Abstract
The cfiA gene, encoding an imipenem-hydrolyzing metallo-β-lactamase produced by Bacteroides fragilis, and insertion-like elements were detected by PCR amplification with B. fragilis strains isolated in Japan. The cfiA gene was found in 1.9 and 4.1% of the imipenem-susceptible B. fragilis isolates collected from 1987 to 1988 and from 1992 to 1994, respectively. Insertion-like elements adjacent to the cfiA gene were found in all nine metallo-β-lactamase-producing imipenem-resistant strains tested but not in nine cfiA-positive strains with no detectable metallo-β-lactamase activity.
Bacteroides fragilis is an anaerobic bacterium most frequently isolated from suppurative anaerobic infections and exhibits a broad spectrum of resistance to antimicrobial agents (17). Nationwide surveys in Japan and the United States showed imipenem to be very active against B. fragilis (2, 4). However, the emergence of resistance to imipenem among B. fragilis strains has been reported (1, 3, 5). It has been suggested that the production of an imipenem-hydrolyzing metallo-β-lactamase contributes to imipenem resistance among B. fragilis strains (1, 10).
The metallo-β-lactamase produced by B. fragilis is encoded by the cfiA gene (22), which has also been called the ccrA gene (15). A recent study demonstrated that an insertion element (IS), IS1186, located immediately upstream of the cfiA gene promoted the expression of this carbapenemase gene (13) as well as other insertion elements (14). Podglajen et al. suggested that a one-step mutation can allow the silent cfiA gene to be expressed (12). If so, B. fragilis strains carrying the silent cfiA gene would be expected to be eradicated in clinical settings before mutation occurs.
The aim of this study was to investigate the distribution of the cfiA gene among B. fragilis strains in Japan and to analyze the relationships between susceptibility to imipenem, metallo-β-lactamase production, and the presence of the cfiA gene adjacent to IS-like elements. A one-step mutation of cfiA-positive, imipenem-susceptible B. fragilis strains was also tested.
B. fragilis clinical strains used were placed into one of three groups. (i) The first group consisted of 21 stock strains, including 7 imipenem-resistant strains (MIC, ≥256 μg/ml, 4 strains; 32 μg/ml, 1 strain; and 16 μg/ml, 2 strains) from our laboratory, which were collected between 1986 and 1994 from various hospitals in Japan, and 13 imipenem-susceptible strains (MIC, 4 μg/ml, 1 strain; 1 μg/ml, 5 strains; and 0.5 μg/ml, 7 strains), and 1 imipenem-intermediate strain (MIC, 8 μg/ml), which were collected before 1987. (ii) The second group included 162 isolates, collected between 1987 and 1988, from a central clinical laboratory in Tokyo, Japan. (iii) The third group consisted of 124 isolates collected at Gifu University Hospital, Gifu, Japan, between 1992 and 1994.
Susceptibility was tested by an agar dilution method (8). Imipenem of known potency was obtained from Banyu Pharmaceutical, Tokyo, Japan.
Metallo-β-lactamase activity was assayed by both a spectrophotometric technique (1) and a biological method. For the biological assay, a 2-day culture of B. fragilis on modified Gifu anaerobe medium (GAM) agar (Nissui Pharmaceutical, Tokyo, Japan) was suspended in Anaerobe Broth MIC medium (Difco Laboratories, Detroit, Mich.). The cell suspension of 106 CFU/ml was mixed with the same volume of 200 mM 3-(N-morpholino)propanesulfonic acid–potassium hydroxide buffer (pH 7.2) containing imipenem at a final concentration of 6.3 μM or with imipenem solution supplemented with 2 mM EDTA. The mixture was incubated anaerobically for 18 h at 37°C. Imipenem alone and a mixture of imipenem and EDTA were incubated in parallel as controls.
To measure the remaining imipenem bioactivity, blank paper disks (Toyo-roshi, Tokyo, Japan) were impregnated with 30 μl of the mixture and placed on Antibiotic Medium 3 (Difco) plus 1.5% agar which was seeded with Bacillus subtilis MB-32 as an indicator strain. Plates were read for the presence of inhibition zones after overnight aerobic incubation at 37°C.
