Abstract
The persistency of COVID-19 in the world and the continuous rise of its variants demand new treatments to complement vaccines. Computational chemistry can assist in the identification of moieties able to lead to new drugs to fight the disease. Fullerenes and carbon nanomaterials can interact with proteins and are considered promising antiviral agents. Here, we propose the possibility to repurpose fullerenes to clog the active site of the SARS-CoV-2 protease, Mpro. Through the use of docking, molecular dynamics, and energy decomposition techniques, it is shown that C60 has a substantial binding energy to the main protease of the SARS-CoV-2 virus, Mpro, higher than masitinib, a known inhibitor of the protein. Furthermore, we suggest the use of C70 as an innovative scaffold for the inhibition of SARS-CoV-2 Mpro. At odds with masitinib, both C60 and C70 interact more strongly with SARS-CoV-2 Mpro when different protonation states of the catalytic dyad are considered. The binding of fullerenes to Mpro is due to shape complementarity, i.e., vdW interactions, and is aspecific. As such, it is not sensitive to mutations that can eliminate or invert the charges of the amino acids composing the binding pocket. Fullerenic cages should therefore be more effective against the SARS-CoV-2 virus than the available inhibitors such as masinitib, where the electrostatic term plays a crucial role in the binding.
Keywords: C60, C70, masitinib, Mpro, SARS-CoV-2, COVID-19, drug repurposing, MM-GBSA, inhibitors, nanobio interface
1. Introduction
The onslaught of COVID-19 waves and the continuous rise of variants of the SARS-CoV-2 virus [1] demand new treatments to complement vaccinations. Endocytic entry into host cells, RNA replication and transcription, translation and proteolytic processing of viral proteins, virion assembly, and release of new viruses through exocytic mechanisms are all potentially targetable processes in the coronavirus life cycle [2].
Among the viral proteins, only a few are essential in the life cycle of the virus. The main protease, known as Mpro or 3CLpro, plays a critical function in viral replication and transcription and represents the main target for medicinal chemistry [3,4]. This enzyme breaks down the polyprotein chain coded by the RNA of the virus into functional proteins, which the virus needs to construct itself and proliferate [3,4]. Disrupting this important part of the virus’s self-replication engine blocks the infection.
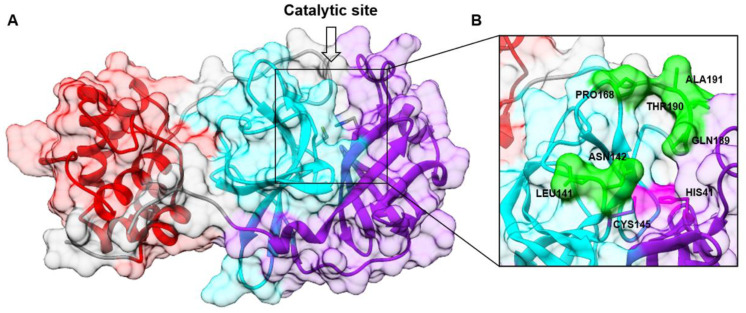
Just a few weeks after the first COVID-19 outbreak, the crystallographic structure of Mpro was determined and deposited under the PDB code 6LU7 [3]. Mpro is composed of 306 amino acids, characterized by 3 distinct domains (domains I, II, and III) [3]. Domain I (residues 8–101) and domain II (residues 102–184) have a similar fold composed of antiparallel ß-barrel structures. Domain III (residues 201–303), instead, consists of a cluster of five α-helices, responsible for protein dimerization. The active site is placed in a cleft between domains I and II (Figure 1). The catalytic residues Cys145 and His41 are buried in this cavity that can accommodate four substrate residues in positions P1′ through P4 and is flanked by residues from both domains I and II (Figure 1B). The catalytic dyad may be activated by a proton transfer from Cys145 to His41, possibly triggered by substrate binding or occurring in a transition state during the attack by the sulfur on the carbonyl carbon atom of the scissile peptide bond. It was suggested that a water molecule might complete the catalytic triad by mediating crucial interactions between His41 and other important conserved residues, such as His164 and Asp187 [4].
Figure 1.
(A) Mpro structure (PDB ID 7JU7) shown in ribbons and surface representation: domain I (residues 8–101) in purple, domain II (residues 102–184) in cyan, and domain III (residues 201–303) in red. (B) A magnification of the catalytic site showing the catalytic dyad (His41 and Cys145) in purple and residues that lie within the binding pocket in green.
Due to the immediate availability of the Mpro crystal structure, structure-based drug discovery (SBDD) techniques were promptly used to expedite the rational identification of potential Mpro inhibitors [5,6,7] or to drive the repurposing of known molecules [8,9,10,11,12,13,14]. Many protease inhibitors of the human immunodeficiency virus (HIV) were identified as possible anti-COVID candidates [15].
Fullerenes and carbon nanomaterials are able to interact with peptides [16,17] and proteins [18,19,20,21,22,23,24,25,26] and, in general, are considered promising antiviral agents [27,28,29,30,31,32]. The idea of using C60 as an inhibitor of the HIV protease dates back to 1993 [33]. C60 inhibits the protein thanks to its size and unusual spherical shape [33]. The buckyball fits snugly into the substrate binding pocket, blocks the active site, and prevents the HIV polypeptide chain from entering [33]. C60 is, however, insoluble in water, and fullerene derivatives were designed and then synthesized for use in a physiological environment [34,35,36,37,38,39,40,41,42,43,44]. The structure–activity relationship between functionalized fullerenes and HIV protease inhibition showed the importance of positioning the derivative moieties in a well-defined geometry on the fullerene cage [34,35,36,37,38,39,40,41,42,43,44].
The idea of repositioning C60 for the inhibition of SARS-CoV-2 Mpro is a natural consequence [45,46,47]. In this work, we compare (i) the binding energy of C60 with the HIV protease and with SARS-CoV-2 Mpro to understand the efficiency of the repurposing, and (ii) the performances of C60 and masitinib [48], a known inhibitor of SARS-CoV-2 Mpro, to verify the possibility of using C60 derivatives as effective Mpro inhibitors. We further propose, for the first time, the use of C70 as an innovative scaffold for the inhibition of SARS-CoV-2 Mpro.
