Abstract
Ambrosia beetles are insect vectors of important plant diseases and have been considered as a threat to forest ecosystems, agriculture, and the timber industry. Several factors have been suggested as promoters of the pathogenic behavior of ambrosia beetles; one of them is the nature of the fungal mutualist and its ability to establish an infectious process. In Mexico, Xylosandrus morigerus is an invasive ambrosia beetle that damages many agroecosystems. Herein, two different isolates from the X. morigerus ambrosia beetle belonging to the Fusarium genus are reported. Both isolates belong to the Fusarium solani species complex (FSSC) but not to the Ambrosia Fusarium clade (AFC). The two closely related Fusarium isolates are pathogenic to different forest and agronomic species, and the morphological differences between them and the extracellular protease profile suggest intraspecific variability. This study shows the importance of considering these beetles as vectors of different species of fungal plant pathogens, with some of them even being phylogenetically closely related and having different pathogenic abilities, highlighting the relevance of the fungal mutualist as a factor for the ambrosia complex becoming a pest.
Keywords: ambrosia fungi, symbiote fungi, Fusarium, Xylosandrus morigerus, phytopathogen
1. Introduction
An insect–fungus mutualism is an interaction that implies a reciprocal influence where each species provides mutual benefits such as dispersal, protection and nutrition [1]. The supply of nutrients by a partner can be direct, by serving them as food, or indirect, by providing digestible compounds or detoxifying a food source. They protect each other against environmental variations, competitors and/or natural enemies, and the dispersal aspect is clearly a benefit for the fungus, since it is a sessile organism and uses the insect as vector for its spores or propagules [1]. Fungiculture is the best-known mutualistic interaction between insect and fungi. This activity is performed by fungus-farming ants (Hymenoptera: Formicidae: Myrmicinae: Attini) [2], fungus-farming termites (Blattodea: Termitidae: Macrotermitinae) [3], the stingless bee Scaptotrigona depilis (Hymenoptera: Apidae: Meliponini) [4,5] and bark and ambrosia beetles (Coleoptera: Curculionidae: Scolytinae and Platypodinae) [6].
Bark and ambrosia beetles (Curculionidae: Scolytinae) have been adopted as a model of evolutionary ecology and phytopathology since they have emerged as a threat to forests and agricultural areas. Among these beetles, there are 16 hypothesized origins of fungus farming [7,8], with 63 genera in 10 tribes [9], and members of the tribe Xyleborini are considered to be strict fungus farmers, e.g., the genera Euwallacea, Xyleborus, Xyleborinus, and Xylosandrus [8]. Ambrosia beetles are xylem-borers of dead and stressed trees and feed primarily on cultivated co-evolved fungi, from which they acquire nutrients such as amino acids, vitamins and sterols; at the same time, fungus grows as mycelium on the walls of their galleries. Most of the ambrosia beetle species transport their food (fungi) in the mycetangium or gut [9,10,11], and it is proposed that the mycetangium may enforce the fidelity to the fungal mutualist [12].
The fungal mutualists of ambrosia beetles are reported to be Ascomycetes—belonging to the orders Ophiostomatales, Microascales, Hypocreales, and Saccharomycetales—and Basidiomycetes, of the orders Russulales and Polypolares [7]. Some species of the genera Ambrosiella (Microascales), Raffaelea (Ophiostomatales), Geosmithia (Hypocreales) and Ambrosiozyma (Saccharomycetales) are the best-known ambrosia fungi [6,7,11]; however, several interactions with Fusarium (Hypocreales), a genus that encloses several plant pathogens, have been documented [13].
Until now, the Fusarium species, described as a nutritional mycangial mutualist of ambrosia beetles, belongs phylogenetically to the Ambrosia Fusarium Clade (AFC), which was first described within the F. solani species complex (FSSC) as a monophyletic lineage, and which includes several phylogenetically distinct species [14,15,16,17,18,19]. These species from the AFC are obligate mutualists of Euwallacea ambrosia beetles (Coleoptera: Scolytinae) and it has been suggested that at least seven Euwallacea species are engaged in this obligate mutualism with at least 16 ambrosia fusaria species [19]. However, there is a constant stream of new reports about Fusarium species associated with ambrosia beetle species [20,21,22]. The data from these suggest that the Fusarium species could be common mutualists from others closely related species belonging to the Xileborini tribe but is clearly non-exclusive from beetles of the Euwallacea genus. For example, some species belonging to FSSC have been isolated from different species of the Xylosandrus genus, e.g., X. germanus [23], X. crassiusculus [24] and X. compactus [25,26].
The Xylosandrus genus includes at least 54 species, such as X. morigerus, X. crassiusculus, X. germanus, X. compactus and X. curtulus [27,28], and it has been suggested that members of this genus should be considered potential quarantine pests [29]. In particular, X. morigerus is defined as an ecological generalist [29]; thus, it can establish itself in new areas and become invasive, damaging agriculture and/or forestry areas under certain conditions. The damage to new environments, such as orchard and urban landscapes, is due to its capacity to attack live but weakened trees [30] and perform long-term attacks [6,30,31], since Xylosandrus is attracted to ethanol, which is produced by affected trees during biotic and abiotic stress, generating a continued infestation [30], and also for its association with phytopathogenic fungi [31,32]. The Ambrosiella species is considered to be the main fungal mutualist of Xylosandrus spp. [33,34]; however, Ambrosiella rarely behaves as a phytopathogen. Nevertheless, the acquisition of fungi from the environment modifies the beetle mycobiome [35] and, through lateral transmission, Xylosandrus spp. can associate with plant pathogens.
Analyses are currently underway to determine how the acquired plant pathogen impacts the health of an infested tree. An approach to address this matter is pre-invasion assessment [36], a phenotypical analysis of the symbiotes/mutualists, focusing on the behavior of the pathogen in potential hosts. Here, we report the identity of two fungal isolates from X. morigerus captured in Veracruz, Mexico. We determined the pathogenic capacity of both isolates in different possible hosts, giving insights into the complex Xylosandrus–Fusarium and its phytosanitary implications in some environments.
Understanding this interaction will increase the knowledge of the ecology of the ambrosia complexes and improve the management strategies designed to prevent and control the damage that they can cause.
2. Materials and Methods
2.1. Fungal Isolation
INECOL_BM-04 and INECOL_BM-06 were isolated from laboratory-reared ambrosia beetles, Xylosandrus morigerus. The beetles used to start the colony were collected in 2015 at Jaguaroundi Ecological Park, a protected natural area located in Coatzacoalcos, Veracruz (N 18.10931, W 94.36044). Ambrosia beetles were captured using ethanol-baited traps similar to those described by [37]. To build these traps, the neck of a two-liter polyethylene terephthalate (PET) plastic bottle was removed and joined with adhesive tape to the neck of another two-liter PET plastic bottle that had a cut frame of 11 × 20 cm on the side. A 50 mL Falcon tube with 96% ethanol was tied to the wall of the upper bottle and a cotton cord served as a wick to release the ethanol. In the lower bottle, moistened paper towels were placed to avoid the dehydration of captured beetles. Five of these traps were hung on trees at 5 PM and removed at 7 AM the next morning.
Female beetles were sorted and placed in an artificial culture media [38] for laboratory rearing and incubated at 26°C in total darkness. After 30 days, the colonies were dissected, and 3 females were collected, mounted and morphologically identified using the taxonomic keys of [39]. Female specimens of X. morigerus were surface sterilized by vortexing them twice in a 96% ethanol solution for 1 min; then, 10 X. morigerus beetles were aseptically segmented into two parts: head/thorax and abdomen. The surfaces of heads/thorax segments were washed by vortexing in a 1.5 mL microcentrifuge tube with 500 μL of 96% ethanol for 30 s and rinsed three times with sterile distilled water. The beetle tissue was ground with a sterile plastic micropistil in a 1.5 mL microcentrifuge tube containing 50 µL of sterile distilled water. Twenty-five microliters of the suspension was spread on a Petri dish with potato dextrose agar (PDA, 39 g/L Sigma-Aldrich, St. Luis, MO, USA) in duplicate and the cultures were incubated at 28 ± 1 °C in darkness. The colonies that showed differences in morphology were selected and placed individually in Petri dishes containing PDA (Sigma-Aldrich, St. Luis, MO, USA), which were incubated at 28 ± 1 °C in darkness for 14 days. These isolates were purified by single conidial culture in water-agar medium (2% agar) and incubated at 28 ± 1 °C for 1 to 3 days in darkness. A single colony was cultured on PDA (Sigma-Aldrich, St. Luis, MO, USA) and incubated at 28 ± 1 °C for 7 days in darkness. Two morphologically different isolates, INECOL_BM-04 and INECOL_BM-06, were selected for further characterization.
The isolates are maintained in the Internal Collection of the Molecular Biology Laboratory at the Department of Advanced Molecular Studies at Ecology Institute (INECOL).
2.2. Culture Conditions
The Fusarium spp. isolates INECOL_BM-04 and INECOL_BM-06 were cultured by 5 mm diameter mycelium plugs on PDA (Sigma-Aldrich, St. Luis, MO, USA) at 25 ± 1 °C in darkness.
