 1.9 | 91.2 | 1.9 | 5.0 |
| III | C34 | 0.5 | 90.6 | 2.4 | 6.5 |
| III | C26 | 8.1 | 89.4 | 0.0 | 2.5 |
| III | C82 | 3.2 | 89.1 | 2.4 | 5.3 |
| IV | C27 | 1.5 | 89.8 | 2.2 | 6.4 |
| IV | C37 | 5.4 | 87.0 | 2.6 | 5.0 |
| IV | C40 | 5.3 | 90.8 | 1.0 | 2.8 |
| V | C43 | 2.5 | 89.2 | 1.7 | 5.4 |
| V | C48 | 30.7 | 60.4 | 1.7 | 4.7 |
| V | C56 | 21.1 | 73.7 | 1.3 | 3.2 |
These results are from pilot experiments.
The trimethylsterol fraction is a mixture of lanosterol and eburicol.
TABLE 4.
Yield of cells and ergosterol content of a 100-ml culture under stationary growth conditions
| Method (units of measurement) | Avg yield ± SDa (itraconazole IC50 [μM])
|
||
|---|---|---|---|
| C43 | C48 | C56 | |
| Coulter counter (108 cells) | 201 ± 24 (0.03) | 127 ± 10b (0.07) | 111 ± 6b (0.1) |
| UV analysis (μg of Δ5,7-diene sterol) | 1,068 ± 125 (0.02) | 1,049 ± 169 (0.1) | 921 ± 81 (0.12) |
| Sterol/cell ratio (fg/cell) | 53 ± 8 | 82 ± 9 | 83 ± 9 |
Based on at least six measurements.
P < 0.05 as determined by Wilcoxon statistical analysis.
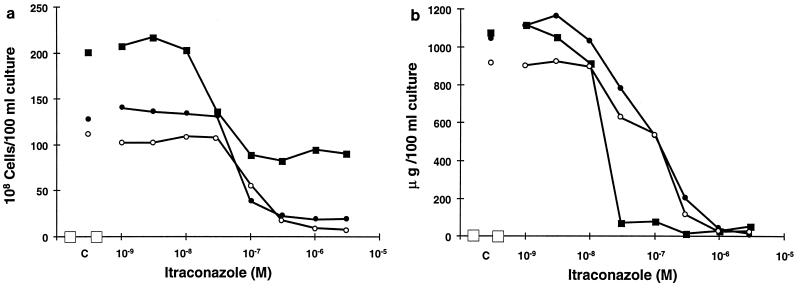
FIG. 1.
Effects of itraconazole on C. albicans C43 (■), C48 (●), and C56 (○) after 24 h in shaken culture in CYG medium. (a) Growth measured by Coulter counter. (b) Ergosterol content measured by determining the A281 of a heptane extract obtained from C43 (■), C48 (●), and C56 (○). The results shown are averages of at least three independent experiments.
FIG. 2.
Histograms of the forward scatter of cultures of C. albicans C43 (a and b), C48 (c and d), and C56 (e and f) grown for 24 h in CYG medium under control conditions (a, c, and e) or after exposure to 3 μM itraconazole (b, d, and f). The single cells are found in region S, whereas region M represents multicellular clusters.
