Summary
High ambient temperatures have adverse impacts on crop yields. Although a few plant thermosensors have been reported, these sensors directly or indirectly impact PIF4-controlled transcriptional regulation. Moreover, high temperatures also trigger a number of post-transcriptional alternative splicing events in plants and even in animals. Here, we show that LAMMER kinase AFC2 in Arabidopsis controls high-temperature-triggered alternative splicing. Plants without AFC2 exhibited distorted splicing patterns at a high ambient temperature. Further investigations revealed that high temperatures triggered alternative splicing in the majority of PIF4 target genes as a means of desensitizing PIF4 signaling. Consistently, the afc2 mutants exhibited more exaggerated high ambient temperature responses in a PIF4-dependent manner. AFC2 directly phosphorylated the serine/arginine-rich protein splicing factor RSZ21, and AFC2 kinase activity decreased with increasing temperature, indicating that the AFC2 itself may sense temperature changes. In summary, we report that alternative splicing is a safe-guard mechanism when plants encounter high temperature.
Subject areas: Molecular biology, Molecular mechanism of gene regulation, Plant biology
Graphical abstract
Highlights
-
•
AFC2 controls alternative splicing under high ambient temperature
-
•
AFC2 negatively regulates thermomorphogenesis
-
•
AFC2 kinase activity is temperature-sensitive
Molecular biology; Molecular mechanism of gene regulation; Plant biology
Introduction
At high ambient temperatures (28°C–29°C), Arabidopsis plants elongate their hypocotyls and petioles, elevate their leaves, and trigger earlier flowering to cool down their leaf surfaces and shorten their life history to adapt (Quint et al., 2016). These morphological changes are collectively termed thermomorphogenesis. Phytochrome-interacting factor (PIF4) is the pivotal transcription factor that regulates thermomorphogenesis. PIF4 directly binds to thousands of target gene promoters, including auxin biosynthesis-related genes, and modulates their expression (Sun et al., 2012). Three types of plant thermosensors (phytochromes, PIF7 mRNA, and ELF3 protein) have been identified that regulate PIF4 transcriptional activity by either interacting with PIF4 or controlling PIF4 mRNA expression (Jung et al., 2016, 2020; Legris et al., 2016; Chung et al., 2020; Silva et al., 2020).
In addition to causing differential gene expression, high ambient temperatures also trigger a number of alternative splicing events (Lee et al., 2013; Pose et al., 2013; Sureshkumar et al., 2016; Jin et al., 2020; Neumann et al., 2020). Alternative splicing either produces distinct mature mRNAs that are then translated into different protein isoforms with different functions (Lee et al., 2013; Pose et al., 2013) or transcribes aberrant mRNAs and causes mRNA decay normally through the nonsense-mediated mRNA decay (NMD) pathway (Chaudhary et al., 2019; Neumann et al., 2020; Lin and Zhu, 2021). For example, high temperature triggers the alternative splicing of FLOWERING LOCUS M (FLM) and downregulates FLM expression through NMD, which consequently de-represses FLOWERING LOCUS T (FT) expression and results in early flowering (Sureshkumar et al., 2016). Interestingly, high-temperature-responsive alternative splicing events also widely occur in animals (Martin Anduaga et al., 2019; Haltenhof et al., 2020), suggesting that they might be an example of convergent evolution during heat adaption in multicellular eukaryotes.
A recent study reported that subtle temperature increases inactivated human or mouse CDC-like kinases (CLKs) and reduced their phosphorylation of serine/arginine-rich proteins (SR proteins), which changed global alternative splicing patterns (Colwill et al., 1996; Haltenhof et al., 2020). SR proteins have both RNA-binding domains and serine/arginine-rich repeat domains, which bind to exonic splicing enhancer area to facilitate the U1 small nuclear ribonucleoprotein (U1 snRNP) recognizing the 5′ splicing site (5′SS) (Lin and Zhu, 2021). The phosphorylation status of SR proteins is crucial for its function (Golovkin and Reddy, 1999). CLK1 also phosphorylated the splicing factor U1-70K and controlled the initial step in spliceosome assembly (Aubol et al., 2021). Interestingly, CLK homologs in other animals (Alligator mississippiensis, Trachemys scripta, and Drosophila melanogaster) also displayed similar high temperature sensitivity, though they differed in their individual growth temperature ranges (Haltenhof et al., 2020). Molecular dynamics simulations have shown that high temperature alters the conformation of the human CLK1 kinase activation segment and suggested that CLKs directly sense temperature through a biophysical response (Haltenhof et al., 2020).
In this study, we revealed that one CLK homolog in Arabidopsis thaliana (AFC2) participated in the high ambient temperature-responsive alternative splicing control and negatively regulated thermomorphogenesis. AFC2 kinase activity was also sensitive to temperature increase, which suggested a conserved temperature-sensing mechanism in both plants and animals.
