Abstract
All known DNA ligases catalyze the formation of a phosphodiester linkage between adjacent termini in double-stranded DNA via very similar mechanisms. The ligase family can, however, be divided into two classes: eubacterial ligases, which require NAD+ as a cofactor, and other ligases, from viruses, archaea, and eukaryotes, which use ATP. Drugs that discriminate between DNA ligases from different sources may have antieubacterial activity. We now report that a group of arylamino compounds, including some commonly used antimalarial and anti-inflammatory drugs and a novel series of bisquinoline compounds, are specific inhibitors of eubacterial DNA ligases. Members of this group of inhibitors have different heterocyclic ring systems with a common amino side chain in which the two nitrogens are separated by four carbon atoms. The potency, but not the specificity of action, is influenced by the DNA-binding characteristics of the inhibitor, and the inhibition is noncompetitive with respect to NAD+. The arylamino compounds appear to target eubacterial DNA ligase in vivo, since a Salmonella Lig− strain that has been rescued with the ATP-dependent T4 DNA ligase is less sensitive than the parental Salmonella strain.
DNA ligation is an important step in a number of cellular processes, including replication, recombination, and repair of damaged DNA (8, 14, 15). As a consequence, DNA ligase is an essential enzyme in every organism. Its key role in living organisms is evidenced by the facts that eukaryotic cells encode up to five isoenzymes (40) and that many viruses encode their own DNA ligase (13, 28). Studies of conditionally lethal and null mutants of DNA ligase genes (2, 8, 22) indicate that the single DNA ligase of bacteria is an essential enzyme. In eukaryotic systems, disruption of LIG1, which encodes the major DNA ligase of proliferating cells, prevents normal cell growth and differentiation (1, 23).
All of the DNA ligases studied to date catalyze the joining of ends of nicked double-stranded DNA by similar processes that involve adenylation of a critical lysine residue within the active site (14, 15). The ligases can be divided, however, into two separate families. In the highly conserved eubacterial family of DNA ligases, NAD+ functions as the adenyl donor (27, 35, 38, 44), while in the much more variable family of ligases from viruses, archaea, and eukaryotes (13, 28, 41, 42), the adenyl moiety is donated by ATP. One exception to this rule is Haemophilus influenzae, which appears to make use of both NAD+-dependent and ATP-dependent ligases (2).
Amino acid sequence comparisons suggest that the NAD+-dependent DNA ligases are phylogenetically unrelated to the ATP-dependent DNA ligases (13). Only one motif (K-X-D-G) is shared by the two families of enzymes (13, 31), and the presence of this motif may reflect convergent rather than divergent evolution (13). The ATP-dependent ligases are more varied in sequence but nevertheless appear to belong to a nucleotidyltransferase superfamily (29). Enzymes within this superfamily are characterized by a set of six short motifs containing conserved residues that are critical for covalent nucleotide transfer (9, 34).
The structure of an ATP-dependent DNA ligase has been reported (34), and very recently the structure of the NAD+-binding domain of a bacterial DNA ligase has also been determined (30). The crystallographic data indicate that while there is some structural similarity between the two families of enzymes, the nature of some of the residues lining the active cleft and the spatial organizations of the two domains of the ligases may differ significantly. The differences between the two families of enzymes and the high degree of conservation of the eubacterial DNA ligases (2, 14) have led to the suggestion that the NAD+-dependent ligase may be a potential target for new bactericidal drugs (30).
A number of effective inhibitors of DNA ligases have previously been described (3, 19, 20), but none of them appears to be capable of discriminating between DNA ligases from different sources (21). We now report the identification of a series of compounds that are the first reported specific inhibitors of Escherichia coli DNA ligase. We also demonstrate that the endogenous eubacterial DNA ligase can be targeted in vivo by some of these compounds, thereby providing the first practical demonstration that DNA ligase inhibitors do indeed represent a novel class of agents with antimicrobial activity.
MATERIALS AND METHODS
Cells and chemicals.
The LT2 wild-type strain of Salmonella typhimurium (22), its DNA ligase-minus (null) derivative which was constructed in the presence of the T4 Lig+ plasmid pBR313/598/8/1b (TT15151 [43]), the conditional lethal (8) mutant strain of E. coli lig-7(ts) supF (TT6508), and its derivative E. coli lig-7(ts) supF (UAG, Ty8) pBR313/598/8/1b (TT8352), harboring the gene for T4 DNA ligase (T4 Lig+ [43]), were all kindly provided by J. R. Roth (University of Utah, Salt Lake City). Doxorubicin and hydroxychloroquine were gifts from Farmitalia-Carlo Erba (Milan, Italy) and Sanofi Winthrop (Newcastle upon Tyne, United Kingdom), respectively. The 2-amino-5-diethylaminopentane was purchased from Aldrich, Castle Hill, Australia. [3H]dTTP was obtained from Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom. Mefloquine was kindly donated by Hoffmann-La Roche, Basel, Switzerland. N,N′- bis[4-((4-(Diethylamino)-1-methylbutyl)amino)-quinolin-6-yl]sebacamide(6bisQ8) and other bisquinolines were synthesized as described previously (25). T4 DNA ligase was purchased from Sigma Chemical Co., St. Louis, Mo. E. coli DNA ligase and exonuclease III were from Boehringer Mannheim, Castle Hill, Australia. Human recombinant DNA ligase I was purified as described elsewhere (7). Due to the difficulty of isolating large quantities of active human DNA ligase, the more extensive studies of inhibition of ATP-dependent ligase were performed with the T4 DNA ligase. Nutrient broth and nutrient agar were supplied by Oxoid Pty. Ltd., West Heidelberg, Australia. All other drugs and chemicals were from Sigma Chemical Co.
DNA ligase assays.
