Abstract
We investigated the association between DICER1 (rs3742330) and DROSHA (rs10719) polymorphisms and pseudoexfoliation glaucoma (PXG) and related clinical phenotypes in a Saudi cohort. In a retrospective case-control study, TaqMan real-time, PCR-based genotyping was performed in 340 participants with 246 controls and 94 PXG cases. The minor (G) allele frequency of rs3742330 in PXG (0.03) was significantly different from that in the controls (0.08) and protective against PXG (odds ratio (OR) = 0.38, 95% confidence interval (CI) = 0.16–0.92), p = 0.017). Similarly, the rs3742330 genotypes showed a significant protective association with PXG in dominant (p = 0.019, OR = 0.38, 95% CI = 0.15–0.92), over-dominant (p = 0.024, OR = 0.39, 95% CI = 0.16–0.95), and log-additive models (p = 0.017, OR = 0.38, 95% CI = 0.16–0.92). However, none remained significant after an adjustment for age, sex, and multiple testing. Rs10719 in DROSHA did not show any significant allelic or genotype association with PXG. However, a protective effect of the GA haplotype in DICER1 and DROSHA and PXG (p = 0.034) was observed. Both polymorphisms showed no significant effect on intraocular pressure and the cup–disk ratio. In conclusion, we report a significant genetic association between variant rs3742330 in DICER1, a gene involved in miRNA biogenesis, and PXG. Further investigation in a larger group of patients of different ethnicities and functional studies are warranted to replicate and validate its potential role in PXG.
Keywords: DICER1, DROSHA, genetics, glaucoma, intraocular pressure, microRNA, polymorphisms, pseudoexfoliation, rs10719, Saudi
1. Introduction
Pseudoexfoliation glaucoma (PXG) is an age-related and more aggressive form of open-angle glaucoma associated with a poor prognosis. PXG is characterized by excessive production and abnormal accumulation or deposition of pseudoexfoliative material, typically in the anterior segment of the eye that obstructs the aqueous flow pathway, leading to increased intraocular pressure (IOP), optic nerve head damage, retinal ganglion cell death (RGC), and subsequent loss of vision [1,2]. Genetic and environmental factors also play an important role in the development and progression of the disease [3,4]. Previous genome-wide studies have identified genetic loci and polymorphisms associated with the disease phenotype [3,5,6]. However, the genetic factors and molecular mechanisms contributing to glaucomatous eye damage are still unclear.
Increasing evidence suggests the critical roles of microRNA (miRNA) in glaucoma [7,8]. miRNAs are small (~22 bp), conserved, noncoding RNAs that act by binding to complementary sequences in the 3′ untranslated region (3′ UTR) of messenger RNAs (mRNAs) to regulate posttranscriptional gene expression or translational repression [9]. miRNAs regulate nearly 30% of human genes and each miRNA can regulate several gene targets [10]. Consequently, miRNAs can influence various pathophysiological processes, such as proliferation, differentiation, migration, and apoptosis, and modulate several disease outcomes as a result [11,12]. Two critical RNase III enzymes, DROSHA and DICER1, are involved in the biogenesis of miRNAs. Following the synthesis of primary miRNAs (pri-miRNAs) by RNase II in the nucleus, DROSHA cleaves pri-miRNAs into a 70 bp stem-loop structure called pre-miRNA. Subsequently, pre-miRNA is transported by the exportin-5 protein to the cytoplasm, wherein DICER1 processes pre-miRNAs into mature miRNAs that are subsequently incorporated into the RNA-induced silencing complex (RISC) to interact with target mRNAs and regulate their expression and function [13].
Differential expression of DICER1 or DROSHA enzymes due to polymorphisms in the miRNA coding genes can have pathological consequences [14,15]. Two commonly investigated variants affecting the miRNA binding and mRNA stability, expression, and function include rs10719 G > A in DROSHA and rs3742330 A > G in DICER1, located in the 3′ UTRs of their respective genes. These polymorphisms have been associated with several complex human diseases, including glaucoma [7,16,17,18,19]. We hypothesize that these polymorphisms via miRNA regulation of mRNA stability or translational repression might trigger downstream changes to influence disease processes (e.g., extracellular matrix (ECM) remodeling and trabecular meshwork (TM) homeostasis) involved in PXG pathogenesis [20]. Thus, we investigated the genetic association of polymorphisms rs3742330 and rs10719 in DICER1 and DROSHA genes, respectively, in PXG patients of Saudi origin.
2. Materials and Methods
2.1. Study Design and Participants
In a retrospective case-control study, the participants were recruited at the King Abdulaziz University Hospital in Riyadh, Saudi Arabia. PXG patients (n = 94) of Saudi origin were diagnosed by glaucoma consultants based on the clinical evidence of exfoliation material on the pupil margin or anterior lens surface, the presence of glaucomatous optic neuropathy with associated visual field loss in one or both eyes, and high untreated IOP (≥21 mm Hg) as per the guidelines of the European Glaucoma Society [21]. The exclusion criteria included secondary glaucoma, optic neuropathies or visual loss unrelated to glaucoma, inability to examine fundus for disk assessment, steroid usage, ocular trauma, and refusal to participate. A group of healthy subjects (n = 246) of the same ethnicity, who were >40 years of age, had normal IOP (without medication), and were free from any form of glaucoma, was included as a control group. Subjects refusing to participate were excluded.
