Abstract
Tubulins are critical for the internal organization of eukaryotic cells, and understanding their emergence is an important question in eukaryogenesis. Asgard archaea are the closest known prokaryotic relatives to eukaryotes. Here, we elucidated the apo and nucleotide-bound x-ray structures of an Asgard tubulin from hydrothermal living Odinarchaeota (OdinTubulin). The guanosine 5′-triphosphate (GTP)–bound structure resembles a microtubule protofilament, with GTP bound between subunits, coordinating the “+” end subunit through a network of water molecules and unexpectedly by two cations. A water molecule is located suitable for GTP hydrolysis. Time course crystallography and electron microscopy revealed conformational changes on GTP hydrolysis. OdinTubulin forms tubules at high temperatures, with short curved protofilaments coiling around the tubule circumference, more similar to FtsZ, rather than running parallel to its length, as in microtubules. Thus, OdinTubulin represents an evolutionary stage intermediate between prokaryotic FtsZ and eukaryotic microtubule-forming tubulins.
The architecture and GTP hydrolysis mechanism of Odinarchaeota tubulin provide insight into the evolution of the microtubule.
INTRODUCTION
The tubulin/FtsZ/CetZ superfamily of proteins polymerizes into filaments for which nucleotide-dependent dynamics and curvature are critical to their functions. The prokaryotic guanosine 5′-triphosphate (GTP)–hydrolyzing FtsZ and CetZ form homo-filaments, which adopt straight and curved conformations (1–4). These filaments are part of the ring systems that constrict during prokaryotic cell division (5). The eukaryotic microtubule-forming tubulins have resulted from a series of gene duplications (6) and have diverged significantly from FtsZ (7) and CetZ (2). The γ-tubulin ring complex patterns the microtubule (8), which typically comprises 13 parallel strands. Each straight strand (protofilament) nucleates from a single subunit of γ-tubulin via incorporation of the obligate α/β-tubulin heterodimers (9). Thus, in the cell, the microtubule nucleation step is separated at microtubule-organizing centers from other assembly and disassembly dynamics. α-Tubulin contains a nonexchangeable, nonhydrolyzing GTP-binding site (N-site), whereas β-tubulin (E-site) and γ-tubulin contain exchangeable and hydrolyzing GTP-binding sites. The switch from straight to curved protofilaments at microtubule + ends, following GTP hydrolysis, results in catastrophe disassembly (10, 11).
The Asgard archaea superphylum has been proposed as the closest prokaryotic relative to eukaryotes (12). Their genomes, which were mainly obtained from metagenomic studies, include genes that have homology to eukaryotic signature protein (ESP)–encoding genes. These ESPs were previously thought to be exclusive to eukaryotes, before the genomic characterization of the first Asgard archaea, Candidatus Lokiarchaeota (13). Thus, Asgard archaea genomes have become valuable resources to understand proto-eukaryotic protein machineries at the functional level, such as the ESPs studied in actin dynamics (14–16) and membrane fusion (17). The Candidatus Odinarchaeota archaeon LCB_4 (Odin) metagenome-assembled genome (MAG; GenBank accession number MDVT00000000.1) includes two genes predicted to be FtsZ homologs (OLS17704.1 and OLS17546.1) and a single gene (OdinTubulin, OLS18786.1) with greater homology to eukaryotic tubulin rather than to prokaryotic FtsZ (12). OdinTubulin is significantly more similar to eukaryotic tubulins than the next closest archaeal tubulin-like sequences from Nitrosoarchaeum (18), BLAST e-values of 10−67 compared with 10−13, respectively. However, the properties of OdinTubulin are unknown.
RESULTS
X-ray structure of OdinTubulin
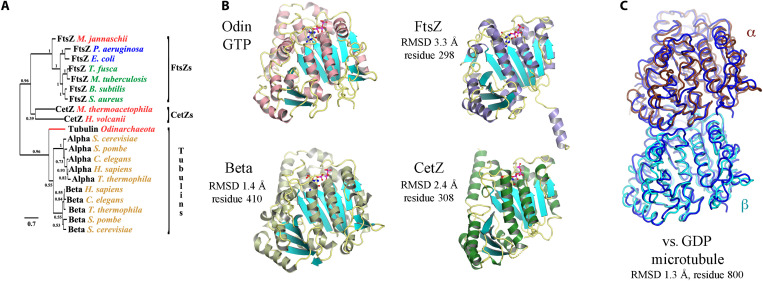
To address whether OdinTubulin is a genuine tubulin at the protein level, we expressed, purified, crystallized, and determined the structure of OdinTubulin in the apo form, and bound to GTP or guanosine diphosphate (GDP) (fig. S1 and table S1 and summarized in table S2). Phylogenetic analysis, using structure alignment, confirmed that OdinTubulin has diverged significantly from FtsZ and CetZ and branches in the same clade as eukaryotic tubulins (Fig. 1A) (12). Structure homology searches revealed that the GTP-bound OdinTubulin, refined at 1.62 Å (Protein Data Bank PDB 7EVB, capital letters refer to structures determined in this study), is most similar to α- and β-tubulins within a microtubule, regardless of the nucleotide state within the microtubule, rather than to nonpolymerized tubulin subunits or to FtsZ or CetZ (Fig. 1B and table S3). OdinTubulin shares ~34 to 35% sequence identity with the human α- and β-tubulins. Within the crystal packing, OdinTubulin subunits are arranged as in a microtubule protofilament (figs. S1 and S2). Superimposition of the OdinTubulin lower subunit (−) onto a eukaryotic microtubule GDP-containing β-tubulin subunit (19) revealed that the position of the OdinTubulin upper subunit (+) aligned closely with the proximal α-tubulin subunit from the microtubule protofilament (Fig. 1C) (19). By contrast, superimposition onto the microtubule structure containing a GTP mimetic in the β-tubulin subunit (11), or onto the β-tubulin subunit from the stathmin-bound curved protofilaments (20), aligned the α-tubulin subunits poorly to the OdinTubulin upper subunit (fig. S3, table S4, and movies S1 to S3).
Fig. 1. The crystal structure of OdinTubulin.
(A) Phylogenetic analysis of OdinTubulin from structure-based sequence alignment in comparison to the prokaryotic cell division proteins, FtsZ and CetZ, and the eukaryotic microtubule-forming tubulins. (B) Comparison of the protomer structures of GTP-bound OdinTubulin (PDB 7EVB) to β-tubulin (PDB 6o2r) (19), CetZ (PDB 4b45) (2), and FtsZ (PDB 1w5a) (4). The matching numbers of residues and RMSD values indicate the relative structural similarities to OdinTubulin. RMSD, root mean square deviation. (C) Superimposition of the two GTP-bound OdinTubulin symmetry-related subunits from the crystal packing (dark blue) onto two subunits of eukaryotic tubulin from the GDP-bound microtubule (PDB 6o2r) (19).
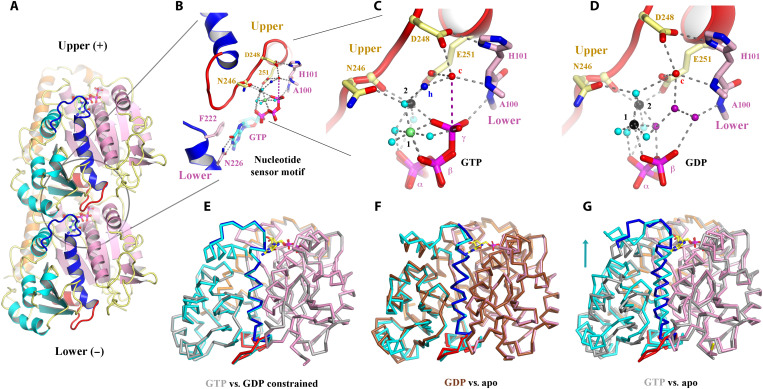
Similar to eukaryotic tubulin, OdinTubulin comprises an N-terminal domain (residues 1 to 202), an intermediate domain (residues 203 to 367), and a C-terminal domain (residues 368 to 424; Fig. 2A), as described for eukaryotic tubulin (9). The α7 helix and its preceding loop (blue) and the α8 helix and its preceding loop (red), which we term the “nucleotide sensor motif,” lie in the intermediate domain, connecting the nucleotide from the lower subunit (−) to the nucleotide in the upper subunit (+) (Fig. 2A and movie S4). In eukaryotes, the α7 helix is known to undergo a translation movement in response to the presence of different nucleotides and protofilament curvature (21). Together, these data indicate that the GTP-bound OdinTubulin crystals contain a straight microtubule-like tubulin protofilament stabilized by native nucleotide binding.
