Abstract
Colorectal cancer (CRC) is the second deadly and the third most common malignancy worldwide. It has been projected that annual new cases of CRC will increase by 63% in 2040, constituting an even greater health challenge for decades to come. This study has linked DEC1 (differentiated embryonic chondrocyte expressed gene 1) to the pathogenesis of CRC. Based on the analysis of patient samples and database data, DEC1 is expressed much higher in CRC than the adjacent normal tissues. CRC patients with higher DEC1 expression have a shorter survival time. The carcinogenesis protocol with azoxymethane/dextran sulfate induces a higher number of tumors with larger sizes in DEC1+/+ than DEC1−/− mice. Overexpression of DEC1 increases the expression of proliferation- and antiapoptosis-related genes, but decreases the level of proapoptotic genes. Mechanistically, this study has shown that DEC1 is functionally looped to the IL-6/STAT3 signaling pathway (interleukin-6/signal transducer and activator of transcription 3). IL-6 induces DEC1, and DEC1 enhances the phosphorylation of STAT3, resulting in increased pSTAT3/STAT3 ratio. DEC1 and STAT3 are present in reciprocal immunocomplexes, pointing to physical interactions (presumably with pSTAT3). These findings establish that DEC1 is a CRC enhancer. The enhancement is achieved largely through the IL-6/STAT3 pathway. The potential of the physical interaction between DEC1 and STAT3 will likely serve as a foundation to develop intervention strategies for CRC prevention and therapy.
Keywords: Differentiated embryonic chondrocyte expressed gene 1 (DEC1), Colorectal cancer (CRC), STAT3, Proliferation, Antiapoptosis
Abbreviations: AOM, azoxymethane; Bcl-2, B cell lymphoma-2; Bcl-XL, B-cell lymphoma-extra-large; BHLH, basic helix-loop-helix; BrdU, 5-bromo-2′-deoxyuridine; Co-IP, co-immunoprecipitation; CDKs, cyclin-dependent protein kinases; CRC, colorectal cancer; DSS, dextran sulfate sodium salt; DEGs, differentially expressed genes; DEC1, differentiated embryonic chondrocyte expressed gene 1; GEO, gene expression omnibus; GEPIA, gene expression profiling interactive analysis; H&E, hematoxylin and eosin; IHC, immunohistochemical staining; IL-6, interleukin-6; KO, knockout; PCNA, proliferating-cell nuclear antigen; STAT3, signal transducer and activator of transcription 3; TUNEL, TdT-mediated dUTP nick-end labeling
Introduction
Colorectal cancer (CRC) is the second deadly and the third most common malignancy. In 2020, there were over 1.9 million new cases with approximately 1.0 million deaths worldwide [1,2]. It has been projected that annual new cases of CRC will increase by 63% in 2040 [1], constituting an even greater health challenge for decades to come. Like many other malignancies, the incidence of CRC is higher among developed countries [3]. While there are many contributing factors, inflammatory bowel disease has been closely linked to an increased risk of CRC [4]. Indeed, inflammation is known to promote the development of colorectal adenoma, which is intimately involved in the pathogenesis of CRC [2].
Several proinflammatory cytokines such as interleukin-6 (IL-6) are increased in CRC patients [5]. IL-6 triggers signaling cascades centered by signal transducer and activator of transcription 3 (STAT3) [6,7]. STAT3 functions are multifaceted and associated with both cancer-promoting and suppressing activities, probably depending on integrated signaling networks in various tissues and cells [8], [9], [10]. For example, STAT3 functions as a tumor suppressor in prostate cancer [9] but a promoter in colon cancer [10]. It is well established that STAT3 activation leads to increased expression of proliferation-related genes, such as cyclin-dependent protein kinases (CDKs) and proliferating-cell nuclear antigen (PCNA) [11]. Likewise, STAT3 activation is associated with increased expression of antiapoptosis-related genes such as survivin, human B cell lymphoma-extra-large (Bcl-XL) and human B cell lymphoma-2 (Bcl-2) [12].
Differentiated embryonic chondrocyte expressed gene 1 (DEC1, BHLHE40) belongs to the superfamily of the basic helix-loop-helix (bHLH) transcription factors [13]. This transcription factor has a broad tissue distribution and is involved in diverse cellular events such as differentiation, proliferation and apoptosis [14]. Physiologically, DEC1 is a clock gene and regulates circadian rhythmicity and metabolic functions [14,15]. Pathologically, DEC1 expression is positively associated with tumor development and invasiveness in such malignancies as breast cancer [16], gastric carcinomas [17] and colon cancer [18]. The level of DEC1 mRNA in colon adenocarcinoma is 20 times as much as that in the adjacent normal tissues [18] with a robust increase in DEC1 protein expression. Overexpression of DEC1 has been shown to increase the expression of survivin, a gene known to enhance cancer survival and chemoresistance [17,19]. However, the precise role of DEC1 in the development of CRC remains to be determined.
This study has used database analysis, patient samples, animal as well as cell models, and demonstrated that DEC1 is a CRC enhancer. The enhancement is achieved through the IL-6/STAT3 pathway. IL-6 induces DEC1, and DEC1 enhances the phosphorylation of STAT3, forming a functional loop. Consistent with increased pSTAT3/STAT3 ratio, DEC1 physically interacts STAT3 (presumably with pSTAT3). The physical interaction likely serves as a foundation to develop intervention strategies for CRC prevention and therapy.
Materials and methods
Participants
Colon cancer patients, five males and five females with an average age of 52.9 (45–62 years of old), were recruited from Sir Run Hospital affiliated to Nanjing Medical University. Histopathological examination was performed after colectomy. The inclusion criteria were: (1) patients were diagnosed and confirmed by multiple approaches including colonoscopy, barium meal and pathological results; (2) patients underwent simple resection of colon cancer; and (3) patients voluntarily joined the study and signed the informed consent form. Exclusion criteria were: (a) patients had an oncological history and concurrent multiple primary tumors; (b) patients had colon cancer resection combined with other operations; and (c) patients had doubtful pathological diagnosis. This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Nanjing Medical University (Reg. No. 2020-602 Nanjing, China), and all participants gave informed consent to the study.
GEPIA database analysis (gene expression profiling interactive analysis)
The website of GEPIA database is: http://gepia.cancer-pku.cn/index.html. The database was analyzed for differential expression of DEC1 between normal and tumor, and the correlation between DEC1 and STAT3.
GEO database analysis (gene expression omnibus)
The dataset of GSE17536, with clinic information and microarray data, was analyzed. Programming language for statistical computing and graphics, commonly referred to as the R language, was used to specify the survival rate at different levels of DEC1 expression, prognostic factors of CRC, DEC1-associated gene expression and the correlation between DEC1 and IL-6 levels. The protein-protein interaction (PPI) network was constructed with the STRING database (https://string-db.org/).
Animals
DEC1+/- mice (C57BL/6J background) were purchased from the Department of Developmental Biology, Research Institute for Radiation Biology and Medicine, Japan. The generation of C57BL/6 mice harboring targeted disruption of the DEC1 gene was described previously [20]. Heterozygous male mice mated with female mice to generate the wild type (DEC1+/+) and DEC1 knockout (DEC1−/−) mice. The genotyping was performed right after birth and confirmed before the experiment to ensure the genotyping accuracy. Tail genetic identification was shown in Fig. S1. Mice were maintained in a pathogen-free animal facility under standard 12:12 h light:dark cycle, and fed with standard rodent chow and water in the Animal Core Facility of Nanjing Medical University (Nanjing, China). This study was approved by the Institutional Animal Care and Use Committee (No. IACUC-1704004) and the Animal Ethical and Welfare Committee (AEWC).
AOM/DSS-induced CRC (azoxymethane/ dextran sulfate sodium salt)
The chemical carcinogenesis protocol of AOM/DSS was used to establish CRC model. Male mice (DEC1+/+ and DEC1−/−) at the age of 6–8 weeks were divided into two groups for each genotype: the control (n = 10), and the AOM/DSS group (n = 20). Mice in the AOM/DSS groups (DEC1+/+ or DEC1−/−) were administered a single intraperitoneal injection of 10 mg/kg AOM (Sigma-Aldrich, St Louis, MO, USA; Cat: A5486-25MG). Thereafter, mice were given three cycles of 2.5% DSS (MP Biomedicals, Santa Ana, USA; Cat: 216011080) through drinking water. Each cycle started with 2.5% DSS for 7 days, followed by a 14-day recovery period (Fig. 1E). Mice in the control groups were given an intraperitoneal injection of the same volume of PBS. After 86 days, mice were anesthetized with chloral hydrate and then euthanized. The large intestine (from cecum to anus, referred to as colon segment for clarity) was collected, and its length was measured. The intestine was cut longitudinally and washed with PBS. The number and volume of tumors were counted, measured and recorded. One part of the tissues was frozen at -80 °C for RNA and protein extraction. The other part was fixed in paraformaldehyde for histological examination as described below.