2.2. Genotyping of rs3742330 in DICER1 and rs10719 in DROSHA
DNA extracted from EDTA blood using a QIAamp DNA blood kit (Cat. # 51306, QIAGEN GmbH, Hilden, Germany) was subjected to genotyping. TaqMan® assays C__27475447_10 and C___7761648_10 (Catalog number: 4351379, Applied Biosystems Inc., Foster City, CA, USA) were used to genotype rs3742330 (A>G) and rs10719 (G>A), respectively, under the recommended conditions on an ABI 7500 Real-Time PCR system (Applied Biosystems Inc., Foster City, CA, USA). Briefly, each PCR reaction was performed in a total volume of 25 µL consisting of a 1× TaqMan® genotyping master mix (Applied Biosystems Inc., Foster City, CA, USA), 1X SNP genotyping assay mix, and 20 ng DNA. Each 96-well plate included two no-template (negative) controls. Amplification conditions included incubation at 95 °C for 10 min, followed by 40 cycles, denaturation at 92 °C for 15 s, and annealing and extension at 60 °C for 1 min. The VIC® and 6-carboxy-fluorescein (FAM) fluorescence levels of the PCR products were measured at 60 °C for 1 min. A fluorescence analysis was performed using automated 2-color allele discrimination software to identify the DICER1 and DROSHA genotypes on a 2-dimensional graph [22].
2.3. In Silico Analysis of rs3742330
The annotation of rs3742330 and its localization to the genomic region was visualized using the UCSC genome browser with the SNPedia option. Regulation and comparative genomics tracks were used to visualize regulatory elements and conservation across species. TargetScanHuman 7.2 was added as a custom track to visualize miRNA targets. The RNA binding partners to the 3′ UTR reference of 30 nucleotides upstream and downstream of rs3742330 were predicted using an RBPmap database (http://rbpmap.technion.ac.il/index.html, accessed on 24 February 2022). Regulatory motif alteration was examined using HaploReg v4.1.
2.4. Statistics
The Hardy–Weinberg equilibrium (HWE) and other allelic and genotype associations were tested by chi-square analysis. The continuous parameters were tested by an independent samples t-test and one-way ANOVA. Regression analysis was performed to evaluate the effect of multiple risk factors such as age, sex, and genotype on PXG outcome. Statistical analysis was performed using SPSS version 22 (IBM Inc., Chicago, IL, USA) and SNPStats online software (https://www.snpstats.net/start.htm, accessed on 30 December 2021). SHEsis online software (Analysis tool for random samples, by YongYong Shi (analysis.bio-x.cn, accessed on 30 December 2021)) was used to examine linkage disequilibrium (LD) and analyze haplotypes. Power analysis was performed using PS software (https://ps-power-and-sample-size-calculation.software.informer.com/3.1/, accessed on 30 December 2021). A two-tailed p < 0.05 was considered statistically significant and a Bonferroni-corrected p-value (0.05/5 = 0.01) was considered where applicable.
3. Results
3.1. Demographic Characteristics and Minor Allele Frequency Distribution
Table 1 shows the demographic characteristics of the participants and minor allele frequency (MAF) distribution of the examined polymorphisms. The cases were significantly older than the controls (p < 0.001). However, the differences in sex distribution between PXG cases compared to controls were nonsignificant. Of 246 controls and 94 PXG cases, genotyping was available for 241 controls for rs3742330 DICER1 polymorphism. The five DNA samples from controls that failed to amplify were excluded from DICER1 analysis. There was no significant deviation from the HWE (p > 0.05). The rs3742330 (G) MAF in PXG (0.03) varied more significantly than the controls (0.08) and was protective against PXG (odds ratio (OR) = 0.38, 95% confidence interval (CI) = 0.16–0.92), p = 0.017). In contrast, the rs10719 (A) allele showed no significant distribution between PXG and the controls. Furthermore, no significant sex-specific allelic association was observed (Table 1).
Table 1.
Demographic characteristics and distribution of minor allele frequency of DICER1 rs3742330 (G) and DROSHA rs10719 (A) polymorphisms in pseudoexfoliation glaucoma cases and control participants.
| Characteristics | Controls | Cases | Odds Ratio (95% Confidence Interval) |
p-Value |
|---|---|---|---|---|
| No. of participants | 246 | 94 | - | - |
| Age in years (SD) | 59.5 (7.2) | 66.4 (9.7) | - | <0.001 1 |
| Men, n (%) | 132 (53.6) | 61 (64.8) | - | - |
| Women, n (%) | 114 (46.3) | 33 (33.5) | - | 0.061 2 |
| Minor Allele Frequency | ||||
| rs3742330 (G) | ||||
| Total | 0.08 | 0.03 | 0.38 (0.16–0.92) | 0.017 2 |
| Men | 0.08 | 0.03 | 0.40 (0.13–1.21) | 0.076 2 |
| Women | 0.08 | 0.03 | 0.34 (0.07–1.55) | 0.120 2 |
| rs10719 (A) | ||||
| Total | 0.43 | 0.45 | 1.08 (0.78–1.50) | 0.630 2 |
| Men | 0.45 | 0.47 | 1.08 (0.71–1.65) | 0.720 2 |
| Women | 0.41 | 0.42 | 1.05 (0.62–1.77) | 0.870 2 |
1 Independent t-test; 2 chi-square analysis. Significant odds ratio and p-value in bold.
3.2. Genotype Association Analysis with PXG
PXG is a complex disease with no apparent genetic mode of inheritance. We used co-dominant, dominant, recessive, over-dominant, and log-additive genetic models to examine the association between DICER1 and DROSHA polymorphisms and the risk of PXG using SNPStat software (Table 2). Rs3742330 genotypes in DICER1 showed a significant association with PXG risk in dominant, over-dominant, and log-additive genetic models. The log-additive model exhibited the best fit with the lowest Akaike information criterion (AIC) and Bayesian information criterion (BIC) values. However, the significance was lost after an adjustment for age, sex, and Bonferroni correction (pcorrection < 0.01). In contrast, rs10719 polymorphism in DROSHA showed no significant association (Table 2). In addition, a sex-stratified genotype analysis for rs3742330 and rs10719 showed no significant association (Table 3 and Table 4).
Table 2.