Fig. 2. Structural implications for GTP hydrolysis.
(A) The OdinTubulin protofilament in the crystal packing (PDB 7EVB). Two subunits of OdinTubulin are depicted. The α7 helix and preceding loop (dark blue) and α8 helix and preceding loop (red) comprise the nucleotide sensor motif, which connect the upper and lower GTP-binding sites (sticks). Secondary structure elements are colored by domain: N-terminal (pink), intermediate (cyan), and C-terminal (orange). The nucleotide sensor motif lies within the intermediate domain (see movie S4). (B) Enlargement of the GTP interactions. Only part of each nucleotide sensor motif is shown for clarity. Selected residues from the upper and lower subunits are labeled in yellow and pink, respectively. (C) Enlargement of the interactions around the GTP γ-phosphate. Black, lime green, and cyan spheres indicate Na+, Mg2+ (numbered in black), and water molecules, respectively. The proposed hydrolytic water is shown as a red sphere and labeled “c,” and water molecule suitably placed to receive the hydrogen ion from the hydrolytic water is labeled “h” in blue. The purple dashed line indicates the route for nucleophilic attack on the GTP γ-phosphate (see movie S5). (D) The same region from the GDP-bound structure (PDB 7EVE). Three water molecules (purple) replace the GTP γ-phosphate, and both cations are assigned as Na+ on the basis of bond distances and crystallization conditions (see movie S5). (E to G) Superimposition of protomer structures. (E) GTP-bound OdinTubulin (gray, PDB 7EVB) overlaid on the constrained GDP-bound structure (colored, PDB 7EVE). (F) The unconstrained GDP-bound OdinTubulin (brown, PDB 7F1B) overlaid on the apo structure (colored, PDB 7EVG). (G) GTP-bound OdinTubulin (gray, PDB 7EVB) overlaid on the apo structure (colored, PDB 7EVG). The arrow highlights the conformational change for the intermediate domain (see movie S7).
GTP binding
Inspection of the nucleotide-binding site revealed that GTP is bound to the “E”-site, with the guanine moiety interacting with the side chains of Phe222 and Asn226 from the N-terminal region of the nucleotide sensor motif, and the γ-phosphate is bound to the main-chain amide nitrogen from Ala100 from the lower subunit (Fig. 2B). The C-terminal region of the nucleotide sensor motif, from the upper subunit, interacts indirectly with the GTP phosphate groups through a bonding network of cations and water molecules (Fig. 2, B and C). There are two GTP-bound cations: the commonly observed ion that bridges the β- and γ-phosphates (“1” in Fig. 2C), and a second ion that directly coordinates Asn246, Glu251, and the GTP γ-phosphate (“2” in Fig. 2C). Cation 2–stabilized Glu251 orders a water molecule (“c” in Fig. 2, B and C), which is also in bonding distance of Asp248 from the upper subunit and to the main-chain amide nitrogen from His101 in the lower subunit. Water “c” is 4.1 Å from the GTP γ phosphorous atom, suitably positioned for straight-line nucleophilic attack for hydrolysis (movie S5). The cation-binding sites (1 and 2) are promiscuous but, in the crystal structures, preferentially bind Mg2+ and K+/Na+, respectively (fig. S4).
Proposed hydrolysis mechanism
The GTP-binding site arrangement indicates a probable hydrolysis mechanism, whereby Asp248 and/or Glu251 activate the hydrolytic water “c.” The activated water will be directed by the main-chain amide nitrogen from His101 and the side chain of Glu251, which may swivel while remaining bound to cation 2, resulting in nucleophilic attack on the γ-phosphorous atom, leading to hydrolysis (Fig. 3A). Another water molecule “h” is suitably placed to receive the hydrogen ion from the hydrolytic water (Figs. 2C and 3A and movie S5). The extended distance of water “c” from the GTP γ-phosphate phosphorous atom (4.1 Å) in the crystal structure may be part of the cooperative conformational changes within a protofilament. We hypothesize that protofilaments initially polymerize as straight protofilaments. Stochastic fluctuations in the water “c” position may then initiate hydrolysis in one or more subunits, which, in turn, can be propagated by the allosteric motion of the nucleotide sensor motif to produce concerted hydrolysis and conformational change throughout the protofilament, perhaps acting as a delay mechanism to allow adequate polymerization before cooperative curving.
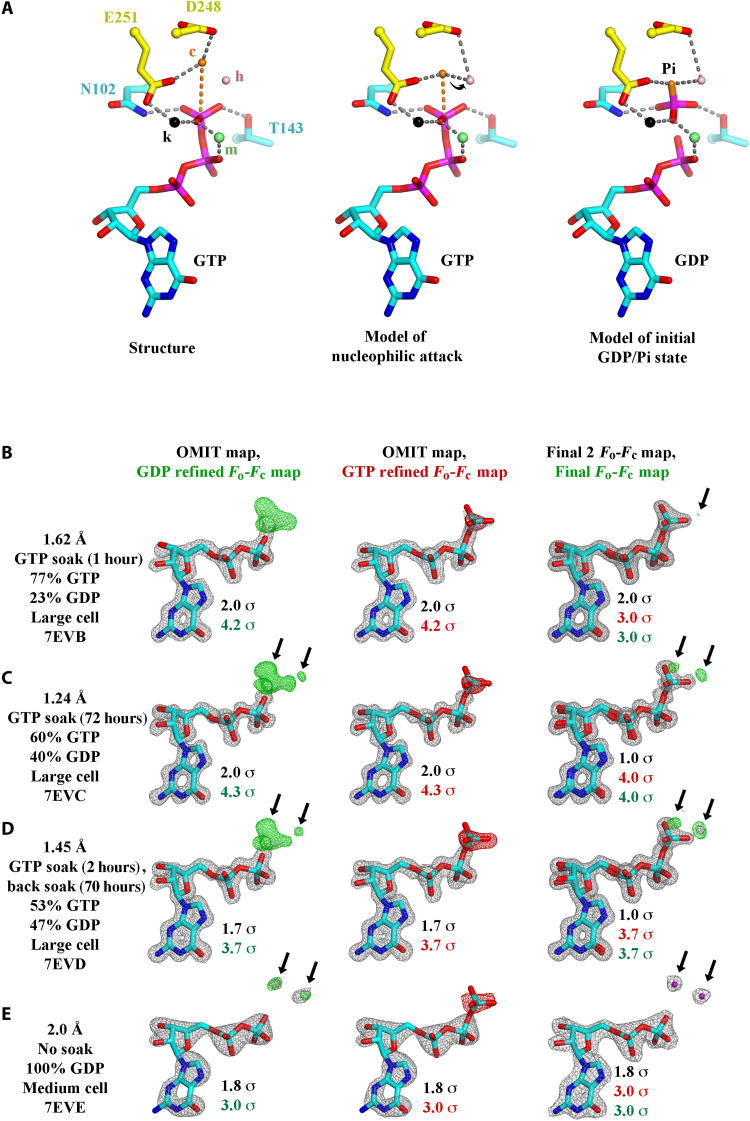
Fig. 3. GTP hydrolysis.
