Abstract
RNA polymerase (RNAP) binding protein A (RbpA) is essential for mycobacterial viability and regulates transcription initiation by increasing the stability of the RNAP-promoter open complex (RPo). RbpA consists of four domains: an N-terminal tail (NTT), a core domain (CD), a basic linker, and a sigma interaction domain. We have previously shown that truncation of the RbpA NTT and CD increases RPo stabilization by RbpA, implying that these domains inhibit this activity of RbpA. Previously published structural studies showed that the NTT and CD are positioned near multiple RNAP-σA holoenzyme functional domains and predict that the RbpA NTT contributes specific amino acids to the binding site of the antibiotic fidaxomicin (Fdx), which inhibits the formation of the RPo complex. Furthermore, deletion of the NTT results in decreased Mycobacterium smegmatis sensitivity to Fdx, but whether this is caused by a loss in Fdx binding is unknown. We generated a panel of rbpA mutants and found that the RbpA NTT residues predicted to directly interact with Fdx are partially responsible for RbpA-dependent Fdx activity in vitro, while multiple additional RbpA domains contribute to Fdx activity in vivo. Specifically, our results suggest that the RPo-stabilizing activity of RbpA decreases Fdx activity in vivo. In support of the association between RPo stability and Fdx activity, we find that another factor that promotes RPo stability in bacteria, CarD, also impacts to Fdx sensitivity. Our findings highlight how RbpA and other factors may influence RNAP dynamics to affect Fdx sensitivity.
Keywords: RNA polymerase, mycobacteria, bacterial transcription, antibiotic action, bacterial genetics, RbpA, fidaxomicin
Abbreviations: BL, basic linker; CD, core domain; DMSO, dimethyl sulfoxide; Fdx, fidaxomicin; NTT, N-terminal tail; RNAP, RNA polymerase; RbpA, RNA polymerase binding protein A; RPo, RNAP-promoter open complex; SID, sigma interaction domain
Mycobacterium tuberculosis is the causative agent of the disease tuberculosis, which resulted in an estimated 1.5 million deaths worldwide in 2019 (https://www.who.int/publications/i/item/9789240013131). New strategies are necessary to fight this global health crisis, including the development of novel therapies. Bacterial transcription is a druggable essential process in M. tuberculosis, demonstrated by the transcription inhibitor rifampicin’s continued status as a cornerstone of tuberculosis treatment. Bacterial transcription is carried out by an RNA polymerase (RNAP) comprised of five subunits (α2ββ’ω), referred to as the core RNAP, and a sixth dissociable subunit (σ) that when bound to core RNAP forms a complex termed the RNAP holoenzyme. Mycobacterial transcription initiation in vivo also requires two additional essential RNAP-interacting proteins, RbpA and CarD (1, 2, 3, 4, 5, 6). RbpA and CarD regulate transcription initiation by binding to the RNAP and modulating the kinetics of RNAP-promoter open complex (RPo) formation and RNAP promoter escape (2, 5, 6, 7, 8, 9, 10).
RbpA is comprised of four structural domains, including the N-terminal tail (NTT), core domain (CD), basic linker (BL), and sigma interaction domain (SID) (4, 10, 11). Most of the characterization of RbpA has focused on the BL and SID. The RbpA SID domain directly interacts with σ region 1.2, σ nonconserved region, and σ region 2.3 in group I (M. tuberculosis σA) and group II (M. tuberculosis σB) σ factors (4, 5, 10, 12, 13, 14). The SID domain is both necessary and sufficient for RbpA to associate with the RNAP holoenzyme (5). An arginine at position 88 in the M. tuberculosis RbpA SID is critical for the interaction with σA and σB (5, 15). The M. tuberculosis RbpA BL contains several positively charged residues, including K73, K74, K76, and R79, that are positioned to interact with the negatively charged DNA phosphate backbone near the upstream edge of RPo (4, 10). Alanine substitution at either R79 in the BL or R88 in the SID has demonstrated that the interactions between RbpA and the RNAP and DNA are necessary for RbpA to increase RPo stability during transcription initiation (5, 10, 14). In vivo, R79A or R88A substitutions in RbpA result in upregulation of some genes and downregulation of other genes, suggesting that the outcome of RbpA activity may be promoter dependent, possibly due to differences in the kinetics of transcription initiation at each promoter (5, 16, 17).
Much less is known about the functions performed by the RbpA NTT and CD. Deletion of the RbpA NTT increases the ability of RbpA to stabilize RPo, and deletion of both the RbpA NTT and CD further increases RPo stability, indicating that both domains antagonize RbpA-mediated stabilization of RPo (5, 10). Structural analysis of RbpA bound to the M. tuberculosis RNAP-σA RPo shows that the RbpA NTT is positioned near the RNA exit channel, possibly contacting the RNAP β switch 3 region (Sw3), β flap, β′ lid, σA region 3.2 (σA3.2, also referred to as the σ “finger” domain), and the β′ zinc binding domain (ZBD), while the RbpA CD is positioned near the RNAP β′ zipper and RNAP β′ ZBD (10, 11). These RNAP structural domains have been characterized to varying levels in Escherichia coli, which lacks RbpA. The RNAP β Sw3 is one of five switch regions that are thought to undergo conformational changes during transcription initiation (18). RNAP β Sw3 is positioned near the template DNA −3 and −4 nucleotides, raising the possibility that RNAP β Sw3 could play a role in DNA template strand positioning (19). The RNAP β flap, which includes the flap tip helix that interacts with σ region 4, is important for positioning σ region 4 for interaction with the −35 element of the promoter (20) and represents a common binding interface for transcription factors that directly interact with σ (21, 22). The RNAP β′ lid separates the RNA/DNA hybrid as part of the RNA exit channel and is required for RPo stability and transcription in E. coli and Thermus aquaticus (23, 24). RNAP σ703.2 plays a role in initiating nucleotide triphosphate binding by positioning the DNA template strand for interaction with −4 and −5 nucleotides of the DNA template strand, which affects abortive transcription and promoter escape (25, 26, 27, 28). Both the RNAP β′ ZBD and β′ zipper facilitate RPo formation on promoters with −35 elements that form weak interactions with σ by making promoter contacts within the spacer region between the −10 and −35 motifs (29, 30).