2. Results
2.1. Determining the C60 Binding Pocket in SARS-CoV-2 Mpro
The crystal structures of SARS-CoV-2 Mpro complexed with different ligands (Figure 2A) showed that noncovalent and covalent inhibitors behave differently. Noncovalent inhibitors interact with the multiple residue system of the substrate binding pocket of SARS-CoV-2 Mpro. Covalent inhibitors mainly interact with the catalytic Cys145 residue after an initial noncovalent interaction in the substrate binding pocket.
Figure 2.
(A) Binding of Mpro ligands N3 (PDB ID 6LU7), masitinib (PDB ID 7JU7), parampanel analogue compound 5 (PDB ID 7L11), MPI4 (PDB ID 7JQ1), ML188 (PDB ID 7L0D), isofloxythepin (PDB ID 7AY7), carmofur (PDB ID 7BUY), UAW243 (PDB ID 6XFN), and boceprevir (PDB ID 6XQU) in Mpro. The protein structure is shown with the ribbon. (B) Identification of the fullerene binding pocket in Mpro.
Using a docking protocol able to identify the fullerene binding pockets of proteins [18,19,49,50,51,52,53,54], we docked the C60 in SARS-CoV-2 Mpro. C60 binds on the substrate binding pocket of Mpro (Figure 2B). The fullerene cage shows a strong shape complementarity with this pocket of Mpro. C60 occupies exactly the same position occupied by known Mpro inhibitors, suggesting an inhibitory activity of the cage.
2.2. Determining the Binding Energy between C60 and SARS-CoV-2 Mpro
The atomistic understanding of the interactions of C60 with SARS-CoV-2 Mpro is crucial for real applications of nanomolecules in medicine [26]. MD simulations represent a powerful tool to investigate such interactions [55]. Starting from the docking pose, 100 ns of molecular dynamics simulations was carried out. To estimate the binding energy between Mpro and C60, an MM-GBSA analysis of the trajectories was performed. C60 lies above His41, giving sandwich-like π–π interactions [56,57], and interacts hydrophobically [56,57] with Met49 and Leu27. Surfactant-like interactions [56,57] with Cys145 and Ser46 are also observed.
The ΔGbinding between Mpro and C60 is −18.8 kcal mol–1. This value is very close to the interaction energy between lysozyme and C60 (−18.5 kcal mol−1) [50], a complex that is experimentally accessible and widely used in nanomedicine [49,58,59,60]. The result demonstrates the feasibility of the exploitation of the C60 molecule in the inhibition of Mpro.
According to the analysis of the binding components of the energy, the driving force for binding (−44.8 kcal mol–1) is represented by van der Waals interactions. Hydrophobic interactions, (Enon-polar), assist the binding, despite the fact that their value (−2.5 kcal mol–1) is far lower than that of the vdW interactions. The contributions of polar solvation (12.8 kcal mol–1) and entropy (15.7 kcal mol−1) are positive and oppose the binding. Because of the rigidity of CNPs, the entropic term is frequently overlooked [45] while studying protein–CNP interactions. However, this factor, which is estimated to be 15.7 kcal mol–1, is energetically significant and should be considered when protein–CNP hybrids are studied. The binding of C60 to the protein cavity produces a significant reduction in amino acid mobility, giving rise to this high value.
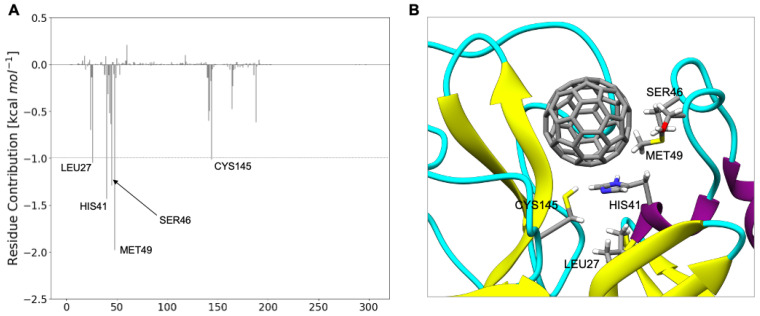
The decomposition analysis of the total binding energy provides the contribution to the binding of each amino acid (Figure 3). The most interacting amino acids (∆Gbinding larger than 1.0 kcal mol−1) are Met49, His41, Ser46, Leu27, and Cys145. Very interestingly, C60 strongly interacts both with the catalytic dyad (Cys145-His 41) and with residues located on the substrate binding pocket (Met49, Ser46, Leu27).
Figure 3.
(A) C60@Mpro interactions. ΔGbinding decomposed per residue. (B) Interaction between Met49, His41, Ser46, Leu27, and Cys145 and C60.
At the same time, C60 shields the catalytic dyad, blocking its catalytic activity, and occupies the substrate binding pocket, impeding the interaction of Mpro with its substrate.
2.3. Comparing the Binding of C60 in SARS-CoV-2 Mpro and HIV Protease
To validate the results and evaluate the reliability of the use of C60 as an Mpro inhibitor, we calculated the interaction of C60 with the HIV protease (ProHIV), using the same protocol adopted to calculate the binding energy between C60 and Mpro (Table 1).
Table 1.
Energy components of ΔGbinding (VDW, Eel, EGB, and Enon-polar) for C60@Mpro and C60@ProHIV complexes. All energies are reported in kcal mol−1.
| Complex | ΔH | VDW | EEl | EGB | Enon-polar | TΔS | ΔGbind |
|---|---|---|---|---|---|---|---|
| C60@Mpro | −34.5 | −44.8 | 0.0 | 12.8 | −2.5 | −15.7 | −18.8 |
| C60@ProHIV | −38.0 | −51.6 | 0.0 | 16.0 | −2.4 | −18.3 | −19.7 |
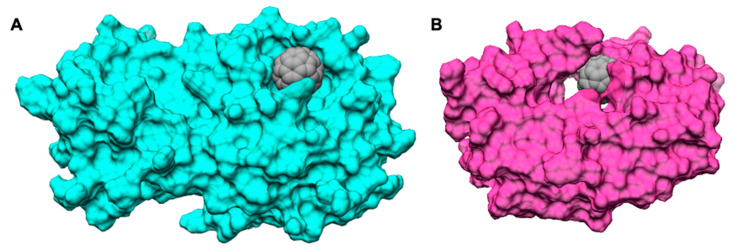
Shape complementarity is the crucial parameter governing the interaction of fullerene with proteins [57]. In ProHIV, as well as in Mpro, C60 fits snugly in the active site (Figure 4), and as a consequence, the total binding energy and the energy components of ΔGbinding between C60 and the two proteases are similar [61].