(A) Proposed mechanism of GTP hydrolysis. Left: Structure 7EVI is shown with the residues and ions that stabilize the GTP γ-phosphate. c, hydrolytic water; h, hydrogen ion receiving water; m, magnesium ion; k, potassium or sodium ion. Middle: To initiate hydrolysis, the hydrolytic water (orange) is required to approach the GTP γ-phosphate phosphorus atom, along the orange dashed line, likely losing a hydrogen ion to the hydrogen ion receiving water (pink), indicated by the arrow. Right: The dissociated γ-phosphate ion will receive a hydrogen ion, possibly from a surrounding water molecule or from Thr143. (B to E) GTP hydrolysis followed by x-ray crystallography. Structures determined (B) 1 or (C) 72 hours after soaking with 10 mM GTP showed a decrease in the GTP:GDP ratio. (D) Back soaking the crystals (70 hours) decreased the ratio further. (E) Structure of a nonsoaked crystal with 100% GDP bound to OdinTubulin arranged in the regular protofilament packing, similar to the microtubule, which is stabilized by a different crystal unit cell (figs. S1 and S2). The maps are contoured at the color-coded sigma levels. Left: The structures were refined with GDP. Middle: Refined with GTP. Right: The structures were refined with final GTP/GDP ratios. Green (+) and red (−) Fo-Fc density indicates the need for more or less atoms, respectively. The arrows indicate the position of two ordered water molecules (purple) that appear following γ-phosphate release after hydrolysis (E, right). The third ordered water molecule (Fig. 2D), which appears after hydrolysis, is bound to the metal ions, occupies a similar position to an oxygen from the GTP γ-phosphate, and does not appear in the difference maps. This water has weaker electron density compared with the other two waters and likely has partial occupancy (movie S5).
A crystal structure containing 100% GDP in the nucleotide-binding site (PDB 7EVE, refined at 2.0 Å) revealed that the phosphate ion is released following hydrolysis and is replaced by three water molecules, and the hydrolytic water–binding site is occupied (Fig. 2D and movie S5), without eliciting notable conformational changes to the OdinTubulin protomer or protofilament structures (Fig. 2E and fig. S1). We propose that the exchange of the covalently bound γ-phosphate for three water molecules, following hydrolysis, results in weakening the binding between the upper and lower OdinTubulin subunits in this region, producing strain in the protofilament.
Conformational changes on GTP hydrolysis
Incubation of GTP-soaked crystals in the presence of Na+ over time resulted in an initial increase in the bound GDP:GTP ratio (3 days; Fig. 3, B to E, and table S2) followed by a blurring of the electron density for the intermediate domain (2 months), indicating a slow structural transition within the crystals. A single GTP-soaked OdinTubulin crystal (5 mM GTP, 1 mM MgCl2, and 0.1 M KCl; 1 hour) was frozen and confirmed to contain a ratio of GTP:GDP of ~9:1 by x-ray crystallography. Subsequently, the crystal was thawed and reequilibrated in crystallization conditions supplemented by 1 mM MgCl2, 0.1 M KCl, and 0.2 mM sodium acetate in the absence of nucleotide (2 weeks at 20°C), before being refrozen and the crystal structure determined (PDB 7F1B, refined at 2.40 Å). The resulting 100% GDP-bound conformation represents a second class of OdinTubulin structure that is similar to the structures that we determined in the apo state (PDB 7EVG, refined at 2.48 Å) or partially bound to background GDP that resulted from the purification protocol (~60%; PDB 7EVH, refined at 2.50 Å; Fig. 2F, figs. S1 and S5, and movie S6). We interpret this alternate OdinTubulin conformation to be that after the structural transitions resulting from GTP hydrolysis to GDP, allowing for release of the nucleotide.
Comparison of the apo/GDP-bound structures with the GTP-bound structures revealed that the intermediate domain, including the nucleotide sensor motif, moves relative to the N-terminal and C-terminal domains (Fig. 2G and movie S7). This conformational change alters the interactions between protomer subunits and likely results in a curving of the protofilament, unless the straight form is stabilized by interprotofilament interactions as observed for the central portion of the microtubule or in the OdinTubulin crystal packing (fig. S2). By contrast, the 100% GDP-bound OdinTubulin (PDB 7EVE) adopts a GTP-like conformation stabilized by a different crystal packing (figs. S1 and S2). We interpret the 7EVE structure to be the GDP-bound strained structure before the conformational change. The role of the nucleotide sensor motif in allosterically linking the occupancy of the nucleotide-binding site between adjacent protomers is likely twofold: first, in ensuring that GTP-bound monomers are preferentially added to a growing filament, and, second, in cooperatively coordinating the conformational change throughout a protofilament following hydrolysis and phosphate release.
Conservation with microtubule-forming tubulins
Because the proposed hydrolytic and ion-binding residues from the nucleotide sensor motif are conserved between OdinTubulin and α-tubulin (Fig. 4A), we propose that GTP hydrolysis in microtubules may proceed via the same mechanism involving two cations and a strained intermediate where the phosphate ion is released following hydrolysis and replaced by three water molecules (Fig. 2D). Support for this mechanism can be found in the structures of eukaryotic tubulins. A water molecule is found bound to α-tubulin Glu254 in the sequestered α/β-tubulin dimer (22), equivalent to the hydrolytic water bound to Glu251 in OdinTubulin (compare Fig. 4, B and C). α-Tubulin Glu254 has been predicted to be involved in catalysis by comparison to the structure of FtsZ (23). The main-chain amide nitrogens of Ala99 and Gly100 (β-tubulin) adopt similar positions to Ala100 and His101 from the lower OdinTubulin subunit, which interact with the GTP γ-phosphate and water “c.”
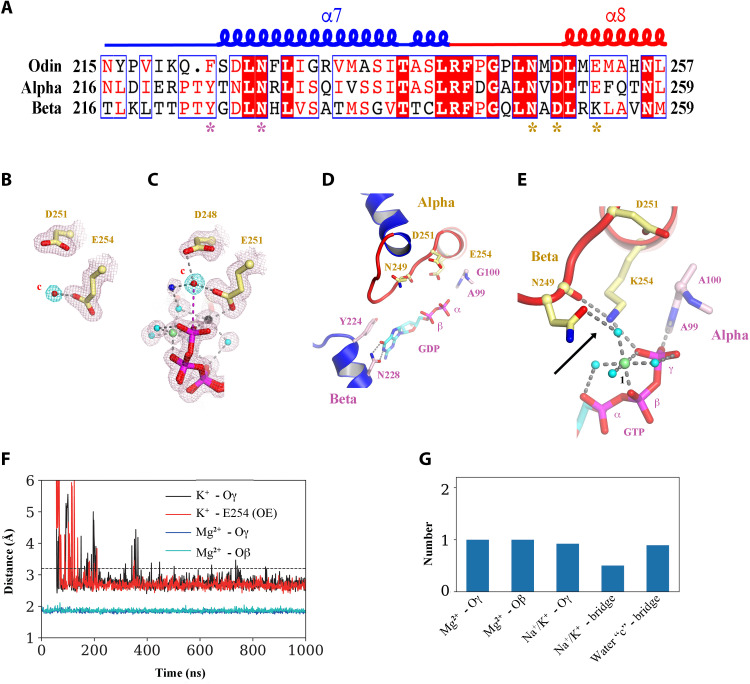
Fig. 4. Similarities in OdinTubulin and microtubule nucleotide interactions.
(A) Conservation in the sequence of the nucleotide sensor motif from OdinTubulin and human α- and β-tubulins. Colored stars below the alignment indicate the residues highlighted in Fig. 2 (B to D) and in (B) to (E). (B) A water molecule is found bound to α-tubulin Glu254 in the sequestered α/β-tubulin dimer (PDB 6s8k), equivalent to (C) the hydrolytic water “c” bound to Glu251 in OdinTubulin. The 2Fo-Fc electron density maps contoured at 1 σ (pink) and the density around potential hydrolytic water (cyan). (D) Subunits within a GDP-bound microtubule E-site (PDB 6o2r) in a similar conformation to Fig. 2B showing structural similarity. (E) β-Tubulin interactions with bound-GTP in the α-tubulin N-site (PDB 6s8k) in a similar orientation to Fig. 2C. The cation-bound, hydrolysis-guiding residue Glu251 from OdinTubulin is substituted by a basic residue (Lys254, indicated by the arrow) in β-tubulin. (F) Coordination of metal ions in molecular dynamics (MD) simulations at the GTP exchangeable site of a microtubule for a representative simulation. A magnesium ion is stably coordinated via oxygen atoms from GTP β-phosphate (cyan) and γ-phosphate (blue) throughout the 1-μs simulation. K+ becomes associated with an oxygen atom from the GTP γ-phosphate (black) and Glu254 (red). Similar results were obtained when Na+ became coordinated at the same site. (G) Occupancies of the metal ions at each site during the simulation. Stable occupancies of the GTP-bound Mg2+ and K+ or Na+ were observed. Approximately half the time, the K+ or Na+ was jointly coordinated by the GTP γ-phosphate and Glu254 (Na+/K+ bridge). Last, a water molecule was located at position “c” between Glu254 and the phosphorous atom of the GTP γ-phosphate (water “c”-bridge).
