Abstract
Drug resistance is always a challenge in conquering breast cancer clinically. Recognition of drug resistance and enhancing the sensitivity of the tumor to chemotherapy is urgent. Herein, a dual-responsive multi-function “Matryoshka" nanosystem is designed, it activates in the tumor microenvironment, decomposes layer by layer, and release gene and drug in sequence. The cell is re-educated by NgBR siRNA first to regain the chemosensitivity through regulating the Akt pathway and inhibit ERα activation, then the drugs loaded in the core are controlled released to killing cells. Carbonized polymer dots are loaded into the nanosystem as an efficient bioimaging probe, due to the GE11 modification, the nanosystem can be a seeker to recognize and evaluate drug-resistance tumors by photoacoustic imaging. In the tumor-bearing mouse, the novel nanosystem firstly enhances the sensitivity to chemotherapy by knockdown NgBR, inducing a much higher reduction in NgBR up to 52.09%, then effectively inhibiting tumor growth by chemotherapy, tumor growth in nude mouse was inhibited by 70.22%. The nanosystem also can inhibit metastasis, prolong survival time, and evaluate tumor drug resistance by real-time imaging. Overall, based on regulating the key molecules of drug resistance, we created visualization nanotechnology and formatted new comprehensive plans with high bio-safety for tumor diagnosis and treatment, providing a personalized strategy to overcome drug resistance clinically.
Keywords: Tumor microenvironment responsive, Small interfering RNA (siRNA), Gene-silencing, Carbonized polymer dots, Multimodal imaging, Sequence released, Drug resistance
Graphical abstract
Highlights
-
•
Knockdown NgBR regulate the Akt pathway and inhibit ERα activate, enhance the sensitivity of chemotherapy.
-
•
Knockdown of NgBR inhibits metastasis and prolongs survival.
-
•
Nanosystem can evaluate drug resistance and kill tumors at the same time.
1. Introduction
Breast cancer has always been a malignant tumor with high morbidity and mortality. In 2020, breast cancer surpassed lung cancer for the first time to become the world's largest cancer (11.7% of the total cases and 24.5% of the female cases) and ranks first in cancer deaths (15.5% of all cancer deaths) [1]. The molecular typing of breast cancer includes progesterone receptor (PR), estrogen receptor (ER), human epidermal growth factor receptor 2 (HER-2), and proliferating cell nuclear antigen (Ki-67), among which ERα-positive breast cancer accounts for the majority (70%–75%) [2]. Studies have shown that the incidence of ERα-positive breast cancer has been increasing and the ERα-negative breast cancer is declining [3,4], and most deaths also occur in ERα-positive breast cancer patients [5,6]. Although increased detection through widespread uptake of mammographic screening can reduce breast cancer mortality to a certain extent [7,8], mammographic screening has limitations of overdiagnosis and overtreatment [9]. Therefore, personalization, early diagnosis, and treatment of ERα-positive breast cancer are future clinical trends.
With the rapid progress of tumors, for advanced malignant tumors, especially breast cancers derived from epithelial tissues, surgical treatment often fails to achieve satisfactory results. Chemotherapy has a high sensitivity for solid tumors, and paclitaxel is a representative of chemotherapy drugs often used for advanced breast cancer and patients with breast conservation needs. However, studies have shown that only 7%–16% of ERα-positive breast cancer patients can achieve pathological complete remission after chemotherapy [2]. The drug resistance of tumors can be roughly divided into the following two reasons: (1) Pharmacological and physiological factors [10], such as the drugs reaching the tumor are insufficient caused by excessive drug excretion or degraded in the circulation, for example, many chemotherapeutic drugs such as paclitaxel are hydrophobic, which makes it difficult to arrive the target region, so we use nanoplatforms to specifically deliver them to target cells, to play their maximum efficacy [11]; (2) cell or tumor self-specific drug resistance, such as drug transporter P- glycoprotein (P-gp) [[12], [13], [14]], which not only prevent drugs enter into cancer cells but also can efflux entered drugs. Second, multiple cell-related signaling pathways [15] can trigger drug resistance. However, it is worth noting that the drug resistance mechanism of ERα-positive breast cancer cells is a little different from other types of breast cancer due to the constant stimulation of estrogen. Lippman ME first proposed a link between ERα expression and chemotherapy [16]. They found that only 12% of breast cancer patients with high ERα expression responded to chemotherapy, while 76% of patients with low or no ERα expression responded significantly to chemotherapy. Subsequently, researchers have found that ERα-positive breast cancer patients have a poor response to various chemotherapy drugs such as paclitaxel and anthracyclines. A large amount of evidence points out that estrogen and ERα interact with various signaling pathways such as human neuroglobin (NGB) [17], transforming growth factor (TGF)-β [18], P53 [19] to promote proliferation, inhibiting apoptosis, stimulating metastasis [20], and angiogenesis [21] while triggering the drug resistance of tumor cells [2,22,23]. Due to the high morbidity and mortality of ERα-positive breast cancer, early recognition of drug resistant tumors and personalized treatment is vital. Although scholars are continuously dedicated to solving drug resistance, but almost focused on preventing the efflux of drugs [24,25], it's to cure the symptoms, not the disease.
Nogo-B receptor (NgBR) provides a new idea to solve the drug resistance dilemma. NgBR was first reported by Miao's team [26], it is a type I receptor with a single transmembrane domain and the specific receptor for Nogo-B. It is highly expressed in a variety of cancer and proliferating vascular endothelial cells [18,[27], [28], [29]], which have aroused the extensive interest of researchers as soon as they were proposed. NgBR is involved in various disease processes, including diabetes [30], hemangioma [28], inflammation [31], and is involved in the occurrence and development of tumors. Although the research on NgBR has not been comprehensive, the role of NgBR in tumors has been gradually revealed: (1) NgBR binds to farnesylated Ras and recruits Ras on the plasma membrane, which is necessary steps of RTKs-mediated Raspberry signaling activation and tumorigenesis [32]; (2) NgBR is associated with a variety of tumor signaling pathways, including Ras/ERK/Snail1 signaling pathway and PI3K/Akt [20]; (3) NgBR is involved in tumor metastasis-related epithelial-mesenchymal transition [18,33]; (4) NgBR is associated with drug resistance in hepatocellular carcinoma [27] and breast cancer [34]. In addition, studies have confirmed that NgBR is also associated with ERα-positive breast cancer and the degree of malignancy of breast cancer [35]. These studies prompted us to explore whether NgBR can be used as a novel molecular therapeutic target to fundamentally solve the drug resistance of ERα-positive breast cancer cells and to evaluate the correlation between NgBR and tumor growth.
Small interfering RNA (siRNA) is milestone progress in cancer therapy, siRNA can directly work on tumor molecular pathways to inhibit tumor proliferation and invasion, and regain chemotherapy sensitivity of cancer cells [36] through RNA interference (RNAi). But due to siRNA can be degraded by the enzymes in the blood and non-specific off-target side effects that may induce immune responses [37], that limits the siRNA entrance to the target cells. Mesoporous organosilica nanoparticles (MONs) are one of the most promising drug and gene delivery nanocarriers due to their structure/chemical tunability, large surface area, matrix degradation, and controllable drug release [[38], [39], [40], [41], [42]]. The large pore size of MONs make siRNA can not only be adsorbed on the surface but also can be co-loaded with small-sized therapeutic agents into the mesopores to achieve long-term stable release. It is generally believed that compared with negatively and neutrally charged nanomaterials, positively charged has a higher cellular uptake rate [43], but on the contrary, cationic are easily aggregated and have an adverse immune response, our study uses “invisible" PEG to break this bottleneck. PEGylated make the nanocarriers stably in the circulatory and PEG acid-detachment make nanoparticle turn positive in the tumor microenvironment (TME) to promote endocytosis, at the same time, nanosystem can protect the siRNA against degradation and a reduction of the off-target effects [44]. Next, we selected the GE11(a polypeptide that specifically targets EGFR) to achieve the active target for the following reasons. First, GE11 increases cellular uptake not only through EGFR-mediated endocytosis [45,46] but also exists in another actin-driven pathway [47]. To reduce the effect of cation PEI, we added a GGGGC chain to the C-terminus of GE11 [48]. Second, the EGFR is closely related to the proliferation and differentiation of tumors and partially co-localizes with NgBR [32]. It can overcome the potential self-defeating issue. Finally, our study found that EGFR has a higher expression in drug resistant breast cancer cells, which lays the foundation for the identification and evaluation of drug resistance tumors by photoacoustic imaging.
Here, we designed a “Matryoshka” strategy that used redox-responsive MONs as core, PEI coated to absorb siRNA, acid-detached PEG as an outer shell, GE11 to achieve the target, co-loaded gene and drug to the target cell. The nanosystem shows the following features for multistaged delivery (Scheme 1): i) GE11 polypeptide for active targeting, increasing the uptake capacity and as a seeker for drug resistant cell [[49], [50], [51]]; ii) PEG outer shell prolongs blood circulation, protecting loaded cargo, and thus enhances tumor accumulation, reduce the off-target effects [52]; iii) Sensitive response of the benzoic acid imine bond to pH induces the nanosystem been “activated” in TME (PEG detachment and charge increase) and improves the uptake of cells, exposure of PEI promote the endosome escapes through the “proton sponge" effect [53], enhances the cytosolic siRNA transport to achieve efficient gene silencing; iv) MONs core exhibit excellent cargo (including nucleic acid and drugs) loading efficiency and post-modification capabilities [54,55], and further degraded by the high GSH of intracellular [38,56] to sequentially release paclitaxel. v) The newly deep red carbonized polymer dots (CPDs) with narrow full width at half maximum (FWHM) loaded to the MONs frame, which has strong absorption and emission capabilities in the deep red-light region with high biocompatibility [57] to achieve real-time and security bio-imaging. Overall, when the nanosystem recognizes drug resistant tumor cells, it is “activated" and the control sequence releases gene and drug, enhancing the sensitivity of chemotherapy through RNAi first, and then releases the drug to kill the cells, significantly improving the therapeutic effect of ERα-positive drug resistant breast cancer.
Scheme 1.
Schematic illustration of the procedures of the “Matryoshka" nanosystem based on TME pH and redox responsive, co-loaded siRNA and drug and control release. The nanosystem recognizes EGFR on the cell membrane and after entering the target cell, the siNgBR enhances the drug sensitivity of cancer cells and paclitaxel efficient killing cells.
2. Results and discussion
2.1. Preparation and characterization of the MONC-PPG nanosystem
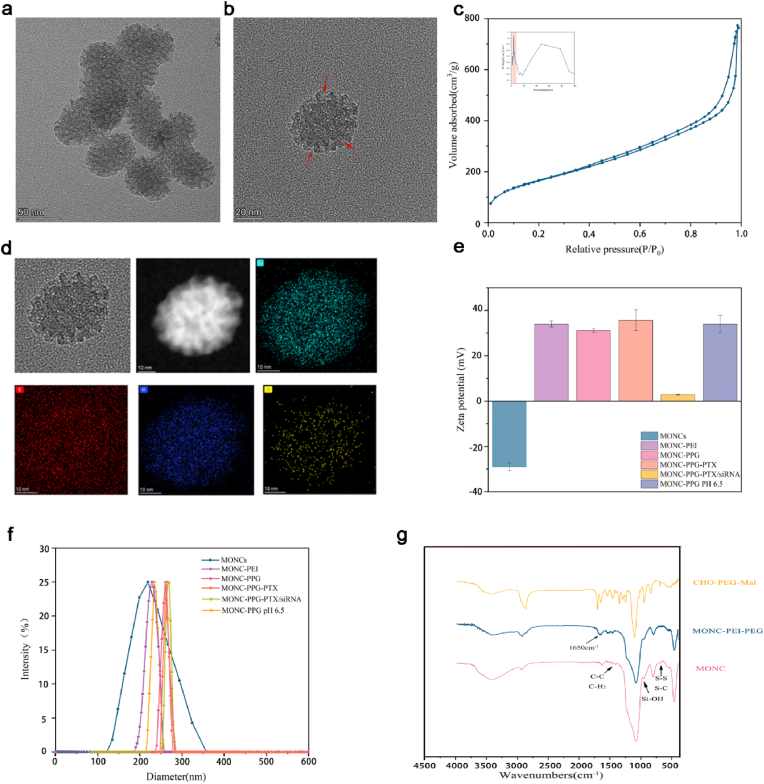
We use the micelle/precursor co-templating assembly (M/P-CA) strategy to prepare MONCs [56]. Transmission electron microscopy (TEM) images show that MONC-CPDs (hereinafter referred to as MONCs) have a typical spherical structure, with clearly pore-like structures, the diameter of 52.07 ± 6.29 nm (Fig. 1a and b). MONCs have a large pore volume (1.16 cm3/g) and pore diameter (7.71 nm) that is enough to load cargo (including small anticancer drugs and large molecular siRNA) (Fig. 1c). An obvious peak centered at 680 nm is observed in CPDs and MONCs when excited by 413 nm, proving the successful connection between MONs and CPDs. The Raman spectroscopy (S–S and S–C tensile vibration at 488 cm−1) (Figure S1c, Supporting Information) and energy dispersive spectroscopy (EDS) element mapping (Fig. 1d) verified the existence of disulfide bonds in MONCs, laid the foundation for its subsequent response.
