Abstract
Studies on animal models with mutations in ameloblastin gene have suggested that the extracellular matrix protein ameloblastin (AMBN) plays important roles in controlling cell-matrix adhesion and ameloblast polarization during amelogenesis. In order to examine the function of AMBN in cell polarization and morphology, we developed an in vitro 3D cell culture model to examine the effect of AMBN and amelogenin (AMEL) addition on ameloblast cell lines. We further used high resolution confocal microscopy to detect expression of polarization markers in response to AMBN addition. Addition of AMBN to the 3D culture matrix resulted in the clustering and elongation (higher aspect ratio) of ALC in a dose dependent manner. The molar concentration of AMEL required to exact this response from ALC was 2.75- times greater than that of AMBN. This polarization effect of ameloblastin was attributable directly to an evolutionary conserved domain within its exon 5-encoded region. The lack of exon 6-encoded region also influenced AMBN-cell interactions but to a lesser extent. The clusters formed with AMBN were polarized with expression of E-cadherin, Par3 and Cldn1 assembly at the nascent cell-cell junctions. The elongation effect was specific to epithelial cells of ameloblastic lineage ALC and LS8 cells. Our data suggest that AMBN may play critical signaling roles in the initiation of cell polarity by acting as a communicator between cell-cell and cell-matrix interactions. Our investigation has important implications for understanding the function of ameloblastin in enamel-cell matrix adhesion and the outcomes may contribute to efforts to develop strategies for enamel tissue regeneration.
Keywords: Ameloblastin, Ameloblast polarization, 3D culture, Enamel, Amelogenin
Introduction
Mature enamel, which is the hardest biomineralized tissue in the vertebrate body, is acellular and contains little to no residual organic matter. The prisms or rods are the basic structural units of prismatic enamel. The prisms run from the dentinoenamel junction to the outer surface and are each made of numerous hydroxyapatite (HAP) crystallites. Present between the arrays of prisms are the interprismatic mineral phase [1,2].
Unlike bone and dentin, enamel has a unique mechanism of mineralization occurring in an extracellular, non-collagenous environment [3]. This dynamic extracellular matrix, which is comprised primarily of amelogenin, followed by ameloblastin and enamelin, is constantly remodeled as amelogenesis progresses from the secretory stage through the transition and maturation stages [4]. The two major matrix modifiers during amelogenesis are matrix metalloproteinase-20 (Mmp20) during the secretory stage and kallikrein-4 (Klk4) during the maturation stage [5]. In humans, defects in enamel formation occurring secondary to genetic abnormalities are collectively referred to as Amelogenesis Imperfecta (AI). Mutations in AMBN and AMELX genes lead to AI [6,7]. Although ameloblastin (AMBN) is not the most abundant enamel matrix protein (EMP), AMBN truncation or loss in mouse models has consequences for enamel development that are far more severe than those caused by truncation or loss of amelogenin. The effects of AMBN truncation include severe cell and mineral abnormalities resulting in an absence of true enamel [8,9].
In mature ectodermal secretory organs like salivary and lacrimal glands and in the ameloblasts, the epithelial cells share common features: elongated shapes with polarized nuclei, close cell-cell adhesion, and specialized intercellular junctions with an apico-basal cell polarity. Ameloblasts undergo dynamic morphological changes during the various stages of amelogenesis. They begin as short cuboidal cells and dramatically transform into tall, columnar, highly polarized cells during the onset of presecretory stage in humans (or secretory stage in mice) [2,10]. They perform important functions including but not limited to the secretion of EMPs during the secretory stage of amelogenesis, followed by mineral ion transport and EMPs reabsorption during the maturation stage. Loss of ameloblast polarity due to the knockout of junctional complex proteins [11–13] or occurring secondary to the absence of certain EMPs (specifically ameloblastin and enamelin) results in enamel microstructural defects [8,14].
AMBN has been specifically immunolocalized in developing rodent incisor ameloblasts to the Tomes’ process [15–17] —a feature of ameloblasts theorized to be responsible for the generation of prismatic enamel structure in higher vertebrates [18]. Given the reported correlation between the presence of Tomes’ processes and the enamel prismatic microstructure and the apparent immunolocalization of AMBN to ameloblast Tomes’ process [15,17] we hypothesize that AMBN indirectly participates in the orchestrated prism formation process by impacting cell morphological changes or maintenance of the Tomes’ processes. Although AMBN is not the predominant EMP, during the early and late bell stage of human tooth development, it has been detected immunohistochemically in the acellular zone of the dental papilla near the inner enamel epithelial cells that are cuboidal in comparison to the highly elongated secretory ameloblasts [19]. Although the exact function of such an expression pattern has not been explained, it was suggested to be part of the well-characterized process of reciprocal induction during odontogenesis [2].Considering the dynamic roles played individually by the cells and the proteins during amelogenesis, we further hypothesize that they undergo specific cooperative cell-matrix interactions that function to pattern the microstructure of enamel through control of cell architecture.
A direct link between ameloblast cell morphology, polarization and the resultant higher-order structures of enamel is yet to be established. The spectrum of abnormalities observed in Ambn mutant animals precludes the possibility of examining the effect of absence of functional AMBN on the cell morphology [8,9]. Developing reliable cell culture models in which AMBN effect on cell behavior can be examined is therefore timely and essential. Upon enamel maturation ameloblasts transform into cells of reduced enamel epithelium which are eventually shed into the oral cavity upon tooth eruption, resulting in the lack of a reliable primary cell source of terminally differentiated ameloblasts [2]. Here, we used ameloblast lineage cells (ALC) that are derived from mouse molars through a spontaneous immortalization process. They have been shown to stably express the ameloblast cell markers and appear cobblestone like in 2D monolayer cultures and are considered to be a suitable in vitro ameloblast cell line [20]. Additionally, LS-8 cells that are immortalized using SV40 were used as an adjunct ameloblast cell line [21]. To examine the effects of exogenous AMBN on the morphology of ameloblasts we developed a novel 3D cell culture model onto which recombinant AMBN and AMEL were added. We evaluated changes in cell clustering and elongation and demonstrated expression and localization of polarization markers as the result of AMBN addition to the 3D gel system.
Results
The effect of AMBN on cell clustering and elongation is greater than AMEL
A three-dimensional on-top type culture system with GFR Geltrex was optimized to examine the role of exogenously supplemented recombinant EMPs AMBN and AMEL on ALC (Fig 1). At the end of 24h of culture in the presence of AMBN and AMEL, the majority of cells exhibited a characteristic pattern that involved the formation of cell clusters (Fig. 1 b, c), as visualized using cell membrane (DiD) and nuclear labeling (DAPI). Sequential Z stacks were recorded to examine the morphology of these cell clusters in 3D in the absence and presence of AMBN (Fig. 1 d–g). The reconstructed images revealed an organized selective elongation of cells in the presence of AMBN, along the Z axis into the depths of the gel (from the surface) (Fig. 1 e, g). Fluorescent imaging revealed a preferential nuclear localization towards one pole of the cells as viewed in 3D reconstructions (Supplementary Fig. S1). In the group of cells cultured in growth-factor reduced (GFR) Geltrex alone (control), the surface cell morphology remained largely devoid of the pattern of clustering seen in the samples with proteins (data not shown). Similarly, the surface cell morphology in the other negative controls (BSA and heat-denatured AMBN) remained largely devoid of this pattern of clustering, with the cells remaining discrete (Fig. 1a). In 3D reconstructions, the discrete cells appeared planar without any significant elongation along the Z axis (Fig. 1 d,f). Interestingly, heat-denatured AMBN was sufficient to reverse the effect of AMBN on ALC (Fig. 1a,d). Within the first 24h of culture, the cells exhibited a 7-fold average increase in the aspect ratio (height to width) in the presence of AMBN and AMEL compared individually to the cells grown in gel only, with BSA or with heat-denatured AMBN (n=30; p < 0.001 each) (Fig. 1h.). This difference in the cell aspect ratio between the test and control groups remained consistent at the end of 72h of culture (n=15; p < 0.001) (Supplementary Fig. S2). Comparison of ALC aspect ratio values in the AMBN and AMEL groups revealed significantly greater cell elongation in the presence of AMBN (n=30; p < 0.001) (Fig. 1h).
Fig. 1.
