Abstract
The active inference framework, and in particular its recent formulation as a partially observable Markov decision process (POMDP), has gained increasing popularity in recent years as a useful approach for modeling neurocognitive processes. This framework is highly general and flexible in its ability to be customized to model any cognitive process, as well as simulate predicted neuronal responses based on its accompanying neural process theory. It also affords both simulation experiments for proof of principle and behavioral modeling for empirical studies. However, there are limited resources that explain how to build and run these models in practice, which limits their widespread use. Most introductions assume a technical background in programming, mathematics, and machine learning. In this paper we offer a step-by-step tutorial on how to build POMDPs, run simulations using standard MATLAB routines, and fit these models to empirical data. We assume a minimal background in programming and mathematics, thoroughly explain all equations, and provide exemplar scripts that can be customized for both theoretical and empirical studies. Our goal is to provide the reader with the requisite background knowledge and practical tools to apply active inference to their own research. We also provide optional technical sections and multiple appendices, which offer the interested reader additional technical details. This tutorial should provide the reader with all the tools necessary to use these models and to follow emerging advances in active inference research.
Keywords: Active inference, Computational neuroscience, Bayesian inference, Learning, Decision-making, Machine learning
Introduction
Active inference, and in particular its recent application to partially observable Markov decision processes (POMDPs; defined below), offers a unified mathematical framework for modeling perception, learning, and decision making (Da Costa, Parr et al., 2020; Friston, Parr, & de Vries, 2017c; Friston, Rosch, Parr, Price, & Bowman, 2018; Parr & Friston, 2018b). This framework treats each of these psychological processes, and their interactions, as interdependent forms of inference. Namely, decision-making agents are assumed to infer the probability of different external states and events in the environment – including their own actions – by combining prior beliefs with sensory input. Unlike ‘passive’, perceptual inference processes (e.g., inferring the presence of an external object based on patterns of light impinging on the retina), the inferences underlying decision-making are ‘active’, in the sense that the agent infers the actions most likely to generate preferred sensory input (e.g., inferring that eating some food will reduce a feeling of hunger). Agents also infer the actions most likely to reduce uncertainty and facilitate learning (e.g., inferring that opening the fridge will reveal available food options). This leads decision-making to favor actions that optimize a trade-off between maximizing reward and information gain. The resulting patterns of perception and behavior predicted by active inference match well with those observed empirically (e.g., see Smith et al., 2021d, 2021c, 2020b; Smith, Kuplicki, Teed, Upshaw, & Khalsa, 2020c; Smith et al., 2021e, 2020e). The neural process theory associated with active inference has also successfully reproduced empirically observed neural responses in multiple research paradigms and generated novel, testable predictions (Friston, FitzGerald, Rigoli, Schwartenbeck, & Pezzulo, 2017a; Schwartenbeck, FitzGerald, Mathys, Dolan, & Friston, 2015; Whyte & Smith, 2020). Due to these and other considerations, this framework has become increasingly influential in recent years within psychology, neuroscience, and machine learning.
Over the last decade there have been many articles that offer either (1) broad intuitions about the workings and potential implications of active inference (e.g., (Badcock, Friston, Ramstead, Ploeger, & Hohwy, 2019; Clark, 2013, 2015; Clark, Watson, & Friston, 2018; Hohwy, 2014; Pezzulo, Rigoli, & Friston, 2015, 2018; Smith, Badcock, & Friston, 2020a)), or (2), technical presentations of the mathematical formalism and how it continues to evolve (e.g., Da Costa, Parr et al., 2020; Friston et al., 2016a, 2017a, 2017c; Hesp, Smith, Allen, Friston, & Ramstead, 2020; Parr & Friston, 2018b). However, for those first becoming acquainted with this field, the former class of articles does not provide sufficient detail to instill a thorough understanding of the framework, leading to potential misunderstanding and potentially inaccurate empirical predictions. At the other extreme, the latter class of articles is highly technical and requires considerable mathematical expertise, familiarity with notational conventions, and the broader ability to translate the mathematical formalism into empirical predictions relevant to a given field of study. This has made the active inference literature less accessible to a broader audience who might otherwise benefit from engaging with it. To date, there are also relatively few materials available for students seeking to gain the practical skills necessary to build active inference models and apply them to their own research aims (although some very helpful material has been prepared by others; e.g., Philipp Schwartenbeck [https://github.com/schwartenbeckph] and Oleg Solopchuk [https://medium.com/@solopchuk/tutorial-on-active-inference-30edcf50f5dc]).
The goal of this paper is to provide an accessible tutorial on the POMDP formulation of active inference that is easy to follow for readers without upper-level undergraduate/graduate-level training in mathematics and machine learning, while simultaneously offering basic mathematical understanding — as well as the practical tools necessary to build and use active inference models for their own purposes. We review the conceptual and formal foundations and provide a step-by-step guide on how to use code in MATLAB (provided in the appendices and supplementary code) to build active inference (POMDP) models, run simulations, fit models to empirical data, perform model comparison, and perform further steps necessary to test hypotheses using both simulated and fitted empirical data (all supplementary code can also be found at: https://github.com/rssmith33/Active-Inference-Tutorial-Scripts; we note here that there is also a recently developed python implementation of active inference that can be found at: https://github.com/inferactively/pymdp). We have tried to assume as little as possible about the reader’s background knowledge in hopes of making these methods accessible to researchers (e.g., psychologists and neuroscientists) without a strong background in mathematics or machine learning. However, we have also included sections that provide additional technical detail, which the pragmatic reader can safely skip over and still follow the practical tutorial aspects of the paper. We have also provided additional material in appendices and supplementary code with:
Definitional material to help the non-expert reader who would like to attempt the technical sections.
Additional mathematical detail for interested readers with a stronger technical background.
Pencil-and-paper exercises that help build an intuition for the behavior of these models.
A stripped down but well commented version of the most commonly used model inversion script (described below) for running simulations, which can serve as a springboard for readers seeking a deeper understanding of the code that implements these models.
Throughout the article, we will refer to the associated MATLAB code, assuming the reader is working through the paper and the code in parallel.
While we assume as little mathematical background as possible, some limited knowledge of probability theory, calculus, and linear algebra will be necessary to fully appreciate some sections of the tutorial. Building models in practice also requires some basic familiarity with the MATLAB programming environment. We realize that this background knowledge is nontrivial. However, to minimize these potential hurdles, we (1) provide thorough explanations when presenting the mathematics and programming (with further expansion within optional technical sections and in Appendix A), (2) include hands-on examples/exercises in the companion MATLAB code, and (3) provide pencil-and-paper exercises (see Appendix B and Pencil_and_paper_exercise_solutions.m code) that readers can work through themselves. In total, this tutorial should offer the reader the necessary resources to:
Acquire a basic understanding of the mathematical formalism.
Build generative models of behavioral tasks and run simulations of both behavioral and neural responses.
Fit models to behavioral data and recover model parameters on an individual basis, which can then be used for subsequent (e.g., between-subjects) analyses.
Our hope is that this will increase the accessibility and use of this framework to a broader audience. Note, however, that our focus is specifically on the POMDP formulation, which models time in discrete steps and treats beliefs and actions as discrete categories (referred to as ‘discrete state–space’ models with ‘discrete time’). This means that we do not cover a number of other topics associated with active inference and the broader free energy principle from which it is derived. For example, we do not cover ‘continuous state–space’ models, which can be used to model perception of continuous variables (e.g., brightness; for a tutorial, see Bogacz (2017) as well as motor control processes (e.g., controlling continuous levels of muscle contraction; see (Adams, Shipp, & Friston, 2013) and Buckley, Sub Kim, McGregor, and Seth (2017)). Nor do we cover ‘mixed’ models, in which discrete and continuous state–space models can be linked — allowing decisions to be translated into motor commands (e.g., see Friston et al. (2017c), Millidge (2019) and Tschantz et al. (2021). We also do not cover work on free energy minimization in self-organizing systems or the basis of the free energy principle in physics. The most thorough technical introduction to the physics perspective can be found in Friston (2019); a less technical (but still rigorous) introduction is presented in Andrews (2020).2 Thus, the focus of this tutorial is somewhat narrow and practical. Our aim is to equip the reader with the understanding and tools necessary to build models in practice and apply them in their own research.
The paper is organized as follows. In Part 1, we introduce the reader to the terms, concepts, and mathematical notation used within the active inference literature, and present the minimum mathematics necessary for a basic understanding of the formalism (as applied in a practical experimental setting). In Part 2, we introduce the reader to the concrete structure and elements of POMDPs and how they are solved. In Part 3, we provide a step-by-step description of how to build a generative model of a behavioral task (a variant on commonly used explore–exploit tasks), run simulations using this model, and interpret the outputs of those simulations. In Part 4, we introduce the reader to learning processes in active inference. In Part 5, we introduce the reader to the neural process theory associated with active inference and walk the reader through generating and interpreting the outputs of neural simulations that can be used to derive empirical predictions. In Part 6, we introduce hierarchical models and illustrate how, based on the neural process theory, they can be used to simulate established electrophysiological responses in a commonly used auditory mismatch paradigm. Finally, in Part 7 we describe how to fit behavioral data to a model and derive individual-level parameter estimates and how they can be used for further group-level analyses.
1. Basic terminology, concepts, and mathematics
1.1. Mathematical foundations: Bayes’ theorem and active inference
The active inference framework is based on the premise that perception and learning can be understood as minimizing a quantity known as variational free energy (VFE), and that action selection, planning, and decision-making can be understood as minimizing expected free energy (EFE), which quantifies the VFE of various actions based on expected future outcomes. To motivate the use and derivation of these quantities, we need to first introduce the reader to Bayesian inference and explore its relation to the notion of active inference. We will cover these foundational principles here. By the end of this subsection, the reader should have a working knowledge of the basic building blocks of active inference. This includes understanding what a model is, how rules within probability theory can be used to perform inference within a model, and how this inference process can be extended to perform action selection.
As an initial note to readers with less mathematical background, a full understanding of the equations presented below will not be necessary to begin building models and applying them to behavioral data. Often, building and working with models in practice is a great way to get an intuitive grasp of the underlying mathematics. So, if some of the equations below have unfamiliar notation and become hard to follow, do not get discouraged. An intuitive grasp of the concepts described in this section will be enough to learn the practical applications in the subsequent sections. That said, we also explain the equations and notation in this section assuming minimal mathematical background.
We start by highlighting that the term ‘active inference’ is based on two concepts. The first is the idea that organisms actively engage with (e.g., move around in) their environments to gather information, seek out ‘preferred’ observations (e.g., food, water, shelter, social support, etc.), and avoid non-preferred observations (e.g., tissue damage, hunger, thirst, social rejection, etc.). The second concept is Bayesian inference, a statistical procedure that describes the optimal way to update one’s beliefs (understood as probability distributions) when making new observations (i.e., receiving new sensory input) based on the rules of probability (for a brief introduction to the rules of probability, see Appendix A). Specifically, beliefs are updated in light of new observations using Bayes’ theorem, which can be written as follows:
| (1) |
Starting on the right-hand side of the equation, the term p(s|m) indicates the probability (p) of different possible states (s) under a model of the world (m). This ‘prior belief’ (the ‘prior’) encodes a probability distribution (‘Bayesian belief’) with respect to s before making a new observation (o). In general, the concept of a ‘state’ is abstract and can refer to anything one might have a belief about. For example, s might refer to the different possible shapes of an object, such as a square vs. a circle vs. a triangle, and so forth. The term p (o|s, m) is the ‘likelihood’ term and encodes the probability within a model that one would make a particular observation if some state were the true state (e.g., observing a straight line is consistent with a square shape but not with a circular shape). The symbol (|) means ‘conditional on’ and is also often read as ‘given’ (e.g., the probability of o given s). The term p (o|m) is the ‘model evidence’ (also called the ‘marginal likelihood’) and indicates how consistent an observation is with a model of the world in general (i.e., across all possible states). Finally, the term p (s|o, m) is the ‘posterior’ belief, which encodes what one’s new belief (i.e., adjusted probability distribution over possible states) optimally should be after making a new observation.
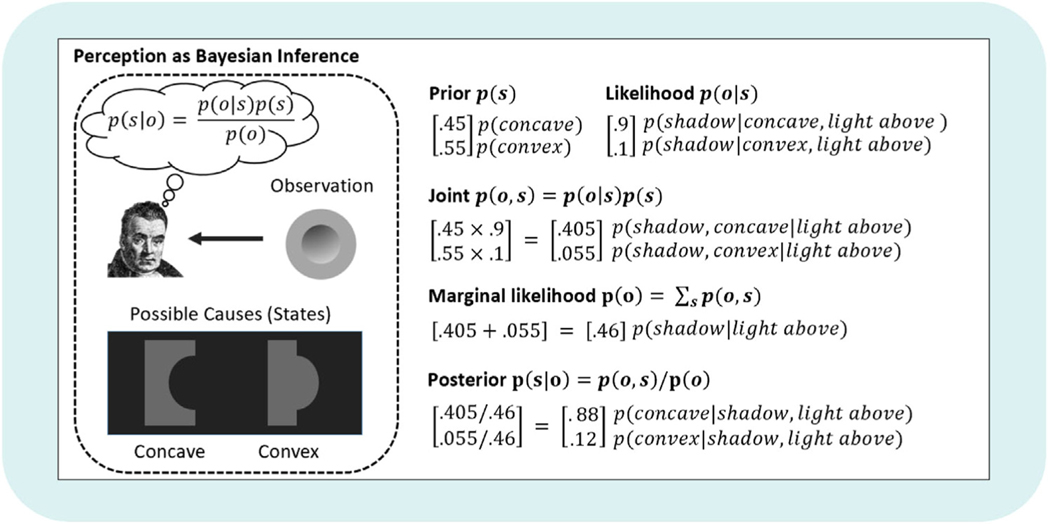
In essence, Bayes rule describes how to optimally update one’s beliefs in light of new data. Specifically, to arrive at a new belief (your posterior), you must: (1) take what you previously believed (your prior), (2) combine it with what you believe about how consistent a new observation is with different possible states (your likelihood), and (3) consider the overall consistency of that observation with your model (i.e., how likely that observation is under any set of possible states included in your model; the model evidence, p (o|m)). The last step (i.e., dividing by p (o|m)) ensures that your posterior belief remains a proper probability distribution that sums to 1 (i.e., it accomplishes ‘normalization’). For a simple numerical example of Bayesian inference in the context of perception, see Fig. 1.
Fig. 1.
Simple example of perception as Bayesian inference (based on Ramachandran, 1988). Please note that, while we have not explicitly conditioned on a model (m) in the expression of Bayes’ rule shown in the left panel (as in the text), this should be understood as implicit (i.e., priors and likelihoods are always model-dependent). In this example, we take a ‘brain’s eye view’ and imagine that we are presented with the shaded gray disk (the ‘observation’) in the left panel of the figure. Due to the shading pattern, the central portion of the disk is typically perceived as concave (as shown in the bottom-left), but it can also be perceived as convex (and typically is perceived as convex if rotated 180o). This is because the brain is equipped with a strong (unconscious) belief that light sources typically come from above. Given this assumption, the apparent shadow in the upper portion of the disk is much more likely to arise from a concave surface. To capture this mathematically, on the right we consider ‘concave’ and ‘convex’ as the two possible hidden states or ‘causes’ of sensory input (i.e., the shadow on the gray disk). We want to know whether the shadow pattern on the disk (observation) is caused by a concave or convex surface. The optimal way to infer the hidden state (concave or convex) is to use Bayes’ theorem. For the sake of this example, assume we believe the chances of observing a concave vs. convex surface in general are almost equal, with a slight bias toward expecting a convex surface (e.g., perhaps we have come across convex surfaces slightly more often in the past; encoded in the prior distribution shown above). The likelihood is a different story. The apparent shadow is much more consistent with a concave surface if light is coming from above (i.e., encoded in the likelihood distribution). To infer the posterior probability, we multiply the likelihood and prior probabilities, giving us the joint distribution. We then sum the probabilities in the joint distribution, yielding the total probability of the observation across the possible hidden states (i.e., the marginal likelihood). Finally, we divide the joint distribution by the marginal likelihood to reach the posterior. The posterior tells us that the most probable hidden state is a concave surface (i.e., corresponding to what is most often perceived). Thus, even though the two-dimensional gray disk alone is equally consistent with a convex or concave surface, the assumption that light is coming from above (encoded in the likelihood) most often leads us to perceive a concave 3-dimensional shape.
In this tutorial, the concept of a model is key. As briefly introduced above, we here focus specifically on generative models, which are models of how observations (sensory inputs) are generated by objects and events outside of the brain that cannot be known directly (typically termed ‘hidden states’ or ‘hidden causes’; e.g., a baseball generating a specific pattern of activation on the retina). In simple generative models (i.e., not yet incorporating action), the necessary variables correspond to those presented within Bayes’ theorem above (although note that conditioning on the model variable m is often left implicit, as we will also do going forward). That is, each model includes a set of possible hidden states (s), priors over those states p(s), a set of possible observations (o; also called ‘outcomes’), and a likelihood that specifies how states generate observations p(o|s). The notion of hidden or unobservable states causing observable outcomes illustrates how inference can be seen as a type of model inversion. Namely, updating one’s beliefs from prior to posterior beliefs is like inverting the likelihood mapping — that is, moving from p(o|s) to p(s|o). In other words, starting with a mapping from causes to consequences and then using it to infer the causes from consequences.
Importantly, models can also include multiple types/sets of states (i.e., different state-spaces). For example, one set of states could encode possible shapes, while another set of states could encode possible object locations. When different sets of states are independent in this way, each set is called a different ‘hidden state factor’. Similarly, models can include multiple types/sets of observable outcomes. For example, one set of possible observations could come from vision, while another set of possible observations could come from audition. When sets of observable outcomes are independent in this way, each set is called a different ‘outcome modality’. Once all sets of possible states and observations are specified, the generative model is defined in terms of the joint distribution p(o, s) — that is, the probability distribution over all possible combinations of states and observations. Based on the product rule in probability theory (see Appendix A), this can be decomposed into the separate terms just mentioned:
| (2) |
If there is only one set of states and observations, this joint distribution is a 2-dimensional distribution. If there are more sets, it becomes a higher-dimensional distribution that, while harder to visualize (and more time consuming to compute), can be treated in the same way.
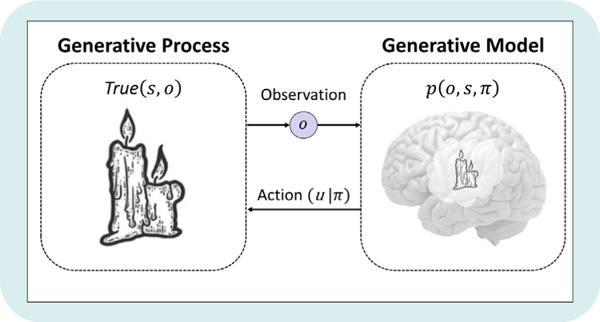
A crucial point to keep in mind at this point is the distinction between a generative model and the generative process (see Fig. 2). A generative model, as discussed above, is constituted by beliefs about the world and can be inaccurate (sometimes referred to as ‘fictive’). In other words, explanations for (i.e., beliefs about) how observations are generated do not have to represent a veridical account of how they are actually generated. Indeed, explanations for sensory data within models are often simpler than the true processes generating those data. In contrast, the generative process refers to what is actually going on out in the world — that is, it describes the veridical ‘ground truth’ about the causes of sensory input. For example, a model might hold the prior belief that the probability of seeing a pigeon vs. a hawk while at a city park is [.9 .1], whereas the true probability in the generative process may instead be [.7 .3]. This distinction is important in practical uses of modeling when one wants to simulate behavior under false beliefs and unexpected observations (e.g., when modeling delusions or hallucinations).
Fig. 2.
Visual depiction of the distinction between the generative process and the generative model, as well as their implicit coupling in the perception–action cycle. The generative process describes the true causes of the observations that are received by the generative model, which inform posterior beliefs about those causes (i.e., perception). In active inference, a generative model also includes policies (π), where each policy is a possible sequence of actions (u) that could be selected. The policy with the highest posterior probability (given preferred outcomes) is typically chosen, which couples the agent back to the generative process by changing the true state of world through action.
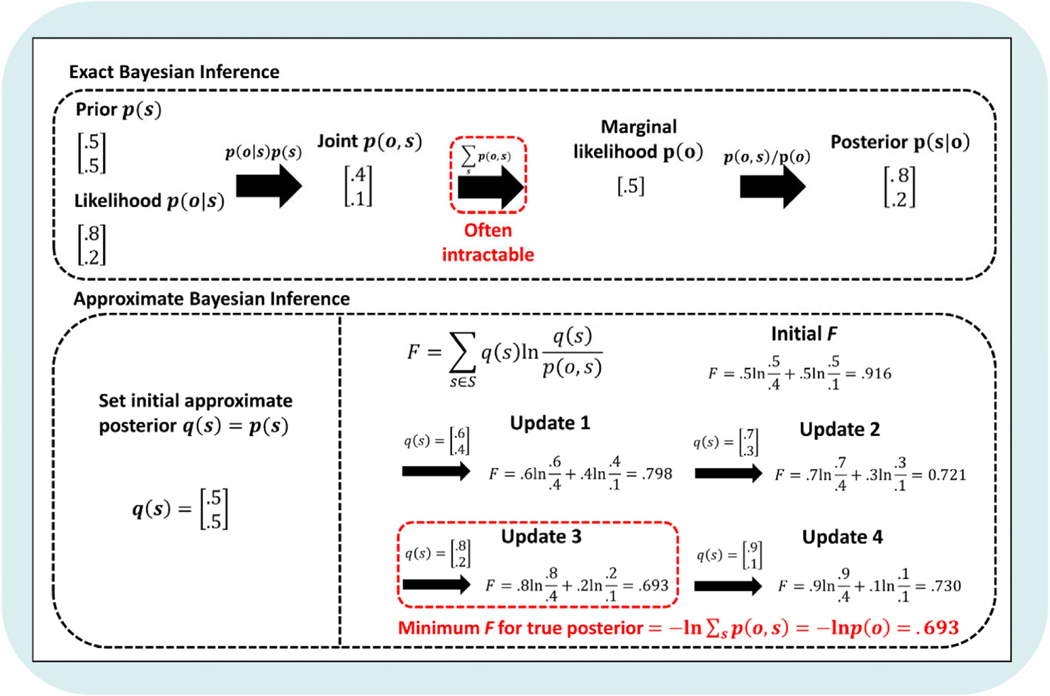
While Bayesian inference represents the optimal way to infer posterior beliefs within a generative model, Bayes theorem is computationally intractable for anything but the simplest distributions. This is because evaluating p(o|m) – the marginal likelihood (denominator) in Bayes’ theorem – requires us to sum the probabilities of observations under all possible states in the generative model (i.e., based on the sum rule of probability; see Appendix A and Fig. 1). For discrete distributions, as the number of dimensions (and possible values) increases, the number of terms that must be summed increases exponentially. In the case of continuous distributions, it requires the evaluation of integrals that do not always have closed-form (analytic) solutions. As such, approximation techniques are required to solve this problem. This is where VFE is crucial, as it provides a computationally tractable quantity that allows for approximate inference. One common way this is explained requires the introduction of an information-theoretic quantity known as self-information or surprisal (often also just called ‘surprise’, but we avoid this term here to minimize confusion with the distinct concept of psychological surprise). Surprisal reflects a deviation between observed outcomes and those predicted by a model. It is typically written as the negative log-probability of that observation, − ln p (o|m), where ln is the natural logarithm. Consistent with the intuitive notion of surprise, lower probability events generate higher surprisal values (e.g., − ln (.5) = 0.69, while − ln(.9) = 0.1). It therefore follows that minimizing surprisal is equivalent to maximizing the evidence an observation provides for a model; i.e., p(o|m). As will be demonstrated in the next section, VFE is always greater than or equal to surprisal, which means that minimizing VFE is also a way to maximize model evidence. This gets around the problem of computational intractability mentioned above and allows for inference of posterior beliefs over states. Fig. 3 provides an example of inference using Bayes’ theorem and using minimization of VFE (see the next subsection for an introduction to the relevant mathematics).
Fig. 3.
Simple example of exact versus approximate Bayesian inference. As with the example in Fig. 1, we are given a prior belief over states p (s) and the likelihood of a new observation p (o|s), and we wish to infer the posterior probability over states given that new observation p (s|o). Exact inference requires the evaluation of the marginal likelihood p (o), which, for anything but the simplest distributions, is either computationally intensive or intractable. Instead, variational inference minimizes VFE (here denoted by F), which scores the difference between an (initially arbitrary) approximate posterior distribution q(s) and a target distribution (here the exact posterior; for an introduction to variational inference, see Appendix A). By iteratively updating the approximate posterior to minimize F (usually via gradient descent, see main text for details), a distribution can be found that approximates the exact posterior. That is, q(s) will approximate the true posterior when it produces a minimum value for F. Here, we have shown an example of iterative updating for the simplest distribution possible to illustrate the concept. As shown in the bottom-left, in this example we start the agent with an approximate posterior distribution q (s) = p (s) = [.5 .5]T — which can be thought of as an initial guess about what the true posterior belief p(s|o) should be after making a new observation (o). We then define a generative model with the joint probability, p (o, s), where the true posterior we wish to find is p (s|o) = [.8 .2]T for the observation o (as calculated using exact inference in the top panel). On the bottom-right, under ‘Initial F’, we first solve for F using our initial q(s). Under ‘Update 1’, we then find (by searching neighboring values) a nearby value for q(s) that leads to a lower value for F, and we repeat this process in ‘Update 2’. In ‘Update 3’, q (s) is equal to p(s|o), which corresponds to finding a minimum value for F (i.e., where the remaining value above zero corresponds to surprisal, − ln p(o)). The fact that F has reached a minimum can be seen in ‘Update 4’, where continuing to change q(s) causes F to again increase in value. Thus, by finding the posterior q (s) over states that generates the minimum for F, that q(s) will also best approximate the true posterior. In the supplementary code, we have included a script VFE_calculation_example.m that will allow you to define your own priors, likelihoods, and observations and calculate F for different q (s) values.
Although Bayesian inference is used to model perception and learning in related frameworks (e.g., predictive coding; see (Bogacz, 2017), the active inference approach covered in this tutorial extends this application of Bayesian inference in two ways. First, it models categorical inference (e.g., the presence of a cat vs. a dog), as opposed to continuous inference (i.e., variables that take a continuous range of values, such as speed, direction of motion, brightness, etc.). Second, it models the inference of optimal action sequences during decision-making (i.e., inferring a probability distribution over possible action options, which can be thought of as encoding the estimated probability of achieving one’s goals if each action were chosen). In planning, possible sequences of actions (called ‘policies’)3 are denoted by the Greek letter pi (π), so the generative model is extended to:
| (3) |
We will return to the prior over policies later. For now, we simply note that active inference models can include additional elements that control (for example) how much randomness is present in decision-making and how habits can be acquired and influence decisions. They can also be extended to include learning. We will return to these extensions in later sections.
To make decisions, an agent requires a means of assigning higher value to one policy over another. This in turn requires that some observations are preferred over others. One of the more (superficially) counterintuitive aspects of active inference is the way it formalizes preferences. This is because there are no additional variables labeled as ‘rewards’ or ‘values’. Instead, preferences are encoded within a specific type of prior probability distribution — which is often called a ‘prior preference distribution’. This distribution is often simply denoted as p(o); however, the term p(o) is also used in other ways, which can be a source of confusion. Therefore, we will instead represent this distribution as , where the variable C denotes the agent’s preferences (Parr, Pezzulo, & Friston, 2022). In this distribution, observations with higher probabilities are treated as more rewarding. Note that this is distinct from priors over states, p(s), which encode beliefs about the true states of the world (i.e., irrespective of what is preferred).
The value of each policy in active inference is also specified within a probability distribution, where a higher value corresponds to a higher probability of being selected. This probability is based on EFE (i.e., lower EFE indicates higher value) and reflects beliefs about how likely each policy is to generate preferred observations (and how effective it is expected to be at maximizing information gain; discussed further below). In one sense, the use of probability distributions to encode preferences and policy values can simply be considered a kind of mathematical ‘trick’ to bring all elements of action selection within the domain of Bayesian belief updating — a kind of planning as inference (Attias, 2003; Botvinick & Toussaint, 2012; Kaplan & Friston, 2018). However, many articles have considered the possibility (or use language suggesting) that the formalism may have deeper implications. Specifically, the active inference literature often discusses how prior preferences may be thought of as encoding the observations that are implicitly ‘expected’ by an organism in virtue of its phenotype (i.e., the observations an organism must seek out to maintain its survival and/or reproduction). For example, consider body temperature. Humans can only survive if body temperatures continue to be observed within the range of 36.5 – 37.5 degrees Celsius. Thus, the human phenotype implicitly entails a high prior probability of making such observations. If a human perceives (i.e., infers) that their body temperature has (or is going to) deviate from ‘expected’ temperatures, they will infer which policies are most likely to minimize this deviation (e.g., seeking shelter when they are cold or expect to become cold). In this sense, body temperatures within survivable ranges (for the human phenotype) are the least ‘surprising’. In formal terms, the variable C in prior preferences can therefore be thought of as standing in for a model of an organism’s phenotype, where this model predicts specific (internal and external) observations consistent with that phenotype and motivates actions expected to maintain those observations.
However, it is important to highlight that this formal treatment of preferences and values as ‘Bayesian beliefs’ (i.e., probability distributions) need not be understood as a psychological description, nor must it be if one wishes to use active inference models in practice. In other words, not all beliefs at the mathematical level of description need to be equated with beliefs at the psychological level; some Bayesian beliefs in the formalism can instead correspond to rewarding or desired outcomes at the psychological level (Smith, Ramstead, & Kiefer, 2022). Similarly, the notion of ‘surprise’ with respect to prior preferences is not equivalent to the conscious experience of surprise; minimizing the type of ‘phenotypic surprise’ discussed in active inference is better mapped onto psychological states associated with achieving one’s goals. Traditional beliefs (in the psychological sense) can instead be identified with other model elements, such as priors over states, p(s), and observations expected given policies, . However, regardless of the way one views the mapping between mathematical and psychological levels of description, active inference can more broadly (and less controversially) be seen as suggesting that the brain just is (or that it ‘implements’ or ‘entails’) a generative model of the body and external environment of the organism.
1.2. Non-technical introduction to solving partially observable Markov decision processes via free energy minimization
In this subsection, we expand on the need for approximate inference in cases where Bayes’ theorem cannot be computed directly and explain how this motivates the use of VFE and EFE. By the end of this subsection, the reader should have a basic understanding of how VFE can be used to perform approximate Bayesian inference within a generative model and of how EFE extends this approach to infer optimal choices.
The specific type of generative model used here is a partially observable Markov decision process (POMDP). A Markov decision process describes beliefs about abstract states of the world, how they are expected to change over time, and how actions are selected to seek out preferred outcomes or rewards based on beliefs about states. This class of models assumes the ‘Markov property’, which simply means that beliefs about the current state of the world are all that matter for an agent when deciding which actions to take (i.e., that all knowledge about past states is implicitly ‘packed into’ beliefs about the current state). The agent then uses its model, combined with beliefs about the current state, to select actions by making predictions about possible future states. To be ‘partially observable’ means that the agent can be uncertain in its beliefs about the state of the world it is in. In this case, states are referred to as ‘hidden’ (as introduced above). The agent must infer how likely it is to be in one hidden state or another based on observations (i.e., sensory input) and use this information to select actions.
In active inference, these tasks are solved using a form of approximate inference known as variational inference (Attias, 2000; Beal, 2003; Markovic, Stojic, Schwoebel, & Kiebel, 2021) for a brief introduction, see Appendix A). Broadly speaking, the idea behind variational inference is to convert the intractable sum or integral required to perform model inversion into an optimization problem that can be solved in a computationally efficient manner. This is accomplished by introducing an approximate posterior distribution over states, denoted q(s), that makes simplifying assumptions about the nature of the true posterior distribution. For example, it is common to assume that hidden states under the approximate distribution do not interact (i.e., are independent), which gives the approximate distribution a much simpler mathematical form. Such assumptions are often violated, but the approximation is usually good enough in practice. Note that, despite its aim of approximating the true posterior , the approximate posterior q(s) is typically not written as being conditioned on observations. This is because it does not directly depend on observations — it is simply an (initially arbitrary) distribution over states that is iteratively updated to match the true posterior distribution as closely as possible (described below).
After introducing q(s), the next step in variational inference is to measure the similarity between this distribution and the generative model, p (o, s), using a measure called the Kullback–Leibler (KL) divergence. We will discuss the KL divergence in more detail in the following (technical) section. For now, it is sufficient to think of the KL divergence as a measure of the dissimilarity between two distributions. It is zero when the distributions match, and it gets larger the more dissimilar the distributions become. VFE corresponds to the surprisal we want to minimize plus the KL divergence between the approximate and true posterior distributions. In variational inference, we systematically update q (s) until we find the value that minimizes VFE, at which point q (s) will approximate the true posterior, . In Fig. 3 we provide a simple example of calculating VFE under different values for q (s) to provide the reader with an intuition for how this works (this example can also be reproduced and customized in the VFE_calculation_example.m script provided in the supplementary code). For ease of calculation, this figure uses the following expression for VFE (note that VFE is denoted by F when presented in equations):
| (4) |
However, this expression does not make it obvious how minimizing VFE will lead q (s) to approximate the true posterior. As discussed further in the technical sections, this can be seen more clearly by algebraically manipulating VFE into the following form, which is more often seen in the active inference literature:
| (5) |
For details on how we move between different expressions for F, see the optional technical section (Section 1.3). Here the term indicates the expected value or ‘expectation’ of a distribution and is equivalent to the term in Eq. (4). It indicates that is evaluated for each value of q (s) and then the resulting values are summed (see numerical example in Fig. 3). Based on this form of the equation, we can see that, because ln p(o) does not depend on q (s), the value of F will become smaller as the value of q (s) approaches the value of the true posterior, — since the former is divided by the latter and the log of one is zero.
