Abstract
Pathogenic Rickettsia are obligate intracellular bacteria and the etiologic agents of many life‐threatening infectious diseases. Due to the serious nature of these infections, it is imperative to both identify the responsive immune sensory pathways and understand the associated immune mechanisms that restrict Rickettsia proliferation. Previous studies have demonstrated that the mammalian complement system is both activated during Rickettsia infection and contributes to the immune response to infection. To further define this component of the mammalian anti‐Rickettsia immune response, we sought to identify the mechanism(s) of complement activation during Rickettsia infection. We have employed a series of in vitro and in vivo models of infection to investigate the role of the classical complement activation pathway during Rickettsia infection. Depletion or elimination of complement activity demonstrates that both C1q and pre‐existing IgM contribute to complement activation; thus implicating the classical complement system in Rickettsia‐mediated complement activation. Elimination of the classical complement pathway from mice increases susceptibility to R. australis infection with both increased bacterial loads in multiple tissues and decreased immune activation markers. This study highlights the role of the classical complement pathway in immunity against Rickettsia and implicates resident Rickettsia‐responsive IgM in the response to infection.
Keywords: C1q, classical complement pathway, complement, IgM, Rickettsia
Pathogenic Rickettsia are extremely dangerous bacteria that parasitize the cytoplasm of the mammalian vasculature. Due to the serious nature of these infections, it is imperative to both identify the responsive immune sensory pathways and understand the associated immune mechanisms that restrict bacterial growth. This manuscript identifies a component of the complement system and pre‐existing IgM as key contributors to immune recognition of these bacteria.
1. INTRODUCTION
Rickettsia are α‐proteobacteria belonging to the family Rickettsiaceae (Chan et al., 2010; Parola et al., 2013). These small, obligate intracellular, Gram‐negative bacteria are transmitted to humans and animals by hematophagous arthropods whereby the bacteria reside primarily in the cytoplasm of vascular cells (Mansueto et al., 2012). Serious to fatal Spotted Fever Group (SFG) Rickettsia infections are associated with Rickettsia rickettsii, the agent of Rocky Mountain spotted fever (RMSF), and Rickettsia conorii, the etiologic agent of Mediterranean spotted fever (MSF). Case mortality rates among people infected with RMSF and MSF can reach 20%–30% in the absence of timely diagnosis and intervention with the appropriate antibiotic (Biggs et al., 2016; Sahni et al., 2013; De Sousa et al., 2003). R. rickettsii is the most pathogenic SFG species and is distributed throughout the Americas (Ribeiro et al., 2021). R. conorii is found throughout southern Europe, northern Africa, and south western Asia (Blanton, 2019). The newly emerging SFG pathogen Rickettsia parkeri is the causative agent of the comparatively less dangerous infection R. parkeri rickettsiosis (Parola et al., 2005; Straily et al., 2016). Of additional note for this manuscript is Rickettsia australis, the agent of Queensland Tick Typhus, which is less closely related to the other SFG Rickettsia but causes a similar disease (Stewart et al., 2017).
During SFG Rickettsia infection, transmission from an infected arthropod vector and proliferation of Rickettsia in endothelial cells induce inflammatory events including the secretion of cytokines and activation of complement proteins. The combination of Rickettsia proliferation and the host immune response are responsible for the destruction of blood vessels resulting in the characteristic maculopapular rash (Blanton, 2019; Mansueto et al., 2012; Sahni et al., 2019). While the bacteria primarily reside within the host cytoplasm, some bacteria exit via the luminal surface of blood vessels into the bloodstream (Walker, 2007). It is during these extracellular phases of infection or during tick transmission that the bacteria are exposed to the potentially deleterious effects of the complement system.
As a component of innate immunity, the complement system is a collection of soluble proteins that circulate through bodily fluids as inactive precursors. Complement recognition of molecular markers on damaged or foreign cells induces a cascade of proteolysis, conformational changes, and macromolecular assembly that ultimately activates effector mechanisms that deposit the lytic membrane attack complex, opsonize the cell, and produce pro‐inflammatory anaphylatoxins (Merle et al., 2015a). Complement activation also has important secondary effects on the immune system by enhancing the innate immune response, prompting development of the adaptive response, and augmenting effectiveness of the adaptive response to infection (Merle et al., 2015b). Complement can be initiated by three largely independent activation pathways: classical, lectin, and alternative pathways (Merle et al., 2015a). The alternative pathway activates through a “tick‐over” mechanism whereby autocatalysis leads to activation and only the existence of cell‐proximal complement regulatory proteins prevents complement activation (Nilsson & Nilsson Ekdahl, 2012). The lectin pathway is stimulated by sugar‐binding proteins that recognize foreign sugars on the surface of an invading organism (Garred et al., 2016; Merle et al., 2015a; Reid, 2018; Thielens et al., 2017). The classical complement pathway is stimulated by deposition of antibodies on a surface. Both IgG and IgM molecules recruit the C1 complex to initiate complement activation (Harboe et al., 2011; Thielens et al., 2017). The three complement activation pathways converge at generation of a C3 convertase enzyme that greatly amplifies complement proteolytic cascade that ultimately results in generation of three effector mechanisms including: recruitment of the C6‐9 proteins to produce a lytic pore that can kill a microorganism, deposition of C3b and C5b results in opsonization to encourage engulfment and digestion the organism by phagocytic cells, and release of soluble proinflammatory anaphylatoxins C3a and C5a (and C4a to lesser extent) that activate and recruit immune cells to the areas of complement activation (Harboe et al., 2011; Merle et al., 2015a; Merle et al., 2015b; Ricklin et al., 2010, 2016).
Complement is primarily a serum‐borne innate immune component and Rickettsia preferentially infect the vascular system of susceptible mammals (Mansueto et al., 2012). Therefore, these entities are likely to have an adversarial relationship in vivo. Recent investigations into the interaction between pathogenic Rickettsia species and the host complement system have elucidated part of this host–pathogen interplay. Clinical findings from patients infected with MSF report increased plasma levels of several complement activation markers (Otterdal et al., 2016). While complement appears to be activated in vivo, Rickettsia have been shown to be inherently resistant to complement‐mediated killing (Fish et al., 2017; Garza et al., 2017; Riley et al., 2014) (Riley et al., 2012). However, the dichotomy between Rickettsia and complement is not purely one dimensional as a recent study has shown that immunity against the pathogenic species R. australis requires a functional complement system (Riley et al., 2018). The complement proteins C3 and C9 are not deposited on the bacteria, so complement efficacy is not dependent on complement‐mediated phagocytosis nor direct killing of the bacteria. Instead, infected C3‐knock‐out mice show a decrease in IFN‐γ production and early IgG production, suggesting that activation of complement influences the remainder of the immune system to produce anti‐Rickettsia effectors (Riley et al., 2018). With the knowledge that complement contributes to the immune response to these obligate intracellular pathogens these questions remain: Could we modulate the complement system to enhance the efficacy of the immune response? Could we employ complement activation during vaccine‐induced immunity?
In this manuscript, we sought to investigate the mechanisms of complement activation in naive mammals. To investigate Rickettsia‐induced complement activation, we have developed different approaches to evaluate the role of C1q in the initiation of the classical pathway during Rickettsia infection and have defined a central role of IgM in this mechanism. Finally, we have evaluated impact of the lack of classical complement activation in vivo by using genetically modified mice. Our findings advance our understanding of the immune response to Rickettsia infection through elucidation of a mechanism of complement recognition of pathogenic Rickettsia and identification of pre‐existing Rickettsia‐responsive IgM in both human and mouse serum.
2. RESULTS
2.1. The classical complement pathway protein C1q contributes to Rickettsia‐mediated complement activation in multiple mammal species
A previous study had shown that the complement system is required for the immune response to Rickettsia infection (Riley et al., 2018). We sought to identify the mechanism of complement activation that occurs during Rickettsia exposure, with the hypothesis that Rickettsia‐mediated complement activation relies on the classical complement activation pathway. The classical complement pathway normally connects antibody deposition on a membrane to complement activation and consists of the C1, C2, and C4 proteins before merging into the common complement cascade at proteolysis of C3 (Dunkelberger & Song, 2010). Of the classical complement proteins, the C1 protein complex (C1q, r, s) is unique to this pathway, so manipulation of C1 proteins only affects the classical pathway without off‐target effects on other complement activation pathways (Reid, 2018).
To evaluate the contribution of the classical complement activation pathway in Rickettsia‐mediated complement activation, we first removed individual complement components from pooled normal human serum (NHS) using antibody‐mediated protein depletion. Using agarose‐conjugated anti‐C1q or anti‐IgM, we were able to specifically reduce the quantity of these proteins in human serum as compared with the control protein, factor B (Laskowski & Thurman, 2018) (Figure 1a). We refer to these depleted serum samples as C1q‐depleted human serum (C1q‐dplHS) and IgM‐depleted human serum (IgM‐dplHS). Analysis of repeated serum depletions indicates that C1q depletion significantly depletes C1q protein (76 ± 22% depletion) but does not significantly deplete IgM. Similarly, IgM depletion depleted IgM protein (49 ± 14% depletion) from serum but does not cross‐deplete C1q (Figure S1a/b).
FIGURE 1.
