Abstract
Culture and susceptibility testing of Helicobacter pylori strains was performed in a large multinational, multicenter randomized clinical trial. Culture was carried out on gastric biopsy samples obtained from 516 patients at entry and had a sensitivity of 99% when the [13C]urea breath test was used as a reference. Susceptibility testing was performed for clarithromycin and metronidazole on 485 strains by an agar dilution method and the epsilometer test (Etest) and for amoxicillin by an agar dilution method only. Resistance to clarithromycin (>1 μg/ml) was found in 3% of the H. pylori strains, with a perfect correlation between Etest and agar dilution methods. Resistance to metronidazole (>8 μl/ml) was found in 27% of the strains by agar dilution, but there were important discrepancies between it and the Etest method. No resistance to amoxicillin was found. The logarithms of the MICs of the three antibiotics against susceptible strains had a distribution close to normal. The impact of resistance was tested in the four arms of the trial. There were not enough clarithromycin-resistant strains to evaluate the impact of resistance on the cure rate of clarithromycin-based regimens. For metronidazole-resistant strains, the impact noted in the clarithromycin-metronidazole arm was partially overcome when omeprazole was added (76% eradication for resistant strains versus 95% for susceptible strains). Secondary resistance to clarithromycin occurred in strains from 12 of 105 patients (11.4%) after the failure of a clarithromycin-based regimen to effect eradication. The detection of point mutations in clarithromycin-resistant strains was performed by a combination of PCR and restriction fragment length polymorphism. Mutations (A2142G and 2143G) were found in all strains tested except one. This study stresses the importance of performing susceptibility tests in clinical trials in order to explain the results of different treatments.
The eradication of Helicobacter pylori has been the subject of numerous clinical trials these last 10 years in order to find the optimum therapy. However, few of these trials have been large multicenter randomized clinical trials, and moreover the culture of H. pylori which allows the testing of antimicrobial susceptibility has rarely been performed (3, 15, 27, 39).
There are several limitations to previous studies of the antimicrobial characteristics of H. pylori strains, which include small sample size, no details about the history of the patients, a geographical distribution limited to specific areas, and inadequate methodology, with few exceptions (13).
Despite the fact that culture is the standard detection method for most pathogenic bacteria, the specific requirements of H. pylori in terms of transport and growth render it difficult to culture. Other methods such as histological detection have even been proposed as the “gold standard” (1). Moreover, the impact of primary resistance on the eradication rate and the occurrence of secondary resistance have been challenged (17, 28) and deserve to be studied further in the context of large trials.
The aim of our study was to perform culture on gastric biopsies from a large, double-blind, randomized, multinational, multicenter clinical trial performed with a homogeneous group of duodenal ulcer patients in remission in Europe (i) to study the distribution of MICs of antibiotics against H. pylori strains, (ii) to compare the results obtained by agar susceptibility testing and epsilometer test (Etest), (iii) to detect the point mutations associated with resistance to macrolides, and (iv) to measure the impact of primary resistance on treatment outcome as well as the occurrence of secondary resistance in H. pylori.
MATERIALS AND METHODS
Detection of H. pylori.
Patients with duodenal ulcer disease at 47 centers in six countries (France, Germany, Ireland, Norway, Sweden, and the United Kingdom) were included in the study. Only patients with no or at the most one previous H. pylori eradication attempt were enrolled in the study. The details of the clinical trial design are provided elsewhere (18).
The antibiotics tested were amoxicillin, clarithromycin, and metronidazole since the drugs used in the trial were either amoxicillin (1 g) plus clarithromycin (500 mg) with (OAC) and without (AC) omeprazole (20 mg) or metronidazole (400 mg) plus clarithromycin (250 mg) with (OMC) and without (MC) omeprazole (20 mg). All of the drugs were given twice daily for 7 days.
At the first visit, patients underwent both a [13C]urea breath test (UBT), as previously described (19), and endoscopy. The UBT kit used was made by Utandningsester Sverize AB (Gôteborg, Sweden), and the cutoff value was 5 δ 0/00. During endoscopy, a screening test (CLO test) was performed and biopsies were taken for culture from the antrum (two biopsies 1 to 2 cm from the pylorus on the anterior and posterior walls) and the corpus (two biopsies approximately 10 cm from the cardia along the greater curvature). They were immediately introduced into a transport medium (Portagerm pylori; bioMérieux, Marcy l’Etoile, France) and sent by courier to a local bacteriology laboratory within 24 h in a cool transport container (Sarstedt, Nümbrecht, Germany). There was one bacteriology laboratory per country except in Ireland, where the biopsies were sent to the United Kingdom.
