Abstract
BACKGROUND
Clinical features from electronic health records (EHRs) can be used to build a complementary tool to predict coronary artery disease (CAD) susceptibility.
OBJECTIVES
The purpose of this study was to determine whether an EHR score can improve CAD prediction and reclassification 1 year before diagnosis, beyond conventional clinical guidelines as determined by the pooled cohort equations (PCE) and a polygenic risk score for CAD.
METHODS
We applied a machine learning framework using clinical features from the EHR in a multiethnic, clinical care cohort (BioMe) comprising 555 CAD cases and 6,349 control subjects and in a population-based cohort (UK Biobank) comprising 3,130 CAD cases and 378,344 control subjects for external validation.
RESULTS
Compared with the PCE, the EHR score improved CAD prediction by 12% in the BioMe Biobank and by 9% in the UK Biobank. The EHR score reclassified 25.8% and 15.2% individuals in each cohort respectively, compared with the PCE score. We observed larger improvements in the EHR score over the PCE in a subgroup of individuals with low CAD risk, with 20% increased discrimination and 34.4% increased reclassification. In all models, the polygenic risk score for CAD did not improve CAD prediction, compared with the PCE or EHR score.
CONCLUSIONS
The EHR score resulted in increased prediction and reclassification for CAD, demonstrating its potential use for population health monitoring of short-term CAD risk in large health systems.
Keywords: biobank, coronary artery disease, electronic health record, machine learning, polygenic risk score, pooled cohort equations, prevention
Early identification of individuals at risk of developing coronary artery disease (CAD) has been a long-standing goal in cardiovascular medicine.1 Risk assessment methods have been designed to predict an individual’s long-term risk to disease. Conventional guidelines use the American College of Cardiology and American Heart Association pooled cohort equations (PCE) to assess prescription of cholesterol-lowering medications (statins) for prevention of atherosclerotic cardiovascular disease (ASCVD).2 Current guidelines recommend that individuals 40–75 years of age with a PCE score ≥7.5%, without clinical ASCVD or diabetes, and with low-density lipoprotein cholesterol 70 to 189 mg/dL will benefit from cholesterol-lowering treatment.3 However, known limitations of the PCE score include both underestimation and overestimation of CAD risk,4–6 as well as biases in certain populations.7–9 Therefore, it is important to identify alternative approaches that complement PCE-based risk prediction and stratification.
Genetic studies have investigated the clinical utility of a polygenic risk score (PRS) for CAD. A recent study reported a 3-fold increased risk of CAD among the top 8% of individuals with a high PRS,10 suggesting that high PRS can potentially be used for CAD risk stratification.11 However, subsequent studies have shown only minimal gains in CAD prediction when PRS is considered along with the PCE.12,13 One study further showed that a high PRS is insensitive to conventional clinical guidelines using the PCE, highlighting that PRS and the PCE capture different axes of CAD risk.14 In addition, PRS for CAD are predominantly derived from European ancestry cohorts and have reduced performance in non-European individuals.15 Thus, the clinical utility of high PRS for CAD remains unclear and is an area of ongoing research.16 Nevertheless, this underscores the need to develop new prediction scores that capture different axes of CAD risk.
Electronic health records (EHRs) enable the evaluation of disease risk for a wide range of conditions. These large data sets provide a rich resource of structured and unstructured medical data that can be used to capture core characteristics of several diseases, making them ideal for clinical risk prediction and stratification. A number of studies have begun to use machine learning (ML)-based approaches to accurately predict complex diseases,17 including CAD.18–21 However, its utility for automated short-term prediction of CAD in hospital settings remains unexplored, particularly in low-risk individuals determined by conventional clinical guidelines. A prediction score built using system-wide EHR and ML can identify residual risk of disease not captured by conventional tools that have been built on a reduced number of traditional risk factors. Such a prediction score can potentially be used as a population health screening tool to identify high-risk patients within large EHR-based health systems.
Here, we test whether an EHR score built using an ML approach on EHR-based clinical features can improve 1-year risk prediction and reclassification of CAD beyond that of a conventional clinical risk score, the PCE, as well as a PRS for CAD. To evaluate this, we used a multiethnic, EHR-linked clinical care cohort in the BioMe Biobank comprising 555 CAD cases and 6,349 control subjects (n = 6,904 total), together with a validation cohort of 3,130 CAD cases and 378,344 controls in the UK Biobank (n = 381,474 total).
METHODS
SAMPLE COHORTS.
We first assessed the predictive performance of clinical features from the EHR, PCE, and PRS in the discovery cohort of the BioMe Biobank, a multiethnic, EHR-linked, clinical care biobank of >30,000 individuals. The study was approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board. We limited the study population to one for which the American College of Cardiology and American Heart Association PCE is designed to guide statin initiation. We selected only individuals 40–79 years of age and eliminated individuals taking cholesterol-lowering medications (statins) and individuals with second-degree relatedness or higher.
UK Biobank, a population-based cohort with EHR and genotype data from 502,505 individuals in the United Kingdom, was used as a validation cohort. Data gathering and preprocessing were performed using the same criteria used in BioMe.
Our primary analysis was performed solely on CAD. A secondary analysis was performed on stroke alone and on ASCVD (CAD, angina, and stroke). More detailed information can be found in the Supplemental Appendix.
CLINICAL FEATURES FROM THE EHR.
