Abstract
Aspergillus species are cosmopolitan and ubiquitous, closely related to human daily life. They are also of food, industrial and medical importance. From the examination of cultures isolated from soil samples collected on tropical islands of China, four new species of the genus were discovered based on phylogenetic analyses and morphological comparisons. Aspergillus xishaensis sp. nov. and A. neoterreus sp. nov. belong to sections Flavipedes and Terrei of subgenus Circumdati, and A. hainanicus sp. nov. and A. qilianyuensis sp. nov. are in sections Cavernicolarum and Nidulantes of subgenus Nidulantes. To accommodate A. hainanicus, a new series Hainanici was proposed. Detailed descriptions and illustrations of the new taxa were provided.
Keywords: Ascomycota, Eurotiales, fungal biodiversity, phylogeny, taxonomy
1. Introduction
Species of Aspergillus P. Micheli ex Haller are cosmopolitan and ubiquitous. Some of them are closely related to human daily life. Strains of A. niger Tiegh. and A. oryzae (Ahlb.) Cohn were used for the fermentation of food for more than two millennia and the manufacturing of food enzymes for over 50 years [1]. Aspergillus niger is also a workhorse and cell factory for the production of citric acid, an organic acid with high economic importance, which is widely used in beverage, food, detergents, cosmetics and pharmaceutical industries [2]. Aflatoxins, produced by A. flavus Link and other aspergilli, are highly toxic secondary metabolites and severely contaminate food supplies of humans and animals, resulting in health hazards and even death [3]. Some black aspergilli were reported to be postharvest pathogens of economically important crops, e.g., A aculeatus Iizuka, A. japonicus Saito and A. uvarum G. Perrone et al. infecting the fruits of grapes [4]. Aspergillosis infections caused by Aspergillus species are of significant morbidity and mortality. Mostly, they are attributed to A. fumigatus Fresen., followed by A. flavus and A. terreus Thom [5].
The genus was originally introduced in 1729 and has more than one thousand names recorded in the database Index Fungorum. According to a recent monographic study, Aspergillus was divided into six subgenera (namely, Aspergillus, Circumdati, Cremei, Fumigati, Nidulantes and Polypaecilum), 27 sections and 75 series, with 446 species accepted [6]. Recently, more than 20 new species were added, e.g., A. kumbius (Pitt) and A. malvicolor A.D. Hocking in sect. Circumdati, A. agricola Pummi Singh et al. and A. burnettii Pitt in section Flavi, A. alboluteus F. Sklenar et al. and A. okavangoensis Visagie and Nkwe in section Flavipedes, A. nanangensis Pitt in section Janorum, A. hydei Doilom and A. vinaceus Ferranti et al. in section Nigri, and A. barbosae A.C.R. Barros-Correia et al. in section Terrei of subgenus Circumdati; A. arizonensis Jurjević et al. and A. banksianus Pitt in section Fumigati of subgenus Fumigati; A. lannaensis N. Suwannarach et al. in section Sparsi, and A. sigarelli B.D. Sun et al. in section Usti of subgenus Nidulantes; A. limoniformis Z.F. Zhang and L. Cai and A. telluris B.D. Sun et al. in sect. Polypaecilum of subgenus Polypaecilum [7,8,9,10,11,12,13,14,15,16,17,18,19]. The increasing number of species reveals the extremely high biodiversity of Aspergillus.
During the examinations of the cultures isolated from sandy soil collected on tropical islands of China, four new species were discovered based on phylogenetic analyses and morphological comparisons. They belong to sections Flavipedes and Terrei of subgenus Circumdati and sections Cavernicolarum and Nidulantes of subgenus Nidulantes, respectively. The detailed descriptions and illustrations of the new taxa are provided.
2. Materials and Methods
2.1. Fungal Materials
Cultures were isolated from sandy soil collected on tropical islands of China in 2015. Dried cultures were deposited in the Herbarium Mycologicum Academiae Sinicae (HMAS), and living ex-type strains were preserved in the China General Microbiological Culture Collection Center (CGMCC).
2.2. Morphological Observations
Morphological characterization was conducted following standardized methods [20]. Four standard growth media were used: Czapek yeast autolysate agar (CYA, yeast extract Oxoid), malt extract agar (MEA, Amresco), yeast extract agar (YES) and potato dextrose agar (PDA). If sporulation failed on the above media, PDA with 3% sea salts (3% NaCl, Psaitong) and oatmeal agar (OA) were further applied. The methods for inoculation, incubation, microscopic examinations and digital recordings followed our previous studies [21,22,23,24].
2.3. Molecular Experiments
DNA was extracted from the cultures grown on PDA for 7 days using the Plant Genomic DNA Kit (DP305, TIANGEN Biotech, Beijing, China). Polymerase chain reaction (PCR) amplifications of the internal transcribed spacer (ITS), beta-tubulin (BenA), calmodulin (CaM) and RNA polymerase II second largest subunit (RPB2) gene regions were conducted with the routine methods [21,22,23,24]. The products were purified and subject to sequencing on an ABI 3730 DNA Sequencer (Applied Biosystems). Although the ITS region is proposed as the universal DNA barcode for fungi, it is not sufficient to distinguish species of Aspergillus. The ITS sequences provided in this study might be helpful for other researchers in case of need.
2.4. Phylogenetic Analyses
Forward and reverse sequences newly generated in this study were assembled using Seqman v. 7.1.0 (DNASTAR Inc., Madison, WI, USA). The assembled sequences were deposited at GenBank. The sequences used for phylogenetic analyses are listed in Table 1 and Table 2. Sequences of the combined loci (BenA, CaM and RPB2) of each of the two subgenera were aligned using MAFFT v. 7.221 [25] and then manually edited and combined in BioEdit v. 7.1.10 [26] and MEGA v. 6.0.6 [27]. The combined datasets of individual subgenera were analyzed to infer their phylogeny. Maximum likelihood (ML) analyses were conducted using RAxML-HPC2 [28] on XSEDE 8.2.12 on CIPRES Science Gateway v. 3.3 [29] with the default GTRCAT model. Bayesian inference (BI) analyses were performed with MrBayes v. 3.2.5 [30]. Appropriate nucleotide substitution models and parameters were determined by Modeltest v. 3.7 [31]. The consensus trees were viewed in FigTree v. 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 3 June 2015). Aspergillus flavus of subgen. Circumdati sect. Flavi served as an outgroup.
Table 1.
Fungal species and sequences used in phylogenetic analyses of Aspergillus subgen. Nidulantes.