2.3. Molecular Identification
The genomic DNA of axenic colonies of INECOL_BM-04 and INECOL_BM-06 was extracted using the protocol described by [40] with minor modifications. The rRNA cluster, consisting of the internal transcribed spacers (ITS1 and ITS2) and the 5.8S rRNA genes, was amplified using ITS4 and ITS5 primers [41], and the PCR mix for the amplification was prepared following the manufacturer’s protocol of the Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, Waltham, MA, USA, Cat. 11304). Amplification was conducted in a MultiGeneTM OptiMax thermocycler with the following conditions: 94 °C for 1 min, followed by 32 cycles of 94 °C for 15 s, 58 °C for 30 s, and 72 °C for 1 min with a final extension step of 72 °C for 5 min. The sequences of the purified PCR product were determined in both directions with the BigDye Terminator v3.1 technology and multicapillary DNA sequencing system AB3730, using the specific primers.
2.4. Molecular Phylogenetic Analysis
The phylogenetic analysis was based on four gene fragments: the translation elongation factor-1 alpha (tef1), the internal transcribed spacer region of rDNA (ITS), the 28S large subunit of the rDNA (LSU) and the RNA polymerase second-largest subunit (rpb2). The sequences of the FSSC species were those used in previous studies [42,43,44]. The sequences of the four loci of INECOL_BM-04 and INECOL_BM-06 were retrieved from the unpublished assembled genomes, provided by Ibarra-Laclette and Sánchez-Rangel from the Ecology Institute (INECOL) at Xalapa, Veracruz-Mexico. The sequences of the four markers for INECOL_BM-04 and INECOL_BM-06 isolates were submitted to GenBank with the following accessions: OM455454, OM455455, OM455456, OM455457, OM455458, OM455459, OM455460 and OM455461.
The sequences were manually curated when necessary and aligned with ClustalX [45]. After performing a heuristic trimming with Trimal [46], the four markers were concatenated in a single sequence representative of each species and the best model for molecular evolution was identified for each marker using the corrected Aikaike Information Criterion with PartitionFinder2 [47]. The phylogenetic tree and its clade credibility values were inferred using MrBayes [48] through a Markov chain Monte Carlo (MCMC) analysis of 2 runs over 1 × 106 generations. The resulting phylogenetic tree was visualized and annotated using the Ggtree [49] and the Treeio [50] packages in R. The four-locus data set and the Bayesian Inference (BI) tree are publicly available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S29320, accessed on 24 January 2022).
2.5. Macroscopic Morphology Examination
Petri dishes with PDA were inoculated with a 5 mm diameter plug taken from the edge of the actively growing one-week-old colony and incubated at 25 ± 1 °C in darkness. Colony morphology was evaluated seven and fourteen days post inoculation (dpi). The photo-documentation was carried out with a Sony Cybershot DSC-W55 camera.
2.6. Colony Radial Growth
Each isolate was cultivated in darkness at 25 ± 1 °C in PDA. The diameter of the colony was measured at the end of 14 days. The assay was performed with five technical replicates. A one-way ANOVA with post hoc Tukey HSD test was performed for statistical analysis.
2.7. Microscopic Analyses
Microscopic characters were investigated in Spezieller Nährstoffarmer Agar (SNA) and Carnation Leaf Agar (CLA) [51] at 25 ± 1 °C under a photoperiod of 12 h light/12 h dark for 10 days. The presence of sporodochia and chlamydospores was evaluated after one month of incubation. Conidia, conidiophores and chlamydospores were examined and documented with a Leica DMI6000 B microscope using a Leica DFC450 C camera and recorded using LAS X software after they were mounted in water. Average, standard deviation (SD) and minimum–maximum values for the size of individual conidial types for each isolate were calculated from measurements of at least 30 randomly selected conidia. The conidia from SNA cultures were collected with sterile distilled water and quantified in a Leica DM750 microscope using a Neubauer chamber. The sporodochia images were acquired with a Zeiss SteREO Discovery.V8 stereomicroscope. For scanning electron microscopy (SEM), 5 mm mycelium plugs were taken from the actively growing edge of a five-day-old colony cultured on PDA (Sigma-Aldrich, St. Luis, MO, USA) and fixed in 2.5% glutaraldehyde for 24 h at 4 °C and washed three times in 0.1 M Sorensen’s phosphate buffer for 5 min each. The samples were dehydrated through a graded ethanol series (30, 40, 50, 60, 70, 80, 90 and 96%); each sample remained for 60 min in each dilution and was then transferred to absolute ethanol three times (30 min each). Dehydrated samples were dried up to the critical point with CO2 in a Polaron-E500 drying apparatus. Dried samples were mounted on aluminum stubs and coated with gold using a Polaron 11-HD sputter-coating unit and observed in an FEI QuantaTM 250 FEG SEM operating at 5 kV [52].
2.8. Pathogenesis Assay in Coffea arabica, Salix lasiolepis, Populus nigra, Citrus sinensis and Citrus latifolia
Coffea arabica cv. Marsellesa trees were purchased from Sociedad de Productores de Café sustentable Aromas de Coatepec SPR de RL in Emiliano Zapata, Veracruz; this organization acquired the coffee seeds from the germplasm bank of Cafetalera Guadalupe Zajú, S.A de C.V., which is Rainforest Alliance certified. C. arabica cv. Oro Azteca trees were kindly donated by Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP). S. lasiolepis and P. nigra trees were acquired from a Las Palmas nursery in Saltillo, Coahuila, Mexico; this nursery is registered at the Secretaría del Medio Ambiente y Recursos Naturales (UMA-VIV-0605-COA) as a Management Unit for Wildlife Conservation. Meanwhile, C. sinensis and C. latifolia trees were bought from the “El Olmo” nursery in Xalapa, Veracruz, Mexico. The age of the trees ranged between one and two years and their height varied from 1.40 to 2.20 m. The trees were maintained in a zenith-type greenhouse under the following conditions: minimum temperature of 19 ± 2 °C, maximum of 27 ± 2 °C and a relative humidity between 50 and 70%. Nutrient solution (Nitro Sol) was periodically applied to all the plants to increase vegetative development.
Stem segments 8 cm in length were cut longitudinally and, together with the leaves of each plant species, were disinfected with a 2% (v/v) chlorine solution for 2 min and washed three times (2 min each) with sterile distilled water. For the pathogenesis assay, a wet chamber system was used, consisting of a 150 mm × 25 mm Petri dish with a cellulose filter paper (Whatman Grade 1) placed at the bottom and saturated with sterile distilled water. Two to three leaves or stems of the plant species were placed in the wet chamber. Before the inoculation, mechanical damage was inflicted with a scalpel at the base of the leaf and the central part of the stem. The plant tissue was inoculated by placing on the damaged site a plug of mycelium from the edge of the actively growing colony in seven-day-old PDA (the plug was carefully placed so the mycelium touched the plant tissue). The wet chamber was sealed and incubated at 25 °C ± 1 under a photoperiod of 8 h/16 h light/darkness for 12 days and for 21 days in the case of C. arabica. The assay was performed with three technical replicates. The lesion area was measured by Image J conducting particle quantification based on image contrast.
2.9. Extracellular Protease Activity Assay
The extracellular protease activity of both Fusarium sp. isolates was evaluated with a milk powder plate assay in six different growth media: PDA (BD-DifcoTM), Minimal Medium (MM) (+C+N), MM without nitrogen source (+C−N), MM without carbon source (−C+N), MM without carbon and nitrogen source (−C−N) and water agar 1.5% (WA). MM and its variants were prepared as described by [51]. A 25% solution of dried skimmed milk was prepared separately and added to the culture media to a final concentration of 3%. Plates were inoculated with a plug of mycelium obtained from the edge of an actively growing colony on PDA and incubated for 7 days at 28 °C. To determine the Lysis Index, the diameter of the protease halo was divided by the diameter of the colony. The assay was performed with three technical replicates. The Heat Map was generated using the GraphPad Prism 8 Software.