In the BioMe Biobank, we considered both categorical and continuous data as clinical features. To predict 1-year risk to CAD (as well as ASCVD and stroke in secondary analysis), only clinical features 1 year before first diagnosis were considered for all cases. To ensure measurements were consistent across timepoints, continuous features 1 year after the most recent entry were removed for all individuals. Age was defined according to the last considered entry. All categorical features available in the BioMe biobank were used including 14,940 International Classification of Diseases (ICD)-9/10 diagnosis codes, and 27,232 medications with repeated prescriptions. For categorical data, presence of a diagnostic code/medication prescription in the EHR was coded as “1”; the absence of a diagnostic code/ medication prescription was coded as “0.” Continuous features with >60% missing values were discarded. In total, 107 laboratory and 9 vital traits, measured as part of routine clinical care, were used in posterior analyses. Individuals with >60% missing values were also discarded. All remaining missing values were imputed using a random forest-based algorithm (missForest version 1.4).22 For continuous features with multiple timepoints in an individual, the median values were considered. After imputation, features with a Pearson’s correlation coefficient >0.90 were removed. In instances where 2 features were highly correlated, the one with the higher added correlation with all other features was removed. Statistical variability for 39 ICD-9/10 diagnosis codes, 37 medications, 31 laboratory results, and 9 vital traits used to train the models can be found in Supplemental Table 1.
In the UK Biobank, we performed 1-year prediction by using EHR data 1 year before first diagnosis for all CAD cases. Similarly, continuous data were restricted to a 1-year margin after the most recent entry. Continuous features and patients with >60% missing values were discarded, and the remaining were imputed. Detailed information on how the PCE and PRS were calculated can be found in the Supplemental Appendix.
ML-BASED APPROACH TO DEVELOP PREDICTIVE MODELS IN BioMe BIOBANK.
We implemented a ML approach to develop the EHR score using clinical features from the EHR. To minimize sampling bias, we repeated the workflow 100 times using different samples for training and testing in each iteration. The ML workflow is described below for a single iteration (Figure 1A). We randomly selected 90% of cases and an equal number of noncomorbid control subjects to create the train set. Given that BioMe is a hospital-based cohort with greater prevalence of disease, only noncomorbid control subjects were used for training to minimize misclassification. Two separate balanced test sets were generated using the remaining 10% of cases and either of the following: 1) noncomorbid control subjects; or 2) a random sample of control subjects. All subsequent steps were performed only on the train set and then applied to the test set to avoid overfitting. We reduced the complexity of the model by performing feature selection23 on the continuous and categorical train set separately to make the prediction task clinically interpretable.24 Nonselected features were then removed from the test set accordingly. Age, sex, and self-reported ethnicity or 10 principal components (10 PCs) of genetic ancestry were used as covariates. Continuous features were scaled within the train set, and the resulting metrics were used to scale the test set accordingly. The model regresses predictions from 3 algorithms to compute a final score, namely random forest,25 gradient boosted trees,26 and support vector machine with polynomial kernel27 using the caret package28 in R (R Foundation for Statistical Computing). A table describing the hyperparameters evaluated for each algorithm can be found in the Supplemental Table 2. For each algorithm, hyperparameters were optimized using an internal 10-fold cross-validation within the train set. The resulting model was then used to predict CAD, ASCVD, and stroke cases in the test set. By randomly selecting samples in each iteration, we obtained performance metrices representative of the entire population while still avoiding overfitting. The reported performance metrics correspond to the mean ± SD of performance metrics across the 100 iterations. Further details regarding testing of different combinations of the EHR score, PCE, and PRS in various models and sampling of the test sets can be found in the Supplemental Methods.
FIGURE 1.
Study Design and Flowchart The BioMe Biobank (A) was used for training and the UK Biobank (B) was used for external validation. Statin indicates cholesterol-lowering medications. AUROC = area under the receiver-operating characteristic curve; GBT = gradient boosted trees; NRI = net reclassification improvement; PCE = pooled cohort equations; RF = random forest; SVM = support vector machines.
ML VALIDATION IN UK BIOBANK.
To predict 1-year risk for disease in the validation cohort of UK Biobank, we trained 100 new models. For each iteration in the previously mentioned training process, common features between UK Biobank and selected features in BioMe were used to train a new model (Figure 1B). Each model was then used to predict all CAD cases in UK Biobank and an equal number of control subjects to ensure a balanced validation set. Continuous features were scaled according to the metrics used to scale the identical features in the train set for each iteration. Reported performance metrics are the mean value and SD across all predictions performed with each model. Further information regarding sampling and PCE and baseline models in UK Biobank can be found in the Supplemental Methods.
STATISTICAL ANALYSES.
Statistical analyses were performed using R software version 3.5.3.29 Predictive performance for the models was measured using the receiver-operating characteristic curve using the R pROC package version 1.14.0.30 Sensitivity, specificity, negative predictive value, positive predictive value, and overall accuracy of the models were calculated using R caret package version 6.0.84. Overall, in-cases and in-control subjects net reclassification improvement (NRI) was calculated using the R nricens package version 1.6.31
RESULTS
BASELINE STUDY POPULATIONS.
The BioMe Biobank comprised a total of 30,825 individuals of diverse ancestries (7,615 African Americans, 10,536 Hispanic Americans, 9,370 European Americans, and 3,304 individuals from other ancestries). Filtering resulted in 555 cases and 6,349 control subjects for use in the ML workflow (Supplemental Table 3). In UK Biobank, there were a total of 502,505 individuals (472,695 White, 9,882 Asian or Asian British, 8,061 Black or Black British, 2,958 Mixed, 1,574 Chinese, 4,558 other ethnic group, and 2,777 not specified). After applying similar filtering criteria as in the BioMe sample (see Methods section), we identified 3,130 CAD cases and 378,344 control subjects in UK Biobank as the validation cohort. In a subset of these individuals, we assessed the predictive performance of the EHR score in individuals who would typically have low predicted CAD risk using the PCE <7.5. We identified 555 CAD cases (114 low-risk) and 6,349 control subjects (3,185 low-risk) in the BioMe Biobank and 3,130 CAD cases (589 low-risk) and 378,344 control subjects (204,143 low-risk) in UK Biobank.