Figure 4.
Surface complementarity between (A) Mpro and (B) ProHIV proteins and the C60 cage.
Since it is known that C60 experimentally works as an HIV protease inhibitor [33,34,35,36,37,38,39,40,41,42,43,44], this comparison validates the idea of repurposing C60 as a SARS-CoV-2 Mpro inhibitor.
2.4. Comparing the Binding of C60 in SARS-CoV-2 Mpro with Masitinib
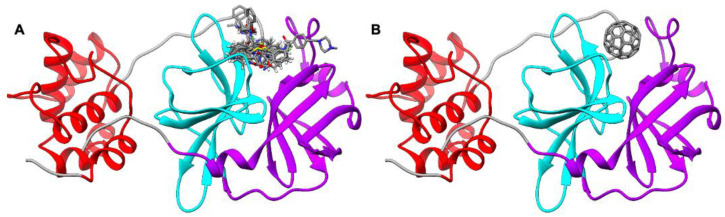
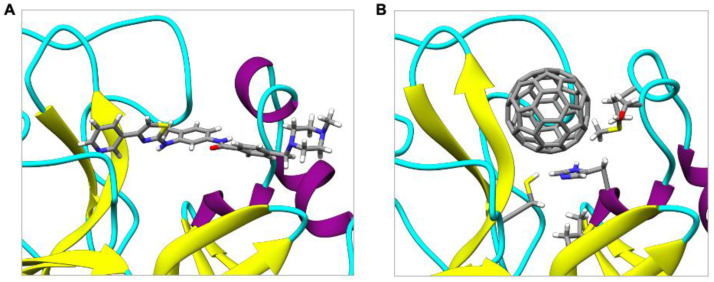
To estimate the potential application of C60 as an inhibitor of Mpro, we calculated the binding energy with a known inhibitor of Mpro, namely, masitinib. Masitinib is an orally bioavailable tyrosine kinase inhibitor, repurposed as an inhibitor of SARS-CoV-2 Mpro [48]. X-ray crystallography and biochemistry experiments showed that masitinib acts as a competitive inhibitor of Mpro [48] and occupies the same binding pocket of C60 (Figure 5). Mice infected with SARS-CoV-2 and subsequently treated with masitinib had a 200-fold decrease in viral titers in the lungs and nose, as well as lower lung inflammation [48].
Figure 5.
Binding of (A) masitinib and (B) C60 with Mpro.
The results show that C60 has a larger binding energy than masitinib, due to two factors that are usually ignored when the interactions between inhibitors and proteins are evaluated, namely, desolvation energy and entropy (Table 2).
Table 2.
Energy components of ΔGbinding (VDW, Eel, EGB, and Enon-polar) for C60@Mpro and masitinib@Mpro complexes. All energies are reported in kcal mol−1.
| Complex | ΔH | VDW | EEl | EGB | Enon-polar | TΔS | ΔGbind |
|---|---|---|---|---|---|---|---|
| C60@Mpro | −34.5 | −44.8 | 0.0 | 12.8 | −2.5 | −15.7 | −18.8 |
| masitinib@Mpro | −41.2 | −51.4 | −18.1 | 34.0 | −5.8 | −24.7 | −16.5 |
Even if vdW and electrostatic terms are larger for masitinib than for C60, binding of the more polar and more flexible masitinib molecule gives larger penalty terms due to the desolvation energy and entropy. C60 is an ideal inhibitor because the terms that generally oppose binding (i.e., desolvation energy and entropy) are minimized by its hydrophobicity and rigidity.
2.5. Determining the Binding Energy between C70 and SARS-CoV-2 Mpro
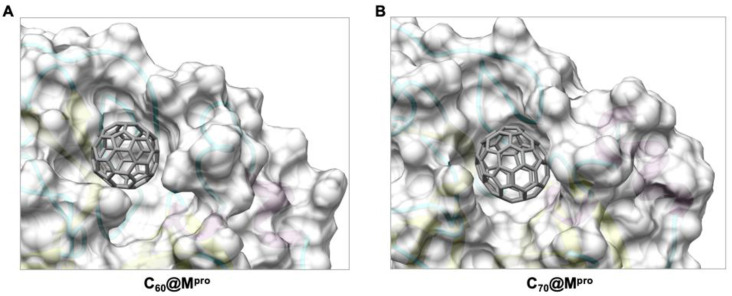
With the increase in the carbon cage size, the binding strength between proteins and fullerenes usually increases [60,62]. It is natural to suppose that C70, in principle, may be a better inhibitor than C60 for Mpro. Docking calculations showed that C70 occupies the same binding pocket (Figure 6).
Figure 6.
Surface representation of the binding cleft of (A) C60 and (B) C70 in Mpro.
The ΔGbinding between C70 and Mpro is considerably higher (−28.0 kcal mol−1) than in the case of C60 (−18.8 kcal mol−1) (Table 3).
Table 3.
