Abstract
Engineered bacteria for therapeutic applications would benefit from control mechanisms that confine the growth of the bacteria within specific tissues or regions in the body. Here we show that the tropism of engineered bacteria can be enhanced by coupling bacterial growth with genetic circuits that sense oxygen, pH or lactate through the control of the expression of essential genes. Bacteria that were engineered with pH or oxygen sensors showed preferential growth in physiologically relevant acidic or oxygen conditions, and reduced growth outside the permissive environments when orally delivered to mice. In syngeneic mice bearing subcutaneous tumours, bacteria engineered with both hypoxia and lactate biosensors coupled through an AND gate showed increased tumour specificity. The multiplexing of genetic circuits may be more broadly applicable for enhancing the localization of bacteria to specified niches.
An emerging focus in synthetic biology is to engineer microorganisms to grow selectively within specific niches of the environment and human body1–9. As genetic programming enables bacteria to sense and respond to physiological conditions in situ, this approach is poised to change existing paradigms for diagnosing and treating diseases such as inflammation10,11, infection12,13 and cancer14–16. A critical consideration of using engineered bacteria for medicine is the need for the containment of microbial growth to disease sites to prevent off-target tissue damage and septic shock17,18. Thus, engineering genetic circuits to confine bacterial growth at specific locations in the human body has the potential to address the challenge of translating next-generation microbial therapies.
To date, the majority of bacterial therapies have relied on the natural tropism of bacteria, defined here as the preferential growth within specific host tissues or microenvironments such as the gastrointestinal tract, skin and tumours10,15,19–21. Although relying on inherent bacterial growth preferences can sometimes control bacteria localization, many bacteria can grow outside of their natural niches—quickly spreading to unintended locations and resulting in off-target effects. Genetic engineering approaches, such as metabolite auxotrophy, dependence on synthetic amino acids and toxin/anti-toxin-based systems, have been used to control bacterial growth22–26. Coupling these techniques with environmentally responsive biosensors can improve the containment of engineered bacteria and prevent unintended spread27–31, but precise localization to specified organs remains a challenge. In this Article, we demonstrate an approach to engineer bacterial tropism with genetic circuits programmed to sense one or multiple distinct physiological signatures allowing for enhanced bacterial growth in predetermined conditions of organ niches (Fig. 1).
Fig. 1 |. Schematic of biosensors for engineered bacteria tropism.
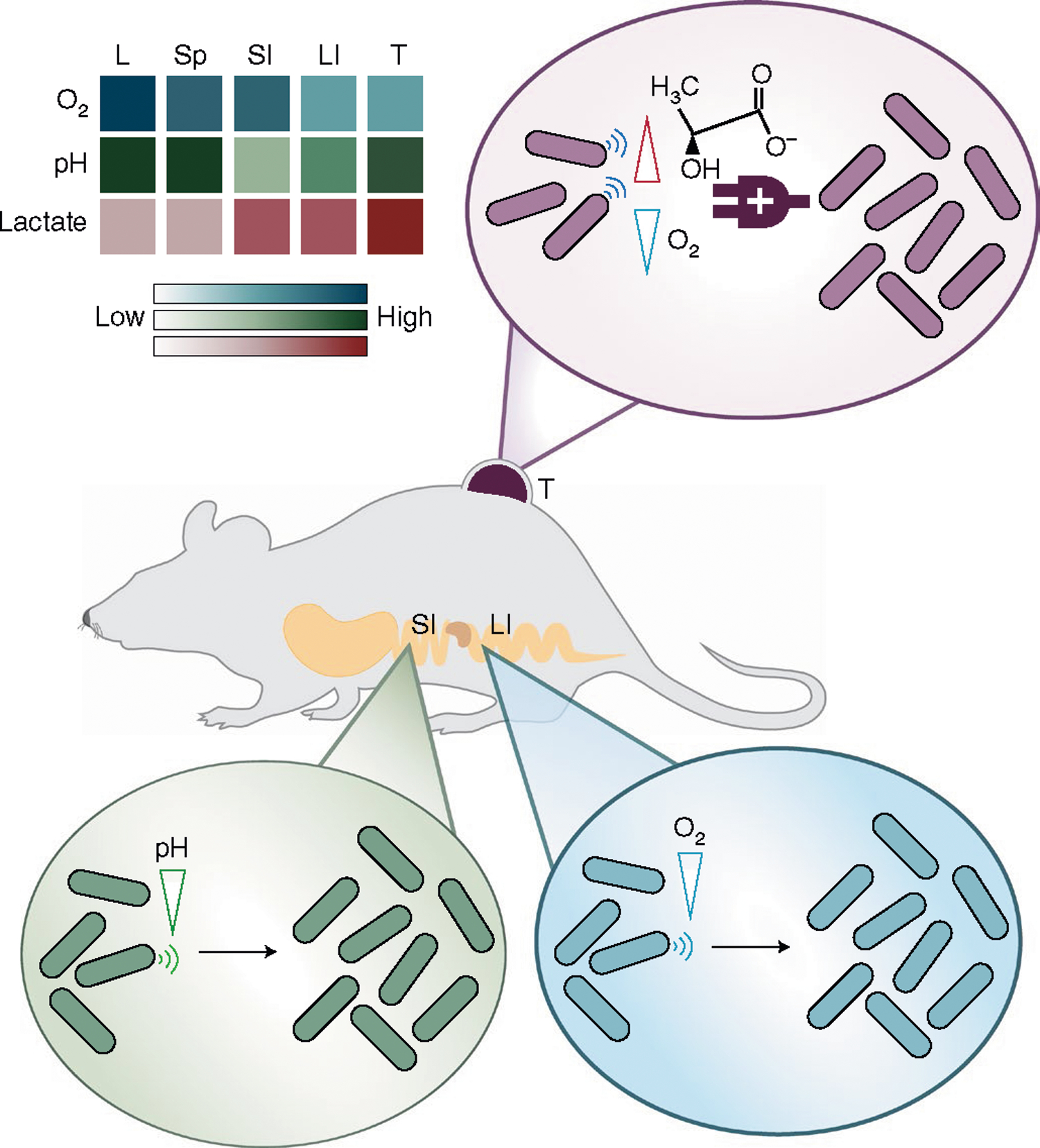
Engineered biosensors detect specific oxygen, lactate and pH levels in organs to enable tropism of bacteria in vivo. Different organs, including the liver (L), spleen (Sp), small intestine (SI), large intestine (LI) and tumour (T), exhibit varying biochemical signatures (Supplementary Table 1). The schematic shows that bacteria programmed to sense pH and oxygen enhance bacterial colonization in the small and large intestine, respectively. Bacteria with multiplexed lactate and oxygen AND logic gate sensing grow selectively within the tumour microenvironment.
Results
Development and characterization of engineered biosensors.
To construct bacterial biosensors that can distinguish unique organ environments, we chose oxygen, pH and lactate as common physiological indicators with substantial previous quantification in mice and humans32–36 (Supplementary Table 1). To sense oxygen, we used a hypoxia-sensing promoter (pPepT) that is primarily regulated by the transcriptional activator, fumarate and nitrate reduction regulatory protein (FNR)28 (Fig. 2a). In the absence of oxygen, FNR binds to a [4Fe–4S]2+ cluster to generate a transcriptionally active homodimer. However, the cluster is degraded in the presence of oxygen, which dissociates the FNR dimer into inactive monomers37. Measuring green fluorescent protein (GFP) expressed under the control of the pPepT promoter on a plasmid, we detected elevated levels of fluorescence in response to hypoxic conditions (Fig. 2a). We next designed an l-lactate biosensor, derived from the native lldPRD operon38–40, to detect lactic acid fermentation by host mammalian cells. This lactate-sensing system was constructed on two plasmids: a lactate-inducible reporter plasmid driving expression of a gene of interest and a repressor plasmid, which produces the repressor LldR that dimerizes to inhibit expression of the reporter gene unless bound to lactate (Fig. 2a). In response to increasing concentrations of l-lactate in the culture medium, we observed a corresponding increase in GFP (Fig. 2a). Finally, we engineered the pH-sensitive promoter pCadC among other systems41,42 that is regulated by a membrane-tethered activator protein (CadC)43–45, which shows increased activity in acidic medium compared with medium at a neutral pH (Fig. 2a).
Fig. 2 |. Design and characterization of hypoxia, lactate and pH biosensors.