EHR MODEL IMPROVES CAD PREDICTION.
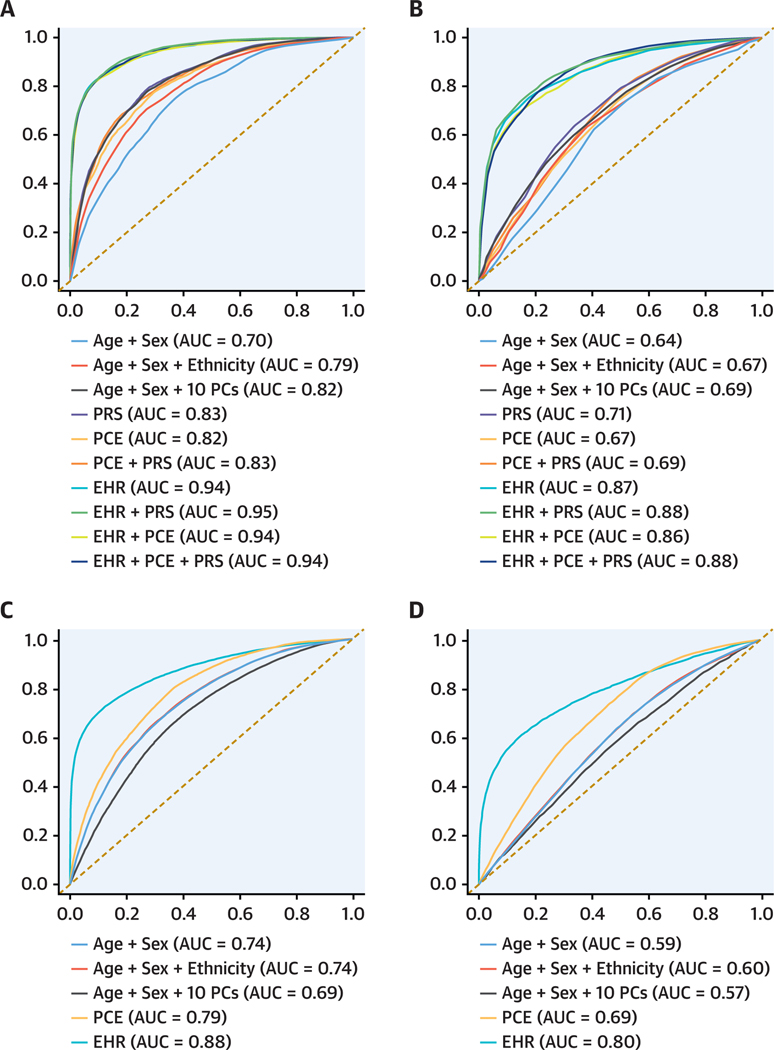
We first assessed the predictive performance of 1-year risk of CAD for the PCE, PRS, and EHR models, both individually and in combination, on 6,904 CAD cases and control subjects in the BioMe cohort. The distribution of PCE and PRS is shown in Supplemental Figure 1. Compared with the baseline model (age + sex + ethnicity), the PCE improved CAD prediction by 3% (area under the receiver-operating characteristic curve [AUROC]: 0.82 for PCE vs AUROC: 0.79 for age + sex + ethnicity) on the noncomorbid (Table 1, Figure 2A) and 4% (AUROC: 0.75 for PCE vs AUROC: 0.71 for age + sex + ethnicity) on the random (Supplemental Table 4A, Supplemental Figure 2A) test set. The addition of PRS did not contribute to either the noncomorbid (AUROC: 0.83 for PRS [age + sex + 10 PCs] vs AUROC: 0.82 for age + sex + 10 PCs) or the random test set (AUROC: 0.79 for PRS vs AUROC: 0.78 for age + sex + 10 PCs).
table 1.