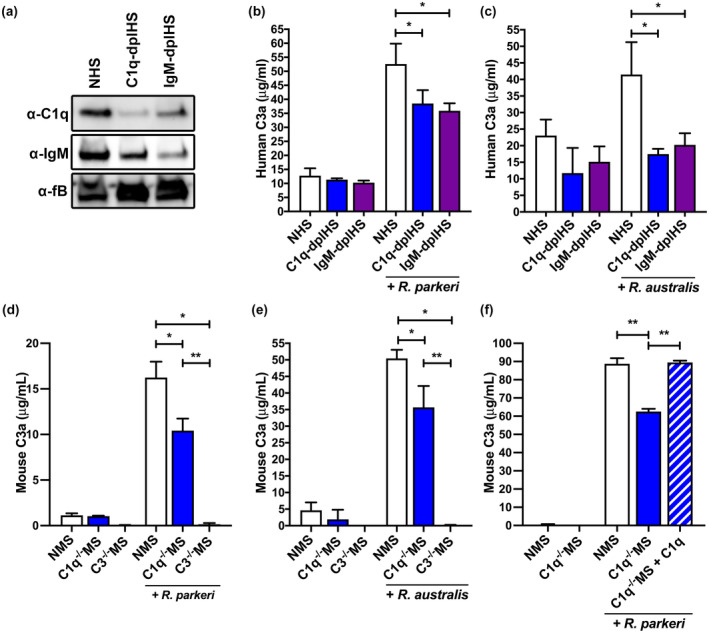
C1q and IgM contribute to Rickettsia‐induced complement activation in serum. (a) Western blot analysis of depletion of C1q and IgM protein in C1q‐depleted human serum (C1q‐dplHS) and IgM‐depleted human serum (IgM‐dplHS) as compared with normal human serum (NHS) with serum protein factor B (fB) as loading control. (b) Human complement activation as measured by production of the anaphylatoxin C3a. NHS (white), C1q‐dplHS (blue), and IgM‐dplHS (purple) have similar initial complement activation markers. After incubation with R. parkeri, both C1q‐dplHS and IgM‐dplHS demonstrated deficient complement activation as compared with NHS. (c) Similar complement activation impairment in C1q‐dplHS and IgM‐dplHS as compared with NHS upon incubation with R. australis. (d) Mouse complement activation as measured by production of the anaphylatoxin C3a. Normal mouse serum (NMS) and C1q−/−MS have similar initial complement activation levels. After incubation with R. parkeri, C1q−/−MS demonstrated deficient complement activation as compared with NMS but greater than negative control C3−/− mouse serum. (e) Similar complement activation defect in C1q−/−MS after incubation with R. australis. (f) Addition of exogenous C1q protein restores complement activation in C1q−/− mouse serum. After incubation with R. parkeri, C1q−/−MS (blue) elaborates a deficient complement response as compared with NMS (white). Addition of C1q protein to C1q−/−MS (patterned) restores complement activation to the level of NMS. *p < .05 by one‐way ANOVA with Dunnet's multiple comparison with NHS control. **p < .05 by one‐way ANOVA with Bonferroni's multiple comparison test
With NHS, C1q‐dplHS, and IgM‐dplHS, we were able to directly assess the contribution of the classical pathway to Rickettsia‐mediated complement activation. To this end, we added extensively purified R. parkeri to each serum and incubated to allow for complement activation. After incubation, the bacteria were removed by centrifugation, and the quantity of the complement activation peptide C3a was determined by ELISA (Merle et al., 2015a). As demonstrated in Figure 1b, each serum sample possessed similar background C3a levels. However, both C1q‐ and IgM‐dplHS were less stimulated by R. parkeri than NHS. Of note, these depleted serum samples still contain residual C1q and IgM proteins, so they may retain limited capacity for classical complement activation. Thus, the observed reduction in complement activation indicates that both human C1q and IgM contribute to C3a production and implicates the classical complement activation pathway in R. parkeri‐mediated complement activation.
To determine if results obtained with R. parkeri can be applied to the remainder of the Rickettsia genus, we choose to assess complement activation with more distantly related Rickettsia species, R. australis (Parola et al., 2013). Incubation of each serum type with purified R. australis produced similar results to those obtained with R. parkeri. As shown in Figure 1c, each serum had low background levels of C3a prior to incubation with R. australis. However, both C1q‐ and IgM‐dplHS had significantly reduced C3a production as compared with NHS. Thus, classical complement activation pathway contributes to Rickettsia‐mediated complement activation in human serum.
To assess the role of classical complement activation in other susceptible mammalian species, we switched the source of serum to another Rickettsia‐susceptible mammal. We acquired serum from naïve WT, C3−/− (Wessels et al., 1995), and C1q−/− (Fonseca et al., 2017) mice which we refer to as NMS, C3−/−MS, and C1q−/−MS, respectively. The serum from these genetically modified mice is different from the depleted human serum employed in Figure 1a–c in that there is no production of the specific complement protein being analyzed. C3 is the central complement protein with broad influence on the entire complement system, so we employed C3−/−MS as a negative control for complement activation. Conversely C1q is unique to the classical pathway, so C1q−/−MS only lack classical complement activation (Merle et al., 2015a). The genetic identity of the source animals was confirmed by PCR (Figure S1c) and to eliminate the possibility that C1q removal is directly toxic to the bacteria, we incubated R. parkeri in NMS, C1q−/−MS, and NMS supplemented with anti‐Rickettsia antibody and assessed survival of the bacteria by limiting dilution titration. Rickettsia are inherently resistant to complement‐mediated killing in NMS, but addition of a specific anti‐Rickettsia antibody overcomes this bacterial phenotype (Figure S1d). C1q−/− serum is unable to kill R. parkeri, indicating that the lack of classical complement activation in mouse serum does not affect the survival of Rickettsia in serum.
NMS, C3−/−MS, and C1q−/−MS were incubated with extensively purified R. parkeri to allow for complement activation. After removal of the bacteria by centrifugation, the quantity of the complement activation peptide C3a was measured. As shown in Figure 1d, NMS and C1q−/−MS possessed low background amounts of C3a, and C3−/−MS did not possess any C3a. After incubation with R. parkeri, C1q−/−MS produced significantly less C3a than NMS, indicating that classical complement activation contributes to Rickettsia‐mediated complement activation in murine serum. To validate this murine serum phenotype, we also assessed complement activation with R. australis. As shown in Figure 1e, the classical complement protein C1q also contributes to R. australis‐mediated complement activation in murine serum.
Together, these results establish that the classical pathway contributes to complement activation in both human and mouse serum and with both R. parkeri and R. australis. Overall, these results reveal that (a) the classical complement pathway contributes to Rickettsia‐mediated complement activation, (b) complement activation is associated with IgM, as depletion of IgM leads to a decrease in complement activation, and (c) these results apply to two Rickettsia species in serum from two different animal species.
2.2. Exogenous C1q protein restores Rickettsia‐mediated complement activation in C1q‐deficient mouse serum
The data from Figure 1 demonstrates that the classical pathway contributes to Rickettsia‐mediated complement activation in two Rickettsia species and in two mammals. To validate that C1q mutant mammals did not possess any off‐target phenotypes and to determine if reduced complement activation is truly C1q specific, we used recombinant C1q protein to reconstitute the classical complement pathway in C1q−/−MS. R. parkeri was incubated with NMS, C1q−/−MS, or C1q−/−MS supplemented with 5 µg/ml C1q. As shown in Figure 1f, all serum samples had low pre‐activation C3a concentrations, but incubation with R. parkeri resulted in a significantly lower C3a concentration in C1q−/−MS as compared with NMS control. However, supplementation of 5 µg/ml C1q to C1q−/−MS reconstituted full complement activation (Sandholm et al., 2019). These results confirm that C1q contributes to Rickettsia‐mediated complement initiation.
2.3. Presence of broadly reactive anti‐Rickettsia IgM in human serum
The previous figures demonstrate that the C1 complex, specifically C1q, contributes to complement recognition of Rickettsia. The major function of C1q is to connect antibody deposition to complement activation (Gadjeva et al., 2008). Since C1q contributed to complement activation in human and murine serum, we hypothesized that Rickettsia‐reactive IgM may be present in naïve mammalian serum. To this end, we depleted IgM from pooled NHS. Western blotting analysis showed that the level of human IgM in the depleted serum (IgM‐dplHS) was reduced as compared with the level in mock‐depleted serum (NMS) with the ubiquitous serum protein factor B as a loading control (Figure 2a).
FIGURE 2.