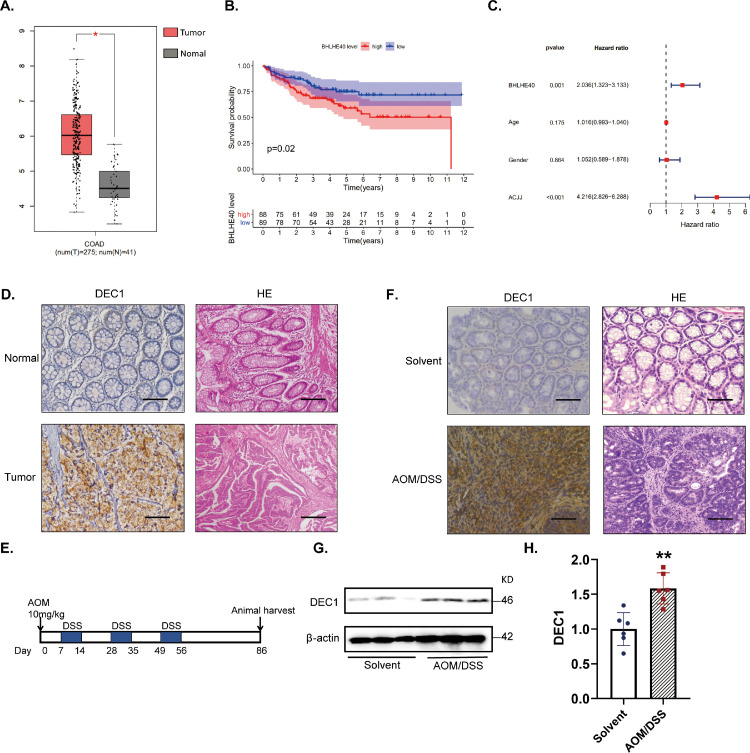
Fig. 1.
Increased DEC1 expression in human and AOM/DSS-induced CRC tissues (A) GEPIA database showed a higher DEC1 expression in human colorectal cancer than that in normal tissues. (B) GEO database showed that patients with low DEC1 expression had a higher survival probability. (C)Multivariate analysis of prognostic factors of colorectal cancer was elucidated based on the GEO. (D) DEC1 in human colorectal cancer by using IHC (N = 10). Scale bar = 40 μm. (E) Schedule of colorectal cancer induced by AOM/DSS in mice. (F) DEC1 expression in AOM/DSS-induced CRC mice through IHC (N = 6 in each group). Scale bar = 40 μm. (G-H) DEC1 protein level in AOM/DSS-induced CRC mice by Western blotting (t-text, t = 4.392, df = 10, p = 0.0014. N = 6 in each group). Data are expressed as mean ± SD. * p < 0.05, ** p < 0.01 vs. normal or solvent.
Histological analysis
For hematoxylin and eosin staining (H&E), paraformaldehyde-preserved tissues were embedded in paraffin and sections (5 μm) were made. Tissue sections were stained with H&E and examined by a pathologist who was blinded to the experimental groups. The severity of hyperplasia score was assigned as previously described [21]. For immunohistochemical staining (IHC), sections were deparaffinized followed by rehydration with ethanol at a series of decreased concentrations. Antigen retrieval was performed with 0.01 M preheated citrate buffer (pH 6.0, 90 °C) for 15 min. The sections were incubated overnight at 4 °C with, respective primary antibodies (e.g., Ki67) diluted in PBS. Slides were incubated with appropriate secondary antibody for 1 h at room temperature and then visualized with 3,3′-Diaminobenzidine (Maxim, Fuzhou, China; Cat: DAB-0031) as a substrate. For BrdU incorporation assay (5-bromo-2′-deoxyuridine,) mice were injected at 10 mg/kg body weight with BrdU (Sigma-Aldrich, St Louis, MO, USA; Cat: B5002) 2 h before euthanization. Tissues were fixed in paraformaldehyde.
Quantitative real-time PCR
Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, USA; Cat:15596-026). Reverse transcription was performed with the HiScript II Q RT SuperMix kit (Vazyme, Nanjing, China; Cat: R222-01). Quantitative real-time PCR was performed with SYBR Green Master in a 7300 Real-time PCR System (Applied Biosystems). The PCR amplification was performed with commercial kits (Vazyme, Nanjing, China; Cat: Q331-02) according to the manufacturer's instructions. The qPCR primer sequences are: β-actin forward, 5′-ATGCTCCCCGGGCTGTAT-3′, β-actin reverse, 5′-CATAGGAGTCCTTCTGACCCATTC-3′, IL-6 forward, 5′-CTGCAAGAGACTTCCATCCAGTT-3′, IL-6 reverse, 5′-GAAGTAGGGAAGGCCGTGG-3′.
TUNEL assay (TdT-mediated dUTP nick-end labeling)
The TUNEL method was used to detect apoptotic changes. Paraffin-embedded tissue sections (5 μm) were processed for 15 min with 0.01 M preheated citrate buffer (pH 6.0, 90 °C). The samples were then incubated with TUNEL reaction buffer at 37 °C for 1 h. The slides were washed for three times and then incubated for 30 min with fluorescein isothiocyanate (FITC)-conjugated anti-fluorescein antibody (KeyGEN, Nanjing, China; Cat: KGA7052). TUNEL-positive cells were quantified by a fluorescent microscope (Acquisition software: DP2-BSW).
Cell culture and treatment
Human colon cancer cell lines including DLD-1, SW480 and HCT116 were purchased from ATCC (Manassas, VA, USA). Cells were cultured in 1640 medium supplemented with 10% FBS at 37 °C in a humidified incubator containing 5% CO2. The medium was changed daily. For IL-6 treatment, cells were seeded in 6-well plates for overnight, and then treated with IL-6 (0, 5, 10, 25, 50 ng/ml) for 24 h. Thereafter, the cells were harvested.
DEC1 overexpression and knockdown/sg-DEC1/cas9 experiments
For DEC1 overexpression, DLD-1 cells were seeded in a 6-well plate at the density of 1 × 106 cells/well and then transfected by GenJet TM in vitro DNA Transfection Reagent II (SignaGen, Maryland, USA;Cat:SL100489). The transfection mixtures contained 1 μg Flag-CMV2 vector or Flag-DEC1 plasmid. The transfection medium was replaced with the regular medium 6 h thereafter. The transfected cells were used for subsequent experiments as described below.
For DEC1 knockdown experiment, DLD-1 cells were lentivirally transduced to LV-CON or LV-DEC1-RNAi (Genechem, Shanghai, China). After 48 h, cells were cultured in media containing 5 μg/ml of puromycin to select the stable cells for two weeks. For DEC1 sg-RNA/cas9 experiment, DLD-1 cells were transfected with vector control or lentivirus encoding sgDEC1/cas9 (Genechem, Shanghai, China). Later, cells were cultured in media containing puromycin (5 µg/mL) for two weeks. The cells were designated as sg-CON (control, polyclonal cells) and sg-DEC1 (sg-DEC1/cas9, polyclonal cells) in this study, respectively.
Immunofluorescence
The transfected or transduced DLD-1 cells (chamber slides) were washed three times with PBS, treated with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. The slides were treated with 5% bovine serum albumin (Applygen, Beijing, China; Cat: P1621-25) for 1 h to block non-specific binding sites and then incubated with the primary antibodies (STAT3 or DEC1 antibody) at 4 °C for overnight. The secondary antibodies (goat anti-mouse Rhodamine TRITC or goat anti-rabbit FITC) were incubated for 1 h, followed by treatment with DAPI (Biogot, Nanjing, China; Cat: BD5010) for 15 min. All images were taken on a Laser scanning confocal or fluorescence microscopy.
Co-immunoprecipitation (Co-IP) assay
DLD-1 cells were seeded in 60 mm dishes for overnight and then treated with IL-6 (50 ng/ml) or the same volume of PBS for 24 h. Cell lysates were prepared in ice-cold RIPA buffer (1 ml). Cell lysates (500 μg) were incubated with anti-STAT3 or anti-DEC1 antibody (1 μl) at 4 °C for overnight. The immune complexes were precipitated with 20 μl of Protein A/G Plus-Sepharose beads (Santa Cruz, Dallas, USA, Cat: sc-2003) at 4 °C for 4 h. The beads were washed for 8 times with ice-cold buffer (50 mM Tris/HCl, pH 7.4 and 150 mM NaCl). The immuno-precipitates were analyzed for the presence of DEC1 or STAT3 by Western blotting with anti-DEC1 and anti-STAT3 antibodies. The cell lysates (5 μg), prior to immunoprecipitation, were also analyzed for the levels of DEC1 and STAT3.
Western blotting
Colon homogenates or cell lysates (30 μg) were resolved by 7.5% SDS-PAGE (sodium dodecyl sulfate-polyacrylamide gel electrophoresis) and transferred electrophoretically to PVDF membranes. The membranes were incubated for 1 h with 5% bovine serum albumin to block non-specific binding sites and then with primary antibodies at 4 °C for overnight. Thereafter, the membranes were incubated with the secondary antibody conjugated with horseradish peroxidase, which was detected with the chemiluminescent detection system (Vazyme, Nanjing, China; Cat: E412-01). The chemiluminescent signal was captured by Kodak Image Station 2000.
Statistical methods
Data were expressed as mean ± SD. Student's t-test, one-way ANOVA and two-way ANOVA were used to analyze data through the GraphPad Prism 8 software. Statistical significance was indicated when p values were less than 0.05.