Bacterial DNA was obtained by heating cells for 10 min at 95°C. The primers for detection of the cfiA gene and IS-like elements and the predicted size of PCR products with primer sets are listed in Table 1. PCR amplification was run for 35 cycles consisting of 20 s at 95°C and 2 min at 64°C as described elsewhere (9). Southern hybridization was performed as described previously (7). Oligonucleotide probe GBI-3 was used for a PCR product with GBI-1 and GBI-2 primers, and oligonucleotide probe GBI-2 was used for an amplicon with GBI-3 and GBI-4 primers (Table 1).
TABLE 1.
Sequences of oligonucleotide primers and probes
| Genetic element and oligonucleotide | Sequence (5′–3′) | Positiona | Usage | Amplicon (predicted size) |
|---|---|---|---|---|
| cfiA gene | ||||
| GBI-1 | CCCAACTCTCGGACAAAGTG | 624–643 | Forward primer | GBI-1–GBI-2 (340 bp) |
| GBI-2 | AGTGAATCGGTGAATCCATG | 944–963 | Reverse primer and probe for a GBI-3 and GBI-4 primer set | |
| GBI-3 | CGAACCAGATGACGATAGAC | 891–910 | Forward primer and probe for a GBI-1 and GBI-2 primer set | GBI-3–GBI-4 (358 bp) |
| GBI-4 | ACGATCTGCTTGGTATGCTC | 1229–1248 | Reverse primer | GBI-1–GBI-4 (625 bp) |
| IS | ||||
| Gb | CGCCAAGCTTTGCCTGCCATTAT | Upstream of cfiA | Forward primer | G-E (approx. 2 kbp) |
| Eb | CTTCGAATTCGGCGAGGGATACATAA | Inside of cfiA | Reverse primer |
Four cfiA-positive and four cfiA-negative imipenem-susceptible strains were tested for a one-step mutation resulting in imipenem resistance. A 48-h culture of each of these strains was suspended in Anaerobe Broth MIC medium at a concentration of 109 CFU/ml. A 100-μl aliquot of cell suspension was spread on modified GAM agar containing 16 μg of imipenem per ml and incubated anaerobically for 72 h at 37°C. Ten colonies on each agar plate, if available, were subcultured on modified GAM agar and subjected to imipenem susceptibility testing as described above.
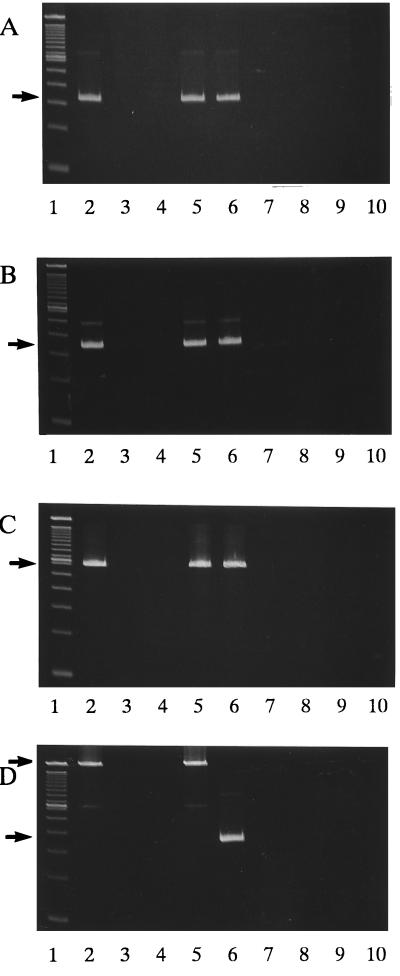
To detect the cfiA gene, PCR amplification with three primer sets (GBI-1 and GBI-2, GBI-3 and GBI-4, and GBI-1 and GBI-4) was carried out. A positive PCR test was detected in seven imipenem-resistant laboratory stock strains of B. fragilis which produced detectable levels of metallo-β-lactamase by spectrophotometric assay or bioassay; in one imipenem-susceptible strain, which produced no detectable metallo-β-lactamase; and in one imipenem-intermediate strains, which generated no detectable metallo-β-lactamase. Twelve other imipenem-susceptible strains, which had no detectable metallo-β-lactamase, had a negative PCR test. Representative PCR results are shown in Fig. 1A to C. The results of the Southern hybridization agreed with those of the PCR assay (data not shown). All seven imipenem-resistant strains were PCR positive for IS-like elements; a PCR product of approximately 2 kbp in size was generated (Fig. 1D, lanes 2 and 5). One imipenem-susceptible strain, which gave a positive PCR test for cfiA, was PCR negative for IS-like elements with an amplicon of approximately 400 bp (Fig. 1D, lane 6), a DNA size which indicates that there is no IS-like element immediately upstream of cfiA.
