Abstract
Our previous studies have shown that cholesterol-conjugated, peptide-based pan-coronavirus (CoV) fusion inhibitors can potently inhibit human CoV infection. However, only palmitic acid (C16)-based lipopeptide drugs have been tested clinically, suggesting that the development of C16-based lipopeptide drugs is feasible. Here, we designed and synthesized a C16-modified pan-CoV fusion inhibitor, EK1-C16, and found that it potently inhibited infection by SARS-CoV-2 and its variants of concern (VOCs), including Omicron, and other human CoVs and bat SARS-related CoVs (SARSr-CoVs). These results suggest that EK1-C16 could be further developed for clinical use to prevent and treat infection by the currently circulating MERS-CoV, SARS-CoV-2 and its VOCs, as well as any future emerging or re-emerging coronaviruses.
Keywords: palmitic acid, lipopeptide, Omicron, β-coronavirus, entry inhibitor
1. Introduction
Coronaviruses (CoVs) comprise a group of RNA viruses that can cause human or animal infection. Seven coronaviruses can infect humans, thus being named human CoVs (HCoVs), including severe acute respiratory syndrome coronavirus (SARS-CoV), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Middle East respiratory syndrome coronavirus (MERS-CoV), HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1 [1]. Five belong to the β-CoV genus (i.e., sarbecoviruses), including SARS-CoV, SARS-CoV-2, MERS-CoV, HCoV-OC43, and HCoV-HKU1. Some SARSr-CoVs from bat (e.g., WIV1, Rs3367, and RsSHC014) also belong to the β-CoV genus.
SARS-CoV, SARS-CoV-2, and MERS-CoV, which belong to the group of highly pathogenic CoVs, possess high infectivity and transmissibility and can cause serious disease after infecting humans [2,3]. Since the end of 2019, when coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 was first reported, SARS-CoV-2 and its variants have infected about 3.6 billion individuals and caused more than 5.6 million deaths worldwide (https://covid19.who.int (accessed on 10 January 2022)). SARS-CoV-2 variants of concern (VOC), including Alpha, Beta, Gamma, Delta, and Omicron, have seriously compromised the clinical efficacy of many vaccines and antibody therapies [4,5,6], making it more difficult to control the COVID-19 pandemic.
In addition, low-pathogenic HCoVs in the β-CoV genus (HCoV-OC43 and HCoV-HKU1) usually cause the common cold in humans (mild upper respiratory tract infections) [7], but sometimes pneumonia in children, the elderly, or immunocompromised adults [8], calling for the development of more effective and broad-spectrum antivirals against both high- and low-pathogenic HCoVs [9,10,11].
An HCoV infects the host target cell through either a cytoplasmic or endosomal membrane fusion pathway. Each of these fusion processes occurs after the interaction of the receptor-binding domain (RBD) in the viral spike protein and cellular receptor, and proteolysis of spike protein mediated by transmembrane protease serine 2 (TMPRSS-2) on the cell surface or cathepsin L in the endosome. RBD-receptor interaction can be by RBD-specific antibodies and some mini-protein inhibitors [12], while the proteolytic function of TMPRSS-2 or cathepsin L can be inhibited by TMPRSS-2 inhibitors (e.g., camostat and nafamostat) or cathepsin L inhibitors (e.g., K11777), respectively [13].
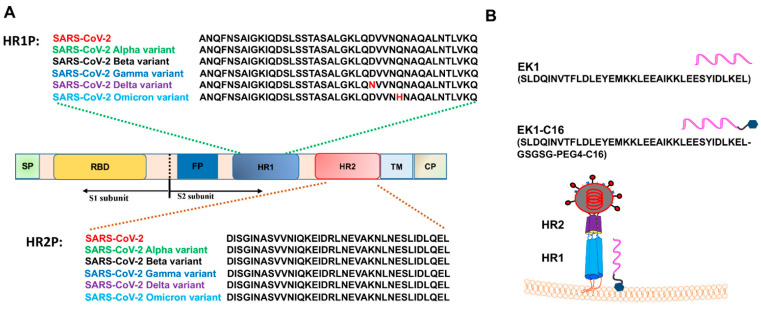
The six-helix bundle (6-HB) fusion core structure formed by HR1 and HR2 domains of SARS-CoV-2 is key for mediating membrane fusion. Previous studies confirmed its stability (Figure 1A), suggesting that the 6-HB fusion core is an important target for the development of pan-CoV fusion inhibitors against SARS-CoV-2 and its variants [14,15]. We and others demonstrated that peptides derived from the HR2 domain of SARS-CoV-2, such as 2019-nCoV-HR2P, IPB01, and SARS-CoV-2-HRC, could potently inhibit SARS-CoV-2 infection by interacting with the HR1 domain of SARS-CoV-2 S protein to block the formation of 6-HB fusion core between viral HR1 and HR2 domains [14,15,16]. In particular, our previously developed pan-CoV fusion inhibitor EK1 is effective against infection by SARS-CoV-2 D614G and its VOCs [17,18,19]. Later, we found that cholesterol- and 25-hydroxycholesterol-conjugated EK1 peptides, such as EK1C4 [17], EKL1C [20], and EK1P4HC [21], exhibited much improved antiviral activity against SARS-CoV-2, its VOCs, and other HCoVs, including SARS-CoV, MERS-CoV, HCoV-229E, HCoV-NL63, and HCoV-OC43, as well as bat SARSr-CoV WIV1, SARSr-CoV Rs3367, and SARSr-CoV SHC014. However, we note that no cholesterol-conjugated peptide drug is currently in clinical use, indicating the difficulty of developing clinically applicable cholesterol-based lipopeptide drugs. Interestingly, however, several palmitic acid-based lipopeptide drugs have been studied in clinical trials [22,23], suggesting the feasibility of their development.