The positioning of the RbpA NTT and CD near multiple different structural and functional domains of the RNAP-σA holoenzyme implies that the RbpA NTT and CD could impact RNAP activity through a number of mechanisms. However, it is unclear what contacts between the RbpA NTT/CD and the RNAP mediate the antagonism of RPo stability. In addition, structural studies indicate that the RbpA NTT is positioned in the RNAP-σA holoenzyme complex in such a way that it contributes to the binding site for the antibiotic fidaxomicin (Fdx) (11), which is used to treat Clostridium difficile infections. Fdx inhibits transcription initiation by binding the RNAP and blocking the closing of the RNAP clamp that occurs during RPo formation (11, 31). Deletion of the RbpA NTT decreases sensitivity of M. tuberculosis RNAP to Fdx in vitro and in vivo (11), which is proposed to be due to the loss of RbpA’s contribution to the RNAP-Fdx binding interface. However, given that RbpA NTT also decreases RPo stability (5) and is predicted to interact with σA3.2, which is known to affect Fdx activity (11, 32), it is possible that RbpA may impact Fdx activity by additional mechanisms. In this study, we interrogate the roles played by residues within the NTT in RbpA-dependent Fdx sensitivity and find that the amino acids predicted by the structural studies to interact with Fdx do partially contribute to Fdx activity in vitro. However, we also find that RbpA’s impact on Fdx activity in vivo extends beyond the role of the NTT in binding the antibiotic, revealing a dominant contribution for RNAP conformation in Fdx sensitivity.
Results
RbpA E17 and R10 synergize to promote Fdx activity against M. tuberculosis RNAP-σAin vitro
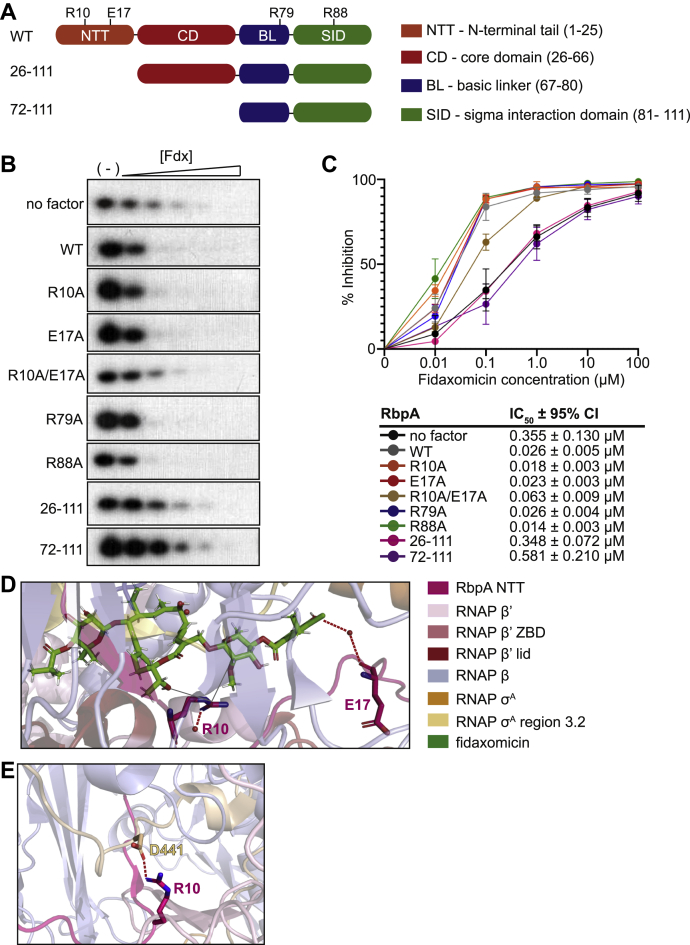
In vitro assays that monitor the production of a 3-nucleotide product as a proxy of RPo stability have shown that addition of Fdx to M. tuberculosis RNAP-σA holoenzymes reduces the amount of RPo formed following the subsequent addition of NTPs and a DNA template harboring the M. tuberculosis rrnAP3 promoter (11). We used this assay with a range of Fdx concentrations to calculate the concentration of Fdx that inhibits 50% of RPo (IC50) formed by RNAP-σA on the rrnAP3 promoter in the presence or absence of different RbpA variants. Addition of WT RbpAMtb (RbpAMtbWT) to the RNAP-σA holoenzyme increases the sensitivity of the RNAP-σA holoenzyme to Fdx in this assay, and this is dependent on the presence of the NTT (deleted in the RbpAMtb26–111 and RbpAMtb72–111 mutants) (11) (Fig. 1, A–C). Deletion of both the RbpA NTT and CD resulted in an IC50 within the confidence interval of the IC50 when the NTT alone was deleted (Fig. 1, B and C), indicating that the presence of the RbpA CD does not affect Fdx activity against M. tuberculosis RNAP-σA in vitro. In contrast, an R88A substitution in the RbpA SID that weakens the interaction between RbpA and the RNAP resulted in an IC50 that was lower and outside the confidence interval, compared to RbpAMtbWT, suggesting that domains outside of the NTT could increase Fdx sensitivity (Fig. 1C). Importantly, a saturating concentration of RbpA protein was used in these assays, and therefore, the different effects of RbpA variants on Fdx sensitivity in this assay should not be a result of altered proportions of RbpA-bound RNAP-σA complexes.
Figure 1.
