Abstract
Acute lymphoblastic leukemia (ALL) is the most common cancer during childhood, representing about 30–35% of cases. Its etiology is complex and not fully understood. ALL is influenced by genetic variants, and their frequencies (Fq) vary in different ethnic groups, which consequently could influence the epidemiology of this cancer worldwide. The aim of this study was to investigate the correlation between the genetic variants and their impacts on incidence (IC), mortality (MT), and prevalence (PV) rates of ALL in different world populations. Methods: Sixty variants were selected from the literature with Genome Wide Association studies (GWAS). Information regarding allele Fq was selected from the 1000 Genomes platform. Epidemiological data were taken from the Global Burden of disease visualisations (GBD) Compare website. Statistical analyses were calculated in RStudio v.3.5.1 software. Results: Four variants demonstrated significant results in correlations with epidemiological data for ALL. The PAX5 gene variant (rs2297105) had an indirect relationship with PV and IC of ALL, showing that an increased Fq of this variant is related to low rates of both. An increased Fq of rs915172 in EPB4IL2 gene was also correlated with a lower IC of ALL. The rs1048943 of the CYP1A1 gene and the rs3088440 polymorphism of the CDKN2A gene were shown to have a direct proportional relationship with MT rate, showing that an increased Fq of these variants correlates with a worse prognosis worldwide. Conclusion: Our study points out four important variants for understanding the IC, PV, and MT rates for ALL. The ascertainment of these data may help to choose molecular markers to investigate the susceptibility and prognosis of ALL.
Keywords: acute lymphoblastic leukemia, susceptibility, epidemiological data
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignant cancer in childhood, representing about 30–35% of cases. The leukemogenesis process can occur in both B and T lymphoid lineages, which can invade the bone marrow, peripheral blood, and tissues [1].
Incidence and lethality rates of childhood ALL vary among countries [2]. In 2017, the global incidence of ALL was 1.04/100,000, while the global mortality was 0.72/100,000. In this analysis, the American continent had the highest incidence and mortality rates (1.45/100,000 and 1.03/100,000, respectively) [3].
ALL’s etiology is complex and not fully understood, however, it is known to be associated with both endogenous and exogenous factors. Confirmed risk factors include congenital cytogenetic disorders and exposure to various environmental factors, such as certain viral infections. However, these factors can explain only about 10% of all cases, while the remaining continue with their origins unknown [4,5].
Recent Genome Wide Association Studies (GWAS) have identified different genetic variants associated with higher risk for ALL, including single nucleotide polymorphisms (SNPs) in important genes, suggesting this type of leukemia could relate to a polygenic susceptibility [6,7,8,9]. However, there is no consensus in these studies in worldwide populations.
Another factor associated with the incidence of ALL is the ethnic characteristics of different populations [7]. This disparity observed between populations occurs due to the presence of genetic polymorphisms and intrinsic environmental factors to which these groups are exposed [10]. Some studies have already highlighted Amerindian ancestry and mixed populations as risk factors for developing of toxicity in ALL [9,11,12,13]. Therefore, our study aimed to investigate the correlation between genetic polymorphisms involved in biological pathways for ALL and their impacts on incidence, mortality, and prevalence rates in different world populations.
2. Materials and Methods
2.1. Variant Selection and Genetic Data Collection
Polymorphisms were selected from the literature by searching the PubMed platform with the following descriptors: “Acute lymphoblastic leukemia”, “GWAS”, “risk”, “susceptibility”, “population”, which led to 46 articles. Inclusion criteria were: Variants found in GWAS and meta-analysis studies, variants associated with protection, susceptibility, or risk to ALL. We obtained 60 polymorphisms described for the development of acute lymphoblastic leukemia (Supplementary Table S1). Among these, 56 were excluded because they did not significantly correlate with acute lymphoblastic leukemia.