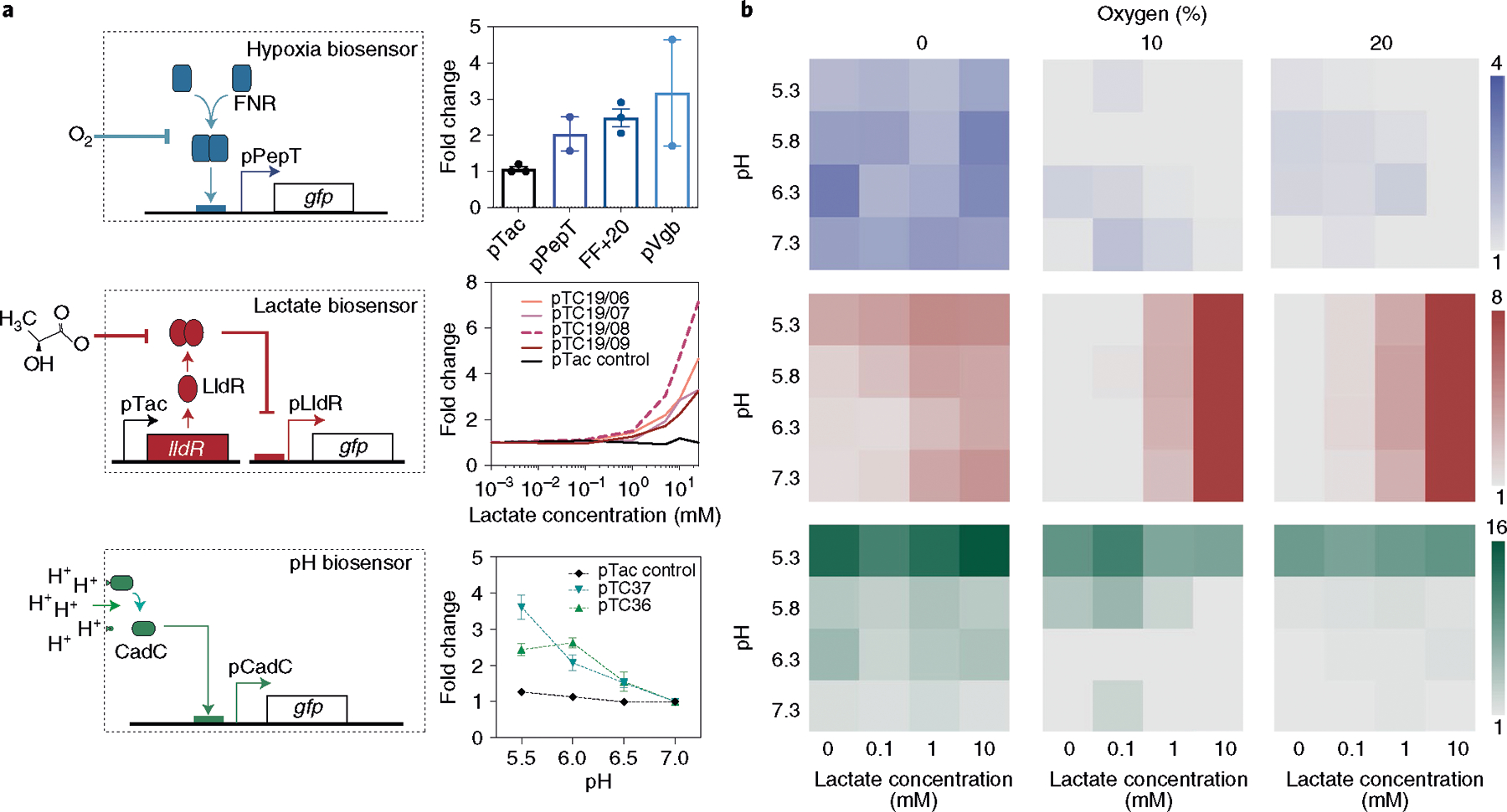
a, Left: each biosensor architecture consists of modified native bacteria promoters to sense specific environmental changes and express GFP. The hypoxia biosensor uses the pPepT promoter (or others) that relies on dimerized FNR to drive gene expression under low oxygen levels. The lactate biosensor contains constitutive production of an LldR repressor, which derepresses and activates the pLldR promoter in the presence of lactate. Finally, the pH biosensor, which is based on the pCadC system, is regulated by the membrane-tethered transcriptional factor CadC. Right: biosensor strains were grown in a specified environment (0% and 20% oxygen, 0–10 mM lactate, and pH 5.5, 6, 6.5 and 7) for 16 h. The fold change was calculated as the ratio of fluorescence in the induced to uninduced state. n = 3 biological replicates. Data are mean ± s.e.m. (Supplementary Table 2). b, Three biosensors (pPepT, pTC1908 and pTC37; Supplementary Table 2) were chosen from the variants and each was grown in pH 5.3–7.3, 0 mM, 0.1 mM, 1 mM and 10 mM of lactate, and under 0%, 10% and 20% oxygen for 16 h to characterize their induction in intersecting environmental conditions. All of the strains were cultured and assayed for fluorescence signal compared with a control baseline condition with oxygen at 20%, lactate concentration at 0 mM and pH 7.3. n = 3 biological replicates. Data are mean. The raw data and s.e.m. values are provided in Supplementary Table 5.
We next built a library of biosensor variants by tuning genetic parameters to establish systems that specifically activate in contrasting environmental conditions. For hypoxia biosensors, we tested three distinct hypoxia promoters (pPepT, FF+20 and pVgb)28,46,47 and demonstrated that hypoxia induces gene expression downstream of each promoter when compared to a constitutive promoter (Fig. 2a). For lactate biosensors, we varied the origin of replication as a knob to tune the copy numbers of the plasmids encoding the promoter (pLldR) and the regulatory protein (LldR). Using this approach, several biosensor variants showed notable activation under increasing lactate concentrations with varied sensitivity and dynamic ranges (Fig. 2a and Supplementary Fig. 1). Finally, for pH biosensors, we also constructed a library of biosensor variants by changing the plasmid copy numbers and we observed biosensor activation within the physiological pH range (Fig. 2a and Supplementary Fig. 2).
We chose to further characterize a single biosensor variant for oxygen, lactate and pH conditions. The variants pPepT, pTC1908 and pTC37 were selected owing to their activation under physiological concentrations that are similar to naturally occurring levels in various organs, such as the gastrointestinal tract and solid tumours48,49. Here, the hypoxia biosensor pPepT demonstrated low basal expression, which is suitable for preventing non-specific activation (Supplementary Fig. 3a). The lactate biosensor variants generally demonstrated low basal expression, and we also chose pTC1908 on the basis of its relatively high dynamic range (Supplementary Fig. 1). Specifically, pTC1908 is composed of a middle-copy-number reporter plasmid and its basal expression is quenched by the LldR repressor on a high-copy-number plasmid. For the pH biosensor, we chose pTC37, engineered on a single low-copy-number plasmid as it exhibited robust sensing at lower pH ranges (Supplementary Fig. 2a). As bacteria are often subjected to multiple environmental conditions simultaneously in vivo, we also tested the cross-reactivity of the biosensors in overlapping environmental conditions. Here we observed functional activity of biosensors exposed to multiple conditions, suggesting that these biosensors could be used in a combinatorial manner (Fig. 2b).
Engineering biosensors as containment strains to control bacterial growth.
We next sought to restrict bacterial replication in response to distinct biochemical signatures by expressing essential genes under the control of the biosensor promoters. This coupled design allows for potentially larger fold differences in bacterial number between environmental conditions despite the relatively low dynamic range of an individual biosensor. We chose an essential gene (asd) that is required in lysine, threonine and methionine biosynthesis. Supplementation by diaminopimelic acid (DAP), a bacteria-specific amino acid, allows for growth in asd-knockout strains28. Importantly, DAP cannot be produced or metabolized from the host cellular environment, which provides an ideal strategy for biocontainment in vivo. We knocked out asd from the genome of Escherichia coli (Supplementary Fig. 4a) and placed the gene under the control of the three biosensor promoters. As the initial circuits showed high levels of basal expression, we reduced this expression by constructing a library of variants with a range of gene copy numbers and introducing additional gene regulators (Fig. 3a). In brief, we first decreased the plasmid copy number to lower basal gene expression level (colE1, p15a or sc101 plasmid origin of replications and a single genome integration). We also tuned expression levels by introducing transcriptional (ribosomal-binding site (RBS) strength and antisense RNA) and translational (protein degradation tag) controls (Fig. 3a). A complete list of modifications and sequences is provided in Supplementary Tables 2 and 4.
Fig. 3 |. Engineering biosensor-dependent containment circuits and multiplexing for AND logic gate growth in vitro.
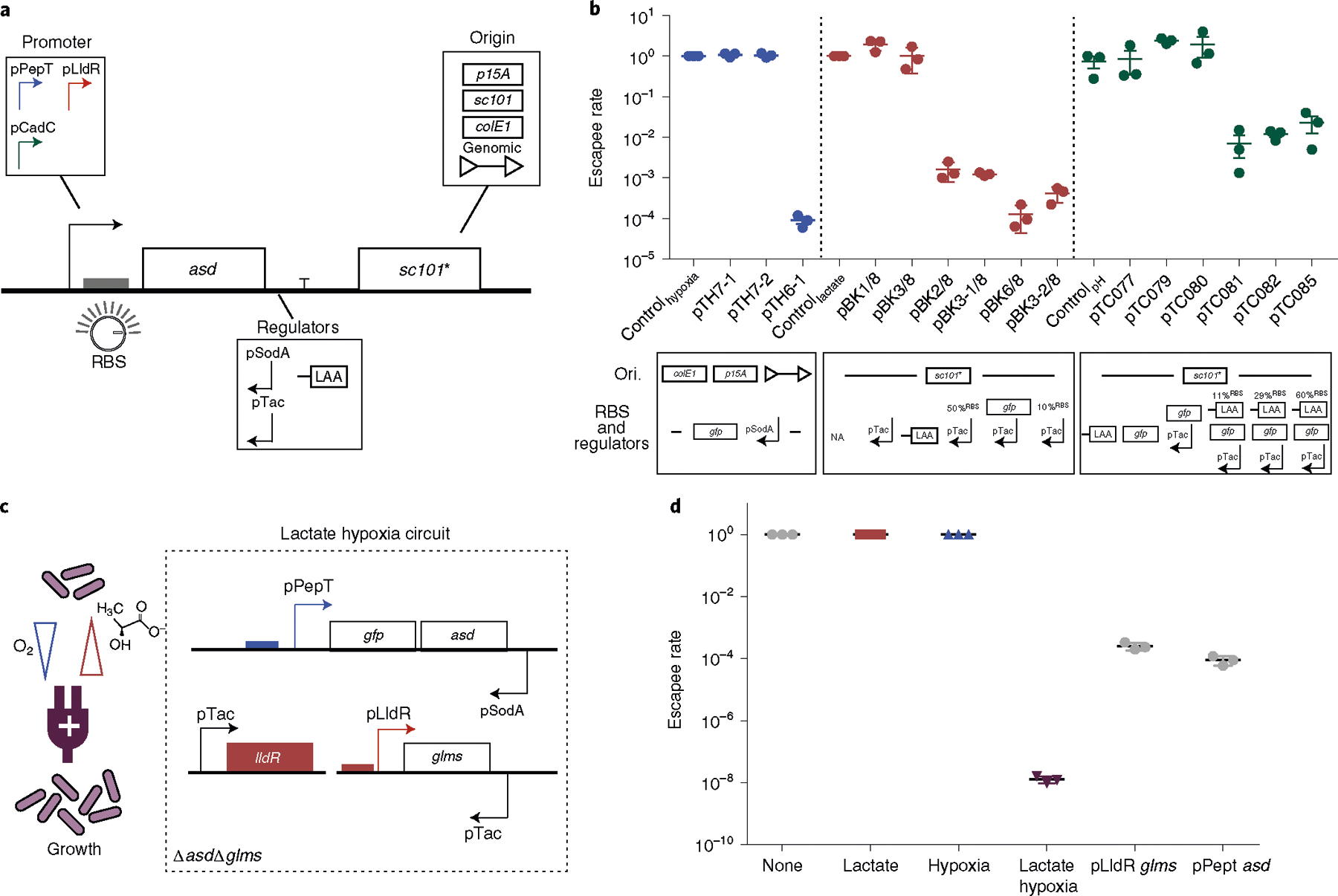
a, Design of a modular containment circuit that includes a biosensor promoter (pPepT, pLldR or pCadC) driving an essential gene (asd). To tune the sensitivity and reduce noise, additional regulators such as antisense promoters (pTac or pSodA), origin of replication (colE1, p15A, sc101* or genome integration), RBS or protein degradation tag (LAA) were used. b, Top: characterization of biocontainment variants on the basis of the escapee rate, defined as the ratio between colonies grown in non-permissive (normoxic, 0 mM lactate and pH 7) and permissive (hypoxic, 10 mM lactate and pH 6) conditions. All variants were cultured for 12–16 h with a starting density of 107 c.f.u. per ml and plated on LB agar plates with added supplements, after which colonies were counted the next day. n = 3 biological replicates. Data are mean ± s.e.m. All variants drove an essential gene along with additional genetic parts such as anti-sense, tuned RBS strength or a degradation tag (LAA). Blue, red and green indicate the hypoxia-, lactate- and pH-driven containment circuit, respectively (Supplementary Tables 2 and 4). Bottom: design of the circuit variants specifying changes in origin of replication (Ori.), RBS and regulators for each construct. NA indicates no modifications in RBS and regulators. c, Schematic of the lactate–hypoxia AND logic gate circuit, which consists of both the hypoxia promoter pPepT driving an essential gene (asd) and the lactate biosensor pLldR driving a second essential gene (glms). d, The escapee rates of the lactate–hypoxia AND logic gate circuit. The engineered bacteria were cultured under the four different conditions (none, 10 mM lactate supplemented, hypoxic conditions and a combination of 10 mM lactate with hypoxic conditions). The escapee rate was calculated as the ratio of non-permissive (no inducers, lactate only or hypoxic only) to permissive (both lactate and hypoxia) conditions. Two single circuits were grown in either lactate or hypoxic conditions. n = 3 biological replicates. Data are mean ± s.e.m. Samples were grown for 16 h and then plated on LB agar supplemented with DAP and d-glucosamine. Colonies were counted after incubating at 37 °C overnight.