Figure 1.
The amino acid sequences of HR1 and HR2 core regions of SARS-CoV-2 and its VOC, and design of EK1-C16 lipopeptide. (A) The amino acid sequences of HR1 and HR2 core regions of SARS-CoV-2 and its VOCs, including Alpha, Beta, Gamma, Delta, and Omicron variants. (B) Design of EK1-C16 lipopeptide and putative mechanism of potent antiviral activity of EK1-C16 lipopeptide. The C16 group of EK1-C16 can bind tightly with the cellular membrane of target cells, promoting the membrane-bound EK1-C16 peptide entering the endosome to inhibit the viral entry into the cytoplasm for replication, while lipid-free peptides only inhibit cytoplasm membrane fusion [24,25].
Therefore, in this study, we designed and synthesized a palmitic acid (C16)-modified EK1 lipopeptide by adding a C16 group at the C-terminus of EK1 peptide, termed EK1-C16 (Figure 1B). We found that EK1-C16 could potently inhibit infection by SARS-CoV-2 wild-type strain (D614G) and its VOCs, including Alpha, Beta, Gamma, Delta, and Omicron, as well as other β-CoVs, including SARS-CoV, MERS-CoV, HCoV-OC43, and bat SARSr-CoV WIV1 and SARSr-CoV Rs3367. These results suggest that EK1-C16 is a potent lipopeptide-based pan-CoV fusion inhibitor with promise as an antiviral candidate with efficacy in preventing and treating infection by current circulating MERS-CoV and SARS-CoV-2 and its variants, as well as any future emerging or re-emerging coronaviruses.
2. Materials and Methods
2.1. Cell Lines, Plasmids, Peptides, and Viruses
HEK293T and Vero-E6 cell lines were obtained from the American Type Culture Collection (ATCC). Caco2, RD, and Huh-7 cell lines were obtained from the cell bank of the Chinese Academy of Science. All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS).
HIV-backbone plasmid (pNL4-3.Luc.R-E) and other plasmids coding the spike protein of coronaviruses (pAAV-IRES-GFP-SARS-CoV-2-D614G-spike, pAAV-IRES-GFP-MERS-CoV-spike, pcDNA3.1-SARS-CoV-2-spike, pcDNA3.1-SARS-CoV-2-B.1.1.7-spike, pcDNA3.1-SARS-CoV-2-P.1-spike, pcDNA3.1-SARS-CoV-2-B1.351-spike, pcDNA3.1-SARS-CoV-2-B1.617.2-spike, pcDNA3.1-SARS-CoV-2-B.1.1.529-spike, pcDNA3.1-SARS-CoV-spike, pcDNA3.1-MERS-CoV-spike, pcDNA3.1-Bat-CoV-WIV1-spike, pcDNA3.1-Bat-CoV-Rs3367-spike, and pcDNA3.1-VSV-G) were all preserved in our laboratory.
EK1-C16 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL-GSGSG-PEG4-C16) and EK1 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL) were synthesized by Chengdu Shengnuo Biotechnology Co., Ltd. Authentic SARS-CoV-2 WT strain (nCoV-SH01, GenBank number: MT121215.1) was isolated and preserved in the Biosafety Level 3 (BSL-3) Facility of Fudan University. Authentic SARS-CoV-2 Omicron (hCoV-19/Hong Kong/HKU-344/2021) was isolated from a patient and maintained in the Biosafety Level 3 (BSL-3) Facility of the University of Hong Kong (HKU). HCoV-OC43 (VR-1558) was obtained from ATCC.