Within the active inference framework, the task of both perception and learning is to minimize VFE in order to find (approximately) optimal posterior beliefs after each new observation. Perception corresponds to posterior state inference after each new observation, while learning corresponds to more slowly updating the priors and likelihood distributions in the model over many observations (which facilitates more accurate state inference in the long run). It is important to note, however, that minimizing VFE is not simply a process of finding the best-fitting approximation on every trial. Sensory input is inherently noisy, and simply finding the best-fitting posterior on each trial would lead to fitting noise, which would result in exaggerated and metabolically costly updates. In statistics, this is known as overfitting. Fortunately, VFE minimization naturally avoids this problem. Put into words, VFE measures the complexity of a model minus the accuracy of that model. Here, the term ‘accuracy’ refers to how well a model’s beliefs predict sensory input (i.e., the goodness of fit), while the term ‘complexity’ refers to how much beliefs need to change to maintain high accuracy when new sensory input is received (i.e., VFE remains higher if beliefs need to change a lot to account for new sensory input). Perception therefore seeks to find the most parsimonious (smallest necessary) changes in beliefs about the causes of sensory input that can adequately explain that input.
Analogously, the task of action selection and planning is to select policies that will bring about future observations that minimize VFE. The problem is, of course, that future outcomes have not yet been observed. Actions must therefore be selected such that they minimize expected free energy (EFE). Crucially, EFE scores the expected cost (i.e., a lower value indicates higher reward) minus the expected information gain of an action. This means that decisions that minimize EFE seek to both maximize reward and resolve uncertainty. When beliefs about states are very imprecise or uncertain, actions will tend to be information-seeking. Conversely, selected actions will tend to be reward-seeking when confidence in beliefs about states is high (i.e., when there is no more uncertainty to resolve and the agent is confident about what to do to bring about preferred outcomes).
However, as we will see later, if the magnitude of expected reward is sufficiently high (i.e., if a preference distribution is highly precise), actions that minimize EFE can become ‘risky’ — in that they seek out reward in the absence of sufficient information (i.e., reward value outweighs information value). In general, the imperative to minimize EFE is especially powerful in accounting for commonly observed behaviors in which, instead of seeking immediate reward, organisms first gather information and then maximize reward once they are confident about states of the world (e.g., turning on a light before trying to find food). It can also capture interesting behaviors that occur in the absence of opportunities for reward, where organisms appear to act simply out of ‘curiosity’ (Barto, Mirolli, & Baldassarre, 2013; Oudeyer & Kaplan, 2007; Schmidhuber, 2006). Further, variations in the precision of preferences during EFE minimization can capture interesting individual differences in behavior, as exemplified in the example of ‘risky’ behavior just mentioned. Note that this crucial aspect of active inference effectively addresses the ‘explore–exploit dilemma’ (discussed further below), because the imperatives for exploration (information-seeking) and exploitation (reward-seeking) are just two aspects of expected free energy, and whether exploratory or exploitative behaviors are favored in a given situation depends on current levels of uncertainty and the level of expected reward.
As we shall see in later sections, the posterior over policies can be informed by both VFE and EFE. For now, we simply note that this is because VFE is a measure of the free energy of the present (and implicitly the past), while EFE is a measure of the free energy of the future. This is important, because while some policies may lead to a minimization of free energy in the future, they may not have led to a minimization of free energy in the past (and are therefore suboptimal policies when evaluated overall). In other words, EFE scores the likelihood of pursuing (i.e., the value assigned to) a particular course of action based upon expected future outcomes, while VFE reflects the likelihood of (i.e., the value assigned to) a course of action based upon past/present outcomes. This means the posterior distribution over policies is a function of both VFE and EFE, where these quantities respectively furnish retrospective and prospective policy evaluations. At the psychological level, one can intuitively think of VFE as asking ‘how good has this action plan turned out so far?’, while EFE asks ‘how good do I expect things to go if I continue to follow this action plan?’.
When viewed from the perspective of potential neuroscientific applications, another crucial benefit of VFE and EFE is that they can be computed in a biologically plausible manner. This has inspired neural process theories that specify ways in which sets of neuron-like nodes (e.g., neuronal populations), with particular patterns of synapse-like connection strengths, can implement perception, learning, and decision-making through the minimization of these quantities. These neural process theories postulate several neuronal populations whose activity represents:
Categorical probability distributions (a special case of the multinomial distribution) over the possible states of the world. For those without background in probability theory, these distributions assign one probability value to each possible interpretation of sensory input, and all these probability values must add up to a value of 1. In other words, this assumes the world must be in this state or that state, but not both at the same time, and assigns one probability value to the world being in each possible state. (Note: in other papers you may come across the notation Cat(x), which simply indicates that a distribution x is a categorical distribution).
Prediction-errors, which signal the degree to which sensory input is inconsistent with current beliefs. Prediction errors drive the system to find new beliefs – that is, adjusted probability distributions – so that they are more consistent with sensory input, and therefore minimize these error signals.
Categorical probability distributions over possible policies the agent might choose.
These neural process theories also include simple, coincidence-based learning mechanisms that can be understood in terms of Hebbian synaptic plasticity (i.e., which involve adjusting the strength of the connections between two neurons when both neurons are activated simultaneously; (Brown, Zhao, & Leung, 2010). They further incorporate message passing algorithms (discussed in detail below) that can model the connectivity and firing rate patterns of neuronal populations organized into cortical columns. Such theories afford precise quantitative predictions that can be tested using neuroimaging and other electrophysiological measures of neuronal activation during specific experimental paradigms.
1.3. Technical introduction to variational free energy and expected free energy (optional)
In the previous subsections we introduced two quantities, VFE and EFE, which were described in largely qualitative terms. In this subsection, we consider the formal details behind the above descriptions. By the end of this subsection, the reader should have a working understanding of the different ways that VFE and EFE are often expressed in the literature, how these expressions are derived, and the theoretical insights that each expression provides. Readers without strong mathematical background can safely skip much of this section (if so desired) and move on to the next section without significant loss of understanding. That is, they should still be able to follow the rest of the paper. For those who choose to read this section, we provide accessible explanations of each equation in the hope that as many people as possible will be able to follow and learn from the material.
Before formally defining VFE and EFE, it will be helpful to first familiarize the reader with the relevant mathematical machinery. We will first expand on the KL divergence (; also sometimes called relative entropy), which was briefly introduced in the previous subsection. To remind the reader, this is a measure of the similarity, or dissimilarity, between two distributions. The KL divergence between two distributions, q(x) and p(x), is written as follows:
| (6) |
This equation states that the KL divergence is found by taking each value of x in range X (for which p and q assign values), calculating the value of the right-hand quantity, and then summing the resulting values (for a concrete numerical example, see the calculation of F in Fig. 3). From the perspective of information theory, the KL divergence can be thought of as scoring the amount of information one would need to reconstruct p(x) given full knowledge of q(x). In this context, ‘information’ is measured in a quantity called nats because it depends on the natural log, as opposed to log base 2 where information would be quantified in bits.
As mentioned above, because calculating model evidence is generally not possible, we instead minimize VFE, which is constructed to be an upper bound on negative (log) model evidence. As we noted above, this is also called ‘surprisal’ in information theory: − ln p (o). In other words, by minimizing VFE, one can minimize the negative model evidence — or, more intuitively, one can maximize model evidence (i.e., by finding beliefs in a model for which observations provide the most evidence).
Before turning to the formal definition of VFE, it will be helpful to clarify some notational conventions and concepts. Specifically, using the sum rule of probability we can express model evidence in terms of our generative model as: . That is, p (o) is the sum of the probabilities of observations for every combination of states and policies in the model. Next, one can multiply and divide the joint distribution, , by the (initially arbitrary) approximate distribution . By definition, multiplying and then dividing by does not change the value of this distribution. However, this trick ends up being quite useful as we will see. For mathematical convenience, one can take the negative logarithm of the resulting term, leading to:
| (7) |
As briefly mentioned in the previous subsection, denotes the expected value or expectation of a distribution. This can be thought as a kind of weighted average, where each value of one distribution (here ) is multiplied by the associated value in another distribution (here , and each of the resulting values is summed to get the expected value of the latter distribution. Note that, although written as a summation, the calculation of F in Fig. 3 represents a numerical example. Readers should note the formal similarity between the KL divergence and the expectation value. This is no accident, as the KL divergence is simply the expected difference of the log of two distributions, where the expectation is taken with respect to the distribution in the numerator. Note that, because ln , the distributions in the numerator are often swapped around, as we have done below. In the active inference literature, it is very common to move between KL divergence notation and expectation notation. As such, it can be useful to keep the following identities in mind:
| (8) |
With a clear idea of expectation values and the KL divergence now in mind, we move onto the definition of VFE. Specifically, leveraging the mathematical result referred to as Jensen’s inequality (Kuczma & Gilányi, 2009) – which states that the expectation of a logarithm is always less than or equal to the logarithm of an expectation – we arrive at the following inequality:
| (9) |
On the right-hand side of this equation is VFE, which is defined in terms of the KL divergence – that is, expected difference of the respective logs – between the generative model and the approximate posterior distribution . When the approximate posterior distribution and the generative model match, VFE is equal to zero (i.e., when . To understand the equality on the left-hand side, consider that the expectation, , entails summing over all values of s and π; that is, . This removes s and π from the expression in both the denominator and the numerator, leaving − ln p (o). From this, we can see that, by minimizing VFE, we minimize an upper bound on negative log model evidence (here, with respect to states under each policy). This means that VFE will always be greater than or equal to − ln p (o), which entails that by minimizing the value of VFE, the model evidence p (o) will either increase or remain the same (i.e., it will be maximized, as the logarithm is a monotonically increasing function).
Therefore, all that is needed to perform approximate Bayesian inference in perception, learning, and decision-making is a tractable approach to finding the value of s (i.e., the approximate posterior distribution over s) that minimizes VFE. This can be accomplished by performing a gradient descent on VFE. Gradient descent is a technique that starts by picking some initial value for s and then calculates VFE for this value. It then calculates VFE for neighboring values of s and identifies the neighboring values for which VFE decreases most. It then samples from values of s that neighbor those values and continues to do so iteratively until a minimum VFE value is found (i.e., where VFE no longer decreases for any neighboring values). At this point, an approximation to the optimal beliefs for s has been found (given some set of observations o). On a final terminological note, because VFE is a function that is defined in terms of probability distributions, which are themselves functions, VFE is sometimes referred to as a ‘functional’, which is simply the mathematical term for a function of a function.
Note that in active inference we calculate VFE with respect to each available policy individually (denoted by Fπ). This is because different policies, through their impact on hidden states in the generative process, make certain observations more likely than others. For example, consider a situation where I believe there is a chair to my left and a table to my right. Conditional on having chosen to look left, it is more likely that I will observe a chair than a table. This means that observing the chair acts as evidence that I have chosen the policy of looking left. Because observations provide evidence for policies in this way, both the approximate posterior , and the generative model , are conditioned on policies. This may be useful in some cases where, for example, one is considering the possibility that an agent could have false beliefs about the actions they are carrying out or could be surprised when their intended policy does not match the true observed actions. Going forward, VFE will be presented in terms of Fπ (as shown in the following paragraph).
As we will discuss below, one way in which the brain may accomplish gradient descent on VFE during perception is through the minimization of prediction error. The reason for this can be brought out by doing some algebraic rearrangement to express VFE as a measure of complexity minus accuracy (i.e., as touched upon informally in the previous subsection):
| (10) |
| (L2) |
| (L3) |
| (L4) |
The first line expresses VFE in terms of the expected log difference (KL divergence) between the approximate posterior and the generative model. In the second line we use log algebra to express the division as a subtraction (). In the third line we use the product rule of probability ( to take the likelihood term out of the first expectation term. The fourth line re-expresses the third line, but uses more compact notation for the first term and drops the dependency on policies in the second term (i.e., because we assume here that the likelihood mapping does not depend on policies). The first term in line 4 is the KL divergence between prior and posterior beliefs. This value will be larger if one needs to make larger revisions to one’s beliefs, which is the measure of complexity introduced earlier. A greater complexity means there is a greater chance of changing beliefs to explain random aspects of one’s observations, which can reduce the future predictive power of a model (analogous to ‘overfitting’ in statistics). The second term in line 4 reflects predictive accuracy (i.e., the probability of observations given model beliefs about states). The brain will therefore minimize VFE if it minimizes prediction error (maximizing accuracy) while not changing beliefs more than necessary (minimizing complexity).
Another common way that VFE is expressed is in terms of placing a bound on surprisal:
| (11) |
This equation rearranges line 1 in Eq. (10) (again using the product rule: ) to show that VFE is always greater than or equal to surprisal (i.e., is an upper bound on surprisal) with respect to a policy (i.e., greater than − ). In machine learning, the sign of VFE is usually switched, so that it becomes an evidence lower bound, also known as an ELBO. Maximizing the ELBO is a commonly used optimization approach in machine learning (Winn & Bishop, 2005).
From a slightly different perspective, the gradients of VFE leveraged in active inference can also be expressed as a mixture of prediction errors (i.e., when thought of more broadly as differences to be minimized). This is because complexity is the average difference between posterior and prior beliefs, while accuracy is the difference between predicted and observed outcomes. This therefore also licenses a description of active inference as prediction error minimization (Burr & Jones, 2016; Clark, 2017; Fabry, 2017; Hohwy, Paton, & Palmer, 2016), corresponding to the minimization of these two differences (minimizing VFE through prediction error minimization is described in more detail in Sections 2.4 and 5).
However, active inference is not solely concerned with minimizing prediction error in perception. It is also a model of action selection. When inferring optimal actions, one cannot simply consider current observations, because actions are chosen to bring about preferred future observations. As described informally above, this means that, to infer optimal actions, a model must predict sequences of future states and observations for each possible policy, and then calculate the expected free energy (EFE) associated with those different sequences of future states and observations. As a model of decision-making, EFE also needs to be calculated relative to preferences for some sequences of observations over others (i.e., how rewarding or punishing they will be). In active inference, this is formally accomplished by equipping a model with prior expectations over observations, p(o|C), that play the role of preferences.4 For an initial intuition of how this works, consider two possible policies that correspond to two different sequences of states and observations, where one sequence of observations is preferred more than the other. Since ‘preferred’ here formally translates to ‘expected by the model’, then the policy expected to produce preferred observations will be the one that maximizes the accuracy of the model (and hence minimizes EFE). This means that the probability (or value) of each policy can be inferred based on how much expected observations under a policy will maximize model accuracy (i.e., match preferred observations). When preferred observations are treated as implicit expectations definitive of an organism’s phenotype (e.g., those consistent with its survival, such as seeking warmth when cold, or water when thirsty) this has also been described as ‘self-evidencing’ (Hohwy, 2016).
To score each possible policy in this way, EFE (denoted Gπ in equations) can be expressed as follows:
| (12) |
| (L2) |
| (L3) |
| (L4) |
The first line expresses EFE as the expected difference between the approximate posterior and the generative model. Note that because EFE is calculated with respect to expected outcomes that (by definition) have not yet occurred, observations enter the expectation operator Eq as random variables (i.e., otherwise it is identical in form to the expression for VFE in Eq. (10)). The second line uses the product rule of probability, to rearrange EFE into two terms that can be associated with information-seeking and reward-seeking. To make this clear, the third line does two things. First, it replaces the true posterior () with an approximate posterior (). Second, it drops the conditionalization on π in the second term and instead conditions on the variable C described above that encodes preferences (i.e., ). This is a central move within active inference. Namely, is used to encode preferred observations, and the agent seeks to find policies expected to produce those observations. The agent’s preferences can be independent of the policy being followed, which allows us to drop the conditionalization on π. As mentioned earlier, in most papers on active inference prior preferences are simply written as ; however, to clearly distinguish this from the ln p (o) term within VFE (i.e., where o is an observed variable), we write the term here as explicitly conditioned on C (Parr et al., 2022).
The first term on the right-hand side of line 3 is commonly referred to as the epistemic value, or the expected information gain of a state when it is conditioned on expected observations. The second term is commonly referred to as pragmatic value, which, as just mentioned, scores the agent’s preferences for particular observations. To make the intuition behind epistemic value more apparent, the fourth line flips the terms inside the first expectation so that it becomes prefixed with a negative sign (i.e., ). Because the epistemic value term is subtracted from the total, it is clear that to minimize EFE overall an agent must maximize the value of this term by selecting policies that take it into states that maximize the difference between prior and posterior beliefs; that is, maximize the difference between and . In other words, the agent is driven to seek out observations that reduce uncertainty about hidden states (Parr & Friston, 2017a). For example, if an agent were in a dark room, the mapping between hidden states and observations would be entirely ambiguous, so it would be driven to maximize information gain by turning on a light before seeking out preferred observations (i.e., as it would be unclear how to bring about preferred outcomes before the light was turned on).
Another very common expression of EFE in the active inference literature is:
| (13) |
For a full description of how you get from line 1 of Eq. (12) to this decomposition, see Appendix A. The first term on the right-hand side of this equation scores the anticipated difference (KL divergence) between (1) beliefs about the probability of some sequence of outcomes given a policy, and (2) preferred outcomes (i.e., those expected a priori within the model). This term is sometimes referred to as ‘risk’ (or expected complexity), but it can more intuitively be thought of as beliefs about the probability of reward for each choice one could make. That is, the lower the expected divergence between preferred outcomes and those expected under a policy, the higher the chances of attaining rewarding outcomes if one chose that policy. The second term one the right-hand side of the equation is the expected value of the entropy (H) of the likelihood function, where . Entropy is a measure of the dispersion of a distribution, where a flatter (lower precision) distribution has higher entropy. A higher-entropy likelihood means there are less precise predictions about outcomes given beliefs about the possible states of the world. This term is therefore commonly referred to as a measure of ‘ambiguity’. Policies that minimize ambiguity will try to occupy states that are expected to generate the most precise (i.e., most informative) observations, because those observations will provide the most evidence for one hidden state over others. Putting the risk and ambiguity terms together means that minimizing EFE will drive selection of policies that maximize both reward and information gain (for simple numerical examples of calculating the risk and ambiguity terms, see discussion of ‘outcome prediction errors’ in Section 2.4). Typically, seeking information will occur until the model is confident about how to achieve preferred outcomes, at which point it will choose reward-seeking actions. Importantly, as briefly mentioned earlier, the expression for EFE above entails that stronger (more precise) preferences for one outcome over others will have the effect of down-weighting the value of information, leading to reduced information-seeking (and vice-versa if preferences are too weak or imprecise). This affects how a model resolves the ‘explore–exploit dilemma’ (Addicott, Pearson, Sweitzer, Barack, & Platt, 2017; Friston et al., 2017b; Parr & Friston, 2017a; Schwartenbeck et al., 2019; Wilson, Geana, White, Ludvig, & Cohen, 2014) — that is, the difficult judgement of whether or not one ‘knows enough’ to trust their beliefs and act on them to seek reward or whether to first act to gather more information (see example simulations below). For a more detailed description and step-by-step derivation of the most common formulations of EFE in the active inference literature, see Appendix A.
2. Building and solving POMDPs
2.1. Formal POMDP structure
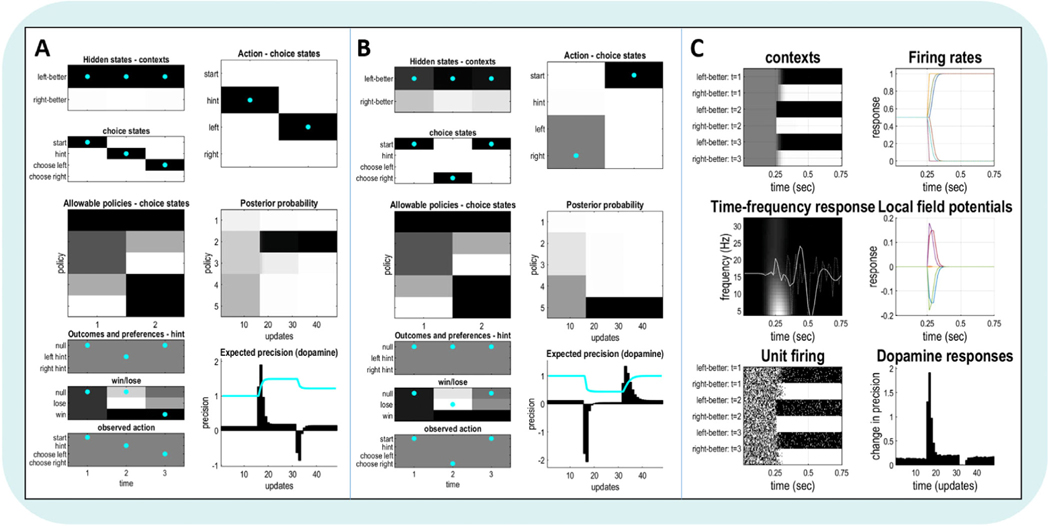
In this first subsection, we introduce the reader to the abstract structure and elements of an active inference POMDP, which is the standard modeling approach in active inference research at present. In a POMDP, one is given a specific type of generative model, including observations, states, and policies, and the goal is to infer posterior beliefs over states and policies when conditioning on observations. By the end of this subsection, the reader should be able to identify and interpret each type of variable in these models and understand the role they play in performing inference. A warning: upon initial exposure, gaining a full understanding of this abstract structure can feel daunting. However, after we put together a model of a concrete behavioral task (in Section 3), this structure – and how to practically use it – tends to become much clearer. The task we will model is an ‘explore–exploit’ task similar to commonly used multi-armed bandit tasks employed in computational psychiatry research (see Fig. 4). In this variant, there are two slot machines with unknown probabilities of paying out. A participant can simply guess, resulting in either a large reward or no reward, or they can ask for a hint (which may or may not be accurate). If they get it right after taking the hint, they receive a smaller reward. This allows for competition between an information-seeking drive and a reward-seeking drive. This task will be described in detail in Section 3, but we will use parts of this broad-strokes description below to exemplify uses of the more abstract elements making up POMDPs.
Fig. 4.
Depiction of the explore–exploit task example. Note that the states and outcomes shown on the right are only examples. Table 1 and Section 3 list all states, outcomes, and policies required to build a generative model for this task.
The term POMDP denotes two major concepts. As described above, the first is partial observability, which means that observations may only provide probabilistic information about hidden states (e.g., observing a hint may indicate that one or another slot machine is more likely to pay out). The second is the Markov property, which simply means that, when making decisions, all relevant knowledge about distant past states is implicitly included within beliefs about the current state. This assumption can be violated, but it allows modeling to be more tractable and is ‘good enough’ in many cases. When dealing with violations of the Markovian assumption – such as when modeling memory – it is necessary to model several interconnected Markovian processes that evolve over different timescales. We will see related examples in later sections covering both hierarchical models and how the parameters of a POMDP can be learned through repeated observations. Here we start with a simple, single-level POMDP where Markovian assumptions are not violated. In the presentation below, note that vectors (i.e., single rows or columns of numbers) are denoted with italics, while matrices (i.e., multiple rows and columns of numbers) are not italicized and denoted with bold.
A POMDP includes both trials and time points (tau; τ) within each trial (sometimes called ‘epochs’). An important thing to note here is that τ indexes the time points about which agents have beliefs. This is distinct from the variable t, which denotes the time points at which each new observation is presented. This is a common (and understandable) source of confusion for those new to the active inference literature (perhaps exacerbated by the fact that t and τ look so similar). To appreciate the need for this distinction, consider cases in which an observation in the present can change one’s beliefs about the past. For example, imagine that you start out in one of two rooms (a green room or a blue room), but you do not know what color the walls are. Later, when you open your eyes and find out the room is painted blue, you will change your belief now about where you were earlier before you opened your eyes (i.e., you had been in the blue room the whole time). In a formal model, this would be a case in which beliefs about one’s state at time τ = 1 change after making a new observation at time t = 2. Thus, the inclusion of both t and τ in active inference entails that the agent updates its beliefs about states at all time points τ with new observations at each time point t. This allows for retrospective inference, as in the previous example, as well as for prospective inference, in which an agent updates beliefs about the future (e.g., τ = 3), when making new observations in the present (e.g., t = 2). This would be the case in the explore–exploit task example, where observing a hint at one time point could update beliefs about which slot machine will be better at the next time point. This is an important distinction to keep in mind when trying to understand simulation results (e.g., in terms of working memory for the past and future; i.e., postdiction and prediction).
In practice, this is accomplished by having entries of 0 for all elements of an observation vector when t < τ. To illustrate this formally, we will use the simpler example of being in one of two rooms described above. In this case, there will be a ‘color’ observation modality where observations could be ‘blue’ or ‘green’ (i.e., a vector with one element for each color). At time t = 1, the observed color for time τ = 2 has not yet occurred. So, at t = 1:
If blue were then observed at t = 2, the observation for τ = 2 would be updated to:
This vector would then remain unchanged for all future time points t > 2 (i.e., the observation is never ‘forgotten’ once it has taken place). The same thing would then occur for all subsequent observations (e.g., at t = 1 and 2; but if green were observed at t = 3 then the vector would be updated to and remain that way for t > 3, etc.). This allows beliefs about states for all time points to be updated at each time point t when these observation vectors are updated.
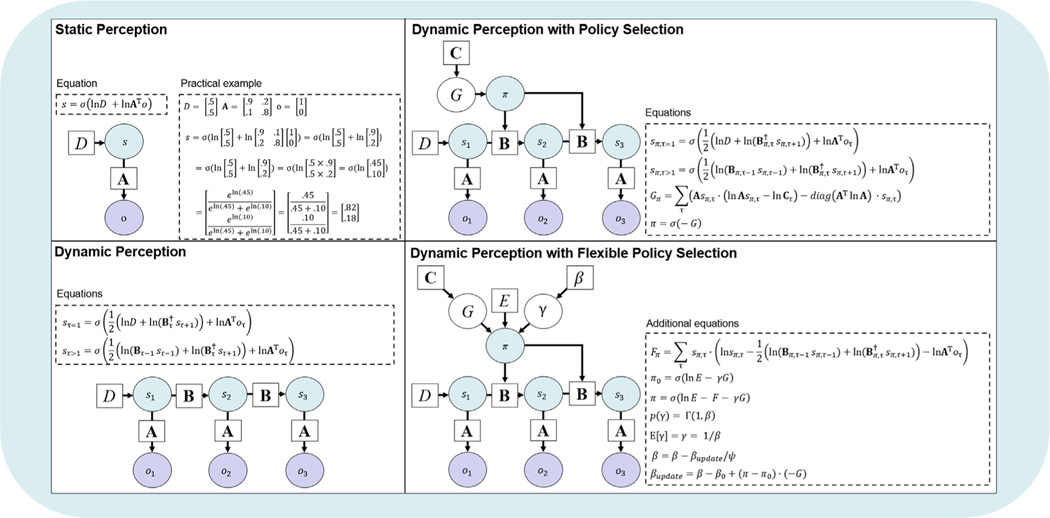
Having now clarified time indexing, we will move on to other model elements. At the first time point in a trial , the model starts out with a prior over categorical states, , encoded in a vector denoted by D – one value per possible state (e.g., which slot machine is more likely to pay out). When there are multiple state factors, there will be one D vector per factor. As touched on above, multiple state factors are necessary to account for multiple types of beliefs one can hold simultaneously. One common example is holding separate beliefs about an object’s location and its identity. In the explore–exploit task example, this could include beliefs about which slot machine is better and beliefs about available choice states (e.g., the state of having taken the hint).
At each subsequent time point, the model has prior beliefs about how one state will evolve into another depending on the chosen policy, , encoded in a ‘transition matrix’ denoted by — one column per state at τ and one row per state at . If transitions for a given state factor are identical across policies, they can be represented by a single matrix. When transitions for a state factor are policy-dependent, there will be one matrix per possible action (i.e., one for each possible state transition under a policy). In other words, the combination of a policy and a time specifies a transition matrix (i.e., encoding the action that would be taken under that policy at that point in time; described further below). In the explore–exploit task example, this could include transitioning to the state associated with getting the hint or transitioning to the state associated with selecting one of the two machines (depending on the policy).
The likelihood function, , is encoded in a matrix denoted by A — one column per state at τ and one row per possible observation at τ. When there are multiple outcome modalities, there will be one A matrix per outcome modality. As touched upon above, multiple outcome modalities are necessary to account for parallel channels of sensory input (e.g., one for possible visual inputs and one for possible auditory inputs). In the explore–exploit task example, this could include one modality for observing the hint and another modality for observing reward vs. no reward.
Preferred outcomes, ln , are specified using a matrix denoted by C — one column per time point and one row per possible observation. When there are multiple outcome modalities, there will be one C matrix per modality. In the explore–exploit task example, this could encode a strong prior preference for a large reward, a moderate preference for a small reward, and low preference for no reward.
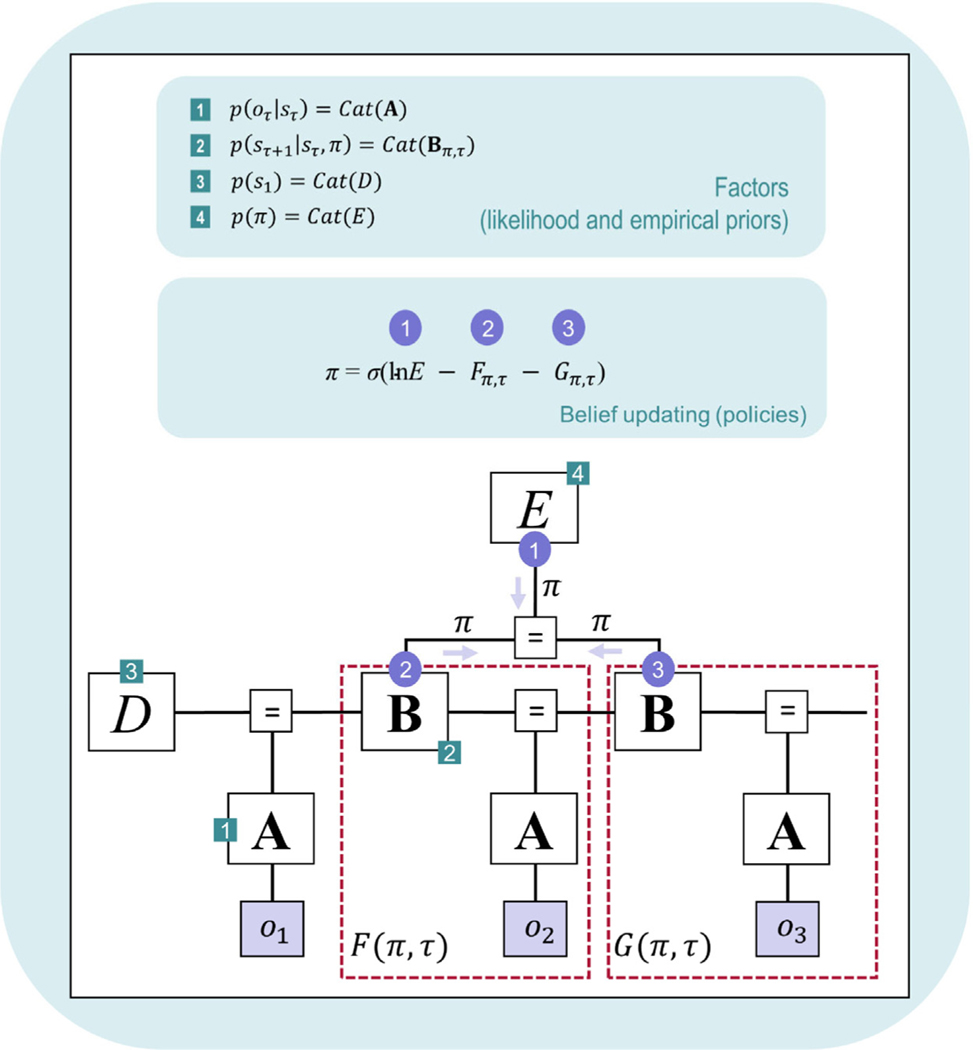
Prior beliefs about policies p (π) are encoded in a (column) vector E (one row per policy) — increasing the probability that some policies will be chosen over others (i.e., independent of observed/expected outcomes). This can be used to model the influence of habits. For example, if an agent has chosen a particular policy many times in the past, this can lead to a stronger expectation that this policy will be chosen again. In the explore–exploit task example, E could be used to model a simple choice bias in which a participant is more likely to choose one slot machine over another (independent of previous reward learning). However, it is important to distinguish between this type of prior belief and the initial distribution over policies from which actions are sampled before making an observation (π0). As explained further below (and in Table 2), this latter distribution depends on E, G, and γ, where the influences of habits and expected future outcomes each have an influence on initial choices.
Table 2.