Association analysis of polymorphisms rs3742330 in DICER1 and rs10719 in DROSHA with the risk of pseudoexfoliation glaucoma cases compared to controls under different genetic models.
| SNP Number | Genetic Model 1 | Genotype | Controls n (%) |
Cases n (%) |
Odds Ratio (95% Confidence Interval) | p-Value 2 | AIC 3 | BIC 4 | p-Value 2,5 |
|---|---|---|---|---|---|---|---|---|---|
| rs3742330 | Co-dominant | A/A | 204 (84.7) | 88 (93.6) | 1.00 | 0.055 | 397.9 | 409.3 | 0.170 |
| A/G | 36 (14.9) | 6 (6.4) | 0.39 (0.16–0.95) 6 | ||||||
| G/G | 1 (0.4) | 0 (0.0) | 0.00 (0.00-NA) | ||||||
| Dominant | A/A | 204 (84.7) | 88 (93.6) | 1.00 | 0.019 | 396.2 | 403.8 | 0.077 | |
| A/G-G/G | 37 (15.3) | 6 (6.4) | 0.38 (0.15–0.92) | ||||||
| Recessive | A/A-A/G | 240 (99.6) | 94 (100.0) | 1.00 | 0.420 | 401.0 | 408.6 | 0.380 | |
| G/G | 1 (0.4) | 0 (0.0) | 0.00 (0.00-NA) | ||||||
| Over-dominant | A/A-G/G | 205 (85.1) | 88 (93.6) | 1.00 | 0.024 | 396.6 | 404.2 | 0.098 | |
| A/G | 36 (14.9) | 6 (6.4) | 0.39 (0.16–0.95) | ||||||
| Log-additive | - | - | - | 0.38 (0.16–0.92) 7 | 0.017 | 396.0 | 403.6 | 0.068 | |
| rs10719 | Co-dominant | G/G | 82 (33.3) | 32 (34.0) | 1.00 | 0.530 | 405.6 | 417.1 | 0.250 |
| A/G | 116 (47.1) | 39 (41.5) | 0.86 (0.50–1.49) | ||||||
| A/A | 48 (19.5) | 23 (24.5) | 1.23 (0.65–2.34) | ||||||
| Dominant | G/G | 82 (33.3) | 32 (34.0) | 1.00 | 0.900 | 404.9 | 412.6 | 0.350 | |
| A/G-A/A | 164 (66.7) | 62 (66.0) | 0.97 (0.59–1.60) | ||||||
| Recessive | G/G-A/G | 198 (80.5) | 71 (75.5) | 1.00 | 0.320 | 403.9 | 411.6 | 0.350 | |
| A/A | 48 (19.5) | 23 (24.5) | 1.34 (0.76–2.35) | ||||||
| Over-dominant | G/G-A/A | 130 (52.9) | 55 (58.5) | 1.00 | 0.350 | 404.0 | 411.7 | 0.098 | |
| A/G | 116 (47.1) | 39 (41.5) | 0.79 (0.49–1.29) | ||||||
| Log-additive | - | - | - | 1.08 (0.78–1.50) | 0.630 | 404.7 | 412.3 | 0.930 |
1 Tested by SNPStat; 2 chi-square analysis; 3 AIC, Akaike information criterion; 4 BIC, Bayesian information criterion; 5 adjusted for age and sex; 6 A/A vs. A/G p-value = 0.032; 7 best-fit model p-value. Significant odds ratio and p-value in bold. Bonferroni-corrected p-value is 0.01.
Table 3.
Association analysis of rs3742330 polymorphism in DICER1 with pseudoexfoliation glaucoma cases in men and women.
| Group | Genetic Model 1 | Genotype | Control n (%) |
Cases n (%) |
Odds Ratio (95% Confidence Interval) | p-Value 2 | AIC 3 | BIC 4 | p-Value 2,5 |
|---|---|---|---|---|---|---|---|---|---|
| Men | Co-dominant | A/A | 109 (85.2) | 57 (93.4) | 1.00 | 0.190 | 240.4 | 250.1 | 0.430 |
| A/G | 18 (14.1) | 4 (6.6) | 0.42 (0.14–1.32) | ||||||
| G/G | 1 (0.8) | 0 (0.0) | 0.00 (0.00–NA) | ||||||
| Dominant | A/A | 109 (85.2) | 57 (93.4) | 1.00 | 0.087 | 238.8 | 245.3 | 0.280 | |
| A/G-G/G | 19 (14.8) | 4 (6.6) | 0.40 (0.13–1.24) | ||||||
| Recessive | A/A-A/G | 127 (99.2) | 61 (100.0) | 1.00 | 0.380 | 241.0 | 247.4 | 0.380 | |
| G/G | 1 (0.8) | 0 (0.0) | 0.00 (0.00–NA) | ||||||
| Over-dominant | A/A-G/G | 110 (85.9) | 57 (93.4) | 1.00 | 0.120 | 239.3 | 245.7 | 0.350 | |
| A/G | 18 (14.1) | 4 (6.6) | 0.43 (0.14–1.33) | ||||||
| Log-additive | - | - | - | 0.40 (0.13–1.21) | 0.076 | 238.6 | 245.1 | 0.240 | |
| Women | -- | A/A | 95 (84.1) | 31 (93.9) | 1.00 | 0.120 | 157.6 | 163.6 | 0.120 |
| A/G | 18 (15.9) | 2 (6.1) | 0.34 (0.07–1.55) | ||||||
| G/G | 0 (0.0) | 0 (0.0) | - |
1 Tested by SNPStat; 2 chi-square analysis; 3 AIC, Akaike information criterion; 4 BIC, Bayesian information criterion; 5 adjusted for age and sex. Note: No rare homozygous genotype G/G was observed among women.
Table 4.