DNA joining activity was measured by determining the extent to which nicked [3H] poly(dA-dT) became resistant to degradation by exonuclease III, using a modification of the method of Modrich and Lehman (18). The E. coli DNA ligase reaction mixture (50 μl) contained 30 mM Tris-HCl (pH 8.0), 4 mM MgCl2, 1.2 mM EDTA, 1 mM dithioerythritol, 0.026 mM NAD, 50 μg of bovine serum albumin/ml, and 2 nmol of [3H]poly(dA-dT) (ca. 60 cpm/pmol). The bacteriophage T4 DNA ligase and human recombinant DNA ligase I reaction mixtures (50 μl) contained 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2 1 mM dithioerythritol, 1 mM ATP, and 2 nmol of [3H]poly(dA-dT) (ca. 60 cpm/pmol). Upon addition of enzyme, the reaction mixture was incubated for 30 min at 37°C, then boiled for 3 min. After the reaction mixture was chilled, 50 μl of a solution containing 80 mM Tris-HCl (pH 8.0), 5 mM dithioerythritol, 30 μg of bovine serum albumin/ml, and 15 U of exonuclease III was added. After incubation for 30 min at 37°C, 80-μl samples were spotted onto Whatman GF/C filters and the filters were batch washed three times with 5% (wt/vol) trichloroacetic acid, two times with ethanol, and once with acetone. At the concentrations used in the present work, none of the test compounds interfered with the action of the exonuclease III or caused breakdown of the ligated polymer upon boiling. For experiments examining the effect of NAD+ on the inhibition of DNA ligase activity by chloroquine (CQ), NAD+ (0.026 to 1 mM) was included in the E. coli DNA ligase reaction mixture.
Drug-DNA interactions.
The interaction of CQ (50 μM) with DNA (100 μM) in 30 mM Tris-HCl, pH 8.0, was determined by monitoring the absorbance of CQ at 343 nm with a Shimadzu spectrophotometer as described by Cohen and Yielding (4).
Growth inhibition assays.
For studies of inhibition of growth in liquid medium, log-phase cultures of S. typhimurium were incubated at 37°C in the presence of CQ as suspensions in either saline (0.85% NaCl) or nutrient broth. After treatment, cell cultures were diluted appropriately and plated on nutrient agar. The resulting colonies were counted after overnight incubation of the plates at 37°C. For studies of growth inhibition on solid medium, log-phase cell cultures were deposited as small drops (10 μl of the appropriate dilution) on nutrient agar plates supplemented with the required amount of CQ and the plates were incubated overnight at either 37°C (in the case of the S. typhimurium strains) or 30°C (in the case of the temperature-sensitive E. coli strains).
RESULTS
Doxorubicin inhibits ATP-dependent and NAD+-dependent DNA ligases with similar efficiencies.
The antitumor drug doxorubicin has previously been shown to be a potent inhibitor of ATP-dependent DNA ligases (19). Using a slightly modified assay of poly(dA-dT) joining activity, we confirmed that doxorubicin inhibits the ATP-dependent DNA ligase of bacteriophage T4 and showed that it inhibits the NAD+-dependent DNA ligase of E. coli with a similar potency (Table 1).
TABLE 1.
Drug concentrations required to inhibit the activities of DNA ligases from E. coli, T4 bacteriophage, and humans (type I) by 50%, and specificity factors
| Druga | IC50 (μM) for:
|
SFa | ||
|---|---|---|---|---|
| E. coli ligase | T4 ligase | Human ligase I | ||
| Doxorubicin | 1.3 ± 0.3 | 2 ± 0.4 | 1.8 ± 0.4 | 1.5 |
| CQ | 53 ± 3 | >1,500 | 720 ± 80 | >28 |
| Hydroxychloroquine | 63 ± 6 | >2,000 | NDb | >31 |
| Quinacrine | 1.5 ± 0.2 | 23 ± 5 | ND | 16 |
| 6bisQ0 | 2.6 ± 0.4 | 14 ± 2 | 16 ± 2 | 5.4 |
| 6bisQ6 | 2.2 ± 0.3 | 18.2 ± 2 | 14.8 ± 3 | 8.3 |
| 6bisQ8 | 1.8 ± 0.2 | 16.8 ± 2 | 12.8 ± 2 | 9.3 |
| 8bisQ0 | 10.2 ± 2 | 33 ± 3 | 49 ± 9 | 3.2 |
| 8bisQ4 | 9 ± 2 | 44 ± 5 | 43 ± 7 | 4.9 |
| N,N′-bis[4-Chloroquinolin-8-yl]succinamide | >2,000 | >2,000 | >2,000 | ND |
| Quinine | >2,000 | >2,000 | >2,000 | ND |
| Cinchonidine | >5,000 | >5,000 | >5,000 | ND |
| 2-Amino-5-diethylaminopentane | 2,100 ± 180 | >16,000 | ND | >7.6 |
| Spermidine | 5,000 ± 580 | 23,000 ± 3,800 | ND | 4.6 |
| Putrescine | 9,200 ± 950 | >20,000 | ND | >2.2 |
SF, specificity factor (ratio of IC50s of drug for T4 and E. coli DNA ligases).
ND, not determined.
Arylamino compounds are specific inhibitors of E. coli DNA ligase.
