Abstract
Embryonal rhabdomyosarcoma (ERMS) is the most common subtype of rhabdomyosarcoma (RMS). Amongst RMS subtypes, ERMS is associated with a favorable outcome with an overall survival of 70% at 5 years for localized disease. The molecular profile of ERMS is heterogeneous, including mostly point mutations in various genes. Therapeutic strategies have remained relatively consistent irrespective of the molecular abnormalities. In this study, we focus on a homogeneous RAS/RAF mutated ERMS subset and correlate with clinicopathologic findings. Twenty-six cases (16 males and 10 females) were identified from screening 98 ERMS, either by targeted DNA sequencing (MSK-IMPACT) or by Sanger sequencing. Fourteen (54%) cases had NRAS mutations, 6 (23%) had KRAS mutations, 5 (19%) had HRAS mutations and 1 case (4%) had BRAF mutation. Median age at diagnosis was 8 years (range 1-70) with two-thirds occurring in the children. Tumor sites varied with H&N and GU sites accounting for 62% of cases. RAS isoform hot spot mutations predominated: NRAS p.Q61K (57%), KRAS p.G12D (67%) and HRAS (codons 12, 14 and 61). Additional genetic abnormalities were identified in 85% of the RAS-mutated cases. At last follow-up, 29% patients died of disease and 23% were alive with disease. The 3-year and 5-year survival rates were 75% and 61% respectively. In conclusion, RAS mutations occur in 27% of ERMS, with NRAS mutations encompassing half of the cases. Overall RAS-mutant RMS do not correlate with age or site, but most tumors show an undifferentiated and spindle cell morphology.
Keywords: NRAS, KRAS, HRAS, Embryonal Rhabdomyosarcoma
INTRODUCTION
Rhabdomyosarcomas (RMS) are the most common soft tissue sarcomas in children, accounting for 5-8% of all pediatric malignancies, with a small subset occurring in adults as well. There are now 4 subtypes of RMS recognized in the latest WHO classification of soft tissue tumors,1 mainly defined based on their genetic profiles, including fusion-negative ERMS, PAX3/7-FOXO1 fusion-positive alveolar RMS (ARMS), spindle and sclerosing RMS with either MYOD1 mutations or various gene fusions, and pleomorphic RMS. While ARMS and MYOD1-mutant spindle and sclerosing RMS follow a highly aggressive clinical course, the prognosis of ERMS has significantly improved in recent years with an overall survival of 70% at 5 years for patients presenting with localized disease. Based on large genomic studies,2-4 the molecular landscape of ERMS has emerged as driven by a wide range of causative mutations, with most frequent single nucleotide point mutations detected within one of the Ras genes (NRAS > KRAS > HRAS), a receptor tyrosine kinase (FGFR4 ≫ ERBB2), or the catalytic component (PIK3CA) of the phosphoinositide-3 kinase (PI3K) complex.3 As the most common molecular subset of ERMS appears to be driven by an oncogenic mutation in one of the Ras isoforms, the aim of this study is to investigate the potential clinical and pathologic correlates associated with this genotype, as well as its pattern of additional co-existing mutations and impact on survival and response to standard therapy protocols.
MATERIALS AND METHODS
Patient Selection
Archival and consultation material from adult and pediatric patients with a diagnosis of ERMS where additional testing identified RAS / RAF mutations was retrieved from the pathology files at Memorial Sloan Kettering Cancer Center. Overall, 93 cases of ERMS were identified in which next-generation targeted DNA sequencing (MSK-IMPACT) was performed. In order to increase the study group, archival material from an additional cohort of 5 ERMS, which preceded the clinical MSK-IMPACT testing (2010-2014), was studied by targeted DNA PCR and Sanger sequencing for hot spot mutations in the 3 RAS genes. Overall, 26 ERMS were identified harboring RAS mutations, with 22 cases being detected by MSK-IMPACT and 4 by Sanger sequencing. The pathologic features of these cases were re-reviewed and analyzed for cell morphology, presence of rhabdomyoblasts, pleomorphism or anaplasia, mitotic activity, and necrosis. The diagnosis of RMS was confirmed by positivity for desmin (Ventana, DE-R-11, RTU) and myogenin (Ventana, F5D, RTU) in all cases. Clinical Risk Group was assessed based on current guidelines5, 6 and follow-up data was collected by review of clinical records. The study was approved by the Institutional Review Board at MSKCC (IRB# 02-060).