Furthermore, in the 3.3-Å cryo–electron microscopy (EM) structure of GDP-bound E-site, microtubule (19) adopts a similar conformation around the nucleotide (compare Figs. 2B and 4D). However, identification of low–molecular weight species, such as water molecules, has not been possible at the resolution of the microtubule EM maps. In the sequestered α/β-tubulin dimer (22), the hydrolysis-activating residue Glu251 from OdinTubulin is substituted by a basic residue (Lys254) in β-tubulin, which places a positive charge in the same location as cation 2 (compare Figs. 2C and 4E). This not only results in a lack of hydrolytic activity by the β-tubulin in the N-site but also indicates that a positive charge is acceptable, in eukaryotic tubulins, in bridging the carbonyl of Asn249 (Asn246 in OdinTubulin; Fig. 2C) and the GTP γ-phosphate (Fig. 4E).
To add weight to the prediction that there is a common mechanism of hydrolysis between OdinTubulin and microtubules, we carried out molecular dynamics (MD) simulations on restrained α/β-tubulin subunits at the E-site interface in a background of magnesium, sodium, and potassium ions. In the 1-μs simulation, a magnesium ion stably associated with the oxygen atoms from the β- and γ- GTP phosphates (Fig. 4F), similarly to the OdinTubulin crystal structure (Fig. 2C). Furthermore, a potassium or sodium ion quickly became within bonding distance of oxygen atom of the GTP γ-phosphate and a carboxyl atom of Glu254 (Fig. 4F), similar to cation 2 in the OdinTubulin structure (Fig. 2C). We measured the occupancy of the ions at these sites during the simulations (Fig. 4G). The Mg2+- and K+/Na+-binding sites, in interacting with GTP, were fully occupied during the simulations. Approximately, 40% of the time, a K+/Na+ occupied the bridging site between oxygen atom of the GTP γ-phosphate and a carboxyl atom of Glu254 (Fig. 4G). Last, a water molecule was observed at 100% occupancy in bridging Glu254 (Glu251 in OdinTubulin) and the phosphorous atom of the GTP γ-phosphate (water “c”—bridge; Figs. 2C and 4G). Together, the conservation in the important GTP hydrolysis residues, which bind cations and the proposed hydrolytic water, combined with the occupancy of these sites in MD simulations, reinforces the hypothesis that GTP hydrolysis proceeds through a similar mechanism in OdinTubulin and microtubules that involves two cations.
OdinTubulin filament assembly
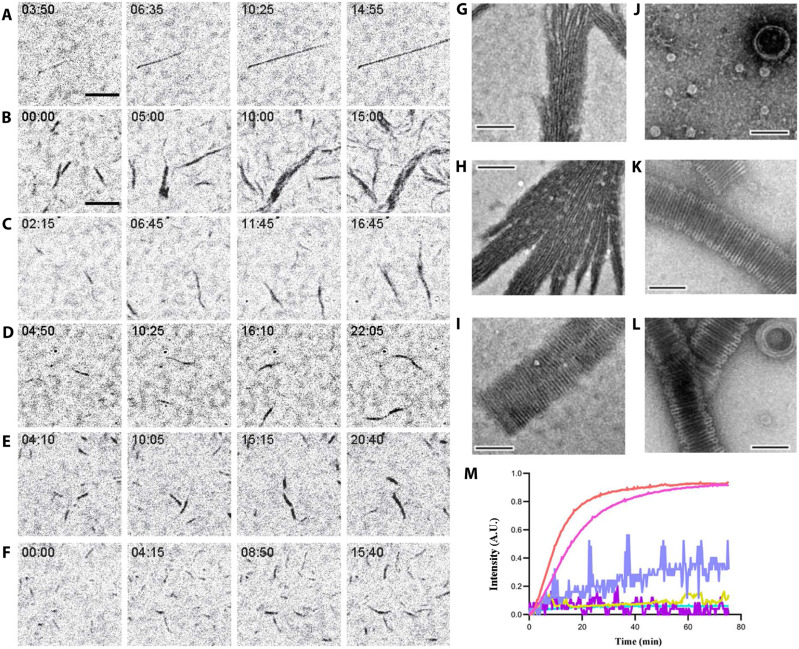
We observed the dynamics of OdinTubulin polymerization by interference reflection microscopy (IRM) (24). Unlike the sparse, straight, smoothly elongating microtubules (Fig. 5A and movies S8 and S9), OdinTubulin tended to form wider bundles of filaments, under similar conditions (Fig. 5B and movies S8 and S9). The elongation proceeded with a high rate of nucleation via polymerization and filament annealing. Lower concentrations of OdinTubulin led to shorter more uniform filaments, which could be assembled under a variety of cation conditions (Fig. 5, C to F, and movies S8 and S9).
Fig. 5. Polymerization of OdinTubulin.
(A to F) IRM. (A) Elongation of tubulin (15 μM) into microtubules in 100 mM K-Pipes (pH 6.9), 0.5 mM MgSO4, 0.5 mM EGTA, 10% glycerol, and 0.7 mM GTP. (B to D) Polymerization of OdinTubulin at 6, 2, and 1 μM, respectively, under the same solution conditions. (E) Polymerization of OdinTubulin (0.5 μM) in 100 mM K-Pipes (pH 6.9), 100 mM NaCl, 0.5 mM EGTA, and 0.7 mM GTP or (F) supplemented with 0.5 mM MgSO4. Scale bar, 5 μm. (G to L) EM of negatively stained samples. Two morphologies of filaments were observed. Bundles of straight filaments appeared in solutions of monovalent cations. (G) 100 mM NaCl, 2 mM EGTA with background K+, or (H) 100 mM KCl and 2 mM EGTA. Tubules were observed in the presence of divalent cations. (I) 100 mM NaCl, 2 mM MgSO4, and 2 mM EGTA with background K+. Bundled straight filaments were occasionally observed under these conditions (not shown). The temperature dependence of tubule assembly was monitored in 100 mM KCl, 2 mM MgSO4, and 2 mM EGTA. (J) 4°C, (K) 37°C, and (L) 80°C. Scale bar, 100 nm. (M) Light scattering profiles for native and H393D mutant OdinTubulin (8 μM) on polymerization. GTP (0.7 mM) control (cyan), H393D without (purple) or with GTP (mustard), and native OdinTubulin without (lilac) or with GTP at 30°C (pink) or 37°C (orange). A.U., arbitrary units.
Observation of negatively stained polymers by EM indicated that two forms of polymer could be assembled. In the absence of Mg2+, bundles of straight protofilaments assembled in solutions containing the monovalent cations, K+ or Na+ (Fig. 5, G and H). Inclusion of Mg2+ with a high concentration of Na+ led to a mixture of bundled protofilaments and tubules (Fig. 5I). By contrast, Mg2+/K+ solutions, in the absence of Na+, produced exclusively tubules (Fig. 5K). Because K+/Na+ and Mg2+ are able to bind to the GTP in OdinTubulin protofilaments (fig. S4), but hydrolysis is slow in the presence of Na+ and absence of Mg2+ (Fig. 3, B to E), we interpret the straight bundles to form from polymerized OdinTubulin protofilaments before significant GTP hydrolysis, whereas the tubules likely are formed simultaneously with GTP hydrolysis, allowing a structural transition to a curved morphology.
Next, light scattering was used to monitor the polymerization. OdinTubulin (8 μM) was observed to increase light scattering in the presence of Mg2+-containing polymerization buffer without GTP; however, the signal was noisy (Fig. 5M). Including GTP (0.7 mM) in the Mg2+-containing polymerization buffer led to a smooth increase in light scattering over a period of 30 min, reaching a steady state, consistent with orderly polymerization (Fig. 5M), and indicating that the nucleotide is required for polymerization.