Energy components of ΔGbinding (VDW, Eel, EGB, and Enon-polar) for C60@Mpro and C70@Mpro complexes. All energies are reported in kcal mol−1.
| Complex | ΔH | VDW | EEl | EGB | Enon-polar | TΔS | ΔGbind |
|---|---|---|---|---|---|---|---|
| C60@Mpro | −34.5 | −44.8 | 0.0 | 12.8 | −2.5 | −15.7 | −18.8 |
| C70@Mpro | −45.6 | −59.2 | 0.0 | 16.8 | −3.2 | −17.6 | −28.0 |
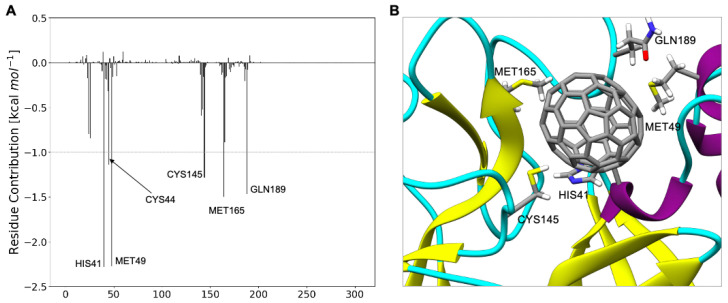
This increase is due to a substantial increase in the van der Waals interactions, which grow from −44.8 kcal mol−1 for C60 to −59.2 kcal mol−1 for C70. Van der Waals and hydrophobic interactions are −47.3 kcal mol−1 in C60 and −62.4 kcal mol−1 in C70, with a net increase of 15.1 kcal mol−1. In C70, the increase compensates the small energy penalties due to higher entropic and desolvation terms (28.5 kcal mol−1 in C60 and 34.4 kcal mol−1 in C70, with a net increase of 5.9 kcal mol−1). The measure of shape complementarity between the fullerene cage and the protein is usually taken as a quick way to estimate the stabilizing interactions. This is also true in this case considering that the variation in the solvent-accessible surface area (∆SASA) upon binding is 347.2 Å2 for C60@Mpro and 444.4 Å2 for C70@Mpro, as evident also in Figure 6. The larger C70 engages stronger interactions with a larger number of the amino acids that make up the binding pocket (Figure 7). The interactions of C60 with the catalytic dyad of His41 and Cys145 are −1.4 kcal mol−1 and −1.0 kcal mol−1, respectively. The interactions of C70 with the same residues are −2.3 kcal mol−1 and −1.3 kcal mol−1, with an increase of 0.9 kcal mol−1 and 0.3 kcal mol−1. In addition, C70 also strongly interacts with Met165 (−1.5 kcal mol−1) and Gln189 (−1.5 kcal mol−1), residues that were identified as the hot spot for the activity of Mpro inhibitors [63].
Figure 7.
(A) C70@Mpro interactions. ΔGbinding decomposed per residue. (B) Interaction between His41, Cys44, Met49, Cys145, Met165, and Gln189 and C70.
2.6. Evaluating the Binding Enrgy of Masitinib, C60, and C70 to SARS-CoV-2 Mpro in Different Protonation States of the Catalytic Dyad
The protonation state of histidine [64,65,66,67], and in particular of the catalytic dyad (His41-Cys145), plays a crucial role in Mpro activity, stability, and protein–ligand interactions. Very recently, neutron crystallographic studies [65,67] provided direct visualization of the hydrogen atoms’ locations in the SARS-CoV-2 Mpro enzyme. If in the bound state His41 is in a neutral form [65], in the ligand-free structure, the catalytic site of Mpro adopts a zwitterionic form where Cys145 is deprotonated and negatively charged and His41 is doubly protonated and positively charged [67]. Since the free energy of activation for the initial proton transfer from Cys145 to His41 is very low [68,69] and the relative energies of the zwitterionic and neutral states are very close [68,69], we reinvestigated the binding of masitinib, C60, and C70 to Mpro in both protonation states of the catalytic dyad (Table 4).
Table 4.
Comparison of the binding energy of masitinib, C60, and C70 with Mpro in the two protomeric states, namely, His41-Cys145 and His41+-Cys145−. All energies are reported in kcal mol−1.
| Complex | His41-Cys145 ΔGbinding | His41+-Cys145-ΔGbinding |
|---|---|---|
| masitinib@Mpro | −16.5 | −5.1 |
| C60@Mpro | −18.8 | −24.3 |
| C70@Mpro | −28.0 | −29.4 |
The protonation state of the catalytic dyad strongly affects the binding of masitinib, and in the zwitterionic form (His41+-Cys145−), the ∆Gbinding is markedly reduced (going from −15.5 kcal mol−1 to −5.1 kcal mol−1). The electrostatic term decreases from −18.1 kcal mol−1 for the doubly neutral dyad to −11.0 kcal mol−1 for the zwitterionic dyad, due to the different distributions of the charges in the binding site. This term is unaffected in the fullerene binding because it does not have net charges. Owing to increased vdW interactions, ∆Gbinding of fullerenes increases in the case of the zwitterionic form of the dyad. It goes from −44.8 kcal mol−1 to −54.6 kcal mol−1 for C60 and from −59.2 kcal mol−1 to −60.9 kcal mol−1 for C70.
3. Conclusions
In summary, we demonstrated the possibility of repurposing fullerenes as inhibitors of SARS-CoV-2 Mpro. We calculated and compared the binding energies of (a) C60 and C70 to Mpro, (b) C60 to the HIV protease, and (c) masitinib to Mpro. The results indicate the feasibility of the repurposing. Fingerprint analysis showed the role of π–π, hydrophobic, and surfactant-like interactions in the binding of the fullerenes to Mpro. Fullerenes interact with the His41-Cys145 dyad, blocking the catalytic activity. At the same time, they occupy the substrate binding pocket, impeding the interaction of Mpro with its substrate.
Fullerenes are ideal inhibitors because, in these molecules, the terms that generally oppose the binding (i.e., desolvation energy and entropy) are minimized by their hydrophobicity and rigidity.
Shape complementarity is crucial to govern the interaction of fullerenes with proteins, and also in this case, the larger C70 interacts more strongly than C60 with the binding site of Mpro. C70 engages stronger interactions with more residues that form the binding pocket. In particular, C70 strongly interacts with the Met165 and Gln189 residues that were identified as the hot spot for the activity of Mpro inhibitors.
Fullerenes are insensitive to mutations that perturb the electrostatic characteristics of the binding site. They interact even more strongly with Mpro when different protonation states of the catalytic dyad are considered.