RbpA E17 and R10 synergize to promote Fdx activity against M. tuberculosis RNAP-σAin vitro.A, schematic of RbpA’s four domain structure including the location of the substituted residues, R10, E17, R79, R88, and the two M. tuberculosis truncation mutants, RbpA 26–111 lacking the NTT, and RbpA 72–111 lacking the NTT and CD. B, representative gels showing Fdx (0 μM, 0.01 μM, 0.1 μM, 1.0 μM, 10 μM, and 100 μM) inhibition of M. tuberculosis RNAP-σA production of three nucleotide transcripts alone or in complex with RbpAMtbWT, RbpAMtbR10A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMtbR79A, RbpAMtbR88A, RbpAMtb26–111, or RbpAMtb72–111 from a linear dsDNA template containing positions −80 to +70 of M. tuberculosis rrnAP3 (relative to the +1 transcription start site). C, dose–response curves of the experiments shown in (A). The curves are generated from at least four replicates from at least two different experiments. Percent inhibition at each Fdx concentration included in the plots compared to no drug is depicted as the mean ± SD. The IC50 for each replicate was calculated by nonlinear regression analysis with four-parameter (EC50, Hill Slope, top and bottom curve plateaus) fitting of log transformed Fdx concentration versus normalized response, with the mean IC50 and 95% confidence interval listed in the table. D, structural modeling of Fdx binding pocket on the RbpA-bound M. tuberculosis RNAP-σA from PDB structure 6BZO. Fdx and RNAP residues involved in the RNAP-Fdx binding interface are shown with PyMol stick representation while the rest of the structure is shown with PyMol cartoon representation. Polar interactions are indicated by red dashed lines, and potential van der Waals interactions are shown as gray lines. E, structural modeling of RbpA-bound M. tuberculosis RNAP-σA from PDB structure 6C04. RbpA R10 and RNAP σA D441 are shown with PyMol stick representation while the rest of the structure is shown with PyMol cartoon representation. The polar interaction between RbpA R10 and RNAP σA D441 is indicated by the red dashed line. CD, core domain; Fdx, fidaxomicin; NTT, N-terminal tail; RbpA, RNA polymerase binding protein A; RNAP, RNA polymerase.
Structural studies predicted that the NTT contributes contacts with Fdx when the antibiotic is bound to the M. tuberculosis RNAP-σA holoenzyme (PDB: 6BZO), specifically through a water-mediated interaction between RbpA E17 and Fdx (Fig. 1D) (11). To determine whether the predicted interaction between Fdx and RbpA E17 underpins NTT-dependent Fdx activity, we calculated the IC50 of Fdx in the presence of RbpAMtbWT versus an RbpAMtbE17A mutant protein. The activity of Fdx against the M. tuberculosis RNAP-σA in the presence of RbpAMtbE17A was nearly equal to Fdx activity against the M. tuberculosis RNAP-σA in the presence of RbpAMtbWT, indicating that alterations in the size and charge of the amino acid side chain at RbpA NTT position 17 do not impact Fdx activity against the M. tuberculosis RNAP-σA (Fig. 1, B and C).
The structure in Boyaci et al. (11) also highlights potential van der Waals interactions between RbpA R10 and Fdx in the RNAP-σA holoenzyme bound to double stranded forked DNA (PDB: 6BZO) (Fig. 1D); however, given the distance between RbpA R10 and Fdx, one would predict this to be a weak interaction. In a separate structure of RbpA bound to M. tuberculosis RNAP-σA in complex with two double-stranded forked DNA molecules that mimics the RPo (PDB: 6C04), the RbpA R10 positively charged side chain is positioned within 2.4 Å of the negatively charged side chain of σA3.2 D441, forming a polar interaction (11) (Fig. 1E). Fdx activity against E. coli RNAP-σ70 holoenzyme lacking σ703.2 is attenuated approximately 20-fold (32), indicating that σ703.2 contributes to Fdx inhibition of the E. coli RNAP. Therefore, if RbpA R10 interacts with σA3.2, this may also affect Fdx activity. To examine whether RbpA R10 contributes to M. tuberculosis RNAP-σA Fdx sensitivity, we measured Fdx IC50 against the M. tuberculosis RNAP-σA in the presence of RbpAMtbR10A. Similar to the RbpAMtbE17A mutant, we observed no change in Fdx IC50s against the M. tuberculosis RNAP-σA in the presence of RbpAMtbR10A compared to RbpAMtbWT (Fig. 1, B and C), indicating that the R10 residue is not required for RbpA NTT-dependent Fdx activity. To determine the effect of disrupting the contacts made by the both RbpA E17 and R10, we measured the Fdx IC50 against M. tuberculosis RNAP-σA in the presence of RbpAMtbR10A/E17A. Mutating both the R10 and E17 residues resulted in an approximately 3-fold increase in the Fdx IC50 compared to RbpAMtbWT, although this was still at least 5-fold lower than RbpA mutants lacking the entire NTT (RbpAMtb26–111 and RbpAMtb72–111) (Fig. 1, B and C). These data indicate that loss of one of these residues increases the importance of the other for Fdx activity, but additional mechanisms also contribute to NTT-dependent Fdx activity in vitro.