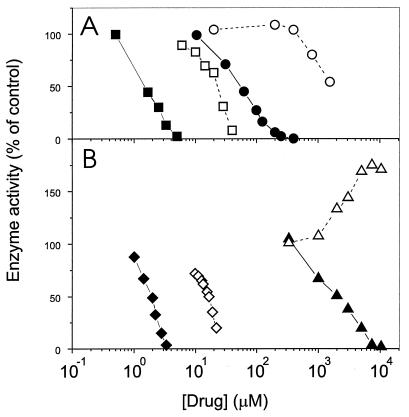
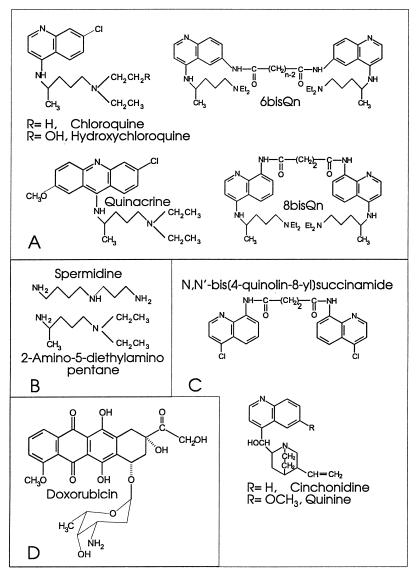
In the course of a search for specific inhibitors, we have identified several representative arylamino compounds that inhibit NAD+-dependent ligases preferentially (Fig. 1 and Table 1). CQ, a known antimalarial and DNA-binding agent (4, 26), is a much more effective inhibitor of the NAD+-dependent DNA ligase of E. coli than of either the bacteriophage T4 DNA ligase or the human recombinant (7) DNA ligase I (Fig. 1A and Table 1). The specificity factor, i.e., the ratio of the 50% inhibitory concentration (IC50) for the T4 ligase to the IC50 for the E. coli ligase, is >28. The closely related quinoline derivative hydroxychloroquine, which is widely utilized as a nonsteroidal anti-inflammatory drug (17), was also shown to be a specific inhibitor of the E. coli DNA ligase (Table 1). The quinoline ring structure does not seem to be necessary for activity, since quinacrine, a 9-aminoacridine that binds to DNA even more tightly than CQ (4, 26), was also a more potent inhibitor of the E. coli DNA ligase (Fig. 1A and Table 1). Members of a series of novel bisquinoline compounds (25) were also examined. 6bisQ8, which has an eight-carbon linker (25), showed good activity against the NAD+-dependent E. coli ligase while retaining reasonable specificity of action (Fig. 1B and Table 1). Two other members of this 6-linked bisquinoline series, for which the number of carbons in the linker was either zero or six, showed very similar inhibitory characteristics. A member of a related 8-linked bisquinoline series, N,N′- bis[4-((4-(diethylamino)-1-methylbutyl)amino)-quinoline-8- yl]adipamide (8bisQ4 [25]) (Fig. 2A), also showed good activity and some specificity (Table 1). Thus, we have identified a number of arylamino compounds that are efficient inhibitors of a bacterial DNA ligase but are less active against ATP-dependent ligases from viral and human sources.
FIG. 1.
Inhibition of DNA joining activity of DNA ligases. The circularization of [3H]poly(dA-dT) substrate by the DNA ligase of E. coli (NAD+ dependent) (filled symbols) or the DNA ligase of the T4 bacteriophage (ATP dependent) (open symbols) was monitored in the presence of increasing concentrations of quinacrine (squares) or CQ (circles) (A) and of 6bisQ8 (diamonds) or 2-amino-5-diethylaminopentane (triangles) (B). Data represent the averages of values determined in at least two separate experiments.
FIG. 2.
Structures of some compounds examined in this work. (A) Compounds showing potent and specific activity against eubacterial DNA ligase. (B) Simple diamino compounds that retain specific activity against E. coli DNA ligase. (C) Related aminoquinoline compounds that lack specific inhibitory activity against eubacterial DNA ligase. (D) Doxorubicin, a previously identified inhibitor of DNA ligases which lacks specificity against E. coli DNA ligase.
A common structural feature of the compounds that display specificity of action is the presence of an amino side chain (with four carbons separating the nitrogens) attached to a heterocyclic ring system (Fig. 2A). Quinoline compounds that lack the diamino side chain (i.e., quinine, cinchonidine, and a bisquinoline compound with no side chain, N,N′-bis[4-chloroquinolin-8-yl]succinamide [Fig. 2C]) showed no measurable inhibitory activity against either type of DNA ligase (Table 1). We have further shown that the free side chain of CQ, 2-amino-5-diethylaminopentane (Fig. 2B), is sufficient to inhibit the activity of E. coli DNA ligase, albeit at very much higher concentrations. By contrast, 2-amino-5-diethylaminopentane enhanced the activity of the ATP-dependent T4 DNA ligase at concentrations above 2 mM (Fig. 1B). Similarly, putrescine (Fig. 2B), the simplest molecule with a NCCCCN structure, preferentially inhibited E. coli DNA ligase, with no significant inhibition of the T4 ligase being observed at the highest concentration examined (Table 1). Stimulation of ATP-dependent DNA ligases by polyamines has been described previously (24, 36). These data indicate that a structure with four carbon atoms separating two nitrogens is the minimum inhibitory element and that this particular inhibitory moiety can be presented on a variety of scaffolds without losing its capacity for specific inhibition of the eubacterial ligase.
To determine whether inhibition of the E. coli DNA ligase by the range of arylamino compounds under investigation involves a direct interaction with the NAD+ recognition site of the enzyme, we examined the effect of varying the concentration of NAD+ from 0.026 to 1 mM. Using CQ as an inhibitory compound, we found that NAD+ did not relieve the inhibition of the enzyme. For example, 100 μM CQ was associated with 76% ± 5% and 74% ± 4% (means ± standard deviations) inhibition of the E. coli DNA ligase activity in the presence and absence of 1 mM NAD+, respectively. This indicates that the inhibitory effects are unlikely to involve direct binding of the arylamino compounds to the cofactor recognition site of the enzyme.
Effect of protonation state on inhibition of DNA ligases.