Results
Increased DEC1 expression in human and AOM/DSS-induced CRC tissues
We initially used GEPIA and GEO databases to shed light on the role of DEC1 in human CRC. As shown in Fig. 1, CRC tissues (N = 275) expressed a significantly higher level of DEC1 compared with that in the adjacent normal tissues (N = 41) (Fig. 1A). Analysis of the GEO database demonstrated that patients with higher DEC1 expression had a shorter survival time (p < 0.05) (Fig. 1B). Multivariate analysis of microarray data has concluded that DEC1 was an independent prognostic factor for CRC patients (p < 0.05) (Fig. 1C). Consistent with these findings from analyzing databases, immunohistochemistry with patient tissues detected much higher expression of DEC1 in the CRC than the adjacent normal tissues (Fig. 1D). Likewise, significant increases of DEC1 expression in AOM/DSS-induced CRC tissues were detected by both immunohistochemistry and Western blotting (Fig. 1F–H).
Protection of DEC1 deficiency against AOM/DSS-induced CRC in mice
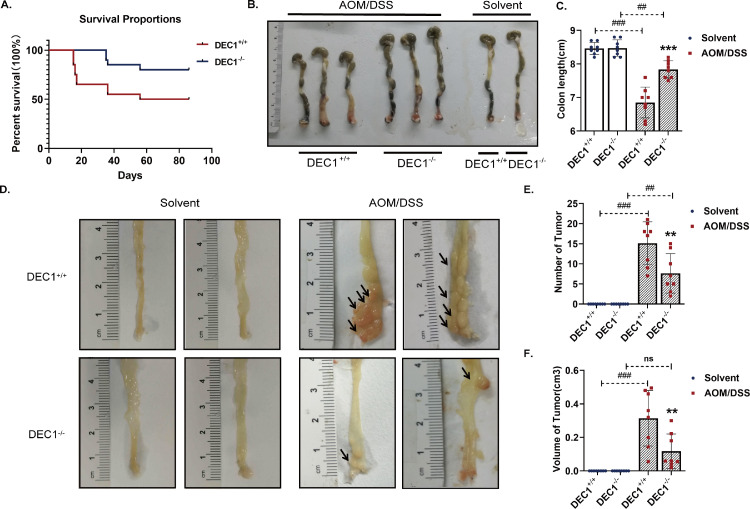
To shed light on an involvement of DEC1 in the development of CRC, DEC1+/+and DEC1−/− mice were treated, initially with AOM followed with DSS, a well-established protocol for CRC induction in animals [22]. As shown in Fig. 2, the first death in the DEC1+/+ group occurred on day 15, whereas the first death in the DEC1−/− group occurred on day 36. At the conclusion of the experiment (86 days), the DEC1−/− group had a mortality rate of 20%, whereas the DEC1+/+ group of 50% (Fig. 2A). Interestingly, DEC1+/+ CRC mice had a much shorter colon than their DEC1−/− counterparts (Fig. 2B, C). Likewise, DEC1+/+ mice developed more tumors with larger sizes than DEC1−/− mice by as many as 2–3 times (Fig. 2D–F). Histologically, tumors in DEC1+/+ mice invaded the basement membrane, whereas those in DEC1−/− mice were much less invasive and largely limited in situ location (Fig. 3A and B). These results suggest that DEC1 promotes but DEC1 deficiency protects against AOM/DSS-carcinogenesis.
Fig. 2.
Protection of DEC1 deficiency against phenotypic changes associated with AOM/DSS-CRC induction in mice. (A) Overall survival in the two types (DEC1+/+, DEC1−/−) CRC mice by using the Kaplan-Meier method (Log-rank test, p = 0.0315). (B, C) The colon length in in the two types (DEC1+/+, DEC1−/−) CRC mice (two-way ANOVA, gene: F (1, 28) = 110.2, p < 0.0001, model: F (1, 28) = 21.77, p < 0.0001, interaction: F (1, 28) = 20.69, p < 0.0001). (D) Representative longitudinal luminal views of colons in the two types (DEC1+/+, DEC1−/−) CRC mice. (E, F) The number (two-way ANOVA, gene: F (1, 28) = 110.2, p < 0.0001, model: F (1, 28) = 21.77, p < 0.0001, interaction: F (1, 28) = 20.69, p < 0.0001) and volume (two-way ANOVA, gene: F (1, 28) = 39.07, p < 0.0001, model: F (1, 28) = 8.140, p = 0.0081, interaction: F (1, 28) = 8.589, p = 0.0081) of tumor nodules in the two types (DEC1+/+, DEC1−/−) CRC mice. N = 8 in each group in this figure, the data are expressed as mean ± SD. ** p < 0.01, *** p < 0.001 vs. DEC1+/+ CRC mice, ## p < 0.01, ### p < 0.001 vs. corresponding solvent mice, ns p > 0.05.
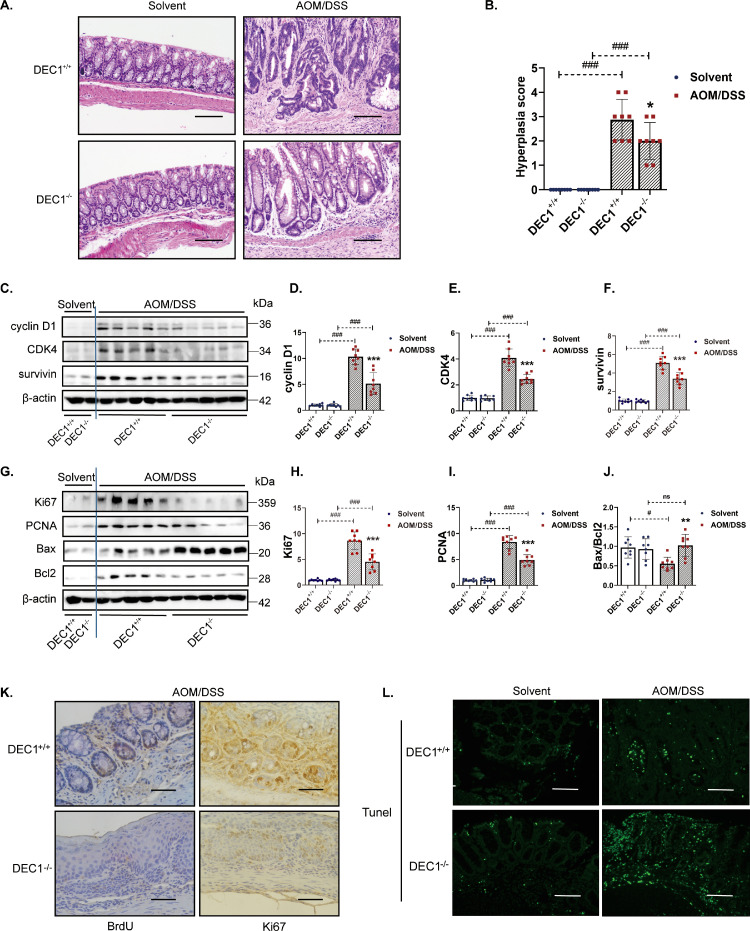
Fig. 3.
Association of DEC1 deficiency with inhibition of colonic tumorous cell proliferation and enhancement of apoptotic changes in AOM/DSS-induced CRC mice. (A) H&E staining of colon sections in the two types (DEC1+/+, DEC1−/−) CRC mice. Scale bar = 200 μm. (B) Quantification of hyperplasia in the two types (DEC1+/+, DEC1−/−) CRC mice (two-way ANOVA, gene: F (1, 28) = 150.0, p<0.0001, model: F (1, 28)=4.831, p=0.0364, interaction: F (1, 28)=4.831, p=0.0364). (C–F) Cyclin D1 (two-way ANOVA, gene: F (1, 28)=215.1, p<0.0001, model: F (1, 28)=31.77, p<0.0001, interaction: F (1, 28)=31.60, p<0.0001), CDK4 (two-way ANOVA, gene: F (1, 28)=249.5, p<0.0001, model: F (1, 28)=32.70, p<0.0001, interaction: F (1, 28)=30.74, p<0.0001) and survivin (two-way ANOVA, gene: F (1, 28)=5.896, p<0.0001, model: F (1, 28)=82.11, p<0.0001, interaction: F (1, 28)=5.340, p<0.0001,) protein levels by Western blotting. (G–J) Ki67 (two-way ANOVA, gene: F (1, 28)=9.258, p<0.0001, model: F (1, 28)=70.36, p<0.0001, interaction: F (1, 28)=9.584, p<0.0001), PCNA (two-way ANOVA, gene: F (1, 28)=363.3, p<0.0001, model: F (1, 28)=33.09, p<0.0001, interaction: F (1, 28)=35.42, p<0.0001), and Bax/Bcl2 (two-way ANOVA, gene: F (1, 28)=5.885, p=0.0220, model: F (1, 28)=3.323, p=0.0790, interaction: F (1, 28)=8.267, p=0.0076) protein levels in the two types (DEC1+/+, DEC1−/−) CRC mice. (K) BrdU and Ki67 in the two types (DEC1+/+, DEC1−/−) CRC mice, scale bar=40μm. (L) TUNEL assay in the two types (DEC1+/+, DEC1−/−) CRC mice, scale bar=40 μm. N=8 in each group in this figure, the data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. DEC1+/+ CRC mice, # p < 0.05, ### p < 0.001 vs. the corresponding solvent mice, ns p > 0.05.