MSK-IMPACT Assay
Details of the MSK-IMPACT assay have been previously published. 7 Briefly, MSK-IMPACT is a comprehensive molecular profiling assay that involves hybridization capture and deep sequencing of all exons and selected introns of up to 468 oncogenes and tumor-suppressor genes, allowing the detection of point mutations, small and large insertions or deletions, and rearrangements. In addition to capturing all coding regions of the genes, the assay also captures >1000 intergenic and intronic single-nucleotide polymorphisms (tiling probes), interspersed homogenously across the genome, aiding the accurate assessment of genome-wide copy number. The assay uses paired tumor / normal DNA for sequencing and analysis. In total, the probes target approximately 1.2 megabases of the human genome.
DNA Extraction and Polymerase Chain Reaction (PCR)
Genomic DNA was extracted from either frozen or formalin-fixed paraffin-embedded tissue, using the phenol/chloroform method or the QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA), respectively. The targeted exon regions of candidate genes (NRAS, KRAS, HRAS) were amplified by the PCR using corresponding forward and reverse primers (Supplementary Table 2). PCR was conducted using the Clontech Advantage 2 PCR Enzyme System kit (Clontech, Mountain View, CA). The PCR products were purified using QIAquick PCR Purification Kit (Qiagen) and confirmed by Sanger sequencing. All mutations were verified bidirectionally.
Statistical Analysis
Statistical analysis was conducted on the SPSS software platform (version 20; IBM, New York, NY). Associations between RAS mutation and clinicopathological factors were assessed by Pearson χ2, Fisher exact or Kruskal-Wallis tests according to whether the variables were categorical or continuous. The overall survival was measured from the date of surgery and examined by Kaplan-Meier analysis and the log-rank test. Two-sided P values < 0.05 were considered significant in all statistical analyses.
RESULTS
Clinicopathologic Features
The clinicopathologic features are summarized in Table 1. Overall, 26 cases (27%) were identified with RAS/RAF mutations. These included 14 cases (54%) with NRAS mutation, 6 cases (23%) with KRAS mutation, 5 cases (19%) with HRAS mutation and 1 case (4%) with BRAF mutation (Supplementary Figure 1). The patients age at diagnosis ranged widely, from 1 to 70 years-old (median: 8 years). Two-thirds of tumors occurred in the pediatric age group, 15 patients (58%), while the remaining 11 (42%) occurred in adults. Sixteen (64%) of the patients were males and 10 (38%) were females. The most common clinical presentation was the intra-abdominal and genitourinary site, occurring in 15 (58%) patients, including 5 involving the pelvis/intra-abdominal, 6 the vagina/uterus/cervix, and 4 occurring paratesticular. The second most common location was the head and neck, occurring in 6 (23%) patients. Five patients occurred in the lower extremity (4) and axilla (1). Among the 15 pediatric tumors, 9 (60%) occurred in the intra-abdominal / genitourinary sites, 4 (27%) tumors occurred in the head and neck locations and 2 (13%) occurred in the soft tissue. Tumors ranged in size from 1.0 to 33.0 cm in greatest dimension (mean 6.3 cm). Twenty-two (85%) of the 26 patients presented with localized disease at the time of presentation.