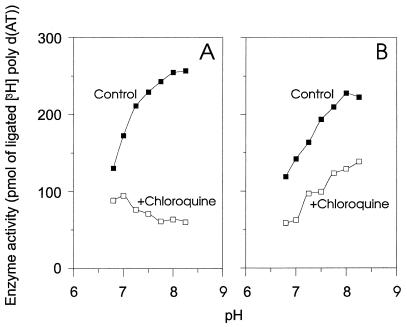
To obtain additional information about the mechanism(s) of inhibition of the DNA ligases by different compounds, we analyzed the inhibitory effects of CQ at different pH values. Chloroquine is a diprotic weak base (pKa1 = 8.1, pKa2 = 10.2), and its protonation state is readily modulated by altering the pH (11). In agreement with a previous report (33), we found that the activities of both the NAD+-dependent and ATP-dependent DNA ligases increased as the pH increased (Fig. 3). For the ATP-dependent ligase, the residual activity (in the presence of 800 μM CQ) also increased with increasing pH (Fig. 3B). This suggests that the degree of protonation of CQ has little effect on the inhibition of the ATP-dependent ligase. By contrast, the residual activity of the NAD+-dependent ligase (in the presence of 110 μM CQ) decreased as the pH was increased from 6.2 to 8.2 (Fig. 3A). The ratio of the activities in the presence and absence of CQ increased from 1.5 to 4.3 (r = 0.972, P = 0.0002). Hence, monoprotonated CQ may be a significantly more effective inhibitor of the NAD+-dependent ligase than diprotonated CQ. These findings suggest that the effect of CQ on the NAD+-dependent ligase involves a mechanism which is quite different from that which is responsible for its weaker effect on the ATP-dependent ligases.
FIG. 3.
Effect of pH on the inhibition of NAD+- and ATP-dependent DNA ligases by chloroquine. (A) The DNA joining activity of E. coli DNA ligase was monitored as a function of pH in the absence of CQ (filled symbols) or in the presence of 110 μM CQ (open symbols). (B) The DNA joining activity of human recombinant (rec.) DNA ligase I was monitored as a function of pH in the absence of CQ (filled symbols) or in the presence of 800 μM CQ (open symbols). poly d(AT), poly(dA-dT).
The DNA-binding properties of the arylamino compounds affect the potency but not the specificity of inhibition of NAD+-dependent DNA ligase.
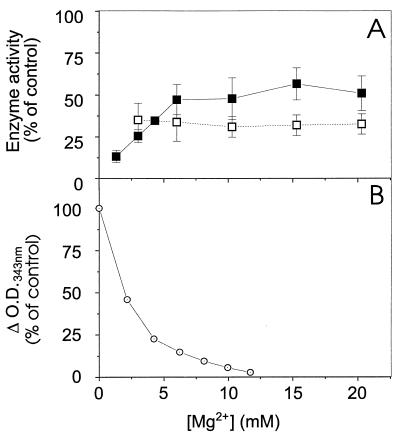
Quinoline antimalarial agents (4, 26) and polyamines (16) have previously been shown to interact with DNA. Moreover, quinacrine, which is a more potent DNA-binding agent than CQ (4, 26), is also a much more potent inhibitor of the E. coli DNA ligase than is CQ (Fig. 1A). This could mean that these inhibitors of DNA ligases exert their activity via an interaction with DNA. To investigate this possibility further, we examined the effect of Mg2+ concentration on the inhibitory effect of CQ. In agreement with a previous report (4), we found that CQ binds maximally to DNA in the absence of divalent cations and shows little binding to DNA at Mg2+ concentrations above 10 mM (Fig. 4B). Indeed, CQ has previously been shown to have little effect on DNA polymerase at Mg2+ concentrations above 2 mM (5). We examined the effect of different Mg2+ concentrations on the activities of the NAD+- and ATP-dependent ligases in the presence of a CQ concentration which gave about 70% inhibition under standard assay conditions (Fig. 4A). For the E. coli DNA ligase, the inhibitory effect of CQ (at 0.1 mM) was weakened at higher Mg2+ concentrations; however, CQ was still partially effective as an inhibitor even at 20 mM Mg2+ (Fig. 4A). The divalent-cation concentration had no effect on the inhibition of the T4 ligase by 2 mM CQ (Fig. 4A). These data provide further support for the suggestion that the molecular basis of the activity of CQ against the NAD+-dependent ligase probably is different from that of its (weaker) effect against the ATP-dependent ligases. The data also indicate that while the ability of CQ and other arylamino inhibitors to bind DNA does contribute to their potency against the NAD+-dependent ligase, DNA binding is not a critical determinant of the specificity of action.
FIG. 4.
Effect of Mg2+ concentration on inhibition of NAD+- and ATP-dependent DNA ligases by chloroquine. (A) The DNA joining activities of E. coli DNA ligase in the presence of 100 μM CQ (filled squares) and of T4 DNA ligase in the presence of 2 mM CQ (open squares) were monitored as a function of increasing Mg2+ concentration. The enzyme activities are presented as percentages of activity in the absence of CQ at the same Mg2+ concentration. (B) The interaction of CQ (50 μM) with DNA (100 μM) was determined by monitoring the change in absorbance of CQ at 343 nm.
Arylamino compounds target eubacterial DNA ligase in vivo.