Temperature dependence of filament formation
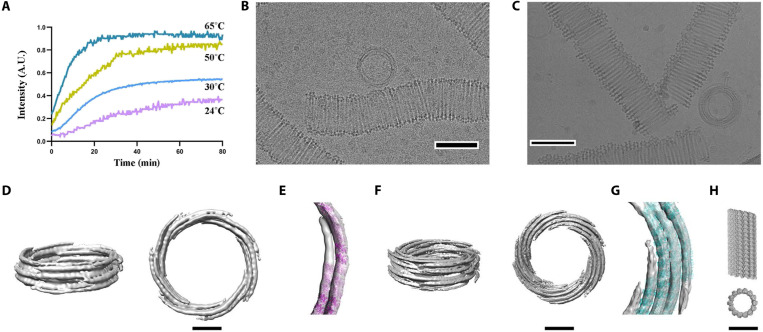
These initial assembly assays (Fig. 5M) revealed a slight difference in the polymerization rates at 30° and 37°C. We confirmed that tubules could be assembled at higher temperatures by EM. OdinTubulin forms tubules with a diameter of ~100 nm in the presence of K+/Mg2+ at 37° or 80°C, with more uniform and thicker-walled structures being formed at the higher temperature (Fig. 5, K and L). Similar to microtubules (25), OdinTubulin tubules were not observed at 4°C, rather immature rings formed that resemble templates for tubule formation (Fig. 5J). The thermostability of the OdinTubulin tubules is consistent with the temperature of the Yellowstone Lower Culex Basin hot spring (~70°C) from where Candidatus Odinarchaeota archaeon LCB_4 MAG was sampled (26). The temperature dependence of polymerization was further investigated by light scattering. The rate of polymerization increased from 24° to 65°C, the temperature limits of the spectrometer (Fig. 6A).
Fig. 6. OdinTubulin tubule architecture.
(A) OdinTubulin (8 μM) polymerization-monitored light scattering at different temperatures. (B) Cryo–electron micrograph of OdinTubulin (40 or 60 μM) polymerized at 37°C and (C) at 80°C, respectively. Scale bar, 100 nm. (D) Two orientations of the 3D reconstruction at 3-nm resolution of OdinTubulin polymerized at 37°C. (E) The crystal structure fitted into the reconstruction. (F) Two orientations of the 3D reconstruction at 4-nm resolution of OdinTubulin polymerized at 80°C and (G) with the fitted model. (H) Two views of the eukaryotic microtubule (11). Scale bars, 25 nm (D to H).
OdinTubulin tubule structure
The OdinTubulin tubules are semi-uniform but display sufficient homogeneity for calculation of low-resolution reconstructions from cryo-EM images (Fig. 6, B to G). The OdinTubulin crystal structure was placed into the EM density maps (Fig. 6, E and G). The OdinTubulin tubules are constructed from two to five layers of short discontinuous curved protofilaments that spiral around the wall of the tubule, approximately perpendicular to the tubule length. By contrast, straight protofilaments run along the length of eukaryotic microtubules (Fig. 6H). Tubules assembled at higher temperature (80°C) appeared more uniform and contained four to five layers (Fig. 6, F and G) relative to tubules assembled at lower temperature (37°C), which typically contained two to three layers (Fig. 6, D and E).
We propose that GTP hydrolysis and phosphate release in OdinTubulin protofilaments lead to curving, enabling assembly into tubules, and that the temperature dependence of polymerization can be understood at two levels. At the monomer level, a two-state thermodynamic equilibrium exists: the enthalpic-favored apo/GDP-bound and the entropically favored GTP-bound conformations. Elevated temperatures bias the monomer conformation toward the GTP-bound state and protofilament assembly. At the protofilament level, GTP hydrolysis and phosphate release rates will likely increase with temperature, favoring protofilament curving and tubule formation. This second mechanism explains the temperature dependence of the tubule geometries. The lower temperature structures (Fig. 6D), above the assembly temperature threshold, may result from incomplete hydrolysis in the protofilaments during tubule assembly, while higher temperature tubules result from complete GTP hydrolysis. Odinarchaeota have yet to be isolated; thus, the physiological role of the OdinTubulin tubules is unknown. Because of their relatively large diameters (~100 nm), we speculate that these tubules may use the straight-to-curved protofilament transition to shape membranes. The ~100-nm diameter of OdinTubulin tubules compares to the ~500-nm diameter of Candidatus Prometheoarchaeum syntrophicum MK-D1, a Lokiarchaeon, the only Asgard archaeon to be isolated to date (15, 27), which can produce long protrusions extending away from the main cell body of diameter ~90 nm. In comparison, fluorescence in situ hybridization images of Lokiarchaeota and Heimdallarchaeota suggest varied morphologies, with some diameters extending beyond 2 μm (28, 29). The dimensions and morphologies of Odinarchaeota are unknown; however, we speculate that OdinTubulin may play a role in subcellular membrane elongation, possibly in forming protrusions. We hypothesize that duplication of an ancient FtsZ/CetZ gene allowed the OdinTubulin protomer and protofilament to evolve and adopt functions outside cell division.
DISCUSSION
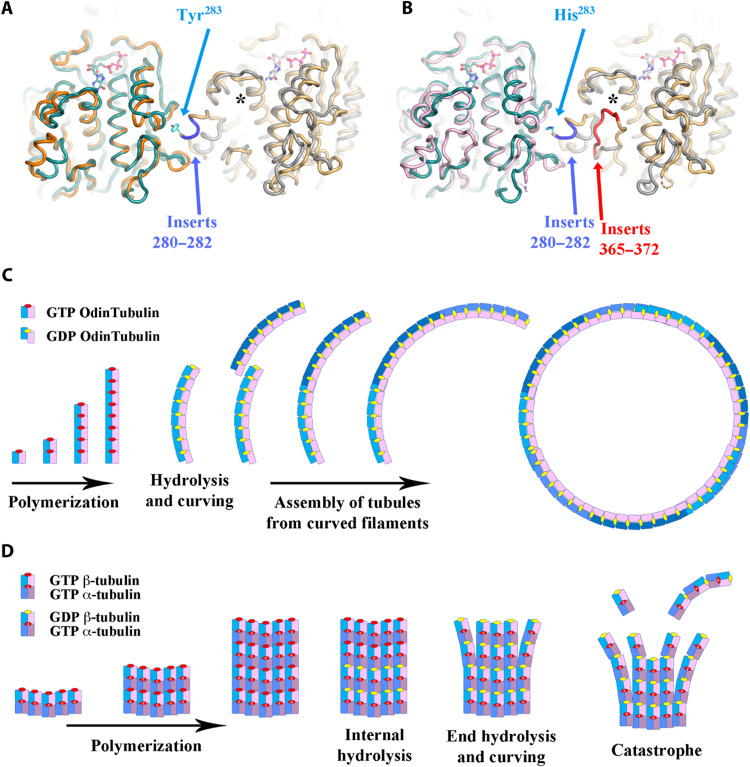
OdinTubulin forms protomers and protofilaments most similar to eukaryotic microtubules, yet assembles into ring systems more similar to FtsZ (3), indicating that OdinTubulin may represent an evolution intermediate between FtsZ and microtubule-forming tubulins. We speculate that segregation of chromosomes of increasing size, and possible enlargement of cell size, during eukaryogenesis, may have necessitated the emergence of stiffer tubules to navigate the increasing cellular distances and/or payloads, providing evolutionary pressure that would favor a switch from a malleable tubulin coil geometry to the stiffer parallel protofilament arrangement seen in microtubules. Such switches in filament suprastructure architecture, using similar protofilament assemblies, have occurred several times during actin-like and tubulin-like filament evolution (6, 30). α- and β-tubulins have a three–amino acid insertion (280 to 282), relative to OdinTubulin, which is involved in assembling the microtubule through interprotofilament interactions (Fig. 7, A and B, and fig. S6). We hypothesize that the evolution of the side-to-side interactions of tubulin protofilaments preceded gene duplication of the prototypical microtubule tubulin gene, because of the presence of the insertion in both α- and β-tubulins. We suggest that the microtubule architecture, composed of straight protofilaments, allowed for the straight-to-curved tubulin protofilament conformational change to be repurposed for catastrophe disassembly (Fig. 7, C and D). Following gene duplication, we hypothesize that divergence of the α- and β-tubulins will have resulted in changes to microtubule dynamics. From structural considerations, loss of GTP hydrolysis at the α-tubulin N-site, because of the glutamic acid–to–lysine substitution in the β subunit (Fig. 4, B and E), would likely result in relatively less strain, cooperativity, and sensing between subunits in the protofilament following GTP hydrolysis in the β subunits. In addition, an α-tubulin insertion (365 to 372), relative to OdinTubulin and β-tubulin, which interacts with the nucleotide sensor motif, may also contribute to reduced plasticity in the α-tubulin subunits. Together, we propose that these adaptations extended the transient stability of the straight form of the protofilaments in the early microtubule. Another gene duplication event allowed the emergence of γ-tubulin as a nucleation complex in a parallel scenario to actin gene duplication in the emergence of the ARP2/3 actin-filament nucleating complex (31).