Multiple RbpA domains impact Fdx activity in vivo
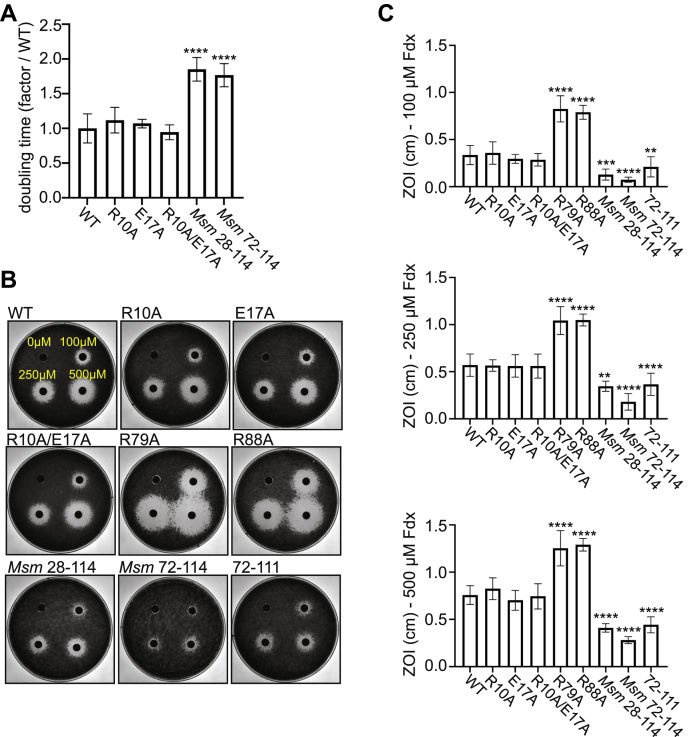
Previous work showed that truncation of the RbpA NTT decreases the sensitivity of Mycobacterium smegmatis to Fdx (11). To investigate the effect of mutations in RbpA on Fdx sensitivity in vivo, we used a strain we previously engineered that expresses rbpAMtbWT at the attB site of M. smegmatis and has the endogenous rbpA gene deleted (5). We then attempted to replace the rbpAMtbWT gene at the attB site in M. smegmatis with alleles encoding each of the RbpA mutants studied in Figure 1 using a gene swapping method (5, 33, 34). We have previously used this approach to generate an M. smegmatis strain expressing rbpAMtb72–111, which has a deletion of both the NTT and CD (Fig. 1A), as its only rbpA allele (5). However, we were unable to generate a viable strain expressing rbpAMtb26–111, which deletes only the NTT (Fig. 1A), in place of rbpAMtbWT. In contrast, we were able to replace the rbpAMtbWT allele with the M. smegmatis allele rbpAMsm28–114, which has previously been used to study the NTT in M. smegmatis (10, 11). Similar to our previous report with the M. smegmatis strain expressing RbpAMtb72–111 (5), RbpAMsm28–114 and RbpAMsm72–114 strains also exhibited a slow growth phenotype (Fig. 2A), confirming that while the NTT and CD are not required for viability in M. smegmatis, they are important domains for RbpA activity. We have also previously shown that M. smegmatis expressing RbpAMtbR88A or RbpAMtbR79A as its only rbpA allele also exhibits a slow growth phenotype due to the importance of RbpA’s interaction with the RNAP and DNA (5). Using the gene swapping approach, we found that the RbpAMtbR10A, RbpAMtbE17A, and RbpAMtbR10A/E17A point mutants could support viability in M. smegmatis and had no effect on growth rate compared to RbpAMtbWT in LB media (Fig. 2A), indicating that these mutations do not affect RbpA’s essential role in M. smegmatis.
Figure 2.
Multiple RbpA domains impact Fdx activity in vivo.A, ratio of the doubling times of M. smegmatis strains expressing RbpAMtbR10A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMsm28–114, or RbpAMsm72–114 as compared to the average doubling time for the strain expressing RbpAMtbWT. The mean ± SD from at least two independent experiments with three replicates per experiment. B, zones of inhibition (ZOI) by Fdx on bacterial lawns of M. smegmatis expressing RbpAMtbWT, RbpAMtbR10A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMtbR79A, RbpAMtbR88A, RbpAMsm28–114, RbpAMsm72–114, or RbpAMtb72–111 as the only copy of rbpA. C, mean radii of ZOI ± SD from at least two experiments with at least three replicates at 100 μM, 250 μM, and 500 μM Fdx is plotted. For A and C, statistical significance of differences was analyzed by ANOVA and Tukey’s multiple comparison test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. All comparisons to RbpAMtbWT were included in the analysis, but only statistically significant comparisons are indicated in the figure. Fdx, fidaxomicin; RbpA, RNA polymerase binding protein A; RNAP, RNA polymerase.
To examine the Fdx sensitivity of each M. smegmatis strain, we used a zone of inhibition assay, similar to previous studies (2, 11). By spreading approximately 2.5 × 108 colony forming units of bacteria on an agar plate and spotting 10 μl of 100, 250, or 500 μM Fdx dissolved in dimethyl sulfoxide (DMSO) onto a disk placed onto the plate, the bacteria form a lawn after incubation at 37 °C for 2 days, and a zone absent of bacterial growth indicates growth inhibition by Fdx. DMSO had no effect on M. smegmatis growth in this assay and did not generate a zone of clearing on its own, whereas incubation of M. smegmatis with Fdx resulted in growth inhibition (Fig. 2B). We compared the radii of the zones of inhibition formed on each M. smegmatis mutant with Fdx and reproduced previous findings that deletion of the RbpA NTT results in resistance to Fdx in vivo (RbpAMtb72–111, RbpAMsm28–114, and RbpAMsm72–114 mutants in Fig. 2, B and C) (11), which is consistent with the in vitro findings (Fig. 1, B and C). In contrast, the RbpAMtbR10A, RbpAMtbE17A, and RbpAMtbR10A/E17A mutants were not more resistant to Fdx in vivo, despite the trend observed in vitro of RbpAMtbR10A/E17A displaying decreased Fdx sensitivity compared to RbpAMtbWT (Figs. 1, B and C and 2, B and C). Strikingly, the M. smegmatis RbpAMtbR79A and RbpAMtbR88A mutants, which have decreased affinity for DNA and the σ factor, respectively, were significantly more sensitive to Fdx treatment (Fig. 2, B and C). These in vivo data highlight the existence of other contributors to RbpA’s effect on Fdx activity that exist in the bacteria but are not recapitulated in the in vitro assay.