Mean Performance Metrics for 1-Year Risk Prediction of CAD
| All | Low-Risk | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Model | AUROC | PPV | NPV | AUROC | PPV | NPV |
|
| ||||||
| BioMe | ||||||
| Age + sex | 0.70 ± 0.04 | 0.68 ± 0.04 | 0.69 ± 0.04 | 0.64 ± 0.12 | 0.60 ± 0.17 | 0.63 ± 0.13 |
| Age + sex + eth | 0.79 ± 0.04 | 0.70 ± 0.04 | 0.72 ± 0.05 | 0.67 ± 0.11 | 0.62 ± 0.11 | 0.64 ± 0.13 |
| Age + sex + 10 PC | 0.82 ± 0.03 | 0.75 ± 0.04 | 0.76 ± 0.04 | 0.69 ± 0.12 | 0.66 ± 0.12 | 0.65 ± 0.12 |
| PRS | 0.83 ± 0.03 | 0.76 ± 0.05 | 0.76 ± 0.04 | 0.71 ± 0.11 | 0.66 ± 0.12 | 0.66 ± 0.11 |
| PCE | 0.82 ± 0.04 | 0.74 ± 0.04 | 0.73 ± 0.05 | 0.67 ± 0.13 | 0.61 ± 0.11 | 0.63 ± 0.15 |
| PCE + PRS | 0.83 ± 0.04 | 0.76 ± 0.04 | 0.74 ± 0.05 | 0.69 ± 0.13 | 0.63 ± 0.13 | 0.63 ± 0.13 |
| EHR | 0.94 ± 0.02 | 0.88 ± 0.04 | 0.85 ± 0.04 | 0.87 ± 0.07 | 0.81 ± 0.10 | 0.78 ± 0.10 |
| EHR + PRS | 0.95 ± 0.02 | 0.88 ± 0.04 | 0.85 ± 0.04 | 0.88 ± 0.07 | 0.81 ± 0.09 | 0.80 ± 0.09 |
| EHR + PCE | 0.94 ± 0.02 | 0.88 ± 0.03 | 0.85 ± 0.04 | 0.86 ± 0.07 | 0.78 ± 0.10 | 0.77 ± 0.10 |
| EHR + PCE + PRS | 0.94 ± 0.02 | 0.88 ± 0.04 | 0.85 ± 0.04 | 0.88 ± 0.07 | 0.79 ± 0.11 | 0.79 ± 0.10 |
| UK Biobank | ||||||
| Age + sex | 0.74 ± 0.02 | 0.67 ± 0.02 | 0.68 ± 0.02 | 0.59 ± 0.02 | 0.57 ± 0.01 | 0.57 ± 0.03 |
| Age + sex + eth | 0.74 ± 0.02 | 0.67 ± 0.02 | 0.68 ± 0.02 | 0.60 ± 0.03 | 0.57 ± 0.02 | 0.58 ± 0.03 |
| Age + sex + 10 PC | 0.69 ± 0.02 | 0.58 ± 0.04 | 0.68 ± 0.02 | 0.57 ± 0.04 | 0.50 ± 0.02 | 0.81 ± 0.02 |
| PCE | 0.79 ± 0.02 | 0.69 ± 0.02 | 0.75 ± 0.03 | 0.69 ± 0.01 | 0.62 ± 0.02 | 0.65 ± 0.03 |
| EHR | 0.88 ± 0.01 | 0.92 ± 0.02 | 0.73 ± 0.02 | 0.80 ± 0.04 | 0.68 ± 0.10 | 0.87 ± 0.02 |
Values are mean ± SD across 100 iterations. Performance metrics for models are shown for BioMe and UK Biobank. Columns correspond to performance metrics in test set from BioMe Biobank and validation set from UK Biobank. Rows correspond to the model being tested.
AUROC = area under the receiver-operating characteristic curve; EHR = electronic health records; eth = ethnicity; NPV = negative predicted value; PC = principal components; PCE = pooled cohort equations; PRS = polygenic risk score; PPV = positive predicted value.
FIGURE 2.
Receiver-Operating Characteristic Curves for 1-Year CAD Risk Prediction Models The models were tested in BioMe (A), low-risk BioMe (B), UK Biobank (C), and low-risk UK Biobank (D) individuals. The y- and x-axes correspond to averaged true positive and false negative rates, respectively, across 100 iterations. The averaged area under the curve is indicated for every model. AUC = area under the curve; EHR = electronic health record; PC = principal component; PCE = pooled cohort equations; PRS = polygenic risk score.
All models with the EHR score had higher predictive performance in both the noncomorbid (AUROC: 0.94–0.95 for various EHR models) and random test sets (AUROC: 0.80 for various EHR models), compared with models without the EHR score (AUROC ≤0.83 in the noncomorbid test sets and AUROC ≤0.79 in the random test sets). The EHR score improved CAD prediction by 12% and 5% when compared with the PCE and 15% and 9% when compared with the baseline model in the noncomorbid and random test sets, respectively. Performance metrics were consistent across ancestries (Supplemental Table 5). Similar results were observed in the validation cohort from UK Biobank where the EHR score contributed the most to CAD prediction in comparison to the PCE with a 9% and 4% improvement in the noncomorbid (Table 1, Figure 2C) and random test sets (Supplemental Table 4C, Supplemental Figure 2C), respectively.
Next, we evaluated whether the EHR model can predict 1-year risk of CAD adequately among low-risk individuals with a PCE score <7.5 in both the BioMe Biobank and UK Biobank. Similar to results observed in all individuals, the PRS contributed little to CAD prediction (Table 1, Figure 2B). In contrast, the EHR model had considerably better predictive performance even among this low-risk group(AUROC:0.87forEHR),with a 20% and 11% improvement over the PCE in the BioMe Biobank and UK Biobank, respectively (Table 1, Figures 2B and 2D). Similar performance in internal cross-validation from the training process was observed in both the all and low-risk individuals’ models (Supplemental Table 6, Supplemental Figure 3), suggesting minimal overfitting.
EHR MODEL IMPROVES RECLASSIFICATION OF CAD RISK.
In addition to prediction, we also compared the ability of the PRS and EHR score to reclassify 1-year CAD risk originating from the PCE score. In line with the discrimination results, the PRS contributed little to reclassifying CAD risk (NRI = 3.8 for PRS vs NRI = 3.4 for age + sex + 10 PCs) (Table 2). On the other hand, the EHR score reclassified a large number of CAD cases and control subjects (NRI = 25.8 and NRI = 5.0 for EHR in the noncomorbid and random test set, respectively) (Table 2, Supplemental Table 7A). In the validation cohort from UK Biobank, the EHR score substantially reclassified 15.2% and 5.5% of cases and control subjects in the noncomorbid and random test set, respectively (NRI = 15.2 and NRI = 5.5, respectively, for EHR) (Table 2).
table 2.