Table 1:
Clinico-pathological features of RAS / RAF mutated RMS
| Case # | Age (yrs) |
Sex | Site | RAS / RAF mutation | Risk Group |
FU duration (months) |
FU status | Treatment |
|---|---|---|---|---|---|---|---|---|
| 1¥ | 1 | F | buttock | NRAS exon3 p.Q61K (c.181 C>A) | High | 15 | DOD | S+C |
| 2 | 2 | M | Para-testicular | NRAS exon3 p.Q61K (c.181C>A) | Low | 48 | NED | S+C |
| 3¤ | 2 | M | soft palate | NRAS exon3 p.Q61K (c.181C>A) | Low | 15 | AWD | S+C |
| 4§ | 2 | F | thigh | NRAS exon3 p.Q61K (c.181C>A) | NA | 19 | NED | C+R+S |
| 5¤ | 3 | M | pelvis | NRAS exon3 p.Q61H (c.183A>T) | Int | 16 | NED | C+R+S |
| 6 | 3 | F | vagina | NRAS exon3 p.Q61K (c.181C>A) | Low | 17 | NED | C+R+S |
| 7 | 5 | M | jaw | NRAS exon2 p.G13V (c.38G>T) | Int | 22 | NED | S+C |
| 8 | 7 | M | Para-testicular | NRAS exon3 p.Q61K (c.181C>A) | Low | 36 | NED | S+C |
| 9¤ | 9 | M | infratemporal fossa | NRAS exon3 p.Q61L (c.182A>T) | Int | 55 | NED | C+R+S |
| 10 | 10 | M | abdomen | NRAS exon3 p.Q61R (c.182A>G) | Int | 3 | DOD | C+S |
| 11§ | 19 | F | vagina | NRAS exon3 p.Q61K (c.181C>A) | Int | 34 | DOD | C+R+S |
| 12 | 34 | F | cervix | NRAS exon3 p.Q61R (c.182A>G) | Int | 2 | AWD | C |
| 13¤ | 62 | M | calf | NRAS exon3 p.Q61K (c.181C>A) | Int | 60 | NED | C+R+S |
| 14 | 70 | M | axilla | NRAS exon3 p.Q61L (c.182A>T) | Int | 35 | NED | C+R+S |
| 15 | 1 | M | pelvis | KRAS exon 2 p.G12D (c.35G>A) | High | 12 | DOD | C+R+S |
| 16¤ | 1 | F | vagina | KRAS exon2 p.G12D (c.35G>A) | Low | 28 | AWD | C+R+S |
| 17 | 2 | M | pelvis | KRAS exon 2 p.G12D (c.35G>A) | Int | 31 | NED | C+R+S |
| 18¥ | 23 | M | Para-testicular | KRAS exon4 p.K117T (c.350A>C) | High | 52 | NED | C+R+S |
| 19 | 44 | F | uterus | KRAS exon2 p.G12A (c.35G>C) | Low | 19 | NED | S+C |
| 20¥ | 62 | F | endometrium | KRAS exon2 p.G12D (c.35G>A) | NA | 4 | DOD | S+C |
| 21¤ | 3 | F | orbit | HRAS exon3 p.Q61K (c.181C>A) | Low | 39 | NED | C+R+S |
| 22¥ | 20 | M | Para-testicular | HRAS exon2 p.G12V (c.35G>T) | Low | 54 | DOD | C+R+S |
| 23 | 22 | F | buccal | HRAS exon 3 p.Q61L | Low | 124 | NED | C+R+S |
| 24¤ | 32 | M | pterygoid fossa | HRAS exon 2 p.G13R | Int | 64 | DOD | C+R+S |
| 25 | 50 | M | thigh | HRAS exon3 p.T50M (c.149C>T) | Int | 5 | AWD | S+C |
| 26¥ | 4 | M | abdomen | BRAF exon15 p.V600E (c.1799T>A) | High | 8 | AWD | C |
F-female, M-male, int-intermediate, NA-not available, DOD-died of disease, AWD-alive with disease, NED-no evidence of disease, C-chemotherapy, S-surgery, R-radiation therapy
Distant recurrence
Local recurrence
Local and distant recurrence
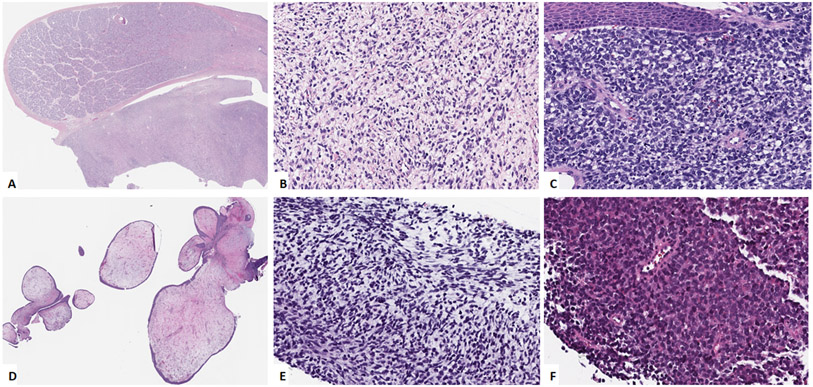
Morphologically, all 26 tumors were classified as ERMS. Ten (10, 40%) cases showed primitive undifferentiated round to ovoid cells in compact sheets (Figure 1A-1C). In focal areas the primitive cells were in a loose, partially myxoid, stroma (Figure 1B). Nine (9, 35%) showed admixture of undifferentiated and spindle cell areas with the spindle cell areas showing cellular spindle cells arranges in fascicles. One case (case # 25) showed a pure spindle cell morphology and one case (case #6) showed a distinctive botryoid morphology (Figure 1D). Rhabdomyoblastic differentiation with strap cells was not a common feature and was noted only in 2 cases. Mitoses ranged from 2 to 25 (mean – 11) per 10 high power fields. Seven cases (27%) showed areas of necrosis.