Effects on RPo stability correlate to sensitivity to Fdx in M. smegmatis in vivo
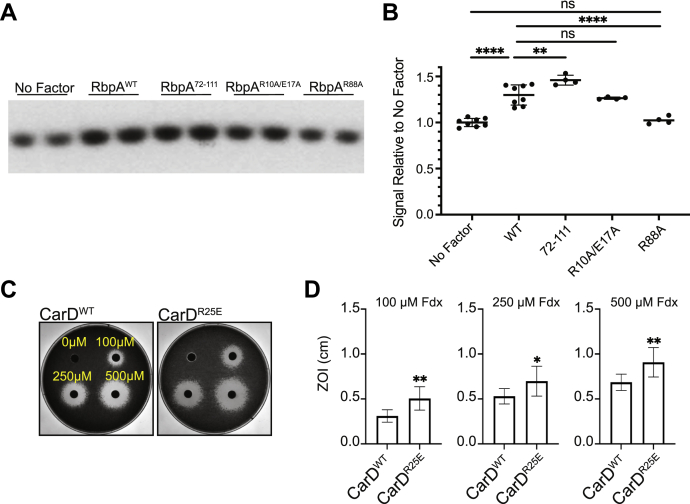
Although the RbpA SID and BL domains are not predicted to contact Fdx in structural models, mutations of residues within the SID (R88A) and BL (R79A) still affected Fdx sensitivity in vivo (Fig. 2, B and C). This suggests that the relationship between RbpA and Fdx sensitivity is not limited to the contribution of specific amino acids within the NTT for Fdx binding to RbpA-bound RNAP-σA. Therefore, we investigated whether RbpA’s functional role during transcription initiation contributed to its effects on Fdx sensitivity. During transcription initiation, RbpA stabilizes RNAP-σA (or σB) RPo (5, 8, 9, 10, 13, 14), which requires binding of the SID to the σ factor and binding of the BL to the DNA (5). In contrast, the NTT and CD antagonize RbpA’s RPo stabilizing activity (5, 10). Using the 3-nucleotide transcription assay to measure RPo stability in the absence of Fdx, we found that addition of RbpAMtbWT to M. tuberculosis RNAP-σA and the rrnAP3 promoter increased RPo stability compared to no factor, and this effect was abolished with the RbpAMtbR88A mutant (Fig. 3, A and B), consistent with previously published stopped flow fluorescence data (5). Addition of RbpAMtb72–111 to M. tuberculosis RNAP-σA and the rrnAP3 promoter increased RPo stability compared to RbpAMtbWT, while addition of the RbpAMtbR10A/E17A mutant showed similar activity as compared to RbpAMtbWT (Fig. 3, A and B), demonstrating that R10 and E17 are not involved in RbpA’s activity on RPo stability.
Figure 3.
RPostability in associated with Fdx sensitivity in vivo.A, representative gels of three nucleotide transcripts produced by M. tuberculosis RNAP-σA alone or in complex with RbpAMtbWT, RbpAMtb72–111, RbpAMtbR10/E17A, or RbpAMtbR88A from a plasmid DNA template containing positions −39 to +4 of M. tuberculosis rrnAP3 relative to the +1 transcription start site. B, ratio of transcript produced as compared to the average of “No Factor” replicates included on the same gel. Results are plotted as individual values with the mean ± SD shown. Statistical significance of differences was determined by ANOVA and Tukey’s multiple comparison test. ‘ns’, not significant; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. C, zones of inhibition (ZOI) by Fdx on bacterial lawns of M. smegmatis expressing CarDMtbWT or CarDMtbR25E as the only copy of carD. D, mean radii of ZOI ± SD from at least two experiments with at least three replicates at 100 μM, 250 μM, and 500 μM Fdx is plotted. Statistical significance was analyzed by two-tailed Welch’s t test. ∗p < 0.05; ∗∗p < 0.01. Fdx, fidaxomicin; RbpA, RNA polymerase binding protein A; RPo, RNAP-promoter open complex.
The effect of these RbpA alleles on RPo stability mirrors the pattern of Fdx sensitivity in vivo, where M. smegmatis strains expressing RbpA alleles conferring higher RPo stability (RbpAMtb72–111) in vitro were less sensitive to killing by Fdx. Conversely, M. smegmatis strains expressing RbpA alleles conferring decreased RPo stability (RbpAMtbR88A) in vitro were more sensitive to killing by Fdx. This correlation led us to further investigate the relationship between RPo stability and Fdx sensitivity. CarD is another essential transcription factor in mycobacteria that functions to stabilize RPo (1, 7, 8, 10, 35, 36). We reasoned that if RPo stability was linked to Fdx activity in vivo, then M. smegmatis strains expressing the CarDMtbR25E mutant allele, which has a weaker affinity for the RNAP and is defective in stabilizing RPo (2, 7, 35) would be more sensitive to Fdx than M. smegmatis expressing CarDMtbWT. Indeed, when we performed the zone of inhibition assays on these strains, we found that the R25E mutation in CarD also increased the sensitivity of M. smegmatis to Fdx (Fig. 3, C and D). In summary, our experiments uncover a relationship between RPo stability and Fdx sensitivity in M. smegmatis (Table 1), suggesting that the role of RbpA for Fdx sensitivity in mycobacteria may involve RbpA’s functional activity during transcription initiation in addition to the role of the RbpA NTT in Fdx binding. In addition, these studies highlight that other factors that regulate RPo stability, such as CarD, could also affect sensitivity to Fdx.
Table 1.
Summary of the effects of RbpA and CarD mutants on fidaxomicin (Fdx) sensitivity and open complex (RPo) stability, compared to WT protein
| RbpA construct | In vitro Fdx sensitivity | In vivo Fdx sensitivity | RPo stability |
|---|---|---|---|
| RbpA72–111 | Decrease | Decrease | Increase |
| RbpAR88A | No change | Increase | Decrease |
| RbpAR10A/E17A | Decreasea | No change | No change |
| CarDR25E | N/A | Increase | Decrease |
The level of decrease in Fdx sensitivity in vitro with RbpAR10A/E17A is intermediate to that of RbpA72–111, when both are compared to RbpAWT.