The following procedure was used in all laboratories. The biopsies were first ground in 0.5 ml of brucella broth in an electric homogenizer. The suspension was then plated on three different media. Two were made in-house: Wilkins-Chalgren agar (Oxoid, Basingstoke, United Kingdom) enriched with 10% human blood and supplemented with vancomycin (10 μg/ml), cefsulodin (5 μg/ml), trimethoprim (5 μg/ml), and actidione (100 μg/ml) and Columbia agar enriched with 10% human blood without antibiotics; the third was a commercial selective medium: pylori agar (bioMérieux). The plates were incubated for 12 days in a microaerobic atmosphere (jars with GasPaks). Colonies suspected to be H. pylori were identified by the presence of urease, catalase, and oxidase activities. The strains were kept frozen at −70°C in brucella broth with 25% glycerol and sent in dry ice by courier at regular intervals to a central laboratory in Bordeaux, France, for further study.
The UBT was repeated 4 and 8 weeks after the end of treatment. Only the patients positive by UBT after treatment underwent a second endoscopy and had a culture for H. pylori performed.
Eradication was defined as the conversion of a positive test at entry (culture or UBT) to two negative UBTs 4 and 8 weeks after cessation of therapy.
Determination of MICs of antibiotics against H. pylori by the agar dilution method.
All of the plates were prepared extemporaneously. McFarland 3 suspensions were prepared in brucella broth from 48-h agar plate cultures and inoculated with a Steers apparatus onto Wilkins-Chalgren agar enriched with 10% sheep blood and containing the antibiotic at the following concentrations: 0.0035 to 4 μg/ml (amoxicillin), 0.0035 to 128 μg/ml (clarithromycin), and 0.03 to 128 μg/ml (metronidazole). The plates were immediately incubated at 37°C in a microaerobic atmosphere (jars with GasPaks) and read after 48 h. The MIC was defined as the lowest concentration with complete growth inhibition.
The quality control strains used were CCUG 38770, CCUG 38771, and CCUG 38772, proposed in Europe for this purpose. In the absence of standard breakpoints, the breakpoints used to define a resistant strain in this study were based on a previous publication (18) and on our experience in clinical trials. They were the following: metronidazole, >8 μg/ml (11); clarithromycin, >1 μg/ml (0.25 to 1 μg/ml, intermediate). No breakpoint was predefined for amoxicillin.
Determination of MICs of antibiotics against H. pylori by the Etest.
Clarithromycin and metronidazole were tested. McFarland 3 suspensions were prepared in brucella broth from 48-h agar plate cultures; 0.5 ml was used to flood Wilkins-Chalgren agar plates enriched with 10% human blood but without antibiotics, and the excess was removed. After the plates were dried, Etest strips were placed on them and they were incubated for 48 h in a microaerobic atmosphere (jars with GasPaks) without anaerobic preincubation and read according to the supplier’s recommendations.
Detection of point mutations associated with clarithromycin resistance by PCR-RFLP.
One pair of primers was used to amplify one fragment of the peptidyltransferase region of the 23S rRNA. The sequences of the primers were based on the published sequence of the 23S rRNA gene of H. pylori (GenBank accession no., U27270). A primer extending from position 1820 to 1839 (5′-CCA CAG CG CTC AG-3′) and a reverse primer from position 2244 to 2225 (5′-CTC CAT AAG AGC CAA AGC CC-3′) were used to amplify a fragment of 425 bp. PCR amplification of DNA was performed in a final volume of 50 μl containing 1 μg of H. pylori genomic DNA, 67 mM Tris-HCl (pH 8.8), 16 mM (NH4)2SO4, 0.01% Tween 20, 1.5 mM MgCl2, a 0.2 mM concentration of deoxynucleoside triphosphate mixture, 0.2 μM concentrations of primers, and 1 U of Taq DNA polymerase. Each reaction mixture was overlaid with 50 μl of mineral oil. The cycling program was 1 cycle at 94°C for 5 min; 40 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min; and a final elongation step at 72°C for 7 min. The two reactions resulted in a single fragment of the expected size.
Advantage was taken of the occurrence of restriction sites for specific enzymes to perform PCR-restriction fragment length polymorphism (RFLP) as a rapid method of detection of the mutation. The method used to detect the mutations without sequencing involved restriction of the PCR products. Ten microliters of amplicon (425 bp) was treated with enzyme BsaI or BbsI (New England Biolabs, Beverly, Mass.). The fragments were incubated for 24 h at 50°C for BsaI and at 37°C for BbsI in order to detect the restriction site occurring when the mutation was A→G at position 2143 and at position 2142, respectively.