The binding of fullerenes to Mpro, based as it is on shape complementarity, i.e., vdW interactions, is aspecific. As such, it is not sensitive to mutations that eliminate or invert the charges of the amino acids composing the binding pocket. Indeed, for some inhibitors, such as masinitib, the electrostatic terms play a crucial role. As a consequence, even a punctual modification of the catalyic site may strongly affect their binding, and therefore their activity. Fullerenes therefore appear to be ideal moieties to exploit in the identification of potential new drugs against the SARS-CoV-2 protease and other viral proteins.
4. Materials and Methods
4.1. System Setup
The crystal structures of the SARS-CoV-2 main proteases (PDB ID 6LU7, 7JU7, and 7L10) and the HIV protease (PDB ID 1ZTZ) were downloaded from the Protein Data Bank (PDB). The crystal structure of PDB ID 7JU7 was used for the docking calculations.
The Amber ff14SB force field [70] was used to model the Mpro and ProHIV proteins. C60 and C70 carbon atoms were modeled as uncharged Lennard-Jones particles by using sp2 carbon parameters taken from the ff14SB force field [70]. Masitinib was parametrized by calculating the partial atomic charges using the restraint electrostatic potential method (RESP) at the HF/6-31G* level of theory. The corresponding parameters were then generated by the standard procedure reported for antechamber, as implemented in Amber16 [71].
4.2. Docking
Docking models were obtained using the PatchDock algorithm [72] that computes the shape complementarity between two entities (ligand and receptor), minimizing the number of steric clashes. PatchDock (i) assigns concave, convex, or flat patches to the ligand and receptor surface, (ii) matches concave–convex/flat–flat, and generates a set of candidate transformations. (iii) Each transformation is then ranked by the shape complementarity and the atomic desolvation energy of the complex (scoring functions). Root mean square deviation clustering avoids the generation of redundant solutions.
4.3. MD Simulations
The docking structures were minimized by 5000 steps of steepest descent minimization, followed by 5000 steps of the conjugate gradient algorithm. The minimized structures underwent a 1 ns equilibration step and were heated from 0 to 300 K (Langevin thermostat). Periodic boundary conditions (PBC) and particle mesh Ewald summation were used throughout (with a cut-off radius of 10 Å for the direct space sum). The MD simulations were carried out using an explicit solvent (TIP3P water model). Sodium counterions were included to exactly neutralize the charge of the system. After the equilibration, a production MD simulation of 100 ns was carried out for every system at 300 K. Amber 16 was used to run all the simulations [71].
4.4. Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) Analysis
A total of 5000 frames were extracted from MD simulations and used for the MM-GBSA analysis. An infinite cut-off was used for all the interactions. The electrostatic contribution to the solvation free energy was calculated with the Generalized Born (GB) model, as implemented in MMPBSA.py [73]. The nonpolar contribution to the solvation free energy was determined with solvent-accessible surface-area-dependent terms. To obtain an estimate of the binding entropy, the normal modes for the complex, receptor, and ligand were calculated, and the results were averaged using the PTRAJ program (Normal Mode Analysis) via MMPBSA.py [73].
Author Contributions
Conceptualization, M.C.; methodology, M.C and T.D.M.; investigation, T.D.M. and E.J.M.; data curation, T.D.M.; supervision, M.C.; writing—original draft, T.D.M. and M.C.; writing—review and editing, T.D.M., E.J.M., F.Z. and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All data in this study can be requested from the corresponding authors (tainah.marforio2@unibo.it, T.D.M.; matteo.calvaresi3@unibo.it, M.C.).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li J., Lai S., Gao G.F., Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 2.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Kneller D.W., Phillips G., O’Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:7–12. doi: 10.1038/s41467-020-16954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., Xie X., Jin Z., Peng J., Liu F., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Z., Yang L., Zhao X.E. Co-crystallization and structure determination: An effective direction for anti-SARS-CoV-2 drug discovery. Comput. Struct. Biotechnol. J. 2021;19:4684–4701. doi: 10.1016/j.csbj.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38:379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar A., Mandal K. Repurposing an Antiviral Drug against SARS-CoV-2 Main Protease. Angew. Chem. Int. Ed. 2021;60:23492–23494. doi: 10.1002/anie.202107481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta A., Zhou H.X. Profiling SARS-CoV-2 Main Protease (MPRO) Binding to Repurposed Drugs Using Molecular Dynamics Simulations in Classical and Neural Network-Trained Force Fields. ACS Comb. Sci. 2020;22:826–832. doi: 10.1021/acscombsci.0c00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuzikov M., Costanzi E., Reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., et al. Identification of Inhibitors of SARS-CoV-2 3CL-Pro Enzymatic Activity Using a Small Molecule in Vitro Repurposing Screen. ACS Pharmacol. Transl. Sci. 2021;4:1096–1110. doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasan M., Parvez M.S.A., Azim K.F., Imran M.A.S., Raihan T., Gulshan A., Muhit S., Akhand R.N., Ahmed S.S.U., Uddin M.B. Main protease inhibitors and drug surface hotspots for the treatment of COVID-19: A drug repurposing and molecular docking approach. Biomed. Pharmacother. 