2.2. Authentic SARS-CoV-2 WT Strain Inhibition
Wild-type SARS-CoV-2 live virus inhibition assay was performed in the BSL-3 Facility, Fudan University. Briefly, peptides were first incubated with SARS-CoV-2 (100 TCID50) for 30 min and then added into the Vero-E6 cell line seeded in a 96-wall plate. After 1 h incubation, the supernatants containing peptide and SARS-CoV-2 were changed for fresh DMEM containing 5% FBS. After 48 h culture, Vero-E6 cells infected with SARS-CoV-2 were fixed with 4% paraformaldehyde, followed by 0.2% Triton X-100 treatment. Next, an immunofluorescence assay was performed to detect the nucleocapsid protein of SARS-CoV-2 in Vero-E6 cells [26]. The SARS-CoV-2 nucleocapsid antibody (1:200, Sino Biological, Beijing, China) was used as a primary antibody, the Alexa Fluor 488 goat anti-rabbit IgG (1:100, Thermo Fisher) was used as a secondary antibody, and DAPI (Thermo Fisher, Waltham, MA, USA) was used to stain the nucleus.
2.3. Authentic SARS-CoV-2 Omicron Variant Inhibition Assay
The inhibitory activity of peptides against SARS-CoV-2 isolate Omicron variant infection was assessed at HKU. Briefly, a diluted peptide was first incubated with 0.01 MOI Omicron variant (hCoV-19/Hong Kong/HKU-344/2021; GISAID accession number EPI_ISL_7357684) for 60 min. Next, this peptide–virus mixture was added into Vero-E6-TMPRSS2 cells which were seeded in a 96-well plate. After 72 h culture, CPE was observed and scored as 100% inhibition or 0% inhibition.
2.4. Package of Coronavirus Pseudovirus
Coronavirus PsVs were produced as previously reported. Briefly, HEK293T cells were seeded in a 6-well plate 24 h before transfection. Upon transfection, HIV backbone plasmid (pNL4-3.Luc.R-E) and spike-expressing plasmid, such as pcDNA3.1-SARS-CoV-2-spike, were co-transfected into HEK293T cells by Vigofect (Vigorous Biotechnology, Beijing, China). At 10 h post-transfection, cellular supernatants containing transfection reagent were changed for fresh DMEM containing 5% FBS. After another 36–48 h culture, cell supernatants containing PsV particles were collected and stored at −80 °C.
2.5. Coronavirus Pseudovirus Inhibition Assay
The inhibitory activity of peptides against pseudovirus infection was assessed as previously reported [18]. In brief, a serially diluted peptide was first incubated with pseudovirus for 30 min, and then this peptide–pseudovirus mixture was added into Caco2 cells seeded in a 96-well plate. After a 12 h culture, culture supernatants were discarded, and fresh DMEM was added. After another 36 h culture, luciferase assay (Promega, Madison, WI, USA) was performed to measure luciferase activity according to the manufacturer’s instructions. Inhibition curves were produced with GraphPad Prism 8 software, and IC50 values were calculated.
2.6. Authentic HCoV-OC43 Inhibition Assay
The inhibitory activity of peptides against authentic HCoV-OC43 infection was measured as previously reported [17]. A diluted peptide was first incubated with HCoV-OC43 (100 TCID50) for 30 min, and the peptide–virus mixture was added to the RD cell line seeded in a 96-well plate. The CCK-8 assay was used to assess cell viability by observing CPE of HCoV-OC43, and an inhibition curve was produced by GraphPad Prism 8 software.
2.7. Cell–Cell Fusion Inhibition Assay
A cell–cell fusion inhibition assay was performed as previously reported [17]. Briefly, HEK293T cells were transfected with plasmid pAAV-IRES-GFP-SARS-CoV-2-spike (or pAAV-IRES-GFP-MERS-CoV-spike, pAAV-IRES-GFP-HCoV-OC43-spike) to obtain 293T cells expressing GFP and SARS-CoV-2 spike protein (or MERS-CoV spike protein, HCoV-OC43-spike). A diluted peptide was then incubated with these transfected HEK293T cells for 30 min and added into target cells seeded in a 96-well plate. Two hours later, fusion status was observed using fluorescence microscopy.
2.8. Cytotoxicity Assay
The cytotoxicity of peptides was assessed as previously reported [27]. Briefly, a diluted peptide was co-incubated with RD cells seeded in a 96-well plate for 12 h. Next, the culture medium containing peptides was replaced with fresh DMEM. After another 36 h culture, the CCK-8 assay was used to assess cell viability.
2.9. Statistical Analysis
The inhibition curves and IC50 values of peptide inhibitors were all produced by GraphPad Prism 8 software.