Cell proliferation and survival are two major mechanisms for tumor expansion [23,24] We next determined the expression of genes related to these cellular events. As shown in Fig. 3C–F, tumor tissues from DEC1−/− CRC mice, compared those from DEC1+/+ CRC mice, had much lower levels of proliferation-proteins (cyclin D1 and CKD4) and anti-apoptosis protein (survivin). Similar changes were detected on the expression of Ki67 and PCNA (Fig. 3G–I), two other genes related to cell proliferation [25]. The higher expression of Ki67 in the tumors from DEC1+/+ CRC tissues was confirmed by immunohistochemistry (Fig. 3K). The expression of Bax was upregulated in DEC1−/− CRC tissues but downregulated in DEC1+/+ CRC, and the opposite was true with Bcl2 (Fig. 3G and J). Bax and Bcl-2 belong the Bcl-2 gene family [26,27]. However, Bax is proapoptotic, whereas Bcl-2 is antiapoptotic [26]. The decreased Bax/Bcl2 ratio in DEC1+/+ CRC tissues, compared with that in DEC1−/− CRC, suggests that DEC1 deficiency protected against AOM/DSS-carcinogenesis through proliferation-inhibition and apoptotic induction. Overall, the increased proliferation and decreased apoptosis were confirmed by BrdU incorporation assay and TUNEL staining (Fig. 3K and L).
DEC1 expression is positively related to STAT3 activation
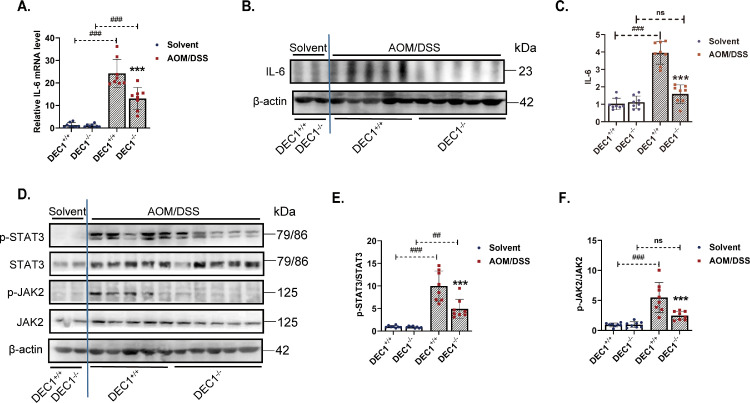
Aberrantly elevated STAT3 activity has been observed in more than 70% human cancer [28] including CRC [29]. IL-6 is known to trigger the activation of STAT3 [30,31]. Next, we tested whether DEC1 impacts STAT3 signaling in CRC. As shown in Fig. 4A, the level of IL-6 increased in both DEC1+/+ and DEC1−/− CRC tissues. However, the increase in DEC1+/+ CRC mice was twice as much as that in their DEC1−/− counterparts. Similar trends of changes were detected on IL-6 protein expression but with a greater magnitude of changes (Fig. 4B and C). The pSTAT3/STAT3 ratio (Fig. 4D and E) and the p-JAK2/JAK2 ratio (Fig. 4D and F) were significantly lower in DEC1−/− than DEC1+/+ CRC tissues. Phosphorylated STAT3 and JAK2 are active signaling molecules [30]. Decreased ratios in DEC1−/− CRC mice suggest that DEC1 deficiency attenuates IL-6/JAK2-STAT3 signal pathways and inhibits CRC development.
Fig. 4.
Positive correlation of DEC1 expression with STAT3 activation. (A) IL-6 mRNA level in the two types (DEC1+/+, DEC1−/−) CRC mice through real-time PCR (two-way ANOVA, gene: F (1, 28)=151.4, p<0.0001, model: F (1, 28)=15.99, p=0.0004, interaction: F (1, 28) =14.75, p=0.0006. N=8 in each group). (B, C) IL-6 protein level in the two types (DEC1+/+, DEC1−/−) CRC mice by Western blotting (two-way ANOVA, gene: F (1, 28)=15.06, p=0.0006, model: F (1, 28)=67.24, p<0.0001, interaction: F (1, 28) = 14.61, p=0.0007. N=8 in each group). (D–F) The p-STAT3/STAT3 (two-way ANOVA, gene: F (1, 28)=15.06, p=0.0006, model: F (1, 28)=67.24, p<0.0001, interaction: F (1, 28) = 14.61, p=0.0007. N=8 in each group) and p-JAK2/JAK2 (two-way ANOVA, gene: F (1, 28)=41.35, p<0.0001, model: F (1, 28)=9.604, p=0.0044, interaction: F (1, 28)=10.36, p=0.0032) in the two types (DEC1+/+, DEC1−/−) CRC mice. N=8 in each group in this figure, and the data are expressed as mean ± SD. *** p < 0.001 vs. DEC1+/+ CRC mice, ## p<0.01, ### p<0.001 vs. corresponding solvent mice, ns p > 0.05.
To gain clinical relevance, we analyzed the GEO database for the connection of DEC1 and IL-6. The data were normalized by eliminating batch differences (p < 0.05). As many as 61 genes were differentially expressed genes (DEGs) with DEC1. These DEGs were used to generate a Heatmap and Volcano plot by R languages. As shown in Fig. S2A-B, IL-6 (an activator of STAT3) was one of the DEC1-associated DEGs. There was a strong correlation between DEC1 and IL-6 expression (Fig. S2C). As expected, a strong correlation between DEC1 and STAT3 was detected (Fig. S2D). These findings conclude that DEC1 is an integrated player of the IL-6/STAT3 signaling in the development of CRC.
DEC1 promotes STAT3 phosphorylation and increases its target gene expression
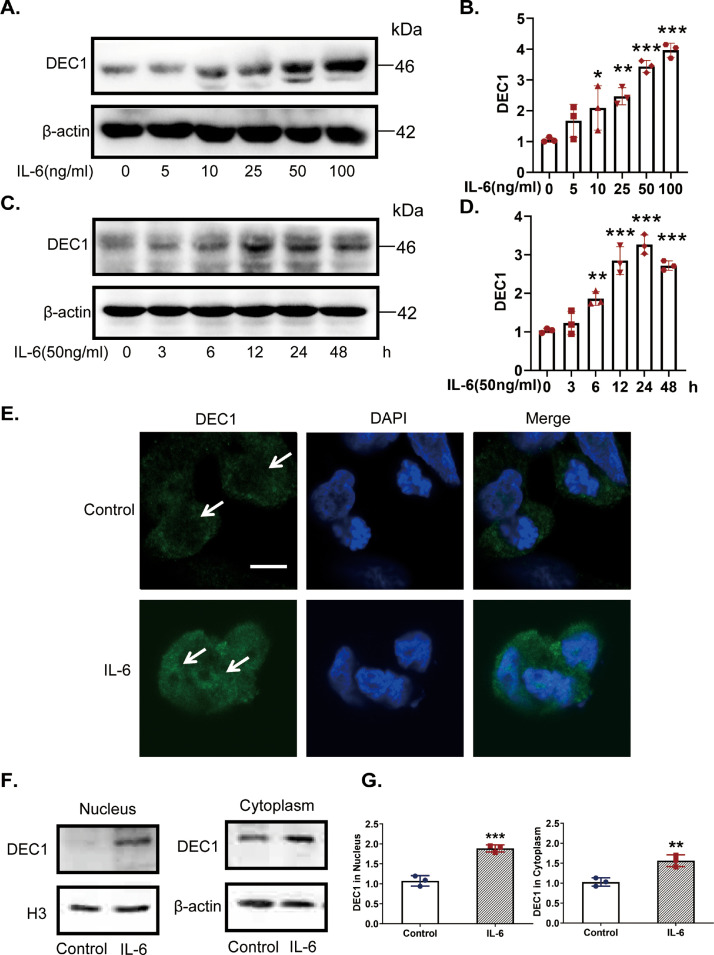
We next performed a set of experiments to provide mechanistic insight. First, we tested whether IL-6 induces DEC1 expression and whether the induction varies depending on a human colorectal cancer cell line (DLD-1, HCT116 and SW480 cells). As shown in Figs. 5A–D and S3A–D, IL-6 induced DEC1 expression in a dose- and time-dependent manner, and similar magnitude of the induction was detected among all three cell lines. The induced expression was confirmed by immunofluorescence assay with a strong presence of DEC1 in the nucleus (Fig. 5E). To further specify the relative induction between the cytoplasm and nucleus, subcellular factions were prepared and analyzed by Western blotting. As shown in Fig. 5F and G, DEC1 was induced by 90% in the nucleus and 54% in the cytoplasm. These results suggest that IL-6 induces DEC1 expression and enhances its nuclear translocation.
Fig. 5.