Fig. 1.
Preparation and characterization of MONC-PPG-siRNA/PTX nanosystem. TEM image of a) MONs and b) obvious pore in MONs, the red arrows in the images indicate the pore of MONs. c) N2 adsorption-desorption isotherm and (inset of c) the corresponding pore size distribution of MONs. d) TEM image and element mapping of MONs. e) Zeta potential of the nanosystem in deionized water (pH 7.4 or 6.5). The data are shown as the mean ± s.d. (n = 3). f) Size distribution of the nanosystem measured by dynamic light scattering (DLS). g) FI-IR spectrum of CHO-PEG-mal, MONCs and MONC-PPG.
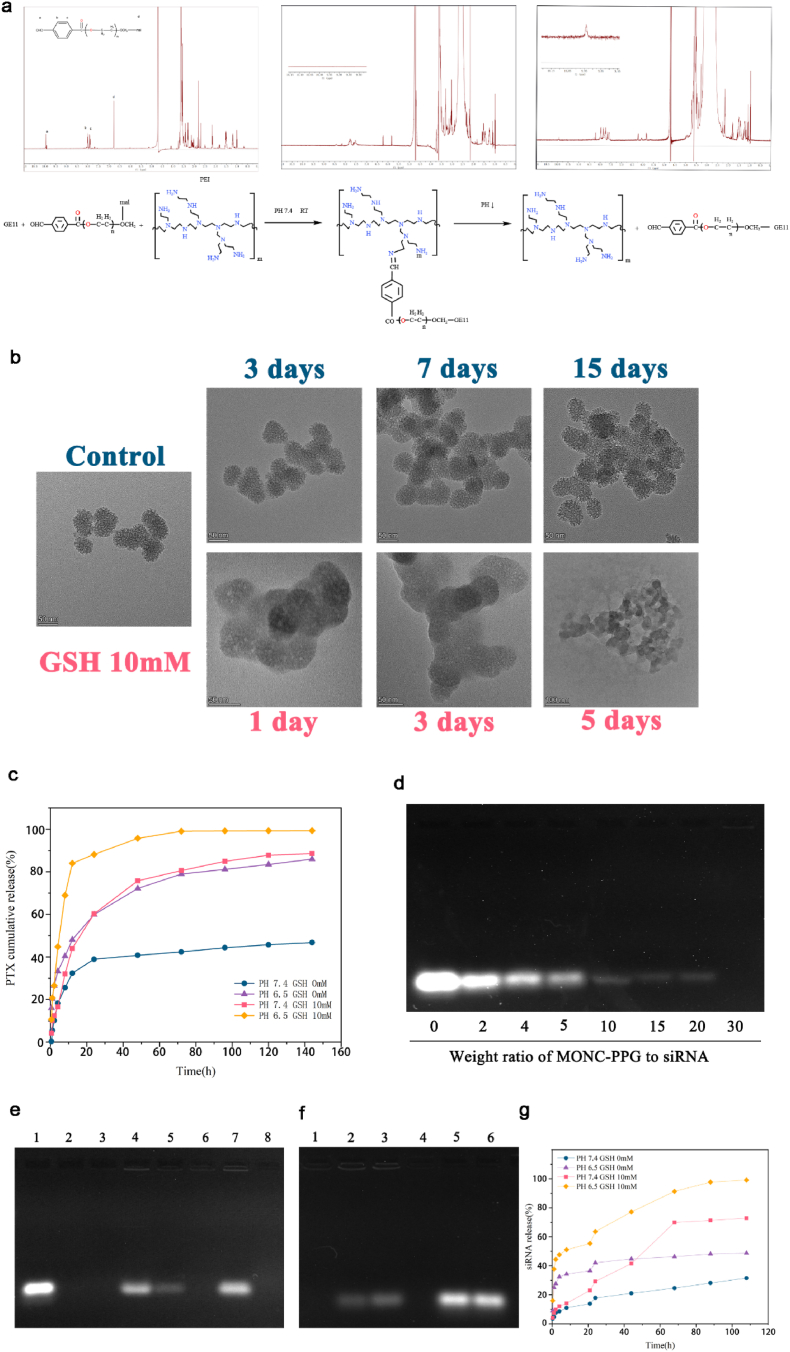
Subsequently, we modified MONCs with PEI, PEG and GE11 stepwise, and monitored the successful modification through dynamic light scattering (DLS, Fig. 1f), Zeta potential (Fig. 1e), Fourier transform infrared (FT-IR, Figure 1g), 1H NMR spectrum (Fig. 2a), and thermogravimetric analysis (TGA, Figure S1e, Supporting Information). TEM shows that the MONCs core was surrounded by a polymer (Figure S1a, b Supporting Information) and its hydrodynamic diameter and zeta potential also changed with modification (Table 1 and Fig. 1e and f). The MONC-PPG has good colloidal stability under physiological conditions at least 3 days (Table 1). FT-IR spectrum shows the formation of the benzoic imine linker between CHO-PEG-mal and PEI as denoted by an absorption peak at 1650 cm−1 (Fig. 1g). In the 1H NMR spectrum (Fig. 2a), the aldehyde proton peak (10.01 ppm) and maleimide peak (6.7 ppm) of CHO-PEG-mal disappeared after connection, indicating that the PEI and GE11 were successfully connected with CHO-PEG-mal. TGA curves (Figure S1e, Supporting Information) of MONCs, MONC-PEI and MONC-PPG show that the weight loss percentages of functional MONCs gradually increase following functional polymer decoration. We also tested by flow cytometry that the connection efficiency of GE11 to the maleimide of PEG can be as high as 94.3% (Figure S1f, Supporting Information).
Fig. 2.
A) Schematic diagram of the low pH-triggered benzoic imine bond disassociation PEG detachment from PEI (down) and 1H-NMR spectra of CHO-PEG-mal and PEI-PEG-GE11 in D2O at pH 7.4 and 6.5(up). The aldehyde proton peaks (10.01 ppm) reappeared on PH 6.5 after 30min. b) TEM image of in vitro degradation of the MONs under 10 mM GSH at 37°C. c) The cumulative releasing curve of PTX from MONC-PPG-PTX in buffer solutions at different GSH concentrations (0 and 10 mM) and PH (7.4 and 6.5). d) Gel retardation electrophoresis of MONC-PPG-PTX/siRNA complex nanoparticles. Lane 1: naked siRNA; complex nanoparticles of MONC-PPG-PTX/siRNA prepared at different MRs of 2, 4, 5, 10, 15, 20, 30, respectively. e) RNase protection assay. Lane 1: naked siRNA; lane 2: naked siRNA + RNase A; lane 3: MONC-PPG-PTX/siRNA complex; lane 4: MONC-PPG-PTX/siRNA + RNase A + heparin; lane 5: MONC-PPG-PTX/siRNA + RNase A + heparin + RNase A + heparin (Secondary elution experiment); lane 6: MONC-PPG-PTX/siRNA + RNase A; Lane 7: MONC-PPG-PTX/siRNA + heparin; lanes 8: MONC-PPG-PTX/siRNA + heparin + RNase A. f) Releases of siRNA from MONC-PPG-PTX/siRNA with a different environment. Lanes 1,4: MONC-PPG-PTX/siRNA under PH 7.4 and GSH 0 mM; lane 2: MONC-PPG-PTX/siRNA under PH 6.5 and GSH 0 mM; lane 3: MONC-PPG-PTX/siRNA under PH 7.4 and GSH 10 mM; lane 5: MONC-PPG-PTX/siRNA under PH 6.5 and GSH 10 mM; lane 6: naked siRNA. g) The cumulative release of siRNA from MONC-PPG-PTX/siRNA under different environments.
Table 1.
Characterization of nanosystem prepared in study.
| Groups | Particle size(nm) | PDI | Zeta potential(mV) |
|---|---|---|---|
| MONCs | 216.41 ± 11.68 | 0.136 | −29.00 ± 1.69 |
| MONC-PEI | 226.98 ± 8.75 | 0.114 | 33.98 ± 1.37 |
| MONC-PPG | 259.81 ± 15.99 | 0.039 | 31.12 ± 0.95 |
| MONC-PPG-PTX | 262.39 ± 15.71 | 0.090 | 35.64 ± 4.61 |
| MONX-PPG-PTX/siRNA | 267.35 ± 19.03 | 0.050 | 2.81 ± 0.23 |
| MONC-PPG PH 6.5 | 232.01 ± 13.21 | 0.046 | 34.00 ± 3.85 |
| MONC-PPG 3days | 266.71 ± 12.62 | 0.060 | 32.79 ± 2.31 |
2.2. The sequence response to pH and redox of MONC-PPG nanosystem
Next, we verify the sequential degradation of the MONC-PPG nanosystem. As is well-known, PEGylation can increase the stability of nanoparticles in the circulatory and protect cargo, but it also inhibits the endocytosis and lysosomal escape, which is also called the “PEG dilemma" [53]. We used a mild-acid degradable benzoic imine bond to bypass the dilemma (Figure S2, Supporting Information). 1H NMR shows (Fig. 2a) after PEI-PEG incubation 30min under pH 6.5, the aldehyde proton peak reappeared but was weak than mal-PEG-CHO, indicating the dissociation of PEG begun after 30min under acid conditions. The change of particle size (Fig. 1e) also proves it. The degradation of the MONCs core lasted for 5 days, using 10 mM GSH to simulate the intracellular microenvironment. Glutathione is a reducing agent containing active side group, which is about 3 orders of magnitude higher in the cell (2–10 mM) than in the extracellular (2–20 μM) [38,58,59]. The TEM (Fig. 2b) shows that due to the high surface area of MONCs, GSH can penetrate deep into the pores to cleave the disulfide bond, MONCs have been degraded from the first day and have undergone surface and overall erosion processes and completely decomposed at 5 d, the control group without GSH did not show obvious degradation even after 14 d, which also shows the stability of the MONCs system. In addition, it is worth mentioning that our research process is carried out under static conditions, while in a living environment, the balance between oxidized (GSSG) and reduced (GSH) glutathione can be achieved by peroxidase/reductase [60,61], so the degradation rate of nanoparticles will be higher as the concentration of GSH is maintained. Based on the above analysis, we have successfully prepared the MONC-PPG nanosystem. The PEG in the outer layer quickly detaches from the PEI in the acidic environment. After entering the cell, the high GSH causes the MONs core to decompose, achieving the purpose of decomposing layer by layer.
2.3. PTX and siRNA loading and release with dual-response
Next, the MONC-PPG system is co-loaded with drugs and genes. Paclitaxel is an effective anticancer drug with poor water-soluble, MONC's mesoporous structure can reduce the drug particle size and increase the specific surface area [62], which would increase the dissolution rate of PTX. When the weight ratio of MONC-PPG to PTX is 3:1, the drug loading rate can be as high as 24.14 ± 1.87% and have the highest drug entrapment efficiency (94.56%), so we choose 3:1 for the following research (Table S3, Supporting Information). More importantly, the release of PTX is pH and GSH dependent (Fig. 2c). When MONC-PPG-PTX is under TME condition (pH 6.5 and GSH 10 mM), the cumulative release of PTX can reach 99% and continue to act on cells within 72 h. It is worth noting that PTX also exhibited a partial pH-dependent release when only in an acidic environment, inferring that due to the large pore size of MONs, with the “protective layer” PEG detachment, the small molecular drug loaded in the pore may be released accordingly. Subsequently, as the MONs core were decomposed, the remaining PTX loaded in was further completely released to exert its full effect. The dual-responsive of MONC-PPG reduced the ineffective release of PTX during the circulatory, full release after reaching the target region to maximize its effectiveness.
SiRNA was loaded into MONC-PPG by electrostatic adsorption. Fig. 2d shows the siRNA condensation capability for MONC-PPG-PTX at different mass ratios (MR), the band of siRNA disappeared at MR of 30, suggesting enough siRNA-loading (33.3 μg/mg) of MONC-PPG-PTX to achieve the RNAi in vivo. RNases A protection assay (Fig. 2e) showed that MONC-PPG-siRNA was first treated with RNases A and then replaced with heparin to appear siRNA (lane 4), the secondary replacement with heparin (lane 5) still has siRNA in the MONC-PPG, which proves that MONC-PPG can effectively protect siRNA from enzymatic degradation, that is the prerequisite of RNAi in the vivo. More importantly, MONC-PPG efficiently releases siRNA when it reaches the target region (acidic reducing environment). When MONC-PPG-siRNA is under physiological conditions (lanes 1, 4), siRNA bands cannot be detected. When adjusted to pH 6.5 and 10 mM GSH (lanes 5), a clear and bright band appears on the gel, but when only with high GSH or low pH (lanes 2 and 3 respectively), just a part of siRNA has been released (Fig. 2f). The cumulative release curve of siRNA under different conditions shows siRNA is almost completely released at GSH 10 mM and pH 6.5 (Fig. 2g). Interestingly, we found that under the acidic condition, the siRNA cumulative release curve appeared a plateau after 2 h. 20 h later, the group with high GSH showed a tendency to burst first and then gradually release and did not exist GSH group only released a small amount after this plateau. Suggesting that due to our control of the pore size (7.7 nm), siRNA adsorbed on the surface was released quickly at the initial stage, then with the MONCs core was cracked under the action of GSH, the remaining siRNA was released for more than 80 h. Such a special structure makes MONC-PPG different from traditional transfection, it releases siRNA only when gets the target region and achieve continue to releases siRNA so that RNAi can act on cells for a long time to effectively knockdown protein and synergy PTX to killing cells (Figure S3, Supporting Information).
