Abstract
Hepatotoxicity is one of the most frequently observed adverse effects resulting from exposure to a xenobiotic. For example, in pharmaceutical research and development it is one of the major reasons for drug withdrawals, clinical failures, and discontinuation of drug candidates. The development of faster and cheaper methods to assess hepatotoxicity that are both more sustainable and more informative is critically needed. The biological mechanisms and processes underpinning hepatotoxicity are summarized and experimental approaches to support the prediction of hepatotoxicity are described, including toxicokinetic considerations. The paper describes the increasingly important role of in silico approaches and highlights challenges to the adoption of these methods including the lack of a commonly agreed upon protocol for performing such an assessment and the need for in silico solutions that take dose into consideration. A proposed framework for the integration of in silico and experimental information is provided along with a case study describing how computational methods have been used to successfully respond to a regulatory question concerning non-genotoxic impurities in chemically synthesized pharmaceuticals.
Keywords: In Silico, Computational Toxicology, Organ toxicity, In Silico Toxicology Protocols, Hepatotoxicity, Liver toxicity, Hazard Identification, Risk Assessment, QSAR, Expert Alerts, Read-across
1. Introduction
Development of alternative approaches for the evaluation of organ toxicity is driven by the ongoing general paradigm shift in toxicology that aims at identifying, developing, and applying more sustainable and practical methods that can limit animal testing [1–6]. Recently, the concept of new approach methodologies (NAM) has been introduced to indicate any technology, methodology, approach, or combination thereof that can be used as (replacement, reduction or refinement) alternatives to animal testing (e.g., in silico, in chemico and in vitro methods) supporting chemical hazard and risk assessment [7,8]. NAMs may also involve the use of in vivo methods such as those with phylogenetically lower animals or those that help to replace, reduce, and refine animal usage [9]. In silico methods that aim at predicting the toxicity of chemicals from their structure play an important role in NAM workflows and are capable of providing information to complement and ultimately enhance the reliability of the human health hazard assessment of chemicals. They can contribute to an understanding of the structural and mechanistic basis underlying toxicity and give insights for the development of testing strategies or for an overall weight of evidence (WoE) evaluation [10].
In silico toxicology (IST) methods include category formation (i.e., chemical grouping), read-across, expert rule-based systems (i.e., structural alerts), and statistical-based systems (i.e., Quantitative Structure-Activity Relationship (QSAR)) [10]. Other computational approaches used in toxicology practice include dose-response and time-response models, biokinetic models and uncertainty factors (e.g., assessment, extrapolation, and risk factor) models [11]. Mechanistic understanding is recognized as a key element for the successful use of NAMs for chemical safety assessment [12] and, as such, the development of predictive in silico approaches needs to take into consideration the increasing knowledge of the underlying biological pathways leading to adverse outcomes and the links between chemistry and the biological activity that triggers these pathways.
Efforts are underway to establish IST protocols, namely standardized approaches for the prediction of toxicity from a chemical structure. Their objective is the definition of in silico assessment methodologies for various endpoints using principles that ensure that results can be generated, recorded, communicated, archived, and then evaluated in a uniform, consistent, and reproducible manner [10]. IST protocols build on structured hazard assessment frameworks as shown in the genetic toxicology and skin sensitization protocols [13,14] or as discussed in relation to the development of IST protocols for carcinogenicity [15].
The present work focuses on hepatotoxicity and aims at building a robust and pragmatic framework upon which IST protocols for organ toxicity can then be established; toxicity to other major target organs (heart, kidney, lung and nervous system) is discussed elsewhere [16,17]. This manuscript outlines a series of potential applications of the protocols, then summarizes general concepts concerning the state-of-the-science and organization of knowledge related to organ toxicity, and reviews liver toxicity to provide context to the endpoints requiring prediction. This introductory review is aimed to provide a framework for applying computational models in the context of existing knowledge on liver toxicity and the underlying mechanisms. Current in vivo and in vitro methods are discussed as this information is essential to incorporate within the weight of evidence in any hazard assessment as well as supporting the development of IST methods. The current advances in the understanding of the molecular pathways and events underlying pathogenesis of hepatotoxicity are discussed alongside the identification of challenging issues and limitations for the development of in silico models. A proposed IST framework for liver toxicity is presented and its use illustrated with a regulatory submission case study describing a response to a question from a regulatory authority.
1.1. Examples of potential protocol use
The use of in silico methods for the assessment of potential organ toxicity of chemicals is of interest for different industry sectors (e.g., pharmaceuticals, cosmetics, agrochemicals), where these approaches can be employed in the context for both product development and regulatory purposes (see Table 1). It is essential any eventual IST protocol addresses issues important to its adoption in these different situations.
Table 1.
Illustrative cases of application of in silico methods (and corresponding IST protocol) for liver toxicity.
| Context | Discussion |
|---|---|
| Non-genotoxic impurities in chemically synthesized pharmaceuticals | In the regulatory context of pharmaceuticals, International Council for Harmonization of Technical Requirements for Pharmaceuticals for Human Use (ICH), ICH Q3A and ICH Q3B guidelines address qualification of non-genotoxic drug impurities. Qualification is a process of acquiring and evaluating data that establishes the biological safety of an individual impurity or a given impurity profile at the level(s) specified. This process should result in a rationale for establishing impurity acceptance criteria that includes safety considerations [18,19]. According to these guidelines, when non-genotoxic impurities (NGIs) above the qualification threshold lack impurity-specific safety data, additional safety testing should be considered. Practices vary across different regulatory regions, but a recent reflection paper on the qualification of NGIs by the European Medicines Agency (EMA) discusses the use of an integrated risk assessment, where alternative strategies including (Q)SAR, Threshold of Toxicological Concern (TTC), read-across, and in vitro approaches can collect impurity-specific safety information that can be used to decide whether the NGI may be considered safe at the specified level [20]. More specifically, the EMA reflection paper considers the use of in silico methods (e.g., QSAR) to evaluate the pharmacological and toxicological properties of the NGI, acknowledging both the lack of standardized protocols and the worldwide efforts to develop such protocols [10]. Several factors, including the intended use and route of administration of the pharmaceutical and existing knowledge on similar substances, drive the selection of endpoints of interest for the assessment. In silico toxicology predictions of endpoints related to organ toxicity are beneficial and may be included in the safety profile and embedded WoE approach and/or used as a flag for further investigations and testing. |
| Extractables & Leachables | Toxicological risk assessment of extractables and leachables for medicinal products is becoming an important issue in the regulatory framework [21]. Industry has to evaluate potential toxic or harmful substances that can migrate into a product from the packaging materials such as drug delivery systems, and medical device components. Indeed, chemical contamination can also occur from processing equipment or container enclosures. In silico toxicity predictions of organ-toxicity may assist hazard assessment in a WoE approach for data-poor substances; they can also provide key information in the initial assessment that identifies the higher-risk compounds that should be tracked during testing. |
| Industrial chemicals | Organs such as the heart, brain, liver, kidneys, pancreas, spleen, immune system, and lungs are investigated in repeated-dose toxicity studies, that are required under REACH (Registration, Evaluation, Authorisation and Restriction of Chemical Substances) regulation depending on tonnage bands. In this regulatory context, IST predictions of specific target organ toxicity may be integrated into WoE considerations to support available in vivo studies [22]. Under Regulation (EC) No 1272/2008 on the Classification, labelling and packaging of substances and mixtures (CLP Regulation), information from repeated-dose toxicity studies are mainly used for classification for specific organ toxicity, and IST tools can provide further evidence on hazardous effects on certain organs [23]. Appropriate IST approaches may be integrated in the priority designation under the Toxic Substances Control Act (TSCA), where the principle of the 3Rs (replacement, refinement, and reduction) relating to the use of animals is supported [24]. |
| Biocides | Legislation on biocides usually requires information on repeated-dose toxicity and in this context the assessment of organ toxicity may be supported by the use of IST methods. Such approaches, together with all alternative methods, are generally encouraged in this context to reduce animal experiments [25]. |
| Plant Protection Products (PPP) | Several initiatives have been undertaken by the European Food Safety Authority (EFSA) to investigate the use of alternative methods for the assessment of pesticides, including the evaluation of the relevance of IST methods for metabolites and degradants [26]. Indeed, few metabolites and degradation products of pesticides are tested for toxicity, although they may be of concern for human health. The limited availability of QSAR models for chronic toxicity endpoints (e.g., hepatic and urinary tract toxicities, nephrotoxicity) has encouraged the use of read-across as alternatives within a WoE framework in this context. |
| Cosmetics | Some regulatory frameworks, such as the EU Cosmetics Regulation No. 1223/2009 [27], prohibit testing on animals for cosmetics. As such, alternative approaches including IST methods are promoted for the safety assessment of cosmetics-related substances [28]. Repeated-dose toxicity of cosmetic ingredients is a crucial endpoint in this context and it entails assessment of organ toxicity which can be investigated by means of IST (grouping, read-across, and (Q)SAR) together with in vitro data and physiologically-based pharmacokinetic (PBPK) modelling [29–32]. |
| Food | Use of alternative methods including in silico approaches may provide valuable means to assist safety assessment of food-related chemicals, where, for example, threshold levels (e.g., NOAEL) are generally used to derive Acceptable Daily Intake (ADI) levels. In silico methods may contribute to the assessment within a WoE rationale as discussed by the European Food Safety Authority (EFSA) [33]. The FDA’s predictive toxicology roadmap supports QSAR programs and development of new computational approaches [34]. As such, in silico models can be used as predictive tools to examine target organ effects, including hepatotoxicity, cardiotoxicity, and nephrotoxicity of food toxicants, providing, for example, relevant mechanistic insights on adverse effects [35]. |
| Occupational | Derivation of Occupational Exposure Limit (OEL) values entail the collection of data on toxicokinetics and toxicological endpoints relevant to worker exposure including specific target organ toxicity. In silico methods for the prediction of organ toxicity can be integrated into a WoE approach to help establish the point of departure (PoD) (e.g., Benchmark Dose (BMD) or no-observed-adverse-effect-level (NOAEL)) that is relevant for deriving exposure limit values (i.e., observed-effect-levels (OELs)) [36]. In silico methods can also complement the toxicological profile of data poor-substances in support of the overall risk assessment/management in cases where OEL are absent such as in the context of the Occupational Exposure Banding (OEB) process [37]. |
| Product Development | In the pharmaceutical industry secondary pharmacology is used for early hazard identification where compounds are routinely tested against in vitro off-target panels to assess their promiscuity [38,39]. The targets of these panels usually have an established linkage to adverse drug reactions (ADR); notably, the number of off-target effects related to liver injury is very limited. In silico toxicity predictions, either based on the prediction of off-target interactions, or inferred from other methods such as structural alerts [40], may be useful for flagging organ toxicity and prompting scientists to monitor the corresponding liability as the candidate advances through discovery. In silico methods can also provide relevant information to elucidate the structural basis underlying specific organ toxicity or to understand the corresponding biological mechanisms of such toxicity. In the preclinical phase of drug development, predictions of organ toxicity can be used to adjust the testing strategy and possibly adapt the in vivo protocols. Subsequently, they can assist in the understanding of the observations from preclinical studies (e.g., liver pathology) throughout the different durations of the studies: acute, sub-chronic, chronic, and eventually lifetime (2-years). During drug development, the information and data provided by in silico approaches may also assist in establishing the human relevance of the observed adverse effects. |
| Mixtures | NAMs are considered valuable approaches to support the hazard assessment of mixtures following exposure via air, water, food, consumer products, materials and goods [41]. NAMs may provide information to address different data gaps such as the limited number of toxicity data for mixtures assessed as a whole, or the lack of information (e.g. chemical hazard, dose-response, mode of action) needed for component-based approaches [42]. In this context, QSAR can assist the hazard and risk assessment of chemical mixtures with predictions of toxicological effects for the individual components or supporting the grouping of chemicals. Grouping is based on the mode of action of the chemicals, a key step in the assessment of mixtures, where in silico methods may inform on similarities of the toxicity profiles of the chemicals, such as common target organ toxicity profiles [42]. |
1.2. Organ toxicity
Adverse effects can be characterized in terms of their nature, target organ, potency, and mode of action (MoA) and these factors may differ greatly across toxicants [43]. Chemicals elicit local and/or systemic effects with most xenobiotics inducing main adverse effects in one (or several) target organ(s). In general, susceptibility of the target organ or a high concentration of the toxicant (the parent compound or metabolites) in the organ will influence the site of action. The toxic effects can be reversible or irreversible: low concentration and/or a short duration of exposure often induce reversible effects, while higher concentrations and/or longer durations of exposure may induce irreversible effects.
Adverse effects in acute toxicity and in chronic toxicity are generally brought about by distinct mechanisms [44,45]. Prolonged exposure and low doses of a chemical agent may elicit a variety of adverse effects arising from the perturbation of different biological pathways [44]. Understanding the effects on a specific target organ in terms of the individual mechanisms underpinning chronic toxicity provide a suitable framework for the development of in silico models (and alternative models in general) capable of predicting the dose at which a chemical elicits an adverse effect based upon chronic exposure.
Mitochondrial dysfunction can lead to both acute or chronic injury and may impact different organs and tissues; organs containing high levels of mitochondria, such as liver, kidney, heart, and nervous system are most susceptible to mitochondrial dysfunction [46,47].
1.3. Adverse Outcome Pathway (AOP)
Existing knowledge about a biological pathway can be organized within the AOP conceptual framework [48–51]. Such a framework comprises a logical sequence of KEs (key events) triggered by a MIE (Molecular Initiating Event) that arises from an initial interaction between a stressor and a biological target(s) (e.g., DNA binding, protein oxidation, receptors) [52,53]. KEs lead to structural and functional changes ultimately culminating in the AO (adverse outcome) relevant to the human organism and the human population.
The AOP framework can be combined with the AEP (Aggregate Exposure Pathway) framework that extends the organization of the relationship between AOs and chemistry. An AEP links the introduction of a stressor from sources, fate, and transport through environmental media, patterns of external exposure, and biokinetic processes leading to target site exposures [54–56].
AEP and AOP together serve as basis for developing a mechanistically-informed IATA (Integrated Approaches to Testing and Assessment). A mechanistically-informed IATA uses information derived from appropriate combinations of alternative approaches (including in silico and in vitro) that target KEs within well-defined AOPs, to aid hazard and risk assessments by guiding minimal but well-informed higher tier testing. To facilitate the advances of mechanistically-informed approaches, AOPs are being collected in the AOP-Wiki, i.e., a collaborative platform for the development of AOPs overseen by an expert group from the Organisation for Economic Co-operation and Development (OECD) [57,58]. One current area of development is to make AOPs quantitative for practical risk assessment, which is a considerable undertaking given the lack of available data [59–61].
1.4. Target organ toxicity models
Studies of concordance of the toxicity of pharmaceuticals reported that 70% of human-relevant toxicities are detected in animal experimentation [62], though the prediction of safety (absence of events) is difficult [63,64]. Importantly, extrapolation of human risk from in vivo animal testing must account for species differences in toxic responses; it was observed that the affected organ influences the translational relevance [65,66]. It can also be noted that human adverse drug reactions (ADRs) are mostly linked to toxicities to liver, heart, and neurological organs [65–68], as these are areas known for high rates of metabolism (e.g., CYP, transporters).
In the context of pharmaceuticals, in vitro models for the assessment of target organ toxicity are routinely applied and play an important role, especially in early drug development [66,69]. The in vitro systems take into account specific physiological conditions of the organ (e.g., electrophysiology, metabolism, proliferation, and specific homeostasis) and usually provide mechanism-based biological observations.