Matrix formulation of equations used for inference.
| Model update component | Update equation | Explanation | Model-specific description for explore-exploit task (described in detail Section 3) |
|---|---|---|---|
| Updating beliefs about initial states expected under each allowable policy. |
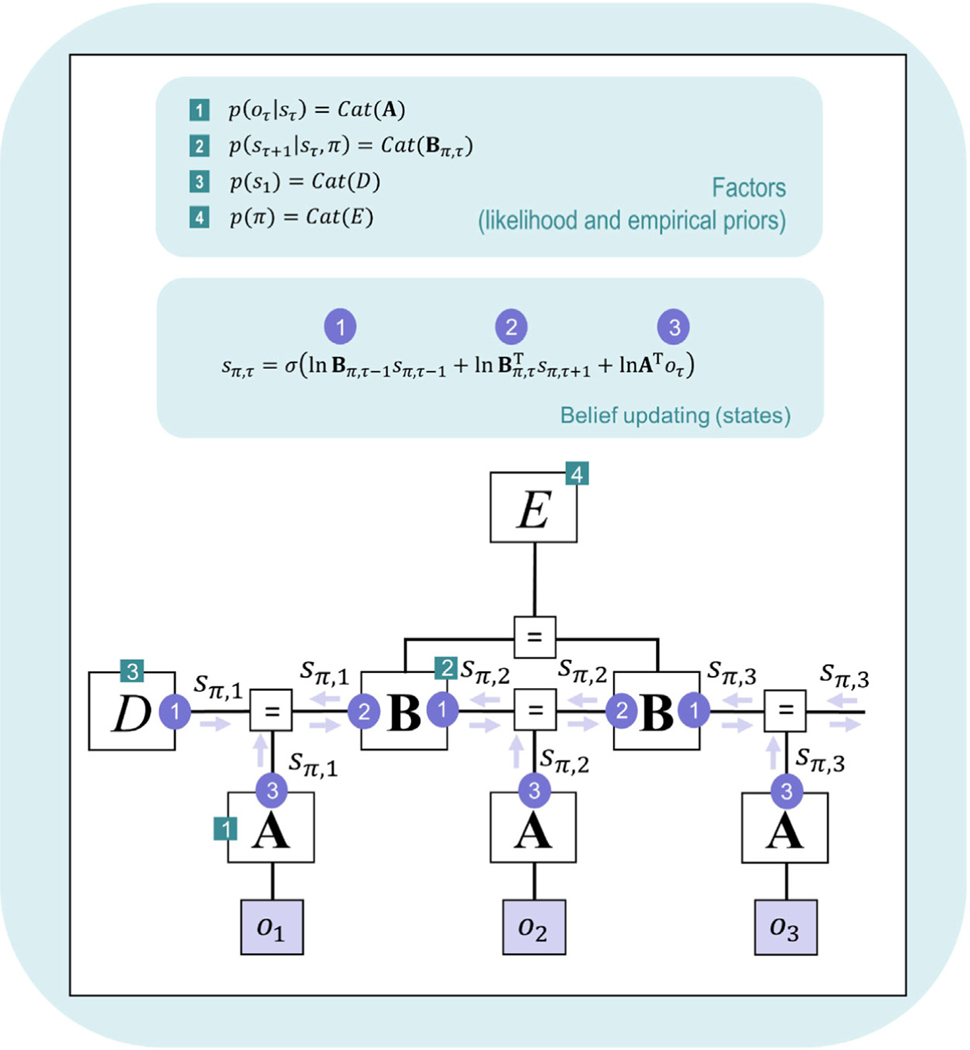
|
First equation: The variable is the state prediction error with respect to the first time point in a trial. Minimizing this error corresponds to minimizing VFE (via gradient descent) and is used to update posterior beliefs over states. The term () corresponds to prior beliefs in Bayesian inference, based on beliefs about the probability of initial states, D, and the probability of transitions to future states under a policy, . The term corresponds to the likelihood term in Bayesian inference, evaluating how consistent observed outcomes are with each possible state. The term ln corresponds to posterior beliefs over states (for the first time point in a trial) at the current update iteration. Second Equation: We move to the solution for the posterior by setting , solving for ln , and then taking the softmax (normalized exponential) function (denoted σ) to ensure that the posterior over states is a proper probability distribution with non-negative values that sums to 1. This equation is described in more detail in the main text. A numerical example of the softmax function is also shown in Appendix A. |
Updating beliefs about: 1. Whether the left vs. right slot machine is more likely to pay out on a given trial. 2. The initial choice state (here, always the ‘start’ state). |
| Updating beliefs about all states after the first time point in a trial that are expected under each allowable policy. |
|
First equation: The variable is the state prediction error with respect to all time points in a trial after the first time point. Minimizing this error corresponds to minimizing VFE (via gradient descent) and is used to update posterior beliefs over states. The term () corresponds to prior beliefs in Bayesian inference, based on beliefs about the probability of transitions from past states, , and the probability of transitions to future states, , under a policy. The term ln corresponds to the likelihood term in Bayesian inference, evaluating how consistent observed outcomes are with each possible state. Second Equation: As in the previous row, we move to the solution for the posterior, , by setting , solving for ln , and then taking the softmax function (σ). This equation is described in more detail in the main text. |
Updating beliefs about: 1. Whether the left vs. right slot machine is more likely to pay out on a given trial. 2. Beliefs about choice states after the initial time point (here, this depends on the choice to take the hint or to select one of the slot machines). |
| Probability of selecting each allowable policy |
|
The initial distribution over policies before making any observations (π0), and the posterior distribution over policies after an observation (π). The initial distribution is made up of the learned prior over policies encoded in the E vector (reflecting the number of times a policy has previously been chosen) and the expected free energy of each allowable policy (G). The posterior distribution is determined by E, G, and the variational free energy (F) under each policy after making a new observation. The influence of G is also modulated by an expected precision term (γ), which encodes prior confidence in beliefs about G (described further in the main text; also see Fig. 9). See row 1 for an explanation of the function of the σ symbol. We note, however, that incorporation of E, F, and/or γ when computing π is a modeling choice. These need not be included in all cases (e.g., see top-left portion of Fig. 5; also see Da Costa, Parr et al., 2020). In some contexts, one might choose to include some of these terms but not others, or to only include G. This depends on the research question. (e.g., E will be useful if task behavior is influenced by habits, while can be useful when there are many possible deep policies to choose from). See the row in this table on ‘Expected free energy precision’ for more details about inference over policies when are included. This is also discussed further in the main text. |
Updating overall beliefs about whether the best course of action is to take the hint and/or to choose the left vs. right slot machine. |
| Expected free energy of each allowable policy |
|
The first equation reproduces the ‘risk + ambiguity’ expression for the expected free energy of each policy (Gπ) that is explained in the main text. The second equation shows this same expression in terms of the elements in the POMDP model used in this tutorial (i.e., in matrix notation). Expected free energy evaluates the value of each policy based on their expected ability to: (1) generate the most desired outcomes, and (2) minimize uncertainty about hidden states. Achieving the most desired outcomes corresponds to minimizing the KL divergence between preferred observations, , and the observations expected under each policy, . Minimizing uncertainty corresponds to minimizing the expected entropy of the likelihood . Note that the diag() function simply takes the diagonal elements of a matrix and places them in a row vector. This is simply a convenient method for extracting and operating on the correct matrix entries to calculate the entropy, , of the distributions encoded within each column in A. For simple numerical examples of calculating the risk and ambiguity terms, see discussion of ‘outcome prediction errors’ in Section 2.4. |
The ‘risk’ term - - drives the agent to select the slot machine expected to be most likely to pay out. If the value of winning money in Cτ is high enough (i.e., if is sufficiently precise), this will deter the agent from choosing to ask for the hint. The ‘ambiguity’ term - - drives the agent to minimize uncertainty by asking for the hint. |
| Marginal free energy of each allowable policy |
|
The first equation shows the marginal (as opposed to variational) free energy, which is now used in the most recent implementations of active inference. The second equation shows this same expression in terms of the elements in the POMDP model used in this tutorial (i.e., in matrix notation). Marginal free energy has a sightly different form than the expressions for VFE that are also shown in the text (and which have been used in many previous papers in the active inference literature). This updated form improves on certain limitations of the message passing algorithms derived from minimization of VFE (see Section 2.3; also see (Parr, Markovic, Kiebel, & Friston, 2019). Marginal free energy evaluates the evidence that inferred states provide for each policy (based on new observations at each time point). See the first two rows in this table on updating beliefs about states for an explanation of how each term in the equation relates to Bayesian inference. |
This would encode the amount of surprise (given a choice of policy) when observing a hint or a win/loss after selecting a specific slot machine. |
| Expected free energy precision |
Iterated to convergence: |
The β term, and its prior value β0, is a hyperparameter on the expected free energy precision term (γ). Specifically, β is the ’rate’ parameter of a gamma distribution (Γ) with a ‘shape’ parameter value of 1. The expected value of this distribution, , is equal to the reciprocal of β. Note that we use the non-italicized γ to refer to the random variable and use the italicized γ to refer to the scalar value of that variable. This scalar is what is subsequently updated based on the equations shown here. The γ term controls the precision of G, based on the agent’s confidence in its estimates of expected free energy. This confidence changes when new observations are consistent or inconsistent with G. More specifically, y modulates the influence of G on policy selection based upon a G prediction error (Gerror). This is calculated based on the difference between the initial distribution over policies (π0) and the posterior distribution after making a new observation (π). The difference between these terms reflects the extent to which new observations (scored by F) make policies more or less likely. If the vector encoding the posterior over policies increases in magnitude in comparison to the prior, and still points in the same direction, the difference vector between the posterior and the prior will point in the same direction as the — G vector (i.e., less than a 90° angle apart; see Fig. 9). If so, the value of γ will increase, thereby increasing the impact of G on policy selection. In contrast, if the difference vector between the posterior and the prior does not point in the same direction as the — G vector (i.e., greater than a 90° angle apart), γ will decrease and thereby reduce the impact of G on policy selection (i.e., as the agent’s confidence in its estimates of expected free energy has decreased). Note that the ft update term mediating these updates technically corresponds to the gradient of free energy with respect to . The subsequent update in the value of γ is such that G contributes to the posterior over policies in an optimal manner. β and Gerror are often discussed in relation to dopamine in the active inference literature. Note that β0 is the initial prior (which is not updated), and β is the initial posterior, which is subsequently updated to provide a new estimate for γ = 1/β. The variable ψ is a step size parameter that reduces the magnitude of each update and promotes stable convergence to final values of γ. For a derivation of these equations, see Appendix in Sales, Friston, Jones, Pickering, and Moran (2019). |
A higher value for β would reduce an agent’s confidence in the best policy based on the values in G. This might lead the agent to select a slot machine more randomly or based to a greater extent on its past choices (i.e., if it has a precise prior over policies in the vector E). |
Table note: The term denotes the transpose of with normalized columns (i.e., columns that sum to 1). Note that you may commonly see the dot (·) notation used in the active inference literature to denote transposed matrix multiplication, such as , which means (we use the latter notation here). When A matrices have more than two dimensions (i.e., when they are tensors), the transpose is applied to the two-dimensional matrix associated with each value of the other dimensions. The σ symbol indicates a softmax operation (for an introduction see Appendix A), which transforms vector values to make up a proper probability distribution (i.e., with non-negative values that sum to 1). Italicized variables indicate vectors (or single numbers [scalars] in the case of β and γ). Bold, non-italicized variables indicate matrices. Subscripts indicate conditional probabilities; e.g., ).
Each allowable action (u) is encoded as a possible state transition (one of several B matrices that can be chosen for a state factor). In this case, each possible action is encoded in a vector U, and the possible sequences of actions (where each allowable sequence defines a policy) are encoded in a matrix denoted by V (one row per time point, one column per policy, and a third dimension for state factor). In the explore–exploit task example, U could include the choice to take the hint and the choice to select each of the two machines, while V could include the possible action sequences, such as, for example, taking the hint and then choosing the left machine vs. taking the hint and then choosing the right machine. Note that the possible actions encoded in the vector U are also sometimes referred to as ‘control states’ in the active inference literature.
As we have done in previous sections, the free energy and expected free energy for each policy are denoted by vectors F and G, respectively. The degree to which G controls policy selection is modulated by a further parameter γ (a single number; i.e., a scalar). This parameter is a precision estimate for the expected free energy over policies. It can be thought of as encoding a prior belief about the confidence with which policies can be inferred (i.e., how reliable beliefs about the best policy are expected to be). It is often called the ‘prior policy precision’ parameter; however, it is important to note that this is not the same thing as the precision of posterior beliefs over policies (π). This is because π also depends on the vectors E (habits) and F (shown in Table 2 further below) — which means, for example, that π could be precise even if γ were low (Hesp et al., 2020). For this reason, it is better to think of γ as an ‘expected free energy (G) precision’ parameter as opposed to a policy precision parameter per se. If no habits are present (i.e., if E is a flat distribution), lower γ values lead to more randomness in policy selection. In the presence of strong habits, lower γ values increase how much habits influence policy selection, because the influence of G is reduced relative to E (see equation for π in Table 2 further below). You can simulate these dynamics yourself by specifying values for γ, E, F, and G within the EFE_Precision_Updating.m code provided in the supplementary code.
In some models we will discuss below with multi-step policies, the prior value for γ is updated into a posterior γ estimate (via updates to its hyperparameter β). Table 2 and Fig. 9 explain this in detail. Briefly, the value of this precision parameter is increased after each observation if the variational free energy over policies is consistent with the expected free energy over policies prior to that observation. If these free energies are inconsistent, this precision is decreased (i.e., the agent becomes less confident in its estimates of G). However, it is important to note that there are situations in which policies are only considered from the current time step into the future (such as ‘shallow’ or ‘short-sighted’ policies that, at each time step, only ‘look ahead’ one time step to consider the immediate consequences of different actions). In such cases, previous observations do not inform the relative probabilities of policies (i.e., they are just ‘reset’ at each time step) — and the expected precision reduces to the prior value for γ (and is not updated). (Note: below, and in the supplementary code, we show how ‘shallow’, one-step policies vs. ‘deep’, multi-step policies can be included by specifying policies with the variable U vs. V).
Fig. 9.
Illustration of a geometric interpretation of the factors contributing to updates in expected free energy precision estimates (γ). In these examples we include two policies and specify the values of priors over policies (E), expected free energy over policies (G), and the variational free energy over policies following a new observation (F). Based on these, a prior and posterior over policies (π0 and π) are computed (calculations shown in each of the top panels). Updates are then computed for β, where γ = 1/β, constrained by a fixed prior value β0 and a step size parameter (ψ) that promotes convergence to a stable posterior across iterations. The direction and magnitude of the resulting update in γ is driven by the dot product between the difference vector for prior and posterior policy beliefs (π − π0) and the −G vector. This dot product can be thought of as a prediction error (Gerror) reflecting the level of (dis)agreement between expected free energy and the variational free energy of a subsequent observation. The plots in the top panels show cases where γ is positively updated (left) and negatively updated (right). In the case on the left, the two vectors (π − π0 and −G) point in a similar direction (i.e., less than 90° apart), which represents a way to visualize how new observations (through F) provide evidence for the reliability of G (leading to an increase in its precision weighting γ). In the case on the right, the vectors are greater than 90° apart, providing evidence against the reliability of G (leading to a decrease in its precision weighting γ). Note that, for reasons of clarity, the endpoints of the vectors shown here are not the actual values of π − π0 and −G; they instead correspond to scaled values of these vectors, which makes them similar in length and more clearly illustrates the angle separating them. The middle panels show 16 iterations of γ updating, as is done per time point (i.e., observation) in a trial in the supplementary code (and in the standard SPM routines) until a stable posterior estimate is reached. This is similar to prediction error minimization dynamics for the state and outcome predictions errors described earlier. The bottom panels show the rate of change in γ, which bears some similarity to prediction error responses. The updates shown in this figure have been associated with dopamine in the neural process theory accompanying active inference. These simulations can be reproduced using the EFE_Precision_Updating.m code provided in the supplementary code.
All the model variables are summarized in Table 1. Solutions for inference in the POMDP are shown in Table 2 at the end of this section. In each of these tables we also provide a description of the way each model element can be used to implement the explore–exploit task example, which we build in Section 3.
Table 1.
Model variables.
| Model variable* | General definition | Model specification for explore-exploit task (described in detail Section 3) |
|---|---|---|
| oτ | Observable outcomes at time τ. | Outcome modalities: 1. Hints (no hint, hint-left, hint-right) 2. Reward (start, lose, win) 3. Observed behavior (start, take hint, choose left, choose right) |
| sτ | Hidden states at time τ. One vector of possible state values for each state factor (i.e., each independent set of states; e.g., visual vs. auditory states). | Hidden state factors: 1. Context (left machine is better vs. right machine is better) 2. Choices (start, take hint, choose left, choose right) |
| π | A vector encoding the distribution over policies reflecting the predicted value of each policy. Each policy is a series of allowable actions in a vector U, where actions correspond to different state transitions (i.e., different matrices) that can be chosen by the agent for each state factor. Policies are chosen by sampling from this distribution. | Allowable policies include the decision to: 1. Stay in the start state 2. Get the hint and then choose the left machine 3. Get the hint and then choose the right machine 4. Immediately choose the left machine (and then return to the start state) 5. Immediately choose the right machine (and then return to the start state) |
| A matrix: | A matrix encoding beliefs about the relationship between hidden states and observable outcomes at each time point τ (i.e., the probability that specific outcomes will be observed given specific hidden states at specific times). Note that in the POMDP structure typically used in the active inference literature (and which we describe in this tutorial), the likelihood is assumed to remain constant across time points in a trial, and hence will not differ at different values for τ (although one could adjust this if so desired). The likelihood is also assumed to be identical across policies, and so there is no indexing with respect to π. When there is more than one outcome modality, there is one A matrix per outcome modality. When there is more than one state factor, these matrices become high-dimensional and are technically referred to as tensors. For example, a second state factor corresponds to a third matrix dimension, a third state factor corresponds to a fourth matrix dimension, and so forth. |
Encodes beliefs about the relationship between: 1. Probability that the hint is accurate in each context 2. Probability of reward in each context 3. Identity mapping between choice states and observed behavior |
| matrix: | A matrix encoding beliefs about how hidden states will evolve over time (transition probabilities). For states that are under the control of the agent, there are multiple matrices, where each matrix corresponds to one action (state transition) that the agent may choose at a given time point (if consistent with an allowable policy). When there is more than one hidden state factor, there is one or more matrices per state factor (depending on policies). | Encodes beliefs that: 1. Context does not change within a trial 2. Transitions from any choice state to any other are possible, depending on the policy. |
| C matrix: | A matrix encoding the degree to which some observed outcomes are preferred over others (technically modeled as prior expectations over outcomes). When there is more than one outcome modality, there is one C matrix per outcome modality. Rows indicate possible observations; columns indicate time points. Note that each column of values in C is passed through a softmax function (transforming it into a proper probability distribution) and then log-transformed (using the natural log). Thus, preferences become log-probabilities over outcomes. | Encodes the stronger preference for wins than losses. Wins are also more preferred at the second time point than the third time point. |
| D vector: p(s1) | A vector encoding beliefs about (a probability distribution over) initial hidden states. When there is more than one hidden state factor, there is one D vector per state factor. | The agent begins in an initial state of maximal uncertainty about the context state (prior to learning), but complete certainty that it will start in the ‘start’ choice state. |
| E vector: p(π) | A distribution encoding beliefs about what policies will be chosen a priori (a prior probability distribution over policies, implemented as a vector assigning one value to each policy), based on the number of times different actions have been chosen in the past. | The agent has no initial habits to choose one slot machine or another (prior to learning). |
While, for consistency, we have used the standard notation found in the active inference literature, it is important to note that it does not always clearly distinguish between distributions and the possible values taken by random variables under those distributions. For example, π refers to the distribution over policies, but when used as a subscript it indexes each individual policy (e.g., indicates a distinct matrix for each different policy). This same convention holds for s and o.
2.2. Graphical models
In many papers in the active inference literature, POMDPs are represented using graphical models. We will now discuss these representations and what their role is in active inference. By the end of this subsection, the reader should be able to interpret graphical models and understand the different benefits they provide.
Graphical models, such as the graphs shown in Figs. 5–7, are a useful method for visually depicting how variables in a model depend on one another. When models include probability distributions over variables, these graphs can be used to represent the conditional relationships between these variables. These types of probabilistic graphical models are particularly useful in the context of active inference because they provide a clear visual summary of the computational architecture of the models, and the way (biologically plausible) message passing algorithms (described below) can be used to update beliefs. Here we consider two types of graphical models – Bayesian networks (or ‘Bayes nets’, see Fig. 5; and for an introduction, see chapter 8 of (Bishop, 2006) and Forney-style (normal) factor graphs (Dauwels, 2007; Loeliger, 2004). For readers interested in a more detailed introduction to the use of Forney-style factor graphs in active inference, we recommend the excellent tutorial introduction by de Vries and Friston (2017).
Fig. 5.
Bayesian network representations of state estimation (perception) and policy selection. Each graph depicts a generative model of the causes of observations, which can be inverted to perform inference. Top left Static perception with a worked example. Variables: s = states, o = observations, A = likelihood mapping between states and outcomes, D = initial state priors. This is equivalent to exact Bayesian inference. Bottom left: Dynamic perception. Transition matrices (Bτ) have been added to describe (beliefs about) how states change over time. Subscripts for observations and states correspond to time point in a trial (denoted by tau; τ). Importantly, when τ > 1, the Bτ matrix from the previous τ (i.e., Bτ−1) functions as an empirical prior, playing the same role as the D vector at τ = 1. Top right: Dynamic perception with policy selection. Each policy (π) entails a different sequence of actions, which corresponds to different transitions between states (i.e., different Bπ,τ matrices). Based on expected free energy (G; which in turn depends on prior preferences, C), the highest probability will be assigned to policies expected to minimize uncertainty over states and maximize the probability of preferred observations. Bottom right: Dynamic perception with flexible policy selection. This final model includes an expected free energy precision term γ = E[γ], where p (γ) corresponds to a gamma distribution (Γ) with a ‘shape’ parameter equal to 1 and a ‘rate’ parameter specified by β. Note that the non-italicized γ in the generative model is a random variable, whereas the italicized γ is a statistic (expected value) of the gamma distribution (i.e., a fixed scalar) that is updated based on the equations shown in the figure. The value of γ encodes the agent’s confidence in policy selection and adjusts the contribution of G to the posterior distribution over policies (π). This precision value is also optimized by updating the value of β after new observations, based on the variational free energy (F) over policies associated with those observations. In short, when a new observation is inconsistent with prior beliefs about policies (π0; i.e., based on G), the agent assigns a lower expected precision (γ) to G when arriving at posteriors over policies (π; see Fig. 9 and Table 2 for more details). A prior over policies (E) is also included, which can be used to model habit formation. A lower γ (i.e., less confidence in model-based beliefs about G) also entails a stronger influence of the habits encoded in E on policy selection. Note that the dependency of π on F has been omitted from the graphical depiction of the generative model in this panel. See main text for further variable descriptions. See Table 2 for further explanation of these equations.
Fig. 7.
This figure reproduces the same graph shown in Fig. 6 to illustrate the link between message passing and policy selection in active inference. Top: As in Fig. 6, these equations specify the factors that constitute the factorized generative model, and the numbers in the green squares highlight the correspondence between the equations and the factors represented by the normal factor graph below. Middle Belief update equation for inferring the posterior over policies. Purple numbers indicate the correspondence between terms within the update equation and the messages passed between each factor shown in the factor graph. Bottom: Normal factor graph representation of message passing in the context of inference over policies. Red dotted lines show partition functions of the graph, which are used to construct the free energy approximations to the probability of current observations conditioned on policies, − ln p (oτ|π) ≈ Fπ,τ, and the expected probability of future observations conditioned on policies, −Eq(oτ>t,sτ>t|π) [ln p (oτ>t|π)] ≈ Gπ,τ. The factors Fπ,τ and Gπ,τ then become the messages (shown by the purple arrows) sent from the two transition probability factors (Bπ,τ−1 and Bπ,τ) that converge on the equality constraint node (connecting the Bπ,τ nodes and the E node). When combined with the message sent from E, and after the application of a softmax function, this becomes the posterior over policies (adapted from Friston et al. (2017c), Parr and Friston (2018a)). As noted in the text, this representation of inference over policies as message passing is heuristic and only meant as an analogy to message passing with respect to posteriors over states. This is because it is not carried out iteratively (i.e., the posterior is arrived at using a single iteration), the messages are not bidirectional, and Fπ,τ and Gπ,τ are not factors. This graphical representation also cannot illustrate all dependencies with respect to Gπ,τ . This is because Gπ,τ depends on two different types of predicted future observations – p (oτ>t|π) and p (oτ>t|C) — only the first of which is depicted here (i.e., with respect to o3). For a proposed scheme for carrying out iterative message passing with respect to inference over policies, see Champion, Grześ, and Bowman (2021).
When depicting active inference models with Bayes nets (Fig. 5), the circles (‘nodes’) correspond to variables (e.g., observations, hidden states, and policies), while the arrows connecting nodes (‘edges’) show the dependencies between variables represented by each node. For example, the arrows in Fig. 5 going from the ‘sτ node’ (i.e., states at time τ) to the ‘oτ node’ (i.e., outcomes at time τ) means that the value of oτ depends on the value of sτ. This entails that if one knows something about observations, then one can infer something about the hidden states that cause them (i.e., the hidden states that generate those observations in the generative model). Readers familiar with Bayesian networks will note that the form of the graphical model shown in Fig. 5 is slightly unusual, as squares denoting the factors that mediate the conditional relationships have been placed on top of the edges (e.g., the A and matrices).
This graphical model serves as a concise visual depiction of the relationships between model elements covered in detail in the previous subsection. It illustrates how observations at each time step (purple) are generated by hidden states (green) via a mapping encoded by the likelihood matrix A. The matrix is shown as mediating the dependencies between states at different time points (i.e., encoding beliefs about how states change over time). The probability over states at the first time point is shown to depend on the D vector. State transitions () are shown to depend upon policies (π). The probability distribution over policies in turn depends on learned priors over policies (E) and the EFE of each allowable policy (G). The EFE is shown to depend on the prior distribution over observations (C), which encodes the agent’s preferences for some observations over others (i.e., this dependency entails that policies with the lowest EFE will be those expected to generate the most preferred observations). The influence of EFE on policies also depends upon its precision term (γ), which encodes confidence in current EFE estimates. This in turn depends on the value of β (an initial prior over γ that can subsequently be updated; see Table 2 for a description). To help readers gain an intuition for inference using graphical models, Fig. 5 builds up a full POMDP starting from a graphical representation of perception at a single time point using Bayes theorem (with a worked example). It then adds the evolution of states over time, followed by their dependence on policies, and the dependence of policies on the variables just described.
The defining characteristic of a generative model such as that shown in Fig. 5 is that it can be used to generate data (i.e., observations). The conditional dependencies depicted in the Bayesian network in the bottom-right of this figure show how observations are generated by a POMDP. Starting at the top of the network, a policy (π) is first selected via a softmax (normalized exponential) function (σ) of the aforementioned variables (for an introduction to the softmax function, see Appendix A). The initial distribution over policies prior to receiving an observation is denoted , while the posterior over policies after receiving an observation also incorporates the VFE: . Next, policy-dependent transition probabilities encoded in the matrix (or the D vector at ) generate hidden states, which in turn generate observations at each time point. The likelihood (A) matrix determines which observations are generated by each hidden state.
Recall that to perform inference we must invert the generative model (i.e., infer the most likely states and policies given each new observation). This is where normal factor graphs (Fig. 6) are crucial, as they can be leveraged to both derive and visualize a suite of message passing algorithms (see below) for Bayesian inference. Normal factor graphs are made up of square nodes and edges (lines connecting square nodes). Square nodes can be thought of as functions (i.e., factors or conditional probability distributions; see below) that take in some input (e.g., sufficient statistics5 of beliefs over states or observations) and transform that information in some way to produce an output (e.g., the sufficient statistics of the conditional probabilities specified by the factor). These inputs and outputs are called ‘messages’ and are represented by the edges connecting the square nodes. When an edge connects to only one square node it is called a half edge, and it only carries messages to and from that node. When an edge connects two square nodes, this indicates that these nodes exchange messages and that each contributes to the value represented on that edge. When three square nodes exchange messages, the graph is adjusted to contain three edges (one connected to each square node) that converge onto an equality node (a small square node with an ‘=‘ sign), which combines the messages in a specific manner (described in the following technical section). As described further below, the messages represented on each edge correspond to the variables represented by the circular nodes in the Bayes’ net depiction in Fig. 5, while the square nodes in a factor graph correspond directly to the square nodes in this figure. Note that, in some cases, the edges in factor graphs are also supplemented by including circle nodes to represent (the sufficient statistics of) hidden variables (as in Fig. 5), but we depict them without circle nodes in Figs. 6 and 7 to give the reader some familiarity with this commonly presented form. Also note that, unlike Bayes net depictions, normal factor graphs have undirected edges, which highlights the bidirectional nature of message passing (see light purple arrows in the bottom portion of Fig. 6).
Fig. 6. Top:
Equations specifying the factors that constitute the factorized generative model. Numbers in the green squares highlight the correspondence between the equations and the factors in the generative model that are represented within the normal factor graph in the bottom panel. Here, Cat() indicates a categorical distribution. Middle: Belief update equation for approximate posteriors over states that is derived from variational message passing (note the difference between this message passing scheme and the marginal message passing approach described in the text). Purple numbers indicate the correspondence between terms within the update equation and the messages passed between each factor shown in the factor graph in the bottom panel. Bottom: Normal factor graph representation of the factorized POMDP. In contrast to the Bayes net representation shown in Fig. 5, nodes (large white boxes) represent factors, whereas the edges (lines connecting each box) represent the sufficient statistics of approximate posteriors, which are passed as messages between factors (i.e., edges represent the common variables that participate in the factors they connect, such as posteriors over states under each policy for each time point, ). Source: Adapted from Friston et al. (2017c).
More technically, normal factor graphs represent a factorization of the generative model. Recall that generative models are formally defined as the joint probability distribution over observations, states, and policies of the POMDP across time, . Factorization means that this joint probability can be defined as the product of several conditionally independent distributions. In POMDPs, the factorization assumes that each state only depends on the state at the previous time step and policy (i.e., the so-called Markov property). This is described by the following equation, which shows a factorization of the joint distribution into prior distributions over states and policies, and the distributions representing the likelihood and state transitions.
| (14) |
| (L2) |
| (L3) |
For the unfamiliar reader, please note that the symbol indicates taking the product of each of the distributions to the right of it for each time point τ to the final time point T. Line 2 shows the form of the distribution conditioned on policies. Line 3 rewrites line 2 in matrix form by replacing each categorical distribution with the above-described matrices/vectors whose columns contain the parameters of the respective distributions. Specifically, , and . Here and are vectors of zeros with a one placed in the element corresponding to the state/observation of interest. Their role is simply to select out the elements of the A or B matrix corresponding to the relevant state-outcome pair or current state-previous state pair. Once in matrix form, it is easy to see the direct correspondence between the factorized distribution shown in the equation above and the factors included in the normal factor graph in Fig. 6. Each of these distributions is associated with a factor node. Each edge represents the probability distribution over the variable that needs to be inferred (i.e., the approximate posteriors over states sπ,τ, and policies π). Edges connect factors that exchange messages with the same variables (e.g., D, A, and are all connected by the variable ).
2.3. Technical section on variational and marginal message passing (optional)
In this subsection we will introduce variational message passing (Winn & Bishop, 2005), which is foundational to the way active inference performs approximate inference of posteriors over states. By the end of this subsection, the reader should understand the general steps for performing variational message passing and how they can be carried out using the factor graph representation of POMDPs in active inference. For readers with less mathematical background, this section can also be safely skipped without compromising the ability to understand the rest of the tutorial. Although, as always, we have made efforts to fully explain all equations. For readers with specific interest in this topic, we also note here that more recent implementations of active inference have used a refined algorithm – called marginal message passing – that is more robust to problems of overconfidence that arise with variational message passing (i.e., where posterior beliefs can become too precise too quickly; see Parr et al. (2019). However, understanding marginal message passing requires us to first understanding variational message passing. Therefore, we will focus on this approach here, and return to how it has been refined at the end of the section.
To invert the model (i.e., condition on observations to infer approximate posteriors over states and policies) via the minimization of VFE, some simplifying assumptions need to be made (i.e., since exact inference is intractable in most real-word cases). Variational message passing is based on the mean-field approximation, which assumes that the approximate posterior factorizes into the product of (independent) distributions (Bishop, 2006). This approximation often works well in practice, but it has the limitation of ignoring possible pairwise (or more complex) interactions between variables. In the POMDPs under discussion here, the mean-field approximation assumes that the approximate posterior factorizes into a prior distribution over policies and the distributions over states expected under each policy at each time point:
| (15) |
Note that, by convention, approximate posterior distributions are denoted with the variable q. Also again recall that T corresponds to the final time point in a trial, such that this posterior distribution is over the values of states across time points under each policy — and this distribution itself evolves over time with each new observation. This means that an observation at a later time can change posterior beliefs about states at earlier times (i.e., retrospective inference).
With this factorization in hand, we can employ variational message passing to infer the approximate posterior at each edge of the graph, and then combine them into a global posterior using the equation just presented. Variational message passing can be summarized in terms of the following steps:
Initialize the values of the approximate posteriors for all hidden variables (i.e., all edges) in the graph.
Fix the value of observed variables (here, ).
Choose an edge (V) corresponding to the hidden variable you want to infer (here, ).
Calculate the messages, , which take on values sent by each factor node connected to V.
Pass a message from each connected factor node N to V (often written as ).
- Update the approximate posterior represented by V according to the following rule: . The arrow notation here indicates messages from two different factors arriving at the same edge.
- Normalize the product of these messages so that corresponds to a proper probability distribution.
- Use this new to update the messages sent by connected factors (i.e., for the next round of message passing).
Repeat steps 4–6 sequentially for each edge.
Steps 3–7 are then repeated until the difference between updates converges to some acceptably low value (i.e., resulting in stable posterior beliefs for all edges).