3. Results
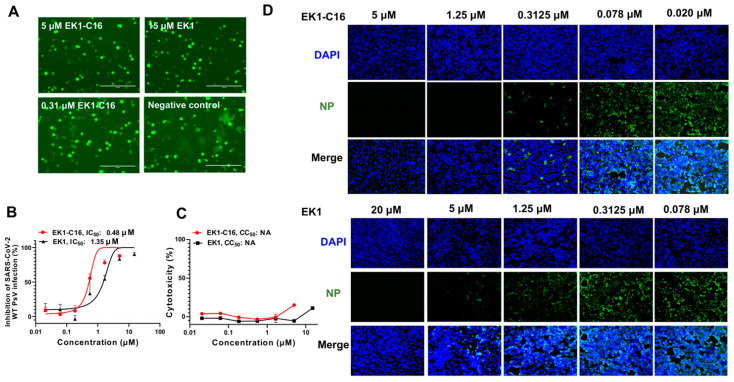
3.1. EK1-C16 Potently Inhibited Infection of SARS-CoV-2 Wild-Type (WT) Strain
After designing and synthesizing the EK1-C16 lipopeptide, we first tested its inhibitory activity against SARS-CoV-2 D614G S-mediated cell–cell fusion and infection of the pseudotyped SARS-CoV-2 WT strain (Wuhan-Hu-1). We found that EK1-C16 at high (5.0 μM) and low (0.31 μM) concentrations could suppress SARS-CoV-2 S-mediated cell–cell fusion (Figure 2A). It also effectively inhibited SARS-CoV-2 WT pseudovirus (PsV) infection in Caco2 cells in a dose-dependent manner with an IC50 (half maximal inhibitory concentration) of 0.48 μM (Figure 2B). We then assessed the potential cytotoxicity of EK1-C16 using the CCK-8 assay. At the concentration of 5 μM, it exhibited no significant cytotoxicity (Figure 2C). Next, we used an authentic SARS-CoV-2 inhibition assay to determine the inhibitory activity of EK1-C16 against infection of authentic SARS-CoV-2 WT strain (nCoV-SH01, GenBank number: MT121215.1); an immunofluorescence assay was used to detect SARS-CoV-2 N protein expression. As shown in Figure 2D, EK1-C16 at 0.31 μM could effectively inhibit authentic SARS-CoV-2 WT infection. While EK1 at 0.31 μM showed no significant inhibitory activity, it did inhibit authentic SARS-CoV-2 infection at 5.0 μM, suggesting that EK1-C16 is more effective than EK1 in inhibiting authentic SARS-CoV-2 infection.
Figure 2.
EK1-C16-mediated inhibition of SARS-CoV-2 infection. (A) EK1-C16-mediated inhibition of SARS-CoV-2 D614G S-meditated cell–cell fusion. (B) EK1-C16-mediated inhibition of SARS-CoV-2 WT (Wuhan-Hu-1) PsV infection. (C) Cytotoxicity of EK1-C16 to RD cells was tested by using a CCK-8 assay. (D) EK1-C16-mediated inhibition of authentic SARS-CoV-2 WT (nCoV-SH01) infection. Samples were tested in triplicate, and the experiment was repeated at least twice.
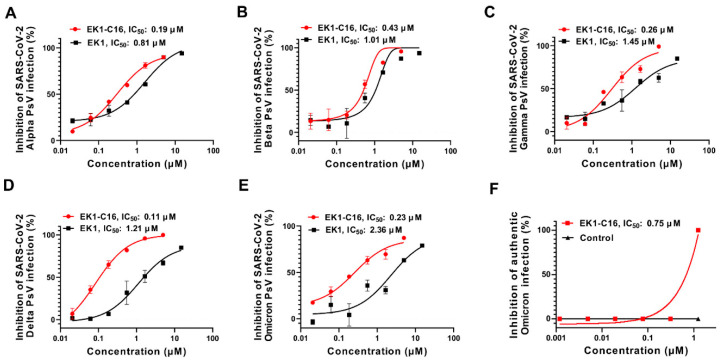
3.2. EK1-C16 Inhibited Infection of SARS-CoV-2 VOCs, Including Omicron
SARS-CoV-2 variants are constantly emerging. Some show increased infectivity and transmissibility, as well as reduced sensitivity to neutralization of therapeutic antibodies and vaccine-elicited sera. Here, we assessed the inhibitory activity of EK1-C16 against these SARS-CoV-2 VOCs. As shown in Figure 3A–E, EK1-C16 could effectively inhibit infection by pseudotyped SARS-CoV-2 VOC Alpha, Beta, Gamma, Delta, and Omicron with IC50 values of 0.19, 0.43, 0.26, 0.11, and 0.23 μM, respectively, which are about 3- to 10-fold more potent than that of SARS-CoV-2 WT. We further determined the inhibitory activity of EK1-C16 against infection of the authentic Omicron variant in Vero-E6-TMPRSS-2 cells by detecting the cytopathic effect (CPE) at 72 h post-infection. We found that EK1-C16 could effectively inhibit authentic Omicron infection with an IC50 value of 0.75 μM (Figure 3F).
Figure 3.
EK1-C16-mediated inhibition of infection by SARS-CoV-2 VOCs. EK1-C16-mediated inhibition of infection by pseudotyped SARS-CoV-2 Alpha (A), Beta (B), Gamma (C), Delta (D), and Omicron (E), and by authentic Omicron variant (F). The samples were tested in triplicate, and the experiment was repeated at least twice.