2021;140:111742. doi: 10.1016/j.biopha.2021.111742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pinzi L., Tinivella A., Caporuscio F., Rastelli G. Drug Repurposing and Polypharmacology to Fight SARS-CoV-2 Through Inhibition of the Main Protease. Front. Pharmacol. 2021;12:636989. doi: 10.3389/fphar.2021.636989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macip G., Garcia-Segura P., Mestres-Truyol J., Saldivar-Espinoza B., Ojeda-Montes M.J., Gimeno A., Cereto-Massagué A., Garcia-Vallvé S., Pujadas G. Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition. Med. Res. Rev. 2022;42:744–769. doi: 10.1002/med.21862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu W., Shyr Z., Lo D.C., Zheng W. Viral proteases as targets for coronavirus disease 2019 drug development. J. Pharmacol. Exp. Ther. 2021;378:166–172. doi: 10.1124/jpet.121.000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di Giosia M., Nicolini F., Ferrazzano L., Soldà A., Valle F., Cantelli A., Marforio T.D., Bottoni A., Zerbetto F., Montalti M., et al. Stable and Biocompatible Monodispersion of C 60 in Water by Peptides. Bioconjug. Chem. 2019;30:808–814. doi: 10.1021/acs.bioconjchem.8b00916. [DOI] [PubMed] [Google Scholar]
- 17.Rozhin P., Charitidis C., Marchesan S. Self-assembling peptides and carbon nanomaterials join forces for innovative biomedical applications. Molecules. 2021;26:4084. doi: 10.3390/molecules26134084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calvaresi M., Zerbetto F. Baiting proteins with C60. ACS Nano. 2010;4:2283–2299. doi: 10.1021/nn901809b. [DOI] [PubMed] [Google Scholar]
- 19.Calvaresi M., Zerbetto F. Fullerene sorting proteins. Nanoscale. 2011;3:2873–2881. doi: 10.1039/c1nr10082c. [DOI] [PubMed] [Google Scholar]
- 20.De Leo F., Magistrato A., Bonifazi D. Interfacing proteins with graphitic nanomaterials: From spontaneous attraction to tailored assemblies. Chem. Soc. Rev. 2015;44:6916–6953. doi: 10.1039/C5CS00190K. [DOI] [PubMed] [Google Scholar]
- 21.Calvaresi M., Zerbetto F. The Devil and Holy Water: Protein and Carbon Nanotube Hybrids. Acc. Chem. Res. 2013;46:2454–2463. doi: 10.1021/ar300347d. [DOI] [PubMed] [Google Scholar]
- 22.Marchesan S., Prato M. Under the lens: Carbon nanotube and protein interaction at the nanoscale. Chem. Commun. 2015;51:4347–4359. doi: 10.1039/C4CC09173F. [DOI] [PubMed] [Google Scholar]
- 23.Di Giosia M., Valle F., Cantelli A., Bottoni A., Zerbetto F., Fasoli E., Calvaresi M. High-throughput virtual screening to rationally design protein—Carbon nanotube interactions. Identification and preparation of stable water dispersions of protein—Carbon nanotube hybrids and efficient design of new functional materials. Carbon N. Y. 2019;147:70–82. doi: 10.1016/j.carbon.2019.02.043. [DOI] [Google Scholar]
- 24.Di Giosia M., Marforio T.D., Cantelli A., Valle F., Zerbetto F., Su Q., Wang H., Calvaresi M. Inhibition of α-chymotrypsin by pristine single-wall carbon nanotubes: Clogging up the active site. J. Colloid Interface Sci. 2020;571:174–184. doi: 10.1016/j.jcis.2020.03.034. [DOI] [PubMed] [Google Scholar]
- 25.Rozhin P., Abdel Monem Gamal J., Giordani S., Marchesan S. Carbon Nanomaterials (CNMs) and Enzymes: From Nanozymes to CNM-Enzyme Conjugates and Biodegradation. Materials. 2022;15:1037. doi: 10.3390/ma15031037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Costanzo L., Geremia S. Atomic details of carbon-based nanomolecules interacting with proteins. Molecules. 2020;25:3555. doi: 10.3390/molecules25153555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reina G., Peng S., Jacquemin L., Andrade A.F., Bianco A. Hard Nanomaterials in Time of Viral Pandemics. ACS Nano. 2020;14:9364–9388. doi: 10.1021/acsnano.0c04117. [DOI] [PubMed] [Google Scholar]
- 28.Serrano-Aroca Á., Takayama K., Tuñón-Molina A., Seyran M., Hassan S.S., Pal Choudhury P., Uversky V.N., Lundstrom K., Adadi P., Palù G., et al. Carbon-Based Nanomaterials: Promising Antiviral Agents to Combat COVID-19 in the Microbial-Resistant Era. ACS Nano. 2021;15:8069–8086. doi: 10.1021/acsnano.1c00629. [DOI] [PubMed] [Google Scholar]
- 29.Mallakpour S., Azadi E., Hussain C.M. Fight against COVID-19 pandemic with the help of carbon-based nanomaterials. New J. Chem. 2021;45:8832–8846. doi: 10.1039/D1NJ01333E. [DOI] [Google Scholar]
- 30.Innocenzi P., Stagi L. Carbon-based antiviral nanomaterials: Graphene, C-dots, and fullerenes. A perspective. Chem. Sci. 2020;11:6606–6622. doi: 10.1039/D0SC02658A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sengupta J., Hussain C.M. Carbon nanomaterials to combat virus: A perspective in view of COVID-19. Carbon Trends. 2021;2:100019. doi: 10.1016/j.cartre.2020.100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A., Ahsan O., Wei D.Q., Ansari J.K., Najmi M.H., Muhammad K., Waheed Y. Computational evaluation of abrogation of hbx-bcl-xl complex with high-affinity carbon nanotubes (Fullerene) to halt the hepatitis b virus replication. Molecules. 2021;26:6433. doi: 10.3390/molecules26216433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman S.H., DeCamp D.L., Kenyon G.L., Sijbesma R.P., Srdanov G., Wudl F. Inhibition of the HIV-1 Protease by Fullerene Derivatives: Model Building Studies and Experimental Verification. J. Am. Chem. Soc. 1993;115:6506–6509. doi: 10.1021/ja00068a005. [DOI] [Google Scholar]