Recently, the use of high-content imaging assays based on human cell lines from different organs (hepatocytes, neurons, cardiomyocytes and endothelial cells) was investigated to derive NAM-based point of departures (NAM-based PODs) for risk characterization [70]. Such NAM-based PODs are being explored as conservative surrogates for in vivo values [70,71]. In the future, it is hoped that generic screens based on high-content image-based assays such as the CellPainting assay are able to give safety-relevant information on a broad scale and at practically applicable low cost [72].
A higher level of complexity is introduced with the development of in vitro three-dimensional (3D) models and microphysiological systems to address the composite endpoints associated with target organ toxicity [73,74]. In 3D cell culture techniques, the cell environment can be manipulated to mimic the in vivo environment and provide more accurate data on toxicity [75]. Techniques such as the “organ-on-a-chip” and organoids aim at replicating the complex attributes of the organ [76,77].
By modeling the inter-relationship between cells, tissues and possibly organs, such models may hopefully predict adverse effects either as single organ systems or as part of an integrated, multi-organ system. Multi-organ toxicity is difficult to accurately address with in vivo models and is not reflected in the isolated single cells from a single organ [78,79].
1.5. Molecular targets
Adverse drug reactions (ADRs) usually have an established relationship to undesired interactions of the drug or drug candidate with proteins (off-targets) other than its therapeutic target [38,39,80–83]. The pharmaceutical industry generally explores such secondary effects by means of in vitro high throughput screens against a large number of unintended targets (receptors, ion channels, enzymes, transporters) with the aim of limiting off-target interactions and thus reducing liabilities leading to toxicity [84,85].
An industry-regulatory consensus on the list of off-targets is currently lacking. One example is a minimal safety panel of 44 targets by Bowes and co-workers that links the biological targets to specific ADRs as, for example, cardiovascular system (CVS), central nervous system (CNS), pulmonary, and renal effects (liver is not on the list). Two molecular targets (i.e., Muscarinic Acetylcholine M1 Receptor and Muscarinic Acetylcholine M3 Receptor) specifically associated with potential adverse effects occurring in the liver are included in another pharmacology screening battery [39].
1.6. Toxicogenomics
Toxicogenomic studies are mainly exploited to gather mechanistic information or as predictive tools. Predictive toxicogenomics is based on databases of genomic profiles resulting from the response of a biological model following exposure to reference chemicals (i.e., chemicals acting with a known mode of action); the toxicogenomic signatures can then be used to predict or classify the toxicological behavior of an unknown chemical based on its toxicogenomic profile [86,87]. Databases such as DrugMatrix [88], Open TG-GATEs (Toxicogenomics Project-Genomics Assisted Toxicity Evaluation System) [89] and the Comparative Toxicology Database (CTD) [90] directly link toxicity to gene expression data, and can be exploited to develop QSAR models useful to investigate organ toxicity [91]. Such databases can also be used to explore multi-organ toxicity [92]. Results from this type of analysis may heavily depend on how the data examined was generated though, as well as the precise methods and parameters chosen for its subsequent processing and interpretation.
1.7. ADME and toxicokinetics
Understanding internal exposures in humans is deemed essential for a successful application and use of alternative methods. This implies understanding the concentrations or exposure patterns at which an adverse effect is observed (e.g., in an alternative framework) in relation to the corresponding internal concentrations at the target tissue of the test chemical in humans [93].
More specifically, mechanistically-based paradigms arising from alternative methods (e.g., in vitro and in silico) provide predictions on the intrinsic toxicological effects on a given tissue that need to be combined with information on chemical exposure to evaluate the relevance of the findings to human safety. The integrated action of ADME (adsorption, distribution, metabolism, excretion) processes will determine whether the biologically active form of a chemical (i.e., the parent compound or its metabolite(s)) reaches the target tissue at a certain concentration for a specific duration to potentially elicit an adverse effect [94].
Information on systemic exposure as derived from ADME and toxicokinetic investigations are used to interpret toxicological findings supporting, for example, the identification of the circulating moieties (parent substance/metabolites) or the extrapolation of animal toxicity data to humans [95–101].
Lists of models and software for the prediction of ADME properties are available in the literature [102–105]. QSARs for the prediction of metabolism include CYP inhibitor/substrate predictions, with some of these models deriving the site of metabolism (SOM) (i.e., the structural fragment of the xenobiotic where the metabolic reaction occurs) [105,106]. Expert system approaches are also available to predict biotransformation pathways from the molecular structure of the parent chemical [107,108]. A downside of these approaches is the generation of an excessive number of metabolites from a multitude of potential pathways and their human relevancy [107]. It can be noted that predicting the relative likelihood of such events is generally easier than the prediction of the absolute likelihood of a transformation, which is however needed to understand the fate of a molecule in an organism quantitatively.
Compartmental TK models and PB-TK (physiologically based-toxicokinetic) models quantitatively describe the temporal change in the concentration of xenobiotics and/or their metabolites in biological matrices (e.g., blood, tissue, urine, alveolar air) of the exposed organism [109–111]. They provide an estimate of the internal concentrations of the test chemical, and their toxicological applications include extrapolations across species, in vitro to in vivo extrapolation, route to route extrapolation, high to low dose extrapolation, and intra-species extrapolations [112–114].
2. Background to Liver toxicity
The liver is a primary target of toxicity following oral exposure to drugs and, in general, to xenobiotics. Liver susceptibility arises from its portal location and key role in xenobiotic metabolism [115,116]. Orally-ingested xenobiotics are absorbed in the gastrointestinal tract, thus making this organ particularly vulnerable to chemical-induced injury, most likely due to the high concentrations of xenobiotics to which it is exposed [117]. The liver is also the principal site of xenobiotic metabolism involving biotransformation that may result in the formation of highly reactive metabolites (or otherwise bioactive) which may cause adverse effects [118–120].
In the context of pharmaceutical toxicology, liver damage caused by drugs is referred to as DILI (Drug Induced Liver Injury) [121–124] and is one of the most frequent causes for drug attrition and market withdrawals [125,126]. DILI is a result of complex mechanisms, which include both compound-related adverse effects on the liver and patient-specific factors such as the genetic background or factors related to lifestyle [127].
Two types of DILI are described in the literature [128,129], intrinsic DILI and idiosyncratic DILI. The former is reproducible and dose-dependent and, in general, can be predicted. The latter is not fully understood: it only affects a limited number of people, is difficult to predict, usually develops at therapeutic doses, and lacks a clear dose dependency.
With the liver being a key organ for the interpretation of acute [130] and repeated-dose toxicity [44,131,132], hepatotoxicity appears to be a concern related to many other types of chemicals, including herbal agents or nutritional supplements, industrial chemicals, agrochemicals, biocides, cosmetics, chemicals present in food, and substances of abuse [133].
2.1. Processes and endpoints
Chemical-induced hepatotoxicity is referred to as acute or chronic (see Fig. 1), depending on the duration of the liver injury [134]; it results in impaired hepatocyte function and viability that are observed in different histopathological patterns including necrosis, apoptosis, fibrosis, steatosis, and cholestasis [135]. Necrosis, i.e., cell death caused by unregulated biological events, is commonly observed in both acute and chronic liver diseases, which can progress to fibrosis, characterized by the deposition replacement of parenchymal tissue with collagen, proteoglycans, and glycoproteins [135,136]; deposition of collagen throughout the liver results in cirrhosis, which, in turn, may lead to liver failure. Apoptosis, that is the normal physiological process of regulated cell death, can also be induced or suppressed by xenobiotics. Steatosis, also known as fatty liver disease, refers to the abnormal accumulation of triglycerides in hepatocytes and in severe cases it is accompanied by inflammation and hepatocellular necrosis. A condition related to steatosis is phospholipidosis characterized by phospholipid accumulation within the lysosomes of hepatocytes. Cholestasis is the disruption of bile production or flow. Certain liver injuries are characterized by local inflammation contributing to the progression of the injury [137].
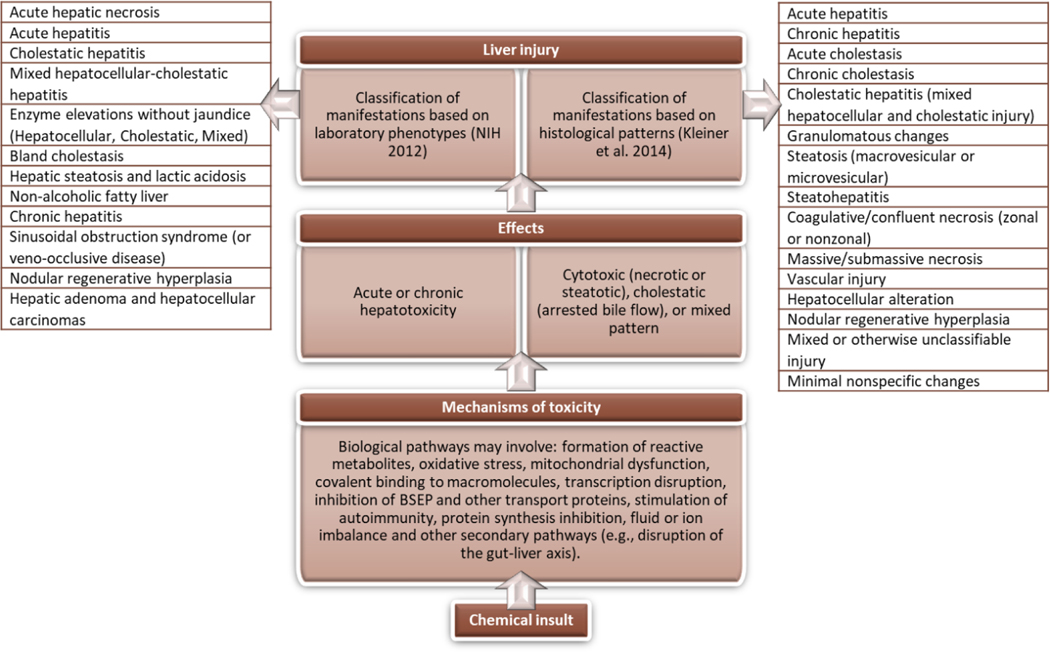
Figure 1.
Chemical insult leading to diverse manifestations of hepatotoxicity. Chemical-induced hepatotoxicity may involve different biological pathways and can be classified in different ways, as for example based on clinical laboratory phenotypes [141] or histological patterns [144].
Acute hepatotoxicity is manifested in various forms and can include hepatic necrosis, hepatocellular steatosis and/or degeneration, or acute cholestasis. Manifestations of chronic hepatotoxicity include hepatitis, steatosis, fibrosis, and phospholipidosis [138,139].
Chemically-induced hepatotoxicity is broadly categorized as cytotoxic (necrotic or steatotic), cholestatic (arrested bile flow), or a mixed pattern [115,138]; hepatocytes (i.e., the most abundant liver cell type) may be targeted resulting in hepatocellular liver injury; impairment of excretion of bile leads to cholestatic or mixed hepatocellular/cholestatic injury [133].
Different classifications of liver injury exist (Fig. 1.) such as the classification according to the various clinical laboratory phenotypes resembling the corresponding liver disease [140,141], or the classification based on histological patterns reported by the Drug-Induced Liver Injury Network (DILIN) [142–144]. As noted by Cronin and co-workers these “classic toxicology” classifications based on clinical or histopathological criteria cover the main effects to the liver, comprising a mixture of mechanisms and observations [44].
Hepatotoxicity data from histopathology-related findings as originating from preclinical toxicity study reports for regulatory submissions can be collated into groups of histopathology terms related to similar findings (and, possibly, potential mechanisms); general clusters are identified: tissue damage, inflammatory changes, structural alteration, and accumulative lesions with each general cluster separating into more specific groups as shown in Table 2 [145]. The same hierarchical organization is applied to other target organs (i.e., heart and kidney) [16].
Table 2.
The hierarchical organization used to group histopathology terms of similar findings (and mechanism) for hepatotoxicity; findings were extracted from preclinical toxicity study reports for regulatory submissions [145].
| LIVER TOXICITY | |
|---|---|
| General clusters | Specific clusters |
| Tissue damage | Necrosis |
| Degeneration | |
| Inflammatory changes | Inflammation |
| Infiltration | |
| Structural alterations | Proliferation |
| Hyperplasia | |
| Hypertrophy | |
| Accumulative lesions | Steatosis |
| Vacuolation | |
Primary mechanisms underpinning hepatotoxicity include formation of reactive metabolites, oxidative stress, mitochondrial dysfunction, covalent binding to macromolecules, transcription disruption, inhibition of the Bile Salt Exporter pump (BSEP) protein and other transport proteins, stimulation of autoimmunity, protein synthesis inhibition, fluid or ion imbalance [124,133,146–148], and other secondary mechanisms such as those linked to the disruption of the gut-liver axis [149]. Chemicals can trigger innate or adaptive immune responses leading to liver injury [124,150], and an immune mediated mechanism involving reactive metabolites has been described for idiosyncratic DILI [151,152]. The different general pathways underlying pathogenesis of liver injury may work together to promote toxicity: mitochondrial dysfunction, oxidative stress, and alterations in intrahepatic bile acid and/or lipid homeostasis [128]. The latter effect commonly is associated with nuclear receptor activation and mitochondrial toxicity [153]. A three-step working model of liver injury pathogenesis has been proposed [138,154], where the first step starts with an initial cellular injury promoted by the chemical or its metabolites via direct cell stress, direct mitochondrial inhibition and/or specific immune reactions; the second step is induction of mitochondrial dysfunction, that leads to the third step, cell death (necrosis or apoptosis). Current information on the mechanisms related to hepatotoxicity is being organized in the AOP-Wiki, and Table S1 (available in the supplementary material) lists several AOPs as defined in this repository [57,58]. As mentioned above, quantitative aspects of AOPs for DILI still require further work in the future.
2.2. In vivo and in vitro methods
In the drug development process, hepatotoxicity assessment relies on in vivo toxicity studies in rodent and other animal species or clinical trials. Animal models must be critically evaluated, as interspecies differences, especially in metabolism and disposition of xenobiotics, are crucial aspects in the context of hepatotoxicity [155]. Differences in hepatic transporters between species may affect the resulting toxic effects, even if the production and elimination of toxic metabolites are similar [156]. Ex vivo models (liver tissue slices and isolated perfused liver) may also be used to complement the in vivo models [157].
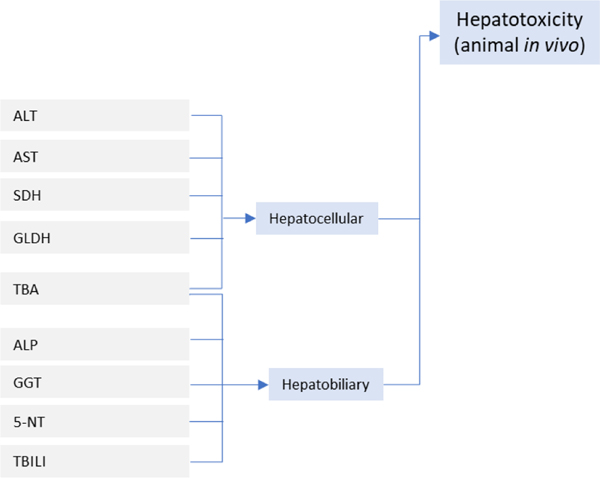
Common biomarkers associated with liver toxicity can be grouped into hepatocellular and hepatobiliary [158]; hepatocellular biomarkers are: ALT (alanine aminotransferase), AST (aspartate aminotransferase), SDH (sorbitol dehydrogenase), GLDH (glutamate dehydrogenase), and TBA (total bile acids); hepatobiliary biomarkers are: ALP (alkaline phosphatase), GGT (gamma-glutamyltransferase), 5-NT (5’-nucleotidase), TBILI (total bilirubin), and TBA. Some biomarkers have been specifically associated with mitochondrial injury, a primary factor in hepatotoxicity, and these are: alanine aminotransferase (isoenzyme ALT2), cytochrome c, GLDH, carbamoyl-phosphate synthetase 1 (CPS1), mitochondrial DNA (mtDNA), and long-chain acylcarnitines [159].