| Section | Series | Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|---|---|
| Aenei | Aenei | A. aeneus Sappa 1954 | CBS 128.54 T | Somalia | forest soil | EF652474 | EF652298 | EF652386 | EF652210 |
| A. bicolor M. Chr. and States 1978 | CBS 425.77 T | USA | soil | EF652511 | EF652335 | EF652423 | EF652247 | ||
| Bispori | Bispori | A. bisporus Kwon-Chung and Fennell 1971 | CBS 707.71 T | USA | soil | EF661208 | EF661121 | EF661139 | EF661077 |
| Cavernicolarum | Cavernicolarum | A. californicus Frisvad et al. 2011 | CBS 123895 T | USA | chaparral of Adenostoma fasciculatum | FJ531153 | FJ531180 | FJ531128 | MN969065 |
| A. cavernicola Lörinczi 1969 | CBS 117.76 T | Romania | on walls of cave | EF652508 | EF652332 | EF652420 | EF652244 | ||
| A. kassunensis Baghd. 1968 | CBS 419.69 T | Syria | soil | EF652461 | EF652285 | EF652373 | EF652197 | ||
| A. subsessilis Raper and Fennell 1965 | CBS 502.65 T | USA | desert soil | EF652485 | EF652309 | EF652397 | EF652221 | ||
| Egyptiaci | A. egyptiacus Moub. and Moustafa 1972 | CBS 656.73 T | Egypt | sandy soil | EF652504 | EF652328 | EF652416 | EF652240 | |
| Hainanici | A. hainanicus X.C. Wang and W.Y. Zhuang, sp. nov. | ZC79 T | China: Hainan | sandy soil | OM414846 | OM475626 | OM475630 | OM475634 | |
| Nidulantes | Aurantiobrunnei | A. aurantiobrunneus Raper and Fennell 1965 | CBS 465.65 T | Australia | canvas haversack for respirator | EF652465 | EF652289 | EF652377 | EF652201 |
| Multicolores | A. multicolor Sappa 1954 | CBS 133.54 T | Somalia | forest soil | EF652477 | EF652301 | EF652389 | EF652213 | |
| Nidulantes | A. nidulans (Eidam) G. Winter 1884 | CBS 589.65 T | Belgium | unknown | EF652427 | EF652251 | EF652339 | EF652163 | |
| Speluncei | A. spelunceus Raper and Fennell 1965 | CBS 497.65 T | USA | soil and dead Orthoptera | EF652490 | EF652314 | EF652402 | EF652226 | |
| Stellati | A. stellatus Curzi 1934 | CBS 598.65 T | Panama | soil | EF652426 | EF652250 | EF652338 | EF652162 | |
| Unguium | A. unguis (Émile-Weill and L. Gaudin) Thom and Raper 1934 | CBS 132.55 T | USA | shoe leather | EF652443 | EF652267 | EF652355 | EF652179 | |
| Versicolores | A. amoenus M. Roberg 1931 | CBS 111.32 T | Germany | fruit of Berberis sp. | EF652480 | JN853946 | JN854035 | JN853824 | |
| A. austroafricanus Jurjević et al. 2012 | CBS 145748 T | South Africa | soil | JQ301891 | JN853963 | JN854025 | JN853814 | ||
| A. creber Jurjević et al. 2012 | CBS 145749 T | USA | air | JQ301889 | JN853980 | JN854043 | JN853832 | ||
| A. cvjetkovicii Jurjević et al. 2012 | CBS 599.65 T | USA | soil | EF652440 | EF652264 | EF652352 | EF652176 | ||
| A. fructus Jurjević et al. 2012 | CBS 584.65 T | USA | fruit of date palm | EF652449 | EF652273 | EF652361 | EF652185 | ||
| A. griseoaurantiacus Visagie et al. 2014 | CBS 138191 T | Micronesia | house dust | KJ775553 | KJ775086 | KJ775357 | KU866988 | ||
| A. hongkongensis C.C. Tsang et al. 2016 | CBS 145671 T | China: Hong Kong | nails of Homo sapiens | AB987907 | LC000552 | MN969320 | LC000578 | ||
| A. jensenii Jurjević et al. 2012 | NRRL 58600 T | USA | soil | JQ301892 | JN854007 | JN854046 | JN853835 | ||
| A. pepii Despot et al. 2016 | CBS 142028 T | Croatia | air | KU613368 | KU613371 | KU613365 | n.a. | ||
| A. protuberus Munt.-Cvetk. 1968 | CBS 602.74 T | former Yugoslavia | rubber coated electric cables | EF652460 | EF652284 | EF652372 | EF652196 | ||
| A. puulaauensis Jurjević et al. 2012 | CBS 145750 T | USA: Hawaii | dead hardwood | JQ301893 | JN853979 | JN854034 | JN853823 | ||
| A. qilianyuensis X.C. Wang and W.Y. Zhuang, sp. nov. | ZC101 T | China: Hainan | sandy soil | OM414847 | OM475627 | OM475631 | OM475635 | ||
| A. subversicolor Jurjević et al. 2012 | CBS 145751 T | India | green berries of coffee | JQ301894 | JN853970 | JN854010 | JN853799 | ||
| A. sydowii (Bainier and Sartory) Thom and Church 1926 | CBS 593.65 T | France | unknown | EF652450 | EF652274 | EF652362 | EF652186 | ||
| A. tabacinus Nakaz. et al. 1934 | CBS 122718 T | unknown | tobacco | EF652478 | EF652302 | EF652390 | EF652214 | ||
| A. tennesseensis Jurjević et al. 2012 | CBS 145752 T | USA | toxic dairy feed | JQ301895 | JN853976 | JN854017 | JN853806 | ||
| A. venenatus Jurjević et al. 2012 | CBS 145753 T | USA | toxic dairy feed | JQ301896 | JN854003 | JN854014 | JN853803 | ||
| A. versicolor (Vuill.) Tirab. 1908 | CBS 583.65 T | unknown | unknown | EF652442 | EF652266 | EF652354 | EF652178 | ||
| Ochraceorosei | Funiculosi | A. funiculosus G. Sm. 1956 | NRRL 4744 T | Nigeria | loam soil | EF661223 | EF661112 | EF661175 | EF661078 |
| A. lannaensis N. Suwannarach et al. 2021 | SDBR-CMUO8 T | Thailand | soil | MW588211 | MW219783 | MW219781 | MW219785 | ||
| Ochraceorosei | A. ochraceoroseus Bartoli and Maggi 1979 | CBS 550.77 T | Côte d’Ivoire | forest soil | EF661224 | EF661113 | EF661137 | EF661074 | |
| Raperorum | Raperorum | A. ivoriensis Bartoli and Maggi 1979 | CBS 551.77 T | Côte d’Ivoire | forest soil | EF652441 | EF652265 | EF652353 | EF652177 |
| A. raperi Stolk and J.A. Mey. 1957 | CBS 123.56 T | Congo | soil | EF652454 | EF652278 | EF652366 | EF652190 | ||
| Silvatici | Silvatici | A. silvaticus Fennell and Raper 1955 | CBS 128.55 T | Ghana | soil | EF652448 | EF652272 | EF652360 | EF652184 |
| Sparsi | Biplani | A. biplanus Raper and Fennell 1965 | CBS 468.65 T | Costa Rica | soil | EF661210 | EF661116 | EF661130 | EF661036 |
| Conjuncti | A. conjunctus Kwon-Chung and Fennell 1965 | CBS 476.65 T | Costa Rica | soil | EF661179 | EF661111 | EF661133 | EF661042 | |
| Implicati | A. implicatus Persiani and Maggi 1994 | CBS 484.95 T | Côte d’Ivoire | forest soil | FJ491656 | FJ491667 | FJ491650 | MN969078 | |
| Sparsi | A. sparsus Raper and Thom 1944 | CBS 139.61 T | Costa Rica | soil | EF661181 | EF661125 | EF661173 | EF661071 | |
| Usti | Calidousti | A. calidoustus Varga et al. 2008 | CBS 121601 T | Netherlands | bronchoalveolar lavage fluid of Homo sapiens | HE616558 | FJ624456 | HE616559 | MN969061 |
| Deflecti | A. deflectus Fennell and Raper 1955 | CBS 109.55 T | Brazil | soil | EF652437 | EF652261 | EF652349 | EF652173 | |
| Monodiorum | A. monodii (Locq.-Lin.) Varga et al. 2011 | CBS 435.93 T | Chad | dung of Agnus | FJ531150 | FJ531171 | FJ531142 | MN969082 | |
| Usti | A. ustus (Bainier) Thom and Church 1926 | CBS 261.67 T | USA | culture contaminant | EF652455 | EF652279 | EF652367 | EF652191 | |
| outgroup | A. flavus Link 1809 | CBS 569.65 T | South Pacific | cellophane | AF027863 | EF661485 | EF661508 | EF661440 |
GenBank accession numbers in bold indicate the newly generated sequences.
Table 2.
Fungal species and sequences used in phylogenetic analyses of Aspergillus subgen. Circumdati.