Sex-stratified association analysis of polymorphism rs10719 in DROSHA with pseudoexfoliation glaucoma cases.
| Group | Genetic Model 1 | Genotype | Control n (%) |
Cases n (%) |
Odds Ratio (95% Confidence Interval) | p 2 | AIC 3 | BIC 4 | p 2,5 |
|---|---|---|---|---|---|---|---|---|---|
| Men | Co-dominant | G/G | 40 (30.3) | 20 (32.8) | 1.00 | 0.450 | 245.2 | 255.0 | 0.230 |
| A/G | 66 (50) | 25 (41.0) | 0.76 (0.37–1.54) | ||||||
| A/A | 26 (19.7) | 16 (26.2) | 1.23 (0.54–2.80) | ||||||
| Dominant | G/G | 40 (30.3) | 20 (32.8) | 1.00 | 0.730 | 244.7 | 251.2 | 0.470 | |
| A/G-A/A | 92 (69.7) | 41 (67.2) | 0.89 (0.46–1.71) | ||||||
| Recessive | G/G-A/G | 106 (80.3) | 45 (73.8) | 1.00 | 0.310 | 243.8 | 250.3 | 0.230 | |
| A/A | 26 (19.7) | 16 (26.2) | 1.45 (0.71–2.96) | ||||||
| Over-dominant | G/G-A/A | 66 (50.0) | 36 (59.0) | 1.00 | 0.240 | 243.4 | 250.0 | 0.094 | |
| A/G | 66 (50.0) | 25 (41.0) | 0.69 (0.38–1.28) | ||||||
| Log-additive | - | - | - | 1.08 (0.71–1.65) | 0.720 | 244.7 | 251.2 | 0.830 | |
| Women | Co-dominant | G/G | 42 (36.8) | 12 (36.4) | 1.00 | 0.970 | 162.5 | 171.5 | 0.800 |
| A/G | 50 (43.9) | 14 (42.4) | 0.98 (0.41–2.35) | ||||||
| A/A | 22 (19.3) | 7 (21.2) | 1.11 (0.38–3.23) | ||||||
| Dominant | G/G | 42 (36.8) | 12 (36.4) | 1.00 | 0.960 | 160.6 | 166.5 | 0.510 | |
| A/G-A/A | 72 (63.2) | 21 (63.6) | 1.02 (0.46–2.28) | ||||||
| Recessive | G/G-A/G | 92 (80.7) | 26 (78.8) | 1.00 | 0.810 | 160.5 | 166.5 | 0.900 | |
| A/A | 22 (19.3) | 7 (21.2) | 1.13 (0.43–2.93) | ||||||
| Over-dominant | G/G-A/A | 64 (56.1) | 19 (57.6) | 1.00 | 0.8800 | 160.5 | 166.5 | 0.590 | |
| A/G | 50 (43.9) | 14 (42.4) | 0.94 (0.43–2.06) | ||||||
| Log-additive | - | - | - | 1.05 (0.62–1.77) | 0.870 | 160.5 | 166.5 | 0.610 |
1 Tested by SNPStat; 2 chi-square analysis; 3 AIC, Akaike information criterion; 4 BIC, Bayesian information criterion; 5 adjusted for age and sex.
3.3. Linkage and Haplotype Analysis
The two polymorphisms were tested for LD and haplotype analysis using the SHEsis platform. The standardized LD coefficient D’ value between rs3742330 and rs10719 was 0.06 and r2 = 0.00, indicating that these polymorphisms are not in LD. Additionally, the haplotype distribution did not significantly affect the risk of PXG (X2 = 6.357, df = 3, p = 0.095). However, haplotype GA showed a significant protective effect (p = 0.034, OR = 0.20, 95% CI = 0.04–1.03, Table 5).
Table 5.
Haplotype analysis of DICER1 rs3742330 and DROSHA rs10719 polymorphisms.
| Haplotypes 1 | Controls, Frequency | Cases, Frequency | p-Value | Odds Ratio (95% Confidence Interval) |
|---|---|---|---|---|
| AA | 0.38 | 0.44 | 0.179 | 1.26 (0.89–1.77) |
| AG | 0.53 | 0.52 | 0.821 | 0.96 (0.68–1.35) |
| GA | 0.04 | 0.01 | 0.034 | 0.20 (0.04–1.03) |
| GG | 0.04 | 0.02 | 0.334 | 0.59 (0.21–1.71) |
1 Tested by SHEsis in the order of rs3742330 and rs10719. Overall chi-square = 6.357, df = 3, p = 0.095. Significant p-value and odds ratio in bold.
3.4. Regression Analysis and Genotype Influence on Clinical Parameters
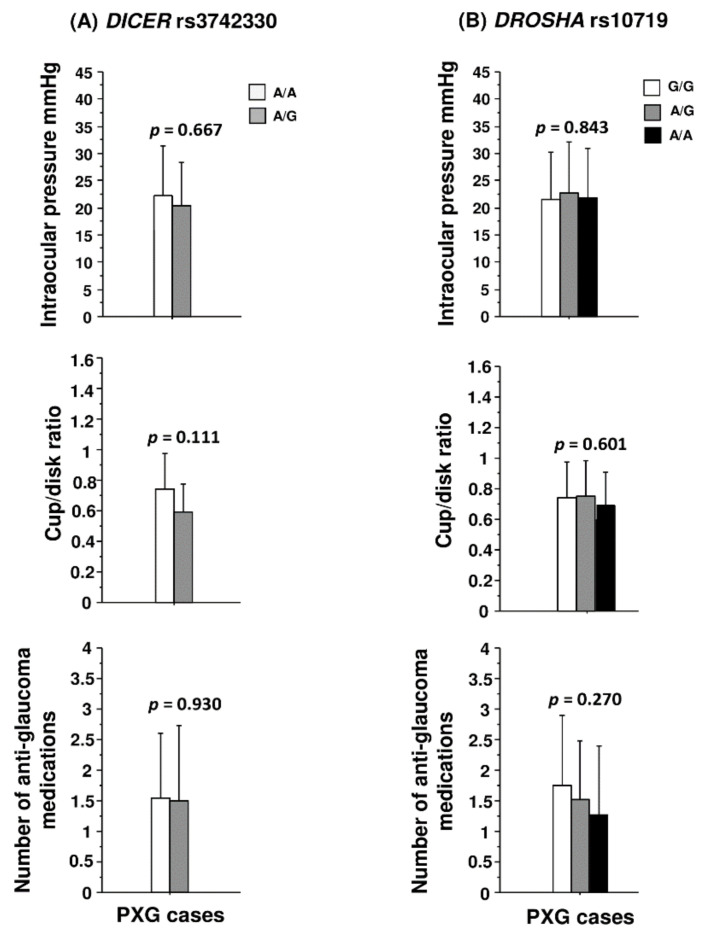
A binary logistic regression analysis was performed to assess the influence of multiple risk factors such as age, sex, and genotypes of rs3742330 and rs10719 on PXG outcome. Except for age (p < 0.001), no other variable showed a significant contribution to PXG risk (Table 6). In addition, neither polymorphism showed a significant genotype effect on clinical markers such as IOP, the cup–disk ratio, and the number of antiglaucoma medications (Figure 1).