Limitations in reproducing human specific metabolism and drug transport processes, and limited representation of the liver tissue heterogeneity can lead to failure of in vitro models to predict hepatotoxicity. The current two-dimensional (2D) in vitro assays, that are based on cell lines such as HepG2 or differentiated HepaRG, are effective at modeling some components of hepatocyte biology but they fail to represent liver tissue heterogeneity (as well as some aspects of its metabolism) and therefore have a limited capacity to predict hepatotoxicity [160]. 3D in vitro liver models are emerging technologies holding great potential to understand the molecular mechanisms of hepatotoxicity and screen for compounds that cause liver injury [161–165].
2.3. In silico methods
Several in silico models for potential use in predicting human hepatotoxicity from molecular structure have been described in the literature [145,166–170,133,171–176] including advanced modelling based on deep learning algorithms [177–179] and prediction models that combine structural descriptors and in vitro ToxCast assay data for the prediction of in vivo organ toxicity [180–184]. Different reviews have thoroughly summarized and discussed available models for this endpoint [126,133,185–190]. Models specifically addressing a particular mechanism, such as mitochondrial dysfunction may also provide insights into liver toxicity [191].
Annotation schemes of liver toxicity information, especially related to DILI, are being developed using various sources [133], such as the FDA adverse event reporting system (AERS) that has been employed to develop a database characterized by well-defined hepatotoxicity-related preferred terms that is suitable for QSAR modelling [192]. Performance of DILI models is expected to improve with well-annotated hepatotoxicity data [125,133,193]. However, agreement on standardized DILI labels does not currently exist [174].
Available models can be grouped according to the following factors (see Table 3): a) methodology used (statistical-based or expert rule-based); b) endpoint being modeled (general hepatotoxicity or a specific aspect such as steatosis or a molecular initiating event); c) source of data for the training set (in vitro data, human data, animal data). Hewitt and coworkers noted that [188]: a) most of the available models aim at predicting the negative/positive general hepatotoxicity regardless of specific mechanism/toxicity observations; b) most models are developed using in vivo data; c) the number of models focusing on more specific endpoints (e.g., liver serum enzymes, hepatic steatosis, cholestasis, jaundice) are becoming more frequent. Some models based on in vivo data are discussed below to exemplify some of the issues underlying the development of in silico models for hepatotoxicity.
Table 3.
Classification of in silico models based on methodology, endpoint, and source of data (adapted from Hewitt and Przybylak [188])
| Methodology: | • Statistical-based • Expert rule-based |
| Endpoint: | • General hepatotoxicity • Specific endpoints • Molecular initiating events (MIEs) |
| Source of data: | • In vitro • In vivo (human) • In vivo (animal) |
The DILI model by Hong and coworkers is based on the Decision Forest machine learning algorithm to model three-classes of DILI severity (most-DILI, less-DILI, no-DILI) with a training set (more than 700 drugs) derived from the FDA’s Liver Toxicity Knowledge Base [194,195]. Notably, the authors recognize that the inclusion of a three-class DILI severity classification as compared to a DILI and no-DILI classification is a key factor positively affecting model performance.
Mulliner and coworkers [196] modeled general hepatotoxicity (negative/positive) by means of a Support Vector Machine (SVM) approach using a dataset from the pharmaceutical domain with more than 3,700 compounds. The modeling algorithms were specifically selected by the authors to account for the complex endpoints that incorporate different modes of action. Additional models were developed for other liver toxicity findings that were hierarchically organized: clinical chemistry, morphological findings, hepatocellular clinical chemistry, hepatobiliary clinical chemistry, hepatocellular morphological findings, and hepatobiliary morphological findings. These more specific models were separately developed both for human and preclinical data. As noted by the authors, the choice of the hierarchical organization of the endpoints was driven by the data set coverage and imbalance of positive versus negative compounds; indeed, specific individual findings such as cytolytic hepatitis would be heavily imbalanced containing only a few positive compounds and thus being unfeasible for modeling.
IST modelling of target organ toxicity by Amberg and coworkers shows that appropriate hierarchical organization of histopathology data (see Table 2) leads to good predictivity of the toxicity findings [145]. The development of these prediction models (i.e., structural alerts, fragment-based, molecular descriptor-based machine learning approaches) indeed requires a key preparation step to collate similar findings (and mechanisms) into suitable clusters. The same approach is applied to model toxicity for different target organs (i.e., liver, kidney, heart).
Read-across is also another approach to predict hepatotoxicity, where the importance of a mechanistically-driven rationale is demonstrated in a recent read-across study based on a large hepatotoxicity dataset of more than 4000 compounds collated from the public literature [197].
The in silico models discussed herein belong to the cheminformatics approach, that is the prediction of hepatotoxicity from chemical structure. As noted by Béquignon and coworkers [198], other types of in silico approaches other than cheminformatics have been employed for liver toxicity prediction and include quantitative adverse outcome pathways (qAOPs), metabolomics, pharmacokinetic-pharmacodynamics (PK-PD) modeling, dynamical pathway modeling with ordinary differential equation (ODE) models, and multi-scale approaches modeling DILI with systems biology approaches.
2.4. Draft assessment framework for liver toxicity
In the present work, mechanisms and toxicological effects are preliminarily combined into a high-level summary demonstrating a potential hazard assessment framework applicable in the context of liver toxicity prediction. The proposed draft assessment framework builds on the mechanistic vision of hepatotoxicity (see Fig. 2) and is shown in Fig. 3. Its structure combines information from in vitro approaches (e.g., biological responses from receptor-based assays), results from in vivo experiments and human data. The protocol structure may also be expanded to consider various exposure scenarios (e.g., environmental, drug, consumer, accidental). Other information, potentially organized and described in other protocols, needs to feed the hepatotoxicity protocol. These include information associated with ADME, the gastrointestinal tract, and the immune system.
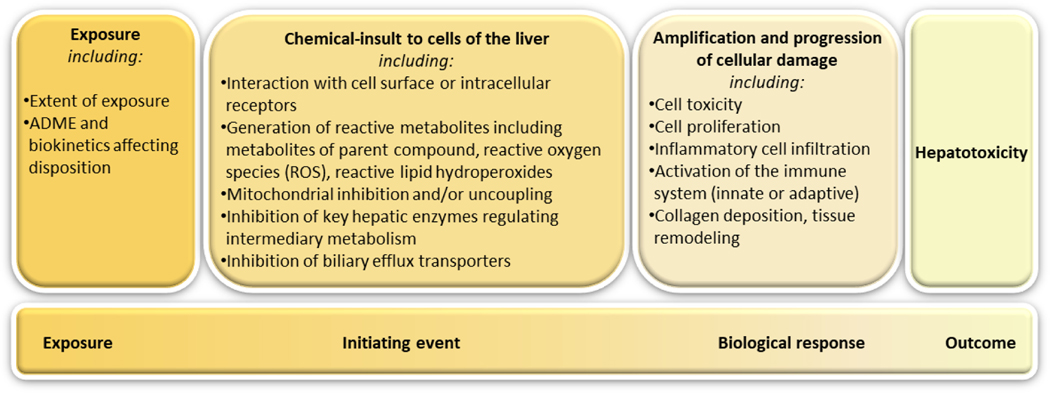
Figure 2.
A general schematic framework adapted from Kenna and co-workers [133] outlining the mechanisms underlying dose-dependent hepatotoxicity. The chemical insult is affected by exposure (e.g., the extent of exposure) and by ADME and biokinetic processes (e.g., balance between bioactivation and detoxification of reactive intermediates). Chemical insults to cells may lead to different biological responses, which may be protective to limit and control the cell damage, or they may be amplified and progress to toxicity.
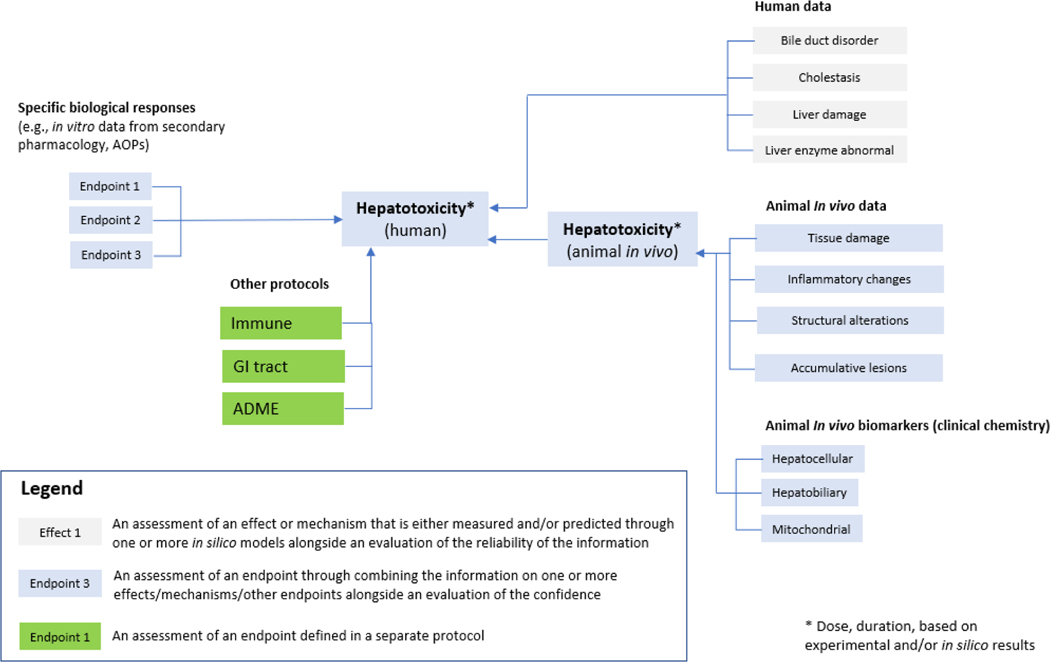
Figure 3.
High level summary of potential hazard assessment framework for liver toxicity. This draft framework combines different sources of information: human data, animal in vivo data, and in vitro data. In vitro data captures specific biological responses associated with hepatotoxicity such as data from secondary pharmacology or data from other relevant in vitro assays. In silico models may be integrated into the assessment of the different endpoints associated with specific biological responses, human data or animal in vivo data. A spectrum of major endpoints and sub-endpoints are included in the draft assessment framework.
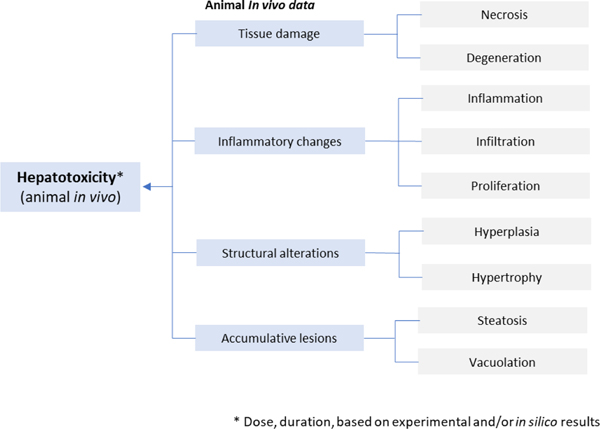
The detailed breakdown of specific types of animal in vivo data (histopathology) shown in Fig. 4. is based on work by Sanofi [145,166], where histopathology-related findings from preclinical toxicity study reports for regulatory submissions (see Table 2) are organized into groups of histopathology terms linked to similar findings (and, possibly, similar mechanisms). Common in vivo biomarkers are associated with hepatocellular- and hepatobiliary-related effects as shown in Fig. 5.. Biomarkers associated with mitochondrial dysfunction (e.g., ALT2, cytochrome, GLDH, CPS1, mtDNA and long-chain acylcarnitines [159]) may also be considered.
Figure 4.
Relevant toxicological effects from histopathology in animal studies (i.e. animal in vivo data) in relation to liver toxicity [145].
Figure 5.
In vivo biomarkers relevant to investigate liver toxicity in animal studies (i.e., animal in vivo data) [158]. Biomarkers associated with mitochondrial dysfunction (e.g., ALT2, cytochrome, GLDH, CPS1, mtDNA and long-chain acylcarnitines [159]) may also be considered.
The assessment framework integrates results of several other biological responses. For example, it may receive input from ligand-binding assays associated with hepatotoxicity, or from in vitro assays measuring mitochondrial dysfunction or, in general, from in vitro assays detecting KEs or MIEs of AOPs (e.g. cholestasis, fibrosis, steatosis, steatohepatitis). It should be noted that the number of molecular targets linked to liver toxicity is limited, with the AbbVie’s molecular pharmacology screening battery including only two molecular targets (i.e., Muscarinic Acetylcholine M1 Receptor and Muscarinic Acetylcholine M3 Receptor) that are specifically associated with potential hepatic ADRs (among other potential target organs) [39].
2.5. Case study
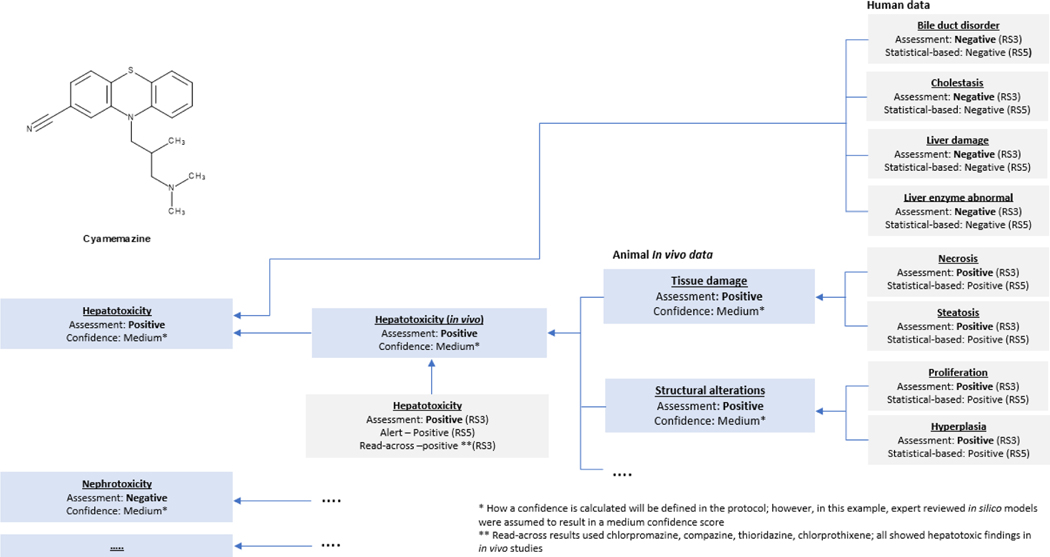
The following case study illustrates the use of the in silico framework for the prediction of organ toxicities. It outlines a response to a regulatory request for the qualification of non-genotoxic impurities of the drug cyamemazine (shown in Fig. 6.). This request was received after submission of the mutagenic impurities assessment according ICH M7 [199] for cyamemazine impurities. The ANSM (i.e., French Health Authority) requested further assessments for specific impurities which were above the qualification threshold and therefore in scope of ICH Q3A/B comprising cyamemazine amide, a degradation product shown in Fig. 6., which was classified in the first submission as Class 5 (i.e., no structural alerts, or alerting structure with sufficient data to demonstrate lack of mutagenicity or carcinogenicity) and non-mutagenic according ICH M7. An in silico analysis assessing the risks of the main biological functions was requested. This was interpreted to include an in silico request for target organ toxicity in the context of the “Reflection paper on the qualification of non-genotoxic impurities” [20]. It is noted, however, that regulatory acceptance of these methods differs across regulatory jurisdictions.