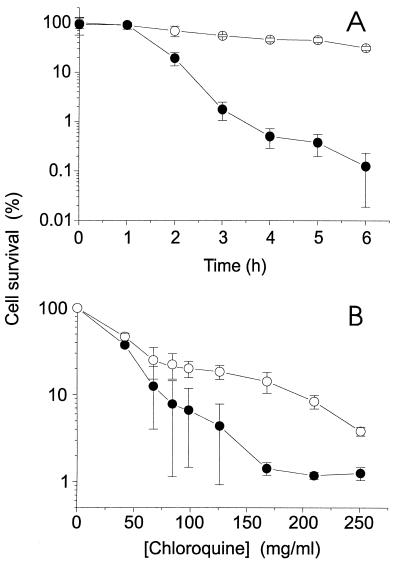
To determine whether these arylamino compounds might have potential as lead structures for novel eubactericidal agents, we examined the abilities of the arylamino compounds to inhibit the growth of bacteria. We used CQ for these studies since it showed the highest specificity factor in vitro. CQ was shown to exert both cytostatic and cytotoxic effects. When an aliquot of a log-phase culture of a wild-type S. typhimurium strain (LT2) was transferred to medium containing CQ, the colony-forming ability of the culture decreased with time (Fig. 5A). Cell survival of this wild-type strain of Salmonella was decreased to 50% of control levels by a CQ concentration of about 25 mg/ml (Fig. 5B). A cytostatic effect of CQ was observed when log-phase S. typhimurium LT2 cultures were plated on solid medium containing CQ (Fig. 6a and b, right sides of plates). At CQ concentrations above 15 mg/ml, the growth of the bacterial colonies was substantially inhibited, indicating that CQ may be able to target the DNA ligase in vivo. The concentration of CQ that was required to inhibit bacterial growth in vivo was very high compared with the IC50 of CQ for the E. coli DNA ligase (i.e., 53 ± 3 μM or 27 μg/ml [Fig. 1A]), presumably due to poor uptake of CQ by bacteria (32).
FIG. 5.
Cytotoxicity of chloroquine against S. typhimurium in liquid culture. (A) Log-phase wild-type S. typhimurium LT2 (filled circles) or S. typhimurium TT15151 (lig-2::Mu dJ/pBR313/598/8/1b [T4 Lig+] AMPr [43]) (open circles) was incubated at 37°C in the presence of chloroquine at 42 mg/ml. At the indicated time points, aliquots were removed and plated on nutrient agar after appropriate dilution. (B) Log-phase wild-type S. typhimurium LT2 and strain TT15151 cells were incubated at 37°C in the presence of increasing concentrations of CQ. After 1 h, aliquots were removed and plated on nutrient agar after appropriate dilution. The resulting colonies were counted after overnight incubation at 37°C. Data are mean values ± standard errors of the means. Student’s t-test analysis indicates a significant difference between the growth of the two strains of bacteria in the presence of 125 and 250 mg of CQ/ml, with P values of <0.01, <0.01, <0.001, and <0.02, respectively.
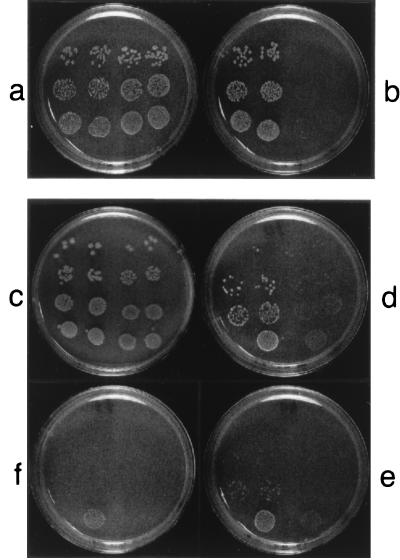
FIG. 6.
Chloroquine inhibition of bacterial growth on a solid medium. Log-phase wild-type S. typhimurium LT2 cells (right sides of plates) or S. typhimurium TT15151 (Lig−/T4 Lig+) cells (left sides of plates) were plated at dilution ratios of 10−3 (bottom rows), 10−4 (middle rows), and 10−5 (top rows) on nutrient agar containing no drug (a) or 15 mg of CQ/ml (b), and the plates were incubated overnight at 37°C. Log-phase cells of a conditional lethal mutant strain of E. coli, TT6508 [lig-7(ts) supF] (right sides of plates) or of its lig-7(ts) supF (UAG, Ty8) pBR313/598/8/1b derivative, E. coli TT8352, harboring the gene for T4 DNA ligase (T4 Lig+ [43]) (left sides of plates), were plated at dilution ratios of 5 × 10−3, 5 × 10−4, 5 × 10−5, and 5 × 10−6 (from bottom to top rows) on nutrient agar containing no drug (c), 30 mg of CQ/ml (d), 40 mg of CQ/ml (e), or 45 mg of CQ/ml (f), and the plates were incubated overnight at 30°C.
To investigate the specificity of action of CQ in vivo, we compared the sensitivity of the wild-type strain of Salmonella (LT2) with that of a DNA ligase-minus (null) derivative (TT15151) which had been “rescued” with a plasmid (pBR313/598/8/1b) encoding the T4 DNA ligase gene. The wild-type LT2 strain was significantly more sensitive to CQ than the mutant strain, which expresses only the ATP-dependent T4 DNA ligase (Fig. 5). Similarly, when the two Salmonella strains were plated on solid medium in the presence of CQ, wild-type bacteria expressing an NAD+-dependent ligase were found to be significantly more sensitive to CQ than bacteria expressing the ATP-dependent T4 DNA ligase (Fig. 6).
We also examined the sensitivity of the two Salmonella strains to the bisquinoline compound 6bisQ8, which shows specific activity against the E. coli DNA ligase. The wild-type Salmonella strain was sensitive to 6bisQ8 at 2.3 mg/ml, while the Lig−(T4 Lig+) partial diploid was resistant to 6bisQ8 at this concentration (data not shown). By contrast, the two Salmonella strains were equally sensitive to quinine and cinchonidine, which show no specificity of action against the NAD+-dependent DNA ligase, and the Lig−(T4 Lig+) strain was slightly more sensitive to doxorubicin (data not shown). The apparent resistance of the Lig−(T4 Lig+) strain to these arylamino compounds is not due to overexpression of the T4 ligase, since the wild-type and rescued strains have previously been shown to possess similar overall levels of DNA ligase activity (22). The difference in the sensitivities of the two bacterial strains to CQ is relatively modest, indicating that there are other targets for CQ in the bacteria. Further studies using more-readily permeating arylamino compounds would be useful to confirm the importance of DNA ligase as a target in vivo. Nonetheless, these studies indicate that, under the conditions of these experiments, CQ and similar compounds can inhibit bacterial growth by directly targeting the eubacterial DNA ligase.