Table 6.
Binary logistic regression analysis to determine the effect of age, sex, and polymorphisms rs3742330 (DICER1) and rs10719 (DROSHA) on the risk of pseudoexfoliation glaucoma.
| Group Variables |
B | SE | Wald | Odds Ratio (95% Confidence Interval) |
p |
|---|---|---|---|---|---|
| Age | 0.100 | 0.017 | 35.755 | 1.10 (1.07–1.14) | 0.000 |
| Sex | 0.392 | 0.273 | 2.068 | 1.48 (0.86–2.52) | 0.150 |
| rs3742330 | 2.350 | 0.309 | |||
| A/G | −0.729 | 0.476 | 2.349 | 0.48 (0.19–1.22) | 0.125 |
| G/G | - | - | - | - | 1.000 |
| rs10719 | 3.002 | 0.223 | |||
| G/A | −0.410 | 0.305 | 1.801 | 0.66 (0.36–1.20) | 0.180 |
| A/A | 0.127 | 0.364 | 0.121 | 1.13 (0.55–2.31) | 0.728 |
| Constant | −7.236 | 1.079 | 44.981 | 0.001 | 0.000 |
Significant p-value in bold.
Figure 1.
Genotype effect of (A) rs3742330 (DICER1) and (B) rs10719 (DROSHA) on glaucoma specific clinical indices in pseudoexfoliation glaucoma (PXG). Note: No rare homozygous rs3742330 G/G genotype was observed in pseudoexfoliation glaucoma cases. p-value is based on one-way ANOVA test.
3.5. Potential Significance of rs3742330
The genomic region containing rs3742330 and its associated neighboring features is shown in Supplementary Figure S1. An in silico analysis showed that the region contains multiple sites for miRNAs, transcription factors, and other regulatory elements (Figure S1A). In addition, the variant may alter the binding of certain RNA-binding proteins (Figure S1B) and regulatory motifs (Figure S1C), suggesting that the region is significant for mRNA stability and gene expression. However, rs3742330 is classified as benign in ClinVar.
4. Discussion
In this paper, we report for the first time a moderate allelic association of variant rs3742330 (G) in DICER1 with PXG in a Saudi cohort. The polymorphism rs3742330 in DICER1 was associated with a decreased risk of PXG (OR of 0.38). Although no homozygous G/G genotype was observed in the PXG patients, the heterozygous A/G genotype also conferred significant protection compared to A/A in different genetic models. However, it did not survive correction for multiple testing.
Emerging evidence suggests that polymorphism(s) in DICER1 may alter the biological functions of miRNAs and play an essential role in the pathogenesis of various diseases [16,17,18,19]. In accordance with our findings, Chatzikyriakidou et al. reported a different DICER1 variant, rs1057035 (C>T), which conferred protection (OR of 0.69) in patients with pseudoexfoliation syndrome [7]. Using the LDlink analysis (https://ldlink.nci.nih.gov/?tab = ldpop, accessed on 30 December 2021) to predict linkage across the 1000 Genomes database, we noted that variants rs1057035 and rs3742330 in DICER1 were not in LD (r2 = 0.032); yet, this study lends further support to the protective association of the DICER1 variant observed in our patient cohort.
Dicer has an essential role in development and angiogenesis [23,24]. Germ-line mutations in DICER1 are associated with DICER1 syndrome [25]. DICER1 carriers exhibit ocular abnormalities including optic nerve damage and retinal changes [25]. Genetic manipulation of dicer1 in mice has been shown to cause retinal degeneration, suggesting its role in cell survival [26,27]. Differential expressions of DICER1 have been associated with various types and stages of cancers, albeit with contradictory findings. The increased expression has been linked to a good prognosis in lung, breast, and ovarian cancer as opposed to a poor prognosis in colorectal and prostate cancer [28,29,30]. These studies suggest that DICER1 may have varied roles in different diseases. The underlying potential mechanism(s) linking DICER1 polymorphism rs3742330 to PXG pathogenesis is unknown. Rs3742330 polymorphism has been related to dysregulation of DICER1 mRNA, wherein the polymorphic A/G and G/G genotypes harbored lower levels of DICER1 mRNA [31,32]. Altered DICER1 levels (due to polymorphism) may directly affect enzyme function or can indirectly influence disease pathogenesis via differential regulation of miRNA expression.