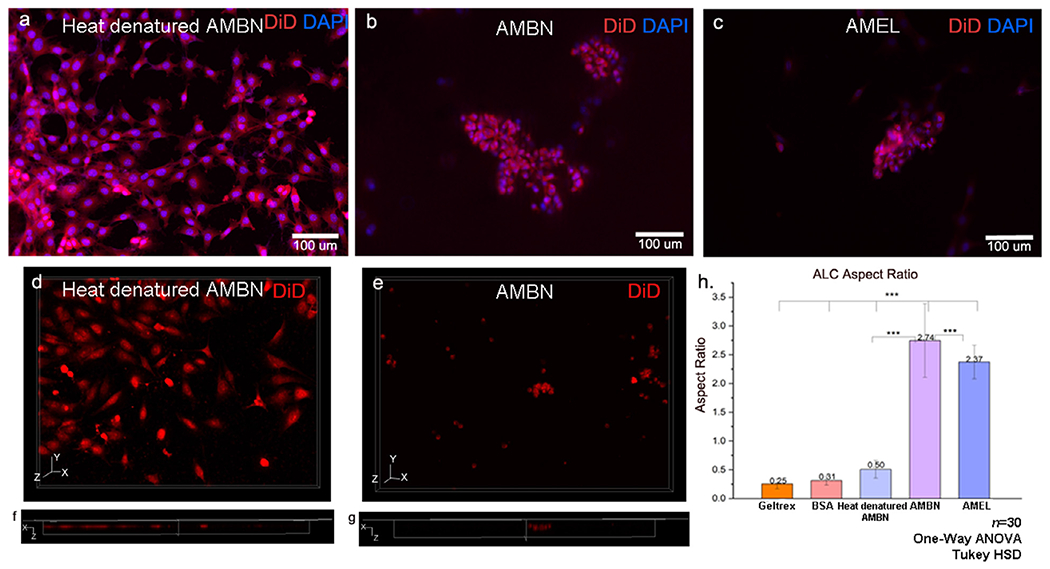
ALC form 3D clusters of elongated cells in the presence of amelogenin and ameloblastin in GFR-Geltrex. XY surface morphology of the cells in the presence of (a) heat denatured AMBN (negative control), (b) AMBN, (c) AMEL. Note the characteristic clustering of cells in b and c whereas cells remain discrete in a. (d, e) three-dimensional reconstruction of Z stacks from control and test groups and (f, g) orthogonal axial sections of the Z stacks. Note the presence of flat cells without significant height in Z direction in (f) negative control. (g) elongated cells within the clusters in AMBN. ALC aspect ratio comparison between control, BSA, heat denatured AMBN and tests (AMBN and AMEL). * One-way ANOVA; Tukey HSD; p<0.05; ** p<0.01; *** p<0.001.
The effects of growth factors and gel stiffness on clustering and elongation
To examine the possibility of the gel substrate impacting cell morphology in 3D culture, and to eliminate the role of residual growth factors in culture substrates, type I collagen and transglutaminase crosslinked gelatin (ColTgel) were used as adjunct gel substrates. Upon comparing the ALC aspect ratio values between GFR Geltrex and type I collagen gels, it was observed that there was no statistically significant difference in the ALC aspect ratio between these two substrates indicating the elongation caused by AMBN does not dependent upon the presence of growth factors (n=15; p > 0.05) (Supplementary Fig. S2).
When cultured in ‘stiff’ (35-47kPa) or ‘soft’ (0.9-1.5kPa) ColTgel, the same level of cell elongation in response to AMBN was observed only after 72h. However, with the medium-stiffness gel (14-20 kPa), the ALC aspect ratio was enhanced in the presence of AMBN after 24h of culture. (Supplementary Fig. S3). Given the observation that substrate composition does not have a significant effect on ALC elongation in 3D culture, except the timing, all further experiments were carried out using GFR Geltrex gel alone where the effect can be seen after 24 hours.
AMBN and AMEL do not have a synergistic effect on elongation of ALC
The optimized 3D-on-top type culture system with GFR Geltrex was used to explore the possibility of a combined synergistic effect of AMEL and AMBN on ALC. This was accomplished by combining recombinant AMEL and AMBN, and by using purified porcine enamel matrix extract. In the AMEL+AMBN group, the two proteins were introduced at an exaggerated AMEL: AMBN molar ratio of 2:1 (1.98 μM AMEL: 0.48 μM AMBN) keeping the protein concentrations consistent with previous experiments presented in Fig 1. ALC cell surface morphology in both of the combination groups (recombinant and native) were comparable to those cultured with full-length AMBN (Fig. 2 a–c), with the cells forming the characteristic clusters. Upon measuring the aspect ratio of the cell clusters after 24h in 3D culture, it was observed that neither the recombinant nor the native groups exhibited a significant increase in the ALC aspect ratio compared to the culture with AMBN alone (Fig. 2.d). The ALC aspect ratio with recombinant AMBN was 1.24-fold greater than with AMEL+AMBN (n=30; p < 0.01) and 1.39-fold greater than with native AMEL+AMBN (n=30; p < 0.001) (Fig. 2d). The aspect ratios of ALC in the combination groups were not statistically significantly different from those in the presence of AMEL alone (n=30; p > 0.05).
Fig. 2.
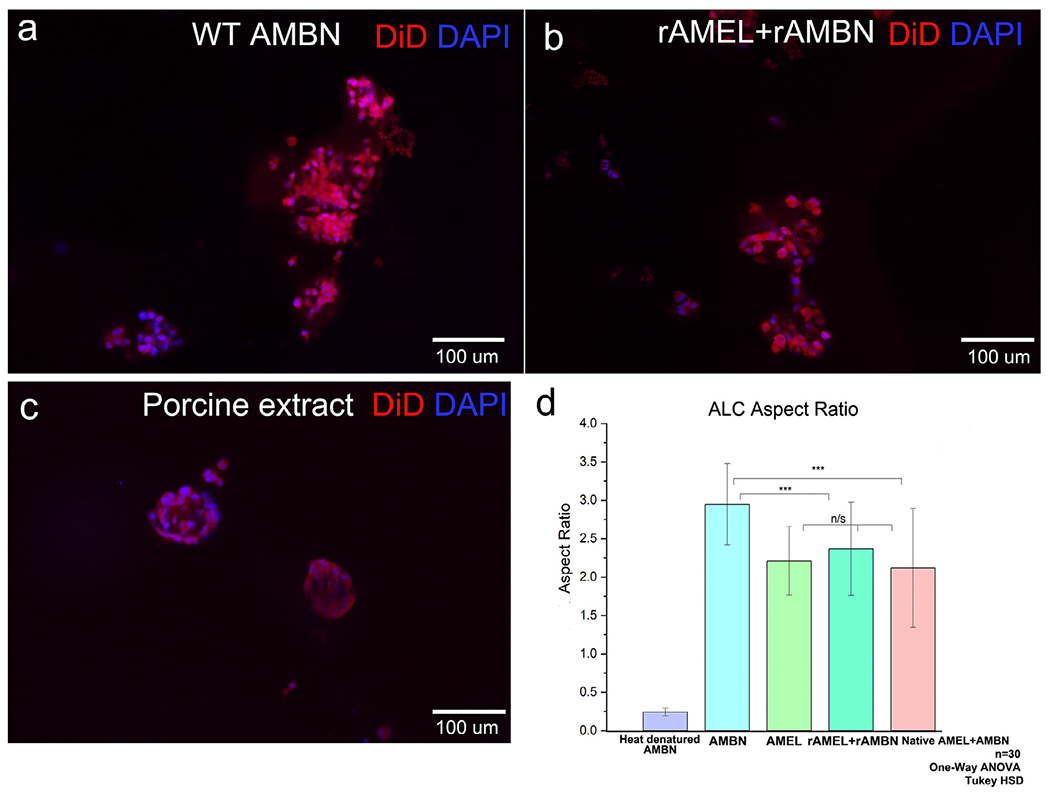
Combination of native and recombinant AMEL (rM179) and AMBN in GFR-Geltrex and their effect on cell elongation. Fluorescent microscope images of the clusters labeled with DiD (membrane) and DAPI (nucleus); (a) recombinant AMBN only, (b) recombinant AMEL+ recombinant AMBN, (c) native AMBN and AMEL isolated from developing porcine enamel, (d) aspect ratio measurements of the clusters. Statistical analysis performed using One-way ANOVA and Tukey HSD; p<0.05; ** p<0.01; *** p<0.001.
The effect of AMBN on elongation is dose-dependent and specific to ALC and LS8
To examine the specificity of AMBN’s effect on ameloblast cells we first examined whether ALC elongation is dependent upon the dosage of AMBN. We then examined the elongation of kidney tubular epithelial cells (TCMK-1) (negative control) and ameloblast-like LS-8 (positive control) cells in response to AMBN. LS8 cells represent an earlier stage of amelogenesis when compared to ALC cells [22]. The effects of varying concentrations of AMBN were analyzed by repeating the 3D ALC aspect ratio measurements with AMBN at 5 μg/ml (0.12 μM), 10 μg/ml (0.24 μM), 15 μg/ml (0.36 μM) and 20 μg/ml (0.48 μM) concentrations. ALC clustering and elongation along the Z axis occurred in the presence of AMBN with a concentration as low as 0.12 μM, with cells recording an average aspect ratio of 2.27 +/− 0.46 (n=30). One-way ANOVA revealed a statistically significant difference in ALC aspect ratio between the 4 different concentrations of ameloblastin tested (n=30; p < 0.001). Furthermore, a direct correlation between AMBN concentration and ALC aspect ratio was extrapolated (at concentrations higher than 10 μg/ml) with a correlation co-efficient of 0.92 and R2 of 0.85 (n=30) (Fig. 3a). The ALC aspect ratio that was obtained in the presence of AMEL (2.37+/−0.29) at 0.99 μM molar concentration was achieved with AMBN at 0.36 μM concentration (Fig. 2). Similar to the ALC, the LS-8 cells cultured in the presence of AMBN formed clusters of cells unlike the discreet cells in the control (Geltrex alone and heat-denatured AMBN). Measurements of cell aspect ratio revealed a significant increase in cell elongation with AMBN compared to the controls (n=15; p < 0.001) (Supplementary Fig. S4).