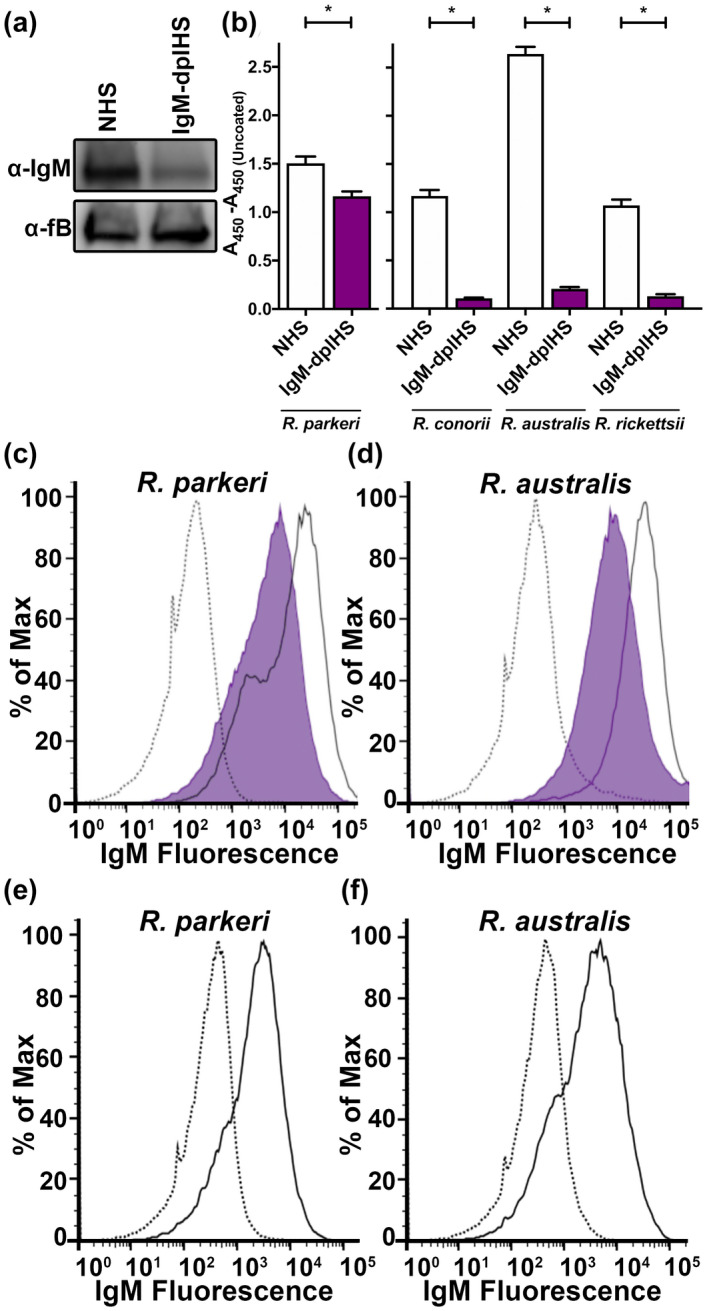
Presence broadly reactive of anti‐Rickettsia IgM in serum. (a) Western blot analysis of IgM in normal human serum (NHS) and IgM‐depleted human serum (IgM‐dplHS) with serum factor B (fB) as loading control. (b) Deposition of human IgM from NHS (white) and IgM‐dplHS (purple) onto R. parkeri‐coated ELISA plate or R. conorii, R. australis, and R. rickettsii‐coated ELISA plate. The absorbance from PBS (uncoated) wells is subtracted from all values. (c) Flow cytometric analysis of human IgM deposition on R. parkeri bacilli from PBS (dotted line), NHS (solid line), and IgM‐dplHS (purple). The increase in fluorescence from NHS as compared with PBS demonstrates the presence of Rickettsia‐reactive IgM. (d) Flow cytometric analysis of human IgM recognition of R. australis from PBS (dotted line), NMS (solid line), and IgM‐dplHS (purple). (e) Flow cytometric analysis of mouse IgM deposition on R. parkeri bacilli from PBS (dotted line) and normal mouse serum (NMS, solid line). (f) Flow cytometric analysis of mouse IgM deposition on R. australis bacilli from PBS (dotted line) and NMS (solid line) *p < .05 by one‐way ANOVA with Bonferroni's multiple comparison test
To query for the presence of Rickettsia‐reactive IgM in human serum, we first analyzed IgM recognition of Rickettsia by ELISA. Equal amounts of whole R. parkeri, R. conorii, R. australis, and R. rickettsii were used to coat ELISA plates. Subsequently, NHS or IgM‐dplHS was added to the wells to allow for IgM recognition of the immobilized Rickettsia. After washing, IgM deposition was determined with an anti‐IgM antibody and ELISA analysis. As shown in Figure 2b, coating of the ELISA well with R. parkeri or other Rickettsia antigens increased IgM deposition as compared with uncoated wells, indicating that there are Rickettsia‐reactive IgM antibodies in human serum. Importantly, specific depletion of IgM in IgM‐dplHS reduced IgM deposition, demonstrating that the change in absorption is specifically due to IgM deposition. These results indicate that human serum contains broadly Rickettsia‐reactive IgM that can recognize R. parkeri, R. conorii, R. australis, and R. rickettsii.
To verify the presence of IgM and to determine if these antibodies recognize the surface of Rickettsia bacilli, we sought to identify IgM deposition on purified bacteria by flow cytometry. To this end, paraformaldehyde‐fixed R. parkeri (Figure 2c) and R. australis (Figure 2d) were incubated with PBS, NHS, or IgM‐dplHS. After washing, IgM deposition was analyzed using fluorescent‐labeled anti‐IgM antibodies. As shown in Figure 2c, incubation of R. parkeri in NHS (solid line) increases IgM deposition on the bacterial surface as compared with PBS (dotted). Additionally, depletion of IgM from human serum (IgM‐dplHS, shaded) reduces IgM deposition on the bacterial surface, indicating that the fluorescent signal is IgM specific. Comparison of fluorescence from IgM‐dplHS and NHS indicates SE Dmax = 43.45% and T(x) = 3,156, indicating robustly significant differences in histogram profiles (Roederer & Hardy, 2001; Roederer et al., 2001). This result is confirmed with analysis R. australis (Figure 2d) with an increase in fluorescence associated with NHS incubation that is reduced in IgM‐dplHS. Comparison of fluorescence from IgM‐dplHS and NHS indicates SE Dmax = 69.51% and T(x) = 4,899, indicating robust significant difference in histogram profiles. As such, NHS contains IgM that recognizes both R. parkeri and R. australis.
To confirm that the existence of Rickettsia‐reactive IgM from naïve serum is not human specific, we assessed deposition of IgM from normal mouse serum. As shown in Figure 2e–f, incubation of R. parkeri or R. australis with normal B57BL6/J serum (solid line) resulted in an increase in fluorescence associated with IgM deposition as compared with PBS control (dotted line). Therefore, both naïve human and mouse serum contain IgM that is reactive to both R. parkeri and R. australis.
2.4. Anti‐Rickettsia IgM deposition is required for classical complement activation
Having demonstrated the presence of broadly reactive anti‐Rickettsia IgM in human and mouse sera (Figure 2), we next sought to evaluate the connection between IgM and C1q‐dependent complement activation. Classical complement activation normally proceeds sequentially from antibody deposition to C1 deposition (Merle et al., 2015a). As IgM depletion from human serum decreases complement activation (Figure 2), we hypothesized that depletion of IgM from human serum should also decrease the quantity of C1q accumulation on the bacterial surface. To this end, R. australis were incubated with PBS (dotted line), NHS (solid line), or IgM‐dplHS (purple shaded). After removal of the serum and washing, the bacteria were stained with fluorescent antibodies to both C1q and IgM. First, incubation of the bacteria in NHS (solid line) leads to both IgM and C1q accumulation on the bacterial surface as compared with PBS (dotted) (Figure 3a,b). However, depletion of IgM from human serum (IgM‐dplHS) leads to both decreased IgM (Figure 3b) and C1q (Figure 3a) on the bacterial surface. Therefore, C1q deposition on Rickettsia is dependent on the amount of IgM fixation on the bacteria and IgM recognition of the Rickettsia surface leads to classical complement activation.
FIGURE 3.
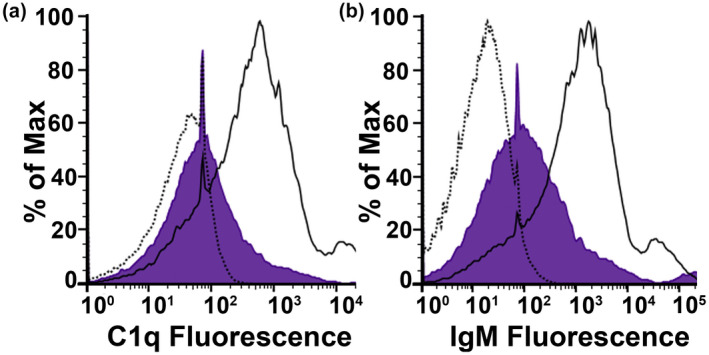
Classical complement activation occurs through IgM recognition of Rickettsia. (a) Flow cytometric analysis of C1q deposition on R. parkeri after incubation in NHS (solid line), IgM‐dplHS (purple), or PBS (dotted line). Decreased quantity of IgM in human serum leads to decreased complement activation and C1q on the bacterial surface. (b) Flow cytometric analysis of IgM deposition on R. parkeri after incubation in NHS (solid line), IgM‐dplHS (purple), and PBS (dotted line). IgM recognition of Rickettsia is demonstrated through increase in fluorescence from NHS as compared with PBS. Signal specificity is confirmed through IgM‐dplHS
2.5. The classical complement activation pathway contributes to control of Rickettsia infection
Heretofore, we have explored the interaction between Rickettsia and serum‐borne complement and established that both IgM and the classical pathway contribute to Rickettsia‐mediated complement activation in ex vivo serum samples. However, biological relevance of classical complement activation in the immune response to infection remains unanswered. To address this question, we queried C57BL/6J mice (WT) and mice containing a targeted mutation in the c1q gene (C1q−/−) for susceptibility to R. australis infection (Feng et al., 1993). C1q−/− mice lack exon 3 of the complement component 1, q subcomponent, alpha polypeptide gene leading to incomplete assembly of the C1 complex and ablation of the entire classical complement activation pathway (Fonseca et al., 2017). WT and C1q−/− mice were intravenously infected with 0.5 minimum lethal doses of R. australis strain Cutlack (Riley et al., 2018).
