Abstract
Conventional approaches to isolate and characterize nanobodies are laborious. We combine phage display, multivariate enrichment, next-generation sequencing, and a streamlined screening strategy to identify numerous anti–severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) nanobodies. We characterize their potency and specificity using neutralization assays and hydrogen/deuterium exchange mass spectrometry (HDX-MS). The most potent nanobodies bind to the receptor binding motif of the receptor binding domain (RBD), and we identify two exceptionally potent members of this category (with monomeric half-maximal inhibitory concentrations around 13 and 16 ng/ml). Other nanobodies bind to a more conserved epitope on the side of the RBD and are able to potently neutralize the SARS-CoV-2 founder virus (42 ng/ml), the Beta variant (B.1.351/501Y.V2) (35 ng/ml), and also cross-neutralize the more distantly related SARS-CoV-1 (0.46 μg/ml). The approach presented here is well suited for the screening of phage libraries to identify functional nanobodies for various biomedical and biochemical applications.
High-throughput mining of camelid immune repertoires identified broad and potent nanobodies from an immunized alpaca.
INTRODUCTION
Camelids, including llamas and alpacas, express unique immunoglobulins composed of just heavy chains (1). The antigen-binding variable fragment is a single domain that can be expressed recombinantly as a 15-kDa antibody fragment called VHH or nanobody. For applications where the functions of an Fc domain are not required, nanobodies have many advantages over their full-size antibody counterparts. Nanobodies can be produced at high quantities much more cost-effectively than monoclonal antibodies. Their small size and single-gene nature allow for easy cloning, modification, and functionalization and also permit better tissue penetration and, for imaging purposes, closer proximity of fluorophores or radioisotopes to the antigen. As a result, nanobodies have applications in cell biology (2), structural biology (3), cancer research (4), and immunology (5). In addition, nanobodies are ideal neutralizing molecules or perturbants of viruses. Potent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–neutralizing nanobodies target the receptor binding domain (RBD) of the spike protein. They neutralize the virus by blocking either angiotensin-converting enzyme 2 (ACE2) receptor interactions (6, 7) or other mechanisms such as the triggering of conformational changes (8). One affinity-matured nanobody fused to a human immunoglobulin G is currently in clinical development for treatment of SARS-CoV-2 infections (9).
Antigen-specific nanobodies are typically isolated from large immune libraries (10, 11) or, more recently, also from synthetic libraries (12, 13). Typically, these libraries are screened using robust phage display or yeast display techniques. To encompass the larger diversity of nonimmune and synthetic libraries, screening typically starts with ribosome display. Often, these screens only yield a handful of useful binders. Other screens using lentiviral nanobody libraries allow direct phenotypic readouts that are more productive and limit the time-consuming functional testing of individual identified binders (14). However, phenotypic readouts cannot be implemented in all cases. To address this gap, we combined the robust and versatile phage display with rapid high-throughput functional testing, both bridged by next-generation sequencing (NGS) and enrichment analysis. Applying this approach, we identify a panel of potent SARS-CoV-2–neutralizing nanobodies and provide detailed methodological descriptions, enabling easy implementation in other nanobody discovery workflows.
RESULTS
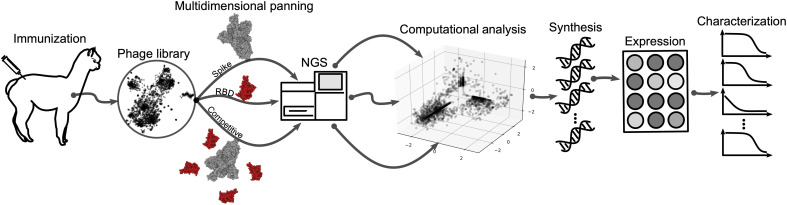
To generate a library of nanobodies against SARS-CoV-2, we immunized one alpaca four times with prefusion-stabilized SARS-CoV-2 spike protein, while the last three immunizations additionally included the RBD (Fig. 1). For each immunization, both proteins were injected separately into different flanks of the animal. Four days after the last immunization, we isolated peripheral blood mononuclear cells, amplified nanobody-specific regions, and constructed a phagemid library. We performed three parallel phage selections on proteins C-terminally immobilized on magnetic beads. The first screen was performed with recombinant spike protein (“S”), the second screen with RBD (“RBD”), and the third screen with immobilized spike in the presence of nonimmobilized RBD to deplete RBD-specific nanobodies from the phage pool (“ScRBD”). Thus, the RBD and ScRBD pannings should enrich for nonoverlapping sets of nanobodies, and the S panning should enrich for the union of these.
Fig. 1. Strategy overview.
A nanobody phage library is constructed from the immune repertoire of an immunized alpaca. This undergoes multiple independent panning steps, each enriching for distinct epitope targets. The original library and the enriched population from each panning are deeply sequenced. Computational enrichment analysis characterizes the repertoire and aids in the selection of nanobody variants for synthesis, expression, and downstream characterization.
Enrichment analysis by NGS
Phage pools were sequenced by Illumina, both before enrichment and after each panning step. The premise of our approach was that when the starting library contains a large number of distinct variants with highly variable frequencies [as is expected of immune repertoires (15)], the final frequency after panning is dominated by the starting frequency of a variant. The enrichment, the increase in frequency due to panning, which should be a better proxy for binding affinity, can be overwhelmed by the starting frequency. This suggests that the traditional approach of picking colonies after panning (11) may miss potent nanobodies with low starting frequencies.
We calculate the enrichment for each variant as the log ratio of the frequency after panning over before panning, regularized with a pseudocount (16) (to accommodate variants that are only observed after panning). To base our choices on enrichment, we need to know how reliable the estimates of enrichment are. Here, we exploit the fact that the library construction step includes a primer with degenerate bases, allowing us to consider independent versions of each variant that differ at a synonymous position. A correlation plot of the enrichment for two versions of each variant shows excellent agreement for RBD and ScRBD but weak agreement for S (see fig. S1). Possible explanations for this reduced correlation could be due to S offering more targets to the nanobody repertoire, increasing the competition and perhaps the stochasticity, or the introduction of a bottleneck at the start of the panning step, causing some variants to drop out. Regardless of the explanation, this indicates that enrichment calculated from the S panning may provide a less reliable signal of enrichment than RBD and ScRBD pannings.
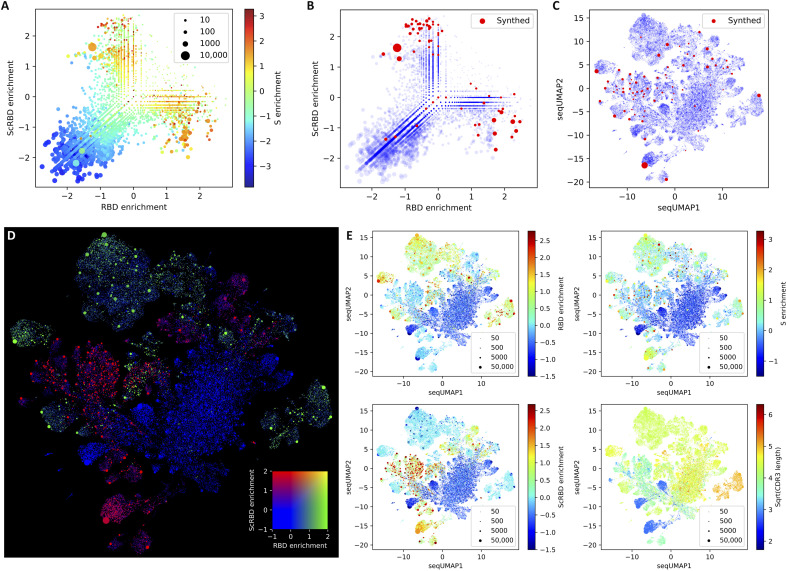
For further analysis, the cloning primer regions were ignored to provide total counts for all versions of each variant from which the final enrichment metrics were calculated. Figure 2A shows the RBD against the ScRBD enrichment. As intended by the panning design, there were very few sequence variants showing enrichment in both of these panning steps. Further, we color by S enrichment, which tends to be higher when either RBD or ScRBD enrichment is high, but the unreliability of the S enrichment identified by the barcode analysis is also visible at this level, especially for variants that were smaller in the baseline library.
Fig. 2. Multivariate repertoire analysis.
Panel (A) shows the enrichment across three parallel panning runs, with RBD panning on the x axis, spike with competing soluble RBD (“ScRBD”) on the y axis, and colored by spike (S). This both shows that a large number of unique nanobodies are enriched and confirms that RBD and ScRBD enrichment is mutually exclusive (by the lack of points in the top-right quadrant). Variants selected for further screening are shown both upon the enrichment plot (B), showing that we generally favored selection of the most enriched nanobodies, and upon a seqUMAP embedding of the nanobody sequences (C), which embeds nanobody sequences into two dimensions such that closely related variants are neighbors, allowing us to visualize nanobody “sequence space.” This facilitates the selection of nanobodies in a way that is sensitive to their relatedness and their enrichment metrics and shows that we avoided selecting nanobodies that were too closely related. Panel (D) shows which regions of seqUMAP space are targeting RBD (green), are targeting the non-RBD parts of spike (red), and are not SARS-CoV-2 specific at all (blue). The fact that the color cluster strongly shows that nanobodies with similar sequences are enriched under similar conditions, and the lack of double enrichment (yellow) in RBD and ScRBD confirms that these two panning runs enriched for mutually exclusive variants. Panel (E) shows RBD, S, and ScRBD enrichment separately and the CDR3 lengths (number of amino acids, square root transformed) for all nanobody variants overlaid on the seqUMAP plot.
Visualizing nanobody repertoire VDJ space
When selecting nanobody variants, a key dimension is their relatedness. For screening purposes, nanobodies with similar VDJ sequences should be avoided, but later, it might be useful to screen further candidates related to any promising hits. This would be aided by a way of visualizing sequence relatedness. One standard approach for visualizing a set of sequences would be a phylogeny or clustering dendrogram, but these are unwieldy for such large sequence datasets, often requiring multiple sequence alignments and behaving poorly for regions of problematic homology, which are especially common in nanobody complementarity-determining region 3 (CDR3s). Here, we adapt the uniform manifold approximation and projection [UMAP; (17)]—an approach that is now standard in the single-cell RNA sequencing literature and popular in many other domains but is not commonly used to visualize sequence data, possibly because of technical challenges. We circumvent these issues using kmer sequence embeddings (18) and call the resulting approach “seqUMAP,” which embeds sequences into two-dimensional (2D) space, such that closely related sequences are neighbors.
Figure 2 (C to E) shows seqUMAP embeddings of the entire nanobody repertoire, overlaid with different data for each variant. Where enrichment is plotted (especially for RBD and ScRBD, but less so for S), there is a notable spatial association with enrichment, showing that genetic relatedness strongly predicts whether a variant is enriched. The mutual exclusivity of RBD and ScRBD panning is recapitulated in Fig. 2D, where entire regions of VDJ space are enriched exclusively in one or the other, but not both.
We selected 72 nanobodies from across enrichment and VDJ space, shown in Fig. 2 (B and C). Because less is known about anti-spike nanobodies that do not target the RBD, we biased our selection to include approximately twice as many ScRBD-enriched candidates as RBD-enriched candidates.