As could be expected from the MIC determinations according to NCCLS guidelines, itraconazole was most active against the C43 isolate. Maximal inhibition of up to 40% of control growth was obtained at 0.1 μM (0.07 μg · ml−1) itraconazole. No further decrease in growth was observed at higher itraconazole concentrations, a phenomenon known as residual or trailing growth (41). For strains C48 and C56, 1 μM itraconazole reduced growth to 15 and 8% of the control, respectively, indicating that strains C48 and C56 showed a more complete growth inhibition profile under these experimental conditions. No enhanced clustering of cells was found in the C48 and C56 isolates after exposure to 3 μM itraconazole compared to C43 (Fig. 2). On the contrary, a higher proportion of single cells was found in the two resistant isolates. The MICs according to the NCCLS methodology, shown in Table 2, are far more discriminatory for resistant strains C48 and C56 (16 and 32 times higher, respectively) relative to the C43 value than the 50% inhibitory concentrations (IC50s) for growth found under the ergosterol synthesis experimental conditions. The very high level of residual growth observed only for the C43 isolate could explain this difference. Indeed, the NCCLS method was designed to minimize residual growth after exposure to azoles in order to get as clear-cut MICs as possible. As shown in Fig. 1b, ergosterol content in the C43 isolate was reduced at lower itraconazole concentrations and the change from high to low sterol content occurred over just 1 dilution step. By contrast, reduction of ergosterol content from maximal to minimal levels in C48 and C56 occurred over 4 to 5 dilution steps. The IC50s were, respectively, 0.02, 0.1, and 0.12 μM. The less steep slope of the inhibition curve transitions in the two resistant isolates, C48 and C56, resulted in even greater differences when inhibition of ergosterol content to 10% of control levels was taken as the endpoint. To reduce ergosterol below 10% of control levels for C43, 0.025 μM itraconazole was sufficient, whereas in the case of C48 and C56, respectively, 0.6 and 0.7 μM, concentrations 24 and 28 times higher, were needed, respectively.
The products of ergosterol biosynthesis inhibition in the three strains, measured by incorporation of radioactively labeled acetate, showed considerable differences (Table 5). For isolate C43, 90% of the radioactivity incorporated into sterols was found in the ergosterol fraction; 50% inhibition of ergosterol synthesis was achieved at 0.002 μM itraconazole. Gas chromatography-mass spectrometry analysis identified 3,6-diol as the major 14-methylated accumulation product in cells exposed to 0.03 μM itraconazole, the lowest concentration giving maximal inhibition. Other sterols found were 14α-methyl-ergosta-5,7,22,24(28)tetraene-3βol, eburicol, lanosterol, obtusifoliol, and 14-methyl-fecosterol. In this strain, only minor quantities of obtusifolione, a 3-ketosteroid, were found. With isolates C48 and C56, 50% inhibition of ergosterol synthesis was reached at 0.05 and 0.04 μM itraconazole, respectively. In the absence of azole antifungals, both isolates accumulated significant amounts of 3,6-diol. The radioactivity incorporated into 3,6-diol increased slightly when these isolates were incubated in the presence of low itraconazole concentrations. After exposure to 0.3 μM itraconazole, in addition to the accumulating sterols described above, substantial quantities of obtusifolione were formed. This indicated that, in these isolates, itraconazole interfered with the 4-demethylation process, as well as with cytochrome P-450-catalyzed 14α-demethylation. Accumulation of obtusifolione after exposure to itraconazole was not observed in any of the 13 other strains studied (data not shown).
TABLE 5.
Sterol compositions of C. albicans isolates
| Sterol(s) | Avg % of [14C]acetate incorporation into total sterolsa
|
|||||
|---|---|---|---|---|---|---|
| C43
|
C48
|
C56
|
||||
| Control | 0.03 μM itraconazole | Control | 0.3 μM itraconazole | Control | 0.3 μM itraconazole | |
| Ergosterol | 89.2 | 0.6 | 60.4 | 10.4 | 73.7 | 17.3 |
| 3,6-Diol | 2.5 | 51.4 | 30.7 | 33.1 | 21.1 | 27.2 |
| Eburicol, lanosterol | 5.4 | 18.9 | 4.7 | 20.9 | 3.2 | 16 |
| Obtusifoliol | 1.7 | 13 | 1.7 | 13.9 | 1.3 | 15.3 |
| 14α-Methyl-fecosterol | 1.7 | 2.7 | ||||
| 14α-Methyl-ergosta-5,7,22,24(28)-tetraene-3βol | 12.7 | 6.6 | 1.3 | |||
| Obtusifolione | 1 | 3.5 | 15 | 1 | 20 | |
Results shown are averages of at least three independent experiments. The itraconazole IC50s for ergosterol synthesis were 0.002, 0.05, and 0.04 μM in strains C43, C48, and C56, respectively.