To assess the impact of these modifications, we measured circuit function as the ratio of bacterial growth in non-permissive environments (normoxia, 0 mM lactate or neutral pH) to permissive conditions (0% oxygen, 10 mM lactate or pH 6), which represents the ‘escapee rate’ for characterizing the containment capability of the engineered bacteria, as has been used in other publications24,25,29. We identified several strain variants with low escapee rates that could grow selectively under permissive conditions (Fig. 3b). For the hypoxia containment circuit (pTH6-1), we genomically integrated the construct containing an additional gene (gfp) upstream of asd to reduce asd gene expression levels, followed by an antisense normoxia promoter, pSodA (Supplementary Fig. 3b), to reduce basal expression of asd. This optimized circuit design achieved selective bacterial growth under hypoxic conditions (Fig. 3b and Supplementary Table 2). For the lactate containment circuit (pBK3-2/8), lowering RBS strength and building on the low-copy plasmid were required for lactate-dependent bacterial growth (Fig. 3b and Supplementary Table 2). Finally, for the pH-containment circuit (pTC085), in addition to a reduced RBS strength, we also included a degradation tag downstream of asd to lower basal gene expression (Fig. 3b and Supplementary Table 2). The proportions of bacteria that grew in non-permissive conditions (escapee rate) were less than 10−4, 10−3 and 10−2 for the hypoxia, lactate and pH containment circuits, respectively. We further sampled oxygen, lactate and pH levels and demonstrated that these containment strains grew in ranges similar to biosensor activation (Supplementary Fig. 5). Taken together, we demonstrated engineered bacterial growth driven by three different environmental cues.
Multiplexing biosensors to improve specificity.
As a single physiological condition can be similar in two organs, for example, hypoxia in both the large intestine and in tumours, integration of multiple signals can enable further distinguishing of organ locations. Thus, to further enhance the specificity of our biocontainment circuits, we designed an AND logic gate circuit that permits bacterial replication only in the presence of two different environmental conditions. To do this, we engineered an additional containment strain with the lactate biosensor driving expression of glms, an essential gene orthogonal to asd that encodes a glucosamine-6-phosphate synthase that can be rescued with d-glucosamine50 (Supplementary Fig. 4b). This engineered lactate containment circuit exhibited selective growth in the presence of 10 mM lactate with a 2.6 × 10−4 escapee rate (Fig. 3d and Supplementary Fig. 6a). Before combining with the hypoxia containment circuit, we tested the lactate containment strain under hypoxic conditions and consistently observed lactate-dependent growth (Supplementary Fig. 6b). We next combined the glms-based lactate containment circuit with the asd-based hypoxia containment circuit in a double-knockout strain (Fig. 3c). This AND logic gate system grew when cultured under both low oxygen and high lactate, while we measured no colonies up to the limit of detection (LOD, 102 c.f.u. per ml) for hypoxic- or lactate-only culture conditions (Fig. 3d and Supplementary Fig. 7). Importantly, the AND logic gate system demonstrated ~104 improvement in escapee rate (10−8) compared with single biosensor containment circuits. These findings highlight that using multiplexed biosensor-based logic gate circuit architecture can improve growth specificity.
To further examine the functionality of single- and multi-input biocontainment circuit architectures, we developed a mathematical model to simulate bacteria growth regulated by one or more biosensor promoters. To do this, we described the rate of reporter protein (GFP) production governed by different biosensor regulators (activators for hypoxia and pH biosensors, repressors for lactate biosensor) with a system of ordinary differential equations (Supplementary Materials). We simulated bacterial growth on the basis of biosensor activation (Extended Data Fig. 1), and explored multiplexed environmental biocontainment using various biosensor combinations (Supplementary Fig. 8). We observed that the relatively low dynamic range of the promoters, when coupled to essential gene production and tuned, was sufficient to drive bacterial growth and result in substantial differences in bacterial number based on environmental inputs in the physiological ranges of interest (Supplementary Fig. 8 and Extended Data Fig. 2). While some biosensors displayed slight differences in activation kinetics, our model predicted the growth of bacteria in various specified environmental combinations over longer time scales in in vitro experimental observations (Supplementary Fig. 9), and provides a generalizable model of circuit architectures to explore beyond our immediate system and applications.
Bacterial biosensors and containment strains alter bacterial population levels in physiological environments.
As a step towards in vivo characterization of biosensors, we first assessed the capability of bacterial biosensors to sense metabolic activity of mammalian cell cultures in vitro (Fig. 4a). We cultured cell lines from various origins, including colorectal, lung, lymph nodes and breast, over 5 d and measured the levels of lactate and pH in the culture medium after collection (Supplementary Fig. 10). Subsequently, we cultured biosensor-containing bacteria (pPepT, pTC1908, pTC37) in the collected cell medium supernatant and measured the fluorescence from the biosensor strains after 16 h. We observed a concomitant increase in the bacterial fluorescent signal as lactate concentrations increased and the pH level decreased (Fig. 4b,c). To confirm that the biosensor activity is independent of cell origin, we assayed medium supernatant samples from six different lung cancer cell lines and a lung fibroblast cell line51. We found that the biosensor showed an increase in fluorescent signal in only the cancer cell lines that produced ~10 mM lactate, suggesting that the activation range of our lactate biosensor is within in vivo tumour levels (Supplementary Fig. 11 and Supplementary Table 1). To characterize the hypoxia biosensor in culture medium, we incubated the bacteria with cell medium in a culture chamber with varying oxygen levels (0%, 10%, 20%). We observed consistent increases in the fluorescence signal following decreasing oxygen levels across cell lines (Fig. 4d). Collectively, these results demonstrate that our engineered bacteria can sense and enhance growth in distinct biochemical signatures found in host environments.
Fig. 4 |. Engineered biosensors respond to physiological cues.
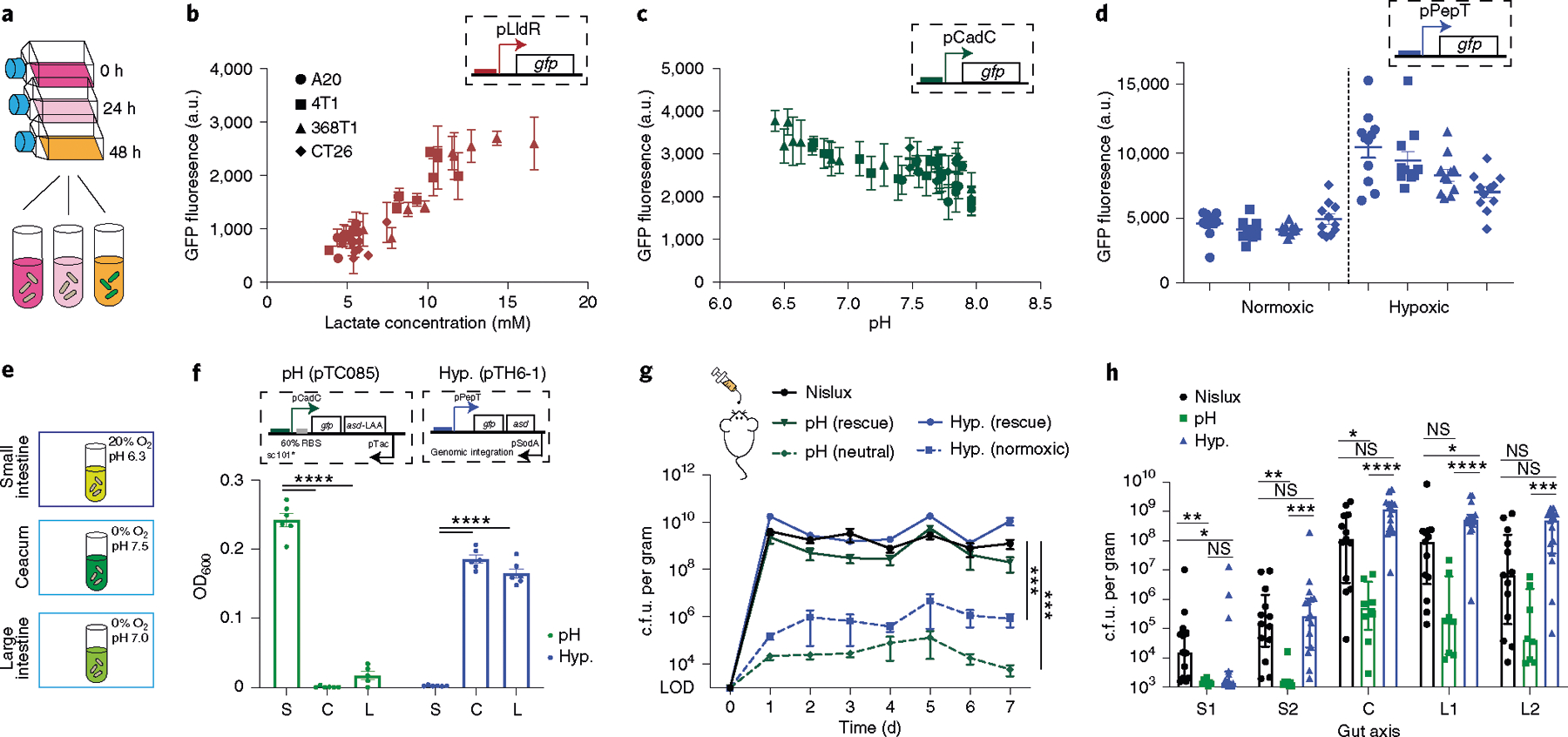
a, Cell culture medium supernatant from four cancer cell lines (A20, CT26, 368T1 and 4T1) was collected twice a day over 5 d and then cultured with our three bacterial biosensors (pTC1908, pTC37 and pPepT). b–d, Fluorescence activation of lactate (b; red), pH (c; green) and hypoxia (d; blue) biosensors when cultured in the collected supernatant overnight at 37 °C. The hypoxia biosensor cultured in cell medium supernatant was grown under conditions with or without oxygen. The GFP signal from the hypoxia biosensor was normalized to the constitutive promoter control. n = 3 biological replicates. Data are mean ± s.e.m. e, Bacteria strains were cultured under the following gut-compartment-relevant pH and O2 concentrations: small intestine (S; pH 6.3 and 20% oxygen); caecum (C; pH 7.5 and 0% oxygen); and large intestine (L; pH 7 and 0% oxygen). f, End-point optical density at 600 nm (OD600) of bacteria grown in gut-mimicking conditions with a starting density of 106 cells per ml for 12–16 h. n = 6 (pTC085) and n = 6 (pTH6-1) biological replicates. Data are mean ± s.e.m. Statistical analysis was performed using two-way ANOVA with Tukey’s multiple-comparisons test; ****P < 0.0001; Hyp, hypoxia g, Fresh faecal pellets were collected every day for 7 d, homogenized and plated on LB agar plates with an antibiotics (Ab.) selection with or without DAP. n = 7 (Nislux), n = 7 (pH (pTC085)) and n = 8 (hypoxia (pTH6-1)) biological replicates per time point. Data are mean ± s.e.m. Statistical analysis was performed using ANOVA with Tukey’s multiple-comparisons test; ***P = 0.0005. LOD, 103 c.f.u. per gram. h, Mice were euthanized at the end of the experiment (day 7), and the gastrointestinal tract was sectioned into five regions (upper small intestine track (S1); lower small intestine track (S2); caecum (C); upper large intestine track (L1); lower large intestine track (L2)). The regions were homogenized and plated on LB agar plates with antibiotic selection and DAP. Colonies were counted the next day. Absolute c.f.u. per gram of recovered bacteria from each gut compartment. n = 13 (Nislux), n = 8 (pH (pTC085)) and n = 15 (hypoxia (pTH6-1)) biological replicates. Data are median ± interquartile range. Statistical analysis was performed using Kruskal–Wallis ANOVA; *P < 0.04, **P < 0.009, ***P = 0.0003, ****P < 0.0001; NS, not significant. LOD, 103 c.f.u. per gram.
