Abstract
The resistance mechanisms to azole antifungal agents were investigated in this study with two pairs of Candida glabrata clinical isolates recovered from two separate AIDS patients. The two pairs each contained a fluconazole-susceptible isolate and a fluconazole-resistant isolate, the latter with cross-resistance to itraconazole and ketoconazole. Since the accumulation of fluconazole and of another unrelated substance, rhodamine 6G, was reduced in the azole-resistant isolates, enhanced drug efflux was considered as a possible resistance mechanism. The expression of multidrug efflux transporter genes was therefore examined in the azole-susceptible and azole-resistant yeast isolates. For this purpose, C. glabrata genes conferring resistance to azole antifungals were cloned in a Saccharomyces cerevisiae strain in which the ATP binding cassette (ABC) transporter gene PDR5 was deleted. Three different genes were recovered, and among them, only C. glabrata CDR1 (CgCDR1), a gene similar to the Candida albicans ABC transporter CDR genes, was upregulated by a factor of 5 to 8 in the azole-resistant isolates. A correlation between upregulation of this gene and azole resistance was thus established. The deletion of CgCDR1 in an azole-resistant C. glabrata clinical isolate rendered the resulting mutant (DSY1041) susceptible to azole derivatives as the azole-susceptible clinical parent, thus providing genetic evidence that a specific mechanism was involved in the azole resistance of a clinical isolate. When CgCDR1 obtained from an azole-susceptible isolate was reintroduced with the help of a centromeric vector in DSY1041, azole resistance was restored and thus suggested that a trans-acting mutation(s) could be made responsible for the increased expression of this ABC transporter gene in the azole-resistant strain. This study demonstrates for the first time the determinant role of an ABC transporter gene in the acquisition of resistance to azole antifungals by C. glabrata clinical isolates.
Patients with advanced human immunodeficiency virus infection develop opportunistic infections due to the decrease in their immunity. Oropharyngeal candidiasis (OPC) caused by Candida albicans is a very common opportunistic infection in these patients and is treated mainly with azole antifungal agents, particularly with fluconazole. Treatment failures have been observed following the repeated use of this agent in relapses of OPC (21, 37, 45, 55). Different laboratories have reported that C. albicans isolates sampled sequentially during fluconazole treatment showed decreased susceptibility to fluconazole compared to that of the isolates sampled at the time of the first episode of infection (6, 30, 41, 53). Clinical resistance to fluconazole has been correlated with in vitro resistance of the yeasts recovered from patients undergoing antifungal therapy (54). This phenomenon has been also documented in other yeast species, including Candida glabrata (4, 16), Candida tropicalis (28), and Candida krusei (57, 58), and in Cryptococcus neoformans (27).
The increasing number of azole-resistant isolates recovered in many institutions during the past decade has motivated studies with the aim of understanding their mechanisms of resistance at the molecular level. Until now, C. albicans isolates have provided a major source for the discovery of mechanisms of azole resistance. Recent findings have shown that increased azole efflux is an important mechanism of resistance in yeast clinical isolates. In azole-resistant C. albicans isolates, increased azole efflux has been correlated with the upregulation of multidrug efflux transporter genes from two distinct families, the ATP binding cassette (ABC) transporters (CDR1 and CDR2) and the major facilitators (C. albicans MDR1) (3, 27, 30, 50, 53, 60). Deletion of CDR1 in C. albicans leads to azole hypersusceptibility and increased fluconazole accumulation (51). Decrease in azole affinity of the target enzyme of these antifungals, i.e., the cytochrome P450 lanosterol demethylase (called CYP51A1, or ERG11), has also been explained at the molecular level. Mutations in the genes encoding CYP51A1 (CYP51A1) have been detected in azole-resistant yeasts. These mutations resulted, in some cases, in amino acid substitutions with the probable effect of altering the binding properties of azoles and thus contributing to a decrease in azole susceptibility in clinical yeast isolates (49). Another mechanism of azole resistance originates from alterations in the ergosterol biosynthesis pathway, often resulting in the absence of ergosterol. This feature renders cells affected by this mechanism cross-resistant to amphotericin B. A few C. albicans clinical isolates possess this property and have been found to accumulate 14α-methylergosta-8,24(28)-dien-3β,6α-diol, which is indicative of a defect in Δ5,6 sterol desaturase (24, 35). The above-mentioned resistance mechanisms can combine with each other in C. albicans and complicate the analysis of such isolates, since it is difficult to establish the role of an individual mechanism in the decrease in azole susceptibility (48). Dissection of resistance mechanisms by the use of genetics in C. albicans clinical isolates resistant to azole antifungal agents has not been reported yet. This lack of important information arises from the difficulties of developing reliable genetic systems for this diploid organism.
Historically, C. albicans accounted for 70 to 80% of organisms isolated in patients infected by fungal species. However, recent data report a population shift toward non-C. albicans yeast species, such as C. glabrata, C. tropicalis, or C. krusei (15). Among the non-C. albicans species, C. glabrata has emerged as an important nosocomial pathogen. Berrouane et al. (7) reported that among Candida species, the proportion of C. glabrata infections in the Iowa University Hospitals from 1988 to 1994 increased significantly, while it remained unchanged for other yeast species and even decreased for C. albicans. Other investigators have noted similar increases in the frequency of infections caused by C. glabrata, mostly in conjunction with the use of azoles (34, 39, 40). We also observed that C. glabrata was often recovered from cultures originating from AIDS patients with OPC. C. glabrata is known to be less susceptible to fluconazole than most of the C. albicans fluconazole-susceptible isolates. Rex et al. (43) reported that the minimal inhibitory concentration inhibiting 50% of the yeast population investigated (MIC50) of fluconazole for 31 C. glabrata isolates was 16 μg/ml, while the MIC50 was 0.25 μg/ml for 129 C. albicans isolates. In several patients who responded poorly to fluconazole therapy, we noticed that C. glabrata isolates could persist and that their susceptibility to azoles was decreased. Since mechanisms of resistance to azoles have been less intensively investigated in non-C. albicans species such as C. glabrata, we addressed here the molecular basis of resistance in two pairs of isolates taken from two different AIDS patients with documented azole antifungal treatment failure. We first isolated C. glabrata azole resistance genes by complementation of hypersusceptibility of a Saccharomyces cerevisiae ABC transporter mutant. From the three different azole resistance genes isolated, only CgCDR1, which resembles the C. albicans ABC transporter CDR genes, was upregulated in the azole-resistant C. glabrata isolates from these two patients. By introducing a genetic marker in an azole-resistant clinical isolate, not only could evidence for the participation of this gene in azole resistance be obtained, but the nature of the mutation or mutations implicated in the upregulation of CgCDR1 could be predicted.
MATERIALS AND METHODS
Strains and media.
The yeast strains used in this study are listed in Table 1. They were grown at 30°C on yeast extract-peptone-dextrose (YEPD) complex medium containing 2% glucose, 1% Bacto peptone (Difco Laboratories, Detroit, Mich.), and 0.5% yeast extract (Difco). YEPD agar plates contained 2% agar (Difco) as a supplement. Yeast nitrogen base (YNB [Difco]) with 2% glucose and 2% agar (Difco) with appropriate amino acids and bases was used as a selective medium after transformation of S. cerevisiae YKKB-13 and C. glabrata. Agar plates containing 50 μg of 5-fluoroorotic acid (5-FOA) per ml were made for the introduction of the ura3 genetic marker in YNB selective medium with 50 μg of uridine per ml. Escherichia coli DH5α (19) was used as a host for plasmid constructions and propagation and was grown on standard media.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| S. cerevisiae YKKB-13 | MATa ura3-52 lys2-801(Am) ade2-101(Oc) trp1-Δ63 his3-Δ200 leu2-Δ1 Δpdr5::TRP1 | 8 |
| C. glabrata | ||
| ATCC 90030 | ATCC reference strain | |
| DSY528 | Clinical isolate, azole susceptible from patient 1 | This study |
| DSY530 | Clinical isolate, azole resistant from patient 1 | This study |
| DSY562 | Clinical isolate, azole susceptible from patient 2 | This study |
| DSY565 | Clinical isolate, azole resistant from patient 2 | This study |
| DSY1029 | ura3 derivative of DSY565 | This study |
| DSY1041 | Δcgcdr1::hisG-URA3-hisG, derived from DSY1029 | This study |
| DSY1056 | Δcgcdr1::hisG, derived from DSY1041 | This study |
| DSY671 | ura3 | 61 |
| DSY1033 | Δcgcdr1::hisG-URA3-hisG, derived from DSY671 | This study |
| DSY1067 | Δcgcdr1::hisG, derived from DSY1033 | This study |
| DSY1717 | DSY1041 transformed with pDS670 | This study |
| DSY1718 | DSY1033 transformed with pDS670 | This study |
Fluconazole and rhodamine 6G accumulation.