Fig. 3.
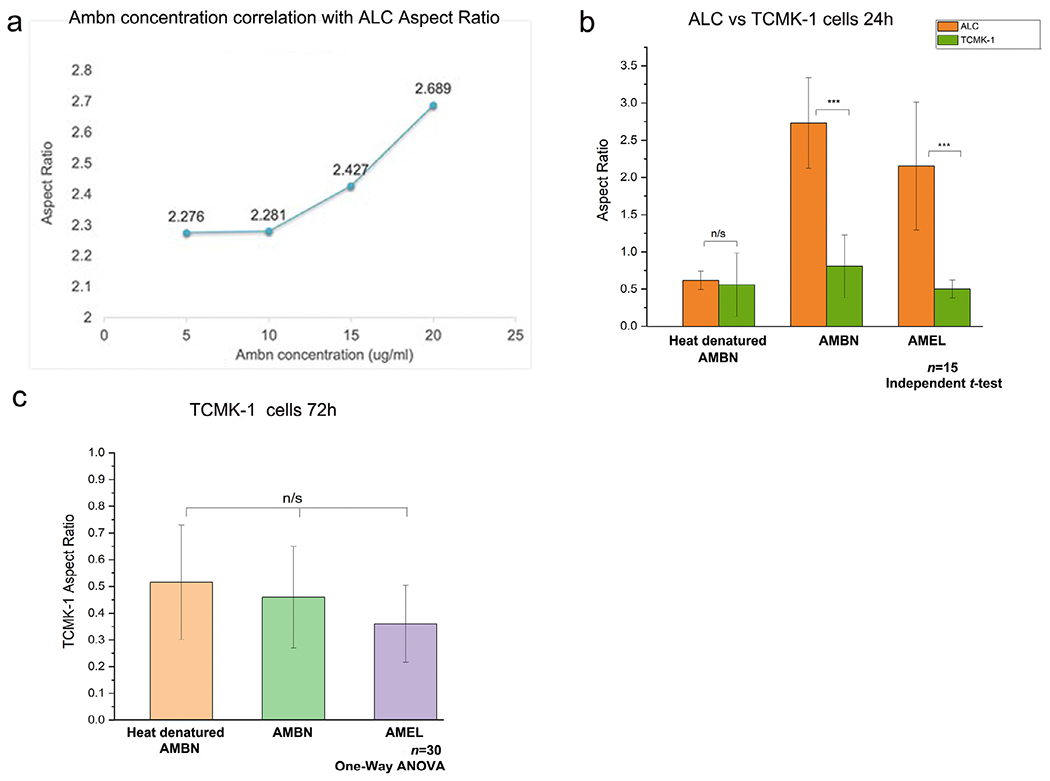
Specificity and dose dependency of AMBN effect on ALC in 3D GFR-Geltrex. (a) Curve representing the linear correlation between wild type recombinant AMBN concentration and ALC aspect ratio. Aspect ratio values corresponding to 5 μg/ml (2.27+/−0.46); 10 μg/ml (2.28+/−0.38); 15 μg/ml (2.42+/− 0.35); 20 μg/ml (2.68 +/− 0.47). (b) Comparison of ALC and TCMK-1 aspect ratio in the presence of control and test proteins at 24h, (c) TCMK-1 cell aspect ratio in the presence of test and control proteins at 72h.
However, the kidney tubular epithelial cells (TCMK-1) did not mirror the effects of AMBN on ALC or LS-8. Examining TCMK-1 cells at both time-points of 24h (Fig 3b) (n=15) and 72h (Fig 3c) (n=30) following initiation of 3D culture revealed no statistically significant difference in aspect ratio between the test (AMBN, AMEL) and control (heat-denatured AMBN) groups (p > 0.05) (Fig. 3b–c). In contrast to ALC and LS-8, TCMK-1 cell surface morphology remained planar in all three groups (Fig. 3c).
Elongated ALC clusters display polarized distributions of junctional complex proteins
Polarization of ALC cells was confirmed with actin labeling using Alexa Fluor 488 conjugated Phalloidin under a confocal laser scanning microscope (Supplementary Fig. S5). Given the clustering of cells observed in the presence of EMPs and the well-characterized role of E-cadherin-mediated initiation and maintenance of epithelial cell polarity [23], junctional complex protein immunolabeling was carried out in 3D. The specific polarization markers that were chosen were E-cadherin (Alexa 555 conjugated), Par3 and Claudin-1 (Cldn1) alongside membrane markers including anti-beta actin antibody and DiD. Double and triple labeling were carried out with relevant combinations of these markers to examine their distribution patterns within the cell clusters. Representative confocal laser scanning images reconstructed from sequential Z stacks confirmed that the clusters formed in the presence of AMBN were composed of multiple individual cells (Fig. 4a–d). Within these cell clusters, membrane labeling using DiD revealed a symmetric, uniform lipid distribution along the outline of the cell (Fig 4c&d), whereas E-cadherin immunolabeling was restricted to the basal side of the membrane relative to the nucleus (Fig. 4 d, e, h white arrows). The same preferential localization pattern of E-cadherin was observed using anti-actin antibody (Fig. 5). The co-localization patterns of DiD with Anti-E-cadherin and Anti-actin with Anti-E-cadherin in the elongated cells were restricted to one pole of the cell, as indicated by asterisks in Fig. 4 e &h and Fig. 5a &d. This asymmetric E-cadherin labeling pattern along the Z axis of elongated cell clusters was confirmed using a series of 2D images taken at varying Z positions (Supplementary Fig. S6). Taken together, these findings suggest that the immunolabeling range (along the Z axis) for E-cadherin was significantly lower than that of DiD and Actin. In the planar cells that were present in the group cultured with heat-denatured AMBN, no such asymmetric distribution patterns of junctional complex markers were observed (Fig. 6). Par3, a polarity protein that is known to accumulate at nascent cell-cell junctions along with E-cadherin, co-localized along the same side of the cell membrane that was labeled with E-cadherin (Fig. 7 a, b asterisks). Tight junctional protein marker Claudin-1 revealed a similar asymmetric distribution pattern (Fig. 8d green arrow). Antibody controls for these experiments using secondary antibodies alone are shown in Supplementary Fig. S7.
Fig. 4.
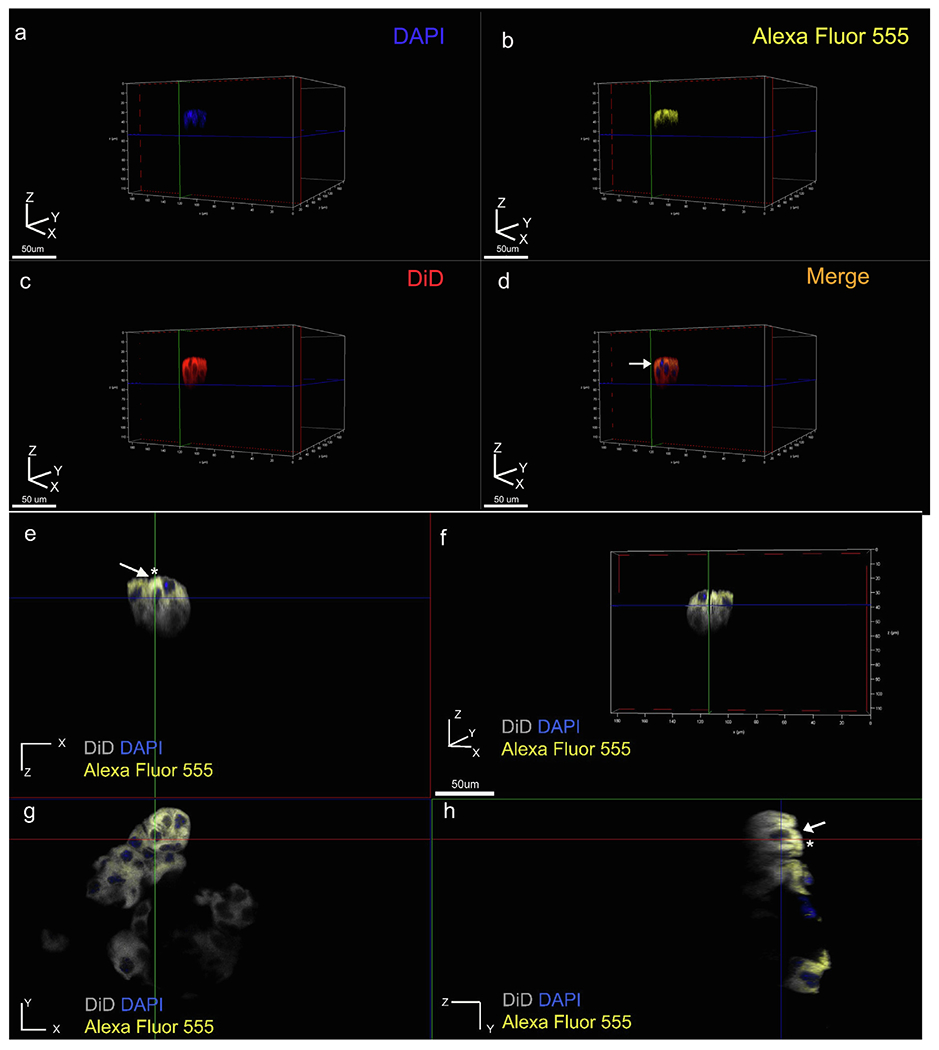
Elongated clusters of cells display polarized distribution of E-cadherin. (a-d) Three-dimensional volume rendering of a representative cell cluster viewed individual channel wise (a-c), and merged (d). Membrane labelled in red (DiD), nucleus in blue (DAPI) and E-cadherin in yellow (Alexa Fluor 555 conjugated primary antibody (yellow), (e-h) two-dimensional sectioning of representative cluster where e, g, h represents XZ, XY and ZY plane sections respectively, (f) three-dimensional volume rendering of the cluster viewed using all three channels. Membrane labeling with DiD (grey pseudo color), nucleus (blue), and E-cadherin (yellow). Membrane-cadherin co-localization is restricted to one pole of the cell along the Z axis (white arrows and asterisks in e, h).