Figure 1: Morphologic spectrum of NRAS-mutated RMS.
A, B. (case 2, 2/M) Paratesticular tumor (A) showing primitive appearing cells in a loose stroma (B). C. Soft palate lesion (case 3, 2/M) composed of undifferentiated cells with round to irregular nuclei arranged in sheets, involving submucosa. D. Vaginal tumor (case 5, 3/F) showing botryoid morphology with polypoid nodulear growth. E. Jaw lesion (case 6, 5/M) showing primitive ovoid to spindle cells in a myxoid background. F. Calf tumor (case 11, 62/M) showing undifferentiated small cells arranged in sheets.
Immunohistochemically, all except one case showed diffuse cytoplasmic positivity for desmin. Myogenin showed focal nuclear positivity in all cases except one. MyoD1 staining was performed on 8 cases and showed focal nuclear positivity in all of the cases.
In all cases, FISH studies performed at the initial diagnosis did not show evidence of FOXO1 gene fusions.
Clinical risk-group data was available on 24 of 26 cases in the study. Nine patients were classified in the low-risk group, 11 in the intermediate-risk group, while 4 in the high-risk group. (Table 1)
NRAS mutations in RMS predominantly occur in males with codon 61 being the most common site of mutation.
Ten of 14 (71%) NRAS-mutated tumors occurred in children less than 18 years of age. The remaining 4 cases occurring in patients aged 19, 34, 62 and 70 years. Nine cases occurred in males and 5 in females. NRAS Q61K was the most common mutation identified in 8 cases, NRAS Q61L and NRAS Q61R mutations were identified in 2 cases each, and the other 2 cases showed NRAS Q61H and NRAS G13V mutations. The tumors were located mainly in the abdomino-pelvic / genitourinary sites (7 cases), with 4 cases occurring in the soft tissues and 3 cases in the head and neck. More than half the cases showed morphology of primitive undifferentiated cells with round to ovoid nuclei in sheets. In focal areas, the tumor cells were arranged in a partially myxoid background (Figure 1A-1C, 1F). Three cases showed a combination of undifferentiated cells with areas of spindle cells (Figure 1E). One case showed a botryoid morphology (Figure 1D).
KRAS G12D is the most common KRAS mutation in RMS
KRAS mutations in ERMS were identified in 3 children and 3 adults, with equal gender distribution. KRAS G12D hot spot mutation occurred in 4 of the 6 cases. The remaining 2 cases showed KRAS G12A and KRAS K117T mutations. All tumors were located in the genitourinary sites including pelvis, paratesticular, vaginal and uterus. Three of the tumors show hybrid morphology with undifferentiated round cell areas and areas with spindle cell morphology. The other three tumors show sheets of primitive undifferentiated cells (Figure 2A, 2B).
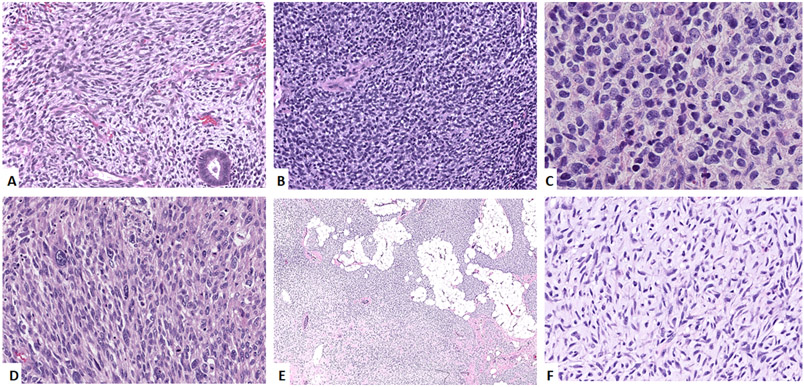
Figure 2: Morphologic spectrum of KRAS- (A-C), HRAS- (D) and BRAF-mutated (E, F) ERMS.