In summary, based on the “layer-by-layer" decomposition of the nanosystem, the gene and drug are gradually released, indicating that the MONC-PPG nanosystem can reduce the useless release of cargo in circulation and fully release in target cells. The optimal sequence of RNAi first to reverse drug resistance and then chemotherapeutics to kill cells have formed, while the long-term release of siRNA provides sufficient support for continuous treatment. The content of each component of the nanosystem is shown in Figure S4 (Supporting Information).
2.4. Cellular uptake, subcellular co-localization, and lysosomal escape of MONC-PPG
First, we used flow cytometry to study the ability of the nanosystem to deliver siRNA into cells. The traditional transfection reagent lipo2000 was used as a control, to compare the ability of the MONC-PPG and MONC-PP to deliver siRNA into cells. The results show that MONC-PPG has the highest gene delivery (reach more than 99% in 2h), while the MON-PP is 81%, both higher than 59% of lipo2000 (Figure S5a, Supporting Information). To observe more intuitively, we used CLSM to observe the cellular uptake of GE11-targeted MONC-PPG and non-targeted MONC-PP under different pH conditions (Figure S5b-c, Supporting Information). Due to the loaded of CPDs, the MONC-PPG emits red light under CLSM, without reloading the fluorescent probe like the previous study. In the neutral environment (pH 7.4), MONC-PPG quickly (within 30 min) binds to the ligand on the cell membrane, showing red fluorescence signals around the cell. In contrast, MONC-PP showed obvious endocytosis after incubating with cells after 1 h. The charge reversal in TME is an important way for the MONC-PPG nanosystem to achieve long circulation and promote endocytosis simultaneously, so, we adjusted the pH to simulate the TME (pH 6.5) to further analyze the influence of detachable-PEG for endocytosis. The results showed that under short-term (10min), MONC-PPG exhibited significantly stronger cell binding capacity than MONC-PP both in a neutral or acid environment. In an acidic environment after 30 min, according to the result of 1H-NMR, PEG began to dissociate, the charge of the nanosystem began to be positive. Compared with in the neutral environment, both the MONC-PPG and the MONC-PP system showed enhanced endocytosis, especially the MONC-PP, which illustrates the importance of charge reversal for promoting endocytosis [51,[63], [64], [65]].
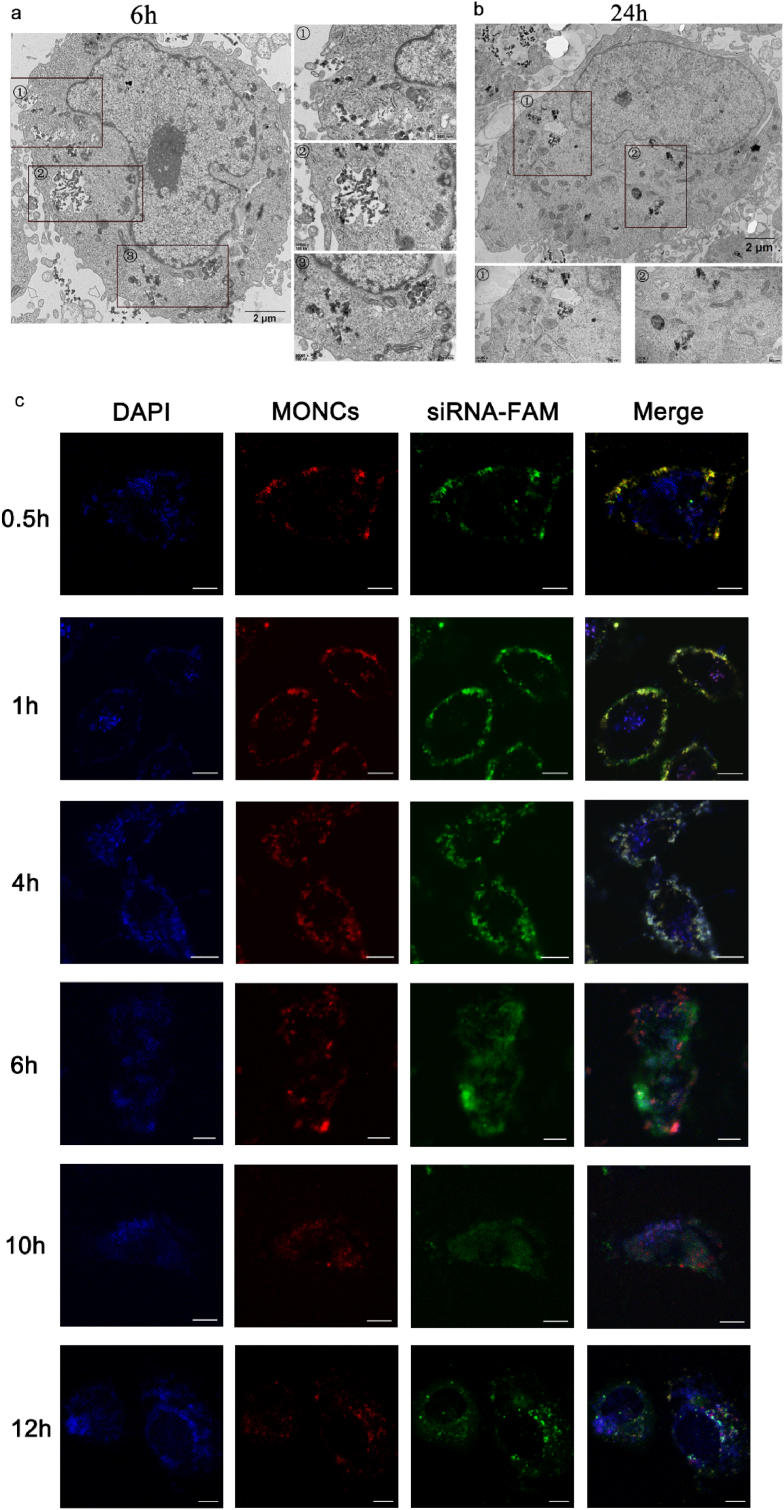
Next, the subcellular localization of MONC-PPG in the cell was detected through bioTEM and CLSM. First, MONC-PPG was incubated with MCF-7/PTX cells for 6 h and observed with bioTEM (Fig. 3a and b). Mesoporous structured nanoparticles and their aggregates were found in both vesicles and cytoplasm. 3a① showed a typical event of prephagocytosis, that cell membrane protrusions around some nanoparticles. When MONC-PPG is endocytosed into the cell, phagosomes are formed (3a②). At the same time, we can also see that some nanoparticles have escaped from lysosomes into the cytoplasm, and degradation has begun (3a③). This lysosome escape and degradation are more obvious at 24h (Fig. 3b). Furthermore, we used CLSM to intuitively observe the co-localization of the MONC-PPG-siRNA system with lysosomes and siRNA release (Fig. 3c). After 30min incubation, the siRNA and MONC-PPG were co-localized on the cell membrane, MONC-PPG-siRNA began to be endocytosed by the lysosome after 1h. Lysosomal escape has begun after 6h, CLSM shows that some green fluorescence (siRNA) separated from red (MONCs) and blue (lysosome), and in the cytoplasm. After 12h, they were completely separated. Our research is the first to describe the entire process that after MONC-PPG-siRNA enters cells, confirms that MONC-PPG nanosystem can directly deliver the siRNA into the target cell, making a lysosomal escape and releasing siRNA into the cytoplasm, reduce intracellular wastage and minimizing off-target effects.
Fig. 3.
BioTEM images of MCF-7/PTX cells after incubation with MONC-PPG for 6h(a) and 24h(b). c) CLSM images of MCF-7/PTX cells after the incubation with MONC-PPG-siRNA for various times (0.5, 1, 4, 6, 10, 12h). Lysosomes stained with Lysotracker BLUE (blue fluorescence), MONC-PPG shows red fluorescein due to CPDs, siRNA labeled with green fluorescein FAM, respectively. scale bar:10 μm.
2.5. Gene silencing efficiency of MONC-PPG-siNgBR and the mechanism of reversing drug resistance in ERα-positive breast cancer cell
First, we compared the transfection efficiency of the traditional transfection Lipofectamine 2000 and MONC-PPG-siRNA. The MONC-PPG-siNgBR knockdown efficiency is much higher than lipo2000, 48h with a 90.56% downregulation (Fig S6a, b, Supporting Information).
Previous studies have also shown that NgBR is highly expressed in tumor sections of ERα-positive breast cancer patients who received chemotherapy [2]. We found that NgBR expression was significantly upregulated in drug resistant cells (MCF-7/PTX) compared with their parental cells (MCF-7) (Figure S6c, d Supporting Information). RT-PCR assays further confirmed the result (Figure S6e, Supporting Information). Based on this, we focus on the mechanism of knocking down NgBR to make ERα-positive breast cancer regain chemosensitivity. According to our research, MONC-PPG-siNgBR may regulate the drug resistance of ERα-positive breast cancer in various ways (Scheme 2).
Scheme 2.
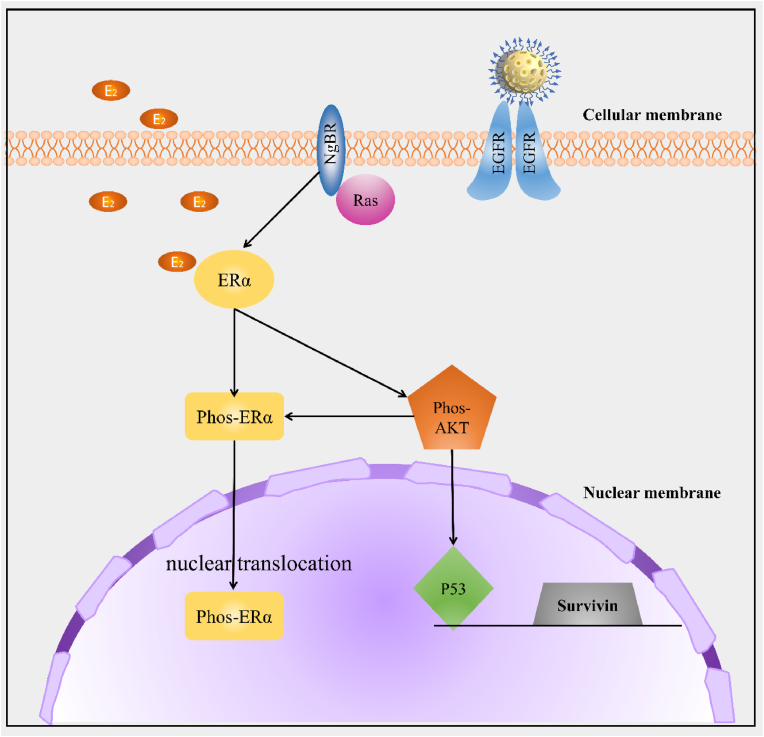
The proposed working model elucidates the roles of MONC-PPG-siNgBR in regulating the drug resistance of ERα positive breast cancer cells. EGFR is used for nanosystem recognize specific cells. Knocking-down NgBR regulates the phosphorylation of ERα and inhibits nuclear translocation. On the other hand, NgBR influence the Akt phosphorylation and downstream molecules.
It is well known that estradiol-induced proliferation and anti-apoptosis are the main causes of drug resistance [22,23]. Phosphorylation ERα (phos-ERα S118) is the manifestation of ERα activation [66] and a confirmation signal of ERα-mediated drug resistance [67]. So, we explored whether NgBR is involved in the process of activating ERα (Fig. 4a and b). The expression of phos-ERα increased by 1.72 times after estrogen stimulation, and with NgBR knockdown by MONC-PPG-siNgBR, it decreases to 0.39 times. Interestingly, we found that NgBR knockdown does not cause a decrease of total-ERα protein, but significantly reduces the phos-ERα. Then, we use immunofluorescence to observe the cell expression of phos-ERα, the image shows that the estrogen stimulates strengthen the ERα S118 nuclear translocation, and knockdown NgBR reduced the nuclear translocation, the ERα S118 located in the nucleus is significantly reduced and can be seen in the cytoplasm(Figure S7, Supporting Information), remind that NgBR may be related to the nuclear translocation of phosphorylated ERα, which is the key step that initiates the transcription of ERα genes.
Fig. 4.
A) Western blot analysis of MCF-7/PTX cell after various treatments, with or without stimulate by E2 for 48 h. b) Quantitative analysis of the normalized protein expression (using Image J software). The data are shown as the mean ± s.d. (n = 3). ∗∗∗p < 0.001. RT-PCR analysis of NgBR(c) and survivin(d) mRNA levels after treatment with MONC-PPG-siNgBR 48h. The data are presented as the mean ± s.d.(n = 3).