3. Results
3.1. Fungal Isolates INECOL_BM-04 and INECOL BM-06 Belong to the FSSC but Not to the AFC
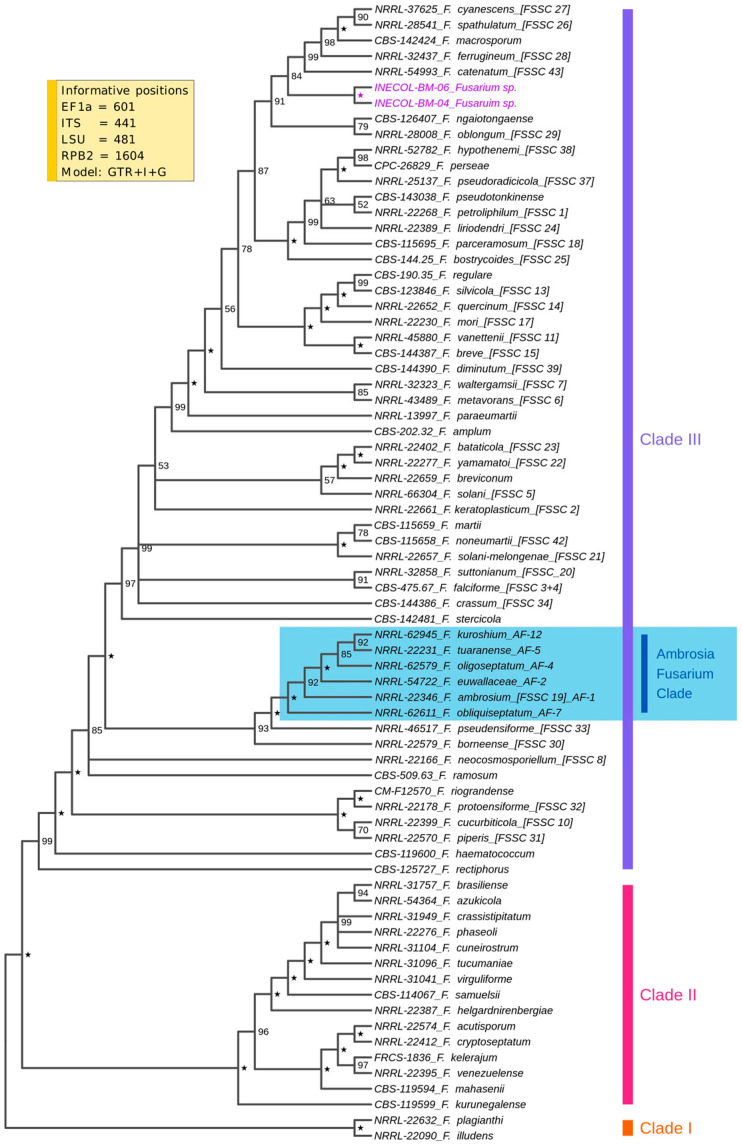
A BLAST analysis of the ITS sequence of both isolates against the ITS database of NCBI showed 96.13% and 96.30% identity to Fusarium solani CBS 140079 for INECOL_BM-04 and INECOL_BM-06, respectively. These ITS sequences also compared by BLAST against the Fusarioid-ID database [42] were similar to those of FSSC 12a NRRL 46705, with 98.58% identity for INECOL_BM-04 and 98.75% identity for INECOL_BM-06. Based on the results described above, these isolates were considered as members of the genus Fusarium belonging to the FSSC. To improve and establish their phylogenetic relationship with other species from FSSC, a multilocus sequence analysis based on the tef1, ITS, LSU and rpb2 sequences was performed. The Bayesian inferred phylogeny included 3127 bp characters from these four loci and from a total of 73 strains belonging to the three clearly distinguishable subclades from FSSC (Figure 1). The multilocus sequence typing revealed that both INECOL isolates belong to clade 3 but not to AFC, which is made up of F. ambrosium AF-1, F. euwallaceae AF-2, F. oligoseptatum AF-4, F. tuaranense AF-5, F. obliqueseptatum AF-7 and F. kuroshium AF-12. However, both INECOL isolates formed a highly supported clade showing the close relatedness among them and are related to the clade represented by F. macrosporum CBS 142424, F. spathulatum NRRL 28541 (FSSC 26), F. cyanescens CBS 518.82 (FSSC 27), F. ferrugineum NRRL 32427 (FSSC 28) and F. catenatum NRRL 54993 (FSSC 43).
Figure 1.
Cladogram of the phylogenetic relationship of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06 with members of the three clades of FSSC. Phylogeny constructed through Bayesian inference using a combined data set of four gene markers’ (tef1, ITS, LSU and the second-largest subunit of RNA polymerase II (rpb2)) sequences. The representative species of clades I, II and III are those proposed by [53]. Numerical designations referring to the informal nomenclature for phylogenetic species of FSSC (e.g., FSSC 1) are provided. The AFC is indicated. Support values from the Bayesian inference are indicated at the nodes. A star in the node indicates 100% support. INECOL isolates are highlighted.
3.2. The Two Closely Related Fusarium sp. INECOL_BM-04 and INECOL_BM-06 Isolated from X. morigerus Are Phenotypically Different
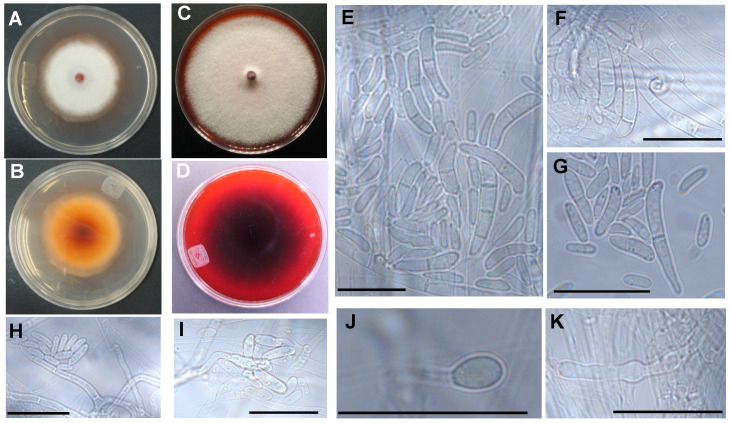
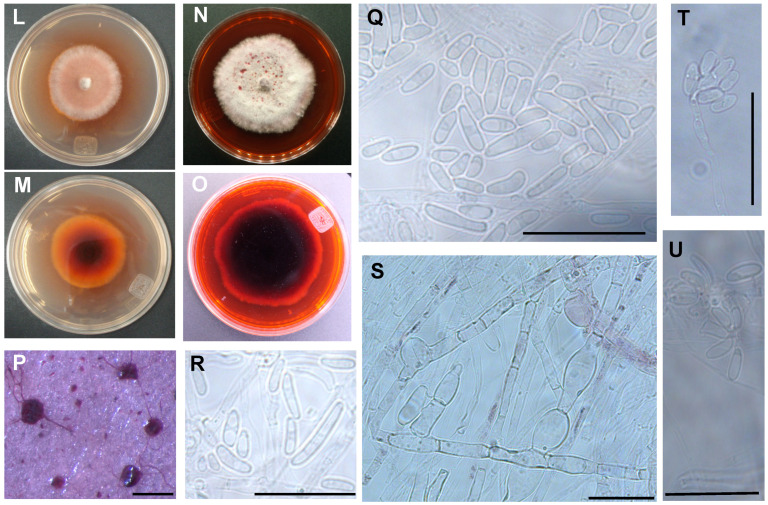
Fusarium sp. INECOL_BM-04 and INECOL_BM-06 showed differences in the morphology of their colonies. After 7 days of incubation in PDA, Fusarium sp. INECOL_BM-04 developed a dense white mycelium and the colony presented a smooth margin (Figure 2A,B). Meanwhile, Fusarium sp. INECOL_BM-06 showed aerial white mycelium in the periphery with a lower density in comparison with Fusarium sp. INECOL_BM-04. The colony also presented concentric rings of purplish mycelium and secreted a reddish compound shown by the pigmentation of the agar (Figure 2L,M). Both isolates presented reddish pigmentation in the reverse of the colony, although in different amounts (Figure 2B,M). At 14 days of incubation, Fusarium sp. INECOL_BM-04 developed a dense cottony mycelium with pale cream-white tonalities. The colony presented a smooth margin, and a strong moldy odor was detected (Figure 2C,D). Meanwhile, Fusarium sp. INECOL_BM-06 showed aerial white mycelium with a higher density and irregular margin of the colony in comparison with Fusarium sp. INECOL_BM-04. The colony presented reddish exudate droplets and a strong moldy odor (Figure 2N,O). Both isolates secreted an intense violet compound shown by the pigmentation of the agar (Figure 2D,O). The colony radial growth was statistically different since Fusarium sp. INECOL_BM-04 and INECOL_BM-06 presented mean colony diameters of 86.62 ± 1.77 mm and 66.51 ± 3.05 mm, respectively, at the end of 14 days of incubation.
Figure 2.
Morphological characteristics of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL BM-06 isolated from X. morigerus. Colony morphology in PDA at 7 and 14 days of incubation of Fusarium sp. INECOL_BM-04: (A,C) colony surface; (B,D) colony undersurface, and Fusarium sp. INECOL_BM-06; (L,N) colony surface; (M,O) colony undersurface. Microscopic characters of Fusarium sp. INECOL_BM-04: conidia (E–G), chlamydospores (J,K), and aerial conidiophores (H,I) in SNA or CLA; and Fusarium sp. INECOL_BM-06: sporodochia in PDA (P), conidia (Q,R), chlamydospores (S) and aerial conidiophores (T,U) in SNA or CLA. Photographs were taken at 10 dpi. Scale bar = 25 µm; scale bar in p = 1 mm.