A. Uterine tumor (case 19, 44/F) showing primitive-appearing spindle cells involving the endometrium. B. Vaginal tumor (case 16, 1/F) showing undifferentiated cells with round nuclei in sheets. C. Paratesticular mass (case 22, 20/M) showing sheets of undifferentiated round cells. D. Thigh lesion (case 25, 50/M) showing a sarcomatoid morphology with spindle to pleomorphic cells in a fascicular pattern. E, F Abdominal tumor (case 26, 4/M) showing a cellular tumor infiltrating adipose tissue. Higher power image (F) showing primitive spindle cells in a collagenous to myxoid stroma.
ERMS with HRAS mutations have heterogeneous clinical and pathologic features.
The 5 cases with HRAS mutations occurred in 3 males and 2 females, affecting 1 child and 4 adults. The mutations that were identified were HRAS G12V, HRAS G13R, HRAS Q61K, HRAS Q61L and HRAS T50M. Three tumors were located in the head and neck location with the other two located in paratesticular and thigh. Two tumors showed morphology of undifferentiated round cells, one of which had focal rhabdomyoblastic differentiation. Two other tumors showed hybrid morphology of primitive undifferentiated cells and areas with spindle cell features. One of the tumors show a pure spindle cell morphology with spindle cells arranged in fascicles. Scattered cells in this case showed large pleomorphic nuclei (Figure 2C, 2D).
One case showed a BRAF V600E mutation and occurred in the abdomen of a 4-year-old boy and showed primitive undifferentiated cells in a loose myxoid stroma (Figure 2E, 2F).
Additional co-existing genetic abnormalities identified.
Next-generation sequencing identified additional genetic alterations in 17 of the 20 (85%) cases studied. These are listed in Supplementary Table 1. The recurrent additional genetic alterations included 4 cases each with TP53, NF1, or BCOR mutations, 2 cases each with MYC amplification, RAD21 amplification, AKT3, PIK3CA, PTEN and FGFR4 mutations. Other non-recurrent genetic alterations identified are listed in Supplementary Table 1.
Follow-up
Clinical follow-up was available on all patients (Table 1) and ranged from 2 to 124 months (mean – 29.5 months). Fifteen patients were managed with a combination of chemotherapy, radiation therapy and surgery, 9 received surgery and chemotherapy and 2 recent cases are being treated with chemotherapy. Seven (7) patients had local recurrence, 5 patients had distant recurrence and 2 patients had local and distant recurrences. At last follow up, 13 patients (52%) had no evidence of disease (NED), 6 patients (23%) were alive with disease (AWD) and 7 patients (28%) had died of disease (DOD) at 2 to 64 months following diagnosis. The 3-year and 5-year survival rates for the entire cohort were 75% and 61% respectively, while for patients presenting with localized disease, the 3-year and 5-year survival rates were 81% and 65% respectively. No significant differences in survival were noted among patients with different RAS mutations. (Supplementary Figure 2)
Among the NRAS mutated group, 3 of the patients died of disease at 3, 15 and 34 months following diagnosis; 3 were alive with disease and 8 had no evidence of disease. Among the KRAS mutated group, 2 patients died of disease at 4 and 12 months following diagnosis, 1 was alive with disease at 28 months and 3 had no evidence of disease. Among the HRAS mutated group, 2 patients died of disease at 54 and 64 months following diagnosis, 1 was alive with disease at 5 months and 2 had no evidence of disease. The BRAF mutated patient was alive with disease at 8 months following diagnosis.