Figure 6.
Chemical structure of cyamemazine and cyamemazine amide
The in silico hazard assessment of cyamemazine amide (alongside other newly specified cyamemazine degradation products) above the qualification threshold was conducted using different in silico prediction systems, database searches for read-across, and expert knowledge. This hazard assessment was intended to better assess the risk of the degradation products on main biological functions, covering the main toxicological endpoints mutagenicity, clastogenicity, hepatotoxicity, nephrotoxicity, cardiotoxicity, developmental toxicity, phototoxicity and ocular toxicity.
The first step in the toxicity hazard assessment was to perform a search in public or internal databases to determine if experimental toxicity data were already available on the identified impurities. Structure-based assessments were performed using two complementary in silico (quantitative) structure-activity relationship (Q)SAR systems to predict the toxic potential of impurities, including both commercial systems alongside in-house QSAR models. Fig. 7. summarizes the in silico predictions, read-across results, and final expert review for the hepatotoxicity hazard assessment of the drug substance (Active Pharmaceutical Ingredient, API) cyamemazine. Fig. 8. summarizes the in silico predictions and read-across results for the degradation impurity cyamemazine amide. Notably, a similar analysis was performed on other organ systems. The in silico results are mapped onto a hazard assessment framework based on the outline proposed in Fig. 3. Impurities are classified by expert review into different classes of toxicological concern based on the hazard assessment of these data for each endpoint. The final assessment follows the main principles that are proposed and standardized in the IST protocols by Myatt and co-workers [10] including the use of reliability scores (RS) to represent the quality of each hazard assessment. This assessment ( see Fig. 7 and Fig. 8) comprises an expert review on the QSAR outcome which increased the reliability of the results to RS3 (i.e., reliability score following expert review), according to the reliability score outlined by Myatt and co-workers [10].
Figure 7.
Framework for assessment of organ toxicity for cyamemazine.
Figure 8.
Framework for assessment of organ toxicity for cyamemazine amide
The cyamemazine amide is present at only low levels (i.e., at levels slightly above the ICH Q3 qualification threshold of 0.15% [18]) as compared to the drug substance, which means that its contribution to the overall safety profile is limited as compared to the parent compound unless additional alerts are introduced. Therefore, the corresponding hazard assessment mainly focused on the deviating and different substructures of the degradation product as compared to the drug substance and on assessing if these differences in the substructure were predicted to have an increased, decreased, or similar hazard compared to cyamemazine. It should be noted that this is a hazard assessment and does not predict any safety risk for the known safe doses and exposures.
The results of the in silico predictions, both from expert alert and statistical models, are very similar for the cyamemazine amide and the API cyamemazine. Based on a weight-of-evidence paradigm that accounted for the different information available, the hazard of the impurity was evaluated, leading to the conclusion that the toxicity hazard was very similar to the API’s toxicity hazard (it needs to be kept in mind that both types of predictions are based on currently available data, so data coverage of in silico systems is of crucial importance). The in silico predictions of the hepatotoxicity models, for example, show a lower prediction probability in most models, signifying that the hepatotoxic hazard is even less than the API cyamemazine. From a structural point of view, the amide substructure was the only difference between the API and cyamemazine amide and was not predicted as an additional toxicophore likely to increase the hazard for the investigated endpoints. The degradation impurity cyamemazine amide was indeed predicted to have a similar or relatively decreased toxicity hazard when compared to the API cyamemazine for the endpoints of interest.
3. Discussion
New approach methodologies based on a more mechanistic vision are emerging at a fast rate for supporting hepatotoxicity assessment of chemicals; high expectations call for novel strategies that would be less dependent on in vivo testing and where integration of the different approaches (including in silico methods) may improve prediction of liver toxicity [200]. In this context, it should be noted that a mechanistic-based classification of liver injury is challenging, because of the number and complexity of biological pathways underpinning hepatotoxicity and because there is still limited understanding of these mechanisms [44,137,201].
Development of in silico models to predict liver toxicity poses many hurdles, as highlighted in the literature by Cronin and co-workers [44,188]. The first issue concerns the poor understanding of chemically-induced hepatotoxicity mechanisms and pathogenesis, which limits the development of mechanistically-based in silico models (as well as current in vitro panels for this purpose). This issue in turn affects the performance of the models.
The second issue arises from the availability of heterogeneous and sparse (and often biased) experimental data, such as pre-clinical studies or clinical reports of adverse drug reactions, resulting in pools of data that come from different assays and protocols. Biases from the assay side may arise from the source and precise composition of biological and chemical samples used for data generation (as well as the detailed assay protocol); biases from the clinical side may arise from differences between individuals (differences associated with genetic or other origin), as well as, in particular for post-marketing data, reporting biases and other confounding factors. Data from both the assay and the clinical side will hence generally be of variable quality and typically may not reflect potency. When these datasets are used as training sets, they will compromise the predictive power of the resulting computational models, since any model is only as good as the data that goes into it. As noted by Mulliner and coworkers [196], the specific liver toxicity endpoint that is often modeled is not present in the available databases, but it is described by a collection of observations (e.g., liver necrosis, increase of transaminases, bilirubin). The third issue concerns the complex mechanisms occurring in the liver, such as metabolism or defense mechanisms. These complex biological processes are difficult to embed in the computational approaches aimed at predicting toxicity. The fourth issue arises from the limited chemical space that the current models can cover, as the available experimental data are skewed towards the pharmaceutical domain. Finally, it should be noted that in silico models that predict dose/timepoints are limited in part due to technical limitations and the lack of properly annotated data. Models that do not build on dose/timepoints data fail to differentiate between those chemicals that have the potential to cause injury but never reach exposures and durations where damage is observed clinically, and those chemicals that lack the potential to cause injury altogether. Accounting for the time and exposure concept may shed light on mechanisms leading to liver injury as some negative compounds could have been classified as negative just because they did not reach the exposures or durations required for the damage to be observed. Classification algorithms for hepatotoxicity (positive/negative prediction) restrict the usability of the corresponding in silico models, as these models lack the ability to provide information on the doses producing the adverse effects or prediction of the severity of the effects. Predicting a quantitative measure of potency would be ideal. A practical and effective approach to modelling quantitative data would be to group the responses into multiple dose ranges based on a specific duration of exposure.
Recent advances in science show an increasing understanding of the molecular pathways and events underlying pathogenesis of hepatotoxicity. This knowledge serves as a robust basis to improve the predictions of hepatotoxicity within a mechanistic framework possibly using the AOP scenario [44,138,202–204]. The mechanistically-driven analysis carried out in the current position paper has resulted in the proposal of a practical and extensible framework building on the IST protocol project for integrating information on organ toxicity (both experimental and in silico) along with an example of how this approach was successfully used to assess non-genotoxic impurities. Definition of endpoints and their relationships in a mechanistically-informed framework constitutes the basis for pragmatic use and integration of in silico models built on various types of data.
Based on the draft framework proposed in Fig. 3, future development of the IST protocol applicable to hepatotoxicity will include:
enumeration of the different in silico approaches as well as experimental data to be associated with the effects and biological responses defined in the framework; these effects or responses that can be measured (or predicted) are shown as gray boxes in Fig. 3, 4 and 5.
definition of major and sub-endpoints (shown in the framework as blue boxes in Fig. 3, 4 and 5. alongside clear definition of the rules and principles for combining the information on effects/biological responses into these major and sub- endpoints to result in an overall assessment and an evaluation of the confidence of this assessment;
incorporation of an expert review process of all major and sub-endpoint assessments, including specific guidelines for incorporating internal and external knowledge and formats for documenting the results.
4. Conclusion
In silico prediction of organ toxicity is poised to play a significant role in future research and development supporting the move toward mechanistic-based assessments of toxicity and application of the replace, refine, and reduce (3Rs) principles to whole animal toxicity testing. Although much work is needed for the integration of in silico models in robust NAM workflows or for their use in defined approaches (that could replace the current testing paradigm in the assessment of target organ toxicity), such models can still play an important role in specific scenarios such as the assessment of non-genotoxic impurities, prioritization of testing strategies, or screening of chemicals. The development of robust and transparent in silico toxicology protocols, based on suitable underlying data, paves the way for a more extensive use of these methods, taking advantage of the underlying standardizations. Protocols must provide a defensible assessment of the overall confidence (based on the reliability and completeness of the information provided, and its relevance to the toxicological endpoint) in addition to information on toxic concentrations and timepoints to be effectively used as part of any hazard assessment.
The hazard assessment framework organizes and classifies available and emerging experimental approaches and the underlying endpoints that are going to be integrated for the evaluation of liver toxicity. The output will serve as a basis for the development of an IST protocol that standardizes the use of in silico approaches for the prediction of hepatotoxicity from chemical structure. The resulting standardization catalyzes the acceptability of both the in silico methods and the corresponding predictions by end users, colleagues, collaborators, and regulators as well as provides a means to support a more transparent analysis of the results.
Supplementary Material
Summary of the biological mechanisms and processes underpinning hepatotoxicity
Description of experimental approaches to support the prediction of hepatotoxicity
Discussion of the role of in silico approaches highlighting challenges to the adoption of these methods
Proposed framework for the integration of in silico and experimental information
Acknowledgements
Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R44ES026909. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Arianna Bassan: Conceptualization, Writing - Original Draft, Writing - Review & Editing. Vinicius M. Alves: Writing - Review & Editing. Alexander Amberg: Writing - Review & Editing.Lennart T. Anger: Writing - Review & Editing. Scott Auerbach: Writing - Review & Editing. Lisa Beilke: Writing - Review & Editing. Andreas Bender: Writing - Review & Editing. Mark T.D. Cronin: Writing - Review & Editing. Kevin P. Cross: Writing - Review & Editing. Jui-Hua Hsieh: Writing - Review & Editing. Nigel Greene: Writing - Review & Editing. Raymond Kemper: Writing - Review & Editing. Marlene T. Kim: Writing - Review & Editing. Moiz Mumtaz: Writing - Review & Editing. Tobias Noeske: Writing - Review & Editing. Manuela Pavan: Writing - Review & Editing. Julia Pletz: Writing - Review & Editing. Daniel P. Russo: Writing - Review & Editing. Yogesh Sabnis: Writing - Review & Editing. Markus Schaefer: Writing - Review & Editing. David T. Szabo: Writing - Review & Editing. Jean-Pierre Valentin: Writing - Review & Editing. Joerg Wichard: Writing - Review & Editing. Dominic Williams: Writing - Review & Editing. David Woolley: Writing - Review & Editing. Craig Zwickl: Writing - Review & Editing. Glenn J. Myatt: Conceptualization, Writing - Review & Editing, Funding acquisition.
Declaration of Competing Interest
Disclaimer
CDC Disclaimer
The findings and conclusions in this article are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry.
FDA disclaimer
This article reflects the views of the authors and should not be construed to represent FDA’s views or policies. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Supplementary data
(see supplementary file)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hartung T, Toxicology for the twenty-first century, Nature. 460 (2009) 208–212. 10.1038/460208a. [DOI] [PubMed] [Google Scholar]
- [2].ICCVAM, Interagency Coordinating Committee on the Validation of Alternative Methods (ICCVAM) Authorization Act of 2000, 42 U.S.C. 285l-3, Public Law; 106–545, 106th Congress, 2000. https://ntp.niehs.nih.gov/iccvam/docs/about_docs/pl106545.pdf. [Google Scholar]
- [3].Judson R, Houck K, Martin M, Knudsen T, Thomas RS, Sipes N, Shah I, Wambaugh J, Crofton K, In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme, Basic Clin. Pharmacol. Toxicol. 115 (2014) 69–76. 10.1111/bcpt.12239. [DOI] [PubMed] [Google Scholar]
- [4].Krewski D, Acosta D, Andersen M, Anderson H, Bailar JC, Boekelheide K, Brent R, Charnley G, Cheung VG, Green S, Kelsey KT, Kerkvliet NI, Li AA, McCray L, Meyer O, Patterson RD, Pennie W, Scala RA, Solomon GM, Stephens M, Yager J, Zeise L, Toxicity testing in the 21st century: a vision and a strategy, J. Toxicol. Environ. Health, Part B. 13 (2010) 51–138. 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].National Academies of Sciences, Engineering, and Medicine, Using 21st Century Science to Improve Risk-Related Evaluations, National Academies Press, Washington, DC, 2017. 10.17226/24635. [DOI] [PubMed] [Google Scholar]
- [6].Rusyn I, Arzuaga X, Cattley RC, Corton JC, Ferguson SS, Godoy P, Guyton KZ, Kaplowitz N, Khetani SR, Roberts R, Roth RA, Smith MT, Key Characteristics of Human Hepatotoxicants as a Basis for Identification and Characterization of the Causes of Liver Toxicity, Hepatology. (2021) hep.31999. 10.1002/hep.31999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].US EPA, Strategic Plan to Promote the Development and Implementation of Alternative Test Methods within the TSCA Program., U.S. Environmental Protection Agency, Office of Chemical Safety and Pollution Prevention, Washington, DC, 2018. https://www.epa.gov/sites/production/files/2018-06/documents/epa_alt_strat_plan_6-20-18_clean_final.pdf. [Google Scholar]