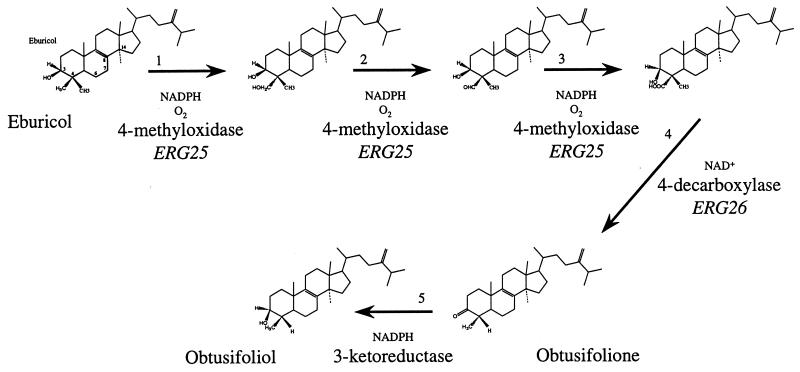
ERG25 and ERG3 sequence analysis.
Obtusifolione is a substrate for the 3-ketoreductase stage in the five-step 4-demethylation process, as shown in Fig. 3. So far, ERG25 is the only gene identified in C. albicans in this process and its product, the 4-methyloxidase, catalyzes at least three of the five reactions required (1). Gachotte et al. identified the ERG26 gene catalyzing the decarboxylase step in S. cerevisiae (10). Only the 3-ketoreductase reaction has not yet been assigned to a gene. To investigate whether mutations in the ERG25 gene, specific to both resistant isolates, contributed to the inhibition of the 3-ketoreduction, we sequenced the ERG25 alleles of the three isolates, as those of two unrelated strains (C. albicans ATCC 44858 and B59630). All isolates from patient V tested were heterozygotes in the 5′-untranslated region for the presence of four or five repeats of the sequence ATTT starting at position 67 in the sequence with EMBL accession no. AF051914. The sequences obtained from the reference strains were identical; strain B59630 contained four ATTT repeats, as found in the GenBank sequence obtained from C. albicans CAI-8, whereas strain ATCC 44848 contained five ATTT repeats. The ERG25 sequences of the two unrelated strains, B59630 and ATCC 44848, were identical to the published sequence, whereas five heterozygotic but silent mutations, A412A/C, A421A/G, A640A/T, A661A/G, and G706G/T, were found to be identical in C43, C48, and C56. This sequence analysis brings additional support for the relatedness of the strains investigated here. The Δ5,6-desaturase is encoded by ERG3. It is hypothesized that 3,6-diol is the presumed product of attempted Δ5,6-desaturation of 14-methylated precursors (14). Because accumulation of this sterol was seen under control conditions, we sequenced the gene in all three isolates. The 1,380-nucleotide amplification products were identical for all three isolates. Compared to the sequence of C. albicans B311 deposited at GenBank, one silent heterozygotic substitution (A1223A/G) and one T1438C mutation, leading to a conserved change of valine into an alanine at position 352, were found. Because this mutation was the same in all three isolates, it probably does not contribute to the accumulation of 3,6-diol.
FIG. 3.
The C4-demethylation process during ergosterol biosynthesis. Reactions are shown to remove the first C4-methyl group. To remove the second methyl group, the entire process is repeated.