As oxygen and acidity levels decrease along the longitudinal axis of the intestine49, we examined whether our engineered strains could lead to enhanced biocontainment. We transformed pH and oxygen biosensors driving asd gene expression into a probiotic bacterium, E. coli Nissle 1917, which is currently used for oral administration in humans with gastrointestinal disorders52,53. To test the biocontainment circuits under physiological conditions, we first grew the pH and hypoxia containment strains in media matching gut environments of the small intestine, caecum and large intestine in vitro (Fig. 4e). The pH containment strain demonstrated highly specific growth in the acidic small intestine environment, whereas the hypoxia containment strain showed preferential growth in the hypoxic caecum and large intestine environment (Fig. 4f). To test biocontainment in vivo, we collected faecal samples over several days after oral delivery of bacteria. For both the hypoxia and pH containment strains, approximately 100-fold to 10,000-fold less bacteria from faecal samples grew during the course of one week compared with the control strain (labelled Nislux, E. coli Nissle 1917 with integrated luxCDABE cassette; Fig. 4g). However, with DAP supplementation in the agar, containment strains were rescued to similar colony-forming units (c.f.u.) as the control strain. These results suggest that the reduced bacterial numbers outside the host occurred through engineered auxotrophy.
We subsequently examined the bacterial distribution along the axis of the gut by measuring c.f.u. from homogenized tissue along five regions of the gastrointestinal tract. Generally, more bacteria were found in the distal colon (caecum and large intestine) for all strains, as expected from previous studies49,54–56 (Fig. 4h and Supplementary Fig. 12a–c). Compared to the control Nislux strain, fewer hypoxia-containment bacteria (pTH6-1) were found in the S1 region of the small intestine probably due to high oxygen, and similar bacteria counts were found in the large intestine regions, as expected (Fig. 4h). The pH containment strain (pTC085) showed lower levels of bacteria overall compared with the Nislux control strain. This was potentially due to reduced growth rates observed for pH values of >6 (Supplementary Fig. 13), which correspond to gastrointestinal tract measurements (Supplementary Fig. 14). To analyse relative bacterial profiles across the gastrointestinal tract, we scaled by c.f.u. per gram measurements of S1 for each strain (Supplementary Fig. 12d). Compared with the Nislux control strain, we found an approximately tenfold relative enrichment in hypoxia-dependent bacteria in the large intestine compared with in the small intestine. By contrast, we observed a decrease in relative bacteria levels in the distal colon of the gastrointestinal tract for pH-dependent bacteria compared with the control Nislux strain. To confirm that the altered biodistribution of pTC085 was not solely due to a reduced growth rate in general or low viability of this strain, we created a strain that constitutively expresses asd in the same Nissle Δasd background (pTC012). Here we observed that pTC012 had similar viability and maximal growth rate to that of pTC085 (as well as Nislux) at pH 6, but no pH-growth dependence (Supplementary Figs. 13e and 15). Although the single containment strains demonstrated selective growth in gut environment conditions in vitro, we did not observe specific targeting in vivo. To explain this, we constructed a mathematical model by factoring in the escapee rates we observed for these strains. This model was able to recapitulate the shift in bacteria distribution along the gut axis (Supplementary Fig. 16) with a corresponding input of oxygen and pH values (Supplementary Fig. 14 and Supplementary Table 1). Thus, although engineered strains could preferentially grow in the simulated environmental conditions of different gut compartments, single sensors were not sufficient to achieve targeting or overall profile shifts in the gastrointestinal tract in vivo, potentially due to their high escapee rates. These results provided further motivation for the need for combining sensors to improve specificity by reducing escapee rates.
Combined hypoxia–lactate containment strain improves tumour specificity.
Aside from modulation of bacterial growth in the healthy gastrointestinal tract, we next tested whether multiplexed containment circuits can enhance specificity in diseased sites such as tumours. In this context, bacteria such as Salmonella enterica serovar Typhimurium have been commonly studied as a cancer therapy due to their increased growth in tumour environments57. While S. Typhimurium has been reported to colonize tumours, it can also reach and survive in the liver and spleen28,58–60, suggesting the need to improve safety by reducing off-target bacterial load. We sought to improve on the natural tropism of S. Typhimurium for tumours by integrating multiple unique metabolic signatures from the tumour microenvironment, such as hypoxia, high lactate and low pH levels48,61–63 (Supplementary Table 1).
We first used a recently designed three-dimensional (3D) bacteria spheroid coculture system that recapitulates properties of the tumour microenvironment, including oxygen and nutrient gradients, mammalian cell metabolism and local 3D growth of the bacterial population in tumours64 (Fig. 5a). Attenuated S. Typhimurium ELH1301 was transformed with biosensor plasmids (Supplementary Fig. 17) and cocultured with the tumour spheroid system. Once in the spheroid core, we observed an increase in the total fluorescence signal from bacteria carrying hypoxia, lactate or pH biosensors driving GFP (Fig. 5b,c,d and Supplementary Fig. 18 and 19). We consistently measured the highest reporter signals in the centre of the spheroid, reflecting the expected biochemical gradients in the spheroid core64–66. The combined lactate–hypoxia AND logic gate containment strain showed comparable tumour-colonizing capabilities as the control strain (ELH1301) in tumour spheroids (Fig. 5e). To verify that the containment strain requires the expression of both essential genes, we constructed strains in which one biosensor drives a single essential gene in a double-knockout strain (ELH1301 ΔasdΔglms). After co-culture, these strains could not effectively colonize tumour spheroids (Supplementary Fig. 20), indicating that bacteria required both essential genes to grow.
Fig. 5 |. The multiplexed biosensor achieves enhanced specificity of bacteria tumour colonization.