Results
In A. thaliana, three LAMMER kinases (At3g53570 (AFC1), At4g24740 (AFC2), and At4g32660 (AFC3)) were CLK homologs (Figure S1A). Among them, AFC2 was reported to phosphorylate SR proteins two decades ago (Golovkin and Reddy, 1999), but the biological functions of AFC2 are still unknown. Phylogenetic studies have shown that AFC2-related kinases expanded during plant terrestrialization. Only one AFC2-related gene was identified in the water-living Chlorophyta (Chlamydomonas reinhardtii), but more than one ortholog was found in all the land-grown plant lineages in our investigations (Figure S2). More interestingly, the diversified AFC members did not exist before the appearance of early angiosperms (Amborella trichopoda), suggesting that AFC2-related LAMMER kinases might be necessary for plants to conquer the land (Figure S2).
AFC2 shows sequence similarity to mammalian CLKs, especially in their kinase activation segment regions (Figure S1A). The amino acids arginine 342 (R342) and histidine 343 (H343) in human CLK1 are sensitive to temperature increases and are necessary for its kinase activity (Haltenhof et al., 2020). Our results showed that mutation of their analogs in Arabidopsis AFC2 (AFC2R284K or AFC2H285K) inhibited their phosphorylation on RS-containing zinc finger protein 21 (RSZ21, one SR substrate protein for AFC2) (Figure S1B). These results confirmed that these two conserved amino acids in AFC2 were also necessary for AFC2 kinase activity.
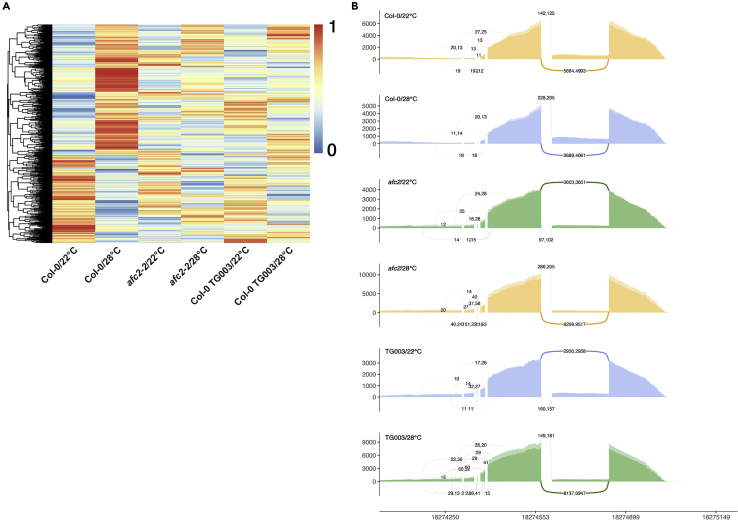
Next, we compared differential alternative splicing events at high temperature (28°C) in wild-type plants (Col-0) and AFC2-defective plants. We included both afc2-2 loss-of-function mutants and TG003 (CLK inhibitor (Ninomiya et al., 2011; Haltenhof et al., 2020))-treated plants as AFC2-defective plants in this study. Clustering the alternative splicing events from our RNA-seq data showed that in the absence of functional AFC2 (in afc2-2 mutants or TG003-treated Col-0 samples), the high-temperature-triggered alternative splicing events were almost completely eliminated (Figures 1A and S3; Table S1). Five typical alternative splicing types were uncovered in the high-temperature-treated Col-0 samples. Intron-retained and alternative 3′ splice sites were the two major forms (Figure S3). We randomly selected one intron-retained gene (At4g39260) for a more detailed investigation. The high-temperature treatment enhanced intron retention in Col-0 based on their read distributions, but these patterns did not occur in either the afc2-2 mutants or the TG003-treated Col-0 samples (Figure 1B). These results indicated that AFC2 was required for high-temperature-responsive alternative splicing.
Figure 1.
AFC2 is required for high-temperature-responsive alternative splicing
(A) Heatmap showing the patterns of exon inclusion levels from 2456 shared alternative splicing events among treatments. The mean exon inclusion level value was used in the clustering and was calculated according to the exon inclusion levels in two biological duplicates.
(B) Sashimi plots illustrating the read distributions of At4g39260 in different samples. The curved lines represent the reads spanning the junctions of two exons. Numbers separated by a comma indicate the exact reads from two independent RNA-seq experiments.
See also Figures S1–S4; Tables S1 and S2.
Further analysis of the RNA-seq dataset found that there were few overlaps between differentially alternative spliced genes (DASGs) and differentially expressed genes (DEGs). In Col-0, high temperature elicited 1903 DEGs and 2971 DASGs. However, only 219 genes were both DEGs and DASGs, which suggest that 92.6% (2752 in 2971 DASGs) of the alternatively spliced genes were not differentially expressed at high temperature (Figure S4). In addition, 15.3% (421 in 2752 DASGs, Table S2) of them were reported to be putative PIF4 target genes (Figure S4), according to previously published high-quality PIF4 chromatin immunoprecipitation (ChIP)-seq data (Oh et al., 2012). Interestingly, a couple of genes encoding auxin signaling components (including AUXIN RESPONSE FACTOR 4 (ARF4), ARF6, INDOLEACETIC ACID-INDUCED PROTEIN 16 (IAA16), IAA29, TOPLESS) or auxin transporters (PIN-LIKES 5 (PILS5), LIKE AUXIN RESITANT 1 (LAX1)) were among the 421 genes (Table S2). These results indicated that PIF4 directly controlled these auxin signaling-related gene expressions; however, their expression levels were not significantly changed most likely due to alternative splicing. In fact, similar to shade avoidance syndrome, plants need a brake to optimize their hypocotyl elongation to balance optimal growth with lodging (Romero-Montepaone et al., 2021). Therefore, we assumed that alternative splicing was a means to attenuate the PIF4-regulated transcriptome and avoid the overactivation of thermomorphogenesis.