Increases of DEC1 expression and nuclear translocation in DLD-1 cells in response to IL-6. (A–D) IL-6 increases DEC1 expression, both in a dose- and time-dependent manner. DLD-1 cells were treated with IL-6 (0, 5, 10, 25, 50 and 100 ng/ml) for 24 h (one-way ANOVA, F (5, 12) = 21.87, p<0.0001. 0 vs. 10, p=0.0331, 0 vs. 25, p=0.0043, 0 vs. 50, p<0.0001, 0 vs. 100, p<0.0001), or were treated with IL-6 (50 ng/ml) for 0, 3, 6, 12, 24 or 48 h (one-way ANOVA, F (5, 12) = 45.57, p<0.0001. 0 vs. 6, p=0.0043, 0 vs. 12, p<0.0001, 0 vs. 50, p<0.0001, 0 vs. 100, p<0.0001). After that, the DEC1 expression was analyzed by Western blotting. (E) Effect of IL-6 on the DEC1 translocation with immunofluorescence analysis. DLD-1 cells were treated with IL-6 (50 ng/ml) for 24 h, and DEC1 protein translocation was monitored by immunofluorescence analysis. Scale bar=10 μm. (F, G) IL-6 induced DEC1 expression and translocation with plasmolysis. DLD-1 cells were treated with IL-6 for 24 h. DEC1 or β-actin cytoplasm protein (t-text, t=5.123, df=4, p=0.0069) and DEC1 or H3 nuclear protein expression (t-text, t=8.994, df=4, p=0.0008) were detected by Western blotingt. All experiments were repeated at least three times, and the data are expressed as mean ± SD. *p<0.05, ** p < 0.01, *** p < 0.001 vs. control or 0 h.
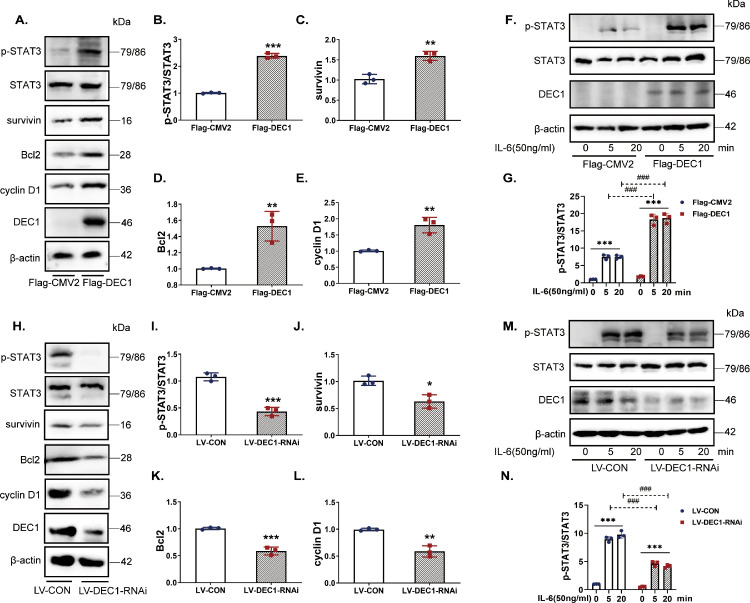
Second, we tested whether DEC1 directly regulates the expression and phosphorylation of STAT3. Overexpression and knockdown/knockout (RNAi and sgRNA/cas9) strategies were used to provided complementary information. The efficiency of DEC1 overexpression or knockdown was shown in Fig. 6A (line 6) and in Fig. 6H (line 6). Interestingly, neither overexpression nor knockdown or sg-RNA/cas9 of DEC1 altered the expression of STAT3 (mRNA and total protein) (Fig. 6A, H and S4). Nevertheless, the overexpression alone significantly increased the p-STAT3/STAT3 ratio (Fig. 6A and B) and the expression of target genes such as survivin, Bcl-2 and cyclin D1 (Fig. 6A and C–E). Conversely, knockdown of DEC1 downregulated the expression of these genes (Figs. 6H–L, S5A). In addition, overexpression of DEC1 increased the ratio of p-STAT3/STAT3 induced by IL-6 and the opposite was true with DEC1 knockdown (Fig. 6F, G, M and N). These findings suggest that DEC1 enhances the phosphorylation of STAT3 and upregulates its target genes.
Fig. 6.
Activation of STAT3 and increased expression of its target genes by DEC1 in DLD1 cells. (A-E) The overexpression of DEC1 increased the protein levels of p-STAT3/STAT3(t-text, t=25.82, df=4, p<0.0001), survivin (t-text, t=6.132, df=4, p=0.0036), Bcl2(t-text, t=4.953, df=4, p=0.0077) and cyclin D1(t-text, t=5.653, df=4, p=0.0048). (F, G) The p-STAT3/STAT3 level induced by IL-6 (two-way ANOVA, gene: F (1, 12)=311.5, p<0.0001, IL-6: F (2, 12)=320.4, p<0.0001, interaction: F (2, 12)=62.18, p<0.0001) in the DEC1 overexpression cells. (H–L) The knockdown of DEC1 decreased the protein levels of p-STAT3/STAT3 (t-text, t=10.36, df=4, p=0.0005), survivin (t-text, t=4.368, df=4, p=0.0120), Bcl2 (t-text, t=9.589, df=4, p=0.0007), and cyclin D1 (t-text, t=6.604, df=4, p=0.0027). (M, N) The p-STAT3/STAT3 level induced by stattic (two-way ANOVA, gene: F (1, 12)=365.7, p<0.0001, IL-6: F (2, 12)=511.5, p<0.0001, interaction: F (2, 12)=72.79, p<0.0001) in the DEC1 knockdown stable cells. All the experiments were repeated at least three times, and the data are expressed as mean ± SD. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. control (PBS) or 0 min, and ### p < 0.001 vs. the corresponding time of Flag-CMV2 or LV-CON.
DEC1 promotes CRC cell survival and proliferation via STAT3 signaling
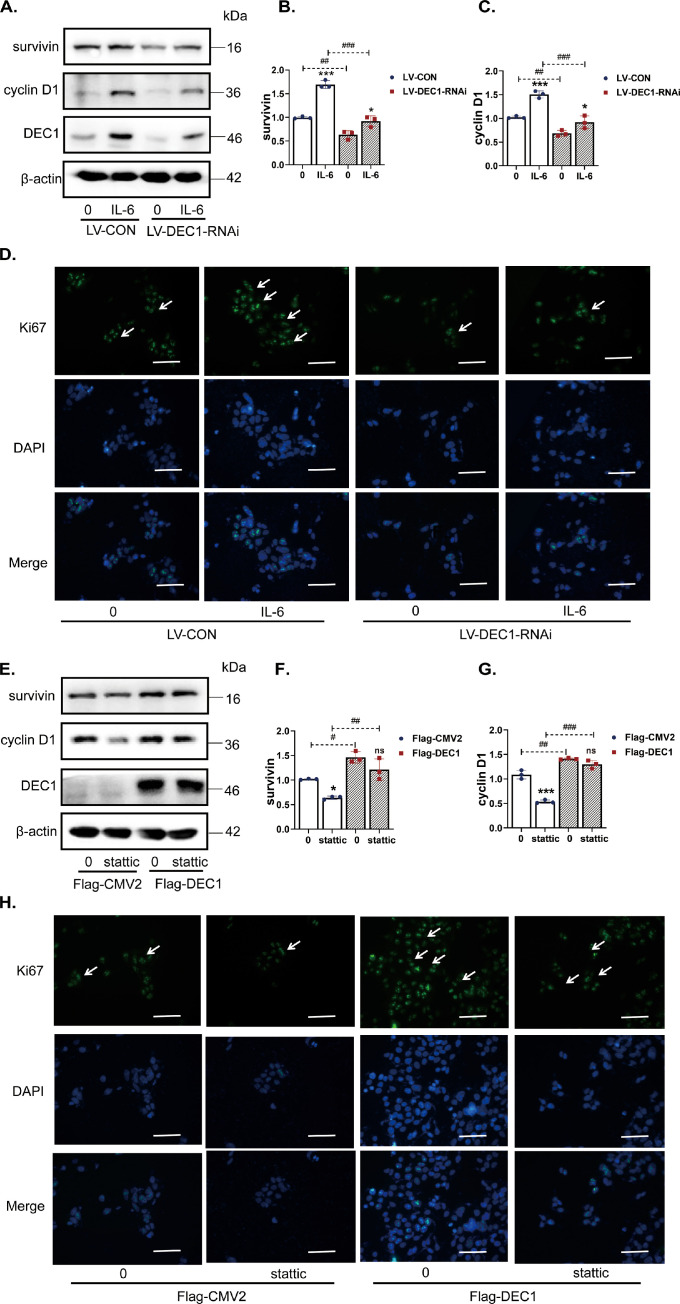
To make a functional connection of DEC1 with STAT3 signaling, we tested the effect of DEC1 on the expression of genes related to proliferation or survival in the presence or absence of the STAT3 activator IL-6 or the STAT3 inhibitor stattic [32]. Once again, DLD-1 cells were used with DEC1 overexpression or knockdown/knockout (RNAi or sgRNA/cas9). For the knockdown experiment, DLD-1-WT (LV-CON) and DLD-1-KD (LV-DEC1-RNAi, stable cells) were seeded in a 6-well plate at the density of 1 × 106 cells/well and treated with IL-6 (50 ng/ml) for 24 h. As shown in Fig. 7A–C (lanes 1 and 2), the protein expression of both survivin and cyclin D1 significantly decreased in LV-DEC1-RNAi compared with that in LV-CON cells. The decrease was more profound in IL-6 treated cells. Similar results were detected with sg-CON and DEC1 sgRNA/cas9 (Fig. S5B). In addition, IL-6 significantly increased Ki67 expression, whereas knockdown of DEC1 reduced the increase as shown by immunofluorescence assay (Fig. 7D). These results suggest that knockdown or sgRNA/cas9 of DEC1 alone is sufficient to reduce the protein expression of genes related to proliferation or survival and to counteract the effect of IL-6 on the expression of these genes.