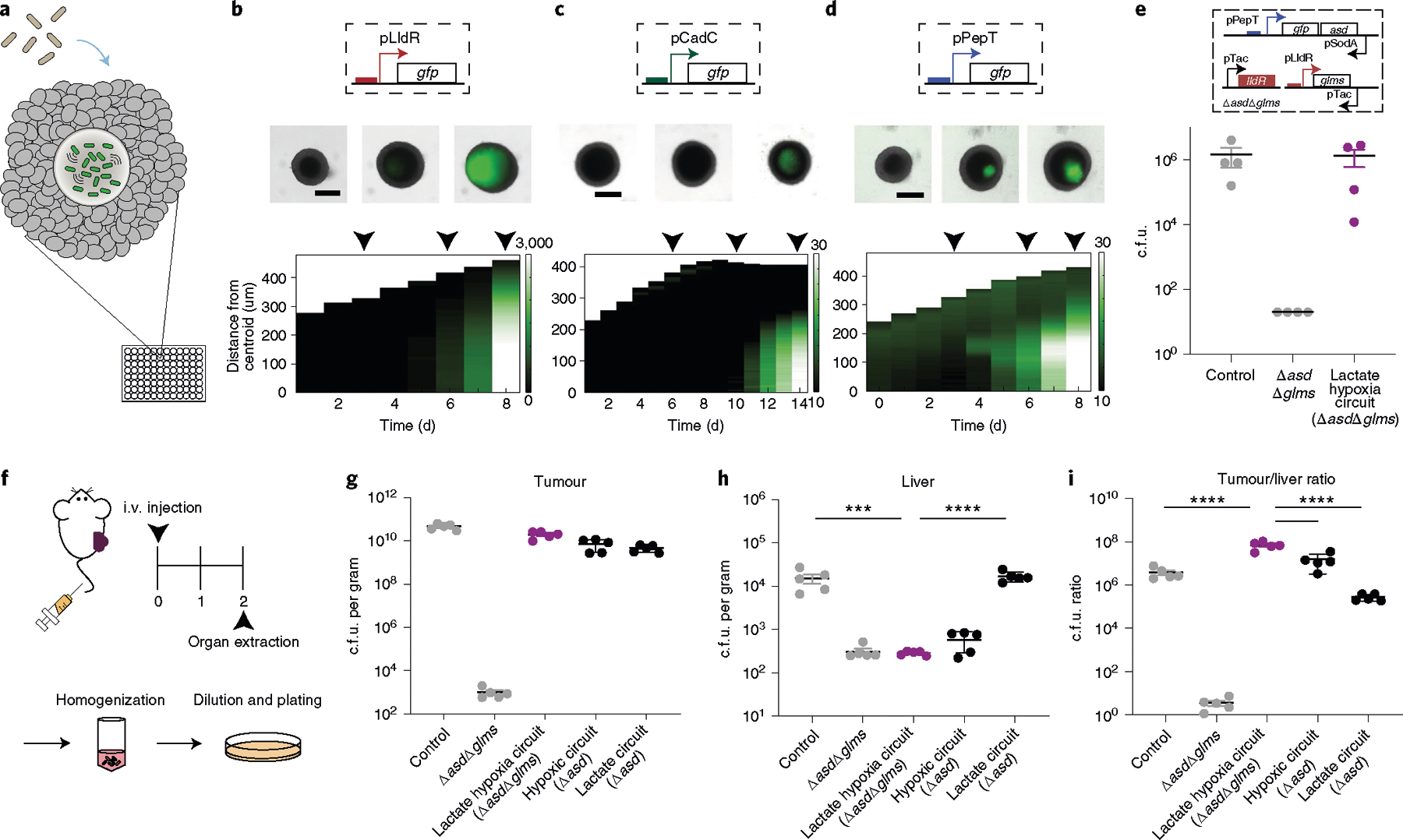
a, Engineered bacterial biosensors were cocultured in tumour spheroids and monitored for biosensor activation. b–d, Top: representative images of biosensors in tumour spheroids from n = 3 biological replicates (see Supplementary Fig. 19 for replicates). The black arrows indicate the day that the image was taken (top). Scale bars, 200 μm. Bottom: corresponding space–time diagram demonstrating radially averaged fluorescence intensity of lactate (pTC1908) (b), pH (pTC037) (c) and hypoxia (pPepT) (d) biosensors. The white boundary indicates the edge of the spheroid. e, Recovered colony counts of control strain S. Typhimurium ELH1301, ELH1301 ΔasdΔglms and the hypoxia–lactate circuit tested in tumour spheroid technology. All strains were cocultured in spheroids for 6 d, after which the spheroid was homogenized and the samples were plated on LB agar plates. n = 3 biological replicates. Data are mean ± s.e.m. LOD, 20 c.f.u. f, BALB/c mice (n = 5 per group) were implanted subcutaneously with 5 × 106 CT26 cells in one hind flank. When the tumour volumes were 100–150 mm3, the mice were intravenously administered with ELH1301 (control), double knockout ELH1301 (ΔasdΔglms), the lactate–hypoxia circuit in ΔasdΔglms, hypoxic only (Δasd) or lactate only (Δasd) ELH1301. After 2 d, the tumour, liver and spleen were homogenized and plated on LB agar plates with supplements. i.v., intravenous. g,h, Bacteria colonizing tumour (g) and liver (h) tissues were quantified and counted after 1 d. n = 5 biological replicates. Data are mean ± s.e.m. Statistical analysis was performed using one-way ANOVA with Bonferroni’s multiple-comparisons test; ****P < 0.0001, ***P = 0.0002. Tumour and spleen LOD, 103 c.f.u. per gram; liver LOD, 102 c.f.u. per gram. i, The tumour:liver ratio of bacterial c.f.u. per gram was calculated on the basis of recovered c.f.u. from extracted organs. n = 5 biological replicates. Data are mean ± s.e.m. Statistical analysis was performed using one-way ANOVA with Bonferroni’s multiple-comparisons test; ****P < 0.0001. Tumour LOD, 103 c.f.u. per gram; liver LOD, 102 c.f.u. per gram.
We next intravenously injected our tumour-containment strain (ELH1301 ΔasdΔglms with the lactate–hypoxia AND gate) in a subcutaneous syngeneic mouse tumour model (Fig. 5f). To measure improvement in tumour localization by the containment strain, we administered positive control (ELH1301 with no circuit) and negative control (ELH1301 ΔasdΔglms) strains that set the maximum and minimum bacterial load we can expect from the organs after systemic delivery, respectively. Organs and tumours were collected and homogenized to assess bacterial colonization 2 d after injection by c.f.u. enumeration. As expected, we observed that the lactate–hypoxia AND gate containment strain was able to colonize tumours to a similar level as the control strain (Fig. 5g). These results suggest that the containment strain was able grow in the tumour environment that provides permissive lactate and oxygen levels. In comparison, the recovered c.f.u. of the containment strain was lowered in the spleen and liver compared with 104–105 c.f.u. per gram bacterial load of the control strain (Fig. 5h and Supplementary Fig. 21a). As the double-knock-out (ΔasdΔglms) negative control strain was found to be at similar levels as the containment strain in the spleen and liver, we reasoned that the non-tumour environment was unable to induce essential gene expression and permit growth. On the basis of these results, we conclude that multiplexed bacteria decreased off-target colonization in the spleen and liver with a demonstrated increase in tumour-targeting specificity (Fig. 5i and Supplementary Fig. 21b). Finally, we showed that the AND gate strain exhibits a tumour/liver ratio that is significantly higher than the hypoxia-only and lactate-only circuit strains, demonstrating increased tumour specificity with multiplexing containment circuits in vivo (Fig. 5g,h,i).
To test whether observed differences in bacterial growth were dependent on environmental signatures in situ, we grew engineered bacteria in extracted organs ex vivo (tumour, liver and spleen). No bacterial growth was observed in any homogenized organs that no longer possessed native physiological environments that were permissive for growth (20% oxygen, <1 mM lactate; Supplementary Fig. 22). By contrast, we observed a significant increase in bacterial number when we modulated these conditions for activation by spiking in 10 mM lactate and incubating at 0% oxygen conditions (Supplementary Fig. 22). Similar levels of bacterial growth were observed when rescued with essential supplements (DAP and d-glucosamine). These results indicate that the in vivo growth tropism of the engineered bacteria was independent of organ types and was primarily driven by the local hypoxia and lactate levels.
Discussion
Our engineered system demonstrates the proof-of-concept that bacterial growth distributions can be enhanced towards conditions of the mouse gastrointestinal tract and tumours harbouring distinct environmental signatures. By tuning and multiplexing biosensors responsive to physiological cues to construct biocontainment circuits, we demonstrated enhanced tropism of microbial growth.
One critical design of our biocontainment circuit is the coupling of bacterial growth to environmentally responsive promoters. Although inducible promoters have been used widely to highly express genes of interest with low basal expression, many native promoters possess relatively low dynamic ranges67–69. This presents a challenge for in vivo microbial applications as insufficient or high basal expression of diagnostic or therapeutic payloads leads to lack of efficacy and safety. Here we coupled engineered promoter–biosensor machinery with bacterial growth through the expression of essential genes to address this limitation. Owing to the low gene expression needed for essential genes, minimal promoter activity was sufficient to enable bacterial growth at specified environmental conditions. We observed that, coupled with the signal, amplification through replication successfully achieved >1,000-fold differences in bacterial number using three independent biosensors. In terms of specificity, the ideal promoter would possess threshold-like activation in the desired environmental condition compared with a control. However, most natural and engineered promoters display a more graded response, as we observed in our biosensor variants. Nonetheless, amplification of the promoter signal through coupling to essential genes produces an improved profile due to the nonlinear amplification of exponential growth.
While the amplification through a single biosensor-growth coupling results in condition-specific growth, we ultimately see that single sensors could be combined with others to improve the escapee rate, leading to more precise organotropism. For example, although in vitro data support preferential growth of pH containment strains at levels found in small intestine compared with the caecum and large intestine, the escapee rate of this single strain is near 10−2, implying that the strain will produce escapees at a high frequency, which could then lose pH dependence and grow in unwanted regions. We therefore engineered the system to incorporate multiple biosensors and containment circuits to further enhance tropism. By using orthogonal biosensor–essential-gene pairs, bacterial growth can be regulated with multiple environmental signatures in an AND logic gate manner. This is particularly important for systemic bacterial delivery, whereby microorganisms have access to multiple organs and can induce severe toxicity after off-target colonization. As a proof-of-concept, we showed an improvement in bacterial tumour specificity by multiplexing oxygen and lactate biosensors. Although similar oxygen or lactate conditions can individually be found in other organs, the use of two environmental signatures increased the specificity to the tumour, resulting in reduction in off-target colonization. Using a computational model, we demonstrated that coupling of multiple biosensors can indeed lead to improved tropism in relevant physiological range. Furthermore, this model allows for future predictions as to the expected tropism enhancements and generalizability of multiplexed containment circuit designs.
Looking forwards, the approach described here can enable future precision targeting of specific physiological regions. For example, we envision the use of the multiplexed biosensors to control the expression of therapeutic payloads to enhance efficacy and avoid off-target toxicity. More precise activation using multiplexed biosensors can be used to differentiate microenvironment of various tumour types, potentiating the use of bacteria as precision diagnostic devices70,71. Furthermore, we propose generalizing this approach to other bacterial species and targeting other organs for biomedical research and translation to human disease. As translation of engineered microorganisms for various technological applications continues, robust and precise engineering of bacterial localization to desired environments will provide a useful method to improve biocontainment and safety to target specific sites for the local delivery of treatment options.
Methods
Host strains and culturing.
ELH1301 was provided by E. Hohmann. E. coli Nissle 1917 was obtained from the Bhatia lab15. Full strain information is provided in Supplementary Table 3. All bacteria were cultured in LB media (Sigma-Aldrich) with appropriate antibiotic selection (100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, 25 μg ml−1 chloramphenicol) at 37 °C.
Plasmids and biosensor library constructions.
Plasmids were constructed using Gibson Assembly or using standard restriction digest and ligation cloning and transformed into Mach1 competent cells (Invitrogen). The biosensors were constructed by synthesizing promoters from IDT, except for the pPepT, pLldR and pCadC promoters, which were obtained using colony PCR from E. coli Nissle 1917. Promoters were cloned in front of the sfGFP gene of a previously used ColE1 pTD103 sfGFP plasmid72. To construct biocontainment circuits, the essential gene asd or glms was added after the sfGFP gene. To tune the circuit sensitivity, gene copy numbers (colE1, p15a or sc101 replication origins and single genome integration), antisense promoters, RBSs and protein-degradation tags were engineered by cloning each segment using synthesized DNA followed by Gibson assembly. A detailed table of biosensor plasmids is provided (Supplementary Table 2).
Chromosomal gene deletion and integration in bacteria.
