Abstract
Cell-free system has emerged as a powerful platform with a wide range of in vitro applications and recently has contributed to express metabolic pathways for biosynthesis. Here we report in vitro construction of a native biosynthetic pathway for L-4-nitrotryptophan (L-4-nitro-Trp) synthesis using an Escherichia coli-based cell-free protein synthesis (CFPS) system. Naturally, a nitric oxide (NO) synthase (TxtD) and a cytochrome P450 enzyme (TxtE) are responsible for synthesizing L-4-nitro-Trp, which serves as one substrate for the biosynthesis of a nonribosomal peptide herbicide thaxtomin A. Recombinant coexpression of TxtD and TxtE in a heterologous host like E. coli for L-4-nitro-Trp production has not been achieved so far due to the poor or insoluble expression of TxtD. Using CFPS, TxtD and TxtE were successfully expressed in vitro, enabling the formation of L-4-nitro-Trp. After optimization, the cell-free system was able to synthesize approximately 360 μM L-4-nitro-Trp within 16 h. Overall, this work expands the application scope of CFPS for study and synthesis of nitro-containing compounds, which are important building blocks widely used in pharmaceuticals, agrochemicals, and industrial chemicals.
Keywords: Cell-free protein synthesis, NO synthase, P450 enzyme, L-4-nitrotryptophan, In vitro biosynthesis, Synthetic biology
1. Introduction
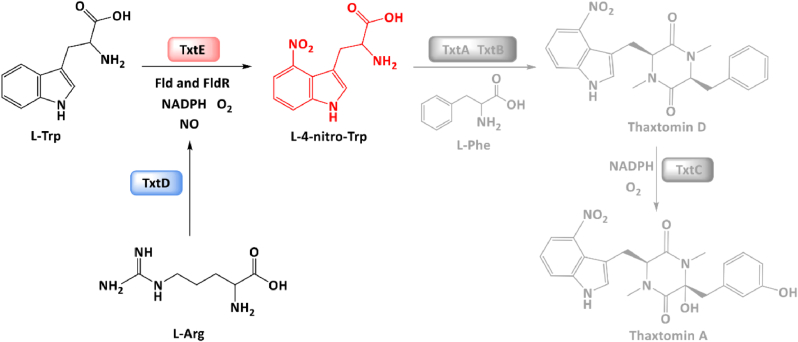
Nitro group (-NO2) containing compounds, especially the nitroaromatic chemicals, are widely used as explosives, dyes, pharmaceuticals, pesticides, and antibiotics [[1], [2], [3]]. Among these compounds, nitro amino acids have been found in microorganisms serving as building blocks for the synthesis of bioactive natural products [3,4]. For example, the unnatural amino acid L-4-nitrotryptophan (L-4-nitro-Trp) is an essential precursor for the biosynthesis of thaxtomins A-D, which are nonribosomal peptides produced by various Streptomyces strains (e.g., S. scabies, S. acidiscabies, and S. turgidiscabies) [5,6]. The predominant member thaxtomin A has the potent phytotoxicity in plants by inhibiting cellulose biosynthesis, thus destroying the integrity of the cell wall and the development of normal cells through cytokinesis [7,8]. As a result, thaxtomin A can be potentially developed as an herbicide in agriculture [9]. Its herbicidal performance has been demonstrated and the unique 4-nitroindole structure acts as an essential moiety for the phytotoxicity [10]. In addition, L-4-nitro-Trp also has been used as an intermediate in organic reactions for synthesizing chemicals like triboc-6-boronopinacol tryptophan (TB-6-BT), which has potential for tumor treatment together with other drugs [11].
In chemistry, nitro groups required for the 4-nitration of l-tryptophan (L-Trp) can be first generated by the mixed-acid reaction of sulfuric and nitric acids, and then used to substitute C4 of L-Trp via electrophilic substitution [1,2]. However, this method requires complex reaction conditions and lacks regioselectivity [2]. By contrast, biological systems can produce many target compounds in a mild way without the use of relative harsh chemical reagents and conditions. Naturally, L-4-nitro-Trp can be synthesized by two biocatalysts: a nitric oxide (NO) synthase (TxtD) and a cytochrome P450 enzyme (TxtE), which are involved in the thaxtomin A biosynthetic pathway (Fig. 1) [[12], [13], [14]]. Briefly, TxtD first generates NO from the substrate l-arginine (L-Arg). Then, TxtE catalyzes the direct regiospecific 4-nitration of L-Trp using NO and O2 as cosubstrates to form L-4-nitro-Trp. Interestingly, TxtE is one of only two known enzymes enabling direct nitration of a C–H bond in bacterial natural product biosynthesis [15]. Due to the unique catalytic function of TxtE, it has been engineered and developed as promising nitration biocatalysts for biotechnological applications, for example, the generation of L-4-nitro-Trp analogs [[16], [17], [18]]. While TxtE can be overexpressed and purified to implement in vitro catalysis, recombinant expression of TxtD in a heterologous host like E. coli was not successful. Therefore, NO can only be generated by adding expensive and unstable NO donors (e.g., NOC-5) for in vitro biocatalytic nitration [17,18]. So far, there is no report by using both native enzymes (TxtD and TxtE) together to synthesize L-4-nitro-Trp in vitro. Recently, an isozyme NO synthase from Bacillus subtilis was used to reconstitute an artificial biosynthetic pathway with the coexpression of TxtE to produce L-4-nitro-Trp and its analogs in E. coli [19]. However, overexpression of NO synthase in vivo may generate a high level of NO in cells, leading to cytotoxicity on the producers [13,15]. Therefore, a new strategy for L-4-nitro-Trp synthesis by coexpression of native enzyme partners TxtD and TxtE is highly desirable.
Fig. 1.
TxtD (NO synthase) and TxtE (P450 enzyme) catalyze the biosynthesis of L-4-nitro-Trp. Note that L-4-nitro-Trp is a precursor for thaxtomin A biosynthesis involved two nonribosomal peptide synthetases (TxtA and TxtB) and another cytochrome P450 enzyme (TxtC).
In recent years, cell-free systems as complements to living cell systems have been well-developed, showing a broad range of applications in biotechnology and synthetic biology [[20], [21], [22], [23], [24]]. Crude extract based cell-free protein synthesis (CFPS) systems were mainly used for in vitro protein production in the early days. Now, the CFPS systems have been evolved into promising and powerful platforms for biomanufacturing by reconstitution of cell-free expressed enzymes. For example, several studies have demonstrated the rapid, high-yield synthesis of valuable chemicals relevant to energy, food, pharmaceuticals, and materials [[25], [26], [27], [28]]. Because of the absence of cell walls, cell-free reactions are open that enables easy and flexible manipulation, monitoring, and optimization. Recently, CFPS-based systems have been used in an aqueous-organic biphasic system for cascade biotransformation by construction of an artificial enzymatic pathway [29]. If the full metabolic pathway is long with multiple steps (e.g., 6–7 enzymes), cell-free systems can also be designed with different reaction modules for enzyme expression, followed by combining the enzyme-enriched modules for efficient biocatalysis [30]. Moreover, there are no living, intact cells in CFPS reactions. Therefore, cell-free systems are more tolerant to toxic molecules as compared to living systems, which has been showcased with the synthesis of styrene, a cellular toxic compound, at high titers using CFPS [31].