Fig. 7. Microtubule adaptations in tubulin.
Structural superimposition of two OdinTubulin subunits (green and gray) onto two laterally related microtubule subunits: (A) β-tubulins (orange and yellow) or (B) α-tubulins (pink and yellow). The microtubule inserts 280 to 282 (blue), relative to OdinTubulin, create a protrusion used for interprotofilament contacts, centered around Tyr283 and His283 in β- and α-tubulins, respectively. The second α-tubulin inserts 365 to 372 (red) interact with the nucleotide sensor motif, indicated by an asterisk. (C and D) Cartoons depicting how the straight-to-curved protofilament transition is used in the two tubule systems. (C) OdinTubulin polymerizes as straight protofilaments, which curve on GTP hydrolysis. The curved protofilaments assemble into tubules. End on view. (D) α/β-Tubulin heterodimers assemble as tubules of typically 13 protofilaments (5 are shown). GTP hydrolysis in the central region of the tubule causes strain; however, the microtubule architecture prevents curving. GTP hydrolysis at the microtubule + ends leads to curving and dissociation, via the loss of curved stretches of protofilaments or monomers. GTP hydrolysis occurs in alternate subunits, reducing the strain and cooperativity within microtubule protofilaments, relative to OdinTubulin, leading to longer more stable protofilaments in the straight form. Side view.
It is notable from most phylogenetic analyses that Odinarchaeota are more distant from eukaryotes relative to other Asgard archaea, which do not have tubulin genes (12). Further discovery of Asgard lineages may clarify this tubulin gene distribution. At the present level of genome sequencing, horizontal transfer of the tubulin gene appears to be the most likely explanation for tubulin’s exclusivity to Odinarchaeota and eukaryotic lineages. The structural data presented here place some limitations on any such gene transfer. The transfer would be required to occur in an ancestor of Odinarchaeota to or from a proto-eukaryotic organism, potentially another Asgard archaea, before the switch to microtubule geometry and subsequent gene duplication. Thus, we propose that tubulin originated prior to eukaryogenesis and the switch to microtubule filament morphology was an important adaption during eukaryogenesis.
Further evidence for OdinTubulin representing a record of the prototypical tubulin before the evolution into microtubule-forming tubulins can be found in sequence analysis. Comparison of a hybrid human α/β-tubulin sequence, which includes the interface residues at the E-site from both α-tubulin and β-tubulin, increased the identity with OdinTubulin from 34 to 35% for α-tubulin and β-tubulin to 37.5% for the hybrid sequence, indicating that OdinTubulin represents a reasonable model of eukaryotic tubulin before the gene duplications (fig. S6). Thus, tubulin is an example in evolution, in which gene duplication coupled with sequence variation, without significant structural change to the core protein component (the tubulin protomer), gave rise to a novel complex protein machine, the microtubule. The microtubule is essential to eukaryotic chromosome segregation, and its emergence was likely a key event in eukaryogenesis. In summary, OdinTubulin appears to have the characteristics of a primordial tubulin before the transition into the eukaryotic microtubule-forming tubulins.
MATERIALS AND METHODS
Protein expression and purification
The Escherichia coli codon-optimized gene encoding the Odinarchaeota tubulin protein (OLS18786.1) was synthesized and placed in the pSY5 vector, which encodes an N-terminal human rhinovirus (HRV) 3C protease cleavage site and 8-histidine tag (14). The OdinTubulin mutation (H393D) was introduced using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) according to the manufacturer’s protocol. Plasmids were transformed into E. coli (DE3), the cells grown to a density of OD600 (optical density at 600 nm) = 0.8, and the protein expressed by induction with 0.5 mM isopropyl-d-1-thiogalactopyranodside (IPTG) at 18°C overnight. After centrifugation, cell pellets were resuspended in binding buffer [20 mM Hepes, 500 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP) (pH 7.5)], supplemented with Triton X-100 (0.01%), protease inhibitor cocktail (Set III, EDTA-free; Calbiochem), and benzonase (2 μl of 10,000 U/μl; Merck), or in EM buffer [100 mM Pipes, 500 mM NaCl, 50 mM imidazole, 10 mM MgSO4, and 2 mM EGTA (pH 6.9)]. Cells were lysed using an ultrasonic cell disrupter Vibra-Cell (Sonics). The protein was purified from the clarified supernatant using a Ni-NTA affinity chromatography column (HisTrap FF, GE Healthcare) with binding buffer and eluted through on column cleavage with HRV 3C protease. Affinity-purified protein was further purified by size exclusion chromatography (16/60 Superdex 75 PG, GE Healthcare) in the gel filtration buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM TCEP; or for EM samples: 100 mM Pipes (pH 6.9), 150 mM NaCl, 1 mM MgSO4, 2 mM EGTA, and 50 μM GTP]. Pure protein–containing fractions were identified by SDS–polyacrylamide gel electrophoresis, pooled and concentrated with 2000– to 10,000–molecular weight cutoff (MWCO) Vivaspin concentrators (Vivascience), and flash frozen in liquid nitrogen in small aliquots or used freshly.
Crystallization, structure determination, model building, and refinement
Native OdinTubulin and H393D crystallization trials, at 5 to 15 mg/ml in 20 mM Hepes (pH 7.5), 150 mM NaCl, and 1 mM TCEP, were performed using the sitting drop or vapor diffusion methods with a precipitant solution (1:1) at 293 K. Native OdinTubulin crystals were formed in 0.1 M bis-tris, pH 7.5, 25% (w/v) PEG 3350. These crystals diffracted x-rays poorly to 4 Å; however, the resulting dataset was amenable to successful molecular replacement using the Sus scrofa β-tubulin structure as a search model (PDB 6o2r, chain K) (19). Mutational analysis, of crystal contacts, identified a single–amino acid substitution (H393D) that had improved diffraction to 2.5 Å but did not alter protofilament packing; however, the nucleotide-binding site showed partial occupancy (7EVH; fig. S7 and table S1). Soaking of these crystals with GTP increased the diffraction limit and sharpened the electron density suitable for unambiguous structure determination (table S1). Crystals were frozen in the mother liquor. X-ray data were collected on RAYONIX MX-300 HS charge-coupled device detector on beamline Taiwan Photon Source (TPS) 05A [National Synchrotron Radiation Research Center (NSRRC), Taiwan, Republic of China] at λ = 1.0 Å or on BL41XU (λ = 1.0 Å) of SPring-8 on a Pilatus 6M detector.
Data were indexed, scaled, and merged following the standard protocols (16). Molecular replacement and refinement were carried out using PDB 6o2r chain K as the search model using standard methods to solve the 1.62-Å structure 7EVB (table S1) (16). The identity of the bound nucleotide was assessed by refinement of GTP, GDP, or a combination of GTP and GDP in the nucleotide-binding site. Subsequent soaks and alternate crystal structures are detailed in tables S1 and S2.