Fig. 5.
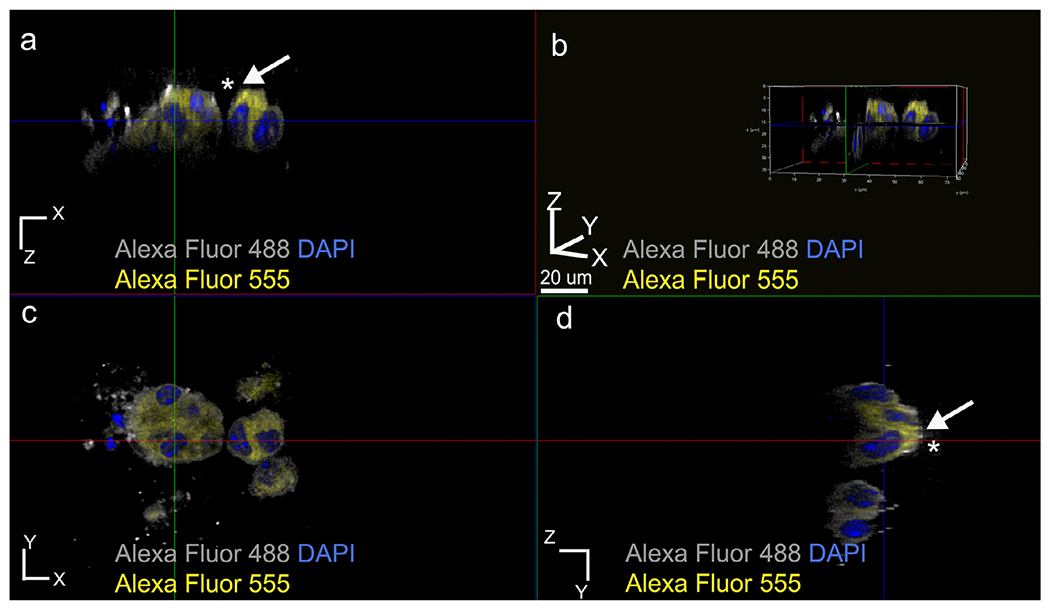
Membrane labeling using anti-actin antibody to confirm the asymmetrical E-cadherin distribution pattern within ALC clusters formed in the presence of WT recombinant AMBN. (a) XZ plane section, (b) three-dimensional volume rendering of the cluster, (c) XY plane section and (d) YZ plane section. White arrows with asterisks in a and d indicate membrane-actin colocalization pattern. Grey pseudo color for Alexa Fluor 488 actin and yellow Alexa fluor 555 conjugated anti-E-cadherin.
Fig. 6.
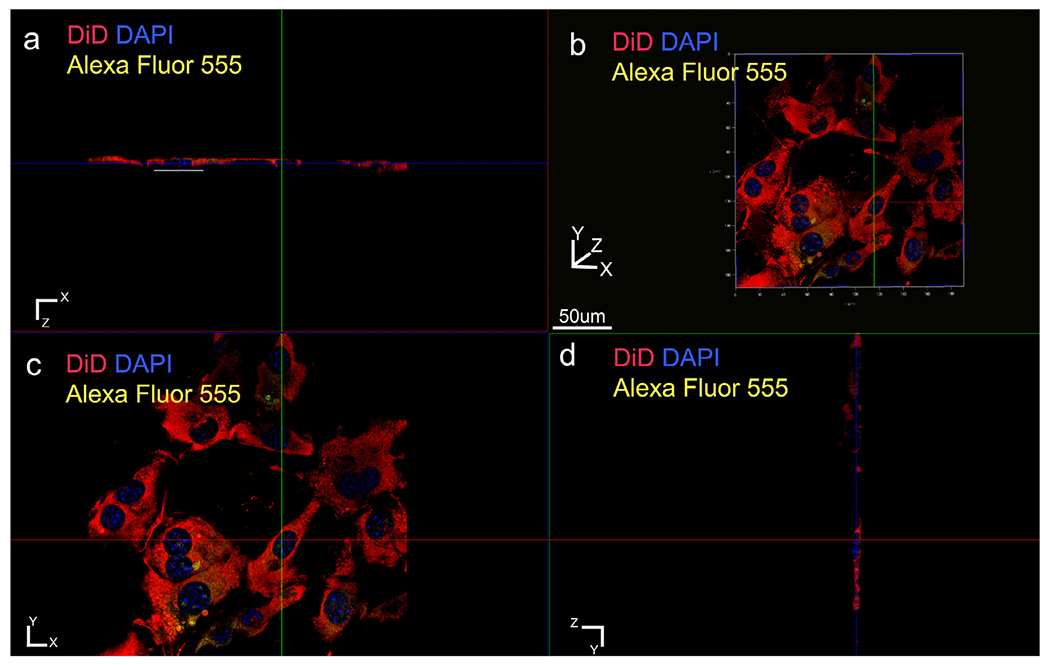
E-cadherin and membrane labeling in the control (heat denatured ameloblastin). The cells are strikingly different with negligible elongation along the Z direction (white dashed line in a). (a) XZ plane section, (b) three-dimensional volume rendering, (c) XY plane section, (d) YZ plane section. Membrane labeled with DiD (red); nucleus with DAPI (blue) and E-cadherin with Alexa Fluor 555 conjugated primary antibody (yellow).
Fig. 7.
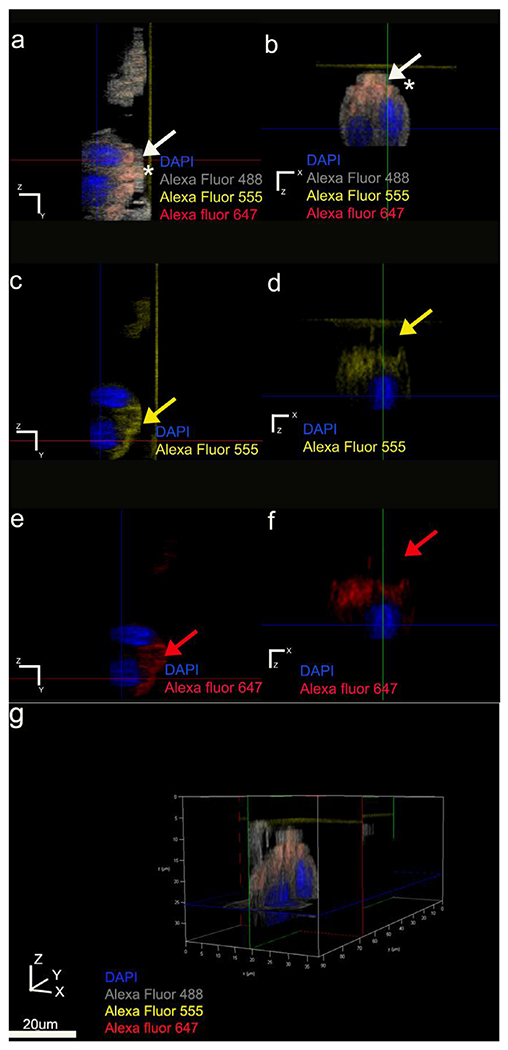
Asymmetrical immunolabeling pattern of polarity protein Par3. Par3 localization within the clusters restricted to the same pole of the cell membrane as E-cadherin. Anti-actin antibody used as the symmetrical membrane marker. Actin represented with grey pseudo color; Par3 in red, E-cadherin in yellow and nucleus in blue. (a, b) Par3 and E-cadherin colocalize (represented in orange) basal to the nucleus position indicated by white arrows and asterisks. (c, d) YZ and XZ plane sections of Alexa Fluor 555 E-cadherin channel respectively. (e, f) YZ and XZ plane sections of Alexa Fluor 647 Par-3 channel respectively. (g) three-dimensional volume rendering of the cluster viewed using all channels.
Fig. 8.