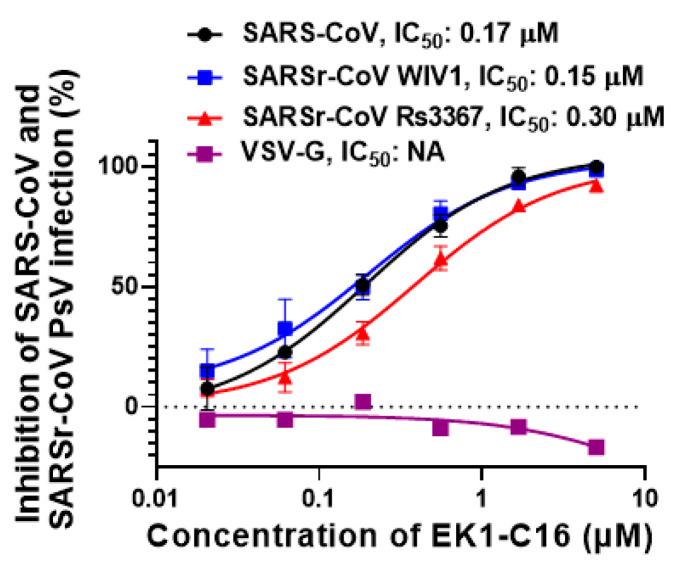
3.3. EK1-C16 Broadly Inhibited Infection by Other Sarbecoviruses
SARS-CoV has the potential to re-emerge in the future, while bat SARSr-CoVs may cause emerging SARS-like infectious diseases in the future [28]. To prepare for these emerging or re-emerging coronavirus infectious diseases, it is essential to develop broad-spectrum antivirals. Here, we assessed the inhibitory activity of EK1-C16 against infection by pseudotyped SARS-CoV and bat SARSr-CoVs. We found that EK1-C16 could potently inhibit SARS-CoV PsV infection with an IC50 of 0.17 μM and bat SARSr-CoV WIV1 and Rs3367 infection with IC50 of 0.15 and 0.3 μM, respectively (Figure 4). In contrast, EK1-C16 exhibited no significant inhibitory activity against VSV-G PsV infection at a concentration as high as 5.0 μM (Figure 4), suggesting that the antiviral activity of EK1-C16 is specific for coronaviruses.
Figure 4.
EK1-C16 can broadly inhibit sarbecoviruses. EK1-C16 can inhibit SARS-CoV, WIV1, and Rs3367 PsV, but it has no inhibitory activity against VSV-G PsV-mediated infection, indicating its specificity for sarbecoviruses. Each peptide inhibitor was tested in duplicate, and experiments were repeated twice.
3.4. EK1-C16 Inhibited MERS-CoV Infection
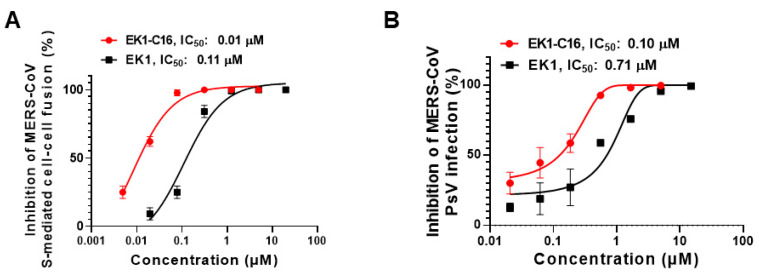
Another highly pathogenic HCoV in human circulation is MERS-CoV. Although its infectivity and transmissibility are much lower compared to SARS-CoV-2, its case-fatality rate is as high as 34% [29]. Therefore, it is also essential to develop antivirals against MERS-CoV infection. Here, we first assessed the inhibitory activity of EK1-C16 against MERS-CoV S-mediated membrane fusion. We found that it could potently inhibit MERS-CoV S-mediated cell–cell fusion with an IC50 of 0.012 μM (Figure 5A), indicating nearly 10-fold more efficacy than that of EK1 peptide. Next, we evaluated the inhibitory activity of EK1-C16 against MERS-CoV PsV infection in Caco2 cells and found that it inhibited MERS-CoV PsV infection with an IC50 of 0.10 μM, about sixfold more potent than that of EK1 (Figure 5B). These results suggest that EK1-C16 could be further developed as a candidate antiviral for the prevention and treatment of MERS-CoV infection.
Figure 5.
EK1-C16 can broadly inhibit MERS-CoV infection. (A) Inhibitory activity of EK1-C16 against MERS-CoV spike protein-mediated cell–cell fusion. (B) Inhibitory activity of EK1-C16 against MERS-CoV PsV. Samples were tested in triplicate, and the experiment was repeated twice.