- [8].OECD, Overview of concepts and available guidance related to integrated approaches to testing and assessment (IATA), OECD Publishing, Paris, 2020. https://www.oecd.org/chemicalsafety/risk-assessment/concepts-and-available-guidance-related-to-integrated-approaches-to-testing-and-assessment.pdf. [Google Scholar]
- [9].Avila AM, Bebenek I, Bonzo JA, Bourcier T, Davis Bruno KL, Carlson DB, Dubinion J, Elayan I, Harrouk W, Lee S-L, Mendrick DL, Merrill JC, Peretz J, Place E, Saulnier M, Wange RL, Yao J, Zhao D, Brown PC, An FDA/CDER perspective on nonclinical testing strategies: Classical toxicology approaches and new approach methodologies (NAMs), Regulatory Toxicology and Pharmacology. 114 (2020) 104662. 10.1016/j.yrtph.2020.104662. [DOI] [PubMed] [Google Scholar]
- [10].Myatt GJ, Ahlberg E, Akahori Y, Allen D, Amberg A, Anger LT, Aptula A, Auerbach S, Beilke L, Bellion P, Benigni R, Bercu J, Booth ED, Bower D, Brigo A, Burden N, Cammerer Z, Cronin MTD, Cross KP, Custer L, Dettwiler M, Dobo K, Ford KA, Fortin MC, Gad-McDonald SE, Gellatly N, Gervais V, Glover KP, Glowienke S, Van Gompel J, Gutsell S, Hardy B, Harvey JS, Hillegass J, Honma M, Hsieh J-H, Hsu C-W, Hughes K, Johnson C, Jolly R, Jones D, Kemper R, Kenyon MO, Kim MT, Kruhlak NL, Kulkarni SA, Kümmerer K, Leavitt P, Majer B, Masten S, Miller S, Moser J, Mumtaz M, Muster W, Neilson L, Oprea TI, Patlewicz G, Paulino A, Lo Piparo E, Powley M, Quigley DP, Reddy MV, Richarz A-N, Ruiz P, Schilter B, Serafimova R, Simpson W, Stavitskaya L, Stidl R, Suarez-Rodriguez D, Szabo DT, Teasdale A, Trejo-Martin A, Valentin J-P, Vuorinen A, Wall BA, Watts P, White AT, Wichard J, Witt KL, Woolley A, Woolley D, Zwickl C, Hasselgren C, In silico toxicology protocols, Regul. Toxicol. Pharmacol. 96 (2018) 1–17. 10.1016/j.yrtph.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Raies AB, Bajic VB, In silico toxicology: computational methods for the prediction of chemical toxicity, WIREs Comput. Mol. Sci. 6 (2016) 147–172. 10.1002/wcms.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mahony C, Ashton RS, Birk B, Boobis AR, Cull T, Daston GP, Ewart L, Knudsen TB, Manou I, Maurer-Stroh S, Margiotta-Casaluci L, Müller BP, Nordlund P, Roberts RA, Steger-Hartmann T, Vandenbossche E, Viant MR, Vinken M, Whelan M, Zvonimir Z, Cronin MTD, New ideas for non-animal approaches to predict repeated-dose systemic toxicity: Report from an EPAA Blue Sky Workshop, Regul. Toxicol. Pharmacol. 114 (2020) 104668. 10.1016/j.yrtph.2020.104668. [DOI] [PubMed] [Google Scholar]
- [13].Hasselgren C, Ahlberg E, Akahori Y, Amberg A, Anger LT, Atienzar F, Auerbach S, Beilke L, Bellion P, Benigni R, Bercu J, Booth ED, Bower D, Brigo A, Cammerer Z, Cronin MTD, Crooks I, Cross KP, Custer L, Dobo K, Doktorova T, Faulkner D, Ford KA, Fortin MC, Frericks M, Gad-McDonald SE, Gellatly N, Gerets H, Gervais V, Glowienke S, Van Gompel J, Harvey JS, Hillegass J, Honma M, Hsieh J-H, Hsu C-W, Barton-Maclaren TS, Johnson C, Jolly R, Jones D, Kemper R, Kenyon MO, Kruhlak NL, Kulkarni SA, Kümmerer K, Leavitt P, Masten S, Miller S, Moudgal C, Muster W, Paulino A, Lo Piparo E, Powley M, Quigley DP, Reddy MV, Richarz A-N, Schilter B, Snyder RD, Stavitskaya L, Stidl R, Szabo DT, Teasdale A, Tice RR, Trejo-Martin A, Vuorinen A, Wall BA, Watts P, White AT, Wichard J, Witt KL, Woolley A, Woolley D, Zwickl C, Myatt GJ, Genetic toxicology in silico protocol, Regul. Toxicol. Pharmacol. 107 (2019) 104403. 10.1016/j.yrtph.2019.104403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Johnson C, Ahlberg E, Anger LT, Beilke L, Benigni R, Bercu J, Bobst S, Bower D, Brigo A, Campbell S, Cronin MTD, Crooks I, Cross KP, Doktorova T, Exner T, Faulkner D, Fearon IM, Fehr M, Gad SC, Gervais V, Giddings A, Glowienke S, Hardy B, Hasselgren C, Hillegass J, Jolly R, Krupp E, Lomnitski L, Magby J, Mestres J, Milchak L, Miller S, Muster W, Neilson L, Parakhia R, Parenty A, Parris P, Paulino A, Paulino AT, Roberts DW, Schlecker H, Stidl R, Suarez-Rodrigez D, Szabo DT, Tice RR, Urbisch D, Vuorinen A, Wall B, Weiler T, White AT, Whritenour J, Wichard J, Woolley D, Zwickl C, Myatt GJ, Skin sensitization in silico protocol, Regul. Toxicol. Pharmacol. 116 (2020) 104688. 10.1016/j.yrtph.2020.104688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Tice RR, Bassan A, Amberg A, Anger LT, Beal MA, Bellion P, Benigni R, Birmingham J, Brigo A, Bringezu F, Ceriani L, Crooks I, Cross K, Elespuru R, Faulkner D, Fortin MC, Fowler P, Frericks M, Gerets HHJ, Jahnke GD, Jones DR, Kruhlak NL, Lo Piparo E, Lopez-Belmonte J, Luniwal A, Luu A, Madia F, Manganelli S, Manickam B, Mestres J, Mihalchik-Burhans AL, Neilson L, Pandiri A, Pavan M, Rider CV, Rooney JP, Trejo-Martin A, Watanabe-Sailor KH, White AT, Woolley D, Myatt GJ, In silico approaches in carcinogenicity hazard assessment: current status and future needs, Comput. Toxicol. (2021) Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bassan A, Alves VM, Amberg A, Anger LT, Beilke L, Bender A, Bernal A, Cronin M, Hsieh J-H, Johnson C, Kemper R, Mumtaz M, Nelson L, Pavan M, Pointon A, Pletz J, Ruiz P, Russo DP, Sabnis Y, Sandhu R, Schaefer M, Stavitskaya L, Szabo DT, Valentin J-P, Woolley D, Zwickl C, Myatt GJ, In silico approaches in organ toxicity hazard assessment: current status and future needs for predicting heart, kidney and lung toxicities, (2021) Unpublished results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Crofton K, Bassan A, Behl M, Chushak Y, Fritsche E, Gearhart J, Marty S, Mumtaz M, Pavan M, Ruiz P, Shaffer T, Sachana M, Selvam R, Stavitskaya L, Szabo D, Tice R, Wilson D, Woolley D, Myatt GJ, Current status and future needs for a neurotoxicity hazard assessment framework that integrates in silico approaches, (2021) Unpublished results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].ICH, ICH Q3A (R2) Impurities in new drug substances, European Medicines Agency, 2006. https://database.ich.org/sites/default/files/Q3A%28R2%29%20Guideline.pdf.
- [19].ICH, ICH Q3B (R2) Impurities in new drug products, European Medicines Agency, 2006. https://database.ich.org/sites/default/files/Q3B%28R2%29%20Guideline.pdf.
- [20].EMA, Reflection paper on the qualification of non-genotoxic impurities, 2018. https://www.ema.europa.eu/en/qualification-non-genotoxic-impurities.
- [21].Broschard TH, Glowienke S, Bruen US, Nagao LM, Teasdale A, Stults CLM, Li KL, Iciek LA, Erexson G, Martin EA, Ball DJ, Assessing safety of extractables from materials and leachables in pharmaceuticals and biologics - Current challenges and approaches, Regul. Toxicol. Pharmacol. 81 (2016) 201–211. 10.1016/j.yrtph.2016.08.011. [DOI] [PubMed] [Google Scholar]
- [22].ECHA, Guidance on information requirements and chemical safety assessment Chapter R.7a: endpoint specific guidance. Version 6.0, Publications Office of the EU, 2017. 10.2823/337352. [DOI] [Google Scholar]
- [23].ECHA, Non-animal approaches Current status of regulatory applicability under the REACH, CLP and Biocidal Products regulations, Publications Office of the EU, 2017. 10.2823/000784. [DOI] [Google Scholar]
- [24].US EPA, Low-Priority Substances under TSCA, US EPA. (2019). https://www.epa.gov/assessing-and-managing-chemicals-under-tsca/low-priority-substances-under-tsca (accessed April 16, 2021). [Google Scholar]
- [25].ECHA, Guidance on the Biocidal Products Regulation Volume III: human health, assessment & evaluation (Parts B+C) Version 4.0, Publications Office of the EU, 2017. 10.2823/143042. [DOI] [Google Scholar]
- [26].JRC, Applicability of QSAR analysis to the evaluation of the toxicological relevance of metabolites and degradates of pesticide active substances for dietary risk assessment - Prepared by European Commission Joint Research Centre, Institute for Health & Consumer Protection, Ispra, Italy, EFSA Supporting Publications. 7 (2010) EN-50, 311. 10.2903/sp.efsa.2010.EN-50. [DOI] [Google Scholar]
- [27].EC, Regulation (EC) No 1223/2009 of the European Parliament and of the Council of 30 November 2009 on cosmetic products, OJ. L 342 (2009) 59–209. http://data.europa.eu/eli/reg/2009/1223/oj. [Google Scholar]
- [28].Gellatly N, Sewell F, Regulatory acceptance of in silico approaches for the safety assessment of cosmetic-related substances, Comput. Toxicol. 11 (2019) 82–89. 10.1016/j.comtox.2019.03.003. [DOI] [Google Scholar]
- [29].COSMOS, Final Report Summary - COSMOS (Integrated In Silico Models for the Prediction of Human Repeated Dose Toxicity of Cosmetics to Optimise Safety) | FP7 | CORDIS | European Commission, (2015). https://cordis.europa.eu/project/id/266835/reporting (accessed May 7, 2021).
- [30].Rogiers V, Benfenati E, Bernauer U, Bodin L, Carmichael P, Chaudhry Q, Coenraads PJ, Cronin MTD, Dent M, Dusinska M, Ellison C, Ezendam J, Gaffet E, Galli CL, Goebel C, Granum B, Hollnagel HM, Kern PS, Kosemund-Meynen K, Ouédraogo G, Panteri E, Rousselle C, Stepnik M, Vanhaecke T, von Goetz N, Worth A, The way forward for assessing the human health safety of cosmetics in the EU - Workshop proceedings, Toxicology. 436 (2020) 152421. 10.1016/j.tox.2020.152421. [DOI] [PubMed] [Google Scholar]
- [31].Schultz TW, Przybylak KR, Richarz A-N, Mellor CL, Bradbury SP, Cronin MTD, Read-across of 90-day rat oral repeated-dose toxicity: A case study for selected 2-alkyl-1-alkanols, Comput. Toxicol. 2 (2017) 28–38. 10.1016/j.comtox.2017.02.005. [DOI] [Google Scholar]
- [32].Schultz TW, Cronin MTD, Lessons learned from read-across case studies for repeated-dose toxicity, Regul. Toxicol. Pharmacol. 88 (2017) 185–191. 10.1016/j.yrtph.2017.06.011. [DOI] [PubMed] [Google Scholar]
- [33].EFSA Scientific Committee, Hardy A, Benford D, Halldorsson T, Jeger MJ, Knutsen HK, More S, Naegeli H, Noteborn H, Ockleford C, Ricci A, Rychen G, Schlatter JR, Silano V, Solecki R, Turck D, Benfenati E, Chaudhry QM, Craig P, Frampton G, Greiner M, Hart A, Hogstrand C, Lambre C, Luttik R, Makowski D, Siani A, Wahlstroem H, Aguilera J, Dorne J-L, Dumont AF, Hempen M, Martínez SV, Martino L, Smeraldi C, Terron A, Georgiadis N, Younes M, Guidance on the use of the weight of evidence approach in scientific assessments, EFSA J. 15 (2017) e04971. 10.2903/j.efsa.2017.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].FDA, FDA’s Predictive Toxicology Roadmap, FDA. (2020). https://www.fda.gov/science-research/about-science-research-fda/fdas-predictive-toxicology-roadmap (accessed May 20, 2020).
- [35].Gosslau A, Assessment of food toxicology, Food Sci. Hum. Wellness. 5 (2016) 103–115. 10.1016/j.fshw.2016.05.003. [DOI] [Google Scholar]
- [36].ECHA, Appendix to Chapter R.8: Guidance for preparing a scientific report for health-based exposure limits at the workplace. Version 1.0, Publications Office of the EU, 2019. 10.2823/333736. [DOI] [Google Scholar]
- [37].NIOSH, Technical report: The NIOSH occupational exposure banding process for chemical risk management, 2019. 10.26616/NIOSHPUB2019132. [DOI]
- [38].Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S, Reducing safety-related drug attrition: the use of in vitro pharmacological profiling, Nat. Rev. Drug Discovery. 11 (2012) 909–922. 10.1038/nrd3845. [DOI] [PubMed] [Google Scholar]
- [39].Lynch JJ, Van Vleet TR, Mittelstadt SW, Blomme EAG, Potential functional and pathological side effects related to off-target pharmacological activity, J. Pharmacol. Toxicol. Methods. 87 (2017) 108–126. 10.1016/j.vascn.2017.02.020. [DOI] [PubMed] [Google Scholar]
- [40].Rognan D, The impact of in silico screening in the discovery of novel and safer drug candidates, Pharmacol. Ther. 175 (2017) 47–66. 10.1016/j.pharmthera.2017.02.034. [DOI] [PubMed] [Google Scholar]
- [41].Drakvik E, Altenburger R, Aoki Y, Backhaus T, Bahadori T, Barouki R, Brack W, Cronin MTD, Demeneix B, Hougaard Bennekou S, van Klaveren J, Kneuer C, Kolossa-Gehring M, Lebret E, Posthuma L, Reiber L, Rider C, Rüegg J, Testa G, van der Burg B, van der Voet H, Warhurst AM, van de Water B, Yamazaki K, Öberg M, Bergman Å, Statement on advancing the assessment of chemical mixtures and their risks for human health and the environment, Environ. Int. 134 (2020) 105267. 10.1016/j.envint.2019.105267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bopp SK, Kienzler A, Richarz A-N, van der Linden SC, Paini A, Parissis N, Worth AP, Regulatory assessment and risk management of chemical mixtures: challenges and ways forward, Crit. Rev. Toxicol. 49 (2019) 174–189. 10.1080/10408444.2019.1579169. [DOI] [PubMed] [Google Scholar]
- [43].Lu FC, Kacew S, Lu FC, Lu’s basic toxicology: fundamentals, target organs, and risk assessment, Fourth Edition, CRC Press, London, 2002. 10.1201/9781003026976. [DOI] [Google Scholar]
- [44].Cronin MTD, Enoch SJ, Mellor CL, Przybylak KR, Richarz A-N, Madden JC, In silico prediction of organ level toxicity: Linking chemistry to adverse effects, Toxicol. Res. 33 (2017) 173–182. 10.5487/TR.2017.33.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Leblanc GA, Acute Toxicity, in: Hodgson E. (Ed.), A Textbook of Modern Toxicology, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2004: pp. 213–224. 10.1002/0471646776.ch11. [DOI] [Google Scholar]
- [46].Porceddu M, Buron N, Rustin P, Fromenty B, Borgne-Sanchez A, In Vitro Assessment of Mitochondrial Toxicity to Predict Drug-Induced Liver Injury, in: Chen M, Will Y. (Eds.), Drug-Induced Liver Toxicity, Humana, New York, NY, 2018: pp. 283–300. 10.1007/978-1-4939-7677-5_14. [DOI] [Google Scholar]
- [47].Will Y, Dykens JA, eds., Mitochondrial Dysfunction Caused by Drugs and Environmental Toxicants, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2018. 10.1002/9781119329725. [DOI] [Google Scholar]
- [48].Ankley GT, Bennett RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder PK, Serrrano JA, Tietge JE, Villeneuve DL, Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment, Environ. Toxicol. Chem. 29 (2010) 730–741. 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- [49].OECD, Guidance Document for The Use Of Adverse Outcome Pathways In Developing Integrated Approaches To Testing And Assessment (IATA) Series on Testing & Assessment No. 260, OECD Environment, Health and Safety Publications, Paris, 2016. 10.1787/44bb06c1-en. [DOI] [Google Scholar]
- [50].Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M, Adverse outcome pathway (AOP) development I: strategies and principles, Toxicol. Sci. 142 (2014) 312–320. 10.1093/toxsci/kfu199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, Ottinger MA, Vergauwen L, Whelan M, Adverse outcome pathway development II: best practices, Toxicol. Sci. 142 (2014) 321–330. 10.1093/toxsci/kfu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Allen TEH, Goodman JM, Gutsell S, Russell PJ, A History of the Molecular Initiating Event, Chem. Res. Toxicol. 29 (2016) 2060–2070. 10.1021/acs.chemrestox.6b00341. [DOI] [PubMed] [Google Scholar]
- [53].Allen TEH, Goodman JM, Gutsell S, Russell PJ, Defining Molecular Initiating Events in the Adverse Outcome Pathway Framework for Risk Assessment, Chem. Res. Toxicol. 27 (2014) 2100–2112. 10.1021/tx500345j. [DOI] [PubMed] [Google Scholar]
- [54].Hines DE, Edwards SW, Conolly RB, Jarabek AM, A case study application of the Aggregate Exposure Pathway (AEP) and Adverse Outcome Pathway (AOP) frameworks to facilitate the integration of human health and ecological end points for Cumulative Risk Assessment (CRA), Environ. Sci. Technol. 52 (2018) 839–849. 10.1021/acs.est.7b04940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tan Y-M, Leonard JA, Edwards S, Teeguarden J, Paini A, Egeghy P, Aggregate exposure pathways in support of risk assessment, Curr. Opin. Toxicol. 9 (2018) 8–13. 10.1016/j.cotox.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tan C, Edwards S, Teeguarden J, Egeghy P, Leonard J, The Aggregate Exposure Pathway (AEP): A conceptual framework for advancing exposure science research and transforming risk assessment, Orlando, FL, 2016. https://cfpub.epa.gov/si/si_public_record_report.cfm?Lab=NERL&dirEntryId=335346. [Google Scholar]
- [57].AOP Knowledgebase, AOPwiki, (2021). https://aopwiki.org/ (accessed March 5, 2021).