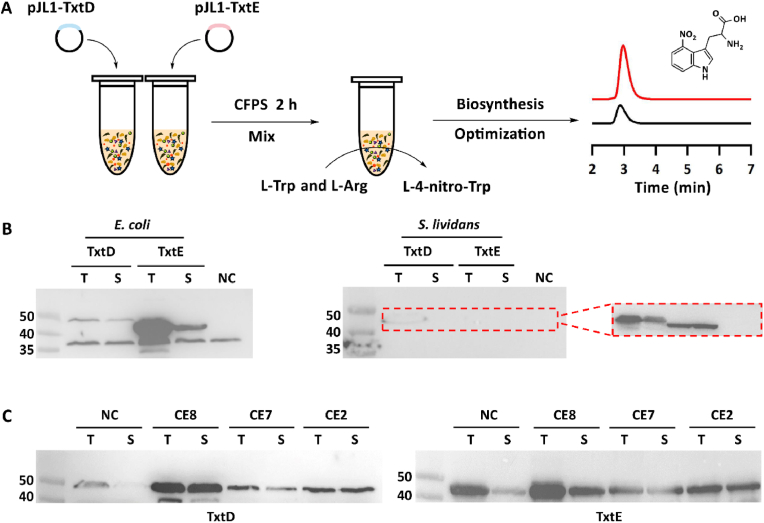
In this work, we aim to reconstitute the biosynthetic route for L-4-nitro-Trp synthesis by in vitro expressed TxtD and TxtE using an E. coli-based CFPS system (Fig. 2A). To begin, we evaluate the effects of different CFPS systems and cell-free reaction formats on the expression of both enzymes (TxtD and TxtE). Then, cell-free expressed TxtD and TxtE without purification can catalyze the formation of L-4-nitro-Trp. Finally, the key parameters for in vitro L-4-nitro-Trp biosynthesis are optimized for enhanced production. Overall, our cell-free system is robust to synthesize the NO synthase (TxtD) and P450 enzyme (TxtE) with biocatalytic activity. Especially, our data suggest that CFPS is efficient for the expression of TxtD, which is hardly expressed in vivo using the heterologous host E. coli [19]. Looking forward, we envision that cell-free biosynthetic systems will serve as promising platforms for the rapid synthesis of value-added molecules such as the nitro-containing compounds of industrial and agricultural interest.
Fig. 2.
Cell-free expression of TxtD and TxtE for L-4-nitro-Trp biosynthesis. (A) Schematic diagram of CFPS reactions and optimization for enhanced production. (B) Western-blot analysis of cell-free expressed TxtD (45.6 kDa) and TxtE (45.7 kDa) with the anti-His antibody. TxtD and TxtE were expressed in E. coli and S. lividans CFPS systems, respectively. In the E. coli system, the band between 35 and 40 kDa was an unknown protein derived from the cell lysates as it also appeared in the NC sample. In the S. lividans system, the bands of TxtD and TxtE were not visible as shown in the red line frame when they were imaged (high-resolution mode imaging program, UVP Chemstudio Touch 815, analytikjena) together with the E. coli CFPS expressed proteins. When the S. lividans CFPS samples were imaged separately using the ultrahigh-sensitivity mode imaging program, TxtD and TxtE bands could be visualized as shown in the right side red line frame. T, total protein; S, soluble protein; NC, negative control without plasmids in the CFPS reaction. (C) Western-blot analysis of TxtD (left) and TxtE (right) expressed in E. coli CFPS reactions with different enriched molecular chaperones. NC, negative control – cell extracts (CE) without chaperones; CE8 with chaperones: DnaK-DnaJ-GrpE + GroES-GroEL; CE7 with chaperones: GroES-GroEL; CE2 with chaperones: GroES-GroEL-tig; T, total protein; S, soluble protein. Using protein band density analysis by ImageJ, the solubility of TxtD was determined to be 28% (NC), 86% (CE8), 42% (CE7), and 96% (CE2); the solubility of TxtE was determined to be 27% (NC), 67% (CE8), 76% (CE7), and 88% (CE2). Percent solubility (%) = soluble protein/total protein.
2. Materials and methods
2.1. Strains and plasmids
E. coli DH5α was used for molecular cloning and plasmid propagation. E. coli BL21 Star(DE3) was used for preparation of cell extracts. The genes of txtD (1203 bp) and txtE (1221 bp) were amplified by PCR from the genome of Streptomyces acidiscabies and cloned into the vector pJL1. The primers used for PCR amplification are shown in Table 1. Plasmid pJL1-sfGFP was digested with NdeI and SalI to obtain a linearized backbone, which was ligated with the genes txtD and txtE, respectively, using a recombinant ligation kit (Beyotime, China). The resulting plasmids for cell-free expression were named as pJL1-TxtD and pJL1-TxtE. The plasmids of molecular chaperones (pG-KJE8, pGro7, and pG-Tf2) were obtained from TaKaRa (Beijing, China). pG-KJE8 (dnaK-dnaJ-grpE and groES-groEL), pGro7 (groES-groEL), and pG-Tf2 (groES-groEL-tig) were transformed into of E. coli BL21 Star(DE3), respectively, for the preparation of cell extracts containing chaperone proteins.
Table 1.
Primers used for PCR amplification.
| Genes | Primer sequences (5’→3′) |
|---|---|
| txtD | |
| Forward | catcatcatcaccatatgacctccgaagtcgctctgggccctt |
| Reverse | tgttagcagccggtcgacttactggtgggggtagaagttggggcgct |
| txtE | |
| Forward | catcatcatcaccatatgaccgtcccctcgccgctcgccga |
| Reverse | tgttagcagccggtcgacttagcggaggctgagcggcaggga |
2.2. Preparation of cell extracts
E. coli BL21 Star(DE3) cells were grown in 1 L 2 × YTPG medium (10 g yeast extract, 16 g tryptone, 5 g NaCl, 7 g K2HPO4, 3 g KH2PO4, and 18 g glucose, pH 7.2) in 2.5 L baffled Ultra Yield™ flasks (Thomson Instrument Company, USA). After inoculation with 20 mL overnight preculture (initial OD600 of 0.05–0.1), the cells were incubated in the shaker at 220 rpm and 34 °C. When the OD600 reached 0.6–0.8, cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to express T7 RNA polymerase. To prepare cell extracts consisting of chaperones, all cultures were induced with 0.8 mM IPTG. Note that the final concentration of IPTG was reduced to 0.8 mM according to the manufacturer's manual (Chaperone Plasmid Set, Code No. 3340, TaKaRa). In addition, 1 mg/mL l-arabinose and 1 ng/mL tetracycline were added to induce pG-KJE8, 1 mg/mL l-arabinose was added to induce pGro 7, and 1 ng/mL tetracycline was added to induce pG-Tf2. Then, cells were harvested at an OD600 of 3.0 by centrifugation at 5,000 g and 4 °C for 15 min. Afterward, cell pellets were washed three times with cold S30 Buffer (10 mM Tris-acetate, 14 mM magnesium acetate, 60 mM potassium acetate, and 2 mM dithiothreitol (DTT)). After that, the pellet was resuspended in S30 Buffer (1 mL per gram of wet cell biomass) and lysed by sonication (10 s on/off, 50% of amplitude). The lysate was then centrifuged twice at 12,000 g and 4 °C for 10 min. The resulting supernatant was flash frozen in liquid nitrogen and stored at −80 °C until use.
