Abstract
Human TMEM175, a noncanonical potassium (K+) channel in endolysosomes, contributes to their pH stability and is implicated in the pathogenesis of Parkinson’s disease (PD). Structurally, the TMEM175 family exhibits an architecture distinct from canonical potassium channels, as it lacks the typical TVGYG selectivity filter. Here, we show that human TMEM175 not only exhibits pH-dependent structural changes that reduce K+ permeation at acidic pH but also displays proton permeation. TMEM175 constitutively conducts K+ at pH 7.4 but displays reduced K+ permeation at lower pH. In contrast, proton current through TMEM175 increases with decreasing pH because of the increased proton gradient. Molecular dynamics simulation, structure-based mutagenesis, and electrophysiological analysis suggest that K+ ions and protons share the same permeation pathway. The M393T variant of human TMEM175 associated with PD shows reduced function in both K+ and proton permeation. Together, our structural and electrophysiological analysis reveals a mechanism of TMEM175 regulation by pH.
Human TMEM175, a noncanonical potassium channel, conducts protons but displays reduced K+ permeation at lower pH.
INTRODUCTION
Lysosomes function as degradation stations and signaling hubs, which play vital roles in regulating cell growth, cell differentiation, cell metabolism, and immune responses (1). An acidic environment is required for maintaining the physiological functions of lysosomes. Vacuolar-type H+-adenosine triphosphatase (V-ATPase) is the primary driving force for establishing and maintaining the acidic pH of lysosomes by actively pumping in protons. In contrast, many channels and transporters in the membrane of lysosomes contribute to lysosomal pH homeostasis by providing counterions (2). The potassium (K+) channel named as transmembrane protein 175 (TMEM175) has been shown to mediate voltage-independent K+ conductance in endosomes and lysosomes, regulate the organelle membrane potential, and maintain pH stability of lysosomes in human cells (3). Knocking out TMEM175 abolished K+ conductance across the lysosomal membrane and led to overacidification of the lysosomal lumen under fed condition, more alkaline lysosomes under starved condition, and abnormal organelle fusion during autophagy (3). TMEM175 homologs in bacteria and archaea also form K+-permeable channels on the cell membrane, although their biological functions remain unknown (3). Hence, TMEM175 represents a previously unidentified potassium channel family that exists in all three domains of life.
Human TMEM175 (hsTMEM175) is associated with Parkinson’s disease (PD), first evident in a genome-wide association study (4). Subsequently, the most notable missense mutation, M393T, and a less frequent mutation, Q65P, have been characterized in molecular and cellular context in hsTMEM175 (4–11). Abnormal lysosomal degradation has been implicated in the pathogenesis of PD. For example, impaired activity of the lysosomal hydrolase glucocerebrosidase (GBA) substantially increased the risk of PD (12). In hsTMEM175 knockout cells, the activity of a few lysosomal hydrolases, including GBA, was significantly reduced (5), with impaired autophagosome clearance by lysosomes (5, 6). Thus, hsTMEM175 missense mutations impaired degradation of α-synuclein oligomers to increase the risk of PD, making hsTMEM175 a potential therapeutic target for treating PD (5, 6).
Recent progress in structural studies of TMEM175 has significantly advanced our understanding of the assembly, gating, and ion selectivity of TMEM175. Crystal structures of TMEM175 homologs from two bacterial species, Chamaesiphon minutus (cm) and Marivirga tractuosa (mt), were solved in closed conformations, which revealed homotetramers with six transmembrane (TM) helices for each monomer (13, 14). Cryo–electron microscopy (cryo-EM) structures of human TMEM175 at pH 8.0 revealed that hsTMEM175 assembles as a dimeric pseudotetramer, highly resembling the bacterial TMEM175 (15). hsTMEM175 at pH 8.0 adopts both open state and closed state with bound potassium ions in the ion permeation pore. Structural comparison between closed and open states of hsTMEM175 suggested that the conformational changes in the pore-lining helices may account for channel gating. Further structural analysis combined with electrophysiological recordings revealed a potential mechanism of TMEM175 ion selectivity by which the hydrophobic gate residues of isoleucine and key pore-lining hydrophilic residues together contribute to potassium selectivity (14, 15). Despite the tremendous progress, whether and how hsTMEM175 is regulated by pH, important questions for an endolysosomal channel, remain unexplored.
Using whole-cell electrophysiological recording, we found that lowering pH reduces potassium conductivity of TMEM175. In line with this, cryo-EM structures of hsTMEM175 showed that TMEM175 displays an open conformation at neutral pH and a closed conformation at lower pH. We further discovered that hsTMEM175 also generates proton currents that become apparent at low pH. Together with molecular dynamics simulation and structure-guided mutagenesis, we deduce that K+ and protons share a common permeation pathway. Collectively, our data reveal an unanticipated role for hsTMEM175 in balancing both pH and potassium homeostasis in lysosomes.
RESULTS
Reduced K+ binding and conductance at low pH
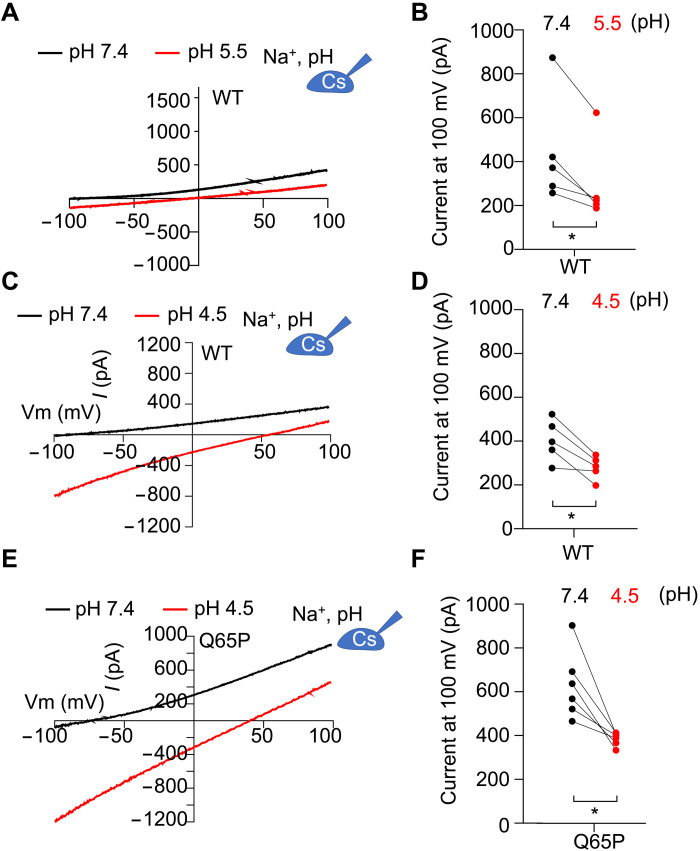
To probe whether and how pH regulates the channel activity of hsTMEM175, we used whole-cell electrophysiological recording in human embryonic kidney (HEK) 293T cells expressing hsTMEM175. As pH undergoes variations in a wide range from 7.4 to 4.5 in earlier endosomes to lysosomes, we assessed ion conductance of hsTMEM175 at pH 7.4, 5.5, and 4.5, respectively, in bath solution. The pH 7.4 represents the earlier endosomal lumen pH, while pH 5.5 and 4.5 mimic the lysosomal pH. We found that the outward current amplitude at 100 mV, representing K+ permeation, was significantly reduced at both pH 5.5 and 4.5 (Fig. 1, A to D, and fig. S1A), which is in line with our structural observation that the central ion permeation pathway is more restricted at a lower pH. We also observed that Q65P, a human variant protective against PD (7), displayed larger currents at pH 7.4 than wild type (WT), thus representing a gain-of-function mutant, and exhibited a more apparent reduction in K+ permeation at pH 4.5 (Fig. 1, E and F, and fig. S1, B and C). Together, our data revealed a previously unrecognized regulation of hsTMEM175 conductivity by pH.
Fig. 1. K+ conductivity of human TMEM175 regulated by pH.
(A) Representative current-voltage (I-V) curves obtained in whole-cell configuration from HEK293T cells expressing hsTMEM175 WT at pH 7.4 and 5.5. (B) Averaged currents measured at either 100 or −100 mV from (A) for both hsTMEM175 WT at pH 7.4 and 5.5 (N = 5), showing the difference between pH 7.4 and pH 4.5. *P < 0.05. (C) Representative I-V curves obtained in whole-cell configuration from HEK293T cells expressing hsTMEM175 WT at pH 7.4 and pH 4.5. (D) Averaged currents measured at 100 mV from (C) for both hsTMEM175 WT at pH 7.4 and pH 4.5 (N = 5), showing the difference between pH 7.4 and pH 4.5. *P < 0.05. (E) Representative I-V curves obtained in whole-cell configuration from HEK293T cells expressing hsTMEM175 Q65P mutant at pH 7.4 and pH 4.5. (F) Averaged currents at 100 mV of hsTMEM175 Q65P at pH 7.4 and pH 4.5 (N = 5), showing the difference at pH 7.4 or pH 4.5. *P < 0.05.