Fluconazole accumulation testing was performed in duplicate with 3H-labelled fluconazole (Amersham Life Science, Little Chalfont, Buckinghamshire, United Kingdom) as described previously (53), but with a single incubation time of 20 min. Rhodamine 6G (Sigma, Fluka Chemie AG, Buchs, Switzerland) accumulation testing was performed with yeast cells grown to the logarithmic phase in 14 ml of sterile polystyrene tubes with 2 ml of YEPD at 30°C under constant agitation. Rhodamine 6G labelling of cells was performed in 1 ml of YEPD with 107 cells and containing 10 μM rhodamine 6G. The mixture was incubated for 30 min at 30°C, after which it was stopped by placing the tubes on ice. These conditions have been optimized for minimal incubation time and maximal rhodamine 6G accumulation (data not shown). The reaction mixture was then diluted 40-fold in cold sterile 0.1 M phosphate-buffered saline (PBS) at pH 7.0 and then directly subjected to flow cytometry in a FACScan fluorescence-activated cell sorter (FACS) (Becton Dickinson, San Jose, Calif.). Fluorescence was measured at an excitation wavelength of 488 nm and an emission wavelength of 515 nm (F1 detector). The sheath fluid was Isotone II. Data were acquired for 1,500 cells with the FACScan Lysis II software. Rhodamine 6G efflux was determined with 107 cells previously loaded by incubation with 10 μM rhodamine 6G at 30°C in YEPD. Cells were washed three times with YEPD medium at 4°C to remove excess rhodamine 6G, and efflux was started by incubation at 30°C in the same medium. The decrease in fluorescence of loaded cells was then recorded at regular time intervals.
Drug susceptibility tests.
Tests of susceptibility to azole antifungals were performed by broth microdilution assay according to the National Committee for Clinical Laboratory Standards (NCCLS) protocol M27-A (33) with RPMI-1640 medium (Difco) and incubation at 35°C for 48 h. Endpoint readings were recorded with a microplate reader (Bio-Rad, Hercules, Calif.), and the azole concentration yielding at least 50% growth inhibition compared to the growth in drug-free medium was defined as the MIC. Amphotericin B susceptibility was measured according to growth in Antibiotic Medium 3 broth (Difco) as recommended previously (42).
Susceptibility to different compounds of the C. glabrata isolates and of S. cerevisiae strains containing C. glabrata drug resistance genes was also tested qualitatively by spotting serial dilutions of yeast cultures onto complex YEPD medium agar plates. This provides an easy visualization of growth differences between different yeast strains. Since S. cerevisiae does not grow well in the RPMI medium described in the NCCLS protocol M27-A, the use of the qualitative plate assay for drug susceptibility is more adequate. The following drugs were solubilized in dimethyl sulfoxide: ketoconazole and itraconazole (Janssen Pharmaceuticals, Beerse, Belgium), 4-nitroquinoline-N-oxide (Sigma), and benomyl (Riedel-de-Haën, Seelze, Germany). Fluconazole (Pfizer UK, Sandwich, United Kingdom), cycloheximide, fluphenazine, and crystal violet (Sigma) were dissolved in water. Each plate contained 15 ml of agar. The drugs were diluted in the corresponding solvents to achieve the concentrations used in YEPD plates. Preliminary tests were performed to optimize drug concentrations in YEPD plates so that growth differences between the different S. cerevisiae and C. glabrata strains used in this study could be observed. To perform the susceptibility tests, yeasts were grown overnight at 30°C with constant shaking in YEPD liquid medium. The cultures were diluted to 2 × 107 cells per ml in 0.9% NaCl. Five microliters of this suspension and of serial dilutions of each yeast culture was spotted onto each type of plate and incubated for 48 h at 30°C.
Ergosterol biosynthesis inhibition.
Ergosterol biosynthesis inhibition assays were performed with cellular extracts from C. glabrata strains. Cellular extracts were from cells grown in 50 ml of YEPD medium and were obtained by mechanical disruption with glass beads (0.3 to 0.5 mm in diameter) in phosphate buffer (0.1 M sodium phosphate [pH 7.5]). The extracts were centrifuged at 10,000 × g for 10 min at 4°C. The assays were performed with increasing fluconazole concentrations by the method described by Vanden Bossche et al. (56). Each assay mixture contained 5 mg of cellular proteins from the individual strain which was measured by the Bradford assay (Bio-Rad, Hercules, Calif.).
Isolation of DNA and RNA.
The small-scale isolation of DNA and RNA from C. glabrata was performed from cultures grown to the logarithmic growth phase in YEPD medium at 30°C under constant shaking. One milliliter of each culture was centrifuged in Eppendorf tubes at 4°C. After a washing step with TE (10 mM Tris-HCl [pH 7.5], 1 mM EDTA), yeast DNA was extracted by adding 0.3 g of glass beads (0.3 to 0.5 mm in diameter), 200 μl of a breaking buffer (2% Triton X-100, 1% sodium dodecyl sulfate [SDS], 10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl), and 200 μl of phenol-chloroform-isoamyl alcohol (24:24:1). After 1 min of vortexing in a Mini-Beadbeater (Biospec Products, Inc., Bartlesville, Okla.), the tubes were centrifuged at maximum speed for 10 min in a microcentrifuge, and the supernatant was reextracted with chloroform-isoamyl alcohol (24:1). Nucleic acids were then precipitated with 20 μl of 3 M sodium acetate (pH 5.0) and 400 μl of ethanol at −20°C, washed with 70% ethanol, and resuspended in 50 μl of TE. For the extraction of RNA, the yeast cell pellet was mixed with 0.3 g of glass beads, 300 μl of RNA extraction buffer (0.1 M Tris-HCl [pH 7.5], 0.1 M LiCl, 10 mM EDTA, 0.5% SDS) and 300 μl of phenol-chloroform-isoamyl alcohol (24:24:1). After 1 min of vortexing in the Mini-Beadbeater, the aqueous phase was reextracted with phenol-chloroform-isoamyl alcohol, and RNA was precipitated with 600 μl of ethanol at −20°C for 1 h. The RNA pellet was resuspended in 50 μl of diethyl pyrocarbonate-treated H2O, and the concentration was measured spectrophotometrically at A260 and A280. RNA was stored at −80°C.
Typing of C. glabrata clinical isolates.
Two different methods were utilized for the typing analysis of the C. glabrata clinical isolates. The first method was based on restriction fragment length polymorphism (RFLP) with HinfI as restriction enzyme as described by others (5, 11). The second method was based on the use of repetitive probes Cg6 and Cg12 as described previously (29). In this method, 5 μg of C. glabrata genomic DNA, prepared as described above, was cut with EcoRI. The digested DNA was electrophoresed in a 0.7% agarose gel. Transfer was made by vacuum blotting on GeneScreen Plus membranes (DuPont NEN, Boston, Mass.) after depurination of the DNA with HCl. The Cg6 and Cg12 repetitive probes were prepared from λ phage DNA by liquid lysate with the Qiagen λ kit (Qiagen, Chatsworth, Calif.). The probes were labelled with [32P]dATP by nick translation with a nick translation system (Gibco BRL, Life Technologies, Inc., Rockville, Md.). After prehybridization for 30 min, membranes were hybridized overnight at 65°C in 5× SSPE (1× SSPE is 0.15 M NaCl with 10 mM NaH2PO4 and 1 mM EDTA) containing 5% dextran sulfate and 0.3% SDS. The membranes were washed four times for 30 min each at 48°C in 2× SSPE containing 0.2% SDS. Fuji RX film was used for visualization of hybridization patterns.
Yeast transformations.
S. cerevisiae and C. glabrata were transformed by the lithium acetate procedure as described by Sanglard et al. (51).
Cloning of C. glabrata azole resistance genes.
The C. glabrata gene library was constructed in the plasmid YEp24 (GenBank accession no. L09156). C. glabrata genomic DNA from strain ATCC 90030 was partially digested with Sau3A to obtain fragment lengths ranging from 6 to 9 kb. The fragments were purified by agarose gel electrophoresis and ligated to YEp24 previously digested with BamHI. The gene library was amplified in E. coli DH5α. C. glabrata genes conferring resistance to azole antifungals were cloned by complementation of hypersusceptibility to fluconazole of the S. cerevisiae strain YKKB-13. First, about 80,000 Ura+ clones were selected after transformation of YKKB-13 with the C. glabrata genomic library. The Ura+ clones were then pooled in separate aliquots, part of which were spotted onto YEPD medium containing 5 and 10 μg of fluconazole per ml, while the remaining cells were kept frozen at −80°C.
Plasmid rescue from S. cerevisiae.
Episomal plasmids from the parent vector YEp24 were rescued from S. cerevisiae transformants by electroporation in E. coli. Yeast cells were grown in selective YNB medium to late log phase, and total DNA was extracted as outlined above. One microliter of the DNA suspension (50 μl) was electroporated into E. coli DH5α, and ampicillin-resistant clones from each transformant were analyzed by restriction enzyme analysis.
Construction of C. glabrata vectors.
To enable the replication of YEp24-derived plasmids in C. glabrata and particularly of pNB126, the CgCEN and CgARS sequences contained in pCgACU-5 were subcloned in these vectors. pCgACU-5 (a kind gift of K. Kitada) was generated by inserting CgURA3 into the XhoI site of pCgAC-5 (25). The presence of the CgCEN and CgARS sequences in plasmids contributes to their stable replication in C. glabrata in one copy per cell. The CgCEN and CgARS sequences were recovered from pCgACU-5 by digestion with SalI and XhoI and subcloned into the single SalI site of pNB126 to generate pDS670. pDS670 contains CgCDR1 and could transform successfully ura3 mutants from both S. cerevisiae and C. glabrata.
Disruption of CgCDR1.
For the disruption of CgCDR1 in C. glabrata, a 4.9-kb fragment from pNB126 was cloned into the same sites of pMTL21 (10), yielding pDS458. An 800-bp fragment was removed from pDS458 by digestion with BglII, thus creating a deletion in CgCDR1. The 3.7-kb BamHI-BglII fragment from pNKY51, which contained a hisG-URA3-hisG cassette with the S. cerevisiae URA3 gene, was inserted in the BglII-treated pDS458, thus yielding pDS460. Digestion of pDS460 with PvuII and XhoI, the latter cutting in the polylinker site of pMTL21, generated a fragment of 5.3 kb which was used to transform DSY1029 by lithium acetate.