For unfamiliar readers, the ‘∝’ symbol in step 6 denotes proportionality, meaning that the ratio between variables is always constant. We can change from the proportionality sign to an equals (‘=‘) sign by explicitly introducing a constant (k) into the equation, so x ∝ y becomes . For probability distributions, the constant is the normalization factor that ensures a distribution sums to 1. Also note that, while the arrows above each μ in step 6 are used to distinguish messages conveyed from two different factor nodes onto the same edge, the factor graphs in active inference models require three factor nodes to exchange messages. As mentioned earlier, when more than two factors exchange messages, this requires edges from each factor node to converge onto an equality node. In this case, the message conveyed to each edge is the product of the messages from the other connected factors: .
For hidden states , each message conveys the exponentiated expected log value of each factor , where denotes the function represented by each factor (Dauwels, 2007). For observed variables, the message simply conveys the known value of the factor, which is easily calculated (e.g., in the POMDPs considered in this tutorial, the message is simply ATo). When combined, these messages allow for approximation of the posterior represented by the associated edge. The posterior at each edge is normalized by applying a softmax function prior to the next round of message passing.
Using these update rules, we arrive at the following update equations for approximate posteriors over states in our POMDP models, which we will call messages 1–3 (i.e., performed in log-space; represented by purple circles on the factor graph at each location in Fig. 6 where edges meet factors):
Note the straightforward relation between these messages and Bayes’ theorem. As depicted in Fig. 6, message 1 corresponds to the prior from the previous time point (denoted by the right-facing arrow). Message 2 corresponds to prior information from the future time point (denoted by the left-facing arrow; e.g., allowing retrospective inference about the state at time point 1 after receiving an observation at time point 2). Message 3 corresponds to the likelihood of an observation at the current time point (no arrow notation; here assumed to be the same for all values of π). So, for example, if we take the edge corresponding to the posterior for sπ,2 (in the middle of the graph), this posterior will then correspond to integrating priors ( and ) with the likelihood (A) and then normalizing to convert back to a proper probability distribution (i.e., as in Bayes’ theorem). When adopting the matrix notation for these messages, belief updating can be written as:
| (16) |
Note that is replaced by the prior over initial states D for . As described in the following section, these updates (or their marginal message passing counterparts) can also be reformulated in terms of prediction errors that illustrate the biological plausibility of this message passing scheme. To help the reader get a concrete sense of the dynamics of message passing, we have provided some simple example code within the accompanying MATLAB scripts (Message_passing_example.m).
Inferring policies can be thought of as making use of an analogous process. Although it should be emphasized that posterior inference over policies in the current implementation (spm_MDP_VB_X_tutorial.m) is not explicitly done in this manner. Still, we will stay with the message passing notation for didactic purposes. Recall that, under active inference, policies are selected based on their (expected) ability to generate preferred observations and maximize information gain. In addition, recall from the previous two sections that preferred observations are formally treated as being more probable a priori. State transitions under a policy can then be seen as more probable if they maximize the probability of current observations, ln , and the expected probability of future observations conditioned upon the policy in question, . Notice that future observations are here treated as random variables that need to be inferred (i.e., because they have not yet been given to the model). Also notice the similarity between this and the expression for EFE. This similarity is no accident, as we shall see shortly (Parr & Friston, 2018a). Inferring these distributions requires us to evaluate partition functions of the normal factor graph. This means summing over the variables (i.e., probabilities) represented by the edges enclosed in the red dotted lines in Fig. 7. This operation is also sometimes called ‘closing the box’ (Loeliger, 2004). For example, to obtain the probability of current observations conditioned upon policies, and that of expected future observations conditioned upon policies, we must evaluate the following summations:
| (17) |
| (18) |
As we have seen, however, such summations are often intractable. Instead, we evaluate the free energy functionals VFE and EFE, as they approximate the required probabilities (as we saw in Section 1) and can be computed efficiently:
| (19) |
| (20) |
The posterior over policies can then be computed in a similar manner as the posterior over states. Specifically, we can express the messages sent from the and matrix factor nodes, and the E vector factor node, to the edges representing the posterior over policies as follows (messages 1–3 shown in Fig. 7).
| (21) |
Here again, messages from past and future time points are denoted with right-pointing and left-pointing arrows (respectively), while the message conveying priors over policies is denoted without arrow notation. Once these messages are passed, if we normalize the result by applying a softmax function, we arrive at an expression for the posterior over policies that (suppressing the precision term γ) corresponds to the equation shown in Table 2:
| (22) |
It is important to note, however, that unlike the state inference process shown in Fig. 6, there is no need for iterative message passing in this case. A single round of message passing is equivalent to the equation above. Thus, while inference over policies can be heuristically viewed in terms of message passing (for illustrative consistency with variational message passing in state inference), it need not be described in this manner (and there are differences; e.g., the messages are not bidirectional).
At this juncture, we return to the more recent development of marginal message passing that was mentioned at the beginning of this section. Because variational message passing and the mean-field approximation have known limitations, this improved algorithm has been adopted, and is incorporated into the most recent software implementation (spm_MDP_VB_X.m; as well as in the tutorial version included as supplementary code: spm_MDP_VB_X_tutorial.m). Briefly, marginal message passing represents a type of compromise between the computational efficiency of variational message passing and another widely used algorithm – called belief propagation – that is more computationally expensive but can perform exact (as opposed to approximate) inference under suitable conditions (for details, see Parr et al., 2019). A full explanation of marginal message passing is beyond the scope of this tutorial, as it first requires a more thorough introduction to both variational message passing and belief propagation. Here, we chose to introduce the reader to the mean-field approximation in combination with variational message passing due to its simplicity and wide usage within the active inference literature, and to provide the interested reader with a foundation to build from when pursuing more details on these topics elsewhere.
However, the major resulting adjustment under marginal message passing is that the posterior over states becomes:
| (23) |
As above, note that is replaced by the prior over initial states D for . The result of adding the to scale the influence of transition beliefs () is that the precision of transition probabilities is reduced. This prevents overestimation of the precision of posteriors — something that can occur with variational message passing. Also note that denotes the transpose of with normalized columns (i.e., columns that sum to 1). As presented here, this modification may come across somewhat ad hoc. However, as with variational message passing, the update equations for marginal message passing can be derived in a principled manner (described in Parr et al., 2019).
2.4. Prediction error formulation
One strength of active inference is that it comes equipped with a biologically plausible instantiation in terms of prediction-error minimization. In this subsection, we will introduce the reader to the different types of prediction errors described in the active inference literature and how their minimization affords state inference and policy selection. We will also provide explicit numerical examples. By the end of this subsection, the reader should understand the basis of these prediction errors, the role of the different terms within their respective equations, and how they relate to VFE and EFE.
There are two types of prediction errors described in active inference – ‘state’ and ‘outcome’ prediction errors – based on the equations for Fπ and Gπ, respectively (see Table 2). State prediction errors drive belief updating with respect to states and are based on message passing algorithms. Outcome prediction errors drive policy selection. They are not based on an explicit message passing algorithm, but they illustrate how Gπ can be formulated within the same type of biologically plausible error-minimization scheme. We will now cover each of these types of prediction errors in turn.
State prediction errors track how Fπ changes over time as beliefs about states sπ,τ are updated (i.e., reductions in Fπ correspond to reductions in state prediction error). These prediction errors are based on the marginal message passing scheme described in the previous technical section. For those who skipped this section, please briefly review Eq. (23), which corresponds to message passing between the square nodes and edges in the factor graph for an active inference POMDP (shown in the bottom panel of Fig. 6). As we now describe, it is this equation that can be reformulated in terms of a state prediction-error signal that the brain seeks to minimize in order to infer posteriors over states sπ,τ, using three types of messages (i.e., message 1: ln , message 2: ln , and message 3: ln ). This is part of a more general mapping proposed between the variables included in active inference and both neuronal and synaptic activity.
In the proposed mapping, firing rates in specific neuronal populations represent the continually updated posteriors over states sπ,τ – corresponding to edges in the factor graph shown in Fig. 6 or circle nodes in the Bayes’ net depiction in Fig. 5. Patterns of synaptic connection strengths implement factors (i.e., the square nodes within graphs), such as the A and matrices, that implement functions and transform the incoming messages encoded within firing rates; see Parr and Friston (2018a). To simulate neuronal dynamics, one can set up an ordinary differential equation, based on marginal message passing, that performs a gradient descent on VFE by introducing the state prediction error (επ,τ) as an auxiliary variable. This prediction error scores the difference between the log prior probability of each hidden state (i.e., the posterior from the previous time step) and the log probability of each hidden state following a round of message passing (i.e., when a new observation has been received). As described in the previous section, with each observation there will be many rounds (iterations) of message passing (i.e., the message passing equation will be repeated many times) until posterior beliefs over states converge to a stable value. All of this is conditioned upon a specific policy (denoted by the subscript π), because the agent is trying to infer the states it will occupy if it chooses one policy vs. another. To arrive at empirical predictions about measurable neural responses, we can then substitute in a ‘depolarization’ or ‘voltage’ variable, vπ,τ, to stand in for the log posterior over states; . The resulting state prediction error equation and belief updating are then written as follows.
State Prediction Errors
| (24) |
| (25) |
| (26) |
For those who skipped the technical section, note that in Eq. (24) denotes the transpose of with normalized columns (i.e., columns that sum to 1). In this equation, the combination of the two B matrices (combined with state beliefs) correspond to priors, whereas the A matrix (combined with observations) corresponds to the likelihood. The arrow notation indicates updates to the value of a variable at each iteration. Eq. (25) states that the change in level of depolarization vπ,τ with each iterative update corresponds to the prediction error επ,τ. Note that this error term corresponds to the rate of change in VFE; . The updated value of is subsequently put through a softmax function (σ) in Eq. (26) to return an updated posterior distribution over states . The key aspect of this set of update equations is that the value of continues to change (i.e., the equations are continually repeated) until the value of the state prediction error term επ,τ is minimized. In other words, the equations are set up such that they change the value of (in the direction of steepest descent) until this produces the lowest value of , at which point the resulting value of will correspond to an approximate posterior over states. This is because is the attracting fixed point, meaning that the system tends to evolve towards , and that once it reaches this value it will remain there. This leaves us with a biologically plausible prediction-error minimization scheme that can perform posterior inference over states and can be instantiated in a relatively simple neural network (see Section 5; for more details, see Parr and Friston (2018a). That is, by finding posterior beliefs over states (on the far right of Eq. (24)) that minimize , Fπ is minimized and becomes a stable posterior belief.
As described in more detail below (and elsewhere; Da Costa, Parr, Sengupta, & Friston, 2021), the variable is used to model the average voltage or membrane potential of a neural population (i.e., by taking either positive or negative values), where one population is assumed to encode information about each state factor (i.e., the probability of each state within that factor for each time τ under each policy). The state variable then corresponds to the firing rates of that population, which are driven by their membrane potential. This is because is the softmax (σ) of the voltage and therefore, similar to a firing rate, takes only non-negative values (i.e., between 0 and 1). This follows from the assumption made in mean-field models of neural dynamics that the average firing rate of a population can be treated as a sigmoid function of the average membrane potential (Breakspear, 2017; Da Costa et al., 2021). Local field potentials (LFPs) and event-related potentials (ERPs) in electroencephalography (EEG) research are then modeled as the time derivative (rate of change) in the firing rates .
To make these equations more concrete, consider a worked example with the following generative model entailed by a policy, combined with a specific observation and initialized value of the approximate posterior:
As can be seen here, the likelihood (A) matrix indicates that outcome 1 (row 1) is more likely (i.e., p = .8) under state 1 (column 1). Under the policy being considered, the agent believes it will most likely remain in its current state in the first state transition (i.e., p = .9 and .8 on the diagonal in ; columns indicate states at time τ − 1, rows indicate state at time τ) and more likely to move to state 2 during the second state transition (i.e., p = .8 and .7 in the bottom row in ). Further, the agent observes outcome 1 at time and has a prior expectation that both states are equally likely ( and v). Here we will also set , which is a common initialization at the beginning of a trial. However, the values of these three variables will often not remain equal after new observations as a trial progresses. In this case, the error signal will be:
This error signal will then update beliefs over states through the depolarization variable vπ,τ as follows:
Notice that in this example the variational update (i.e., single step of gradient descent) results in a negative value for the state prediction error term (i.e., . As can be seen, this shifts the approximate posterior such that it will better minimize prediction error in the next variational update (i.e., here increasing the probability of occupying state 1).
In contrast to state prediction errors, the outcome prediction errors mentioned above track how Gπ changes over time as beliefs about policies are updated (i.e., reductions in Gπ correspond to reductions in outcome prediction error). In other words, when this type of prediction error is minimized, policies are identified that minimize both uncertainty over states (i.e., ambiguity) and the expected difference between predicted and preferred outcomes. However, as noted above, it is important to clarify that, unlike state prediction errors, outcome prediction errors are not directly tied to the message passing schemes described above — and the currently available routines in SPM for performing active inference do not explicitly calculate outcome prediction errors. The current implementation instead calculates Gπ directly. When this prediction error formulation has been presented in previous literature, it has largely been for illustrative purposes with respect to demonstration of biological plausibility (Parr & Friston, 2018a). However, calculating outcome predictions errors could feasibly be added if one were interested in modeling the associated neuronal responses predicted by this aspect of the process theory (for one recently proposed scheme for inferring policies through message passing, see Champion et al. (2021). In contrast, the current routines do calculate state prediction errors, which can be used without modification for purposes of empirical prediction.
The update equation for outcome prediction error is as follows.
Outcome Prediction Errors:
| (27) |
This prediction error is best understood as a mixture of two types of expected predictions errors. The first term, , corresponds to the expected difference between preferred outcomes (i.e., the probability distribution encoding preferences over outcomes specified by Cτ) and the outcomes expected under a policy (i.e., corresponds to the observations expected under a policy, oπ,τ). This can therefore be thought of as the expected prediction error (under each policy) with respect to the observations predicted by prior preferences. The second term, , corresponds to how much observations are expected to update beliefs if adopting a particular policy (i.e., it is the entropy term, where lower entropy entails greater information gain). Therefore, as with state prediction error, minimizing this term minimizes uncertainty — but in this case it is uncertainty with respect to policies. Note that the diag() function simply takes the diagonal elements of a matrix and places them in a row vector. Note also that, unlike with state prediction errors, we have not used the update (←) notation for outcome prediction errors. This is because, in the current formulation of active inference, outcome prediction errors are not iteratively minimized; they are simply computed once for each policy. Those who read the technical section on VFE and EFE will recognize these two terms as the matrix forms of the risk , and ambiguity terms in EFE.
Again, to make this more concrete we provide a worked example of each term under two possible policies, assuming the following variable values:
In other words, the agent prefers outcome 1 (row 1), and the likelihood (A) matrix indicates that state 1 (column 1) is more likely to generate outcome 1 (i.e., p = .9). Further, state beliefs under policy 1 entail a higher probability of being in state 1 (i.e., p = .9) than state beliefs under policy 2 (; i.e., p = .5). We can first calculate the risk (reward-seeking) term within the outcome prediction error for each policy:
Policy 1:
Policy 2:
Note that a negligibly small number (here, e−16) is added to the values in Cτ because ln (0) is undefined. As expected, because the approximate posterior over states for policy 1 makes the generation of preferred observations more likely, policy 1 has lower values for the risk term (i.e., leading to lower outcome prediction error, all else being equal).
Moving onto the ambiguity (information-seeking term), consider another example with the following variables:
In this case, the likelihood (A) matrix indicates that state 2 (column 2) has a more precise distribution than state 1 (column 1). In other words, observations are expected to provide more precise information about states when in state 2. As such, we expect outcome prediction errors to drive selection of the policy that will lead the agent toward state 2. In this case, policy 2 assigns a higher probability to state 2 (i.e., = .9 in row 2). We can confirm this by calculating the ambiguity term for each policy as follows:
Policy 1:
Policy 2:
As expected, because the outcomes generated by state 1 are more ambiguous (i.e., less informative), and policy 2 assigns a higher probability to state 2 than policy 1, policy 2 better minimizes ambiguity.
It is important to stress that the risk and ambiguity terms for outcome prediction errors work synergistically, and one often has policies that minimize both risk and ambiguity. As can be seen in the outcome prediction error equation above, subtracting the ambiguity term from the risk term corresponds to adding (i.e., note the double negative) the entropy of the likelihood mapping under a policy to the risk of the policy, which we have calculated separately here. This drives selection of policies that maximize both reward- and information-seeking by minimizing the overall resulting error.
While these example calculations may appear somewhat involved (even for a single policy), an intuitive way to think about these two prediction errors is that minimizing state prediction error maximizes confidence in posterior beliefs, while minimizing outcome prediction error maximizes confidence in how to achieve goals or desires. To reproduce the worked examples above and allow the reader to calculate state and outcome prediction errors under different model parameters, we have provided the Prediction_error_example.m script in the supplementary code.
3. Building specific task models
3.1. Explore-exploit task
To make the structure of a POMDP more concrete, in this and subsequent sections we will build models of specific behavioral tasks commonly used in empirical studies. This will provide the reader with the necessary tools to build their own models and use them in both simulation work and empirical studies. This will also allow us to concretely demonstrate some of the unique resources offered by active inference when modeling behavior in a simple reinforcement learning context. To be sure, in many task contexts (e.g., when there is no uncertainty about states) active inference models can perform similarly to reinforcement learning models, and they do not always generate optimal behavior (Da Costa, Sajid, Parr, Friston, & Smith, 2020; Markovic et al., 2021).6 However, as discussed above, when tasks involve various types of uncertainty (e.g., about task condition or reward probabilities), active inference offers a unique approach for modeling information-seeking behavior that can lead to superior performance in some cases (Markovic et al., 2021; Sajid, Ball, Parr, & Friston, 2021). Another resource offered by active inference, even when it performs similarly to other types of models, is its associated neural process theory (i.e., describing how neural signaling might implement variational or marginal message passing; see Section 5). The task models we build in this tutorial will further allow us to illustrate how active inference can be used to make testable empirical predictions about neural responses. As we will see, because active inference models integrate perception, learning, and decision-making within a single model architecture, this affords the generation of predictions about neural responses across a wide range of perceptual tasks in addition to reinforcement learning and decision-making tasks.
In this subsection, we will build a model of the explore–exploit task briefly described in the previous section. Every step we outline in this section for building the explore–exploit task model is laid out in the accompanying MATLAB code (Step_by_Step_AI_Guide.m). This code is included in the supplementary code files and can also be found at: https://github.com/rssmith33/Active-Inference-Tutorial-Scripts. While going through this section, we encourage the reader to work through this code in parallel. Here we will use non-bold italics when presenting the general mathematical notation and subsequently show the associated MATLAB syntax in bold.
In the beginning of the explore–exploit task, the participant is told that on each trial one machine will tend to pay out more often, but they will not know which one. They are also told that the better machine will not always be the same on each trial. They can choose to select one right away and possibly win $4. Or they can choose to press a button that gives them a hint about which slot machine is better on that trial. However, if they choose to take the hint, they can only win $2 if they pick the correct machine. Over many trials, the participant can learn which slot machine tends to pay out more often and either make safe or risky choices (i.e., take the hint or not).
To model this task, it can be helpful to start by specifying the sets of possible hidden states (state factors). In this case, one state factor corresponds to whether the left or right slot machine is more likely to win (‘left-better context’, ‘right-better context’). The second state factor corresponds to the choice state (‘start state’, ‘asking for the hint’, ‘choosing the left machine’, ‘choosing the right machine’). Moving to MATLAB code, we can set these up by specifying the priors over initial states with a set of vectors D, with one vector per state factor (i.e., where the factor number is specified in brackets). The general structure for these vectors is:
In this case, we specify:
Note that, to match MATLAB syntax, we use the apostrophe (‘) to indicate a transpose. This says that the participant begins with the belief that the ‘left-better’ and ‘right-better’ contexts have equal probability (left and right entries, respectively), and with a fully precise belief that he/she will start the trial in the start state (from left to right: ‘start’, ‘get hint’, ‘choose left’, and ‘choose right’ states).
It is important to briefly note, however, that things change slightly if we wish to simulate learning as opposed to just inference, because we need to separate the generative process from the generative model. In this case, capital D stands for initial state probabilities in the generative process, while lowercase d stands for the initial state priors in the generative model (which are learned). For example, one could specify and , which would mean the true context is ‘left-better’ but the agent believes each context is equally likely. In the supplementary code accompanying this section, we do this as a way of controlling which context we want to simulate. The same capital vs. lowercase letter convention holds for all other matrices/vectors used here. We return to this in the section on learning further below (Section 4).
Moving forward, we must next specify the (in this case three) sets of possible observations (outcome modalities). Here, the first set of observations corresponds to the hint (‘no hint’, ‘machine-left hint’, ‘machine-right hint’). The second set of observations corresponds to decision outcomes (‘start’, ‘lose’, and ‘win’). Finally, the participant also observes their own behavior; namely, the observed action (‘start’, ‘asking for the hint’, ‘choosing the left machine’, ‘choosing the right machine’). This last outcome modality can be important in active inference models because choice states must be inferred just like any other state. Observing their own behavior therefore allows a participant to be more confident about whether their intended actions have been successfully carried out. In MATLAB, we can set these up by specifying the likelihood (A) matrices. There will always be one A matrix for each outcome modality. Rows correspond to outcomes, columns correspond to the states in the first state factor, and there is an additional dimension for each additional state factor (note that, as mentioned in Table 1, the A matrices are more correctly referred to as tensors if they include more than two dimensions). The general structure is therefore:
A {outcome modality} (outcome, factor 1, factor 2,
. . ., factor N) = [matrix]
In this case, only the ‘get hint’ state (state 2) in state factor 2 generates a hint observation, so for the third dimension we specify a ‘2’ as follows:
Here, columns from left to right correspond to the ‘left-better’ and ‘right-better’ states, while rows from top to bottom correspond to the ‘no hint’, ‘machine-left hint’, ‘machine-right hint’ observations. This matrix indicates that the hint is accurate with a probability of 1 (100% accuracy). For example, a ‘machine-left hint’ observation (row two) will be generated by the ‘left-better context’ (column one) with probability = 1. Here each column must add up to 1.
For the other dimensions of state factor 2 (i.e., matrix dimension 3):
for i = 1, 3, 4 :
end
This indicates that all other choice states will generate the ‘no hint’ observation (i.e., a hint will never be observed in those states).
For the second outcome modality, the ‘start’ and ‘get hint’ states generate ‘start’ observations (row 1):
for i = 1, 2
end
We will now specify the probability of winning in the ‘left-better context’ vs. the ‘right-better context’ depending on choice state. First, we specify that choosing the left machine (i.e., transitioning to the ‘choose left machine’ state; state 3 in factor 2) will lead to a win 80% of the time (row 3) if in the ‘left-better context’ (column 1) and lead to a win 20% of the time if in the ‘right-better context’ (column 2), with inverse probabilities for a loss (row 2), and a probability of 0 of continuing to observe the ‘start’ observation (row 1):
We will then specify that the probabilities of winning are reversed if choosing the right machine (i.e., transitioning to the ‘choose right machine’ state; state 4 in factor 2):
Remember, the first column is the context where the left machine is better, and the second column is the context where the right machine is better. It is the third dimension that corresponds to choice states (in this case, choice state 3 and 4, corresponding to choosing the left vs. right machine).
Finally, for the third observation modality (observed action), states simply map 1-to −1 to outcomes (across all other state combinations):
This simply allows the individual to infer what their choice was with complete certainty (rows top to bottom: ‘start’, ‘asking for the hint’, ‘choosing the left machine’, ‘choosing the right machine’ observations).
Now that we have the likelihood, the next step is to specify the (policy-dependent) state transition (Bπ,τ) matrices. The general structure for these matrices is:
Here the vector U contains indices specifying the action number assigned to each matrix, where each policy π subsequently specifies a sequence of these action numbers (described further below).
Because we have two state factors, we need two sets of matrices. The first matrix is for the context factor. In this case, because a context remains the same within each trial, this is simply an identity matrix that says states at time τ (columns) remain the same at τ + 1 (rows):
Columns from left to right and rows from top to bottom both correspond to the ‘left-better’ and ‘right-better’ states. There is only one ‘action’ (possible transition from each state) for this factor, so the third dimension is a 1 and has a length of 1; i.e., there is no B {1} (:, :, 2).
In contrast, the second state factor is choice state, where four different transitions (actions) are possible at each time step. In this case, each matrix below indicates that one could move from any state to the chosen state:
In other words, action 1, B {2} (:, :, 1), entails moving to the ‘start’ state from any other state, action 2, B {2} (:, :, 2), entails moving to the ‘get hint’ state from any other state, and so forth. The third dimension labels these as action numbers 1, 2, 3, and 4.
Next, we need to specify preferences over each set of outcomes (C), with one matrix per outcome modality. Here, rows indicate observations (same order as in the corresponding A matrices) and columns indicate time points in a trial from left to right. In other words:
In this case, the model has no direct preference for getting a hint (i.e., preferences for outcome modality 1: C {1}) or for observing the choice of a particular action (i.e., preferences for outcome modality 3: C {3}). So:
Here, columns from left to right indicate τ = 1, 2, 3 in the trial. The model does have preferences for winning and losing (outcome modality 2), which are specified as follows:
This indicates that, at the second and third time points, a win (row 3) has a value of 4 and 2, respectively. Remember, the value is less at the third time point (third column) because the individual wins less money if they instead choose to take the hint at the second time point (i.e., and thus continues to observe the ‘start’ outcome in row 1 for this outcome modality). The values of −1 in row 2 indicate a preference against observing a loss at time points 2 and 3.
Note that we only initially specify the C matrix values in this form for convenience. These preference distributions are passed through a softmax (normalized exponential) function (σ) such that each column (i.e., the preference distribution for each time τ) in the C matrix encodes a proper probability distribution of non-negative values that sums to 1, at which point a natural log is applied. This means that the values are transformed into log-probabilities as follows (i.e., less negative indicates more preferred):
For example, in the case of the preferences for Cwin specified above:
To be clear, in the line immediately above, the softmax is applied to each column separately (i.e., corresponding to the preference distribution over outcomes at each time point).
Next, we need to specify allowable policies. There are three time points in a trial for this task, which means a policy will consist of two actions. If we want to include ‘shallow’ policies, where the model only looks one step ahead, we need to specify a set of vectors U that index each action (as already referred to above). Technically, this set of vectors is specified as a matrix including one row, one column for each allowable action, and a third dimension specifying each state factor. Thus, the structure is:
Entries for U(:, :, 2) allow all four possible transitions (actions) between choice states (factor 2) at each time point. The entries for U(:, :, 1) are all ones because there is only one possible ‘action’ – that is, one transition matrix – for state factor 1. There still needs to be four ones within U(:, :, 1) to match the number of actions in U(:, :, 2). In other words, each overall action option corresponds to the combined entries in a given column for both state factors.7
If we instead want to include ‘deep’ policies, where the simulated participant plans ahead until the end of the trial, this means that we need to specify one column for each allowable policy (with each entry indicating an action number) in a matrix V, with one row per time point and a third dimension specifying each state factor. Thus, the general structure is:
In this case, we might reasonably include five policies:
As with U, all policies here do not change state factor 1 at either time point (hence, all entries are ones8). For state factor 2, we have included policies in which the model chooses to remain in the ‘start state’ (i.e., choosing action 1 twice; column 1), chooses to take the hint (action 2) and then select either of the slot machines (i.e., actions 3 or 4; columns 2–3), or decides to choose a slot machine right away (columns 4–5; note, these policies subsequently return to state 1, since it is not possible to win twice in one trial). We will use deep policies in the simulations below.
If so desired, one can also specify a fixed prior over policies E to incorporate a bias or ‘habit’ to select some policies over others. This is simply a column vector with one entry per policy that encodes the probability of that policy. Here we will not include such a bias, which means that E will simply be a flat distribution over our 5 allowable policies in V:
Finally, there are several scalar (single-value) parameters one can set. One parameter is beta (β), which is the prior on the expected free energy precision term γ discussed above (which encodes the precision estimate for the expected free energy over policies). A low β value (around 1) indicates high expected precision, whereas higher values (e.g., 3, 5, 10) indicate lower expected precision. Higher β values will increase randomness in policy selection and also make policy selection more influenced by habits encoded in the E vector (for an example of these dynamics, see Smith, Khalsa, & Paulus, 2021b. This follows from the fact that it implies less confidence that model beliefs will generate preferred outcomes (Hesp et al., 2020). Another parameter is alpha (α), which is a standard ‘inverse temperature’ (or ‘action precision’) parameter that controls randomness (e.g., motor stochasticity) in action selection under a chosen policy (higher values indicate less randomness; typical range is between around 1 – 32, but very high values can be chosen to remove choice stochasticity).
| (28) |
Here, sigma (σ) indicates a softmax function that transforms the quantity on the right into a proper probability distribution that sums to 1 (see Appendix A for more detail). Both β and α must be positive numbers. Here we will make β = 1 and α = 32, specifying reasonable amounts of indeterminacy in action selection.
3.2. Running and plotting simulations
We have now specified a generative model and are ready to run single-trial simulations. To do so in MATLAB, we will assign each of our variables to a structure called mdp (for Markov decision process). Concretely, this means assigning mdp.D = D, mdp.V = V, mdp.beta = beta, and so forth for all the scalars, vectors, and matrices constructed above. We can then run this structure through the standard active inference estimation function spm_MDP_VB_X.m (available in the DEM toolbox of the most recent versions of SPM academic software: http://www.fil.ion.ucl.ac.uk/spm/). This just means entering:
However, because SPM software is often updated, we include a specific version for this tutorial. So here you should run:
This function will simulate behavior based on an POMDP structure (i.e., it runs the equations in Table 2), and the output MDP (capital letters) structure will contain the simulation results. As we have specified it here, it assumes the generative process and generative model are identical (see section on learning below where we remove this assumption). It will thus generate outcomes based on the generative process and simulate the subsequent inference and decision dynamics within the generative model when observing those outcomes. Because the above-mentioned simulation script is quite complex, we also direct readers interested in the details of how the belief updating scheme is implemented to the Simplified_simulation_script.m script included in the supplementary code, which is a stripped down (but heavily commented) version of the standard model inversion scheme used in spm_MDP_VB_X.m. For clarity, this additional tutorial script inverts the same generative model of the explore–exploit task introduced above.
Single-trial behavior can be plotted with some default plotting routines. The primary single-trial plotting routine available in SPM can be run in MATLAB by entering:
This plotting routine can also take additional optional inputs:
Gf: state factors to plot.
Gg: outcome modalities to plot.
For example, spm_MDP_VB_trial(MDP, 1: 2, 2: 3) would plot the first two state factors and the second and third outcome modalities.
At this point, the reader is encouraged to set the variable Sim in the first section of the accompanying tutorial code (i.e., Step_by_Step_AI_Guide.m, line 51) to Sim = 1 and then click ‘Run’, which will run the model and this plotting script. Before running this script, remember to make sure SPM12 is installed and that the ‘DEM’ folder within the SPM folder structure is added as a path in MATLAB (...spm12\toolbox\DEM).
Based on the current model specification, a representative plot of simulation results is shown in Fig. 8A. This and similar plots are generated from specific output fields in the MDP structure (Table 3 describes each output field). The two panels in the top-left of Fig. 8A show posteriors over states at the end of the trial (i.e., the states the model believes it was in at each time point τ when at the last time point t). Here time goes from left to right, darker indicates higher probability, and the cyan dots denote the true states. Here, the model believes it was in the ‘left-better context’ and that it chose to take the hint and then chose the left slot machine. The top-right shows the action probabilities and true actions. Here the agent is highly confident that taking the hint and choosing the left machine were the best choices (and cyan dots indicate that these were also the actual actions taken). The left-middle panel just shows the different possible two-step action-sequences specified in the model (from left to right). Note that lighter shades in this panel just indicate higher action numbers (e.g., action 1 is black, action 2 is dark gray, etc.). The right-middle panel shows the evolution of the posterior distribution over policies over time (from left to right). Here, it can be seen that at the second time point the model became highly confident in policy 2 (i.e., the ‘take the hint and then choose the left slot machine’ policy). The three panels in the bottom-left show the outcomes (cyan dots) and preference distributions. The first ‘hint’ modality shows that the model received the hint at time point 2. The plot is gray because there is no preference for one observation over others. This is also the case for the third ‘observed action’ plot, which simply confirms what the model chose. The second ‘win/lose’ modality shows that a win was observed at the third time point. The preference distribution here indicates the strong preference for the win at time points 2 and 3 (darker value), and a preference not to lose (lighter value). The ‘null’ (starting) outcome in the top row is an intermediate gray at time point 2 (no preference for or against this outcome); the distribution becomes darker gray at the third time point because the value of the win at time point 3 was relatively less (i.e., $2 vs. $4) and so the overall distribution over outcomes at the third time point is less precise.
Fig. 8.