2.3. Cell-free protein synthesis (CFPS) and Western-blot analysis
A standard 15 μL CFPS reaction was incubated at 30 °C with the following reagents: 12 mM magnesium l-glutamate, 10 mM ammonium l-glutamate, 130 mM potassium l-glutamate, 1.2 mM ATP, 0.85 mM each of GTP, UTP, and CTP, 34 μg/mL folinic acid, 170 μg/mL of E. coli tRNA mixture, 2 mM each of 20 standard amino acids, 0.33 mM nicotinamide adenine dinucleotide (NAD), 0.27 mM coenzyme A (CoA), 1.5 mM spermidine, 1 mM putrescine, 4 mM sodium oxalate, 33 mM phosphoenolpyruvate (PEP), 13.3 μg/mL plasmid, and 27% (v/v) of cell extract.
After CFPS reactions, all cell-free expressed proteins were analyzed by Western-blot. To do this, the reaction mixture was first centrifuged at 4 °C and 12,000 g for 10 min, and the supernatant was collected as soluble fractions. Then, 10 μL of total protein (without centrifugation) and 10 μL of soluble protein (supernatant) samples were mixed with 10 μL of 2x loading buffer, respectively, and heated at 98 °C for 10 min. Afterward, 10 μL of each heated sample was loaded to SDS-PAGE gel for protein separation, followed by wet transferring to PVDF membrane (Bio-Rad) with 1x transfer buffer (25 mM Tris-HCl, 192 mM glycine, and 20% methanol, pH 8.3). Then, the PVDF membrane was blocked with Protein Free Rapid Blocking Buffer (EpiZyme) for 30 min at room temperature. After washing thrice with 1x TBST buffer (10 mM Tris-HCl, 150 mM NaCl, and 0.1% Tween 20, pH 7.5) for each 5 min, 1:10000 TBST buffer-diluted His-Tag Mouse Monoclonal Antibody (Proteintech) solution was added to the membrane and incubated for 1 h at room temperature. After washing again with 1x TBST buffer thrice for each 5 min, 1:10000 TBST buffer-diluted HRP-Goat Anti-Mouse IgG (H + L) Antibody (Proteintech) solution was added to the membrane and incubated for another 1 h at room temperature. After the last washing with TBST thrice for each 5 min, the PVDF membrane was visualized using Omni ECL reagent (EpiZyme) under UVP ChemStudio (analytikjena). Finally, protein band densities were analyzed by ImageJ, which were then used to determine the percent solubility (%) of target proteins (soluble protein/total protein).
2.4. Cell-free biosynthesis of L-4-nitro-Trp
To synthesize L-4-nitro-Trp, two enzymes of TxtD (EC:1.14.14.47; UniProt Entry No. Q6XXZ5; an oxidoreductase) and TxtE (UniProt Entry No. A0A124C0F9; a monooxygenase) were employed to construct the biosynthetic pathway in vitro. Cell-free synthesis of L-4-nitro-Trp was carried out at a 180 μL reaction volume in 2 mL microcentrifuge tubes. The reactions were incubated at 30 °C with shaking at 600 rpm, including the protein synthesis phase and the L-4-nitro-Trp formation phase. For protein expression, the final concentrations of all CFPS reaction components were the same as described in the section of “2.3. Cell-free protein synthesis (CFPS) and Western-blot analysis”. The production of L-4-nitro-Trp was performed by two approaches. In the first one-step method, TxtD and TxtE were coexpressed in one pot with a reaction volume of 180 μL, allowing for simultaneously the enzyme expression and the L-4-nitro-Trp formation. The total reaction time was 16 h for this production format. In the second two-step method, TxtD and TxtE were initially expressed separately and the volume of each CFPS reaction was 90 μL. After protein expression for 5 h, the two reactions were mixed for another 16 h incubation for L-4-nitro-Trp production (note that after mixing the total reaction volume was 180 μL). Unless otherwise noted, the substrates and related cofactors were added to cell-free reactions with the following final concentrations: 1 mM L-Arg, 1 mM L-Trp, 1 mM Fld-FldR (a molar ratio of 1:1), and 0.5 mM NADPH. When cell-free biosynthesis finished, L-4-nitro-Trp was extracted and analyzed by LC-MS (see “2.5. L-4-nitro-Trp extraction and quantification” for details).
2.5. L-4-nitro-Trp extraction and quantification
After cell-free biosynthesis, an equal volume of 100% methanol (i.e., 180 μL methanol per reaction) was added to the reaction mixture to terminate the cell-free reaction. The resulting solution was mixed thoroughly for 1 min by vortex (VORTEX-GENIE2, Scientific Industries). Afterward, the mixture was centrifuged for 15 min at 21,000 g and the resulting supernatant was directly used for product analysis by LC-MS. All experiments were repeated three times.
L-4-nitro-Trp quantification was performed using an Agilent 1260 LC-MS System equipped with an Agilent Proshell 120 EC-C18 column (3.0 × 50 mm, 2.7 μm) coupled with a ThermoFisher Orbitrap Elite system. For L-4-nitro-Trp detection, 2 μL of each sample was injected and separated at a flow rate of 0.3 mL/min and 40 °C column temperature. Water (A) and acetonitrile (B) each containing 0.1% formic acid were used as mobile phases with a linear gradient program (10–90% solvent B) for 15 min to separate chemicals. A pre-wash phase of 10 min with 10% solvent B was added at the beginning of each run. For MS/MS analysis, MS was acquired under a Full-Scan mode in a mass range of m/z 150–1000. Fragmentation was introduced by high energy collision-induced dissociation (HCD) technique with normalized collision energy at 35 eV. MS/MS resolution was 15,000 and the scan range was 50–300.