2.2. Frequency Information on Analyzed Variants
Variants’ frequency information was obtained from the 1000 Genomes Project platform (Available online: https://www.internationalgenome.org/ (Accessed on 5 October 2021), available in Phase 3 Release of the 1000 Genomes Database
Specific frequencies were collected for each ethnic group described in the platform: African (AFR), American Mixed (AMR), Asian (EAS), European (EUR), and South Asian (SAS).
2.3. Epidemiological Data Collection
Data on incidence (IC), prevalence (PV), and mortality (MT) rates of ALL in global populations were obtained from the Global Burden of disease visualisations (GBD) Compare website, created by the Institute for Health Metrics and Evaluation (http://vizhub.healthdata.org/gbd-compare, accessed on 28 January 2022). We collected the epidemiological rates from all countries analyzed in 1000 Genomes database (Supplementary Table S2).
2.4. Statistical Analysis and Plots
Correlation analysis between frequencies of genetic variants and epidemiological rates was calculated by Pearson’s correlation coefficient. All analyses were performed in RStudio v.3.5.1 software.
3. Results
A total of 60 polymorphisms were analyzed regarding their association with IC, MT and/or PV rates of ALL. Among these, four (rs2297105, rs915172, rs1048943, and rs3088440) showed significance in the correlation analyses. Information about these variants is available in Table 1.
Table 1.
Information about the significant variants investigated.
| Position | Gene | Variant ID | Location | Impact Predicted by SNPeff |
Variable |
|---|---|---|---|---|---|
| 9: 37020625 | PAX5 | rs2297105 | Intronic | Modifier | Incidence and prevalence |
| 6: 130863630 | EPB41L2 | rs915172 | Splice site region | Low | Incidence |
| 15: 74720644 | CYP1A1 | rs1048943 | Non-synonymous coding | Moderate | Mortality |
| 9: 21968160 | CDKN2A | rs3088440 | 3′-UTR | Modifier | Mortality |
Abbreviations: Genetic variant annotation and functional effect prediction toolbox (SNPeff).
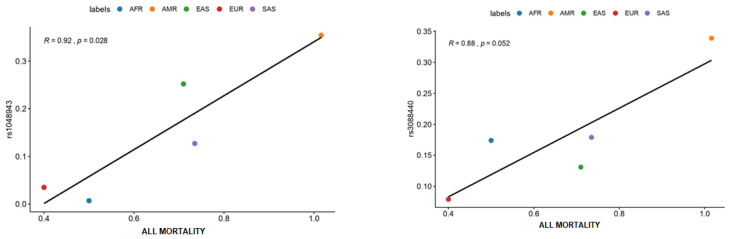
Figure 1 shows that two variants–rs1048943 of the CYP1A1 gene and rs3088440 of the CDKN2A gene–were positively correlated with mortality rates. Both images show homogeneity in our findings, since the higher the frequency of the variant allele in these polymorphisms, the higher were mortality rates (showed as per million of individuals). Based on this correlation, AMR individuals have higher frequencies of the variant alleles, and, consequently, have higher rates of ALL mortality. On the other hand, European populations have the lowest frequency of these variants, also having the lowest mortality rates.
Figure 1.
Mortality rates of ALL. Abbreviations: African (AFR), American Mixed (AMR), Asian (EAS), European (EUR), and South Asian (SAS), Acute lymphoblastic leukemia (ALL).
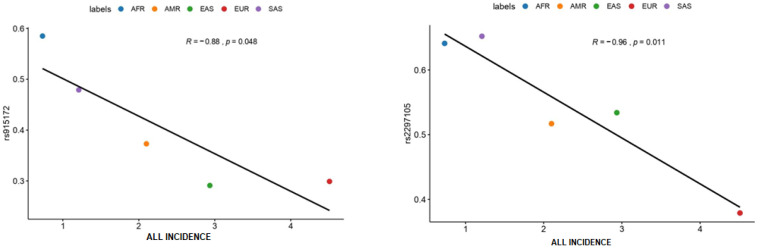
Regarding incidence (Figure 2), we found that both rs2297105 of the PAX5 gene and rs915172 of EPB41L2 gene are negatively correlated with ALL incidences. AFR and SAS have the highest frequencies of both these variants, also having the lowest rates of ALL incidences, while EUR has the lowest frequency and the highest incidence. This finding may indicate a possible protective factor regarding these variants.