To obtain further information about the role of DNA ligase as a target for CQ in vivo, we compared the sensitivity of the conditional lethal (8) mutant strain of E. coli lig-7(ts) supF, TT6508, with that of its E. coli lig-7(ts) supF (UAG, Ty8) pBR313/598/8/1b derivative, TT8352, harboring the gene for T4 DNA ligase (T4 Lig+ [43]). The parental E. coli lig-7(ts) strain TT6508 was sensitive to CQ when grown at the permissive temperature of 30°C on solid medium (Fig. 6c to f, right sides of plates). The T4 Lig+ strain TT8352, which expresses both a temperature-sensitive NAD+-dependent and an ATP-dependent DNA ligase, showed a somewhat increased level of resistance to CQ (Fig. 6c to f, left sides of plates). The less dramatic level of resistance to CQ of the E. coli strains harboring the T4 Lig+ plasmid [compared with the Lig−(T4 Lig+) Salmonella strain] may be due to the presence of the endogenous NAD+-dependent enzyme. This endogenous enzyme may compete for binding to substrate or for interaction with proteins of the replicative apparatus (22).
The enhanced resistance of the bacterial strains expressing an ATP-dependent DNA ligase strongly suggests that the NAD+-dependent DNA ligase of eubacteria is a target of the arylamino compounds in vivo.
DISCUSSION
Eubacteria possess an NAD+-dependent DNA ligase enzyme that is quite distinct from the ATP-dependent ligase enzymes employed by other organisms. The crystal structure of an ATP-dependent DNA ligase from bacteriophage T7 is available, having been solved at 2.6 Å resolution (34). The T7 enzyme contains a core structure conserved in all ATP-dependent ligases (28). The T7 DNA ligase has a markedly asymmetric structure, comprising two domains divided by a deep basic cleft, which presumably accommodates the DNA duplex. The ATP-binding site is located at the base of this cleft (34, 41). ATP binding appears to be stabilized by a number of interactions with the protein, in particular with Lys-34, which is positioned adjacent to the α-phosphate group of the bound ATP (34).
Recently, structural information relating to the NAD+-dependent ligase from Bacillus stearothermophilus has also become available (30, 39). This enzyme also appears to consist of two domains; however, these domains are more biochemically independent than the two domains of the ATP-dependent DNA ligase. The crystal structure of the N-terminal domain of B. stearothermophilus has been solved at 2.8 Å resolution (30). Despite the lack of sequence homology with ATP-dependent ligases, the central core of this domain shows significant structural homology with the equivalent region of the ATP-dependent enzyme, and some key residues in the adenylation domain are retained in equivalent positions. Nonetheless, there appear to be a number of structural differences, since the ATP-dependent enzyme needs to undergo a transition from an open to a closed conformation for adenylation to occur. This requires the participation of both the N-terminal and C-terminal domains. By contrast, the N-terminal domain of the NAD+-dependent enzyme can be efficiently adenylated in the absence of the C-terminal domain (30). Thus, the fine structures and topologies of the cofactor-binding cavities may be quite different. The arylamino compounds identified in this study presumably interfere specifically with the NAD+-dependent DNA ligase by interacting with residues that lie adjacent to the adenylation site, since our results indicate that CQ does not compete directly for binding to the NAD+-binding site.
We have shown that a diamino side chain (NCCCCN) is necessary for the specific inhibitory activity of arylamino compounds but that this moiety can be presented on either a quinoline or an acridine nucleus. The differential interactions of polyamines (24, 36) with the two families of DNA ligases may have similar molecular bases. Hydroxylation of the terminal tertiary amine of the diaminopentane side chain (as in hydroxychloroquine) did not appear to affect the potency of inhibition, but quinolines bearing amino alcohol side chains (e.g., quinine and cinchonidine) were extremely weak inhibitors. The DNA-binding ability of the ring system, to which the diamino side chain is attached, appears to enhance the potency of an inhibitor but is not a critical determinant of its specificity.
Eubacteria would be expected to be sensitive to significant, or even partial, inhibition of their DNA ligase activities. Studies of temperature-sensitive DNA ligase mutants have revealed some important effects of defective DNA ligase genes on cellular growth and DNA repair (8). Two mutants (one with a lig-7(ts) mutation and the other with a lig4 mutation) that express DNA ligase at 15 to 20% of wild-type levels, as assessed by their ability to sustain phage replication, were studied in some detail (8). These DNA ligase mutants showed both DNA replication and repair defects when grown at nonpermissive temperatures (8), suggesting that DNA ligase is a suitable target for novel antibacterial agents.
While some of the aminoquinoline compounds identified in this study (e.g., CQ, quinacrine, and 6bisQ8) were reasonably potent inhibitors of NAD+-dependent DNA ligase in vitro, they were >1,000-fold less active in vivo. This is presumably due to the fact that basic drugs, such as CQ, are actively excluded by bacteria (32), so the intracellular concentration of the aminoquinolines is likely to be very low. If the aminoquinolines could be modified to enhance uptake, they might have potential as bactericidal drugs.
We suggest that inhibition of DNA ligase can account for the previously reported antibacterial activity of CQ against both spore outgrowth and vegetative multiplication in Bacillus species (32). Ligase inhibition may also help to explain the ability of CQ to induce frameshift mutations in E. coli and S. typhimurium (37). By contrast, it seems unlikely that inhibition of DNA ligase underlies the antimalarial activities of CQ and quinacrine, since partial sequencing of a Plasmodium falciparum DNA ligase gene indicates that it is a typically eukaryotic ATP-dependent enzyme (14a).