Proton conductance and selectivity by hsTMEM175
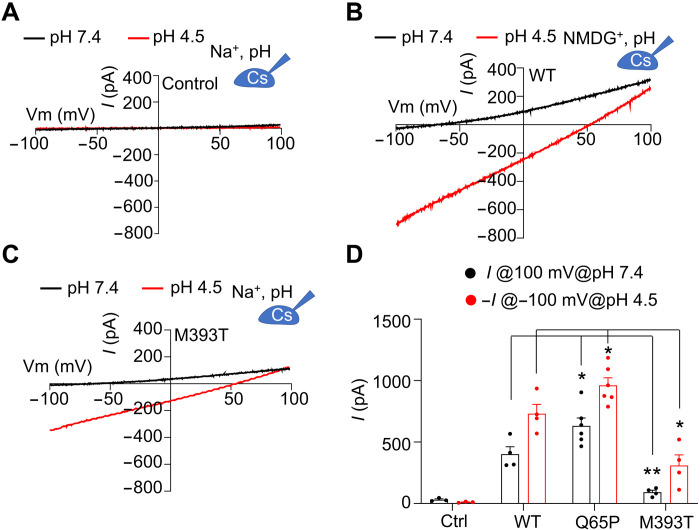
While characterizing the pH-regulated K+ conductance by TMEM175, we unexpectedly observed that pH 4.5 evoked significant inward currents at −100 mV in hsTMEM175 WT (Fig. 1C). As a control, cells without expression of hsTMEM175 exhibited no such inward current at pH 4.5 (Fig. 2A), suggesting that these currents are mediated by hsTMEM175.
Fig. 2. Proton conductance by TMEM175 WT and PD-associated mutants.
(A) Representative I-V curves obtained in whole-cell configuration from HEK293T cells without expressing hsTMEM175 at pH 7.4 and pH 4.5. The pipette solution contains 150 mM Cs+, and 150 mM Na+ was used in the bath solution. (B) Representative I-V curves from HEK293T cells expressing WT hsTMEM175 at pH 7.4 and pH 4.5, with 150 mM NMDG+ in the bath solution. (C) Representative I-V curves obtained in whole-cell configuration from HEK293T cells expressing hsTMEM175 M393T mutant at pH 7.4 and pH 4.5. (D) Averaged currents at 100 and −100 mV of control, hsTMEM175 WT, Q65P, and M393T at pH 7.4 and pH 4.5 (N = 5), showing the difference at pH 7.4 or pH 4.5. *P < 0.05; **P < 0.01.
Considering the high concentration of Na+ in the bath solution and low concentration of Cl− in both pipette and bath solutions, the inward currents could be induced by influx of either extracellular Na+ or protons. To distinguish these two possibilities, we substituted Na+ in the bath solution with equal molar of N-methyl-d-glucamine (NMDG+), a large impermeant cation, and found that the inward current was little affected (Fig. 2B), suggesting that these currents were not carried by Na+ but perhaps by protons. To further validate our conclusion, we performed pH imaging using the membrane-permeable pHrodo Green dye to monitor the proton influx mediated by hsTMEM175 in HEK293T cells. As predicted, both WT and a gain-of-function mutant, Q360L, conducted protons, leading to a transient increase in fluorescence, indicating acidification of transfected cells when exposed to a pH 4.5 solution (fig. S2, A to C). Nontransfected control cells and cells transfected with a loss-of-function mutant, I271W, did not display significant change in fluorescence, indicating little pH alteration (fig. S2, A to C). These data further support that hsTMEM175 is capable of conducting protons. We predict that in lysosomes, hsTMEM175 acts to prevent luminal overacidification from the constitutive inward proton pumping by V-ATPases; hence, its loss of TMEM175 function could lead to more acidic lysosomes. In line with this, the lysosomal lumen in TMEM175 knockout or knockdown cells is more acidic than those of WT cells under fed condition (5).
M393T mutant impairs proton conductance
Earlier studies have identified two TMEM175 mutants that are linked to PD pathogenesis. Of these mutants, M393T was found to promote PD pathogenesis, while Q65P is protective against PD. A recent study proposed that these mutants influence PD pathogenesis through modulating the K+ conductance (16). Compared with WT, M393T mutant decreases the K+ conductance, while Q65P increases the K+ conductance (Fig. 2, C and D, and fig. S1, B and C). Thus, the perturbation of lysosomal K+ homeostasis may be one factor contributing to the PD pathogenesis. We also observed that M393T also impairs the proton conductance of TMEM175 and Q65P promotes the proton conductance (Fig. 2, C and D). We propose that modulation of proton conductance by these mutants may also play an important role in PD pathogenesis because proton conductance of TMEM175 is directly related to lysosomal pH stability, which is critical for the function of lysosomal hydrolases and the degradation of α-synuclein. Therefore, the PD-associated, loss-of-function mutation M393T may also result in lysosomal overacidification and jeopardize the function of lysosomal enzymes such as GBA. Because GBA requires a narrow pH range of 4.7 to 5.5 for optimal activity (17), overacidification may lead to reduced GBA activity, which, in turn, may result in the accumulation of α-synuclein and PD pathogenesis.
Proton conductivity and selectivity by TMEM175
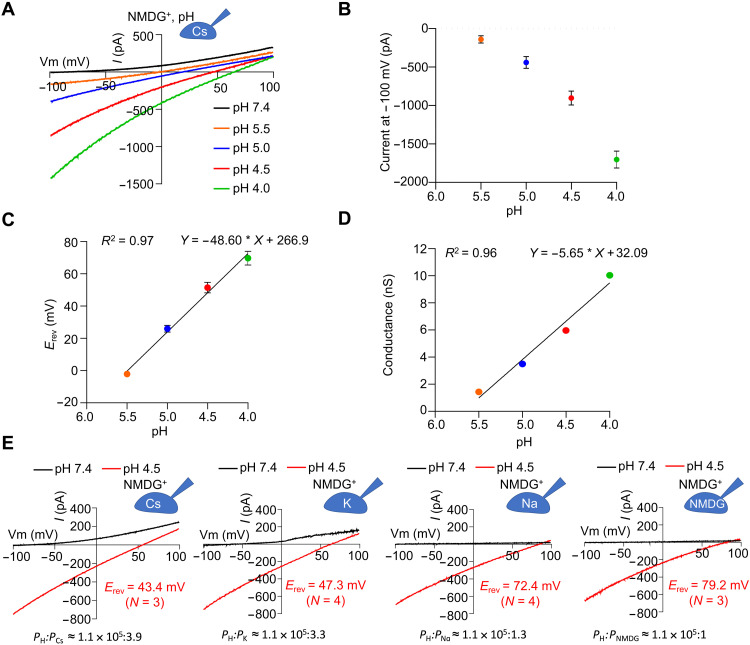
To further characterize proton permeation by hsTMEM175, we recorded the inward currents at a series of pH from 7.4 to 4.0 and found that the current amplitude at −100 mV increased as the pH decreased (Fig. 3, A and B). We then plotted the reversal potential (Erev) as a function of proton concentration or pH (Fig. 3C). For a selective ion channel, the Goldman-Hodgkin-Katz equation predicts a linear relationship between Erev and the log of the ion concentration gradient for the permeant ion. Here, a linear regression was fitted to the Erev-pH relationship with correlation coefficient of 0.97 (Fig. 3C), supporting that the inward currents we observed were carried by protons. Furthermore, we found a 53.6-mV shift in Erev for a 10-fold change in proton concentration (from pH 5.5 to 4.5), similar to the 58-mV shift in Erev predicted by the Nernst equation for a selective ion channel.
Fig. 3. Proton conductance and selectivity of human TMEM175.
(A) Representative I-V curves of hsTMEM175 WT at various pH as indicated showing the drift of the currents at −100 and +100 mV. (B) Averaged currents at −100 mV of hsTMEM175 WT from (A) as a function of pH values. Data are presented as means ± SEM for n = 4 (pH 5.5) and n = 3 (other pH). (C) Erev as a function of pH values from experiments in (A). Data are presented as means ± SEM for n = 4 (pH 5.5) and n = 3 (other pH). (D) Conductance at −100 mV as a function of pH values derived from (B) and (C). (E) Representative I-V curves of hsTMEM175 WT at pH 7.4 and pH 4.5, showing the ion selectivity for proton over potassium, sodium, cesium, and NMDG+ ions.
To test whether the proton current is gated or constitutive, we plotted the apparent conductance (current/voltage) at the applied voltage of −100 mV as a function of pH (Fig. 3D). The driving force or voltage experienced by the channel was taken as the difference between the applied voltage (−100 mV) and the measured Erev. The plotted proton conductance as a function of pH was well fit by a linear regression with a correlation coefficient of 0.96 (Fig. 3D), suggesting that hsTMEM175 is constitutively active and is not gated by pH over this pH range.