| Section | Series | Species | Strain | Locality | Substrate | ITS | BenA | CaM | RPB2 |
|---|---|---|---|---|---|---|---|---|---|
| Flavipedes | Flavipedes | A. ardalensis A. Nováková et al. 2015 | CBS 134372 T | Spain | soil | FR733808 | HG916683 | HG916725 | HG916704 |
| A. capensis Visagie et al. 2014 | CBS 138188 T | South Africa | house dust | KJ775550 | KJ775072 | KJ775279 | KP987020 | ||
| A. flavipes (Bainier and R. Sartory) Thom and Church 1926 | NRRL 302 T | France | dung of dog | EF669591 | EU014085 | EF669549 | EF669633 | ||
| A. iizukae Sugiy 1967 | CBS 541.69 T | Japan | core sample from stratigraphic drilling | EF669597 | EU014086 | EF669555 | EF669639 | ||
| A. micronesiensis Visagie et al. 2014 | CBS 138183 T | Micronesia | house dust | KJ775548 | KJ775085 | KP987067 | KP987023 | ||
| A. neoflavipes Hubka et al. 2015 | CBS 260.73 T | Thailand | forest soil | EF669614 | EU014084 | EF669572 | EF669656 | ||
| A. okavangoensis Visagie and Nkwe 2021 | CBS 147420 T | Botswana | bat guano contaminated soil in cave | MW480880 | MW480788 | MW480706 | MW480790 | ||
| A. suttoniae J.P.Z. Siqueira et al. 2018 | FMR 13523 T | USA | sputum of Homo sapiens | LT899487 | LT899536 | LT899589 | LT899644 | ||
| A. templicola Visagie et al. 2014 | CBS 138181 T | Mexico | church dust | KJ775545 | KJ775092 | KJ775394 | KP987017 | ||
| A. urmiensis Arzanlou et al. 2016 | CBS 139558 T | Iran | soil | KP987073 | KP987041 | KP987056 | KP987030 | ||
| A. xishaensis X.C. Wang and W.Y. Zhuang, sp. nov. | ZC108 T | China: Hainan | sandy soil | OM414848 | OM475628 | OM475632 | OM475636 | ||
| Terrei | Terrei | A. alabamensis Balajee et al. 2009 | CBS 125693 T | USA | wound of Homo sapiens | KP987071 | KP987049 | EU147583 | KP987018 |
| A. aureoterreus Samson et al. 2011 | CBS 503.65 T | USA | soil | EF669580 | EF669524 | EF669538 | EF669622 | ||
| A. citrinoterreus J. Guinea et al. 2015 | CBS 138921 T | Spain | sputum of Homo sapiens | KP175260 | LN680657 | LN680685 | MN969155 | ||
| A. floccosus (Y.K. Shih) Samson et al. 2011 | CBS 116.37 T | China: Hubei | waste cloth | KP987086 | FJ491714 | KP987066 | KP987021 | ||
| A. heldtiae Visagie 2020 | PPRI 4229 T | South Africa | seed of Pennisetum glaucum | MK450656 | MK450981 | MK451518 | MK450809 | ||
| A. hortae (Langeron) C.W. Dodge 1935 | CBS 124230 T | Brazil | ear of Homo sapiens | KP987087 | FJ491706 | KP987054 | KP987022 | ||
| A. neoafricanus Samson et al. 2011 | CBS 130.55 T | Ghana | soil | EF669585 | EF669516 | EF669543 | EF669627 | ||
| A. neoterreus X.C. Wang and W.Y. Zhuang, sp. nov. | ZC111 T | China: Hainan | sandy soil | OM414849 | OM475629 | OM475633 | OM475637 | ||
| A. pseudoterreus S.W. Peterson et al. 2011 | CBS 123890 T | Argentina | soil | EF669598 | EF669523 | EF669556 | EF669640 | ||
| A. terreus Thom 1918 | CBS 601.65 T | USA | soil | EF669586 | EF669519 | EF669544 | EF669628 | ||
| Flavi | Flavi | A. flavus Link 1809 | CBS 569.65 T | South Pacific | cellophane | AF027863 | EF661485 | EF661508 | EF661440 |
GenBank accession numbers in bold indicate the newly generated sequences.
3. Results
3.1. Phylogenetic Analysis
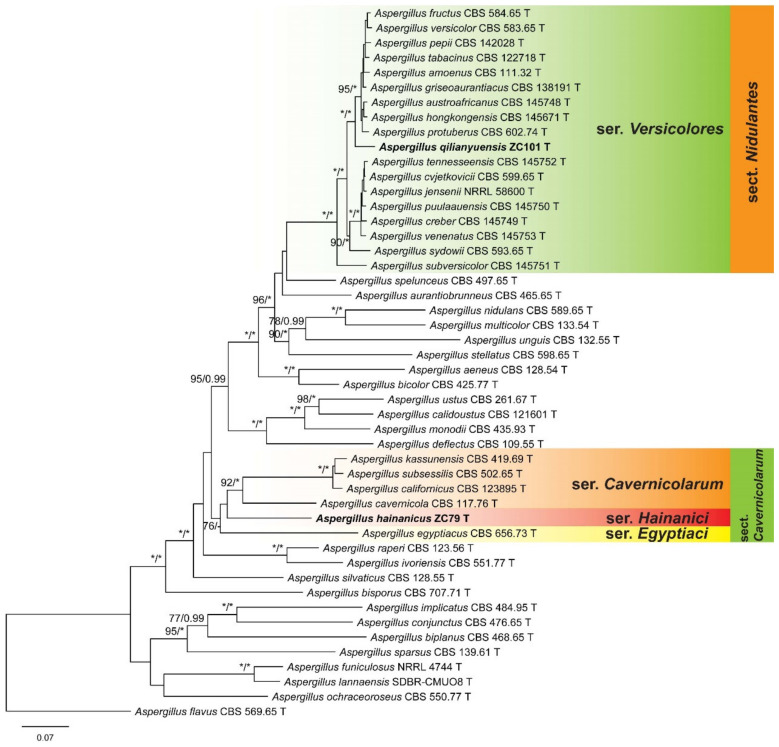
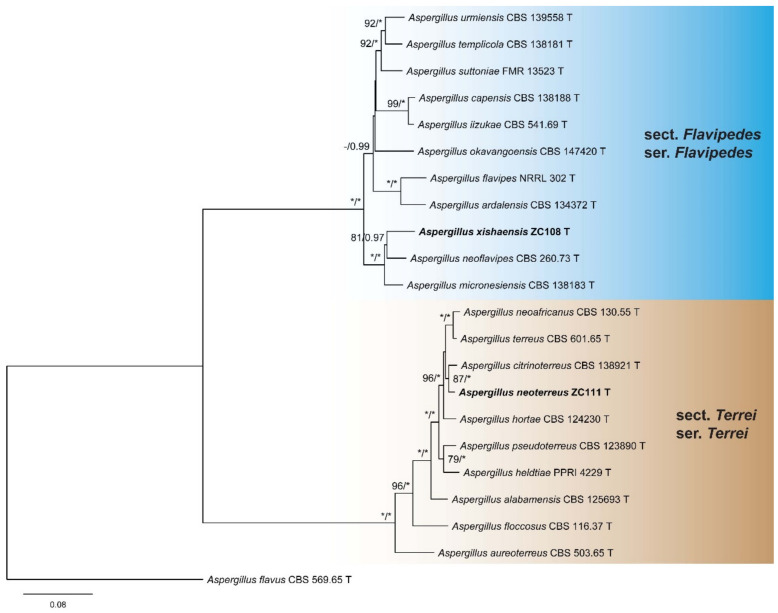
To determine the positions of the isolates, two combined datasets (BenA + CaM + RPB2) of Aspergillus subgenera Nidulantes and Circumdati were compiled and analyzed. The detailed characteristics of the datasets are listed in Table 3. In the phylogeny of Aspergillus subg. Nidulantes (Figure 1), the strains ZC79 and ZC101 were located in sect. Cavernicolarum and Nidulantes, respectively. The strain ZC79 was sister to the species of ser. Cavernicolarum and Egyptiaci, and a new series was proposed as ser. Hainanici to accommodate it. The strain ZC 101 formed a distinct lineage in ser. Versicolores. As shown in the phylogenetic tree of Aspergillus subg. Circumdati (Figure 2), the strain ZC108 was a member of sect. Flavipedes ser. Flavipedes, and clustered with A. micronesiensis and A. neoflavipes. The strain ZC111 was revealed to be affiliated to sect. Terrei ser. Terrei, and shared a close relationship with A. citrinoterreus.
Table 3.
Detailed characteristics of datasets of Aspergillus.
| Subgenus | Locus | No. of Seq. | Length of Alignment (bp) | No. of Variable Sites | No. of Parsimony-Informative Sites | Model for BI |
|---|---|---|---|---|---|---|
| Nidulantes | BenA | 48 | 528 | 292 | 235 | |
| CaM | 48 | 829 | 477 | 408 | ||
| RPB2 | 47 | 1014 | 429 | 377 | ||
| combined | 48 | 2371 | 1198 | 1020 | TIM + I + G | |
| Circumdati | BenA | 22 | 541 | 273 | 199 | |
| CaM | 22 | 589 | 286 | 218 | ||
| RPB2 | 22 | 998 | 301 | 216 | ||
| combined | 22 | 2128 | 860 | 633 | TIM + I + G |
Full names of the used models: TIM + I + G (transition model with invariable sites and gamma distribution).
Figure 1.
ML phylogeny of Aspergillus subgen. Nidulantes inferred from combined BenA, CaM and RPB2 dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. *Asterisk denotes 100% bootstrap or 1.00 posterior probability.