Figure 2.
ALL Incidence Rates. Abbreviations: African (AFR), American Mixed (AMR), Asian (EAS), European (EUR), and South Asian (SAS), Acute lymphoblastic leukemia (ALL).
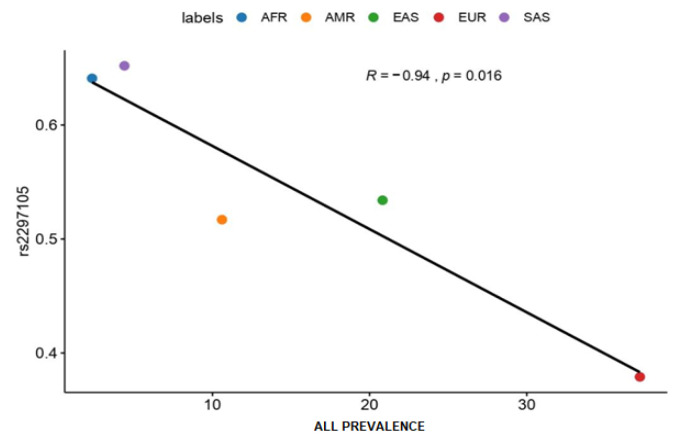
Regarding prevalence rates (Figure 3), only one polymorphism showed a statistically significant inverse correlation (rs2297105 of the PAX5 gene). According to these results, the EUR and EAS populations have lower frequencies of these variant alleles, which grant them higher ALL prevalence rates.
Figure 3.
ALL Prevalence Rates. Abbreviations: African (AFR), American Mixed (AMR), Asian (EAS), European (EUR), and South Asian (SAS), Acute lymphoblastic leukemia (ALL).
4. Discussion
A large fluctuation in ALL’s epidemiological rates is observed worldwide [1]. One of the risk factors that may contribute to this variability is the inherent genetic factors of each ethnicity [7,10,11]. Several genomic studies with a large number of patients show that genetic variants may act as risk factors for ALL in populations with different patterns of ethnic composition [13,14].
This study gathered the IC, PV, and MT rates found in countries compounding the 1000 Genomes database and correlated these data with the allele frequencies of genetic variants, which have already been associated with susceptibility for ALL GWAS studies. Our results show a strong correlation of four genetic variants (rs2297105, rs915172, rs1048943, and rs3088440) in four genes (PAX5, EPB41L2, CYP1A1, and CDKN2A) with PV, IC, and MT rates.
The PAX5 gene encodes the paired box protein PAX5, a member of the family of transcription factors essential for B-cell differentiation since it activates specific genes and represses those that compromise hematopoietic lineages [15]. Thus, PAX5 is one of the proteins with a high frequency of genetic alterations in B-cell ALL. Such alterations are the results of deletions, translocations, or mutations. About one-third of cancers are consequences of PAX5 gene modifications, and the complete loss of this gene is related to the highest aggressiveness of B-cell ALL [16,17].
Our results show that a higher frequency of the polymorphism rs2297105 in the PAX5 gene is associated with a lower IC and a lower PV of ALL. The rs2297105 variant in the PAX5 gene showed fluctuations between frequencies of the polymorphism in the different populations, as we found high frequency in the African population (0.641) and low frequency in Europeans (0.379). However, the frequency of rs2297195 was inversely associated with the prevalence and incidence of ALL, as the European population showed higher prevalence and incidence of ALL when compared to the African population. This polymorphism has been associated as a susceptibility locus for ischemic or hemorrhagic stroke [14] and glucocorticoid resistance in ALL [18].