DISCUSSION
RAS-ERK (RAS-RAF-MEK-ERK) pathway is a signal transduction cascade affecting cell proliferation and survival. 8, 9 Various growth factors bind to their receptors and activate RAS which, in turn, activates RAF and results in MEK phosphorylation. Phosphorylated MEK then phosphorylates ERK which activates various cell cycle-related proteins. This pathway has been implicated in the oncogenesis of various hematological and solid tumors. Activating RAS mutations occur in approximately 30% of human cancers. 10, 11 The RAS family, including HRAS, KRAS, and NRAS, belongs to the small G protein superfamily and involves the Ras/MAPK (mitogen-activated protein kinase) pathway, which transduces the extracellular input to the intracellular compartment through the GDP/GTP-regulated switches. The cancer-related somatic mutations in RAS members often occur at amino acids 12, 13, or 61, which are the conserved sites for modulating GDP/GTP binding and hydrolysis. There is increasing evidence that Ras proteins have isoform-specfic biological functions and tumorigenic effects, possibly attributed to the C-terminal hypervariable region, and that different mutation codons might induce different transformation potential.11 Different tumor types often display specificity in RAS gene involvement. For example, HRAS mutations are prevalent in bladder and kidney carcinoma, while KRAS mutations predominate in colorectal, pancreatic, endometrial, lung, and cervical cancers, and NRAS lesions are more commonly observed in melanoma and liver carcinoma as well as in lymphoid and myeloid malignancies.12
The RAS/MEK/ERK pathway also plays a major role in RMS 13 as it can trigger uncontrollable proliferation of cancer cells and prolong their survival time. 14-16 Mutations in RAS in zebrafish have been shown to lead to development of ERMS. 17, 18 Previous studies have shown that RAS gene mutations can be identified in 5-30% of all fusion negative RMS patients. 2, 12, 19-22 Shern et al. observed some age-specific correlations in RAS mutated RMS patients with HRAS mutations occurring in infants, KRAS mutations in toddlers and NRAS mutations in adolescents.4
In our study, RAS / RAF mutations were seen in 27% of 98 ERMS studied. Similar to prior studies 2-4, 12, 22, NRAS gene mutations were the most prevalent, accounting for half of the cases. KRAS and HRAS gene mutations occurred at a similar rate, comprising a quarter of RAS mutant ERMS. BRAF mutation was identified in only one case, indicating that RAS gene mutations far outnumber the RAF gene mutations in RMS. Mutations in all of the genes were missense mutations and occurred predominantly in the well-known hotspots described in various cancers. Codon 61 was the most common site of NRAS mutations, seen in 13 of the 14 NRAS-mutated tumors. One tumor showed a codon 13 mutation, another hotspot for NRAS gene. All except one of the KRAS mutations were seen in codon 12. Only one case showed codon 117 mutation, a rare site of KRAS mutations. Similarly, all except one HRAS mutations were identified in hot spot regions of codons’ 61, 12 and 13. One case showed instead an infrequent site mutation in codon 50 mutation. The BRAF mutated RMS occurred in the hot spot codon 600. The significance of the rare non-hotspot RAS mutations such as KRAS p.K117T and HRAS p. T50M, with respect to being ‘driver’ or ‘passenger’ mutations, remains uncertain.
Most of the RAS / RAF mutated tumors in our study analyzed by next-generation sequencing platform showed additional genetic alterations. TP53 gene alterations were the most common additional genetic alterations seen in 4 cases. Interestingly, a number of co-mutated genes such as NF1, FGFR4, PIK3CA, AKT3, etc, are part of the RAS / RAF / AKT pathway, which raises an important question as to the significance of these different mutations in the pathogenesis of ERMS. These results are in keeping with those reported by other groups.2-4 Three cases also showed evidence of coexistent loss of function mutations in BCOR gene, which has been previously described. 3
HRAS mutations have also been reported in malignant ectomesenchymoma (MEM), a rare multi-phenotypic sarcoma that occurs in pediatric age group and composed of mesenchymal component in the form of rhabdomyosarcoma and an intimately admixed neuroectodermal component in the form of ganglioneuroblastoma or ganglioneuroma. In a recent study by Huang et al. 23 6 of the 7 malignant ectomesenchymoma cases showed HRAS mutations, 4 showing p.G13R mutation and 2 showing p.Q61L mutation. Most of the tumors had an ERMS like morphology. In unsupervised hierarchical clustering based on gene expression data, the MEM cases grouped tightly with the RMS cases. We have since seen 3 additional cases of MEM, all with HRAS mutations (2 with G13R and 1 with Q61L) and, similar to the prior study, were mostly located in the GU area. In the study from Huang et al.23, none of the cases with available follow-up had died of disease. The 3 additional cases we have seen also had no evidence of disease at last follow-up. This suggests that, with similar therapy, MEM have a better prognosis than RAS-mutated ERMS. The shared clinicopathologic, molecular and gene expression findings suggest that MEM has a close relationship to ERMS.
The impact of RAS / RAF mutations on prognosis of ERMS is uncertain, based on current literature. Although some studies suggest that RAS mutations are significantly associated with intermediate- and high-risk ERMS 24, other studies have shown that RAS gene mutations do not portend a poor prognosis.4 In our study, half (48%) of the RAS / RAF mutated RMS patients were either alive with disease or had died of the disease in spite of multimodal management including surgery, chemotherapy and radiation therapy. For patients presenting with localized disease, the 3-year and 5-year survival rates were 81% and 65% respectively. Although comparable, this is marginally lower than the reported 70% 5-year survival of ERMS patients presenting with localized disease. The presence and type of additional genetic alterations, as identified by next-generation sequencing molecular studies, did not seem to have a consistent impact on either survival or response to multimodal management including chemotherapy, radiation therapy and surgery.