Discussion
Prior studies on RbpA have focused almost exclusively on the SID interaction with σ factor and the BL interaction with DNA, leaving the NTT and CD largely uncharacterized. Structural studies have provided tremendous insight into the potential interactions between the NTT and CD with multiple RNAP-σA holoenzyme domains as well as the antibiotic Fdx (10, 11, 37). Herein, we test the prediction that RbpA R10 and E17 contribute contacts with the antibiotic Fdx that are important for RbpA’s NTT-dependent activity against M. tuberculosis RNAP-σA. We find that in vitro, combined mutation of both residues affects the IC50 of Fdx activity against the M. tuberculosis RNAP-σA (Fig. 1, B and C); however, it is still not clear whether RbpA R10 and E17 promote RbpA NTT-dependent Fdx activity through direct interaction with Fdx or through an alternative mechanism. Maintenance of partial Fdx activity against M. tuberculosis RNAP-σA bound by RbpAMtbR10A/E17A in vitro indicates that additional RbpA NTT residues, or perhaps the entire structural domain, mediate RbpA NTT-dependent Fdx activity. In addition, the RbpAMtbR10A/E17A mutant did not alter Fdx sensitivity in M. smegmatis (Fig. 2), indicating that those residues play less of a role in Fdx activity in vivo. The R88A substitution that weakens RbpA’s interaction with the RNAP in vivo (5), and thus would be expected to decrease M. smegmatis sensitivity to Fdx since less RbpA would be associated with RNAP-σA, also unexpectedly increased M. smegmatis sensitivity to Fdx. Taken together, these observations reveal differences in the effects of RbpA mutants on Fdx sensitivity in vitro compared to in vivo and support a model where RbpA can impact Fdx activity independent of its direct contacts with the antibiotic.
These discrepancies between the measured sensitivities in vitro versus in vivo may be due in part to the limited scope of the in vitro assay used here and in previous studies to probe Fdx activity (11), where Fdx is added to RbpA and RNAP-σA holoenzyme before DNA addition. Whereas in the cell, RNAP-σA holoenzyme could be bound to DNA prior to Fdx binding. This limitation may bias the in vitro assay toward identifying the factors that affect Fdx binding to free RbpA-RNAP-σA holoenzyme complex. In particular, our in vivo results support an association between effects on RPo stability and Fdx sensitivity. Our work indicates that RPo stability is a newly characterized way that RbpA contributes to Fdx activity. During transcription initiation, RPo stabilization involves closing of the RNAP clamp module around downstream nucleic acid as the transcription bubble is formed (38). Structural studies indicate that Fdx inhibits transcription initiation by trapping the mycobacterial transcription initiation complex in an open-clamp conformation (11). In addition, Fdx is predicted to be unable to bind the closed-clamp conformation (11). Therefore, mycobacterial transcription factors such as RbpA and CarD that favor RPo formation (7, 8, 10) may impact Fdx sensitivity by reducing the lifetime of open-clamp RNAP complexes that Fdx can bind. Conversely, Fdx has also been shown to decrease the affinity of CarD to RNAP in vitro (39). CarD has a lower affinity to the open-clamp RNAP complex compared to the closed-clamp RNAP complex (RPo) (7). Thus, it is possible that Fdx lowers the fraction of CarD bound to RNAP-promoter complexes by reducing the amount of RPo formed at equilibrium. This work highlights the need to biochemically understand Fdx activity against the diversity of RNAP complexes that exist within the bacteria.
In addition to the initiation complexes formed following RNAP-σA binding to DNA, one could envision other factors that exist in vivo and not in vitro that could impact Fdx activity. The in vitro assays of Fdx activity also exclude RNAP holoenzymes containing alternative σ factors and additional RNAP interacting proteins present in the bacteria. Fdx has been shown to be more active at inhibiting the E. coli RNAP-σs holoenzyme compared to the E. coli RNAP-σ70 holoenzyme (32), suggesting that the presence of alternative σ factor–bound holoenzymes may also explain some discrepancies between our in vitro and in vivo findings. In addition to these direct effects on RNAP, truncation of the RbpA NTT and CD results in global dysregulation of gene expression in M. smegmatis (5, 10), which could also affect sensitivity to Fdx. Therefore, the effect of RbpA on Fdx activity in vivo is likely multifactorial. As such, analysis of RbpA mutants with substitutions in conserved residues within the NTT that are predicted to contact different domains in the RNAP-σA holoenzyme revealed diverse effects of RbpA on the Fdx sensitivity of M. smegmatis (Fig. S1). The impact of these mutants on transcription initiation is unknown, but further investigation into this area could shed more light on how association of RbpA on transcription initiation complexes contributes to antibiotic susceptibility.
Collectively, our results demonstrate that the RbpA NTT domain is a significant contributor to the Fdx sensitivity of the mycobacterial transcription machinery, consistent with previous studies. However, we also discover that the role for RbpA involves more than simply providing amino acids to the Fdx binding site. Our data support a model where multiple RbpA domains, including the NTT, can impact Fdx sensitivity through modulation of transcription initiation kinetics. Our studies reveal a role for another factor that also regulates RPo stability, CarD, in Fdx sensitivity. Fdx is currently used to treat infections caused by C. difficile, a bacterium that does not encode an RbpA homolog but does encode CarD and other factors that will regulate transcription by modifying RPo lifetime (1). Therefore, these studies also shed light on pathways that can be targeted to improve Fdx activity in the clinic.