The EPB41L2 is a member of the 4.1 proteins, encoded by the EPB41 (erythrocyte membrane protein band 4.1-like 2) genes, which are components of the cortical cytoskeleton underlying the cell membrane [19]. The EPB41 family form nodes in the cell cortex, connecting further components of the cortical cytoskeleton–like spectrins, actin, and transmembrane adhesion proteins, receptors, and transporters [20]. Polymorphisms in the EPB41L2 gene has been associated with childhood acute lymphoblastic in genome-wide association study in Korea [6].
In our study, the rs915172 variant of the EPB41L2 gene showed an association with ALL incidences, as the lower the frequency of this variant, the higher the incidence for ALL. In our study, the rs915172 variant of the EPB41L2 gene showed an association with ALL incidences, as the lower the frequency of this variant, the higher the incidence for ALL. We found the highest frequency in the African population (0.585), which was associated with lower incidence of ALL, while the opposite happened with the European population, which showed a lower frequency of the SNP (0.299) and a higher incidence of ALL. This result may suggest the polymorphism as a protective factor for susceptibility to ALL. This is the first study to correlate this specific polymorphism with a pathological condition, for which further clinical studies are required to clarify their relationship.
The CYP1A1 gene is a member of the CYP family that participates in the metabolism and elimination of exogenous chemical compounds. Genetic alterations can modify enzyme activity and compromise the metabolism of mutagenic substances. Polymorphisms in these enzymes positively influence susceptibility to ALL in children [21]. Most studies investigating the role of this gene in leukemia susceptibility are done in Oriental populations [22]. A meta-analysis has associated the presence of this polymorphism with an increased risk for developing ALL and Acute Myeloid Leukemia in Caucasian individuals [23].
Our findings also include a positive correlation between the rs1048943 polymorphism of the CYP1A1 gene with ALL mortality rates, given that, the higher the frequency of this variant, the higher the ALL mortality rates. The highest frequency was found in the American population (0.354), and the lowest in European (0.035).
The cyclin-dependent kinase 2A inhibitor (CDKN2A) encodes two proteins important for cell cycle regulation: p14ARF, which binds to p53 and inhibits MDM2, and p16INK4a, which is a cyclin-dependent kinase inhibitor. Both act as tumor suppressors [24]. Mutations in the CDKN2A gene have been observed in several types of cancers, such as melanoma-like, pancreatic, intestinal, prostate, and gastric lymphoma [25]. Deletions in the CDKN2A/B gene have been correlated with a worse prognosis of ALL in patients of all ages, and a missense variant associated with the unique risk of ALL in European and Hispanic children has been identified, conferring a threefold increased risk for ALL [26,27].
In our study, the rs3088440 variant in the CDKN2A/B gene was positively associated with MT rate, so the higher the frequency of the variant allele, the higher the numbers of ALL’s incidence and mortality. The highest frequency was found in the American population (0.354), and the lowest in European (0.127).
In the present study, it was not possible to stratify the populations investigated according to the socioeconomic level of the patients and/or their levels of access to medical resources. This limitation made it possible to include unique ethnic groups with relevant data for the analyses.
5. Conclusions
Large genome-wide studies are important for identifying variants associated with susceptibility to complex diseases. However, the results found may differ among populations worldwide due to fluctuation in allele frequencies of these variants among different ethnic groups. Our study aimed to correlate data on genetic variants associated with ALL with the epidemiology rates of this disease.
Our results point to four important variants (rs2297105, rs915172, rs1048943, and rs3088440) for understanding the incidence, prevalence, and mortality rates associated with this cancer. The rs1048943 and rs3088440 variants were positively associated with ALL mortality, and both variants were more frequent in the American population and less frequent in the European population.