The significance of RAS mutations from a therapeutic aspect is not certain. Most of our patients were treated with standard rhabdomyosarcoma regimens. A recent study 25 showed that a novel RAF/MEK inhibitor CH5126766 causes G1 cell cycle arrest in RAS-mutated RMS cell lines. More recently, inhibitors specifically targeting the KRAS p. G12C (AMG-510, MRTX-849, and JNJ-74699157)26 have been developed with AMG-510 obtaining FDA approval for treatment of adult patients with KRAS p. G12C mutated locally advanced or metastatic non-small cell lung cancer. This holds great promise in treatment of other tumors with KRAS p. G12C mutations. Additional studies are needed to study the efficacy of these targeted drugs in patients with RAS-mutated RMS.
In conclusion, RAS mutations are noted in approximately a quarter of all ERMS. NRAS gene mutations constitute one-half of these mutations, while KRAS and HRAS gene mutations account for the remaining cases. Most of the RAS-mutant RMS showed additional genetic alterations including mutations in genes affecting the RAS/RAF/ERK pathway. Although RAS mutated ERMS in our study appeared to have similar survival rates to that reported in all ERMS, additional larger studies are necessary to determine the true prognostic impact of RAS/RAF mutations. Although there was no statistically significant correlation, most RAS-mutant ERMS occurred in children and in the H&N and GU anatomic locations. In fact, all KRAS mutant ERMS were located in the GU area. All except two ERMS characterized by a RAS-positive genotype harbored an undifferentiated and/or spindle cell morphology lacking the typical strap or rhabdomyoblastic type cells.
Supplementary Material
Supplementary Figure 2: Kaplan-Meyer survival plot depicting probability of survival based on the type of RAS mutation. No statistically significant correlation was noted.
Supplementary Figure 1: Schematic Oncoprint representation of the RAS-mutated RMS with clinical features and follow-up data.
ACKNOWLEDGEMENTS
The authors would like to thank Bruce Crilly for preparation of composite figures.
Supported in part by:
P50 CA 140146-01 (CRA), P50 CA217694 (NA, CRA), P30 CA008748 (NA, CRA), Kristin Ann Carr Foundation (CRA), Cycle for survival (LW, CRA)
Footnotes
Conflict of Interest Disclosures: None.
REFERENCES
- 1.WHO Classification of Tumours Editorial Board. WHO classification of tumours: soft tissue and bone tumours. 5th ed. Lyon, France: International Agency for Research on Cancer; 2020. [Google Scholar]
- 2.Seki M, Nishimura R, Yoshida K, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nature communications. Jul 3 2015;6:7557. doi: 10.1038/ncomms8557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer discovery. Feb 2014;4(2):216–31. doi: 10.1158/2159-8290.CD-13-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shern JF, Selfe J, Izquierdo E, et al. Genomic Classification and Clinical Outcome in Rhabdomyosarcoma: A Report From an International Consortium. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. Jun 24 2021:JCO2003060. doi: 10.1200/JCO.20.03060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malempati S, Hawkins DS. Rhabdomyosarcoma: review of the Children's Oncology Group (COG) Soft-Tissue Sarcoma Committee experience and rationale for current COG studies. Pediatric blood & cancer. Jul 15 2012;59(1):5–10. doi: 10.1002/pbc.24118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hibbitts E, Chi YY, Hawkins DS, et al. Refinement of risk stratification for childhood rhabdomyosarcoma using FOXO1 fusion status in addition to established clinical outcome predictors: A report from the Children's Oncology Group. Cancer Med. Oct 2019;8(14):6437–6448. doi: 10.1002/cam4.2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. May 2015;17(3):251–64. doi: 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. May 14 2007;26(22):3279–90. doi: 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- 9.Matallanas D, Birtwistle M, Romano D, et al. Raf family kinases: old dogs have learned new tricks. Genes Cancer. Mar 2011;2(3):232–60. doi: 10.1177/1947601911407323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. Sep 1 1989;49(17):4682–9. [PubMed] [Google Scholar]