Dicer knockdown results in a marked global reduction in miRNA levels [33,34]. miRNAs are tissue-specific and expressed in ocular tissues related to glaucoma [35]. Several studies have highlighted the significant role of miRNAs and their plausible underlying mechanisms in different types of glaucoma, including PXG [8,36,37,38,39,40]. Rao et al. reported the overexpression of 12 miRNAs in PXG, of which miR-122-5p, miR-124-3p, and miR-424-5p targeted TGFβ1, fibrosis/ECM, and proteoglycan metabolism pathways [39]. These signaling pathways are known mediators of ECM secretion and deposition and are strongly implicated in glaucoma [41]. Interestingly, using MicroSNiPer [42], we observed that the DICER1 rs3742330 (G) variant might enhance the binding of hsa-miR-124-3p and probably cause a decrease in DICER1 gene expression, which may impact the DICER1 enzyme’s function and/or sequentially affect miRNA expression. Similarly, Zenkel et al. reported downregulation of the miR-29 family in iridal and ciliary tissue specimens from donor eyes with pseudoexfoliation syndrome [43]. In vitro studies have reported the miR-29 family as an essential modulator of ECM genes under chronic oxidative stress conditions and TGFß stimulation in the human TM cells [44,45]. miR-1260b, mainly involved in the process of hypoxia and apoptosis, was reported to be most abundantly expressed in PXG [38]. Hindle et al. reported 20 circulatory miRNAs targeting neuroinflammation, nitric oxide, and neurotrophin–tropomyosin-related kinase (TRK) signaling pathways [40], which have all been strongly implicated in the pathophysiology of glaucoma [46,47]. In another study, Drewry et al. identified dysregulated miRNAs (miR-122-5p, miR-3144-3p, miR-320a, miR-320e, and miR-630) in PXG involved in focal adhesion, tight junctions, and TGFß signaling [37].
Oxidative stress, inflammation, breakdown of the blood–aqueous barrier, a decrease in clusterin, and genetic predisposition (e.g., lysyl oxidase-like-1 (LOXL1) polymorphisms) are among the pathologic mediators of the abnormal elastotic process in PXG [1]. Taken together, we speculate that polymorphism rs3742330 in DICER1 may alter DICER levels and, via miRNA regulation, may be associated with PXG by influencing processes or pathways discussed above (e.g., TRK signaling, TGFß signaling, oxidative stress genes, tight junctions, and apoptosis) that might affect ECM remodeling and TM homeostasis or RGC survival. We are interested in discovering whether the DICER1 polymorphism modulates the PXG risk by affecting DICER enzyme function and/or via RNA interference. The in silico analysis suggested that the genomic region of rs3742330 located in the 3′ UTR of DICER1 may be important for mRNA transcript stability and post-transcriptional regulation of gene expression. However, there is no direct evidence supporting this role. Additionally, the effect of gene–gene and/or gene–environment interactions, or linkage with a causal variant in PXG, cannot be ruled out. Further molecular and in vitro studies are needed to validate these hypotheses.
In contrast, rs10719 polymorphism in DROSHA, another critical enzyme involved in miRNA biogenesis [33], did not show any allelic or genotype association with PXG or clinical markers (e.g., IOP and the cup–disk ratio), indicating that this variant may not have a significant role in PXG. However, the role of other variants in this gene cannot be ruled out. In addition, haplotype analysis of DICER1 and DROSHA polymorphisms indicated that haplotype GA was protective against PXG. However, it is difficult to ascertain whether the protective effect observed in our study is attributable to a real haplotype effect or reflects a strong LD with any other variant(s) not included in this study.
The results of the study have certain limitations. It should be noted that the significant association of the DICER1 variant was restricted to the unadjusted models of analysis when corrections for multiple testing and confounding variables were included, indicating that the association may be dependent on other gene–gene or gene–environmental interactions [48]. It also needs to be emphasized that the study was exploratory, performed in a single ethnic group of patients, and provides no functional evidence. In addition, the results are limited by sample size, particularly considering the low frequency of the variant observed in this cohort of Arab ethnicity, and the possibility of chance association cannot be ruled out. Based on the observed allele frequencies and an α level of 0.05, the estimated power was 0.73 per allele for rs3742330 (DICER1) and >0.9 per allele for rs10719 (DROSHA) to detect an OR of two. However, a much larger sample size would be required to detect an odds ratio of 1.5 or less, which is commonly seen in genetic association studies.
In conclusion, we report a genetic association between a potentially functional polymorphism rs3742330 in DICER1, a gene involved in miRNA biogenesis, and PXG patients of Saudi origin, suggesting a need to further investigate the involvement of epigenetic pathways as modulators in disease development and progression. Further replication in a large population-based sample cohort of different ethnicities and functional studies are warranted to validate the potential role of DICER1 variant rs3742330 in PXG.
Acknowledgments
We would like to thank the Vice Deanship of Scientific Research Chair, Glaucoma Research Chair in Ophthalmology at King Saud University. We also thank Abdulrahman Al-Mosa for his clinical assistance during the study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13030489/s1, Figure S1: The in silico analysis of rs3742330 in DICER1 showing (A) The genomic region containing the polymorphism on chromosome 14q32.13 and its neighboring features, such as miRNA binding sites, transcription binding sites, regulatory elements, eQTLs, and conservation among species obtained using the UCSC genome browser. (B) The altered RNA binding protein partners predicted using RBP map database. (C) Altered regulatory motifs as detected by HaploReg v4.1.