Enrichment predicts binding
The 72 selected nanobodies were synthesized and cloned into a nanobody expression vector. Nanobodies were expressed in a 96-deep-well plate with a culture volume of 1 ml. Expressed nanobodies were retrieved from the periplasm by osmotic shock, and the periplasmic extract was analyzed by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining. A band corresponding to the nanobody was visible for most periplasmic extracts at ~15 kDa, alongside other bands typically of higher molecular weight (fig. S3). To get an estimate of the expression efficiency of the different nanobodies, we quantified the band intensity. For most nanobodies, expression efficiency and purity were sufficient to analyze binding specificity by enzyme-linked immunosorbent assay (ELISA) (spike- or RBD-coated) and antiviral activity by pseudotyped virus (PSV) neutralization assays.
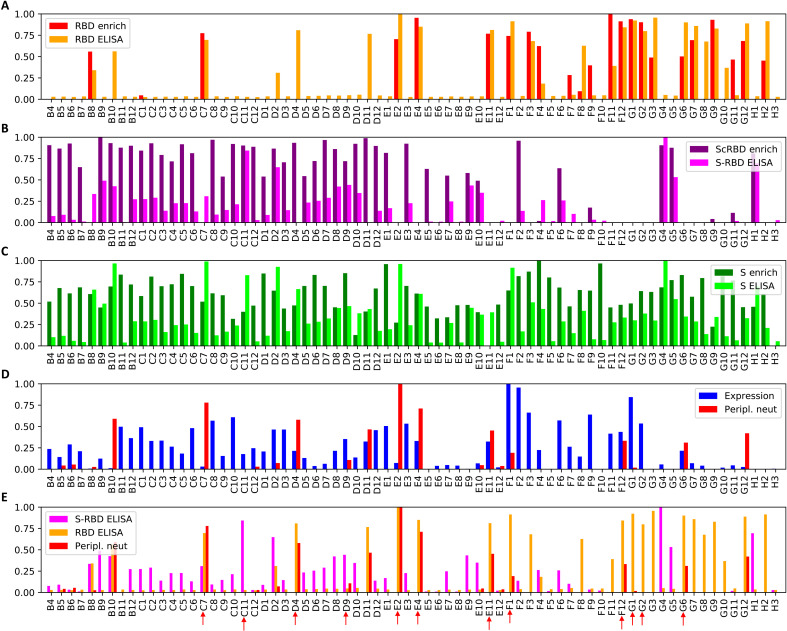
Figure 3 shows the enrichment metrics from the NGS data and the ELISA values for all selected nanobodies. We pair the ScRBD enrichment with the difference between the spike and RBD ELISA values. With only a few exceptions, nanobodies that were enriched for a particular target show ELISA signal for that target, with correlation coefficients of r = 0.72 for RBD (P < 10−5) and r = 0.66 for ScRBD (P < 10−5). The correlation for S was not significant, which is mostly because both spike and RBD targets exhibit S ELISA signal, reducing the variance, but may be due, in part, to the less reliable enrichment estimates for S than for RBD or ScRBD.
Fig. 3. Rapid nanobody screening.
Seventy-two nanobodies, selected from the multivariate analysis, were synthesized and expressed, and the crude periplasmic extract was screened for expression, binding, and neutralization. All values are normalized to the maximum value across nanobodies. Panels (A), (B), and (C) depict, for each nanobody, the enrichment calculated from the NGS data and the corresponding periplasmic extract ELISA, for RBD, ScRBD, and S, respectively. For panel (B), the S-RBD ELISA signal is the RBD optical density at 450 nm (OD450) subtracted from the S OD450 and should only be strongly positive when a nanobody binds the spike outside of the RBD. Together, panels (A) and (B) show that, for the vast majority of nanobody variants, the enrichment analysis is strongly predictive of whether the nanobody targets RBD or not. Panel (D) shows (log-domain) PSV neutralization IC50s and nanobody expression measured from the periplasmic extract. Panel (E) shows RBD ELISA, S-RBD ELISA, and PSV neutralization together, with nanobodies selected for subsequent investigation are highlighted with red arrows.
Neutralization
To identify nanobodies capable of neutralizing SARS-CoV-2, we used a high-throughput pseudotyped-virus neutralization assay, directly assessing neutralization by periplasmic extracts over four serial threefold dilutions. We observed a common baseline signal for inhibition at low dilutions relative to wells without periplasmic extract, which we therefore subtracted from all measurements. This screen identified a number of nanobodies displaying potent neutralizing capacity. Normalized log PSV neutralization titers are shown in Fig. 3 (D and E), alongside normalized expression results and ELISA values.
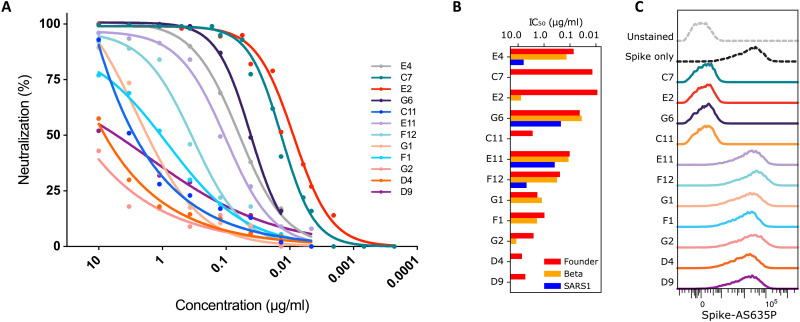
We selected 11 candidates for downstream analysis, including the most potently neutralizing RBD-specific nanobodies and several non-RBD binders, which were then expressed and purified. Neutralizing antibody titers of purified nanobodies were highly correlated with the preliminary periplasm screens, with nanobodies C7 and E2 displaying exceptional potency with median inhibitory concentrations (IC50s) in the range of 0.01 μg/ml (Fig. 4A). Note that E4 is identical to the “Fu2” nanobody that was isolated via more traditional colony picking from the same immunized animal and is extensively described elsewhere (19). Two of the selected non-RBD–specific nanobodies (C11 and D9) were also capable of neutralization, albeit weakly, with D9 exhibiting a maximal inhibition plateau at approximately 50% neutralization.
Fig. 4. Neutralization by nanobody monomers.
Eleven candidate nanobodies selected from the rapid screen, were profiled by pseudotyped lentivirus assay for neutralization activity. (A) Neutralization curves. (B) Neutralization IC50s against the SARS-CoV-2 founder variant (Wu-Hu-1), the Beta variant of concern (first described in South Africa, 501Y.V2/B.1.351), and the more distantly related SARS-CoV-1 (SARS1). (C) Intensity of labeled spike in a flow cytometry assay, showing whether a nanobody can prevent fluorescent spike protein from binding to HEK293T-hACE2 target cells, clarifying the mechanism of neutralization. Previously characterized nanobody Fu2/E4 is included in panels (A) and (B).
A subset of nanobodies are broadly neutralizing
Variants of concern (20–22) are rapidly rising in frequency. Some of these exhibit mutations that confer escape from prior immunity and from many existing monoclonal antibody therapy candidates (23). Given this context, one approach to addressing this problem is to attempt to discover broadly neutralizing biologics. Figure 4B shows that many of the identified nanobodies are sufficiently broad to neutralize both the SARS-CoV-2 “founder” variant and the Beta variant of concern (B.1.351/501Y.V2), sometimes without any reduction in potency. The most potently neutralizing nanobodies, C7 and E2, lack any meaningful cross-neutralization, but G6 is exceptionally potent against Beta (IC50 = ~35 ng/ml). Furthermore, two nanobodies (E11 and G6) show substantial cross-neutralization of SARS-CoV-1, which is a far more distantly related member of the Betacoronavirus genus, suggesting the targeting of a more conserved epitope.
Nanobodies do not need to bind the RBD or block ACE2 receptor interaction to neutralize SARS-CoV-2
Nanobodies have been shown to neutralize SARS-CoV-2 by various mechanisms, including direct competition with ACE2 (6, 7), locking of RBDs in an ACE2-inaccessible conformation (24), dimerizing spikes and agglutinating virions (19), or triggering the postfusion conformation and shedding of S1 (8). To test whether identified nanobodies interfere with the binding of the spike protein to the human ACE2 (hACE2) receptor, we performed a flow cytometry–based competitive binding assay. hACE2-expressing human embryonic kidney (HEK) 293T cells were stained with fluorescently labeled prefusion-stabilized spike trimers, either alone or preincubated with saturating amounts of the different nanobodies (Fig. 4C). Preincubation of fluorescently labeled spike protein with the most potent nanobodies, C7 and E2, completely abolished staining of the hACE2-positive cells. Similarly, G6 effectively prevented spike binding to ACE2. Unexpectedly, one nanobody (C11) that binds to a non-RBD epitope was still capable of preventing spike binding to ACE2-expressing cells. In contrast, other tested nanobodies did not reduce staining, suggesting that they neutralize SARS-CoV-2 by mechanisms other than blocking ACE2 interaction.
RBD-specific nanobodies bind with subnanomolar affinity
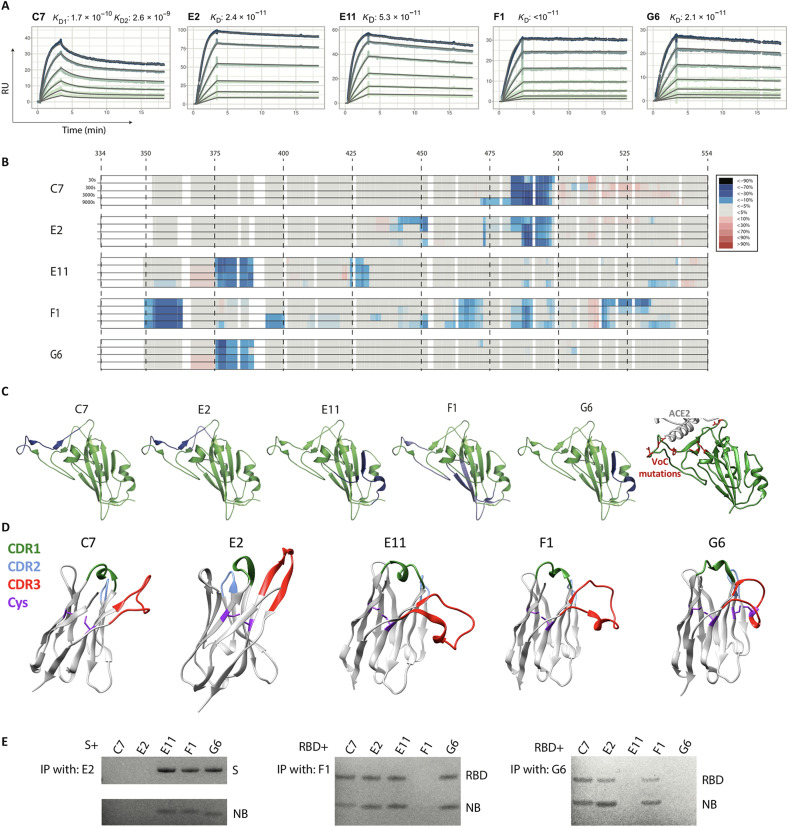
Binding affinities can affect neutralization potential. We determined the binding kinetics of five RBD-specific nanobodies by surface plasmon resonance (SPR). We observed high affinities in the picomolar range for all tested nanobodies (C7, E2, E11, F1, and G6) (Fig. 5A). F1, despite being only moderately neutralizing, showed the highest affinity, with a barely detectable dissociation rate in our assay, and an estimated dissociation constant to the RBD of <10−11. For the nanobody C7, there was a poor fit of the 1:1 Langmuir model, and the elution profile during size exclusion purification indicated the tendency for natural dimer- and multimerization. The heterogeneous binding model fit the C7 SPR sensorgrams well, confirming that two binding events are observed simultaneously: binding of C7 to the RBD and binding of C7 to C7. Overall, for these five nanobodies, binding affinities do not straightforwardly correlate with neutralization, highlighting the importance of the epitope location.