Northern and Southern blots.
DNA was separated by conventional 1% agarose gel electrophoresis in TAE buffer (40 mM Tris-acetate [pH 7.5], 1 mM EDTA). For RNA electrophoresis, RNA samples were resuspended in a loading buffer (50% formamide, 100 mM morpholinepropanesulfonic acid [MOPS] [pH 7.0], 6.4% formaldehyde, 5% glycerol, 5% of a water solution saturated with bromophenol blue), denatured at 85°C for 5 min, and separated by 1% agarose gel electrophoresis. The agarose was melted in a buffer containing 0.1 M MOPS, 0.6 M formaldehyde, and 10 μg of ethidium bromide per ml. The electrophoresis buffer was 0.1 M MOPS (pH 7.0). After completion of electrophoresis, RNA was visualized under UV light, and the position of the rRNA was determined. Both Northern and Southern blots were performed by vacuum blotting onto GeneScreen Plus membranes (DuPont NEN, Boston, Mass.). Membranes containing RNA were baked under vacuum for 2 h at 80°C. Membranes were prehybridized at 42°C with a buffer consisting of 50% formamide, 1% SDS, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 10% dextran sulfate, and 100 μg of salmon sperm DNA per ml. 32P-DNA-labelled probes were generated by random priming (14) and added to the hybridization solution overnight. Washing steps were performed at high stringency identical to those recommended by the supplier (DuPont NEN, Boston, Mass.). Stripping of probes off Northern blots for sequential hybridizations was achieved by boiling the membranes for 10 min in TE buffer with 0.1% SDS.
The DNA probes used in Southern and Northern blots were as follows: CgCDR1, 1.8-kb XbaI-BamHI fragment from pNB126; CgYAP1, 2.3-kb PstI-NruI fragment from pNB125; and CgMDR1, 1.2-kb EcoRV fragment from pNB124. The CgERG11 and CgURA3 probes were generated from fragments amplified by PCR from genomic DNA. The primers for CgURA3 (GenBank accession no. L13661) were 5′ CTCGAGAACCAATTGCATCA 3′ and 5′ CTAGCTTCCTATTGGATATG 3′ and amplified a 900-bp fragment. The primers for CgERG11 (GenBank accession no. L40389) were 5′ ATGTCCACTGAAAACACTTCTTTG 3′ and 5′ CTAGTACTTTTGTTCTGGATGTCT 3′ and amplified the 1.6-kb CgERG11 open reading frame (ORF).
Quantifications of Northern blot bands were performed by scanning the hybridized membranes in an Instant Imager (Packard Instrument Company, Meriden, Conn.). Signals were integrated by the software supplied by the manufacturer and normalized to the corresponding values of an internal standard. In the case of C. glabrata, the internal standard was the CgURA3 probe.
DNA sequencing.
Sequence data were obtained with DNA fragments subcloned from pNB124, pNB125, and pNB126 into pBluescript (Stratagene GmbH, Zürich, Switzerland). Sequence data were generated on both DNA strands by using reverse or universal primers and customized primers by automated sequencing in a Li-Cor 4200 sequencer (Li-Cor, Inc., Lincoln, Nebr.).
RESULTS
Origin and azole susceptibility of C. glabrata isolates.
For the study of mechanisms of resistance to azoles, C. glabrata clinical isolates were selected retrospectively from a collection of yeasts recovered from AIDS patients with OPC. The C. glabrata strains were chosen from two different patients with recurrent OPC and were first selected on the basis of decreased susceptibility to azole antifungals.
Patient 1 had his first OPC episode in October 1990 and was diagnosed with AIDS. C. albicans was isolated initially from the patient’s oral cavity in December 1993 and was subsequently found in recurrent episodes of OPC. In April 1994, the first C. glabrata strain (DSY528) was isolated. At that time, patient 1 had received a cumulative dose of 17.3 g of fluconazole, and his CD4 count in blood was 4 cells per mm3. After treatment with 400 mg of fluconazole for 7 consecutive days, OPC was still persistent 47 days later. DSY530 was isolated at this time and showed a reduced susceptibility to fluconazole compared to DSY528 (Table 2). Patient 2 had his first clinically documented episode of OPC in November 1991 and was diagnosed with AIDS. C. albicans was first isolated from the oral cavity in February 1993. In May 1995, C. glabrata DSY562 was isolated for the first time together with C. albicans. The patient had received a cumulative dose of 4.1 g of fluconazole, and his CD4 count in blood was 34 cells per mm3. After two courses of treatment with 200 mg of fluconazole for 7 consecutive days each, OPC was still persistent 50 days later, and the C. glabrata strain DSY565 was isolated, which showed reduced susceptibility to fluconazole compared to that of DSY562 (Table 2). In both patients, C. glabrata isolates were isolated in mixed culture with C. albicans. The less susceptible C. glabrata isolate of each patient will be designated in this study as azole resistant, without reference to the clinical MIC breakpoints proposed by Rex et al. (44).
TABLE 2.
Antifungal drug susceptibility of C. glabrata isolates taken from two AIDS patients with OPC
| Isolate | MIC (μg/ml)
|
Time elapsed between samplings (days) | |||
|---|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | Amphotericin B | ||
| DSY528 | 4 | 0.062 | 0.125 | 0.5 | |
| DSY530 | 32 | 0.5 | 1 | 0.5 | 47 |
| DSY562 | 4 | 0.031 | 0.125 | 0.5 | |
| DSY565 | 128 | 2 | 4 | 0.5 | 50 |
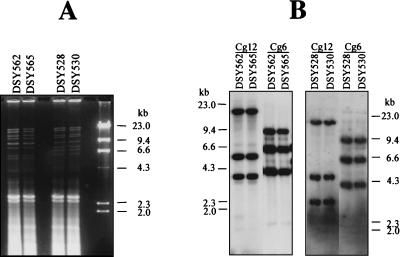
The C. glabrata strains from these two patients were typed by two different methods. The results shown in Fig. 1 demonstrate that identical banding patterns for the two C. glabrata strains from a given patient could be obtained by these methods and thus suggest that the azole-susceptible and azole-resistant isolates from these two patients were related to each other. This implies that no strain replacement occurred during azole therapy in these patients, but rather that azole resistance developed from the original azole-susceptible isolates. The stability of the resistance phenotype did not change after more than 50 consecutive passages (over 500 generations) in drug-free medium. Thus, azole resistance in the azole-resistant isolates could be due to genome alterations rather than to transient adaptation to azole antifungals.
FIG. 1.
Typing of C. glabrata clinical isolates used in this study. (A) Restriction enzyme analysis of C. glabrata genomic DNA digested with HinfI. Restriction fragments were separated by 1% agarose gel electrophoresis and stained with ethidium bromide. (B) Profiles of band patterns revealed by hybridization with the repetitive element probes Cg6 and Cg12 as described by Lockhart et al. (29). Molecular size standards were depicted on each photograph.
Fluconazole and rhodamine 6G accumulations in C. glabrata isolates.
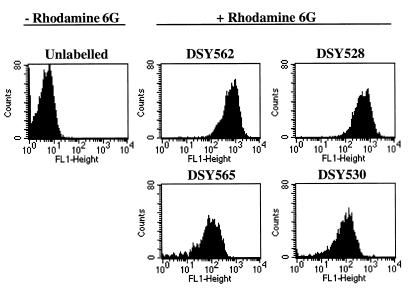
We performed several experiments in order to determine the mechanisms of azole resistance in the azole-resistant isolates. First, no changes in amphotericin B susceptibility were observed in the four clinical strains used in this study (Table 2). Therefore, since some alterations in the ergosterol biosynthetic pathway are coupled with amphotericin B resistance (17, 35), it is likely that no alterations in this pathway were occurring in these strains. In agreement with this hypothesis was the detection of this sterol in ergosterol biosynthesis inhibition assays. Second, fluconazole concentrations at which the ergosterol biosynthesis was inhibited in cellular extracts by 50% (IC50) did not vary significantly between azole-susceptible and azole-resistant isolates (Table 3). This led us to deduce that an alteration in the affinity of the C. glabrata CYP51A1 proteins to azoles was not the cause of azole resistance in these isolates. We therefore tested the accumulation of two different substances in the azole-susceptible and azole-resistant isolates. As shown in Table 3, the azole-resistant isolates were accumulating less fluconazole than their azole-susceptible parents. The accumulation of fluconazole was reduced by factors of 3.8 and 3.1 in DSY565 and DSY530 compared to those in DSY562 and DSY528, respectively. Failure in drug accumulation was not restricted to a single compound, since intracellular levels of rhodamine 6G were also affected in the azole-resistant isolates (Fig. 2). The decreases in rhodamine 6G accumulation were 6.1- and 5.1-fold in DSY565 and DSY530 compared to those in DSY562 and DSY528, respectively. Failure in accumulating several unrelated drugs in the azole-resistant isolates was reminiscent of the effect of multidrug efflux transporters observed in C. albicans, and therefore we exploited the possibility that multidrug efflux transporter genes were upregulated in the azole-resistant C. glabrata isolates.
TABLE 3.
Fluconazole accumulation and inhibition of ergosterol biosynthesis by fluconazole in C. glabrata isolates
| Isolate | Amt of [3H]fluconazole/2 × 107 cells (cpm) | IC50 of fluconazole for ergosterol biosynthesis (nM) |
|---|---|---|
| DSY528 | 300 ± 20 | 55 ± 10 |
| DSY530 | 95 ± 20 | 65 ± 8 |
| DSY562 | 313 ± 22 | 40 ± 12 |
| DSY565 | 81 ± 5 | 50 ± 8 |
| DSY1041 | 457 ± 7 | NDa |
ND, not determined.