Author Contributions
A.A.K.: concept, investigation, methodology, analysis, writing draft, review and editing, and supervision; T.A.A., T.S.: investigation, data curation, and manuscript review and editing; R.R.: analysis, resources, and manuscript review and editing; E.A.O. and F.A.A.: data curation, resources, and manuscript review and editing; G.P.L.: analysis and data interpretations, resources, and manuscript review and editing; S.A.A.-O.: concept, resources, funding acquisition, project administration, and manuscript review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by King Saud University through the Vice Deanship of Scientific Research Chair and Glaucoma Research Chair in Ophthalmology (GRC-2022).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (IRB) Committee of the College of Medicine, King Saud University (IRB protocol number # 08–657).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data supporting the conclusions of this article are all presented within the report.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vazquez L.E., Lee R.K. Genomic and proteomic pathophysiology of pseudoexfoliation glaucoma. Int. Ophthalmol. Clin. 2014;54:1–13. doi: 10.1097/IIO.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma: A review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorleifsson G., Magnusson K.P., Sulem P., Walters G.B., Gudbjartsson D.F., Stefansson H., Jonsson T., Jonasdottir A., Stefansdottir G., Masson G., et al. Common sequence variants in the loxl1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 4.Dewundara S., Pasquale L.R. Exfoliation syndrome: A disease with an environmental component. Curr. Opin. Ophthalmol. 2015;26:78–81. doi: 10.1097/ICU.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aung T., Ozaki M., Lee M.C., Schlotzer-Schrehardt U., Thorleifsson G., Mizoguchi T., Igo R.P., Jr., Haripriya A., Williams S.E., Astakhov Y.S., et al. Genetic association study of exfoliation syndrome identifies a protective rare variant at loxl1 and five new susceptibility loci. Nat. Genet. 2017;49:993–1004. doi: 10.1038/ng.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aung T., Ozaki M., Mizoguchi T., Allingham R.R., Li Z., Haripriya A., Nakano S., Uebe S., Harder J.M., Chan A.S., et al. A common variant mapping to cacna1a is associated with susceptibility to exfoliation syndrome. Nat. Genet. 2015;47:387–392. doi: 10.1038/ng.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatzikyriakidou A., Founti P., Melidou A., Minti F., Bouras E., Anastasopoulos E., Pappas T., Haidich A.B., Lambropoulos A., Topouzis F. Microrna-related polymorphisms in pseudoexfoliation syndrome, pseudoexfoliative glaucoma, and primary open-angle glaucoma. Ophthalmic. Genet. 2018;39:603–609. doi: 10.1080/13816810.2018.1509352. [DOI] [PubMed] [Google Scholar]
- 8.Rong R., Wang M., You M., Li H., Xia X., Ji D. Pathogenesis and prospects for therapeutic clinical application of noncoding rnas in glaucoma: Systematic perspectives. J. Cell Physiol. 2021;236:7097–7116. doi: 10.1002/jcp.30347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal micrornas. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Schickel R., Boyerinas B., Park S.M., Peter M.E. Micrornas: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 12.Ardekani A.M., Naeini M.M. The role of micrornas in human diseases. Avicenna J. Med. Biotechnol. 2010;2:161–179. [PMC free article] [PubMed] [Google Scholar]
- 13.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of microrna biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aghajanova L., Giudice L.C. Molecular evidence for differences in endometrium in severe versus mild endometriosis. Reprod. Sci. 2011;18:229–251. doi: 10.1177/1933719110386241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nothnick W.B., Swan K., Flyckt R., Falcone T., Graham A. Human endometriotic lesion expression of the mir-144-3p/mir-451a cluster, its correlation with markers of cell survival and origin of lesion content. Sci. Rep. 2019;9:8823. doi: 10.1038/s41598-019-45243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borghini A., Vecoli C., Mercuri A., Turchi S., Andreassi M.G. Individual and joint effects of genetic polymorphisms in microrna-machinery genes on congenital heart disease susceptibility. Cardiol. Young. 2021;31:965–968. doi: 10.1017/S1047951120004874. [DOI] [PubMed] [Google Scholar]
- 17.Wang M., Gu J., Shen C., Tang W., Xing X., Zhang Z., Liu X. Association of microrna biogenesis genes polymorphisms with risk of large artery atherosclerosis stroke. Cell Mol. Neurobiol. 2021:1–7. doi: 10.1007/s10571-021-01057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Cao A.L., Dong C. Rs10719 polymorphism located within drosha 3’-untranslated region is responsible for development of primary hypertension by disrupting binding with microrna-27b. Med. Sci. Monit. 2017;23:911–918. doi: 10.12659/MSM.897607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cardoso J.V., Medeiros R., Dias F., Costa I.A., Ferrari R., Berardo P.T., Perini J.A. Drosha rs10719 and dicer1 rs3742330 polymorphisms in endometriosis and different diseases: Case-control and review studies. Exp. Mol. Pathol. 2021;119:104616. doi: 10.1016/j.yexmp.2021.104616. [DOI] [PubMed] [Google Scholar]
- 20.McDonnell F., O’Brien C., Wallace D. The role of epigenetics in the fibrotic processes associated with glaucoma. J. Ophthalmol. 2014;2014:750459. doi: 10.1155/2014/750459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.European Glaucoma Society Terminology and Guidelines for Glaucoma, 5th edition. Br. J. Ophthalmol. 2021;105:1–169. doi: 10.1136/bjophthalmol-2021-egsguidelines. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Amero K.K., Kondkar A.A., Mousa A., Azad T.A., Sultan T., Osman E.A., Al-Obeidan S.A. Analysis of toll-like receptor rs4986790 polymorphism in saudi patients with primary open angle glaucoma. Ophthalmic. Genet. 2017;38:133–137. doi: 10.3109/13816810.2016.1151900. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 24.Yang W.J., Yang D.D., Na S., Sandusky G.E., Zhang Q., Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J. Biol. Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 25.Huryn L.A., Turriff A., Harney L.A., Carr A.G., Chevez-Barrios P., Gombos D.S., Ram R., Hufnagel R.B., Hill D.A., Zein W.M., et al. Dicer1 syndrome: Characterization of the ocular phenotype in a family-based cohort study. Ophthalmology. 