Fig. 5. RBD-targeting affinities and epitope mapping.
Five RBD-targeting nanobodies were selected for affinity characterization by SPR and for epitope mapping by hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS). (A) Picomolar affinities (170 to <10 pM) for all tested nanobodies. RU, resonance units; KD, dissociation constant. (B) HDX-MS signal across the RBD sequence, which is mapped onto the RBD structure in (C), together revealing three distinct epitope classes. (C) Right-most: The positions of all RBD mutations occurring in variants Alpha, Beta, Gamma, Delta, Kappa, Epsilon, Eta, Iota, and Lambda are shown in red. VoC, variant of concern. (D) AlphaFoldv2 (31, 32) predictions of the structure for these five nanobodies, highlighting the CDRs, and the cysteine pairs. Unlike the others, G6 shows an additional predicted disulfide bond between cysteines in the CDR3 and in FR2. (E) Immunoprecipitation (IP) competition analysis supporting the three epitope classes. C7 and E2, the two most potent neutralizers of SARS-CoV-2, target an epitope at the ACE2 interface, which explains both their potency and their lack of cross-reactivity. Nanobody (NB) G6 targets an epitope that is well conserved across the founder virus of SARS-CoV-2, the Beta variant, and SARS-CoV-1 (where there is only a single substitution relative to SARS-CoV-2), explaining its cross-reactivity, with E11 having a similar epitope. F1 has by far the largest epitope, which potentially explains the very low dissociation rate.
RBD-specific nanobodies bind to different epitopes on the RBD
To determine the binding sites of five RBD-binding nanobodies, we used hydrogen/deuterium exchange coupled to mass spectrometry (HDX-MS). This technology provides a powerful means to study protein dynamics and interactions in solution (25, 26). In the case of epitope mapping, it can be viewed as a comparison of deuterium uptake between two states of a protein (unbound and bound), where the interaction leads to a change in conformational stability and/or solvent accessibility. In brief, nanobody-RBD complexes and RBD were exposed to deuterated H2O, resulting in HDX, followed by denaturation and digestion with pepsin. Peptides were then analyzed by liquid chromatography–MS (LC-MS). Comparison of bound and unbound RBD then allows to identify areas with altered HDX. All the investigated nanobodies were found to exhibit clear and defined interaction sites on the RBD (Fig. 5, B and C). The structural resolution as defined by the degree of overlap of the peptides generated by pepsin digestion, i.e., the peptide map overlap, and the kinetics of the uptake is shown for respective state in data file S1. Nanobodies E2 and C7 appear to have a common strong binding site spanning amino acids 487 to 496 where a strong protection was observed, with a unique weaker site for E2 spanning 441 to 452 and an extension of the strong binding site for C7 to include 471 to 486. F1 was the nanobody that introduced the most deuteration protection on the RBD, with a strong interaction not only at amino acids 352 to 361 but also at 392 to 400, 462 to 470, 483 to 492, and 514 to 530, explaining its exceptional affinity. When annotating these sites on a 3D structure of the RBD, it appears that F1 engages a large continuous surface. Last, for G6 and E11, a common strong interaction site was observed spanning amino acids 375 to 387 and a unique E11 site at residues 423 to 431. G6, but not E11, has an additional cysteine pair connecting the CDR3 and Framework region 2 (FR2) (Fig. 5D). Although all nanobodies had at least one strong interaction site, it should be noted that from the HDX data, it is not possible to discriminate between a direct interaction surface and a possible conformational change in the RBD introduced by nanobody binding when several interaction sites are observed. We confirmed the three distinct epitopes by immunoprecipitation-based competition assay (Fig. 5E).
Epitopes in the RBD have been broadly classified into four classes based on overlapping epitopes frequently targeted by antibodies isolated from convalescent humans (27). Nanobodies C7 and E2 were mapped to a class 2–like epitope, consistent with their ability to compete with ACE2 for binding to RBD and the inability to neutralize the Beta variant harboring E484K. Nanobodies E11 and G6 were mapped to epitopes overlapping that of the monoclonal antibody COV2-2677 (28, 29), consistent with that of a class 4 antibody [that also more broadly includes the antibody CR3022 (30)]. However, the ability of G6 but not E11 to prevent spike binding to ACE2 suggests that it may use a different angle of approach. F1 appears to have a unique mode of recognition that does not map to any previously well-described epitopes.
Therapeutic potential of nanobodies
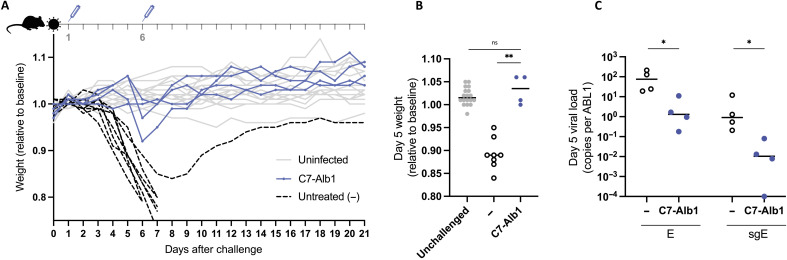
To evaluate the therapeutic potential of neutralizing nanobodies for the treatment of SARS-CoV-2 infection, we used transgenic mice that express hACE2 under the control of the cytokeratin-18 promoter (K18-hACE2 mice) (33). These mice are highly susceptible to SARS-CoV-2 infection and experience weight loss following infection that correlates with pathology and disease severity (34). One major limitation to the therapeutic application of nanobodies is their short half-life in vivo. We therefore conjugated C7 to a nanobody specific for albumin (Alb1) that has been demonstrated to increase serum half-life (35). Mice were challenged with 86 plaque-forming units (PFU) of SARS-CoV-2 (2.4 × 106 RNA genome copies) and subsequently treated with 320 μg of C7-Alb1 [in 160 μl of phosphate-buffered saline (PBS)] intraperitoneally (ip) on days 1 and 6 after infection. Untreated control mice experienced substantial weight loss, beginning around 4 days after challenge. Both weight loss and viral load in oropharyngeal swabs were significantly lower for mice treated with C7-Alb1 compared to untreated mice (Fig. 6). Although three animals treated with C7-Alb1 experienced some transient weight loss between days 5 and 6, weight loss was significantly reduced compared to untreated mice (Fig. 6, A and B). Furthermore, oropharyngeal viral loads in C7-Alb1–treated mice were approximately 100-fold lower than in untreated mice on day 5 (Fig. 6C). All but one of the untreated control mice succumbed to infection and had to be euthanized by day 7. However, all mice treated with C7-Alb1 survived, demonstrating the therapeutic efficacy of this neutralizing nanobody for SARS-CoV-2 infection.
Fig. 6. A half-life–extended nanobody heterodimer rescues K18-hACE2 mice from a lethal SARS-CoV-2 challenge.
K18-hACE2 mice were challenged with 86 PFU (2.4 × 106 genome copies) of SARS-CoV-2, produced in Calu-3 cells, and weight was monitored over time (A). Four mice received 320 μg of C7-Alb1 ip at days 1 and 6 after challenge. The mean weight of each mouse from days 0 to 2 served as the baseline, and the weight loss relative to this baseline is shown. Uninfected mice are shown in gray, and untreated infected mice are shown in black. (B) Weight loss at day 5 after infection is shown for treated (C7-Alb1) and untreated (−) mice and compared to cohoused mice that were not challenged (unchallenged). ns, not significant (P > 0.05). **P < 0.01. (C) Viral load for both genomic (E) and subgenomic (sgE) RNA in oropharyngeal swabs taken at day 5 is shown for treated (C7-Alb1) and untreated (−) mice. *P < 0.05.
Rapid click chemistry–based dimerization to identify potent homo- and heterodimer combinations
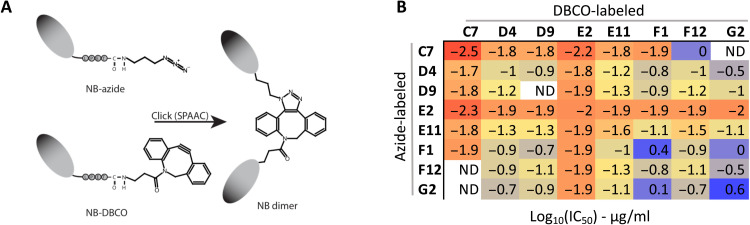
Nanobody dimerization can subtantially increase potency (36, 37). Combining nanobodies with distinct specificity can, in addition, limit viral escape (8). While molecular structures can be useful to select specific nanobody combinations, detailed structural information is not always readily available for larger pools of nanobodies. To identify potent dimers, we developed a screen to rapidly generate and test different nanobody combinations. We used sortase A functionalization and click chemistry to generate nanobody homo- and heterodimers as described in detail previously (36). In brief, using sortase A, nanobodies were C-terminally functionalized with a dibenzocyclooctyne (DBCO) or an azide. Labeled nanobodies were then mixed and incubated to permit sprain-promoted azide-alkyne cycloaddition (SPAAC) and C- to C-terminal fusion of azide- and DBCO-labeled nanobodies (Fig. 7A). Successful dimerization for all tested combinations was confirmed by SDS-PAGE analysis (fig. S4), with dimerization efficiencies ranging between 55 and 70%. Given that we were screening for substantial potency increases relative to the monomers, dimer reactions were not purified further before testing for their neutralization potential (Fig. 7B). The two most potent nanobodies C7 and E2 proved to be very potent in combination with any other nanobody. D9 and E11 proved to be good in combination with most other nanobodies. D4 and F12 displayed improved potency to varying degrees in combinations with the other nanobodies. F1 and G2 only performed well when combined with other potent neutralizers, and homo- or heterodimer combinations of these two appeared similar or worse than the monomeric versions.
Fig. 7. Rapid screening for potent nanobody dimer pairs.
C- to C-terminal–fused nanobody dimers were generated using a combination of sortase A functionalization and click chemistry (A). (B) A heatmap of the neutralization IC50s against SARS-CoV-2 founder virus for crude homo- and heterodimer reaction products. Neutralization curves for select purified homo- and heterodimers are shown in fig. S2. ND, not determined.
For more precise concentration and neutralization value determination, homo- and heterodimers incorporating G6, E11, and E2 were produced and purified at a larger scale (fig. S2). All dimers proved extremely potent. An E2 homodimer with a solubility enhancing polyethylene glycol 11 (PEG11) linker neutralized SARS-CoV-2 with an IC50 of approximately 0.7 ng/ml (23 pM). E11 and G6 homo- and heterodimers neutralized with similar potency between 10 and 20 ng/ml (333 to 667 pM). In conclusion, we show that high-throughput generation and screening of nanobody dimers can facilitate the rapid identification of extremely potent and synergistic combinations.
Lineage-level analysis
Nanobodies do not exist as independent sequences but as lineages arising from common VDJ recombination events that expand and undergo somatic hypermutation. Anti-spike, single-domain antibodies from the same lineage are expected to bind the same epitope but with varying affinities, and so identifying lineages can help guide both screening selection and downstream searches for optimized candidates. The grouping of sequence variants into lineage is not directly observed and must be inferred from the sequences themselves. While the analysis of antibody lineages is growing in popularity in the antibody repertoire sequencing community, it has not been widely adopted for nanobodies, which may be, in part, due to the poor availability of germline sequence data. Many strategies for grouping lineages take all sequences with the same V and J germline assignments and then cluster these on the basis of the distance between their CDR3 (38). One of the benefits of relying on V and J assignments is reduced computational burden. The number of pairwise CDR3 comparisons is quadratic in the number of variants that must be compared, and splitting into groups by V and J assignment markedly reduces the number of comparisons.