Statistical analysis.
Differences in eradication rates were tested with the Fisher exact test. The influence of the metronidazole MICs on the eradication probability in the OMC group was tested by a stepwise procedure based on a permutation test for trend. The underlying assumption for the procedure was that the probability of treatment failure, as a function of the MIC, is nondecreasing, i.e., that higher MICs are not associated with higher success probability. In the first step, the permutation test, with the logarithms of the MICs as scores, was applied to all OMC data to test for a trend towards lower eradication rates for higher MICs. In case of statistical significance, it was concluded that (at least) the highest MIC (128 μg/ml) was associated with a decreased eradication rate and the stepwise procedure was then continued. In the second step, the trend analysis was repeated excluding the strains for which the MIC was 128 μg/ml. A MIC of 64 μg/ml was also interpreted as a significant result giving a decreased probability of eradication. The stepwise procedure was continued in this way, gradually excluding the highest MICs, until the first nonsignificant result was encountered. The procedure was then stopped. This stepwise procedure is a kind of closed test (20) that strongly protects the overall type I error rate.
RESULTS
There were 539 patients randomized in the study of whom 514 were included in the intention to treat analysis and 449 were included in the per protocol analysis.
A comparison between UBT and culture was made for only 529 patients at entry, since one test was missing in 10 cases. With UBT as the reference, the sensitivity of culture was 99%.
There were 485 strains available for determination of the MICs. The strains were isolated from Sweden (203), Norway (93), Germany (85), the United Kingdom and Ireland (60), and France (44). The distribution of the MICs of clarithromycin, metronidazole, and amoxicillin against H. pylori determined by the agar dilution method are presented in Fig. 1 to 3, respectively. Only 14 strains (3%) were categorized as intermediate or resistant to clarithromycin, while 131 strains (27%) were resistant to metronidazole. The distribution of metronidazole resistance rates varied between countries from 16 to 42%, the highest rate occurring in Norway (Table 1). The results of the Etest were compared to those of the agar dilution method for clarithromycin and metronidazole (Table 2). There was a perfect correlation for clarithromycin. In contrast, there was a large proportion of discrepancies (22%) for metronidazole: the Etest led to higher MICs, and more strains were categorized as resistant. When the dilution steps were compared (Fig. 4), it was found that the percentage of agreement within plus or minus 1 dilution was 57% and within plus or minus 2 dilutions was 74%.
FIG. 1.
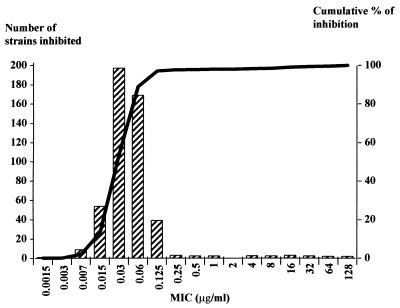
Distribution of the MICs of amoxicillin against 485 strains of H. pylori. MIC at which 50% of the strains were inhibited (MIC50), 0.03 μg/ml; MIC90, 0.125 μg/ml; range, 0.035 to 0.5 μg/ml.
FIG. 3.
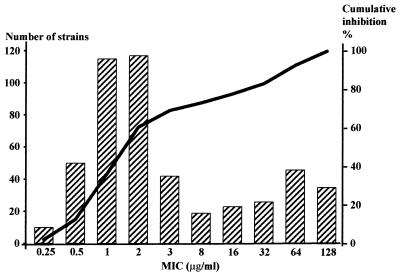
Distribution of the MICs of metronidazole against 485 strains of H. pylori. MIC at which 50% of the strains were inhibited (MIC50), 2 μg/ml; MIC90, 64 μg/ml; range, 0.25 to 128 μg/ml.
TABLE 1.
Distribution of primary resistance of H. pylori to antibiotics in different countries determined by the agar dilution method
TABLE 2.
Comparison of the MIC results for clarithromycin and metronidazole against H. pylori by agar dilution and Etest
| Drug and Etest result | No. of strains with indicated agar dilution result
|
||
|---|---|---|---|
| Susceptible | Resistant | Total | |
| Clarithromycin | |||
| Susceptible | 460 | 0 | 460 |
| Resistant | 0 | 14 | 14 |
| Total | 460 | 14 | 474 |
| Metronidazole | |||
| Susceptible | 261 | 37 | 298 |
| Resistant | 72 | 99 | 171 |
| Total | 333 | 136 | 469 |
FIG. 4.