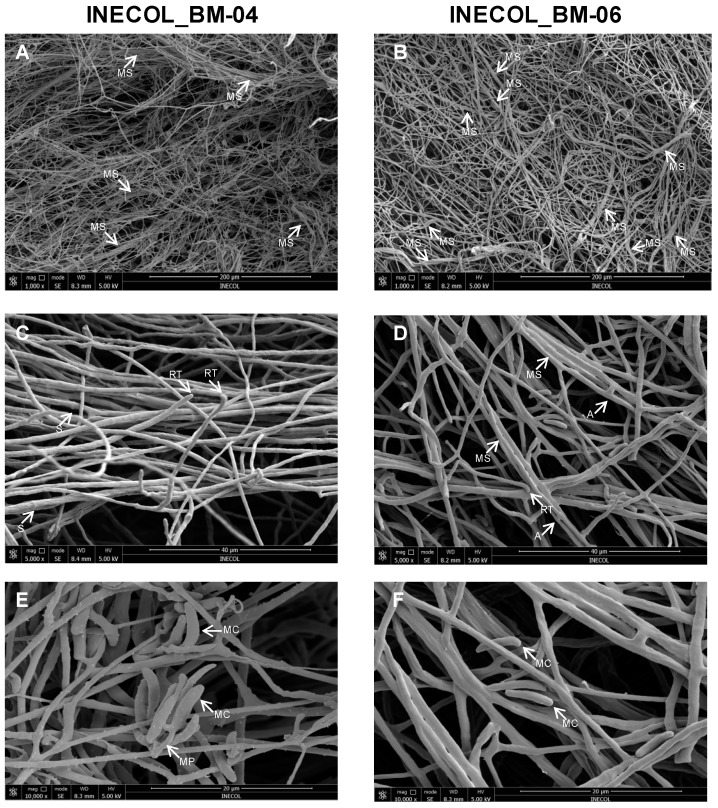
Microscopic characters were evaluated on SNA, CLA and PDA. Both isolates have septate branched hyphae with rounded tips (Figure 3); chlamydospores formed but were scarce, intercalary in or the terminal of the hyphae, mostly globose, single, hyaline, and smooth walled (Figure 2J,K,S). Purple sporodochia were observed in Fusarium sp. INECOL-BM-06 (Figure 2P) and absent in Fusarium sp. INECOL_BM-04 at 6 weeks of incubation in PDA. Aerial conidiophores were scarce, harboring aseptate microconidia-forming false heads (Figure 2H,I,T,U). There were also conidiophores arising from the substrate mycelium with a simple phialide-forming aseptate microconidia in Fusarium sp. INECOL-BM-06 (Figure 2Q,R) and 1–3 septate conidia in Fusarium sp. INECOL_BM-04 (Figure 2E–G). Aseptate microconidia were indistinguishable from aerial or substrate mycelium conidiophores. Microconidia from Fusarium sp. INECOL-BM-04 were hyaline and obovoid, reniform, oval or allantoid shaped, measuring 6.6 − (9.6 ± 1.8) − 17 × 2.1 − (3.1 ± 0.6) − 5.3 µm; in contrast, Fusarium sp. INECOL-BM-06 had hyaline-, obovoid-, oval-, fusiform- and allantoid-shaped and 5.6 − (9.1 ± 1.8) − 20.1 × 2.3 − (3.1 ± 0.4) − 4.2 µm-sized microconidia. The 1-3 septate conidia from Fusarium sp. INECOL_BM-04 were scarce in comparison with aseptate conidia and 15.8 − (22.2 ± 5) − 34.3 × 1.1 − (4.3 ± 0.8) − 6.6 µm sized. Fusarium sp. INECOL_BM-04 had higher production of conidia in comparison to Fusarium sp. INECOL_BM-06, 3.13 × 106 conidia (±1,102,065.94) and 1.39 × 106 conidia (±302,108.148) per colony, respectively. By means of SEM, a myceliar organization was more evident, composed of loosely organized hyphal filaments, hyphal aggregates, such as myceliar strands, and some interconnects by anastomosis (Figure 3); our observations suggest that Fusarium sp. INECOL_BM-06 develop a more robust network since the myceliar strands and anastomosis are more evident in this species (Figure 3).
Figure 3.
Microscopic features of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06, evaluated by scanning electron microscopy (SEM). (A,B) Micrographs taken at 1000× magnification. (C,D) Micrographs taken at 5000× magnification. (E,F) Micrographs taken at 10,000× magnification. MS: myceliar strand; S: septum; RT: rounded tip; A: anastomosis; MP: monophialide; MC: microconidia.
Based on the molecular and morphological characters, these two isolates associated with Xylosandrus morigerus can be considered to be closely related, belonging to the Fusarium genus, specifically to the FSSC but not to AFC.
3.3. Phytopathogenicity Screening Shows Differences in Virulence among Fusarium spp. Associated with Xylosandrus morigerus
To probe whether Fusarium spp. isolated from X. morigerus exhibits phytopathogenic behavior, we implemented various pathosystems using a wide range of hosts, including agronomical species, principally those that are important in Veracruz, Mexico, such as Coffea arabica, which is a known host of X. morigerus, and species of Citrus, as well as forest species that are potential hosts of X. morigerus. Both isolates provoked disease symptoms in all the plant species used; however, we observed important differences between assays and the Fusarium isolates.
3.3.1. Pathogenesis Assays in Coffea arabica
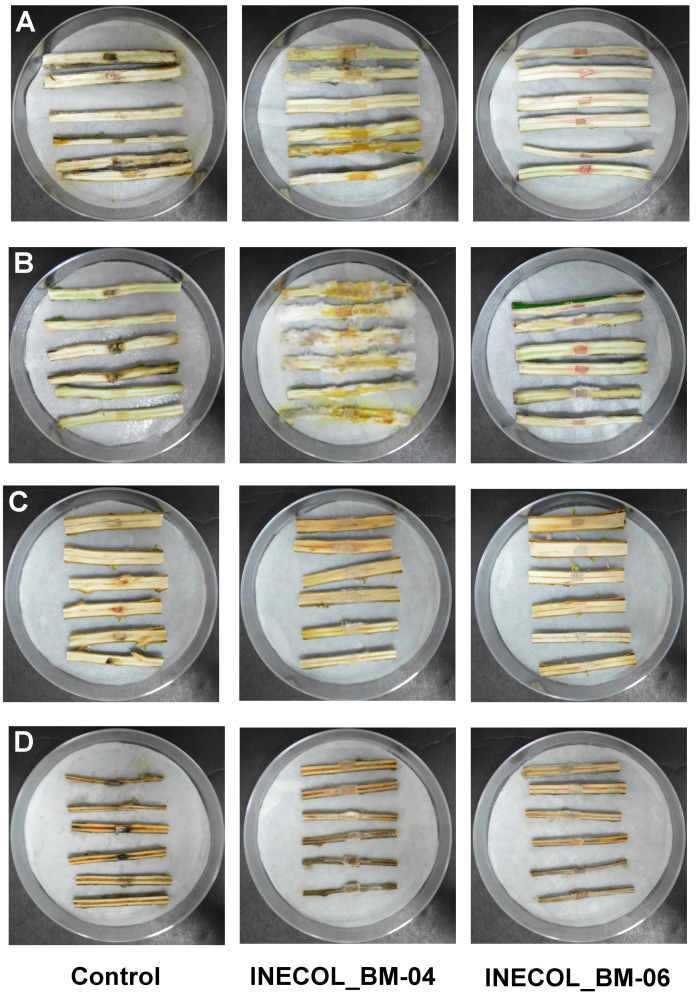
Leaves and stems of C. arabica cv. Marsellesa and cv. Oro azteca were challenged with Fusarium spp. (Figure 4). Both isolates provoked infection symptoms of necrosis, discreetly; however, the damage in leaves of C. arabica cv. Marsellesa inoculated with INECOL_BM-04 was statistically higher in comparison with those inoculated with INECOL_BM-06, with 4.20% (±1.25) and 2.53% (±0.79) of leaf surface being affected by each isolate. Meanwhile, the tissue damage provoked by both isolates in C. arabica cv. Oro azteca was similar, Fusarium sp. INECOL_BM-04 affected 3.67% (±0.65) and Fusarium sp. INECOL_BM-06 damaged 3.35% (±0.70) of the leaf surface (Figure S1). On the other hand, the stems seem to be resistant to Fusarium spp., since no differences were detected in comparison with control stems after 21 dpi (Figure 4).
Figure 4.
Pathogenicity screening of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06 isolates in leaves and vascular tissue of Coffea arabica. (A) C. arabica cv. Marsellesa (B) C. arabica cv. Oro azteca. Photographs were taken at 21 dpi.
3.3.2. Pathogenesis Assays in Important Citrus Species, Citrus latifolia and Citrus sinensis
In leaves of the agronomical species C. latifolia and C. sinensis, both isolates triggered clear infection symptoms characterized by chlorosis in the zone of the principal vein and brownish necrosis at 12 dpi. Citrus sinensis is more susceptible to Fusarium spp. associated with X. morigerus infection since the symptoms were more pronounced in this citrus species (Figure 5A,B). Interestingly, in C. latifolia, the infection by Fusarium sp. INECOL_BM-04 progressed significantly faster than the infection by Fusarium sp. INECOL_BM-06, and the expansion of tissue damage triggered by Fusarium sp. INECOL_BM-04 was approximately four times more than that damage caused by Fusarium sp. INECOL_BM-06 (Figure S1). In C. sinensis, no significant differences were observed in the expansion of tissue damage (Figure S1), but the intensity of symptoms differed; the leaves inoculated with Fusarium sp. INECOL_BM-04 show a bigger necrosed area than those inoculated with Fusarium sp. INECOL-BM-06; however, they present chlorosis, also considered a symptom of infection (Figure 5A,B). Regarding the infection of stems, only Fusarium sp. INECOL_BM-04 caused rot, indicated by an amber discoloration; this symptom was more pronounced in C. sinensis. Additionally, we clearly observed the presence of white cottony mycelium over the stem tissue. Fusarium sp. INECOL_BM-06 did not produce a significant fungal mass nor disease symptoms in both citrus species (Figure 6A,B).
Figure 5.
Pathogenicity screening of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06 in leaves of citrus and forest species. (A) Citrus latifolia. (B) Citrus sinensis. (C) Salix lasiolepis. (D) Populus nigra. The photographs were taken at 12 dpi for (A–D) and 8 dpi for (B).
Figure 6.
Pathogenicity screening of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06 in vascular tissue of citrus and forest species. (A) Citrus latifolia. (B) Citrus sinensis. (C) Salix lasiolepis. (D) Populus nigra. The photographs were taken at 12 dpi.