Next, we explored the impact of NgBR knockdown on ERα-related signaling pathways. P53 is a tumor suppressor and its inactivation is one of the most common events in cancer. ERα can directly bind to P53 [68] and inhibit its function [69], which may also be important for the development of drug resistance. Next, ERα and p53 are bound to the survivin gene promoter, inhibiting the inhibitory effect of p53 on survivin, which gives ERα-positive breast cancer cells the ability to inhibit apoptosis. Survivin is participated in the division and apoptosis and affects the drug resistance of tumor cells [14,70]. As the result shows, with estrogen stimulate, the expression of NgBR of MCF-7/PTX increased by 1.07 times, the corresponding expression of phos-Akt increased by 1.22 times, survivin was 1.12 times, and p53 was 0.32 times (Fig. 4a and b). After using MONC-PPG-siRNA, the expression of phos-Akt reduced to 0.25 times, p53 increased 3.97 times higher than before. Correspondingly, survivin was reduced to 0.09 times. RT-PCR shows the same result (Fig. 4c and d). It verified that NgBR is highly correlated with the expression of p53 and survivin through the Akt pathway [2,20,32,71], i indicating that knockdown NgBR can fundamentally enhance the sensitivity of ERα-positive breast cancer cells to chemotherapy and influence the cell apoptosis.
In summary, on the one hand, knockdown NgBR not only reduces the activation of ERα but also inhibits the nuclear translocation of ERα S118, on the other hand, NgBR knockdown increases the expression of P53 (a tumor suppressor) and reduces the survivin induced by estrogen through the Akt pathway, suggesting that NgBR plays an important role in the drug resistance of ERα-positive breast cancer (Scheme 2). Last, we found compared with Lipofectamine 2000, the gene silencing efficiency of the MONC-PPG-siRNA system is more stable and efficient.
2.6. The bio-safety and therapy efficiency of MONC-PPG-PTX/siNgBR nanosystem of drug resistant ERα-positive breast cancer cell
2.6.1. The bio-safety and hemolytic activities of MONC-PPG
CCK-8 and Annexin V-PI dual staining were used to detect the bio-safety of the nanosystem. MONCs and MONC-PPG will not arouse cytotoxicity, but when MONCs are only modified PEI, it has cytotoxicity when the concentration is higher than 200 μg/mL (Figure S8, Supporting Information), maybe the direct exposure of cationic PEI makes MONC-PEI have biological toxicity, but PEG-modified can effectively reduce biological toxicity.
We also test the membrane biological property of the nanosystem [72,73]. As it is displayed in Figure S9 (Supporting Information), the MONCs and MONC-PPG nanosystem exhibited comparable hemolysis with the control group, indicating good biocompatibility. But when MONCs were only modified with PEI (MONC-PEI), the hemolysis further increased. In conclusion, PEGylation can effectively reduce the biological toxicity caused by cationic PEI, and ensure safety while enhancing the circulating half-life.
2.6.2. The therapy effect of the MONC-PPG-siNgBR/PTX to drug resistant ERα-positive breast cancer cell
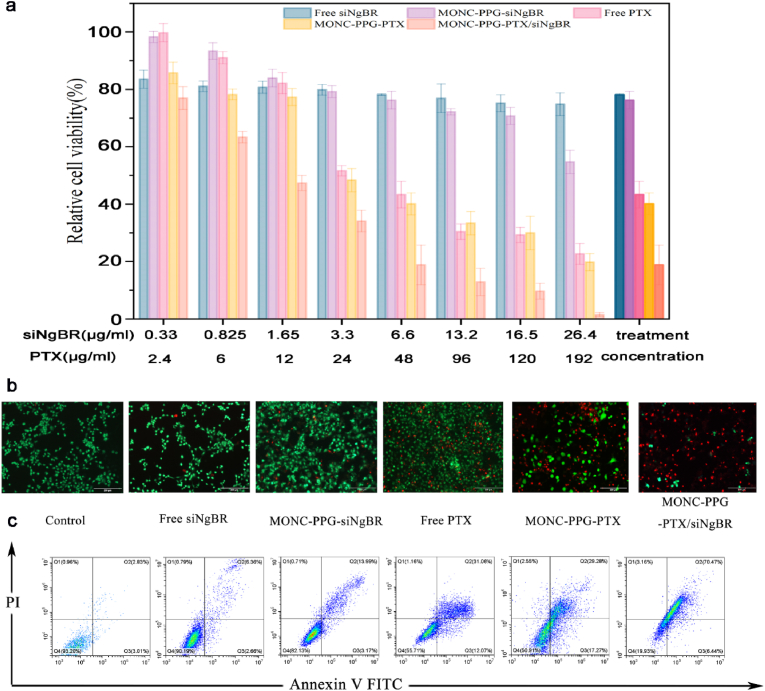
Next, we detect cell apoptosis to prove whether knockdown NgBR can regain the chemotherapy sensitivity of MCF-7/PTX. The single gene therapy group (including free siNgBR and MONC-PPG-siNgBR group) has milder killing effects on cells (the percentage of apoptotic cells is 9.02% and 17.16%, respectively). Based on the previous analysis, we can infer that knockdown NgBR has an inhibitory effect on the proliferation of tumor cells due to related to a variety of tumor factors. The chemotherapeutic group showed moderate cell killing ability. This is due to the resistance of MCF-7/PTX to paclitaxel, PTX fails to exert the best cytotoxicity. The performance of the MONC-PPG-PTX/siNgBR group illustrates the important role of knockdown NgBR in reversing drug resistance. The sequential degradation of the MONC-PPG-PTX/siNgBR makes the gene silent and chemotherapeutic drugs can be played in sequence, making its therapeutic effect far superior to the other groups, apoptosis rate can reach 76.91% at the same therapeutic concentration (Fig. 5). MONC-PPG-PTX/siNgBR provides the possibility for personalized treatment of drug resistant cancer.
Fig. 5.
Combination therapy of MCF-7/PTX cell in vitro. a) Cell viability of MCF-7/PTX after incubation with different concentrations of PTX, siRNA, and MONC-PPG-PTX/siNgBR for 48 h. The data are presented as the mean ± s.d. b) CLSM images of CAM- and PI-costained MCF-7/PTX cells after various treatments (scale bar: 200 μm). c) Flow cytometry analysis of apoptosis after various treatments.
2.7. Photoacoustic performances and biodistribution of MONC-PPG
Due to the complexity and heterogeneity of tumors, personalized treatment is a trend. A potential way to achieve it is theranostics, which combines treatment with real-time diagnosis to provide an opportunity to monitor the effectiveness of treatment and adjust the treatment window over time [[74], [75], [76]]. Due to its high depth penetration and spatial/temporal resolution, photoacoustic (PA) imaging can guide drug release, localization, and diagnosis [77,78]. Carbon dots have great potential in PA bioimaging with low-cost sources and low cytotoxicity but are limited by visible light excitation [79,80]. The CPDs introduced in our research overcome these obstacles, it has ultra-high deep red/near-infrared emissivity, narrow full width at half maximum (FWHM) [57], and greatly improve the imaging contrast, it is of great significance as a laser emitting material.
We scan the full wavelength of CPDs and MONC-PPG from 680 nm to 970 nm under the PA imaging system and choose 680 nm as the excitation wavelength (Figure S10a, Supporting Information). As shown in Fig. 6a, the PA signal of CPDs and MONC-PPG are both dose-dependent with an excellent linear relationship. MONC-PPG can get satisfactory imaging within treatment concentration (200 μg/mL), so MONC-PPG can ensure the biological safety, imaging, and treatment effect at the same time, is an ideal carrier for imaging and therapeutic, enriches its clinical applications. Next, we injected MONC-PPG into MCF-7/PTX tumor-bearing mice via the tail vein. The accumulation of MONC-PPG in the tumor region varies with time-dependent, reaching a peak at 6 h post-injection and then gradually decreasing. After 24 h, some PA signals remained in the tumor (Fig. 6c), the quantitative analysis showed a similar trend (Fig. 6b). The above results show that the MONC-PPG nanosystem can effectively accumulate in the tumor and has an excellent PA imaging performer, which has a huge potential for clinical transformation.
Fig. 6.
In vivo multiple imaging and corresponding quantification analysis of MONC-PPG nanosystem. a) In vitro PA imaging intensities of CPDs(red) and MONC-PPG (black) with various concentrations. b) Quantification analysis of normalized PA imaging signal within the tumor region at corresponding time points. The data are presented as the mean ± s.d. n = 3. c) In vivo PA images of MCF-7/PTX tumor-bearing mice after intravenous injection of MONC-PPG. In vivo fluorescent imaging of MCF-7/PTX tumor-bearing nude mice with intravenous injection of d) MONC-PPG or e) MONC-PP at different time points.
The tumor accumulation profiles after intravenous injection of MONC-PPG and non-targeted MONC-PP as controls were tracked by fluorescence imaging. Compared with MONC-PP (Fig. 6d), MONC-PPG more specifically accumulate in the tumor regions (Fig. 6e) and quantitative analysis shows that the fluorescence intensity of MONC-PPG in the tumor region is significantly stronger than MONC-PP (Figure S10b, c, Supporting Information). Same as PA imaging, the tumor accumulation has the most aggregation after 6h intravenous injection.
Summing up the above, compared with MONC-PP only through the EPR effect, MONC-PPG can more efficiently accumulate in the tumor with the dual-action of the active targeting of the GE11 and the passively EPR effect [42]. Indicating that the MONC-PPG nanosystem can pass by the blood-tumor barrier (BTB) and deliver the cargo loaded into the target cells, minimizing unnecessary drug consumption and the off-target effect of genes.
2.8. The ability of the MONC-PPG nanosystem evaluate drug resistance of breast cancer in vivo
In the progression of tumors, the key regulatory molecules of tumor cells will be heterogeneous change, for malignant tumors, especially drug resistant tumors, use visualization strategies to assist tumor diagnosis, evaluate its progress and treatment response is a new standard. GE11 can efficiently and specifically recognize EGFR on the surface of tumor cell membranes. Our research shows that drug resistant cells (MCF-7/PTX) have higher EGFR expression levels than their parental cells (MCF-7) (Fig. 7 a-d). Therefore, we have constructed tumor models of MCF-7 and MCF-7/PTX respectively and injected MONC-PPG. The results showed (Fig. 7 e, f) that the drug resistant tumor has a significantly stronger PA signal than MCF-7. To sum up, the MONC-PPG nanosystem is expected to be a possible way for early diagnosis of tumor drug resistance and subsequent construction of a personalized diagnosis and treatment system.
Fig. 7.
Western blot and quantitative analysis (using Image J software) of MCF-7/PTX and MCF-7 cells (a, b) and tumor model tissue (c,d). The data are shown as the mean ± s.d. (n = 3). ∗∗∗p < 0.001. e) In vivo PA images of MCF-7/PTX and MCF-7 tumor-bearing mice after intravenous injection of MONC-PPG 6 h. f) Quantification analysis of PA imaging signal within the tumor region. The data are presented as the mean ± s.d. (n = 3).
2.9. Biological safety and anti-tumor efficacy of MONC-PPG-PTX/siNgBR in MCF-7/PTX tumor models
2.9.1. The biological safety of the MONC-PPG nanosystem
Biological safety is necessary for clinical transformation. MONC-PPG-PTX/siNgBR and MONC-PPG were injected into healthy mice through the tail vein every three days, PBS as a control. The process lasted 28 days. The mice's weight stably (Figure S11a, Supporting Information) and biochemical blood indexes in each group showed negligible fluctuation (Figure S11b, Supporting Information), demonstrating the high safety of MONC-PPG-PTX/siNgBR. Besides, H&E staining for major organs has no obvious pathological damage and inflammatory damage (Figure S11c, Supporting Information). Suggesting that MONC-PPG nanosystem have no toxicity regardless of long-term or short-term studies, supporting its potential in vivo application.
2.9.2. Anti-tumor efficacy of MONC-PPG-PTX/siNgBR in MCF-7/PTX tumor models
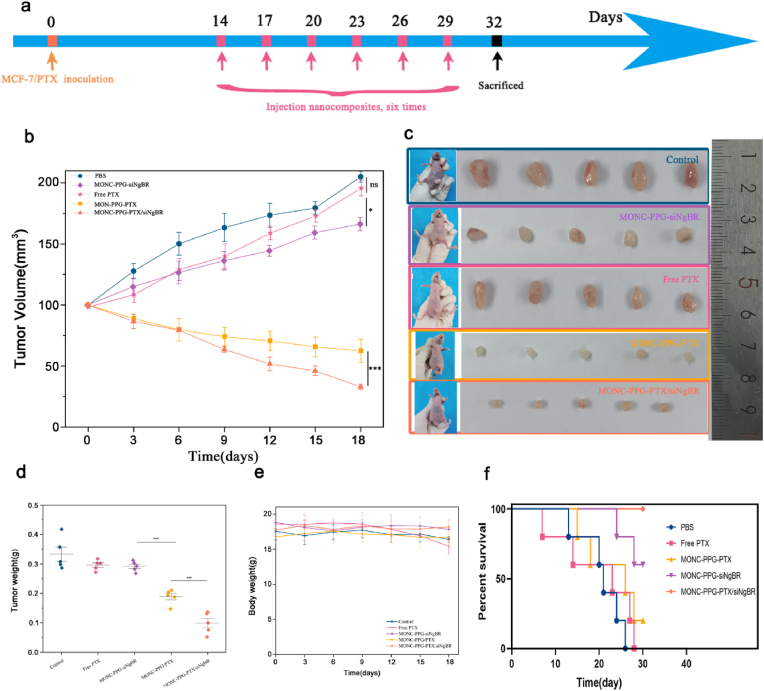
The personalized therapeutic efficacy of MONC-PPG-PTX/siNgBR was evaluated in the MCF-7/PTX xenograft mouse model (Fig. 7). The data showed that the tumor growth curve between the control and the free PTX group did not show a significant difference, this may be that the tumor model is formed by PTX-resistant cells and the degradation of free PTX in the circulation. So, it is urgent to reverse drug resistance and target delivery drugs. The tumor curve of the MONC-PPG-siNgBR group showed an inhibitory trend compared with the control group (P < 0.05). The MONC-PPG-PTX group achieved moderate growth inhibition. Compared with free PTX, MONC-PPG-PTX can achieve more killing effect because that MONC-PPG can target deliver PTX into the tumor cells, but due to the drug resistance of cells, the tumor-killing of MONC-PPG-PTX has not yet reached a satisfactory level. The MONC-PPG-PTX/siNgBR anti-tumor efficacy was further enhanced, the tumor was reduced to only 15.9% of the control group (P < 0.001). The excellent therapeutic effect is thanks to the synergistic effect of RNAi and chemotherapeutics.