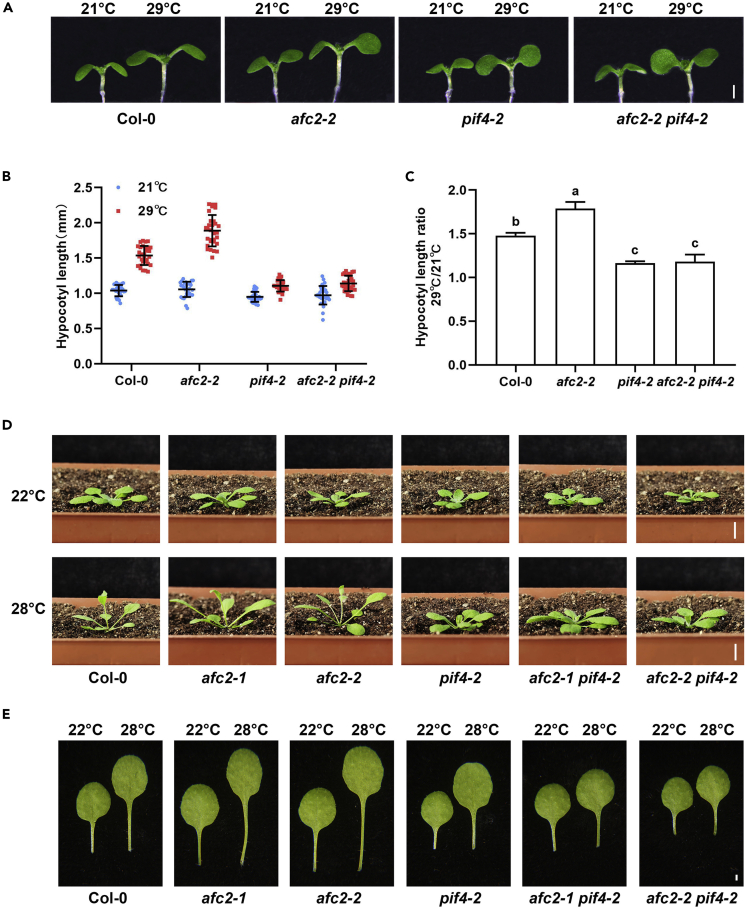
To test this hypothesis, we observed thermomorphogenesis phenotypes in afc2 mutants. In the seedling stages, two independent alleles of afc2 mutants (afc2-1 and afc2-2) displayed hypersensitivity to high temperature (Figures S5A–S5C). Meanwhile, the leaf upward growth (Figures S5D–S5F) and petiole elongations (Figures S5G and S5H) in afc2 mutants were also more exaggerated than those in the wild-type controls. Next, we generated afc2 pif4 double mutants (afc2-1 pif4-2 or afc2-2 pif4-2) and found that loss of PIF4 in the afc2 background successfully suppressed their high-temperature hypersensitivity in either hypocotyl elongation (Figures 2A–2C) or leaf petiole elongation (Figures 2D and 2E). Because PIF4-triggered auxin biosynthesis is the major cause of thermomorphogenesis, we also took advantage of auxin biosynthesis inhibitors to study whether blocking auxin production could rescue afc2 hypersensitivity. Kynurenine (Kyn) and 4-phenoxyphenylboronic acid (PPBo) were reported to specifically inhibit two auxin biosynthesis enzymes (TAA1 or YUCCA), respectively (He et al., 2011; Kakei et al., 2015). Kyn and PPBo treatment suppressed hypocotyl elongation in all the mutants in our tests, which suggested that the longer hypocotyl elongations in afc2 mutants relied on auxin biosynthesis (Figure S6). Taken together, our results showed that plants exhibited hypersensitivity to high temperature in a PIF4-dependent manner in the absence of AFC2.
Figure 2.
AFC2 negatively controls thermomorphogenesis
(A) Hypocotyl phenotypes of the indicated genotypes grown at 29°C (scale bar = 1 mm).
(B) Quantification analysis of hypocotyl lengths of Col-0, afc2-2, pif4-2, and afc2-2 pif4-2 grown at 21°C or 29°C. Data are represented as mean ± SD n = 30.
(C) The hypocotyl elongation ratio was calculated from the results shown in (B). Significant differences are indicated by letters, p ≤ 0.01.