Mean Net Reclassification Improvement for 1-Year Risk Prediction of CAD
| All | Low-Risk | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Model | Overall | Cases | Control Subjects | Overall | Cases | Control Subjects |
|
| ||||||
| BioMe | ||||||
| Age + sex | −9.6 ± 7.8 | −1.8 ± 7.0 | −7.8 ± 7.1 | −0.9 ± 28.2 | 4.2 ± 26.1 | −5.1 ± 23.3 |
| Age + sex + eth | −4.6 ± 7.2 | 1.1 ± 6.4 | −5.7 ± 5.7 | 2.1 ± 26.9 | 4.4 ± 24.3 | −2.3 ± 19.4 |
| Age + sex + 10 PC | 3.4 ± 7.7 | 4.4 ± 6.3 | −1.0 ± 6.1 | 5.1 ± 24.1 | 0.7 ± 23.4 | 4.4 ± 19.2 |
| PRS | 3.8 ± 7.7 | 3.7 ± 6.4 | 0.2 ± 6.2 | 6.9 ± 26.6 | 2.2 ± 24.0 | 4.7 ± 19.6 |
| PCE + PRS | 2.7 ± 5.5 | 0.6 ± 5.2 | 2.1 ± 4.7 | 4.2 ± 23.6 | 4.2 ± 23.0 | 0.0 ± 17.2 |
| EHR | 25.8 ± 8.5 | 12.4 ± 7.5 | 13.4 ± 5.6 | 34.4 ± 27.1 | 14.0 ± 23.5 | 20.4 ± 18.4 |
| EHR + PRS | 26.1 ± 8.7 | 12.4 ± 7.7 | 13.7 ± 5.1 | 35.6 ± 26.9 | 15.6 ± 23.6 | 19.7 ± 17.3 |
| EHR + PCE | 25.1 ± 7.7 | 12.2 ± 7.0 | 12.9 ± 5.1 | 31.3 ± 26.9 | 14.8 ± 23.0 | 18.5 ± 18.0 |
| EHR + PCE + PRS | 25.6 ± 7.7 | 12.7 ± 7.1 | 12.9 ± 4.8 | 33.7 ± 25.5 | 16.1 ± 22.0 | 17.6 ± 17.8 |
| UK Biobank | ||||||
| Age + sex | −7.9 ± 2.8 | −8.9 ± 4.0 | 1.0 ± 4.1 | −13.0 ± 4.7 | −8.8 ± 11.4 | −4.2 ± 8.9 |
| Age + sex + eth | −7.7 ± 2.7 | −8.3 ± 3.4 | 0.6 ± 3.8 | −12.8 ± 6.0 | −6.8 ± 11.0 | −6.0 ± 8.4 |
| Age + sex + 10 PC | −15.1 ± 3.8 | −35.4 ± 4.7 | 20.3 ± 3.7 | −38.3 ± 45.2 | −66.4 ± 11.2 | 27.6 ± 43.5 |
| EHR | 15.2 ± 4.1 | −13.5 ± 4.7 | 28.7 ± 3.9 | 9.3 ± 53.5 | −14.6 ± 14.7 | 24.1 ± 49.3 |
Values are mean ± across 100 iterations. Performance metrics for models are shown for BioMe and UK Biobank. Columns correspond to overall and by-class Net Reclassification Improvement in test set from BioMe Biobank and validation set from UK Biobank. Rows correspond to the model being tested.
Abbreviations as in Table 1.
In low-risk individuals with a PCE score <7.5, we observed modest reclassification when adding PRS to the PCE model (NRI = 4.2 for PCE + PRS) (Table 2). In the BioMe Biobank and UK Biobank, the EHR model provided notable improvement with 34.4% and 9.3% of individuals being reclassified in each cohort, respectively (Table 2).
EHR MODEL LEVERAGES CLINICAL RISK OVER ESTIMATION AMONG CASES AND CONTROL SUBJECTS.
We next evaluated the predictive performance of the EHR model in CAD cases and control subjects separately. Among cases, we observed 14% and 23% higher positive predictive value (probability of being true positives) and 12% and −13% change in sensitivity (percentage of cases identified) for the EHR score compared with the PCE score in the BioMe Biobank and the UK Biobank, respectively (Table 1, Supplemental Table 8). Among control subjects in BioMe, we observed 12% and −2% change in negative predictive value (probability of being true negatives) and 13% and 29% higher specificity (percentage of controls identified) for the EHR score compared with the PCE score in the BioMe Biobank and UK Biobank, respectively.