Fig. 7.
Effect of DEC1 knockdown or overexpression on the expression of STAT3 in response to IL-6 or static. (A–C) Protein levels of survivin (two-way ANOVA, gene: F (1, 8)=137.0, p<0.0001, IL-6: F (1, 8)=103.6, p<0.0001, interaction: F (1, 8) = 17.79, p=0.0029) and cyclin D1 induced by IL-6(two-way ANOVA, gene: F (1, 8)=90.58, p<0.0001, IL-6: F (1, 8)=54.79, p<0.0001, interaction: F (1, 8)=6.525, p=0.0339) in the DEC1 knockdown stable cells. (D) Ki67 induced by IL-6 in the DEC1 knockdown stable cells. (E–G) Protein levels of survivin (two-way ANOVA, gene: F (1, 8) = 50.19, p=0.0001, stattic: F (1, 8)=19.10, p=0.0024, interaction: F (1, 8) = 0.8092, p=0.3946) and cyclin D1 induced by stattic (two-way ANOVA, gene: F (1, 8) = 211.8, p<0.0001, stattic: F (1, 8)=79.50, p<0.0001, interaction: F (1, 8)=35.00, P=0.0004) in the DEC1 overexpression cells. (H) Ki67 level induced by stattic in the DEC1 overexpression cells. Scale bar=40 μm. All the experiments were repeated at least three times, and the data are expressed as mean ± SD. * p < 0.05, *** p < 0.001 vs. 0, and # p < 0.05, ## p < 0.01, ### p < 0.001 vs. the corresponding LV-CON or Flag-CMV2.
For the overexpression experiment, DLD-1 cells were seeded in a 6-well plate at the density of 1 × 106 cells/well and transfected with Flag-DEC1 construct (800 ng/well) or the corresponding vector for 24 h. The transfected cells were treated with stattic (5 μM) for 24 h. Contrary to DEC1 knockdown or sgRNA/cas9, DEC1 overexpression increased the expression of survivin, cyclin D1 and Ki67 (Fig. 7E–G). Stattic significantly decreased survivin, cyclin D1 and Ki67 expression in vector-transfected cells (Fig. 7E–H), whereas the overexpression of DEC1 abolished the decrease (Fig. 7E–H). These findings suggest that DEC1 enhances colorectal cancer cell survival and proliferation with an involvement of STAT3 signaling.
DEC1-STAT3 physical interaction
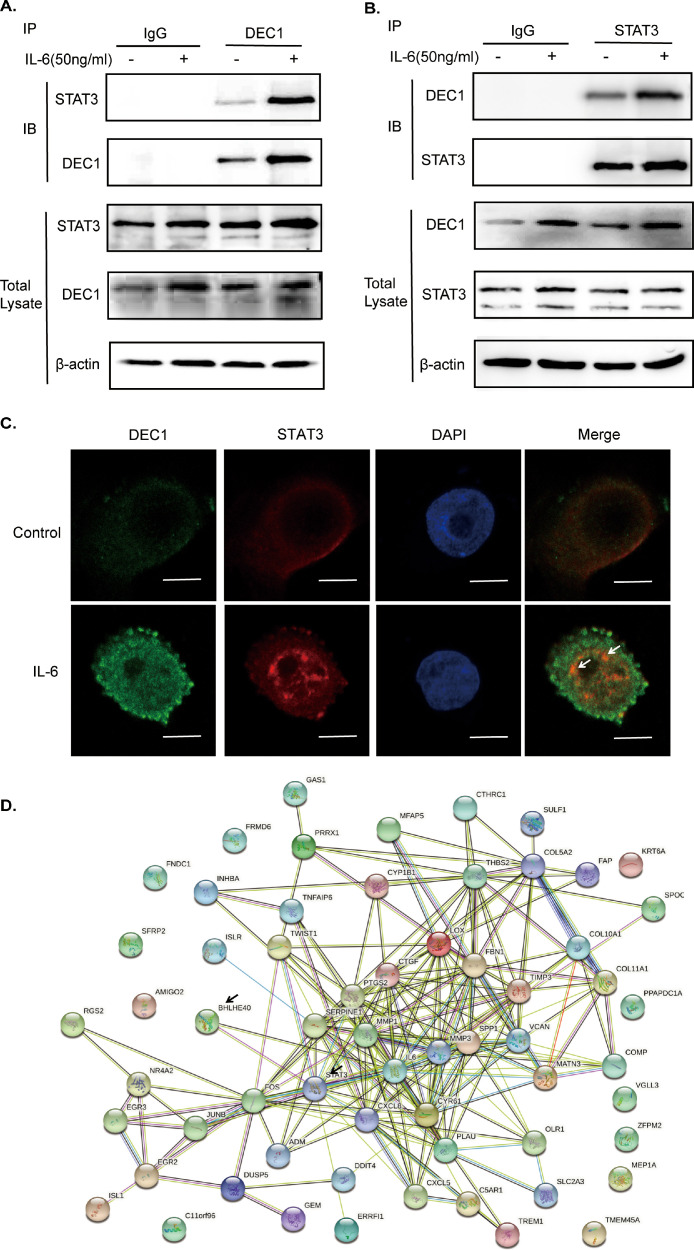
To explore the mechanism underlying how DEC1 and STAT3 signaling functionally interact, we tested whether DEC1 directly interacts with STAT3 as we have shown that DEC1 increased the pSTAT3/STAT3 ratio but did not cause changes on its mRNA and protein expression (Figs. 6A and H, S4). First, we performed co-immunoprecipitation with lysates from cells treated with IL-6. The lysates were immunoprecipitated with IgG, anti-DEC1 or anti-STAT3 antibody. The precipitates were then detected by Western blotting with anti-STAT3 or anti-DEC1 antibody. As shown in Fig. 8, STAT3 was presented in the immunocomplex precipitated with the anti-DEC1 antibody (Fig. 8A). Reciprocally DEC1 was presented in the immunocomplex precipitated with anti-STAT3 antibody (Fig. 8B). IL-6 significantly increased the DEC1-STAT3 interaction (Fig. 8A and B). Second, we performed immunofluorescence assay to test for their co-localization. DLD-1 cells were treated with PBS or IL-6 for 24 h and fixed with 4% paraformaldehyde. As expected, DEC1 and STAT3 were colocalized (Fig. 8C) and IL-6 increased DEC1 and STAT3 with greater presence in the nucleus (Fig. 8C). Finally, the PPI (protein-protein interaction) network analysis revealed that DEC1 and STAT3 could have protein interactions by using the STRING database (https://string-db.org/) (Fig. 8D). These findings suggest that DEC1 enhances CRC cell survival and proliferation, likely through direct interaction with STAT3 (pSTAT3).
Fig. 8.
Interaction of DEC1 with STAT3. (A, B) Cells were treated with IL-6 (50 ng/ml) or PBS for 24 h. Cell lysates were incubated with an antibody against DEC1, STAT3 or non-immune IgG. The immunoprecipitated complex was analyzed through using antibodies against STAT3 or DEC1. The input was analyzed by Western blotting for DEC1, STAT3 or GAPDH. (C) Co-localization of DEC1 and STAT3 through using immunofluorescence. DLD-1 cells were treated with IL-6 (50 ng/ml) or PBS for 24 h. DEC1 (green), STAT3 (red), and DAPI (blue) were analyzed with immunofluorescence staining. An over-lapping subcellular distribution (yellow) was presented in merged images. Scale bar=10 μm. (D) DEC1 and STAT3 have interaction in protein-protein interaction network through using the STRING database (https://string-db.org/). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
CRC is the second deadly and the third most common malignancy worldwide [1]. It has been projected that annual new cases of CRC will increase by 63% in 2040 [1], constituting an even greater health challenge for decades to come. The pathogenesis of CRC is driven by the increased proliferation/survival in the superficial epithelium [33,34]. However, the precise mechanisms remain to be determined for such increases during the development of CRC. Several transcriptional factors, such as P63, Sox2 and HIF-1α (hypoxia inducible factor), have been linked to play critical roles in the epithelial proliferation [35]. In this study, we have used complementary approaches and demonstrated that DEC1, a well-established transcription factor [13], enhances cell proliferation and survival related to CRC pathogenesis in both clinical and chemicals-induced conditions.