(D) Representative images showing the rosette leaf elevation angles in 20-day-old Col-0, afc2-1, afc2-2, afc2-1 pif4-2, and afc2-2 pif4-2 plants grown at the indicated temperatures (scale bar = 5 mm).
(E) Representative images showing the petiole lengths of the fifth or sixth rosette leaves in 20-day-old Col-0, afc2-1, afc2-2, afc2-1 pif4-2, and afc2-2 pif4-2 plants grown at the indicated temperatures.
See also Figures S5 and S6.
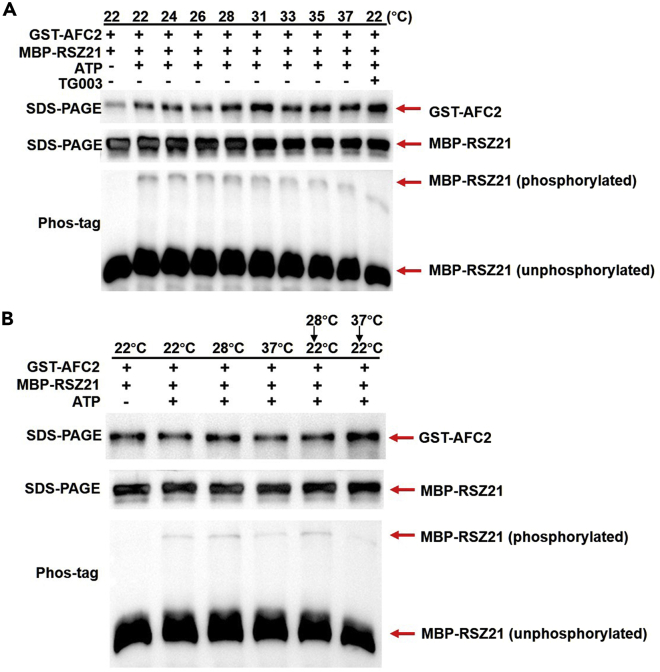
Finally, we tested whether AFC2 proteins were able to sense high temperature in an in vitro kinase assay. We expressed and purified both RSZ21 substrates and AFC2 kinase from E. coli and mixed them with ATP to react at different temperatures (ranging from 22°C to 37°C) before loading onto the gel. In the presence of Phos-tag reagents, the shifted bands (representing phosphorylated RSZ21) were clear at lower temperatures but were weak when the temperature increased (Figure 3A). Studies in mammals reported that the reduced human CLK1 activity at high temperatures could be reversed when the reactions were shifted back to a normal temperature range (Haltenhof et al., 2020). However, the reduced AFC2 kinase activity at 37°C could not be reversed when the reactions were shifted back to 22°C (Figure 3B). This different result is reminiscent of the totally distinct behaviors of plants and animals living in fluctuating temperatures. Animal behaviors (such as sex determination in reptiles) are reversible when the temperature returns to normal, but already-elongated hypocotyls in plants cannot shorten when the temperature decreases. Therefore, we hypothesized that plant AFC2 proteins lost this reversibility during evolution. Future crystal structure comparisons of mammalian CLKs with AFC2 may provide more direct answers to this hypothesis.
Figure 3.
High temperature reduces AFC2 kinase activity
(A) In vitro kinase assay performed at the indicated temperatures. GST-AFC2 kinase and MBP-RSZ21 substrates were incubated at the indicated temperatures with or without ATP for 30 min. Kinase activity was monitored by immunoblotting on Phos-tag SDS-PAGE gels, while the loading controls were detected by immunoblotting on regular SDS-PAGE gels. The shifted MBP-RSZ21 bands on the Phos-tag SDS-PAGE gel represented phosphorylated forms, while the unshifted bands indicated unphosphorylated forms. GST-AFC2 proteins were detected with anti-GST antibody, and MBP-RSZ21 proteins were detected with anti-MBP antibody.
(B) Kinase activity reversibility assay. GST-AFC2 kinase and MBP-RSZ21 substrates were initially incubated at the indicated temperatures with or without ATP for 30 min and then stopped by adding sample loading buffer. Two reactions performed at 28°C or 37°C were not stopped but were transferred to 22°C for additional 30 min to test kinase activity reversibility. Kinase activities were then monitored as described in (A).
Discussion
Taken together, these results illustrate that the conserved CLK kinase in plants (AFC2) was necessary for controlling plant alternative splicing. Although thermo-responsive hypocotyl elongation is an effective way for plants to cope with high temperature, unlimited elongation causes lodging risk and requires more energy allocations for growth. Therefore, plants already develop several brakes to restrict PIF4 activity. For example, transcription factors HECATEs (HEC1 and HEC2) directly interact with PIF4 and inhibit its transcriptional activity (Lee et al., 2021). In this study, we revealed that alternative splicing was also an efficient way to attenuate PIF4-regulated thermomorphogenesis. afc2 mutants displayed a more exaggerated thermomorphogenesis phenotype, indicating that the manipulation of AFC2 could also enhance plant heat tolerance. It is noteworthy that targeted therapy on spliceosomes or their related components is already a topic of great interest in biomedical research (Hsu et al., 2015; Zhang et al., 2020a; Bowling et al., 2021). Our work suggests that AFC2-controlled spliceosome phosphorylation could be an entry point for future plant spliceosome-based breeding.