Figure 2.
ML phylogeny of Aspergillus subgen. Circumdati inferred from combined BenA, CaM and RPB2 dataset. Bootstrap values ≥70% (left) or posterior probability values ≥0.95 (right) are indicated at nodes. *Asterisk denotes 100% bootstrap or 1.00 posterior probability.
3.2. Taxonomy
Series Hainanici X.C. Wang and W.Y. Zhuang, ser. nov.
Fungal Names: FN570966.
Etymology: Named after Aspergillus hainanicus.
Type: Aspergillus hainanicus X.C. Wang and W.Y. Zhuang.
In Aspergillus subgen. Nidulantes sect. Cavernicolarum.
Diagnosis: Series Hainanici belongs to subgen. Nidulantes sect. Cavernicolarum and is sister to series Cavernicolarum and Egyptiaci (Figure 1). Colonies no growth at 37 °C; conidia en masse greyish black; conidiophores biseriate; stipes short, thick walls, brown; vesicles globose to subglobose; metulae cylindrical to obovate, covering almost a half surface of the vesicle; phialides flask-shaped; conidia large, subglobose, strongly echinulate.
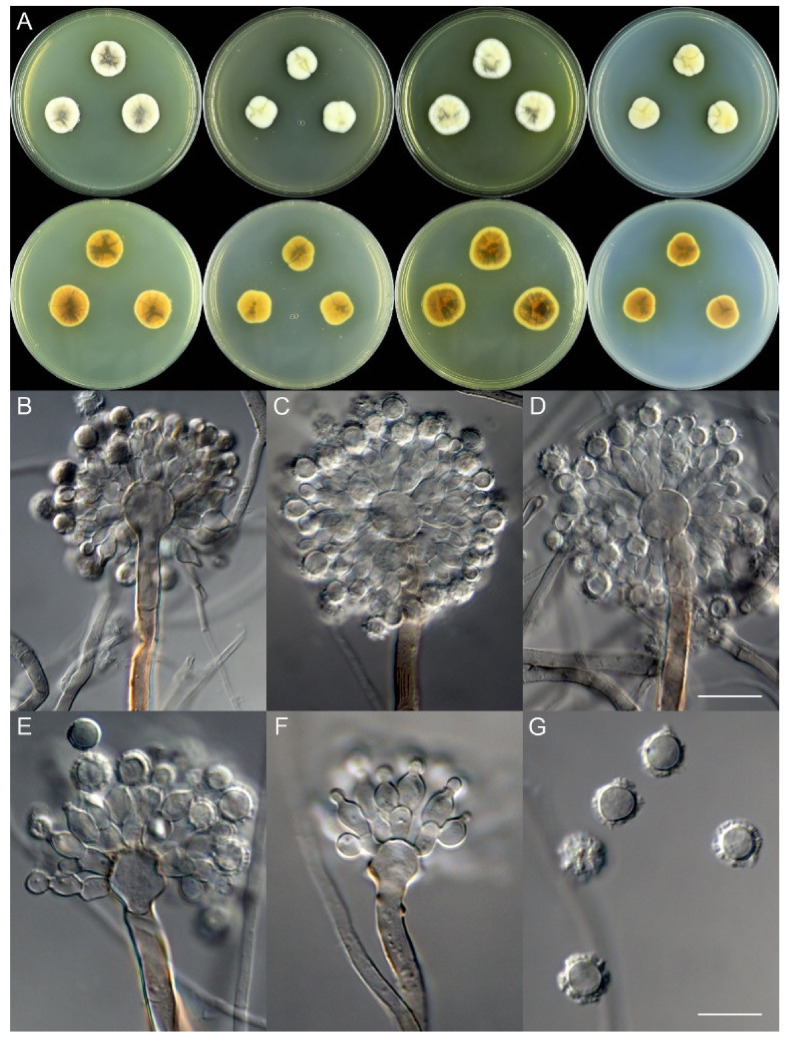
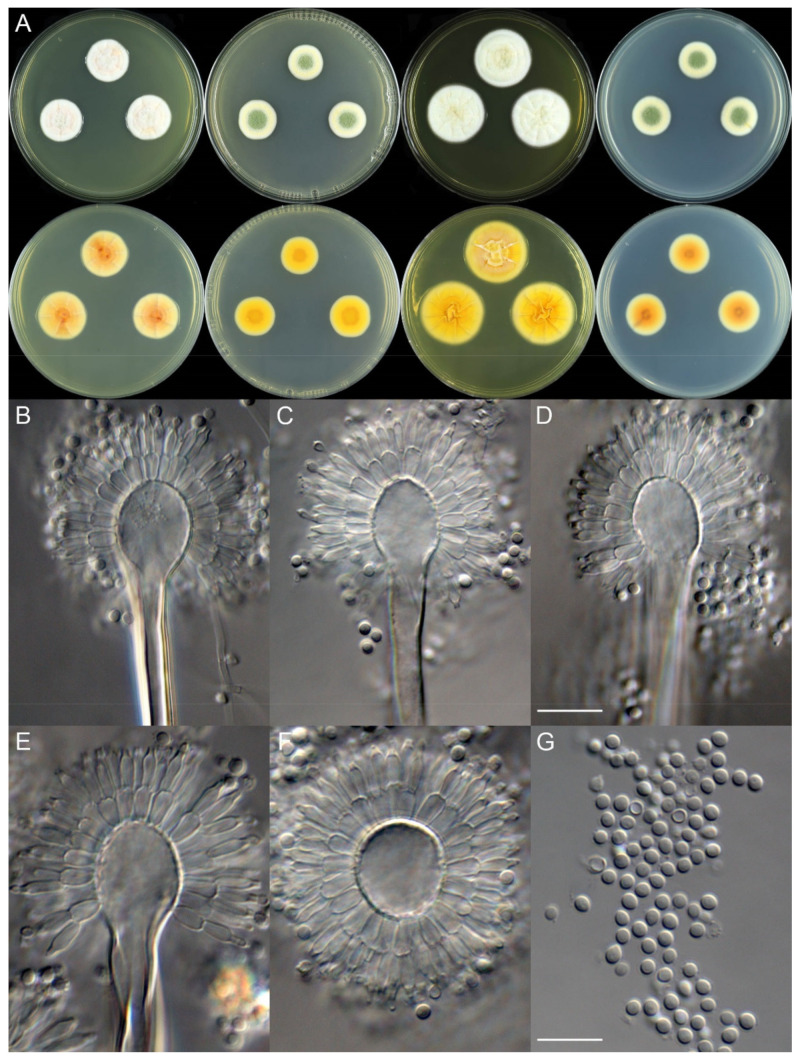
Aspergillus hainanicus X.C. Wang and W.Y. Zhuang, sp. nov. Figure 3.
Figure 3.
Colonial and microscopic morphology of Aspergillus hainanicus (ZC79). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES and PDA; bottom row left to right, reverse CYA, MEA, YES and PDA); (B–F) Conidiophores; (G) Conidia. Bars: (D) = 15 µm, applies to (B,C); (G) = 10 µm, applies to (E,F).
Fungal Names: FN570967.
Etymology: The specific epithet refers to the type locality.
In Aspergillus subgen. Nidulantes sect. Cavernicolarum ser. Hainanici.
Typification: CHINA. Hainan Province, Sansha City, Xisha District, Xisha Islands, Xuande Islands, Yongxing Island, 16°50′4″ N 112°20′49″ E, in sandy soil (phosphorous lime soil) under unidentified plants, 29 March 2015, Ye-Wei Xia, culture, Kai Chen, ZC79 (holotype HMAS 247855, ex-type strain CGMCC 3.20888).
DNA barcodes: ITS OM414846, BenA OM475626, CaM OM475630, RPB2 OM475634.