Polymerization assay
Polymerization of native OdinTubulin or H393D (8 μM) was induced by the addition of GTP (0.7 mM) in K-Pipes buffer [100 mM Pipes (pH 6.9), 0.5 mM EGTA, 0.5 mM MgSO4, and 10% (v/v) glycerol], total volume of 100 μl at various temperatures. Absorption at 340 nm was used to measure an increase in light scattering consistent with polymerization in 96-well, clear, flat-bottomed plates (Corning, Nunc). The plates were equilibrated (30 min) to the appropriate temperatures before the assays. Changes in absorbance were monitored with an Infinite M Nano+ plate reader (Tecan).
Interference reflection microscopy
Glass coverslips and slides were cleaned for 30 min in Hellmanex III (2% in water) at 60°C with sonication and rinsing in ultrapure water. Coverslips were dried using nitrogen gas flow. In vitro polymerization assays were performed using flow chambers with dimensions of 3 × 20 × 0.07 mm (width × length × height) that were assembled with double-sided tape as the spacer from 20 × 20–mm cover slip and slide. Brain tubulin elongation assay: Seeds (in the mix) were elongated with a mix containing 15 μM tubulin at 30°C in IRM buffer: 100 mM K-Pipes (pH 6.9), 0.5 mM MgSO4, 0.5 mM EGTA supplemented with 0.7 mM GTP, an oxygen scavenger cocktail [20 mM DTT, glucose (3 mg/ml), catalase (20 μg/ml), and glucose oxidase (100 μg/ml)], and 0.25% methyl cellulose (1500 centipoises, Sigma-Aldrich). OdinTubulin was similarly polymerized by addition of 0.7 mM GTP in the IRM buffer at the protein concentrations and cation conditions indicated in Fig. 5.
Nonlabeled microtubules and nonlabeled OdinTubulin filaments were imaged with IRM on an epifluorescence microscope (Eclipse Ti2, Nikon). The samples were illuminated with a SOLA Light Engine (Lumencor) through a cube equipped with a monochromatic filter at 520 nm, a 50/50 dichroic mirror, and a 60× numerical aperture 1.49 total internal reflection fluorescence (TIRF) objective. The microscope stage was kept at 30°C using a warm stage controller [LCI (Live Cell Instrument)]. Images and movies were captured using an Orca flash 4LT camera (Hamamatsu) every 5 s for 30 min.
Sequence and structure analyses
Tubulin, FtsZ, and CetZ structures were aligned using PDB codes 1rlu, 1w5e, 2vam, 2vaw, 3zid, 4b45, 4e6e, 4ffb, 5jco, 5mjs, 5n5n, 5ubq, 5w3j, 6e88, 6rvq, and 6unx. The resulting alignment was subjected to phylogenetic analysis using the published methods (16). Structure comparisons were carried out using the Dali server (http://ekhidna2.biocenter.helsinki.fi/dali/).
MD simulations
The all-atom MD simulations were performed by the software GROMACS 2019 (32). The Amber ff14SB force field (33) and TIP3P (34) water model were used for the protein molecule and solvent. The force field parameters for the GTP were taken from (35). The atomic coordinates of the tubulin subunits were taken from the PDB (code 6o2r) (19). The protein atoms were solvated in the truncated octahedral water boxes (with 28,413 water molecules). Na+, K+, and Cl− were added to neutralize the systems and to simulate the salt concentrations of [NaCl] = 10 mM and [KCl] = 150 mM. Covalent bonds involving hydrogen atoms were restrained by the LINCS algorithm (36). The system was first minimized by 50,000 steps using the steepest descent method and then equilibrated for 0.1 ns in the canonical ensemble (NVT) and another 1.0 ns in the isothermal-isobaric ensemble (NPT). After the equilibration simulations, the production simulations with the length of 1.0 μs were conducted. The temperature and pressure were controlled at 298.0 K and 1.0 atm, respectively. We performed seven independent simulations with different initial atom velocities. In calculating the occupancies, snapshots of the first 200 ns in each of the MD trajectories were omitted. MDTraj was used for analysis (37).
Negative staining of Odin tubulin
Negatively stained GTP-induced tubules were observed using a Hitachi-H7600 transmission electron microscope (Institute for Advanced Research, Nagoya University). Thawed OdinTubulin protein was diluted to a final concentration of 40 to 60 μM in prewarmed polymerization K-Pipes buffer as previously described for eukaryotic tubulin (38) and polymerized for 10 to 20 min using 2 mM GTP (Sigma-Aldrich). The mixture (2.5 μl) was applied onto glow-discharged grid STEM100Cu elastic carbon grids (Ohkenshoji Co. Ltd.) and absorbed for 1 min. The sample was blotted with filter paper, and then, 2% uranyl acetate solution (5 μl) was applied. After 1 min, the grids were blotted again and allowed to dry overnight.
Cryo-EM grid preparation
Molybdenum R1.2/1.3 and R2/2 200-mesh grids with a holey carbon support film (Quantifoil, Jena, DE) were glow discharged for 40 s under high vacuum shortly before sample application. OdinTubulin protein (40 to 60 μM) was polymerized for 10 to 20 min at 37° and 80°C using an adapted protocol for eukaryotic tubulin (38) by adding temperature-equilibrated K-Pipes buffer and 2 mM GTP (Sigma-Aldrich). The polymerized sample (2.5 μl) was applied to the glow-discharged grids, blotted for 0.5 to 1.5 s, and plunge frozen with an EM GP plunge freezer (Leica Microsystems) operated at room temperature at 90% humidity.
Cryo-EM data acquisition and image processing
Frozen molybdenum cryo-EM grids were initially screened on a JEOL JEM-3000SFF electron microscope (Cellular and Structural Physiology Institute, Nagoya University) operated at 200 kV at minimal dose system. Images were recorded using a K2 Summit camera with exposure settings of 1.35 Å per pixel size. Grids were subsequently imaged on a Titan Krios (FEI), at the Institute for Protein Research, Osaka University, equipped with the Field Emission Gun FEG operated at 300 kV and a minimal dose system. Imaging was performed using the EPU software (FEI) or SerialEM software (Nexperion) attached to the Titan Krios. Images of OdinTubulin incubated at 37°C were recorded at nominal magnification of 47,000, without using objective aperture, nominal defocus range of −2.0 to −2.6 μm with a dose rate of 40.05 e−/Å2, and exposure time of 2.42 s. Images were recorded using a Falcon III detector (FEI) at a pixel size of 1.45 Å per pixel and a frame rate of 60 frames per individual images. For incubation at 80°C, two sessions of data collection at a magnification of 64,000, without objective aperture, defocus range of −2.0 to −2.4 μm with a dose rate of 50 e−/Å2, and exposure time of 5.21 seconds, were used. Images were recorded with a K3 summit detector (FEI) in counting mode at a pixel size of 1.11 Å per pixel and a frame rate of 58 frames per image.
A total of 1217 (for 37°C) and 7600 (for 80°C) raw movies were collected and processed in RELION 3.0/3.1.1 (39). Drift was motion corrected with MotionCor2 (40), and the CTF for each micrograph was estimated with CTFFind-4.1 (41). Micrographs with good observed CTF estimations were selected for further processing. Tubules were picked manually using EMAN2 e2helixboxer (42) and extracted in RELION 3.0/3.1.1 with a 4 × 4 binning (box size of 250 × 250 pixels). Particles from two-dimensional (2D) classes displaying clear and similar structure and radius were selected. The helical pitch of the coil was determined using the 2D classes. A 3D classification was performed with a wide range of initial helical parameters using the helical pitch of the coil as a restriction. The 3D classes that had consistent projections with 2D classes and top views of the filaments were selected. Initial 3D reference models were prepared using RELION toolbox kit cylinder. Two rounds of 3D classification were performed. 3D refinement was performed with a reference model low pass filtered at 40 Å with solvent mask. Particles were reextracted with a 2 × 2 binning (box size of 500 × 500 pixels), and the final 3D refinement was performed.
Model fitting
In the crystals, the protofilaments are unable to bend because of the crystal packing. However, superimposing the apo-OdinTubulin (7EVG) onto two adjacent subunits from the GTP-bound protofilament (7EVB), via the C-terminal domains, led to a curved model that could be fitted into the EM density.