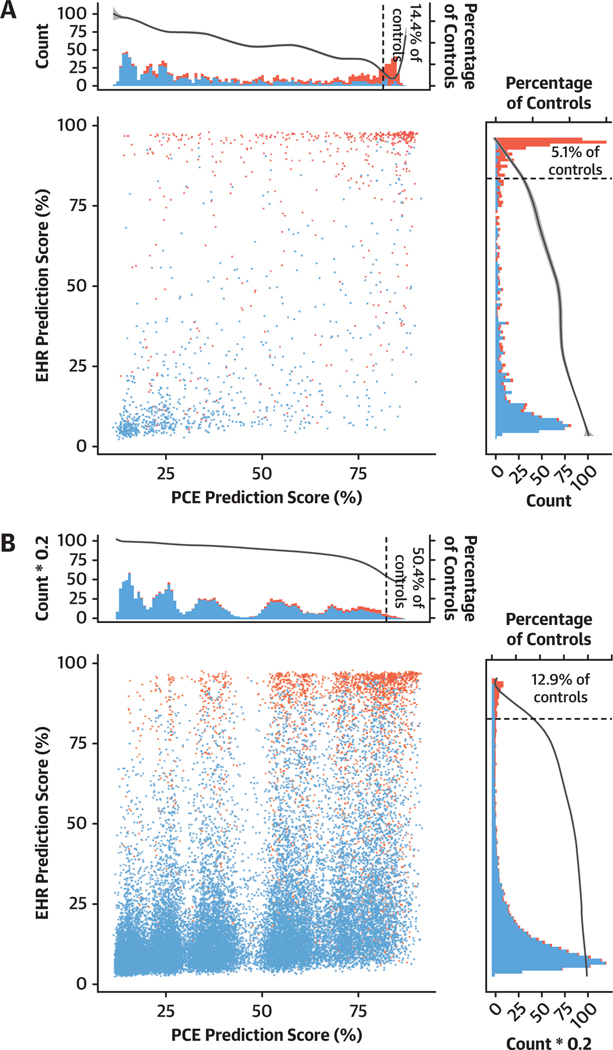
Previous studies have reported that the PCE score tends to overestimate CAD risk.4,5,7,8 We investigated this finding by examining the proportion of false positives (control subjects with overestimated clinical risk) in the top 15% of EHR and PCE scores. We observed 5.1% and 12.9% of false positives in the top 15% of the EHR scores compared with 14.4% and 50.4% of false positives in the top 15% of the PCE scores in the BioMe Biobank and UK Biobank, respectively (Figure 3). These results suggest that the EHR score can correctly reclassify CAD risk. Notably, a larger difference in UK Biobank indicates increased reclassification in settings where PCE can overestimate clinical risk for CAD.
FIGURE 3.
Change in Prediction Score Between PCE model and EHR Model The models were tested in BioMe (A) and UK Biobank (B) individuals. Dot plot shows the averaged prediction of the EHR and PCE models for every tested individual in the BioMe and UK Biobank cohorts. Red and blue indicates actual cases and control subjects, respectively. Top and right histograms show the distribution of averaged PCE and EHR prediction scores, respectively. The secondary y-axes in the histograms show the percentage of control subjects predicted in a 1% wide bin of the prediction score. Dotted vertical line indicates the 85% threshold. Abbreviations as in Figure 2.
NONTRADITIONAL CLINICAL FEATURES IN THE EHR SCORE CONFER PREDICTIVE POWER FOR CAD.
Given that the EHR score maximally predicts CAD risk, we examined the most important features in training the EHR score. When training either the model encompassing all individuals or those with low-risk with PCE <7.5%, we observed that only 1 (age) of the 10 most important features was a risk factor used to calculate the PCE score (Supplemental Figure 4). Other important features included primary essential hypertension (I10), hemoglobin A1c, low-density lipoprotein cholesterol, estimated glomerular filtration rate, and red blood cell distribution width, among others. Although we considered 116 features in the EHR model, we observed that only a small number of features contributes substantially to CAD prediction, and most features are not known risk factors for CAD (Supplemental Figure 5).
PREDICTION OF ASCVD AND STROKE.
In secondary analysis, we further assessed the predictive performance of 1-year risk of ASCVD and stroke for the PCE and EHR models. By applying the same filtering criteria, 715 cases and 6,359 control subjects for ASCVD and 237 cases and 7,268 control subjects for stroke were used in the BioMe Biobank.
Both the ASCVD and stroke prediction models showed similar performance compared with the CAD EHR score. Both the ASCVD and stroke models improved prediction by 16% when compared with the PCE score (AUROC: 0.93 for EHR vs 0.77 for PCE and AUROC: 0.94 for EHR vs 0.78 for PCE, respectively) (Supplemental Tables 9 and 10, Supplemental Figures 6 and 7).
DISCUSSION
Our findings have several implications for risk prediction of CAD. First, our study is consistent with others showing that the addition of PRS does not improve prediction power for CAD as measured by discrimination metrics like AUROC.
Second, the addition of EHR-based clinical features conferred 12% improvement in 1-year risk prediction of CAD, when compared with the PCE. We observed similar improvements for risk prediction of ASCVD and stroke. Many of the features in the EHR model were nontraditional, suggesting that additional disease risk can be captured using alternative sources of clinical information. An EHR score can potentially serve as an automated health screening tool enabling systematic identification of high-risk individuals for ASCVD based on their prior medical history in the EHR (Central Illustration). The clinical use case for this score would be to identify high-risk individuals (who are flagged as low risk by traditional scores), optimizing prevention and care using embedded pathways. However, development of this algorithm is just the starting point; there are several steps before such approaches become part of routine clinical care. First, there needs to be prospective validation to ensure that calibration and discrimination power are within acceptable parameters,17 input features in the model are explained adequately,32 and the performance is consistent across key subgroups including gender and race/ethnicity.33 Second, the model and associated clinical decision support need to be tested in a randomized fashion by recognized guidelines (including SPIRIT-AI [Standard Protocol Items: Recommendations for Interventional Trials-Artificial Intelligence]34) to ensure that such alerts are leading to patient benefit and not harm. Finally, after clinical implementation, the model needs to be monitored prospectively to ensure issues such as data set shift do not occur; if they do occur, model retraining to ensure acceptable performance needs to be performed.35
CENTRAL ILLUSTRATION.