3.3.3. Pathogenesis Assays of Salix lasiolepis and Populus nigra
The pathogenicity tests in leaves of S. lasiolepis and P. nigra exhibited differences in both pathogenicity and host susceptibility (Figure 5C,D). In S. lasiolepis, both isolates provoked statistically similar effects as the mean percentage of the tissue damage was 13% (±2.73 for Fusarium sp. INECOL-BM-04 and ±2.46 for Fusarium sp. INECOL-BM-06) (Figure S1) and the main symptom was necrosis in the inoculation zone and beyond (Figure 5C). In contrast, for P. nigra, a statistically significant difference between the pathogenicity of both isolates was noticed, as Fusarium sp. INECOL-BM-06 triggered 19.04% (±5.85) tissue damage in comparison with the 7.30% (±2.13) tissue damage provoked by Fusarium sp. INECOL_BM-04 (Figure S1). The necrosis area was accompanied by an accentuated chlorosis in the P. nigra leaves infected with Fusarium sp. INECOL_BM-06; on the other hand, the leaves infected with Fusarium sp. INECOL_BM-04 did not show evident chlorosis symptoms (Figure 5D). With respect to the vascular tissue infection, in both plants there was a development of mycelium (Figure 6C,D); the inoculated stems of S. lasiolepis and P. nigra showed slight necrosis in vascularity in comparison with their respective controls, being accentuated in those inoculated with Fusarium sp. INECOL_BM-04 (Figure 6C,D); however, there are no significant differences in tissue damage.
3.4. Extracellular Protease Activity Is Slightly Different among Fusarium spp. Associated with Xylosandrus morigerus
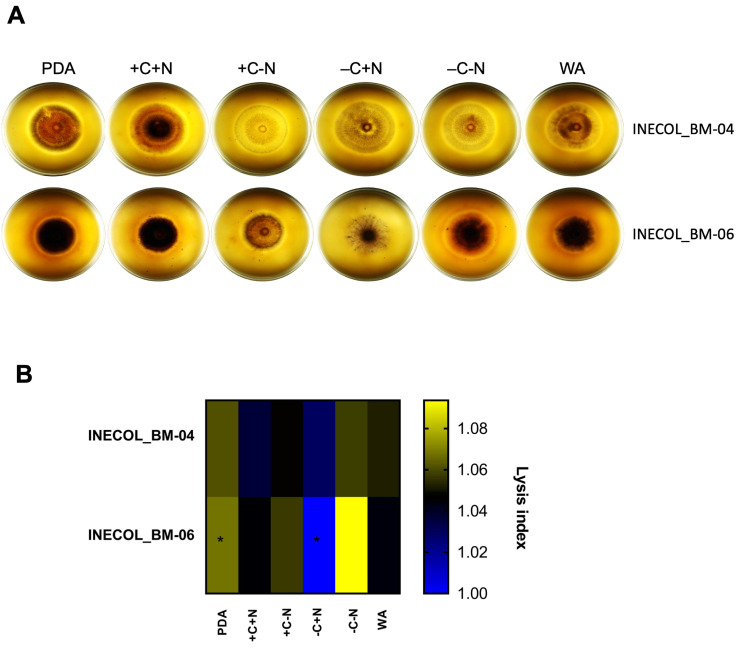
Secreted proteases contribute to fungal virulence either for the degradation of host tissue, facilitating penetration, or for the acquisition of nutrients. Protease secretion, observed as a lysis halo in the plates, was evaluated and compared in different nutritional conditions in both Fusarium isolates associated with X. morigerus (Figure 7A). Fusarium sp. INECOL_BM-04 secretes proteases in all the tested conditions and the quantities of secreted proteases seem to be similar; however, it secretes higher amounts of proteases in the C-rich condition (PDA) and in the absence of C and N sources (−C−N) (Figure 7B);. A smaller amount of secreted proteases was found in both culture medium with the presence of an N source, +C+N and −C+N (Figure 7B). In contrast, Fusarium sp. INECOL_BM-06 is clearly the null lysis halo in the absence of a C source (−C+N) compared with the rest of conditions (Figure 7A,B); thus, growing in the −C+N condition has a repressive effect on protease secretion in this species, while the absence of both C and N sources (−C−N) and the C-rich medium PDA have an inductive effect, followed by the +C−N condition (Figure 7A,B). For both isolates, higher amounts of secreted proteases were observed when there was no N source.
Figure 7.
Extracellular Protease activity of Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06. (A) Protease activity of Fusarium spp. associated with X. morigerus indicated by clearing zones in different culture media supplemented with low-fat powder milk 3%. (B) Heat Map of Protease secretion index (normalized by colony diameter) of Fusarium spp. associated with X. morigerus grown in different culture media supplemented with low-fat powder milk 3%. Heat Map shows the average of three technical replicates. Data obtained 7 dpi. (* p-value < 0.05 obtained by one-way ANOVA with Bonferroni multiple comparison calculation). Lysis index = 1 indicates no Lysis.
4. Discussion
Bark and ambrosia beetles have emerged as study models in phytopathology, since invasive species have changed their behavior by attacking living and healthy trees, provoking serious phytosanitary problems [30,31,32,54]. In addition, many examples of tree diseases provoked by the fungal mutualist are reported [31,32]. Therefore, it is important to gain a comprehensive understanding of the contribution of the associated fungi, in terms of their pathogenicity, for the development of plant disease.
Xilosandrus morigerus should be considered as a potential quarantine pest; its introduction and establishment in non-native habitats are facilitated by its regular inbreeding, males mating with their sisters within the parental gallery before dispersal; haplodiploidy, females that are not inseminated by a brother before leaving the nest can mate with a haploid son produced by an unfertilized egg; and by a broad host range stimulated by their mutualist fungus [29,55]. Xilosandrus morigerus has a nearly pantropical distribution. In Latin America, its distribution stretches from Veracruz, Mexico, to Brazil; this ambrosia beetle is able to attack a wide variety of host plants, and therefore it is considered as an important pest of crop and ornamental trees. It is called a “brown coffee twig borer” since it is a well-known pest of Coffea canephora and C. arabica but it can also attack Persea americana, Theobroma cacao, and Salix humboldtiana, among others [55].
For this work, specimens of laboratory-reared X. morigerus were used. Originally, the beetles were captured in the nature reserve Jaguaroundi Park for wildlife conservation in Veracruz State, Mexico. The phylogenetic analysis, based on four genetic markers, ITS, LSU, tef1 and rpb2, showed that the Fusarium spp. associated with X. morigerus belong to the FSSC and not to the AFC. The fact that INECOL isolates form an independent clade suggests that they can be classified as conspecific isolates of a new Fusarium species within FSSC associated with X. morigerus; however, future work needs to be carried out in order to define new species and it will be interesting analyze a higher number of isolates to evaluate the extent of genetic variation within the species [56,57].
Fusarium spp. associated with X. morigerus INECOL_BM-04 and –06 are phylogenetically related to Fusarium species, which are little described and morphologically different. Only F. macrosporum CBS 142424 is related to plants since it was isolated from C. sinensis crown; the rest are related to animals. F. spathulatum NRRL 28541 (FSSC 26), F. cyanescens CBS 518.82 (FSSC 27) and F. ferrugineum NRRL 32427 (FSSC 28) were isolated from human tissues; F. catenatum NRRL 54993 (FSSC 43) was isolated from Stegostoma fasciatum (zebra shark) tissues [58]. Fusarium species have been identified as symbiotes of Xylosandrus species and pathogenic for the plant host based on pathogenic tests or because the recovery of the fungus was from infected plant tissue. In Hawaii, X. compactus inoculated F. solani in Acacia koa but virulence assays showed that F. solani was not pathogenic [59,60,61]. In Italy, F. solani was isolated from entry holes, galleries and stained woody tissues of Quercus ilex attacked by X. compactus, and a pathogenicity test showed the symptoms exhibited by the naturally infected plants [62]; in a National Park in Lazio, X. crassiusculus was trapped when attacking Ceratonia siliqua, and F. solani was isolated from necrotic tissue observed in the proximity of the entry holes [24]. In addition, Fusarium spp. were found to be associated with X. compactus in the Mediterranean maquis [63]. In New York apple orchards, X. germanus was claimed to be the agent that caused damage; F. solani was isolated from unsterilized insect bodies, internal contents and infested wood surrounding the entry hole, but the authors attributed the wilting symptoms observed in the trees to the tunneling of the insects [23]. Interestingly, our results suggest that other Fusarium species, especially those belonging to FSSC, aside from F. solani could be mutualists of Xylosandrus spp.
Our analyses showed that even Fusarium spp. associated with X. morigerus INECOL_BM-04 and –06 are closely genetically related isolates, and they exhibit a particular pathogenic profile and specific morphological traits; these differences can be explained by intraspecific trait variation that can be driven at the gene expression regulation level in response to specific conditions [64,65]. The possible variances in gene regulation can impact how the pathogen responds to a specific host and reprograms the expression of its pathogenic arsenal, i.e., secreted proteases, toxins, and secondary metabolites, among others. Here, we compared the extracellular protease activity of both Fusarium isolates associated with X. morigerus. Specifically, the nutrition status was shown to regulate the expression of secreted proteases. For both isolates, higher amounts of secreted proteases were found in the absence of an N source. However, nutrition heterogeneity, as denoted by the profiles of protease activity among them, can impact the virulence in a species-, strain- and host-dependent manner, as reported for Aspergillus nidulans and others [66,67,68]. On the other hand, it is interesting that some phenotypical differences are evident in habitual culture conditions such as PDA, which suggests that some regulatory differences among the strains are sharp and do not need special conditions highlight; thus, the gene expression could be regulated differentially in both strains “normally”, and this has repercussions on the phenotype.