The essential genes asd and glms were deleted using the λ-Red recombination system73. Linear DNA with ploxp-cmR-loxp template were PCR amplified using pkD3 plasmid and electroporated into bacteria carrying pKD46 plasmid. Bacteria were recovered and plated with supplement DAP and d-glucosamine. Chromosomal deletions of the essential genes were verified by PCR and sequencing. To integrate biosensor circuits into the bacterial genome, the CRIM plasmid system was employed74. Plasmid pAH162 carrying a tetracycline resistance gene was used to integrate the construct at φ80 sites73. Hypoxia promoter driving asd gene was cloned into the plasmid followed by genomic integration. Integration was verified using PCR and sequencing.
Characterization of biosensors in vitro.
Each bacterial strain was grown in liquid culture overnight, and then used to inoculate induction experiments. For hypoxia biosensors, each variant was cultured overnight in normoxic and hypoxic conditions. Hypoxia was achieved by growing bacteria in BD GasPak EZ anaerobic pouches and static culture at 37 °C. For lactate biosensors, each lactate biosensor variant strain was grown in 6 wells of a 24-well Qiagen Block overnight in LB broth with the relevant antibiotics and at lactic acid concentrations of 0, 0.1, 1, 5 and 10 mM. For pH biosensors, each variant was cultured in 96-well plates with a pH ranging from around 4.4–8. Negative controls of untransformed Mach1 and Nissle cells, along with a positive control of pTac sfGFP, a strain that expresses sfGFP using the synthetic Tac promoter, were grown in the same conditions. All cultures were started at a bacterial OD600 of 0.1. After 16–20 h of growth, fluorescence and absorbance data were collected using the Tecan Infinite MicroPlate reader.
Characterization of biocontainment circuit in vitro.
All containment strains were grown in LB medium overnight with supplements (DAP and/or d-glucosamine) added. Cultures were then washed three times with PBS to remove residual supplements, followed by serial tenfold dilutions ten times into LB with inducers (10 mM lactate, pH 5.5, 6, 7 or cultured under hypoxia). All variants were cultured for 12–16 h and plated on LB agar plates with supplements, and c.f.u values were calculated the next day.
Biosensor in vitro characterization data analysis.
All biosensor in vitro fluorescence signals were calculated by dividing raw GFP pixel intensity by the OD600 value, both obtained from plate reader data. The background fluorescence signal (no plasmid control of the same strain) was subtracted. For the hypoxia biosensor, the fluorescence signal was normalized to constitutive promoter (pTac) control, to account for protein folding maturation under hypoxia, as has been observed our group and others47,75–79 (Supplementary Fig. 3c). The fold change of each biosensor was quantified as the ratio between normalized fluorescence signal of induced and non-induced conditions. All triplicate values were averaged. For Fig. 1c, the baseline condition was at 20% oxygen concentration, 0 mM lactate concentration and pH 7.3.
Mammalian cell culture.
The 393T5, 373T1, 802T1, 482T1, 368T1 and 393T1 cell lines were provided by T. Jacks. All other cell lines were obtained from ATCC. Mammalian cells were cultured in DMEM/F-12 medium with GlutaMAX supplement (Gibco; for 393T5, 373T1, 802T1, 482T1, 368T1 and 393T1) or RPMI 1640 medium (Gibco; for CT26, 4T1, A20 and 368T1) and supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin–streptomycin (CellGro); human lung fibroblasts were cultured in Fibroblast Growth Kit-Low serum (ATCC PCS-201-041) with 2% FBS and placed inside a tissue culture incubator at 37 °C maintained at 5% CO2. Full cell line information is provided in Supplementary Table 2.
Bacterial culture with monolayer cell supernatants.
All cell lines were seeded in 9 identical wells on 6-well plates in 3 ml of DMEM/10% FBS with sodium bicarbonate or RPMI/10% FBS medium without buffer at an initial cell count of 100,000. Twice daily over the next 5 d, in 8–12 h intervals, the entire contents of each well were removed and transferred to a 15 ml tube. The sample was centrifuged at 200 r.c.f. for 5 min, and the medium supernatant was collected into another tube, which was frozen at −80 °C to preserve the lactate concentration at the time of collection. After collection, each monolayer cell supernatant was separately incubated with the 3 biosensors, negative control and positive control. Hypoxia biosensor was tested with varying levels of oxygen in a culturing chamber connected to the CO2–O2 controller (Okolab). All bacteria were grown for 12–16 h and assayed for fluorescence and absorbance using the Tecan Infinite MicroPlate reader.
Bacteria coculture with tumour spheroids.
Tumour spheroids were generated by seeding cells in round-bottom ultra-low attachment 96-well plate (Corning). Each well contained 2,500 CT26 cells in 100 μl of RPMI/10% FBS medium without antibiotics. The plate was centrifuged at 3,000 r.c.f. for 5 min to aggregate cells at the bottom of the plate and placed inside a tissue culture incubator for 4 d before coculture with bacteria.
Bacteria were cultured in a 37 °C shaker overnight to reach stationary phase before use. S. Typhimurium (106 c.f.u.) was inoculated into wells containing 4-day-old mature tumour spheroids and placed back into the tissue culture incubator. After 2 h of bacteria inoculation, medium was removed. Tumour spheroids were washed with 200 μl of PBS repeatedly, while leaving spheroids at the bottom of plate. After washing, 200 μl of medium containing 2.5 μg ml−1 gentamicin (Gibco) was added, and tumour spheroids were monitored for growth. Acquisition of spheroid still images was performed with EVOS FL Auto 2 Cell Imaging Systems. The scope and accessories were programmed using the Celleste Imaging Analysis software.
Bacterial colonization quantification using colony counts.
Spheroids containing bacteria were repeatedly washed with 200 μl of PBS, while leaving spheroids at the bottom of plate. After washing, spheroids were resuspended in 100 μl of PBS and homogenized using mechanical dissociation with sterile tips and repeated pipetting. Destruction of spheroids was confirmed using microscopy. Serial tenfold dilutions of the samples were inoculated on appropriate agar plates.
GFP average fluorescence and radial histograms for spheroids.
To measure the spatiotemporal dynamics of bacteria invading tumour spheroids, we first found a threshold brightness value for each transmitted light (TL) image to distinguish the dark spheroid from the light background. Scikit-image implementations of two popular thresholding methods were used: the minimum method80 for images taken daily, and Yen’s method81 for other images. The largest region within the resulting threshold-based image mask was identified as the tumour spheroid, and the mean intensity of sfGFP fluorescence within this region was calculated. To compute radial histograms, mean sfGFP fluorescence for many thin annuli with variable mean radii was measured and centred on the centroid of the spheroid mask region.
Animal gastrointestinal models.
All animal experiments were approved by the Institutional Animal Care and Use Committee (Columbia University, protocol AC-AAAN8002). Animal experiments were performed on BALB/c mice (aged 4–6 weeks; Taconic Biosciences) after pretreatment with antibiotics (ampicillin and neomycin) for 7–10 d before oral gavage of bacterial strains82. Faecal matter was collected daily and plated on selective plates with or without DAP to quantify containment. Animals were euthanized at the end point of 7 d, and the whole gut was excised, separated into five sections (S1, S2, C, L1 and L2), weighed and then homogenized to plate on DAP-supplemented plates for colony counting the next day.
Measurements of gastrointestinal bacteria distribution.
Recovered c.f.u. values were all normalized by weight of organ in grams as described previously10,83 (Supplementary Figs. 14 and 18). Absolute c.f.u. per gram of individual mice was divided by the respective c.f.u. per gram from the S1 region to compute the relative bacterial distribution along the gastrointestinal axis (Fig. 5c and Supplementary Fig. 14).
Animal tumour models.
All animal experiments were approved by the Institutional Animal Care and Use Committee (Columbia University, protocol AC-AAAN8002). The protocol requires animals to be euthanized when tumour burden reaches 2 cm in diameter or under veterinary staff recommendation. Mice were randomized into various groups in a blinded manner.
Animal experiments were performed on female BALB/c mice (aged 6–8 weeks; Taconic Biosciences) with bilateral subcutaneous hind flank tumours from CT26 colorectal cells. The concentration for implantation of the tumour cells was 5 × 107 cells per ml in RPMI medium (no phenol red). Cells were injected at a volume of 100 μl per flank, with each implant consisting of 5 × 106 cells. Tumours were grown to an average of approximately 150 mm3 before bacterial injections. Tumour volume was quantified using calipers to measure the length, width and height of each tumour (V = L × W × H).
Bacterial administration for in vivo experiments.
Bacterial strains were grown overnight in LB medium containing the appropriate antibiotics. A 1:100 dilution into medium with antibiotics was started the day of injection and grown until an OD600 of approximately 0.4. Bacteria were centrifuged and washed three times with sterile PBS before injection into mice. Oral gavage and intravenous injections of bacteria were performed at a concentration of 5 × 1010 and 5 × 108 cells per ml in PBS respectively and administered at a total volume of 100 μl.
Biodistribution.
After 2 d of bacterial injection, mice were euthanized to collect the tumours, spleen and liver. Subsequently, the organs were weighed and homogenized using a gentleMACS tissue dissociator (Miltenyi Biotec; C-tubes). Homogenates were serially diluted and plated on LB agar plates at 37 °C overnight. Colonies were counted and computed as c.f.u. per gram of tissue. Tumour-targeting capability was calculated by comparing the tumour:spleen and tumour:liver ratios of the control ELH1301 and engineered lactate hypoxia AND gate circuit strains. LOD values were determined by the lowest c.f.u. detectable per dilution of sample.
Ex vivo organ measurements.
pH was determined using a precalibrated microprocessor-based pH metre with a spear ending (Oakton Instruments). Gut pH measurements were taken from mice pretreated with antibiotics and after oral gavage. Measurements were then taken with the gastrointestinal tract cut open and the lumen exposed to measure the pH level of lumen with the pH metre in contact with the lumen surface. The tumour, liver and spleen were extracted, weighed and homogenized using the gentleMACS tissue dissociator (Miltenyi Biotec; C-tubes) in 5 ml PBS containing 15% glycerol. pH measurements were taken from homogenates with compensation for added PBS. A pH reading was taken immediately after the animals were euthanized to minimize the influence of post-mortem time on the pH. The samples were covered by a probe tip and a stable reading was acquired. The pH metre was washed with distilled water between measurements. Lactate assays were also performed on the extracted organs. The standard protocol using the lactate colorimetric assay kit II (BioVision) was performed, and we compensated for dilution from the added PBS by calculation.
Mathematical model.