Here, we adopt a germline-naive lineage assignment strategy. Rather than V and J assignment, we use proximity in seqUMAP space to define a search neighborhood over which to compare CDR3s (see Materials and Methods for a description of how to do this efficiently) and merge variants into lineages.
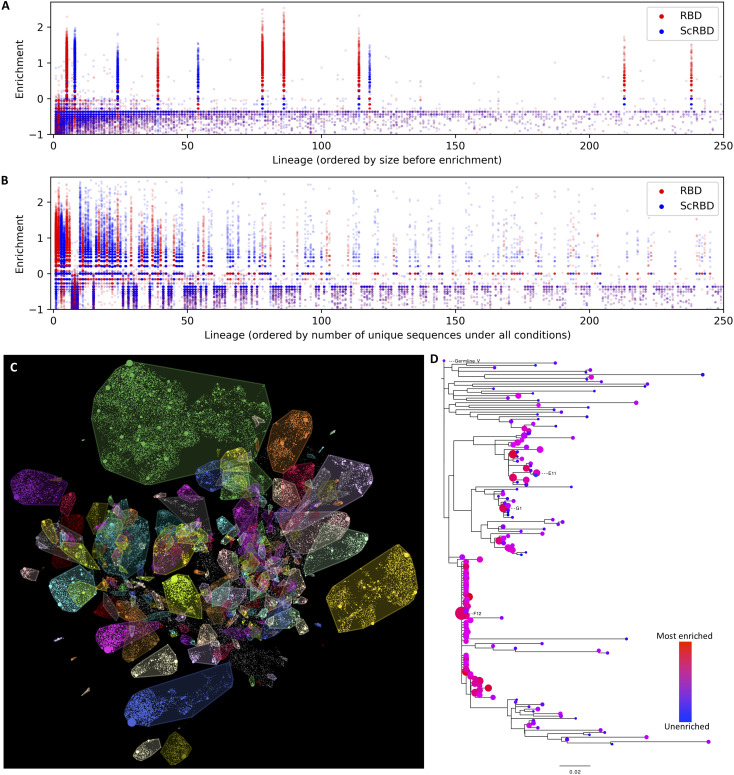
Figure 8 explores the lineage structure of this nanobody dataset. In some cases, multiple nanobodies were screened from the same lineage. For example, Fig. 8D shows that three nanobodies—E11, G1, and F12—arise from distinct clades of a single lineage. These three nanobodies are up to 17 amino acids apart from each other, exhibit a 20-fold difference in potency between the most and least potent, and can cross-neutralize the Beta variant and SARS-CoV-1. This lineage would be an excellent candidate for further screening—if another order of magnitude improvement were found in a variant on this lineage, this would be the most potent cross-neutralizing nanobody in the dataset.
Fig. 8. Nanobody lineages.
Grouping nanobody variants by their inferred lineage, sharing a common original VDJ rearrangement, allows us to interrogate the clonality of the alpaca immune response and the consistency of the enrichment and to identify other candidate nanobody variants that might have improved properties. (A and B) The enrichment of variants, organized by lineage, under RBD and ScRBD enrichment conditions. Similar to that at the variant level, enrichment at the lineage level is mutually exclusive, suggesting that lineages are each restricted to a single target. The lineage ordering in (A) shows that, before panning enrichment, only 11 of the largest 250 lineages are spike specific, and (B) shows how this shifts with a single round of panning. (C) Lineages overlaid on the seqUMAP embeddings and (D) the phylogeny for a subset of variants from a single lineage containing nanobodies E11, G1, and F12. Neutralization data show a 20-fold difference in potency between E11 and G1.
DISCUSSION
The use of nanobodies or nanobody-like proteins has increased greatly across a variety of applications in recent years. Although NGS-enabled analysis of immune repertoires is common for studying the elicitation and maturation of conventional antibodies (38–42), this approach is not yet standard for camelid repertoires nor commonly used for nanobody discovery. Here, we describe a rapid and straightforward approach to nanobody discovery that exploits the fact that, once established, libraries can be expanded indefinitely. This allows enrichment against multiple targets, which can be different subcomponents or variants of the same antigen. Multiplexed NGS of the starting library and of each distinct enrichment step provides us with massively parallel information about the affinity of individual nanobodies against each target. In contrast to conventional panning and colony picking, this approach relies on enrichment metrics instead of postpanning abundance, which enables the identification of high-affinity nanobodies even when they exist at low abundance in the baseline library, and are not sufficiently enriched to be sampled during traditional colony picking (see fig. S5). Another benefit is that, as this approach relies on enrichment metrics, it only requires a single panning round.
This could be expanded further to more complex antigens such as cell surfaces or different conformational states of proteins. Serendipitous discoveries have identified a number of nanobodies that recognize specific conformational states of proteins (43, 44). By panning, for example, against different conformational states and identifying nanobodies that are enriched in one but not the other, our approach could provide the means to rationally isolate nanobodies with such conformation specificity.
This high-throughput approach provides joint enrichment and genetic information, and the genetic information allows grouping the nanobodies into lineages that will share functional properties. While their affinity may differ, nanobodies from the same lineage will target the same epitope. The advantages of a lineage-based analysis are twofold: (i) It avoids redundancy arising from testing multiple highly related nanobodies in the initial screen, and (ii) once a promising lineage has been identified, information from across the lineage can be leveraged to identify or further optimize nanobodies with improvements in a particular function.
We demonstrate the utility of this approach using a nanobody library obtained from an alpaca immunized with recombinant SARS-CoV-2 spike proteins, isolating a number of extremely potent, neutralizing nanobodies. These included nanobodies capable of neutralizing SARS-CoV-2 in the low picomolar range and broadly neutralizing nanobodies capable of cross-neutralizing variants of concern and SARS-CoV-1.
Nanobodies E2 and C7 are among the most potent monomeric neutralizing nanobodies isolated to date, consistent with targeting of a “class 2” epitope (27) overlapping the ACE2-binding site, which is highly represented among potently neutralizing antibodies isolated to date (45, 46). Several neutralizing nanobodies have been evaluated against the Beta variant, and the frequent resistance to neutralization suggests that this is a frequent target for potently neutralizing nanobodies as well. Sziemel et al. (47) characterized three potent neutralizing nanobodies, and while one nanobody was similarly potent against Beta, the other two neutralizing nanobodies failed to cross-neutralize this variant. Furthermore, a single mutation (E484K) was sufficient to recapitulate this escape. Similarly, in another nanobody library, the Beta variant mutations (and E484K alone) were sufficient to almost completely abolish binding by two (of seven) of the most potently neutralizing nanobodies (48).
F1, while not a potent neutralizer, shows exceptional binding properties that are of substantial benefit for diagnostic applications, where favorable binding may improve the ability to detect small quantities of SARS-CoV-2 antigen. The binding to a unique and conserved epitope, with associated cross-reactivity to the Beta variant, is also desirable, because diagnostic applications need to maintain sensitivity in a landscape dominated by these variants of concern.
G6, E11, and E4 are almost as potent as E2/C7 but exhibit robust cross-neutralization of the Beta variant (B.1.351/501Y.V2). Other prevalent or antibody evasive variants of concern, such as Delta and Mu, also do not harbor mutations in the epitopes identified for G6 and E11, suggesting that these may provide broad cross-variant neutralization. This property is becoming progressively more critical as variants of concern continue to emerge. For G6 and E11, this breadth is further supported by their neutralization of SARS-CoV-1, which has a single amino acid mutation in the epitope region common to G6 and E11 and one additional mutation in the E11-specific epitope region. Among sarbecoviruses that have the potential to use ACE2 [lacking two common deletions in the RBD that prevent ACE2 use (49)], the overwhelming majority of these are identical in the G6 epitope to either SARS-CoV-1 or SARS-CoV-2, and because G6 neutralizes SARS-CoV-2 and tolerates the substitution in SARS-CoV-1, it is likely to exhibit substantial pan-sarbecovirus neutralization for ACE2 using sarbecoviruses.
We further show the therapeutic potential of nanobodies in a transgenic mouse model. A half-life–extended C7 construct (C7-Alb1) administered following infection was able to rescue all treated animals from a near-universally fatal SARS-CoV-2 challenge dose. C7-Alb1 administered on days 1 and 6 after infection substantially reduced pathology, demonstrated by significantly reduced oropharyngeal viral loads and weight loss.
To be applied as therapeutics or prophylactics, nanobodies may need to undergo “humanization” to reduce their immunogenicity. One potential strategy is to replace the nanobody framework regions with those from related human immunoglobulin genes (50). With G6 in particular, the extreme variation in the CDR1 and CDR2 of enriched members of the G6 lineage suggests that binding may be completely dominated by the CDR3, suggesting that humanization may be achievable without affecting neutralization potency. With its minimal binding footprint, a suitably humanized G6 construct would have exceptional potential as a broadly neutralizing therapeutic or prophylactic against SARS-CoV-2 and its many emerging variants.
MATERIALS AND METHODS
Protein and probes
The plasmid for the expression of the SARS-CoV-2 prefusion-stabilized spike (51) was a gift from the McLellan Lab. The plasmid was used for transient transfection of FreeStyle 293F cells using the FreeStyle MAX reagent (Thermo Fisher Scientific). The spike trimer was purified from filtered supernatant on Strep-Tactin XT resin (IBA Lifesciences) or Ni–nitrilotriacetic acid (Ni-NTA) resin and purified by size exclusion chromatography on a Superdex 200.
The RBD was cloned upstream of a sortase A motif (amino acid sequence LPETG) and a 6xHIS tag. The plasmid was used for transient transfection of FreeStyle 293F cells as described above. The protein was purified from filtered supernatant on His-Pur Ni-NTA resin followed by size exclusion chromatography on a Superdex 200.
Nanobodies were cloned in the pHEN plasmid with a C-terminal sortase motif (LPETG) and a 6xHIS tag. BL21 Escherichia coli were transformed with this plasmid, expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at optical density at 600 nm = 0.6, and cells were grown overnight at 30°C. Nanobodies were retrieved from the periplasm by osmotic shock and purified on Ni-NTA resin and size exclusion chromatography.
The albumin binding nanobody Alb1 was described earlier (35), and the sequence was obtained from WO/2006/122787. Sortase A 5M was produced as described before (36) in BL21 E. coli and purified by Ni-NTA and size exclusion chromatography.
Fluorescent spike ectodomain was generated by first attaching DBCO–N-hydroxysuccinimidyl ester to the spike trimer in a 3:1 molar ratio before attaching Abberior STAR 635P azide by click chemistry. The final product was purified from unreacted DBCO and fluorophore on a PD-10 desalting column.
The biotinylated RBD was generated using sortase A and amine-PEG3-biotin as a nucleophile. The reaction was performed with 50 μM RBD, 5 μM sortase A 5M, and 8 mM amine-PEG3-biotin for 6 hours at 4°C. Sortase A and unreacted RBD were removed on Ni-NTA resin, and excess nucleophile was removed by two consecutive purifications on PD-10 desalting columns. The biotinylated nanobodies were generated using sortase with a reaction of 100 μM nanobody, 5 μM sortase A 5M, and 8 mM amine-PEG3-biotin for 2 hours at 25°C. Sortase and unreacted nanobody were removed on Ni-NTA resin, and the nanobodies were purified by size exclusion chromatography or PD-10 desalting columns.