However, local tumor inhibition is not the only goal clinically, long-term survival is also an important consideration. The long-term survival of mice after different treatments shows (Fig. 8f) that the MONC-PPG-PTX/siNgBR has the longest survival time, surviving over 30 days after the treatment. Interestingly, the MONC-PPG-siNgBR group is the second-longest survival time only to the MONC-PPG-PTX/siNgBR, with an average of 28.4 days, and 60% of the mice survived over 30 days, significantly better than other groups. The above suggests that the MONC-PPG-PTX/siNgBR treatment system is not only greatly inhibits the growth of tumors, but also significantly prolongs the survival time of mice with a better prognosis. It is the first study to discover the possible correlation between NgBR protein and the survival time, which is consistent with previous studies confirming that NgBR is closely related to the poor prognosis of breast cancer patients [2,34].
Fig. 8.
The therapeutic efficacy of MONC-PPG nanosystem in vivo. a) Schematic illustration of therapeutic experiments in MCF-7/PTX models. The arrows indicate the timing of tail-vein injected MONC-PPG nanosystem (pink). b) Tumor growth curve of the different treatment groups in vivo. Tumor size measurement began on the 14th day and continued every three days until the 32nd day (the first recording time as 0 days). n = 5 mice per group. The data are presented as the mean ± s.d. c) Images of tumor tissues after sacrifice and picture of mice before sacrifice are shown. d) The tumor weight after sacrifice. e) As described in d), the body weight curve of mice during the whole treatment process. f) Time-dependent survival curve. ∗p<0.05 ∗∗∗p < 0.001.
2.10. The mechanism of MONC-PPG-PTX/siNgBR reverse drug resistance and inhibit metastasis in vivo
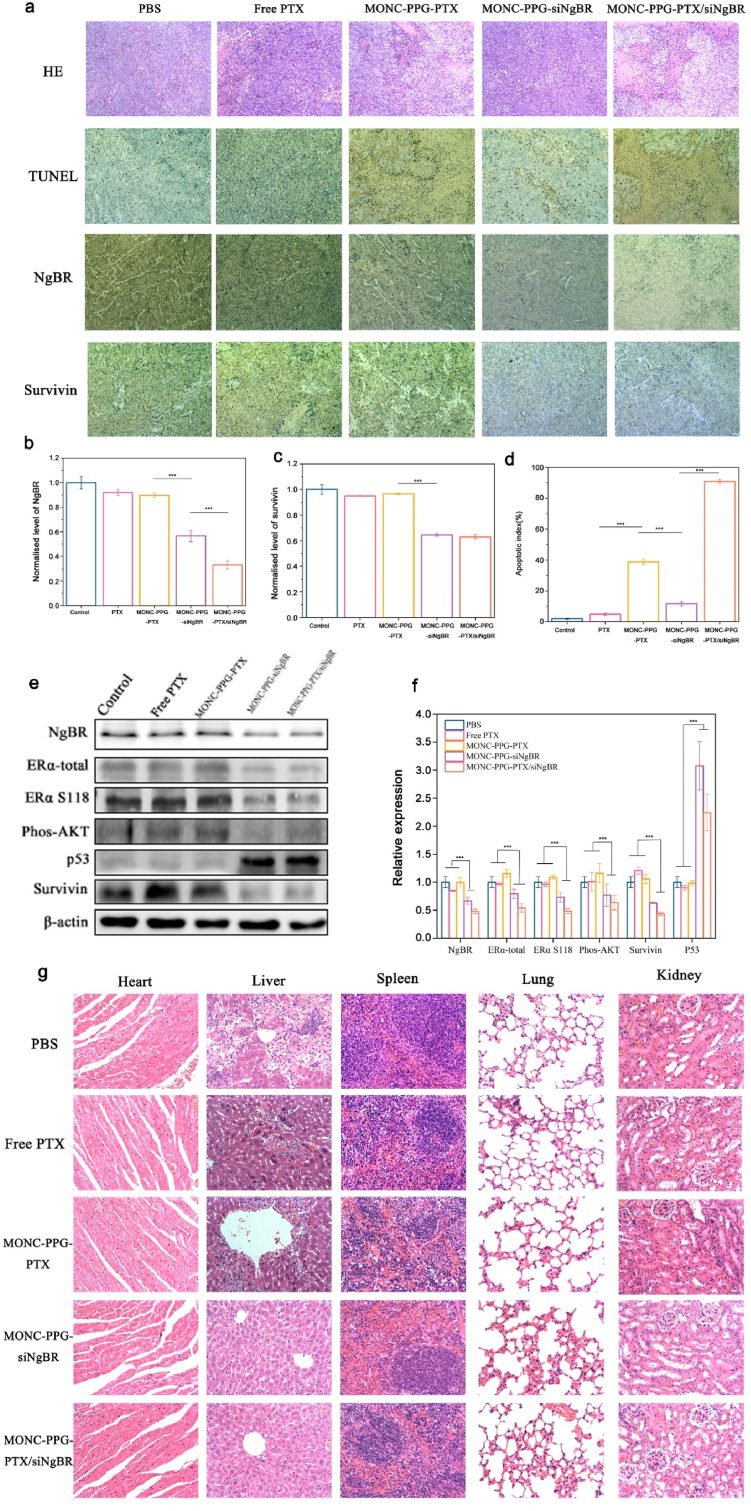
Next, we performed IHC and TUNEL assay (Fig. 9a) and quantitative analysis (Fig. 9 b-d) of different treatment groups. NgBR is the high expression in the control group and was significantly down-regulated in the MONC-PPG-siNgBR and MONC-PPG-PTX/siNgBR groups, which were about 0.56 times and 0.33 times that of the control group, respectively. With NgBR knockdown, the survivin also changed, the reduction in the MONC-PPG-siNgBR group and the MONC-PPG-PTX/siNgBR group were 0.64 times and 0.62 times that of the control group, respectively, indicating that the MONC-PPG system can successfully deliver siRNA to the target cells and achieve RNAi effects. Subsequently, we performed TUNEL assay analysis (Fig. 9d), showing that the proportion of nuclear apoptosis in the MONC-PPG-PTX/siNgBR group was the highest in all groups, indicating that the MONC-PPG-PTX/siNgBR has the best tumor-killing effects.
Fig. 9.
The MONC-PPG-PTX/siRNA nanosystem regains drug sensitivity through knockdown NgBR protein in vivo. a) H&E, TUNEL, NgBR and survivin staining of tumor sections after various treatments. b-d) Quantification of positive tumor cells are shown as the mean ± s.d. (n = 3). ∗∗∗p < 0.001. e) Western blot of proteins in tumors tissue and f) quantitative analysis of the normalized protein expression in tumors (using Image J software). G) H&E staining of major organs after treatment. Scale bars,17.11 μm.
The western bolt analysis (Fig. 9e and f) shows consistent results with immunohistochemistry. Compared with the group not loaded with siNgBR, the group loaded with siNgBR showed a significant decrease in the expression level of NgBR protein (the MONC-PPG-siNgBR group and the MONC-PPG-PTX/siNgBR group were 0.66 and 0.48 times that of the control group, respectively). Correspondingly, the knockdown of NgBR reduced phos-Akt and survivin, increased P53 in the corresponding, which is consistent with the in vitro results, also prove that the MONC-PPG nanosystem can deliver nucleic acids to the target, making it fully functional. Slightly different from the in vitro results, with the NgBR knockdown not only reduced the expression of activated ERα (MONC-PPG-siNgBR group and MONC-PPG-PTX/siNgBR group was 0.73 and 0.48 times that of the control group, respectively), but the total ERα also decreased (the MONC-PPG-siNgBR group and MONC-PPG-PTX/siNgBR group were 0.79 and 0.53 times that of the control group, respectively). This may be the short stimulus of estrogen in vitro, the total ERα did not have enough time to change [2], while the in vivo experiment performed a total of six injections, which lasted 18 days, the continuous knockdown of NgBR and estrogen stimulus gave the total ERα sufficient time to change, further indicating that NgBR may be closely related to ERα.
Next, we performed HE staining of organs in each group after treatment (Fig. 9g). We find that in the control group, free PTX group and MONC-PPG-PTX group, metastatic lesions were seen in the liver, and they were more obvious around the central vein. Corresponding pathological changes occurred in the spleen. More interesting is that the organs of the MONC-PPG-siNgBR and the MONC-PPG-PTX/siNgBR group have no metastasis, inflammation, or other pathological changes. Indicating that the drug resistant ERα-positive breast cancer may have a higher degree of malignancy, making it more prone to metastasis, knocking down NgBR can not only regain chemotherapy sensitivity but may also inhibit tumor metastasis. Previous studies have also shown that NgBR may be involved in a variety of tumor metastasis-related mechanisms [21,32,81], including epithelial-mesenchymal transition (EMT) and tumor angiogenesis. Combining the previous results, we can try to infer that the reason MONC-PPG with siNgBR groups have a longer survival time is that on the one hand, the growth of local tumors is inhibited, on the other hand, NgBR knockdown may inhibit tumor metastasis and prolong survival. Synthesize our research results, because of the high malignancy and rapid growth of drug resistant ERα-positive breast cancer, the low responsiveness to chemotherapeutic further hinders its clinical treatment. NgBR is related to a variety of tumor factors, knock it down can decrease tumor proliferation factors and increase apoptosis factors, inhibiting the tumor process and regaining chemotherapy sensitivity. At the same time, NgBR maybe has an inhibitory effect on metastasis.
To sum up, MONC-PPG-PTX/siNgBR nanosystem combines multimodal imaging and RNAi with chemotherapy, it can as a “pioneer” to find drug resistant cells, evaluate its drug resistance, and to be a “killer”, sequentially releases genes and drugs in the tumor region, effectively and specific kills tumor, and with high bio-safety. At the same time, our study also revealed that NgBR knockdown may be related to the metastasis of drug resistant ERα-positive breast cancer. In summary, the establishment of the MONC-PPG-PTX/siNgBR nanosystem provides the possibility to achieve personalized diagnosis and treat of drug resistant breast cancer.
3. Conclusion
In this study, we have developed a “Matryoshka" multi-function nanosystem as a recognizer, chemosensitizer, and cell-killers for drug resistant breast cancer. The outermost layer is GE11 polypeptide to specifically recognize drug resistant cells. The “invisible" PEG increases the long-circulation and protects cargos and detached after arriving tumor region. Then the PEI is exposed, making a dual-enhanced cellular uptake and lysosomal escape to promote the release of siRNA into the cytoplasm. Last, the MONs core is responsive to the intracellular reductive microenvironment for the controlled releasing of the drugs encapsulated in its pores. In the breast tumor model, MONC-PPG-PTX/siNgBR can directly deliver gene and drug into tumor region, distinguishing and evaluating drug resistant tumors and then decompose layer by layer and release gradually, the siNgBR continuous release to effectively down-regulate the NgBR to re-educate the drug resistant cells regain sensitivity, and then the drugs followed release due to core dissociation. The combination therapy effectively suppresses tumor progression and inhibits metastasis, prolonging the survival time. Furthermore, we loaded CPDs to track the nanoparticles for multimodal imaging and evaluate drug resistance tumors. It is the first research that combines high-efficiency PA agent CPDs with MONs to recognize drug resistance and then focus on enhancing chemotherapy sensitivity and inhibiting metastasis after directly knockdown NgBR. Based on our data, this strategy may provide a potential way for clinical translational therapy and promote personalized treatment.
MONC-PPG nanosystem can be a smart and bio-safe nanocarrier for protecting cargo while circulating in the blood, increasing intracellular internalization, and maximizing the effect of genes and drugs while simultaneously achieving real-time imaging. The nanosystem is easy to modify and can be a personalization therapy means for various tumors, providing conditions for bioengineering and clinical transformation. However, our study also has some limitations. We focused on the therapeutic effect of MONC-PPG-PTX/siNgBR on tumors but did not study its metabolism in the body. Although MONs are relatively safe, avoiding unnecessary cumulative damage is required for clinical translation. Second, although the dual-response release of MONC-PPG prevents a big part of the ineffective drug and gene release, there is still a small amount of early release in a neutral environment, seeking a more “closed” gatekeeper is our next goal.