On the other hand, the rs9151172, rs2297105 variants were inversely related to the incidence rates of ALL, with higher frequency in the African population and lower frequency in the European population. The ascertainment of these data may help to choose molecular markers to investigate the susceptibility and prognosis of ALL.
Acknowledgments
We acknowledge funding from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FAPESPA (Fundação Amazônia de Amparo a Estudos e Pesquisa), UFPA (Universidade Federal do Pará), and PROPESP (Pró-reitoria de Pesquisa e Pós-graduação) from UFPA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jpm12030370/s1, Table S1: Studied Genes and References; Table S2: Epidemiological Data on Global Population.
Author Contributions
Conceptualization, L.W.M.S.V., J.C.G.R. and N.P.C.d.S.; methodology, J.C.G.R.; formal analysis, J.C.G.R.; investigation, M.R.F., L.W.M.S.V., L.P.A.G., A.L.d.A. and B.B.d.S.; resources, A.V.W., S.M.F. and R.M.R.B.; writing—original draft preparation, M.R.F., L.W.M.S.V., J.C.G.R., L.P.A.G. and A.L.d.A.; writing—review and editing, M.R.F., L.W.M.S.V., L.P.A.G., A.L.d.A. and N.P.C.d.S.; supervision, N.P.C.d.S.; project administration, S.E.B.d.S., P.P.d.A. and R.M.R.B.; funding acquisition, S.E.B.d.S. and P.P.d.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), FAPESPA(Fundação Amazônica de Amparo a Estudos e Pesquisas), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and UFPA (Universidade Federal do Pará). These funding agencies played no role in the study design, data collection and analysis, or the decision to publish, or the preparation of the manuscript.
Institutional Review Board Statement
The study was approved by the Ethics Committee of Research Center of Oncology/Federal University of Pará (Protocol nº 3.419.333/2019).
Informed Consent Statement
Not applicable for this study. All data used were obtained from secondary sources available on the 1000 Genomes Project platform/database and GBD Compare website.
Data Availability Statement
All relevant data will be shared as Supporting Information files if the manuscript is accepted for publication.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Malard F., Mohty M. Acute Lymphoblastic Leukaemia. Lancet. 2020;395:1146–1162. doi: 10.1016/S0140-6736(19)33018-1. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Institute for Health Metrics and Evaluation (IHME) University of Washington; Seattle, WA, USA: 2015. [(accessed on 15 June 2021)]. GBD Compare. Available online: http://vizhub.healthdata.org/gbd-compare. [Google Scholar]
- 4.Li S.Y., Ye J.Y., Liang E.Y., Zhou L.X., Yang M. Association between MTHFR C677T Polymorphism and Risk of Acute Lymphoblastic Leukemia: A Meta-Analysis Based on 51 Case-Control Studies. Med. Sci. Monit. 2015;21:740–748. doi: 10.12659/MSM.892835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gharbi H., Ben Hassine I., Soltani I., Safra I., Ouerhani S., Bel Haj Othmen H., Teber M., Farah A., Amouri H., Toumi N.H., et al. Association of Genetic Variation in IKZF1, ARID5B, CDKN2A, and CEBPE with the Risk of Acute Lymphoblastic Leukemia in Tunisian Children and Their Contribution to Racial Differences in Leukemia Incidence. Pediatr. Hematol. Oncol. 2016;33:157–167. doi: 10.3109/08880018.2016.1161685. [DOI] [PubMed] [Google Scholar]