DISCUSSION
This study highlights the paradox that itraconazole reduces the growth of resistant isolates C48 and C56 more profoundly, relative to control growth, than that of parent susceptible isolate C43 under the conditions of the present tests in vitro. However, the onset of growth inhibition in the resistant isolates occurs at higher itraconazole concentrations, presumably because intracellular azole content is reduced as a result of enhanced expression of both CDR1 and CDR2 in these strains. We hypothesize that the accumulation of obtusifolione as a result of itraconazole exposure, which is an unusual phenomenon in C. albicans, contributes to the more complete inhibition of growth. Accumulation of obtusifolione induced by itraconazole has been described in Histoplasma capsulatum (35, 42) and Cryptococcus neoformans (37). In both species, itraconazole inhibits growth completely, to the baseline level. Obtusifolione is the substrate for the 3-ketoreductase stage in the five-step 4-demethylation process, as shown in Fig. 3. So far, ERG25 is the only gene identified in C. albicans in this process and its product catalyzes at least three of the five reactions required. In S. cerevisiae, the gene coding for the sterol decarboxylase was recently identified as ERG26 (YGL001c) and found to be essential in cells with no other deficiency in the sterol biosynthetic pathway (10). The authors of this study concluded that the accumulation of the toxic oxygenated sterol intermediates prevented growth and that ERG26 was not involved in the 3-ketoreductase reaction. Unfortunately, only limited sequence information about a putative C. albicans ERG26 homolog is available. In addition, no accumulation of such carboxylic acid sterols was observed in the C. albicans strains studied. For these reasons, ERG26 was not further analyzed. Because in the 14α-demethylation process all three chemical steps necessary for the removal of the 14-methyl group are catalyzed by a single cytochrome P-450 (ERG11 or CYP51), the possibility exists that Erg25p is also involved in the 3-ketoreductase reaction. No mutations in ERG25 were found in our isolates, and thus, the ERG25 gene product is not likely to participate in the obtusifolione accumulation. This sequence analysis, however, does not conclusively eliminate the role of ERG25 in the 3-ketoreductase-catalyzed reaction. Indeed, because Erg25p is a membrane-bound enzyme, the possibility exists that indirect effects such as a different sterol composition, which influences membrane fluidity, induce conformational changes in the enzyme in the resistant strains, rendering it sensitive to itraconazole. The resistant isolates differed from the sensitive strain in sterol composition in both the quantity of ergosterol and in 3,6-diol content. It seems likely that the whole ergosterol synthetic pathway is upregulated in the resistant strains. The higher ergosterol content in the resistant isolates correlates with the twofold higher transcription signal for ERG11 described in these isolates by Sanglard et al. (27). The unusual accumulation of 3,6-diol observed under control conditions in the resistant isolates studied here was previously described by Shimokawa et al. in a 14α-demethylase mutant (32). We could not link the 3,6-diol accumulation to mutations in the ERG3 sequence, whose product is hypothesized to be involved in 3,6-diol formation. An alternative explanation for the accumulation is that it arises as a result of higher concentrations of 14-methylated substrates which could be generated by an upregulation of early steps in the ergosterol pathway followed by insufficient cytochrome P-450 activity to remove the 14α-methyl group from all of the substrate molecules available. The less active 14α-demethylase enzyme could exist in the azole-resistant strains investigated here. In fact, Sanglard et al. (30) identified two point mutations, G129A and G464S, in C56 ERG11 that differed from the C43 sequence. They elegantly showed that these point mutations specifically diminished the affinity of the proteins for azole antifungals and demonstrated their functionality by heterologous overexpression in an S. cerevisiae strain. However, these mutations might have affected other features of these proteins, particularly their functional integrity.
From this study, it can be concluded that, as already shown by others (8, 20), multiple changes are found in clonally related clinical isolates. The observed azole resistance is probably due to the combination of multiple factors that often evolve sequentially during treatment. This complicates the interpretation of mechanistic resistance studies and highlights a need to obtain multiple clonal isolates from a patient at frequent intervals.
ACKNOWLEDGMENTS
We thank H. Schreuders, T. Verhulst, A. Schijfs, and L. Van Nuffel for their excellent technical assistance and B. van den Hazel for interesting discussions.