FIG. 1.
PCR amplification for detection of the cfiA gene with primers GBI-1 and GBI-2 (A), GBI-3 and GBI-4 (B), and GBI-1 and GBI-4 (C) and for detection of IS-like element with primers G and E (D). Lane 1, 100-bp DNA ladder (Gibco BRL); lanes 2 and 5, metallo-β-lactamase-producing, imipenem-resistant B. fragilis strains; lanes 3, 4, and 6 to 9, detectable metallo-β-lactamase-negative, imipenem-susceptible strains; lane 10, negative control without DNA sample. Arrows indicate 340-bp (A), 358-bp (B), 625-bp (C), ca. 2-kbp (D), and ca. 400-bp (D) amplicons. Lanes 2, 5, and 6 were PCR positive for the cfiA gene. Lanes 2 and 5 were positive for IS-like elements immediately upstream of the cfiA gene.
Prevalence of cfiA, susceptibility to imipenem, metallo-β-lactamase production, and carriage of IS-like elements were studied in two cohorts of B. fragilis strains (Table 2). Based on the results from the stock strains mentioned above, a primer set of GBI-1 and GBI-4 was used to detect cfiA. All cfiA-positive strains were subjected to a test for metallo-β-lactamase production by both spectrophotometric assay and bioassay.
TABLE 2.
Distribution of the cfiA gene and IS-like element and metallo-β-lactamase production among clinical isolates of B. fragilisa
| No. of strains tested (yr isolated) | Susceptibility to imipenem | cfiA gene | IS-like element | Metallo-β-lactamase production | No. of productive strains |
|---|---|---|---|---|---|
| 162 (1987–1988) | R | + | + | + | 2 |
| I | + | − | − | 1 | |
| S | + | − | −b | 3 | |
| S | − | −c | ND | 156 | |
| 124 (1992–1994) | R | − | − | − | 1 |
| I | − | − | ND | 1 | |
| S | + | − | −b | 5 | |
| S | − | −c | ND | 117 |
R, resistant with MICs of ≥16 μg/ml; I, intermediate with a MIC of 8 μg/ml; S, susceptible with MICs of ≤4 μg/ml; +, positive; −, negative; ND, not done.
One strain was tested.
Ten strains were tested.
Imipenem resistance was found in 2 (1.2%) of 162 strains recovered between 1987 and 1988 and 1 (0.8%) of 124 strains isolated between 1992 and 1994. Two resistant isolates collected between 1987 and 1988 had the cfiA gene and IS-like elements and produced metallo-β-lactamase, whereas one resistant strain (MIC of imipenem, 32 μg/ml) isolated between 1992 and 1994 was cfiA- and IS-negative and showed no detectable metallo-β-lactamase activity. The cfiA gene was detected in 1.9% of the 159 imipenem-susceptible strains isolated between 1987 and 1988 and in 4.1% of the 122 imipenem-susceptible strains recovered between 1992 and 1994. Regardless of the susceptibility to imipenem, the cfiA gene was found in 6 (3.7%) of the 162 strains isolated between 1987 and 1988 and 5 (4.0%) of the 124 strains isolated between 1992 and 1994.
Although tiny colonies were found after eight imipenem-susceptible strains were cultured on imipenem-supplemented agar plates, recovered colonies (irrespective of cfiA carriage) developed no resistance to imipenem by susceptibility testing and produced no detectable metallo-β-lactamase.
In this study of two cohorts of B. fragilis strains, the prevalence of the cfiA gene was 3.7 and 4.0%, respectively. Of imipenem-susceptible B. fragilis strains, 1.9% of the first cohort and 4.1% of the second cohort carried the cfiA gene. These results are relatively similar to those obtained in previous studies from France showing that approximately 3% of 500 randomly selected strains of B. fragilis were cfiA positive (14) and that a silent cfiA gene was found in 1.6% of the isolates (12, 13). The similarities derived from geographically distinct surveys suggest that the prevalence of cfiA-positive strains among B. fragilis may be relatively constant in each country.