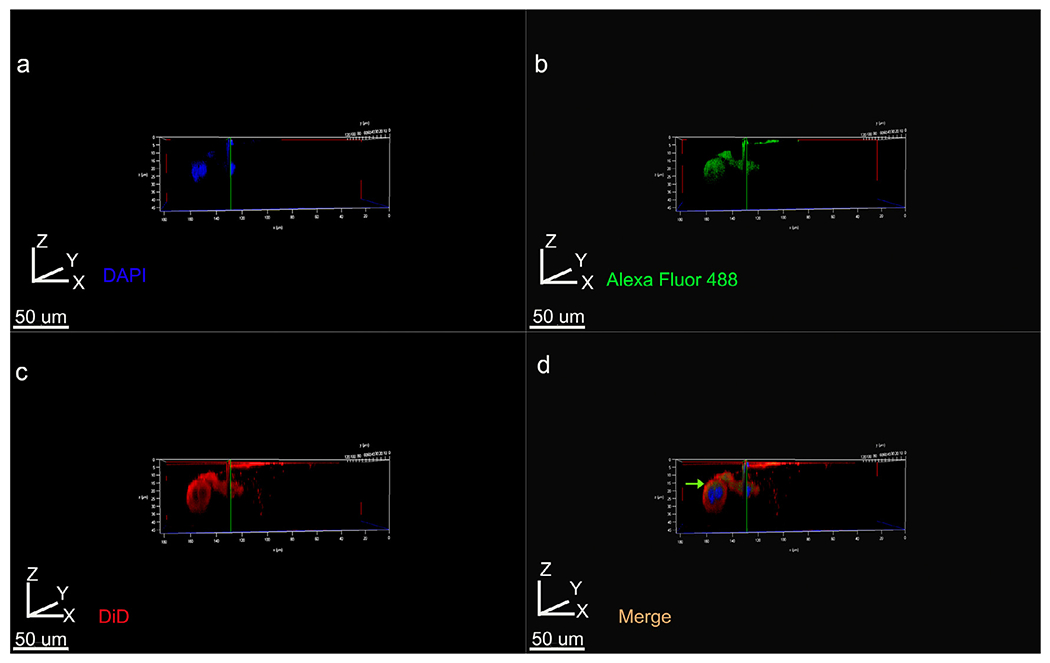
3D volume rendering of Z stacks showing tight junctional protein Claudin-1 distribution within ALC clusters labeled with DiD for cell membrane. (a) DAPI, (b) Cldn1 labeling with Alexa Fluor 488 conjugated secondary, (c) DiD, (d) merged imaged using all three channels. Cldn1 labeling is restricted to one pole of the cell and is not detectable on the below the nucleus (green arrow in d).
The exon 5-encoded sequence of AMBN is involved in ALC clustering and elongation
Recent in vitro and cell culture data from our laboratory highlighted the potential of the exon 5-encoded region of ameloblastin to interact with ameloblast cells [24]. In an attempt to examine the function of this domain in cell clustering and elongation, we generated a recombinant AMBN protein lacking the sequence encoded by exon 5 (AMBNΔ5). In order to examine the influence of adjacent amino acids we also generated a protein lacking the sequence encoded by exon 6 (AMBNΔ6) [25]. The loss of these abovementioned regions has significant effects on enamel development [8] as documented by evidence from animal models. ALC cultured with AMBNΔ5 exhibited none of the characteristics of cell clustering seen when cultured with full-length AMBN (Fig. 9b). Measurements of cell aspect ratio in the presence of AMBNΔ5 confirmed the absence of cell elongation, with cells remaining planar compared to those cultured with full-length AMBN (n=30; p < 0.001) (Fig. 9d). These planar cells did not display any asymmetry in the distribution patterns of junctional complex proteins (data not shown). The distinct planar morphologies observed with AMBNΔ5 was similar to what was observed with negative controls (Geltrex alone, BSA and heat-denatured AMBN). With AMBNΔ6, a heterogenous cell population consisting of some clusters interspersed with planar cells was observed. Although the ability of ALC to cluster was not completely lost with the removal of exon 6 encoded sequence, the aspect ratios of the cells were significantly lower compared to those cultured with full-length AMBN (n=30; p < 0.05) (Fig. 9d).
Fig. 9.
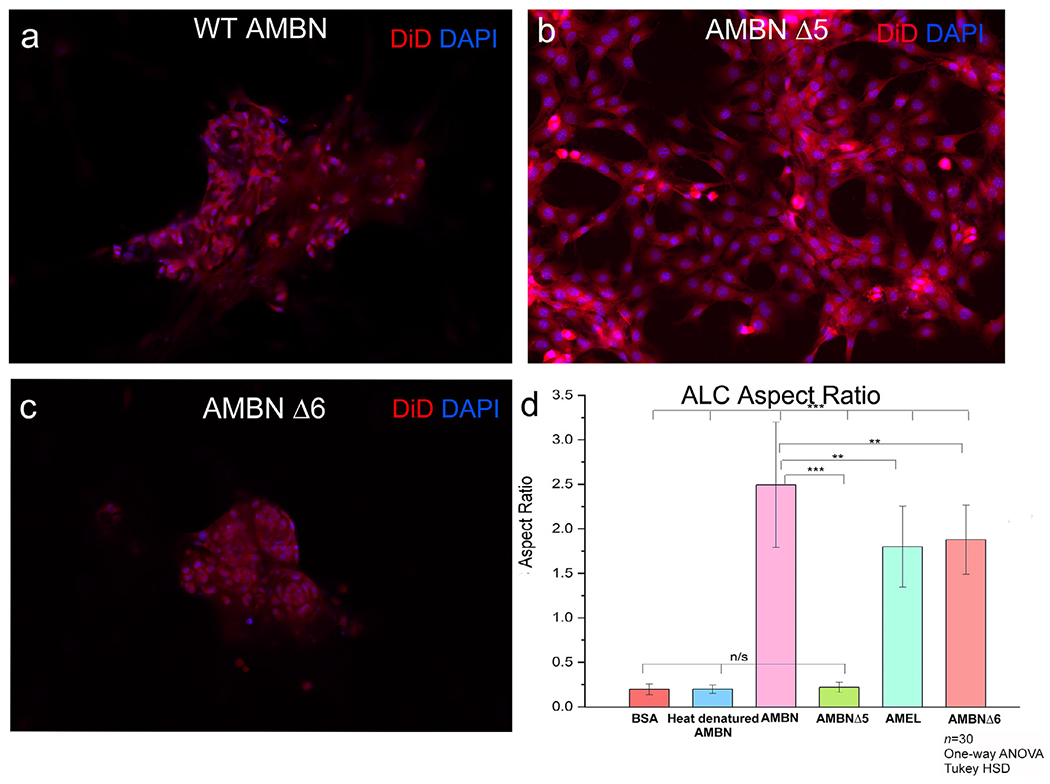
ALC ALC morphology in the presence of AMBN mutants lacking exon 5 and exon 6 encoded regions (AMBNΔ5 and AMBNΔ6). Fluorescent microscope images of DiD (membrane) and DAPI (nucleus) labeled cell clusters in the presence of: (a) recombinant wild type AMBN, (b) AMBNΔ5 showing the complete reversal of this clustering phenomena in the case of the AMBNΔ5 mutant, (c) AMBNΔ6 showing some cells remaining planar while others elongating in the presence of this mutant. (d) aspect ratio measurements of ALC cells in the presence of mutants. One-way ANOVA; Tukey HSD; p<0.05; ** p<0.01; *** p<0.001.
Discussion
We used ameloblast-like cell lines ALC and LS-8 to demonstrate that enamel extracellular matrix proteins AMEL and AMBN altered ameloblast cell morphology and polarization status when cultured in a 3D gel matrix. We observed that enamel epithelial cell lines cultured in this manner in the presence of ameloblastin formed clusters of elongated cells with asymmetrical distribution of cell polarity and tight junctional proteins. This effect of ameloblastin was stronger than amelogenin and was attributable directly to a domain within its exon 5-encoded region. The lack of exon 6-encoded region also influenced AMBN-cell interactions but to a lesser extent.
In general, 3D microenvironments allow for a better replication of in vivo conditions [26] in terms of controlled cell proliferation [27] viability [28] and morphology [29]. Although primary enamel epithelial cells have been previously cultured in 3D, [28,30] we aimed at generating a model that would allow non-uniform contact surfaces in 3D. This is in striking contrast with the earlier models in which the cells were embedded in the entirety of the gel for longer periods of time. The rationale behind generating this 3D-on-top type model was to ensure that the bioactive molecules could be presented along only one face of the cell (along the Z axis). Although the Geltrex substrates were largely devoid of growth factors, the stiffness of the gel can impact the cell surface receptors and result in cells assuming certain morphologies [31]. Furthermore, substrate stiffness can affect the diffusion rates of added bioactive molecules. To optimize experimental conditions and to examine the effect of mechanical properties of the gel substrate, all three commercially available densities of the gelatin substrate were tested. In the stiffest and softest of the three, with strengths of 35-47kPa and 0.9-1.5kPa respectively, only after 72h did ALC respond to AMBN exhibiting morphological changes. We therefore elected to work with GFR Geltrex (protein concentration of 9-12 mg/ml), which produced a more rapid cellular response and allowed us to collect data after 24 hours.