REFERENCES
- 1.Bard M, Bruner D A, Pierson C A, Lees N D, Biermann B, Frye L, Koegel C, Barbuch R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodey G P. Resistance to antimicrobial agents revisited. Curr Opin Infect Dis. 1997;10:419–421. [Google Scholar]
- 3.De Muri G P, Hostetter M K. Resistance to antifungal agents. Pediatr Clin N Am. 1995;42:665–685. doi: 10.1016/s0031-3955(16)38984-2. [DOI] [PubMed] [Google Scholar]
- 4.Denning D W, Baily G G, Hood S V. Azole resistance in Candida. Eur J Clin Microbiol Infect Dis. 1997;16:261–280. doi: 10.1007/BF01695630. [DOI] [PubMed] [Google Scholar]
- 5.Doignon F, Aigle M, Ribereau-Gayon P. Resistance to imidazoles and triazoles in Saccharomyces cerevisiae as a new dominant marker. Plasmid. 1993;30:224–233. doi: 10.1006/plas.1993.1054. [DOI] [PubMed] [Google Scholar]
- 6.Dupont B. Azole antifungal agents: emerging and inherent resistance. Curr Opin Infect Dis. 1995;8:424–427. [Google Scholar]
- 7.Ebert E, Gaudin J, Muecke W, Ramsteiner K, Vogel C, Fuhrer H. Inhibition of ergosterol biosynthesis by etaconazole in Ustilago maydis. Z Naturforsch. 1983;38C:28–34. [Google Scholar]
- 8.Franz R, Kelly S L, Lamb D C, Kelly D E, Ruhnke M, Morschhäuser J. Multiple molecular mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frosco M, Barrett J F. Importance of antifungal drug-resistance: clinical significance and need for novel therapy. Exp Opin Investig Drugs. 1998;7:175–198. doi: 10.1517/13543784.7.2.175. [DOI] [PubMed] [Google Scholar]
- 10.Gachotte D, Barbuch R, Gaylor J, Nickel E, Bard M. Characterization of the Saccharomyces cerevisiae ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) involved in sterol biosynthesis. Proc Natl Acad Sci USA. 1998;95:13794–13799. doi: 10.1073/pnas.95.23.13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartman P G, Sanglard D. Inhibitors of ergosterol as antifungal agents. Curr Pharm Design. 1997;3:177–208. [Google Scholar]
- 12.Hitchcock C A, Barrett-Bee K J, Russell N J. The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J Gen Microbiol. 1986;132:2421–2431. doi: 10.1099/00221287-132-9-2421. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E M, Warnock D W. Azole drug resistance in yeast. J Antimicrob Chemother. 1995;36:751–755. doi: 10.1093/jac/36.5.751. [DOI] [PubMed] [Google Scholar]
- 14.Joseph-Horn T, Hollomon D W, Loeffler J, Kelly S. Cross-resistance to polyene and azole drugs in Cryptococcus neoformans. Antimicrob Agents Chemother. 1995;39:1526–1529. doi: 10.1128/aac.39.7.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph-Horn T, Hollomon D W. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol Lett. 1997;149:141–149. doi: 10.1111/j.1574-6968.1997.tb10321.x. [DOI] [PubMed] [Google Scholar]
- 16.Kelly S L, Lamb D C, Corran A J, Baldwin B C, Kelly D E. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun. 1995;197:428–432. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 17.Kelly S L, Lamb D C, Kelly D E, Manning N J, Loeffler J, Hebart H, Schumacher U, Einsele H. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol Δ5,6-desaturation. FEBS Lett. 1997;400:80–82. doi: 10.1016/s0014-5793(96)01360-9. [DOI] [PubMed] [Google Scholar]
- 18.Kolaczkowski M, Goffeau A. Active efflux by multidrug transporters as one of the strategies to evade chemotherapy and novel practical implications of yeast pleiotropic drug resistance. Pharmacol Ther. 1997;76:219–242. doi: 10.1016/s0163-7258(97)00094-6. [DOI] [PubMed] [Google Scholar]
- 19.Lamb D C, Kelly D E, Schunck W-H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Ribot J, McAtee R K, Lee L N, Kirkpatrick W R, White T C, Sanglard D, Paterson T F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marichal P, Vanden Bossche H. Mechanisms of resistance to azole antifungals. Acta Biochim Pol. 1995;42:509–516. [PubMed] [Google Scholar]