Limitations of the study
Although we showed here that AFC2 phosphorylated RSZ21, the detailed relationships between RSZ21 phosphorylation status and splicing control are still unknown. We will generate phosphorylation mimic forms or un-phosphorylation mimic forms of RSZ21 transgenic plants and compare their RNA binding and spliceosome assembling in future to dissect this question. On another side, as a kinase, AFC2 may have more substrates besides SR proteins. We are going to prepare AFC2 overexpression plants and perform immunoprecipitation-coupled mass spectrometry (IP-MS) to identify novel AFC2 substrates.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| GST-Tag Antibody | Affinity Biosciences | Cat# T0007; RRID: AB_2839414 |
| Anti-MBP Monoclonal Antibody | New England Biolabs | Cat# E8032S; RRID: AB_1559730 |
| Bacterial and virus strains | ||
| DH5α competent cell | Vazyme | Cat# C502-02 |
| BL21 (DE3) competent cell | Vazyme | Cat# C504-02 |
| Rosetta (DE3) competent cell | TOLOBIO | Cat# CC96139-01 |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium hypochlorite | SINOPHARM | Cat# 80010428 |
| Triton X-100 | Sigma-Aldrich | Cat# V900502 |
| Murashige and Skoog Basal Medium (MS) | Sigma-Aldrich | Cat# M5519 |
| Sucrose | Sigma-Aldrich | Cat# V900116-500G |
| Agar | Sigma-Aldrich | Cat# A1296-1KG |
| Phos-tag Acrylamide | Wako | Cat# 304-93521 |
| Manganese (II) chloride tetrahydrate (MnCl2) | Sigma-Aldrich | Cat# V900197-500G |
| Sodium dodecyl sulfate | Sigma-Aldrich | Cat# V900859-500G |
| Ammonium persulfate substitute (APS substitute) | Beyotime | Cat# ST005-10g |
| Glycine | Sigma-Aldrich | Cat# V900144-5KG |
| Methanol | SINOPHARM | Cat# 10014118 |
| Ethylenediaminetetraacetic acid (EDTA) | Sigma-Aldrich | Cat# V900106-500G |
| Glutathione Sepharose beads | Cytiva | Cat# 17075601 |
| Amylose resin | New England Biolabs | Cat# E8021S |
| Phenylmethylsulphonyl fluoride (PMSF) | Sigma-Aldrich | Cat# P7626 |
| L-Glutathione reduced | Sigma-Aldrich | Cat# V900436-25G |
| D-(+)-Maltose monohydrate | Sigma-Aldrich | Cat# V900435-100G |
| Trizma base | Sigma-Aldrich | Cat# V900483-5KG |
| Magnesium chloride hexahydrate (MgCl2) | Sigma-Aldrich | Cat# V900020-500G |
| dithiothreitol (DTT) | Beyotime | Cat# ST040-5g |
| adenosine triphosphate (ATP) | Coolaber | Cat# CA1261-5G |
| L-Kynurenine (Kyn) | Sigma-Aldrich | Cat# K8625 |
| TG-003 | Sigma-Aldrich | Cat# T5575 |
| 4-Phenoxyphenylboronic acid (PPBo) | Sigma-Aldrich | Cat# 480142 |
| TRIzol | Invitrogen | Cat# 15596018 |
| Critical commercial assays | ||
| Mut Express MultiS Fast Mutagenesis Kit V2 | Vazyme | Cat# C215-01 |
| Deposited data | ||
| RNA-seq dataset | National Genomics Data Center (https://ngdc.cncb.ac.cn) | CRA001128 and CRA003933 |
| The Arabidopsis genome | TAIR10 | http://ensemblgenomes.org |
| Experimental models: Organisms/strains | ||
| Arabidopsis afc2-1 | SALK_118114 | N/A |
| Arabidopsis afc2-2 | SALK_043601 | N/A |
| Arabidopsis pif4-2 | Ma et al., 2016 | N/A |
| Arabidopsis afc2-1 pif4-2 | This paper | N/A |
| Arabidopsis afc2-2 pif4-2 | This paper | N/A |
| Oligonucleotides | ||
| GST-AFC2 Forward: TGGGATCCCCGAATTCATGGAGCGT GTGCATGAATTC |
This paper | N/A |
| GST-AFC2 Reverse: GTCGACCCGGGAATTCTCTTCTCCTTGCGAAAAAC | This paper | N/A |
| MBP-RSZ21 Forward: TATCGTCGACGGATCCATGACGAGGGTTTATGTC | This paper | N/A |
| MBP-RSZ21 Reverse: CAGGGAATTCGGATCCCACCCCATTGGCATATGG | This paper | N/A |
| GST-AFC2R284K Forward: ACCaaaCATTATAGGGCACCAGAAGTCATTTT | This paper | N/A |
| GST-AFC2R284K Reverse: GCCCTATAATGtttGGTTGATACAATGTAGGTTTGGTCC | This paper | N/A |
| GST-AFC2H285K Forward: ACCAGAaaaTATAGGGCACCAGAAGTCATTTTAGG | This paper | N/A |
| GST-AFC2H285K Reverse: GCCCTATAtttTCTGGTTGATACAATGTAGGTTTGG | This paper | N/A |
| Recombinant DNA | ||
| GST-AFC2 | This paper | N/A |
| MBP-RSZ21 | This paper | N/A |
| GST-AFC2R284K | This paper | N/A |
| GST-AFC2H285K | This paper | N/A |
| Software and algorithms | ||
| ImageJ | NIH | http://rsb.info/nih/gov/ij |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ziqiang Zhu (zqzhu@njnu.edu.cn).