3. Results and discussion
3.1. Selection of CFPS systems
Recently, with the demand for rapid synthesis of proteins in vitro, several new CFPS systems have been developed from different types of organisms (e.g., Bacillus subtilis, Pseudomonas putida, Streptomyces lividans, Vibrio natriegens, and Pichia pastoris) [[32], [33], [34], [35], [36], [37]], which can better mimic their cellular physicochemical environment for protein expression. For example, the Streptomyces-based CFPS system shows benefits for the expression of high GC-content genes with enhanced protein solubility [34]. To select a suitable cell-free system for TxtD and TxtE expression, we started to evaluate two CFPS systems derived from E. coli and Streptomyces lividans [27,34]. We chose these two platforms on the basis of two main reasons. First, the E. coli-based CFPS system has been well established with high protein productivity. Second, the genes of txtD and txtE are originated from Streptomyces with high GC-content that should be favored by the Streptomyces CFPS system.
Initially, two strains E. coli BL21 Star(DE3) and S. lividans TK24 were used for cell extract preparation to perform CFPS reactions. For protein expression, the plasmids pJL1-TxtD and pJL1-TxtE were used as DNA templates. The reaction temperatures for E. coli and Streptomyces CFPS systems were set at 30 °C and 23 °C, respectively, the optimal temperature for each system as reported previously [29,34]. Then, individual expression of the two proteins in both cell-free systems was carried out for 8 h to eliminate the effect of reaction time on protein synthesis. After CFPS reactions, the samples were analyzed by Western-blot. The results indicated that TxtD and TxtE could be expressed in both CFPS systems (Fig. 2B). However, most of the proteins (especially TxtE) were formed in insoluble fractions in the E. coli BL21 Star(DE3)-based CFPS reactions. By contrast, while the protein solubility was increased in the Streptomyces CFPS system, the total protein expression levels were significantly lower than those in the E. coli system. This result is in agreement with the previous report that Streptomyces CFPS system can enhance the solubility of Streptomyces-originated proteins without chaperone supplementation [34], perhaps as a result of the similarity of cellular physicochemical and biological environments among different Streptomyces species. Recently, more efforts have been performed to improve the productivity of Streptomyces CFPS [35,38]. Hopefully, further enhanced Streptomyces cell-free systems will be used as a potential platform for the expression of many other GC-rich gene clusters directly amplified from Streptomyces genomes for natural product biosynthesis. In this work, based on the total protein expression levels, we decided to choose the high-yield E. coli cell-free system for TxtD and TxtE expression but firstly with the main focus on the improvement of protein solubility.
Molecular chaperones have been demonstrated to help facilitate protein folding and block protein aggregation, which are often coexpressed in vivo to improve the solubility of target proteins [[39], [40], [41]]. To take advantage of chaperones, we next aimed to prepare cell extracts enriched with chaperone proteins that might be beneficial for enhancing the solubility of cell-free expressed proteins in vitro. To this end, we first transformed different molecular chaperone plasmids into the chassis strain E. coli BL21 Star(DE3), which was then used for cell extract preparation. Using chaperones-enriched cell extracts (CE8: DnaK-DnaJ-GrpE + GroES-GroEL; CE2: GroES-GroEL-tig; CE7: GroES-GroEL), TxtD and TxtE were expressed separately and then analyzed by Western-blot. As shown in Fig. 2C, CE8 and CE2 performed better and expressed notably higher soluble proteins than CE7 and the control cell extracts without any chaperones. For example, according to the protein band density analysis by ImageJ, the percent solubility of TxtD expressed in CE8 and CE2 was 86% and 96%, respectively, both of which are higher than that of TxtD expressed in CE7 (42%). In addition, CE8-based CFPS reactions generated the most amount of soluble TxtD and TxtE among all tested chaperones. The amount of soluble TxtD and TxtE in CE8-based CFPS reactions was 233% and 115% higher, respectively, as compared to the CE2-based protein expression. Our results demonstrated that chaperones indeed helped increase protein solubility albeit to different levels in CFPS reactions. Overall, the combination of DnaK and GroEL chaperone systems performed the best in our work, which is in agreement with previous reports that these two chaperone systems can significantly increase the solubility of aggregation-prone proteins expressed in cell-free systems [42]. Since the property of each protein is naturally different, how to choose chaperones for the soluble expression of proteins (if only insoluble without chaperones) is in principle dependent, and we believe that CFPS systems might be a robust platform for rapid evaluation. Taken together, our cell-free platform enables soluble expression of TxtD and TxtE by supplying reasonable chaperones. This is significant because (i) soluble expression of TxtD in vivo was difficult as reported previously [19], but it can be easily expressed in CFPS with high solubility. Moreover, our results might also suggest a reasonable solution for others to express soluble TxtD proteins in vivo by coexpression of chaperones; and (ii) most importantly, soluble TxtD and TxtE synthesized in the cell-free system should be active to catalyze the formation of the target compound L-4-nitro-Trp. Finally, CE8-based cell extracts were selected for the following experiments because of its capability for high-level expression of soluble proteins in vitro. Meanwhile, we analyzed the protein synthesis profiles of TxtD and TxtE over 8 h in the E. coli CFPS system using CE8 cell extracts. The results indicated that both proteins could be synthesized for 8 h, although approximately 80% of the two proteins were synthesized during the initial 3–4 h (Fig. 3).
Fig. 3.
Protein synthesis profile over 8 h in E. coli CFPS reactions using CE8 cell extracts. Western-blot analysis of (A) TxtD and (B) TxtE. Time courses of (C) TxtD and (D) TxtE synthesis. The protein band densities in (A, B) were analyzed by ImageJ and then used for calculating the relative expression in (C, D). The protein band densities of TxtD and TxtE at 8 h were set as 100%, respectively, for the calculation of relative expression (%).