To assess the permeability of hsTMEM175 to protons relative to other cations, we recorded hsTMEM175-mediated currents with a voltage ramp protocol from −100 to 100 mV in bath solution at pH 4.5 and pipette solution containing 150 mM Na+, Cs+, K+, or NMDG+ at pH 7.4 (Fig. 3E). The reversal potential was measured for each recording, and using the Goldman equation that defines the relationship among ion concentration gradients, reversal potential, and permeability, we calculated permeability ratios of hsTMEM175 for protons relative to Na+ or NMDG+ at greater than 105-fold, and relative to K+ or Cs+ at ~0.5 × 105–fold (Fig. 3E and fig. S3, A to D). Thus, even with a physiological concentration of protons versus Na+ or K+ in lysosomal lumen at 10−4- to 10−3-fold less, the high selectivity of hsTMEM175 for protons over other cations makes it possible for protons to permeate in the presence of the competing ions.
Overall structures of human TMEM175 at different pH
To elucidate potential pH-induced conformational changes of TMEM175, we collected cryo-EM data on samples of TMEM175 purified in the detergent lauryl maltose neopentyl glycol (LMNG) and the lipid cholesteryl hemisuccinate (CHS) (fig. S4A) and determined structures of TMEM175 under different conditions by applying C2 symmetry, including the pH 6.8 structure in NaCl (3.2-Å resolution), the pH 7.4 structure in NaCl (3.4-Å resolution), the pH 7.4 structure in KCl (3.4-Å resolution), and the pH 5.5 structure in KCl (3.5-Å resolution) (figs. S4 and S5, A to C, and table S1). Because the K+ density in the center of the channel does not obey C2 symmetry, we also determined the pH 7.4 structure in KCl and the pH 5.5 structure in KCl without imposing the symmetry at 3.6- and 3.9-Å resolution, respectively (fig. S5, D and E).
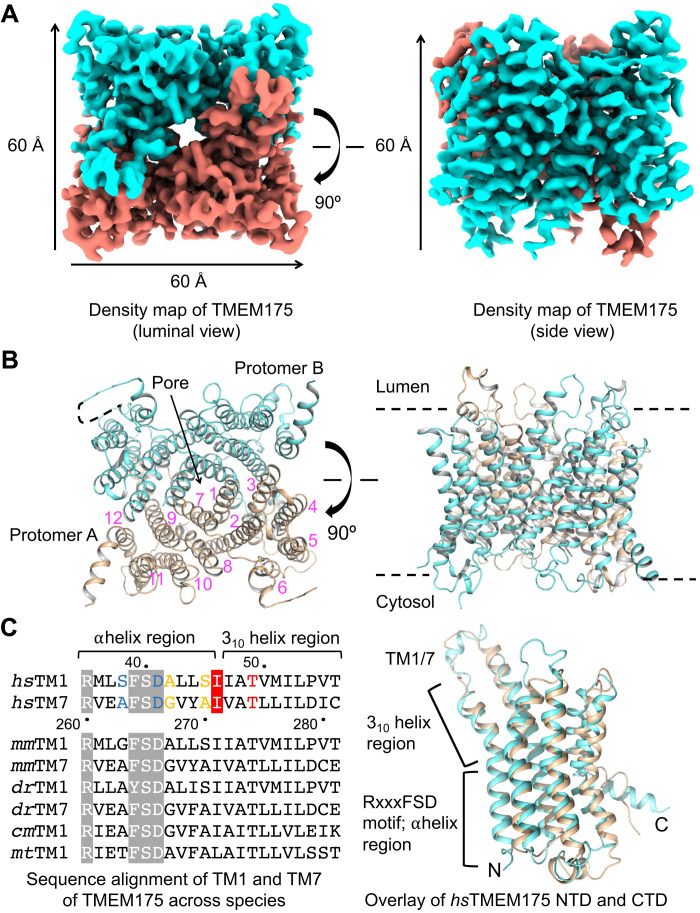
Human TMEM175 assembles into a dimer, with each subunit subdivided into an N-terminal domain (NTD) and a C-terminal domain (CTD) of six TM helices each (Fig. 4, A and B; and fig. S6, A to C). The NTD and CTD are highly structurally homologous with a pairwise overall root mean square deviation (RMSD) of 2.4 Å (Fig. 4C). Thus, the dimeric hsTMEM175 channel has a pseudo fourfold symmetry (Fig. 4B). From a top view, the structure has an inner compact core that forms the pore, which is composed of TM1 to TM3 of the NTD and TM7 to TM9 of the CTD interacting with the neighboring subunit (fig. S6, B and C). In contrast, TM4 to TM6 of the NTD and TM10 to TM12 of the CTD that form the outer portion of the channel do not participate in intersubunit interactions (fig. S6, B and C). The overall structures of hsTMEM175 at different pH are highly similar, with pairwise RMSDs of 0.3 to 0.6 Å.
Fig. 4. The assembly mechanism of human TMEM175.
(A) 3D cryo-EM density of human TMEM175. Dimensions of the TMEM175 dimer are ~60 × 60 × 60 Å. (B) Ribbon diagrams of the human TMEM175 structure with the subunits colored in cyan and wheat, respectively. The 12 helices in one of the two subunits are labeled by numbers in magenta. (C) Sequence alignment of TM1 and TM7 across species and structural comparison of human TMEM175 NTD and CTD. In the aligned sequences, the conserved RxxxFSD motif is highlighted in gray, and the conserved gate residue is highlighted in red. Residues involved in K+ coordination are colored in blue, yellow, and red for hydrophilic layers 3, 2, and 1, respectively.
Our structures of hsTMEM175 are also highly similar to the published hsTMEM175 structures determined at pH 8.0 in LMNG (15), with overall RMSDs of 0.7 and 1.0 Å, respectively, for our pH 7.4 structure (in KCl) against the open-state structure [Protein Data Bank (PDB) ID 6WC9, in KCl] and the closed-state structure (PDB ID 6WCA, in KCl). However, both 6WC9 and 6WCA lack TM5 and TM6 in comparison to our structures (fig. S6D). In our pH 7.4 structure (in KCl), we observed extra density at a pocket formed by TM2, TM6, and TM9 (fig. S6E), which we built as a cholesterol molecule either copurified endogenously or as part of CHS present in our purification condition. This bound molecule likely stabilized the TM5 and TM6 parts of hsTMEM175 (fig. S6E).
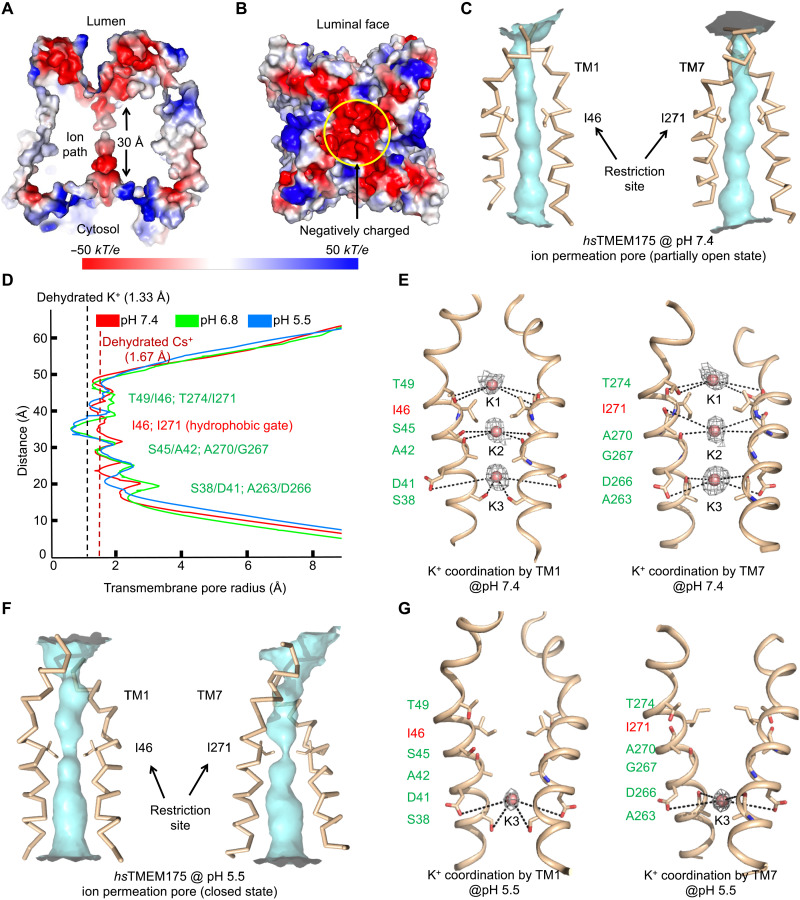
The ion permeation pore with three K+-binding sites
The dimeric or pseudotetrameric hsTMEM175 has a central pore for ion permeation that is lined by TM1 and TM7, which share 40% sequence identity (Fig. 4, B and C). A conserved RxxxFSD signature motif in these helices partly faces the pore (3, 13) (Fig. 4C and fig. S6C). Electrostatic surface analysis showed that a large patch of the luminal funnel just outside the ion permeation pore is highly negatively charged (Fig. 5, A and B), which may effectively generate an attractive potential for positively charged ions. When we calculated the pore radii along the ion permeation pathway, we found that the hsTMEM175 structure at pH 7.4 is in an open state with a pore wide enough to allow K+ to pass through based on the radius of a dehydrated K+ (Fig. 5, C and D). Of note, our TMEM175 structure at pH 7.4 suggests that the pore diameter is not wide enough to permit Cs+ permeation, which is puzzling, given electrophysiological data that demonstrate TMEM175 permeability is greater for Cs+ than for K+ (3). Local conformational changes within TMEM175 expressed in cells may account for this difference.