Differences of dilution steps for MICs determined by Etest and the agar dilution method (n = 469).
The impact of primary resistance of H. pylori on the clinical outcome in the different treatment groups was determined for 470 patients. For clarithromycin, it was difficult to evaluate this impact because of the low number of resistant strains. However, none of the resistant strains was eradicated in the two arms whose patients received only the antibiotics without omeprazole. For metronidazole, there was a clear impact of resistance on the cure of the infection, which was partly overcome by the addition of omeprazole. The eradication rate increased from 76 (95% confidence interval [CI], 58 to 89%) to 91% (95% CI, 82 to 96%) when it was added (P < 0.007). In the OAC group, considered a control group, no difference between strains susceptible and resistant to metronidazole was observed.
Metronidazole MICs of 128, 64, and 32 μg/ml were responsible for a significantly higher risk of failure (P < 0.001, P = 0.001, and P < 0.05, respectively) but not 16 μg/ml (P = 0.06).
The failure of OAC was not associated with a higher MIC of amoxicillin against H. pylori. With an MIC of 0.03 μg/ml, there was one failure among 39 cases (2.5%); with an MIC of 0.06 μg/ml, there were three failures among 27 cases (11%); and with an MIC of 0.125 μg/ml, there was one failure among 12 cases (8%). The distribution of failures was similar in the groups which did not receive amoxicillin.
Fifteen percent of the patients had received previous eradication therapy: 57 patients had received amoxicillin, 22 had received macrolides, and 15 had received metronidazole. An impact of this previous therapy was found only for macrolides (P = 0.001).
Secondary resistance of H. pylori to clarithromycin, defined as a strain which converted from below (at inclusion) to above (at the posttreatment control) the defined breakpoint, occurred in 12 of 105 patients (11.4%) receiving clarithromycin; secondary resistance to metronidazole occurred in 12 patients receiving metronidazole. In addition, for the patients receiving amoxicillin and clarithromycin alone, there were discrepancies between the results obtained with metronidazole: nine susceptible strains became resistant to metronidazole, and six resistant strains became susceptible to metronidazole.
PCR-RFLP to detect point mutations associated with clarithromycin resistance was performed for 10 strains which were resistant pretreatment and for the 12 strains which became resistant after treatment. The distribution of the mutation according to the pre- or posttreatment isolation is presented in Table 3. Only one pretreatment strain did not have one of the two mutations sought for; this strain possibly had an A2142C mutation. For those strains not eradicated, the same mutation was found before and after treatment. A mixed population of resistant wild-type bacteria was only found in one case, both before and after treatment. The median MIC was 16 μg/ml for the strains with the A2143G mutation and 64 μg/ml for those with the A2142G mutation.
TABLE 3.
Distribution of point mutations associated with clarithromycin resistance in H. pylori strains isolated pre- and posttreatment
| Point at which strain resistance noted | No. of strains showing:
|
||
|---|---|---|---|
| BsaI restriction (A2143G) | BbsI restriction (A2142G) | No restriction with BsaI and BbsI | |
| Pretreatment | 7 | 2 | 1 |
| Posttreatment | 4 | 8 | |
DISCUSSION
In contrast to what has been previously published and recommended (1), we found that culture, which is a prerequisite for antimicrobial susceptibility testing (23), is also an accurate way to diagnose H. pylori infection. Despite the fact that there were 47 centers involved in this study, the recovery rate was higher than 90% and even slightly better than the sensitivity of the UBT. The most probable explanation for this high yield was the use of a rigorous and standardized protocol, identical in the five laboratories involved in performing the bacterial culture. The critical points were the grinding of biopsies, the use of several kinds of agar media including two which were freshly prepared, and the follow-up readings of the plates over a 12-day period. Moreover, a transport system which maintained the biopsies at a low temperature, avoided dessication and contact with air (26a), and assured delivery within 24 h was used.
We have consistently used Wilkins-Chalgren agar enriched with human blood for culture for the past 10 years following an initial comparative study of media (25). It has provided the best results in clinical practice compared to other media, but these findings have never been formally published (22). Human serum is considered the best growth supplement (38). Cefsulodin is a good selective agent for gram-negative bacilli including Pseudomonas aeruginosa (16) and has been proposed in a commercially available selective supplement (6). The improved growth capacity of fresh media has been stressed by others (12), as well as the use of selective and nonselective media (33).