3.5. EK1-C16 Inhibited HCoV-OC43 Infection
Apart from the above highly pathogenic HCoVs, some HCoVs with low pathogenicity, such as HCoV-OC43, continue to circulate widely in humans during the winter months and cause upper and respiratory tract illness and common cold-like symptoms [7,8]. HCoV-OC43 infection may also be associated with acute exacerbation of chronic obstructive pulmonary disease (AECOPD) and pneumonia in all age groups with immunocompromised conditions [8]. Therefore, it is also important to develop antivirals against HCoVs showing low pathogenicity [9,10]. Accordingly, in this study, we first measured the inhibitory activity of EK1-C16 against HCoV-OC43 S-mediated cell–cell fusion and found that EK1-C16 can potently inhibit HCoV-OC43 S-mediated cell–cell fusion with an IC50 value of 0.01 μM, which is about a 28-fold improvement compared to EK1 (Figure 6A). Next, we measured the inhibitory activity of EK1-C16 on the authentic HCoV-OC43 infection in RD cells. As shown in Figure 6B, EK1-C16 exhibited highly effective in inhibiting HCoV-OC43 infection with IC50 of 0.07 μM, about 22-fold more potent than that of EK1, indicating that EK1-C16, if well-developed, can also be used as a prophylactic or therapeutic against low pathogenic HCoV infection.
Figure 6.
EK1-C16 can broadly inhibit HCoV-OC43 infections. (A) Inhibitory activity of EK1-C16 against HCoV-OC43 S-mediated cell–cell fusion. (B) Inhibitory activity of EK1-C16 against authentic HCoV-OC43 infection. The samples were tested in triplicate and the experiment was repeated once.
4. Discussion
The outbreak of COVID-19 sparked the development of a broad spectrum of antivirals, including therapeutic monoclonal antibodies, protein-, peptide- and small-molecule compound-based inhibitors, against SARS-CoV-2 infection [12,30,31]. However, the newly emerged SARS-CoV-2 VOCs, such as Omicron, have shown increasing resistance to some developed antiviral treatments and, even more concerning, SARS-CoV-2 RBD-specific neutralizing antibodies and vaccines being used worldwide [6,32,33,34,35]. The presence of SARSr-CoVs in bats may cause future outbreaks of SARS-like infectious diseases [28]. Thus, the growing list of SARS-CoV-2 VOCs and other emerging sarbecoviruses calls for the urgent development of antivirals with broad applicability and improved anti-coronavirus activity. It should be noted that MERS-CoV is still circulating in the Middle East region [36]. Several cases of SARS-CoV-2 and MERS-CoV co-infection were identified in Saudi Arabia [37], and both SARS-CoV-2 and MERS-CoV could infect type-II alveolar cells [38]. Furthermore, co-infection of immunocompromised individuals—for instance, by SARS-CoV-2 Omicron or MERS-CoV—could lead to a new species through genetic recombination [29]. Such an event could potentially increase the transmissibility of the current Omicron variant and reduce, even further, the sensitivity to SARS-CoV-2 neutralizing antibodies, while gaining a higher case-fatality rate (CF) of MERS-CoV. Such a scenario would spell disaster in countries with a low COVID-19 vaccination rate. Therefore, it is essential to develop highly effective pan-CoV therapeutics or prophylactics [10].
Our previous studies have shown that the HR1 domain is an important target for the development of potent and broad-spectrum HCoV fusion inhibitors [27,39]. We found that EK1 peptide targeting the HR1 domain of divergent HCoVs could broadly and effectively inhibit infection of all HCoVs and bat SARSr-CoVs tested [27]. Our cholesterol-conjugated EK1 lipopeptide, EK1C4, showed significant improvement in its inhibitory activity against SARS-CoV-2 and its VOCs, including Omicron [17,19]. However, while no cholesterol-based lipopeptides are currently in clinical use, some C16-based lipopeptide drugs are in clinical trials, showing the practicality of developing a C16-conjugated lipopeptide drug, as we have herein reported.
Specifically, our C16-conjugated, lipopeptide-based pan-CoV fusion inhibitor, EK1-C16, effectively inhibited infection by SARS-CoV-2 WT and its VOCs, including Omicron, with the highest transmissibility and lowest sensitivity to SARS-CoV-2 neutralizing antibodies. EK1-C16 lipopeptide is also highly effective against infection by SARS-CoV and bat SARSr-CoVs, MERS-CoV, and HCoV-OC43. Similarly, some small-molecule antivirals targeting the conserved region of other viral proteins of SARS-CoV-2, such as remdesivir and molnupiravir targeting viral RdRp and nirmatrelvir targeting Mpro, also exhibit broad-spectrum anti-HCoV activity and can potently inhibit infection from the Omicron variant [35,40]. Combinations of EK1-based peptides with these inhibitors targeting the conserved regions of other viral proteins are expected to have synergistic antiviral activity against infection of SARS-CoV-2 variants and other HCoVs.
Taken collectively, these results suggest that EK1-C16 is a highly promising candidate for development as a potent and broad-spectrum anti-HCoV drug for the prevention and treatment of infection by current and future SARS-CoV variants, as well as emerging and re-emerging coronaviruses.