Experimental procedures
Media and bacterial strains
All M. smegmatis strains were derived from mc2155 and grown at 37 °C in LB medium supplemented with 0.5% dextrose, 0.5% glycerol, and 0.05% Tween 80. M. smegmatis strains expressing RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E RbpAMtbR10A, RbpAMtbS15A, RbpAMtbE17A and RbpAMtbR10A/E17A, RbpAMtbR79A, RbpAMtbR88A, RbpAMtb26–111, RbpAMtb72–111, RbpAMsm28–114, and RbpAMsm72–114 were engineered using pMSG430 plasmids that express each rbpA allele from a constitutive Pmyc1-tetO promoter and integrated into the attB site of the M. smegmatis ΔrbpA attB::tet-rbpA strain previously described (5, 33, 34). The primers used to make RbpA strains are in Table S1. RbpAMtbR79A, RbpAMtbR88A, RbpAMtb26–111, and RbpAMtb72–111 have been previously described in (5). The M. smegmatis ΔrbpA attB::tet-rbpA strains expressing RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E, RbpAMtbR10A, RbpAMtbS15A, RbpAMtbE17A, RbpAMtbR10A/E17A, RbpAMsm28–114, RbpAMsm72–114, RbpAMtbR79A, and RbpAMtbR88A were named csm455, csm461, csm456, csm457, csm458, csm451, csm462, csm450, csm498, csm510, csm511, csm322, and csm314, respectively.
Protein preparation for biochemical assays
Plasmids containing the M. tuberculosis H37Rv genomic DNA encoding the different M. tuberculosis RNAP holoenzyme subunits were a gift from Jayanta Mukhopadhyay (Bose Institute) (40). Expression and purification were carried out in accordance with the methods described previously (5). Recombinant M. tuberculosis RbpA proteins were purified from E. coli as previously described using the pET-SUMO vector (primers used to make RbpA constructs for protein purification are in Table S2) (5). RbpA was stored at −80 °C in 150 mM NaCl, 20 mM Tris pH 8.0, and 1 mM β-mercaptoethanol. M. tuberculosis RNAP-σA holoenzyme was stored at −80 °C in 50% glycerol, 10 mM Tris pH 7.9, 200 mM NaCl, 0.1 mM EDTA, 1 mM MgCl2, 20 μM ZnCl2, and 2 mM DTT.
Fdx zone of inhibition
M. smegmatis cultures were grown to OD600 = 0.4 to 0.8. Based on the approximation that OD600 = 1.0 is equivalent to 5 × 108 mycobacteria, 2.5 × 108 cells were collected, resuspended in 100 μl of LB, and plated on LB agar plates. Whatman filter paper disks were applied to the plates, and 10 μl of 100 μM, 250 μM, or 500 μM Fdx (Selleck Chemicals) resuspended in DMSO or DMSO alone were added to the Whatman filter paper disks. The plates were incubated at 37 °C for 48 h, and the zones of inhibition were measured. The zone of inhibition for each replicate at each drug concentration is the average of four measurements approximately 90o apart.
3-Nucleotide in vitro transcription assay
For the Fdx studies in Figure 1, a linear 150 bp dsDNA template containing the M. tuberculosis rrnAP3 promoter was prepared by annealing and extending 85-mer oligonucleotide primers (Integrated DNA Technologies) with a 20 nucleotide overlap ranging from nucleotides 1,471,577 to 1,471,726 in the M. tuberculosis H37Rv genome (9) and HPLC purified as previously described (7). For the RPo stability assays in Figure 3, a plasmid DNA template containing the M. tuberculosis rrnAP3 promoter from the −39 to +4 positions relative to the +1 transcription start site, ranging from nucleotides 1,471,618 to 1,471,660 in the M. tuberculosis H37Rv genome, was used. Plasmid DNA was isolated by Midi-prep (Qiagen) and cleaned by alcohol precipitation. For all 3-nucleotide transcription assays, RbpA, M. tuberculosis RNAP-σA holoenzyme, and dsDNA template were incubated at 37 °C for 10 min. Reactions were initiated by adding 2.5 μl of a substrate mixture containing GpU, UTP, and 32P radiolabeled UTP and incubating at 37 °C for 10 min to allow for production of a 3-nucleotide product in 20 μl reactions that included a final concentration of 2 μM RbpA (saturating concentration based on (5, 8), 100 nM M tuberculosis RNAP-σA holoenzyme, 10 nM dsDNA template, 1 mM DTT, 0.1 mg/ml BSA (NEB), 200 μM GpU, 20 μM UTP, 0.2 μl of 32P radiolabeled UTP, 75 mM NaCl, 10.1 mM MgCl2, 2 μM ZnCl2, 18 mM Tris pH 8.0, 0.01 mM EDTA, 5% glycerol, and 0.1 mM β-mercaptoethanol. Reactions were stopped with 2X formamide stop buffer (98% [vol/vol] formamide, 5 mM EDTA and 0.05% w/v bromophenol blue). Reaction products were resolved by 22% polyacrylamide-urea gel electrophoresis and exposure to autoradiography film. Products were quantified using ImageJ. Dose–response curves were carried out the same way with the exception that Fdx was added to RbpA and M. tuberculosis RNAP-σA holoenzyme, incubated for 10 min at 37 °C, at which point linear dsDNA template was added and allowed to incubate at 37 °C for 15 min before initiating the reactions with the substrate mixture. The in vitro transcription reaction conditions are slightly different than those used in previously published work (11), including different salts in the buffers, different type of holoenzyme preps, and a different dsDNA template, all likely contributing to overall differences in the Fdx IC50 values. Nonetheless, the trends between samples are consistent between this manuscript and previously published work, and therefore, the different reaction conditions do not change the data interpretations or conclusions.
Data availability
All data are contained in the manuscript and the supporting information file.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Gregory Harrison, Michael Nehls, Rachel Kinsella, and Skyler Hendrix for reviewing the manuscript and for helpful discussions.