- 6.Han S., Lee K.M., Park S.K., Lee J.E., Ahn H.S., Shin H.Y., Kang H.J., Koo H.H., Seo J.J., Choi J.E., et al. Genome-Wide Association Study of Childhood Acute Lymphoblastic Leukemia in Korea. Leuk. Res. 2010;34:1271–1274. doi: 10.1016/j.leukres.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Hsu L.I., Briggs F., Shao X., Metayer C., Wiemels J.L., Chokkalingam A.P., Barcellos L.F. Pathway Analysis of Genome-Wide Association Study in Childhood Leukemia among Hispanics. Cancer Epidemiol. Biomark. Prev. 2016;25:815–822. doi: 10.1158/1055-9965.EPI-15-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vijayakrishnan J., Studd J., Broderick P., Kinnersley B., Holroyd A., Law P.J., Kumar R., Allan J.M., Harrison C.J., Moorman A.V., et al. Genome-Wide Association Study Identifies Susceptibility Loci for B-Cell Childhood Acute Lymphoblastic Leukemia. Nat. Commun. 2018;9:1340. doi: 10.1038/s41467-018-03178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pui C.H., Nichols K.E., Yang J.J. Somatic and Germline Genomics in Paediatric Acute Lymphoblastic Leukaemia. Nat. Rev. Clin. Oncol. 2019;16:227–240. doi: 10.1038/s41571-018-0136-6. [DOI] [PubMed] [Google Scholar]
- 10.Quiroz E., Aldoss I., Pullarkat V., Rego E., Marcucci G., Douer D. The Emerging Story of Acute Lymphoblastic Leukemia among the Latin American Population—Biological and Clinical Implications. Blood Rev. 2019;33:98–105. doi: 10.1016/j.blre.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 11.De Carvalho D.C., Wanderley A.V., dos Santos A.M.R., Moreira F.C., de Sá R.B.A., Fernandes M.R., Modesto A.A.C., de Souza T.P., Cohen-Paes A., Leitão L.P.C., et al. Characterization of Pharmacogenetic Markers Related to Acute Lymphoblastic Leukemia Toxicity in Amazonian Native Americans Population. Sci. Rep. 2020;10:10292. doi: 10.1038/s41598-020-67312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chuah B., Goh B.C., Lee S.C., Soong R., Lau F., Mulay M., Dinolfo M., Lim S.E., Soo R., Furuie T., et al. Comparison of the Pharmacokinetics and Pharmacodynamics of S-1 between Caucasian and East Asian Patients. Cancer Sci. 2011;102:478–483. doi: 10.1111/j.1349-7006.2010.01793.x. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y.L., Hung C.C., Chen J.S., Lin K.H., Jou S.T., Hsiao C.C., Sheen J.M., Cheng C.N., Wu K.H., Lin S.R., et al. IKZF1 Deletions Predict a Poor Prognosis in Children with B-Cell Progenitor Acute Lymphoblastic Leukemia: A Multicenter Analysis in Taiwan. Cancer Sci. 2011;102:1874–1881. doi: 10.1111/j.1349-7006.2011.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamada Y., Sakuma J., Takeuchi I., Yasukochi Y., Kato K., Oguri M., Fujimaki T., Horibe H., Muramatsu M., Sawabe M., et al. Identification of Six Polymorphisms as Novel Susceptibility Loci for Ischemic or Hemorrhagic Stroke by Exome-Wide Association Studies. Int. J. Mol. Med. 2017;39:1477–1491. doi: 10.3892/ijmm.2017.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J., Li T., Zhao L., Liang X., Fu X., Wang J., Shang Z., Huang W., Zhou J. Identification of Significant Pathways Induced by PAX5 Haploinsufficiency Based on Protein-Protein Interaction Networks and Cluster Analysis in Raji Cell Line. BioMed Res. Int. 2017;2017:5326370. doi: 10.1155/2017/5326370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smeenk L., Fischer M., Jurado S., Jaritz M., Azaryan A., Werner B., Roth M., Zuber J., Stanulla M., Boer M.L., et al. Molecular Role of the PAX 5- ETV 6 Oncoprotein in Promoting B-cell Acute Lymphoblastic Leukemia. EMBO J. 2017;36:718–735. doi: 10.15252/embj.201695495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isidro-Hernández