The arylamino compounds identified in this study appear to be the first examples of specific inhibitors of eubacterial DNA ligases identified to date. Our data indicate that a structure with four carbon atoms separating two nitrogens is the minimum element with specific inhibitory activity. This class of inhibitors is therefore unusual in that a small inhibitory moiety can be presented on a variety of scaffolds without losing its specificity of action. We believe that the arylamino compounds identified in this work will prove useful as tools for further studies of mechanisms of bacterial mutagenesis and DNA ligase action. In addition, we believe that the identification of even more-potent and more-specific inhibitors of NAD+-dependent DNA ligases will allow the detection and development of potent, broad-spectrum, eubactericidal drugs. If compounds that are efficiently accumulated by bacteria can be identified, they may well prove to be of assistance in our ongoing struggle with bacteria that show resistance to some of the currently available and clinically useful antibacterial agents.
ACKNOWLEDGMENTS
The ligase mutants and plasmid-containing strains were kindly provided by J. R. Roth, University of Utah.
G.C. acknowledges a short-term travel grant from CNR. This work was partially supported by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Bentley D, Selfridge J, Millar J K, Samuel K, Hole N, Ansell J D, Melton D W. DNA ligase I is required for fetal liver erythropoiesis but is not essential for mammalian cell viability. Nat Genet. 1996;13:489–491. doi: 10.1038/ng0896-489. [DOI] [PubMed] [Google Scholar]
- 2.Cheng C, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by Haemophilus influenzae. Nucleic Acids Res. 1997;25:1369–1374. doi: 10.1093/nar/25.7.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciarrocchi G, Lestingi M, Fontana M, Spadari S, Montecucco A. Correlation between anthracycline structure and human DNA ligase inhibition. Biochem J. 1991;279:141–146. doi: 10.1042/bj2790141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen S N, Yielding K L. Spectrophotometric studies of the interaction of chloroquine with deoxyribonucleic acid. J Biol Chem. 1965;240:3123–3131. [PubMed] [Google Scholar]
- 5.Cohen S N, Yielding K L. Inhibition of DNA and RNA polymerase reactions by chloroquine. Proc Natl Acad Sci USA. 1965;54:521–527. doi: 10.1073/pnas.54.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doherty A J, Wigley D B. Functionals domains of an ATP-dependent DNA ligase. J Mol Biol. 1999;285:63–71. doi: 10.1006/jmbi.1998.2301. [DOI] [PubMed] [Google Scholar]
- 7.Gallina A, Rossi F, Milanesi G, Rossi R, Montecucco A, Ciarrocchi G. Characterization of human DNA ligase I expressed in a baculovirus-insect cell system. Biochem J. 1995;312:593–597. doi: 10.1042/bj3120593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gottesman M M, Hicks M L, Gellert M. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol. 1973;77:531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- 9.Hakansson K, Doherty A J, Shuman S, Wigley D B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 10.Ho C K, Van Etten J L, Shuman S. Characterization of an ATP-dependent DNA ligase encoded by Chlorella virus PBCV-1. J Virol. 1997;71:1931–1937. doi: 10.1128/jvi.71.3.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irvin J L, Irvin E M. Spectrophotometric and potentiometric evaluation of apparent acid dissociation exponents of various 4-aminoquinolines. J Am Chem Soc. 1947;69:1091–1099. doi: 10.1021/ja01197a034. [DOI] [PubMed] [Google Scholar]
- 12.Johnston L H, Nasmyth K A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 13.Kletzin A. Molecular characterisation of a DNA ligase gene of the extremely thermophilic archaeon Desulfurolobus ambivalens shows close phylogenetic relationship to eukaryotic ligases. Nucleic Acids Res. 1992;20:5389–5396. doi: 10.1093/nar/20.20.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg A, Baker T. DNA replication. W. H. New York, N.Y: Freeman and Co.; 1992. [Google Scholar]
- 14a.La Greca, N., G. Ciarrocchi, and L. Tilley. Unpublished observations.
- 15.Lindahl T, Barnes D E. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–258. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 16.Liquori A M, Costantino L, Crescenzi V, Elia V, Giglio E, Puliti R, De Sanctis Savino M, Vitagliano V. Complexes between DNA and polyamines: a molecular model. J Mol Biol. 1967;24:113–122. [Google Scholar]
- 17.Mackenzie A H. Antimalarial drugs for rheumatoid arthritis. Am J Med. 1983;75:48–58. doi: 10.1016/0002-9343(83)90474-6. [DOI] [PubMed] [Google Scholar]
- 18.Modrich P, Lehman I R. Enzymatic joining of polynucleotides. IX. A simple and rapid assay of polynucleotide joining (ligase) activity by measurement of circle formation from linear deoxyadenylate-deoxythymidylate copolymer. J Biol Chem. 1970;245:3626–3631. [PubMed] [Google Scholar]
- 19.Montecucco A, Pedrali-Noy G, Spadari S, Zanolin E, Ciarrocchi G. DNA unwinding and inhibition of T4 DNA ligase by anthracyclines. Nucleic Acids Res. 1988;16:3907–3918. doi: 10.1093/nar/16.9.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montecucco A, Fontana M, Focher F, Lestingi M, Spadari S, Ciarrocchi G. Specific inhibition of human DNA ligase adenylation by a distamycin derivative possessing antitumor activity. Nucleic Acids Res. 1991;19:1067–1072. doi: 10.1093/nar/19.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montecucco A, Lestingi M, Rossignol J M, Elder R H, Ciarrocchi G. Lack of discrimination between DNA ligases I and III by two classes of inhibitors, anthracyclines and distamycins. Biochem Pharmacol. 1993;45:1536–1539. doi: 10.1016/0006-2952(93)90057-4. [DOI] [PubMed] [Google Scholar]