- 34.Sijbesma R., Srdanov G., Wudl F., Castoro J.A., Wilkins C., Friedman S.H., Decamp D.L., Kenyon G.L. Synthesis of a Fullerene Derivative for the Inhibition of HIV Enzymes. J. Am. Chem. Soc. 1993:6510–6512. doi: 10.1021/ja00068a006. [DOI] [Google Scholar]
- 35.Schinazi R.F., Sijbesma R., Srdanov G., Hill C.L., Wudl F. Synthesis and virucidal activity of a water-soluble, configurationally stable, derivatized C60 fullerene. Antimicrob. Agents Chemother. 1993;37:1707–1710. doi: 10.1128/AAC.37.8.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura E., Tokuyama H., Yamago S., Shiraki T., Sugiura Y. Biological activity of water-soluble fullerenes. Structural dependence of DNA cleavage, cytotoxicity, and enzyme inhibitory activities including HIV-protease inhibition. Bull. Chem. Soc. Jpn. 1996;69:2143–2151. doi: 10.1246/bcsj.69.2143. [DOI] [Google Scholar]
- 37.Friedman S.H., Ganapathi P.S., Rubin Y., Kenyon G.L. Optimizing the Binding of Fullerene Inhibitors of the HIV-1 Protease through Predicted Increases in Hydrophobic Desolvation. J. Med. Chem. 1998;41:2424–2429. doi: 10.1021/jm970689r. [DOI] [PubMed] [Google Scholar]
- 38.Luca Marcorin G., Da Ros T., Castellano S., Stefancich G., Bonin I., Miertus S., Prato M. Design and synthesis of novel [60] fullerene derivatives as potential HIV aspartic protease inhibitors. Org. Lett. 2000;2:3955–3957. doi: 10.1021/ol000217y. [DOI] [PubMed] [Google Scholar]
- 39.Bosi S., Da Ros T., Spalluto G., Balzarini J., Prato M. Synthesis and anti-HIV properties of new water-soluble bis- functionalized[60]fullerene derivatives. Bioorg. Med. Chem. Lett. 2003;13:4437–4440. doi: 10.1016/j.bmcl.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 40.Marchesan S., Da Ros T., Spalluto G., Balzarini J., Prato M. Anti-HIV properties of cationic fullerene derivatives. Bioorg. Med. Chem. Lett. 2005;15:3615–3618. doi: 10.1016/j.bmcl.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 41.Troshina O.A., Troshin P.A., Peregudov A.S., Kozlovskiy V.I., Balzarini J., Lyubovskaya R.N. Chlorofullerene C60Cl6: A precursor for straightforward preparation of highly water-soluble polycarboxylic fullerene derivatives active against HIV. Org. Biomol. Chem. 2007;5:2783–2791. doi: 10.1039/b705331b. [DOI] [PubMed] [Google Scholar]
- 42.Strom T.A., Durdagi S., Ersoz S.S., Salmas R.E., Supuran C.T., Barron A.R. Fullerene-based inhibitors of HIV-1 protease. J. Pept. Sci. 2015;21:862–870. doi: 10.1002/psc.2828. [DOI] [PubMed] [Google Scholar]
- 43.Yasuno T., Ohe T., Kataoka H., Hashimoto K., Ishikawa Y., Furukawa K., Tateishi Y., Kobayashi T., Takahashi K., Nakamura S., et al. Fullerene derivatives as dual inhibitors of HIV-1 reverse transcriptase and protease. Bioorg. Med. Chem. Lett. 2021;31:127675. doi: 10.1016/j.bmcl.2020.127675. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi T., Yasuno T., Takahashi K., Nakamura S., Mashino T., Ohe T. Novel pyridinium-type fullerene derivatives as multitargeting inhibitors of HIV-1 reverse transcriptase, HIV-1 protease, and HCV NS5B polymerase. Bioorg. Med. Chem. Lett. 2021;49:128267. doi: 10.1016/j.bmcl.2021.128267. [DOI] [PubMed] [Google Scholar]
- 45.Hurmach V.V., Platonov M.O., Prylutska S.V., Scharff P., Prylutskyy Y.I., Ritter U. C60 fullerene against SARS-CoV-2 coronavirus: An in silico insight. Sci. Rep. 2021;11:17748. doi: 10.1038/s41598-021-97268-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suárez M., Makowski K., Lemos R., Almagro L., Rodríguez H., Herranz M.Á., Molero D., Ortiz O., Maroto E., Albericio F., et al. An Androsterone-H2@C60 hybrid: Synthesis, Properties and Molecular Docking Simulations with SARS-Cov-2. Chempluschem. 2021;86:972–981. doi: 10.1002/cplu.202000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skariyachan S., Gopal D., Deshpande D., Joshi A., Uttarkar A., Niranjan V. Carbon fullerene and nanotube are probable binders to multiple targets of SARS-CoV-2: Insights from computational modeling and molecular dynamic simulation studies. Infect. Genet. Evol. 2021;96:105155. doi: 10.1016/j.meegid.2021.105155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drayman N., DeMarco J.K., Jones K.A., Azizi S.A., Froggatt H.M., Tan K., Maltseva N.I., Chen S., Nicolaescu V., Dvorkin S., et al. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science. 2021;373:931–936. doi: 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Calvaresi M., Arnesano F., Bonacchi S., Bottoni A., Calò V., Conte S., Falini G., Fermani S., Losacco M., Montalti M., et al. C60@Lysozyme: Direct observation by nuclear magnetic resonance of a 1:1 fullerene protein adduct. ACS Nano. 2014;8:1871–1877. doi: 10.1021/nn4063374. [DOI] [PubMed] [Google Scholar]
- 50.Calvaresi M., Bottoni A., Zerbetto F. Thermodynamics of Binding between Proteins and Carbon Nanoparticles: The Case of C60@Lysozyme. J. Phys. Chem. C. 2015;119:28077–28082. doi: 10.1021/acs.jpcc.5b09985. [DOI] [Google Scholar]
- 51.Calvaresi M., Furini S., Domene C., Bottoni A., Zerbetto F. Blocking the passage: C60 geometrically clogs K+ channels. ACS Nano. 2015;9:4827–4834. doi: 10.1021/nn506164s. [DOI] [PubMed] [Google Scholar]
- 52.Trozzi F., Marforio T.D., Bottoni A., Zerbetto F., Calvaresi M. Engineering the Fullerene-protein Interface by Computational Design: The Sum is More than its Parts. Isr. J. Chem. 2017;57:547–552. doi: 10.1002/ijch.201600127. [DOI] [Google Scholar]