Materials availability
Strains, plasmids, and Arabidopsis lines used in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Arabidopsis thaliana
All plant materials used in this study were in Columbia (Col-0) background. afc2-1 (SALK_118114) and afc2-2 (SALK_043601) were obtained from the Arabidopsis Biological Resource Center. afc2-1 pif4-2 and afc2-2 pif4-2 double mutants were generated through genetic crossing and identified by PCR-based genotyping. The seeds were first sterilized with 10% sodium hypochlorite and 0.1% Triton X-100 for 7 min and were then washed with sterilized water five times and placed on Murashige and Skoog (MS) medium (4.4 g/L MS salt, 1% sucrose pH 5.8, 0.8% agar).
Method details
Phenotypes
For the hypocotyl observations, plates were kept at 4°C for 3 days, held at 21°C for 3 days to stimulate germination and transferred to 29°C or kept at 21°C for additional 4 days. The light conditions were a 16 h light/8 h dark photoperiod and 70 μmol m−2 s−1 white light. For the leaf angle or petiole observations in adult plants, plates were kept at 4°C for 3 days, held at 22°C for 3 days to stimulate germination and transferred to 28°C or kept at 22°C for additional 4 days. Observations were transferred to soil for additional 13 days at the indicated temperatures. All quantifications were measured with ImageJ software (https://imagej.nih.gov/ij/).
Sequence alignments and phylogenetic analysis
Protein sequences of mouse CLK1, human CLK1, mouse CLK4 and human CLK4 were kindly provided by Dr. Florian Heyd. Arabidopsis AFC sequences were obtained from the Arabidopsis Information Resource (http://www.arabidopsis.org). Amino acid alignments were performed with the ClustalW algorithm in DNAMAN software (Lynnon Biosoft, Canada). For phylogenetics, we selected the Chlamydomonas reinhardtii, Physcomitrella patens, Selaginella moellendorffii, Amborella trichopoda, Arabidopsis thaliana and Oryza sativa genomes to represent charophytes, bryophytes, lycophytes, and the ancestors of living angiosperms, dicotyledons and monocotyledons. Phylogenetic trees were generated with an online tool (PhyloGenes, http://www.phylogenes.org) (Zhang et al., 2020b).
In vitro phosphorylation assay
Full-length coding sequences of AFC2 or RSZ21 were cloned and constructed into pGEX-5X-1 or pMAL-c5X vectors, respectively. Mutated versions of AFC2 (AFC2R284K, AFC2H285K) were generated through a Mut Express MultiS Fast Mutagenesis Kit V2 (Vazyme). GST or MBP fusion proteins were expressed in E. coli (DE3) cells and further purified with either glutathione sepharose 4B (GE Healthcare) or amylose resin (New England Biolabs). For the kinase assay, 20 μL of AFC2 kinase and RSZ21 substrates were incubated in 100 μL of kinase reaction buffer (25 mM Tris-HCl (pH 7.5), 12 mM MgCl2, 1 mM dithiothreitol) at the indicated temperatures with or without 1 mM adenosine triphosphate (ATP) supplement. The reactions were stopped by adding SDS sample loading buffer. After boiling and centrifugation, the samples were separated on either regular 10% SDS-PAGE gel or 8% Phos-tag SDS-PAGE gel (8% SDS-PAGE with 75 μM Phos-tag (Wako) and 100 μM MnCl2 reagents). After being transferred onto a nitrocellulose membrane (Pall), proteins were detected with either anti-GST (Affinity Biosciences) or anti-MBP antibodies (New England Biolabs) according to standard immunoblot protocols.