Author Contributions
Q.W., S.J., K.-Y.Y. and L.L. conceived, planned, and supervised the experiments; Q.L., J.F.-W.C., W.X., L.W., F.J., J.Z., J.P., G.Z. and S.X. performed the experiments; Q.L., W.X. and S.X. analyzed the data and wrote the draft, while Q.W., S.J. and K.-Y.Y. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study is supported by the National Natural Science Foundation of China (82161138002 and 92169112 to S.J.; 82002142 to S.X.); National Key Research and Development Program of China (2021YFC2300703 to L.L.); Program of Shanghai Academic/Technology Research Leader (20XD1420300 to L.L.); the Health and Medical Research Fund (COVID1903010–Project 7 to J.F.-W.C.), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region; Health@InnoHK, Innovation and Technology Commission, the Government of the Hong Kong Special Administrative Region (to K.-Y.Y.); the National Program on Key Research Project of China (2020YFA0707500 and 2020YFA0707504 to J.F.-W.C.); and Sanming Project of Medicine in Shenzhen, China (SZSM201911014 to K.-Y.Y.). The funding sources had no role in the study design, data collection, analysis, interpretation, or writing of the report.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data of this paper are available on request from the corresponding author.
Conflicts of Interest
S.J., L.L., S.X., and Q.W. are the inventors in the patent or patent application covering the peptide EK1 or EK1-C16. J.F.-W.C. has received travel grants from Pfizer Corporation Hong Kong and Astellas Pharma Hong Kong Corporation Limited and was an invited speaker for Gilead Sciences Hong Kong Limited and Luminex Corporation. The other authors declare no conflicts of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chen B., Tian E.K., He B., Tian L., Han R., Wang S., Xiang Q., Zhang S., El Arnaout T., Cheng W. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020;5:89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 6.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 7.Jiang S., Du L. Effect of Low-Pathogenic Human Coronavirus-Specific Antibodies on SARS-CoV-2. Trends Immunol. 2020;41:853–854. doi: 10.1016/j.it.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh E.E., Shin J.H., Falsey A.R. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J. Infect. Dis. 2013;208:1634–1642. doi: 10.1093/infdis/jit393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao M., Su X., Jiang S. Broad-Spectrum Anti-coronavirus Vaccines and Therapeutics to Combat the Current COVID-19 Pandemic and Future Coronavirus Disease Outbreaks. Stem Cell Rep. 2021;16:398–411. doi: 10.1016/j.stemcr.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L., Su S., Yang H., Jiang S. Antivirals with common targets against highly pathogenic viruses. Cell. 2021;184:1604–1620. doi: 10.1016/j.cell.2021.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Xia S., Zhu Y., Lu L., Jiang S. Pan-coronavirus fusion inhibitors as the hope for today and tomorrow. Protein Cell. 2021;12:84–88. doi: 10.1007/s13238-020-00806-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu F., Xiang R., Deng X., Wang L., Yu Z., Tian S., Liang R., Li Y., Ying T., Jiang S. Receptor-binding domain-specific human neutralizing monoclonal antibodies against SARS-CoV and SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:212. doi: 10.1038/s41392-020-00318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson C.B., Farzan M., Chen B., Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022;23:3–20. doi: 10.1038/s41580-021-00418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y., Yu D., Yan H., Chong H., He Y. Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J. Virol. 2020;94:e00635-20. doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Outlaw V.K., Bovier F.T., Mears M.C., Cajimat M.N., Zhu Y., Lin M.J., Addetia A., Lieberman N.A.P., Peddu V., Xie X., et al. Inhibition of Coronavirus Entry In Vitro and Ex Vivo by a Lipid-Conjugated Peptide Derived from the SARS-CoV-2 Spike Glycoprotein HRC Domain. mBio. 2020;11:e01935-20. doi: 10.1128/mBio.01935-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia S., Lan Q., Zhu Y., Wang C., Xu W., Li Y., Wang L., Jiao F., Zhou J., Hua C., et al. Structural and functional basis for pan-CoV fusion inhibitors against SARS-CoV-2 and its variants with preclinical evaluation. Signal Transduct. Target. Ther. 2021;6:288. doi: 10.1038/s41392-021-00712-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia S., Chan J.F., Wang L., Jiao F., Chik K.K., Chu H., Lan Q., Xu W., Wang Q., Wang C., et al. Peptide-based pan-CoV fusion inhibitors maintain high potency against SARS-CoV-2 Omicron variant. Cell Res. 2022 doi: 10.1038/s41422-022-00617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., Xu W., Liu Z., Wang C., Xia S., Lan Q., Cai Y., Su S., Pu J., Xing L., et al. A highly potent and stable pan-coronavirus fusion inhibitor as a candidate prophylactic and therapeutic for COVID-19 and other coronavirus diseases. Acta Pharm. Sin. B. 2021 doi: 10.1016/j.apsb.