Our study suggests that metallo-β-lactamase production is clearly related to the presence of the cfiA gene and IS-like elements immediately upstream of the metallo-β-lactamase gene. Gene activation by IS elements in B. fragilis is being identified: for example, IS21 (21) activation of the cepA gene (20); IS4351, IS942, and IS1186 (13, 16) activation of the ccrA or cfiA gene; IS4351 activation of the ermF gene (18, 19); and IS1170 and IS1169 activation of nimC and nimD (23).
In this study, one strain of B. fragilis (MIC of imipenem, 32 μg/ml) lacked production of metallo-β-lactamase. By contrast, Bacteroides distasonis (6) and Enterobacter cloacae (11) have been shown to have other imipenem resistance mechanisms, including reduced outer membrane permeability and the production of other types of β-lactamase, such as serine β-lactamase. Further studies are needed to determine the other resistance mechanism(s) of B. fragilis strains against imipenem.
The intraspecific transfer of imipenem resistance in a B. fragilis strain associated with the production of an imipenem-hydrolyzing metallo-β-lactamase has been previously reported (2). However, this earlier study has been the sole report of plasmid-mediated transmission of metallo-β-lactamase. Activation of the silent cfiA gene by one-step mutation was not confirmed in this study. Taken together, our data suggest that neither transfer of imipenem resistance by a plasmid nor spontaneous mutation leading to resistance seems to be a common way for B. fragilis to acquire resistance to imipenem.
Our study did not prove the conversion of cfiA gene-harboring imipenem-susceptible strains to imipenem resistance by a single mutation. This failure may be due to the lack of the IS element necessary for imipenem resistance within the strains tested.
The PCR assay described here, in combining detection of the cfiA gene and of IS-like elements immediately upstream of the cfiA gene, may be a useful tool to monitor the prevalence of metallo-β-lactamase-mediated imipenem-resistant B. fragilis strains.
Acknowledgments
This study was supported in part by a grant for the “Study of Drug-Resistant Bacteria,” founded by the Ministry of Health and Welfare, Japan, in 1996.
REFERENCES
- 1.Bandoh K, Muto Y, Watanabe K, Katoh N, Ueno K. Biochemical properties and purification of metallo-β-lactamase from Bacteroides fragilis. Antimicrob Agents Chemother. 1991;35:371–372. doi: 10.1128/aac.35.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bandoh K, Watanabe K, Muto Y, Tanaka Y, Kato N, Ueno K. Conjugal transfer of imipenem resistance in Bacteroides fragilis. J Antibiot. 1992;45:542–547. doi: 10.7164/antibiotics.45.542. [DOI] [PubMed] [Google Scholar]
- 3.Cuchural G J, Jr, Malamy M H, Tally F P. β-Lactamase-mediated imipenem resistance in Bacteroides fragilis. Antimicrob Agents Chemother. 1986;30:645–648. doi: 10.1128/aac.30.5.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cuchural G J, Jr, Tally F P, Jacobus N V, Aldridge K, Cleary T, Finegold S M, Hill G, Iannini P, O’Keefe J P, Pierson C, Crook D, Russo T, Hecht D. Susceptibility of the Bacteroides fragilis group in the United States: analysis by site of isolation. Antimicrob Agents Chemother. 1988;32:717–722. doi: 10.1128/aac.32.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hedberg M, Edlund C, Lindqvist L, Rylander M, Nord C E. Purification and characterization of an imipenem hydrolyzing metallo-β-lactamase from Bacteroides fragilis. J Antimicrob Chemother. 1992;29:105–113. doi: 10.1093/jac/29.2.105. [DOI] [PubMed] [Google Scholar]
- 6.Hurlbut S, Cuchural G J, Tally F P. Imipenem resistance in Bacteroides distasonis mediated by a novel β-lactamase. Antimicrob Agents Chemother. 1990;34:117–120. doi: 10.1128/aac.34.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jotwani R, Kato N, Kato H, Watanabe K, Ueno K. Detection of Bacteroides fragilis in clinical specimens by polymerase chain reaction amplification of the neuraminidase gene. Curr Microbiol. 1995;31:215–219. doi: 10.1007/BF00298376. [DOI] [PubMed] [Google Scholar]