Fig. 5. Ion permeation pore of human TMEM175.
(A) Cross section through the pore showing the surface electrostatic potential of the pore and the pore entrances at the luminal and cytosolic sides (pH 6.8 structure model). (B) The surface electrostatic potential of the luminal face of the channel. (C) The ion permeation pore of hsTMEM175 at pH 7.4 formed by TM1 and TM7 with restriction residues in sticks representing an open conformation. (D) The pore radii of pH 5.5 (blue), pH 6.8 (green), and pH 7.4 (red) hsTMEM175 structures along the pore axis calculated using the HOLE program (44). The conserved gate residues are labeled in red, and residues coordinating K+ are colored in green. (E) K+ sites coordinated by hydrophilic residues lining the ion permeation pore (pH 7.4). The conserved gate residues are labeled in red, and residues coordinating the three K+ ions are labeled in green. Cryo-EM density for the K+ ions are contoured at 2.1 σ and shown in gray. (F) The ion permeation pore of hsTMEM175 at pH 5.5 formed by TM1 and TM7 with restriction residues in sticks, representing a closed conformation for K+ permeation. (G) One K+ site coordinated by hydrophilic residues lining the ion permeation pore (pH 5.5). The conserved gate residues are labeled in red, and residues coordinating the K+ site are labeled in green. Cryo-EM density for the K+ ions are contoured at 1.3 σ and shown in gray.
The cryo-EM map of hsTMEM175 at pH 7.4 in KCl displayed three additional densities in the ion permeation pore along the pseudo fourfold axis (Fig. 5E), which were absent from the structure in NaCl. These densities were unambiguous under different sigma cutoffs (fig. S7A) and were interpreted as three single potassium ions (K1–K3) spaced at 6.5 to 6.7 Å apart from one another (Fig. 5E). K1–K3 are surrounded by main-chain carbonyl oxygen atoms or side-chain hydroxyls from three hydrophilic layers in the ion permeation pathway (Fig. 5E). The hydroxyl groups of T49 on TM1 and of T274 on TM7 and the carbonyl oxygen atoms of I46/I271 gating residues are in position to interact with K1. The side-chain hydroxyl of S45 and the carbonyl oxygen of A42 on TM1, and the carbonyl oxygen atoms of G267 and A270 on TM7 are adjacent to K2. The hydroxyl group of S38 and the carboxylate of D41 on TM1, and the carbonyl oxygen of A263 and the carboxylate of D266 on TM7 interact with K3 (Fig. 5E). The distances between the oxygen ligands in our structures and K+ (~4 to 6 Å) are longer than those needed for direct coordination, suggesting the involvement of water molecules. In line with the published structures (15), we also propose that the gate for hsTMEM175 is formed by the conserved hydrophobic residues I46 and I271 situated between K1 and K2 (Fig. 5, D and E).
Our observed K1–K3 sites in an open state of hsTMEM175 are different from the published structure 6WC9 at pH 8.0 in open state (15). Seven K+ sites were discerned in 6WC9, and the comparison with our pH 7.4 structure is shown (fig. S7B). When we placed our observed K+ sites into the cryo-EM density that corresponds to 6WC9 [Electron Microscopy Data Bank (EMDB) ID EMD-21603], these sites localized away from the density (fig. S7C), suggesting that the difference in K+ sites could not be explained by the current resolution of our structure. Of note, it appears that our three K+ sites are more aligned with the hydrophilic layers in the permeation path (fig. S7B). Because conformations of the side chains lining the pore are structurally conserved between our structure and 6WC9 with only minor conformational differences (fig. S7B), we have not identified a reason for the difference in the identified K+ sites.
pH-induced structural changes
To elucidate potential pH-induced structural changes, we compared the hsTMEM175 structure at pH 7.4 in KCl (open conformation) with those at pH 6.8 in NaCl and at pH 5.5 in KCl, both of which are in a closed conformation. Unlike the pH 7.4 structure, calculation of pore radii along the ion permeation pathway revealed a distinct narrowing to ~0.8 Å at residue I46 of TM1 and residue I271 of TM7 in the pH 6.8 and 5.5 structures (Fig. 5F). Because 0.8 Å is below the radius of a dehydrated K+, we reasoned that the pH 5.5 structure should represent a closed conformation of the channel that reduced the permeation of K+. The pH 5.5 structure also appeared to have reduced K+ binding with only one K+ site (K3) visible, and this site has weaker density than that in the pH 7.4 structure (fig. S7, A, D, and E). No density was seen at the K1 and K2 sites, and the potential protonation of pore-lining residues may lead to the reduced K+ binding at the K1 and K2 sites (fig. S7E). This observation is different from the closed conformation structure 6WCA obtained at pH 8.0, in which five K+ sites were observed (fig. S7F). When we placed the K3 site into the cryo-EM density corresponding to 6WCA, it localized away from the density (fig. S7G), suggesting that there is a real difference between these two closed conformations. It is intriguing how both open and closed conformations were observed at pH 8.0 in the published study (15), while we identified these two different conformations at two different pH.
K+ gating and selectivity by TMEM175
To validate the importance of the observed residues in K+ coordination and/or gating, we expressed WT and mutant hsTMEM175 proteins in HEK293T cells and tested their ion permeability and selectivity using whole-cell recording. We first tested the role of the three hydrophilic layer residues and found that their mutations reduced hsTMEM175-dependent currents (fig. S8, A and B), suggesting the significance of these residues in K+ conductance. Consistently, S45A mutation was also shown to reduce K+ conductance by Hite and co-workers (15). In contrast, Brunner et al. (14) showed that S45A is critical for K+ selectivity. Despite this discrepancy, these studies underscore the functional significance of the hydrophilic residues on either side of the gating residues (14, 15). We then tested the role of the Ile residues at the constriction gate and showed that the I46M/I271M mutant also displayed markedly reduced current (fig. S8C), likely due to the occlusion of the permeation pathway by the larger Met side chains. In contrast, the I46N/I271N mutant retained permeation but lost selectivity for K+ with a PK/PNa of about 1.4, in comparison with our measured PK/PNa of about 10.2 for WT hsTMEM175 (fig. S8D) that is in a similar range as obtained previously (3). Our findings support a model in which residues of S45, I46/I271, and T49/T274 form a functional unit to couple gating and potassium selectivity (fig. S8E). Notably, two groups also showed the functional significance of hydrophilic residues and the gating residues in channel gating and ion selectivity, which provided another piece of evidence to support our model (14, 15).
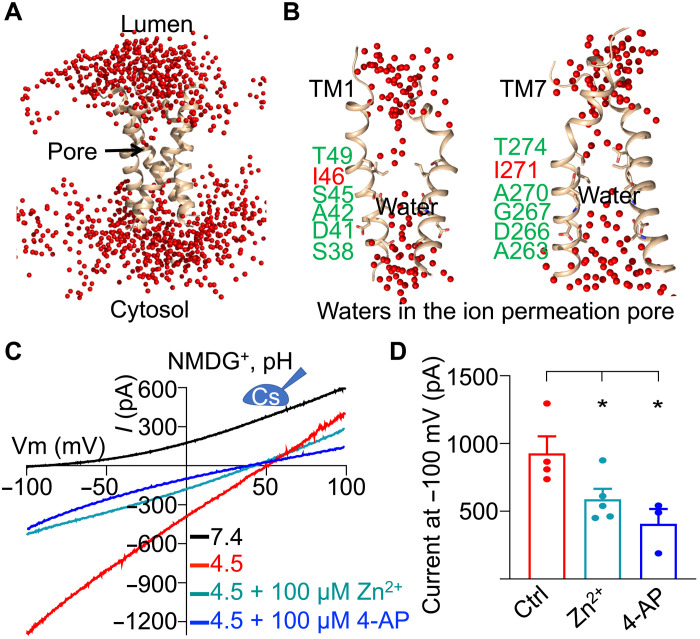
Putative proton permeation pathway
Proton movement often occurs through a “hopping” mechanism, in which water and hydrophilic residues form a “wire” along the proton path (18–20). To test whether water could play a role for hsTMEM175, we subjected the hsTMEM175 structure in KCl at pH 7.4 to an all-atom molecular dynamics simulation in a hydrated system containing solvated K+ and Cl− and 1-palmitoyl-2-oleoyl-glycerol-3-phosphocholine (POPC) lipids. The water profile obtained at 40 ns revealed that the most water-accessible part of the structure is along the K+ permeation pathway (Fig. 6, A and B). Many water molecules were observed along this pathway except exactly at the hydrophobic gate, which had a water gap (Fig. 6B). Similar water permeation pattern was also observed for the proton channel OPTOP1 (20), suggesting that TMEM175 may share the same mechanism of proton conduction used by OTOP1. These data also indicate that proton and K+ may share the same permeation path and that hydrophilic residues lining or surrounding the pore may be involved in proton flux.