Alpaca library generation and phage selection
The alpaca nanobody library used in this study was already described and used here (19). In brief, one adult female alpaca (Funny) at Preclinics, Germany, was immunized four times in a 60-day immunization schedule. Each immunization consisted of two injections. For the first immunization, 200 μg of prefusion stabilized spike and 200 μg of S1 + S2 domain (Sino Biological) were used. In remaining immunizations, each consisted of one injection with 200 μg of RBD and one injection with 200 μg of prefusion-stabilized spike, both produced in FreeStyle 293F cells as described above. The animal study protocol was approved by the Preclinics animal welfare officer commissioner and registered under the registration no. 33.19-42502-05-17A210 at the Lower Saxony State Office for Consumer Protection and Food Safety—LAVES and is compliant with the Directive 2010/63/EU on animal welfare.
The nanobody phage library (size = 7.2 × 107) was generated as described by Hanke et al. (6). Phage display was performed on biotinylated RBD immobilized on streptavidin magnetic beads (Dynabeads M-280, Invitrogen) or strep-tagged spike immobilized on Strep-Tactin XT magnetic beads (IBA Lifesciences). To select for non-RBD binders, the phage enrichment on the spike protein was performed in the presence of nonimmobilized RBD.
NGS and enrichment analysis
Nanobody phage libraries were sequenced at baseline and, for each selection strategy, after each of two rounds of panning, as described by Hanke et al. (6). Briefly, phage libraries are amplified by polymerase chain reaction (PCR) and sequenced on an Illumina MiSeq instrument (2 × 300). Only metrics (e.g., enrichment) from the first panning round were used for downstream analysis, as an interim analysis showed that there was no clear ongoing enrichment during the second round. This is possibly because binding nanobodies were already at a saturating frequency after the first panning round.
The establishment of a nanobody phage library described above overwrites the N-terminal region of the VHH with a cloning primer, which includes ambiguous nucleotides. Here, we used this to investigate the internal consistency of each panning step. Sequencing primers were trimmed, and reads were collapsed by identity, retaining the frequency of each sequence. We performed an interim frequency analysis that compares the frequency of the two most frequent “versions” of each variant, where versions have identical sequences after the cloning primer but distinct ambiguities in the cloning primer sequence (considering the three N-terminal ambiguities to prevent bias driven by priming effects). If a panning run is consistent in which variants are enriched, then there should be agreement between two versions of each variant. Figure S1 shows that this is true of RBD and ScRBD panning but much less so of S panning. For downstream analysis, the cloning primer region was excluded when calculating variant frequencies.
To reduce sequence error and the volume of data for downstream analysis, we exclude all nonfunctional sequences (with early stop codons) and all variants that do not occur at least three times across all datasets. Enrichment is estimated as the change in frequency between pre- and postpanning sequence datasets in the log10 domain. To avoid undefined log ratios, we use pseudocount regularization when estimating frequencies. Enrichment is defined as: log10(Fpost + ε) − log10(Fpre + ε), where Fx is the proportion of reads from dataset x and ε is a small constant, which we choose to be the reciprocal of the number of unique variants across the entire dataset.
Antibody repertoires exhibit complex relatedness patterns, due to the nature of VDJ recombination, and subsequent somatic hypermutation. To visualize the relatedness of sequences, we use UMAP (17). UMAP first constructs a neighbor graph in the original high-dimensional space and then searches for a low-dimensional embedding that best preserves the similarity (measured by cross-entropy loss) between the original neighbor graph and the graph implied by the low-dimensional embedding. UMAP does not natively work on sequences (especially not unaligned sequences), and so we first embed sequences in a high-dimensional “kmer” space, in which the squared Euclidean distance between closely related sequences well approximates their Levenshtein distance (18), especially for sequences that are closely related. As is popular in single-cell RNA sequencing applications of UMAP, we first project the high-dimensional kmer representations into an intermediate space by principal components analysis (PCA) and then apply UMAP to these PCA coordinates. The approach (which we call seqUMAP) is implemented in the Julia language and will be described in detail in a forthcoming manuscript. We apply seqUMAP to embed the set of 68,123 functional VDJ sequences into two dimensions.
Lineages
Antibody repertoires are made up of distinct lineages. Within a lineage, antibodies share the same ancestor but differ by somatic hypermutation. Clustering sequences into lineages typically relies on accurate V and J gene assignments, as the search for lineages is, for computational reasons, constrained to occur within sequences that share the same V and J germline gene. However, alpaca V gene databases appear to be incomplete, so we preferred an alternative strategy for lineage calling.
We begin with our seqUMAP embedding of all of the nanobody variants. We construct a “k-d tree” (52) of the seqUMAP coordinates of all variants, exploiting this space-partitioning data structure (52) to efficiently define a neighbor graph for all points within a set radius (0.4 here) of each other in seqUMAP space. The CDR3 is the strongest signal for lineage membership, and we consider each edge in G and prune that edge if the CDR3s are too dissimilar. For efficient comparison of CDR3s, we again rely on kmer embeddings and a “kmer distance,” which approximates a length-normalized Levenshtein distance (18). For CDR3s that are the same length, we prune edges where the kmer distance is greater than 12.5%, and for CDR3s of unequal length, we only allow up to 10% kmer distance. Most lineage calling strategies do not allow for any CDR3 length variation, which is partly for computational considerations, as this avoids a large number of pairwise alignment comparisons, but our kmer approach avoids any additional computational cost of comparing CDR3s of different lengths. Because CDR3 length variation is less common, and because allowing CDR3s of different lengths to be considered as the same lineage can lead to overly permissive lineage merging, we use a stricter distance requirement for CDR3s of unequal length. After CDR3-based pruning edges in G, the connected components of G define our single-domain antibody lineages.
This lineage calling relies on consistent CDR3 calling. We used an alignment-based strategy to identify CDR3s. We constructed a multiple sequence alignment of all amino acid variants, defined the CDR3 as the portion of the alignment after the canonical cysteine to the end of the conserved DYW, and projected these regions back into nucleotide space, as the comparisons described above occur on CDR3 nucleotide sequences.
With the radii used for the analysis of our 68,123 variant sequences, the exploitation of the seqUMAP neighborhood reduces the number of pairwise CDR3 comparisons from 4.6 billion to 5.3 million, and the entire algorithm completes in around 15 s on a standard laptop.
Although the alpaca germline database is incomplete, it can serve as a sanity check on our lineage segmentation algorithm, which uses no germline assignment information. We assigned germline alleles using IMGT HighV-QUEST (53) and counted how often two random members of the same lineage have concordant V and J allele assignments. To quantify agreement, we sample a random lineage with a size of >2, then sample two unique sequences from the lineage (without replacement), and see if their germline assignments match (in case of ambiguous allele assignment, which happens in, for example, 7% of the nanobodies for V assignment, we allow a match when any of the calls are in common). Repeating this 1 million times and counting the proportions that are concordant give us an agreement metric. For V alleles, this pairwise agreement is 96.3%, and 98.7% for J alleles, which is fairly high considering that the algorithm is blind to these assignments, and we also expect the assignments to be sometimes incorrect. Inspecting pairs of nanobodies that were called, by our algorithm, to be within the same lineage but with discordant V allele calls, we then examined how diverged their CDR3 nucleotide sequences were (by edit distance). The modal value is zero. Around half of the pairs with different V calls have identical CDR3s, and the average CDR3 distance is just 2.2 nucleotides, which does not suggest that the lineage assignment is incorrect. Inspecting these pairs suggests two main explanations: (i) Somatic hypermutations in the V region are likely causing assignment to incorrect alleles (possibly due to the correct candidates not being part of the germline set) and (ii) chimeric variants that can be introduced during the establishment of the original nanobody library, which can have the first part of a sequence from one nanobody and the rest from another (note that these can be functional nanobodies, so we do not aim to exclude them). In this latter case, there is no correct answer to the lineage assignment question, because part of the sequence comes from the same lineage, and part of the sequence comes from a different one.
Cloning and expression of candidates
Selected nanobody sequences were ordered as eBlocks from Integrated DNA Technologies (IDT) with 20–base pair overhangs for Gibson assembly into a pHEN6 plasmid digested with Pst I and BstE II restriction enzymes. The plasmids then encoded for nanobodies followed by a sortase A motif (LPETG) and a HIS tag. The Gibson assembly was performed in a 96-well plate, and 2 μl of the assembly was directly used to transform BL21 E. coli and grown overnight in LB medium and a 96-well plate covered with AirPore tape sheets in a 37°C shaking incubator (>250 rpm). From this “master” plate, 20 μl of culture was used to start an expression plate (1 ml of LB per well in a 96-deep well plate). After 4-hour incubation at 37°C, nanobody expression was induced by addition of 1 mM IPTG (final concentration), and cells were grown at 30°C overnight with shaking. Cells in the expression plate were pelleted and resuspended in 100 μl of TES buffer [200 mM tris (pH 8), 0.65 mM EDTA, and 0.5 M sucrose] for 1 hour. Resuspended cells were then diluted in 300 μl of 0.25× TES buffer overnight. Cells were centrifuged, and periplasmic extracts were collected and directly used for expression level quantification, ELISA, and neutralization assays.
For nanobody candidates that were analyzed in more detail, plasmids from the original Gibson assembly were amplified in DH5α and verified by Sanger sequencing before being produced in larger quantities and purified by Ni-NTA affinity and size exclusion chromatography.
Neutralization assay
Pseudoviruses were generated by cotransfection of HEK293T cells with plasmids encoding firefly luciferase, a lentiviral packaging plasmid (Addgene, catalog no. 8455), and a plasmid encoding the spike protein (with a C-terminal truncation) from SARS-CoV (Addgene, catalog no. 170447), SARS-CoV-2 (54), or SARS-CoV-2 B.1.351 variant (Beta) (23). Medium was changed 12 to 16 hours after transfection, and PSVs were harvested at 48 and 72 hours, clarified by centrifugation, and stored at −80°C until use. PSVs sufficient to generate 100,000 relative light units (RLUs) were incubated with serial dilutions of nanobody for 60 min at 37°C. A total of 15 000 HEK293T-hACE2 cells were then added to each well, and the plates were incubated for 48 hours at 37°C. Luminescence was measured using Bright-Glo (Promega) on a GM 2000 luminometer (Promega) with an integration time of 0.3 s. Neutralizing antibody ID50 (median inhibitory dose) titers were calculated in Prism 9 (GraphPad Software) by fitting a four-parameter logistic curve bounded between 0 and 100 and interpolating the concentration/dilution where RLUs were reduced by 50% relative to control wells in the absence of nanobody.
For periplasmic extract PSV neutralization, low-level background inhibition was evident. For the rapid screen, we subtracted this background value from all IC50s for the results described in Fig. 3 and for candidate selection.
Flow cytometry
HEK293T-hACE2 cells were trypsinized and fixed in 4% formaldehyde in PBS for 20 min. Cells were stained with spike–Abberior STAR 635P not premixed or premixed with target nanobody or control nanobody. Fluorescence was quantified using a BD FACSCelesta and the FlowJo software package.
Surface plasmon resonance
Binding kinetics were determined by SPR using a Biacore 2000. All experiments were performed at 25°C in a running buffer of 10 mM Hepes, 150 mM NaCl (pH 7.4), and 0.005% Tween 20 (v/v). Site-specifically biotinylated RBD was immobilized on streptavidin sensor chips (Series S Sensor Chip SA, GE Healthcare) to a level of ~200 resonance units. A twofold dilution series of the nanobodies was injected at a flow rate of 30 μl/min (association, 180 s; dissociation, 900 s), and the immobilized RBD was regenerated using 0.1 M glycine buffer (pH 2) twice for 10 s. Data were analyzed using BIAevaluation Software and fitted using the 1:1 Langmuir model with mass transfer, except for C7 where we used the heterogeneous ligand model to account for the self-dimerization of this nanobody.