Electronic Health Record Clinical Features Predict Short-Term Coronary Artery Disease Risk
Clinical features from the electronic health record can predict 1-year coronary artery disease risk with a discrimination power of 0.94 AUROC and reclassify 25.8% of predictions coming from the Pooled Cohort Equations. AUROC = area under the receiver-operating characteristic curve; CAD = coronary artery disease; EHR = electronic health record; NRI = net reclassification improvement; PCE = Pooled Cohort Equations.
Third, improvement of the EHR score over the PCE was particularly notable in individuals with low predicted risk for CAD with PCE <7.5. The good prediction performance of the EHR score in this subgroup indicates that it can be used for CAD prediction in a subgroup of individuals for which the PCE has neither granularity nor power to estimate clinical risk.
Finally, we observed that the use of the EHR score showed higher confidence in predicting CAD cases and control subjects separately. This is particularly relevant because the PCE overestimates clinical risk for CAD,5 indicating these can be used as complementary scores. Higher confidence in calling CAD cases and identifying control subjects by the EHR score corrects for misclassified CAD risk by the PCE. Moreover, the EHR score tackles short-term risk prediction, whereas the PCE was developed for long-term risk prediction. Consideration of these scores together can help mitigate overestimation stemming from the PCE alone while informing different aspects of CAD risk.
STUDY LIMITATIONS.
First, CAD cases were identified using ICD codes that could result in misclassification of cases and/or control subjects.36,37 However, prior studies have replicated known genetic associations with CAD status using these codes. Second, restricting the data to 1 year before diagnosis resulted in a small sample size of low-risk cases. Even though sampling bias was accounted for by training 100 models, the small sample size can compromise discrimination power of the EHR score. Third, the single nucleotide variant weights considered for this analysis were derived from a GWAS meta-analysis using a majority (77%) of participants with European ancestry. These were used to calculate PRS for CAD in individuals from diverse ancestries in BioMe, which could limit the discrimination capacity of the PRS.38 However, the low predictive power of the PRS on CAD is consistent with previous reports.12,13 Fourth, given that BioMe is a hospital-based cohort, noncomorbid control subjects were necessarily used for training, which likely decreased EHR score performance in the random test set, highlighting the importance of selecting a clean set of control subjects in the training set. Fifth, the study cohort was limited to BioMe Biobank participants with genotype data to compute the PRS. A broader population not linked to genetic data can be used in future studies, as the EHR score was built on clinical features derived from routine clinical care. Sixth, by building the EHR, we may miss sources of information not recorded in the EHR (eg, nutrition, physical activity, emotional well-being) that may be informative for CAD prediction.
CONCLUSIONS
An EHR score confers greater discriminative power for CAD compared with the PRS and PCE. Appropriate calibration of an EHR score can correct the widely reported overestimation of CAD risk originating from the PCE. The use of clinical features from the EHR can complement conventional clinical risk scores for clinical risk prediction and stratification of CAD.
Supplementary Material
COMPETENCY IN SYSTEMS-BASED PRACTICE:
A score derived from clinical data extracted from the EHR can improve 1-year CAD risk prediction and reclassification beyond conventional guideline-based calculators or polygenic risk scores.
TRANSLATIONAL OUTLOOK:
Further research is needed to assess the pragmatic utility of the EHR score for population-based short-term CAD risk assessment in large-scale health care systems.
Acknowledgments
FUNDING SUPPORT AND AUTHOR DISCLOSURES
Mr Forrest is supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) (T32-GM007280). Dr Nadkarni is supported by a career development award from the NIH (K23-DK107908) and by NIH grants (R01-DK108803, U01-HG007278, U01-HG009610, and U01-DK116100); is scientific cofounder, consultant, advisory board member, and equity owner of Renalytix AI; is a scientific cofounder and equity holder for Pensieve Health; has served as a consultant for Variant Bio; has received grants from Goldfinch Bio; and has received personal fees from Renalytix AI, BioVie, Reata, AstraZeneca, and GLG Consulting. Dr Do is supported by the National Institute of General Medical Sciences of the NIH (R35-GM124836) and the National Heart, Lung, and Blood Institute of the NIH (R01-HL139865 and R01-HL155915); has received grants from AstraZeneca; has received grants and nonfinancial support from Goldfinch Bio; is a scientific cofounder, consultant, and equity holder for Pensieve Health; and has served as a consultant for Variant Bio. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
ABBREVIAT IONS AND ACRONYMS
- ASCVD
atherosclerotic cardiovascular disease
- AUROC
area under the receiver-operating characteristic curve
- CAD
coronary artery disease
- EHR
electronic health record
- ML
machine learning
- NRI
net reclassification improvement
- PCE
Pooled Cohort Equations
- PRS
polygenic risk score
Footnotes
Andrew DeFilippis, MD, MSc, served as Guest Associate Editor for this paper. Athena Poppas, MD, served as Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
REFERENCES
- 1.Zamorano JL, del Val D. Predictive models of atherosclerotic cardiovascular disease: in search of the philosopher’s stone of cardiology. J Am Coll Cardiol. 2016;67:148–150. [DOI] [PubMed] [Google Scholar]
- 2.Goff DC Jr, Lloyd-Jones DM, Bennett G, et al. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63(25 Pt B):2935–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone NJ, Robinson JG, Lichtenstein AH, et al. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J Am Coll Cardiol. 2014;63(25 Pt B):2889–2934. [DOI] [PubMed] [Google Scholar]
- 4.Ridker PM, Cook NR. Statins: new American guidelines for prevention of cardiovascular disease. Lancet. 2013;382:1762–1765. [DOI] [PubMed] [Google Scholar]