- 11.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. May 15 2012;72(10):2457–67. doi: 10.1158/0008-5472.CAN-11-2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinelli S, McDowell HP, Vigne SD, et al. RAS signaling dysregulation in human embryonal Rhabdomyosarcoma. Genes Chromosomes Cancer. Nov 2009;48(11):975–82. doi: 10.1002/gcc.20702 [DOI] [PubMed] [Google Scholar]
- 13.Shukla N, Ameur N, Yilmaz I, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. Feb 1 2012;18(3):748–57. doi: 10.1158/1078-0432.CCR-11-2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ciccarelli C, Vulcano F, Milazzo L, et al. Key role of MEK/ERK pathway in sustaining tumorigenicity and in vitro radioresistance of embryonal rhabdomyosarcoma stem-like cell population. Molecular cancer. Feb 20 2016;15:16. doi: 10.1186/s12943-016-0501-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langenau DM, Keefe MD, Storer NY, et al. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes & development. Jun 1 2007;21(11):1382–95. doi: 10.1101/gad.1545007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salem B, Hofherr S, Turner J, Doros L, Smpokou P. Childhood Rhabdomyosarcoma in Association With a RASopathy Clinical Phenotype and Mosaic Germline SOS1 Duplication. J Pediatr Hematol Oncol. Nov 2016;38(8):e278–e282. doi: 10.1097/MPH.0000000000000566 [DOI] [PubMed] [Google Scholar]
- 17.Kashi VP, Hatley ME, Galindo RL. Probing for a deeper understanding of rhabdomyosarcoma: insights from complementary model systems. Nat Rev Cancer. Jul 2015;15(7):426–39. doi: 10.1038/nrc3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storer NY, White RM, Uong A, et al. Zebrafish rhabdomyosarcoma reflects the developmental stage of oncogene expression during myogenesis. Development. Jul 2013;140(14):3040–50. doi: 10.1242/dev.087858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Takita J, Hiwatari M, et al. Mutations of the PTPN11 and RAS genes in rhabdomyosarcoma and pediatric hematological malignancies. Genes Chromosomes Cancer. Jun 2006;45(6):583–91. doi: 10.1002/gcc.20322 [DOI] [PubMed] [Google Scholar]
- 20.Dolgikh N, Hugle M, Vogler M, Fulda S. NRAS-Mutated Rhabdomyosarcoma Cells Are Vulnerable to Mitochondrial Apoptosis Induced by Coinhibition of MEK and PI3Kalpha. Cancer Res. Apr 15 2018;78(8):2000–2013. doi: 10.1158/0008-5472.CAN-17-1737 [DOI] [PubMed] [Google Scholar]
- 21.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. Nov 15 1989;49(22):6324–7. [PubMed] [Google Scholar]
- 22.Casey DL, Wexler LH, Pitter KL, Samstein RM, Slotkin EK, Wolden SL. Genomic Determinants of Clinical Outcomes in Rhabdomyosarcoma. Clin Cancer Res. Mar 1 2020;26(5):1135–1140. doi: 10.1158/1078-0432.CCR-19-2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang SC, Alaggio R, Sung YS, et al. Frequent HRAS Mutations in Malignant Ectomesenchymoma: Overlapping Genetic Abnormalities With Embryonal Rhabdomyosarcoma. Am J Surg Pathol. Jul 2016;40(7):876–85. doi: 10.1097/PAS.0000000000000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, Stewart E, Shelat AA, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer cell. Dec 9 2013;24(6):710–24. doi: 10.1016/j.ccr.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa N, Kikuchi K, Yagyu S, et al. Mutations in the RAS pathway as potential precision medicine targets in treatment of rhabdomyosarcoma. Biochem Biophys Res Commun. May 7 2019;512(3):524–530. doi: 10.1016/j.bbrc.2019.03.038 [DOI] [PubMed] [Google Scholar]
- 26.Veluswamy R, Mack PC, Houldsworth J, et al. KRAS G12C-Mutant Non-Small Cell Lung Cancer: Biology, Developmental Therapeutics, and Molecular Testing. J Mol Diagn. 2021. May;23(5):507–520.doi: 10.1016/j.jmoldx.2021.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 2: Kaplan-Meyer survival plot depicting probability of survival based on the type of RAS mutation. No statistically significant correlation was noted.
Supplementary Figure 1: Schematic Oncoprint representation of the RAS-mutated RMS with clinical features and follow-up data.