3.2. Construction of L-4-nitro-Trp biosynthetic pathway in vitro
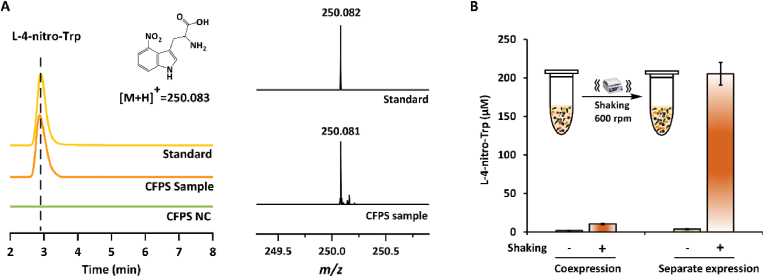
Having demonstrated the soluble expression of TxtD and TxtE in CFPS, we next sought to use the two enzymes to construct the L-4-nitro-Trp biosynthetic pathway in vitro. We initially compared the biosynthesis in two ways. First, TxtD and TxtE were coexpressed in one-pot for L-4-nitro-Trp biosynthesis. Second, CFPS reactions were performed to express TxtD and TxtE separately and then the reactions were mixed to reconstitute the full metabolic pathway. After cell-free biosynthesis, the samples were analyzed by LC-MS. The results suggested that L-4-nitro-Trp could be successfully detected in both reaction approaches with two enzymes either coexpressed or separately expressed and then mixed (see Fig. 4A for a representative LC-MS analysis). During the process of L-Trp nitration, NO and O2 are required by TxtE as cosubstrates [13]. Given that NO can be generated from L-Arg by the pathway-specific NO synthase TxtD, the amount of O2 in the reaction mixture is hypothesized to be another important factor for the biocatalysis. Therefore, we attempted to supply sufficient O2 in CFPS reactions, which might be helpful for the formation of L-4-nitro-Trp. To do this, all reactions were shaken at 600 rpm during the biosynthesis. As shown in Fig. 4B, the reaction format significantly impacted the product formation. On the one hand, the two-stage biosynthesis (i.e., enzymes first expressed for 5 h and then mixed for biocatalysis) worked much better than the one-pot reaction by coexpression of TxtD and TxtE. On the other hand, shaking could remarkably improve the productivity. For example, the product concentration in the two-stage reaction was increased by more than 50 times as compared to the control without shaking. Overall, with shaking the yield of L-4-nitro-Trp in the two-stage biosynthesis was >20-fold higher than the one-pot coexpression reaction. Taken together, our data suggested that O2 is a limiting factor and shaking can provide more O2 for the enzymatic (TxtE) catalysis. In addition, the low L-4-nitro-Trp production in the one-pot reaction is likely due to resource limitations (e.g., energy and substrates) caused by coexpression of two enzymes. This constraint can be circumvented by separate expression of TxtD and TxtE in advance and then mixed them for product synthesis. Therefore, next we set out to use the two-stage reaction format with shaking to further optimize the production of L-4-nitro-Trp in vitro.
Fig. 4.
Cell-free biosynthesis of L-4-nitro-Trp. (A) LC-MS analysis of L-4-nitro-Trp produced by the CFPS reaction. (B) Effects of different cell-free reaction formats on L-4-nitro-Trp production. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
3.3. Optimization of L-4-nitro-Trp biosynthesis
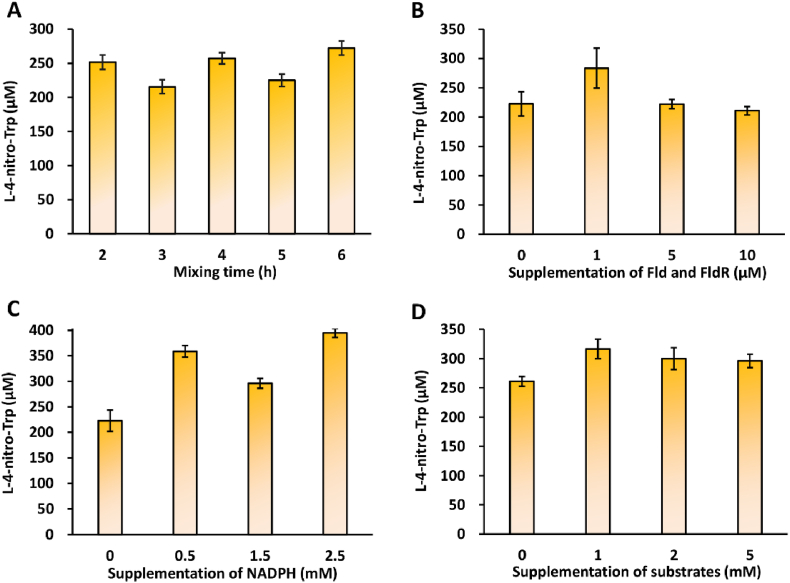
To optimize cell-free biosynthesis of L-4-nitro-Trp, we wanted to evaluate some key factors that might influence the efficiency of enzymatic catalysis. Since enzymes are crucial biocatalysts for the product formation, we first investigated the effect of enzyme expression time before mixing them on product synthesis. To do this, cell-free expression of TxtD and TxtE was carried out separately ranging from 2 to 6 h and then the reactions were mixed for another 16 h incubation. We started the evaluation from 2 h because the expression levels of the two enzymes before 2 h were low that might not be sufficient to implement the biosynthesis (Fig. 3). As shown in Fig. 5A, the L-4-nitro-Trp yields in different groups were comparable, demonstrating that at least 2-h of CFPS was sufficient to synthesize enough enzymes for the catalysis. Further increase of CFPS reaction time did not significantly improve the product yield, perhaps as a result of other limiting factors in the reaction. This result is similar to a previous report that 2-h CFPS reaction was the best for enzyme expression before mixing and the resultant bioconversion [30].
Fig. 5.
Optimization of L-4-nitro-Trp production. (A) Effect of cell-free enzyme (TxtD and TxtE) expression time before mixing them on product synthesis. Effects of supplementation of (B) Fld-FldR (molar ratio of 1:1), (C) NADHP, and (D) substrates (L-Arg and L-Trp, molar ratio of 1:1) on L-4-nitro-Trp formation. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
Like many other microbial cytochrome P450 enzymes, TxtE requires an electron transfer system for its catalytic activity, which can be supported by, for example, the surrogate redox partners of spinach ferredoxin and ferredoxin reductase [13]. In addition, previous studies also have demonstrated that two flavoproteins flavodoxin (Fld) and NADPH-flavodoxin reductase (FldR) from E. coli are capable of transferring electrons to different source P450 enzymes [[43], [44], [45]]. Because our CFPS system is based on E. coli cell lysates, the endogenous Fld-FldR can directly serve as an electron transfer system for supporting the activity of cell-free expressed TxtE to synthesize L-4-nitro-Trp (Fig. 4A). However, we were curious to know if the TxtE activity could be further boosted by supplementation of purified Fld-FldR proteins to the cell-free reactions. We, therefore, added different amounts of purified Fld and FldR with a molar ratio of 1:1 to the mixed reactions. We found that when the cell-free reaction was supplemented with 1 μM of Fld-FldR, the yield of L-4-nitro-Trp could be increased by 27% as compared to the reaction without Fld-FldR addition (Fig. 5B). However, adding more Fld-FldR proteins (5 and 10 μM) slightly reduced the product yields. Meanwhile, we wanted to supplement NADPH, which is also required by TxtE for its reaction [13], to the cell-free system and explore the effect of NADPH concentration on product formation. As shown in Fig. 5C, with the addition of NADPH, the L-4-nitro-Trp yields obviously increased and 2.5 mM NADPH gave rise to the highest yield of nearly 400 μM. We also observed that more than 200 μM of L-4-nitro-Trp were produced without supplementing NADPH. This is because E. coli cell lysates in the cell-free system can provide endogenous NADPH for supporting the activity of TxtE, which is in line with the previous report using E. coli cells for TxtE expression and catalysis [19].