- 8.Kato N, Kato H, Tanaka-Bando K, Watanabe K, Ueno K. Comparison of in vitro activities of DU-6859a and other fluoroquinolones against Japanese isolates of anaerobic bacteria. Clin Infect Dis. 1996;23(Suppl. 1):S31–S35. doi: 10.1093/clinids/23.supplement_1.s31. [DOI] [PubMed] [Google Scholar]
- 9.Kato N, Kato H, Watanabe K, Ueno K. Association of enterotoxigenic Bacteroides fragilis with bacteremia. Clin Infect Dis. 1996;23(Suppl 1):S83–S86. doi: 10.1093/clinids/23.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 10.Khushi T, Payne D J, Fosberry A, Reading C. Production of metal dependent β-lactamases by clinical strains of Bacteroides fragilis isolated before 1987. J Antimicrob Chemother. 1996;37:345–350. doi: 10.1093/jac/37.2.345. [DOI] [PubMed] [Google Scholar]
- 11.Lee E H, Nicolas M H, Kitzis M D, Pialoux G, Collatz E, Gutmann L. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob Agents Chemother. 1991;35:1093–1098. doi: 10.1128/aac.35.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podglajen I, Breuil J, Bordon F, Gutmann L, Collatz E. A silent carbapenemase gene in strains of Bacteroides fragilis can be expressed after a one-step mutation. FEMS Microbiol Lett. 1992;70:21–29. doi: 10.1016/0378-1097(92)90557-5. [DOI] [PubMed] [Google Scholar]
- 13.Podglajen I, Breuil J, Collatz E. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol Microbiol. 1994;12:105–114. doi: 10.1111/j.1365-2958.1994.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 14.Podglajen I, Breuil J, Casin I, Collatz E. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J Bacteriol. 1995;177:5270–5275. doi: 10.1128/jb.177.18.5270-5275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen B A, Gluzman Y, Tally F P. Cloning and sequencing of the class B β-lactamase gene (ccrA) from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother. 1990;34:1590–1592. doi: 10.1128/aac.34.8.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasmussen B A, Kovacs E. Identification and DNA sequence of a new Bacteroides fragilis insertion sequence-like element. Plasmid. 1991;25:141–144. doi: 10.1016/0147-619x(91)90027-t. [DOI] [PubMed] [Google Scholar]
- 17.Rasmussen B A, Bush K, Tally F P. Antimicrobial resistance in Bacteroides. Clin Infect Dis. 1993;16(Suppl. 4):S390–S400. doi: 10.1093/clinids/16.supplement_4.s390. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen J L, Odelson D A, Macrina F L. Complete nucleotide sequence and transcription of ermF, a macrolide-lincosamide-streptogramin B resistance determinant from Bacteroides fragilis. J Bacteriol. 1986;168:523–533. doi: 10.1128/jb.168.2.523-533.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen J L, Odelson D A, Macrina F L. Complete nucleotide sequence of insertion element IS4351 from Bacteroides fragilis. J Bacteriol. 1987;169:3573–3580. doi: 10.1128/jb.169.8.3573-3580.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers M B, Parker A C, Smith C J. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob Agents Chemother. 1993;37:2391–2400. doi: 10.1128/aac.37.11.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers M B, Bennett T K, Payne C M, Smith C J. Insertional activation of cepA leads to high-level β-lactamase expression in Bacteroides fragilis clinical isolates. J Bacteriol. 1994;176:4376–4384. doi: 10.1128/jb.176.14.4376-4384.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson J S, Malamy M H. Sequencing the gene for an imipenem-cefoxitin-hydrolyzing enzyme (CfiA) from Bacteroides fragilis TAL2480 reveals strong similarity between CfiA and Bacillus cereus β-lactamase II. J Bacteriol. 1990;172:2584–2593. doi: 10.1128/jb.172.5.2584-2593.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinh S, Haggoud A, Reysset G, Sebald M. Plasmids pIP419 and pIP421 from Bacteroides: 5-nitroimidazole resistance genes and their upstream insertion sequence elements. Microbiology. 1995;141:927–935. doi: 10.1099/13500872-141-4-927. [DOI] [PubMed] [Google Scholar]