FIG. 2.
Rhodamine 6G accumulation in C. glabrata clinical isolates. Cells were labelled with rhodamine 6G and analyzed by FACS as indicated in Materials and Methods. FACS histograms are given for the unlabelled control (DSY562) and for each yeast strain labelled with rhodamine 6G. The mean fluorescence values for DSY562 and DSY565 were 691 and 113, respectively, and those for DSY528 and DSY530 were 566 and 110, respectively. Accumulation experiments with rhodamine 6G were repeated five times with these strains. DSY565 and DSY530 mean fluorescence reached 16% ± 1.8% and 12% ± 3% of the values obtained with DSY562 and DSY528, respectively.
Cloning of C. glabrata azole resistance genes.
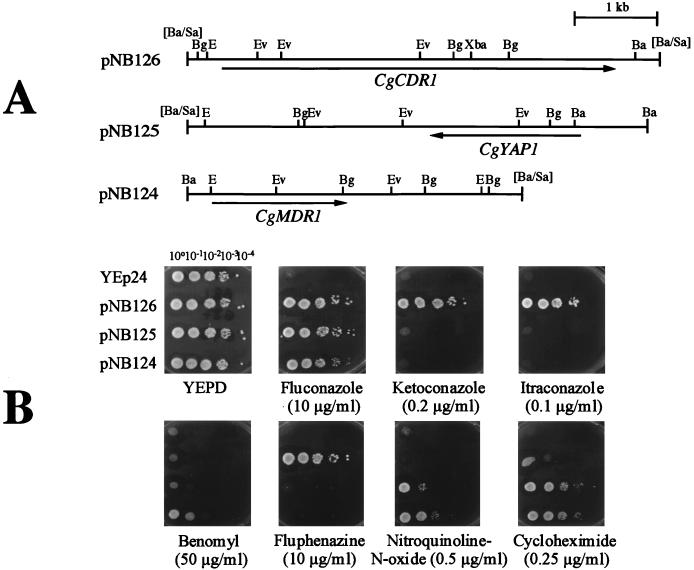
Given the observations mentioned above, we first attempted to isolate multidrug efflux transporter genes from C. glabrata, since at the time these studies were initiated, no C. glabrata gene encoding a transporter was available. We used a strategy consisting of isolation of C. glabrata genes which could confer azole resistance to an S. cerevisiae multidrug efflux transporter mutant. Since the absence of ABC transporter Pdr5p in S. cerevisiae yields hypersusceptibility to azole antifungals, this genetic background was suitable to isolate genes involved in azole resistance in C. glabrata. From a total of about 80,000 Ura+ S. cerevisiae transformants, more than 200 individual yeasts able to grow on media containing 5 and 10 μg of fluconazole per ml were selected. Each fluconazole-resistant clone was then tested for different drug resistance profiles by using agar plates containing 10 and 25 μg of fluconazole per ml, 0.1 and 1 μg of itraconazole per ml, 0.2 and 2 μg of ketoconazole per ml, 0.25 μg of cycloheximide per ml, 10 μg of fluphenazine per ml, 50 μg of benomyl per ml, 0.5 μg of crystal violet per ml, and 0.5 μg of nitroquinoline-N-oxide per ml. Among the 200 fluconazole-resistant clones, only 20 were specifically resistant to fluconazole among the azole drugs tested while remaining resistant to cycloheximide, benomyl, and nitroquinoline-N-oxide. The other yeast transformants were able to grow on medium containing different azole antifungals and other drugs, such as cycloheximide and fluphenazine. Plasmids rescued from these isolates were extracted and separated by gel electrophoresis. To group these plasmids into different categories, C. albicans probes for CDR1, MDR1, and CAP1, which were genes recovered by the same functional complementation (50), were used in low-stringency hybridizations of Southern blots of the C. glabrata plasmids (data not shown). Three different classes of plasmids were recovered, each containing a DNA fragment hybridizing with these probes. From each of these groups, a single plasmid was chosen for further restriction map and sequence analysis. Figure 3A shows the restriction maps of these three plasmids (pNB124, pNB125, and pNB126) with their respective azole resistance genes. pNB124 contained a gene with similarity to C. albicans MDR1 (formerly named BENr), a gene belonging to the class of major facilitator multidrug transporters. This gene was named CgMDR1 (52). pNB125 contained a gene with similarity to the S. cerevisiae YAP1 gene, a transcription factor with a leucine dimerization motif. This gene was therefore named CgYAP1 (52). pNB126 contained a gene with similarity to fungal ABC transporters such as the S. cerevisiae PDR5 gene (8), the C. albicans CDR1 and CDR2 genes (50, 53) or the recently cloned CgPDH1 gene (32). This C. glabrata gene was named CgCDR1 for Candida drug resistance gene, by analogy with the C. albicans CDR genes. Figure 3B shows the drug resistance profiles obtained because of the presence of each plasmid in YKKB-13. All plasmids could confer resistance to fluconazole, but only pNB126 containing CgCDR1 conferred resistance to different azole antifungals and fluphenazine. pNB124 and pNB125 conferred similar drug resistance profiles, but only pNB124 containing CgMDR1 was able to confer slight resistance to benomyl.
FIG. 3.
Cloning of azole resistance genes in C. glabrata. (A) Restriction maps of C. glabrata insert DNA from plasmids pNB124, pNB125, and pNB126 conferring azole resistance in the S. cerevisiae Δpdr5 mutant YKKB-13. The plasmid backbone is YEp24. Sau3A-digested C. glabrata DNA was introduced at the BamHI site of YEp24 in the construction of the gene library. The transcription directions of the ORF of each azole resistance gene deduced from nucleotide sequencing are shown by arrows. Ba, BamHI; Bg, BglII; E, EcoRI; Ev, EcoRV; Xba, XbaI; [Ba/Sa], BamHI site of YEp24 destroyed by insertion of a genomic Sau3A site. (B) Drug resistance profiles of S. cerevisiae YKKB-13 transformed with plasmids pNB124, pNB125, and pNB126. Yeast strains were spotted in serial dilutions onto YEPD medium containing the drug at the indicated concentration. Plates were incubated at 30°C for 48 h.
Expression of azole resistance genes in C. glabrata.
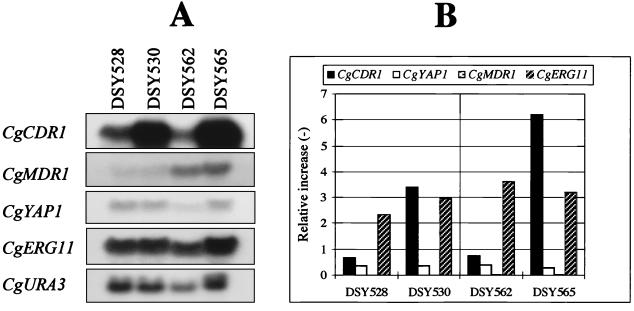
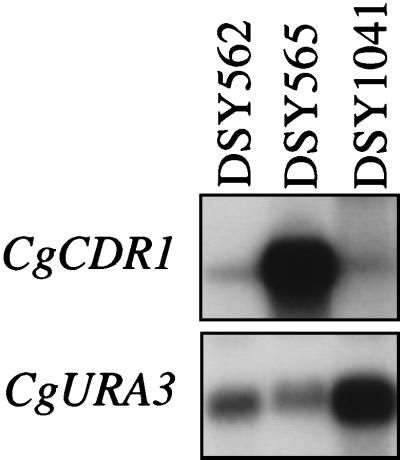
We next performed Northern blot analysis of total RNA extracted from C. glabrata clinical isolates DSY562, DSY565, DSY528, and DSY530 with DNA probes for each of the azole resistance genes. This experiment was aimed to show a possible correlation between the expression of an azole resistance gene and azole resistance in the clinical isolates. The CgURA3 gene was used as an internal control for the standardization of signals. The CgERG11 gene was also included as a probe in this analysis, since it was reported by others that CgERG11 overexpression can moderately establish azole resistance in C. glabrata (31). Figure 4A shows that only CgCDR1 expression was elevated in both azole-resistant isolates from the two different patients. The relative increases in CgCDR1 expression obtained in DSY565 and DSY530 were eight- and fivefold compared to those in DSY562 and DSY528, respectively (Fig. 4B). The expression of the other genes was not significantly affected by azole resistance. CgMDR1 expression was increased in both DSY528 and DSY530 compared to the level in DSY562 and DSY565, but indicated that it did not vary between azole-susceptible and azole-resistant isolates from the same patient.
FIG. 4.
Expression of azole resistance genes in C. glabrata clinical isolates. (A) Northern blot analysis. Five micrograms of RNA from each strain was separated and blotted as indicated in Materials and Methods. The blotted membrane was probed sequentially with 32P-labelled probes for CgCDR1, CgYAP1, CgMDR1, CgERG11, and CgURA3. Labelled DNA probes are described in Materials and Methods. Washed membranes were exposed to Fuji RX films for 1 to 10 h. (B) Quantification of hybridization signals detected by each 32P-labelled probe. Signals given by each probe were counted with an Instant Imager and normalized to the counts obtained with the CgURA3 probe. The values of the normalized counts for CgMDR1 were too small to be reported in the figure. These values were 0.0093 for DSY528 and 0.012 for DSY530, and 0.037 for DSY562 and 0.026 for DSY565.