Next, we sought to investigate the effect of substrate (L-Arg and L-Trp) concentration on the synthesis of L-4-nitro-Trp. To do so, we mixed two CFPS reactions with expressed TxtD and TxtE and added substrates (the molar ratio of L-Arg and L-Trp is 1:1) to the reaction mixtures at different final concentrations from 1 to 5 mM. Our data indicated that the product yields were similar with different substrate concentrations, although 1 mM of L-Arg and L-Trp (1:1) gave rise to the highest production (316 μM), which is only 20% higher than the reaction without substrate supplementation (Fig. 5D). This suggested that L-Arg and L-Trp, which were added at 2 mM for protein expression in CFPS reactions, were not completely consumed and the remaining amino acids were sufficient to synthesize a notable amount of L-4-nitro-Trp (261 μM) under the current conditions.
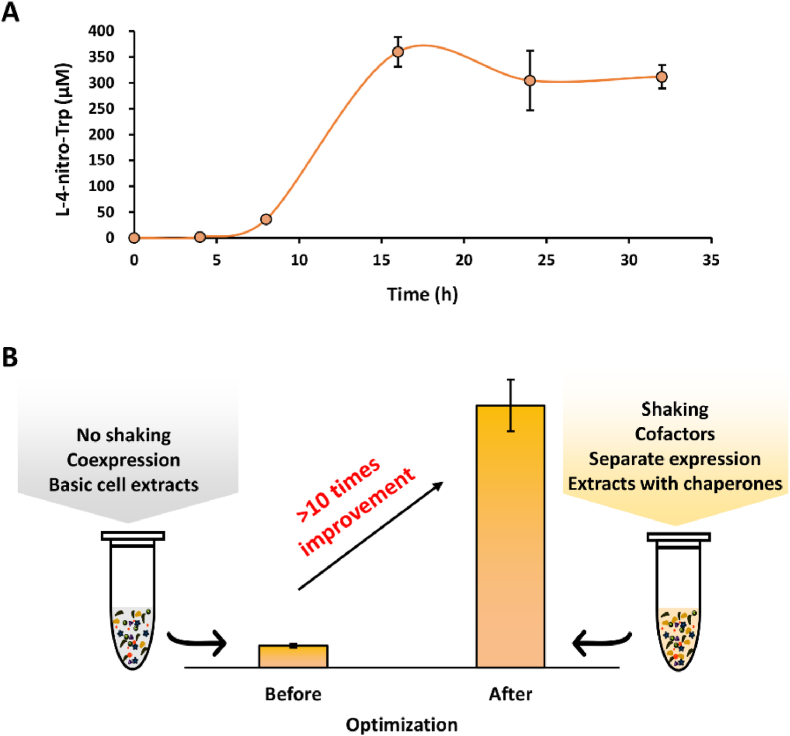
Finally, using the optimized cell-free biosynthesis system - note that 0.5 mM of NADPH (but not 2.5 mM) was used in the reaction as this concentration was sufficient to support high production (Fig. 5C), we tested the time course of product formation by measuring L-4-nitro-Trp concentrations at different time points (Fig. 6A). After TxtD- and TxtE-enriched CFPS were mixed, the cell-free reactions were carried out for 32 h in total. The results indicated that product synthesis in the initial 8 h was slow. While the reason remains unclear, this is likely in part due to the following possibilities: (i) the redox balance between enzymatic reactions and cofactors (e.g., Fld-FldR, NADPH, etc.) is not fully achieved yet [12,13]; (ii) TxtD needs more time to generate a sufficient amount of NO for TxtE to catalyze the nitration of L-Trp; and (iii) O2 might also be a limited substrate for TxtE at the beginning, which has to be slowly transferred into the reaction mixture by shaking. The product accumulation rate rapidly increased during the next 8 h with a maximum yield of ∼360 μM at 16 h. Subsequently, the yields slightly decreased, which is probably due to the degradation of L-4-nitro-Trp by potential E. coli endogenous enzymes (e.g., tryptophanase and nitroreductase) as observed in the cell-based production system [19].
Fig. 6.
Cell-free biosynthesis of L-4-nitro-Trp in the optimized reaction system. (A) Time course of L-4-nitro-Trp production within 32 h. (B) Comparison of L-4-nitro-Trp production before and after system optimization. Values show means with error bars representing standard deviations (S.D.) of at least three independent experiments.
Overall, our final, optimized cell-free system was capable of synthesizing nearly 360 μM of L-4-nitro-Trp, which is more than 10 times higher than the initial system without optimization (Fig. 6B). Our cell-free biosynthesis system has several key features. First, CFPS enables the active expression of TxtD in this work, which cannot be expressed in full-length soluble form in vivo [12,19]. This is in agreement with other in vivo “difficult-to-express” proteins like metalloproteins, which, by contrast, can be highly and actively expressed in CFPS [46]. In addition to the E. coli-based CFPS system, many other CFPS platforms [[32], [33], [34], [35], [36], [37]] may be also available for the expression of enzymes with high (soluble) yields and high activity to reconstitute the related biosynthetic pathways. Second, cell-free systems are flexible to be optimized due to their open reaction environment. For example, the reaction format and cofactor supplementation can be readily manipulated to achieve high productivity. Previous studies also have demonstrated the flexibility of cell-free systems by designing modular CFPS reactions, expressing enzymes in separate modules, and eventually mixing cell-free modules for long metabolic pathway construction and product formation [30,47]. Third, cell-free systems without the use of living cells are tolerant to toxic compounds such as the cellular toxic NO if it is highly accumulated in cells. This robustness feature of cell-free systems has also been shown previously with the production of cellular toxic styrene as well as the application of CFPS in an aqueous-organic biphasic system for cascade biotransformation [29,31]. Taken together, we believe that cell-free systems can be rationally selected, designed, and optimized to increase the biosynthesis performance and as a result lead to high productivity.
4. Conclusion
With the development of cell-free biotechnology, cell-free systems are emerging as powerful platforms not only for protein synthesis but also for many synthetic biology applications. In this context, cell-free strategies are creating a new frontier for next-generation biosynthesis through the rapid and rational construction of in vitro metabolic pathways [[25], [26], [27], [28], [29], [30], [31]]. To further expand the scope (type) of proteins and small high-value compounds that can be produced by cell-free systems, here we demonstrated cell-free biosynthesis of a nitro-containing compound L-4-nitro-Trp by in vitro reconstitution of the native biosynthetic pathway. Specifically, an E. coli-based CFPS system enriched with molecular chaperones was used to express active NO synthase (TxtD) and P450 enzyme (TxtE) for L-4-nitro-Trp biosynthesis. After optimization, the cell-free system was able to synthesize about 360 μM of L-4-nitro-Trp, which is a >10-fold improvement as compared to the initial system without optimization. Looking forward, we anticipate that cell-free biosynthetic platforms will play an important role in the production of various valuable compounds as showcased recently, including, for example, fine chemicals, pharmaceutical intermediates, food-related amino acids, and complex natural products [[25], [26], [27], [28], [29], [30], [31]]. In addition, sustainable production of such compounds in large amounts might also be possible in the near future as the E. coli CFPS system has been shown to scale up linearly from 250 μL to 100 L reaction volumes [48]. Taken together, given the flexibility of cell-free systems, biosynthesis and biomanufacturing beyond the cell holds tremendous potential to create cost-effective, efficient, and high-yield in vitro biosynthetic factories for the production of chemicals, therapeutics, and materials.