A reference method for studying the susceptibility of H. pylori to clarithromycin was recommended by the National Committee for Clinical Laboratory Standards (NCCLS) (29, 34) after this study was performed. The NCCLS method also uses agar dilution but differs on the following points: Wilkins-Chalgren agar was used instead of Mueller-Hinton agar; 10% instead of 5% blood was used; there was a larger bacterial inoculum (109 instead of 107 to 108 CFU/ml), which was prepared in brucella broth from a 48-h plate instead of in saline from a 72-h plate; and a shorter incubation time (48 instead of 72 h) was used. The higher inoculum was used in order to detect possible resistant mutant colonies and to be able to read the plates within 48 h. The agar dilution method, which is considered to be the reference method for performing susceptibility tests for most other bacteria, was also used to test metronidazole and amoxicillin.
This is the first time that a frequency distribution of the logarithms of the MICs for the three antibiotics has been obtained on such a large random sample of strains cultured from duodenal ulcer disease patients who are possibly representative of those in northern Europe.
The MIC distribution for amoxicillin against H. pylori was normal, with the highest MIC at 0.5 μg/ml. It is necessary to seek a resistance mechanism, such as a modification in penicillin binding proteins, before categorizing these strains as resistant. We found no resistant strains, in contrast to the findings of van Zwet et al. (35), or tolerant strains, as recently identified by Dore et al. (7).
The MIC distribution for clarithromycin against H. pylori was normal for 97% of the strains. A point mutation on the 23S rRNA gene (30, 37) was detected in all resistant strains except one by PCR-RFLP, leading one to think that an A2142C mutation could be present. The proportion of resistant strains was very low in this study. This may reflect the limited use of macrolides in countries from the northern part of Europe, especially Scandinavia. However, the resistance rate was also surprisingly low in France, which is contrary to results obtained in several other studies involving hundreds of strains, where it was in the range of 10% (24). Another explanation may be that strains isolated from patients with duodenal ulcer disease are more likely to be susceptible to macrolides. Further studies should be performed to address this question.
The MIC distribution of metronidazole against H. pylori showed a normal distribution for part of the strains. A tendency for a second mode was observed for strains with MICs higher than 8 μg/ml. The resistance mechanism of H. pylori to metronidazole is not well known. It may concern the enzymes involved in the reduction of the nitro group, but alternate pathways may exist, and therefore the observed MICs would be the result of a complex phenomenon (32). The breakpoint of 8 μg/ml has been proposed for metronidazole resistance based on studies using bismuth-based triple therapies (11), and this figure was applied in this study. This breakpoint was valid for the MC group but not for the OMC group. The stepwise test indicated that the threshold of 32 μg/ml would be more appropriate if the latter regimen is used.
In contrast to clarithromycin resistance, resistance to metronidazole (>8 μg/ml) was found in 27% of the strains, with a marked variation between the different countries. Surprisingly, the frequency of resistance was only 16% in France; in previous studies the frequency was double this value. In Norway it was 42%. The reason for this high resistance rate in Norway is unclear.
The Etest has proven to be an accurate method to test the susceptibilities of fastidious organisms, including H. pylori, to antibiotics. However, there is some concern regarding the value of Etest results for testing the effect of metronidazole on this bacterium. While we found an excellent agreement between the agar dilution method and the Etest for clarithromycin, there was an unacceptable discrepancy rate of 22% for metronidazole. The reason is unclear, but in this study, the plates were not preincubated in an anaerobic atmosphere (4). On the other hand, for the agar dilution test, the plates were prepared extemporaneously and therefore they were more likely to have a constant redox potential than the plates used for the Etest. In previously performed studies, the correlation was indeed far from perfect: the percentage of agreement within plus or minus 1 dilution was 89 (31) and 83% (8), but only 56% in the study of Cederbrant (5). Contrary to these results, Hirschl et al. found the Etest to be accurate and precise (14).
The clinical relevance of H. pylori resistance to antibiotics is an important aspect to consider (26). Unfortunately, it was not possible to draw any conclusions for clarithromycin because of the low number of resistant strains encountered. With regard to metronidazole, there was an important impact of the resistance to this drug when it was used only with clarithromycin but the addition of omeprazole had a beneficial effect. The reason is not clear since metronidazole activity is not supposed to be pH dependent and, even more importantly, the diffusion of the drug in the gastric lumen decreases when the pH increases (9, 36). Therefore, it is most likely that the beneficial effect involves clarithromycin, whose activity is pH dependent, and/or a synergism between the two compounds. In another study, where amoxicillin was given instead of clarithromycin with metronidazole and lansoprazole, there was a lower cure rate (2).