Epitope mapping by HDX-MS
All chemicals were from Sigma-Aldrich, pH measurements were made using a SevenCompact pH meter equipped with an InLab Micro electrode (Mettler-Toledo), and, before all measurements, a four-point calibration (pH 2,4,7, and 10) was made. The HDX-MS analysis was made using automated sample preparation on a LEAP H/D-X PAL platform interfaced to an LC-MS system, comprising an UltiMate 3000 Micro LC coupled to an Orbitrap Q Exactive Plus MS.
HDX was performed on an RBD, 0.6 mg/ml without and with nanobody (E2, C7, F1, G6, and E11), in PBS (pH 7.5) (Sigma-Aldrich, #D8537). All HDX-MS was done in one continuous run, with runs of the apo state made in between the nanobody runs. In total, eight replicates were made for the apo state. E2, C7, F1, and E11 were run in triplicate, and G6 in duplicate. Samples were run. Apo state samples constituted 2.5 μl of RBD and 2.5 μl of PBS, the interaction analysis samples constituted 2.5 μl of RBD mixed with 2.5 μl of ligand, and the samples were diluted with 30 μl of 10 mM PBS (pH 7.46) or HDX labeling buffer of the same composition prepared in D2O [pH(read) 7.10]. The HDX labeling was carried out for t = 0, 30, 300, 3000, and 9000 s at 20°C. The labeling reaction was quenched by dilution of 30 μl of labeled sample with 30 μl of 1% trifluoroacetic acid, 0.4 M Tris(2-carboxyethyl)phosphine (TCEP), and 4 M urea (pH 2.5) at 1°C, and 60 μl of the quenched sample was directly injected and subjected to online pepsin digestion at 4°C (in-house immobilized pepsin column, 2.1 mm by 30 mm). The online digestion and trapping were performed for 4 min using a 0.1% formic acid (FA) with a flow of 50 μl/min (pH 2.5). The peptides generated by pepsin digestion were subjected to online SPE on a PepMap300 C18 trap column (1 mm by 15 mm) and washed with 0.1% FA for 60 s. Thereafter, the trap column was switched in-line with a reversed-phase analytical column, Hypersil GOLD, with a particle size of 1.9 μm (1 mm by 50 mm); separation was performed at 1°C using a gradient of 5 to 50% B over 8 min and then from 50 to 90% B for 5 min; and the mobile phases were 0.1% FA (A) and 95% acetonitrile/0.1% FA (B). Following the separation, the trap and the column were equilibrated at 5% organic content until the next injection. The needle port and the sample loop were cleaned three times after each injection with mobile phase 5% MeOH/0.1% FA, followed by 90% MeOH/0.1% FA, and a final wash of 5% MeOH/0.1% FA. After each sample and blank injection, the pepsin column washed by injecting 90 μl of pepsin wash solution with 1% FA/4 M urea/5% MeOH. To minimize carryover, a full blank was run between each sample injection. Separated peptides were analyzed on a Q Exactive Plus MS, equipped with a heated electrospray ionization (HESI) source operated at a capillary temperature of 250°C with a sheath gas of 12 au, an auxiliary gas of 2 au, and a sweep gas of 1 au. For HDX analysis, MS full scan spectra were acquired at 70,000 resolution, automatic gain control of 3 × 106, maximum injection time of 200 ms, and scan range of 300 to 2000. For identification of generated peptides, separate undeuterated samples were analyzed using data-dependent tandem MS with higher-energy C-trap dissociation (HCD) fragmentation.
A summary of the HDX experimental detail is reported in file S1. The MS and HDExaminer analysis files have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (reference ID: 30395289).
HDX-MS data analysis
PEAKS Studio X Bioinformatics Solutions Inc. (Waterloo, Canada) was used for peptide identification after pepsin digestion of undeuterated samples. The search was done on a FASTA file with only the RBD sequence, and search criteria was a mass error tolerance of 15 parts per million and a fragment mass error tolerance of 0.05 Da, allowing for fully unspecific cleavage by pepsin. Peptides identified by PEAKS with a peptide score value of log P > 25, and no modifications were used to generate a peptide list containing peptide sequence, charge state, and retention time for the HDX analysis. HDX data analysis and visualization were performed using HDExaminer version 3.1.1 (Sierra Analytics Inc., Modesto, US). Each nanobody RBD complex state was analyzed and compared to its four closest (in time) apo state runs. The analysis was made on charge states 1 to 6 for each peptide, allowed only for EX2, and the two first residues of a peptide were assumed unable to hold deuteration. Because of the comparative nature of the measurements, the deuterium incorporation levels for the peptic peptides were derived from the observed relative mass difference between the deuterated and nondeuterated peptides without back-exchange correction using a fully deuterated sample (55). As a full deuteration experiment was not made, full deuteration was set to 75% of maximum theoretical uptake. The presented deuteration data are the average of all high and medium confidence results. The allowed retention time window was ±30 s. Heatmaps settings were uncolored proline, and heavy smoothing and the difference heatmaps were drawn using automatically calculated significance based on replicate variance. The spectra for all time points were manually inspected; low-scoring peptides, obvious outliers, and any peptides where retention time correction could not be made consistent were removed. As bottom-up labeling HDX-MS is limited in structural resolution by the degree of overlap of the peptides generated by pepsin digestion, the peptide map overlap is shown for respective state in file S1.
Nanobody competition assay
Immunoprecipitations for competition assays were performed with 8 μg of C-terminally biotinylated nanobody on M-280 streptavidin magnetic beads (Dynabeads, Invitrogen) and 10 μg of spike or RBD preincubated with 10 μg of the indicated (HIS-tagged) nanobodies. Bound spike or RBD was eluted in 0.2 M glycine (pH 2.2) and analyzed by SDS-PAGE and Coomassie staining.
SARS-CoV-2 challenge experiments
K18-hACE2 transgenic mice were purchased from the Jackson Laboratory and maintained as a hemizygous line. Experiments were conducted in biosafety level 3 facilities at the Comparative Medicine department (KM-F) at Karolinska Institutet. Ethics for studies of virus infection and therapeutic intervention were obtained from the Swedish Board of Agriculture (10513-2020). Mice were administered nanobodies as described in the Therapeutic potential of nanobodies section and challenged intranasally with 86 PFU SARS-CoV-2 in 40 μl of PBS following isoflurane sedation. Oropharyngeal sampling was performed on day 5 under light anesthesia with isoflurane. Weight and general body condition were monitored daily until weight drop started, whereupon mice were monitored twice daily. During the experiment, weight loss, changes in general health, breathing, body movement and posture, piloerection, and eye health were monitored. Mice were euthanized when they reached 20% weight loss or when movement was greatly impaired and/or they experienced difficulty breathing that was considered to reach a severity level of 0.5 on Karolinska Institutet’s veterinary plan for monitoring animal health. The weight loss in response to infection was highly reproducible. In Fig. 6, data from 50% of the untreated, challenged animals are historical controls from previous experiments performed under identical conditions. This challenge experiment was run at the same time as that from Hanke et al. (19), and the control mice were shared between both.
RNA extraction and reverse transcription quantitative PCR for SARS-CoV-2 detection
Viral RNA was isolated from buccal swabs collected 5 days after infection and stored in 500 μl of TRIzol reagent (Invitrogen). Total RNA extractions from buccal swab samples were performed using an adapted TRIzol manufacturer’s protocol with a 45-min precipitation step at −20°C. RNA pellets were resuspended in 20 μl of ribonuclease-free water.
Reverse transcription PCR (RT-PCR) reactions were performed using 4 μl of resuspended RNA in a 20-μl reaction volume using the SuperScript III one step RT quantitative PCR (RT-qPCR) system with Platinum Taq Polymerase (Invitrogen) with 400 nM concentrations of each primer and 200 nM probe. Primers and probes for the CoV-E gene target were as previously described (56). The following primers and probes for the ABL1 target were adapted from Ishige et al. (57) to enable detection of the murine homolog: ABL1_ENF1003_deg (5′-TGGAGATAACACTCTCAGCATKACTAAAGGT-3′), ABL1_ENR1063 (5′-GATGTAGTTGCTTGGGACCCA-3′), and ABL1_ENPr1043_deg (5′-HEX-CCATTTTTSGTTTGGGCTTCACACCATT-BHQ1–3′). The CoV-E and ABL1 TaqMan assays were run in multiplex. Detection of the subgenomic CoV-E target was adapted from Wölfel et al. (58) using a leader/E gene junction-specific forward primer: sgEjunc_SARSCoV2_F (5′-CGATCTCTTGTAGATCTGTTCTCTAAACG-3′). All oligonucleotides were synthesized by Eurofins Genomics.
Thermal cycling conditions for all assays consisted of RT at 55°C for 10 min, denaturation at 95°C for 3 min, and 45 cycles of 95°C for 15 s and 58°C for 30 s. Reactions were carried out using a CFX96 Connect Real-Time PCR Detection System (Bio-Rad) following the manufacturer’s instructions. To generate standard curves, a synthetic DNA template gBlock (IDT) was transcribed using the mMessage mMachine T7 Transcription Kit (Invitrogen) and serially diluted. To reduce sampling-related variability, SARS-CoV-2 RNA copies were normalized by ABL1 copies, and this ratio was used for comparisons. ABL1 copies were not significantly different between groups.
Generation of nanobody dimers by sortase-mediated functionalization and click chemistry
Nanobodies were functionalized site specifically on the C terminus using sortase A 5M with either an azide or a DBCO and subsequently dimerized by Cu-free SPAAC reaction as described earlier (36). Briefly, nanobodies at concentrations ranging from 75 to 205 μM were incubated with 5 μM sortase A, 8 mM DBCO-amine (Sigma-Aldrich, #761540), or 10 mM 3-azido-1-propanamine (Sigma-Aldrich, #762016) in 50 mM tris (pH 7.5), 150 mM NaCl, and 10 mM CaCl2 for 3 hours at 25°C. Unreacted nanobody, sortase A, and excess nucleophile were removed using Ni-NTA resin and Zeba spin desalting columns (2 ml; 7000–molecular weight cutoff; Thermo Fisher Scientific, #89890). Dimers were generated with a SPAAC reaction by incubating 30 μg of DBCO-functionalized and 30 μg of azide-functionalized nanobodies in a 96-well plate for 72 hours at 4°C. Reaction products were analyzed by SDS-PAGE (4 to 12% NuPAGE Bis-Tris, Life Technologies) and Coomassie G-250 staining. The relative amount of dimers in the gels (fig. S4) was quantified using ImageJ and an E4 homodimer as a reference. Some homodimers, when produced at larger scale (fig. S2), were generated by combining azide-functionalized nanobody with bis-PEG11-DBCO to increase solubility. These constructs were purified by size exclusion chromatography.
Acknowledgments
We gratefully thank J. Voss, D. Huang, and J. Bloom for reagents. We acknowledge P. Moore and the NICD (South Africa) for providing a B.1.351 spike plasmid, which was generated using the funding from the South African Medical Research Council. pCMV-dR8.2 dvpr was a gift from B. Weinberg (Addgene plasmid no. 8455; http://n2t.net/addgene:8455; RRID: Addgene_8455). pBOBI-FLuc was a gift from D. Nemazee (Addgene plasmid no. 170674; http://n2t.net/addgene:170674; RRID: Addgene_170674). We thank J. Klingstrӧm for providing Calu-3 cells and sharing the infectious SARS-CoV-2 isolate. We thank M. Kaduk for referring us to the AlphaFoldv2 implementation.