CRediT authorship contribution statement
Xintong Tian: Investigation, Methodology, Visualization, Writing – original draft. Wan-Qiu Liu: Investigation, Methodology. Huiling Xu: Methodology, Validation. Xiangyang Ji: Methodology. Yushi Liu: Methodology. Jian Li: Conceptualization, Supervision, Funding acquisition, Resources, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 31971348 and 32171427) and the Natural Science Foundation of Shanghai (No. 19ZR1477200). J.L. also acknowledges the starting grant from ShanghaiTech University.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Ju K.S., Parales R.E. Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev. 2010;74:250–272. doi: 10.1128/MMBR.00006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan G., Yang M. Recent advances in the synthesis of aromatic nitro compounds. Org Biomol Chem. 2013;11:2554–2566. doi: 10.1039/c3ob27354g. [DOI] [PubMed] [Google Scholar]
- 3.Winkler R., Hertweck C. Biosynthesis of nitro compounds. Chembiochem. 2007;8:973–977. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 4.Tomita H., Katsuyama Y., Minami H., Ohnishi H. Identification and characterization of a bacterial cytochrome P450 monooxygenase catalyzing the 3-nitration of tyrosine in rufomycin biosynthesis. J Biol Chem. 2017;292:15859–15869. doi: 10.1074/jbc.M117.791269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kers J.A., Cameron K.D., Joshi M.V., Bukhalid R.A., Morello J.E., Wach M.J., Gibson D.M., Loria R. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol. 2005;55:1025–1033. doi: 10.1111/j.1365-2958.2004.04461.x. [DOI] [PubMed] [Google Scholar]
- 6.Loria R., Bignell D.R.D., Moll S., Huguet-Tapia J.C., Joshi M.V., Johnson E.G., Seipke R.F., Gibson D.M. Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces. Antonie Leeuwenhoek. 2008;94:3–10. doi: 10.1007/s10482-008-9240-4. [DOI] [PubMed] [Google Scholar]
- 7.Fry B.A., Loria R. Thaxtomin A: evidence for a plant cell wall target. Physiol Mol Plant Pathol. 2002;60:1–8. [Google Scholar]
- 8.Loria R., Kers J., Joshi M. Evolution of plant pathogenicity in Streptomyces. Annu Rev Phytopathol. 2006;44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PubMed] [Google Scholar]
- 9.King R.R., Lawrence C.H., Gray J.A. Herbicidal properties of the thaxtomin group of phytotoxins. J Agric Food Chem. 2001;49:2298–2301. doi: 10.1021/jf0012998. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H., Wang Q., Ning X., Hang H., Ma J., Yang X., Lu X., Zhang J., Li Y., Niu C., Song H., Wang X., Wang P.G. Synthesis and biological evaluations of a series of thaxtomin analogues. J Agric Food Chem. 2015;63:3734–3741. doi: 10.1021/jf506153t. [DOI] [PubMed] [Google Scholar]
- 11.Chio C.M., Huang Y.C., Chou Y.C., Hsu F.C., Lai Y.B., Yu C.S. Boron accumulation in brain tumor cells through boc-protected tryptophan as a carrier for boron neutron capture therapy. ACS Med Chem Lett. 2020;11:589–596. doi: 10.1021/acsmedchemlett.0c00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kers J.A., Wach M.J., Krasnoff S.B., Widom J., Cameron K.D., Bukhalid R.A., Gibson D.M., Crane B.R., Loria R. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 13.Barry S.M., Kers J.A., Johnson E.G., Song L., Aston P.R., Patel B., Krasnoff S.B., Crane B.R., Gibson D.M., Loria R., Challis G.L. Cytochrome P450-catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat Chem Biol. 2012;8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson E.G., Krasnoff S.B., Bignell D.R.D., Chung W.C., Tao T., Parry R.J., Loria R., Gibson D.M. 4-Nitrotryptophan is a substrate for the non-ribosomal peptide synthetase TxtB in the thaxtomin A biosynthetic pathway. Mol Microbiol. 2009;73:409–418. doi: 10.1111/j.1365-2958.2009.06780.x. [DOI] [PubMed] [Google Scholar]
- 15.Caranto J.D. The emergence of nitric oxide in the biosynthesis of bacterial natural products. Curr Opin Chem Biol. 2019;49:130–138. doi: 10.1016/j.cbpa.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 16.Dodani S.C., Kiss G., Cahn J.K.B., Su Y., Pande V.S., Arnold F.H. Discovery of a regioselectivity switch in nitrating P450s guided by molecular dynamics simulations and Markov models. Nat Chem. 2016;8:419–425. doi: 10.1038/nchem.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuo R., Zhang Y., Huguet-Tapia J.C., Mehta M., Dedic E., Bruner S.D., Loria R., Ding Y. An artificial self-sufficient cytochrome P450 directly nitrates fluorinated tryptophan analogs with a different regio-selectivity. Biotechnol J. 2016;11:624–632. doi: 10.1002/biot.201500416. [DOI] [PubMed] [Google Scholar]
- 18.Zuo R., Zhang Y., Jiang C., Hackett J.C., Loria R., Bruner S.D., Ding Y. Engineered P450 biocatalysts show improved activity and regio-promiscuity in aromatic nitration. Sci Rep. 2017;7:842. doi: 10.1038/s41598-017-00897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zuo R., Ding Y. Direct aromatic nitration systems for synthesis of nitrotryptophans in Escherichia coli. ACS Synth Biol. 2019;8:857–865. doi: 10.1021/acssynbio.8b00534. [DOI] [PubMed] [Google Scholar]
- 20.Swartz J.R. Expanding biological applications using cell-free metabolic engineering: an overview. Metab Eng. 2018;50:156–172. doi: 10.1016/j.ymben.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Liu W.Q., Zhang L., Chen M., Li J. Cell-free protein synthesis: recent advances in bacterial extract sources and expanded applications. Biochem Eng J. 2019;141:182–189. [Google Scholar]
- 22.Silverman A.D., Karim A.S., Jewett M.C. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21:151–170. doi: 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 23.Copeland C.E., Langlois A., Kim J., Kwon Y.C. The cell-free system: a new apparatus for affordable, sensitive, and portable healthcare. Biochem Eng J. 2021;175:108124. [Google Scholar]