To describe the behaviour of biosensors, we developed a system of ordinary differential equations that describe the dynamic expression of GFP (y) in response to varying levels of environmental lactate (L; equation (1)), oxygen (O2; equation (2)) and pH (H+; equation (3)), regulated by transcriptional factors LldR, FNR and CadC, respectively. Each biosensor’s simplified equation is derived from mass action equations describing the binding of transcription factors to DNA operators with regulation by lactate, oxygen and pH, on the basis of previous approaches for describing promoter activity through repressor and activator occupancy84,85. For simplicity we assumed that the timescales for binding and transcription reactions are substantially faster than translation86–88. Bacterial growth is modelled using the logistic equation with Nmax as the maximum population size, γ is the degradation rate and μ is the growth rate (equation (4)). For containment circuits, we assumed that protein concentrations produced from the asd and glms genes follow GFP concentrations from the below biosensor equations. For the general AND gate containment circuit, we assumed that the growth rate is proportional to the product of glms and asd expression (equation (5); Supplementary Fig. 4d,e).
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
Model parameter values.
We chose model parameters based on fitting experimental GFP biosensor data (Supplementary Fig. 8). We saw GFP activation with increase of L (lactate concentration) and H+ (protons) and decrease of O2 (oxygen level). Other parameters used include the following: αy (production rate), 1,750–3,000; γy (degradation rate), 0.01–1; K1(lldR dimer binding affinity to plldR promoter), 200; KL (Lactate binding affinity to lldR dimer), 200; K2 (FNR dimer binding affinity to pPepT promoter), 0.5; (O2 binding affinity to FNR dimer), 0.4; K3 (CadC binding affinity to pCadC promoter), 0.7; KH (H+ interaction with CadC transcription activator), 10−6; Nmax(maximum population), 1,000; γ (rate of bacterial degradation), 2.5; a (gene expression to growth rate proportionality constant), 1.3 × 10−7 for more than one essential gene, 4.6 × 10−4 for single essential gene. The parameter a was derived from experimental data of comparing supplement concentration to bacterial growth (Supplementary Fig. 4d,e).
Statistical analysis.
Statistical tests were performed using either GraphPad Prism 7.0 (Student’s t-test and analysis of variance (ANOVA)) or Microsoft Excel. The details of the statistical tests are indicated in the respective figure legends. When data were approximately normally distributed, values were compared using a Student’s t-test, one-way ANOVA for a single variable or a two-way ANOVA for two variables with Bonferroni correction for multiple comparisons. Mice were randomized into different groups before the experiments.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw fluorescence and OD600 data in Fig. 2b, and the raw c.f.u. counts and measured weights in Supplementary Fig. 12 are provided as Supplementary Information. Additional data are available from the corresponding author on request.
Code availability
The MATLAB code used in this study is available from the corresponding author on request.
Extended Data
Extended Data Fig. 1 |. Computational modelling of biosensors.
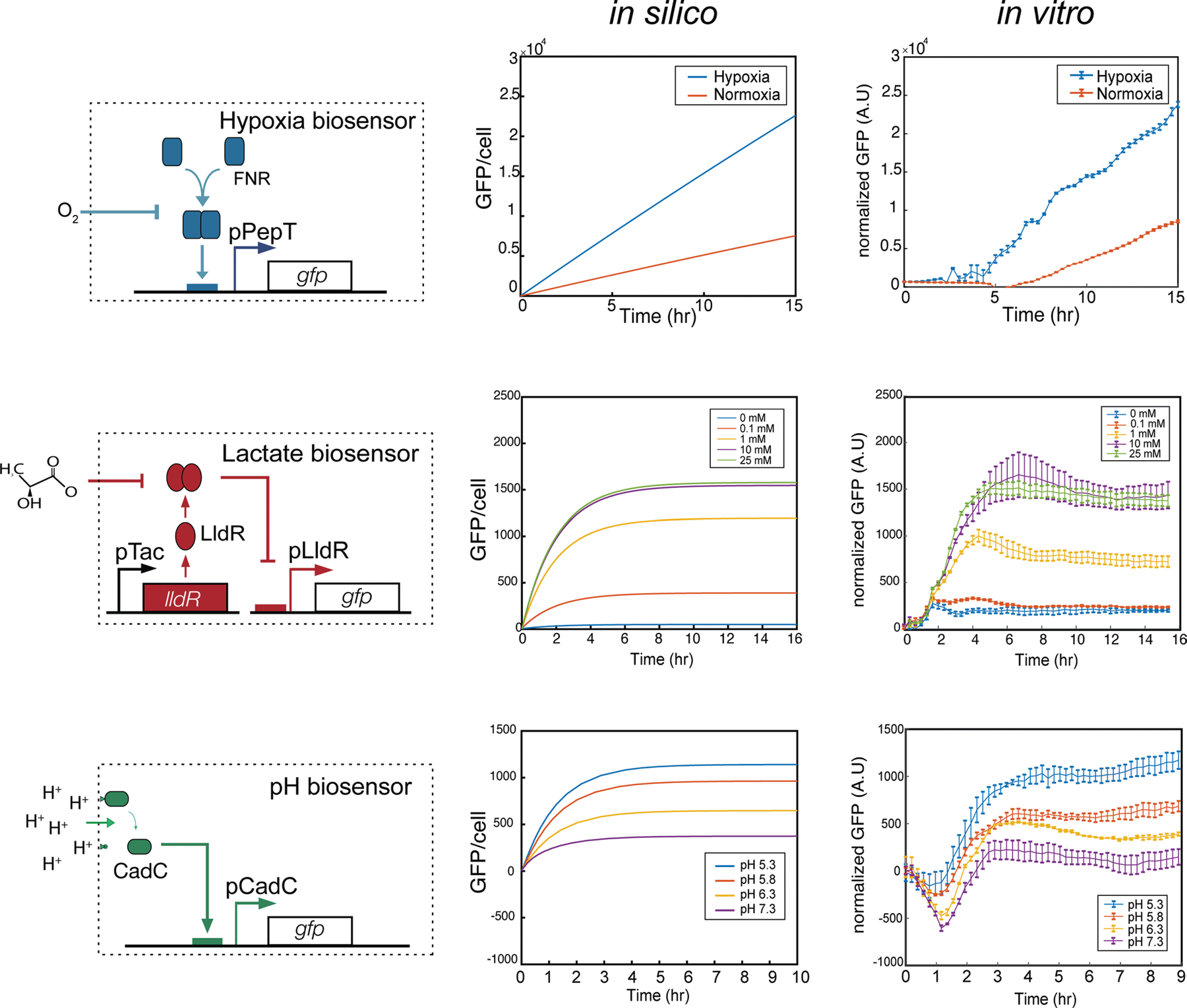
Biosensors are modelled under regulation of transcription activators (FNR, CadC) or repressors (LldR) with varying environmental conditions. (Left) biosensor circuit schematics as shown in Fig. 1 with (middle) modeling in silico predictions compared to (right) in vitro experimental results (n = 3, ±S.E.M). See supplementary materials for detailed equations and parameters used in this study. Lactate and pH biosensor in vitro GFP fluorescent result is normalized by OD. Hypoxia biosensor in vitro GFP fluorescence was measured in an anaerobic chamber and are normalized by data from constitutive promoter pTac.
Extended Data Fig. 2 |. Computational simulation of three 2-input AND-gate containment circuits.
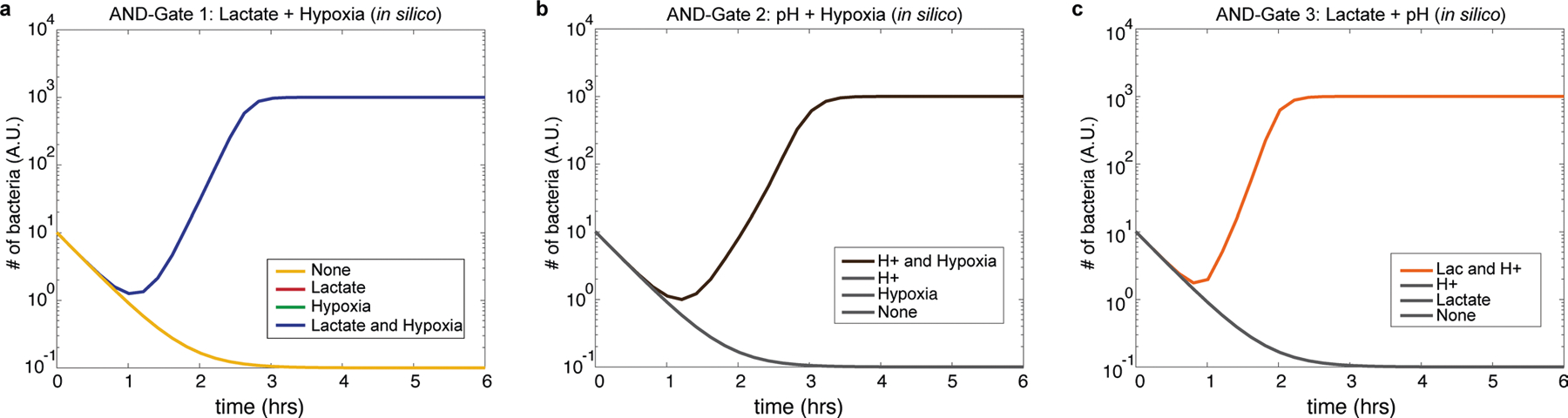
a, Computational modelling of lactate hypoxia AND gate containment strain growth over time in permissive (high lactate and hypoxia) and non-permissive (none, high lactate or hypoxia only) conditions. b, Growth simulation of pH hypoxia AND gate containment strain in permissive (low pH and hypoxia) and non-permissive (none, low pH or hypoxia only) conditions. c, Growth simulation of lactate pH AND gate containment strain in permissive (high lactate and low pH) and non-permissive (none, high lactate or low pH only) conditions. Lines not shown are covered by overlapping lines.
Supplementary Material
Acknowledgements
We thank J. Zhang and W. Mather for technical assistance and the members of the Danino laboratory for reading the manuscript. This work was supported by the DoD Idea Development Award (LC160314), DoD Era of Hope Scholar Award (BC160541), Honjo International Foundation Scholarship (to T.H.) and NIH F99CA253756 (to T.H.).
Footnotes
Competing interests
T.C., T.H. and T.D. have filed a provisional patent application (number 62/930,665) with the US Patent and Trademark Office related to this work.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41551-021-00772-3.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41551-021-00772-3.