Three pre‐identified mice were removed from the experiment at day 3 post‐infection to assess immune activation status. To estimate the level of complement activation in vivo, we used ELISA methods to quantify serum C3a concentration. C3a is a soluble peptide that is produced in vivo during proteolytic processing of the central complement component C3 (Merle et al., 2015a). As shown in Figure 4a, R. australis‐infected C1q−/− animals exhibited significantly reduced C3a as compared with WT mice, demonstrating decreased complement activation in C1q−/− mice. This finding merges in vitro data indicating that the classical pathway is employed with the in vivo immune response to infection.
FIGURE 4.
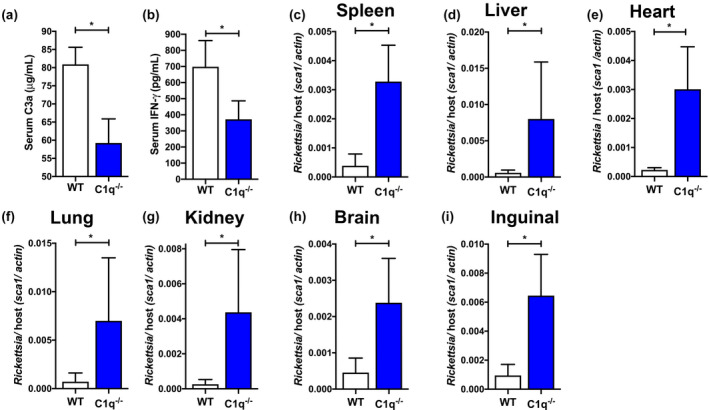
Classical complement deficient animals elaborate a deficient immune response with increased Rickettsia australis burden. (a) Reduced complement activation as assessed by serum C3a at 3 days post R. australis infection in C1q−/− mice (blue) as compared with wild type mice (WT, white). (b) Reduced serum IFN‐γ at 3 days port infection in C1q−/− (blue) as compared with WT mice (white). (c–i) Bacterial loads in mouse organs at 6 days post R. australis infection. Quantity of Rickettsia is expressed as quantitative PCR ratio of R. australis sca1 DNA to murine actin DNA. C1q−/− mice have higher Rickettsia loads than WT in (c) spleen, (d) liver, (e) heart, (f) lung, (g) kidney, (h) brain, and (i) inguinal lymph node. *p < .05 by students t‐test
While complement is classically associated with direct antimicrobial mechanisms, Rickettsia are inherently resistant to complement mediated killing (Fish et al., 2017; Garza et al., 2017; Riley et al., 2012, 2014, 2018). However, the complement system can have major effects on the remainder of the immune system, likely through production of the proinflammatory anaphylatoxins (Merle et al., 2015a). Presence of anaphylatoxins contributes to both local and system proinflammatory phenotypes, including production of other cytokines (Jiménez‐Reinoso et al., 2017; Mathern & Heeger, 2015; Merle et al., 2015b; West et al., 2018). Since IFN‐γ production correlates strongly with the effective immune response to rickettsial infections (Sahni, 2009; Walker, 2007; Walker et al., 2000), we assessed the level of this cytokine in the infected mice. As shown in Figure 4b, the level of IFN‐γ was significantly reduced in C1q−/− mice as compared with WT. This finding demonstrates that C1q deficiency correlates with reduced serum IFN‐γ and suggests a relationship between classical complement and the innate immune response to infection.
Finally, to examine if the classical pathway has a direct effect on Rickettsia pathogenesis, we examined bacterial DNA content in different organs from infected mice. Pre‐identified mice were removed from each group at 6 days post‐infection and major organs were extracted for analysis. A single chromosomal DNA target was amplified for R. australis (sca1) and Mus musculus (actin), and the bacterial load was determined by calculating the ratio of Rickettsia to murine DNA. As shown in Figure 4c–i, the quantity of R. australis was significantly increased in C1q−/− organs as compared with the WT in multiple tissues, including heart (Figure 4c), lung (Figure 4d), liver (Figure 4e), spleen (Figure 4f), kidney (Figure 4g), brain (Figure 4h), and inguinal lymph node (Figure 4i). Together, these data demonstrate that the classical pathway contributes to in vivo complement activation, IFN‐γ production, and control of R. australis proliferation.
3. DISCUSSION
Herein, we report the contribution of the classical pathway to Rickettsia‐induced complement activation. Both in vitro and in vivo experiments demonstrate the importance of the classical pathway in two different animal serum types and against two Rickettsia species. We demonstrate that Rickettsia‐specific IgM molecules exist in naïve murine and human serum, and that these antibodies contribute to complement activation. Finally, we employed in vivo models of R. australis infection to demonstrate that the classical complement activation pathway contributes to both immune activation and control of Rickettsia infection. These results indicate that pre‐existing IgM and the classical complement pathway contribute to the innate immune response and ultimately aid in the control of Rickettsia infection.
Previous reports have demonstrated that Rickettsia bacilli are potent complement activators in vitro and in vivo (Chan et al., 2011; Garza et al., 2017; Riley et al., 2012, 2018). We utilized serum from two different mammalian species to establish that the classical pathway contributes to in vitro complement activation. Both C1q‐free murine serum and C1q‐depleted human serum elaborated incomplete complement activation (Figure 1). These phenotypes were validated with two pathogenic Rickettsia species, so we could reasonably surmise that these phenotypes hold true for other susceptible mammals and other pathogenic Rickettsia species. Another important outcome is that we never observed complete ablation of complement activation in serum lacking the classical pathway. This is expected for C1‐depleted human serum that still retains some C1q but was more unexpected with mouse serum that completely lacks a functioning classical pathway. Therefore, our data does not eliminate the possibility that the other two complement activation pathways, the alternative and lectin pathways, also contribute to Rickettsia‐induced complement activation. These findings led us to query the biological relevance of classical complement activation in animal models of infection.
Mice lacking an essential component of the classical complement pathway, C1q, were more susceptible to intravenous R. australis infection than wild type counterparts (Figure 4). The first phenotype associated with C1q deficiency was reduced complement activation at 3 days post infection. This finding demonstrates that the classical complement activation pathway contributes to Rickettsia‐mediated complement activation in vivo. This reduced complement activation correlated with reduced serum IFN‐γ at 3 days post infection. IFN‐γ and IFN‐stimulated genes have been shown to significantly contribute to the effective immune response in many different models of Rickettsia infection with IFN‐γ gene knockout mice being 100× more susceptible to R. australis infection that WT (Bechelli et al., 2016; Burke et al., 2020; Colonne et al., 2013; Fang et al., 2012; Jordan et al., 2009; Kondo et al., 2020; Rumfield et al., 2020; Walker et al., 2001) In humans and mice, a major source of IFN‐γ is NK cells (Ferlazzo & Morandi, 2014; Min et al., 2014), and NK cells are activated to produce IFN‐γ during Rickettsia infection (Billings et al., 2001; Carl et al., 1986; Fang et al., 2012; Jordan et al., 2009). NK cells express surface receptors to detect complement activation, including C3aR, C5aR, C5L2, CR3, CR3, and CR4, suggesting that NK cells are capable of detecting the small fragments (C3a and C5a) and the large fragments (C3b and its metabolites) (Fusakio et al., 2011; Min et al., 2014). Therefore, our observation of reduced serum IFN‐γ in C1q−/− mice hints at a complement/ NK cell/ IFN‐γ cross‐talk that requires further investigation.
Classical pathway deficiency also resulted in increased Rickettsia burden in all tested organs at 6 days post infection (Figure 4). Therefore the classical pathway has a role in Rickettsia clearance as has been previously reported with Borrelia burgdorferi (Zhi et al., 2018), Streptococcus pneumoniae (Brown et al., 2002), Francisella tularensis (Schwartz et al., 2012), and Haemophilus influenza (Langereis et al., 2019). C1q‐mediated pathogen clearance appears to be associated with anaphylatoxin production or potentially with direct action by the enhancement of phagocytosis or antibody inhibitory effect (Boyle et al., 2015; Schwartz et al., 2012). However, we still have not identified the molecular or cellular mechanisms of complement efficacy during Rickettsia infection. Based on previous results, it is unlikely that complement‐enhanced phagocytosis or direct bacterial killing contribute to control of Rickettsia infection, as these systems are dispendible for the effective immune response to infection (Riley et al., 2018). Therefore, investigating how complement enhances the immune response to Rickettsia infection remains a high priority.
Potentially the most impactful finding in this report is the presence of Rickettsia‐reactive IgM in both pooled human and mouse serum (Figure 2). Therefore, an important question moving forward is: are these IgM antibodies truly innate germline‐encoded Rickettsia‐specific antibodies? We have considered the remote chance that a human serum donor had recently experienced a Rickettsia infection and possesses antigen‐specific antibodies that could activate the classical complement pathway. However, we know the lifelong history of the murine serum donors (Figure 2e–f), so we can be sure that each mouse serum is truly naïve, in that they had never seen Rickettsia antigen prior to experimental analysis. These mice had never been exposed to Rickettsia antigens, yet still possessed Rickettsia‐reactive IgM. A previous study has reported the production of poly reactive IgM in mice infected with another Rickettsiales, Ehrlichia muris; however, these IgM were reported not detected in normal sera from C57BL/6 mice (Jones et al., 2012). Thus, we can conclude that classical pathway‐mediated complement activation is truly an innate immune mechanism. Therefore, our data suggest that these are natural IgM antibodies as defined by Baumgarth et al. as “truly antigen‐independent elaboration of IgM” (Baumgarth, 2017).