Acknowledgments
We thank the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology, and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology, Taiwan, ROC, and the SPring-8 Synchrotron, Japan. EM screening and data collection were supported by the Japan Agency for Medical Research and Development (AMED) grant number JP20am0101074 (A.O.) and the Collaborative Research Program of the Institute for Protein Research, Osaka University (CENCR-20-20). We thank E. Balıkçı for technical support.
Funding: This work was supported by JST CREST, Japan, grant number JPMJCR19S5 (S.A., A.N., and R.C.R); by the Japan Society for the Promotion of Science (JSPS), grant number JP20H00476; and by the Moore-Simons Project on the Origin of the Eukaryotic Cell, grant number GBMF9743. K.F. is supported by ELSI-First Logic Astrobiology Donation Program.
Author contributions: C.A., S.A., A.N., L.B., and R.C.R. conceived the experiments and analyzed the data. C.A., S.A., and L.T.T. performed biochemical experiments. C.A., L.T.T., and R.C.R. conducted x-ray experiments. J.G. performed IRM experiments, and W.L. conducted MD simulations. S.A., A.N., A.O., M.H., and K.H. conducted EM experiments. A.N., K.F., L.B., and R.C.R. supervised the work. R.C.R. wrote the manuscript. All authors edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The atomic coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 7EVB-D, 7EVG-L, and 7F1A-B. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Tables S1 to S4
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S9
REFERENCES AND NOTES
- 1.Wagstaff J. M., Tsim M., Oliva M. A., García-Sanchez A., Kureisaite-Ciziene D., Andreu J. M., Löwe J., A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. MBio 8, e00254-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duggin I. G., Aylett C. H. S., Walsh J. C., Michie K. A., Wang Q., Turnbull L., Dawson E. M., Harry E. J., Whitchurch C. B., Amos L. A., Löwe J., CetZ tubulin-like proteins control archaeal cell shape. Nature 519, 362–365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popp D., Iwasa M., Erickson H. P., Narita A., Maeda Y., Robinson R. C., Suprastructures and dynamic properties of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 285, 11281–11289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliva M. A., Cordell S. C., Lowe J., Structural insights into FtsZ protofilament formation. Nat. Struct. Mol. Biol. 11, 1243–1250 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Errington J., Daniel R. A., Scheffers D.-J., Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67, 52–65 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunning P. W., Ghoshdastider U., Whitaker S., Popp D., Robinson R. C., The evolution of compositionally and functionally distinct actin filaments. J. Cell Sci. 128, 2009–2019 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Löwe J., Amos L. A., Crystal structure of the bacterial cell-division protein FtsZ. Nature 391, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Zheng Y., Wong M. L., Alberts B., Mitchison T., Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Nogales E., Wolf S. G., Downing K. H., Structure of the αβ tubulin dimer by electron crystallography. Nature 391, 199–203 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Ravelli R. B. G., Gigant B., Curmi P. A., Jourdain I., Lachkar S., Sobel A., Knossow M., Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428, 198–202 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Zhang R., LaFrance B., Nogales E., Separating the effects of nucleotide and EB binding on microtubule structure. Proc. Natl. Acad. Sci. U.S.A. 115, E6191–E6200 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaremba-Niedzwiedzka K., Caceres E. F., Saw J. H., Backstrom D., Juzokaite L., Vancaester E., Seitz K. W., Anantharaman K., Starnawski P., Kjeldsen K. U., Stott M. B., Nunoura T., Banfield J. F., Schramm A., Baker B. J., Spang A., Ettema T. J. G., Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Spang A., Saw J. H., Jorgensen S. L., Zaremba-Niedzwiedzka K., Martijn J., Lind A. E., van Eijk R., Schleper C., Guy L., Ettema T. J. G., Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521, 173–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akıl C., Robinson R. C., Genomes of Asgard archaea encode profilins that regulate actin. Nature 562, 439–443 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Akıl C., Kitaoku Y., Tran L. T., Liebl D., Choe H., Muengsaen D., Suginta W., Schulte A., Robinson R. C., Mythical origins of the actin cytoskeleton. Curr. Opin. Cell Biol. 68, 55–63 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Akıl C., Tran L. T., Orhant-Prioux M., Baskaran Y., Manser E., Blanchoin L., Robinson R. C., Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea. Proc. Natl. Acad. Sci. U.S.A. 117, 19904–19913 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neveu E., Khalifeh D., Salamin N., Fasshauer D., Prototypic SNARE proteins are encoded in the genomes of Heimdallarchaeota, potentially bridging the gap between the prokaryotes and eukaryotes. Curr. Biol. 30, 2468–2480.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Yutin N., Koonin E. V., Archaeal origin of tubulin. Biol. Direct 7, 10 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eshun-Wilson L., Zhang R., Portran D., Nachury M. V., Toso D. B., Löhr T., Vendruscolo M., Bonomi M., Fraser J. S., Nogales E., Effects of α-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. U.S.A. 116, 10366–10371 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prota A. E., Bargsten K., Zurwerra D., Field J. J., Díaz J. F., Altmann K.-H., Steinmetz M. O., Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 339, 587–590 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Aldaz H., Rice L. M., Stearns T., Agard D. A., Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature 435, 523–527 (2005). [DOI] [PubMed] [Google Scholar]
- 22.La Sala G., Olieric N., Sharma A., Viti F., de Asis Balaguer Perez F., Huang L., Tonra J. R., Lloyd G. K., Decherchi S., Díaz J. F., Steinmetz M. O., Cavalli A., Structure, thermodynamics, and kinetics of plinabulin binding to two tubulin isotypes. Chem 5, 2969–2986 (2019). [Google Scholar]
- 23.Downing K. H., Nogales E., Crystallographic structure of tubulin: Implications for dynamics and drug binding. Cell Struct. Funct. 24, 269–275 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Mahamdeh M., Simmerts S., Luchniak A., Schäffer E., Howard J., Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. J. Microsc. 272, 60–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borisy G. G., Olmsted J. B., Klugman R. A., In vitro aggregation of cytoplasmic microtubule subunits. Proc. Natl. Acad. Sci. U.S.A. 69, 2890–2894 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker B. J., Saw J. H., Lind A. E., Lazar C. S., Hinrichs K.-U., Teske A. P., Ettema T. J. G., Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat. Microbiol. 1, 16002 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Imachi H., Nobu M. K., Nakahara N., Morono Y., Ogawara M., Takaki Y., Takano Y., Uematsu K., Ikuta T., Ito M., Matsui Y., Miyazaki M., Murata K., Saito Y., Sakai S., Song C., Tasumi E., Yamanaka Y., Yamaguchi T., Kamagata Y., Tamaki H., Takai K., Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577, 519–525 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salcher M. M., Andrei A.-Ş., Bulzu P.-A., Keresztes Z. G., Banciu H. L., Ghai R., Visualization of Lokiarchaeia and Heimdallarchaeia (Asgardarchaeota) by fluorescence in situ hybridization and catalyzed reporter deposition (CARD-FISH). mSphere 5, –e00686-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avcı B., Brandt J., Nachmias D., Elia N., Albertsen M., Ettema T. J. G., Schramm A., Kjeldsen K. U., Spatial separation of ribosomes and DNA in Asgard archaeal cells. ISME J. 16, 606–610 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang S., Ghoshdastider U., Narita A., Popp D., Robinson R. C., Structural complexity of filaments formed from the actin and tubulin folds. Commun. Integr. Biol. 9, e1242538 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson R. C., Turbedsky K., Kaiser D. A., Marchand J. B., Higgs H. N., Choe S., Pollard T. D., Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindahl E., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 33.Maier J. A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K. E., Simmerling C., ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 35.Meagher K. L., Redman L. T., Carlson H. A., Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 24, 1016–1025 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 37.McGibbon R. T., Beauchamp K. A., Harrigan M. P., Klein C., Swails J. M., Hernández C. X., Schwantes C. R., Wang L.-P., Lane T. J., Pande V. S., MDTraj: A modern open library for the analysis of molecular dynamics trajectories. Biophys. J. 109, 1528–1532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond D. R., Kain S., Newcombe A., Hoey C., Katsuki M., Cross R. A., Purification of tubulin from the fission yeast Schizosaccharomyces pombe. Methods Mol. Biol. 777, 29–55 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J., EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 to S4
Movies S1 to S9