Analysis of nucleotide and amino acid sequences of CgCDR1.
Since only CgCDR1 expression was enhanced in azole-resistant isolates, this gene was submitted to closer analysis. Figure 5 shows the nucleotide sequence of a 5.3-kb fragment isolated from pNB126 starting from the Sau3A cloning site on YEp24 and ending at the single BamHI site of the same plasmid. An uninterrupted ORF of 4,500 bp encoding a protein with 1,499 amino acids was detected from the most upstream ATG codon. The 5′ region starting from this ATG codon displayed typical features of yeast promoters: an adenosine at position −3 and a TATA box at position −83 could be distinguished (Fig. 5). We noticed the presence of four putative Pdr1p and Pdr3p binding sites in the 5′ flanking region of CgCDR1. The positions of these sites were as follows:−516GTCCACGGAA−507−387TTCCACGGAA−378 −228TTCCACGGGA−219 −134CTCCACGGGA−125
FIG. 5.
Nucleotide and deduced amino acid sequences of the CgCDR1 gene and its encoded polypeptide. Nucleotides and amino acids are numbered to the left and to the right, respectively. The sequence starts from a Sau3A site and ends with a BamHI site. The CgCDR1 ORF is indicated below the nucleotide sequence, and its position in the pNB126 restriction map corresponds to the arrow shown in Fig. 3A. In the deduced protein sequence, putative membrane-spanning domains are underlined, and the Walker A and B ATP-binding motifs and ABC signatures are boxed in the N- and C-terminal regions. A TATA box in the 5′ flanking region is underlined. Putative Pdr1p and Pdr3p binding sites in the 5′ flanking region are boxed. The CgCDR1 sequence is available in the GenBank database under accession no. AF109723.
The consensus of the Pdr1p and Pdr3p binding sites in the S. cerevisiae PDR5 gene was TTCCG(C/T)GGAA. Nucleotides conserved among the Pdr1p and Pdr3p binding sites are shown in boldface letters. Palindromes are underlined. These sites (and conserved nucleotide sequences) have been shown to be important in the regulation of ABC transporter genes in S. cerevisiae by the transcription factors Pdr1p and Pdr3p (23). These sites may therefore play a similar role in the regulation of CgCDR1.
The CgCDR1 ORF encodes a protein of 169 kDa (CgCdr1p) and displays a structure and domain organization typical of membrane proteins of the ABC superfamily (Fig. 5). A blast search of the CgCDR1 ORF against the entire GenBank database gave the highest score with PDR5 from S. cerevisiae. C. glabrata CgCdr1p is 75% identical to Pdr5p. CgCdr1p is composed of two homologue halves, each comprising a C-hydrophilic domain and a N-hydrophobic domain. The hydropathy plot of CgCdr1p identified six putative transmembrane domains for each of the hydrophobic domains (Fig. 5). Each hydrophilic domain included ATP-binding motifs found in ATP-binding cassette domains. In the N-terminal ABC domain, the Walker A (LGRPGSGCTTLL) and B (QCWDNATRGLD) motifs and the ABC signature (SGGERKRVSIAE) could be recognized. In the C-terminal ABC domain, both the Walker A (GASGAGKTTLL) and Walker B (VFLDEPSGLD) regions are present, but an atypical ABC signature (NVEQRKRLTIGV) is present where the usual SXGQ/E is replaced by NVEQ. Recently Miyazaki et al. (32) reported the nucleotide sequence of an ABC transporter gene from C. glabrata called PDH1. Pdh1p, however, was not identical to CgCdr1p, as revealed by pairwise alignment (data not shown). Both proteins showed 73% identity, which was most prominent in the ABC signatures and Walker A and B domains.
Construction of CgCDR1 mutants in C. glabrata.
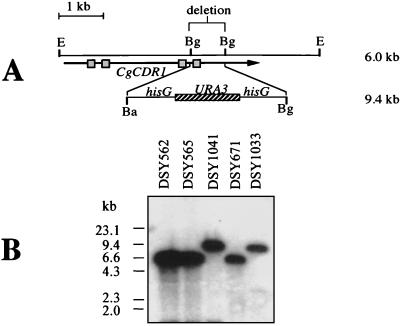
Given the overexpression of CgCDR1 in the azole-resistant clinical isolates, we addressed the involvement of this ABC transporter in azole resistance by the deletion of this gene in one azole-resistant isolate. First, an auxotrophic mutant for uracil was generated by positive selection of resistance to 5-FOA. 5-FOA-resistant colonies in S. cerevisiae are usually deficient for the URA3 gene, and this selection was applied successfully to C. glabrata. 5-FOA-resistant colonies from DSY565 were obtained with a low reversion frequency (>10−8). The identity of the ura3 auxotrophy could be confirmed, since, by transformation of one 5-FOA-resistant DSY565 derivative (DSY1029) with a C. glabrata plasmid containing the CgURA3 gene, Ura+ colonies were obtained (data not shown). DSY1029 was further utilized for the deletion of CgCDR1. A linear fragment which contained the hisG-URA3-hisG cassette from pNKY51 (2) replacing an internal deletion of CgCDR1 (Fig. 6A) was used to transform DSY1029. Ura+ colonies were selected, and analysis of the CgCDR1 locus in one of the Ura+ transformants (DSY1041) confirmed that the correct gene replacement had occurred (Fig. 6B). The wild-type CgCDR1 6-kb EcoRI fragment observed in both DSY562 and DSY565 was not observed in DSY1041 and was replaced by a 9.4-kb band expected from the insertion of the hisG-URA3-hisG cassette. To verify if the deletion made in CgCDR1 affected its expression, total RNA from DSY1041 was analyzed by Northern analysis with a CgCDR1 probe. As shown in Fig. 7, a weak signal could still be detected in RNA from DSY1041, but with a slightly reduced migration when compared to the CgCDR1 mRNA signals observed in DSY562 and DSY565. The intensity of this signal was, however, much reduced compared to that of the CgCDR1 mRNA signal in DSY565. Therefore, deletion of CgCDR1 in DSY565 affected the expression of this gene to a large extent. The weak signal still observed in DSY1041 could be attributed to the formation of aberrant CgCDR1 mRNA or to the presence of additional mRNA(s) from an additional ABC transporter gene or genes hybridizing weakly with the CgCDR1 probe.
FIG. 6.
Disruption of CgCDR1 in C. glabrata. (A) Genomic map of the CgCDR1 locus before and after internal deletion of a 0.8-kb BglII site. The CgCDR1 ORF is represented by an arrow, on which boxes give the location of the N- and C-terminal Walker A and B domains. The BglII deletion destroys the C-terminal domain. The size of the wild-type EcoRI genomic fragment containing CgCDR1 is about 6 kb. The insertion of the hisG-URA3-hisG cassette after deletion of the 0.8-kb BglII fragment yields a product of about 9.4 kb. (B) Southern blot of genomic DNA from C. glabrata isolates. Genomic DNA was extracted from the clinical strains DSY562 and DSY562, from the laboratory strain DSY671, and from the CgCDR1 deletion mutants DSY1041 and DSY1033, both of which were derived from DSY565 and DSY671, respectively. The CgCDR1 probe is described in Materials and Methods. Signals revealed by the labelled probe correspond to those expected from the genomic restriction map. The origin of each DNA is indicated.
FIG. 7.
Expression of CgCDR1 in C. glabrata clinical strains and from the CgCDR1 deletion mutant DSY1041. Total RNA was extracted from each strain and separated by gel electrophoresis. Membranes were probed sequentially with CgCDR1 and CgURA3 labelled probes (same as described in the legend to Fig. 4) and after washing were exposed to Fuji RX films.
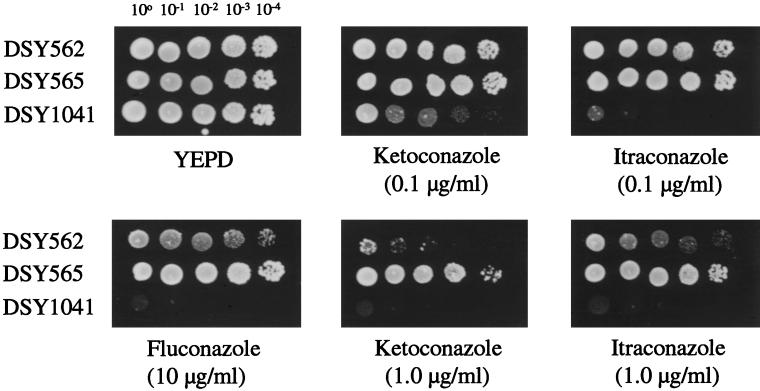
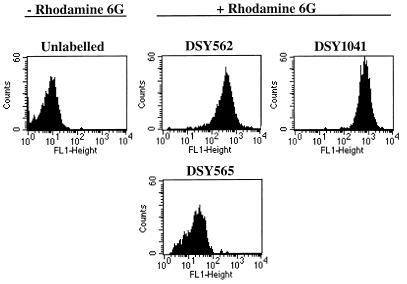
DSY1041 was subjected to azole susceptibility tests and drug accumulation experiments in parallel to DSY562 and DSY565. As shown in Fig. 8, the deletion of CgCDR1 in DSY1041 had a significant effect on the susceptibility to azoles. DSY1041 could not grow at drug concentrations permissive for DSY565 in medium containing fluconazole, itraconazole, or ketoconazole. DSY1041 was also more susceptible to azole antifungal agents in this assay than the azole-susceptible isolate DSY562, as revealed by the difference in growth between both yeasts in the azole-containing YEPD medium tested. Since CgCDR1 was still expressed in DSY562 as opposed to DSY1041, this result could, however, be expected. MICs of azole antifungals for DSY1041 measured with the NCCLS standard protocol in RPMI medium were lower than those for DSY565 and were in the range of those measured for DSY562 (Table 4). We noticed that the MICs of fluconazole were lower for DSY1041 than for DSY562, when other incubation media were used, in particular YNB (data not shown). Intracellular levels of fluconazole and rhodamine 6G were restored in DSY1041 compared to those in DSY565 and reached or exceeded the levels found in DSY562. As shown in Table 3, fluconazole accumulation was increased by 5.6-fold in DSY1041 compared to that in DSY565. The accumulation of rhodamine 6G in DSY1041 was increased by a factor of 16.5 compared to that in DSY565 (Fig. 9). We also observed that the efflux of rhodamine 6G in DSY1041 loaded with this substance was much reduced compared to that in DSY565 or DSY562 (data not shown). Taken together, these results showed that the deletion of CgCDR1 in the azole-resistant isolate DSY565 dramatically affected azole resistance and drug accumulation and thus support the idea that the expression of CgCDR1 was the major cause of azole resistance in this isolate.