- 22.Park U E, Olivera B M, Hughes K T, Roth J R, Hillyard D R. DNA ligase and the pyridine nucleotide cycle in Salmonella typhimurium. J Bacteriol. 1989;171:2173–2180. doi: 10.1128/jb.171.4.2173-2180.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrini J H J, Xiao Y, Weaver D T. DNA ligase I mediates essential functions in mammalian cells. Mol Cell Biol. 1995;15:4303–4308. doi: 10.1128/mcb.15.8.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poso H, Kuosmanen M. Spermidine and spermine stimulate the activity of T4-DNA ligase. Biochem Biophys Res Commun. 1983;117:217–222. doi: 10.1016/0006-291x(83)91563-2. [DOI] [PubMed] [Google Scholar]
- 25.Raynes K, Galatis D, Cowman A F, Tilley L, Deady L W. Synthesis and activity of some antimalarial bisquinolines. J Med Chem. 1995;38:204–206. doi: 10.1021/jm00001a026. [DOI] [PubMed] [Google Scholar]
- 26.Scaria P V, Craig J C, Shafer R H. Differential binding of the enantiomers of chloroquine and quinacrine to polynucleotides: implications for stereoselective metabolism. Biopolymers. 1993;33:887–895. doi: 10.1002/bip.360330604. [DOI] [PubMed] [Google Scholar]
- 27.Shark K B, Conway T. Cloning and molecular characterization of the DNA ligase gene (lig) from Zymomonas mobilis. FEMS Microbiol Lett. 1992;75:19–26. doi: 10.1016/0378-1097(92)90450-3. [DOI] [PubMed] [Google Scholar]
- 28.Shuman S. Vaccinia virus DNA ligase: specificity, fidelity and inhibition. Biochemistry. 1995;34:16138–16147. doi: 10.1021/bi00049a029. [DOI] [PubMed] [Google Scholar]
- 29.Shuman S, Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 30.Singleton M R, Hakanson K, Timson D J, Wigley D B. Structure of the adenylation domain of an NAD+-dependent DNA ligase. Structure. 1999;7:35–42. doi: 10.1016/s0969-2126(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 31.Smith G L, Chan Y S, Kerr S M. Transcriptional mapping and nucleotide sequence of a vaccinia virus gene encoding a polypeptide with extensive homology to DNA ligases. Nucleic Acids Res. 1989;17:9051–9062. doi: 10.1093/nar/17.22.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith K T, Dawes W. The preferential inhibition of Bacillus subtilis spore outgrowth by chloroquine. Arch Microbiol. 1989;152:251–257. doi: 10.1007/BF00409659. [DOI] [PubMed] [Google Scholar]
- 33.Spadari S, Ciarrocchi G, Falaschi A. Purification and properties of a polynucleotide ligase from human cell cultures. Eur J Biochem. 1971;22:75–78. doi: 10.1111/j.1432-1033.1971.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 34.Subramanya H S, Doherty A J, Ashford S R, Wigley D B. Crystal structure of an ATP-dependent DNA ligase from bacteriophage T7. Cell. 1996;85:607–615. doi: 10.1016/s0092-8674(00)81260-x. [DOI] [PubMed] [Google Scholar]
- 35.Takahasi M, Yamaguchi E, Uchida T. Thermophilic DNA ligase. Purification and properties of the enzyme from Thermus thermophilus HB8. J Biol Chem. 1984;259:10041–10047. [PubMed] [Google Scholar]
- 36.Teraoka H, Tsukada K. Activation of mammalian DNA ligase by polyamines. Biochem Biophys Res Commun. 1980;95:638–643. doi: 10.1016/0006-291x(80)90833-5. [DOI] [PubMed] [Google Scholar]
- 37.Thomas S M, Silburn K A, MacPhee D G. Frameshift mutagenesis by chloroquine in Escherichia coli and Salmonella typhimurium. Mutat Res. 1987;192:233–237. doi: 10.1016/0165-7992(87)90062-5. [DOI] [PubMed] [Google Scholar]
- 38.Thorbjarnardottir S H, Jonsson Z O, Andresson O S, Kristijansson J K, Eggertsson G, Palsdottir A. Cloning and sequence analysis of the DNA ligase-encoding gene of Rhodothermus marinus and overproduction, purification and characterization of two thermophilic DNA ligases. Gene. 1995;161:1–6. doi: 10.1016/0378-1119(95)00286-f. [DOI] [PubMed] [Google Scholar]
- 39.Timson D J, Wigley D B. Functional domains of an NAD+-dependent DNA ligase. J Mol Biol. 1999;285:73–83. doi: 10.1006/jmbi.1998.2302. [DOI] [PubMed] [Google Scholar]
- 40.Tomkinson A E, Mackey Z B. Structure and function of mammalian DNA ligases. Mutat Res. 1998;407:1–9. doi: 10.1016/s0921-8777(97)00050-5. [DOI] [PubMed] [Google Scholar]
- 41.Tomkinson A E, Totty N F, Ginsburg M, Lindahl T. Location of the active site for enzyme-adenylate formation in DNA ligases. Proc Natl Acad Sci USA. 1991;88:400–404. doi: 10.1073/pnas.88.2.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss B, Richardson C C. Enzymatic breakage and joining of deoxyribonucleic acid. I. Repair of single-strand breaks in DNA by an enzyme system from Escherichia coli infected with T4 bacteriophage. Proc Natl Acad Sci USA. 1967;57:1021–1028. doi: 10.1073/pnas.57.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson G G, Murray N E. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterisation of the recombinants. J Mol Biol. 1979;132:471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- 44.Zimmerman S B, Little J W, Oshinsky C K, Gellert M. Enzymatic joining of DNA strands: a novel reaction of diphosphopyridine nucleotide. Proc Natl Acad Sci USA. 1967;57:1841–1848. doi: 10.1073/pnas.57.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]