RNA extraction and RNA-sequencing (RNA-seq) analysis
For RNA extraction, the seedlings were grown at 22°C for 3 days and then transferred to 28°C or kept at 22°C for an additional 4 days. TG003-treated Col-0 samples were directly grown on MS medium with 30 μM TG003 (Sigma-Aldrich) at 22°C for 3 days and then transferred to 28°C or kept at 22°C for an additional 4 days. RNA was extracted with TRIzol reagent (Invitrogen). For each treatment, two independent samples were collected as biological duplicates. Further RNA quality checks, library construction and sequencing were performed in GENEWIZ (Suzhou, China). The raw reads were first checked by FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) and then trimmed and filtered by removing adapters, low-quality reads (Qphred <20 reads and short reads) and unpaired reads using Trim Galore (www.bioinformatics.babraham.ac.uk/projects/trim_galore). The Arabidopsis TAIR10 genome was downloaded from Ensembl (Release 49) (http://ensemblgenomes.org/). The clean reads were mapped to the Arabidopsis TAIR10 genome using HISAT2 v2.2.1 (Kim et al., 2015) with the default parameters. The mapped reads in sequence alignment/map (SAM) format were sorted and converted to the binary version of the SAM file (BAM format) using SAMtools version 0.1.19 (Li et al., 2009). The FPKM value of each gene was calculated using StringTie v2.1.4 and the Ballgown package in R (Kim et al., 2015). Differential expression was tested based on the mapped read counts in R using DESeq2 (Love et al., 2014). Significantly differentially expressed genes (DEGs) were identified with the threshold log2 (fold change) ≥1 and the adjusted p value (Padj) ≤ 0.05. Differential alternative splicing events were identified by using rMATS v4.1.0 with the parameters “-t paired -len 150 -c 0.0001 -novelSS” (Shen et al., 2014). rMATS is a useful tool for performing robust and flexible analyses of alternative splicing based on exon inclusion levels (ψ). Events with p < 0.05 and exon inclusion level (ψ) change >0.05 were identified as significant differential alternative splicing events. The shared genes among the treatments were filtered, and FPKM values were clustered. A heatmap was generated with the heatmap package in R. The alternative splicing patterns in each treatment were also analyzed using the exon inclusion levels of the shared alternative splicing events in these groups. The alternative splicing events in representative genes were visualized using ggsashimi (Garrido-Martin et al., 2018).
Data accessibility
The raw sequence data reported in this paper have been deposited in the National Genomics Data Center (https://ngdc.cncb.ac.cn) under accession numbers CRA001128 (Col-0 22°C, Col-0 28°C) and CRA003933 (afc2-2 22°C, afc2-2 28°C, TG003 22°C, TG003 28°C).
Quantification and statistical analysis
Graph presentation and statistical analysis were carried out using the GraphPad Prism 8 software (GraphPad Prism Software Inc., San Diego, CA, USA). Differences among hypocotyl length ratios were determined by Student’s t test.
Acknowledgments
We thank Dr. Florian Heyd for sharing the mammalian CLK protein sequences. This work was supported by the National Natural Science Foundation of China (31970256), the Fok Ying Tong Education Foundation (161023), and the Qing Lan project. The graphical abstract was created with BioRender.com.
Author contributions
Z. Zhu conceived this project, designed experiments, and wrote the manuscript. J.L. initiated this project, extracted RNA for sequencing, made constructs, and prepared plant materials. J. L. and J.S. performed kinase assays and characterized phenotypes. Z. Zhang analyzed the RNA-seq data. B. Z. provided computing platforms and commented on phylogenetics. All authors analyzed the data and contributed to figure preparations.
Declaration of interests
The authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104051.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the Lead Contact upon reasonable request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon reasonable request.
References
- Aubol B.E., Wozniak J.M., Fattet L., Gonzalez D.J., Adams J.A. CLK1 reorganizes the splicing factor U1-70K for early spliceosomal protein assembly. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2018251118. e2018251118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling E.A., Wang J.H., Gong F., Wu W., Neill N.J., Kim I.S., Tyagi S., Orellana M., Kurley S.J., Dominguez-Vidana R., et al. Spliceosome-targeted therapies trigger an antiviral immune response in triple-negative breast cancer. Cell. 2021;184:384–403.e21. doi: 10.1016/j.cell.2020.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary S., Jabre I., Reddy A.S.N., Staiger D., Syed N.H. Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 2019;24:496–506. doi: 10.1016/j.tplants.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Chung B.Y.W., Balcerowicz M., Di Antonio M., Jaeger K.E., Geng F., Franaszek K., Marriott P., Brierley I., Firth A.E., Wigge P.A. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants. 2020;6:522–532. doi: 10.1038/s41477-020-0633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Pawson T., Andrews B., Prasad J., Manley J.L., Bell J.C., Duncan P.I. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Garrido-Martin D., Palumbo E., Guigo R., Breschi A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006360. e1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. An SC35-like protein and a novel serine/arginine-rich protein interact with Arabidopsis U1-70K protein. J. Biol. Chem. 1999;274:36428–36438. doi: 10.1074/jbc.274.51.36428. [DOI] [PubMed] [Google Scholar]