- 5.Kavousi M, Leening MJG, Nanchen D, et al.Comparison of application of the ACC/AHA Guidelines, Adult Treatment Panel III Guidelines, and European Society of Cardiology Guidelines for Cardiovascular Disease Prevention in a European cohort. JAMA. 2014;311:1416–1423. [DOI] [PubMed] [Google Scholar]
- 6.Yeboah J, Polonsky TS, Young R, et al. Utility of nontraditional risk markers in individuals ineligible for statin therapy according to the 2013 American College of Cardiology/American Heart Association Cholesterol Guidelines. Circulation. 2015;132:916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muntner P, Colantonio LD, Cushman M, et al. Validation of the atherosclerotic cardiovascular disease pooled cohort risk equations. JAMA. 2014;311:1406–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFilippis AP, Young R, McEvoy JW, et al. Risk score overestimation: the impact of individual cardiovascular risk factors and preventive therapies on the performance of the American Heart Association-American College of Cardiology Atherosclerotic Cardiovascular Disease risk score in a modern multi-ethnic cohort. Eur Heart J.2016;38:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rana JS, Tabada GH, Solomon MD, et al. Accuracy of the atherosclerotic cardiovascular risk equation in a large contemporary, multiethnic population. J Am Coll Cardiol. 2016;67:2118–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khera AV,Chaffin M,Aragam KG,et al.Genomewidepolygenicscoresforcommondiseasesidentify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weale ME, Riveros-Mckay F, Selzam S, et al. Validation of an integrated risk tool, including polygenic risk score, for atherosclerotic cardiovascular disease in multiple ethnicities and ancestries. Am J Cardiol. 2021;148:157–164. [DOI] [PubMed] [Google Scholar]
- 12.Elliott J, Bodinier B, Bond TA, et al. Predictive accuracy of a polygenic risk score–enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosley JD, Gupta DK, Tan J, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aragam KG, Dobbyn A, Judy R, et al. Limitations of contemporary guidelines for managing patients at high genetic risk of coronary artery disease. J Am Coll Cardiol. 2020;75:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikilitas O, Schaid DJ, Kosel ML, et al. Predictive utility of polygenic risk scores for coronary heart disease in three major racial and ethnic groups. Am J Hum Genet. 2020;106:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotter JI, Lin HJ. An outbreak of polygenic scores for coronary artery disease. J Am Coll Cardiol. 2020;75:2781–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44–56. [DOI] [PubMed] [Google Scholar]
- 18.Ward A, Sarraju A, Chung S, et al. Machine learning and atherosclerotic cardiovascular disease risk prediction in a multi-ethnic population. NPJ Digit Med. 2020;3:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alaa AM, Bolton T, Di Angelantonio E, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS One. 2019;14:e0213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Feng Q, Wu P, et al. Learning from longitudinal data in electronic health record and genetic data to improve cardiovascular event prediction. Sci Rep. 2019;9:717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agrawal S, Klarqvist MDR, Emdin C, et al. Selection of 51 predictors from 13,782 candidate multimodal features using machine learning improves coronary artery disease prediction. Patterns (N Y). 2021;2(12):100364. 10.1016/j.patter.2021.100364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stekhoven DJ, Bühlmann P. MissForest—nonparametric missing value imputation for mixedtype data. Bioinformatics. 2012;28:112–118. [DOI] [PubMed] [Google Scholar]
- 23.Kursa MB, Rudnicki WR. Feature selection with the Boruta package. J Stat Soft. 2010;36:1–13. [Google Scholar]
- 24.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N Engl J Med. 2019;380: 1347–1358. [DOI] [PubMed] [Google Scholar]
- 25.Liaw A, Wiener M. Classification and Regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- 26.Chen T, Guestrin CE. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016: 785–794. [Google Scholar]
- 27.Karatzoglou A, Smola A, Hornik K, Zeileis A. kernlab—an S4 package for kernel methods in R. J Stat Soft. 2004;11:1–20. [Google Scholar]
- 28.Kuhn M. Building predictive models in R using the caret package. J Stat Soft. 2008;28. [Google Scholar]
- 29.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. Accessed February 14, 2022. https://www.r-project.org/ [Google Scholar]
- 30.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and Sþ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12: 77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue E. nricens: NRI for risk prediction models with time to event and binary response data. CRAN; 2018. [Google Scholar]
- 32.Cutillo CM, Sharma KR, Foschini L, et al. Machine intelligence in healthcare—perspectives on trustworthiness, explainability, usability, and transparency. NPJ Digital Medicine. 2020;3:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veinot TC, Mitchell H, Ancker JS. Good intentions are not enough: how informatics interventions can worsen inequality. J Am Med Inform Assoc. 2018;25:1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cruz Rivera SLX, Chan A, et al. , for the SPIRIT-AI and CONSORT-AI Working Group, SPIRIT-AI and CONSORT-AI Steering Group, SPIRIT-AI and CONSORT-AI Consensus Group. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat Med. 2020;26:1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finlayson SG, Subbaswamy A, Singh K, et al.The clinician and dataset shift in artificial intelligence. N Engl J Med. 2021;385:283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khokhar B, Jette N, Metcalfe A, et al. Systematic review of validated case definitions for diabetes in ICD-9-coded and ICD-10-coded data in adult populations. BMJ Open. 2016;6(8): e009952. 10.1136/bmjopen-2015009952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCormick N, Bhole V, Lacaille D, AvinaZubieta JA. Validity of diagnostic codes for acute stroke in administrative databases: a systematic review. PLoS One. 2015;10: e0135834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.