Fig. 6. Proton permeation pathway of human TMEM175.
(A) Selected water distribution in and around the hsTMEM175 structure from the molecular dynamics simulation. TM1 and TM7 of hsTMEM175 are shown in a ribbon diagram, water molecules are displayed as red spheres, and the ion permeation pore is indicated by an arrow. (B) Water molecules in the ion permeation pore of TMEM175. (C) Representative I-V curves of hsTMEM175 WT at conditions as indicated, showing the inhibition of TMEM175 conductivity by Zn2+ and 4-AP. (D) Averaged currents at −100 mV of hsTMEM175 WT from (C) at pH 4.5 in the absence and presence of 100 μM Zn2+ or 100 μM 4-AP. Data are presented as means ± SEM for n = 5.
To test this hypothesis, we first used Zn2+, which has been shown to inhibit the potassium conductance of TMEM175 and also nonspecifically inhibit various proton channels including OTOP1 and Hv1 that use the mechanism of water hopping for conducting protons (3, 21, 22). We found that the proton current of hsTMEM175 was significantly inhibited by Zn2+ (Fig. 6, C and D). We further used 4-aminopyridine (4-AP), a selective pore blocker of K+ channels that has been shown to block K+ conductance of hsTMEM175 (3), and found that 4-AP also significantly suppressed the proton current of hsTMEM175 (Fig. 6, C and D). Collectively, these data suggest that protons may share the same permeation pathway with potassium ions.
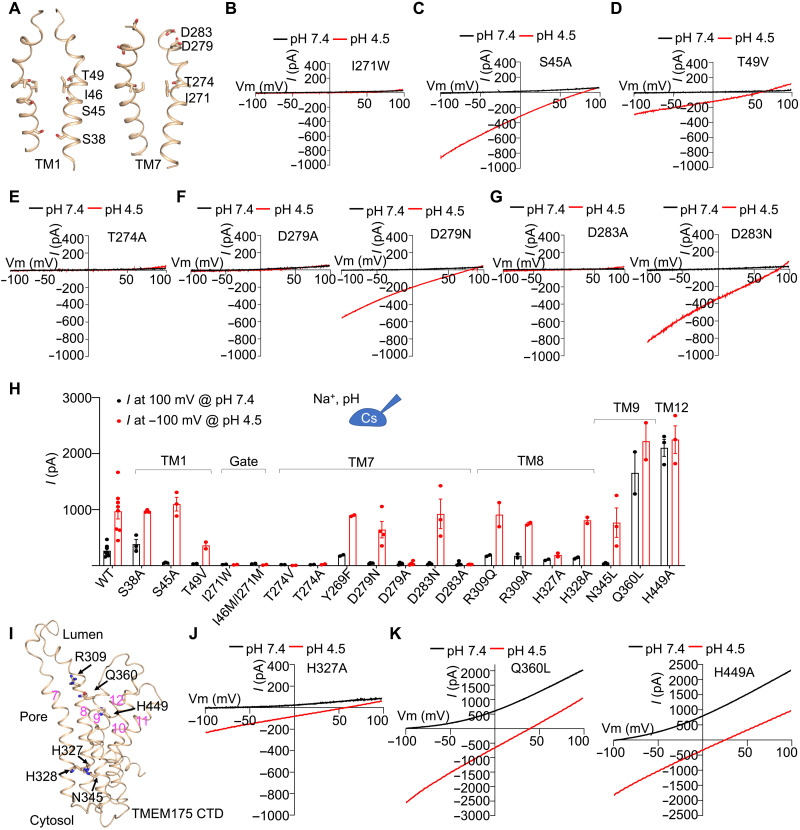
Residues critical for proton permeation
To functionally identify residues important for proton permeation, we first mutated gating residues and the hydrophilic residues lining the ion permeation pore of hsTMEM175 (Fig. 7A) and used the whole-cell recording system in HEK293T cells expressing hsTMEM175. Mutations on the gating residues, I271W and I46M/I271M, blocked both proton and K+ conductance (Fig. 7B and fig. S9, A and B). We speculated that the bulky hydrophobic side chains may have changed water penetration and interrupted the water wire for proton conductance. We then examined pore-lining hydrophilic residues, including S38, S45, and T49 on TM1, and T274, D279, and D283 on TM7 (Fig. 7, C to H, and fig. S9C). On TM1, the S38A mutant showed normal proton and K+ conductance, S45A blocked K+ but not proton conductance, and T49V abolished K+ conductance and reduced proton conductance (Fig. 7, C and D, and fig. S9C). In contrast, all alanine mutations on TM7, including T274A, T274V, D279A, and D283A, abolished both proton and K+ conductance (Fig. 7, E to G). Mutations of D279N and D283N specifically abolished K+ conductance but had little or mild effects on proton conductance (Fig. 7, F and G), suggesting that the hydrophilic nature of the side chains sufficiently contributes to proton conductance. Thus, our data suggest that pore-lining hydrophilic residues on TM1 and TM7 are involved in both K+ and proton conductance and that proton permeation mainly occurs via hoping through the water wire and perhaps hydrophilic residues.
Fig. 7. Residues critical for potassium ion and proton permeation.
(A) Pore-lining residues in hsTMEM175 that we tested in this study for functional involvement in proton and K+ conductance. (B) Representative I-V curves of hsTMEM175 mutant I271W at pH 7.4 and pH 4.5, showing the abolishment of K+ and proton currents. (C) Representative I-V curves of hsTMEM175 mutants S45A at pH 7.4 and pH 4.5, showing abolished K+ conductance but normal proton conductance. (D) Representative I-V curves of hsTMEM175 mutants T49V at pH 7.4 and pH 4.5, showing abolished K+ conductance and reduced proton conductance. (E) Representative I-V curves of hsTMEM175 mutant T274A at pH 7.4 and pH 4.5, showing the abolishment of both proton and K+ currents. (F and G) Representative I-V curves of hsTMEM175 mutants D279A, D279N, D283A, and D283N at pH 7.4 and pH 4.5, showing their effects on K+ and proton conductance. (H) Averaged currents at 100 mV of pH 7.4 or −100 mV of pH 4.5 for hsTMEM175 WT and mutants. Data are presented as means ± SEM for n ≥ 3. (I) Residues on TM8, TM9, and TM12 that were examined for the regulation of proton conductance. (J) Representative I-V curves of hsTMEM175 mutant H327A, showing the deficiency in K+ and proton conductance. (K) Representative I-V curves of hsTMEM175 mutants Q360L and H449A, showing enhancement of K+ and proton conductance.
We next explored the functional contributions of several hydrophilic residues surrounding the ion permeation pore where water molecules tend to accumulate. These include R309, H327, and H328 on TM8; N345 and Q360 on TM9; and H449 on TM12 (Fig. 7I). Among the seven mutants that we generated, R309Q, R309A, H327A, H328A, and N345L displayed reduced or abolished K+ conductance, but only H327A also had reduced proton current (Fig. 7, H and J; and fig. S9, D to G). These mutations might induce local or global protein conformational changes that affect the pore configuration, consistent with the assumption that K+ conductance depends on an intact and precise pore conformation, whereas proton conductance may simply require a watery pore supported by a lining of hydrophilic residues. Q360L and H449A exhibited substantially enhanced conductance for both potassium and protons, perhaps through allosteric effects (Fig. 7, H and K, and fig. S1, B and C). With the larger currents, these activating mutants further consolidate the pH dependence of K+ conductance and demonstrate the proton conductance in hsTMEM175 (Fig. 7, H and K).