- 22.Marichal P, Vanden Bossche H, Odds F C, Nobels G, Warnock D W, Vincent Timmerman, Van Broeckhoven C, Fay S, Mose-Larsen P. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Reference method for broth dilution susceptibility testing of yeasts. Tentative standard M27-A. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 24.Odds F C, Vranckx L, Woestenborghs F. Antifungal susceptibility testing of yeasts: evaluation of technical variables for test automation. Antimicrob Agents Chemother. 1995;39:2051–2060. doi: 10.1128/aac.39.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odds F C. Should resistance to azole antifungals in vitro be interpreted as predicting clinical non-response? Drug Resist Updates. 1998;1:11–15. doi: 10.1016/s1368-7646(98)80209-4. [DOI] [PubMed] [Google Scholar]
- 26.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 30.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P450 lanosterol 14α-demethylase (CYP51) from azole-resistant Candida albicans clinical isolates contributing to the resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanglard D, Ischer F, Calabrese D, de Micheli M, Bille J. Multiple resistance mechanisms to azole antifungals in yeast clinical isolates. Drug Resist Updates. 1998;1:255–265. doi: 10.1016/s1368-7646(98)80006-x. [DOI] [PubMed] [Google Scholar]
- 32.Shimokawa O, Kato Y, Kawano K, Nakayama H. Accumulation of 14α-methyl-ergosta-8,24(28)-diene-3β,6α-diol in 14α-demethylation mutants of Candida albicans: genetic evidence for the involvement of 5-desaturase. Biochim Biophys Acta. 1989;1003:15–19. doi: 10.1016/0005-2760(89)90092-1. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Bossche H, Willemsens G, Cools W, Lauwers W, Le Jeune L. Biochemical effects of miconazole on fungi. II. Inhibition of ergosterol biosynthesis in Candida albicans. Chem Biol Interact. 1978;21:59–78. doi: 10.1016/0009-2797(78)90068-6. [DOI] [PubMed] [Google Scholar]
- 34.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Verhoeven H, Coene M-C, Lauwers W, Janssen P A J. Interaction of azole derivatives with cytochrome P450 systems in yeast, fungi, plants and mammalian cells. Pestic Sci. 1987;21:289–306. [Google Scholar]
- 35.Vanden Bossche H, Marichal P, Gorrens J, Bellens D, Coene M-C, Lauwers W, Le Jeune L, Moereels H, Janssen P A J. Mode of action of antifungals of use in immunocompromised patients. In: Vanden Bossche H, MacKenzie D W R, Cauwenbergh G, Van Cutsem J, Drouhet E, Dupont B, editors. Mycoses in AIDS patients. New York, N.Y: Plenum Press; 1990. pp. 223–243. [Google Scholar]
- 36.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanden Bossche H, Marichal P, Le Jeune L, Coene M-C, Gorrens J, Cools W. Effects of itraconazole on cytochrome P-450-dependent sterol 14α-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob Agents Chemother. 1993;37:2101–2105. doi: 10.1128/aac.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanden Bossche H, Marichal P, Odds F C. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 39.Vanden Bossche H. Mechanisms of antifungal resistance. Rev Iberoam Micol. 1997;14:44–49. [PubMed] [Google Scholar]
- 40.Vanden Bossche H, Dromer F, Improvisi L, Lozano-Chiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36(Suppl. 1):119–128. [PubMed] [Google Scholar]
- 41.Warnock D W, Johnson E M. Antifungal drug susceptibility testing. Curr Opin Infect Dis. 1997;10:444–448. [Google Scholar]
- 42.Wheat J, Marichal P, Vanden Bossche H, Le Monte A, Connolly P. Hypothesis on the mechanism of resistance to fluconazole in Histoplasma capsulatum. Antimicrob Agents Chemother. 1997;41:410–414. doi: 10.1128/aac.41.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White T C, Bowden R A, Marr K A. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]