References
- 1.Riglar DT & Silver PA Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol 16, 214–225 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Ruder WC, Lu T & Collins JJ Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Khalil AS & Collins JJ Synthetic biology: applications come of age. Nat. Rev. Genet 11, 367–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitada T, DiAndreth B, Teague B & Weiss R Programming gene and engineered-cell therapies with synthetic biology. Science 359, eaad1067 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garbeva P, van Veen JA & van Elsas JD Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu. Rev. Phytopathol 42, 243–270 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Hidalgo P, Maymon M, Pule-Meulenberg F & Hirsch AM Engineering root microbiomes for healthier crops and soils using beneficial, environmentally safe bacteria. Can. J. Microbiol 65, 91–104 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Vijay K, Murmu M & Deo SV Bacteria based self healing concrete—a review. Constr. Build. Mater 152, 1008–1014 (2017). [Google Scholar]
- 8.Lawson CE et al. Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol 17, 725–741 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao Y et al. Programmable assembly of pressure sensors using pattern-forming bacteria. Nat. Biotechnol 35, 1087–1093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riglar DT et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol 35, 653–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daeffler KN et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol 13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang IY et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun 8, 15028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao N, Cubillos-Ruiz A, Cameron DE & Collins JJ Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med 10, eaao2586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Din MO et al. Synchronized cycles of bacterial lysis for in vivo delivery. Nature 536, 81–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danino T et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med 7, 289ra284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson JC, Clarke EJ, Arkin AP & Voigt CA Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol 355, 619–627 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Low KB et al. Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol 17, 37–41 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Cao Z, Cheng S, Wang X, Pang Y & Liu J Camouflaging bacteria by wrapping with cell membranes. Nat. Commun 10, 3452 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mimee M, Tucker AC, Voigt CA & Lu TK Programming a human commensal bacterium, bacteroides thetaiotaomicron, to sense and respond to stimuli in the murine gut microbiota. Cell Syst. 1, 62–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatsuji T et al. A commensal strain of Staphylococcus epidermidis protects against skin neoplasia. Sci. Adv 4, eaao4502 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dang LH, Bettegowda C, Huso DL, Kinzler KW & Vogelstein B Combination bacteriolytic therapy for the treatment of experimental tumors. Proc. Natl Acad. Sci. USA 98, 15155–15160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JW, Chan CTY, Slomovic S & Collins JJ Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol 14, 530–537 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Mandell DJ et al. Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stirling F et al. Rational design of evolutionarily stable microbial kill switches. Mol. Cell 68, 686–697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan CT, Lee JW, Cameron DE, Bashor CJ & Collins JJ ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat. Chem. Biol 12, 82–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao M et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella Typhimurium. Proc. Natl Acad. Sci. USA 102, 755–760 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A & Shapiro MG Tunable thermal bioswitches for in vivo control of microbial therapeutics. Nat. Chem. Biol 13, 75–80 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Yu B et al. Explicit hypoxia targeting with tumor suppression by creating an ‘obligate’ anaerobic Salmonella Typhimurium strain. Sci. Rep 2, 436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallagher RR, Patel JR, Interiano AL, Rovner AJ & Isaacs FJ Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res. 43, 1945–1954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S et al. Coupling spatial segregation with synthetic circuits to control bacterial survival. Mol. Syst. Biol 12, 859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Barahona M & Buck M A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens. Bioelectron 40, 368–376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Santis V & Singer M Tissue oxygen tension monitoring of organ perfusion: rationale, methodologies, and literature review. Br. J. Anaesth 115, 357–365 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C & Kieda C Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J. Cell. Mol. Med 15, 1239–1253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen LW et al. Etiology and therapeutic approach to elevated lactate levels. Mayo Clin. Proc 88, 1127–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bakker J, Nijsten MW & Jansen TC Clinical use of lactate monitoring in critically ill patients. Ann. Intensive Care 3, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aoi W & Marunaka Y Importance of pH homeostasis in metabolic health and diseases: crucial role of membrane proton transport. BioMed. Res. Int 2014, 598986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crack JC et al. Signal perception by FNR: the role of the iron-sulfur cluster. Biochem. Soc. Trans 36, 1144–1148 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Aguilera L et al. Dual role of LldR in regulation of the lldPRD operon, involved in l-lactate metabolism in Escherichia coli. J. Bacteriol 190, 2997–3005 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goers L et al. Whole-cell Escherichia coli lactate biosensor for monitoring mammalian cell cultures during biopharmaceutical production. Biotechnol. Bioeng 114, 1290–1300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weghoff MC, Bertsch J & Muller V A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ. Microbiol 17, 670–677 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Xu Y et al. An acid-tolerance response system protecting exponentially growing Escherichia coli. Nat. Commun 11, 1496 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stirling F et al. Synthetic cassettes for pH-mediated sensing, counting, and containment. Cell Rep. 30, 3139–3148 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Schlundt A et al. Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator. Sci. Rep 7, 1051 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viala JP et al. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PLoS ONE 6, e22397 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee YH, Kim JH, Bang IS & Park YK The membrane-bound transcriptional regulator CadC is activated by proteolytic cleavage in response to acid stress. J. Bacteriol 190, 5120–5126 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan RM et al. Bacterial delivery of a novel cytolysin to hypoxic areas of solid tumors. Gene Ther 16, 329–339 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Mengesha A et al. Development of a flexible and potent hypoxia-inducible promoter for tumor-targeted gene expression in attenuated Salmonella. Cancer Biol. Ther 5, 1120–1128 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Petrova V, Annicchiarico-Petruzzelli M, Melino G & Amelio I The hypoxic tumour microenvironment. Oncogenesis 7, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donaldson GP, Lee SM & Mazmanian SK Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 14, 20–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K et al. A novel balanced-lethal host-vector system based on glmS. PLoS ONE 8, e60511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winslow MM et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonnenborn U Escherichia coli strain Nissle 1917—from bench to bedside and back: history of a special Escherichia coli strain with probiotic properties. FEMS Microbiol. Lett 363, fnw212 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Wassenaar TM Insights from 100 years of research with probiotic E. coli. Eur. J. Microbiol Immunol 6, 147–161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheth RU et al. Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol 37, 877–883 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yasuda K et al. Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 17, 385–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI & Borisy GG Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hohmann EL, Oletta CA & Miller SI Evaluation of a phoP/phoQ-deleted, aroA-deleted live oral Salmonella Typhi vaccine strain in human volunteers. Vaccine 14, 19–24 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Zheng JH et al. Two-step enhanced cancer immunotherapy with engineered Salmonella Typhimurium secreting heterologous flagellin. Sci. Transl. Med 9, eaak9537 (2017). [DOI] [PubMed] [Google Scholar]
- 59.Zhao M et al. Monotherapy with a tumor-targeting mutant of Salmonella Typhimurium cures orthotopic metastatic mouse models of human prostate cancer. Proc. Natl Acad. Sci. USA 104, 10170–10174 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman RM Tumor-targeting Salmonella Typhimurium A1-R: an overview. Methods Mol. Biol 1409, 1–8 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Chen LQ et al. Evaluations of tumor acidosis within in vivo tumor models using parametric maps generated with acido CEST MRI. Mol. Imaging Biol 17, 488–496 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Romero-Garcia S, Moreno-Altamirano MM, Prado-Garcia H & Sanchez-Garcia FJ Lactate contribution to the tumor microenvironment: mechanisms, effects on immune cells and therapeutic relevance. Front. Immunol 7, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonveaux P et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Invest 118, 3930–3942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harimoto T et al. Rapid screening of engineered microbial therapies in a 3D multicellular model. Proc. Natl Acad. Sci. USA 116, 9002–9007 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walenta S, Doetsch J, Mueller-Klieser W & Kunz-Schughart LA Metabolic imaging in multicellular spheroids of oncogene-transfected fibroblasts. J. Histochem. Cytochem 48, 509–522 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Riffle S, Pandey RN, Albert M & Hegde RS Linking hypoxia, DNA damage and proliferation in multicellular tumor spheroids. BMC Cancer 17, 338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wan X et al. Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol 15, 540–548 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Chen Y et al. Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun 9, 64 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hicks M, Bachmann TT & Wang B Synthetic biology enables programmable cell-based biosensors. ChemPhysChem 21, 132–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Runa F et al. Tumor microenvironment heterogeneity: challenges and opportunities. Curr. Mol. Biol. Rep 3, 218–229 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stanta G & Bonin S Overview on clinical relevance of intra-tumor heterogeneity. Front. Med 5, 85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Danino T, Mondragon-Palomino O, Tsimring L & Hasty J A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haldimann A & Wanner BL Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol 183, 6384–6393 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Craggs TD Green fluorescent protein: structure, folding and chromophore maturation. Chem. Soc. Rev. 38, 2865–2875 (2009). [DOI] [PubMed] [Google Scholar]
- 76.Coralli C, Cemazar M, Kanthou C, Tozer GM & Dachs GU Limitations of the reporter green fluorescent protein under simulated tumor conditions. Cancer Res. 61, 4784–4790 (2001). [PubMed] [Google Scholar]
- 77.Misra T et al. A genetically encoded biosensor for visualising hypoxia responses in vivo. Biol. Open 6, 296–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee JY et al. A novel chimeric promoter that is highly responsive to hypoxia and metals. Gene Ther. 13, 857–868 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Leventhal DS et al. Immunotherapy with engineered bacteria by targeting the STING pathway for anti-tumor immunity. Nat. Commun 11, 2739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prewitt JM & Mendelsohn ML The analysis of cell images. Ann. N. Y. Acad. Sci 128, 1035–1053 (1966). [DOI] [PubMed] [Google Scholar]
- 81.Yen J-C, Chang F-J & Chang S A new criterion for automatic multilevel thresholding. IEEE Trans. Image Process 4, 370–378 (1995). [DOI] [PubMed] [Google Scholar]
- 82.Carvajal-Aldaz DG et al. Simultaneous delivery of antibiotics neomycin and ampicillin in drinking water inhibits fermentation of resistant starch in rats. Mol. Nutr. Food Res 61, 1600609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ho CL et al. Engineered commensal microbes for diet-mediated colorectal-cancer chemoprevention. Nat. Biomed. Eng 2, 27–37 (2018). [DOI] [PubMed] [Google Scholar]
- 84.Alon U An Introduction to Systems Biology: Design Principles of Biological Circuits 3–19 (Chapman & Hall: /CRC, 2007). [Google Scholar]
- 85.Stricker J et al. A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cookson NA et al. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol. Syst. Biol 7, 561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen Y, Kim JK, Hirning AJ, Josic K & Bennett MR Synthetic biology. Emergent genetic oscillations in a synthetic microbial consortium. Science 349, 986–989 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S & Cohen SN Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl Acad. Sci. USA 99, 9697–9702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its Supplementary Information. The raw fluorescence and OD600 data in Fig. 2b, and the raw c.f.u. counts and measured weights in Supplementary Fig. 12 are provided as Supplementary Information. Additional data are available from the corresponding author on request.