- [58].Pittman ME, Edwards SW, Ives C, Mortensen HM, AOP-DB: a database resource for the exploration of Adverse Outcome Pathways through integrated association networks, Toxicol. Appl. Pharmacol. 343 (2018) 71–83. 10.1016/j.taap.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Spinu N, Cronin MTD, Enoch SJ, Madden JC, Worth AP, Quantitative adverse outcome pathway (qAOP) models for toxicity prediction, Arch. Toxicol. 94 (2020) 1497–1510. 10.1007/s00204-020-02774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Perkins EJ, Gayen K, Shoemaker JE, Antczak P, Burgoon L, Falciani F, Gutsell S, Hodges G, Kienzler A, Knapen D, McBride M, Willett C, Doyle FJ, Garcia-Reyero N, Chemical hazard prediction and hypothesis testing using quantitative adverse outcome pathways, ALTEX. 36 (2019) 91–102. 10.14573/altex.1808241. [DOI] [PubMed] [Google Scholar]
- [61].Perkins EJ, Ashauer R, Burgoon L, Conolly R, Landesmann B, Mackay C, Murphy CA, Pollesch N, Wheeler JR, Zupanic A, Scholz S, Building and applying quantitative adverse outcome pathway models for chemical hazard and risk assessment, Environ Toxicol Chem. 38 (2019) 1850–1865. 10.1002/etc.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Olson H, Betton G, Robinson D, Thomas K, Monro A, Kolaja G, Lilly P, Sanders J, Sipes G, Bracken W, Dorato M, Van Deun K, Smith P, Berger B, Heller A, Concordance of the toxicity of pharmaceuticals in humans and in animals, Regul. Toxicol. Pharmacol. 32 (2000) 56–67. 10.1006/rtph.2000.1399. [DOI] [PubMed] [Google Scholar]
- [63].Bailey J, Thew M, Balls M, Predicting human drug toxicity and safety via animal tests: can any one species predict drug toxicity in any other, and do monkeys help?, ATLA, Altern. Lab. Anim. 43 (2015) 393–403. 10.1177/026119291504300607. [DOI] [PubMed] [Google Scholar]
- [64].Kenna JG, Ram R, Chapter 3.2 - Safety Assessment of Pharmaceuticals, in: Balls M, Combes R, Worth A. (Eds.), The History of Alternative Test Methods in Toxicology, Academic Press, 2019: pp. 167–176. 10.1016/B978-0-12-813697-3.00020-2. [DOI] [Google Scholar]
- [65].Beilmann M, Boonen H, Czich A, Dear G, Hewitt P, Mow T, Newham P, Oinonen T, Pognan F, Roth A, Valentin J-P, Van Goethem F, Weaver RJ, Birk B, Boyer S, Caloni F, Chen AE, Corvi R, Cronin MTD, Daneshian M, Ewart LC, Fitzgerald RE, Hamilton GA, Hartung T, Kangas JD, Kramer NI, Leist M, Marx U, Polak S, Rovida C, Testai E, Van der Water B, Vulto P, StegerHartmann T, Optimizing drug discovery by Investigative Toxicology: Current and future trends, ALTEX. 36 (2019) 289–313. 10.14573/altex.1808181. [DOI] [PubMed] [Google Scholar]
- [66].Weaver RJ, Valentin J-P, Today’s challenges to de-risk and predict drug safety in human “Mind-the-Gap,” Toxicol. Sci. 167 (2019) 307–321. 10.1093/toxsci/kfy270. [DOI] [PubMed] [Google Scholar]
- [67].Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, Pangalos MN, Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework, Nat. Rev. Drug Discovery. 13 (2014) 419–431. 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- [68].Sacks LV, Shamsuddin HH, Yasinskaya YI, Bouri K, Lanthier ML, Sherman RE, Scientific and regulatory reasons for delay and denial of FDA approval of initial applications for new drugs, 2000–2012, JAMA. 311 (2014) 378–384. 10.1001/jama.2013.282542. [DOI] [PubMed] [Google Scholar]
- [69].Will Y, McDuffie JE, Olaharski AJ, Jeffy BD, eds., Drug Discovery Toxicology: From Target Assessment to Translational Biomarkers, John Wiley & Sons, Inc, Hoboken, New Jersey, 2016. https://www.wiley.com/en-ar/Drug+Discovery+Toxicology%3A+From+Target+Assessment+to+Translational+Biomarkers-p-9781119053330. [Google Scholar]
- [70].Chen Z, Liu Y, Wright FA, Chiu WA, Rusyn I, Rapid hazard characterization of environmental chemicals using a compendium of human cell lines from different organs, ALTEX - Alternatives to Animal Experimentation. 37 (2020) 623–638. 10.14573/altex.2002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Paul Friedman K, Gagne M, Loo L-H, Karamertzanis P, Netzeva T, Sobanski T, Franzosa JA, Richard AM, Lougee RR, Gissi A, Lee J-YJ, Angrish M, Dorne JL, Foster S, Raffaele K, Bahadori T, Gwinn MR, Lambert J, Whelan M, Rasenberg M, Barton-Maclaren T, Thomas RS, Utility of in vitro bioactivity as a lower bound estimate of in vivo adverse effect levels and in risk-based prioritization, Toxicol. Sci. 173 (2020) 202–225. 10.1093/toxsci/kfz201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Bray M-A, Singh S, Han H, Davis CT, Borgeson B, Hartland C, Kost-Alimova M, Gustafsdottir SM, Gibson CC, Carpenter AE, Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes, Nat. Protoc. 11 (2016) 1757–1774. 10.1038/nprot.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Peterson NC, Mahalingaiah PK, Fullerton A, Di Piazza M, Application of microphysiological systems in biopharmaceutical research and development, Lab Chip. 20 (2020) 697–708. 10.1039/C9LC00962K. [DOI] [PubMed] [Google Scholar]
- [74].Pridgeon CS, Schlott C, Wong MW, Heringa MB, Heckel T, Leedale J, Launay L, Gryshkova V, Przyborski S, Bearon RN, Wilkinson EL, Ansari T, Greenman J, Hendriks DFG, Gibbs S, Sidaway J, Sison-Young RL, Walker P, Cross MJ, Park BK, Goldring CEP, Innovative organotypic in vitro models for safety assessment: aligning with regulatory requirements and understanding models of the heart, skin, and liver as paradigms, Arch. Toxicol. 92 (2018) 557–569. 10.1007/s00204-018-2152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jensen C, Teng Y, Is it time to start transitioning from 2D to 3D cell culture?, Front. Mol. Biosci. 7 (2020) 33. 10.3389/fmolb.2020.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Esch EW, Bahinski A, Huh D, Organs-on-chips at the frontiers of drug discovery, Nat Rev Drug Discov. 14 (2015) 248–260. 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lancaster MA, Knoblich JA, Organogenesis in a dish: Modeling development and disease using organoid technologies, Science. 345 (2014) 1247125–1247125. 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- [78].Esch MB, Smith AST, Prot J-M, Oleaga C, Hickman JJ, Shuler ML, How multi-organ microdevices can help foster drug development, Adv. Drug Delivery Rev. 69–70 (2014) 158–169. 10.1016/j.addr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wang YI, Carmona C, Hickman JJ, Shuler ML, Multi-organ microphysiological systems for drug development: strategies, advances and challenges, Adv. Healthcare Mater. 7 (2018). 10.1002/adhm.201701000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Deaton AM, Fan F, Zhang W, Nguyen PA, Ward LD, Nioi P, Rationalizing secondary pharmacology screening using human genetic and pharmacological evidence, Toxicol. Sci. 167 (2019) 593–603. 10.1093/toxsci/kfy265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ietswaart R, Arat S, Chen AX, Farahmand S, Kim B, DuMouchel W, Armstrong D, Fekete A, Sutherland JJ, Urban L, Machine learning guided association of adverse drug reactions with in vitro target-based pharmacology, EBioMedicine. (2020) 102837. 10.1016/j.ebiom.2020.102837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Urban L, Whitebread S, Hamon J, Mikhailov D, Azzaoui K, Screening for Safety-Relevant Off-Target Activities, in: Peters J-U (Ed.), Polypharmacology in Drug Discovery, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2012: pp. 15–46. 10.1002/9781118098141.ch2. [DOI] [Google Scholar]
- [83].Whitebread S, Dumotier B, Armstrong D, Fekete A, Chen S, Hartmann A, Muller PY, Urban L, Secondary pharmacology: screening and interpretation of off-target activities – focus on translation, Drug Discovery Today. 21 (2016) 1232–1242. 10.1016/j.drudis.2016.04.021. [DOI] [PubMed] [Google Scholar]
- [84].Jenkinson S, Schmidt F, Rosenbrier Ribeiro L, Delaunois A, Valentin J-P, A practical guide to secondary pharmacology in drug discovery, J. Pharmacol. Toxicol. Methods. 105 (2020) 106869. 10.1016/j.vascn.2020.106869. [DOI] [PubMed] [Google Scholar]
- [85].Valentin J-P, Guillon J-M, Jenkinson S, Kadambi V, Ravikumar P, Roberts S, Rosenbrier-Ribeiro L, Schmidt F, Armstrong D, In vitro secondary pharmacological profiling: an IQ-DruSafe industry survey on current practices, J. Pharmacol. Toxicol. Methods. 93 (2018) 7–14. 10.1016/j.vascn.2018.07.001. [DOI] [PubMed] [Google Scholar]
- [86].Alexander-Dann B, Pruteanu LL, Oerton E, Sharma N, Berindan-Neagoe I, Módos D, Bender A, Developments in toxicogenomics: understanding and predicting compound-induced toxicity from gene expression data, Mol. Omics. 14 (2018) 218–236. 10.1039/c8mo00042e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Stavitskaya L, Aubrecht J, Kruhlak NL, Chemical Structure-Based and Toxicogenomic Models, in: Graziano MJ, Jacobson-Kram D. (Eds.), Genotoxicity and Carcinogenicity Testing of Pharmaceuticals, Springer, Cham, 2015: pp. 13–34. 10.1007/978-3-319-22084-0_2. [DOI] [Google Scholar]
- [88].Ganter B, Snyder RD, Halbert DN, Lee MD, Toxicogenomics in drug discovery and development: mechanistic analysis of compound/class-dependent effects using the DrugMatrix ® database, Pharmacogenomics. 7 (2006) 1025–1044. 10.2217/14622416.7.7.1025. [DOI] [PubMed] [Google Scholar]
- [89].Igarashi Y, Nakatsu N, Yamashita T, Ono A, Ohno Y, Urushidani T, Yamada H, Open TG-GATEs: a large-scale toxicogenomics database, Nucleic Acids Res. 43 (2015) D921–D927. 10.1093/nar/gku955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Davis AP, Grondin CJ, Johnson RJ, Sciaky D, King BL, McMorran R, Wiegers J, Wiegers TC, Mattingly CJ, The Comparative Toxicogenomics Database: update 2017, Nucleic Acids Res. 45 (2017) D972–D978. 10.1093/nar/gkw838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Low Y, Uehara T, Minowa Y, Yamada H, Ohno Y, Urushidani T, Sedykh A, Muratov E, Kuz’min V, Fourches D, Zhu H, Rusyn I, Tropsha A, Predicting drug-induced hepatotoxicity using QSAR and toxicogenomics approaches, Chem. Res. Toxicol. 24 (2011) 1251–1262. 10.1021/tx200148a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].An YR, Kim JY, Kim YS, Construction of a predictive model for evaluating multiple organ toxicity, Mol. Cell. Toxicol. 12 (2016) 1–6. 10.1007/s13273-016-0001-6. [DOI] [Google Scholar]
- [93].Sewell F, Aggarwal M, Bachler G, Broadmeadow A, Gellatly N, Moore E, Robinson S, Rooseboom M, Stevens A, Terry C, Burden N, The current status of exposure-driven approaches for chemical safety assessment: A cross-sector perspective, Toxicology. 389 (2017) 109–117. 10.1016/j.tox.2017.07.018. [DOI] [PubMed] [Google Scholar]
- [94].Lehman-McKeeman LD, Ruepp SU, Chapter 2 - Biochemical and Molecular Basis of Toxicity, in: Wallig MA, Haschek WM, Rousseaux CG, Bolon B. (Eds.), Fundamentals of Toxicologic Pathology, Third Edition, Academic Press, 2018: pp. 15–33. 10.1016/B978-0-12-809841-7.00002-2. [DOI] [Google Scholar]
- [95].Barton HA, Pastoor TP, Baetcke K, Chambers JE, Diliberto J, Doerrer NG, Driver JH, Hastings CE, Iyengar S, Krieger R, Stahl B, Timchalk C, The acquisition and application of Absorption, Distribution, Metabolism, and Excretion (ADME) data in agricultural chemical safety assessments, Crit. Rev. Toxicol. 36 (2006) 9–35. 10.1080/10408440500534362. [DOI] [PubMed] [Google Scholar]
- [96].EC, Guidelines for setting specific concentration limits for carcinogens in Annex I of directive 67/548/EEC Inclusion of potency considerations, Publications Office of the EU, Luxembourg, 1999. https://op.europa.eu/s/o9up. [Google Scholar]
- [97].EFSA, Modern methodologies and tools for human hazard assessment of chemicals, EFSA J. 12 (2014) 3638. 10.2903/j.efsa.2014.3638. [DOI] [Google Scholar]
- [98].US EPA, Guidelines for Carcinogen Risk Assessment, Washington, DC, 2005. https://www.epa.gov/sites/production/files/2013-09/documents/cancer_guidelines_final_3-25-05.pdf. [Google Scholar]
- [99].ICH, Note for guidance on toxicokinetics: the assessment of systemic exposure in toxicity studies S3A, 1995. https://database.ich.org/sites/default/files/S3A_Guideline.pdf.