- 53.di Giosia M., Valle F., Cantelli A., Bottoni A., Zerbetto F., Calvaresi M. C60 bioconjugation with proteins: Towards a palette of carriers for all pH ranges. Materials. 2018;11:691. doi: 10.3390/ma11050691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bologna F., Mattioli E.J., Bottoni A., Zerbetto F., Calvaresi M. Interactions between Endohedral Metallofullerenes and Proteins: The Gd@C60–Lysozyme Model. ACS Omega. 2018;3:13782–13789. doi: 10.1021/acsomega.8b01888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ganazzoli F., Raffaini G. Classical atomistic simulations of protein adsorption on carbon nanomaterials. Curr. Opin. Colloid Interface Sci. 2019;41:11–26. doi: 10.1016/j.cocis.2018.11.008. [DOI] [Google Scholar]
- 56.Marforio T.D., Calza A., Mattioli E.J., Zerbetto F., Calvaresi M. Dissecting the supramolecular dispersion of fullerenes by proteins/peptides: Amino acid ranking and driving forces for binding to c60. Int. J. Mol. Sci. 2021;22:11567. doi: 10.3390/ijms222111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Di Giosia M., Zerbetto F., Calvaresi M. Incorporation of Molecular Nanoparticles Inside Proteins: The Trojan Horse Approach in Theranostics. Acc. Mater. Res. 2021;8:594–605. doi: 10.1021/accountsmr.1c00065. [DOI] [Google Scholar]
- 58.Di Giosia M., Bomans P.H.H., Bottoni A., Cantelli A., Falini G., Franchi P., Guarracino G., Friedrich H., Lucarini M., Paolucci F., et al. Proteins as supramolecular hosts for C60: A true solution of C60 in water. Nanoscale. 2018;10:9908–9916. doi: 10.1039/C8NR02220H. [DOI] [PubMed] [Google Scholar]
- 59.Soldà A., Cantelli A., Di Giosia M., Montalti M., Zerbetto F., Rapino S., Calvaresi M. C60@lysozyme: A new photosensitizing agent for photodynamic therapy. J. Mater. Chem. B. 2017;5:6608–6615. doi: 10.1039/C7TB00800G. [DOI] [PubMed] [Google Scholar]
- 60.Di Giosia M., Soldà A., Seeger M., Cantelli A., Arnesano F., Nardella M.I., Mangini V., Valle F., Montalti M., Zerbetto F., et al. A Bio-Conjugated Fullerene as a Subcellular-Targeted and Multifaceted Phototheranostic Agent. Adv. Funct. Mater. 2021;31:2101527. doi: 10.1002/adfm.202101527. [DOI] [Google Scholar]
- 61.Martínez-Ortega U., Figueroa-Figueroa D.I., Hernández-Luis F., Aguayo-Ortiz R. In Silico Characterization of Masitinib Interaction with SARS-CoV-2 Main Protease. ChemMedChem. 2021;16:2339–2344. doi: 10.1002/cmdc.202100375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X., Yang S.T., Wang H., Wang L., Hu W., Cao A., Liu Y. Influences of the size and hydroxyl number of fullerenes/fullerenols on their interactions with proteins. J. Nanosci. Nanotechnol. 2010;10:6298–6304. doi: 10.1166/jnn.2010.2623. [DOI] [PubMed] [Google Scholar]
- 63.Luo S., Huang K., Zhao X., Cong Y., Zhang J.Z.H., Duan L. Inhibition mechanism and hot-spot prediction of nine potential drugs for SARS-CoV-2 Mproby large-scale molecular dynamic simulations combined with accurate binding free energy calculations. Nanoscale. 2021;13:8313–8332. doi: 10.1039/D0NR07833F. [DOI] [PubMed] [Google Scholar]
- 64.Pavlova A., Lynch D.L., Daidone I., Zanetti-Polzi L., Smith M.D., Chipot C., Kneller D.W., Kovalevsky A., Coates L., Golosov A.A., et al. Inhibitor binding influences the protonation states of histidines in SARS-CoV-2 main protease. Chem. Sci. 2021;12:1513–1527. doi: 10.1039/D0SC04942E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kneller D.W., Phillips G., Weiss K.L., Zhang Q., Coates L., Kovalevsky A. Direct Observation of Protonation State Modulation in SARS-CoV-2 Main Protease upon Inhibitor Binding with Neutron Crystallography. J. Med. Chem. 2021;64:4991–5000. doi: 10.1021/acs.jmedchem.1c00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chan H.T.H., Moesser M.A., Walters R.K., Malla T.R., Twidale R.M., John T., Deeks H.M., Johnston-Wood T., Mikhailov V., Sessions R.B., et al. Discovery of SARS-CoV-2 Mpropeptide inhibitors from modelling substrate and ligand binding. Chem. Sci. 2021;12:13686–13703. doi: 10.1039/D1SC03628A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kneller D.W., Phillips G., Weiss K.L., Pant S., Zhang Q., O’Neill H.M., Coates L., Kovalevsky A. Unusual zwitterionic catalytic site of SARS-CoV-2 main protease revealed by neutron crystallography. J. Biol. Chem. 2020;295:17365–17373. doi: 10.1074/jbc.AC120.016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramos-Guzmán C.A., Ruiz-Pernía J.J., Tuñón I. Unraveling the SARS-CoV-2 Main Protease Mechanism Using Multiscale Methods. ACS Catal. 2020;10:12544–12554. doi: 10.1021/acscatal.0c03420. [DOI] [PubMed] [Google Scholar]
- 69.Arafet K., Serrano-Aparicio N., Lodola A., Mulholland A.J., González F.V., Świderek K., Moliner V. Mechanism of inhibition of SARS-CoV-2 MprobyN3peptidyl Michael acceptor explained by QM/MM simulations and design of new derivatives with tunable chemical reactivity. Chem. Sci. 2021;12:1433–1444. doi: 10.1039/D0SC06195F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maier J., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Case D.A., Betz R.M., Botello-Smith W., Cerutti D.S., Cheatham I.T.E., Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., et al. Amber 16. University of California; San Francisco, CA, USA: 2016. [Google Scholar]
- 72.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data in this study can be requested from the corresponding authors (tainah.marforio2@unibo.it, T.D.M.; matteo.calvaresi3@unibo.it, M.C.).