4. Materials and methods
4.1. Synthesis of mesoporous organosilica nanoparticle-carbonized polymer dots (MON-CPDs)
MONs were synthesized according to the method (M/P-CA strategy) reported by Shi's group [56]. In brief, cetyltrimethylammonium chloride (CTAC, 2 g) and carbonized polymer dots (CPDs,1 mg) were dissolved in 20 mL water at 95 °C under stirring. Add triethanolamine (TEA, 0.08 g) to the system after stirring for 10min. After 30 min, the solution involving 1 g tetraethyl orthosilicate (TEOS) and 1.3 g bis[3-(triethoxysilyl)propyl]tetrasulfide (BTES) was added dropwise and stirred for 4 h. Centrifugation and washed three times to remove the residual reactants. Then, the products were extracted twice for 12 h with ethanol (with 10% v/v HCL) at 78 °C by refluxing to remove the template CTAC. After centrifugation, washing, and drying, the CPD-loaded mesoporous organosilica nanoparticles (MONCs) are obtained.
4.2. Preparation of MONC-PEI-PEG
Synthesis of MONC-PEI: Dissolving 25 mg MONCs in 30 mL ethanol, and then added dropwise to the solution containing PEI (0.2 mL, 100 mg/mL aqueous solution) and ethanol (2 mL). The mixture was stirred for 24 h, then washed with ethanol and water to remove unreacted PEI, and dispersed in 30 mL of water for further use.
Synthesis of MONC-PEI-PEG-mal: It is simply mixing CHO-PEG-mal with MONC-PEI at a weight ratio of 2:1(WPEI: WCHO-PEG-mal) at pH 7.4 for 1 h. The product (MONC-PP-mal) was centrifuged and washed for use.
4.3. Preparation of MONC-PP-GE11
The thiol-maleimide reaction is formed through the cys bond of GE11 and the mal bond of the PEG. The MONC-PP-mal synthesized in the previous step was resuspended in water, 10 mg MONC-PP-mal mix with 0.12 μmol GE11 at a concentration of 5 mg/mL, shaken (300 rpm) for 2 h then incubated overnight at 4 °C. Then the unreacted peptides in the solution were removed by centrifugation (10,000 rpm, 10 min) and wash 3 times.
4.4. Paclitaxel loading and release
PTX was loaded into MONC-PPG by the adsorption equilibrium method [82]. MONC-PPG (30 mg) and PTX were dissolved in 4 mL dichloromethane solution with a mass ratio of 3:1(MONC-PPG: PTX), and then ultrasonic dispersion uniformity. Stirring for 24 h in a closed container under 37 °C, 500 rpm. Then the solvent was evaporated to 1 mL, then centrifugation at 10,000 rpm to remove supernatant, and washed. The MONC-PPG-PTX was dried under a vacuum to remove trace dichloromethane.
To determine the drug loading efficiency, 1 mg MONC-PPG-PTX were added in 2 mL methanol, ultrasonic to release the PTX, then centrifuged at 10,000 rpm for 15 min. The PTX was determined by HPLC.
The release of PTX in vitro was performed in PBS at pH 6.5, 7.4 and GSH concentration of 0 mM or 10 mM, and monitored over time. MONC-PPG-PTX was re-suspended in 2 mL PBS (contain 30% alcohol and 0.05% tween80) at 37 °C with gentle agitation. At predetermined times centrifuged (13000 rpm; 10 min) the solution and the supernatant were removed and replaced with the same volume of fresh buffer. The supernatant was measured using HPLC. The accumulative release of PTX was calculated using the formula:
4.5. Preparation of MONC-PPG-NgBR siRNA
Briefly, 1 mg of MONC-PPG was dispersed in 1 mL RNases free water in a centrifuge tube and ultrasonicated for 10 min. After sterilizing 1 h, 1 mL NgBR siRNA solution (10 nM, dissolved in RNases free water) was added. The mixture was continuously shaken at 150 rpm for 2 h under RT. The MONC-PPG-siRNA was collected by centrifugation at 11,000 rpm for 5 min and washed with RNases free water several times, the obtained products were re-dispersed into 1 mL RNases free water.
4.6. Gel retardation assay
The loading capacity of siRNA on MONC-PPG-PTX evaluate by gel retardation assay, siRNA and MONC-PPG-PTX were combined with various mass ratios and incubated for 2 h to prepared MONC-PPG-PTX/siRNA. Electrophoresis was carried out at 80 V for 30 min. Naked siRNA was used as control. The siRNA bands were visualized using the C400 Visible Fluorescence Imaging System.
Release siRNA in vitro with condition of pH 7.4 or 6.5 and GSH 10 mM or 0 mM. Suspend MONC-PPG-siRNA in 1 mL RNases free water and shaker at 100 rpm in 37 °C. At different time points, the suspension was centrifuged (10000 rpm; 5min), the supernatant was taken out and replaced with the same volume of fresh buffer. The supernatant was measured with Nanodrop 2000.
4.7. RNases A protection assay
For RNases A protection assay, 10 μL naked siRNA and MONC-PPG-siRNA complexes containing 0.4 μg siRNA were incubated with 40 ng RNases A at 37 °C for 1 h. RNases A was inhibited by adding RNases inhibitor according to manufacturer's instruction. Then added 5 μL 150 mg/mL heparin solution and incubated at 50 °C for 2 h to replace the RNA. The siRNA released from the complexes were examined by 1.5% agarose gel electrophoresis.
4.8. Cell culture
The human ERα positive breast cancer cell line MCF-7 was obtained from the Type Culture Collection Committee of the Chinese Academy of Sciences (Beijing, China) and the PTX resistant cell line MCF-7/PTX was purchased from Keygen Biotech (Nanjing, China) and cultured in RPMI 1640 medium containing 10% fetal bovine serum, 100 U/mL penicillin G sodium, 100 μg/mL streptomycin sulfate and 0.5 μg/mL PTX(only MCF-7/PTX). Cancer cells were maintained at 37 °C in a humidified and 5 % CO2 incubator.
4.9. Cellular uptake and colocation of MONC-PPG-siRNA
The cellular uptake of MONC-PPG was investigated by CLSM and flow cytometry. MCF-7/PTX were seeded in 6-well plates at a density of 2 × 105 cells per well. After 24 h, they were treated with 100 μg/mL MONC-PPG for 0.5, 1, 2, and 6 h at 37 °C for flow cytometry. As for CLSM, MCF-7/PTX was treated with MONC-PPG or MONC-PP for 10min, 30min, 1 h, 2 h, and 6 h under pH 7.4 or pH 6.5 for further observation.
For colocation, cells were seeded in glass bottom cell culture dish at a density of 2 × 105 cells per well and incubated for 24 h. Then treated with 100 μg/mL MONC-PPG-siRNA-FAM for 0.5 h, 1 h, 4 h, 6 h, 10 h and 12 h at 37 °C.After incubation, remove the medium and washed with PBS. Cells were stained with LysoTracker-blue for 2 h and CLSM for record the fluorescent images.
4.10. Transfection and western blot analysis
To extract total cellular protein, MCF-7/PTX and MCF-7 were seeded 24 h, they were treated with 100 μg/mL MONC-PPG-siNgBR for 6 h, then washed with PBS. Re-add fresh medium or medium containing 17β-estradiol (E2) for 48 h. Then extract protein for WB. At the same time, siRNA was transfected according to the instructions of Lipofectamine 2000 (Invitrogen, Eugene, OR, USA) for control. Western blot analysis was performed according to standard protocols, β-actin was used as an endogenous control. All antibodies are listed in Table S2 (Supporting information).
4.11. Cellular viability and apoptosis
After treating the cells with different treatments, CCK-8 reagent was added to cell culture medium and measured at 450 nm. Apoptosis was analyzed with a FITC -Annexin V Apoptosis Detection Kit (Dojindo) and Calcein-AM/propidium iodide (PI) double staining.
4.12. Treatment experiments in animal models
Six weeks old female BALB/c nude mice (15-18 g) were purchased from Beijing Experimental Animal Center. All experiments and procedures performed on mice were based on the guidelines of the Institutional Animal Care and Use Committee.
Female nude mice bearing MCF-7/PTX tumor were developed by subcutaneously injecting 3 × 107 cancer cells into the right mammary fat pad. When tumors grew to a volume of 100 mm3, mice were randomly assigned into 5 groups (n = 5). Mice were injected with PBS, MONC-PPG-siRNA, free PTX, MONC-PPG-PTX and MONC-PPG-PTX/siRNA at the dose of 0.4 mg/kg PTX and 60 μg/kg siRNA via tail vein, respectively. The procedure was repeated six times every 3 days. Recording the body weight and tumor volume. At the end of treatments, mice were sacrificed and tumors were excised and photographed. Tumor volume was calculated using the following equation:
The tumors were sliced into the paraffin sections for further research. First, for immunohistochemistry (IHC) staining assay to detect the expression of NgBR and survivin in vivo. Second, terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick and labeling (TUNEL) staining was performed to evaluate the apoptosis condition of tumors. Third, the tumors and the major organs including heart, liver, spleen, lungs and kidney were collected for hematoxylin and eosin (H&E) staining to examine the histological changes. Last, extract protein from tumor tissue for WB detection.
4.13. Biodistribution and PA imaging of MONC-PPG
The biodistribution of MONC-PPG was monitored by FL imaging (λex/λem = 640 nm/705 nm). Tumor-bearing mice were injected with MONC-PPG or MONC-PP (0.2 mg/mL, 100 μL). though tail vein. After different time points (pre-, 0.5, 2, 6 and 24 h post-injection), fluorescence image and intensity were collected and analyzed by IndiGo software.
PA performance of MONC-PPG was evaluated by Vevo LAZR Photoacoustic Imaging System. First, full wavelength scan(680–970 nm) to detect maximum absorbance. Then, detection of photoacoustic signals at different concentrations (0.05, 0.1, 0.2, 0.5, 1 and 2 mg/mL) to mapping curve of PA values. As for PA performance in vivo, tumor-bearing mice were i. v. injected with MONC-PPG solution (0.2 mg/mL, 100 μL) and PA images were acquired at various time points after injection.
4.14. Statistical analysis
All the experiments were carried out three times in parallel at least. The data were presented in the ± SD form. The student's t-test and one-way ANOVA were used for statistical analysis: ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. No animals were excluded from the analysis.
Credit author statement
Xinzhi Xu: Methodology, Software, Investigation, Writing – original draft Chunxiang Jin: Conceptualization, Resources, Writing – review & editing, Kai Zhang: Formal analysis, Formal analysis, Writing – review & editing, Yang Cao: Investigation, Supervision, Junjun Liu: Methodology, Visualization, Yue Zhang: Investigation, Writing – original draft, Haitao Ran: Resources, Supervision, Ying Jin: Resources, Validation
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 82003173), the Jilin Province Department of Finance(Grant Nos. 2019SCZ060.)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100245.
Contributor Information
Xinzhi Xu, Email: xuxz91@126.com.
Chunxiang Jin, Email: jincx722@jlu.edu.cn.
Haitao Ran, Email: rht66@163.com.