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan Q., Wang C., Zhou J., Wang L., Jiao F., Zhang Y., Cai Y., Lu L., Xia S., Jiang S. 25-Hydroxycholesterol-Conjugated EK1 Peptide with Potent and Broad-Spectrum Inhibitory Activity against SARS-CoV-2, Its Variants of Concern, and Other Human Coronaviruses. Int. J. Mol. Sci. 2021;22:11869. doi: 10.3390/ijms222111869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmon-Céron D., Durier C., Desaint C., Cuzin L., Surenaud M., Hamouda N.B., Lelièvre J.D., Bonnet B., Pialoux G., Poizot-Martin I., et al. Immunogenicity and safety of an HIV-1 lipopeptide vaccine in healthy adults: A phase 2 placebo-controlled ANRS trial. Aids. 2010;24:2211–2223. doi: 10.1097/QAD.0b013e32833ce566. [DOI] [PubMed] [Google Scholar]
- 23.Lévy Y., Lacabaratz C., Lhomme E., Wiedemann A., Bauduin C., Fenwick C., Foucat E., Surenaud M., Guillaumat L., Boilet V., et al. A Randomized Placebo-Controlled Efficacy Study of a Prime Boost Therapeutic Vaccination Strategy in HIV-1-Infected Individuals: VRI02 ANRS 149 LIGHT Phase II Trial. J. Virol. 2021;95:e02165-20. doi: 10.1128/JVI.02165-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J.E., Gallagher T. Lipidation increases antiviral activities of coronavirus fusion-inhibiting peptides. Virology. 2017;511:9–18. doi: 10.1016/j.virol.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K.K., Pessi A., Gui L., Santoprete A., Talekar A., Moscona A., Porotto M. Capturing a fusion intermediate of influenza hemagglutinin with a cholesterol-conjugated peptide, a new antiviral strategy for influenza virus. J. Biol. Chem. 2011;286:42141–42149. doi: 10.1074/jbc.M111.254243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Y., Xu W., Gu C., Cai X., Qu D., Lu L., Xie Y., Jiang S. Griffithsin with A Broad-Spectrum Antiviral Activity by Binding Glycans in Viral Glycoprotein Exhibits Strong Synergistic Effect in Combination with A Pan-Coronavirus Fusion Inhibitor Targeting SARS-CoV-2 Spike S2 Subunit. Virol. Sin. 2020;35:857–860. doi: 10.1007/s12250-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.K., Wang Q., Du L., Tan W., Wilson I.A., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su S., Li W., Jiang S. Developing pan-beta-coronavirus vaccines against emerging SARS-CoV-2 variants of concern. Trends Immunol. 2022;43:170–172. doi: 10.1016/j.it.2022.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang S., Hillyer C., Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang R., Yu Z., Wang Y., Wang L., Huo S., Li Y., Liang R., Hao Q., Ying T., Gao Y., et al. Recent advances in developing small-molecule inhibitors against SARS-CoV-2. Acta Pharm. Sin. B. 2021 doi: 10.1016/j.apsb.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium W.H.O.S.T., Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., Kieny M.P., et al. Repurposed Antiviral Drugs for Covid-19—Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., Iwatsuki-Horimoto K., Chiba S., Halfmann P., Nagai H., et al. Efficacy of Antibodies and Antiviral Drugs against Covid-19 Omicron Variant. N. Engl. J. Med. 2022 doi: 10.1056/NEJMc2119407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan A., Nafisah S.B., Mzahim B., Aleid B., Almatrafi D., Assiri A., Jokhdar H. MERS-CoV in the COVID-19 era: Update from Saudi Arabia, 2019-2020. East. Mediterr. Health J. 2021;27:1109–1113. doi: 10.26719/emhj.21.049. [DOI] [PubMed] [Google Scholar]
- 37.Elhazmi A., Al-Tawfiq J.A., Sallam H., Al-Omari A., Alhumaid S., Mady A., Al Mutair A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Middle East Respiratory Syndrome Coronavirus (MERS-CoV) coinfection: A unique case series. Travel Med. Infect. Dis. 2021;41:102026. doi: 10.1016/j.tmaid.2021.102026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sajini A.A., Alkayyal A.A., Mubaraki F.A. The Recombination Potential between SARS-CoV-2 and MERS-CoV from Cross-Species Spill-over Infections. J. Epidemiol. Glob. Health. 2021;11:155–159. doi: 10.2991/jegh.k.201105.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—A target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vangeel L., Chiu W., De Jonghe S., Maes P., Slechten B., Raymenants J., Andre E., Leyssen P., Neyts J., Jochmans D. Remdesivir, Molnupiravir and Nirmatrelvir remain active against SARS-CoV-2 Omicron and other variants of concern. Antiviral Res. 2022;198:105252. doi: 10.1016/j.antiviral.2022.105252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data of this paper are available on request from the corresponding author.