Colony diam.: 7 days, 25 °C (unless stated otherwise): CYA 18–20 mm; CYA 37 °C no growth; MEA 16–17 mm; YES 21–22 mm; PDA 16–17 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, concave at centers, protuberant at margins, radially sulcate; margins narrow, entire; mycelia white and then buff; texture velutinous; sporulation sparse; conidia en masse greyish black; soluble pigments light brown; exudates tiny, hyaline and clear; reverse yellow to orange, but black at centers and with black sectors. On MEA 25 °C, 7 days: Colonies irregular, protuberant; margins narrow, entire; mycelia white and then cream to light yellow; texture velutinous; sporulation absent; soluble pigments light brown; exudates tiny, hyaline and clear; reverse buff, yellow to orange, but black at centers. On YES 25 °C, 7 days: Colonies nearly circular or irregular, protuberant at centers, radially sulcate; margins narrow, fimbriate; mycelia white; texture velutinous; sporulation sparse; conidia en masse greyish black; soluble pigments greenish-brown; exudates absent; reverse orange to black. On PDA 25 °C, 7 days: Colonies irregular, protuberant; margins narrow, entire; mycelia white and then cream to light yellow; texture velutinous; sporulation absent; soluble pigments yellow; exudates tiny, hyaline and clear; reverse buff, yellow to orange, and with black sectors.
Micromorphology: Conidial heads radiate; stipes short, 55–90 × 4.5–6.0 μm, thick walls, smooth, brown, not septate; vesicles 7.5–13 × 9.0–13 μm, globose to subglobose; biseriate; metulae 5.0–9.0 × 3.0–6.5 μm, cylindrical to obovate, covering almost a half surface of the vesicle; phialides 5.5–8.0 × 3.5–5.0 μm, flask-shaped; conidia 6.0–9.5 μm, subglobose, strongly echinulate.
Note: This species is phylogenetically related to A. californicus, A. cavernicola, A. kassunensis and A. subsessilis of ser. Cavernicolarum and A. egyptiacus of ser. Egyptiaci (Figure 1), but differs from the former four species in its brown stipe and larger, strongly echinulate conidia, and differs from the latter one due to no growth on CYA at 37 °C, slower growth rates on MEA and YES, brown stipe and larger and strongly echinulate conidia (Table 4).
Table 4.
Morphological comparisons of new species and their closely related species.
| Species | CYA 25 °C (mm) | CYA 37 °C (mm) | MEA (mm) | YES (mm) | Conidia Shape | Conidia Wall | Conidia Size (µm) | Reference |
|---|---|---|---|---|---|---|---|---|
| A. cavernicola | 10–12 | no growth | 12–13 | 14–15 | subglobose | smooth to echinulate | 5–6 × 3.5–4.5 | [19] |
| A. californicus | 20–24 | no growth | 19–20 | 26–27 | subglobose to ellipsoidal | smooth to finely roughened | 3–4.5 × 2.5–4.5 | [19] |
| A. kassunensis | 15–16 | no growth | 18–20 | 21–22 | globose | smooth | 2–3 | [19] |
| A. subsessilis | 16–17 | no growth | 12–13 | 18–19 | globose | smooth | 3–4 | [19] |
| A. egyptiacus | 13–20 | 21–24 | 29–30 | 32–45 | globose to subglobose | smooth | 4.5–6.5 × 3.5–6 | [19] |
| A. hainanicus | 18–20 | no growth | 16–17 | 21–22 | subglobose | strongly echinulate | 6–9.5 | This study |
| A. versicolor | 28–36 | 8 | 21–31 | n.a. | spherical to subspherical | finely roughened | 2.5–3.5 | [32] |
| A. qilianyuensis | 21–23 | no growth | 17–20 | 29–30 | subglobose | smooth | 2–3 | This study |
| A. micronesiensis | 22–28 | 17–25 | 20–25 | 35–44 | globose to subglobose | smooth to finely roughened | 2.5–3.5 | [33] |
| A. neoflavipes | 30–33 | 20–22 | 34–35 | n.a. | globose to subglobose | smooth | 2.5–3 | [34] |
| A. xishaensis | 19–22 | 19–21 | 16–20 | 25–29 | globose to subglobose | smooth | 3–4 | This study |
| A. citrinoterreus | 33–35 | n.a. | 23–25 | n.a. | globose to subglobose | smooth | 2–3 × 1.5–3 | [35] |
| A. neoterreus | 26–28 | 57–58 | 21–23 | 37–40 | subglobose to broad ellipsoid | smooth | 2–2.5 | This study |
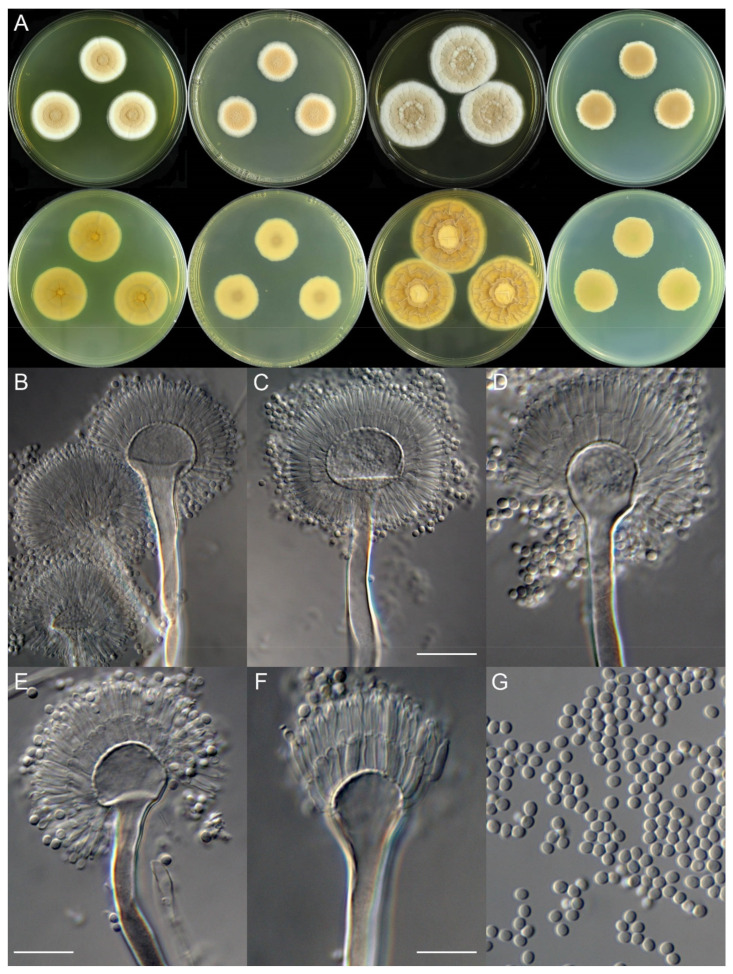
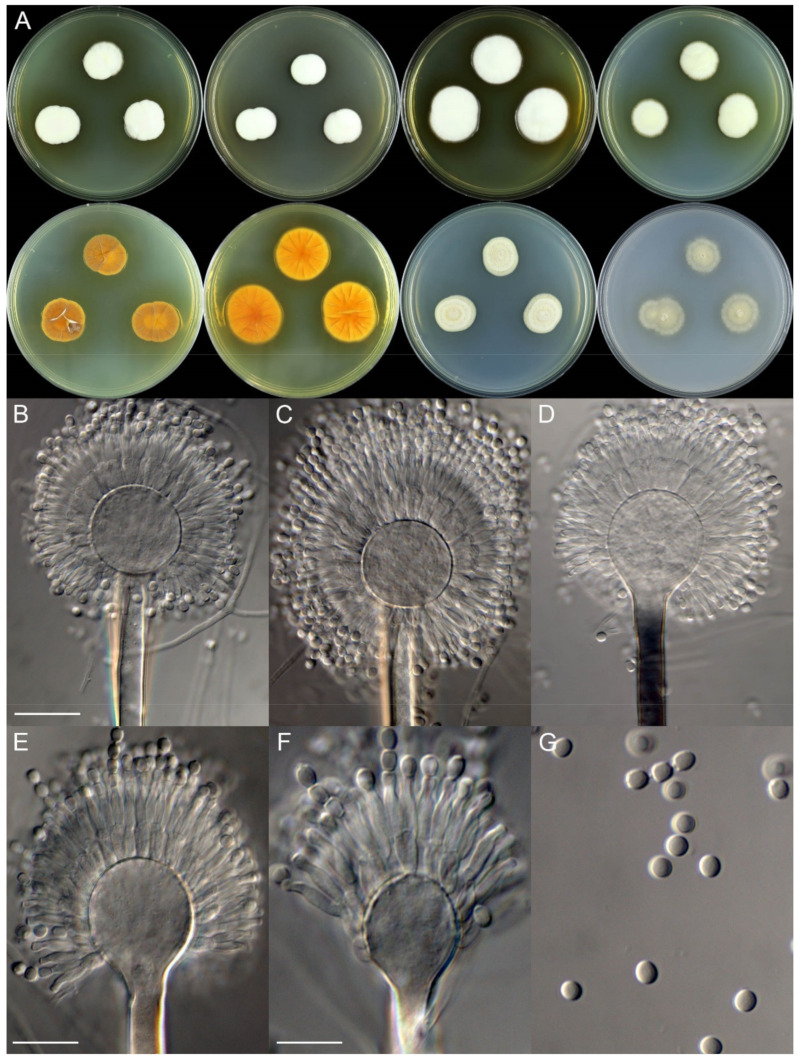
Aspergillus neoterreus X.C. Wang and W.Y. Zhuang, sp. Nov. Figure 4.