As part of an initial approach of pre-invasion assessment, we analyzed the pathogenic behavior of Fusarium sp. INECOL_BM-04 and -06 in the leaves and stems of agricultural and ecological representative species. The pathogenicity assays suggest that both isolates are pathogenic for most of the plant species tested; however, the extent of the lesion for most of the leaves was less than 20% of the leaf surface and there was no greater damage in the vascular tissue. Both cultivars of C. arabica, were agronomically selected for their high yield and resistance to coffee rust, which can contribute to their resistance to Fusarium spp. Meanwhile, the damage in S. lasiolepis and P. nigra was higher, showing mayor susceptibility. Nevertheless, the results suggest that both isolates of Fusarium associated with X. morigerus do not represent a threat to C. arabica, S. lasiolepis and P. nigra, but both isolates, especially Fusarium sp. INECOL_BM-04 can be considered as minor pathogens for Citrus species, given the damaged surface of the leaves and the colonization of the vascular tissue. Interestingly, most of the fungal associates of ambrosia beetles displayed no significant impact on the host during a pre-invasion assessment assay [36] but the possibility of the fungus being more virulent in other conditions was not tested, and so cannot be disregarded.
Traditionally, it has been considered that an ambrosia beetle has a single, primary fungal mutualist, but now it is known that most ambrosia beetles carry, and probably feed on, multiple fungal species [6,69]. Additionally, vector shifts have been reported for some fungal mutualist/symbiotes. In addition to X. glabratus, Raffaelea lauricola was recovered from other ambrosia beetle species, including X. crassiusculus [70]. Even, Fusarium spp. has been associated with other insects, beyond Xylosandrus spp. and Euwallacea spp., and with the decline of forest and agricultural species [71,72]. Given these dynamic associations between beetles and fungal mutualists and varying fidelity, studies to determine the mycobiota of bark and ambrosia beetles, by DNA metabarcoding, have been conducted [20,21,35,73,74]. Altogether, these data show the high diversity of fungal–beetle associations and the probability of being a vector of phytopathogens in diverse ecological niches, even without being the main fungal mutualist. In addition to the factors stated before, the variation in trade level among world regions [75] and the influence of spatial and climatic factors in the abundance and dispersal behavior of invasive ambrosia beetle [76] highlight the relevance of studying ambrosia complexes to determine their potential as forest and agricultural threats and, therefore, the implementation of permanent control and surveillance measures worldwide.
5. Conclusions
In this study, we report the molecular identification and phenotypical characterization of two fungal isolates from the ambrosia beetle Xylosandrus morigerus. The isolates INECOL_BM-04 and INECOL-BM-06 are members of the Fusarium genus, belonging to the Fusarium solani species complex (FSSC) but not to the Ambrosia Fusarium Clade (AFC), with pathogenic potential with regard to forest and agricultural species. The results highlight the possibility of X. morigerus being a vector of phytopathogens.
Acknowledgments
The authors want to thank Mónica Ramírez-Vázquez for providing access to the Advanced Microscopy Unit at INECOL and are also grateful to Greta Hanako Rosas-Saito for her kind and efficient technical support with SEM images. Additionally, we are grateful to Jire A. Muñoz Jaimes for her technical assistance in fungal growth protocols.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8030231/s1, Figure S1: Percentage of tissue damage in leaves of Coffea arabica, Citrus latifolia, Citrus sinensis, Salix lasiolepis and Populus nigra inoculated with Fusarium sp. INECOL_BM-04 and Fusarium sp. INECOL_BM-06.
Author Contributions
Conceptualization, D.S.-R.; formal analysis, N.C.-V., E.I.-L., E.V. and D.S.-R.; investigation, N.C.-V., J.B.R.-H., L.A.M.-R., A.J.P.-L., E.V., A.P.C.-D., L.A.I.-J. and E.D.C.-H.; resources, E.I.-L. and D.S.-R.; writing—original draft preparation, N.C.-V. and D.S.-R.; writing—review and editing, N.C.-V. and D.S.-R.; project administration, D.S.-R.; funding acquisition, D.S.-R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Consejo Nacional de Ciencia y Tecnología (Conacyt) through the Fondo Institucional de Fomento Regional para el Desarrollo Científico, Tecnológico y de Innovación (Fordecyt) Grant 292399 (currently named Programa Presupuestario F003).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequences of the four markers for INECOL_BM-04 and INECOL_BM-06 isolates were submitted to GenBank with the following accessions: OM455454, OM455455, OM455456, OM455457, OM455458, OM455459, OM455460 and OM455461. The four-locus data set and the Bayesian Inference (BI) tree are public available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S29320?x-access-code=7d6edd9c73c50b876654dd26dec6e00e&format=html, accessed on 24 January 2022).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vega F.E., Biedermann P.H.W. On Interactions, Associations, Mycetangia, Mutualists and Symbiotes in Insect-Fungus Symbioses. Fungal Ecol. 2020;44:100909. doi: 10.1016/j.funeco.2019.100909. [DOI] [Google Scholar]
- 2.Lange L., Grell M.N. The Prominent Role of Fungi and Fungal Enzymes in the Ant–Fungus Biomass Conversion Symbiosis. Appl. Microbiol. Biotechnol. 2014;98:4839–4851. doi: 10.1007/s00253-014-5708-5. [DOI] [PubMed] [Google Scholar]
- 3.Da Costa R., Hu H., Li H., Poulsen M. Symbiotic Plant Biomass Decomposition in Fungus-Growing Termites. Insects. 2019;10:87. doi: 10.3390/insects10040087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menezes C., Vollet-Neto A., Marsaioli A.J., Zampieri D., Fontoura I.C., Luchessi A.D., Imperatriz-Fonseca V.L. A Brazilian Social Bee Must Cultivate Fungus to Survive. Curr. Biol. 2015;25:2851–2855. doi: 10.1016/j.cub.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 5.Paludo C.R., Menezes C., Silva-Junior E.A., Vollet-Neto A., Andrade-Dominguez A., Pishchany G., Khadempour L., do Nascimento F.S., Currie C.R., Kolter R., et al. Stingless Bee Larvae Require Fungal Steroid to Pupate. Sci. Rep. 2018;8:1122. doi: 10.1038/s41598-018-19583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hulcr J., Stelinski L.L. The Ambrosia Symbiosis: From Evolutionary Ecology to Practical Management. Annu. Rev. Entomol. 2017;62:285–303. doi: 10.1146/annurev-ento-031616-035105. [DOI] [PubMed] [Google Scholar]
- 7.Biedermann P.H.W., Vega F.E. Ecology and Evolution of Insect–Fungus Mutualisms. Annu. Rev. Entomol. 2020;65:431–455. doi: 10.1146/annurev-ento-011019-024910. [DOI] [PubMed] [Google Scholar]
- 8.Johnson A.J., McKenna D.D., Jordal B.H., Cognato A.I., Smith S.M., Lemmon A.R., Lemmon E.M., Hulcr J. Phylogenomics Clarifies Repeated Evolutionary Origins of Inbreeding and Fungus Farming in Bark Beetles (Curculionidae, Scolytinae) Mol. Phylogenet. Evol. 2018;127:229–238. doi: 10.1016/j.ympev.2018.05.028. [DOI] [PubMed] [Google Scholar]
- 9.Kirkendall L.R., Biedermann P.H.W., Jordal B.H. Bark Beetles. Elsevier; Amsterdam, The Netherlands: 2015. Evolution and Diversity of Bark and Ambrosia Beetles; pp. 85–156. [Google Scholar]
- 10.Huang Y.-T., Skelton J., Hulcr J. Lipids and Small Metabolites Provisioned by Ambrosia Fungi to Symbiotic Beetles Are Phylogeny-Dependent, Not Convergent. ISME J. 2020;14:1089–1099. doi: 10.1038/s41396-020-0593-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Six D.L. Ecological and Evolutionary Determinants of Bark Beetle—Fungus Symbioses. Insects. 2012;3:339–366. doi: 10.3390/insects3010339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skelton J., Johnson A.J., Jusino M.A., Bateman C.C., Li Y., Hulcr J. A Selective Fungal Transport Organ (Mycangium) Maintains Coarse Phylogenetic Congruence between Fungus-Farming Ambrosia Beetles and Their Symbionts. Proc. R. Soc. B Biol. Sci. 2019;286:20182127. doi: 10.1098/rspb.2018.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman J.J. The F Usarium Solani Species Complex: Ubiquitous Pathogens of Agricultural Importance. Mol. Plant Pathol. 2016;17:146–158. doi: 10.1111/mpp.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aoki T., Liyanage P.N.H., Konkol J.L., Ploetz R.C., Smith J.A., Kasson M.T., Freeman S., Geiser D.M., O’Donnell K. Three Novel Ambrosia Fusarium Clade Species Producing Multiseptate “Dolphin-Shaped” Conidia, and an Augmented Description of Fusarium Kuroshium. Mycologia. 2021;113:1089–1109. doi: 10.1080/00275514.2021.1923300. [DOI] [PubMed] [Google Scholar]