Example simulation plots that can be generated with the code included in this tutorial. A detailed walk-through is provided in the main text. Panel A Example simulation of a risk-averse agent performing the explore–exploit task. The agent takes the hint, then chooses the left slot machine, and observes a win. Panel B: Example simulation of a risk-seeking agent performing the explore–exploit task. The agent foregoes the hint and immediately chooses the right slot machine, observes a loss, and then returns to the start state. Panel C: Example neuronal simulations based on the risk-averse agent in Panel A. This illustrates the neuronal firing rates, local field potentials, and dopamine responses predicted by the neural process theory. For all panels, note that darker colors indicate higher probability values for beliefs about states, actions, and policies over time. For outcomes, darker values indicate stronger prior preferences. The ‘allowable policies’ plots in the first two panels simply display the action sequences corresponding to each policy (darker indicates lower numbers, where each number denotes an available action). The dopamine response plots (lower-right plots in each panel) correspond to updates in the expected precision of the EFE distribution over policies (γ); cyan lines indicate γ values while black spikes correspond to their rate of change. In the upper- and lower-left plots (‘contexts’ and ‘firing rates’) of panel C, each column (moving left to right along the x-axis) corresponds to beliefs about context states at the time when an observation was received (t), while rows from top to bottom on the y-axis correspond to the time point for which beliefs are updated (tau; τ). For example, the top-right quadrant corresponds to beliefs at time t = 3 about time τ = 1 (note that, unfortunately, this standard SPM plotting routine inappropriately labels each row with ts instead of τs). Firing rates (upper-right) correspond to the magnitude of posteriors over each state (in this case, the states in the ‘context’ state factor), while local field potentials (middle-right) correspond to their rate of change (in both cases, there is one line plotted for each row in the plots in the upper- and lower-left). See main text for interpretation of time frequency response plots and their motivation. These simulations can be reproduced by running the Sim = 1 option in the supplementary Step_by_Step_AI_Guide.m code (although note that, because outcomes are sampled from probability distributions, results will not be identical each time)
Table 3.
Output fields for spm_MDP_VB_X_tutorial.m simulation script.
| MDP Field | Model Element | Structure | Description |
|---|---|---|---|
| MDP.F | Negative variational free energy of each policy over time. | Rows = policies. Columns = time points. |
Negative variational free energy of each policy at each time point in the trial. For example, if there are 2 policies and 6 time points there will be a 2 × 6 matrix containing the negative variational free energy of each policy at each point in the trial. |
| MDP.G | Negative expected free energy of each policy over time. | Rows = policies. Columns = time points. |
Negative expected free energy of each policy at each time point in the trial. For example, if there are 2 policies and 6 time points there will be a 2 × 6 matrix containing the negative expected free energy of each policy at each point in the trial. |
| MDP.H | Total negative variational free energy over time. | Columns = time points. | Total negative variational free energy averaged across states and policies at each time point. For example, if there are 8 time points there will be a 1 × 8 row vector containing the total negative free energy at each time point. |
| MDP.Fa MDP.Fd MDP.Fb … |
MDP.Fa is the negative free energy of parameter ‘a’ (if learning A matrix). There are also analogous fields if learning other matrices/vectors (e.g., MDP.Fd for learning the parameters of the D vector, etc.). | Columns = one per outcome modality or hidden state factor (i.e., depending on the specific parameters being learned). If the agent is learning parameters of a single vector (e.g., E), this will be a single column. | KL divergence between the parameters of the matrix/vector that is being learned at the beginning of each trial and at the end of each trial. Each column in the vector may represent an outcome modality (i.e., in the case of the A matrix), a hidden state factor (i.e., in the case of the B matrix and D vector), or any other vector (e.g., the E vector). |
| MDP.O | Outcome vectors. | Rows = outcome modalities. Columns = time points. |
Vectors (one per cell) specifying the outcomes for each modality at each time point. Observed outcomes are encoded as 1s, with 0s otherwise. |
| MDP.P | Probability of emitting an action. | Rows = one per controllable state factor. Columns = actions. Third dimension = time point. |
The probability of emitting each particular action, expressed as a softmax function of a vector containing the probability of each action summed over each policy. For example, assume that there are two possible actions, with a posterior over policies of [.4 .4 .2], with policy 1 and 2 leading to action 1, and policy 3 leading to action 2. The probability of action 1 and 2 is therefore [.8 .2]. This vector is then passed through another softmax function controlled by the inverse temperature parameter α, which by default is extremely large (α = 512). Actions are then sampled from the resulting distribution, where higher α values promote more deterministic action selection (i.e., by choosing the action with the highest probability). |
| MDP.Q | Posteriors over states under each policy at the end of the trial. | 1 cell per state factor. Rows = states. Columns = time points. Third dimension = policy number. |
Posterior probability of each state conditioned on each policy at the end of the trial after successive rounds of updating at each time point. |
| MDP.R | Posteriors over policies. | Rows = policies. Columns = time points. |
Posterior over policies at each time point. |
| MDP.X | Overall posteriors over states at the end of the trial. These are Bayesian model averages of the posteriors over states under each policy. | 1 cell per state factor. Rows = states. Columns = time points. |
This means taking a weighted average of the posteriors over states under each policy, where the weighting is determined by the posterior probability of each policy. |
| MDP.un | Neuronal encoding of policies. | 1 cell per policy dimension. Rows = policies. Columns = iterations of message passing (16 per time point). For example, 16 iterations, and 8 time points gives a vector with 128 columns). |
Simulated neuronal encoding of the posterior probability of each policy at each iteration of message passing. |
| MDP.vn | Neuronal encoding of state prediction errors. | 1 cell per state factor. Rows = iterations of message passing (16 per time point). Columns = states. Third Dimension: time point the belief is about (τ). Fourth Dimension: time point the belief is at (t). |
Bayesian model average of state prediction errors at each iteration of message passing (weighted by the posterior probability of the associated policies). |
| MDP.xn | Neuronal encoding of hidden states. | 1 cell per state factor. Rows = iterations of message passing (16 per time point). Columns = states. Third Dimension: time point the belief is about (τ). Fourth Dimension: time point the belief is at (t). |
Bayesian model average of normalized firing rates, which reflect posteriors over states at each iteration of message passing (weighted by the posterior probability of the associated policies). |
| MDP.wn | Neuronal encoding of tonic dopamine, reflecting the current value of γ. | Rows = number of iterative updates (16 per time point). For example, if there were two time points in a trial this would be 1 column with 32 rows. | This reflects the value of the expected precision of the expected free energy over policies (γ) at each iteration of updating. |
| MDP.dn | Neuronal encoding of phasic dopamine responses, reflecting the rate of change in γ. | Rows = number of iterative updates (16 per time point). For example, if there were two time points in a trial this would be 1 column with 32 rows. | This variable reflects the rate of change in the expected precision of expected free energy over policies (γ) at each iteration of updating. |
| MDP.rt | Simulated reaction times. | Columns = time points. | Computation time (i.e., time to convergence) for each round of message passing and action selection. |
As mentioned earlier, however, optimal information-seeking (in the sense of maximizing preferred outcomes in the long-run) depends on having the right balance of reward value and information value. To illustrate this, Fig. 8B shows simulations in which the magnitude of the preference distribution for a win has been multiplied by 2: C {2}(3, :) = [0 8 4]. As can be seen there, the model instead decided to make a guess right away about which machine will win (in this case, choosing right) and unfortunately observed a loss (bottom-left, middle sub-panel). As can be seen in the upper right, its confidence in the left vs. right action is equal (equally gray over each). As can be seen in the upper left, the model’s posterior over states shows high confidence that it was in fact in the ‘left-better’ context at every time point, because (retrospectively) this was most likely the case if it lost after choosing the machine on the right.
Finally, the bottom-right plots in these panels display predictions about dopamine responses in the neural process theory, which we have not yet discussed. These responses are based on changes in confidence in expected free energy estimates after receiving new observations (i.e., the updates to the expected free energy precision parameter γ ; explained in the bottom row of Table 2). In this case, the large ‘dopamine spike’ shown at the second time step is because EFE at that time step favored policy 2 (i.e., taking the hint and then choosing the left machine) and VFE at that time step (based on observations) provided support for policies 2 and 3 (i.e., after observations at the second time step, only the two policies that included taking the hint remained plausible). Because the policy favored by EFE was supported, the precision estimate for EFE increased (corresponding to the positive dopamine spike).
As a numerical example to help offer an intuition for how these updates operate, we can plug the VFE and EFE values at time point 2 in this simulation into the policy distribution and precision update equations shown in Table 2. In these equations, γ is the expected free energy precision term, β0 is the initial prior for this precision at the start of a trial (γ = 1/β), and β is the posterior value that is continuously updated over time by a term we label βupdate. This term technically reflects the gradient of free energy with respect to and is informed by a value scoring the level of (dis)agreement between expected free energy and observed (variational) free energy after making a new observation — which can be thought of as a type of prediction error (Gerror; with proposed associations with emotion; see (Hesp et al., 2020). For the sake of illustration, we can set γ, β0, and β equal to one and specify the distributions over policies as follows9:
As can be seen here, the agent has no habit-like prior expectations over policies (i.e., the E distribution is flat), and the expected free energy over policies (G) favors policy 2 (i.e., entry 2 has the lowest value). The variational free energy after a new observation (F) provides precise evidence for policies 2 and 3 (values much closer to 0, indicating that the new observation is inconsistent with policies 1, 4, and 5). Given this setup, one round of iterative updating would be:
Here we have included a step size parameter of ψ = 2, which reduces the magnitude of each update and promotes stable convergence. Note that, while we have here shown an example of a single round of updating, there will be many rounds of updating to convergence for each new observation. Notice that the increase in the probability of policy 2 and 3 between the prior and posterior over policies is driven by F, which scores the evidence afforded each policy given current observations. Policy 2 and 3 better minimize F and are therefore more plausible. Taking the dot product between the vector encoding the difference between the prior and posterior over policies (π − π0) and the −G vector is equivalent to scaling each element of the difference vector by the associated G value and then summing the results, which in this case creates a positive update. This is because the difference vector is pointing in roughly the same direction as the −G vector. This is apparent in that G initially indicated the highest probability for policy 2, and F also provided evidence for policy 2. As a result, the prediction error (Gerror) is positive and the updated γ value increases the impact of G on the posterior over policies (i.e., because the agent is now more confident in its beliefs about G). In contrast, if the policies favored by G were not supported by F (as in Fig. 8B), the Gerror term would be negative and the updated value of γ would decrease the impact of G on the posterior over policies, as the agent has lost confidence in its beliefs about G. For a derivation of these update equations, see Appendix in (Sales et al., 2019).
Note that the cyan line in the dopamine plot corresponds to the stable expected free energy precision value (γ ; with a hypothesized link to tonic dopamine levels), as opposed to the rate of change in this precision (in black; with a hypothesized link to phasic dopamine responses). To help the reader gain a better intuition for the dynamics of these updates, we have provided supplementary code (EFE_Precision_Updating.m), which allows the reader to specify the number of policies, the values for the vectors E, F, and G, and the value of β0, and then simulate these updates. Fig. 9 also illustrates a helpful geometric interpretation of the factors that determine the direction of β updates. Namely, when the difference vector (π − π0) and the −G vector point in a similar direction (i.e., an angle of less than 90° apart), the dot product of the two will result in an increase in γ. The fact that these vectors point in the same direction is a way to visualize how new observations (through F) provide evidence supporting the reliability of G, and therefore increase its precision weighting. In contrast, when these vectors point in different directions (i.e., the angle separating them is greater than 90°), this suggests that G is less reliable; its precision (γ) is therefore reduced and it contributes less to the posterior distribution over policies (π).
Now that we have gone through an example of these dynamics, an important question concerns the settings in which they may be useful. Here, it is important to highlight that E, F, and β/γ can be viewed as optional elements (e.g., they are not incorporated in the policy selection model within the upper right portion of Fig. 5, or in other examples of active inference (Da Costa, Parr et al., 2020)). For example, incorporating E may not be useful unless modeling a task in which you suspect that participants enter a study with a particular choice bias or that habitual choice behavior could be learned over time. Incorporating F and β/γ updating is only useful in the context of deep policies. As mentioned elsewhere, β/γ can optimize the relative influence of goals and habits (E and G). Among others, one benefit of incorporating F emerges when there are a large number of deep policies to choose from. This is because it allows observations to render some policies highly implausible early in a trial, which has the effect of narrowing the search space for the optimal policy. This typically works in conjunction with an ‘Occam’s window’ parameter that removes policies from the search space if their probability becomes too low relative to the most probable policy (this parameter is specified as mdp.zeta and explained in more detail within the accompanying tutorial code Step_by_Step_Hierarchical_Model.m.
4. Modeling learning
In this section we will discuss how learning is implemented in active inference and how this can be used to model multitrial behavioral data. As a concrete example, we will return to the explore–exploit task model and allow the agent to learn prior beliefs (i.e., within the vector D) about how often the left slot machine and right slot machine tend to pay out. More generally, we will discuss how any set of model parameter values – such as those encoding distributions within the likelihood (A), transition beliefs (Bπ,τ), or priors over policies (E) – can be learned over repeated trials. This is based on updating prior beliefs over these parameters within a class of distributions called Dirichlet distributions. We first provide a technical introduction to the general mathematical foundations of Dirichlet distributions. Then we discuss learning in more intuitive terms, provide numerical examples, and demonstrate how to run simulations in practice.
4.1. Technical introduction to Dirichlet priors (optional)
In this subsection we provide a technical introduction to the Dirichlet distribution used to implement learning in active inference. After completing this section, the reader should have an understanding of how the parameters in a Dirichlet distribution can: (1) act as priors on the categorical distributions used in the POMDP models covered above, and (2) be updated based on posterior beliefs at the end of a trial. For readers less interested in these technical details, this section can be skipped. As mentioned above, we will provide a more intuitive conceptual introduction in the next subsection. We encourage all readers to consider the formal details below, but a complete understanding of this subsection will not be required to follow subsequent sections.
Learning in active inference is formulated in terms of a Dirichlet-categorical model. Specifically, Bayesian inference is performed using a categorical distribution (which was discussed earlier) as the likelihood, and a Dirichlet distribution as the prior. The Dirichlet distribution is a distribution defined over a vector of values that sit on the interval [0, 1], and sum to 1. That is, the values of the vector have the same properties as a probability distribution. As such, the Dirichlet distribution is often described as a distribution over a distribution. In this case, it can be used to encode beliefs about model parameters (e.g., confidence in parameters in the likelihood or transition matrices of a POMDP).
The Dirichlet distribution is used as the prior over the parameters of the categorical distribution because it is the conjugate prior for the categorical distribution. This means that if we multiply a categorical distribution by a Dirichlet distribution, and then normalize to obtain the posterior distribution over the parameters of the categorical distribution, we end up with another Dirichlet distribution — allowing it to be used as a prior in the next round of inference. This allows active inference agents to sequentially update their beliefs about model parameters as they receive new observations. The Dirichlet distribution, denoted Dir (θ|α), is defined as follows:
| (29) |
Where is a normalization constant that ensures the distribution sums to 1, and Γ denotes the gamma function (for a brief introduction to the gamma function, see Appendix A). The variable is a vector of length K containing the parameters of a categorical distribution, and is the set of concentration parameters of the Dirichlet distribution, which satisfy the condition that αk > 0. The gamma function is used in the normalization constant to account for the combinatorics of drawing a random variable from a categorical distribution. That is, it counts the number of ways in which we can place the variable α in one of K mutually exclusive states. Similarly, the categorical distribution is defined as:
| (30) |
Where is a categorical variable that occupies one of K mutually exclusive states (e.g., . Here are the parameters of the distribution and satisfy the conditions , and . The term is the normalization constant.
If we multiply the Dirichlet and categorical distributions to arrive at the posterior distribution over the parameters of the categorical distribution, we obtain the following (ignoring the normalization constant for the sake of brevity):
| (31) |
Notice that this has exactly the same form as the prior defined above, except that we have added a ‘count’ (i.e., xk) of 1 to the concentration parameters corresponding to the observed variable, while a ‘count’ of 0 is added to those corresponding to the non-observed variables. It is the concentration parameters of the Dirichlet distributions in the POMDP structure that are updated during learning. The exact way they are updated depends on the model element in question (e.g., A or B matrix, or D vector) which we discuss in more intuitive terms below. For a more detailed introduction to the Dirichlet-categorical model, see Tu (2014).
4.2. Non-technical continuation on Dirichlet priors
In this subsection we will introduce learning in more concrete and intuitive terms. This will build on what was presented in the previous technical section, but it does not require an understanding of the details presented there. At the end of this subsection, readers should have a practical understanding of what changes in a model during learning and what causes these changes to occur. Although the form of the Dirichlet distribution shown in the previous section can seem complex, the resulting learning process turns out to be quite intuitive. Essentially, it just involves adding counts to a vector or matrix based on posterior beliefs, where larger numbers of counts indicate higher confidence. To illustrate this, we will first consider a Dirichlet distribution for initial state priors over two possible states. As is standard notation in the active inference literature, we represent Dirichlet distributions with the lowercase letters associated with each vector or matrix. For example, we will denote the Dirichlet (Dir) distribution for the initial state prior D as d. Expressed formally:
| (31) |
| (32) |
Here, the concentration parameters for D – denoted by lowercase d = d1, d2 – are the individual parameters that will change during learning. In other words, the process of adding counts mentioned above will apply to the values of these variables. This is based on the following learning equation:
| (34) |
The eta (η) parameter is a learning rate (scalar from 0–1), which controls how much the values in d change after each trial. The omega (ω) parameter is a forgetting rate (scalar from 0–1), which influences how quickly learning in recent trial can ‘overwrite’ the changes in d that occurred in earlier trials. We will return to these below (for now we will assume they are both equal to 1).
To get an intuition for how this ‘learning by counting’ process works, consider a case where you start out with an initial state prior of d = [0.5 0.5]T on the first trial, and your posterior belief at the end of that trial is that you were in state 1 (with probability = 1). In this case, your prior on the second trial would become d = [1.5 0.5]T. In other words, a count of 1 was added to the first entry (i.e., the entry for state 1). If this happened 3 more times, then it would become d = [4.5 0.5]T. In cases of uncertainty, you instead add proportions of counts. For example, if you start out with an initial state prior of d = [1 1]T on the first trial, and your posterior belief at the end of the trial was s = [0.7 0.3]T, then your prior would be updated on the second trial to be d = [1.7 1.3]T. During within-trial inference, these distributions are put through a softmax function so that they retain their same shape but again add up to 1. However, larger numbers indicate greater confidence in the shape of the distribution. This can be seen by comparing d = [1 1]T to d = [50 50]T. While both distributions have the same (in this case flat) shape, it would take many more (new) observations to meaningfully change the shape of the second distribution compared to the first. For example, after one further trial with a precise posterior over state 1, the resulting shape of the distribution d = [2 1]T has changed quite a bit more than d = [51 50]T. This is an important aspect of active learning because it means that the initial (prior) counts determine how ‘open’ an agent is to new experience. Typically, in a novel environment or task, the initial counts are set to very low values (e.g., .25) – so that experience has a substantial effect on an agent’s prior beliefs. This makes inference and planning more context-sensitive, as opposed to an agent with high initial counts who is ‘stuck in its ways’ and would require much more evidence to ‘change its mind’.
As mentioned above, the agent also has a learning rate η. This controls how quickly it gets ‘stuck in its ways’ during learning (this also influences how quickly the agent ceases to select information-seeking policies; see below for more details). For example, if η = 0.5, then an update from d = [1 1]T after inferring state 1 would not lead to d = [2 1]T as shown above. Instead, it would be . Thus, counts (and hence confidence) will increase more slowly after each trial.
As also mentioned above, learning can be further modulated (multiplied) by a forgetting rate (ω). This parameter controls how strongly recent experience is able to ‘overwrite’ what one has learned in the more distant past. A value of ω = 1 indicates no forgetting (i.e., recent experience is unable to overwrite what has been learned previously), while values less than 1 allow increasing levels of forgetting (essentially, with each new observation the agent becomes less confident in what it has previously learned). This is important because, as counts increase during learning, an agent’s beliefs can become rigid and resistant to change, which is suboptimal in changing environments. Higher counts also reduce information-seeking, because the agent is highly confident in its beliefs (described in more detail below), which further hinders the opportunity to learn that (and how) the environment has changed. As such, if an agent believes that the environment is volatile (e.g., that the probabilities of rewards under each policy can change over time), then it is appropriate to adopt a low value for ω (i.e., a high forgetting rate). In other words, a low value for ω can be understood as encoding an agent’s prior belief that the contingencies in the world are unstable. It is worth noting that inferences about changes in the environment (and about the volatility of the environment) could also be implemented in a more principled manner in a hierarchical model (one example of a model with dynamically updated beliefs about environmental volatility is the Hierarchical Gaussian Filter; (Mathys et al., 2014). However, including the simpler forgetting rate parameter described here could be sufficient for modeling task behavior in many cases.
To get an intuition for how this works, glance back at the equation for learning at the end of the previous section (Eq. (34)) and then consider a case where d = [50 50]T and η = 1. If ω = 1, and the agent infers that it is in state 2, then the update will be . In contrast, if ω = 0.1, then the update will be . In this latter case, the agent therefore becomes much less confident in its prior beliefs, and the shape of the posterior (Dirichlet) distribution is changed to a greater degree at the end of that trial.
As a slightly more complex example of learning, the updates for an example A matrix become:
| (35) |
| (36) |
| (37) |
Here, the concentration parameters for A – denoted by lowercase – are the individual parameters in the matrix to be updated. The ⊗ symbol indicates the outer product. This again just involves accumulating (proportions of) counts, modulated by a learning rate and a forgetting rate. But in this case, what is being counted are coincidences between states and observations. For example, assume you have a posterior over states of s = [1 0]T and you made the observation associated with row 1, o = [1 0 0]T. Because you believed you were in state 1 when you observed outcome 1, this indicates that their association in a should increase. That is, a count should be added to a1 (i.e., the intersection of state 1 and outcome 1) before the subsequent trial. If you instead have a posterior over states of s = [0.7 0.3]T and you made the observation associated with row 2, o = [0 1 0]T, this indicates that their association in a should increase proportionally. That is, updates of a3 +0.7 and a4 +0.3 should occur before the next trial. Analogous update rules apply to the other model parameters (B, C, E). This general type of ‘coincidence detection’ learning is analogous to Hebbian synaptic plasticity, where neurons with coincident firing rates increase their synaptic connection strengths (Brown et al., 2010). As discussed further below, the neural process theory associated with active inference proposes that each concentration parameter can therefore be associated with the strength of a synaptic connection.
Another important change when learning is incorporated is that the expected free energy gains an extra term, depending on which parameter is being learned. This is because learning is also based on minimizing expected free energy. For example, if learning A, the equation for expected free energy becomes:
| (38) |
| (39) |
| (40) |
Note that the := symbol just means that two things are defined to be equivalent. The ⨀ symbol indicates the element-wise power (i.e., separately raising each element in a matrix to the power of some number). The term asums is a matrix of the same size as a where each entry within a column corresponds to the sum of the values of the associated column in a. For example, if , then .
Although this updated equation for EFE may appear complex, it simply adds one additional term — often called the ‘novelty’ term (i.e., in Eq. (38), or in the matrix formulation in Eq. (39)). This term scores how much beliefs within the A matrix are expected to change after receiving a new observation. Because the novelty term is a positive value (and subtracted from the total value), this entails that minimizing expected free energy will now also drive information-seeking about parameter values in the A matrix (i.e., as opposed to simply seeking out information about states). In other words, the agent will also seek out observations to increase confidence in its beliefs about . To do this, the agent will seek out state-observation pairings that will maximize the difference in concentration parameters between posterior and prior distributions over A. This difference quantifies the drive or epistemic affordance of finding out ‘what would happen if I do that?’. Although we do not show them explicitly here, similar terms can also be added to the EFE if the agent is learning any of the other matrices or vectors in the model (e.g., learning the transition probabilities in ).
Note that the value of the novelty term is inversely related to concentration parameter values. When the concentration parameters have large values, this term will have a small value, and when the concentration parameters have small values, this term will have a large value. Therefore, when the concentration parameter values are high (i.e., novelty is low), the model will become primarily reward-seeking, as it will be highly confident in its beliefs. In contrast, the agent will be information-seeking when concentration parameter values are low. Analogous dynamics occur when updating concentration parameters for other model parameters.
To make this more concrete, we show a worked example of the novelty term for two A matrices. One with small concentration parameter values (i.e., low confidence in beliefs about the outcomes generated by hidden states), and the other with large concentration parameter values (i.e., high confidence in beliefs about the outcomes generated by hidden states).
Small concentration parameter values (low confidence)
Large concentration parameter values (high confidence):
In both examples, the policy assigns high probability to occupying state 1 (p = .9). The normalized shape of the distribution for each column in A is also the same in both examples. However, the novelty term is larger in the first example where the associated Dirichlet prior a has smaller concentration parameter values (which, when subtracted from the total, will lead to a lower EFE). This means the agent will learn more (i.e., change its beliefs more) when moving to states where it is less confident in its beliefs (encoded as smaller concentration parameter values). To get a more intuitive sense for these computations, you can reproduce these results and adjust the concentration parameter values in the supplementary script EFE_learning_novelty_term.m.
4.3. Simulating learning
In this subsection we will build on the explore–exploit task model we specified above and demonstrate how it can also be used to simulate learning. By the end of this section, the reader should be equipped to run these simulations independently, and to plot and interpret their results. With the explore–exploit task model in place, we only need a few additions to the code. First, a lowercase version of the to-be-learned model element must be created. For example, to enable learning within the A matrix, one must specify an mdp.a with the same dimensions as mdp.A. The same goes for other parameters (mdp.b, mdp.d, etc.). Typically, the initial concentration parameters would be set to low-confidence (i.e., low-magnitude), flat distributions before learning begins; for example: Dir (d) = d {1} = [.25 .25]′. Note here that the generative process continues to correspond to the capital-letter matrices (i.e., which will generate the patterns of observations), while the lowercase-letter matrices are now the generative model. Next, we can specify a learning rate and a forgetting rate by setting mdp.eta and mdp.omega equal to values between 0 and 1. Finally, we need to replicate the mdp structure to include many trials; for example, using code such as:
Then, we can simply run the mdp structure through the spm_MDP_VB_X_tutorial.m function as before. Here, we will simulate two different versions of the task. In the first version, there are 30 trials, and the better slot machine is the same for all trials (left machine). We will allow the simulated agent to learn prior expectations about which context is more likely (i.e., whether the left or right machine tends to lead to wins more often). To enable this type of learning, we will include mdp.d. As shown in the Step_by_Step_AI_Guide.m code, we specify low confidence in initial state priors, d {1} = [.25 .25]′. We also set the learning rate to mdp.eta = .5 and the forgetting rate to mdp.omega = 1 (i.e., no forgetting). We then define a ‘risk-seeking’ (RS) parameter that corresponds to how precise the preference is to win the higher amount of money in the C matrix:
In our first simulation, we set RS = 3 and in our second we set RS = 4. We expect the agent will be less information-seeking, and more risk-seeking, in the latter case. As can be seen in Fig. 10 (top-left), the agent with RS = 3 chooses to take the hint on the first several trials, and slowly begins to forego the hint on later trials (with some choice stochasticity). Unexpected losses, shown in the panel just below, also cause the agent to return to ‘playing it safe’ and again ask for the hint. In contrast, the agent with RS = 4 chooses to take the hint only once (Fig. 10, top-right) and then takes that as sufficient evidence that the left machine must be the better one (and continues to choose this one throughout, despite occasional losses). The lower panels in the figure show simulated event-related potentials (ERPs), simulated dopamine responses, and how beliefs change over time regarding which context is more likely (darker = higher probability). When the Sim variable is set to Sim = 2 in the Step_by_Step_AI_Guide.m code, you can reproduce these simulations (and adjust the RS parameter to other possible values).
Fig. 10.
Simulated learning on the explore–exploit task and predicted neuronal responses. Blue circles in the top panel indicate chosen actions (i.e., the agent’s first choice on each trial); darker shading indicates a higher action probability. Wins/losses, free energies, simulated neuronal responses, and changes in prior beliefs about context over time (darker = stronger prior belief) are shown in the lower panels. See main text for more details. Left: Example simulation of a risk-averse agent (i.e., defined by a moderately precise preference distribution; RS = 3) learning from repeated trials of the explore–exploit task. Here the agent slowly gains confidence that the left machine will always be better and begins to choose that option without taking the hint. This agent often returns to taking the hint after unexpected losses. Right: Example simulation of a risk-seeking agent (i.e., defined by a highly precise preference distribution; RS = 4) learning from repeated trials of the explore–exploit task. This agent only required seeing the hint one time before attempting to pick the correct option directly (and win more money). It did not return to taking the hint with occasional unexpected losses. Note that both agents also show some stochasticity in choice. These simulations can be reproduced by running the Sim = 2 option in the supplementary Step_by_Step_AI_Guide.m code (although note that, because outcomes are sampled from probability distributions, results may not be identical each time).
Next, we simulate a reversal learning paradigm. Here, there are 32 trials in total. Unbeknownst to the agent, in the first 4 trials the left machine will be better, but in the rest of the trials the right machine will be better. Again, we examine an agent with RS = 3 and RS = 4. As shown in Fig. 11, the RS = 3 agent chose to take the hint on all trials in this simulation. In contrast, the RS = 4 agent quickly locked on to the left machine, but it then returned to taking the hint after the reversal. After several trials of again choosing the hint, it becomes confident in directly choosing the right machine in the final trials. When the Sim variable is set to Sim = 3 in the Step_by_Step_AI_Guide.m code, you can reproduce these simulations (and adjust the RS parameter to other possible values).
Fig. 11.
Simulated reversal learning on the explore–exploit task and predicted neuronal responses. Here, the better machine was on the left for the first 4 trials and then on the right for all subsequent trials (without the agent expecting this). Blue circles in the top panel indicate chosen actions (i.e., the agent’s first choice on each trial); darker shading indicates a higher action probability. Wins/losses, free energies, simulated neuronal responses, and changes in prior beliefs about context over time (darker = stronger prior belief) are shown in the lower panels. See main text for more details. Left: Example simulation of a risk-averse agent (i.e., defined by a moderately precise preference distribution; RS = 3), who always chose to take the hint. Right: Example simulation of a risk-seeking agent (i.e., defined by a highly precise preference distribution; RS = 4), who quickly became confident in choosing the left machine without taking the hint. After the unexpected reversal, it decided to again take the hint for many trials before becoming confident in choosing the right machine directly. These simulations can be reproduced by running the Sim = 3 option in the supplementary Step_by_Step_AI_Guide.m code (although note that, because outcomes are sampled from probability distributions, results may not be identical each time).
Each of these examples is meant primarily to give the reader a sense of how to work with these types of simulations. But there are many other parameters that could be manipulated. In the accompanying MATLAB code, the reader can easily re-run these simulations while changing the learning rate, forgetting rate, action precision, or any other parameters in the model. We encourage the reader to do so to get a sense of the unique influences of different parameters on task behavior. For examples of papers that model learning using active inference, see Friston et al. (2016a, 2017b), Schwartenbeck et al. (2019), Smith et al. (2021e), Smith, Parr, and Friston (2019b), Smith, Schwartenbeck, Parr, and Friston (2020d), Smith et al. (2020e) and Tschantz, Seth, and Buckley (2020).
5. Neural process theory
In many active inference papers, one sees figures similar to Fig. 12. These figures typically depict a series of update equations, several columns of ‘ball’ neurons, a specific pattern of synaptic connections, and labels assigning model variables to those neurons and synaptic connections. Such figures are meant to depict one possible neural implementation of active inference, which serves as a concrete illustration of the general biological plausibility of the theory. This type of biological plausibility is considered an important strength of active inference models, due to the resulting ability to make empirical predictions about neurophysiological responses. In turn, these predictions allow one to assess the evidence for distinct message passing algorithms and possible neuronal implementations (Parr et al., 2019). Some of the simulation outputs in Table 3 contain predicted neuronal responses that can be assigned to distinct neuronal populations or synaptic connections (some examples are plotted in Fig. 8). To be clear, some neurophysiological predictions in active inference are not specific to a single neural implementation (i.e., they are based only on the generic prediction error minimization and precision updating equations described above); and these types of predictions have been successfully associated with neural responses observed in previous functional magnetic resonance imaging [fMRI] and EEG studies (e.g., see (Schwartenbeck et al., 2015; Smith et al., 2021e; Whyte, Hohwy, & Smith, 2021; Whyte & Smith, 2020). However, cortical column proposals – such as that shown in Fig. 12 – can also motivate targeted research using methods (such as laminar fMRI) that have been applied to test similar columnar implementations proposed for predictive coding (Stephan et al., 2019). To prepare the reader for understanding – and potentially contributing to – this important area of active inference research, we will walk the reader through Fig. 12 step-by-step. This will also be important when we discuss hierarchical models in the next section (Section 6), in which simulated EEG responses occur over different timescales and are predicted to occur at distinct levels of processing within the brain.
Fig. 12.
Common depiction of the neural process theory associated with active inference. This includes the update equations on the left and an example neural network that could implement them on the right (only exemplar synaptic connections are shown to avoid visual clutter). See main text for an in-depth walk-through. Note that while most equations are presented in the same form as in the main text, we have updated the outcome prediction error (ςπ,τ) equation to include the ‘novelty’ term (Asπ,τ · Wsπ,τ) within Gπ that was introduced in the previous section on learning. We also depict iterative prediction error minimization and precision updating processes using arrows indicating the order of repeated updating until convergence.
In the depicted neural network, each column of neurons (in this case, 3 columns) represents beliefs and prediction errors about each point in time (from left to right, indicated by subscripts for τ = 1, 2, 3). With each new observation, beliefs about all time points (i.e., about the past, present, and future) are updated, corresponding to changes in neural activation across all neurons. The upward arrows from observations (purple nodes at the bottom) to layer 3 (denoted by επ,τ for state prediction errors) are depicted as conveying excitatory (red) observation signals (e.g., sensory input) to granular cells in each cortical column, where these observations can differ at each time point. The receiving (pink) state prediction error neurons calculate their prediction errors by combining observation signals with prediction signals from the state representations in the cyan neurons of layer 2 (supragranular neurons denoted by for state representations). Note that excitatory (red) downward signals from these neurons to layer 3 are conveyed both forward (from the τ = 1, left neurons) and backward (from the τ = 3, right neurons) – indicating both prospective and retrospective predictive influences on state representations about a time point. In contrast, inhibitory (blue) signals are conveyed by these state representation neurons to layer three neurons for the current time point, leading to minimization of prediction error when predictions from state representations match observation signals.