Ying Jin, Email: jinying0216@jlu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. Ca - Cancer J. Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. 2020. [DOI] [PubMed] [Google Scholar]
- 2.Jin Y., Hu W., Liu T., Rana U., Aguilera-Barrantes I., Kong A., Kumar S.N., Wang B., Gao P., Wang X., Duan Y., Shi A., Song D., Yang M., Li S., Han B., Zhao G., Fan Z., Miao Q.R. Nogo-B receptor increases the resistance of estrogen receptor positive breast cancer to paclitaxel. Cancer Lett. 2018;419:233–244. doi: 10.1016/j.canlet.2018.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesa-Eguiagaray I., Wild S.H., Rosenberg P.S., Bird S.M., Brewster D.H., Hall P.S., Cameron D.A., Morrison D., Figueroa J.D. Distinct temporal trends in breast cancer incidence from 1997 to 2016 by molecular subtypes: a population-based study of Scottish cancer registry data. Br. J. Cancer. 2020;123(5):852–859. doi: 10.1038/s41416-020-0938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullooly M., Murphy J., Gierach G.L., Walsh P.M., Deady S., Barron T.I., Sherman M.E., Rosenberg P.S., Anderson W.F. Divergent oestrogen receptor-specific breast cancer trends in Ireland (2004-2013): amassing data from independent Western populations provide etiologic clues. Eur. J. Cancer. 2017;86:326–333. doi: 10.1016/j.ejca.2017.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lumachi F., Brunello A., Maruzzo M., Basso U., Basso S.M. Treatment of estrogen receptor-positive breast cancer. Curr. Med. Chem. 2013;20(5):596–604. doi: 10.2174/092986713804999303. [DOI] [PubMed] [Google Scholar]
- 6.Pennisi A., Kieber-Emmons T., Makhoul I., Hutchins L. Relevance of pathological complete response after neoadjuvant therapy for breast cancer. Breast Cancer. 2016;10:103–106. doi: 10.4137/BCBCR.S33163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.WHO Guidelines Approved by the Guidelines Review Committee, WHO Position Paper on Mammography Screening. World Health Organization Copyright © World Health Organization 2014; Geneva: 2014. [Google Scholar]
- 8.Tabár L., Dean P.B., Chen T.H., Yen A.M., Chen S.L., Fann J.C., Chiu S.Y., Ku M.M., Wu W.Y., Hsu C.Y., Chen Y.C., Beckmann K., Smith R.A., Duffy S.W. The incidence of fatal breast cancer measures the increased effectiveness of therapy in women participating in mammography screening. Cancer. 2019;125(4):515–523. doi: 10.1002/cncr.31840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasad V., Lenzer J., Newman D.H. Why cancer screening has never been shown to "save lives"--and what we can do about it. Br. Med. J. 2016;352:h6080. doi: 10.1136/bmj.h6080. [DOI] [PubMed] [Google Scholar]
- 10.Xie R., Ruan S., Liu J., Qin L., Yang C., Tong F., Lei T., Shevtsov M., Gao H., Qin Y. Furin-instructed aggregated gold nanoparticles for re-educating tumor associated macrophages and overcoming breast cancer chemoresistance. Biomaterials. 2021;275:120891. doi: 10.1016/j.biomaterials.2021.120891. [DOI] [PubMed] [Google Scholar]
- 11.Costoya J., Surnar B., Kalathil A.A., Kolishetti N., Dhar S. Controlled release nanoplatforms for three commonly used chemotherapeutics. Mol. Aspect. Med. 2022;83:101043. doi: 10.1016/j.mam.2021.101043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Chen G., Dai F., Zhang L., Yuan M., Yang D., Liu S., Cheng Y. miR-21 induces chemoresistance in ovarian cancer cells via mediating the expression and interaction of CD44v6 and P-gp. OncoTargets Ther. 2021;14:325–336. doi: 10.2147/OTT.S286639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Phatak V., von Grabowiecki Y., Janus J., Officer L., Behan C., Aschauer L., Pinon L., Mackay H., Zanivan S., Norman J.C., Kelly M., Le Quesne J., Muller P.A.J. Mutant p53 promotes RCP-dependent chemoresistance coinciding with increased delivery of P-glycoprotein to the plasma membrane. Cell Death Dis. 2021;12(2):207. doi: 10.1038/s41419-021-03497-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mirzaei S., Gholami M.H., Hashemi F., Zabolian A., Hushmandi K., Rahmanian V., Entezari M., Girish Y.R., Sharath Kumar K.S., Aref A.R., Makvandi P., Ashrafizadeh M., Zarrabi A., Khan H. Employing siRNA tool and its delivery platforms in suppressing cisplatin resistance: approaching to a new era of cancer chemotherapy. Life Sci. 2021;277:119430. doi: 10.1016/j.lfs.2021.119430. [DOI] [PubMed] [Google Scholar]
- 15.Nie W., Wang B., Mi X., Chen J., Yu T., Miao J., Lin Y., Yang T., Ran M., Hong Z., Liu X., Liang X., Qian Z., Gao X. Co-delivery of paclitaxel and shMCL-1 by folic acid-modified nonviral vector to overcome cancer chemotherapy resistance. Small Methods. 2021;5(5) doi: 10.1002/smtd.202001132. [DOI] [PubMed] [Google Scholar]
- 16.Lippman M.E., Allegra J.C., Thompson E.B., Simon R., Barlock A., Green L., Huff K.K., Do H.M., Aitken S.C., Warren R. The relation between estrogen receptors and response rate to cytotoxic chemotherapy in metastatic breast cancer. N. Engl. J. Med. 1978;298(22):1223–1228. doi: 10.1056/NEJM197806012982203. [DOI] [PubMed] [Google Scholar]
- 17.Fiocchetti M., Fernandez V.S., Montalesi E., Marino M. Neuroglobin: a novel player in the oxidative stress response of cancer cells. Oxid. Med. Cell. Longev. 2019;2019:6315034. doi: 10.1155/2019/6315034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu D., Zhao B., Song Y., Chi X., Fu H., Guan T., Zhang L., Yang X., Hu K., Huang R., Jin X., Miao Q.R., Shao S. Nogo-B receptor is required for stabilizing TGF-beta type I receptor and promotes the TGF-beta1-induced epithelial-to-mesenchymal transition of non-small cell lung cancer. J. Cancer. 2021;12(3):717–725. doi: 10.7150/jca.50483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashour F., Awwad M.H., Sharawy H.E.L., Kamal M. Estrogen receptor positive breast tumors resist chemotherapy by the overexpression of P53 in Cancer Stem Cells. J. Egypt. Natl. Cancer Inst. 2018;30(2):45–48. doi: 10.1016/j.jnci.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Wu D., Zhao B., Qi X., Peng F., Fu H., Chi X., Miao Q.R., Shao S. Nogo-B receptor promotes epithelial-mesenchymal transition in non-small cell lung cancer cells through the Ras/ERK/Snail1 pathway. Cancer Lett. 2018;418:135–146. doi: 10.1016/j.canlet.2018.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B., Ding Y., Zhao X., Han X., Yang N., Zhang Y., Zhao Y., Zhao X., Taleb M., Miao Q.R., Nie G. Delivery of small interfering RNA against Nogo-B receptor via tumor-acidity responsive nanoparticles for tumor vessel normalization and metastasis suppression. Biomaterials. 2018;175:110–122. doi: 10.1016/j.biomaterials.2018.05.034. [DOI] [PubMed] [Google Scholar]
- 22.McKiernan P.J., Smith S.G.J., Durham A.L., Adcock I.M., McElvaney N.G., Greene C.M. The estrogen-induced miR-19 downregulates secretory leucoprotease inhibitor expression in monocytes. J. Innate Immun. 2020;12(1):90–102. doi: 10.1159/000500419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou S., Liu Y., Jin L., Guo P., Liu Q., Shan J., Luo X., He H., Ma W., Zhang T. Estrogen enhances the cytotoxicity of PARP inhibitors on breast cancer cells through stimulating nitric oxide production. J. Steroid Biochem. Mol. Biol. 2021;209:105853. doi: 10.1016/j.jsbmb.2021.105853. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y., Jia L., Wang Q., Hu H., Zhao X., Chen D., Qiao M. pH/redox dual-responsive polyplex with effective endosomal escape for codelivery of siRNA and doxorubicin against drug-resistant cancer cells. ACS Appl. Mater. Interfaces. 2019;11(18):16296–16310. doi: 10.1021/acsami.9b02016. [DOI] [PubMed] [Google Scholar]
- 25.Jiang W., Su L., Ao M., Guo X., Cheng C., Luo Y., Xie Z., Wang X., Wang J., Liu S., Cao Y., Li P., Wang Z., Ran H., Zhou Z., Ren J. Amplified antitumor efficacy by a targeted drug retention and chemosensitization strategy-based "combo" nanoagent together with PD-L1 blockade in reversing multidrug resistance. J. Nanobiotechnol. 2021;19(1):200. doi: 10.1186/s12951-021-00947-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miao R.Q., Gao Y., Harrison K.D., Prendergast J., Acevedo L.M., Yu J., Hu F., Strittmatter S.M., Sessa W.C. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103(29):10997–11002. doi: 10.1073/pnas.0602427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He W., Huang X., Berges B.K., Wang Y., An N., Su R., Lu Y. Artesunate regulates neurite outgrowth inhibitor protein B receptor to overcome resistance to sorafenib in hepatocellular carcinoma cells. Front. Pharmacol. 2021;12:615889. doi: 10.3389/fphar.2021.615889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W., Liu Z., Salato V., North P.E., Bischoff J., Kumar S.N., Fang Z., Rajan S., Hussain M.M., Miao Q.R. NOGOB receptor-mediated RAS signaling pathway is a target for suppressing proliferating hemangioma. JCI Insight. 2021;6(3) doi: 10.1172/jci.insight.142299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong C., Liu Y., Jiang K., Wang H., Qu W., Zhang C., Liang R., Gao Z., Zhao B., Miao Q., Shao S., Wang L. The Nogo-B receptor promotes human hepatocellular carcinoma cell growth via the Akt signal pathway. J. Cell. Biochem. 2018;119(9):7738–7746. doi: 10.1002/jcb.27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y., Hu W., Li Q., Zhao S., Zhao D., Zhang S., Wei Z., Yang X., Chen Y., Li X., Liao C., Han J., Miao Q.R., Duan Y. NGBR is required to ameliorate type 2 diabetes in mice by enhancing insulin sensitivity. J. Biol. Chem. 2021;296:100624. doi: 10.1016/j.jbc.2021.100624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma J., Zeng P., Liu L., Zhu M., Zheng J., Wang C., Zhao X., Hu W., Yang X., Duan Y., Han J., Miao Q.R., Chen Y. Peroxisome proliferator-activated receptor-gamma reduces ER stress and inflammation via targeting NGBR expression. Front. Pharmacol. 2021;12:817784. doi: 10.3389/fphar.2021.817784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao B., Hu W., Kumar S., Gonyo P., Rana U., Liu Z., Wang B., Duong W.Q., Yang Z., Williams C.L., Miao Q.R. The Nogo-B receptor promotes Ras plasma membrane localization and activation. Oncogene. 2017;36(24):3406–3416. doi: 10.1038/onc.2016.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y.K., Xie Y.J., Wu D.C., Long S.L., Tang S., Mo Z.C. NogoB receptor in relevant carcinoma: current achievements, challenges and aims (Review) Int. J. Oncol. 2018;53(5):1827–1835. doi: 10.3892/ijo.2018.4520. [DOI] [PubMed] [Google Scholar]
- 34.Gao P., Wang X., Jin Y., Hu W., Duan Y., Shi A., Du Y., Song D., Yang M., Li S., Han B., Zhao G., Zhang H., Fan Z., Miao Q.R. Nogo-B receptor increases the resistance to tamoxifen in estrogen receptor-positive breast cancer cells. Breast Cancer Res. 2018;20(1):112. doi: 10.1186/s13058-018-1028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R., Tang B.S., Guo J.F. Research advances on neurite outgrowth inhibitor B receptor. J. Cell Mol. Med. 2020;24(14):7697–7705. doi: 10.1111/jcmm.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirzaei S., Mahabady M.K., Zabolian A., Abbaspour A., Fallahzadeh P., Noori M., Hashemi F., Hushmandi K., Daneshi S., Kumar A.P., Aref A.R., Samarghandian S., Makvandi P., Khan H., Hamblin M.R., Ashrafizadeh M., Zarrabi A. Small interfering RNA (siRNA) to target genes and molecular pathways in glioblastoma therapy: current status with an emphasis on delivery systems. Life Sci. 2021;275:119368. doi: 10.1016/j.lfs.2021.119368. [DOI] [PubMed] [Google Scholar]
- 37.Ashrafizadeh M., Hushmandi K., Rahmani Moghadam E., Zarrin V., Hosseinzadeh Kashani S., Bokaie S., Najafi M., Tavakol S., Mohammadinejad R., Nabavi N., Hsieh C.L., Zarepour A., Zare E.N., Zarrabi A., Makvandi P. Progress in delivery of siRNA-based therapeutics employing nano-vehicles for treatment of prostate cancer. Bioengineering (Basel) 2020;7(3) doi: 10.3390/bioengineering7030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu M., Meng Q., Chen Y., Zhang L., Li M., Cai X., Li Y., Yu P., Zhang L., Shi J. Large pore-sized hollow mesoporous organosilica for redox-responsive gene delivery and synergistic cancer chemotherapy. Adv. Mater. 2016;28(10):1963–1969. doi: 10.1002/adma.201505524. [DOI] [PubMed] [Google Scholar]
- 39.Morry J., Ngamcherdtrakul W., Gu S., Reda M., Castro D.J., Sangvanich T., Gray J.W., Yantasee W. Targeted treatment of metastatic breast cancer by PLK1 siRNA delivered by an antioxidant nanoparticle platform. Mol. Cancer Therapeut. 2017;16(4):763–772. doi: 10.1158/1535-7163.MCT-16-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu L., Chen Y., Lin H., Gao S., Chen H., Shi J. Magnesium-Engineered silica framework for pH-accelerated biodegradation and DNAzyme-triggered chemotherapy. Small. 2018;14(35) doi: 10.1002/smll.201800708. [DOI] [PubMed] [Google Scholar]