Funding: L.H. was supported by the David och Astrid Hageléns stiftelse, the Clas Groschinskys Minnesfond, and a Jonas Söderquist’s scholarship. This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 101003653 (CoroNAb; to B.M. and G.M.M.). The work was supported by project grants from the Swedish Research Council (2018-02381 to B.M.) (2018-03914 and 2018-03843 to G.M.M.).
Author contributions: Conceptualization: L.H., G.M.M., and B.M. Formal analysis: L.H., D.J.S., A.P., and B.M. Investigation: L.H., D.J.S., A.P., C.K., L.P.V., V.K., E.U., N.L.S., S.E., and B.M. Methodology: L.H., D.J.S., J.A.-W., S.E., J.M.C., G.M.M., and B.M. Visualization: L.H., D.J.S., and B.M. Resources: L.H., D.J.S., L.P.V., J.M.C., and G.M.M. Supervision: J.M.C., G.M.M., and B.M. Writing–original draft: L.H., D.J.S., and B.M. Writing–review and editing: L.H., D.J.S., J.M.C., G.M.M., and B.M. Funding acquisition: G.M.M. and B.M.
Competing interests: L.H., D.J.S., B.M., and G.M.M. are inventors on a patent application describing SARS-CoV-2 nanobodies, submitted by all four authors. The authors declare that they have no other competing interests.
Data and materials availability: NGS data is deposited at the Sequence Read Archive, under BioProject ID PRJNA814713. A preliminary implementation of the seqUMAP algorithm, in the Julia language for scientific computing, is additionally available at https://github.com/MurrellGroup/SeqUMAP.jl. Other codes for processing NGS nanobody repertoires, as well as postprocessed datasets, are additionally available at https://github.com/MurrellGroup/FuNGS. All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Tables S1 and S2
Figs. S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R., Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993). [DOI] [PubMed] [Google Scholar]
- 2.Nordeen S. A., Andersen K. R., Knockenhauer K. E., Ingram J. R., Ploegh H. L., Schwartz T. U., A nanobody suite for yeast scaffold nucleoporins provides details of the nuclear pore complex structure. Nat. Commun. 11, 6179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uchański T., Masiulis S., Fischer B., Kalichuk V., López-Sánchez U., Zarkadas E., Weckener M., Sente A., Ward P., Wohlkönig A., Zögg T., Remaut H., Naismith J. H., Nury H., Vranken W., Aricescu A. R., Pardon E., Steyaert J., Megabodies expand the nanobody toolkit for protein structure determination by single-particle cryo-EM. Nat. Methods 18, 60–68 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Y. J., Dougan M., Jailkhani N., Ingram J., Fang T., Kummer L., Momin N., Pishesha N., Rickelt S., Hynes R. O., Ploegh H., Nanobody-based CAR T cells that target the tumor microenvironment inhibit the growth of solid tumors in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 116, 7624–7631 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pishesha N., Harmand T., Smeding L. Y., Ma W., Ludwig L. S., Janssen R., Islam A., Xie Y. J., Fang T., McCaul N., Pinney W. 3rd, Sugito H. R., Rossotti M. A., Gonzalez-Sapienza G., Ploegh H. L., Induction of antigen-specific tolerance by nanobody-antigen adducts that target class-II major histocompatibility complexes. Nat. Biomed. Eng. 5, 1389–1401 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Hanke L., Vidakovics Perez L., Sheward D. J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G. B., Hällberg B. M., Murrell B., McInerney G. M., An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 11, 4420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huo J., Le Bas A., Ruza R. R., Duyvesteyn H. M. E., Mikolajek H., Malinauskas T., Tan T. K., Rijal P., Dumoux M., Ward P. N., Ren J., Zhou D., Harrison P. J., Weckener M., Clare D. K., Vogirala V. K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M. L., Tree J. A., Buttigieg K. R., Coombes N., Elmore M. J., Carroll M. W., Carrique L., Shah P. N. M., James W., Townsend A. R., Stuart D. I., Owens R. J., Naismith J. H., Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 27, 846–854 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Koenig P.-A., Das H., Liu H., Kümmerer B. M., Gohr F. N., Jenster L.-M., Schiffelers L. D. J., Tesfamariam Y. M., Uchima M., Wuerth J. D., Gatterdam K., Ruetalo N., Christensen M. H., Fandrey C. I., Normann S., Tödtmann J. M. P., Pritzl S., Hanke L., Boos J., Yuan M., Zhu X., Schmid-Burgk J. L., Kato H., Schindler M., Wilson I. A., Geyer M., Ludwig K. U., Hällberg B. M., Wu N. C., Schmidt F. I., Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science 371, eabe6230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. Schepens, L. van Schie, W. Nerinckx, K. Roose, W. Van Breedam, D. Fijalkowska, S. Devos, W. Weyts, S. De Cae, S. Vanmarcke, C. Lonigro, H. Eeckhaut, D. Van Herpe, J. Borloo, A. F. Oliveira, J. P. Portela Catani, S. Creytens, D. De Vlieger, G. Michielsen, J. C. Zavala Marchan, G. D. Moschonas, I. Rossey, K. Sedeyn, A. Van Hecke, X. Zhang, L. Langendries, S. Jacobs, S. Ter Horst, L. Seldeslachts, L. Liesenborghs, R. Boudewijns, H. J. Thibaut, K. Dallmeier, G. Vande Velde, B. Weynand, J. Beer, D. Schnepf, A. Ohnemus, I. Remory, C. S. Foo, R. Abdelnabi, P. Maes, S. J. F. Kaptein, J. Rocha-Pereira, D. Jochmans, L. Delang, F. Peelman, P. Staeheli, M. Schwemmle, N. Devoogdt, D. Tersago, M. Germani, J. Heads, A. Henry, A. Popplewell, M. Ellis, K. Brady, A. Turner, B. Dombrecht, C. Stortelers, J. Neyts, N. Callewaert, X. Saelens, S. Bert, van S. Loes, N. Wim, R. Kenny, V. B. Wander, F. Daria, D. Simon, W. Wannes, D. C. Sieglinde, V. Sandrine, L. Chiara, E. Hannah, V. H. Dries, B. Jimmy, O. A. Filipa, C. J. P. Portela, C. Sarah, D. V. Dorien, M. Gitte, M. J. C. Zavala, M. G. D., R. Iebe, S. Koen, V. H. Annelies, Z. Xin, L. Lana, J. Sofie, ter H. Sebastiaan, S. Laura, L. Laurens, B. Robbert, T. H. Jan, D. Kai, V. G. Vande, W. Birgit, B. Julius, S. Daniel, O. Annette, R. Isabel, F. C. S., A. Rana, M. Piet, K. S. J. F., R.-P. Joana, J. Dirk, D. Leen, P. Frank, S. Peter, S. Martin, D. Nick, T. Dominique, G. Massimiliano, H. James, H. Alistair, P. Andrew, E. Mark, B. Kevin, T. Alison, D. Bruno, S. Catelijne, N. Johan, C. Nico, S. Xavier, An affinity-enhanced, broadly neutralizing heavy chain–only antibody protects against SARS-CoV-2 infection in animal models. Sci. Transl. Med. 13, eabi7826 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maass D. R., Sepulveda J., Pernthaner A., Shoemaker C. B., Alpaca (Lama pacos) as a convenient source of recombinant camelid heavy chain antibodies (VHHs). J. Immunol. Methods 324, 13–25 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardon E., Laeremans T., Triest S., Rasmussen S. G. F., Wohlkönig A., Ruf A., Muyldermans S., Hol W. G. J., Kobilka B. K., Steyaert J., A general protocol for the generation of Nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zimmermann I., Egloff P., Hutter C. A., Arnold F. M., Stohler P., Bocquet N., Hug M. N., Huber S., Siegrist M., Hetemann L., Gera J., Gmür S., Spies P., Gygax D., Geertsma E. R., Dawson R. J., Seeger M. A., Synthetic single domain antibodies for the conformational trapping of membrane proteins. eLife 7, e34317 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moutel S., Bery N., Bernard V., Keller L., Lemesre E., de Marco A., Ligat L., Rain J.-C., Favre G., Olichon A., Perez F., NaLi-H1: A universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. eLife 5, e16228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt F. I., Hanke L., Morin B., Brewer R., Brusic V., Whelan S. P. J., Ploegh H. L., Phenotypic lentivirus screens to identify functional single domain antibodies. Nat. Microbiol. 1, 16080 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miho E., Roškar R., Greiff V., Reddy S. T., Large-scale network analysis reveals the sequence space architecture of antibody repertoires. Nat. Commun. 10, 1321 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H. Schütze, C. D. Manning, P. Raghavan, Introduction to Information Retrieval (Cambridge Univ. Press, 2008), vol. 39. [Google Scholar]
- 17.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I. W. H., Ng L. G., Ginhoux F., Newell E. W., Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Kumar V., Vollbrecht T., Chernyshev M., Mohan S., Hanst B., Bavafa N., Lorenzo A., Kumar N., Ketteringham R., Eren K., Golden M., Oliveira M. F., Murrell B., Long-read amplicon denoising. Nucleic Acids Res. 47, e104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L. Hanke, H. Das, D. J. Sheward, L. Perez Vidakovics, E. Urgard, A. Moliner-Morro, C. Kim, V. Karl, A. Pankow, N. L. Smith, B. Porebski, O. Fernandez-Capetillo, E. Sezgin, G. K. Pedersen, J. M. Coquet, B. M. Hällberg, B. Murrell, G. M. McInerney, A bispecific monomeric nanobody induces spike trimer dimers and neutralizes SARS-CoV-2 in vivo. Nat. Commun. 13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volz E., Mishra S., Chand M., Barrett J. C., Johnson R., Geidelberg L., Hinsley W. R., Laydon D. J., Dabrera G., O’Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C. V., Boyd O., Loman N. J., McCrone J. T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D. K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D. P.; COVID-19 Genomics UK (COG-UK) consortium, Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N. M., Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 593, 266–269 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E. J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A. J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.-Y., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C. K., Sewell B. T., Lourenço J., Alcantara L. C. J., Pond S. L. K., Weaver S., Martin D., Lessells R. J., Bhiman J. N., Williamson C., de Oliveira T., Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 592, 438–443 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Faria N. R., Mellan T. A., Whittaker C., Claro I. M., da S.Candido D., Mishra S., Crispim M. A. E., Sales F. C., Hawryluk I., McCrone J. T., Hulswit R. J. G., Franco L. A. M., Ramundo M. S., de Jesus J. G., Andrade P. S., Coletti T. M., Ferreira G. M., Silva C. A. M., Manuli E. R., Pereira R. H. M., Peixoto P. S., Kraemer M. U., Gaburo N., da C. Camilo C., Hoeltgebaum H., Souza W. M., Rocha E. C., de Souza L. M., de Pinho M. C., Araujo L. J. T., Malta F. S. V., de Lima A. B., do P. Silva J., Zauli D. A. G., de S. Ferreira A. C., Schnekenberg R. P., Laydon D. J., Walker P. G. T., Schlüter H. M., Santos A. L. P. D., Vidal M. S., Del Caro V. S., Filho R. M. F., Santos H. M. D., Aguiar R. S., Modena J. L. P., Nelson B., Hay J. A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Suchard M. A., Bowden T. A., Pond S. L. K., Wu C.