Note next that each of these state and state prediction error representations are calculated in parallel for each policy (denoted by one example neural column in front of another). The top (red) layer 1 neurons, however, do not have another set of neurons behind them. This is because they perform a Bayesian model average as an overall best guess about states. They do this by taking state representations for each policy, multiplying them by the probability of that policy, and then averaging them to get a final posterior over states. This is accomplished in conjunction with the policy representation (π) neuron on the left (meant to represent a subcortical neural population), the signals from which multiply (via the green modulatory connection) the excitatory (red) signals from the state representation neurons for each policy in layer 2. After each new observation, activity in these policy representation neurons also promotes some actions (u) over others.
Policy representation (π) neurons are in turn activated by neurons encoding habits (E) and inhibited by those encoding expected free energy (G; i.e., greater expected free energy reduces the probability of a policy). The influence of G on π is modulated by the expected free energy precision term (γ) — depicted here as being conveyed by subcortical dopamine neurons. The activity of G neurons is increased by outcome prediction errors (ζπ,τ) in layer 5, multiplied (green connections) by the probability of those outcomes under each policy (oπ,τ, cyan neurons in layer 4; note that these are calculated from Asπ,τ). These outcome prediction errors reflect the expected difference between preferred outcomes and predicted outcomes under each policy (from layer 4; downward excitatory connections between layers 4 and 5), and where those predicted outcomes are in turn based on state representations in layer 2 (excitatory red connections from layer 2 to layers 4 and 5). Note that the matrix W (i.e., calculated based on the synaptic strengths encoded in A) also influences G through interaction with activity in the neurons encoding , but these connections are not explicitly shown to minimize visual clutter.
Based on this description, we assume the reader should be able to follow the equations on the left to identify each associated connection in the network on the right. Note that this figure only shows example connections, assuming two policies and three time points. However, the basic idea is that, if each excitatory connection corresponds to addition, each inhibitory connection corresponds to subtraction, and each modulatory connection corresponds to multiplication, then each of the update equations on the left (in the ‘Belief updating’, ‘Policy inference and expected free energy precision’, ‘Action selection’, and ‘Learning’ boxes) can be implemented in a straightforward manner within a relatively simple neural network. It is worth mentioning that these update equations are not always presented in identical form in such figures across the literature. However, these variations are either algebraically equivalent or have been presented with or without certain elements (e.g., with or without learning, with or without expected free energy precision, etc.), depending on the goals of the paper.
One other aspect of the neural process theory that we will not discuss in detail – but that we would like to briefly point the interested reader toward – pertains to the idea that Bayesian model reduction (i.e., comparing models to find the simplest one that can account for available data) is implemented by homeostatic synaptic adjustment processes during sleep and resting wakefulness (Bucci & Grasso, 2017; Friston et al., 2017b; Hobson & Friston, 2012; Hobson, Hong, & Friston, 2014; Smith et al., 2020d; Tononi & Cirelli, 2014). In short, this literature suggests that, during sleep/rest, the brain can also minimize VFE by finding simpler models (with fewer parameters) that can successfully account for previous experience. This can be accomplished in part via a ‘synaptic downscaling’ process – known to occur during sleep – in which synaptic connections that have gotten stronger during recent learning are subsequently attenuated. Synaptic downscaling can be helpful in removing any small synaptic changes driven by noise (e.g., uninformative coincidences in the presence of multiple stimuli), leaving only the larger synaptic changes needed to account for consistent patterns in recent experience. Although we do not cover it here, we note that the spm_MDP_VB_X.m script (and our tutorial version) does have an additional ‘BMR’ option that can be turned on, which will implement Bayesian model reduction by calling a further SPM script written to simulate this proposed function of sleep/rest (spm_MDP_VB_sleep.m). In this case, the entries in each matrix (e.g., the A matrix) are assumed to represent synaptic connections, and model reduction involves eliminating any changes in the concentration parameters for those matrices during learning that did not improve the explanatory power of the model (i.e., in terms of the accuracy/complexity trade-off).
To facilitate the reader’s ability to use the neural process theory in their own research, the spm_MDP_VB_X_tutorial.m script automatically generates simulated responses for several of the neuronal populations described above (see Table 3 for specific output descriptions). This includes the simulated firing rates of neuronal populations representing posteriors over states (i.e., whose predicted location in the brain would depend on the task). Here, distinct firing rates are generated for the states within each state factor of a model (e.g., in the explore–exploit task, firing rates in one population would encode the probability of the ‘left-better context’, while those in another population would encode the probability of the ‘right-better context’). The script also generates vectors encoding the associated state prediction errors discussed above. Simulated electrophysiological responses are based on the rates of change (derivatives) in firing rates during prediction-error minimization (interpreted as local field potentials [LFPs] or event-related potentials [ERPs], depending on the context). Separate neural populations are postulated to encode posteriors over states and prediction errors for each time point τ (i.e., each of which are updated with new observations over time).
Another set of potentially useful outputs generated by the spm_MDP_VB_X_tutorial.m script corresponds to the VFE of changes in concentration parameters (i.e., higher = greater surprise during learning; see Table 3). Recall here that another important aspect of the neural process theory is that synaptic inputs with different strengths are postulated to carry the prior and conditional probabilities encoded in each of the vectors/matrices that define a model (e.g., the probabilities in the A or Bπ,τ matrices, or in the D or E vectors). More specifically, the value of each entry in particular matrices or vectors can be thought of as corresponding to the strength of a synaptic connection between two neurons. Updating concentration parameters over repeated trials (i.e., over a slower timescale than perception) can therefore be linked to synaptic plasticity — where the specific changes involved depend on the pattern of observations, belief updates, and/or policies chosen on each trial). The VFE of changes in concentration parameters can therefore be used to quantify the magnitude of change in a model on a trial by trial basis, which could be used to identify neural correlates of such changes. The predicted dynamics of within-trial prediction errors and belief updates will also be modulated by these synaptic changes, allowing for predicted time courses that could be used for similar experimental purposes. In addition, synaptic strength changes will also inform the matrix W – dynamically adjusting motivation to seek information about model parameters. Thus, several experimentally useful outputs are provided to test the neural process theory.
An available SPM function for plotting single-trial neural simulations can be run by inputting the following into MATLAB:
Setting the variable Sim in the accompanying tutorial code (i.e., Step_by_Step_AI_Guide.m, line 51) to Sim = 1 will also generate simulation plots using this function.
Based on the current model specification, a representative plot of simulation results is shown in Fig. 8C, based on the single trial depicted in Fig. 8A. The top-left panel depicts the belief updates at each time point t (columns) about each time point τ (rows). As before, darker indicates higher probability. In this case, after presentation of the first observation (t = 1, column 1), the model is fully uncertain about current or future states (each row). After presentation of the second observation (t = 2, column 2), when the model receives the hint, it becomes highly confident that it was, currently is, and will continue to be in the ‘left-better’ state (i.e., based on its transition beliefs that this state does not change within a trial). These beliefs remain stable when it receives the third observation (column 3; i.e., upon observing the expected win). The top-right panel depicts these belief updates (i.e., changes in beliefs over time about the state at each time point; 2 possible states for each of the 3 time points). These belief updates are depicted as traces of changes in neural firing rates, with 2 firing rates per distribution (i.e., encoding the probability of the left- vs. right-better context), resulting in 6 firing rate traces in total (note that, due to overlap, not all 6 traces are clearly visible in this example). The bottom-left plot depicts this same information, but here displayed in terms of a simulated raster plot (i.e., one tick per action potential per neuron in a simulated population). The middle-right panel depicts predicted local field potentials (or event-related potentials), which reflect the rate of change in the simulated firing rates. The middle-left panel depicts the neural responses associated with context state beliefs before (dotted line) and after (solid line) filtering at 4 Hz, superimposed on a time–frequency decomposition of the local field potential (averaged over all simulated neurons). This type of plot has been used in previous work to explain how/why simulated depolarization in specific frequency ranges may coincide with specific stimulus-induced neural responses (Friston et al., 2017a). The bottom-right panel depicts simulated dopamine responses after each new observation (as also depicted in a slightly different way in Fig. 8A). These were covered in detail in Section 3 (see Fig. 9 and the last row of Table 2).
This plotting function also has several options as additional function entries and outputs as follows:
UNITS: a matrix with 2 rows and one or more columns. The first row indicates which hidden state(s) to plot over time for the specified state factor. The second row specifies the time point being represented. For example: UNITS = [1 2 1 2; 1 1 3 3] would plot firing rates for the first two hidden states of the selected state factor over time with regard to beliefs about time points 1 and 3. By default, all units are selected.
FACTOR: a single number denoting which state factor to plot (default = 1).
SPECTRAL: either a 0 or 1 (default = 0). If 1, the top-left plot is replaced by a plot of the power of simulated neural responses in different frequency ranges.
The optional outputs u and v correspond to vectors encoding simulated event-related potentials and firing rates, respectively (for selected units). As was mentioned above, the event-related potentials correspond to the temporal derivative of the firing rates, while the firing rates reflect the magnitude of posterior beliefs over each state at each iteration of marginal message passing.
6. Building hierarchical models
6.1. Hierarchical model structure
Now that we have a clear idea of how to specify a generative model of a behavioral task, how to interpret the relevant outputs, and how we can generate testable predictions regarding neural responses, we will now extend this foundation by building a hierarchical or ‘deep temporal’ model and demonstrating how it can be used to reproduce established neurophysiological results (for examples, see Friston et al., 2018; Parr & Friston, 2017b; Smith, Lane, Parr, & Friston, 2019a; Whyte et al., 2021; Whyte & Smith, 2020). Specifically, we will reproduce the results of experiments examining ERPs in a commonly used auditory mismatch paradigm designed to study the neural basis of perceptual learning and expectation violation. This should help the reader to generalize their understanding by seeing how to build a model with a different structure. It will also demonstrate the versatility and wide range of applications of active inference models. For example, while reinforcement learning models are often used to solve tasks like the slot machine task, such models are not readily applicable to perceptual tasks, like the auditory mismatch paradigm, that do not include rewards or produce notable variability in behavior (e.g., when performance tends to be near ceiling across participants). By the end of this section, the reader should have a solid understanding of both how hierarchical models work and how to implement them.
The steps for building hierarchical models are quite similar to what we have already covered, because this primarily just involves building two models, and then placing one below the other. The further step is figuring out how to link the two models together. This is because, in hierarchical models, the states at the lower level exchange information with states at the higher level in a very specific bidirectional manner (see Fig. 13). First, for each time point in a second-level trial, the hidden states at the second level (; superscript indicates hierarchical level) provide prior beliefs over the initial states of a first-level trial (i.e., for the first first-level trial; for all subsequent first-level trials). In turn, posterior beliefs over initial states at the end of a first-level trial (i.e., ) are treated as observations at a time point in a second-level trial (). This means that the second-level A matrix (likelihood mapping) mediates the ascending and descending messages between hierarchical levels. This structure also entails that the second-level model must operate at a slower timescale than the first-level model, because each observation in the second-level model (i.e., each time point in a second-level trial) corresponds to the results of (i.e., posterior beliefs after) a complete trial in the first-level model. Thus, there are as many first-level trials as there are time points in a second-level trial. For example, if there are four time points in a second-level trial, this means there will need to be a corresponding sequence of four first-level trials (i.e., where each first-level trial could itself have several time points). This is why such models are often called deep temporal models.
Fig. 13.
Top Left: Bayesian network depiction of a 2-level POMDP. Observations depend on hidden states at the first level. In turn, hidden states at the first level depend on hidden states at the second level. Specifically, second-level hidden states provide prior beliefs over initial states at the first level, while posterior beliefs over initial states at the first level are treated as observations by the second level. In the example shown here, the first level has two state transitions per trial. This entails that the first level has two state transitions for every one state transition at the second level. Thus, beliefs at the second level evolve over a slower timescale. Top Right: Example neural network implementing the hierarchical POMDP shown on the left. Bottom: Illustration of the auditory mismatch paradigm for which we build a generative model and run simulations in this section (Section 6). In the model specified in the main text, beliefs about single tones are encoded at the first level, while beliefs about sequences of tones are encoded at the second level. Across trials, the agent then builds up prior expectations through presentation of repeated tones and tone sequences. In line with empirical results, the post-learning model simulations shown in Figs. 14–16 predict earlier ERPs for unexpected tones (local deviations) and later ERPs for unexpected tone sequences (global deviations).
This type of model architecture is essential for capturing perceptual phenomena with nested dynamics, or where objects must be recognized before regularities in the behavior of those objects can be detected. For example, to perceive a baseball flying in a leftward direction, a lower-level model would first need to infer the baseball’s identity and position (i.e., one inferred position per lower-level trial), and a higher-level model would then need to accumulate evidence for a leftward trajectory of motion based on how the baseball’s inferred position changes across several lower-level trials. As another example, to recognize a melody, a lower-level model would be needed to infer the presence of each note, and a higher-level model would then be needed to accumulate evidence for a specific melody, based on a specific sequence of inferred notes over time. A further intuitive example is reading, where the first level infers single words, while the second level infers the narrative entailed by sequences of words (Friston et al., 2018). Note, as soon as we start to use deep or hierarchical generative models, we are essentially relaxing the Markovian assumption by introducing a separation of temporal scales to produce what are known as semi-Markovian models. These are essential for inferring narratives, language, or any deeply structured sequence of state transitions.
Aside from these examples, the hierarchical POMDP setup is quite flexible and can be used to model a wide range of temporally structured phenomena. For example, a policy space could be included at either level alone, or both levels, depending on the target phenomenon to be modeled (e.g., verbal report at a higher level vs. reflexive behavior at a lower level). One could also specify several time points in each lower-level trial, such that higher-level states generate sequences or trajectories of state transitions at the lower level (i.e., within-trial). In previous work, hierarchical models have been used to model working memory, reading, visual consciousness, and emotional awareness, among other phenomena (Friston et al., 2017c, 2018; Hesp et al., 2020; Parr & Friston, 2017b, 2018b; Sandved-Smith et al., 2021a; Smith et al., 2019a; Whyte et al., 2021; Whyte & Smith, 2020). Hierarchical POMDPs also afford further opportunities for simulating neuronal processes. To date, simulations associated with the faster and slower timescales of belief updating have been shown to reproduce an impressive number of task-based electrophysiological findings. For example, empirically observed patterns of ERPs associated with specific cognitive and perceptual processes, such as the P300 and mismatch negativity (MMN), emerge naturally in simulations of different experimental paradigms, which supports the face validity of both the model structure and the neural process theory (e.g., Friston et al., 2017a; Parr & Friston, 2017b; Whyte et al., 2021; Whyte & Smith, 2020).
6.2. Building a hierarchical model
As a concrete, empirically relevant example, we will now demonstrate how one could build a hierarchical model to simulate a simplified version of the auditory mismatch ‘local–global’ paradigm introduced in Bekinschtein et al. (2009); see bottom panel of Fig. 13. In our simplified version of this paradigm, each trial consists of a sequence of four tones (with either low or high frequency), the first three have the same frequency, and the fourth tone either conforms to the predicted pattern (e.g., high-high-high-high; local standard) or violates the predicted pattern by presenting a different tone (e.g., high-high-high-low; local deviation). During EEG, local deviations elicit a mismatch negativity component in ERPs (i.e., a negative component obtained by subtracting ‘local standard’ trials from ‘local deviation’ trials), which appears after approximately 130 ms. Importantly, the sequence of local standard and local deviation trials establishes a global pattern that can itself be confirmed (global standard) or violated (global deviation). For example, this could include several local deviation trials in a row followed by an unexpected local standard trial. Global deviations are known to elicit a P300 ERP component (i.e., a positive component that appears after approximately 300 ms). Unlike other auditory mismatch paradigms, this design also allows local and global violation responses to be dissociated. That is, the factorial design leads to four conditions local standard + global standard, local standard + global deviation, local deviation + global standard, and local deviation + global deviation. For brevity, here we only simulate two of the four possible combinations, local deviation + global deviation, and local deviation + global standard. In our simulated version of the task, we presented an active inference agent with sequences of 4 tones, where each tone could be either low or high, in an analogous manner to the empirical task. We then had the agent report whether the last stimulus on each trial was the same as, or different from, the established pattern.
At this point, the reader is encouraged to open the accompanying MATLAB script and follow along in parallel (Step_by_Step_ Hierarchical_Model.m). As with the previous model, we will start by setting up the priors over initial states for each hidden state factor at the first level:
The specification for D {1} means that there is an equal probability of a high or a low tone (left and right entries, respectively). Note that, because the columns of all the vectors and matrices of the generative process are run through a softmax function in the spm_MDP_VB_X_tutorial.m script, D {1} = [1 1]′ is equivalent to . As the simulation involves learning, we also need to separate the generative process from the generative model by including the lowercase d for the generative model. Here we simply set d = D, as the agent will also start out with the belief that a high and a low tone are equally probable (but with fairly low confidence). However, including d will allow the agent to accumulate concentration parameters (changing the shape of its initial state priors) over trials based on patterns in its observations.
Next, we must specify the likelihood mappings for the first level in the A matrix.
This is simply an identity matrix indicating that the tone states correspond 1-to −1 with tone observations (columns [left to right]: high tone, low tone states; rows [top to bottom]: high tone, low tone observations). However, in the generative model we may want to introduce some noise into tone perception. One convenient way to do this is to first specify:
Then, we can use a softmax function to control the expected precision of the state-observation mapping with a precision parameter:
Note that the exp(−4) is simply a very small number added to A{1} to prevent the possibility of log(0), which is undefined (also note that, while −4 is a reasonable value, other values could be chosen). Depending on the value of the precision parameter (higher = more precise), this will result in a likelihood mapping that specifies different amounts of sensory noise. For example:
Note that, for clarity, this example shows a lower precision than what results from setting precision = 2 in the accompanying tutorial code.
As we are mainly interested in simulating the learning of prior expectations (D vector), we also multiply the concentration parameters within a by 100 (an arbitrary large number) to effectively prevent the learning of these other parameters. This is because we want the level of sensory noise to remain consistent across trials.
Next, we can specify transition probabilities in the B matrix as identity matrices,10 as tones do not change within a lower-level trial.
Here columns (left to right) are high tone and low tone states at time τ, while rows (top to bottom) are high tone and low tone states at time τ + 1. Here there is no need to separate the generative process from the generative model, so we do not specify a separate b matrix.
We do not include preferences or policies at this level, so we now simply assign each variable to the mdp structure. For convenience when later linking this to the higher-level model below, we will denote this structure with an ‘_1’ as follows:
For consistency with how we link first- and second-level models below, we then set MDP_1 = mdp_1 and clear mdp_1.
Now we move on to specifying the second-level model. To keep the variables separate, we will denote each model variable with a ‘_2’ for this level.
Here, we will include one hidden state for each possible sequence of tones:
This indicates that initially there is an equal probability of each tone sequence (left to right: ‘all high’, ‘all low’, ‘high-low’, ‘low-high’). Note again that using four 1s here is just for convenience, as this vector will subsequently be run through a softmax function and each 1 will become a .25.
Here we must also include a second hidden state factor that encodes beliefs about the time point within a trial (e.g., ‘first tone’, ‘second tone’, etc.). This includes (from left to right) time points for 4 tones, a delay period, and then a reporting period:
This indicates that the agent always starts in the ‘time 1’ state.
We also include a reporting state factor, corresponding to the agent either not yet reporting (left entry), reporting ‘same tone’ (middle entry), and reporting ‘different tone’ (right entry) at the end of the trial:
This indicates that the agent always starts in a state of not yet having made a report.
Finally, we allow the agent to build up prior beliefs over time with repeated trials. In this case, because the agent’s beliefs initially match the generative process, we can simply set:
Next, we must specify the likelihood mappings for the second-level A matrix. Because time in trial is a state factor, this becomes somewhat more complex. Specifically, we are now required to specify the type of tone at each time point that is expected under each sequence. To do so, we can specify the matrices as follows. For convenience, we can first specify:
This says that for the first six time points (i = 1: 6), and for all three choice states (j = 1: 3), the first and third sequence states (i.e., ‘all high tones’ and ‘high tones followed by low tone’; columns 1 and 3) are associated with the ‘high tone’ observation (top row), whereas the second and fourth sequence states (i.e., ‘all low tones’ and ‘low tones followed by high tone’; columns 2 and 4) are associated with the ‘low tone’ observation (bottom row). Then, we can adjust this so that the deviation sequences (‘low tones followed by high tone’ and ‘high tones followed by low tone’; columns 3 and 4) are associated with the opposite tone mapping at the fourth time point (i = 4):
The second outcome modality at the higher level (which does not correspond to a lower-level state factor) is feedback about whether a chosen report was correct or incorrect. Here, we need to specify that the agent will observe ‘correct’ feedback (at the final time point) in cases where its report matches the appropriate sequence (row 3), and ‘incorrect’ feedback otherwise (row 2). To do this, we can initially specify that no feedback (‘null’; row 1) will be observed across all time points:
Then we can specify that at time point i = 6, if the agent reports ‘same’ (j = 2), then it will receive correct feedback when it is one of the first two (standard) sequences and incorrect for either of the second two (deviant) sequences:
The we specify the opposite mapping if the agent reports ‘different’ (j = 3):
As with the first-level model, to control the precision of the mapping between first- and second-level states in the generative model, we can use a precision_2 parameter. To do so, we can specify:
This results in a minor amount of noise in the messages passed between levels. As with the first level, we also multiply this parameter by 100 to (effectively) prevent learning.
Next, we must specify the transition matrices for the second level. In this case, the sequence type is stable within a trial, so this should be an identity matrix (columns: states at time τ, rows: states at τ + 1):
Time in trial should progress forward (e.g., ‘Time 1’ should ‘transition to ‘Time 2’, and so forth. As such:
Finally, report states are under control of the agent. In this case, there are three actions:
These three matrices (from 1–3 in dimension 3) correspond to the actions (U) of moving (from any state) to the ‘no report’ state, ‘report same’ state, and ‘report different’ state, respectively. Next, we must specify the allowable sequences of actions (i.e., policies). In this case, we include two policies (two columns) and one row per time point. There are no actions for the first state factor, so (number = action, column = policy, row = time point):
There are also no actions for the second state factor:
For the third state factor, the agent must wait until the last time point and then either select the ‘report same’ or ‘report different’ actions:
Lastly, we must provide the agent with preferences that will motivate accurate reporting. For the first outcome modality (tones), the agent has no preferences (columns = time point, rows [top to bottom] = high tone, low tone observation):
For the second outcome modality (accuracy feedback), the agent prefers to receive ‘correct’ feedback at the last time point (column 6, row 3) and finds ‘incorrect’ feedback to be aversive at the last time point (column 6, row 2):
As already mentioned when building the explore–exploit task model, each column in this matrix is put through a softmax function and then converted into log-probabilities. Having now specified the second-level model, we will place each of these matrices into its own mdp structure:
We then need to connect the lower-level model with the higher-level model as follows:
Next, we need to provide a matrix specifying which outcome modalities at the second level (columns) corresponds to which state factors at the lower level (rows) within a ‘link’ field. Here, the first outcome at the second level (‘tones’) corresponds to the first state factor at the first level:
In this case, the matrix only has a single row because there is only one state factor at the lower level.
Lastly, we need to set the value of the ERP ‘reset’ or ‘decay’ parameter mdp.erp, which at the start of every epoch of gradient descent is used to reset the posterior over states by dividing the posterior by the value of the parameter (i.e., higher values = more resetting). Setting the value of the parameter is entirely up to the discretion of the modeler, depending on assumptions about the particular neurocognitive process under study. In empirical work, this parameter could be fit to observed ERP responses. In the experimental paradigm we simulate here, the tones are played to the participant in quick succession, so we assume that the posterior at each time step carries over and does not decay between presentations. As such, we set mdp.erp = 1. If, however, we were trying to model a task with longer time periods between updating (e.g., a subject navigating a maze), some degree of decay could be appropriate (mdp.erp = 4 is the default value in the current spm_MDP_VB_X.m script).
As before, we can now run this structure through the standard routine to generate simulated behavioral and neuronal responses of an example trial.
To simulate our two conditions of interest, local deviation + global deviation, and local deviation + global standard, we will simulate 10 sequential trials for each condition. For brevity, we will not describe the code that implements this here. Instead, we direct readers to the Step_by_Step_Hierarchical_Model.m script, which has detailed comments describing each of the necessary steps. For both conditions, the first nine trials consist of three high tones and a fourth low tone. On the tenth trial of the local deviation + global standard condition, the trial again consists of three high tones and a fourth low tone. On the tenth trial of the local deviation + global deviation condition, the tenth trial instead consists of four high tones, thereby violating the global regularity. Fig. 14 shows plots of second-level belief updating and policy selection, analogous to the single-level model plots shown in Fig. 8. The model performed at ceiling with 100% accuracy when classifying the last stimulus in the sequence as same or different.
Fig. 14.
Simulated beliefs and behavior in the hierarchical POMDP (analogous to the single-level model plots show in Fig. 8) described in the main task. The top and bottom panels simulate the two task conditions described in the main text both before and after learning (Trial 1 and Trial 10, respectively). These simulations demonstrated that the agent performed the task appropriately. The three panels in the top-left of each plot show posteriors over states at the end of the trial. That is, the states the model believes it was in at each time point τ at the last time point t (i.e., after receiving the last observation). Here, time goes from left to right, darker indicates higher probability, and the cyan dots denote the true states. The top-right panels in each plot show the action probabilities and true actions. The left-middle panel in each plot just shows the different possible action-sequences/policies in the specified model (encoded numerically from left to right). A darker color indicates a lower number. The right-middle panel in each plot shows the progression of posterior beliefs in each policy over time (from left to right, darker = higher confidence). Policies (rows) line up with the action sequences in the plot on the middle-left. The two panels in the bottom-left of each plot display the outcomes in cyan dots and the agent’s preference for each outcome, where darker colors indicate a greater preference (i.e., higher prior probability). Lastly, the bottom-right plot displays predictions about dopamine responses based on the neuronal process theory (i.e., encoding changes in expected precision of EFE; see last row of Table 2 and Fig. 9). In terms of behavior, notice that all models selected the correct actions and received ‘correct’ feedback, as indicated in the lower outcome plot. These simulations can be reproduced using the Step_by_Step_Hierarchical_Model.m script included as supplementary code. Note that, due to random sampling, results may not be identical each time.
The next step is to visualize the resulting simulations in order to make empirical predictions about behavior and neuronal responses.
6.3. Plotting hierarchical models
To visualize the resulting belief updating and simulated neuronal responses for this hierarchical model, we have provided a modified version of the plotting script provided in the freely available SPM routines:
This plotting script shows the simulated firing rates associated with belief updating at each level. It also shows the simulated ERPs (summed first derivative of the firing rates) that would be expected to be generated during the simulated task. Fig. 15 shows the first and tenth trial from each condition (i.e., before and after building up prior beliefs favoring some states over others). Notice that on the first trial of both the local deviation + global deviation condition and the local deviation + global standard condition, the high firing rates at the first level reflect a high posterior confidence in the tone each time it is presented. In contrast, after hearing the first tone, the firing rate is evenly spread between both the ‘high’ and ‘high-low’ hidden states at the second level, reflecting the agent’s uncertainty about which type of sequence is being presented (i.e., since both sequences predict ‘high’ tones for the first three time steps). Note, however, that the fourth tone on the first trial generates an increased firing rate for one of the hidden states, and the firing rate drastically decreases for the other state (depending on whether a ‘high’ or ‘low’ tone is presented). By the tenth trial, the agent has a high prior expectation that it will experience a ‘high-low’ sequence, reflected in the high firing rates for this second-level hidden state from early time points. In the local deviation + global standard condition, this expectation is confirmed, leading to little change in second-level firing rates. Importantly, however, in the local deviation + global deviation condition, this expectation is violated, because the agent is presented with four high tones. This creates a rapid switch in posterior confidence from the ‘high-low’ hidden state to the ‘high’ hidden state at the fourth time step, generating a very strong and rapid shift in second-level beliefs.
Fig. 15.
Simulated ERPs and firing rates extracted from the hierarchical POMDP model of the auditory mismatch paradigm during each task condition (top vs. bottom) both before and after learning (Trial 1 vs. Trial 10). As described in the main text (and illustrated further in Fig. 16), these simulations reproduce ERP results observed in previous empirical studies (e.g., stronger short-latency ERPs in response to local deviations and stronger longer-latency ERPs in response to global deviations). The unit response plots show the posterior probability over states () at each level of the model (as usual, darker colors = higher posterior = higher firing rates). As described in the neuronal process theory section, normalized firing rates are generated by passing the depolarization variable vπ,τ through a softmax function sπ,τ = σ(vπ,τ). The ERP plots show the rate of change (first derivative) of posterior beliefs over states summed over all states at each level of the model (analogous to the aggregate signal measured at the level of the scalp by EEG). Note that each increment of 0.5 along the x-axis corresponds to a trial at the lower level, and to a time point at the higher level (i.e., 6 time points in a higher-level trial, with 6 corresponding lower-level trials). For a detailed description of each plot and its meaning in relation to the task, see the main text. These simulations can be reproduced using the Step_by_Step_Hierarchical_Model.m script included as supplementary code. Note that, due to random sampling, results may not be identical each time.
This brings us to simulated ERPs, which reflect the rate at which posterior beliefs change within each epoch of belief updating, summed over hidden states at each level of the model (i.e., similar to the aggregate signal measured by EEG). At the first level, the repeated presentation of high tones generates small ERPs, whereas deviations from this pattern at the fourth time step (local deviations) generate a larger amplitude mismatch response because posterior beliefs about the tone change rapidly. Importantly, when the local deviation response is subtracted from the local standard response, we can reproduce the classic mismatch negativity effect (MMN; see Fig. 16). At the second level, the repeated occurrence of three high tones and one low tone for the first nine trials creates a strong prior expectation (through the increase in concentration parameters in the D vector) for the ‘high-low’ sequence state. When the model is unexpectedly presented with four high tones on the tenth trial, this expectation violation generates a rapid change in beliefs and a correspondingly large second-level ERP resembling the P300 (see Figs. 15 and 16). Fig. 16 shows custom-made ERP plots, which isolate the contrasts of interest. Again, for the sake of brevity, we will not describe the code that generates this plot in the main text, but direct interested readers to the Step_by_Step_Hierarchical_Model.m script for more details.
Fig. 16.
Custom ERP plots generated from the simulated auditory mismatch paradigm. On the left we have the ‘raw’ first- and second-level ERP waveforms centered on the fourth time step of the tenth trial from each condition. The right side of the figure shows how the subtraction of the deviant trials from the standard trials reproduces both the MMN and P300. At both the first and second level, ‘deviant’ ERPs have a substantially larger amplitude than ‘standard’ ERPs. Note that, although the relative differences in timing between simulated ERPs (and the relative differences in timing between ERPs at different levels of the model) are meaningful, the units we ascribe to time are (usually) somewhat arbitrary. So, for clarity, we have not included any units of time on the x-axis. These simulations can be reproduced using the Step_by_Step_Hierarchical_Model.m script included as supplementary code. Note that, due to random sampling, results may not be identical each time.
7. Fitting models to behavior
So far, we have focused on simulating behavior and establishing the face validity of active inference using canonical examples from the decision-making and electrophysiological literature. In this section, we demonstrate how active inference models can be used in empirical studies. More specifically, we will describe how one can estimate the model parameter values that best explain participant behavior during an experimental task. This approach has been employed in several recent studies that have used active inference models to account for behavior during tasks designed to study a wide range of phenomena — such as attention, risk-taking, approach-avoidance conflict, explore–exploit behavior, and interoception (Mirza, Adams, Mathys, & Friston, 2018; Schwartenbeck et al., 2015; Smith et al., 2021d, 2021c, 2020b, 2020c, 2021e, 2020e). In each of these studies, a model was used to evaluate the prior beliefs that participants most likely held when performing a task (i.e., the prior beliefs that would have generated their behavior in a model). All necessary steps for carrying out this approach are described below, with the goal of preparing the reader to use active inference models in their own empirical studies. By the end of this section, the reader should understand how to fit a model to participant behavior, how to perform a number of diagnostic checks to ensure the validity of parameter estimates, and how to use parameter estimates within group-level Bayesian models.
As stated above, empirical applications of active inference require fitting a task model to participant behavior. When fitting a model to behavior, one would like to find the parameters that maximize the posterior probability of a model given that behavior, p(model|participantbehavior). Assuming a flat prior belief over models, this posterior is proportional to the likelihood term, p(participantbehavior|model). Thus, many fitting approaches (estimation algorithms) try to find the parameters that maximize this likelihood — referred to as maximum likelihood estimation (MLE). In some cases, one might also have reason to expect that certain models are more likely than others a priori. In this case, one can also incorporate an informative prior belief over models, p(model).11 Regardless of the specific approach, the goal of any fitting procedure is to find the set of parameters that would best reproduce/predict the actual behavior of a participant (i.e., with the highest probability) when running simulations using a model (while in some cases also incorporating any prior knowledge one might have).
To do this, one needs to feed the trial-by-trial observations made by participants (e.g., cues, wins/losses, etc.) into the model and look at the actions predicted by the model (i.e., posterior probabilities over actions). One can then compare these predictions to the actions a participant actually chose. Under some sets of parameter values (e.g., prior expectations, precisions, etc.), the model’s predictions may not match behavior well (i.e., the probability of a participant’s actions under the model may be low). However, by searching through different possible combinations of parameter values, the best combination can be found for a given participant. Note, however, that this will only be the best combination possible for that model. This does not mean that the model has high explanatory power (e.g., the best parameter combination for one model might lead to an average action probability of 0.4, while another model might reach 0.7, etc.). This is why it is also important to compare the explanatory power of different models.