FIG. 8.
Azole susceptibility of C. glabrata clinical isolates and of CgCDR1 deletion mutant DSY1041. Yeast strains were spotted in serial dilutions, as indicated, on YEPD medium containing the drug at the corresponding concentration. Plates were incubated at 30°C for 48 h.
TABLE 4.
Azole susceptibility of C. glabrata clinical isolates and of the ABC transporter mutant DSY1041
| Isolate | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Fluconazole | Ketoconazole | Itraconazole | Amphotericin B | |
| DSY562 | 4 | 0.062 | 0.25 | 0.5 |
| DSY565 | 128 | 2 | 4 | 0.5 |
| DSY1041 | 8 | 0.25 | 0.5 | 0.5 |
FIG. 9.
Rhodamine 6G accumulation in C. glabrata clinical isolates and the CgCDR1 deletion mutant DSY1041. Cells were labelled with rhodamine 6G and analyzed by FACS as indicated in Materials and Methods. FACS histograms are given for the unlabelled control (DSY562) and for each yeast strain labelled with rhodamine 6G. Mean values for fluorescence in DSY562, DSY565, and DSY1041 were 300, 47, and 780, respectively.
Rescue of fluconazole resistance phenotype by CgCDR1.
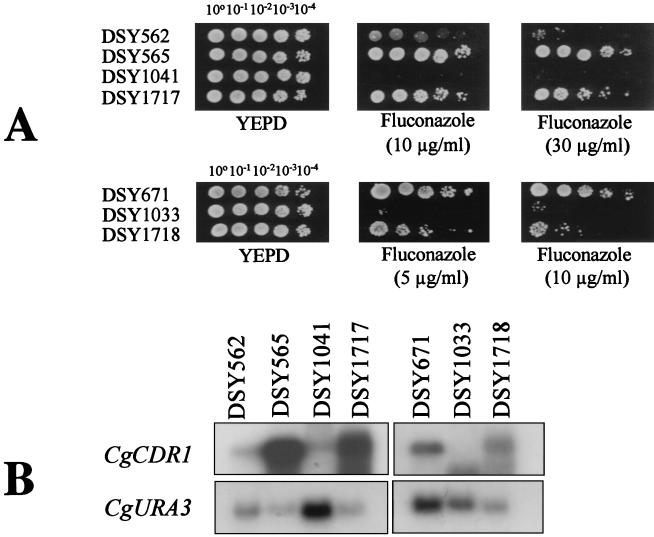
In order to identify the nature of the mutation or mutations responsible for the increase in CgCDR1 mRNA in the azole-resistant clinical isolate DSY565, CgCDR1 isolated from an azole-susceptible C. glabrata strain was reintroduced in the CgCDR1 deletion mutant DSY1053 (the ura3 derivative of DSY1041) with the help of a replicating vector. Since 2μm-derived vectors do not efficiently replicate in C. glabrata, pNB126, which contains CgCDR1 on a 2μm-based vector, was equipped with the CgCEN and CgARS centromeric elements from C. glabrata, thus enabling the replication of the resulting construction (pDS670) in a single copy in C. glabrata (25). As shown in Fig. 10A, the presence of pDS670 in DSY1053 (DSY1717) restored the azole resistance phenotype. This phenotype was followed by a 6.5-fold increase in CgCDR1 mRNA in DSY1717 (Fig. 10B). The same plasmid, pDS670, was used to transform DSY1067, which is the ura3 derivative of DSY1033, a CgCDR1 deletion mutant derived from the azole-susceptible laboratory strain DSY671 (61). The resulting strain, DSY1718, was as susceptible as DSY671 (Fig. 10A). CgCDR1 mRNA levels in DSY1718 were 1.6-fold higher than those in DSY671 and thus were comparable (Fig. 10B). Taken together, these results suggest that the higher CgCDR1 expression levels in DSY1717 should be the consequence of a trans-acting mutation(s) rather than of a cis-acting mutation or mutations on CgCDR1. cis-acting mutations on CgCDR1 from DSY565 would have been revealed if pDS670 had produced similar azole susceptibility and similar CgCDR1 mRNA levels in both DSY1717 and DSY1718. The results obtained in Fig. 10 reflect the pleiotropic drug resistance phenomenon in S. cerevisiae, which is controlled by several mutations of transcription factors acting in trans on several ABC transporter genes (9).
FIG. 10.
Rescue of azole resistance by CgCDR1 in CgCDR1 deletion mutants. (A) Azole susceptibility of C. glabrata strains and mutants transformed with pDS670, which contains CgCDR1, CgCEN, and CgARS elements. Yeast strains were spotted in serial dilutions, as indicated, onto YEPD medium with or without fluconazole at the given concentration. Plates were incubated at 30°C for 48 h. (B) Northern blot analysis of C. glabrata strains and CgCDR1 deletion mutants transformed with pDS670. Five micrograms of total RNA from individual strains was loaded as indicated. The CgCDR1 and CgURA3 probes are described in Materials and Methods. Washed membranes were exposed to Fuji RX films. mRNA signals were quantified as mentioned in the legend to Fig. 4 and indicated that CgCDR1 levels of expression were 8.3-fold and 6.5-fold higher in DSY565 and DSY1717, respectively, than that in DSY562. CgCDR1 expression was 1.6-fold higher in DSY1718 than in DSY671.
DISCUSSION
OPC represents one of the main mucosal infections in AIDS patients. While C. albicans remains the principal agent of OPC, C. glabrata is often recovered from clinical samples in addition to C. albicans. Among 298 human immunodeficiency virus-positive patients monitored in our institution for the occurrence of OPC over a period of 4 years (1990 to 1994), approximately 30% had at least one C. glabrata-positive culture. In our institution, C. glabrata was the second most frequent yeast species recovered after C. albicans. Nguyen et al. (34) also showed that C. glabrata has become the second most frequent yeast species isolated from blood cultures of patients with candidemia. This ranking has been confirmed by more recent studies (39). Gleason et al. (18) reported that C. glabrata was isolated significantly more frequently in patients treated with fluconazole in the surgical intensive care unit of a university hospital. Vennewald et al. (59) noted in their mycological laboratory there was an increase in the occurrence of C. glabrata isolates from 0.5% in 1990 to 22.2% in 1996, mainly originating from high-risk patients in the intensive care unit and the oncology ward. A recent study retrospectively reviewing medical records of patients with hematogenous candidiasis revealed a relative decrease in C. albicans and C. tropicalis infections, but an increase in C. krusei and C. glabrata infections (1). Fluconazole prophylaxis was the single most important determinant for the relative increase in C. krusei and C. glabrata infections. Similar shifts in yeast population favoring the increased appearance of C. glabrata have been reported by others (22, 26). These trends suggest that C. glabrata is becoming increasingly important, and due to the intrinsic low susceptibility of this yeast to azole derivatives, the management of fungal diseases by azole antifungals is rendered more difficult.
We describe here a mechanism of resistance to azole antifungals in two distinct azole-resistant C. glabrata isolates from two different AIDS patients with OPC. The C. glabrata strains used in this study were chosen on the basis of two criteria. First, two pairs of isolates in which changes in azole susceptibility were measured during fluconazole therapy were selected. Second, the isolates of each pair were compared by two typing methods to verify their relatedness. The isolates of the present study satisfied these criteria. We observed retrospectively that the times that elapsed between the recovery of the first azole-susceptible C. glabrata isolate and the azole-resistant isolate were not only short (47 versus 50 days) but were also similar in the two patients. Miyazaki et al. (32) in their investigations of azole resistance in C. glabrata reported an elapsed time of 2 weeks between the sampling of azole-susceptible and the azole-resistant isolates in one AIDS patient under fluconazole therapy. This feature has been also noticed in our collection of C. glabrata isolates and suggests rapid changes in azole susceptibility in C. glabrata strains exposed to azole antifungals. This observation needs to be confirmed by other laboratories.