-H., Ratmann O., Ferguson N. M., Dye C., Loman N. J., Lemey P., Rambaut A., Fraiji N. A., P. S. S. Carvalho M. d., Pybus O. G., Flaxman S., Bhatt S., Sabino E. C., Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wibmer C. K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B. E., de Oliveira T., Vermeulen M., van der Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., von Gottberg A., Cohen C., Morris L., Bhiman J. N., Moore P. L., SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 27, 622–625 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Schoof M., Faust B., Saunders R. A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C. B., Puchades C., Azumaya C. M., Kratochvil H. T., Zimanyi M., Deshpande I., Liang J., Dickinson S., Nguyen H. C., Chio C. M., Merz G. E., Thompson M. C., Diwanji D., Schaefer K., Anand A. A., Dobzinski N., Zha B. S., Simoneau C. R., Leon K., White K. M., Chio U. S., Gupta M., Jin M., Li F., Liu Y., Zhang K., Bulkley D., Sun M., Smith A. M., Rizo A. N., Moss F., Brilot A. F., Pourmal S., Trenker R., Pospiech T., Gupta S., Barsi-Rhyne B., Belyy V., Barile-Hill A. W., Nock S., Liu Y., Krogan N. J., Ralston C. Y., Swaney D. L., García-Sastre A., Ott M., Vignuzzi M., Consortium Q. C. R. G. S. B., Walter P., Manglik A., An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science 370, 1473–1479 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wales T. E., Engen J. R., Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z., Smith D. L., Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnes C. O., Jette C. A., Abernathy M. E., Dam K.-M. A., Esswein S. R., Gristick H. B., Malyutin A. G., Sharaf N. G., Huey-Tubman K. E., Lee Y. E., Robbiani D. F., Nussenzweig M. C., West A. P. Jr., Bjorkman P. J., SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zost S. J., Gilchuk P., Case J. B., Binshtein E., Chen R. E., Nkolola J. P., Schäfer A., Reidy J. X., Trivette A., Nargi R. S., Sutton R. E., Suryadevara N., Martinez D. R., Williamson L. E., Chen E. C., Jones T., Day S., Myers L., Hassan A. O., Kafai N. M., Winkler E. S., Fox J. M., Shrihari S., Mueller B. K., Meiler J., Chandrashekar A., Mercado N. B., Steinhardt J. J., Ren K., Loo Y.-M., Kallewaard N. L., McCune B. T., Keeler S. P., Holtzman M. J., Barouch D. H., Gralinski L. E., Baric R. S., Thackray L. B., Diamond M. S., Carnahan R. H., Crowe J. E. Jr., Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaney A. J., Starr T. N., Gilchuk P., Zost S. J., Binshtein E., Loes A. N., Hilton S. K., Huddleston J., Eguia R., Crawford K. H. D., Dingens A. S., Nargi R. S., Sutton R. E., Suryadevara N., Rothlauf P. W., Liu Z., Whelan S. P. J., Carnahan R. H., Crowe J. E. Jr., Bloom J. D., Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan M., Wu N. C., Zhu X., Lee C.-C. D., So R. T. Y., Lv H., Mok C. K. P., Wilson I. A., A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., Bridgland A., Meyer C., Kohl S. A. A., Ballard A. J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A. W., Kavukcuoglu K., Kohli P., Hassabis D., Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Ovchinnikov, M. Mirdita, M. Steinegger, ColabFold - Making protein folding accessible to all via Google Colab (2021);https://zenodo.org/record/5123297. [DOI] [PMC free article] [PubMed]
- 33.McCray P. B. Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H. P., Halabi C., Sigmund C. D., Meyerholz D. K., Kirby P., Look D. C., Perlman S., Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 81, 813–821 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winkler E. S., Bailey A. L., Kafai N. M., Nair S., McCune B. T., Yu J., Fox J. M., Chen R. E., Earnest J. T., Keeler S. P., Ritter J. H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M. J., Diamond M. S., SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 21, 1327–1335 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roovers R. C., Vosjan M. J. W. D., Laeremans T., el Khoulati R., de Bruin R. C. G., Ferguson K. M., Verkleij A. J., van Dongen G. A. M. S., van Bergen en Henegouwen P. M. P., A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int. J. Cancer 129, 2013–2024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moliner-Morro A., Sheward D. J., Karl V., Vidakovics L. P., Murrell B., McInerney G. M., Hanke L., Picomolar SARS-CoV-2 neutralization using multi-arm PEG nanobody constructs. Biomolecules. 10, 1661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibañez L. I., De Filette M., Hultberg A., Verrips T., Temperton N., Weiss R. A., Vandevelde W., Schepens B., Vanlandschoot P., Saelens X., Nanobodies with in vitro neutralizing activity protect mice against H5N1 influenza virus infection. J Infect Dis 203, 1063–1072 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Phad G. E., Pushparaj P., Tran K., Dubrovskaya V., Àdori M., Martinez-Murillo P., Bernat N. V., Singh S., Dionne G., O’Dell S., Bhullar K., Narang S., Sorini C., Villablanca E. J., Sundling C., Murrell B., Mascola J. R., Shapiro L., Pancera M., Martin M., Corcoran M., Wyatt R. T., Karlsson Hedestam G. B., Extensive dissemination and intraclonal maturation of HIV Env vaccine-induced B cell responses. J. Exp. Med. 217, e20191155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cirelli K. M., Carnathan D. G., Nogal B., Martin J. T., Rodriguez O. L., Upadhyay A. A., Enemuo C. A., Gebru E. H., Choe Y., Viviano F., Nakao C., Pauthner M. G., Reiss S., Cottrell C. A., Smith M. L., Bastidas R., Gibson W., Wolabaugh A. N., Melo M. B., Cossette B., Kumar V., Patel N. B., Tokatlian T., Menis S., Kulp D. W., Burton D. R., Murrell B., Schief W. R., Bosinger S. E., Ward A. B., Watson C. T., Silvestri G., Irvine D. J., Crotty S., Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell 180, 206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havenar-Daughton C., Carnathan D. G., Boopathy A. V., Upadhyay A. A., Murrell B., Reiss S. M., Enemuo C. A., Gebru E. H., Choe Y., Dhadvai P., Viviano F., Kaushik K., Bhiman J. N., Briney B., Burton D. R., Bosinger S. E., Schief W. R., Irvine D. J., Silvestri G., Crotty S., Rapid germinal center and antibody responses in non-human primates after a single nanoparticle vaccine immunization. Cell Rep. 29, 1756–1766.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landais E., Murrell B., Briney B., Murrell S., Rantalainen K., Berndsen Z. T., Ramos A., Wickramasinghe L., Smith M. L., Eren K., de Val N., Wu M., Cappelletti A., Umotoy J., Lie Y., Wrin T., Algate P., Chan-Hui P.-Y., Karita E.; IAVI Protocol C Investigators, IAVI African HIV Research Network, Ward A. B., Wilson I. A., Burton D. R., Smith D., Pond S. L. K., Poignard P., HIV envelope glycoform heterogeneity and localized diversity govern the initiation and maturation of a v2 apex broadly neutralizing antibody lineage. Immunity 47, 990–1003.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umotoy J., Bagaya B. S., Joyce C., Schiffner T., Menis S., Saye-Francisco K. L., Biddle T., Mohan S., Vollbrecht T., Kalyuzhniy O., Madzorera S., Kitchin D., Lambson B., Nonyane M., Kilembe W., Investigators I. A. V. I. P. C.; IAVI African HIV Research Network, Poignard P., Schief W. R., Burton D. R., Murrell B., Moore P. L., Briney B., Sok D., Landais E., Rapid and focused maturation of a VRC01-class HIV broadly neutralizing antibody lineage involves both binding and accommodation of the N276-glycan. Immunity 51, 141–154.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingram J. R., Knockenhauer K. E., Markus B. M., Mandelbaum J., Ramek A., Shan Y., Shaw D. E., Schwartz T. U., Ploegh H. L., Lourido S., Allosteric activation of apicomplexan calcium-dependent protein kinases. Proc. Natl. Acad. Sci. U.S.A. 112, E4975–E4984 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaur H., Hartmann J.-B., Jakob R. P., Zahn M., Zimmermann I., Maier T., Seeger M. A., Hiller S., Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR 73, 375–384 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Yuan M., Liu H., Wu N. C., Lee C.-C. D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T. F., Landais E., Sok D., Jardine J. G., Burton D. R., Wilson I. A., Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes C. O., West A. P. Jr., Huey-Tubman K. E., Hoffmann M. A. G., Sharaf N. G., Hoffman P. R., Koranda N., Gristick H. B., Gaebler C., Muecksch F., Lorenzi J. C. C., Finkin S., Hägglöf T., Hurley A., Millard K. G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P. D., Caskey M., Robbiani D. F., Nussenzweig M. C., Bjorkman P. J., Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell 182, 828–842.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. M. Sziemel, S.-H. Hwa, A. Sigal, G. Tyson, N. Logan, B. J. Willett, P. J. Durcan, Development of highly potent neutralising nanobodies against multiple SARS-CoV-2 variants including the variant of concern B.1.351. bioRxiv:439360 (2021).
- 48.Sun D., Sang Z., Kim Y. J., Xiang Y., Cohen T., Belford A. K., Huet A., Conway J. F., Sun J., Taylor D. J., Schneidman-Duhovny D., Zhang C., Huang W., Shi Y., Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting diverse and conserved epitopes. Nat. Commun. 12, 1–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wells H. L., Letko M., Lasso G., Ssebide B., Nziza J., Byarugaba D. K., Navarrete-Macias I., Liang E., Cranfield M., Han B. A., Tingley M. W., Diuk-Wasser M., Goldstein T., Johnson C. K., Mazet J. A. K., Chandran K., Munster V. J., Gilardi K., Anthony S. J., The evolutionary history of ACE2 usage within the coronavirus subgenus Sarbecovirus. Virus Evol. 7, veab007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincke C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., Conrath K., General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 284, 3273–3284 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C.-L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentley J. L., Multidimensional binary search trees used for associative searching. Commun. ACM. 18, 509–517 (1975). [Google Scholar]
- 53.Li S., Lefranc M.-P., Miles J. J., Alamyar E., Giudicelli V., Duroux P., Freeman J. D., Corbin V. D. A., Scheerlinck J.-P., Frohman M. A., Cameron P. U., Plebanski M., Loveland B., Burrows S. R., Papenfuss A. T., Gowans E. J., IMGT/HighV QUEST paradigm for T cell receptor IMGT clonotype diversity and next generation repertoire immunoprofiling. Nat. Commun. 4, 2333 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers T. F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R. K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M. J., Rawlings S. A., Wu N. C., Yuan M., Smith D. M., Nemazee D., Teijaro J. R., Voss J. E., Wilson I. A., Andrabi R., Briney B., Landais E., Sok D., Jardine J. G., Burton D. R., Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 369, 956–963 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engen J. R., Wales T. E., Analytical aspects of hydrogen exchange mass spectrometry. Annu. Rev. Anal. Chem. 8, 127–148 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brünink S., Schneider J., Schmidt M. L., Mulders D. G., Haagmans B. L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M. P., Drosten C., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 25, 2000045 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishige T., Murata S., Taniguchi T., Miyabe A., Kitamura K., Kawasaki K., Nishimura M., Igari H., Matsushita K., Highly sensitive detection of SARS-CoV-2 RNA by multiplex rRT-PCR for molecular diagnosis of COVID-19 by clinical laboratories. Clin. Chim. Acta 507, 139–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wölfel R., Corman V. M., Guggemos W., Seilmaier M., Zange S., Müller M. A., Niemeyer D., Jones T. C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C., Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2
Figs. S1 and S2
Data file S1