A method employing scaffolds in a 3D culture system using a bioreactor model was recently published [32] where the modification of the cell microenvironment using growth factors and porcine enamel matrix protein crude extracts resulted in the polarization of the incisor cervical loop dental epithelial cells, characterized by the presence of elongated tube-like cell processes. However, in this bioreactor model system, manipulation of the 3D microenvironment did not result in polarization of an immortalized enamel epithelial cell line, HAT-7 cells. In the current study, we primarily used ALC, a spontaneously immortalized ameloblast cell line derived from mouse molars [20]. Although immortalized, ALC have been shown to transform into elongated polarized cells with basolaterally located nuclei when injected into athymic mice with Matrigel [20].
When ALC were cultured in 3D, we observed a characteristic cell clustering in response to the presence of ameloblastin or amelogenin. Even though AMEL also leads to ALC polarization in the 3D culture system, AMBN is more likely to influence ameloblast polarization in vivo (Fig. 1). A similar clustering behavior has been reported in a 2D monolayer culture of osteoblasts with enamel matrix derivative (EMD) extract enriched primarily with amelogenin (but also containing ameloblastin) [33]. EMD extract resulted in the formation of cell clusters with positive immunolabeling for N-cadherin and Connexin-43 at cell-cell junctions with an increase in their protein expression levels [34]. Notably, in our 3D experiments with ALC, the highest cell aspect ratios were achieved in the presence of AMBN alone, with an 85% correlation between AMBN concentration in the gel matrix and ALC aspect ratio. The molar concentration of AMEL required to demonstrate similar cell-cell clustering and elongation was 2.75-times greater than that of AMBN (Fig. 1,2), and when both proteins were added to the culture together, no synergistic effect on cell morphology was observed.
We show that within the first 48h of culture in the presence of ameloblastin, polarization markers such as adherens junction protein E-cadherin, polarity protein Par3 and tight junctional protein Claudin-1 assumed an asymmetrical distribution pattern within the elongated cell clusters. This preferential distribution along the Z axis of the cell clusters suggests the potential initiation of sorting into basal and apical membrane domains within the ALC clusters. This is supported by the co-accumulation of Par3 polarity protein alongside the nascent cell-cell contacts formed by E-cadherin [35]. In fully polarized secretory ameloblasts, Par3 can be selectively immunolocalized to the proximal ameloblast cell membrane [36] . Assembly of tight junctional proteins occurs rapidly after the formation of the initial adhesive cell contacts [37] and establish the axis of polarity by defining the demarcation between basal and apical membranes [23,35,38,39]. Notably, even within the cells that were not significantly elongated, Claudin-1 distribution patterns were asymmetric (Supplementary Fig. S8). Of the three cell polarity markers examined, it was observed that the immunolocalization of Claudin-1 was more cytoplasmic with respect to the positioning of E-cadherin and Par3, which were present along the membrane of ALC in a region located basally from the nuclear position.
E-cadherin is pivotal for the establishment of cell-cell adhesions and the initiation of cell polarity beginning from the cell surface (an outside-in type of signaling) [40]. The dynamics of E-cadherin at the cell surface are stabilized by its association with p120 catenin complex [41,42]. This is necessary for the stabilization of cadherin-catenin complex to prevent its internalization and the subsequent loss of cell polarity. Targeted p120 catenin ablation has also been shown to result in enamel mineralization defects with a loss of ameloblast polarization and morphology [13].
In genetically modified animal models, enamel extracellular matrix protein alteration severely impacts enamel formation. However, of the two major EMPs, ameloblast cell defects are restricted to mutations in Ambn, and are not mirrored by mutations in AmelX [6]. For example, truncation of Ambn (removal of the region encoded by exons 5 and 6) results in the loss of ameloblast polarization, columnar morphology and subsequently attachment to the enamel ECM.
The relationship between AMBN protein and ameloblast cell morphology can be further strengthened by observations concerning the effect of AMBN mutants lacking certain functional domains; AMBNμ5 and AMBNμ6. Within the exon 5-encoded region lies a recently identified, highly conserved 37 amino acid-long amphipathic helix (AH)-forming motif [43] and an AMBN self-assembly domain characterized by a Y/F-x-x-Y/L/F-x-Y/F motif [44]. The self-assembly domain of AMBN is also involved in its co-assembly with AMEL via similar conserved motif [45]. Disruption of AMBN self-assembly domain resulted in perturbations to the rod-interrod architecture of enamel [44]. Synthetic peptides of the evolutionary conserved AH sequence specifically immunolocalized to ALC cell processes, and they directly interacted with epithelial cell membranes, [24] highlighting the pivotal role played by this AH motif in ameloblast cell-matrix interactions. Although the exon 5-encoded region contains the AMBN self-assembly and/or AMEL co-assembly domain, the domain for AMBN-cell interaction seems to be different than that of AMEL-AMBN or AMBN-AMBN interactions [24,44,45]. Current experiments from our laboratory support the notion that AMBN-cell membrane interactions are not dependent on AMBN self-assembly interactions (Kegulian et al., unpublished). In the present study, the lack of synergism between AMBN and AMEL in promoting ALC elongation suggests that AMBN-cell interactions are not dependent upon AMBN interactions with AMEL. AMBN-AMEL interactions however could be important for adhering the cells to the AMEL-rich extracellular matrix.
With the removal of the exon 6-encoded region of Ambn gene, ALC did not lose their potential to cluster entirely, though they assume a much shorter Z height (based on aspect ratio), and in certain regions appear as distinct planar cells. The exon 6-encoded region of ameloblastin holds biological significance, as it is the site of a highly conserved alternative splicing site [17] and has been implicated in a case of human amelogenesis imperfecta (AI) [7]. AI affected patients display a spectrum of abnormalities ranging from severely dysplastic to normal enamel. Strikingly, heat denaturation of full-length AMBN produces a similar effect to exon 5 removal, with a complete lack of the cell effects normally observed in the presence of AMBN, suggesting that assumption of high order structures by ameloblastin can act as a prerequisite for its cell interactions, given its intrinsically disordered nature [44,46].
Although the mechanisms by which cell-cell interaction triggers polarization are common among epithelial cells [38,39], TCMK-1 cells (kidney tubular cells) did not respond to ameloblastin in 3D culture. In 2D monolayer cultures of TCMK-1 cells on AMBN coated plates, these cells were found not to interact specifically with AMBN, based on results from a cell spreading assay [24]. This suggests that the interactions between AMBN and epithelial cell membranes in 3D might be specific to those cells of the ameloblastic lineage (i.e., ALC and LS-8).
In amelogenesis, the inductive potential for terminal differentiation of ameloblasts shifts from the epithelium (during early stages of development) to the dental mesenchyme [47]. Primary human ameloblast cells cultured in 3D Matrigel substrates form similar acinar-like structures, and co-culture with mesenchymal dental pulp cells results in the formation of larger acini with elevated expression levels of ß1 integrin and à2 integrin [10]. In a different model using renal capsule implantation of incisor primordia injected with assembled peptide amphiphiles (PA), it was shown that PA encompassing the cells can act as signaling triggers to initiate the elongation, polarization and formation of enamel pearl-like structures [48]. Taken together, these observations suggest that, when provided with the appropriate signaling factors, ameloblast-like cells possess the potential to form a cluster of cells with increased expression levels of cell-cell and cell-matrix markers.
In summary, our cell culture data suggest that the membrane interaction domains of ameloblastin may play a significant role in triggering the initiation of cell polarity and elongation in enamel epithelial cells, thereby acting as a communicator between cell-matrix and possibly cell-cell interactions. Our investigation has important implications for understanding the function of ameloblastin in cell polarization or enamel-cell matrix adhesion and the outcomes may contribute to efforts to develop strategies for enamel tissue regeneration.
Experimental procedures
Recombinant protein expression and purification
Recombinant full-length ameloblastin (AMBN) and ameloblastin mutants lacking the regions encoded by exon 5 or 6 (AMBNΔ5 and AMBNΔ6, respectively) were expressed and purified from BL21 Escherichia coli cells as per previously published protocols [25]. Full-length recombinant AMBN characterization using SDS-PAGE is shown in Supplementary Fig. S9. Characterization of the recombinant ameloblastin mutants performed using SDS-PAGE electrophoresis was similar to previously published protocols and has been described elsewhere [43]. Recombinant murine amelogenin (rM179) was expressed and purified from BL21 E. coli and was characterized using SDS-PAGE electrophoresis as previously described [45]. Porcine developing enamel matrix was extracted in 0.5M acetic acid and purified as per previously published protocols [45] and characterized with respect to its AMEL and AMBN composition using SDS gel electrophoresis and Western blotting [45]. It was dissolved to reconstitute to working concentrations of 100 μg/ml.