M., Mayado A., Casado-García A., Martínez-Cano J., Palmi C., Fazio G., Orfao A., Ribera J., Ribera J.M., Zamora L., et al. Inhibition of Inflammatory Signaling in Pax5 Mutant Cells Mitigates B-Cell Leukemogenesis. Sci. Rep. 2020;10:19189. doi: 10.1038/s41598-020-76206-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson L., Evans C.A., Matheson E., Minto L., Keilty C., Sanichar M., Case M., Schwab C., Williamson D., Rainer J., et al. Quantitative Proteomic Analysis Reveals Maturation as a Mechanism Underlying Glucocorticoid Resistance in B Lineage ALL and Re-Sensitization by JNK Inhibition. Br. J. Haematol. 2015;171:595–605. doi: 10.1111/bjh.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rangel L., Lospitao E., Ruiz-Sáenz A., Alonso M.A., Correas I. Alternative Polyadenylation in a Family of Paralogous EPB41 Genes Generates Protein 4.1 Diversity. RNA Biol. 2017;14:236–244. doi: 10.1080/15476286.2016.1270003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parra M., Gee S., Chan N., Ryaboy D., Dubchak I., Mohandas N., Gascard P.D., Conboy J.G. Differential Domain Evolution and Complex RNA Processing in a Family of Paralogous EPB41 (Protein 4.1) Genes Facilitate Expression of Diverse Tissue-Specific Isoforms. Genomics. 2004;84:637–646. doi: 10.1016/j.ygeno.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Nida S., Javid B., Akbar M., Idrees S., Adil W., Ahmad G.B. Gene Variants of CYP1A1 and CYP2D6 and the Risk of Childhood Acute Lymphoblastic Leukaemia; Outcome of a Case Control Study from Kashmir, India. Mol. Biol. Res. Commun. 2017;6:77–84. doi: 10.22099/mbrc.2017.4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brisson G.D., Alves L.R., Pombo-De-Oliveira M.S. Genetic Susceptibility in Childhood Acute Leukaemias: A Systematic Review. Ecancermedicalscience. 2015;9:539. doi: 10.3332/ecancer.2015.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu J., Zhao Q., Zhai Y.J., He H.R., Yang L.H., Gao F., Zhou R.S., Zheng J., Ma X.C. Genetic Polymorphisms of CYP1A1 and Risk of Leukemia: A Meta-Analysis. Onco. Targets. Ther. 2015;8:2883–2902. doi: 10.2147/OTT.S92259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang J., Fan J., Wang M., Niu Z., Zhang Z., Yuan L., Tai Y., Chen Z., Song S., Wang X., et al. CDKN2A Inhibits Formation of Homotypic Cell-in-Cell Structures. Oncogenesis. 2018;7:50. doi: 10.1038/s41389-018-0056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao R., Choi B.Y., Lee M.H., Bode A.M., Dong Z. Implications of Genetic and Epigenetic Alterations of CDKN2A (P16INK4a) in Cancer. EBioMedicine. 2016;8:30–39. doi: 10.1016/j.ebiom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walsh K.M., De Smith A.J., Hansen H.M., Smirnov I.V., Gonseth S., Endicott A.A., Xiao J., Rice T., Fu C.H., McCoy L.S., et al. A Heritable Missense Polymorphism in CDKN2A Confers Strong Risk of Childhood Acute Lymphoblastic Leukemia and Is Preferentially Selected during Clonal Evolution. Cancer Res. 2015;75:4884–4894. doi: 10.1158/0008-5472.CAN-15-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X., Liao F., Zhang J., Qin Y., Xu H., Ding Z., Zhang Y., Zhang F. Association of the Independent Polymorphisms in CDKN2A with Susceptibility of Acute Lymphoblastic Leukemia. Biosci. Rep. 2018;38:BSR20180331. doi: 10.1042/BSR20180331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data will be shared as Supporting Information files if the manuscript is accepted for publication.