This study has provided several lines of evidence that support DEC1 as an intimate player in the development of CRC. First, the expression of DEC1 in the tumor tissues from CRC patients significantly increases compared with that in the adjacent normal tissues (Fig. 1A and D), confirming an earlier study with a smaller number of patient samples [18]. Second, CRC patients with higher DEC1 expression have a much shorter survival time (Fig. 1B, C). Third, AOM/DSS-induced CRC mice express significantly higher levels of DEC1 in the colon than the control mice (Fig. 1F–H). Fourth, AOM/DSS induce a fewer number of tumors with smaller sizes in DEC1 knockout than DEC1+/+ mice (Fig. 2E and F). Fifth, the tumors in DEC1+/+ mice are more invasive than those in DEC1−/− mice (Fig. 3A). And finally, upon AOM/DSS induction, the DEC1+/+ group has a mortality rate twice as much as the DEC1−/- group (Fig. 2A).
The enhanced induction of CRC in the DEC1+/+ background by AOM/DSS is likely achieved by promoting cell proliferation, antiapoptosis or both. Indeed, tumors from DEC1+/+ CRC mice, compared those from DEC1−/− CRC mice, express much higher levels of proliferation-proteins: cyclin D1, CKD4, Ki67 and PCNA (Fig. 3C–I) [23,24]. Likewise, the expression of survivin, a major antiapoptotic protein [14], is significantly higher in DEC1+/+ than DEC1−/− CRC mice (Fig. 3C, F). Bax and Bcl-2 are both Bcl-2 gene family members [26] and exhibit inverse changes. The expression of Bax, a pro-apoptotic regulator, is upregulated in DEC1−/− CRC tissues but downregulated in DEC1+/+ CRC, and the opposite is true with Bcl2, an anti-apoptotic protein (Fig. 3G, J) [26,27]. Overall, the increased proliferation and decreased apoptosis, associated with the DEC1+/+ background, are confirmed by BrdU incorporation assay and TUNEL staining (Fig. 3K and L). These results conclude that DEC1 enhances CRC development in response to AOM/DSS through proliferation-promotion and apoptotic inhibition.
The regulated expression of proliferation- and survival-related genes in the animal model is confirmed in cell models including the use of the DLD-1 colorectal cell line. For example, overexpression of DEC1 upregulates the expression of cyclin D1 and Ki67 (Figs. 6A, 7E–H), whereas knockdown and sgRNA/cas9 of DEC1 downregulate the expression of these genes (Figs. 6H, 7A–D, and S5A, B). Similarly, overexpression of DEC1 increases the survivin and Bcl-2 protein levels (Fig. 6A, C and D), whereas knockdown and sgRNA/cas9 of DEC1 decrease them in DLD-1 cells (Figs. 6H, J and K, and S5A). These results are consistent with previous studies [36], [37], [38], [39]. On the other hand, how DEC1 regulates the expression of genes remains to be determined. We have shown that DEC1 exerts transcriptional regulation through E-box or Sp1 elements [17,40]. Interaction with E-box leads to transcriptional repression (e.g., Per) [40], whereas interaction with Sp1 leads to transactivation [17]. Indeed, we have reported that DEC1 increases survivin expression through interaction with SP1 [17]. It is therefore assumed that the observed upregulation of cyclin D1 and Bcl-2 is likely achieved through a DEC1-Sp1 complex.
Several proinflammatory cytokines such as IL-6 are increased in CRC patients [5]. IL-6 triggers signaling cascades centered by STAT3 through phosphorylation [6,11,41]. Interestingly, DEC1 reciprocally enhances the functionality of the STAT3 signaling. For example, tumor tissues from DEC1+/+ CRC mice express higher levels of IL-6 than those from DEC1−/− CRC mice (Fig. 4A–C), but yet IL-6 is shown to robustly induce DEC1 in DLD-1 cells (Fig. 5). The magnitude of induction is higher in the nucleus than the cytoplasm, pointing to an involvement of enhanced nuclear translocation (Fig. 5E). Likewise, overexpression of DEC1 increases the pSTAT3/STAT3 ratio in response to IL-6, and the opposite is true with DEC1 knockdown (Fig. 6F, G, M, N). Similar observations are made on several STAT3 target genes including cyclin D1 (Fig. 7A–C, E–G). Nevertheless, stattic, a specific STAT3 inhibitor [32], reduces the expression of survivin and cyclin D1, but the reduced expression is largely reversed by DEC1 overexpression (Fig. 7E–G). These findings establish that DEC1 is functionally related to the STAT3 signaling pathway.
It is interesting to notice that DEC1 is established as a potent transcription factor [13,17,40], however, this study has demonstrated that overexpression or knockdown/knockout of DEC1 does not alter the expression of STAT3 mRNA and protein (Figs. 6A and H, S4). One of the explanations is that DEC1 regulates the activity of STAT3 by affecting its phosphorylation status, a critical marker for the activity of STAT3 signaling. Indeed, we have shown that DEC1 increases the pSTAT3/STAT3 ratio (Fig. 6F, G, M, N), pointing a mechanism of DEC1 to stabilize pSTAT3. In support of this notion, we and others have shown that DEC1 and STAT3 form protein complex according to protein-protein interaction networks (Fig. 8D) [42,43]. Nevertheless, our coimmunoprecipitation has detected DEC1-STAT3 complexes. The interaction is increased by IL-6 stimulation (Fig. 8A–C), pointing to the preferable interaction between pSTAT3 and DEC1. We have previously shown that IL-6 promotes DEC1 interactions with RXRα, suppressing the activities of dimerization partners PXR and CAR [44], suggesting that protein-protein interactions represent a major mechanism for DEC1 to regulate gene expression.
In summary, we have shown, with database data and patient samples, that DEC1 is expressed much higher in CRC than the adjacent normal tissues. CRC patients with higher DEC1 expression have a shorter survival time. The chemical carcinogenesis protocol with AOM/DSS induces more tumors with larger sizes in DEC1+/+ than DEC1−/− mice. Consistent with the analysis of clinical data and animal samples, overexpression of DEC1 increases the expression of proliferation and survival-related genes, but the opposite is true with apoptotic genes. Critically, this study has shown that IL-6, a potent trigger of STAT3 signaling, induces DEC1, and DEC1 enhances the phosphorylation of STAT3, a critical step toward STAT3 activation. Additionally, we have shown that DEC1 increases the pSTAT3/STAT3 ratio and physically interacts with STAT3 (presumably pSTAT3). These findings establish that DEC1 is an CRC enhancer. The enhancement is achieved with an intimate involvement of the IL-6/STAT3 signaling. These findings provide mechanistic understanding regarding CRC development and metastasis with a potential of developing intervention strategies.
CRediT authorship contribution statement
Enfang Shan: Methodology, Software, Data curation, Writing – original draft. Ying Hao: Visualization, Data curation. Haobin Wang: Software, Formal analysis. Ziheng Zhang: Visualization. Jingwan Hu: Data curation. Guyu Wang: Formal analysis. Wei Liu: Supervision. Bingfang Yan: Investigation, Project administration, Writing – review & editing. Honda Hiroaki: Methodology. Jian Yang: Conceptualization, Project administration, Resources, Funding acquisition, Writing – review & editing.
Competing interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Funding
The work was supported by the Natural Science Foundation of China (Nos: 82073934, 81872937), Nanjing Medical University (NMUB2019024). This study was also supported partially by National Institutes of Health Grants R01EB018748 and 1R21AI153031 (BY).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100783.