Natural antibodies (nAbs) are produced by a group of cells, called B‐1 cells, that are found primarily within spleen and bone marrow, but can also be found in the peritoneal and pleural cavities (Dasgupta et al., 2020). B‐1 secreted IgM has been proposed to play a central role in immunity against multiple pathogens as a mechanism that bridges initial infection to induction of the adaptive response (Brown et al., 2002; Ehrenstein & Notley, 2010; Langereis et al., 2019; Sörman et al., 2014). One action associated with B‐1‐secreted IgM recognition of a pathogen is initiation of complement activation as we have demonstrated in Figure 3. However, natural IgM antibodies may further integrate the immune response to Rickettsia infection. Some natural IgM effector mechanisms are shared with normal antigen‐specific IgM, like Fcμ‐R and Fcα/μ‐R‐mediated endocytosis, but other mechanisms are specific to nAb and B‐1 cells. As an example, soon after infection, body cavity B‐1 cells are rapidly activated to migrate to lymph tissues where they differentiate to cytokine and antibody‐secreting cells (Smith & Baumgarth, 2019). Natural IgM can also promote antigen stimulation of the immune system by mediating deposition of IgM antigen–antibody complexes on follicular dendritic cells (Baumgarth, 2011). Additionally, nAbs can directly influence the adaptive immune response by promoting antigen presentation to stimulate T cells via CD86 (Zhu et al., 2016) and facilitate antigen uptake by B cells to enhance ensuing B‐2 cell‐dependent pathogen‐specific IgG responses (Baumgarth, 2011). Finally, IgM is not the only class of natural antibodies. Natural IgG is a well‐defined component and natural IgA is a burgeoning component of the natural antibody repertoire (Nagele et al., 2013; Panda & Ding, 2015; Panda et al., 2013). Therefore, our identification of anti‐Rickettsia IgM from naïve mammals is likely to influence our understanding of the immune response to infection beyond activation of the complement system. Assessing the contribution of pre‐existing anti‐Rickettsia antibodies to the immune response requires further analysis.
Production of natural IgM by B‐1 cells can serve as an important first‐line defense against pathogens. In the absence of specific antigen, natural antibodies provide broad and rapid protection against pathogens (Baumgarth et al., 2005). Natural IgM are known to demonstrate low‐affinity poly‐reactivity but a high valency that contributes to agglutination of the invading organism (Ehrenstein & Notley, 2010). This broad reactivity is thought to allow animals to recognize diverse invaders as a method to initiate the immune response and dampen proliferation of the pathogen (Boes et al., 1998; Ochsenbein et al., 1999). Interestingly, we have shown that human and mouse serum contain broadly poly‐reactive IgM antibodies that recognize different species of Rickettsia belonging to different clades of the family including R. parkeri, R. conorii, and R. rickettsii, and the more distantly related species, R. australis (Figure 2b). Therefore, it is reasonable to extrapolate that mammalian serum contains IgM that is reactive to many Rickettsia species. This correlates with the ancient nature of the currently identified nIgM antigens, which include phospholipids, glycolipids, glycosylated proteins, and oxidized lipids (Holodick et al., 2017). We therefore hypothesize that mammalian anti‐Rickettsia IgM recognize non‐peptide antigens shared by all Rickettsia species and a residual question that stems from this finding is: what are the rickettsial antigens recognized by these IgM antibodies?
In summary, we have utilized in vitro and in vivo models of Rickettsia infection to elucidate a mechanism of complement activation by these bacteria. We have shown a significant role of C1q binding and IgM in the initiation of complement in human and mouse serum. Mice lacking the essential classical pathway protein, C1q, elaborated reduced complement activation, decreased serum IFN‐γ, and higher bacterial loads in tissues, suggesting a role for the classical complement activation in the immune response against Rickettsia. Together, these data identify an innate immune sensory system that is capable of recognizing Rickettsia and may be responsible for initiating the immune response to Rickettsia infection.
4. MATERIALS AND METHODS
4.1. Ethics statement
All animal experiments were conducted by following protocols approved by the Institutional Biological and Recombinant DNA Safety Committee (IBRDSC) and Institutional Animal Care and Use Committee (IACUC) at the University of Maryland – College Park (R‐NOV‐19‐51). Standards of care and use for all animals conform to all applicable standards and regulations as established by the current version of the Animal Welfare Act and the Guide to the Care and Use of Laboratory Animals. This institution is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC), indicating compliance with the requirements for the proper care and treatment of all vertebrate laboratory animals irrespective of species, location, investigator, use, or funding source. The university has an approved Assurance Statement (D16‐00172) on file with the Office of Laboratory Animal Welfare.
4.2. Cells line and bacterial growth
Vero (ATCC) cells were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% heat‐inactivated fetal bovine serum (Atlanta Biologicals), HyClone MEM Non‐Essential Amino Acid Solution (Thermofisher), and 200 mM L‐Glutamine (Gibco) at 37°C in 5% CO2. R. parkeri Portsmouth, R. australis Cutlack, R. conorii Malish 7, and R. rickettsii Sheila Smith were cultivated in Vero cells, needle‐lysed, and sucrose‐gradient purified as previously described (Ammerman et al., 2008). The rickettsial stocks were aliquoted stored at −80°C until use. Quantity of bacteria was determined by titration as has been previously described (Cardwell & Martinez, 2009).
4.3. Animal husbandry
The following murine models were employed in this study: (a) C57BL/6J; (b) complement component C3‐targeted mutations, B6;129S4‐C3tm1Crr/J mice (C3−/−) (Wessels et al., 1995); (c) complement component C1q‐targeted mutation, B6(Cg)‐ C1qtm1c(EUCOMM)Wtsi (C1q−/−) (Fonseca et al., 2017). Mice used as homozygotes and genotypes were confirmed according to the appropriate genotyping PCR protocol (Jackson Laboratories). All mice were infected at 5–6 weeks of age and were euthanized by 8 weeks old. Wild‐type (WT) C57BL/6J animals were acquired from Jackson Laboratory and were age and sex‐matched to the genetically modified groups.
4.4. Serum samples
Pooled human serum samples described as normal mouse serum (NHS) were purchased from Innovative research. C57BL/6J, B6;129S4‐C3tm1Crr/J mice (C3−/−), and B6(Cg)‐ C1qtm1c(EUCOMM)Wtsi (C1q−/−) blood were isolated by cardiac puncture and processed with Z‐gel (Sarstedt) to recover serum. All serum samples were aliquoted, snap‐frozen, and stored at −80°C. Each thawed sample of sera was used one time.
For C1q depletion: 6 µg of rabbit IgG anti‐human/mouse C1q (Thermofisher) were coupled to Pierce Protein A/G Magnetic Beads (Thermofisher) by overnight incubation at 4°C following the manufacturer instructions. The beads were then centrifuged at 2,000 × g for 2 min and washed by PBS. For IgM depletion, goat IgG anti‐human IgM (Invitrogen) was biotinylated using Antibody Biotinylation kit (Pierce) to obtain a final concentration of 0.8 mg/ml of biotinylated antibody: 8 µl of the biotinylated goat anti‐human IgM were then coupled to Pierce NeutrAvidin Agarose by overnight incubation at 4°C. To obtain a depleted serum, 10% of NHS in PBS were added either to the anti‐C1q+Protein A/G bead, anti‐IgM+NeutrAvidin beads, or mock depleted with free NeutrAvidin beads. The sera were incubated for 1 hr at 4°C with rotation. Samples were then centrifuged at 2,000 × g for 2 min to sediment the beads. Depleted sera were kept on ice and immediately used in the experiments. Depletion was assessed by immunoblotting using rabbit IgG anti‐C1q (Thermofisher), mouse IgG anti‐human IgM (Novusbio), and rabbit IgG anti‐human Factor B (Invitrogen).
4.5. Complement activation and quantification of C3a
To evaluate Rickettsia‐induced complement activation wild type (WT), C3−/−, or C1q−/− mouse serum was diluted to 10% in PBS. 1 × 106 extensively purified R. parkeri or R. australis were added to the different types of sera and incubated at 37°C for 1 hr with rotation. C1q−/− serum was supplemented with 5 µg/ml of mouse C1q protein (Complement tech) where required.
We employed sandwich ELISA to evaluate C3a concentration as a measurement of complement activation. A Maxisorp 96‐well plate (Pierce) was coated with mouse IgG anti‐human C3a/C3a desArg (Novusbio) in bicarbonate buffer (50 mM Na₂CO₃, 50 mM NaHCO₃, pH 9.4) overnight. The plates were then washed three times with wash buffer (PBS‐ 0.5% Tween‐20) and blocked in 1X ELISpot diluent (Invitrogen). Purified human C3a (NMS2096, Invitrogen) was used to generate the standard curve. After washing, biotinylated mouse IgG anti‐human C3a and C3a desArg (Novusbio) were used as a detection antibodies. After washing, streptavidin‐HRP (R&D systems) was added prior to detection with TMB (3,3’,5,5’‐Tetramethylbenzidine, Invitrogen). The reaction was stopped with 2N sulfuric acid. Absorbance was read on Spectra Max M2 spectrometer (Molecular Devices) and the values were expressed as A450nm‐A570nm and graphed against the standard curve.