- Haltenhof T., Kotte A., De Bortoli F., Schiefer S., Meinke S., Emmerichs A.K., Petermann K.K., Timmermann B., Imhof P., Franz A., et al. A conserved kinase-based body-temperature sensor globally controls alternative splicing and gene expression. Mol. Cell. 2020;78:57–69.e4. doi: 10.1016/j.molcel.2020.01.028. [DOI] [PubMed] [Google Scholar]
- He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F., et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu T.Y., Simon L.M., Neill N.J., Marcotte R., Sayad A., Bland C.S., Echeverria G.V., Sun T., Kurley S.J., Tyagi S., et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature. 2015;525:384–388. doi: 10.1038/nature14985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Lin J., Zhu Z. PIF4 and HOOKLESS1 impinge on common transcriptome and isoform regulation in thermomorphogenesis. Plant Commun. 2020;1:100034. doi: 10.1016/j.xplc.2020.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Barbosa A.D., Hutin S., Kumita J.R., Gao M., Derwort D., Silva C.S., Lai X., Pierre E., Geng F., et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585:256–260. doi: 10.1038/s41586-020-2644-7. [DOI] [PubMed] [Google Scholar]
- Jung J.H., Domijan M., Klose C., Biswas S., Ezer D., Gao M., Khattak A.K., Box M.S., Charoensawan V., Cortijo S., et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- Kakei Y., Yamazaki C., Suzuki M., Nakamura A., Sato A., Ishida Y., Kikuchi R., Higashi S., Kokudo Y., Ishii T., et al. Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J. 2015;84:827–837. doi: 10.1111/tpj.13032. [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Ryu H.S., Chung K.S., Pose D., Kim S., Schmid M., Ahn J.H. Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science. 2013;342:628–632. doi: 10.1126/science.1241097. [DOI] [PubMed] [Google Scholar]
- Lee S., Zhu L., Huq E. An autoregulatory negative feedback loop controls thermomorphogenesis in Arabidopsis. PLoS Genet. 2021;17:e1009595. doi: 10.1371/journal.pgen.1009595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M., Klose C., Burgie E.S., Rojas C.C., Neme M., Hiltbrunner A., Wigge P.A., Schafer E., Vierstra R.D., Casal J.J. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Zhu Z. Plant responses to high temperature: a view from pre-mRNA alternative splicing. Plant Mol. Biol. 2021;105:575–583. doi: 10.1007/s11103-021-01117-z. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D., Li X., Guo Y., Chu J., Fang S., Yan C., et al. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proc. Natl. Acad. Sci. U S A. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Anduaga A., Evantal N., Patop I.L., Bartok O., Weiss R., Kadener S. Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. Elife. 2019;8:e4464. doi: 10.7554/eLife.44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A., Meinke S., Goldammer G., Strauch M., Schubert D., Timmermann B., Heyd F., Preussner M. Alternative splicing coupled mRNA decay shapes the temperature-dependent transcriptome. EMBO Rep. 2020;21:e51369. doi: 10.15252/embr.202051369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya K., Kataoka N., Hagiwara M. Stress-responsive maturation of Clk1/4 pre-mRNAs promotes phosphorylation of SR splicing factor. J. Cell Biol. 2011;195:27–40. doi: 10.1083/jcb.201107093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pose D., Verhage L., Ott F., Yant L., Mathieu J., Angenent G.C., Immink R.G., Schmid M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature. 2013;503:414–417. doi: 10.1038/nature12633. [DOI] [PubMed] [Google Scholar]
- Quint M., Delker C., Franklin K.A., Wigge P.A., Halliday K.J., van Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants. 2016;2:15190. doi: 10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- Romero-Montepaone S., Sellaro R., Hernando C.E., Costigliolo-Rojas C., Bianchimano L., Ploschuk E.L., Yanovsky M.J., Casal J.J. Functional convergence of growth responses to shade and warmth in Arabidopsis. New Phytol. 2021;231:1890–1905. doi: 10.1111/nph.17430. [DOI] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U S A. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva C.S., Nayak A., Lai X., Hutin S., Hugouvieux V., Jung J.H., Lopez-Vidriero I., Franco-Zorrilla J.M., Panigrahi K.C.S., Nanao M.H., et al. Molecular mechanisms of evening complex activity in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 2020;117:6901–6909. doi: 10.1073/pnas.1920972117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Qi L., Li Y., Chu J., Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureshkumar S., Dent C., Seleznev A., Tasset C., Balasubramanian S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants. 2016;2:16055. doi: 10.1038/nplants.2016.55. [DOI] [PubMed] [Google Scholar]
- Zhang D., Hu Q., Liu X., Ji Y., Chao H.P., Liu Y., Tracz A., Kirk J., Buonamici S., Zhu P., et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat. Commun. 2020;11:2089. doi: 10.1038/s41467-020-15815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Berardini T.Z., Ebert D., Li Q., Mi H., Muruganujan A., Prithvi T., Reiser L., Sawant S., Thomas P.D., Huala E. PhyloGenes: an online phylogenetics and functional genomics resource for plant gene function inference. Plant Direct. 2020;4:e00293. doi: 10.1002/pld3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data reported in this paper have been deposited in the National Genomics Data Center (https://ngdc.cncb.ac.cn) under accession numbers CRA001128 (Col-0 22°C, Col-0 28°C) and CRA003933 (afc2-2 22°C, afc2-2 28°C, TG003 22°C, TG003 28°C).
Data reported in this paper will be shared by the Lead Contact upon reasonable request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon reasonable request.