2019;126:296–304. doi: 10.1016/j.ophtha.2018.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damiani D., Alexander J.J., O’Rourke J.R., McManus M., Jadhav A.P., Cepko C.L., Hauswirth W.W., Harfe B.D., Strettoi E. Dicer inactivation leads to progressive functional and structural degeneration of the mouse retina. J. Neurosci. 2008;28:4878–4887. doi: 10.1523/JNEUROSCI.0828-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko H., Dridi S., Tarallo V., Gelfand B.D., Fowler B.J., Cho W.G., Kleinman M.E., Ponicsan S.L., Hauswirth W.W., Chiodo V.A., et al. Dicer1 deficit induces alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swahari V., Nakamura A., Deshmukh M. The paradox of dicer in cancer. Mol. Cell Oncol. 2016;3:e1155006. doi: 10.1080/23723556.2016.1155006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karube Y., Tanaka H., Osada H., Tomida S., Tatematsu Y., Yanagisawa K., Yatabe Y., Takamizawa J., Miyoshi S., Mitsudomi T., et al. Reduced expression of dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiosea S., Jelezcova E., Chandran U., Acquafondata M., McHale T., Sobol R.W., Dhir R. Up-regulation of dicer, a component of the microrna machinery, in prostate adenocarcinoma. Am. J. Pathol. 2006;169:1812–1820. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eskandari F., Teimoori B., Rezaei M., Mohammadpour-Gharehbagh A., Narooei-Nejad M., Mehrabani M., Salimi S. Relationships between dicer 1 polymorphism and expression levels in the etiopathogenesis of preeclampsia. J. Cell Biochem. 2018;119:5563–5570. doi: 10.1002/jcb.26725. [DOI] [PubMed] [Google Scholar]
- 32.Huang S.Q., Zhou Z.X., Zheng S.L., Liu D.D., Ye X.H., Zeng C.L., Han Y.J., Wen Z.H., Zou X.Q., Wu J., et al. Association of variants of mirna processing genes with cervical precancerous lesion risk in a southern chinese population. Biosci. Rep. 2018;38:BSR20171565. doi: 10.1042/BSR20171565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.K., Kim B., Kim V.N. Re-evaluation of the roles of drosha, export in 5, and dicer in microrna biogenesis. Proc. Natl. Acad. Sci. USA. 2016;113:E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forbes K., Farrokhnia F., Aplin J.D., Westwood M. Dicer-dependent mirnas provide an endogenous restraint on cytotrophoblast proliferation. Placenta. 2012;33:581–585. doi: 10.1016/j.placenta.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Drewry M., Helwa I., Allingham R.R., Hauser M.A., Liu Y. Mirna profile in three different normal human ocular tissues by mirna-seq. Invest. Ophthalmol. Vis. Sci. 2016;57:3731–3739. doi: 10.1167/iovs.16-19155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molasy M., Walczak A., Szaflik J., Szaflik J.P., Majsterek I. Micrornas in glaucoma and neurodegenerative diseases. J. Hum. Genet. 2017;62:105–112. doi: 10.1038/jhg.2016.91. [DOI] [PubMed] [Google Scholar]
- 37.Drewry M.D., Challa P., Kuchtey J.G., Navarro I., Helwa I., Hu Y., Mu H., Stamer W.D., Kuchtey R.W., Liu Y. Differentially expressed micrornas in the aqueous humor of patients with exfoliation glaucoma or primary open-angle glaucoma. Hum. Mol. Genet. 2018;27:1263–1275. doi: 10.1093/hmg/ddy040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosior-Jarecka E., Czop M., Gasinska K., Wrobel-Dudzinska D., Zalewski D.P., Bogucka-Kocka A., Kocki J., Zarnowski T. Micrornas in the aqueous humor of patients with different types of glaucoma. Graefes Arch. Clin. Exp. Ophthalmol. 2021;259:2337–2349. doi: 10.1007/s00417-021-05214-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao A., Chakraborty M., Roy A., Sahay P., Pradhan A., Raj N. Differential mirna expression: Signature for glaucoma in pseudoexfoliation. Clin. Ophthalmol. 2020;14:3025–3038. doi: 10.2147/OPTH.S254504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hindle A.G., Thoonen R., Jasien J.V., Grange R.M.H., Amin K., Wise J., Ozaki M., Ritch R., Malhotra R., Buys E.S. Identification of candidate mirna biomarkers for glaucoma. Invest. Ophthalmol. Vis. Sci. 2019;60:134–146. doi: 10.1167/iovs.18-24878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhavoronkov A., Izumchenko E., Kanherkar R.R., Teka M., Cantor C., Manaye K., Sidransky D., West M.D., Makarev E., Csoka A.B. Pro-fibrotic pathway activation in trabecular meshwork and lamina cribrosa is the main driving force of glaucoma. Cell Cycle. 2016;15:1643–1652. doi: 10.1080/15384101.2016.1170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barenboim M., Zoltick B.J., Guo Y., Weinberger D.R. Microsniper: A web tool for prediction of snp effects on putative microrna targets. Hum. Mutat. 2010;31:1223–1232. doi: 10.1002/humu.21349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zenkel M., Mößner A., Kruse F.E., Schlötzer-Schrehardt U. Dysregulated expression of fibrogenic micrornas in eyes with pseudoexfoliation syndrome. Invest. Ophthalmol. Vis. Sci. 2012;53:3583. [Google Scholar]
- 44.Luna C., Li G., Qiu J., Epstein D.L., Gonzalez P. Role of mir-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol. Vis. 2009;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- 45.Villarreal G., Jr., Oh D.J., Kang M.H., Rhee D.J. Coordinated regulation of extracellular matrix synthesis by the microrna-29 family in the trabecular meshwork. Investig. Ophthalmol. Vis. Sci. 2011;52:3391–3397. doi: 10.1167/iovs.10-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chitranshi N., Dheer Y., Abbasi M., You Y., Graham S.L., Gupta V. Glaucoma pathogenesis and neurotrophins: Focus on the molecular and genetic basis for therapeutic prospects. Curr. Neuropharmacol. 2018;16:1018–1035. doi: 10.2174/1570159X16666180419121247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wareham L.K., Buys E.S., Sappington R.M. The nitric oxide-guanylate cyclase pathway and glaucoma. Nitric Oxide. 2018;77:75–87. doi: 10.1016/j.niox.2018.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J., Lee J., Oh J.H., Chang H.J., Sohn D.K., Kwon O., Shin A. Dietary lutein plus zeaxanthin intake and dicer1 rs3742330 a > g polymorphism relative to colorectal cancer risk. Sci. Rep. 2019;9:3406. doi: 10.1038/s41598-019-39747-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the conclusions of this article are all presented within the report.