- 41.Cheng Y., Jiao X., Fan W., Yang Z., Wen Y., Chen X. Controllable synthesis of versatile mesoporous organosilica nanoparticles as precision cancer theranostics. Biomaterials. 2020;256:120191. doi: 10.1016/j.biomaterials.2020.120191. [DOI] [PubMed] [Google Scholar]
- 42.Zivojevic K., Mladenovic M., Djisalov M., Mundzic M., Ruiz-Hernandez E., Gadjanski I., Knezevic N.Z. Advanced mesoporous silica nanocarriers in cancer theranostics and gene editing applications. J. Contr. Release. 2021;337:193–211. doi: 10.1016/j.jconrel.2021.07.029. [DOI] [PubMed] [Google Scholar]
- 43.Zhou X., You M., Wang F., Wang Z., Gao X., Jing C., Liu J., Guo M., Li J., Luo A., Liu H., Liu Z., Chen C. Multifunctional graphdiyne-cerium oxide nanozymes facilitate MicroRNA delivery and attenuate tumor hypoxia for highly efficient radiotherapy of esophageal cancer. Adv. Mater. 2021;33(24) doi: 10.1002/adma.202100556. [DOI] [PubMed] [Google Scholar]
- 44.Jia N., Wu H., Duan J., Wei C., Wang K., Zhang Y., Mao X. Polyethyleneimine-coated iron oxide nanoparticles as a vehicle for the delivery of small interfering RNA to macrophages in vitro and in vivo. J Vis Exp. 2019;144 doi: 10.3791/58660. [DOI] [PubMed] [Google Scholar]
- 45.Chen G., Wang Y., Xie R., Gong S. Tumor-targeted pH/redox dual-sensitive unimolecular nanoparticles for efficient siRNA delivery. J. Contr. Release. 2017;259:105–114. doi: 10.1016/j.jconrel.2017.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin H., Pi J., Zhao Y., Jiang J., Li T., Zeng X., Yang P., Evans C.E., Cai J. EGFR-targeting PLGA-PEG nanoparticles as a curcumin delivery system for breast cancer therapy. Nanoscale. 2017;9(42):16365–16374. doi: 10.1039/c7nr06898k. [DOI] [PubMed] [Google Scholar]
- 47.Mickler F.M., Mockl L., Ruthardt N., Ogris M., Wagner E., Brauchle C. Tuning nanoparticle uptake: live-cell imaging reveals two distinct endocytosis mechanisms mediated by natural and artificial EGFR targeting ligand. Nano Lett. 2012;12(7):3417–3423. doi: 10.1021/nl300395q. [DOI] [PubMed] [Google Scholar]
- 48.Li Z., Zhao R., Wu X., Sun Y., Yao M., Li J., Xu Y., Gu J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. Faseb. J. 2005;19(14):1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 49.Tang H., Chen X., Rui M., Sun W., Chen J., Peng J., Xu Y. Effects of surface displayed targeting ligand GE11 on liposome distribution and extravasation in tumor. Mol. Pharm. 2014;11(10):3242–3250. doi: 10.1021/mp5001718. [DOI] [PubMed] [Google Scholar]
- 50.Colzani B., Speranza G., Dorati R., Conti B., Modena T., Bruni G., Zagato E., Vermeulen L., Dakwar G.R., Braeckmans K., Genta I. Design of smart GE11-PLGA/PEG-PLGA blend nanoparticulate platforms for parenteral administration of hydrophilic macromolecular drugs: synthesis, preparation and in vitro/ex vivo characterization. Int. J. Pharm. 2016;511(2):1112–1123. doi: 10.1016/j.ijpharm.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 51.Biscaglia F., Rajendran S., Conflitti P., Benna C., Sommaggio R., Litti L., Mocellin S., Bocchinfuso G., Rosato A., Palleschi A., Nitti D., Gobbo M., Meneghetti M. Enhanced EGFR targeting activity of plasmonic nanostructures with engineered GE11 peptide. Adv. Healthc Mater. 2017;6(23) doi: 10.1002/adhm.201700596. [DOI] [PubMed] [Google Scholar]
- 52.Salimifard S., Karoon Kiani F., Sadat Eshaghi F., Izadi S., Shahdadnejad K., Masjedi A., Heydari M., Ahmadi A., Hojjat-Farsangi M., Hassannia H., Mohammadi H., Boroumand-Noughabi S., Keramati M.R., Jadidi-Niaragh F. Codelivery of BV6 and anti-IL6 siRNA by hyaluronate-conjugated PEG-chitosan-lactate nanoparticles inhibits tumor progression. Life Sci. 2020;260:118423. doi: 10.1016/j.lfs.2020.118423. [DOI] [PubMed] [Google Scholar]
- 53.Han X., Li Y., Xu Y., Zhao X., Zhang Y., Yang X., Wang Y., Zhao R., Anderson G.J., Zhao Y., Nie G. Reversal of pancreatic desmoplasia by re-educating stellate cells with a tumour microenvironment-activated nanosystem. Nat. Commun. 2018;9(1):3390. doi: 10.1038/s41467-018-05906-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su X., Tang Y., Li Y., Wang Z., Tao J., Chen K., Liu Y., Wu J., Wang D., Teng Z. Facile synthesis of monodisperse hollow mesoporous organosilica/silica nanospheres by an in situ dissolution and reassembly approach. ACS Appl. Mater. Interfaces. 2019;11(12):12063–12069. doi: 10.1021/acsami.8b21906. [DOI] [PubMed] [Google Scholar]
- 55.Mai N.X.D., Birault A., Matsumoto K., Ta H.K.T., Intasa-Ard S.G., Morrison K., Thang P.B., Doan T.L.H., Tamanoi F. Biodegradable periodic mesoporous organosilica (bpmo) loaded with daunorubicin: a promising nanoparticle-based anticancer drug. ChemMedChem. 2020;15(7):593–599. doi: 10.1002/cmdc.201900595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu M., Meng Q., Chen Y., Du Y., Zhang L., Li Y., Zhang L., Shi J. Large-pore ultrasmall mesoporous organosilica nanoparticles: micelle/precursor co-templating assembly and nuclear-targeted gene delivery. Adv. Mater. 2015;27(2):215–222. doi: 10.1002/adma.201404256. [DOI] [PubMed] [Google Scholar]
- 57.Liu J., Geng Y., Li D., Yao H., Huo Z., Li Y., Zhang K., Zhu S., Wei H., Xu W., Jiang J., Yang B. Deep red emissive carbonized polymer dots with unprecedented narrow full width at half maximum. Adv. Mater. 2020;32(17) doi: 10.1002/adma.201906641. [DOI] [PubMed] [Google Scholar]
- 58.Montero D., Tachibana C., Rahr Winther J., Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang S., Gao H. Nanoparticles for modulating tumor microenvironment to improve drug delivery and tumor therapy. Pharmacol. Res. 2017;126:97–108. doi: 10.1016/j.phrs.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Hadipour Moghaddam S.P., Saikia J., Yazdimamaghani M., Ghandehari H. Redox-responsive polysulfide-based biodegradable organosilica nanoparticles for delivery of bioactive agents. ACS Appl. Mater. Interfaces. 2017;9(25):21133–21146. doi: 10.1021/acsami.7b04351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu G., Fang Y.Z., Yang S., Lupton J.R., Turner N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004;134(3):489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- 62.Wang T., Liu Y., Wu C. Effect of paclitaxel-mesoporous silica nanoparticles with a core-shell structure on the human lung cancer cell line A549. Nanoscale Res. Lett. 2017;12(1):66. doi: 10.1186/s11671-017-1826-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Genta I., Chiesa E., Colzani B., Modena T., Conti B., Dorati R. GE11 peptide as an active targeting agent in antitumor therapy: a minireview. Pharmaceutics. 2017;10(1) doi: 10.3390/pharmaceutics10010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu W.W., Liu D.Y., Cao Y.C., Wang X.Y. GE11 peptide-conjugated nanoliposomes to enhance the combinational therapeutic efficacy of docetaxel and siRNA in laryngeal cancers. Int. J. Nanomed. 2017;12:6461–6470. doi: 10.2147/IJN.S129946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li K., Pang L., Pan X., Fan S., Wang X., Wang Q., Dai P., Gao W., Gao J. GE11 modified PLGA/TPGS nanoparticles targeting delivery of salinomycin to breast cancer cells. Technol. Cancer Res. Treat. 2021;20 doi: 10.1177/15330338211004954. 15330338211004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee D., Ko Y., Pang C., Ko Y.J., Choi Y.K., Kim K.H., Kang K.S. Estrogenic activity of mycoestrogen (3β,5α,22e)-ergost-22-en-3-ol via estrogen receptor α-dependent signaling pathways in MCF-7 cells. Molecules. 2021;27(1) doi: 10.3390/molecules27010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park S.H., Kim H., Kwak S., Jeong J.H., Lee J., Hwang J.T., Choi H.K., Choi K.C. HDAC3-ERα selectively regulates TNF-α-induced apoptotic cell death in MCF-7 human breast cancer cells via the p53 signaling pathway. Cells. 2020;9(5) doi: 10.3390/cells9051280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teixeira C., Reed J.C., Pratt M.A. Estrogen promotes chemotherapeutic drug resistance by a mechanism involving Bcl-2 proto-oncogene expression in human breast cancer cells. Cancer Res. 1995;55(17):3902–3907. [PubMed] [Google Scholar]
- 69.Konduri S.D., Medisetty R., Liu W., Kaipparettu B.A., Srivastava P., Brauch H., Fritz P., Swetzig W.M., Gardner A.E., Khan S.A., Das G.M. Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc. Natl. Acad. Sci. U. S. A. 2010;107(34):15081–15086. doi: 10.1073/pnas.1009575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babaei M., Abnous K., Taghdisi S.M., Taghavi S., Sh Saljooghi A., Ramezani M., Alibolandi M. Targeted rod-shaped mesoporous silica nanoparticles for the co-delivery of camptothecin and survivin shRNA in to colon adenocarcinoma in vitro and in vivo. Eur. J. Pharm. Biopharm. 2020;156:84–96. doi: 10.1016/j.ejpb.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 71.Wang B., Zhao B., North P., Kong A., Huang J., Miao Q.R. Expression of NgBR is highly associated with estrogen receptor alpha and survivin in breast cancer. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0078083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu C., Zheng J., Ding Y., Meng Y., Tan F., Gong W., Chu X., Kong X., Gao C. Cepharanthine loaded nanoparticles coated with macrophage membranes for lung inflammation therapy. Drug Deliv. 2021;28(1):2582–2593. doi: 10.1080/10717544.2021.2009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun J., Liu J., Gao C., Zheng J., Zhang J., Ding Y., Gong W., Yang M., Li Z., Wang Y., Yang Y., Gao C. Targeted delivery of PARP inhibitors to neuronal mitochondria via biomimetic engineered nanosystems in a mouse model of traumatic brain injury. Acta Biomater. 2022;140:573–585. doi: 10.1016/j.actbio.2021.12.023. [DOI] [PubMed] [Google Scholar]
- 74.Ding Y., Tong Z., Jin L., Ye B., Zhou J., Sun Z., Yang H., Hong L., Huang F., Wang W., Mao Z. An NIR discrete metallacycle constructed from perylene bisimide and tetraphenylethylene fluorophores for imaging-guided cancer radio-chemotherapy. Adv. Mater. 2022;34(7) doi: 10.1002/adma.202106388. [DOI] [PubMed] [Google Scholar]
- 75.Li S., Wu Y., Liu S., Wu T., Liu G., Li T., Chen Z. A multifunctional platinum(IV) and cyanine dye-based polyprodrug for trimodal imaging-guided chemo-phototherapy. J. Mater. Chem. B. 2022;10(7):1031–1041. doi: 10.1039/d1tb02682h. [DOI] [PubMed] [Google Scholar]
- 76.Sun L., Zhou J.E., Luo T., Wang J., Kang L., Wang Y., Luo S., Wang Z., Zhou Z., Zhu J., Yu J., Yu L., Yan Z. Nanoengineered neutrophils as cellular sonosensitizer for visual sonodynamic therapy of malignant tumors. Adv. Mater. 2022 doi: 10.1002/adma.202109969. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z., Fan W., Tang W., Shen Z., Dai Y., Song J., Wang Z., Liu Y., Lin L., Shan L., Liu Y., Jacobson O., Rong P., Wang W., Chen X. Near-infrared semiconducting polymer brush and pH/GSH-responsive polyoxometalate cluster hybrid platform for enhanced tumor-specific phototheranostics. Angew Chem. Int. Ed. Engl. 2018;57(43):14101–14105. doi: 10.1002/anie.201808074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fu Q., Zhu R., Song J., Yang H., Chen X. Photoacoustic imaging: contrast agents and their biomedical applications. Adv. Mater. 2019;31(6) doi: 10.1002/adma.201805875. [DOI] [PubMed] [Google Scholar]
- 79.Humayun M., Wang C., Luo W. Recent progress in the synthesis and applications of composite photocatalysts: a critical review. Small Methods. 2022;6(2) doi: 10.1002/smtd.202101395. [DOI] [PubMed] [Google Scholar]
- 80.Ðorđević L., Arcudi F., Cacioppo M., Prato M. A multifunctional chemical toolbox to engineer carbon dots for biomedical and energy applications. Nat. Nanotechnol. 2022;17(2):112–130. doi: 10.1038/s41565-021-01051-7. [DOI] [PubMed] [Google Scholar]
- 81.Zhao B., Xu B., Hu W., Song C., Wang F., Liu Z., Ye M., Zou H., Miao Q.R. Comprehensive proteome quantification reveals NgBR as a new regulator for epithelial-mesenchymal transition of breast tumor cells. J. Proteonomics. 2015;112:38–52. doi: 10.1016/j.jprot.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang X., Xiong T., Cui M., Li N., Li Q., Zhu L., Duan S., Wang Y., Guo Y. A novel targeted co-delivery nanosystem for enhanced ovarian cancer treatment via multidrug resistance reversion and mTOR-mediated signaling pathway. J. Nanobiotechnol. 2021;19(1):444. doi: 10.1186/s12951-021-01139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.