4.6. IgM ELISA
1 × 106 R. parkeri, R. australis, R. conorii, and R. rickettsii was inactivated with 4% PFA solution (4% paraformaldehyde, 4% sucrose, PBS) These bacteria were used to coat an ELISA plate (Pierce) using bicarbonate coating buffer (50 mM Na₂CO₃, 50 mM NaHCO₃, pH 9.4) overnight. After washing (PBS‐ 0.5% Tween‐20) and blocking (PBS + 1%BSA), 10% corresponding sera or PBS was added and incubated for either 2 hr at room temperature or overnight at 4°C. After washing, biotinylated mouse IgG anti‐human IgM (Novusbio) was added. After washing, streptavidin‐HRP (R&D systems) was added prior to detection with TMB substrate (Invitrogen). The reaction was stopped with 2N sulfuric acid. Absorbance was read on Spectra Max M2 spectrometer (Molecular Devices), and the value was plotted as A450nm. IFN‐γ ELISA assay was performed using Mouse IFN‐γ OptiEIA ELISA set (BD Biosciences) following the manufacturer instructions.
4.7. Immunoblotting
Serum samples were incubated in RIPA buffer (Thermofisher) supplemented with protease inhibitor (Pierce). Total protein was quantified using BSA protein assay kit (Pierce). Proteins were resolved in Mini‐PROTEAN TGX gels (Bio‐Rad) and transferred to polyvinylidene membrane (Immobilion). C1q was detected with rabbit IgG anti‐human/mouse C1q (Invitrogen). Factor B was detected with rabbit IgG anti‐factor B (Invitrogen). Human IgM was detected with mouse IgG anti‐human IgM (Novusbio) with goat anti rabbit‐IgG‐HRP or donkey anti‐mouse‐IgG‐HRP (Invitrogen) as a secondary antibody. Streptavidin‐HRP (R&D systems) and crescendo western HRP substrate (Immobilon) were used prior to acquisition with Chemi‐Imager (Azure Biosystems).
4.8. Flow cytometry
To assess IgM deposition, 1 × 107 R. parkeri or R. australis bacilli were fixed in 4% PFA solution. After washing with PBS‐glycine, bacteria were incubated with 30% of NHS, IgM‐dplHS, or PBS for 1 hr at RT or overnight at 4°C. To assess both human IgM and C1q deposition on Rickettsia bacilli, all samples were treated with rabbit anti‐human C1q (Thermofisher), Alexa Fluor‐546‐conjugated donkey anti‐rabbit‐IgG (Invitrogen) and AlexaFluor‐488‐conjugated goat IgG anti‐human IgM (Invitrogen). Mouse IgM was measured as above but with C57BL6/J serum and AlexaFluor‐488‐conjugated anti‐mouse IgM (clone AF6‐78, Invitrogen). Fluorescence was assessed on a FACSCelesta Cell Analyzer (BD Biosciences) using fluorescein isothiocyanate (FITC) and phycoerythrin (PE) parameters and performed as previously described (Chan et al., 2011; Riley et al., 2014). All samples were analyzed with FloJo software (Tree Star).
4.9. Animal models of infection
Five to 6‐week‐old C57B/6J and C1q−/− were inoculated by retro‐orbital injection with a normally sub‐lethal dose of 5 × 105 R. australis per mouse (Riley et al., 2018). Infected mice were monitored twice daily for signs of disease and daily for weight change. Ten mice from each strain were observed for clinical signs and weight loss (Chan et al., 2011). Overt clinical signs of R. australis infection included ruffled fur, hunched posture, shallow respiration, immobility when touched, and weight loss (Riley et al., 2018). Predesignated mice from each mice strain group were euthanized at 3 and 6‐day post‐infection and were immediately subjected to necropsy. Blood was collected by cardiac puncture and transferred to a Serum Gel Z/1.1 (Starsdedt) and centrifuged to isolate serum. All tissue samples were snap frozen and stored at −80°C until needed.
4.10. DNA extractions and quantitative real‐time PCR (qRT‐PCR) analysis
Genomic DNA was isolated from murine tissues using the PureLinkTM Genomic DNA Mini Kit (Thermofisher). The ratio of R. australis to M. musculus DNA was determined by quantitative PCR utilizing the TaqMan gene amplification master mix (Applied Biosystems) and a CFX‐ Connect, Real‐Time System instrument (BioRad) using sca1_RA_5220F (5′‐TGCAGAACAAGTTTGTTATTACCC), sca1_RA_5465R (5′‐CTACCGCTCCTTGGAACGTTAGACC), and sca1_RA_probe (5′‐56‐FAM/TCGGCTTAA/Zen/GATATGGGAAGT/3IABlFQ). Murine actin primers and probes were previously described (Riley et al., 2018). All unknown samples were graphed against a standard curve of the specific amplicon cloned into pCR2.1. Data are expressed as the ratio of R. australis sca1 to M. musculus actin DNA.
4.11. Serum resistance
R. parkeri (1 × 106 per sample) were incubated with 10% normal mouse sera (NMS), NMS plus 1 µg/ml rabbit anti‐Rickettsia antibody (Rc‐PFA) (Riley et al., 2018), C1q−/− mice serum, or PBS for 1 hr at 37°C with agitation. Samples were transferred to ice to stop complement activation prior to assessing the quantity of input and surviving bacteria, which was determined as has been previously described (Cardwell & Martinez, 2009).
4.12. Statistical analysis
Statistical analysis was performed on Excel and GraphPad PRISM software (Version 8.3). Comparison of serum C3a concentration in serum and serum survival was performed and analyzed by one‐way analysis of variance (ANOVA) with Bonferroni's multiple comparison tests or Dunnett's multiple comparison with control. Analysis of C3a and IFN‐γ concentration in mice serum and DNA tissue DNA content was analyzed by unpaired t‐test. Error bars in bar graphs show the standard deviation from the mean.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
All animal experiments were conducted by following protocols approved by the Institutional Biological and Recombinant DNA Safety Committee (IBRDSC) and Institutional Animal Care and Use Committee (IACUC) at the University of Maryland‐ College Park (R‐NOV‐19‐51). This institution is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC), indicating compliance with the requirements for the proper care and treatment of all vertebrate laboratory animals irrespective of species, location, investigator, use, or funding source. The university has an approved Assurance Statement (D16‐00172) on file with the Office of Laboratory Animal Welfare.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Meiqing Shi and Victoria Verhoeve for helpful comments on the manuscript. Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health award number R21AI156069 and University of Maryland startup funds.
Dahmani, M. , Cook, J.H. , Zhu, J.C. & Riley, S.P. (2021) Contribution of classical complement activation and IgM to the control of Rickettsia infection. Molecular Microbiology, 116, 1476–1488. 10.1111/mmi.14839
Funding information
Research reported in this publication was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health award number R21AI156069 and University of Maryland startup funds.
DATA AVAILABILITY STATEMENT
All data are in the manuscript file.