A consequence of the high eradication rate is a low occurrence of secondary resistance. Indeed, in the group receiving OAC, no secondary resistance was noted. In the group treated with OMC, four strains resistant to clarithromycin and four strains resistant to metronidazole were observed, and even more were observed in the group treated with MC, with five strains resistant to clarithromycin and eight resistant to metronidazole.
The occurrence of resistance to metronidazole in the group receiving AC was unexpected. In fact, the determination of these MICs was repeated and most of them were found to be in the susceptible range. The finding of resistance was probably due to technical problems related to the method of metronidazole testing. Even with agar dilution, the reproducibility is not 100% satisfactory. The recent discovery that mutations of the nitroreductase gene can result in resistance to metronidazole gives us some hope that in the near future molecular tests will be developed (10).
In conclusion, culture of H. pylori proved to be an achievable goal even in a large randomized clinical trial and, as expected, susceptibility testing is important in explaining the results observed. Since the prevalence of primary resistance varies between different areas, it is of paramount importance to include culture and susceptibility testing in future trials. Even though the Etest has proved to be a very satisfactory test for clarithromycin, its use for metronidazole requires further standardization.
FIG. 2.
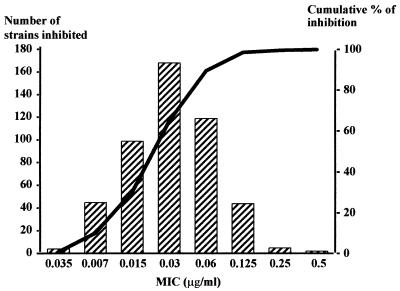
Distribution of the MICs of clarithromycin against 485 strains of H. pylori. MIC at which 50% of the strains were inhibited (MIC50), 0.03 μg/ml; MIC90, 0.125 μg/ml; range, 0.07 to 128 μg/ml.
ACKNOWLEDGMENT
We acknowledge Astra Hässle, Mölndal, Sweden, which financially supported this study.
REFERENCES
- 1.Barthel J S, Everett E D. Diagnosis of Campylobacter pylori infections: the “gold standard” and the alternatives. Rev Infect Dis. 1990;12(Suppl. 1):S107–S114. doi: 10.1093/clinids/12.supplement_1.s107. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard S, Birac C, Lamouliatte H, Forestier S, Mégraud F. Correlation between the MICs of metronidazole on H. pylori strains and the outcome of a lansoprazole-amoxicillin-metronidazole therapy. Gut. 1996;39(Suppl. 2):A05. . (Abstract.) [Google Scholar]
- 3.Buckley M J M, Xia H X, Hyde D M, Keane C T, O’Morain C. Metronidazole resistance reduces efficacy of triple therapy and leads to secondary clarithromycin resistance. Dig Dis Sci. 1997;42:2111–2115. doi: 10.1023/a:1018882804607. [DOI] [PubMed] [Google Scholar]
- 4.Cederbrant G, Kahlmeter G, Ljungh A. Proposed mechanism for metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1992;29:115–120. doi: 10.1093/jac/29.2.115. [DOI] [PubMed] [Google Scholar]
- 5.Cederbrant G, Kahlmeter G, Ljungh A. The Etest for antimicrobial susceptibility testing of Helicobacter pylori. J Antimicrob Chemother. 1993;31:65–71. doi: 10.1093/jac/31.1.65. [DOI] [PubMed] [Google Scholar]
- 6.Dent J C, McNulty C A. Evaluation of a new selective medium for Campylobacter pylori. Eur J Clin Microbiol Infect Dis. 1988;7:555–558. doi: 10.1007/BF01962615. [DOI] [PubMed] [Google Scholar]
- 7.Dore M P, Osato M S, Realdi G, Mura I, Graham D Y, Sepulveda A R. Amoxicillin tolerance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:47–54. doi: 10.1093/jac/43.1.47. [DOI] [PubMed] [Google Scholar]
- 8.Glupczynski Y, Labbé M, Hansen W, Crokahert F, Yourassowsky E. Evaluation of the E test for quantitative antimicrobial susceptibility testing of Helicobacter pylori. J Clin Microbiol. 1991;29:2072–2075. doi: 10.1128/jcm.29.9.2072-2075.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goddard A F, Jessa M J, Barrett D A, Shaw P N, Idström J P, Cederberg C, Spiller R C. Effect of omeprazole on the distribution of metronidazole, amoxicillin and clarithromycin in human gastric juice. Gastroenterology. 1996;111:358–367. doi: 10.1053/gast.1996.v111.pm8690200. [DOI] [PubMed] [Google Scholar]