The preliminary experiments which helped us to determine the cause of resistance in the azole-resistant C. glabrata isolates excluded changes of affinity to azoles in CYP51A1 and also eliminated alteration of sterol biosynthesis, since no differences in amphotericin B susceptibility between azole-susceptible and azole-resistant isolates were measured. Failure of drug accumulation in the azole-resistant isolates was the remaining known mechanism and proved to be present in both azole-resistant isolates. Drug accumulation studies were performed with two different substances, [3H]fluconazole and rhodamine 6G. Rhodamine 6G is a substrate for several ABC transporters and has been used for efflux studies with intact S. cerevisiae cells through the convenient method of flow cytometry (12, 13). The use of rhodamine 6G for measuring accumulation differences by FACS analysis in the C. glabrata isolates of this study has proved to be a very simple method which is much less time-consuming than the method utilizing 3H-labelled fluconazole. By the use of both substrates, not only could the cause of reduced azole susceptibility in isolates DSY530 and DSY565 be explained as a drug accumulation failure, but the nature of this process as an effect due to multidrug transport could be determined as well. FACS analysis of rhodamine 6G-labelled yeast cells can represent a rapid method for the study of azole resistance in a large collection of clinical isolates; however, this method first needs to be validated with more azole-resistant isolates.
The functional cloning of azole resistance genes from C. glabrata greatly assisted the identification of genes involved in azole resistance in the clinical isolates. This strategy had been used already for the cloning of similar genes in C. albicans and revealed six genes, among which CDR1, CDR2, and C. albicans MDR1 were shown to be upregulated in azole-resistant strains (50, 53). In this study, we could identify three different C. glabrata azole resistance genes: a transcription factor (CgYAP1), a multidrug transporter of the major facilitator family (CgMDR1), and a multidrug transporter of the ABC transporter family (CgCDR1). Only the transcription of CgCDR1 was increased in the azole-resistant isolates examined in this study. This effect has also been observed in at least 15 other azole-resistant C. glabrata isolates from AIDS patients of our institution (47), thus suggesting that this mechanism of resistance is common in C. glabrata. Reports on the characterization of azole resistance in C. glabrata from other laboratories have pointed to a failure of drug accumulation as a cause of resistance. Hitchcock et al. (20), Parkinson et al. (38), and Miyazaki et al. (32) reported differences of fluconazole accumulation in azole-resistant C. glabrata isolates due to enhanced drug efflux. Vanden Bossche et al. (56) documented as well a reduced accumulation of fluconazole in another azole-resistant strain as one of the causes of azole resistance. Reduced accumulation of fluconazole in these isolates might have been due to the effect of CgCDR1 upregulation. Other mechanisms of azole resistance have been reported in C. glabrata: Marichal et al. (31) measured an eightfold increase in ERG11 mRNA in an azole-resistant C. glabrata isolate, paralleled by a fourfold increase in gene copy number. Alterations in the ergosterol biosynthesis pathway have not yet been detected in C. glabrata clinical strains. However, laboratory strains with altered sterol profiles have been constructed by deletion of the ERG3 and ERG11 genes (17). Only the simultaneous deletion of both genes yielded mutants with azole and amphotericin B resistance, the latter being consistent with the absence of ergosterol in these cells. Thus, the pathway of azole resistance involving ergosterol biosynthesis may be utilized in clinical strains, but requires multiple gene alterations.
As mentioned above, another ABC transporter gene from C. glabrata called PDH1 has recently been isolated (32). PDH1, which is distinct from CgCDR1, was cloned by using PCR primers matching a consensus in conserved elements among ABC transporters and not by functional complementation as performed in this study. The function of PDH1 as a multidrug transporter has not yet been demonstrated, as opposed to CgCDR1, which is able to confer resistance to multiple drugs when expressed in S. cerevisiae (Fig. 3B). In the report of Miyazaki et al. (32), PDH1 expression was increased between azole-susceptible and azole-resistant C. glabrata isolates, as is the case here for CgCDR1. What is the relevance of both ABC transporter genes for the development of azole resistance in clinical C. glabrata isolates? In the light of our results, since the deletion of CgCDR1 in DSY565 rendered the mutant strain as susceptible to azoles as the susceptible parent isolate (DSY562), it indicates that PDH1 should play a moderate role in the azole resistance of this isolate. This hypothesis is further validated by Northern blot analysis, in which PDH1 mRNA signals were absent in total RNA from DSY562 while present in DSY565, but with a much reduced intensity compared to CgCDR1 mRNA signals (46). PDH1 may, however, be expressed at higher levels in other not-yet-investigated C. glabrata azole-resistant strains, a possibility which remains to be tested. The presence of multiple ABC transporter genes being coordinately upregulated is a feature known in other azole-resistant yeasts, such as C. albicans (50). CDR1 and CDR2, which are two ABC transporter genes with high similarity, are upregulated in azole-resistant isolates, while only CDR1 expression is detected in azole-susceptible strains, thus paralleling the properties of CgCDR1 and PDH1.
The rescue of the azole resistance phenotype by CgCDR1 in the ABC transporter mutant DSY1041 (Fig. 10) is an important result which will aid the further molecular characterization of the azole resistance mechanism in C. glabrata. By identifying trans-acting factors rather than cis-acting factors implicated in the upregulation of CgCDR1 in the azole-resistant isolate DSY565, the role of transcription factors regulating the expression of CgCDR1 becomes more relevant. The presence of putative Pdr1p and Pdr3p binding sites in the promoter region of CgCDR1 suggests strongly that these types of transcription factors might operate in C. glabrata. Interestingly, these binding sites have also been recognized in the promoter region of PDH1 (32). The function of Pdr1p and Pdr3p in S. cerevisiae has been well investigated for the regulation of the ABC transporter gene PDR5. The absence of Pdr1p and Pdr3p binding sites in the promoter of PDR5 decreases the expression of this gene to almost undetectable levels (23). PDR1 and PDR3 gain-of-function alleles have been reported in S. cerevisiae to confer hyperresistance to multiple drugs by increasing the transcription of several target genes, among which is the ABC transporter gene PDR5 (9, 36). A similar situation may occur in C. glabrata. PDR1 and PDR3 homologues are likely to be present in C. glabrata, and gain-of-function alleles of these genes could activate in trans the expression of CgCDR1 or PDH1 in azole-resistant strains.
In summary, this study demonstrates for the first time that azole resistance in clinical C. glabrata isolates is mediated by the upregulation of an ABC transporter gene. It is likely that not-yet-characterized multidrug efflux transporters may participate in this process, a hypothesis currently being addressed in our laboratory. A study of the molecular mechanisms of resistance to azoles in C. glabrata is attractive not only because this yeast has become an important human pathogen in recent years but also because recently developed genetic tools allow the testing of different working hypotheses.
ACKNOWLEDGMENTS
We thank K. Kitada (Nipon Roche Ltd.) and D. Thiele for the gift of plasmids and strains, D. Kaufmann and C. Durussel for assistance with the medical records of patients and management of the yeast collection database at the CHUV, and M. Monod for critical reading of the manuscript.
This work was supported by a grant from the Swiss Research National Foundation (no. 3100-045716) to D.S.