- 15.Aoki T., Smith J.A., Kasson M.T., Freeman S., Geiser D.M., Geering A.D.W., O’Donnell K. Three novel Ambrosia Fusarium Clade species producing clavate macroconidia known (F. floridanum and F. obliquiseptatum) or predicted (F. tuaranense) to be farmed by Euwallacea spp. (Coleoptera: Scolytinae) on woody hosts. Mycologia. 2019;111:919–935. doi: 10.1080/00275514.2019.1647074. [DOI] [PubMed] [Google Scholar]
- 16.Aoki T., Kasson M.T., Berger M.C., Freeman S., Geiser D.M., O’Donnell K. Fusarium oligoseptatum sp. nov., a mycosymbiont of the ambrosia beetle Euwallacea validus in the Eastern U.S. and typification of F. ambrosium. Fungal Syst. Evol. 2018;1:23–39. doi: 10.3114/fuse.2018.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasson M.T., O’Donnell K., Rooney A.P., Sink S., Ploetz R.C., Ploetz J.N., Konkol J.L., Carrillo D., Freeman S., Mendel Z., et al. An Inordinate Fondness for Fusarium: Phylogenetic Diversity of Fusaria Cultivated by Ambrosia Beetles in the Genus Euwallacea on Avocado and Other Plant Hosts. Fungal Genet. Biol. 2013;56:147–157. doi: 10.1016/j.fgb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Lynn K.M.T., Wingfield M.J., Durán A., Oliveira L.S.S., de Beer Z.W., Barnes I. Novel Fusarium mutualists of two Euwallacea species infesting Acacia crassicarpa in Indonesia. Mycologia. 2021;113:536–558. doi: 10.1080/00275514.2021.1875708. [DOI] [PubMed] [Google Scholar]
- 19.O’Donnell K., Sink S., Libeskind-Hadas R., Hulcr J., Kasson M.T., Ploetz R.C., Konkol J.L., Ploetz J.N., Carrillo D., Campbell A., et al. Discordant Phylogenies Suggest Repeated Host Shifts in the Fusarium–Euwallacea Ambrosia Beetle Mutualism. Fungal Genet. Biol. 2015;82:277–290. doi: 10.1016/j.fgb.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Malacrinò A., Rassati D., Schena L., Mehzabin R., Battisti A., Palmeri V. Fungal Communities Associated with Bark and Ambrosia Beetles Trapped at International Harbours. Fungal Ecol. 2017;28:44–52. doi: 10.1016/j.funeco.2017.04.007. [DOI] [Google Scholar]
- 21.Morales-Rodríguez C., Sferrazza I., Aleandri M.P., Dalla Valle M., Speranza S., Contarini M., Vannini A. The Fungal Community Associated with the Ambrosia Beetle Xylosandrus Compactus Invading the Mediterranean Maquis in Central Italy Reveals High Biodiversity and Suggests Environmental Acquisitions. Fungal Biol. 2021;125:12–24. doi: 10.1016/j.funbio.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Skelton J., Jusino M.A., Li Y., Bateman C., Thai P.H., Wu C., Lindner D.L., Hulcr J. Detecting Symbioses in Complex Communities: The Fungal Symbionts of Bark and Ambrosia Beetles Within Asian Pines. Microb. Ecol. 2018;76:839–850. doi: 10.1007/s00248-018-1154-8. [DOI] [PubMed] [Google Scholar]
- 23.Agnello A.M., Breth D.I., Tee E.M., Cox K.D., Villani S.M., Ayer K.M., Wallis A.E., Donahue D.J., Combs D.B., Davis A.E., et al. Xylosandrus Germanus (Coleoptera: Curculionidae: Scolytinae) Occurrence, Fungal Associations, and Management Trials in New York Apple Orchards. J. Econ. Entomol. 2017;110:2149–2164. doi: 10.1093/jee/tox189. [DOI] [PubMed] [Google Scholar]
- 24.Francardi V., Noal A., Francescato S., Pinto R., Bruni A., Loffredi L., Bucini D., Guarnieri D., Bellantuono M., Esposito N., et al. Coexistence of Xylosandrus Crassiusculus (Motschulsky) and X. Compactus (Eichhoff) (Coleoptera Curculionidae Scolytinae) in the National Park of Circeo (Lazio, Italy) Redia-G. Zool. 2017;100:149–155. doi: 10.19263/REDIA-100.17.19. [DOI] [Google Scholar]
- 25.Dixon W., Woodruff R., Foltz J. Black Twig Borer, Xylosandrus Compactus (Eichhoff) (Insecta: Coleoptera: Curculionidae: Scolytinae) Fla. Coop. Ext. Serv. IFAS Univ. Fla. 2003;EENY 311:1–5. [Google Scholar]
- 26.Hara A., Beardsley J. The Biology of the Black Twig Borer, Xylosandrus Compactus (Eichhoff), in Hawaii. Proc. Hawaii Ent. Soc. 1979;1:55–70. [Google Scholar]
- 27.Dole S.A., Beaver R.A. A Review of the Australian Species of Xylosandrus Reitter (Coleoptera: Curculionidae: Scolytinae) Coleopt. Bull. 2008;62:481–492. doi: 10.1649/1108.1. [DOI] [Google Scholar]
- 28.Stephen L.W., Bright D.E. A Catalog of Scolytidae and Platypodidae (Coleoptera), Part 2: Taxonomic Index. Volume B. Gt. Basin Nat. Mem. 1992;13:835–1557. [Google Scholar]
- 29.Andersen H.F., Jordal B.H., Kambestad M., Kirkendall L.R. Improbable but True: The Invasive Inbreeding Ambrosia Beetle Xylosandrus Morigerus Has Generalist Genotypes. Ecol. Evol. 2012;2:247–257. doi: 10.1002/ece3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranger C.M., Schultz P.B., Frank S.D., Chong J.H., Reding M.E. Non-Native Ambrosia Beetles as Opportunistic Exploiters of Living but Weakened Trees. PLoS ONE. 2015;10:e0131496. doi: 10.1371/journal.pone.0131496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hulcr J., Dunn R.R. The Sudden Emergence of Pathogenicity in Insect–Fungus Symbioses Threatens Naive Forest Ecosystems. Proc. R. Soc. B Biol. Sci. 2011;278:2866–2873. doi: 10.1098/rspb.2011.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ploetz R.C., Hulcr J., Wingfield M.J., de Beer Z.W. Destructive Tree Diseases Associated with Ambrosia and Bark Beetles: Black Swan Events in Tree Pathology? Plant Dis. 2013;97:856–872. doi: 10.1094/PDIS-01-13-0056-FE. [DOI] [PubMed] [Google Scholar]
- 33.Harrington T.C., McNew D., Mayers C., Fraedrich S.W., Reed S.E. Ambrosiella Roeperi Sp. Nov. Is the Mycangial Symbiont of the Granulate Ambrosia Beetle, Xylosandrus Crassiusculus. Mycologia. 2014;106:835–845. doi: 10.3852/13-354. [DOI] [PubMed] [Google Scholar]
- 34.Mayers C.G., McNew D.L., Harrington T.C., Roeper R.A., Fraedrich S.W., Biedermann P.H.W., Castrillo L.A., Reed S.E. Three Genera in the Ceratocystidaceae Are the Respective Symbionts of Three Independent Lineages of Ambrosia Beetles with Large, Complex Mycangia. Fungal Biol. 2015;119:1075–1092. doi: 10.1016/j.funbio.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 35.Rassati D., Marini L., Malacrinò A. Acquisition of Fungi from the Environment Modifies Ambrosia Beetle Mycobiome during Invasion. PeerJ. 2019;7:e8103. doi: 10.7717/peerj.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Bateman C., Skelton J., Wang B., Black A., Huang Y.-T., Gonzalez A., Jusino M.A., Nolen Z.J., Freeman S., et al. Pre-Invasion Assessment of Exotic Bark Beetle-Vectored Fungi to Detect Tree-Killing Pathogens. Phytopathology. 2021;112:261–270. doi: 10.1094/PHYTO-01-21-0041-R. [DOI] [PubMed] [Google Scholar]
- 37.Barrera J.F., Villacorta A., Herrera J. Fluctuación Estacional de Las Capturas de La “Broca Del Café” (Hypothenemus Hampei) Con Trampas de Etanol—Metanol e Implicaciones Sobre El Número de Trampas. Entomol. Mex. 2004;3:540–544. [Google Scholar]
- 38.Biedermann P.H.W., Klepzig K.D., Taborsky M. Fungus Cultivation by Ambrosia Beetles: Behavior and Laboratory Breeding Success in Three Xyleborine Species. Environ. Entomol. 2009;38:1096–1105. doi: 10.1603/022.038.0417. [DOI] [PubMed] [Google Scholar]
- 39.Stephen L.W. The Bark and Ambrosia Beetles of North and Central America (Coleoptera: Scolytidae), a Taxonomic Monograph. Gt. Basin Nat. Mem. 1982;6:1–1359. [Google Scholar]
- 40.Tapia-Tussell R., Lappe P., Ulloa M., Quijano-Ramayo A., Cáceres-Farfán M., Larqué-Saavedra A., Perez-Brito D. A Rapid and Simple Method for DNA Extraction from Yeasts and Fungi Isolated from Agave Fourcroydes. Mol. Biotechnol. 2006;33:67–70. doi: 10.1385/MB:33:1:67. [DOI] [PubMed] [Google Scholar]
- 41.White T.J., Bruns T., Lee S.J.W.T., Taylor J. PCR Protocols: A Guide to Methods and Applications. Academic Press; New York, NY, USA: 1990. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; pp. 315–322. [Google Scholar]