Throughout this section, we encourage the reader to follow along in the companion MATLAB script (Step_by_Step_AI_ Guide.m). This can be found in the supplementary code files, as well as at: https://github.com/rssmith33/Active-Inference-Tutorial-Scripts. At the top of this script, if you set Sim = 4 it will perform parameter estimation on a single set of simulated behavioral data from the explore–exploit task model (see the top panels and bottom-left panel of Fig. 17 for example outputs). If you set Sim = 5 it will simulate behavioral data from two models (one with two parameters and one with three parameters; see below) for a few synthetic participants and then perform model comparison. It will also assess how well the estimated parameter values match with the true parameter values used to generate the simulated task behavior (i.e., it will output correlation matrices and associated p-values; bottom-middle panel of Fig. 17). This is important to check (as described below in relation to parameter recoverability) to ensure that parameters can be estimated reliably (this is sometimes referred to as model identifiability). All the code for performing these steps is included and described in the script comments. All examples here will be based on the explore–exploit task model, when simulating behavior during the case of reversal learning used above (see Fig. 11). We will estimate the alpha parameter (α) encoding action precision (i.e., inverse temperature) as well as a risk-seeking parameter (described below). For model comparison, we will also estimate learning rate (η) in a second model.
Fig. 17.
Illustration of different steps in model fitting, model checking, and subsequent analysis methods. Top Left: Single-subject parameter estimates generated by the scripts provided with this tutorial. This example includes estimation of two parameters: alpha (action precision) and RS (risk-seeking). The left subplot shows the trajectory of parameter values as fitting progressed (from red to blue). The middle subplot shows how evidence for the model (given participant data) increased during estimation (via gradient descent on free energy; here shown as ascent on negative free energy). The right subplot depicts how parameters moved from estimation priors to posterior values (pink lines indicate 95% Bayesian confidence intervals). Note that, for clarity, prior values for all parameters in these plots are given a common reference value of 0, such that posterior values are shown as deviations from those priors. For example, the prior values for alpha and RS were 16 and 5, respectively. The plots therefore show that posteriors for alpha deviated to a value below 16 and that those for RS deviated to a value above 5. Top Right: Prior and posterior mean estimates for alpha and RS for a single simulated participant. This shows how posterior estimates for both parameters move toward the true parameter values. Bottom Left: Posterior variances and co-variances for parameters at the single-subject level, here showing that both variances and co-variances were low. These results can be reproduced by running the Sim = 4 option in the supplementary Step_by_Step_AI_Guide.m code. Bottom Middle: Example of a recoverability analysis for alpha in 6 simulated participants. This plot shows the (in this case strong) correlation between alpha values estimated from simulated behavior in each participant and the true parameter values used to generate that simulated behavior. Bottom Right: Output of group-level parametric empirical Bayes’ (PEB) analyses (scripts provided in supplementary code). This example included estimation of three parameters for six simulated participants, where parameters 1, 2, and 3 correspond to alpha (action precision), RS (risk-seeking), and eta (learning rate), respectively. The top row (left to right) shows the evidence (for each parameter) for differences from 0, differences between two groups (simulated as having different RS values, i.e., parameter 2), and relationships to (arbitrarily specified) age values in models assuming effects for all parameters. The bottom row shows analogous results for reduced models that have greater evidence than the full models (here, no group difference in alpha [parameter 1] or eta [parameter 3], and some effects of age on alpha and eta). Recall that these results are based on a very small sample of only 6 simulated participants, which will generally be unreliable. They are for illustration only, and the identified relationships with the arbitrary ‘age’ values should not be taken seriously. Note that we have omitted some additional plots that are also generated. This is because these are the automatic output of scripts originally designed for dynamic causal modeling in neuroimaging, and not all of them are useful in the present context. These results can be reproduced by running the Sim = 5 option in the supplementary Step_by_Step_AI_Guide.m code, while also setting the option PEB = 1 (note that, because outcomes are sampled from probability distributions, results will not be identical each time).
As mentioned above, finding the optimal combination of parameter values to account for a participant’s behavior requires a model fitting (parameter estimation) procedure. There are many different procedures (estimation algorithms) that are available, each with their strengths and limitations. The simplest approach is called a grid search, where each possible combination of parameter values (within some specified range of values) is tried one-by-one, and then the one that best reproduces (e.g., maximizes the likelihood of) behavior is chosen. However, this approach is limited to fairly simple models with a small number of parameters, and it can also lead to overfitting (i.e., finding parameter values that reproduce random aspects of behavior). In more complex models with a larger number of parameters, exhaustive search of the parameter space becomes intractable. Thus, a number of other algorithms have been developed. In this section, we will focus on an estimation technique called variational Bayes (i.e., a specific variant of this approach called variational Laplace; (Friston, Mattout, Trujillo-Barreto, Ashburner, & Penny, 2007)), which is based on exactly the same free energy minimization ideas described above.12 Namely, variational Bayes starts with a prior distribution over parameter values, which acts as the starting value of an approximate posterior (or ‘proposal’) distribution. Via gradient descent on variational free energy, this approximate posterior is adjusted until convergence to a stable minimum value, providing posterior estimates of model parameter values. However, the detailed mathematics underlying this technique are quite advanced and beyond the scope of this tutorial. Therefore, our focus here is on practical applications, with the aim of equipping readers without advanced mathematical training to still be able to make use of such approaches appropriately. We will now go over a set of concrete steps that can be taken to use variational Bayes for model fitting. As we are focusing on the explore–exploit task model, it may help to glance back at the model/task description in earlier sections before moving on.
The first step is to place trial-by trial-observations and participant actions into an mdp structure. In the explore–exploit task model, for each trial this would mean specifying the observations (mdp.o) a participant made for the three outcome modalities at the three time points in each trial, and then specifying the two actions taken (mdp.u). For example, if on trial #1 the participant chose to take the hint and then chose the left machine, this would be:
Here, time goes from left to right. For mdp.u, actions for each state factor correspond to each row from top to bottom. For mdp.o, time also goes from left to right and rows correspond to outcome modalities. The parenthetical ‘(1)’ for the mdp denotes trial #1. Here, for mdp.u, remember there is only one ‘action’ for the first state factor (i.e., the participant does not have control of which machine will most likely lead to a win). So, the top row is simply a 1 for both time points. For the second state factor (choice), the participant first chose the hint (action #2) and then chose the left machine (action #3). For mdp.o, the first row indicates ‘null’ (observation 1), ‘left hint’ (observation #2), and then back to ‘null’. The second row indicates two ‘null’ observations followed by a ‘win’ (observation #3). The last row simply corresponds to the choices (‘start’, ‘take hint’, ‘choose left’). Observations and behavior for each trial need to be inserted in this same way (e.g., mdp(2), mdp(3), etc. until the final trial number).
The second step is to choose the model parameters you want to estimate and which you want to hold fixed. Ultimately, one may want to try estimating different models and/or different numbers of parameters and then compare them to find which best accounts for behavior, as we describe further below. In this example, we will first consider estimating two parameters: learning rate (η) and ‘risk-seeking’ (RS). The latter corresponds to how strong the preference is to win money in the C matrix:
As reviewed above, learning rate scales the size of the ‘count’ that is added to the (Dirichlet) concentration parameters after a new observation. The RS parameter controls the explore–exploit trade-off. In the simulations shown in Figs. 10 and 11, we saw that, as RS values go up, the probability that a participant will take the hint goes down. That is, they will tend to ‘risk it’ and simply guess left or right in hopes of winning the larger amount of money.
When using variational Bayes, the next step is to choose a set of estimation priors. These are not the prior beliefs of a participant, but the initial parameter values that are evaluated during model fitting. Estimation priors include both a prior mean and a prior variance, each of which can be set within the supplementary ‘Estimate_parameters.m’ script, as described further below. A simple way of thinking about the variational Bayes algorithm is that it starts from the chosen estimation priors (i.e., the prior means) and slowly moves away from them to find the combination of parameter values that best explains a participant’s behavior, while also balancing the associated cost of increased model complexity that is incurred when the parameters move too far away from the prior means. More specifically, variational Bayes is accomplished by performing a gradient descent on VFE (i.e., the same process active inference models use to accomplish perception, learning, and action selection). In this case, the gradient descent starts with the prior means, and then evaluates the log-probability of a participant’s actions (e.g., sequence of choices). This log-probability is evaluated using the posterior beliefs about action that the subject would have had, given the priors in question and the outcomes they observed. The scheme then evaluates neighboring parameter values and continues in the direction of increasing likelihood, until a combination is found with no neighboring values that improve the fit.
However, because we seek a VFE minimum, which includes both complexity and accuracy terms, the algorithm will not simply maximize model accuracy. It will also try to minimize how far parameter estimates move away from the mean values of the estimation priors. In other words, parameter estimates for a given participant will represent values that accurately reproduce that participant’s behavior while moving as little as possible from the prior values chosen by the experimenter. Importantly, complexity minimization also depends on the prior variances that are chosen. Setting a small prior variance will lend strong weight to complexity minimization; in contrast, setting a large variance for estimation priors will lead a stronger weight to be placed on maximizing accuracy (but with a greater chance for overfitting). How to set the prior variance can be a crucial choice in variational Bayes, because posterior parameter estimates can in some cases be sensitive to the choice of prior means. When available, prior means can be based on previous studies. However, if previous literature is not available to draw from, one might consider specifying a large prior variance to reflect this uncertainty (in which case the complexity cost will play little-to-no role). This issue of setting appropriate estimation priors, and diagnosing when they may be inappropriate, is one of several important model checking procedures that should be taken to confirm the validity of parameter estimates, which we return to below.
Before moving forward, however, it is important to highlight a specific subtlety in the above scheme. Namely, we are using variational Bayes to estimate the parameters that underwrite variational inference within the brain of a study participant. In other words, there are two generative models: first, a subjective model, which is the POMDP we assume the participant is using to plan their responses. Second, there is an objective model that the experimenter specifies in terms of the estimation priors on parameters of the subjective model. A key conceptual point here is that the parameters of the subjective model can always be interpreted as priors; either explicitly or implicitly in terms of the structure or form of the subjective model. This is important because of something called the complete class theorem (L. D. (Brown, 1981; Wald, 1947). This theorem says that for any pair of reward functions (i.e., preferences) and choice behavior, there exist some prior beliefs that render the choices Bayes optimal. This means that any behavior can be described, under ideal Bayesian assumptions, given the right set of prior beliefs. It is these prior beliefs that provide a theoretically complete characterization of any given participant, in any given experimental paradigm.
After choosing a final set of estimation priors, and obtaining parameter estimates for each participant, it is also important to confirm what is called parameter recoverability (also sometimes referred to as testing whether the model is identifiable or invertible). What this means is that you need to be sure that, if simulated behavior were generated from a model with a given set of parameter values, that the estimation algorithm would provide reasonably precise estimates that approach the parameter values that generated the behavior. For example, assume a given participant’s posterior parameter estimates were RS = 3.2 and α = 3.7 (e.g., using estimation priors of RS = 4 and α = 2). Assessing recoverability would require: 1) putting the values of these posterior parameter estimates into the task model, 2) generating simulated behavior using those values, 3) feeding that simulated behavior into the fitting algorithm, and 4) examining how similar the resulting parameter estimates are to the parameter values that you used to generate the simulated behavior in the first place. If the algorithm returns estimates that move in the right direction from the estimation priors and approach the values that generated the data (e.g., RS = 3.3 and α = 3.5), then we can be more confident that the participant’s estimates are capturing something meaningful and reliable about their decision process. The plot on the top-right of Fig. 17 illustrates a case in which posterior estimates move from estimation priors toward the true values (this plot can be reproduced by setting Sim = 4 when you run the Step_by_Step_AI_Guide.m code). If the fitting algorithm instead returned estimates that were farther away and moved in the wrong direction from the estimation priors (e.g., RS = 4.9 and α = 1.3), this would suggest that the parameter estimates for that participant may not be reliable (e.g., a different combination of parameter values might reproduce their behavior equally well).
Thus, before interpreting parameter estimates in real participants – and using them in subsequent group analyses – it is important to confirm in simulated data that the estimates provided by the fitting algorithm match well with (e.g., are significantly correlated with) the parameters used to generate the simulated behavior in the first place. This should be checked using the combinations of parameter estimates found when fitting a model to the true participant data in a study. For example, if you had three participants with estimated parameter values of [RS,α] = [2.8, 4.1], [3.5, 6.2], [3.8, 8.1], then these three combinations could be used to generate simulated behavior, and this simulated behavior could then be fed into the fitting algorithm to see if it returned results similar to the generative values (e.g., if estimated values were highly correlated with those used to generate the simulated behavior).
It is important to keep in mind, however, that the products of Bayesian model inversion (here, parameter estimation) will, generally speaking, never be the same as the parameters used to generate data. This is because there is usually a simpler way of generating any given set of data, which the model inversion will identify. In other words, in a certain sense there are no ‘true’ parameters — only the best explanation in the sense of Occam’s principle.
If the Sim variable is set to Sim = 5 when you run the Step_by_Step_AI_Guide.m code, simulated behavior will be generated under several parameter values for the explore–exploit task model and it will run correlations between the estimated parameter values and the parameter values that actually generated the simulated behavior. In a representative example simulation, recoverability appeared high for RS (r = .95) and α (r = .94). In a model that also included learning rate (η), this parameter appeared recoverable as well (r = .75). The bottom-middle panel of Fig. 17 shows a scatterplot illustrating the correlation between generative and estimated parameter values for α, which the above-mentioned code will reproduce.
Before assessing parameter recoverability (i.e., model identifiability), however, it is important to first understand more about the concrete steps for fitting. As mentioned above, if you set Sim = 4 in the Step_by_Step_AI_Guide.m script, it will generate simulated explore–exploit task behavior for a single participant. It will feed this simulated behavior into a ‘DCM’ (for Dynamic Causal Modeling) data structure in the appropriate format for model fitting. It then calls the supplementary Estimate_parameters.m script we have provided, which takes the DCM structure as input, runs the variational Bayes routine in SPM12 (spm_nlsi_Newton.m), and calculates the log-likelihood — that is, the sum of the log-probabilities of chosen actions under the model. In our script, the DCM structure includes the following:
Within the Estimate_parameters.m file, you will see locations (starting on line 47) where the script searches for the to-be-fit parameter names. Here, you can enter the estimation priors (means and variances) for each parameter. Note that some parameters (e.g., learning rate) need to be between 0–1. For this reason, they are here transformed into logit-space so that the estimation routine does not assess values outside of that range. Similarly, parameters that can only take on positive values can be transformed into log-space to preclude negative values. For illustration, we have assumed that RS can only take positive values and is therefore log-transformed. Farther down in the script (starting on line 133), these values are re-transformed out of logit- or log-space when they are fed into the model and output as final estimates. Starting on line 188, the log-likelihood function loops through each trial, takes the probability of a participant’s actions in the model (stored in MDP.P; see Table 3), log-transforms it, and then sums the log-probabilities. The closer this value is to 0, the better the model fits the behavior.
When you run the Step_by_Step_AI_Guide.m script (which calls the others automatically) with Sim = 4, a display will also appear (as in the top-left panel of Fig. 17) that shows how the free energy changes over iterations. Note that these values increase because they are negative and will approach 0 as model fit improves. If these values steadily increase, this suggests that fitting is converging to a reliable estimate. If they instead fluctuate up and down inconsistently, this could suggest one or more problems that need to be addressed before model estimates are considered reliable (e.g., poor choice of estimation priors, problems with parameter values remaining in valid ranges, estimates getting stuck in locally [but not globally] optimal values). A failure to converge is usually read as a useful diagnostic that the estimation priors are somehow mis-specified. In other words, if model inversion does not converge gracefully within a few tens of iterations, you may want to think about whether your priors are appropriate — or whether you are trying to fit too many parameters to rather sparse data. This type of model checking is crucial to ensure that parameter estimates are valid and informative.
The estimates resulting from initially chosen prior means can also sometimes offer guidance in this regard. For example, if estimates for all participants tend to move far from the chosen prior means in the same direction, this could suggest that these estimation priors were poorly chosen. If you initially set a prior mean of RS = 1, for example, and then notice that estimates for all participants tend to move up to between 3 and 5, this could indicate that a prior of RS = 4 would be more appropriate (as it appears to be a better prior for the group as a whole) and might help prevent over-weighted complexity costs that could hinder identification of individual differences. It is worth noting that letting data guide the way one chooses estimation priors in this way could be viewed as suspect under some interpretations of Bayesian statistics. However, it is also possible to view estimation priors in variational Bayes as simply being starting values for estimation that can be optimized (and may need to be in cases where some starting values will lead gradient descent to get stuck in suboptimal local minima). In some cases it may be useful to try multiple starting values and then examine whether posterior estimates tend to converge toward similar results. Note also that, although we do not go into detail here, there are also hierarchical ‘empirical Bayes’ methods that can be used to rigorously estimate group-level priors and then re-estimate individual-level parameters based on those group-level priors (Carlin & Louis, 1998; Friston et al., 2016b).
After convergence to the best estimates, the output DCM structure will contain additional fields. The following are relevant to our current use:
DCM.Ep: posterior mean estimates (i.e., expectations) for each parameter.
DCM.Cp: posterior covariance matrix (with posterior parameter variances on the diagonal).
DCM.F: the final free energy value of the best fit model.
The free energy values for each participant will later be used for model comparison. The covariance values (i.e., the off-diagonals in DCM.Cp) should be checked to make sure they are not too high (e.g., > .8), or it would suggest they were not independently estimated and that each estimate may not carry unique/reliable information. This is the same kind of check you would apply to an experimental design — to ensure explanatory variables are orthogonal and the model parameters can be estimated efficiently. The bottom-left panel of Fig. 17 plots an example co-variance matrix (which will be reproduced by the accompanying tutorial scripts; Sim = 4, as with previous plots).
When testing hypotheses about which of several models best explains your data, one must fit each model and then compare how well they each fit behavior. For Bayesian model comparison using variational Bayes, this means comparing the free energies at the group level. One common approach is to use a random effects model, which can be done using the spm_BMS.m function (available within SPM12). This function takes as input a matrix containing the free energies for each participant for each model (one column per model). For the details of this implementation of Bayesian model selection, see (Rigoux, Stephan, Friston, & Daunizeau, 2014) and Stephan, Penny, Daunizeau, Moran, and Friston (2009). An important output of this function is the protected exceedance probability (pxp) of each model. In this case, the model with the highest pxp has the most evidence. Often there will be a clear winner, with pxp = 1 for a single model and 0s for the others. In cases where there is no clear winner (e.g., pxp = [.48 .52]), this will be important to note, as it may reflect insufficient evidence for a ‘best’ model. If parameters in both models are recoverable, one may wish to consider the parameters of each model in further between-subjects analyses (e.g., parameters in one model may have higher explanatory value for specific theoretical questions). Another useful model-checking step that could be informative in this case is to generate simulated datasets from models that include different numbers of parameters (e.g., from models that do vs. do not include a learning rate) and then confirm that the correct model is identified by model comparison (e.g., if the data were generated by a model with only the RS and α parameters, and then several possible models were fit to this data, one would confirm that the model including only these two parameters was identified as having the highest pxp). As mentioned above, setting Sim = 5 in the Step_by_Step_AI_Guide.m script will generate/estimate behavior for a few simulated participants for models with and without the learning rate parameter. It will then do Bayesian model comparison (this will take several minutes). This will output the pxp for each model, as well as the model probabilities and a few other diagnostic outputs we will not cover in detail here; for further information, see Rigoux et al. (2014) and Stephan et al. (2009) as well as the documentation within the spm_BMS.m function. As one example, you could input the free energies for the 2-parameter (F_2_params) and 3-parameter (F_3_params) models as follows:
If the output were pxp = [1 0], this would mean that the probability is equal to 1 that the 2-parameter model is a better fit than the 3-parameter model. In a representative example simulation, the pxp = [.37 .63], weakly favoring the 3-parameter model. After identifying the best model, it is also important to make sure it captures the data well, which could be done by (for example) calculating the average probability of participants’ choices under the winning model or the percentage of trials where participants’ choices were assigned the highest probability. If the winning model still doesn’t capture the data well, this suggests one may need to consider other possible models.
Once (recoverable) parameters for a winning model have been estimated, and we have confirmed that this model captures the data well, one simple approach for group-level analysis would be to analyze the posterior parameter means across participants using standard frequentist analyses. However, one advantage of using variational Bayes is that it also provides information about posterior parameter variances for each individual (i.e., as opposed to only the posterior means). This allows for between-subject Bayesian analyses that take the variances into account. One approach that takes the output DCM structures for each participant as input is parametric empirical Bayes (PEB), described in detail in Friston et al. (2016b) and Zeidman et al. (2019). PEB uses a general linear model and effectively down-weights the contribution of individual subject parameter estimates when those estimates have larger posterior variances (i.e., those with greater uncertainty). PEB can be run using the spm_dcm_peb.m and spm_dcm_peb_bmc.m functions. These functions estimate and compare group models, respectively. In short, they allow a test of whether there is evidence for a model that does include group differences or whether there is more evidence for a model that does not include those differences. When including covariates (e.g., age), it also allows comparing the evidence for or against a relationship between parameters and those covariates. The spm_dcm_peb_review.m function can be used to inspect the results returned from the PEB scripts.
Example code to implement such empirical Bayesian (random effects) analyses is also included in the accompanying MATLAB code Step_by_Step_AI_Guide.m. The code shows examples about (with commented descriptions of) how to set up inputs to PEB. In our example, the code sets several parameters to default values (for more information on these, see Friston et al., 2016b; Zeidman et al., 2019) and then inputs a matrix (M.X) for a general linear model, with a column for the mean, a column separating participants into two groups (here, with low vs. high learning rates), and a column with randomly generated participant age values). This will allow us to assess the evidence for models including effects of group and/or age (and the strength of these effects).
To run PEB, set Sim = 5 and set PEB = 1. This may take some time, as it will first generate and estimate parameters for several (six) simulated participants before feeding them into the PEB scripts (note that the script will save the outputs of Sim = 5 so that this does not need to be repeated each time you want to practice using PEB). In beginning of Section 10 of the script, you can also specify whether you want to use PEB on the 2-parameter or 3-parameter model. When complete, some output plots and the PEB results viewer window will appear. Example outputs are shown in the bottom-right panel of Fig. 17). The figure legend describes how to interpret these outputs.
To review, we have covered several steps for model fitting, model checking, and subsequent analysis. These steps are as follows:
Select estimation priors, including prior means and variances.
- Run variational Bayes on participant data for each model under consideration.
- Check that gradient descent shows smooth convergence toward a free energy minimum (Fig. 17, top-left panel). Note that this is displayed as maximization because negative free energies are used.
Perform Bayesian model comparison to identify the best model, and check that this model captures the data well (e.g., by evaluating the percentage of trials in which it assigns the highest probability to participants’ choices).
- Generate simulated behavior in the best model using the parameter values estimated in your participants, and then run variational Bayes to estimate parameters using this simulated behavior.
- Confirm that posterior estimates approach the parameter values used to generate the simulated behavior (Fig. 17, top-right panel).
- Confirm that posterior co-variances are not high (Fig. 17, bottom-left panel).
- Confirm that generative and estimated parameter values are strongly correlated (Fig. 17, bottom-middle panel).
Use PEB to run regression or group comparison analyses on posterior means and variances at the between-subjects level (Fig. 17, bottom-right panel).
8. Concluding remarks
This concludes the tutorial. For readers seeking more hands-on practice, we have also provided pencil-and-paper exercises in Appendix B (as well as solutions to check your work; see Pencil_and_paper_exercise_solutions.m code). In our experience, doing a few practice problems of this sort, and working with the code, are the best way to gain useful intuitions about the dynamics of these models and how they can be tailored for specific studies. We note that, while we have strived to be comprehensive, there are many new directions in active inference (and associated functionality in the standard SPM routines) incorporating, for example, multi-agent interactions, deep parametric models (e.g., 2nd-order parameters on habits, preferences, precisions, etc.), and mixed models that link POMDPs to continuous motor control processes, among others (e.g., see (Friston et al., 2017c; Hesp et al., 2020)). We hope that working through the materials provided here will offer a ‘launching point’ that will provide the reader with a sufficient foundation to independently extend their work with advances in the field as they emerge.
Supplementary Material
Acknowledgments
The authors would like to thank Lance Da Costa, Thomas Parr, Giuseppe Pagnoni, and Conor Heins for offering useful comments and suggestions during preparation of the manuscript. The authors would also like to acknowledge Samuel Taylor for additional suggestions that improved the clarity of the manuscript.
Funding
R. S. is supported by the William K. Warren Foundation and the National Institute of General Medical Sciences (P20GM121312). C. W. is supported by the University of Cambridge Harding Distinguished Postgraduate Scholars Programme.
Appendix A. Additional mathematical details
Fig. A.1.
Venn diagram depiction of a joint probability distribution and how it is used to infer a conditional probability (such as a posterior probability in Bayesian inference). The total space within the rectangle represents all possible situations. The solid circle corresponds to the subset of all possible situations where a is true. The dashed circle corresponds to the subset of all possible situations where b is true. The overall probability of a relates to how large the circle is for a relative to the total area within the surrounding rectangle (and likewise for the overall probability of b). The blue area where the circles overlap represents the situations where both a and b are true. To infer p(b|a), we simply remove the circle for b (since we are conditioning on a being true). Then we evaluate how much of the circle for a is occupied by the blue region where both a and b are true. If the blue region covers the majority of the circle for a, then p(b|a) will be large, whereas if the blue region only covers a small part of the circle for a, then p(b|a) will be small. Thus, p(b|a) can be thought of as the proportion of situations where a is true in which b is also true.
Introduction to the rules of probability
There are two basic rules in the mathematics of probability that will underpin much of the material covered in this tutorial. The first rule is called the sum rule:
| (A.1) |
Here we use the tilde (∼) symbol to indicate negation. So this states that the probability of a being true, p (a), is equal to the joint probability that both a and b are true, p (a, b), plus the joint probability that a is true and b is false, p (a, ∼ b). If the variable b can take on several values (i.e., more than just true and false), then calculating p (a) requires summing p (a, b) for each possible value of b. Note that p (a) is often referred to as a marginal probability in this context when calculated by summing over all the values of another variable in a joint probability distribution. In the context of Bayesian inference, it is also sometimes referred to as a prior probability when used to describe beliefs about a before making a new observation (described further below).
The second rule is called the product rule:
| (A.2) |
This says that the joint probability of a and b is equivalent to the conditional probability of a given b, p (a|b), multiplied by the probability of b, p (b). Here, the joint probably indicates the likelihood that a and b will occur together (e.g., that it is cloudy and that it is raining). The conditional probability indicates how likely a is if we are told b (e.g., how likely it is to rain if we know that it is cloudy). The marginal probability, p (b), indicates how likely something is to occur in general (e.g., how often it is cloudy overall).
Symmetrically, the product rule says that p (a, b) is also equivalent to the conditional probability of b given a, p (b|a), multiplied by the marginal probability of a, p (a). We can also use the product rule to do standard algebraic manipulations. For example, we can take and then divide both sides by p (a) to get the conditional probability of b given a, p (b|a):
| (A.3) |
We can then derive Bayes’ theorem by simply replacing p (a, b) in Eq. (A.3) with p(a|b)p(b), where this equivalence is shown in Eq. (A.2):
| (A.4) |
In scientific practice, we can represent observational data with the variable a and then use the variable b to represent some theory. In this case, p(b) is referred to as the prior probability of the theory being true before observing the data. Eq. (A.4) then means that if we get some new data a, then we can use Bayes’ theorem to infer whether it increases the probability that some theory b is true. In other words, we can infer whether the posterior probability of the theory given the data, p (b|a), is higher than the prior probability that the theory was true before observing the new data, p(b). This just requires that we know how strongly the theory predicts that new piece of data — typically referred to as the likelihood that the data would be observed if the theory were true, p (a|b).
The use of Venn diagrams is often helpful for gaining intuitions about probabilistic inference. We illustrate this in Fig. A.1 by showing one circle on the left that contains all situations in which a is true (solid outline) and another circle on the right containing all situations in which b is true (dashed outline). The total area within the surrounding rectangle corresponds to all possible situations. Thus, the area outside of both circles corresponds to the situations where neither a or b is true. The set of situations where both a and b are true corresponds to the shaded blue area in the middle where the circles overlap. The overall probability of a corresponds to how large the circle is for a relative to the total area within the surrounding rectangle (and likewise for the overall probability of b). When we infer the posterior probability, p (b|a), we first simply remove the circle for b (i.e., imagine erasing the dashed outline and the separate b variable, since we know from our new data that we are in a situation where a is true). However, we retain the blue shaded region within the circle for a corresponding to when a and b are both true. Then we look at how large the proportion of the circle is for a that corresponds to when b is also true (i.e., how large the blue shaded area is). If this area represents a large portion of the circle for a, then this means the posterior probability, p (b|a), will be high. This proportion is what is captured by (remember this is equivalent to in Bayes’ theorem; see Eq. (A.2)).
A further fact to keep in mind about the rules of probability is that they do not change if other constants are included. For example, the sum rule remains the same if all terms are conditioned
Table A.1.
Prior and conditional probabilities.
| cloudy (p = .7) | not cloudy (p = .3) | |
|---|---|---|
| raining | p (raining|cloudy) = .6 | p (raining|not cloudy) = .01 |
| not raining | p (not raining|cloudy) = .4 | p (not raining|not cloudy) = .99 |
Table A.2.
Joint probabilities.
| cloudy | not cloudy | Marginal probabilities | |
|---|---|---|---|
| raining | p (raining, cloudy) = .7 × .6 = .42 | p (raining, not cloudy) = .3 × .01 = .003 | p (raining) = .42 + .003 = .423 |
| not raining | p (not raining, cloudy) = .7 × .4 = .28 | p (not raining, not cloudy) = .3 × .99 = .297 | p (not raining) = .28 + .297 = .577 |
Table A.3.
Posterior probabilities.
| cloudy | not cloudy | |
|---|---|---|
| raining | p (cloudy|raining) | p (not cloudy|raining) |
| not raining | p (cloudy|not raining) | p (not cloudy|not raining) |
on some other variable c:
| (A.5) |
This also holds for the product rule:
| (A.6) |
As a numerical illustration of these rules, we will let a = “raining” and b = “cloudy” and use Table A.1 to indicate the following prior and conditional probabilities (i.e., the probability of it raining if it is cloudy):
Note that while the values of each column sum to 1, the values of the rows do not, which is why the likelihood term in Bayes’ theorem is not technically considered a proper probability distribution (i.e., if one wanted to treat it as such, the rows would need to be normalized so that they kept the same proportions but did sum to 1).
To get the joint probabilities, we simply multiply each prior by each conditional (i.e., the product rule):
In this case, all the joint probabilities sum to 1, as these four cells (i.e., in the first and second columns) describe all possible outcomes. As shown in the third column, summing across each row gives us the marginal probabilities for each possible value of p (a) (i.e., raining vs. not raining), based on the sum rule. These are the values that end up ‘in the margin’ (which is why they are called marginal probabilities) when summing over the probabilities under each possible value of b (i.e., cloudy vs. not cloudy).
We can then find the posterior probabilities (Table A.3) by dividing the joint probabilities (i.e., the value in each cell within the first and second columns of Table A.2) by the marginal probabilities (i.e., the value of the associated row within the third column in Table A.2):
This tells us, for example, that if you look out your window and see that it is raining, then you can confidently infer that it is cloudy.
A final rule that will be useful to know for this tutorial pertains to logarithmic (log) transformations. Specifically, log transformations of probabilities allow multiplication and division to be expressed in terms of addition and subtraction (respectively). For example, using the natural logarithm (ln), the product rule can be expressed as follows:
| (A.7) |
| (A.8) |
Performing these transformations is often beneficial in practice because it simplifies the necessary computations and can prevent the need to work with very small probabilities (which can happen when many probabilities must be multiplied together in complex real-world problems). Log-probabilities can also be easily converted back into standard probabilities by exponentiating them. That is, , where e ≈ 2.71828 (often called Euler’s number).
This concludes our brief introduction to the rules of probability. Many derivations in this tutorial work primarily based on using these rules in combination with algebraic manipulation.