Figure 4.
Colonial and microscopic morphology of Aspergillus neoterreus (ZC111). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES and PDA; bottom row left to right, reverse CYA, MEA, YES and PDA); (B–F) Conidiophores; (G) Conidia. Bars: (C) = 20 µm, applies to (B); (E) = 12.5 µm, applies to (D); (F) = 10 µm, applies to (G).
Fungal Names: FN570968.
Etymology: The specific epithet refers to the close relationship with A. terreus.
In Aspergillus subgen. Circumdati sect. Terrei ser. Terrei.
Typification: CHINA. Hainan Province, Sansha City, Xisha District, Xisha Islands, Xuande Islands, Qilianyu Islands, Nanshazhou Island, 16°55′46″ N 112°20′55″ E, in sandy soil (phosphorous lime soil) under unidentified plants, 29 March 2015, Ye-Wei Xia, culture, Kai Chen, ZC111 (holotype HMAS 247856, ex-type strain CGMCC 3.20891).
DNA barcodes: ITS OM414849, BenA OM475629, CaM OM475633, RPB2 OM475637.
Colony diam.: 7 days, 25 °C (unless stated otherwise): CYA 26–28 mm; CYA 37 °C 57–58 mm; MEA 21–23 mm; YES 37–40 mm; PDA 20–22 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, slightly protuberant at centers, concentrically sulcate; margins narrow, entire; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse wheat, yellow-brown to khaki; soluble pigments absent; exudates absent; reverse light brown. On CYA 37 °C, 7 days: Colonies nearly circular or irregular, plain, radially sulcate; margins moderately wide, irregular; mycelia white; texture velutinous; sporulation dense; conidia en masse wheat, yellow-brown to khaki; soluble pigments absent; exudates absent; reverse yellow-brown to dark brown. On MEA 25 °C, 7 days: Colonies nearly circular, plain, slightly protuberant at centers; margins wide, entire; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse wheat, yellow-brown to khaki; soluble pigments absent; exudates absent; reverse buff to yellow-brown, but light brown at centers. On YES 25 °C, 7 days: Colonies nearly circular, concave at centers, strongly sulcate; margins wide, fimbriate; mycelia white; texture velutinous; sporulation moderately dense; conidia en masse wheat, yellow-brown to khaki; soluble pigments absent; exudates absent; reverse yellow-brown to light brown. On PDA 25 °C, 7 days: Colonies nearly circular, plain, slightly protuberant at centers; margins narrow, irregular; mycelia white; texture velutinous; sporulation dense; conidia en masse wheat, yellow-brown to khaki; soluble pigments absent; exudates absent; reverse pink-brown, but greenish-brown at centers.
Micromorphology: Conidial heads radiate; stipes 150–225 × 2.5–7.5 μm, thick walls, smooth, hyaline or blackish, not septate; vesicles 11–16.5 × 8.5–27 μm, subglobose to ellipsoid; biseriate; metulae 6.0–7.5 × 2.0–3.0 μm, cylindrical, covering a half to two-thirds the surface of the vesicle; phialides 7.0–8.5 × 1.5–2.0 μm, acerose; conidia 2.0–2.5 μm, subglobose to broad ellipsoid, smooth.
Note: This species is phylogenetically related to A. citrinoterreus (Figure 2) but differs in slower growth rate on CYA and smaller conidia (Table 4).
Aspergillus qilianyuensis X.C. Wang and W.Y. Zhuang, sp. Nov. Figure 5.
Figure 5.
Colonial and microscopic morphology of Aspergillus qilianyuensis (ZC101). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES and PDA; bottom row left to right, reverse CYA, MEA, YES and PDA); (B–F) Conidiophores; (G) Conidia. Bars: (D) = 12.5 µm, applies to (B,C); (G) = 10 µm, applies to (E,F).
Fungal Names: FN570969.
Etymology: The specific epithet refers to the type locality.
In Aspergillus subgen. Nidulantes sect. Nidulantes ser. Versicolores.
Typification: CHINA. Hainan Province, Sansha City, Xisha District, Xisha Islands, Xuande Islands, Qilianyu Islands, Nanshazhou Island, 16°55′46″ N 112°20′55″ E, in sandy soil (phosphorous lime soil) under unidentified plants, 29 March 2015, Ye-Wei Xia, culture, Kai Chen, ZC101 (holotype HMAS 247857, ex-type strain CGMCC 3.20889).
DNA barcodes: ITS OM414847, BenA OM475627, CaM OM475631, RPB2 OM475635.
Colony diam.: 7 days, 25 °C (unless stated otherwise): CYA 21–23 mm; CYA 37 °C no growth; MEA 17–20 mm; YES 29–30 mm; PDA 19–20 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies nearly circular, protuberant, concentrically and radially sulcate; margins narrow, entire; mycelia white and then pink; texture velutinous; sporulation sparse; conidia en masse light greyish green; soluble pigments absent; exudates absent; reverse buff to pink-brown. On MEA 25 °C, 7 days: Colonies nearly circular, slightly protuberant at central areas; margins wide, entire; mycelia white and becoming yellow; texture velutinous; sporulation moderately dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse buff to vivid yellow, but orange-brown at centers. On YES 25 °C, 7 days: Colonies nearly circular, protuberant or concave at centers, concentrically and radially sulcate, deep; margins narrow, entire; mycelia white; texture velutinous; sporulation sparse; conidia en masse light yellow; soluble pigments absent; exudates absent; reverse yellow-brown. On PDA 25 °C, 7 days: Colonies nearly circular, slightly protuberant at central areas; margins wide, entire; mycelia white and then yellow; texture velutinous; sporulation moderately dense; conidia en masse greyish green; soluble pigments absent; exudates absent; reverse buff, yellow-brown to orange-brown.
Micromorphology: Conidial heads radiate; stipes 225–325 × 4.0–8.0 μm, thick walls, smooth, hyaline or blackish, not septate; vesicles 16–20 × 10–18 μm, ellipsoid; biseriate; metulae 5.0–6.0 × 3.0–3.5 μm, cylindrical, covering two-thirds to almost the entire surface of the vesicle; phialides 6.0–8.0 × 2.0–2.5 μm, flask-shaped to acerose; conidia 2.0–3.0 μm, subglobose, smooth.
Note: This species formed a distinct lineage in ser. Versicolores (Figure 1). Morphologically, it differs from the type species of this series, A. versicolor, in slower growth rates on CYA and MEA and smooth and smaller conidia (Table 4).
Aspergillus xishaensis X.C. Wang and W.Y. Zhuang, sp. nov. Figure 6.
Figure 6.
Colonial and microscopic morphology of Aspergillus xishaensis (ZC108). (A) Colony phenotypes (25 °C, 7 days; top row left to right, obverse CYA, MEA, YES and PDA; bottom row left to right, reverse CYA and YES, obverse PDA with 3% NaCl and OA); (B–F) Conidiophores; (G) Conidia. Bars: (B) = 20 µm, applies to (C,D); (E) = 15 µm; (F) = 10 µm, applies to (G).
Fungal Names: FN570970.
Etymology: The specific epithet refers to the type locality.
In Aspergillus subgen. Circumdati sect. Flavipedes ser. Flavipedes.
Typification: CHINA. Hainan Province, Sansha City, Xisha District, Xisha Islands, Xuande Islands, Qilianyu Islands, Nanshazhou Island, 16°55′46″ N 112°20′55″ E, in sandy soil (phosphorous lime soil) under unidentified plants, 29 March 2015, Ye-Wei Xia, culture, Kai Chen, ZC108 (holotype HMAS 247858, ex-type strain CGMCC 3.20890).