DISCUSSION
Our structural studies showed that lower pH caused visible conformational changes in hsTMEM175 in its coordination of K+ in the ion permeation pathway, from three ordered K+ sites (K1–K3) in the pH 7.4 partially open conformation to one weak site (K3) in the pH 5.5 closed conformation. These data, together with electrophysiology, demonstrate that the K+ conductance of hsTMEM175 is regulated by pH. Our observed K+ sites in both the partially open and the closed conformations are different from those in the published hsTMEM175 structures (15). It is unclear how this difference arises. However, as conformations of ion channels are highly sensitive to the environment, the different solubilization conditions that we used in comparison with the published structures may be a potential factor. Functionally, modulation of hsTMEM175 activity by pH could facilitate its role in establishing and maintaining the ionic balance and membrane potential of endolysosomes. When the pH values of these compartments are high, far from the resting pH of 4.5 to 5.5, the hsTMEM175 conductivity is also high, and together with other proteins on endolysosomes may help to quickly lower the pH to the desired values while maintaining the ion homeostasis of endolysosomes. When the pH values of these compartments are already in balance, restricting the K+ conductance of hsTMEM175 activity should reduce any potential ionic disturbance. This K+ channel function of hsTMEM175 appears to be crucial, as the major PD-associated hsTMEM175 mutant M393T exhibits reduced K+ conductance and compromised lysosomal enzyme activity, and the PD-protective hsTMEM175 mutant Q65P shows hyperactivity. A similar observation was found and published by Ren and co-workers (16), and they also made another intriguing discovery that TMEM175 can be gated by directly interacting with AKT1. The molecular mechanisms of how AKT1 interacts and regulates TMEM175 await further investigation.
An entirely unexpected finding from our studies is that hsTMEM175 also functions as a proton channel, constitutively conducting protons down the proton concentration gradient across the organelle membrane. Furthermore, molecular dynamics simulation and structure-guided mutagenesis suggest that protons and K+ may share the same ion permeation pathway, with pore-lining hydrophilic residues important for both proton and K+ permeation. Thus, under acidic pH, the potential competition between proton and K+ ions may also help to explain the reduced K+ binding in our TMEM175 structure at pH 5.5. While previous electrophysiological studies suggested the existence of proton currents in lysosomes (23), no proton channels have so far been identified in lysosomes. Our hsTMEM175 data provide a candidate for such a proton channel, and electrophysiological recordings of TMEM175 currents in lysosomes may further solidify this finding. As V-ATPases actively transport protons into the lysosomal lumen, lysosomes must have evolved a mechanism to prevent the overacidification of their lumen. A role for hsTMEM175 in this function is supported by TMEM175 knockout or knockdown assays, in which the lysosomal lumens of these cells are more acidic than those of WT cells under fed condition (5). On the basis of our measured ion selectivity, hsTMEM175 is actually most selective for protons, ~105-fold more selective than for K+, Cs+, Na+, or NMDG+, and generates a dominant current at low pH even with the much lower proton concentrations relative to lysosomal Na+ or K+ concentrations. Similar to the K+ conductance, the human variants M393T and Q65P also exhibited reduced and enhanced proton conductance, respectively, and the loss-of-function M393T mutation is predicted to result in lysosomal overacidification. Consistently, the PD-associated lysosomal enzyme GBA has an optimal pH range of 4.7 to 5.5 for activity (17), which prompts us to speculate that M393T potentiates PD by dropping the lysosomal pH below this range. Conversely, the Q65P mutant, which protects against PD, drives larger proton currents and may provide optimal lysosomal function. The differential biological functions of proton and K+ permeabilities in hsTMEM175 and their relative physiological and pathophysiological implications remain to be further explored.
MATERIALS AND METHODS
Expression and purification of the full-length human TMEM175
Full-length human TMEM175 gene sequence obtained from a plasmid purchased from the Harvard Plasmid Core was inserted into a BacMam vector with a tobacco etch virus (TEV) protease-cleavable N-terminal maltose-binding protein (MBP) tag using the Gibson Assembly Cloning Kit (New England Biolabs). The resulting construct was expressed in HEK293F cells using the BacMam method (24). In brief, bacmids containing hsTMEM175 were generated using the Bac-to-Bac method according to the manufacturer’s instructions (Invitrogen). The BacMam baculoviruses were obtained by transfecting the bacmids into Sf9 cells, with three generations of virus expansion. Suspension cultures of HEK293F cells in the FreeStyle 293 medium were infected with BacMam baculoviruses at a density of 2 × 106 cells/ml for protein expression. At 10 hours postinfection, 10 mM sodium butyrate was supplemented into infected cultures to boost protein yield. The cultured cells grew for another 48 hours at 30°C before harvesting.
For hsTMEM175 in NaCl at pH 7.4, HEK293F cells were sonicated in buffer A [50 mM Hepes at pH 7.4, 150 mM NaCl, and 1 mM tris(2-carboxyethyl)phosphine (TCEP)] plus 0.5% LMNG, 0.05% CHS, and one tablet of protease inhibitor cocktail (Sigma-Aldrich). The sonicated mixture was further incubated for 3 hours on a nutating mixer. Cell debris was removed by ultracentrifugation at 42,000 revolutions per minute for 1 hour. Supernatant was incubated with amylose resin (New England Biolabs) for 2 hours at 4°C. The resin was then washed with 10 column volumes of wash buffer (buffer A plus 0.1% LMNG and 0.01% CHS). MBP-tagged hsTMEM175 was eluted with wash buffer plus 25 mM maltose, followed by overnight incubation with TEV protease (1:30 treatment, molar ratio) to cleave the MBP tag. hsTMEM175 was further purified away from MBP and TEV by size-exclusion chromatography using a Superdex 200 column (GE Healthcare) in buffer A plus 0.005% LMNG and 0.0005% CHS. Fractions containing hsTMEM175 were pooled and saved for later applications.
For hsTMEM175 in KCl at pH 7.4, the purification procedures were the same as for hsTMEM175 in NaCl except for changing buffer A to buffer B (50 mM Hepes at pH 7.4, 150 mM KCl, and 1 mM TCEP). For hsTMEM175 in NaCl at pH 6.8, buffer C (50 mM Hepes at pH 6.8, 150 mM NaCl, and 1 mM TCEP) was used to substitute buffer A for the entire purification process. For hsTMEM175 in KCl at pH 5.5, buffer D [50 mM 2-(N-morpholino)ethanesulfonic acid (MES) at pH 5.5, 150 mM KCl, and 1 mM TCEP] was used to substitute buffer A for the entire purification process.
Cryo-EM data collection
A 3-μl drop of an hsTMEM175 sample at 1.8 mg/ml was applied to a glow-discharged Quantifoil R1.2/1.3 400-mesh copper grids (Electron Microscopy Sciences), blotted for 4.5 s in 100% humidity at 4°C, and plunged into liquid ethane using an FEI Vitrobot Mark IV. All the grids were screened using an FEI Talos Arctica microscope (Harvard Cryo-EM Center for Structural Biology). The data collection was performed using a 300-kV Titan Krios microscope (FEI) equipped with a K3 direct electron detector (Gatan). For hsTMEM175 at pH 7.4 in NaCl, 5554 micrographs were collected in counting mode, with 50 total frames per movie, 65.0 electrons per Å2 accumulated dose, and 0.825-Å physical pixel size. For hsTMEM175 at pH 7.4 in KCl, 5208 micrographs were collected in counting mode, with 48 total frames per movie, 64.4 electrons per Å2 accumulated dose, and 0.825-Å physical pixel size. For hsTMEM175 at pH 6.8 in NaCl, 3376 micrographs were collected in counting mode, with 50 total frames per movie, 67.7 electrons per Å2 accumulated dose, and 0.825-Å physical pixel size. For hsTMEM175 at pH 5.5 in KCl, 9156 micrographs were collected in counting mode, with 50 total frames per movie, 66.5 electrons per Å2 accumulated dose, and 0.825-Å physical pixel size.
Cryo-EM structure reconstruction
For all datasets, the movie frames were motion corrected and dose weighted by MotionCor2 (25). Initial contrast transfer function (CTF) parameters were estimated by CTFFIND4 (26), and particle picking was carried out using crYOLO (27).
For the pH 6.8 dataset, this resulted in 1,015,093 initial particles. Following successive rounds of two-dimensional (2D) and 3D classification within RELION (28), 142,428 particles were “polished” through the Bayesian polishing process (29). These particles were then subjected to further 2D classification within cryoSPARC (30), after which 141,251 particles were selected (see classification flowchart in fig. S4). The final reconstruction reached 3.2-Å resolution after CTF parameter (beamtilt/trefoil) refinement (30, 31).
For the pH 7.4 dataset in NaCl, particle picking resulted in 1,378,039 initial particles. Following successive rounds of 2D classification within RELION (28), 630,529 particles were polished through the Bayesian polishing process (29). These particles were then subjected to further 2D and 3D classification within cryoSPARC (30), after which 145,051 particles were selected. The final reconstruction reached 3.4-Å resolution after nonuniform refinement.
For the pH 7.4 dataset in KCl, particle picking resulted in 1,111,591 initial particles. Following successive rounds of 2D and 3D classification within RELION (28), 196,835 particles were polished through the Bayesian polishing process (29). These particles were then subjected to further 3D classification within cryoSPARC (30), after which 110,220 particles were selected. The final reconstructions were at 3.6- (C1 map) and 3.4-Å resolution (C2 map) after CTF parameter refinement.
For the pH 5.5 dataset in KCl, particle picking resulted in 790,954 initial particles. Following successive rounds of 2D and 3D classification within RELION (28), good particles were then subjected to further 3D classification within cryoSPARC (30), after which 117,539 particles were selected. The final reconstructions were at 3.9-Å (C1 map) and 3.5-Å resolution (C2 map) after CTF parameter refinement.