REFERENCES
- Ammerman, N.C. , Beier‐Sexton, M. & Azad, A.F. (2008) Laboratory maintenance of Rickettsia rickettsii . Current Protocols in Microbiology, 11, 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth, N. (2011) The double life of a B‐1 cell: self‐reactivity selects for protective effector functions. Nature Reviews Immunology, 11, 34–46. 10.1038/nri2901 [DOI] [PubMed] [Google Scholar]
- Baumgarth, N. (2017) A Hard(y) look at B‐1 cell development and function. The Journal of Immunology, 199, 3387–3394. 10.4049/jimmunol.1700943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth, N. , Tung, J.W. & Herzenberg, L.A. (2005) Inherent specificities in natural antibodies: a key to immune defense against pathogen invasion. Springer Seminars in Immunopathology, 26, 347–362. 10.1007/s00281-004-0182-2 [DOI] [PubMed] [Google Scholar]
- Bechelli, J. , Smalley, C. , Zhao, X. , Judy, B. , Valdes, P. , Walker, D.H. et al. (2016) MyD88 mediates instructive signaling in dendritic cells and protective inflammatory response during Rickettsial infection. Infection and Immunity, 84, 883–893. 10.1128/IAI.01361-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs, H.M. , Behravesh, C.B. , Bradley, K.K. , Dahlgren, F.S. , Drexler, N.A. , Dumler, J.S. et al. (2016) Diagnosis and management of tickborne rickettsial diseases: Rocky Mountain spotted fever, ehrlichioses, and anaplasmosis ‐ United States. MMWR Recommendations and Reports, 65, 1–44. [DOI] [PubMed] [Google Scholar]
- Billings, A.N. , Feng, H.M. , Olano, J.P. & Walker, D.H. (2001) Rickettsial infection in murine models activates an early anti‐rickettsial effect mediated by NK cells and associated with production of gamma interferon. The American Journal of Tropical Medicine and Hygiene, 65, 52–56. 10.4269/ajtmh.2001.65.52 [DOI] [PubMed] [Google Scholar]
- Blanton, L.S. (2019) The Rickettsioses: a practical update. Infectious Disease Clinics of North America, 33, 213–229. 10.1016/j.idc.2018.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boes, M. , Prodeus, A.P. , Schmidt, T. , Carroll, M.C. & Chen, J. (1998) A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. Journal of Experimental Medicine, 188, 2381–2386. 10.1084/jem.188.12.2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle, M.J. , Reiling, L. , Feng, G. , Langer, C. , Osier, F.H. , Aspeling‐Jones, H. et al. (2015) Human antibodies fix complement to inhibit Plasmodium falciparum invasion of erythrocytes and are associated with protection against malaria. Immunity, 42, 580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.S. , Hussell, T. , Gilliland, S.M. , Holden, D.W. , Paton, J.C. , Ehrenstein, M.R. et al. (2002) The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proceedings of the National Academy of Sciences of the United States of America, 99, 16969–16974. 10.1073/pnas.012669199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, T.P. , Engström, P. , Chavez, R.A. , Fonbuena, J.A. , Vance, R.E. & Welch, M.D. (2020) Inflammasome‐mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nature Microbiology, 5, 688–696. 10.1038/s41564-020-0673-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell, M.M. & Martinez, J.J. (2009) The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infection and Immunity, 77, 5272–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl, M. , Martin, E.E. & Dasch, G.A. (1986) Human T helper cells specific for antigens of typhus group rickettsiae enhance natural killer cell activity in vitro . Infection and Immunity, 54, 297–302. 10.1128/iai.54.2.297-302.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y.G.Y. , Riley, S.P. , Chen, E. & Martinez, J.J. (2011) Molecular basis of immunity to rickettsial infection conferred through outer membrane protein B. Infection and Immunity, 79, 2303–2313. 10.1128/IAI.01324-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, Y.G.Y. , Riley, S.P. & Martinez, J.J. (2010) Adherence to and invasion of host cells by spotted fever group Rickettsia species. Frontiers in Microbiology, 1, 1–10. 10.3389/fmicb.2010.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonne, P.M. , Sahni, A. & Sahni, S.K. (2013) Suppressor of cytokine signalling protein SOCS1 and UBP43 regulate the expression of type I interferon‐stimulated genes in human microvascular endothelial cells infected with Rickettsia conorii . Journal of Medical Microbiology, 62, 968–979. 10.1099/jmm.0.054502-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta, S. , Dasgupta, S. & Bandyopadhyay, M. (2020) Regulatory B cells in infection, inflammation, and autoimmunity. Cellular Immunology, 352, 104076. 10.1016/j.cellimm.2020.104076 [DOI] [PubMed] [Google Scholar]
- Dunkelberger, J.R. & Song, W.C. (2010) Complement and its role in innate and adaptive immune responses. Cell Research, 20, 34–50. 10.1038/cr.2009.139 [DOI] [PubMed] [Google Scholar]
- Ehrenstein, M.R. & Notley, C.A. (2010) The importance of natural IgM: scavenger, protector and regulator. Nature Reviews Immunology, 10, 778–786. 10.1038/nri2849 [DOI] [PubMed] [Google Scholar]
- Fang, R. , Ismail, N. & Walker, D.H. (2012) Contribution of NK cells to the innate phase of host protection against an intracellular bacterium targeting systemic endothelium. The American Journal of Pathology, 181, 185–195. 10.1016/j.ajpath.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, H.M. , Wen, J. & Walker, D.H. (1993) Rickettsia australis infection: a murine model of a highly invasive vasculopathic rickettsiosis. The American Journal of Pathology, 142, 1471–1482. [PMC free article] [PubMed] [Google Scholar]
- Ferlazzo, G. & Morandi, B. (2014) Cross‐talks between natural killer cells and distinct subsets of dendritic cells. Frontiers in Immunology, 5, 1–7. 10.3389/fimmu.2014.00159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish, A.I. , Riley, S.P. , Singh, B. , Riesbeck, K. & Martinez, J.J. (2017) The Rickettsia conorii Adr1 interacts with the c‐terminus of human vitronectin in a salt‐sensitive manner. Frontiers in Cellular and Infection Microbiology, 7, 1–12. 10.3389/fcimb.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, M.I. , Chu, S.‐H. , Hernandez, M.X. , Fang, M.J. , Modarresi, L. , Selvan, P. et al. (2017) Cell‐specific deletion of C1qa identifies microglia as the dominant source of C1q in mouse brain. Journal of Neuroinflammation, 14, 1–15. 10.1186/s12974-017-0814-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusakio, M.E. , Mohammed, J.P. , Laumonnier, Y. , Hoebe, K. , Köhl, J. & Mattner, J. (2011) C5a regulates NKT and NK cell functions in sepsis. The Journal of Immunology, 187, 5805–5812. 10.4049/jimmunol.1100338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjeva, M.G. , Rouseva, M.M. , Zlatarova, A.S. , Reid, K.B.M. , Kishore, U. & Kojouharova, M.S. (2008) Interaction of human C1q with IgG and IgM: revisited. Biochemistry, 47, 13093–13102. 10.1021/bi801131h [DOI] [PubMed] [Google Scholar]
- Garred, P. , Genster, N. , Pilely, K. , Bayarri‐Olmos, R. , Rosbjerg, A. , Ma, Y.J. et al. (2016) A journey through the lectin pathway of complement—MBL and beyond. Immunological Reviews, 274, 74–97. 10.1111/imr.12468 [DOI] [PubMed] [Google Scholar]
- Garza, D.A. , Riley, S.P. & Martinez, J.J. (2017) Expression of Rickettsia Adr2 protein in E. coli is sufficient to promote resistance to complement‐mediated killing, but not adherence to mammalian cells. PLoS One, 12, 1–15. 10.1371/journal.pone.0179544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harboe, M. , Thorgersen, E.B. & Mollnes, T.E. (2011) Advances in assay of complement function and activation. Advanced Drug Delivery Reviews, 63, 976–987. 10.1016/j.addr.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Holodick, N.E. , Rodríguez‐Zhurbenko, N. & Hernández, A.M. (2017) Defining natural antibodies. Frontiers in Immunology, 8, 2–9. 10.3389/fimmu.2017.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez‐Reinoso, A. , Marin, A.V. & Regueiro, J.R. (2017) Complement in basic processes of the cell. Molecular Immunology, 84, 10–16. 10.1016/j.molimm.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Jones, D.D. , DeIulio, G.A. & Winslow, G.M. (2012) Antigen‐driven induction of polyreactive IgM during intracellular bacterial infection. The Journal of Immunology, 189, 1440–1447. 10.4049/jimmunol.1200878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, J.M. , Woods, M.E. , Soong, L. & Walker, D.H. (2009) Rickettsiae stimulate dendritic cells through toll‐like receptor 4, leading to enhanced NK cell activation in vivo. Journal of Infectious Diseases, 199, 236–242. 10.1086/595833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, M. , Matsushima, Y. , Mizutani, K. , Iida, S. , Habe, K. & Yamanaka, K. (2020) Transition of serum cytokine concentration in Rickettsia japonica infection. Infectious Disease Reports, 12, 127–131. 10.3390/idr12030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langereis, J.D. , Van Der Pasch, E.S. & De Jonge, M.I. (2019) Serum IgM and C‐reactive protein binding to phosphorylcholine of nontypeable Haemophilus influenzae increases complement‐mediated killing. Infection and Immunity, 87, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, J. & Thurman, J.M. (2018) Factor B. In: Barnum, S. & Schein, T. (Eds.) The complement factsbook: second edition. Elsevier Ltd. pp. 135–146. [Google Scholar]