DNA barcodes: ITS OM414848, BenA OM475628, CaM OM475632, RPB2 OM475636.
Colony diam.: 7 days, 25 °C (unless stated otherwise): CYA 19–22 mm; CYA 37 °C 19–21 mm; MEA 16–20 mm; YES 25–29 mm; PDA 18–22 mm; PDA (3% NaCl) 19–20 mm; OA 19–20 mm.
Colony characteristics: On CYA 25 °C, 7 days: Colonies irregular, protuberant, radially sulcate; margins narrow, entire; mycelia white and then light yellow; texture velutinous; sporulation absent; soluble pigments yellow-brown; exudates absent; reverse yellow-brown to light brown. On CYA 37 °C, 7 days: Colonies nearly circular, protuberant at centers, radially sulcate; margins wide, fimbriate; mycelia white and then light yellow; texture velutinous; sporulation absent; soluble pigments yellow-brown; exudates absent; reverse yellow-brown to dark brown. On MEA 25 °C, 7 days: Colonies nearly circular or irregular, protuberant; margins narrow, entire; mycelia white and then cream; texture velutinous; sporulation absent; soluble pigments yellow-brown; exudates absent; reverse light brown, but buff at margins. On YES 25 °C, 7 days: Colonies nearly circular, protuberant at centers, radially sulcate; margins narrow, entire; mycelia white and then light cream; texture velutinous; sporulation absent; soluble pigments yellow-brown; exudates absent; reverse yellow-brown to orange-brown. On PDA 25 °C, 7 days: Colonies nearly circular or irregular, protuberant; margins narrow, entire; mycelia white and then light cream; texture velutinous; sporulation absent; soluble pigments yellow-brown; exudates greenish-yellow, clear; reverse yellow-brown to light brown. On PDA (3% NaCl) 25 °C, 7 days: Colonies oblong, protuberant; margins moderately wide, entire; mycelia cream; texture velutinous; sporulation dense; conidia en masse white to cream; soluble pigments light yellow-brown; exudates absent; reverse yellow-brown to light brown. On OA 25 °C, 7 days: Colonies nearly circular or irregular, protuberant; margins wide, fimbriate; mycelia cream; texture velutinous; sporulation sparse; conidia en masse white to cream; soluble pigments yellow-brown; exudates absent; reverse light yellow, but light brown at centers.
Micromorphology: Conidial heads radiate; stipes long, 700–1400 × 7.5–10 μm, thick walls, smooth, hyaline or blackish, not septate; vesicles 18–35 × 15–35 μm, globose to broad ellipsoid; biseriate; metulae 7.0–11 × 3.5–4.5 μm, cylindrical, covering two thirds to almost the entire surface of the vesicle; phialides 9.0–11.5 × 2.5–3.0 μm, flask-shaped to acerose; conidia 3.0–4.0 μm, globose to subglobose, smooth.
Note: This species is phylogenetically related to A. micronesiensis and A. neoflavipes (Figure 2) but differs from them in slower growth rates on CYA, MEA and YES and larger conidia (Table 4).
4. Discussion
Aspergillus is a large genus with more than 400 accepted species and more than 1000 names. A comprehensive taxonomic treatment of the genus was recently established on the basis of molecular data and morphological characteristics [6]. Six subgenera, twenty-seven sections and seventy-five series were currently accepted, among which five new sections and seventy-three new series were erected. Based on the above treatment, researchers are able to quickly position their materials to specific ranks of series, sections and subgenera. Three of the four new species described in this study were classified into the known series, except for A. hainanicus, for which the new series Hainanici is proposed. Along with future discovery of new taxa, the current classification system may be updated.
In the Flavi, Fumigati, Nigri and Terrei sections of Aspergillus, some species cause the infectious disease aspergillosis, such as the most frequently occurred and well-known pathogen A. fumigatus [36]. In sect. Nidulantes, A. versicolor (Vuill.) Tirab., a close relative of A. qilianyuensis, was isolated from the skin [37] and nails [38] of humans and also invasively infected multiple organs of dogs [39,40]. Aspergillus hongkongensis C.C. Tsang et al. causes onychomycosis [41]. In subgen. Circumdati, A. citrinoterreus J. Guinea et al. and A. suttoniae J.P.Z. Siqueira et al. were isolated from the sputum of humans [35,42], and A. alabamensis Balajee et al. from the wounds of humans [43]. Whether others of these sections are potentially pathogenic requires future investigation.
Tropical islands represent a unique ecosystem. Due to their extremely isolated location and special environmental conditions, some of them are considered as the world’s biodiversity hotspots. Several species of Aspergillus were recorded from similar geographical origins, such as A. griseoaurantiacus Visagie et al. and A. micronesiensis Visagie et al. from Micronesia [33], and A. puulaauensis Jurjević et al. from Hawaii [32]. The four new species were all derived from the soil samples of the Xisha Islands, which seem to exhibit high species diversity. Further explorations on tropical islands are desperately needed, and we certainly expect to find more new fungi there.
Acknowledgments
The authors would like to thank Tai-Hui Li and Ye-Wei Xia (Guangdong Institute of Microbiology) for providing the soil samples and Kai Chen of this institute for providing cultures for this study.
Author Contributions
Conceptualization, W.-Y.Z. and X.-C.W.; methodology, X.-C.W.; software, X.-C.W.; validation, X.-C.W. and W.-Y.Z.; formal analysis, X.-C.W.; investigation, X.-C.W.; resources, X.-C.W. and W.-Y.Z.; data curation, X.-C.W.; writing—original draft preparation, X.-C.W.; writing—review and editing, W.-Y.Z. and X.-C.W.; visualization, X.-C.W.; supervision, W.-Y.Z.; project administration, W.-Y.Z.; funding acquisition, W.-Y.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This project was supported by the National Natural Science Foundation of China (31750001) and Key Research Program of Frontier Science, Chinese Academy of Sciences (QYZDY-SSW-SMC029).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences newly generated in this study have been submitted to the GenBank database.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Frisvad J.C., Moller L.L.H., Larsen T.O., Kumar R., Arnau J. Safety of the fungal workhorses of industrial biotechnology: Update on the mycotoxin and secondary metabolite potential of Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei. Appl. Microbiol. Biotechnol. 2018;102:9481–9515. doi: 10.1007/s00253-018-9354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behera B.C. Citric acid from Aspergillus niger: A comprehensive overview. Crit. Rev. Microbiol. 2020;46:727–749. doi: 10.1080/1040841X.2020.1828815. [DOI] [PubMed] [Google Scholar]
- 3.Kumar P., Mahato D.K., Kamle M., Mohanta T.K., Kang S.G. Aflatoxins: A global concern for food safety, human health and their management. Front. Microbiol. 2016;7:2170. doi: 10.3389/fmicb.2016.02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solairaj D., Legrand N.N.G., Yang Q.Y., Zhang H.Y. Isolation of pathogenic fungi causing postharvest decay in table grapes and in vivo biocontrol activity of selected yeasts against them. Physiol. Mol. Plant Pathol. 2020;110:101478. doi: 10.1016/j.pmpp.2020.101478. [DOI] [Google Scholar]