- 10.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten S J O, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin C S, Marshall B J, Blincow E D, Wilson D H, Blackbourn S, Philipps M. Prevention of nitroimidazole resistance in Campylobacter pylori by coadministration of colloidal bismuth subcitrate: clinical and in vitro studies. J Clin Pathol. 1988;41:207–210. doi: 10.1136/jcp.41.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hachem C Y, Clarridge J E, Evans D G, Graham D Y. Comparison of agar based media for primary isolation of Helicobacter pylori. J Clin Pathol. 1995;48:714–716. doi: 10.1136/jcp.48.8.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartzen S H, Andersen L P, Bremmelgaard A, Colding H, Arpi M, Kristiansen J, Justesen T, Espersen F, Frimot-Moller N, Bonnevie O. Antimicrobial susceptibility testing of 230 Helicobacter pylori strains: importance of medium, inoculum, and incubation time. Antimicrob Agents Chemother. 1997;41:2634–2639. doi: 10.1128/aac.41.12.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschl A M, Hirschl M M, Rotter M L. Comparison for the determination of the sensitivity of Helicobacter pylori to metronidazole. J Antimicrob Chemother. 1993;32:45–49. doi: 10.1093/jac/32.1.45. [DOI] [PubMed] [Google Scholar]
- 15.Kist M, Strobel S, Fölsch U R, Kirchner T, Hahn E G, van Kleist D H, Klör H U, Dammann H G. Prospective assessment of the impact of primary antimicrobial resistance on cure rates of Helicobacter pylori infection. Gut. 1997;41(Suppl. 1):A90. [Google Scholar]
- 16.Lambert T, Mégraud F, Gerbaud G, Courvalin P. Susceptibility of Campylobacter pyloridis to 20 antimicrobial agents. Antimicrob Agents Chemother. 1986;30:510–511. doi: 10.1128/aac.30.3.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lerang F, Moum B, Haug J B, Tolas P, Breder O, Aubert E, Hoie O, Soberg T, Flaaten B, Farup P, Berge T. Highly effective twice-daily triple therapies for H. pylori infection and peptic ulcer disease: does in vitro metronidazole resistance have any clinical relevance? Am J Gastroenterol. 1997;92:248–253. [PubMed] [Google Scholar]
- 18.Lind T, Mégraud F, Unge P, Bayerdörffer E, O’Morain C, Spiller R, van Zanten S V, Bardhan K D, Hellblom M, Wrangstadh M, Zeijlon L, Cederberg C. The MACH2 study: role of omeprazole in eradication of Helicobacter pylori with 1-week triple therapies. Gastroenterology. 1999;116:248–253. doi: 10.1016/s0016-5085(99)70119-8. [DOI] [PubMed] [Google Scholar]
- 19.Lind T, van Zanten S V, Unge P, Spiller R, Bayerdörffer E, O’Morain C, Bardhan K D, Bradette M, Chiba N, Wrangstadh M, Cederberg C, Idstrom J P. Eradication of Helicobacter pylori using one-week triple therapies combining omeprazole with two antimicrobials: the MACH1 study. Helicobacter. 1996;3:138–144. doi: 10.1111/j.1523-5378.1996.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 20.Marcus R, Peritz E, Gabriel K R. On closed testing procedures with special reference to ordered analysis of variance. Biometrika. 1976;63:655–660. [Google Scholar]
- 21.Mégraud F. Advantages and disadvantages of current diagnostic tests for the detection of Helicobacter pylori. Scand J Gastroenterol. 1996;31(Suppl. 215):57–62. doi: 10.3109/00365529609094536. [DOI] [PubMed] [Google Scholar]
- 22.Mégraud F. Diagnostic bactériologique standard de l’infection àHelicobacter pylori. In: Mégraud F, Lamouliatte H, editors. Helicobacter pylori. Vol. 1. Paris, France: Elsevier; 1996. pp. 249–276. [Google Scholar]
- 23.Mégraud F. How should Helicobacter pylori infection be diagnosed? Gastroenterology. 1997;113:S93–S98. doi: 10.1016/s0016-5085(97)80020-0. [DOI] [PubMed] [Google Scholar]
- 24.Mégraud F. Epidemiology and mechanism of antibiotic resistance in Helicobacter pylori. Gastroenterology. 1998;115:1278–1282. doi: 10.1016/s0016-5085(98)70101-5. [DOI] [PubMed] [Google Scholar]