- 24.Brookwell A., Oza J.P., Caschera F. Biotechnology applications of cell-free expression systems. Life. 2021;11:1367. doi: 10.3390/life11121367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelwick R., Ricci L., Chee S.M., Bell D., Webb A.J., Freemont P.S. Cell-free prototyping strategies for enhancing the sustainable production of polyhydroxyalkanoates bioplastics. Synth Biol. 2018;3:ysy016. doi: 10.1093/synbio/ysy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karim A.S., Dudley Q.M., Juminaga A., Yuan Y., Crowe S.A., Heggestad J.T., Garg S., Abdalla T., Grubbe W.S., Rasor B.J., Coar D.N., Torculas M., Krein M., Liew F.E., Quattlebaum A., Jensen R.O., Stuart J.A., Simpson S.D., Köpke M., Jewett M.C. In vitro prototyping and rapid optimization of biosynthetic enzymes for cell design. Nat Chem Biol. 2020;16:912–919. doi: 10.1038/s41589-020-0559-0. [DOI] [PubMed] [Google Scholar]
- 27.Zhuang L., Huang S., Liu W.Q., Karim A.S., Jewett M.C., Li J. Total in vitro biosynthesis of the nonribosomal macrolactone peptide valinomycin. Metab Eng. 2020;60:37–44. doi: 10.1016/j.ymben.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng J., Yang C., Zhao Z., Xu J., Li J., Li P. Application of cell-free protein synthesis system for the biosynthesis of L-theanine. ACS Synth Biol. 2021;10:620–631. doi: 10.1021/acssynbio.0c00618. [DOI] [PubMed] [Google Scholar]
- 29.Liu W.Q., Wu C., Jewett M.C., Li J. Cell-free protein synthesis enables one-pot cascade biotransformation in an aqueous-organic biphasic system. Biotechnol Bioeng. 2020;117:4001–4008. doi: 10.1002/bit.27541. [DOI] [PubMed] [Google Scholar]
- 30.Yang C., Liu Y., Liu W.Q., Wu C., Li J. Designing modular cell-free systems for tunable biotransformation of L-phenylalanine to aromatic compounds. Front Bioeng Biotechnol. 2021;9:730663. doi: 10.3389/fbioe.2021.730663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grubbe W.S., Rasor B.J., Krüger A., Jewett M.C., Karim A.S. Cell-free styrene biosynthesis at high titers. Metab Eng. 2020;61:89–95. doi: 10.1016/j.ymben.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 32.Kelwick R., Webb A.J., MacDonald J.T., Freemont P.S. Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab Eng. 2016;38:370–381. doi: 10.1016/j.ymben.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., Li J., Jewett M.C. Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth Biol. 2018;3:ysy003. doi: 10.1093/synbio/ysy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Wang H., Kwon Y.C., Jewett M.C. Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol Bioeng. 2017;114:1343–1353. doi: 10.1002/bit.26253. [DOI] [PubMed] [Google Scholar]
- 35.Xu H., Liu W.Q., Li J. Translation related factors improve the productivity of a Streptomyces-based cell-free protein synthesis system. ACS Synth Biol. 2020;9:1221–1224. doi: 10.1021/acssynbio.0c00140. [DOI] [PubMed] [Google Scholar]
- 36.Des Soye B.J., Davidson S.R., Weinstock M.T., Gibson D.G., Jewett M.C. Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth Biol. 2018;7:2245–2255. doi: 10.1021/acssynbio.8b00252. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L., Liu W.Q., Li J. Establishing a eukaryotic Pichia pastoris cell-free protein synthesis system. Front Bioeng Biotechnol. 2020;8:536. doi: 10.3389/fbioe.2020.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H., Yang C., Tian X., Chen Y., Liu W.Q., Li J. Regulatory part engineering for high-yield protein synthesis in an all-Streptomyces-based cell-free expression system. ACS Synth Biol. 2022;11:570–578. doi: 10.1021/acssynbio.1c00587. [DOI] [PubMed] [Google Scholar]
- 39.Thomas J.G., Ayling A., Baneyx F. Molecular chaperones, folding catalysts, and the recovery of active recombinant proteins from E. coli. Appl Biochem Biotechnol. 1997;66:197–238. doi: 10.1007/BF02785589. [DOI] [PubMed] [Google Scholar]
- 40.Nishihara K., Kanemori M., Kitagawa M., Yanagi H., Yura T. Chaperone coexpression plasmids: differential and synergistic roles of Dnak-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol. 1998;64:1694–1699. doi: 10.1128/aem.64.5.1694-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishihara K., Kanemori M., Yanagi H., Yura T. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol. 2000;66:884–889. doi: 10.1128/aem.66.3.884-889.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niwa T., Kanamori T., Ueda T., Taguchi H. Global analysis of chaperone effects using a reconstituted cell-free translation system. Proc Natl Acad Sci USA. 2012;109:8937–8942. doi: 10.1073/pnas.1201380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins C.M., Waterman M.R. Flavodoxin and NADPH-flavodoxin reductase from Escherichia coli support bovine cytochrome P450c17 hydroxylase activities. J Biol Chem. 1994;269:27401–27408. [PubMed] [Google Scholar]
- 44.Jenkins C.M., Waterman M.R. NADPH−flavodoxin reductase and flavodoxin from Escherichia coli: characteristics as a soluble microsomal P450 reductase. Biochemistry. 1998;37:6106–6113. doi: 10.1021/bi973076p. [DOI] [PubMed] [Google Scholar]
- 45.Girhard M., Schuster S., Dietrich M., Dürre P., Urlacher V.B. Cytochrome P450 monooxygenase from Clostridium acetobutylicum: a new α-fatty acid hydroxylase. Biochem Biophys Res Commun. 2007;362:114–119. doi: 10.1016/j.bbrc.2007.07.155. [DOI] [PubMed] [Google Scholar]
- 46.Li J., Lawton T.J., Kostecki J.S., Nisthal A., Fang J., Mayo S.L., Rosenzweig A.C., Jewett M.C. Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol J. 2016;11:212–218. doi: 10.1002/biot.201500030. [DOI] [PubMed] [Google Scholar]
- 47.Dudley Q.M., Karim A.S., Nash C.J., Jewett M.C. In vitro prototyping of limonene biosynthesis using cell-free protein synthesis. Metab Eng. 2020;61:251–260. doi: 10.1016/j.ymben.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Zawada J.F., Yin G., Steiner A.R., Yang J., Naresh A., Roy S.M., Gold D.S., Heinsohn H.G., Murray C.J. Microscale to manufacturing scale-up of cell-free cytokine production − a new approach for shortening protein production development timelines. Biotechnol Bioeng. 2011;108:1570–1578. doi: 10.1002/bit.23103. [DOI] [PMC free article] [PubMed] [Google Scholar]