To the Editor:
Achieving genetic diagnosis is at the core of management for inherited retinal degenerations (IRDs) for prognosis, family planning and access to novel therapies. Genetic panel testing can resolve ~70% of cases [1]. More comprehensive techniques including whole exome and whole genome sequencing (WES/WGS) are employed to explain unresolved pedigrees [2]. Inadvertent discovery of genetic findings unrelated to the testing indication is termed incidental, unsolicited, or secondary findings (SF) [3, 4]. SFs are discovered in 1.7% of WES-screened patients [5]. The American College of Medical Genetics (ACMG) has provided policy guidance regarding the management of SF, mandating pre-test counselling, designating 59 genes where SF have health-relevant actionable outcomes [3]. The ACMG stresses patient/guardian prerogative to opt-out of testing, but advises against clinicians altering the gene testing list at patient/guardian request to avoid SF.
With increased uptake of WES/WGS, SF “are highly likely, if not inevitable” [3, 4] and timing, manner and necessity of SF disclosure for research and clinical testing is debated. The Irish Health Service Executive released a national consent policy in 2019 (https://www.hse.ie/eng/about/who/qid/other-quality-improvement-programmes/consent/national-consent-policy.html), deferring to documented/signed patient preference regarding disclosure of SF; however, this opens a series of ethical dilemmas. Rising use of genotyping for clinical/research purposes, clearer guidance is required regarding SF. Here we outline an exemplar case in the ophthalmic genetics clinic.
A 23-year-old female with clinical retinitis pigmentosa had a negative genetic screen from a panel of 250 IRD-implicated genes [1] and had subsequent ‘trio-WES’ incorporating parental testing. Although an IRD-causative genetic variant was not identified, a PMS2 gene SF pathogenic missense variant, associated with Lynch Syndrome (OMIM#614337), was detected in the patient and mother (Table 1). Pre-WES consent had been given for testing/disclosure of common SF on the diagnostic laboratory’s consent form (https://blueprintgenetics.com/tests/whole-exome-sequencing/whole-exome-family-plus/). PMS2 is associated with increased risk of colon and endometrial cancer risk particularly >50 years. Recommended surveillance from UK Cancer Genetics Group involves 2-yearly colonoscopy (35–75 years), one-off Helicobacter pylori screening (>25 years) and risk-reducing hysterectomy (≥45 years) due to lack of effective endometrial cancer screening measures.
Table 1.
Patient’s WES results.
| Gene | Variant/Protein | Inheritance | Clinical relevance |
|---|---|---|---|
|
PMS2 (7p22.1) OMIM *600259 |
c.137 G>T, p.(Ser46Ile) |
AD |
Lynch Syndrome (OMIM #614337) Colon cancer (12–13% risk) Gastric cancer Small intestine cancer Hepatobiliary cancer Endometrial cancer (13% risk) Ovarian cancer Breast cancer Urinary tract cancer Brain cancer Skin cancer |
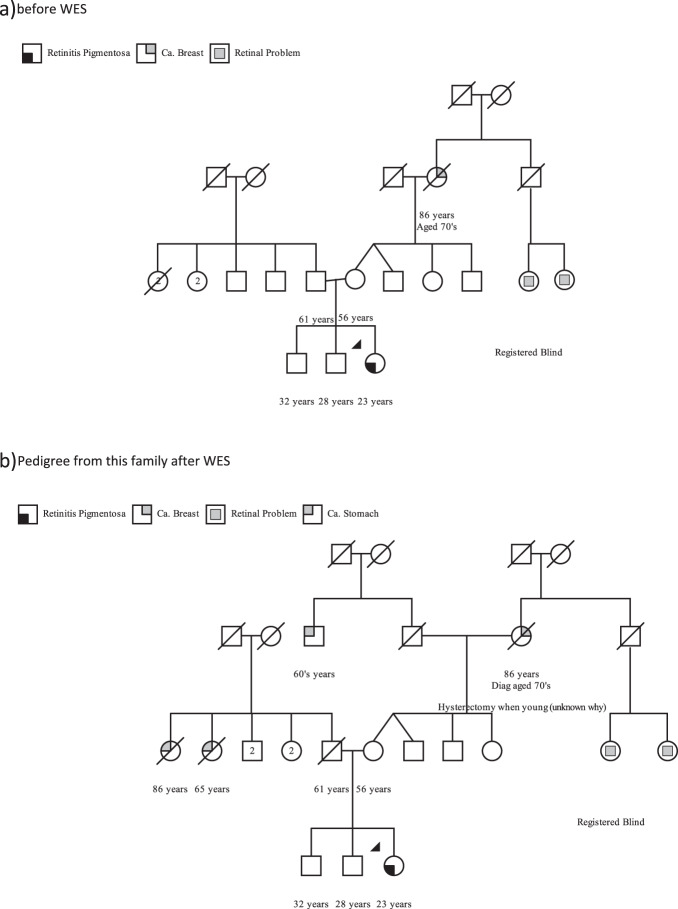
Disclosure of SF may inflict significant psychological burden, particularly in the setting of existing significant sensory disability (e.g., severe IRD). As the genotype-related actionable outcomes were most applicable to the patient’s mother, the mother was contacted first to discuss screening/surgical options. This discussion revealed further cancer history (Fig. 1), echoing published data that re-evaluation of family history post-SF reveals 150% positive family history findings versus first assessment [5].
Fig. 1. Pedigree from this family.
a Before WES. b Pedigree from this family after WES.
This case highlights the need for SF-specific informed pre-consent regarding testing and disclosure [4]. Following negative IRD-panel screening, pre-WES/WGS consent regarding the potential life-altering consequences of SF (mainly cancer-predisposition or cardiac risk genes), must be carried out. A re-evaluation of systemic family medical history should be conducted at this time. If the patient wishes to abstain from SF testing, it is important to discuss what will not be revealed in the analysis: carrier status and analysis of other medically relevant genes at the point of re-consent. A national consensus policy on SF testing/disclosure with focus on standardized consent and multidisciplinary infrastructure for enacting actionable outcomes is needed.
Acknowledgments
Funding
Fighting Blindness Ireland.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Julia Zhu, Email: T5000@mater.ie.
Kirk A J Stephenson, Email: kirkstephenson@hotmail.com.
G. Jane Farrar, Email: jane.farrar@tcd.ie.
Jacqueline Turner, Email: jturner@mater.ie.
James J. O’Byrne, Email: Jamesobyrne@mater.ie
David Keegan, Email: dkeegan@mater.ie.
References
- 1.Whelan L, Dockery A, Wynne N, Zhu J, Stephenson K, Silvestri G, et al. Findings from a genotyping study of over 1000 people with inherited retinal disorders in Ireland. Genes. 2020;11:105. doi: 10.3390/genes11010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, et al. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100:75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalia SS, Adelman K, Bale SJ, Chung WK, Eng C, Evans JP, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19:249–55. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 4.de Wert G, Dondorp W, Clarke A, Dequeker EM, Cordier C, Deans Z, et al. Opportunistic genomic screening. Recommendations of the European Society of Human Genetics. Eur J Human Genet. 2020;29:365–77. [DOI] [PMC free article] [PubMed]
- 5.Hart MR, Biesecker BB, Blout CL, Christensen KD, Amendola LM, Bergstrom KL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med: Off J Am Coll Med Genet. 2019;21:1261. doi: 10.1038/s41436-019-0440-2. [DOI] [PubMed] [Google Scholar]