- [100].OECD, Test No. 417: Toxicokinetics, OECD Publishing, Paris, 2010. 10.1787/9789264070882-en. [DOI] [Google Scholar]
- [101].RIVM, Heringa MB, Brandon EFA, Bessems JG, Bos PMJ, Integration of toxicokinetics and toxicodynamics testing essential for risk assessment, National Institute for Public Health and the Environment, Bilthoven, The Netherlands, 2013. https://www.rivm.nl/bibliotheek/rapporten/055212001.pdf. [Google Scholar]
- [102].Ghosh J, Lawless MS, Waldman M, Gombar V, Fraczkiewicz R, Modeling ADMET, in: Benfenati E. (Ed.), In Silico Methods for Predicting Drug Toxicity, Humana Press, New York, NY, 2016: pp. 63–83. 10.1007/978-1-4939-3609-0_4. [DOI] [PubMed] [Google Scholar]
- [103].Madden JC, In Silico Approaches for Predicting ADME Properties, in: Puzyn T, Leszczynski J, Cronin MT (Eds.), Recent Advances in QSAR Studies, Springer, Dordrecht, 2010: pp. 283–304. 10.1007/978-1-4020-9783-6_10. [DOI] [Google Scholar]
- [104].Mostrag-Szlichtyng A, Worth A, Review of QSAR Models and Software Tools for predicting Biokinetic Properties, Luxembourg, 2010. 10.2788/94537. [DOI] [Google Scholar]
- [105].Shin HK, Kang Y-M, No KT, Predicting ADME Properties of Chemicals, in: Leszczynski J. (Ed.), Handbook of Computational Chemistry, Springer, Dordrecht, Netherlands, 2016: pp. 1–37. 10.1007/978-94-007-6169-8_59-1. [DOI] [Google Scholar]
- [106].Kirchmair J, Williamson MJ, Tyzack JD, Tan L, Bond PJ, Bender A, Glen RC, Computational prediction of metabolism: sites, products, SAR, P450 enzyme dynamics, and mechanisms, J. Chem. Inf. Model. 52 (2012) 617–648. 10.1021/ci200542m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kemper R, Taub M, Bogdanffy M, Metabolism: a determinant of toxicity, in: Hayes A, Kruger C. (Eds.), Hayes’ Principles and Methods of Toxicology, Sixth Edition, CRC Press, London, 2014: pp. 141–214. 10.1201/b17359. [DOI] [Google Scholar]
- [108].Kulkarni SA, Zhu J, Blechinger S, In silico techniques for the study and prediction of xenobiotic metabolism: A review, Xenobiotica. 35 (2005) 955–973. 10.1080/00498250500354402. [DOI] [PubMed] [Google Scholar]
- [109].Clewell III H, Clewell R, Andersen M, Physiologically Based Pharmacokinetic and Toxicokinetic Models, in: Hayes A, Kruger C. (Eds.), Hayes’ Principles and Methods of Toxicology, Sixth Edition, CRC Press, 2014: pp. 247–294. 10.1201/b17359-8. [DOI] [Google Scholar]
- [110].Krishnan K, Andersen M, Physiologically Based Pharmacokinetic and Toxicokinetic Models, in: Hayes A. (Ed.), Principles and Methods of Toxicology, Fifth Edition, CRC Press, London, 2008: pp. 231–292. 10.1201/b14258. [DOI] [Google Scholar]
- [111].van der Merwe D, Gehring R, Buur JL, Chapter 8 - Toxicokinetics in Veterinary Toxicology, in: Gupta RC (Ed.), Veterinary Toxicology (Third Edition), Academic Press, 2018: pp. 133–143. 10.1016/B978-0-12-811410-0.00008-8. [DOI] [Google Scholar]
- [112].Caldwell JC, Evans MV, Krishnan K, Cutting edge PBPK models and analyses: Providing the basis for future modeling efforts and bridges to emerging toxicology paradigms, J. Toxicol. 2012 (2012) 1–10. 10.1155/2012/852384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Mumtaz M, Fisher J, Blount B, Ruiz P, Application of physiologically based pharmacokinetic models in chemical risk assessment, J. Toxicol. 2012 (2012) 1–11. 10.1155/2012/904603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Paini A, Leonard JA, Kliment T, Tan Y-M, Worth A, Investigating the state of physiologically based kinetic modelling practices and challenges associated with gaining regulatory acceptance of model applications, Regul. Toxicol. Pharmacol. 90 (2017) 104–115. 10.1016/j.yrtph.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Sturgill MG, Lambert GH, Xenobiotic-induced hepatotoxicity: mechanisms of liver injury and methods of monitoring hepatic function, Clin. Chem. 43 (1997) 1512–1526. [PubMed] [Google Scholar]
- [116].Zimmerman HJ, Hepatotoxicity: The Adverse Effects of Drugs and Other Chemicals on the Liver, 2nd Edition, Lippincott Williams & Wilkins, Philadelphia, Pennsylvania, 1999. [Google Scholar]
- [117].Pognan F, Detection, Elimination, Mitigation, and Prediction of Drug-Induced Liver Injury in Drug Discovery, in: Chen M, Will Y. (Eds.), Drug-Induced Liver Toxicity, Humana, New York, NY, 2018: pp. 21–43. 10.1007/978-1-4939-7677-5_2. [DOI] [Google Scholar]
- [118].Gu X, Manautou JE, Molecular mechanisms underlying chemical liver injury, Expert Rev. Mol. Med. 14 (2012) e4. 10.1017/S1462399411002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Srivastava A, Maggs JL, Antoine DJ, Williams DP, Smith DA, Park BK, Role of Reactive Metabolites in Drug-Induced Hepatotoxicity, in: Uetrecht J. (Ed.), Adverse Drug Reactions, Springer, Berlin, Heidelberg, 2010: pp. 165–194. 10.1007/978-3-642-00663-0_7. [DOI] [PubMed] [Google Scholar]
- [120].Thompson RA, Isin EM, Ogese MO, Mettetal JT, Williams DP, Reactive Metabolites: Current and Emerging Risk and Hazard Assessments, Chem. Res. Toxicol. 29 (2016) 505–533. 10.1021/acs.chemrestox.5b00410. [DOI] [PubMed] [Google Scholar]
- [121].Chen M, Will Y, eds., Drug-Induced Liver Toxicity, First, Humana, New York, NY, 2018. 10.1007/978-1-4939-7677-5. [DOI] [Google Scholar]
- [122].Ghabril M, Chalasani N, Björnsson E, Drug-induced liver injury: a clinical update, Curr. Opin. Gastroenterol. 26 (2010) 222–226. 10.1097/MOG.0b013e3283383c7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Norman BH, Drug Induced Liver Injury (DILI). Mechanisms and medicinal chemistry avoidance/mitigation strategies, J. Med. Chem. (2020). 10.1021/acs.jmedchem.0c00524. [DOI] [PubMed] [Google Scholar]
- [124].Weaver RJ, Blomme EA, Chadwick AE, Copple IM, Gerets HHJ, Goldring CE, Guillouzo A, Hewitt PG, Ingelman-Sundberg M, Jensen KG, Juhila S, Klingmüller U, Labbe G, Liguori MJ, Lovatt CA, Morgan P, Naisbitt DJ, Pieters RHH, Snoeys J, van de Water B, Williams DP, Park BK, Managing the challenge of drug-induced liver injury: a roadmap for the development and deployment of preclinical predictive models, Nat. Rev. Drug Discovery. 19 (2020) 131–148. 10.1038/s41573-019-0048-x. [DOI] [PubMed] [Google Scholar]
- [125].Chen M, Vijay V, Shi Q, Liu Z, Fang H, Tong W, FDA-approved drug labeling for the study of drug-induced liver injury, Drug Discovery Today. 16 (2011) 697–703. 10.1016/j.drudis.2011.05.007. [DOI] [PubMed] [Google Scholar]
- [126].Kotsampasakou E, Montanari F, Ecker GF, Predicting drug-induced liver injury: The importance of data curation, Toxicology. 389 (2017) 139–145. 10.1016/j.tox.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Ortega-Alonso A, Stephens C, Lucena MI, Andrade RJ, Case characterization, clinical features and risk factors in drug-induced liver injury, Int. J. Mol. Sci. 17 (2016). 10.3390/ijms17050714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Mosedale M, Watkins PB, Drug-induced liver injury: Advances in mechanistic understanding that will inform risk management, Clin. Pharmacol. Ther. 101 (2017) 469–480. 10.1002/cpt.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Roth RA, Ganey PE, Intrinsic versus idiosyncratic drug-induced hepatotoxicity - two villains or one?, J. Pharmacol. Exp. Ther. 332 (2010) 692–697. 10.1124/jpet.109.162651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Prieto P, Graepel R, Gerloff K, Lamon L, Sachana M, Pistollato F, Gribaldo L, Bal-Price A, Worth A, Investigating cell type specific mechanisms contributing to acute oral toxicity, ALTEX. 36 (2019) 39–64. 10.14573/altex.1805181. [DOI] [PubMed] [Google Scholar]
- [131].Horner S, Ryan D, Robinson S, Callander R, Stamp K, Roberts RA, Target organ toxicities in studies conducted to support first time in man dosing: an analysis across species and therapy areas, Regul. Toxicol. Pharmacol. 65 (2013) 334–343. 10.1016/j.yrtph.2013.02.002. [DOI] [PubMed] [Google Scholar]
- [132].Sakuratani Y, Zhang HQ, Nishikawa S, Yamazaki K, Yamada T, Yamada J, Gerova K, Chankov G, Mekenyan O, Hayashi M, Hazard Evaluation Support System (HESS) for predicting repeated dose toxicity using toxicological categories, SAR QSAR Environ. Res. 24 (2013) 351–363. 10.1080/1062936X.2013.773375. [DOI] [PubMed] [Google Scholar]
- [133].Kenna JG, Persson M, Siler SQ, Yu K, Hu C, Chen M, Xu J, Tong W, Will Y, Aleo MD, Liver, in: Will Y, McDuffie JE, Olaharski AJ, Jeffy BD (Eds.), Drug Discovery Toxicology, John Wiley & Sons, Inc, Hoboken, NJ, 2016: pp. 93–129. 10.1002/9781119053248.ch8. [DOI] [Google Scholar]
- [134].David S, Hamilton JP, Drug-induced Liver Injury, US Gastroenterology &Hepatology Review. 6 (2010) 73–80. [PMC free article] [PubMed] [Google Scholar]
- [135].Wallace AD, Meyer SA, Hepatotoxicity, in: Hodgson E. (Ed.), A Textbook of Modern Toxicology, 4th ed, John Wiley & Sons, Hoboken, N.J, 2010: pp. 277–289. [Google Scholar]
- [136].Luedde T, Kaplowitz N, Schwabe RF, Cell death and cell death responses in liver disease: Mechanisms and clinical relevance, Gastroenterology. 147 (2014) 765–783.e4. 10.1053/j.gastro.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ, Mechanisms of hepatotoxicity, Toxicol. Sci. 65 (2002) 166–176. 10.1093/toxsci/65.2.166. [DOI] [PubMed] [Google Scholar]
- [138].Willett C, Rae JC, Goyak KO, Minsavage G, Westmoreland C, Andersen M, Avigan M, Duché D, Harris G, Hartung T, Jaeschke H, Kleensang A, Landesmann B, Martos S, Matevia M, Toole C, Rowan A, Schultz T, Seed J, Senior J, Shah I, Subramanian K, Vinken M, Watkins P, Building shared experience to advance practical application of pathway-based toxicology: Liver toxicity mode-of-action, ALTEX. 31 (2014) 500–519. 10.14573/altex.1401281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zimmerman HJ, Hepatotoxicity DM, Dis.-Mon. 39 (1993) 675–787. [PubMed] [Google Scholar]
- [140].Hoofnagle JH, Serrano J, Knoben JE, Navarro VJ, LiverTox: a website on drug-induced liver injury, Hepatology. 57 (2013) 873–874. 10.1002/hep.26175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].NIH, LiverTox: Clinical and Research Information on Drug-Induced Liver Injury, (2012). http://www.ncbi.nlm.nih.gov/books/NBK547852/ (accessed March 6, 2020).
- [142].Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, Rochon J, DILIN Study Group, Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct, Drug Saf. 32 (2009) 55–68. 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Hoofnagle JH, Drug-induced liver injury network (DILIN), Hepatology. 40 (2004) 773. 10.1002/hep.20445. [DOI] [PubMed] [Google Scholar]
- [144].Kleiner DE, Chalasani NP, Lee WM, Fontana RJ, Bonkovsky HL, Watkins PB, Hayashi PH, Davern TJ, Navarro V, Reddy R, Talwalkar JA, Stolz A, Gu J, Barnhart H, Hoofnagle JH, Drug-Induced Liver Injury Network (DILIN), Hepatic histological findings in suspected drug-induced liver injury: systematic evaluation and clinical associations, Hepatology. 59 (2014) 661–670. 10.1002/hep.26709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Amberg A, Kopanska K, Anger LT, Schaefer M, Spirkl H-P, Stolte M, Durchfeld-Meyer B, Myatt G, Czich A, In silico prediction of organ toxicity - Development of in silico models from in vivo drug histopathology data from regulatory toxicity study reports, The Toxicologist, Supplement to Toxicological Sciences. 174 (2020) Abstract #2050. https://www.toxicology.org/pubs/docs/Tox/2020Tox.pdf. [Google Scholar]
- [146].Bale SS, Vernetti L, Senutovitch N, Jindal R, Hegde M, Gough A, McCarty WJ, Bakan A, Bhushan A, Shun TY, Golberg I, DeBiasio R, Usta BO, Taylor DL, Yarmush ML, In vitro platforms for evaluating liver toxicity, Exp. Biol. Med. 239 (2014) 1180–1191. 10.1177/1535370214531872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Fraser K, Bruckner DM, Dordick JS, Advancing predictive hepatotoxicity at the intersection of experimental, in silico, and artificial intelligence technologies, Chem. Res. Toxicol. 31 (2018) 412–430. 10.1021/acs.chemrestox.8b00054. [DOI] [PubMed] [Google Scholar]
- [148].Pan G, Roles of Hepatic Drug Transporters in Drug Disposition and Liver Toxicity, in: Liu X, Pan G. (Eds.), Drug Transporters in Drug Disposition, Effects and Toxicity, Springer, Singapore, 2019: pp. 293–340. 10.1007/978-981-13-7647-4_6. [DOI] [PubMed] [Google Scholar]
- [149].Albillos A, de Gottardi A, Rescigno M, The gut-liver axis in liver disease: Pathophysiological basis for therapy, J. Hepatol. 72 (2020) 558–577. 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- [150].Jaeschke H, Naisbitt DJ, Immune Mechanisms in Drug-Induced Liver Injury, in: Chen M, Will Y. (Eds.), Drug-Induced Liver Toxicity, Humana, New York, NY, 2018: pp. 511–531. 10.1007/978-1-4939-7677-5_25. [DOI] [Google Scholar]
- [151].Mak A, Uetrecht J, Immune mechanisms of idiosyncratic drug-induced liver injury, J. Clin. Transl. Res. 3 (2017) 145–156. [PMC free article] [PubMed] [Google Scholar]
- [152].Uetrecht J, Mechanisms of idiosyncratic drug-induced liver injury, Adv. Pharmacol. (San Diego, CA, U. S.). 85 (2019) 133–163. 10.1016/bs.apha.2018.12.001. [DOI] [PubMed] [Google Scholar]
- [153].Ipsen DH, Lykkesfeldt J, Tveden-Nyborg P, Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease, Cell. Mol. Life Sci. 75 (2018) 3313–3327. 10.1007/s00018-018-2860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Russmann S, Kullak-Ublick GA, Grattagliano I, Current concepts of mechanisms in drug-induced hepatotoxicity, Curr. Med. Chem. 16 (2009) 3041–3053. 10.2174/092986709788803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Plaa G, Charbonneau M, Plante I, Detection and Evaluation of Chemically Induced Liver Injury, in: Hayes A, Kruger C. (Eds.), Hayes’ Principles and Methods of Toxicology, Sixth Edition, CRC Press, London, 2014: pp. 1445–1488. 10.1201/b17359-33. [DOI] [Google Scholar]
- [156].Morgan SJ, Elangbam CS, Animal Models of Disease for Future Toxicity Predictions, in: Will Y, McDuffie JE, Olaharski AJ, Jeffy BD (Eds.), Drug Discovery Toxicology, John Wiley & Sons, Inc, Hoboken, NJ, 2016: pp. 261–297. 10.1002/9781119053248.ch18. [DOI] [Google Scholar]
- [157].Tuschl G, Hrach J, Hewitt PG, Mueller SO, Application of Short- and Long-Term Hepatocyte Cultures to Predict Toxicities, in: Sahu SC (Ed.), Hepatotoxicity, John Wiley & Sons, Ltd, Chichester, UK, 2007: pp. 139–174. 10.1002/9780470516751.ch7. [DOI] [Google Scholar]
- [158].EMA, Reflection paper on non-clinical evaluation of drug-induced liver injury (DILI), 2010. https://www.ema.europa.eu/en/non-clinical-evaluation-drug-induced-liver-injury-dili.