REFERENCES
- 1.Abisaid D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivarian S. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin Infect Dis. 1997;24:1122–1128. doi: 10.1086/513663. [DOI] [PubMed] [Google Scholar]
- 2.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arias A, Arevalo M P, Andreu A, Rodriguez C, Sierra A. Candida glabrata—in vitro susceptibility of 84 isolates to eight antifungal agents. Chemotherapy. 1996;42:107–111. doi: 10.1159/000239429. [DOI] [PubMed] [Google Scholar]
- 5.Arif S, Barkham T, Power E G, Howell S A. Techniques for investigation of an apparent outbreak of infections with Candida glabrata. J Clin Microbiol. 1996;34:2205–2209. doi: 10.1128/jcm.34.9.2205-2209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barchiesi F, Arzeni D, Delprete M S, Sinicco A, Difrancesco L F, Pasticci M B, Lamura L, Nuzzo M M, Burzacchini F, Coppola S, Chiodo F, Scalise G. Fluconazole susceptibility and strain variation of Candida albicans isolates from HIV-infected patients with oropharyngeal candidosis. J Antimicrob Chemother. 1998;41:541–548. doi: 10.1093/jac/41.5.541. [DOI] [PubMed] [Google Scholar]
- 7.Berrouane Y F, Herwaldt L A, Pfaller M A. Trends in antifungal use and epidemiology of nosocomial yeast infections in a university hospital. J Clin Microbiol. 1999;37:531–537. doi: 10.1128/jcm.37.3.531-537.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bissinger P H, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 9.Carvajal E, Van den Hazel H B, Cybularz-Kolaczkowska A, Balzi E, Goffeau A. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol Gen Genet. 1997;256:406–415. doi: 10.1007/s004380050584. [DOI] [PubMed] [Google Scholar]
- 10.Chambers S P, Prior S E, Barstow D A, Minton N P. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene. 1988;68:139–149. doi: 10.1016/0378-1119(88)90606-3. [DOI] [PubMed] [Google Scholar]
- 11.Cormican M G, Hollis R J, Pfaller M A. DNA macrorestriction profiles and antifungal susceptibility of Candida (Torulopsis) glabrata. Diagn Microbiol Infect Dis. 1996;25:83–87. doi: 10.1016/s0732-8893(96)00083-1. [DOI] [PubMed] [Google Scholar]
- 12.Decottignies A, Grant A M, Nichols J W, de Wet H, McIntosh D B, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J Biol Chem. 1998;273:12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]
- 13.Egner R, Rosenthal F E, Kralli A, Sanglard D, Kuchler K. Genetic separation of FK506 susceptibility and drug transport in the yeast PDR5 ATP-binding cassette multidrug resistance transporter. Mol Biol Cell. 1998;9:523–543. doi: 10.1091/mbc.9.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg A, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 15.Fidel P L, Jr, Vazquez J A, Sobel J D. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin Microbiol Rev. 1999;12:80–96. doi: 10.1128/cmr.12.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortun J, Lopez-San Roman A, Velasco J J, Sanchez-Sousa A, Devicente E, Nuno J, Quereda C, Barcena R, Monge G, Candela A, Honrubia A, Guerrero A. Selection of Candida glabrata strains with reduced susceptibility to azoles in four liver transplant patients with invasive candidiasis. Eur J Clin Microbiol Infect Dis. 1997;16:314–318. doi: 10.1007/BF01695638. [DOI] [PubMed] [Google Scholar]
- 17.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J E. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gleason T G, May A K, Caparelli D, Farr B M, Sawyer R G. Emerging evidence of selection of fluconazole-tolerant fungi in surgical intensive care units. Arch Surg. 1997;132:1197–1201. doi: 10.1001/archsurg.1997.01430350047008. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. A practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–135. [Google Scholar]
- 20.Hitchcock C A, Pye G W, Troke P F, Johnson E M, Warnock D W. Fluconazole resistance in Candida glabrata. Antimicrob Agents Chemother. 1993;37:1962–1965. doi: 10.1128/aac.37.9.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson E M, Warnock D W, Luker J, Porter S R, Scully C. Emergence of azole drug resistance in Candida species from HIV-infected patients receiving prolonged fluconazole therapy for oral candidosis. J Antimicrob Chemother. 1995;35:103–114. doi: 10.1093/jac/35.1.103. [DOI] [PubMed] [Google Scholar]
- 22.Kaben U, Blaschke-Hellmessen R, Hellwig S. Persistence and variability of yeast isolations from hospitalized patients—a comparison of results from Rostock and Dresden, Germany. Mycoses. 1997;40:421–423. doi: 10.1111/j.1439-0507.1997.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 23.Katzmann D J, Hallstrom T C, Mahe Y, Moye-Rowley W S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 24.Kelly S L, Lamb D C, Kelly D E, Loeffler J, Einsele H. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet. 1996;348:1523–1524. doi: 10.1016/S0140-6736(05)65949-1. [DOI] [PubMed] [Google Scholar]
- 25.Kitada K, Yamaguchi E, Arisawa M. Isolation of a Candida glabrata centromere and its use in construction of plasmid vectors. Gene. 1996;175:105–108. doi: 10.1016/0378-1119(96)00132-1. [DOI] [PubMed] [Google Scholar]
- 26.Knoke M, Schulz K, Bernhardt H. Dynamics of Candida isolations from humans from 1992–1995 in Greifswald, Germany. Mycoses. 1997;40:105–110. doi: 10.1111/j.1439-0507.1997.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 27.Lamb D C, Corran A, Baldwin B C, Kwon-Chung J, Kelly S L. Resistant P45051A1 activity in azole antifungal tolerant Cryptococcus neoformans from AIDS patients. FEBS Lett. 1995;368:326–330. doi: 10.1016/0014-5793(95)00684-2. [DOI] [PubMed] [Google Scholar]
- 28.Law D, Moore C B, Joseph L A, Keaney M, Denning D W. High incidence of antifungal drug resistance in Candida tropicalis. Int J Antimicrob Agents. 1996;7:241–245. doi: 10.1016/s0924-8579(96)00328-7. [DOI] [PubMed] [Google Scholar]
- 29.Lockhart S R, Joly S, Pujol C, Sobel J D, Pfaller M A, Soll D R. Development and verification of fingerprinting probes for Candida glabrata. Microbiology. 1997;143:3733–3746. doi: 10.1099/00221287-143-12-3733. [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Ribot J L, McAtee R K, Lee L N, Kirkpatrick W R, White T C, Sanglard D, Patterson T F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marichal P, Vanden Bossche H, Odds F C, Nobels G, Warnock D W, Timmerman V, Vanbroeckhoven C, Fay S, Mosel-Larsen P. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyazaki H, Miyazaki Y, Geber A, Parkinson T, Hitchcock C, Falconer D J, Ward D J, Marsden K, Bennett J E. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob Agents Chemother. 1998;42:1695–1701. doi: 10.1128/aac.42.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Vol. 12. 1995. Tentative standard M27-A, National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 34.Nguyen M H, Peacock J E, Morris A J, Tanner D C, Nguyen M L, Snydman D R, Wagener M M, Rinaldi M G, Yu V L. The changing face of candidemia—emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 35.Nolte F S, Parkinson T, Falconer D J, Dix S, Williams J, Gilmore C, Geller R, Wingard J R. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob Agents Chemother. 1997;41:196–199. doi: 10.1128/aac.41.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nourani A, Papajova D, Delahodde A, Jacq C, Subik J. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol Gen Genet. 1997;256:397–405. doi: 10.1007/s004380050583. [DOI] [PubMed] [Google Scholar]
- 37.Odds F C. Resistance of clinically important yeasts to antifungal agents. Int J Antimicrob Agents. 1996;6:145–147. doi: 10.1016/0924-8579(95)00048-8. [DOI] [PubMed] [Google Scholar]
- 38.Parkinson T, Falconer D J, Hitchcock C A. Fluconazole resistance due to energy-dependent drug efflux in Candida glabrata. Antimicrob Agents Chemother. 1995;39:1696–1699. doi: 10.1128/aac.39.8.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaller M A, Jones R N, Messer S A, Edmond M B, Wenzel R P. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans—frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn Microbiol Infect Dis. 1998;30:121–129. doi: 10.1016/s0732-8893(97)00240-x. [DOI] [PubMed] [Google Scholar]
- 40.Pfaller M A, Messer S A, Houston A, Rangelfrausto M S, Wiblin T, Blumberg H M, Edwards J E, Jarvis W, Martin M A, Neu H C, Saiman L, Patterson J E, Dibb J C, Roldan C M, Rinaldi M G, Wenzel R P. National epidemiology of mycoses survey—a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn Microbiol Infect Dis. 1998;31:289–296. doi: 10.1016/s0732-8893(97)00245-9. [DOI] [PubMed] [Google Scholar]
- 41.Redding S, Smith J, Farinacci G, Rinaldi M, Fothergill A, Rhinechalberg J, Pfaller M. Resistance of Candida albicans to fluconazole during treatment of oropharyngeal candidiasis in a patient with AIDS—documentation by in vitro susceptibility testing and DNA subtype analysis. Clin Infect Dis. 1994;18:240–242. doi: 10.1093/clinids/18.2.240. [DOI] [PubMed] [Google Scholar]
- 42.Rex J H, Cooper C R, Jr, Merz W G, Galgiani J N, Anaissie E J. Detection of amphotericin B-resistant Candida isolates in a broth-based system. Antimicrob Agents Chemother. 1995;39:906–909. doi: 10.1128/aac.39.4.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rex J H, Pfaller M A, Barry A L, Nelson P W, Webb C D. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of nonneutropenic patients with candidemia. Antimicrob Agents Chemother. 1995;39:40–44. doi: 10.1128/aac.39.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Barry A L. Development of interpretive breakpoints for antifungal susceptibility testing—conceptual framework and analysis of in vitro in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 45.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanglard, D. Unpublished data.
- 47.Sanglard, D., and D. Calabrese. Unpublished data.
- 48.Sanglard D, Ischer F, Calabrese D, De Micheli M, Bille J. Multiple resistance mechanisms to azole antifungals in yeast clinical isolates. Drug Resist Updates. 1998;1:255–265. doi: 10.1016/s1368-7646(98)80006-x. [DOI] [PubMed] [Google Scholar]
- 49.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents—characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 51.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanglard D, Ischer F, Bille J. Abstracts of the 37th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1997. Isolation of Candida glabrata genes conferring resistance to azole antifungals and their involvement in the azole resistance of clinical isolates, abstr. C-10; p. 47. [Google Scholar]
- 53.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Troillet N, Durussel C, Bille J, Glauser M P, Chave J P. Correlation between in vitro susceptibility of Candida albicans and fluconazole-resistant oropharyngeal candidiasis in HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1993;12:911–915. doi: 10.1007/BF01992164. [DOI] [PubMed] [Google Scholar]
- 55.Vanden Bossche H, Dromer F, Improvisi I, Lozanochiu M, Rex J H, Sanglard D. Antifungal drug resistance in pathogenic fungi. Med Mycol. 1998;36:119–128. [PubMed] [Google Scholar]
- 56.Vanden Bossche H, Marichal P, Odds F C, Le Jeune L, Coene M-C. Characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1992;36:2602–2610. doi: 10.1128/aac.36.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkateswarlu K, Denning D W, Kelly S L. Inhibition and interaction of cytochrome P450 of Candida krusei with azole antifungal drugs. J Med Vet Mycol. 1997;35:19–25. [PubMed] [Google Scholar]
- 58.Venkateswarlu K, Denning D W, Manning N J, Kelly S L. Reduced accumulation of drug in Candida krusei accounts for itraconazole resistance. Antimicrob Agents Chemother. 1996;40:2443–2446. doi: 10.1128/aac.40.11.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vennewald I, Seebacher C, Roitzsch E. Post-mortem findings in patients with repeatedly mycological demonstration of Candida glabrata. Mycoses. 1998;41:125–132. doi: 10.1111/j.1439-0507.1998.tb00313.x. [DOI] [PubMed] [Google Scholar]
- 60.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou P B, Szczypka M S, Young R, Thiele D J. A system for gene cloning and manipulation in the yeast Candida glabrata. Gene. 1994;142:135–140. doi: 10.1016/0378-1119(94)90368-9. [DOI] [PubMed] [Google Scholar]