- 5.Balajee S.A., Houbraken J., Verweij P.E., Hong S.B., Yaghuchi T., Varga J., Samson R.A. Aspergillus species identification in the clinical setting. Stud. Mycol. 2007;59:39–46. doi: 10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houbraken J., Kocsube S., Visagie C.M., Yilmaz N., Wang X.C., Meijer M., Kraak B., Hubka V., Bensch K., Samson R.A., et al. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Stud. Mycol. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sklenar F., Jurjevic Z., Houbraken J., Kolarik M., Arendrup M.C., Jorgensen K.M., Siqueira J.P.Z., Gene J., Yaguchi T., Ezekiel C.N., et al. Re-examination of species limits in Aspergillus section Flavipedes using advanced species delimitation methods and description of four new species. Stud. Mycol. 2021;99:100120. doi: 10.1016/j.simyco.2021.100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh P., Callicott K.A., Orbach M.J., Cotty P.J. Molecular analysis of S-morphology aflatoxin producers from the United States reveals previously unknown diversity and two new taxa. Front. Microbiol. 2020;11:1236. doi: 10.3389/fmicb.2020.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crous P.W., Wingfield M.J., Chooi Y.H., Gilchrist C.L.M., Lacey E., Pitt J.I., Roets F., Swart W.J., Cano-Lira J.F., Valenzuela-Lopez N., et al. Fungal Planet description sheets: 1042–1111. Persoonia. 2020;44:301–459. doi: 10.3767/persoonia.2020.44.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correia A.C.R.B., Barbosa R.N., Frisvad J.C., Houbraken J., Souza-Motta C.M. The polyphasic re-identification of a Brazilian Aspergillus section Terrei collection led to the discovery of two new species. Mycol. Prog. 2020;19:885–903. doi: 10.1007/s11557-020-01605-4. [DOI] [Google Scholar]
- 11.Gilchrist C.L.M., Lacey H.J., Vuong D., Pitt J.I., Lange L., Lacey E., Pilgaard B., Chooi Y.H., Piggott A.M. Comprehensive chemotaxonomic and genomic profiling of a biosynthetically talented Australian fungus, Aspergillus burnettii sp. nov. Fungal Genet. Biol. 2020;143:103435. doi: 10.1016/j.fgb.2020.103435. [DOI] [PubMed] [Google Scholar]
- 12.Al-Bedak O.A., Moubasher A.H., Ismail M.A., Mohamed R.A. Aspergillus curvatus, a new species in section Circumdati isolated from an alkaline water of Lake Khadra in Wadi-El-Natron, Egypt. Asian J. Mycol. 2020;3:325–334. doi: 10.5943/ajom/3/1/7. [DOI] [Google Scholar]
- 13.Al-Bedak O.A., Moubasher A.H. Aspergillus gaarensis, a new addition to section Circumdati from soil of Lake El-Gaar in Wadi-El-Natron, Egypt. Stud. Fungi. 2020;5:59–65. doi: 10.5943/sif/5/1/5. [DOI] [Google Scholar]
- 14.Boonmee S., Wanasinghe D.N., Calabon M.S., Huanraluek N., Chandrasiri S.K.U., Jones G.E.B., Rossi W., Leonardi M., Singh S.K., Rana S., et al. Fungal diversity notes 1387–1511: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2021;111:1–335. doi: 10.1007/s13225-021-00489-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doilom M., Guo J.W., Phookamsak R., Mortimer P.E., Karunarathna S.C., Dong W., Liao C.F., Yan K., Pem D., Suwannarach N., et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front. Microbiol. 2020;11:585215. doi: 10.3389/fmicb.2020.585215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Z.F., Zhou S.Y., Eurwilaichitr L., Ingsriswang S., Raza M., Chen Q., Zhao P., Liu F., Cai L. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers. 2021;106:29–136. doi: 10.1007/s13225-020-00453-7. [DOI] [Google Scholar]
- 17.Sun B.D., Huang P.P., Wei H.L., Cai W.J., Wang L., Liu S.K., Jiang X.Z., Chen A.J. Aspergillus telluris, a new soil derived species belonging to Aspergillus subgenus Polypaecilum. Phytotaxa. 2020;455:137–151. doi: 10.11646/phytotaxa.455.2.5. [DOI] [Google Scholar]
- 18.Da Silva J.J., Iamanaka B.T., Ferranti L.S., Massi F.P., Taniwaki M.H., Puel O., Lorber S., Frisvad J.C., Fungaro M.H.P. Diversity within Aspergillus niger clade and description of a new species: Aspergillus vinaceus sp. nov. J. Fungi. 2020;6:371. doi: 10.3390/jof6040371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun B.D., Houbraken J., Frisvad J.C., Jiang X.Z., Chen A.J., Samson R.A. New species in Aspergillus section Usti and an overview of Aspergillus section Cavernicolarum. Int. J. Syst. Evol. Microbiol. 2020;70:5401–5416. doi: 10.1099/ijsem.0.004425. [DOI] [PubMed] [Google Scholar]
- 20.Visagie C.M., Houbraken J., Frisvad J.C., Hong S.B., Klaassen C.H., Perrone G., Seifert K.A., Varga J., Yaguchi T., Samson R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014;78:343–371. doi: 10.1016/j.simyco.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X.C., Chen K., Xia Y.W., Wang L., Li T.H., Zhuang W.Y. A new species of Talaromyces (Trichocomaceae) from the Xisha Islands, Hainan, China. Phytotaxa. 2016;267:187–200. doi: 10.11646/phytotaxa.267.3.2. [DOI] [Google Scholar]
- 22.Wang X.C., Chen K., Qin W.T., Zhuang W.Y. Talaromyces heiheensis and T. mangshanicus, two new species from China. Mycol. Prog. 2017;16:73–81. doi: 10.1007/s11557-016-1251-3. [DOI] [Google Scholar]
- 23.Wang X.C., Chen K., Zeng Z.Q., Zhuang W.Y. Phylogeny and morphological analyses of Penicillium section Sclerotiora (Fungi) lead to the discovery of five new species. Sci. Rep. 2017;7:8233. doi: 10.1038/s41598-017-08697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z.K., Wang X.C., Zhuang W.Y., Cheng X.H., Zhao P. New species of Talaromyces (Fungi) isolated from soil in southwestern China. Biology. 2021;10:745. doi: 10.3390/biology10080745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 27.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [Google Scholar]
- 30.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 32.Jurjevic Z., Peterson S.W., Horn B.W. Aspergillus section Versicolores: Nine new species and multilocus DNA sequence based phylogeny. IMA Fungus. 2012;3:59–79. doi: 10.5598/imafungus.2012.03.01.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visagie C.M., Hirooka Y., Tanney J.B., Whitfield E., Mwange K., Meijer M., Amend A.S., Seifert K.A., Samson R.A. Aspergillus, Penicillium and Talaromyces isolated from house dust samples collected around the world. Stud. Mycol. 2014;78:63–139. doi: 10.1016/j.simyco.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubka V., Novakova A., Kolarik M., Jurjevic Z., Peterson S.W. Revision of Aspergillus section Flavipedes: Seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- 35.Guinea J., Sandoval-Denis M., Escribano P., Pelaez T., Guarro J., Bouza E. Aspergillus citrinoterreus, a new species of section Terrei isolated from samples of patients with nonhematological predisposing conditions. J. Clin. Microbiol. 2015;53:611–617. doi: 10.1128/JCM.03088-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Latge J.P., Chamilos G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019;33:e00140-18. doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele A.E. A case of infection with Aspergillus versicolor. Boston Med. Surg. J. 1926;195:536–538. doi: 10.1056/NEJM192609091951105. [DOI] [Google Scholar]
- 38.Torres-Rodriguez J.M., Madrenys-Brunet N., Siddat M., Lopez-Jodra O., Jimenez T. Aspergillus versicolor as cause of onychomycosis: Report of 12 cases and susceptibility testing to antifungal drugs. J. Eur. Acad. Dermatol. Venereol. 1998;11:25–31. doi: 10.1111/j.1468-3083.1998.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S., Corapi W., Quist E., Griffin S., Zhang M. Aspergillus versicolor, a new causative agent of canine disseminated aspergillosis. J. Clin. Microbiol. 2012;50:187–191. doi: 10.1128/JCM.05388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maniam R., Selvarajah G.T., Mazlan M., Lung Than L.T. Pulmonary papillary adenocarcinoma with Aspergillus versicolor infection in a dog. Med. Mycol. Case Rep. 2018;19:25–29. doi: 10.1016/j.mmcr.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsang C.C., Hui T.W., Lee K.C., Chen J.H., Ngan A.H., Tam E.W., Chan J.F., Wu A.L., Cheung M., Tse B.P., et al. Genetic diversity of Aspergillus species isolated from onychomycosis and Aspergillus hongkongensis sp. nov., with implications to antifungal susceptibility testing. Diagn. Microbiol. Infect. Dis. 2016;84:125–134. doi: 10.1016/j.diagmicrobio.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 42.Siqueira J.P.Z., Wiederhold N., Gene J., Garcia D., Almeida M.T.G., Guarro J. Cryptic Aspergillus from clinical samples in the USA and description of a new species in section Flavipedes. Mycoses. 2018;61:814–825. doi: 10.1111/myc.12818. [DOI] [PubMed] [Google Scholar]
- 43.Balajee S.A., Baddley J.W., Peterson S.W., Nickle D., Varga J., Boey A., Lass-Florl C., Frisvad J.C., Samson R.A., the ISHAM Working Group on A. terreus Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot. Cell. 2009;8:713–722. doi: 10.1128/EC.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequences newly generated in this study have been submitted to the GenBank database.