Appendix. Supplementary materials
References
- 1.Xi Y., Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol. 2021;14(10) doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., Martin A. Mismatch repair and colon cancer: mechanisms and therapies explored. Trends Mol Med. 2016;22(4):274–289. doi: 10.1016/j.molmed.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. doi: 10.1038/521S1a. [DOI] [PubMed] [Google Scholar]
- 4.Kim E.R., Chang D.K. Colorectal cancer in inflammatory bowel disease: the risk, pathogenesis, prevention and diagnosis. World J Gastroenterol. 2014;20(29):9872–9881. doi: 10.3748/wjg.v20.i29.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kantola T., Klintrup K., Vayrynen J.P., Vornanen J., Bloigu R., Karhu T., et al. Stage-dependent alterations of the serum cytokine pattern in colorectal carcinoma. Br J Cancer. 2012;107(10):1729–1736. doi: 10.1038/bjc.2012.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang A., Deng S., Chen X., Yu C., Du Q., Wu Y., et al. miR-29a-5p/STAT3 positive feedback loop regulates TETs in colitis-associated colorectal cancer. Inflamm Bowel Dis. 2020;26(4):524–533. doi: 10.1093/ibd/izz281. [DOI] [PubMed] [Google Scholar]
- 7.Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., et al. IL-6 and STAT3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tolomeo M., Cascio A. The multifaced role of STAT3 in cancer and its implication for anticancer therapy. Int J Mol Sci. 2021;22(2):603–627. doi: 10.3390/ijms22020603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pencik J., Schlederer M., Gruber W., Unger C., Walker S.M., Chalaris A., et al. STAT3 regulated ARF expression suppresses prostate cancer metastasis. Nat Commun. 2015;6:7736. doi: 10.1038/ncomms8736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniguchi K., Moroishi T., de Jong P.R., Krawczyk M., Grebbin B.M., Luo H., et al. YAP-IL-6ST autoregulatory loop activated on APC loss controls colonic tumorigenesis. Proc Natl Acad Sci U S A. 2017;114(7):1643–1648. doi: 10.1073/pnas.1620290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y.J., Choi J., Yoon Y.J., Sim Y., Ryu H.W., Oh S.R., et al. 8-Epi-xanthatin induces the apoptosis of DU145 prostate carcinoma cells through signal transducer and activator of transcription 3 inhibition and reactive oxygen species generation. Phytother Res. 2021;35(3):1508–1520. doi: 10.1002/ptr.6918. [DOI] [PubMed] [Google Scholar]
- 13.Qian Y., Jung Y.S., Chen X. Differentiated embryo-chondrocyte expressed gene 1 regulates p53-dependent cell survival versus cell death through macrophage inhibitory cytokine-1. Proc Natl Acad Sci U S A. 2012;109(28):11300–11305. doi: 10.1073/pnas.1203185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao T., Xiong H., Hu X., Hu Y., Wang C., Yang L., et al. DEC1: a potential biomarker of malignant transformation in oral leukoplakia. Braz Oral Res. 2020;34:e52. doi: 10.1590/1807-3107bor-2020.vol34.0052. [DOI] [PubMed] [Google Scholar]
- 15.Sato F., Kohsaka A., Bhawal U.K., Muragaki Y. Potential roles of DEC and Bmal1 genes in interconnecting circadian clock and energy metabolism. Int J Mol Sci. 2018;19(3):781–795. doi: 10.3390/ijms19030781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bi H., Li S., Qu X., Wang M., Bai X., Xu Z., et al. DEC1 regulates breast cancer cell proliferation by stabilizing cyclin E protein and delays the progression of cell cycle S phase. Cell Death Dis. 2015;6:e1891. doi: 10.1038/cddis.2015.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Xie M., Yang J., Yang D., Deng R., Wan Y., et al. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene. 2006;25(23):3296–3306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Zhang H., Xie M., Hu M., Ge S., Yang D., et al. Abundant expression of DEC1/STRA13/SHARP2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J. 2002;367(Pt 2):413–422. doi: 10.1042/BJ20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia Y., Hu R., Li P., Zheng Y., Wang Y., Ma X. DEC1 is required for anti-apoptotic activity of gastric cancer cells under hypoxia by promoting Survivin expression. Gastric Cancer. 2018;21(4):632–642. doi: 10.1007/s10120-017-0780-z. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki K., Miyazaki M., Guo Y., Yamasaki N., Kanno M., Honda Z., et al. The role of the basic helix-loop-helix transcription factor DEC1 in the regulatory T cells. J Immunol. 2010;185(12):7330–7339. doi: 10.4049/jimmunol.1001381. [DOI] [PubMed] [Google Scholar]
- 21.Zaki M.H., Vogel P., Malireddi R.K., Body-Malapel M., Anand P.K., Bertin J., et al. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 2011;20(5):649–660. doi: 10.1016/j.ccr.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song C.H., Kim N., Nam R.H., Choi S.I., Lee H.N., Surh YJ. 17beta-estradiol supplementation changes gut microbiota diversity in intact and colorectal cancer-induced ICR male mice. Sci Rep. 2020;10(1):12283. doi: 10.1038/s41598-020-69112-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y., Gibson S.B. Three dimensions of autophagy in regulating tumor growth: cell survival/death, cell proliferation, and tumor dormancy. Biochim Biophys Acta Mol Basis Dis. 2021;1867(12) doi: 10.1016/j.bbadis.2021.166265. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh P., Vidal C., Dey S., Zhang L. Mitochondria targeting as an effective strategy for cancer therapy. Int J Mol Sci. 2020;21(9):3363–3381. doi: 10.3390/ijms21093363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Shi J., Qi S., Zhang J., Peng D., Chen Z., et al. IL-33 facilitates proliferation of colorectal cancer dependent on COX2/PGE2. J Exp Clin Cancer Res. 2018;37(1) doi: 10.1186/s13046-018-0839-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu W.H., Hua W.J., Qiu W.L., Tseng A.J., Cheng H.C., Lin T.Y. WSG, a glucose-enriched polysaccharide from Ganoderma lucidum, suppresses tongue cancer cells via inhibition of EGFR-mediated signaling and potentiates cisplatin-induced apoptosis. Int J Biol Macromol. 2021;193(Pt B):1201–1208. doi: 10.1016/j.ijbiomac.2021.10.146. [DOI] [PubMed] [Google Scholar]
- 27.El-Far A.H., Godugu K., Noreldin A.E., Saddiq A.A., Almaghrabi O.A., et al. Thymoquinone and costunolide induce apoptosis of both proliferative and doxorubicin-induced-senescent colon and breast cancer cells. Integr Cancer Ther. 2021;20 doi: 10.1177/15347354211035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roeser J.C., Leach S.D., McAllister F. Emerging strategies for cancer immunoprevention. Oncogene. 2015;34(50):6029–6039. doi: 10.1038/onc.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei N., Li J., Fang C., Chang J., Xirou V., Syrigos N.K., et al. Targeting colon cancer with the novel STAT3 inhibitor bruceantinol. Oncogene. 2019;38(10):1676–1687. doi: 10.1038/s41388-018-0547-y. [DOI] [PubMed] [Google Scholar]
- 30.Nagasaki T., Hara M., Nakanishi H., Takahashi H., Sato M., Takeyama H. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br J Cancer. 2014;110(2):469–478. doi: 10.1038/bjc.2013.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai Z., Wang L., Wang X., Zhao B., Zhao W., Bhardwaj S.S., et al. Oxymatrine induces cell cycle arrest and apoptosis and suppresses the invasion of human glioblastoma cells through the EGFR/PI3K/Akt/mTOR signaling pathway and STAT3. Oncol Rep. 2018;40(2):867–876. doi: 10.3892/or.2018.6512. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y.F., Zhuang K.H., Chen B., Li P.W., Zhou X., Jiang H., et al. Expansion and activation of monocytic-myeloid-derived suppressor cell via STAT3/arginase-I signaling in patients with ankylosing spondylitis. Arthritis Res Ther. 2018;20(1) doi: 10.1186/s13075-018-1654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleming M., Ravula S., Tatishchev S.F., et al. Colorectal carcinoma: pathologic aspects. J Gastrointest Oncol. 2012;3(3):153–173. doi: 10.3978/j.issn.2078-6891.2012.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Munro M.J., Wickremesekera S.K., Peng L., Tan S.T., Itinteang T. Cancer stem cells in colorectal cancer: a review. J Clin Pathol. 2018;71(2):110–116. doi: 10.1136/jclinpath-2017-204739. [DOI] [PubMed] [Google Scholar]
- 35.Qian C., Dai Y., Xu X., Jiang Y. HIF-1alpha regulates proliferation and invasion of oral cancer cells through Kv3.4 channel. Ann Clin Lab Sci. 2019;49(4):457–467. [PubMed] [Google Scholar]
- 36.Qian Y., Jung Y.S., Chen X. DeltaNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. J Biol Chem. 2011;286(14):12033–12041. doi: 10.1074/jbc.M110.207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X.M., Lin W., Wang J., Zhang W., Yin A.A., Huang Y., et al. DEC1 expression predicts prognosis and the response to temozolomide chemotherapy in patients with glioma. Mol Med Rep. 2016;14(6):5626–5636. doi: 10.3892/mmr.2016.5921. [DOI] [PubMed] [Google Scholar]
- 38.Sato F., Bhawal U.K., Sugiyama N., Osaki S., Oikawa K., Muragaki Y. Potential role of DEC1 in cervical cancer cells involving overexpression and apoptosis. Clocks Sleep. 2020;2(1):26–38. doi: 10.3390/clockssleep2010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi Y., Liao B., Zheng Z., Yang X., Yang Y., Zhou Y., et al. Downregulation of DEC1 inhibits proliferation, migration and invasion, and induces apoptosis in ovarian cancer cells via regulation of Wnt/beta-catenin signaling pathway. Exp Ther Med. 2021;21(4):372. doi: 10.3892/etm.2021.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Song X., Ma Y., Liu J., Yang D., Yan B. DNA binding, but not interaction with Bmal1, is responsible for DEC1-mediated transcription regulation of the circadian gene mPer1. Biochem J. 2004;382(Pt 3):895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin Y., He Z., Ye J., Liu Z., She X., Gao X., et al. Progress in understanding the IL-6/STAT3 pathway in colorectal cancer. Onco Targets Ther. 2020;13:13023–13032. doi: 10.2147/OTT.S278013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov S.V., Salnikow K., Ivanova A.V., Bai L., Lerman M.I. Hypoxic repression of STAT1 and its downstream genes by a pVHL/HIF-1 target DEC1/STRA13. Oncogene. 2007;26(6):802–812. doi: 10.1038/sj.onc.1209842. [DOI] [PubMed] [Google Scholar]
- 43.Li B., Chu Y., Yan B., Ma X., Liu D., Wang S., et al. Reciprocal expression of differentiated embryonic chondrocyte expressed genes result in functional antagonism in gastric cancer. Dig Dis Sci. 2021 doi: 10.1007/s10620-021-06921-7. [DOI] [PubMed] [Google Scholar]
- 44.Ning R., Zhan Y., He S., Hu J., Zhu Z., Hu G., et al. Interleukin-6 induces DEC1, promotes DEC1 interaction with RXRalpha and suppresses the expression of PXR, CAR and their target genes. Front Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.