Maps used for figures were filtered according to local resolution with b-factor sharpening within cryoSPARC (30). Structural biology applications used in this project (other than cryoSPARC) were compiled and conFig.d by SBGrid (32).
Cryo-EM model building and refinement
The initial model for hsTMEM175 at pH 6.8 in NaCl was created from the structure of cmTMEM175 (PDB ID 5vre) using SWISS-MODEL (33) and fitted as a rigid body into the 3.2-Å cryo-EM map using Chimera (34). The final refined structure of this state was used as the initial models for hsTMEM175 at pH 7.4 in KCl and hsTMEM175 at pH 7.4 in NaCl. All manual adjustments were done using Coot (35), and Phenix (36) was used to refine the models against cryo-EM density maps. All representations of electron densities and structural models were generated using Chimera (34) and PyMOL (37).
Molecular dynamics simulation
The missing residues for the flexible loop between TM6 and TM7 were filled in using MODELLER (38). The structure was protonated with the protonation states of residues at pH 7.0 and immersed into 228 POPC membrane lipids. The system was then solvated by 18,175 water molecules, 6 potassium ions, and 16 chloride ions, resulting in a salt concentration of ~0.01 M. The CHARMM36m force field and TIP3P (transferable intermolecular potential with 3 points) water model were used (39). The modeling and equilibration of the system were performed by the CHARMM-GUI server (40). The graphics processing unit (GPU)–accelerated GROMACS (version 2020.2) was used for the production run (41). The water profile was obtained from 40-ns molecular dynamics simulation at the temperature of 300 K.
Electrophysiology
The hsTMEM175 channel activity in the plasma membrane of HEK293T cells was measured with whole-cell patch-clamp as described previously (13). Specifically, hsTMEM175 WT and mutants cloned into a pLV vector with an N-terminal mScarlet tag were transfected into HEK293T cells using Lipofectamine 3000 (Life Technologies). Around 24 hours following transfection, cells were seeded onto coverslips coated with Matrigel (BD Bioscience). The recording pipette was made from borosilicate glass with an inner diameter of 0.86 mm (Warner Instruments) using a micropipette puller. The pipette resistance was in the range of 3 to 5 megohms when filled with the internal solution. The offset potential was corrected before the gigohm seal was formed. Once the whole-cell configuration was established, membrane capacitance and series resistance were compensated. Currents were recorded using an Axopatch 200B patch-clamp amplifier, filtered at 10 kHz through a low-pass analog filter, and sampled at a rate of 20 kHz with a Digidata 1322A digitizer (Molecular Devices). The current-voltage relation (I-V) curves were generated with a voltage ramp protocol from −100 to 100 mV over 800 ms. For I-V curves at low pH solutions, currents were first monitored in gap-free mode with membrane potential hold at −60 mV during exchange of solutions with low pH. When the peak current amplitude was reached, the same voltage ramp protocol was applied. The pClamp 9 software (Axon Instruments) was used for data acquisition and analysis.
The standard bath solution used in this study contained 150 mM sodium methanesulfonate (Na-MS), 1 mM MgCl2, 1 mM CaCl2, and 10 mM Hepes at pH 7.4 adjusted with tris base. The Na+ was replaced with equal molar K+ or NMDG+ in some cases as indicated in the figure legends. For bath solutions with low pH, ion compositions were kept the same except for the buffers: 10 mM MES (for pH 6 to 5.5) and 10 mM Homo-Pipes (for pH 5 to 4). The standard pipette solution contained 150 mM Cs-MS, 5 mM MgCl2, 10 mM EGTA, and 10 mM Hepes at pH 7.4 adjusted with tris base. For the measurement of proton selectivity, the Cs+ in the pipette solution was replaced with Na+, K+, or NMDG+.
To estimate the ion selectivity of K+ relative to Na+, the I-V curves were obtained in bath solution with either 150 mM Na-MS or K-MS. The relative permeability of Na+ or K+ to Cs+ is given by the Goldman-Hodgkin-Katz equation: PCs/PNa or K = [Na or K]bath/([Cs]pipette*exp(ErevF/RT)). Then the permeability ratio of K+ to Na+ was calculated with the following equation: PK/PNa = ([Na]bath/[K]bath)*exp((Erev (K) − Erev (Na))F/RT), where Erev is the reversal potential, F is Faraday’s constant, R is the gas constant, and T is the absolute temperature. For estimation of H+ selectivity to other cations, the I-V curves were obtained with bath solution containing 150 mM NMDG+ (pH 4.5) and pipette solution containing 150 mM Cs+, Na+, K+, or NMDG+ (pH 7.4). The relative H+ permeability is calculated by PH/PX = ([X]pipette/([H]bath)*exp(ErevF/RT)), where X represents Cs, Na, K, or NMDG. Liquid junction potentials were corrected offline for permeability calculation.
pH imaging
HEK293T cells were transfected via Lipofectamine 3000 (Thermo Fisher Scientific) with WT TMEM175-mCherry, TMEM175-Q360L-mCherry, or TMEM175-I271W-mCherry as described above. Lipofectamine 3000 alone was applied in a fourth condition as a negative control. Transfected cells were plated on Matrigel-coated cell imaging dishes at 37°C. Cells were loaded with an intracellular pH fluorescence indicator pHrodo Green AM according to the protocol described by the manufacturer (Molecular Probes). Live-cell imaging was performed on a Nikon A1R confocal microscope with a Piezo z-drive for fast z-stack acquisition. Transfected and nontransfected cells were identified by the presence or absence, respectively, of fluorescence intensity in the mCherry channel. Regions of interest (ROIs) were determined by the fluorescence in the mCherry channel, and these ROIs were used to determine the average fluorescence intensity in the pHrodo Green channel. The baseline pHrodo Green fluorescence intensity for the mCherry-positive cells were determined for 30 s before recording the fluorescence intensity in response to a pH 4.5 solution (150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, and 10 mM Homo-Pipes with HCl added until desired pH reached). The pHrodo Green fluorescence intensity of each cell was normalized as the difference between the fluorescence intensity F and the baseline intensity divided by the baseline intensity to produce ΔF/F0. Cells that had unstable baseline or terminal fluorescence intensities, dying cells with poor morphology, and recordings that had image shifts during time lapse imaging were excluded from the analysis.
Confocal imaging
The fluorescence images were taken as described previously (42) . In brief, HEK293T cells expressing mScarlet-tagged TMEM175 were first washed with phosphate-buffered saline (PBS) and then fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature. The unreacted PFA was neutralized with 1 M glycine. Then, the cells were mounted in ProLong Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (Molecular Probes, Eugene, OR) and examined on an AIVI spinning disc confocal microscope.
Surface protein biotinylation assay
Biotinylation assays were performed as previously described (43). Briefly, the HEK293T cells transfected with WT TMEM175-mScarlet or mutant were first washed with ice-cold PBS solution. The cells were then incubated with sulfo-NHS-SS-Biotin (0.5 mg/ml in PBS; Pierce, Rockford, IL) at room temperature for 20 min. NH4Cl (1 M) was then used to quench nonreacted biotin. Next, the cells were lysed in CelLytic M lysis buffer (Sigma-Aldrich, St. Louis, MO) supplemented with proteinase inhibitor mixture (Thermo Fisher Scientific, Waltham, MA). Streptavidin beads (Pierce) were then incubated with the cell lysate overnight at 4°C. The captured surface protein by the beads was subjected to Western blot and detected with sScarlet antibody.
Acknowledgments
Funding: Cryo-EM data were collected with the assistance of R. Walsh and S. Sterling at the Harvard Medical School Cryo-Electron Microscopy Center supported by grants from the NIH National Institute of General Medical Sciences (GM103310). T.-M.F. is supported by funding from NIH T32 grant (5T32HL066987-18) and the Ohio State University Startup fund; J.R.H. is supported by funding from NIDCD R01DC013521; and C.S. is supported by a Cancer Research Institute Irvington Postdoctoral Fellowship.
Author contributions: T.-M.F. conceived the project. C.S., T.-M.F., L.W., and S.X. performed molecular cloning, protein purification, and sample preparation. C.S. collected the cryo-EM data, and S.R. performed the cryo-EM data processing. T.-M.F., C.S., and Z.S. built the models. W.Z. and J.W. performed electrophysiological characterizations. C.N.-L. and W.Z. performed the pH imaging assay. J.R.H. supervised the electrophysiological recording experiments and pH imaging assay. W.J.X. performed the molecular dynamic simulation. T.-M.F., H.W., C.S., and W.Z. wrote the manuscript with input from all the authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Accession numbers for the human TMEM175 structures in NaCl at pH 6.8, NaCl at pH 7.4, KCl at pH 7.4, and KCl at pH 5.5 are as follows: 6W8N, 6W8O, 6W8P, and 7LF6 (coordinates of atomic models: Protein Data Bank) and EMD-21575, EMD-21576, EMD-21577, and EMD-23300 (density map: Electron Microscopy Data Bank), respectively.