- 25.Mégraud F, Bonnet F, Garnier M, Lamouliatte H. Characterization of “Campylobacter pyloridis” by culture, enzymatic profile, and protein content. J Clin Microbiol. 1985;22:1007–1010. doi: 10.1128/jcm.22.6.1007-1010.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mégraud F, Doermann H P. Clinical relevance of resistant strains of Helicobacter pylori: a review of current data. Gut. 1998;43(Suppl. 1):S61–S65. doi: 10.1136/gut.43.2008.s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Mégraud F, Lamouliatte H, Bouchard S, Birac C, Villeval F. Abstracts of the European Congress on Clinical Microbiology and Infectious Diseases. 1991. Evaluation of a transport medium: Portagerm pylori for Helicobacter pylori, abstr. 1040. [Google Scholar]
- 27.Misiewicz J J, Harris A W, Bardhan K D, Levi S, O’Morain C, Cooper B T, Kerr G D, Dixon M F, Langworthy H, Piper D. One week triple therapy for Helicobacter pylori: a multicentre comparative study. Gut. 1997;41:735–739. doi: 10.1136/gut.41.6.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moayyedi P, Tompkins D S, Ragunathan P L, Axon A T R. Prevalence of metronidazole in predicting failure of omeprazole, clarithromycin and tinidazole to eradicate H. pylori. Gut. 1996;39(Suppl. 2):A6. . (Abstract.) [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. VIth informational supplement. M100S9 19, 1. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 30.Occhialini A, Urdaci M, Doucet-Populaire F, Bébéar C M, Lamouliatte H, Mégraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piccolomini R, di Bonaventura G, Catamo G, Carbone F, Neri M. Comparative evaluation of the E test, agar dilution, and broth microdilution for testing susceptibilities of Helicobacter pylori strains to 20 antimicrobial agents. J Clin Microbiol. 1997;35:1842–1846. doi: 10.1128/jcm.35.7.1842-1846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith M A, Edwards D I. The influence of microaerophilia and anaerobiosis on metronidazole uptake in Helicobacter pylori. J Antimicrob Chemother. 1995;36:453–461. doi: 10.1093/jac/36.3.453. [DOI] [PubMed] [Google Scholar]
- 33.Tee W, Fairley S, Smallwood R, Dwyer B. Comparative evaluation of three selective media and a nonselective medium for the culture of Helicobacter pylori from gastric biopsies. J Clin Microbiol. 1991;29:2587–2589. doi: 10.1128/jcm.29.11.2587-2589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utrup L J, Flamm R, Osato M, Ferraro M J, Reller L B, Barry A, Bush K, Silliman N. Program and abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C.: American Society for Microbiology; 1998. Susceptibility testing standardization and quality control ranges for Helicobacter pylori, abstr. C-31. [Google Scholar]
- 35.van Zwet A A, Vanderbroucke-Grauls C M J E, ven der Wouden E J, Gerrits M M, Kusters J G. Stable amoxicillin resistance in Helicobacter pylori. Lancet. 1998;352:1595. doi: 10.1016/s0140-6736(98)00064-6. [DOI] [PubMed] [Google Scholar]
- 36.Veldhuyzen van Zanten S J, Pollak P T, Kapoor H, Yeung P K. Effect of omeprazole on movement of intravenously administered metronidazole into gastric juice and its significance in treatment of Helicobacter pylori. Dig Dis Sci. 1996;41:1845–1852. doi: 10.1007/BF02088756. [DOI] [PubMed] [Google Scholar]
- 37.Versalovic J, Shortridge D, Kliber K, Griffy V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westblom T U, Gudipâti S, Madan E, Midkiff B R. Improved growth of Helicobacter pylori using a liquid medium supplemented with human serum. Ital J Gastroenterol. 1991;23(Suppl. 2):48. doi: 10.1007/BF02111882. . (Abstract.) [DOI] [PubMed] [Google Scholar]
- 39.Wurzer H, Rodrigo L, Stamler D, Archambault A, Rokkas T, Skandalis N, Fedorak R, Bazzoli F, Hentschel E, Mora P, Archimandritis A, Mégraud F. Short-course therapy with amoxicillin-clarithromycin triple therapy for 10 days (ACT-10) eradicates Helicobacter pylori and heals duodenal ulcer. ACT-10 Study Group Aliment. Pharmacol Ther. 1997;11:943–952. doi: 10.1046/j.1365-2036.1997.00223.x. [DOI] [PubMed] [Google Scholar]