Introduction to Variational Inference
The typical goal of Bayesian inference is to find the posterior distribution – that is, to infer how the states of the world (s) have changed based on new observations (o). However, this requires one to calculate the marginal likelihood p (o), which often involves intractable sums (or integrals in the case of states/observations with continuous values). The key idea behind variational inference is to convert this inference problem into an optimization problem. To do so, instead of evaluating the marginal likelihood, we optimize an auxiliary distribution q (s) (sometimes called the recognition distribution, or variational posterior) to approximate the true posterior . This is done by using the KL divergence as a measure of the relative difference (in the information-theoretic unit nats – the natural log equivalent of bits) between the two distributions:
| (A.9) |
The KL divergence sums over the states of the two distributions so the output is always greater than or equal to zero. When the recognition distribution and the true posterior match, the KL divergence is zero (i.e., when , . Although, as we do not know the true posterior distribution, this sum also cannot be evaluated. Crucially, however, working from the definition of conditional probabilities, , we can introduce a quantity that can be evaluated directly (noting that ):
| (A.10) |
| (L2) |
| (L3) |
Eq. (A.10) substitutes the alternate definition of the posterior distribution into Eq. (A.9). With some minor rearrangement, we see that the KL divergence between our approximate posterior and the true posterior is now equal to the KL divergence between our approximate posterior and p (o, s) – which can be viewed as a generative model of how states of the world generate observations – plus the log probability of observations (i.e., the log of the marginal likelihood). This is the critical move. Because we are free to specify the generative model, and q (s) is the variable we seek to optimize (and can thus be initially set to an arbitrary value; see below), we have access to both the quantities we need to compute the KL divergence. We now introduce a new quantity, variational free energy (VFE), denoted F, and define it in terms of this KL divergence: . The value of the approximate posterior q (s) that minimizes VFE will then be the q (s) that best approximates the true posterior distribution.
| (A.11) |
Equation A.11 rewrites line 3 of Eq. (A.10), but substitutes in VFE as an explicit variable. From this vantage point, we see that when VFE is minimized the KL divergence between the approximate posterior and the true posterior is also minimized, meaning that the approximate posterior is close to the true posterior. Hence, minimizing VFE allows a tractable means of performing approximate Bayesian inference. One way to do this is using gradient descent. That is, one can start q(s) at an arbitrary value and then test neighboring values to find the one that reduces VFE most. Then one can move to that value, search neighboring values again (etc.), and repeat this process until a value for q(s) is found for which no neighboring values further reduce VFE.
Expected Free Energy
Active inference reverses the usual logic of action selection. Instead of asking ‘which sequence of actions will bring about my preferred outcomes?’, it formally asks, ‘given the assumption that I will achieve my preferred outcomes, what course of action am I most likely to pursue?’ (Millidge, Tschantz, & Buckley, 2021). Within active inference, the answer to this question is the policy (π; i.e., action sequence) that best minimizes a quantity termed expected free energy (EFE). Here we show the most common decompositions of EFE that appear in the active inference literature and describe the workings and the intuition behind each decomposition. EFE is defined in terms of the expected difference between the log of the generative model p (o, s|π) and the log of the approximate posterior given a choice of policy q (s|π). Eq. (A.12) shows the decomposition of the EFE of each policy (Gπ) into terms often referred to as epistemic and pragmatic value (intuitively, expected information gain and reward probability under each policy, respectively). It also shows another common decomposition into terms referred to as risk and ambiguity (similarly corresponding to expected reward and uncertainty minimization under each policy). Note below that, because EFE is calculated with respect to expected outcomes that (by definition) have not yet occurred, observations enter the expectation operator Eq as random variables.
| (A.12) |
| (L2) |
| (L3) |
| (L4) |
| (L5) |
| (L6) |
The second line uses the product rule of probability, , to rearrange EFE into the epistemic and pragmatic value terms described in the main text. In the third line, the dependence on policies is dropped from the pragmatic value term so that it can be used to encode preferences (i.e., this is a key move in active inference). Note that, in most papers on active inference, this is simply written as ; however, to clearly distinguish this from the ln p (o) term within VFE (i.e., where o is an observed variable), we write the pragmatic value term here as explicitly conditioned on a preference variable C (Parr et al., 2022). Line 3 also replaces the true posterior (ln p (s|o,π)) with an approximate posterior (ln q (s|o,π)). Line 4 offers a clearer intuition for epistemic value by flipping the terms inside the first expectation so that it becomes prefixed with a negative sign (i.e., . Because the epistemic value term is now subtracted from the total, it is clear that, to minimize EFE overall, an agent must maximize the value of this term by selecting policies that take it into states that maximize the difference between ln q (s|o,π) and ln q (s|π). In other words, the agent is driven to seek out observations that reduce uncertainty about hidden states (i.e., maximize the change from prior to posterior beliefs after a new observation). For example, if you are in a dark room, then the mapping between hidden states and observations is entirely ambiguous. The best way to minimize uncertainty is to turn a light on.
Moving from the expression in line 3, line 5 uses Bayes rule in the denominator to express the same epistemic imperative, but with the conditional probabilities flipped. Note here that, although algebraically , in this case and refer to the same distribution. The epistemic value terms in lines 3, 4, and 5 are formally equivalent since they each express the mutual information between hidden states and observations. First noting that H[p (x)] denotes the entropy of a distribution p (x), where , mutual information can be written as . This quantity I (x, y) is symmetric and scores the reduction in uncertainty (entropy) about the value of a variable x afforded by knowledge of another variable y. If two variables are independent, mutual information is zero. Finally, line 6 expresses EFE in the form shown in the main text as risk plus ambiguity. The first term, risk, is the KL divergence between the observations expected under a policy and prior preferences. The second term, ambiguity, scores the uncertainty in the likelihood mapping between states and observations. Minimizing EFE thus requires agents to select policies that minimize the difference between expected observations and preferred observations (e.g., seeking warmth when it is cold, maximizing reward, etc.) and to take actions that reduce uncertainty (i.e., ambiguity) about the mapping between hidden states and observations (i.e., another way of expressing the drive to maximize information gain). The work to move between the fifth and the sixth line is somewhat convoluted, so Eq. (A.13) shows it step by step:
| (A.13) |
| (L2) |
| (L3) |
| (L4) |
| (L5) |
| (L6) |
Line 1 of Eq. (A.13) rewrites line 5 of Eq. (A.12), but makes the summations implied by the expectation operators explicit. Line 2 moves back into the same expectation. Line 3 expresses the expectation in the first term in terms of an approximate joint distribution , and in the second term separates out . In the first term of line 4 we evaluate the summation over states in the joint distribution , leaving the fraction inside the log untouched as it does not depend on states. In the second term of line 4 we drop the dependency on policies since the likelihood mapping is constant across choices of policy. In line 5 this then allows us to express in terms of entropy , and rewrite in terms of the KL divergence (defined in the main text) between prior preferences and observations expected under each policy . Finally, because entropy is a negative quantity, we swap the sign between the two terms, leaving us with the canonical form of EFE as risk plus ambiguity in line 6 – where lower risk indicates a higher probability of preferred outcomes under a policy and lower ambiguity indicates more precise (informative) observations expected under a policy.
It is important to highlight here that generative models in active inference also maintain confidence estimates for the model parameters themselves, via a form of distribution called a Dirichlet distribution that encodes priors over these parameters (see main text for an introduction to this type of distribution). Dirichlet distributions contain what are called concentration parameters, where higher concentration parameter values indicate lower uncertainty in the parameters of each distribution. The above expressions of EFE assume that the concentration parameters are saturated (i.e., that they are maximally precise), and hence that there is no uncertainty. However, when there is uncertainty in parameters (as is the case for real organisms), agents must also learn the values for those parameters via the selection of appropriate policies. This means that parameter uncertainty now enters the equation for EFE. For example, Eq. (A.14) shows the form of EFE when the parameters of the likelihood must be learned. Note that this likelihood is termed the A matrix in active inference models (see Table 1).
| (A.14) |
| (L2) |
| (L3) |
| (L4) |
| (L5) |
| (L6) |
Here . Line 1 shows the form of EFE when A is treated as a random variable. Line 2 breaks the approximate posterior and generative model into separate terms using the product rule of probability . It also uses the mean-field approximation – which assumes that the approximate posterior factorizes into the product of independent marginal distributions – to express the approximate joint distribution as . Line 3 approximates line two, replacing the exact posteriors and with approximate posteriors and . Line 4 takes pragmatic value out of the first expectation term and then (as above) conditions on the preference variable C instead of π; i.e., . Line 5 breaks the first expectation in line 4 into two quantities. The first, is the epistemic value term that we saw in the previous expression of EFE without uncertainty in the parameters. As we now have two types of epistemic value, to distinguish them we call the first salience and the second novelty. Salience scores the reduction of uncertainty about states afforded by observations (driving ‘state exploration’ behavior), while novelty scores the reduction in uncertainty about parameters of the generative model afforded by states and observations (driving ‘parameter exploration’ behavior; see (Schwartenbeck et al., 2019). As with line 4 in Eq. (A.12), line 6 here flips the terms inside the first and second expectation to make the salience and novelty terms negative (i.e., such that maximizing these terms brings their value closer to zero). This makes it clearer why maximizing the difference between prior and posterior beliefs – here about both states and parameters – will minimize EFE. The parameters of the model are the sufficient statistics of Dirichlet distributions, which, in the case of learning A, essentially count the number of times a particular categorical state is inferred when a particular outcome is observed (proportional to the posterior probability over each state). Like the epistemic value term in the previous expression of EFE, to minimize EFE here agents must maximize both salience and novelty by seeking out observations that (1) reduce uncertainty about hidden states, and/or (2) reduce uncertainty about parameters. The reduction of uncertainty of the parameters via the maximization of novelty encourages agents to explore novel parts of the state space that are less familiar (i.e., states that have low concentration parameters).
The Softmax Function
The softmax (or normalized exponential) function, denoted by σ, takes a vector x of length k and normalizes the vector such that the elements (1) have a monotonic relationship with the elements of the input vector, and (2) sum to 1 and can thus be treated as a categorical probability distribution over 1 to k mutually exclusive states. Importantly, the vector is weighted by a precision parameter denoted by γ, which controls the extent to which differences between the elements are amplified or dampened by the exponential.
| (A.15) |
For example, for , when .
The Gamma Function
The gamma function (denoted by Γ) is a generalization of the factorial function that, unlike the factorial function (whose domain is restricted to positive integers), is well defined for complex and real valued (i.e., non-integer) inputs (except for the negative integers). For positive, real-valued, and complex numbers, the gamma function is defined by the following definite integral.
| (A.16) |
For the positive integers, the gamma function reduces to the factorial function (i.e., ) but is shifted by 1.
| (A.17) |
| (L2) |
| (L3) |
| (L4) |
To gain an intuition for how the gamma function relates to the factorial function, we will use integration by parts (i.e., to show that .
| (A.18) |
| (L2) |
Because e−x grows faster than xz, the first term is sent to zero leaving us with the following.
| (A.19) |
| (L2) |
If we plug in some examples, we see immediately that this is equivalent to the factorial function.
| (A.20) |
| (L2) |
| (L3) |
In the context of the Dirichlet distribution, the gamma function is used to define a normalization constant that accounts for the combinatorics of the concentration parameters, which are positive real-valued numbers (i.e., they can take non-integer values), most of which are not defined when using the factorial function, which is why the gamma function is employed. We can think of the gamma function as interpolating between the values of the factorial function (i.e., because it is defined with respect to non-negative real numbers, while the factorial function is only defined with respect to the non-negative integers). In other words, when the factorial function and gamma function are given the same inputs, they produce consistent outputs (shifted by one), but the gamma function outputs values between the integer outputs given by the factorial function.
Appendix B. Pencil and paper exercises
The purpose of this supplementary section is to provide a set of exercises that can be worked through using pencil and paper, with the aim of building an intuition for how active inference models operate. The first exercise is a simple example of static perception. The second example extends this by modeling how states (and the observations they generate) change across time, which is an example of a hidden Markov model (HMM). HMMs are the perceptual component of partially observable Markov decision processes (POMDPs). We have not included an example of policy selection, as performing the calculations for even two policies involves too many computations to reasonably expect readers to perform them by hand.
To help the reader build a conceptual bridge between the update equations and their implementation in code, we have also included MATLAB code (Pencil_and_paper_exercise_solutions.m) that has solutions to each of the exercises shown below. Finally, readers should note that, to ensure that the exercises can be solved by hand, we have excluded a key aspect of active inference models as they are usually implemented. Namely, instead of inferring the posterior over hidden states using gradient descent, we use only a single round of marginal message passing. For readers seeking to understand how message passing and policy selection operate in the model inversion procedure implemented in spm_MDP_VB_X.m, please see the stand-alone MATLAB script Simplified_simulation_script.m that is also provided, which is a stripped down, but thoroughly commented, version of the model inversion scheme used in spm_MDP_VB_X.m.
Exercises
Static Perception
For this first example we will keep things as simple as possible.
Example 1
Update Equation
Generative Model and Observation
Model Inversion
Exercise 1
Based on the update equation in Example 1 and the observation listed below, invert the following generative model:
Dynamic Perception
In the second example below we move from a static environment to a dynamic one in which hidden states, and the observations generated by these states, change across time. In addition to including a B matrix that encodes the transition probabilities, we must initialize the approximate posteriors for each tau (τ) before starting model inversion (recall, tau references a time about which one has beliefs, not a time at which one updates beliefs with a new observation). It is also important to note that, because the log of zero is not defined, the model inversion procedure implemented in the code adds a very small number (e−16 = 0.00000011253) to all inputs which turns the log of zero into the log of a very small number. Since we expect readers to be able to do this exercise by hand, we approximate this by adding 0.01 to the input of all logs.
Example 2
Update Equations
*Recall that denotes the transpose of B with normalized columns (i.e., columns that sum to 1). Also note that because the example below only includes 2 time points, only the first and third equations will apply.
Generative Model and Observations
Initialize Approximate Posteriors
Model Inversion: Time Step 1
Model Inversion: Time Step 2
Exercise 2
Using the equations presented in Example 2, and the observations listed below, invert the following generative model. Note again that because this example only includes 2 timepoints, only the first and third equations will apply.
Answers
Answer: Exercise 1
Answer: Exercise 2
Model Inversion: Time Step 1
Model Inversion: Time Step 2
Appendix C. Supplementary data
Supplementary material related to this article can be found online at https://doi.org/10.1016/j.jmp.2021.102632.
Footnotes
This other work appeals to a number of common constructs discussed in the free energy principle literature that are also not covered here, but which the reader may have come across previously. One such construct is a ‘Markov blanket’, which is a mathematical way of describing the boundary that separates the internal states of an organism from the external environment (although note that this term is sometimes used in different ways; see Bruineberg, Dolega, Dewhurst, and Baltieri (2021). Another related construct is a ‘non-equilibrium steady state (NESS) density’, which describes the states an organism must have a high probability of occupying if it is to maintain its existence — where this can be understood as maintaining the integrity of its Markov blanket (i.e., keeping the boundary intact that separates an organism from its environment). The POMDP scheme described in this tutorial does not explicitly appeal to these constructs; however, one can think of an agent’s preferred observations in POMDPs as those that keep it within the high-probability states consistent with its continued existence (i.e., those that would keep its Markov blanket intact).
It is important to note that the term ‘policy’ in active inference is used in a different way than in model-free reinforcement learning. As stated in the main text, a policy in active inference refers to an allowable sequence of actions (e.g., a plan to move to state 1, then to state 2, then to state 3). In contrast, a policy in model-free reinforcement learning typically consists of a mapping from states to actions. That is, a policy specifies the action that should be chosen for each possible state an agent might occupy (e.g., if in state 1, move to state 2; if in state 3, move to state 1, etc.).
Note that in some papers, preferences are formulated over states instead of observations. In this case, one might wonder how an agent can have two priors over states at the same time (one for beliefs and one for preferences). Although the technical details are beyond the scope of this paper, in this case one must think more explicitly in terms of an agent having two models — one of true states of the world and one of preferred states (cast as priors in each model, respectively). Policy selection then attempts to minimize the divergence between the two by bringing true states to match preferred states (for details, see Da Costa, Parr et al., 2020).
Note that, for the categorical distributions we use in this tutorial, the sufficient statistics will correspond to the probability of each possible value of a random variable (e.g., the probability of each possible state or observation).
Although note that, even in task contexts where they perform similarly, active inference models and reinforcement learning models make decisions in a different way. Specifically, while reinforcement learning models seek to maximize a reward signal, active inference models instead seek to reach a target distribution that is treated as rewarding (i.e., the distribution encoding the agent’s preferred observations).
Although not shown in detail here, this also affords the possibility of multidimensional policies in more complex models. For example, if there were multiple possible actions (transition matrices) for two different state factors, one might specify that action 2 for state factor 1 can be chosen together with action 2 for state factor 2, but that action 2 for state factor 1 cannot be chosen together with action 3 for state factor 2 (simply by including a column that has entries of 2 for both state factors but no column that has an entry of 2 for state factor 1 and an entry of 3 for state factor 2).
However, note that they need not all be ones in a more complex model with multidimensional policies, as also described for U in the previous footnote.
Note that the spm_MDP_VB_X.m script (and the tutorial version here) works with negative free energies, and so these F and G values are made negative in the MDP output structure in MATLAB.
As a brief general note, the columns of all matrices are put through a softmax function in the spm_MDP_VB_X_tutorial.m script after the addition of negligibly low values to each entry to prevent the problem that log(0) is undefined. As such, even specification of identity matrices for transition beliefs will not completely rule out the possibility that states could change over time in the face of very strong observational evidence to the contrary.
An example of this approach is maximum a posteriori (MAP) estimation, in which an algorithm tries to find the parameter values (i.e., point estimates) that maximize the value of the posterior as opposed to the likelihood.
Another important class of estimation algorithms uses Markov Chain Monte Carlo (MCMC) methods (Neal, 1993). These methods involve sequentially sampling from a distribution according to specific sets of rules that try to find locations under that distribution with high probability. In this case, the probability of a participant’s actions would be evaluated under a sequence of parameter value combinations (with an initially random starting point) until a sufficient approximation to the true distribution was generated.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Adams R, Shipp S, & Friston K. (2013). Predictions not commands: active inference in the motor system. Brain Structure and Function, 218, 611–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addicott M, Pearson J, Sweitzer M, Barack D, & Platt M. (2017). A primer on foraging and the explore/exploit trade-off for psychiatry research. 42, (pp. 1931–1939). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews M. (2020). The math is not the territory: Navigating the free energy principle (p. 18315). Pittphilsci. [Google Scholar]
- Attias H. (2000). A variational baysian framework for graphical models.
- Attias H. 2003. Planning by probabilistic inference. In Paper presented at the proc. of the 9th int. workshop on artificial intelligence and statistics.
- Badcock PB, Friston KJ, Ramstead MJD, Ploeger A, & Hohwy J. (2019). The hierarchically mechanistic mind: an evolutionary systems theory of the human brain, cognition, and behavior. Cognitive, Affective, & Behavioral Neuroscience, 19(6), 1319–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barto A, Mirolli M, & Baldassarre G. (2013). Novelty or surprise? Frontiers in Psychology, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal MJ (2003). Variational algorithms for approximate bayesian inference. London: University of London. [Google Scholar]
- Bekinschtein T, Dehaene S, Rohaut B, Tadel F, Cohen L, & Naccache L. (2009). Neural signature of the conscious processing of auditory regularities. Proceedings of the National Academy of Sciences of the United States of America, 106, 1672–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CM (2006). Pattern recognition and machine learning. New York: Springer. [Google Scholar]
- Bogacz R. (2017). A tutorial on the free-energy framework for modelling perception and learning. Journal of Mathematical Psychology, 76, 198–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botvinick M, & Toussaint M. (2012). Planning as inference. Trends in Cognitive Sciences, 16(10), 485–488. [DOI] [PubMed] [Google Scholar]
- Breakspear M. (2017). Dynamic models of large-scale brain activity. Nature Neuroscience, 20(3), 340–352. [DOI] [PubMed] [Google Scholar]
- Brown LD (1981). A complete class theorem for statistical problems with finite-sample spaces. The Annals of Statistics, 9(6), 1289–1300. [Google Scholar]
- Brown TH, Zhao Y, & Leung V. (2010). Hebbian plasticity. In Encyclopedia of neuroscience (pp. 1049–1056).
- Bruineberg J, Dolega K, Dewhurst J, & Baltieri M. (2021). The Emperor’s new markov blankets. Behavioral and Brain Research, 1–63. 10.1017/S0140525X21002351, Epub ahead of print. [DOI] [PubMed]
- Bucci A, & Grasso M. (2017). Sleep and dreaming in the predictive processing framework. In Metzinger T, & Wiese W. (Eds.), Philosophy and predictive processing. Johannes Gutenberg-Universität Mainz. [Google Scholar]
- Buckley CL, Sub Kim C, McGregor S, & Seth AK (2017). The free energy principle for action and perception: A mathematical review. Journal of Mathematical Psychology, 81, 55–79. [Google Scholar]
- Burr C, & Jones M. (2016). The body as laboratory: Prediction-error minimization, embodiment, and representation. Philosophical Psychology, 29(4), 586–600. [Google Scholar]
- Carlin BP, & Louis TA (1998). Bayes and empirical bayes methods for data analysis. Boca Raton: Chapman & Hall/CRC. [Google Scholar]
- Champion T, Grześ M, & Bowman H. (2021). Realising active inference in variational message passing: the outcome-blind certainty seeker. Neural Computation. [DOI] [PubMed] [Google Scholar]
- Clark A. (2013). Whatever next? Predictive brains, situated agents, and the future of cognitive science. The Behavioral and Brain Sciences, 36, 181–204. [DOI] [PubMed] [Google Scholar]
- Clark A. (2015). Surfing uncertainty: Prediction, action, and the embodied mind. New York: Oxford University Press. [Google Scholar]
- Clark A. (2017). How to knit your own Markov blanket. In Metzinger TK, & Wiese W. (Eds.), Philosophy and predictive processing. Frankfurt am Main: MIND Group. [Google Scholar]
- Clark JE, Watson S, & Friston KJ (2018). What is mood? A computational perspective. Psychological medicine. (pp. 1–8). [DOI] [PMC free article] [PubMed]
- Da Costa L, Parr T, Sajid N, Veselic S, Neacsu V, & Friston KJ (2020). Active inference on discrete state-spaces: A synthesis. Journal of Mathematical Psychology, 99, Article 102447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa L, Parr T, Sengupta B, & Friston K. (2021). Neural dynamics under active inference: Plausibility and efficiency of information processing. Entropy, 23(4), 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa L, Sajid N, Parr T, Friston KJ, & Smith R. (2020). The relationship between dynamic programming and active inference: the discrete, finite-horizon case. arXiv, arXiv:2009.08111.
- Dauwels J. (2007). On variational message passing on factor graphs. IEEE International Symposium on Information Theory, 2546–2550.
- de Vries B, & Friston KJ (2017). A factor graph description of deep temporal active inference. Frontiers in Computational Neuroscience, 11(95). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry RE (2017). Transcending the evidentiary boundary: Prediction error minimization, embodied interaction, and explanatory pluralism. Philosophical Psychology, 30(4), 395–414. [Google Scholar]
- Friston KJ (2019). A free energy principle for a particular physics. arXiv: 1906.10184.
- Friston KJ, FitzGerald T, Rigoli F, Schwartenbeck P, Doherty JO, & Pezzulo G. (2016a). Active inference and learning. Neuroscience and Biobehavioral Reviews, 68, 862–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, FitzGerald T, Rigoli F, Schwartenbeck P, & Pezzulo G. (2017a). Active inference: A process theory. Neural Computation, 29, 1–49. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Lin M, Frith C, Pezzulo G, Hobson J, & Ondobaka S. (2017b). Active inference, curiosity and insight. Neural Computation, 29, 2633–2683. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Litvak V, Oswal A, Razi A, Stephan KE, van Wijk BCM, et al. (2016b). BayesIan model reduction and empirical Bayes for group (DCM) studies. NeuroImage, 128, 413–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Mattout J, Trujillo-Barreto N, Ashburner J, & Penny W. (2007). Variational free energy and the Laplace approximation. NeuroImage, 34, 220–234. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Parr T, & de Vries B. (2017c). The graphical brain: Belief propagation and active inference. Network Neuroscience, 1, 381–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Rosch R, Parr T, Price C, & Bowman H. (2018). Deep temporal models and active inference. Neuroscience & Biobehavioral Reviews, 90, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesp C, Smith R, Allen M, Friston KJ, & Ramstead MJD (2020). Deeply felt affect: The emergence of valence in deep active inference. Neural Computation, 1–49. [DOI] [PMC free article] [PubMed]
- Hobson J, & Friston K. (2012). Waking and dreaming consciousness: neurobiological and functional considerations. Progress in Neurobiology, 98, 82–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson J, Hong C-H, & Friston K. (2014). Virtual reality and consciousness inference in dreaming. Frontiers in Psychology, 5(1133). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohwy J. (2014). The predictive mind.
- Hohwy J. (2016). The self-evidencing brain. Noû, 50, 259–285. [Google Scholar]
- Hohwy J, Paton B, & Palmer C. (2016). Distrusting the present. Phenomenology and the Cognitive Sciences, 15(3), 315–335. [Google Scholar]
- Kaplan R, & Friston KJ (2018). Planning and navigation as active inference. Biological Cybernetics, 112(4), 323–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczma M, & Gilányi A. (2009). An introduction to the theory of functional equations and inequalities : Cauchy’s equation and Jensen’s inequality (2nd ed.). Basel, Boston, MA: Birkäeuser. [Google Scholar]
- Loeliger HA (2004). An introduction to factor graphs. IEEE Signal Processing Magazine, 21(1), 28–41. [Google Scholar]
- Markovic D, Stojic H, Schwoebel S, & Kiebel S. (2021). An empirical evaluation of active inference in multi-armed bandits. arXiv:2101.08699. [DOI] [PubMed]
- Mathys CD, Lomakina EI, Daunizeau J, Iglesias S, Brodersen KH, Friston KJ, et al. (2014). Uncertainty in perception and the hierarchical Gaussian filter. Frontiers in Human Neuroscience, 8(825). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millidge B. (2019). Combining active inference and hierarchical predictive coding: A tutorial introduction and case study. PsyArXiv.
- Millidge B, Tschantz A, & Buckley CL (2021). Whence the expected free energy?. Neural Computation, 33(2), 447–482. [DOI] [PubMed] [Google Scholar]
- Mirza MB, Adams RA, Mathys C, & Friston KJ (2018). Human visual exploration reduces uncertainty about the sensed world. PLOS ONE, 13, Article e0190429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal RM (1993). Probabilistic inference using Markov chain Monte Carlo methods. Department of Computer Science, University of Toronto. Toronto, ON, Canada. [Google Scholar]
- Oudeyer P-Y, & Kaplan F. (2007). What is intrinsic motivation? a typology of computational approaches. Frontiers in Neurorobotics, 1(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, & Friston KJ (2017a). Uncertainty, epistemics and active inference. Journal of the Royal Society, Interface, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, & Friston KJ (2017b). Working memory, attention, and salience in active inference. Scientific Reports, 7, 14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, & Friston KJ (2018a). The anatomy of inference: Generative models and brain structure. Frontiers in Computational Neuroscience, 12, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, & Friston KJ (2018b). The discrete and continuous brain: From decisions to movement—and back again. Neural Computation, 1–29. [DOI] [PMC free article] [PubMed]
- Parr T, Markovic D, Kiebel S, & Friston KJ (2019). Neuronal message passing using mean-field, bethe, and marginal approximations. Scientific Reports, 9, 1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr T, Pezzulo G, & Friston KJ (2022). Active Inference: The Free Energy Principle in Mind, Brain, and Behavior. MIT Press. [Google Scholar]
- Pezzulo G, Rigoli F, & Friston KJ (2015). Active inference, homeostatic regulation and adaptive behavioural control. Progress in Neurobiology, 134, 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzulo G, Rigoli F, & Friston KJ (2018). Hierarchical active inference: A theory of motivated control. Trends in Cognitive Sciences, 22, 294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran VS (1988). Perceiving shape from shading. Scientific American, 259(2), 76–83. [DOI] [PubMed] [Google Scholar]
- Rigoux L, Stephan KE, Friston KJ, & Daunizeau J. (2014). BayesIan model selection for group studies -revisited. Neuroimage, 84, 971–985. [DOI] [PubMed] [Google Scholar]
- Sajid N, Ball P, Parr T, & Friston K. (2021). Active inference: Demystified and compared. Neural Computation, 1–39. [DOI] [PubMed]
- Sales AC, Friston KJ, Jones MW, Pickering AE, & Moran RJ (2019). Locus coeruleus tracking of prediction errors optimises cognitive flexibility: An active inference model. PLoS Computational Biology, 15(1), Article e1006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandved-Smith L, Hesp C, Mattout J, Friston K, Lutz A, & Ramstead MJ (2021a). Towards a computational phenomenology of mental action: modelling meta-awareness and attentional control with deep parametric active inference. Neuroscience of Consciousness, 2021(2), niab018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhuber J. (2006). Developmental robotics, optimal artificial curiosity, creativity, music, and the fine arts. Journal of Connection Science, 18(2), 173–187. [Google Scholar]
- Schwartenbeck P, FitzGerald T, Mathys C, Dolan R, & Friston KJ (2015). The dopaminergic midbrain encodes the expected certainty about desired outcomes. Cerebral Cortex, 25, 3434–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartenbeck P, Passecker J, Hauser TU, FitzGerald TH, Kronbichler M, & Friston KJ (2019). Computational mechanisms of curiosity and goal-directed exploration. Elife, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Badcock PB, & Friston KJ (2020a). Recent advances in the application of predictive coding and active inference models within clinical neuroscience. Psychiatry and Clinical Neurosciences. [DOI] [PubMed]
- Smith R, Khalsa SS, & Paulus MP (2021b). An active inference approach to dissecting reasons for nonadherence to antidepressants. Biological Psychiatry Cognitive Neuroscience Neuroimaging, 6(9), 919–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Kirlic N, Stewart JL, Touthang J, Kuplicki R, Khalsa SS, et al. (2021d). Greater decision uncertainty characterizes a transdiagnostic patient sample during approach-avoidance conflict: A computational modeling approach. Journal of Psychiatry & Neuroscience, 46(1), E74–E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Kirlic N, Stewart J, Touthang J, Kuplicki R, McDermott T, et al. (2021c). Long-term stability of computational parameters during approach-avoidance conflict in a transdiagnostic psychiatric patient sample. Scientific Reports, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Kuplicki R, Feinstein J, Forthman KL, Stewart JL, Paulus MP, et al. (2020b). A Bayesian computational model reveals a failure to adapt interoceptive precision estimates across depression, anxiety, eating, and substance use disorders. PLoS Computational Biology, 16(12), Article e1008484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Kuplicki R, Teed A, Upshaw V, & Khalsa SS (2020c). Confirmatory evidence that healthy individuals can adaptively adjust prior expectations and interoceptive precision estimates. In Verbelen T, Lanillos P, Buckley C, & De Boom C. (Eds.), Active inference. IWAI 2020, Cham: Springer. [Google Scholar]
- Smith R, Lane RD, Parr T, & Friston KJ (2019a). Neurocomputational mechanisms underlying emotional awareness: Insights afforded by deep active inference and their potential clinical relevance. Neuroscience & Biobehavioral Reviews, 107, 473–491. [DOI] [PubMed] [Google Scholar]
- Smith R, Mayeli A, Taylor S, Al Zoubi O, Naegele J, & Khalsa SS (2021e). Gut inference: A computational modelling approach. Biological Psychology, Article 108152. [DOI] [PMC free article] [PubMed]
- Smith R, Parr T, & Friston KJ (2019b). Simulating emotions: An active inference model of emotional state inference and emotion concept learning. Frontiers in Psychology, 10(2844). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Ramstead MJD, & Kiefer A. (2022). Active inference models do not contradict folk psychology. Synthese, In Press.
- Smith R, Schwartenbeck P, Parr T, & Friston KJ (2020d). An active inference approach to modeling structure learning: Concept learning as an example case. Frontiers in Computational Neuroscience, 14(41). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R, Schwartenbeck P, Stewart JL, Kuplicki R, Ekhtiari H, Investigators T, et al. (2020e). Imprecise action selection in substance use disorder: Evidence for active learning impairments when solving the explore-exploit dilemma. Drug and Alcohol Dependence, 215, Article 108208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan K, Penny WD, Daunizeau J, Moran RJ, & Friston KJ (2009). BayesIan model selection for group studies. Neuroimage, 46(4), 1004–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan KE, Petzschner FH, Kasper L, Bayer J, Wellstein KV, Stefanics G, et al. (2019). Laminar fMRI and computational theories of brain function. Neuroimage, 197, 699–706. [DOI] [PubMed] [Google Scholar]
- Tononi G, & Cirelli C. (2014). Sleep and the price of plasticity: From synaptic and cellular homeostasis to memory consolidation and integration. Neuron, 81, 12–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschantz A, Barca L, Maisto D, Buckley CL, Seth AK, & Pezzulo G. (2021). Simulating homeostatic, allostatic and goal-directed forms of interoceptive control using active inference. bioRxiv, 2021.2002.2016.431365. [DOI] [PubMed]
- Tschantz A, Seth AK, & Buckley CL (2020). Learning action-oriented models through active inference. PLoS Computational Biology, 16(4), Article e1007805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S. (2014). The dirichlet-multinomial and dirichlet-categorical models for bayesian inference. Berkeley, CA: Computer Science Division, UC Berkele; y. [Google Scholar]
- Wald A. (1947). An essentially complete class of admissible decision functions. The Annals of Mathematical Statistics, 54, 9–555. [Google Scholar]
- Whyte C, Hohwy J, & Smith R. (2021). An active inference model of conscious access: How cognitive action selection reconciles the results of report and no-report paradigms. 10.31234/osf.io/mkzx8, PsyArXiv. [DOI] [PMC free article] [PubMed]
- Whyte C, & Smith R. (2020). The predictive global neuronal workspace: A formal active inference model of visual consciousness. Progress in Neurobiology, 2020.2002.2011.944611. [DOI] [PubMed]
- Wilson R, Geana A, White J, Ludvig E, & Cohen J. (2014). Humans use directed and random exploration to solve the explore-exploit dilemma. Journal of Experimental Psychology. General, 143, 2074–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn J, & Bishop CM (2005). Variational message passing. Journal of Machine Learning Research, 6, 661–694. [Google Scholar]
- Zeidman P, Jafarian A, Seghier ML, Litvak V, Cagnan H, Price CJ, et al. (2019). A guide to group effective connectivity analysis, Part 2: Second level analysis with PEB. Neuroimage, 200, 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.