- Mansueto, P. , Vitale, G. , Cascio, A. , Seidita, A. , Pepe, I. , Carroccio, A. et al. (2012) New insight into immunity and immunopathology of Rickettsial diseases. Clinical and Developmental Immunology, 2012, 967852. 10.1155/2012/967852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathern, D.R. & Heeger, P.S. (2015) Molecules great and small: the complement system. Clinical Journal of the American Society of Nephrology, 10, 1636–1650. 10.2215/CJN.06230614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle, N.S. , Church, S.E. , Fremeaux‐Bacchi, V. & Roumenina, L.T. (2015a) Complement system part I ‐ molecular mechanisms of activation and regulation. Frontiers in Immunology, 6, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merle, N.S. , Noe, R. , Halbwachs‐Mecarelli, L. , Fremeaux‐Bacchi, V. & Roumenina, L.T. (2015b) Complement system part II: role in immunity. Frontiers in Immunology, 6, 1–26. 10.3389/fimmu.2015.00257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, X. , Liu, C. , Wei, Y. , Wang, N.A. , Yuan, G. , Liu, D. et al. (2014) Expression and regulation of complement receptors by human natural killer cells. Immunobiology, 219, 671–679. 10.1016/j.imbio.2014.03.018 [DOI] [PubMed] [Google Scholar]
- Nagele, E.P. , Han, M. , Acharya, N.K. , DeMarshall, C. , Kosciuk, M.C. & Nagele, R.G. (2013) Natural IgG autoantibodies are abundant and ubiquitous in human sera, and their number is influenced by age, gender, and disease. PLoS One, 8, e60726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson, B. & Nilsson Ekdahl, K. (2012) The tick‐over theory revisited: Is C3 a contact‐activated protein? Immunobiology, 217, 1106–1110. 10.1016/j.imbio.2012.07.008 [DOI] [PubMed] [Google Scholar]
- Ochsenbein, A.F. , Fehr, T. , Lutz, C. , Suter, M. , Brombacher, F. , Hengartner, H. et al. (1999) Control of early viral and bacterial distribution and disease by natural antibodies. Science, 286, 2156–2159. 10.1126/science.286.5447.2156 [DOI] [PubMed] [Google Scholar]
- Otterdal, K. , Portillo, A. , Astrup, E. , Ludviksen, J.K. , Schjalm, C. , Raoult, D. et al. (2016) Rickettsia conorii is a potent complement activator in vivo and combined inhibition of complement and CD14 is required for attenuation of the cytokine response ex vivo . Clinical Microbiology and Infection, 22, 734. 10.1016/j.cmi.2016.05.010 [DOI] [PubMed] [Google Scholar]
- Panda, S. & Ding, J.L. (2015) Natural antibodies bridge innate and adaptive immunity. The Journal of Immunology, 194, 13–20. 10.4049/jimmunol.1400844 [DOI] [PubMed] [Google Scholar]
- Panda, S. , Zhang, J. , Tan, N.S. , Ho, B. & Ding, J.L. (2013) Natural IgG antibodies provide innate protection against ficolin‐opsonized bacteria. EMBO Journal, 32, 2905–2919. 10.1038/emboj.2013.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola, P. , Paddock, C.D. & Raoult, D. (2005) Tick‐borne rickettsioses around the world: emerging diseases challenging old concepts. Clinical Microbiology Reviews, 18(4), 719–756. 10.1128/CMR.18.4.719-756.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola, P. , Paddock, C.D. , Socolovschi, C. , Labruna, M.B. , Mediannikov, O. , Kernif, T. et al. (2013) Update on tick‐borne rickettsioses around the world: a geographic approach. Clinical Microbiology Reviews, 26, 657–702. 10.1128/CMR.00032-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, K.B.M. (2018) Complement component C1q: historical perspective of a functionally versatile, and structurally unusual, serum protein. Frontiers in Immunology, 9, 1–6. 10.3389/fimmu.2018.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro, C.M. , Carvalho, J.L.B.D. , Bastos, P.A.D.S. , Katagiri, S. , Batalha, E.Y. , Okano, W. et al. (2021) Prevalence of Rickettsia rickettsii in ticks: systematic review and meta‐analysis. Vector‐Borne and Zoonotic Diseases, 21, 557–565. [DOI] [PubMed] [Google Scholar]
- Ricklin, D. , Hajishengallis, G. , Yang, K. & Lambris, J.D. (2010) Complement: a key system for immune surveillance and homeostasis. Nature Immunology, 11, 785–797. 10.1038/ni.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin, D. , Reis, E.S. & Lambris, J.D. (2016) Complement in disease: a defence system turning offensive. Nature Reviews Nephrology, 12, 383–401. 10.1038/nrneph.2016.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, S.P. , Fish, A.I. , Piero, F.D. & Martinez, J.J. (2018) Immunity against the obligate intracellular bacterial pathogen Rickettsia australis requires a functional complement system. Infection and Immunity, 86, 1–15. 10.1128/IAI.00139-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, S.P. , Patterson, J.L. & Martinez, J.J. (2012) The rickettsial OmpB β‐peptide of Rickettsia conorii is sufficient to facilitate factor h‐mediated serum resistance. Infection and Immunity, 80, 2735–2743. 10.1128/IAI.00349-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, S.P. , Patterson, J.L. , Nava, S. & Martinez, J.J. (2014) Pathogenic Rickettsia species acquire vitronectin from human serum to promote resistance to complement‐mediated killing. Cellular Microbiology, 16, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer, M. & Hardy, R.R. (2001) Frequency difference gating: a multivariate method for identifying subsets that differ between samples. Cytometry, 64, 56–64. [DOI] [PubMed] [Google Scholar]
- Roederer, M. , Moore, W. , Treister, A. , Hardy, R.R. & Herzenberg, L.A. (2001) Probability binning comparison: a metric for quantitating multivariate distribution differences. Cytometry, 55, 47–55. [DOI] [PubMed] [Google Scholar]
- Rumfield, C. , Hyseni, I. , McBride, J.W. , Walker, D.H. & Fang, R. (2020) Activation of ASC inflammasome driven by toll‐like receptor 4 contributes to host immunity against rickettsial infection. Infection and Immunity, 88, e00886‐19. 10.1128/IAI.00886-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni, A. , Fang, R. , Sahni, S.K. & Walker, D.H. (2019) Pathogenesis of rickettsial diseases: pathogenic and immune mechanisms of an endotheliotropic infection. Annual Review of Pathology: Mechanisms of Disease, 14, 127–152. 10.1146/annurev-pathmechdis-012418-012800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni, S.K. (2009) Host‐cell interactions with pathogenic Rickettsia species. Future Microbiology, 4, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni, S.K. , Narra, H.P. , Sahni, A. & Walker, D.H. (2013) Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiology, 8, 1265–1288. 10.2217/fmb.13.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandholm, K. , Persson, B. , Skattum, L. , Eggertsen, G. , Nyman, D. , Gunnarsson, I. et al. (2019) Evaluation of a novel immunoassay for quantification of C1q for clinical diagnostic use. Frontiers in Immunology, 10, 1–9. 10.3389/fimmu.2019.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, J.T. , Barker, J.H. , Long, M.E. , Kaufman, J. , McCracken, J. & Allen, L.‐A.‐H. (2012) Natural IgM mediates complement‐dependent uptake of Francisella tularensis by human neutrophils via complement receptors 1 and 3 in nonimmune serum. The Journal of Immunology, 189, 3064–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F.L. & Baumgarth, N. (2019) B‐1 cell responses to infections. Current Opinion in Immunology, 57, 23–31. 10.1016/j.coi.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sörman, A. , Zhang, L. , Ding, Z. & Heyman, B. (2014) How antibodies use complement to regulate antibody responses. Molecular Immunology, 61, 79–88. 10.1016/j.molimm.2014.06.010 [DOI] [PubMed] [Google Scholar]
- De Sousa, R. , Dória Nóbrega, S. , Bacellar, F. & Torgal, J. (2003) Mediterranean spotted fever in Portugal: risk factors for fatal outcome in 105 hospitalized patients. Annals of the New York Academy of Sciences, 990, 285–294. 10.1111/j.1749-6632.2003.tb07378.x [DOI] [PubMed] [Google Scholar]
- Stewart, A. , Armstrong, M. , Graves, S. & Hajkowicz, K. (2017) Rickettsia australis and Queensland tick typhus: a rickettsial spotted fever group infection in Australia. The American Journal of Tropical Medicine and Hygiene, 97, 24–29. 10.4269/ajtmh.16-0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straily, A. , Feldpausch, A. , Ulbrich, C. , Schell, K. , Casillas, S. , Zaki, S.R. et al. (2016) Notes from the field: Rickettsia parkeri Rickettsiosis — Georgia, 2012–2014. MMWR. Morbidity and Mortality Weekly Report, 65, 718–719. [DOI] [PubMed] [Google Scholar]
- Thielens, N.M. , Tedesco, F. , Bohlson, S.S. , Gaboriaud, C. & Tenner, A.J. (2017) C1q: a fresh look upon an old molecule. Molecular Immunology, 89, 73–83. 10.1016/j.molimm.2017.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.H. (2007) Rickettsiae and rickettsial infections: the current state of knowledge. Clinical Infectious Diseases, 45, 39–44. 10.1086/518145 [DOI] [PubMed] [Google Scholar]
- Walker, D.H. , Olano, J.P. & Feng, H.M. (2001) Critical role of cytotoxic T lymphocytes in immune clearance of rickettsial infection. Infection and Immunity, 69, 1841–1846. 10.1128/IAI.69.3.1841-1846.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, D.H. , Popov, V.L. & Feng, H.M. (2000) Establishment of a novel endothelial target mouse model of a typhus group rickettsiosis: evidence for critical roles for gamma interferon and CD8 T lymphocytes. Laboratory Investigation, 80, 1361–1372. 10.1038/labinvest.3780144 [DOI] [PubMed] [Google Scholar]
- Wessels, M.R. , Butko, P. , Ma, M. , Warren, H.B. , Lage, A.L. & Carroll, M.C. (1995) Studies of group B streptococcal infection in mice deficient in complement component C3 or C4 demonstrate an essential role for complement in both innate and acquired immunity. Proceedings of the National Academy of Sciences of the United States of America, 92, 11490–11494. 10.1073/pnas.92.25.11490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, E.E. , Kolev, M. & Kemper, C. (2018) Complement and the regulation of T Cell responses. Annual Review of Immunology, 36, 309–338. 10.1146/annurev-immunol-042617-053245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi, H. , Xie, J. & Skare, J.T. (2018) The classical complement pathway is required to control Borrelia burgdorferi levels during experimental infection. Frontiers in Immunology, 9, 1–12. 10.3389/fimmu.2018.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.‐Y. , Shao, T. , Nie, L.I. , Zhu, L.‐Y. , Xiang, L.‐X. & Shao, J.‐Z. (2016) Evolutionary implication of B‐1 lineage cells from innate to adaptive immunity. Molecular Immunology, 69, 123–130. 10.1016/j.molimm.2015.10.014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All data are in the manuscript file.