Supplementary Materials
This PDF file includes:
Figs. S1 to S9
Table S1
REFERENCES AND NOTES
- 1.Lawrence R. E., Zoncu R., The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21, 133–142 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Xu H., Ren D., Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cang C., Aranda K., Seo Y. J., Gasnier B., Ren D., TMEM175 is an organelle K+ channel regulating lysosomal function. Cell 162, 1101–1112 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Nalls M. A., Pankratz N., Lill C. M., Do C. B., Hernandez D. G., Saad M., De Stefano A. L., Kara E., Bras J., Sharma M., Schulte C., Keller M. F., Arepalli S., Letson C., Edsall C., Stefansson H., Liu X., Pliner H., Lee J. H., Cheng R.; International Parkinson’s Disease Genomics Consortium (IPDGC); Parkinson’s Study Group (PSG) Parkinson’s Research: The Organized GENetics Initiative (PROGENI); 23andMe; GenePD; NeuroGenetics Research Consortium (NGRC); Hussman Institute of Human Genomics (HIHG); The Ashkenazi Jewish Dataset Investigator; Cohorts for Health and Aging Research in Genetic Epidemiology (CHARGE); North American Brain Expression Consortium (NABEC); United Kingdom Brain Expression Consortium (UKBEC); Greek Parkinson’s Disease Consortium; Alzheimer Genetic Analysis Group, Ikram M. A., Ioannidis J. P. A., Hadjigeorgiou G. M., Bis J. C., Martinez M., Perlmutter J. S., Goate A., Marder K., Fiske B., Sutherland M., Xiromerisiou G., Myers R. H., Clark L. N., Stefansson K., Hardy J. A., Heutink P., Chen H., Wood N. W., Houlden H., Payami H., Brice A., Scott W. K., Gasser T., Bertram L., Eriksson N., Foroud T., Singleton A. B., Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinn S., Drolet R. E., Cramer P. E., Wong A. H. K., Toolan D. M., Gretzula C. A., Voleti B., Vassileva G., Disa J., Tadin-Strapps M., Stone D. J., TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc. Natl. Acad. Sci. U.S.A. 114, 2389–2394 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinn S., Blauwendraat C., Toolan D., Gretzula C. A., Drolet R. E., Smith S., Nalls M. A., Marcus J., Singleton A. B., Stone D. J., Functionalization of the TMEM175 p.M393T variant as a risk factor for Parkinson disease. Hum. Mol. Genet. 28, 3244–3254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krohn L., Öztürk T. N., Vanderperre B., Ouled Amar Bencheikh B., Ruskey J. A., Laurent S. B., Spiegelman D., Postuma R. B., Arnulf I., Hu M. T. M., Dauvilliers Y., Högl B., Stefani A., Monaca C. C., Plazzi G., Antelmi E., Ferini-Strambi L., Heidbreder A., Rudakou U., Cochen de Cock V., Young P., Wolf P., Oliva P., Zhang X. K., Greenbaum L., Liong C., Gagnon J. F., Desautels A., Hassin-Baer S., Montplaisir J. Y., Dupré N., Rouleau G. A., Fon E. A., Trempe J. F., Lamoureux G., Alcalay R. N., Gan-Or Z., Genetic, structural, and functional evidence link TMEM175 to synucleinopathies. Ann. Neurol. 87, 139–153 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Zhu X. C., Cao L., Tan M. S., Jiang T., Wang H. F., Lu H., Tan C. C., Zhang W., Tan L., Yu J. T., Association of Parkinson’s disease GWAS-linked loci with Alzheimer’s disease in Han Chinese. Mol. Neurobiol. 54, 308–318 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Jing C. C., Luo X. G., Cui H. G., Li F. R., Li P., Jiang E. Z., Ren Y., Pang H., Screening of polymorphisms located in the FGF20 and TMEM175 genes in North Chinese Parkinson’s disease patients. Genet. Mol. Res. 14, 13679–13687 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Blauwendraat C., Heilbron K., Vallerga C. L., Bandres-Ciga S., von Coelln R., Pihlstrøm L., Simón-Sánchez J., Schulte C., Sharma M., Krohn L., Siitonen A., Iwaki H., Leonard H., Noyce A. J., Tan M., Gibbs J. R., Hernandez D. G., Scholz S. W., Jankovic J., Shulman L. M., Lesage S., Corvol J. C., Brice A., van Hilten J. J., Marinus J.; The 23andMe Research Team, Eerola-Rautio J., Tienari P., Majamaa K., Toft M., Grosset D. G., Gasser T., Heutink P., Shulman J. M., Wood N., Hardy J., Morris H. R., Hinds D. A., Gratten J., Visscher P. M., Gan-Or Z., Nalls M. A., Singleton A. B.; for the International Parkinson’s Disease Genomics Consortium (IPDGC) , Parkinson’s disease age at onset genome-wide association study: Defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 34, 866–875 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Senkevich K., Gan-Or Z., Autophagy lysosomal pathway dysfunction in Parkinson’s disease; evidence from human genetics. Parkinsonism Relat. Disord. 73, 60–71 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Clark L. N., Ross B. M., Wang Y., Mejia-Santana H., Harris J., Louis E. D., Cote L. J., Andrews H., Fahn S., Waters C., Ford B., Frucht S., Ottman R., Marder K., Mutations in the glucocerebrosidase gene are associated with early-onset Parkinson disease. Neurology 69, 1270–1277 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C., Guo J., Zeng W., Kim S., She J., Cang C., Ren D., Jiang Y., The lysosomal potassium channel TMEM175 adopts a novel tetrameric architecture. Nature 547, 472–475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunner J. D., Jakob R. P., Schulze T., Neldner Y., Moroni A., Thiel G., Maier T., Schenck S., Structural basis for ion selectivity in TMEM175 K+ channels. eLife 9, e53683 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh S., Paknejad N., Hite R. K., Gating and selectivity mechanisms for the lysosomal K+ channel TMEM175. eLife 9, e53430 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wie J., Liu Z., Song H., Tropea T. F., Yang L., Wang H., Liang Y., Cang C., Aranda K., Lohmann J., Yang J., Lu B., Chen-Plotkin A. S., Luk K. C., Ren D., A growth-factor-activated lysosomal K+ channel regulates Parkinson’s pathology. Nature 592, E10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karatas M., Dogan S., Spahiu E., Ašić A., Bešić L., Turan Y., Enzyme kinetics and inhibition parameters of human leukocyte glucosylceramidase. Heliyon 6, e05191 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsey I. S., Mokrab Y., Carvacho I., Sands Z. A., Sansom M. S. P., Clapham D. E., An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat. Struct. Mol. Biol. 17, 869–875 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeCoursey T. E., Voltage-gated proton channels: Molecular biology, physiology, and pathophysiology of the H(V) family. Physiol. Rev. 93, 599–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saotome K., Teng B., Tsui C. C. (. A.)., Lee W. H., Tu Y. H., Kaplan J. P., Sansom M. S. P., Liman E. R., Ward A. B., Structures of the otopetrin proton channels Otop1 and Otop3. Nat. Struct. Mol. Biol. 26, 518–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu Y. H., Cooper A. J., Teng B., Chang R. B., Artiga D. J., Turner H. N., Mulhall E. M., Ye W., Smith A. D., Liman E. R., An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047–1050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qiu F., Chamberlin A., Watkins B. M., Ionescu A., Perez M. E., Barro-Soria R., González C., Noskov S. Y., Larsson H. P., Molecular mechanism of Zn2+ inhibition of a voltage-gated proton channel. Proc. Natl. Acad. Sci. U.S.A. 113, E5962–E5971 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cang C., Bekele B., Ren D., The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 10, 463–469 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Goehring A., Lee C. H., Wang K. H., Michel J. C., Claxton D. P., Baconguis I., Althoff T., Fischer S., Garcia K. C., Gouaux E., Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng S. Q., Palovcak E., Armache J.-P., Verba K. A., Cheng Y., Agard D. A., MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Zivanov J., Nakane T., Scheres S. H. W., Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., Sliz P., Collaboration gets the most out of software. eLife 2, e01456 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., Bordoli L., Schwede T., SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E., UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.W. L. DeLano, The PyMOL Molecular Graphics System (2002).
- 38.Fiser A., Sali A., Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B. L., Grubmüller H., MacKerell A. D. Jr., CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. C., GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Zheng W., Yang J. W., Beauchamp E., Cai R., Hussein S., Hofmann L., Li Q., Flockerzi V., Berthiaume L. G., Tang J., Chen X. Z., Regulation of TRPP3 channel function by N-terminal domain palmitoylation and phosphorylation. J. Biol. Chem. 291, 25678–25691 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng W., Yang X., Hu R., Cai R., Hofmann L., Wang Z., Hu Q., Liu X., Bulkley D., Yu Y., Tang J., Flockerzi V., Cao Y., Cao E., Chen X. Z., Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels. Nat. Commun. 9, 2302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Table S1