Cell culture
Mouse ameloblast lineage cells (ALC) were obtained from Professor Toshihiro Sugiyama, Akita University, Japan [20]. Mouse TCMK-1 cells were purchased from ATCC and mouse LS-8 cells were obtained from Professor Malcolm Snead from University of Southern California [21]. The cells were cultured in Dulbecco’s modified Eagle’s Medium (DMEM) (Corning) supplemented with 100 U/L penicillin, 100 mg/ml streptomycin and 10% heat-inactivated fetal bovine serum (FBS) (Corning) in a 5.0% CO2 atmosphere at 37°C until they reached 80% or greater confluence. The 3D cell culture experiments were carried out in Glass-bottomed Multi-well (96-well) Plates (Mattek). Characterization of ALC and LS-8 cells to confirm their innate expression levels of enamel matrix proteins and proteinases was performed using RT-PCR and is described elsewhere [24].
Culture of ALC in growth factor-reduced geltrex
The 3D culture model that was developed for the purposes of this study involved a modification of the 3D-on-top type culture technique [49] that creates differential surfaces for cell contact. Geltrex (Thermofisher) depleted of growth factors using ammonium sulfate precipitation was used as the gel substrate and is referred to here as growth factor-reduced (GFR) Geltrex. Using this modified technique, the test (AMBN, AMEL) and control proteins (heat denatured AMBN, BSA) were restricted to one surface of the cells (along the Z axis), with the opposing Z surface contacting the top 10% Geltrex coat. Briefly, ice-cold Geltrex was added to the wells of the 96-well plate that were pre-coated with 20μg/ml of test (AMBN, AMEL, AMBNΔ5 and AMBNΔ6) or control (bovine serum albumin, heat denatured AMBN and Geltrex alone) proteins and allowed to polymerize into a gel at 37°C for 30 min. The cells were seeded on top of the gel at a density of 3.5 × 104 cells/well [28,49,50]. A 10% Geltrex coat in culture media was added atop the attached cells to complete the 3D-on-top type culture and the cultures were maintained for 24-72h under standard conditions. The 3D cell culture experiment was repeated using TCMK-1 and LS-8 cells.
Immunofluorescent staining
Immunofluorescent staining of 3D cultures was performed following standard protocols for whole-culture fixation [49]. Briefly, cells in the 3D gel were fixed using glutaraldehyde for 30 mins. Cell membranes were labelled with 1,1’-dioctadecyl-3,3,3’,3’-tetramethylindodicarbocyanine perchlorate (DiD) dye overnight at 37°C (1:1000). The cells were then permeabilized using 0.1% Triton-X for 5 min at room temperature (RT) and blocked for 30 mins using 3% FBS or 5% Goat serum. The following primary antibodies were used for junctional complex labeling: Alexa Fluor 555 conjugated mouse monoclonal anti-E-cadherin (1:400; BD Biosciences-610181), rabbit polyclonal anti-Claudin-1 (Cldn1) (1:200; Proteintech-28674-1-AP), rabbit polyclonal anti-Par3 (1:400; Proteintech-11085-1-AP), rabbit polyclonal anti-actin (5 μg/ml; Abcam-ab8227). Unconjugated monovalent anti-mouse Fab fragments (Jackson-715-007-003) were used for mouse-on-mouse labeling and unconjugated monovalent anti-rabbit Fab fragments were used when using two primary antibodies that were raised in the same host [51]. Primary antibody detection was performed using fluorescently conjugated secondary antibodies (1:400) that were incubated for 1h at RT (Alexa Fluor 488 and Alexa Fluor 647; Jackson) Nuclei were counterstained with DAPI (1:1000). Antibody dilutions used were optimized either following titration experiments in 3D or based on the manufacturer’s recommendations.
Aspect ratio measurement and confocal imaging
Fluorescently (DiD) labeled cells were visualized using an inverted fluorescent light microscope (Keyence BZX800) with an objective PlanApo λ NA 0.75 and sequential Z stacks were recorded with a pitch of 0.4 μm. The Z stacks were three-dimensionally reconstructed and analyzed using Keyence Image-Viewer (software version 1.1.1.8). To quantify and differentiate the degree of change in cell morphology in the presence of test and control proteins, a protocol for measuring the cell aspect ratio was developed. The 3D measure tool was used to measure the cell height along the Z axis and the cell width along the XY plane (n=30). Aspect ratio was defined as Aspect ratio = Cell height (Z)/ Cell width (XY) and was calculated for each group from the height and width measurements. Confocal imaging was performed using a Lecia SP8 confocal microscope with an oil immersion objective HCX PL APO CS × 60 (NA 1.4). Sequential Z stacks were recorded with a 0.5 μm pitch and the Z stacks were analyzed using LAS-X version 1.8.1.13759. Detection of Alexa Fluor 555 was performed at 556 to 595 nm (excitation at 561 nm), Alexa Fluor 488 at 502 to 552 nm (excitation at 488 nm), DiD at 645 to 695 nm (excitation at 633 nm).
Cell culture in type I collagen and gelatin substrates
The 3D culture experiments initially carried out using GFR Geltrex were repeated using type I collagen (1.5 mg/ml) (Gibco) and transglutaminase crosslinked gelatin (ColTgel) (101Bio) substrates. In the case of ColTgel, all three commercially available gel densities- soft (0.9-1.4 kPa), medium (14-20 kPa) and stiff (35-47 kPa) were tested. Both gels were cultured following the same 3D-on-top type culture protocol as that of GFR Geltrex gels.
Statistical analysis
Data in Figs. 1–9 were analyzed using Origin 8.0. Values of p less than 0.05 were considered statistically significant. One-way ANOVA tests were used to compare between all the groups and subsequent pairwise comparisons were carried out using Tukey HSD post hoc test.
Supplementary Material
Acknowledgements
We thank Drs. Michael Paine and Malcolm Snead for providing LS-8 cells and for valuable discussions, Dr. Toshihiro Sugiyama for providing ALC cells, Mr. Tach-Vu for technical assistance with fluorescent microscopy, Dr. Rucha Bapat for confocal microscopy training, and Ms. Bridget Samuels for proofreading the manuscript.
Funding
This project was funded by National Institutes of Health–National Institute of Dental and Craniofacial Research grants R01DE013414, R01DE027632 to J. Moradian-Oldak.
Abbreviations:
- AMBN
Ameloblastin
- AMEL
Amelogenin
- AH
Amphipathic Helix
- HAP
Hydroxyapatite
- EMPs
Enamel matrix proteins
- EMD
Enamel Matrix Derivative
- BSA
Bovine serum albumin
- ALC
Ameloblast lineage cells
- Cldn1
Claudin-1
- GFR
Growth factor reduced
Footnotes
Declaration of competing interests
None
References
- [1].Moradian-Oldak J, Protein-mediated enamel mineralization, Front. Biosci 17 (2012) 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nanci A, Ten Cate’s Oral Histology-e-book: development, structure, and function, Elsevier Health Sciences, 2017. [Google Scholar]
- [3].Fincham A, Moradian-Oldak J, Simmer J, The structural biology of the developing dental enamel matrix, J. Struct. Biol 126 (3) (1999) 270–299. [DOI] [PubMed] [Google Scholar]
- [4].Smith CE, Cellular and Chemical Events During Enamel Maturation, Crit. Rev. Oral Biol. Medicine 9 (2) (1998) 128–161. [DOI] [PubMed] [Google Scholar]
- [5].Bartlett J, Simmer J, Proteinases in developing dental enamel, Crit. Rev. Oral Biol. Medicine 10 (4) (1999) 425–441. [DOI] [PubMed] [Google Scholar]
- [6].Hatakeyama J, Fukumoto S, Nakamura T, Haruyama N, Suzuki S, Hatakeyama Y, Shum L, Gibson C, Yamada Y, Kulkarni A, Synergistic roles of amelogenin and ameloblastin, J. Dent. Res 88 (4) (2009) 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ, Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta, Hum. Mol. Genet 23 (20) (2014) 5317–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y, Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts, J. Cell Biol 167 (5) (2004) 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wazen RM, Moffatt P, Zalzal SF, Yamada Y, Nanci A, A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects, Matrix Biol. 28 (5) (2009) 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].He P, Zhang Y, Kim SO, Radlanski RJ, Butcher K, Schneider RA, DenBesten PK, Ameloblast differentiation in the human developing tooth: effects of extracellular matrices, Matrix Biol. 29 (5) (2010) 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Al-Ansari S, Jalali R, Plotkin LI, Bronckers AL, DenBesten P, Zhang Y, Raber-Durlacher JE, de Lange J, Rozema FR, The importance of connexin 43 in enamel development and mineralization, Front. Physiol 9 (2018) 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Guan X, Xu M, Millar SE, Bartlett JD, Beta-catenin is essential for ameloblast movement during enamel development, Eur. J. Oral Sci 124 (3) (2016) 221–227. [DOI] [PubMed] [Google Scholar]