- [159].Beger RD, Bhattacharyya S, Gill PS, James LP, Acylcarnitines as Translational Biomarkers of Mitochondrial Dysfunction, in: Will Y, Dykens JA (Eds.), Mitochondrial Dysfunction Caused by Drugs and Environmental Toxicants, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2018: pp. 383–393. 10.1002/9781119329725.ch24. [DOI] [Google Scholar]
- [160].Collins SD, Yuen G, Tu T, Budzinska MA, Spring K, Bryant K, Shackel NA, Chapter 3 - In Vitro Models of the Liver: Disease Modeling, Drug Discovery and Clinical Applications, in: Tirnitz-Parker JEE (Ed.), Hepatocellular Carcinoma [Internet], Codon Publications, Brisbane (AU), 2019. 10.15586/hepatocellularcarcinoma.2019.ch3. [DOI] [PubMed] [Google Scholar]
- [161].Mittal R, Woo FW, Castro CS, Cohen MA, Karanxha J, Mittal J, Chhibber T, Jhaveri VM, Organ-on-chip models: implications in drug discovery and clinical applications, J. Cell. Physiol. 234 (2019) 8352–8380. 10.1002/jcp.27729. [DOI] [PubMed] [Google Scholar]
- [162].Ramaiahgari SC, Waidyanatha S, Dixon D, DeVito MJ, Paules RS, Ferguson SS, Three-dimensional (3D) HepaRG spheroid model with physiologically relevant xenobiotic metabolism competence and hepatocyte functionality for liver toxicity screening, Toxicol. Sci. 160 (2017) 189–190. 10.1093/toxsci/kfx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Ramaiahgari SC, Ferguson SS, Organotypic 3D HepaRG Liver Model for Assessment of Drug-Induced Cholestasis, in: Vinken M. (Ed.), Experimental Cholestasis Research, Humana, New York, NY, 2019: pp. 313–323. 10.1007/978-1-4939-9420-5_20. [DOI] [PubMed] [Google Scholar]
- [164].Starokozhko V, Groothuis GMM, Judging the value of ‘liver-on-a-chip’ devices for prediction of toxicity, Expert Opin. Drug Metab. Toxicol. 13 (2017) 125–128. 10.1080/17425255.2017.1246537. [DOI] [PubMed] [Google Scholar]
- [165].Usta OB, McCarty WJ, Bale S, Hegde M, Jindal R, Bhushan A, Golberg I, Yarmush ML, Microengineered cell and tissue systems for drug screening and toxicology applications: evolution of in-vitro liver technologies, Technology. 3 (2015) 1–26. 10.1142/S2339547815300012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Amberg A, Anger LT, Stolte M, Hemmerick J, Matter H, Fisk L, Tluczkiewicz I, Pinto-Gil K, López-Massaguer O, Pastor M, In silico prediction of DILI: Extraction of histopathology data from preclinical toxicity studies of the eTOX database for new in silico models of hepatotoxicity, The Toxicologist, Supplement to Toxicological Sciences. 150 (2018) Abstract #2118. https://www.toxicology.org/pubs/docs/Tox/2018Tox.pdf. [Google Scholar]
- [167].Firman JW, Pestana CB, Rathman JF, Vinken M, Yang C, Cronin MTD, A robust, mechanistically based in silico structural profiler for hepatic cholestasis, Chem. Res. Toxicol. 34 (2021) 641–655. 10.1021/acs.chemrestox.0c00465. [DOI] [PubMed] [Google Scholar]
- [168].Fourches D, Barnes JC, Day NC, Bradley P, Reed JZ, Tropsha A, Cheminformatics analysis of assertions mined from literature that describe drug-induced liver injury in different species, Chem. Res. Toxicol. 23 (2010) 171–183. 10.1021/tx900326k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Greene N, Fisk L, Naven RT, Note RR, Patel ML, Pelletier DJ, Developing structure-activity relationships for the prediction of hepatotoxicity, Chem. Res. Toxicol. 23 (2010) 1215–1222. 10.1021/tx1000865. [DOI] [PubMed] [Google Scholar]
- [170].Jain S, Norinder U, Escher SE, Zdrazil B, Combining in vivo data with in silico predictions for modeling hepatic steatosis by using stratified bagging and conformal prediction, Chem. Res. Toxicol. 34 (2021) 656–668. 10.1021/acs.chemrestox.0c00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Liu Z, Shi Q, Ding D, Kelly R, Fang H, Tong W, Translating clinical findings into knowledge in drug safety evaluation - drug induced liver injury prediction system (DILIps), PLoS Comput. Biol. 7 (2011) e1002310. 10.1371/journal.pcbi.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Matthews EJ, Kruhlak NL, Benz RD, Aragonés Sabaté D, Marchant CA, Contrera JF, Identification of structure-activity relationships for adverse effects of pharmaceuticals in humans: Part C: use of QSAR and an expert system for the estimation of the mechanism of action of drug-induced hepatobiliary and urinary tract toxicities, Regul. Toxicol. Pharmacol. 54 (2009) 43–65. 10.1016/j.yrtph.2009.01.007. [DOI] [PubMed] [Google Scholar]
- [173].Matthews EJ, Ursem CJ, Kruhlak NL, Benz RD, Sabaté DA, Yang C, Klopman G, Contrera JF, Identification of structure-activity relationships for adverse effects of pharmaceuticals in humans: Part B. Use of (Q)SAR systems for early detection of drug-induced hepatobiliary and urinary tract toxicities, Regul. Toxicol. Pharmacol. 54 (2009) 23–42. 10.1016/j.yrtph.2009.01.009. [DOI] [PubMed] [Google Scholar]
- [174].Minerali E, Foil DH, Zorn KM, Lane TR, Ekins S, Comparing machine learning algorithms for predicting Drug-Induced Liver Injury (DILI), Mol. Pharmaceutics. 17 (2020) 2628–2637. 10.1021/acs.molpharmaceut.0c00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Rathman J, Yang C, Ribeiro JV, Mostrag A, Thakkar S, Tong W, Hobocienski B, Sacher O, Magdziarz T, Bienfait B, Development of a battery of in silico prediction tools for drug-induced liver injury from the vantage point of translational safety assessment, Chem. Res. Toxicol. 34 (2021) 601–615. 10.1021/acs.chemrestox.0c00423. [DOI] [PubMed] [Google Scholar]
- [176].Ursem CJ, Kruhlak NL, Contrera JF, MacLaughlin PM, Benz RD, Matthews EJ, Identification of structure-activity relationships for adverse effects of pharmaceuticals in humans. Part A: use of FDA post-market reports to create a database of hepatobiliary and urinary tract toxicities, Regul. Toxicol. Pharmacol. 54 (2009) 1–22. 10.1016/j.yrtph.2008.12.009. [DOI] [PubMed] [Google Scholar]
- [177].Asilar E, Hemmerich J, Ecker GF, Image based liver toxicity prediction, J. Chem. Inf. Model. 60 (2020) 1111–1121. 10.1021/acs.jcim.9b00713. [DOI] [PubMed] [Google Scholar]
- [178].Li T, Tong W, Roberts R, Liu Z, Thakkar S, DeepDILI: deep learning-powered drug-induced liver injury prediction using model-level representation, Chem. Res. Toxicol. 34 (2021) 550–565. 10.1021/acs.chemrestox.0c00374. [DOI] [PubMed] [Google Scholar]
- [179].Ma H, An W, Wang Y, Sun H, Huang R, Huang J, Deep graph learning with property augmentation for predicting drug-induced liver injury, Chem. Res. Toxicol. 34 (2021) 495–506. 10.1021/acs.chemrestox.0c00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Liu J, Patlewicz G, Williams AJ, Thomas RS, Shah I, Predicting Organ Toxicity Using in Vitro Bioactivity Data and Chemical Structure, Chem. Res. Toxicol. 30 (2017) 2046–2059. 10.1021/acs.chemrestox.7b00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Liu J, Mansouri K, Judson RS, Martin MT, Hong H, Chen M, Xu X, Thomas RS, Shah I, Predicting hepatotoxicity using ToxCast in vitro bioactivity and chemical structure, Chem. Res. Toxicol. 28 (2015) 738–751. 10.1021/tx500501h. [DOI] [PubMed] [Google Scholar]
- [182].Mahmoud SY, Svensson F, Zoufir A, Módos D, Afzal AM, Bender A, Understanding conditional associations between ToxCast in vitro readouts and the hepatotoxicity of compounds using rule-based methods, Chem. Res. Toxicol. 33 (2020) 137–153. 10.1021/acs.chemrestox.8b00382. [DOI] [PubMed] [Google Scholar]
- [183].Wu L, Liu Z, Auerbach S, Huang R, Chen M, McEuen K, Xu J, Fang H, Tong W, Integrating drug’s mode of action into quantitative structure-activity relationships for improved prediction of druginduced liver injury, J. Chem. Inf. Model. 57 (2017) 1000–1006. 10.1021/acs.jcim.6b00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Xu T, Ngan DK, Ye L, Xia M, Xie HQ, Zhao B, Simeonov A, Huang R, Predictive Models for Human Organ Toxicity Based on In Vitro Bioactivity Data and Chemical Structure, Chem. Res. Toxicol. (2020) acs.chemrestox.9b00305. 10.1021/acs.chemrestox.9b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Brigo A, Muster W, 2.09 - Computational Models to Predict Toxicological Endpoints in Drug Discovery and Strategies for Data Integration, in: Chackalamannil S, Rotella D, Ward SE (Eds.), Comprehensive Medicinal Chemistry III, Elsevier, 2017: pp. 233–258. 10.1016/B978-0-12-409547-2.12321-2. [DOI] [Google Scholar]
- [186].Chen M, Bisgin H, Tong L, Hong H, Fang H, Borlak J, Tong W, Toward predictive models for druginduced liver injury in humans: are we there yet?, Biomarkers Med. 8 (2014) 201–213. 10.2217/bmm.13.146. [DOI] [PubMed] [Google Scholar]
- [187].Ekins S, Progress in computational toxicology, J. Pharmacol. Toxicol. Methods. 69 (2014) 115–140. 10.1016/j.vascn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- [188].Hewitt M, Przybylak K, In Silico Models for Hepatotoxicity, in: Benfenati E. (Ed.), In Silico Methods for Predicting Drug Toxicity, Humana Press, New York, NY, 2016: pp. 201–236. 10.1007/978-1-4939-3609-0_11. [DOI] [PubMed] [Google Scholar]
- [189].Hong H, Zhu J, Chen M, Gong P, Zhang C, Tong W, Quantitative Structure–Activity Relationship Models for Predicting Risk of Drug-Induced Liver Injury in Humans, in: Chen M, Will Y. (Eds.), Drug-Induced Liver Toxicity, Humana, New York, NY, 2018: pp. 77–100. 10.1007/978-1-4939-7677-5_5. [DOI] [Google Scholar]
- [190].Przybylak KR, Cronin MTD, In silico models for drug-induced liver injury - current status, Expert Opin. Drug Metab. Toxicol. 8 (2012) 201–217. 10.1517/17425255.2012.648613. [DOI] [PubMed] [Google Scholar]
- [191].Enoch S, Mellor C, Nelms M, Structure-activity modeling of mitochondrial dysfunction, in: Will Y, Dykens JA (Eds.), Mitochondrial Dysfunction Caused by Drugs and Environmental Toxicants, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2018: pp. 25–34. 10.1002/9781119329725.ch3. [DOI] [Google Scholar]
- [192].Zhu X, Kruhlak NL, Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data, Toxicology. 321 (2014) 62–72. 10.1016/j.tox.2014.03.009. [DOI] [PubMed] [Google Scholar]
- [193].Thakkar S, Li T, Liu Z, Wu L, Roberts R, Tong W, Drug-induced liver injury severity and toxicity (DILIst): binary classification of 1279 drugs by human hepatotoxicity, Drug Discovery Today. 25 (2020) 201–208. 10.1016/j.drudis.2019.09.022. [DOI] [PubMed] [Google Scholar]
- [194].Chen M, Borlak J, Tong W, A Model to predict severity of drug-induced liver injury in humans, Hepatology. 64 (2016) 931–940. 10.1002/hep.28678. [DOI] [PubMed] [Google Scholar]
- [195].Hong H, Thakkar S, Chen M, Tong W, Development of decision forest models for prediction of drug-induced liver injury in humans using a large set of FDA-approved drugs, Sci. Rep. 7 (2017) 17311. 10.1038/s41598-017-17701-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Mulliner D, Schmidt F, Stolte M, Spirkl H-P, Czich A, Amberg A, Computational models for human and animal hepatotoxicity with a global application scope, Chem. Res. Toxicol. 29 (2016) 757–767. 10.1021/acs.chemrestox.5b00465. [DOI] [PubMed] [Google Scholar]
- [197].Zhao L, Russo DP, Wang W, Aleksunes LM, Zhu H, Mechanism-driven read-across of chemical hepatotoxicants based on chemical structures and biological data, Toxicol. Sci. 174 (2020) 178–188. 10.1093/toxsci/kfaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Béquignon OJM, Pawar G, van de Water B, Cronin MTD, van Westen GJP, Computational Approaches for Drug-Induced Liver Injury (DILI) Prediction: State of the Art and Challenges, in: Reference Module in Biomedical Sciences, Elsevier, 2019: p. B9780128012383115000. 10.1016/B978-0-12-801238-3.11535-1. [DOI] [Google Scholar]
- [199].ICH, M7 (R1) Assessment and control of DNA reactive (mutagenic) impurities in pharmaceuticals to limit potential carcinogenic risk, European Medicines Agency, 2017. https://database.ich.org/sites/default/files/M7_R1_Guideline.pdf. [Google Scholar]
- [200].Sistare FD, Mattes WB, LeCluyse EL, The promise of new technologies to reduce, refine, or replace animal use while reducing risks of drug induced liver injury in pharmaceutical development, ILAR J. 57 (2016) 186–211. 10.1093/ilar/ilw025. [DOI] [PubMed] [Google Scholar]
- [201].Shehu AI, Ma X, Venkataramanan R, Mechanisms of drug-induced hepatotoxicity, Clinics in Liver Disease. 21 (2017) 35–54. 10.1016/j.cld.2016.08.002. [DOI] [PubMed] [Google Scholar]
- [202].Gijbels E, Vinken M, An update on adverse outcome pathways leading to liver injury, Appl. In Vitro Toxicol. 3 (2017) 283–285. 10.1089/aivt.2017.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Kim MT, Huang R, Sedykh A, Wang W, Xia M, Zhu H, Mechanism profiling of hepatotoxicity caused by oxidative stress using antioxidant response element reporter gene assay models and big data, Environ. Health Perspect. 124 (2016) 634–641. 10.1289/ehp.1509763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Vinken M, Adverse Outcome Pathways as Tools to Assess Drug-Induced Toxicity, in: Benfenati E. (Ed.), In Silico Methods for Predicting Drug Toxicity, Humana Press, New York, NY, 2016: pp. 325–337. 10.1007/978-1-4939-3609-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.