- 42.Crous P.W., Lombard L., Sandoval-Denis M., Seifert K.A., Schroers H.-J., Chaverri P., Gené J., Guarro J., Hirooka Y., Bensch K., et al. Fusarium: More than a Node or a Foot-Shaped Basal Cell. Stud. Mycol. 2021;98:100116. doi: 10.1016/j.simyco.2021.100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geiser D.M., Al-Hatmi A., Aoki T., Arie T., Balmas V., Barnes I., Bergstrom G.C., Bhattacharyya M.K.K., Blomquist C.L., Bowden R., et al. Phylogenomic Analysis of a 55.1 Kb 19-Gene Dataset Resolves a Monophyletic Fusarium That Includes the Fusarium Solani Species Complex. Phytopathology. 2021;111:1064–1079. doi: 10.1094/PHYTO-08-20-0330-LE. [DOI] [PubMed] [Google Scholar]
- 44.Sandoval-Denis M., Lombard L., Crous P.W. Back to the Roots: A Reappraisal of Neocosmospora. Persoonia-Mol. Phylogeny Evol. Fungi. 2019;43:90–185. doi: 10.3767/persoonia.2019.43.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X Version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 46.Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 48.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 49.Yu G., Lam T.T.-Y., Zhu H., Guan Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Mol. Biol. Evol. 2018;35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang L.-G., Lam T.T.-Y., Xu S., Dai Z., Zhou L., Feng T., Guo P., Dunn C.W., Jones B.R., Bradley T., et al. Treeio: An R Package for Phylogenetic Tree Input and Output with Richly Annotated and Associated Data. Mol. Biol. Evol. 2020;37:599–603. doi: 10.1093/molbev/msz240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blackwell Publishing . In: The Fusarium Laboratory Manual. Leslie J.F., Summerell B.A., editors. Blackwell Publishing; Ames, IW, USA: 2006. [Google Scholar]
- 52.Bozzola J.J., Russell L.D. Electron Microscopy: Principles and Techniques for Biologists. Jones and Bartlett; Location, UK: 1999. Jones and Bartlett Series in Biology. [Google Scholar]
- 53.O’Donnell K. Molecular Phylogeny of the Nectria Haematococca-Fusarium Solani Species Complex. Mycologia. 2000;92:919. doi: 10.2307/3761588. [DOI] [Google Scholar]
- 54.Gugliuzzo A., Criscione G., Tropea Garzia G. Unusual Behavior of Xylosandrus Compactus (Coleoptera: Scolytinae) on Carob Trees in a Mediterranean Environment. Insects. 2019;10:82. doi: 10.3390/insects10030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Invasive Species Compedium. CAB International; Wallingford, UK: 2022. CABI Xylosandrus Morigerus. [Google Scholar]
- 56.Summerell B.A. Resolving Fusarium: Current Status of the Genus. Annu. Rev. Phytopathol. 2019;57:323–339. doi: 10.1146/annurev-phyto-082718-100204. [DOI] [PubMed] [Google Scholar]
- 57.Maharachchikumbura S.S.N., Chen Y., Ariyawansa H.A., Hyde K.D., Haelewaters D., Perera R.H., Samarakoon M.C., Wanasinghe D.N., Bustamante D.E., Liu J.-K., et al. Integrative Approaches for Species Delimitation in Ascomycota. Fungal Divers. 2021;109:155–179. doi: 10.1007/s13225-021-00486-6. [DOI] [Google Scholar]
- 58.Sandoval-Denis M., Crous P.W. Removing Chaos from Confusion: Assigning Names to Common Human and Animal Pathogens in Neocosmospora. Persoonia Mol. Phylogeny Evol. Fungi. 2018;41:109–129. doi: 10.3767/persoonia.2018.41.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daehler C.C., Dudley N. Impact of the Black Twig Borer, an Introduced Insect Pest, on Acacia Koa in the Hawaiian Islands. Micronesica Suppl. 2002;6:35–53. [Google Scholar]
- 60.Dudley N., James R., Sniezko R., Yeh A. Pathogenicity of Four Fusarium Species on Acacia Koa Seedlings. Forest Health Protection. Numbered Report 07-04. 2007. [(accessed on 28 January 2022)]. Available online: https://www.researchgate.net/profile/Richard-Sniezko/publication/228784173_Pathogenicity_of_four_Fusarium_species_on_Acacia_koa_seedlings/links/02e7e52c737caeb912000000/Pathogenicity-of-four-Fusarium-species-on-Acacia-koa-seedlings.pdf.
- 61.Dudley N., James R.L., Sniezko R., Yeh A. Investigating Koa Wilt and Dieback in Hawai’i: Pathogenicity of Fusarium species on Acacia koa seedlings. Native Plants J. 2007;8:259–266. doi: 10.2979/NPJ.2007.8.3.259. [DOI] [Google Scholar]
- 62.Bosso L., Senatore M., Varlese R., Ruocco M., Garonna A., Bonanomi G., Mazzoleni S., Cristinzio G. Severe Outbreak of Fusarium Solani on Quercus Ilex Vectored by Xylosandrus Compactus. J. Plant Pathol. 2012;94:99. doi: 10.4454/JPP.V95I4SUP.036. [DOI] [Google Scholar]
- 63.Vannini A., Contarini M., Faccoli M., Valle M.D., Rodriguez C.M., Mazzetto T., Guarneri D., Vettraino A.M., Speranza S. First Report of the Ambrosia Beetle Xylosandrus Compactus and Associated Fungi in the Mediterranean Maquis in Italy, and New Host–Pest Associations. EPPO Bull. 2017;47:100–103. doi: 10.1111/epp.12358. [DOI] [Google Scholar]
- 64.Gazengel K., Lebreton L., Lapalu N., Amselem J., Guillerm-Erckelboudt A.-Y., Tagu D., Daval S. PH Effect on Strain-Specific Transcriptomes of the Take-All Fungus. PLoS ONE. 2020;15:e0236429. doi: 10.1371/journal.pone.0236429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rowe H.C., Kliebenstein D.J. All Mold Is Not Alike: The Importance of Intraspecific Diversity in Necrotrophic Plant Pathogens. PLoS Pathog. 2010;6:e1000759. doi: 10.1371/journal.ppat.1000759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Adnan M., Zheng W., Islam W., Arif M., Abubakar Y., Wang Z., Lu G. Carbon Catabolite Repression in Filamentous Fungi. Int. J. Mol. Sci. 2017;19:48. doi: 10.3390/ijms19010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ries L.N.A., Beattie S., Cramer R.A., Goldman G.H. Overview of Carbon and Nitrogen Catabolite Metabolism in the Virulence of Human Pathogenic Fungi. Mol. Microbiol. 2018;107:277–297. doi: 10.1111/mmi.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ries L.N.A., Steenwyk J.L., de Castro P.A., de Lima P.B.A., Almeida F., de Assis L.J., Manfiolli A.O., Takahashi-Nakaguchi A., Kusuya Y., Hagiwara D., et al. Nutritional Heterogeneity Among Aspergillus Fumigatus Strains Has Consequences for Virulence in a Strain- and Host-Dependent Manner. Front. Microbiol. 2019;10:854. doi: 10.3389/fmicb.2019.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biedermann P.H.W., Klepzig K.D., Taborsky M., Six D.L. Abundance and Dynamics of Filamentous Fungi in the Complex Ambrosia Gardens of the Primitively Eusocial Beetle Xyleborinus Saxesenii Ratzeburg (Coleoptera: Curculionidae, Scolytinae) FEMS Microbiol. Ecol. 2013;83:711–723. doi: 10.1111/1574-6941.12026. [DOI] [PubMed] [Google Scholar]
- 70.Carrillo D., Duncan R.E., Ploetz J.N., Campbell A.F., Ploetz R.C., Peña J.E. Lateral Transfer of a Phytopathogenic Symbiont among Native and Exotic Ambrosia Beetles. Plant Pathol. 2014;63:54–62. doi: 10.1111/ppa.12073. [DOI] [Google Scholar]
- 71.Alvidrez-Villarreal R., Hernández-Castillo F.D., Garcia-Martínez O., Mendoza-Villarreal R., Rodríguez-Herrera R., Aguilar C.N. Isolation and pathogenicity of fungi associated to ambrosia borer (Euplatypus segnis) found injuring pecan (Carya illinoensis) wood. Agric. Sci. 2012;3:405–416. doi: 10.4236/as.2012.33048. [DOI] [Google Scholar]
- 72.Gil Z., Bustillo A., Gomez D., Marin P. Corthylus n. sp. (Coleoptera: Scolytidae), pest of alder in Rio Blanco basin of Colombia. Rev. Colomb. Entomol. 2004;30:171–178. [Google Scholar]
- 73.Kostovcik M. The Ambrosia Symbiosis Is Specific in Some Species and Promiscuous in Others: Evidence from Community Pyrosequencing. ISME J. 2015;9:126–138. doi: 10.1038/ismej.2014.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miller K.E., Inward D.J.G., Gomez-Rodriguez C., Baselga A., Vogler A.P. Predicting the Unpredictable: How Host Specific Is the Mycobiota of Bark and Ambrosia Beetles? Fungal Ecol. 2019;42:100854. doi: 10.1016/j.funeco.2019.07.008. [DOI] [Google Scholar]
- 75.Lantschner M.V., Corley J.C., Liebhold A.M. Drivers of Global Scolytinae Invasion Patterns. Ecol. Appl. 2020;30:e02103. doi: 10.1002/eap.2103. [DOI] [PubMed] [Google Scholar]
- 76.Williams G.M., Ginzel M.D. Spatial and Climatic Factors Influence Ambrosia Beetle (Coleoptera: Curculionidae) Abundance in Intensively Managed Plantations of Eastern Black Walnut. Environ. Entomol. 2020;49:49–58. doi: 10.1093/ee/nvz125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequences of the four markers for INECOL_BM-04 and INECOL_BM-06 isolates were submitted to GenBank with the following accessions: OM455454, OM455455, OM455456, OM455457, OM455458, OM455459, OM455460 and OM455461. The four-locus data set and the Bayesian Inference (BI) tree are public available in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S29320?x-access-code=7d6edd9c73c50b876654dd26dec6e00e&format=html, accessed on 24 January 2022).