- [13].Bartlett JD, Dobeck JM, Tye CE, Perez-Moreno M, Stokes N, Reynolds AB, Fuchs E, Skobe Z, Targeted p120-catenin ablation disrupts dental enamel development, PLoS One 5 (9) (2010) e12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hu JC-C, Hu Y, Lu Y, Smith CE, Lertlam R, Wright JT, Suggs C, McKee MD, Beniash E, Kabir ME, Enamelin is critical for ameloblast integrity and enamel ultrastructure formation, PLoS One 9 (3) (2014) e89303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krebsbach PH, Lee SK, Matsuki Y, Kozak CA, Yamada KM, Yamada Y, Full-length sequence, localization, and chromosomal mapping of ameloblastin a novel tooth-specific gene, J. Biol. Chem 271 (8) (1996) 4431–4435. [DOI] [PubMed] [Google Scholar]
- [16].Uchida T, Murakami C, Wakida K, Satoda T, Dohi N, Takahashi O, Synthesis, secretion, degradation, and fate of ameloblastin during the matrix formation stage of the rat incisor as shown by immunocytochemistry and immunochemistry using region-specific antibodies, J. Histochem. Cytochem 45 (10) (1997) 1329–1340. [DOI] [PubMed] [Google Scholar]
- [17].MacDougall M, Simmons D, Gu TT, Forsman-Semb K, Kärrman Mårdh C, Mesbah M, Forest N, Krebsbach PH, Yamada Y, Berdal A, Cloning, characterization and immunolocalization of human ameloblastin, Eur. J. Oral Sci 108 (4) (2000) 303–310. [DOI] [PubMed] [Google Scholar]
- [18].Robinson C, Kirkham J, Shore RC, Dental enamel formation to destruction, CRC press; 2017. [Google Scholar]
- [19].Panneer Selvam S, Ponniah I, Expression of ameloblastin in the human tooth germ and ameloblastoma, Oral Dis 24 (8) (2018) 1538–1544. [DOI] [PubMed] [Google Scholar]
- [20].Nakata A, Kameda T, Nagai H, Ikegami K, Duan Y, Terada K, Sugiyama T, Establishment and characterization of a spontaneously immortalized mouse ameloblast-lineage cell line, Biochemical and biophysical research communications 308 (4) (2003) 834–839. [DOI] [PubMed] [Google Scholar]
- [21].Chen L, Couwenhoven R, Hsu D, Luo W, Snead M, Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ, Arch. Oral Biol 37 (10) (1992) 771–778. [DOI] [PubMed] [Google Scholar]
- [22].Sarkar J, Simanian EJ, Tuggy SY, Bartlett JD, Snead ML, Sugiyama T, Paine ML, Comparison of two mouse ameloblast-like cell lines for enamel-specific gene expression, Front. Physiol 5 (2014) 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Drubin DG, Nelson WJ, Origins of cell polarity, Cell 84 (3) (1996) 335–344. [DOI] [PubMed] [Google Scholar]
- [24].Su J, Bapat R, Visakan G, Moradian-Oldak J, An Evolutionarily Conserved Helix Mediates Ameloblastin-Cell Interaction, J. Dent. Res (2020) 0022034520918521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Su J, Bapat RA, Moradian-Oldak J, The expression and purification of recombinant mouse ameloblastin in E. coli, Odontogenesis, Springer; 2019, pp. 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baker BM, Chen CS, Deconstructing the third dimension–how 3D culture microenvironments alter cellular cues, J. Cell Sci 125 (13) (2012) 3015–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Duval K, Grover H, Han L-H, Mou Y, Pegoraro AF, Fredberg J, Chen Z, Modeling physiological events in 2D vs. 3D cell culture, Physiology 32 (4) (2017) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [28].Li W, Machule D, Gao C, DenBesten PK, Growth of ameloblast-lineage cells in a three-dimensional Matrigel environment, Eur. J. Oral Sci 114 (2006) 159–163. [DOI] [PubMed] [Google Scholar]
- [29].Kozlowski M, Gajewska M, Majewska A, Jank M, Motyl T, Differences in growth and transcriptomic profile of bovine mammary epithelial monolayer and three-dimensional cell cultures, J. Physiol. Pharmacol 60 (Suppl 1) (2009) 5–14. [PubMed] [Google Scholar]
- [30].Földes A, Sang-Ngoen T, Kádár K, Rácz R, Zsembery Á, DenBesten P, Steward MC, Varga G, Three-Dimensional Culture of Ameloblast-Originated HAT-7 Cells for Functional Modeling of Defective Tooth Enamel Formation, Front. Pharmacol 12 (2021) 1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Fang JY, Yang Z, Han B, Switch of macrophage fusion competency by 3D matrices, Sci. Rep 10 (1) (2020) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pandya M, Lyu H, Luan X, Diekwisch TG, Polarized, amelogenin expressing ameloblast-like cells from cervical loop/dental pulp co-cultures in bioreactors, Stem Cells Dev (2021) (ja). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].G S, A C, J AC, Persson E, Brodin A, Rydhag L, Hammarstrom L, Formulation of enamel matrix derivative for surface coating, Kinetics and cell colonization, J. Clin. Periodontol 24 (1997) 678. [DOI] [PubMed] [Google Scholar]
- [34].Miron RJ, Hedbom E, Ruggiero S, Bosshardt DD, Zhang Y, Mauth C, Gemperli AC, Iizuka T, Buser D, Sculean A, Premature osteoblast clustering by enamel matrix proteins induces osteoblast differentiation through upregulation of connexin 43 and N-cadherin, PLoS One 6 (8) (2011) e23375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen J, Zhang M, The Par3/Par6/aPKC complex and epithelial cell polarity, Exp. Cell Res 319 (10) (2013) 1357–1364. [DOI] [PubMed] [Google Scholar]
- [36].Inai T, Sengoku A, Hirose E, Iida H, Shibata Y, Differential expression of the tight junction proteins, claudin-1, claudin-4, occludin, ZO-1, and PAR3, in the ameloblasts of rat upper incisors, The Anatomical Record: Advances in Integrative Anatomy and Evolutionary Biology 291 (5) (2008) 577–585. [DOI] [PubMed] [Google Scholar]
- [37].Gumbiner B, Generation and maintenance of epithelial cell polarity, Curr. Opin. Cell Biol 2 (5) (1990) 881–887. [DOI] [PubMed] [Google Scholar]
- [38].Wang AZ, Ojakian GK, Nelson WJ, Steps in the morphogenesis of a polarized epithelium. II. Disassembly and assembly of plasma membrane domains during reversal of epithelial cell polarity in multicellular epithelial (MDCK) cysts, J. Cell Sci 95 (1) (1990) 153–165. [DOI] [PubMed] [Google Scholar]
- [39].Wang AZ, Ojakian GK, Nelson WJ, Steps in the morphogenesis of a polarized epithelium. I. Uncoupling the roles of cell-cell and cell-substratum contact in establishing plasma membrane polarity in multicellular epithelial (MDCK) cysts, J. Cell Sci 95(1) (1990) 137–151. [DOI] [PubMed] [Google Scholar]
- [40].Desai RA, Gao L, Raghavan S, Liu WF, Chen CS, Cell polarity triggered by cell-cell adhesion via E-cadherin, J. Cell Sci 122 (7) (2009) 905–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lohia M, Qin Y, Macara IG, The Scribble polarity protein stabilizes E-cadherin/p120-catenin binding and blocks retrieval of E-cadherin to the Golgi, PLoS One 7 (11) (2012) e51130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Qin Y, Capaldo C, Gumbiner BM, Macara IG, The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin, The Journal of cell biology 171 (6) (2005) 1061–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Su J, Kegulian NC, Bapat RA, Moradian-Oldak J, Ameloblastin binds to phospholipid bilayers via a helix-forming motif within the sequence encoded by exon 5, ACS omega 4 (2) (2019) 4405–4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wald T, Spoutil F, Osickova A, Prochazkova M, Benada O, Kasparek P, Bumba L, Klein OD, Sedlacek R, Sebo P, Intrinsically disordered proteins drive enamel formation via an evolutionarily conserved self-assembly motif, Proceedings of the National Academy of Sciences 114 (9) (2017) E1641–E1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bapat RA, Su J, Moradian-Oldak J, Co-immunoprecipitation reveals interactions between amelogenin and ameloblastin via their self-assembly domains, Front. Physiol 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Wald T, Bednárová L, Osička R, Pachl P, Šulc M, Lyngstadaas SP, Slaby I, Vondrášsek J, Biophysical characterization of recombinant human ameloblastin, Eur. J. Oral Sci 119 (2011)261–269. [DOI] [PubMed] [Google Scholar]
- [47].Mina M, Kollar E, The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium, Arch. Oral Biol 32 (2) (1987) 123–127. [DOI] [PubMed] [Google Scholar]
- [48].Huang Z, Newcomb CJ, Bringas P Jr, Stupp SI, Snead ML, Biological synthesis of tooth enamel instructed by an artificial matrix, Biomaterials 31 (35) (2010) 9202–9211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee GY, Kenny PA, Lee EH, Bissell MJ, Three-dimensional culture models of normal and malignant breast epithelial cells, Nat. Methods 4 (4) (2007) 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hoffman MP, Kibbey MC, Letterio JJ, Kleinman HK, Role of laminin-1 and TGF-beta 3 in acinar differentiation of a human submandibular gland cell line (HSG), J. Cell Sci 109 (8) (1996) 2013–2021. [DOI] [PubMed] [Google Scholar]
- [51].Goodpaster T, Randolph-Habecker J, A flexible mouse-on-mouse immunohistochemical staining technique adaptable to biotin-free reagents, immunofluorescence, and multiple antibody staining, J. Histochem. Cytochem 62 (3) (2014) 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