Author contributions
J. P., D. X. Z., and C. L. S. conceptualization; J. P., D. X. Z., and C. L. S. methodology, J. P. and D. X. Z., validation; J. P. and D. X. Z., formal analysis; J. P., D. X. Z., A. J. F., D. J., and A. R. M. investigation; J. P., D. X. Z., and C. L. S. writing-original draft; J. P., D. X. Z., A. J. F., D. J., A. R. M., E. A. G., and C. L. S. writing-review & editing, J. P., D. X. Z., and C. L. S. visualization; E. A. G. and C. L. S. resources; E. A. G. and C. L. S. supervision; E. A. G. and C. L. S. funding acquisition; C. L. S. project administration.
Funding and additional information
This work was supported by the NIH NIGMS grant GM107544 awarded to E. A. G. and C. L. S., NIH NIGMS grant GM134362 awarded to E. A. G., and a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award awarded to C. L. S. J. P. is supported by NIH NIGMS grant GM007067 and a Stephen I. Morse Graduate Fellowship. D. X. Z. is supported by NIH NIAID grant T32A1007172. D. J. is supported by the Gary K. Ackers Fellowship and Elliot L. Elson Education and Training Fellowship. This publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Ursula Jakob
Supporting information
A, alignment of RbpA proteins from actinobacterial species. Conserved RbpA NTT residues targeted in this analysis are indicated by an asterisk, and the four RbpA structural domains are indicated. B, structural modeling of RbpA NTT interactions with the RNAP from PDB structure 6BZO. RbpA NTT residues targeted in this analysis and the M. tuberculosis RNAP-σA residues positioned to interact with RbpA are shown with PyMol stick representation while the rest of the structure is shown with PyMol cartoon representation. Polar interactions are indicated by red dashed lines. C, zones of inhibition (ZOI) by Fdx on bacterial lawns of M. smegmatis expressing RbpAMtbWT, RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E, or RbpAMtbS15A as the only copy of rbpA. Mean radii of ZOI ± SD from at least two experiments with at least two replicates at 100 μM, 250 μM, and 500 μM Fdx is plotted. D, doubling times of M. smegmatis strains expressing RbpAMtbWT, RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E, or RbpAMtbS15A normalized to the average doubling time of M. smegmatis expressing RbpAMtbWT. The mean ± SD from at least two independent experiments with at least two replicates per experiment. For both (C) and (D), statistical significance of differences was analyzed by ANOVA and Tukey’s multiple comparison test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. All comparisons to RbpAMtbWT were included in the analysis, but only statistically significant comparisons are indicated in the figure. RbpAMtbWT growth rate and ZOI data included in this figure are the same data included in Figure 2, A and C. Fdx, fidaxomicin; NTT, N-terminal tail; RbpA, RNA polymerase binding protein A; RNAP, RNA polymerase.
References
- 1.Stallings C.L., Stephanou N.C., Chu L., Hochschild A., Nickels B.E., Glickman M.S. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garner A.L., Weiss L.A., Manzano A.R., Galburt E.A., Stallings C.L. CarD integrates three functional modules to promote efficient transcription, antibiotic tolerance, and pathogenesis in mycobacteria. Mol. Microbiol. 2014;93:682–697. doi: 10.1111/mmi.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bortoluzzi A., Muskett F.W., Waters L.C., Addis P.W., Rieck B., Munder T., Schleier S., Forti F., Ghisotti D., Carr M.D., O’Hare H.M. Mycobacterium tuberculosis RNA polymerase-binding protein A (RbpA) and its interactions with sigma factors. J. Biol. Chem. 2013;288:14438–14450. doi: 10.1074/jbc.M113.459883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubin E.A., Tabib-Salazar A., Humphrey L.J., Flack J.E., Olinares P.D.B., Darst S.A., Campbell E.A., Paget M.S. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl. Acad. Sci. U. S. A. 2015;112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prusa J., Jensen D., Santiago-Collazo G., Pope S.S., Garner A.L., Miller J.J., Manzano A.R., Galburt E.A., Stallings C.L. Domains within RbpA serve specific functional roles that regulate the expression of distinct mycobacterial gene subsets. J. Bacteriol. 2018;200 doi: 10.1128/JB.00690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu D.X., Garner A.L., Galburt E.A., Stallings C.L. CarD contributes to diverse gene expression outcomes throughout the genome of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 2019;116:13573–13581. doi: 10.1073/pnas.1900176116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rammohan J., Manzano A.R., Garner A.L., Stallings C.L., Galburt E.A. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 2015;43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rammohan J., Ruiz Manzano A., Garner A.L., Prusa J., Stallings C.L., Galburt E.A. Cooperative stabilization of Mycobacterium tuberculosis rrnAP3 promoter open complexes by RbpA and CarD. Nucleic Acids Res. 2016;44:7304–7313. doi: 10.1093/nar/gkw577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen D., Manzano A.R., Rammohan J., Stallings C.L., Galburt E.A. CarD and RbpA modify the kinetics of initial transcription and slow promoter escape of the Mycobacterium tuberculosis RNA polymerase. Nucleic Acids Res. 2019;47:6685–6698. doi: 10.1093/nar/gkz449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hubin E.A., Fay A., Xu C., Bean J.M., Saecker R.M., Glickman M.S., Darst S.A., Campbell E.A. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife. 2017;6 doi: 10.7554/eLife.22520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyaci H., Chen J., Lilic M., Palka M., Mooney R.A., Landick R., Darst S.A., Campbell E.A. Fidaxomicin jams mycobacterium tuberculosis RNA polymerase motions needed for initiation via RBPA contacts. Elife. 2018;7 doi: 10.7554/eLife.34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y., Morichaud Z., Chen S., Leonetti J.P., Brodolin K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σ A-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012;40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y., Morichaud Z., Perumal A.S., Roquet-Baneres F., Brodolin K. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014;42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perumal A.S., Vishwakarma R.K., Hu Y., Morichaud Z., Brodolin K. RbpA relaxes promoter selectivity of M. tuberculosis RNA polymerase. Nucleic Acids Res. 2018;46:10106–10118. doi: 10.1093/nar/gky714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabib-Salazar A., Liu B., Doughty P., Lewis R.A., Ghosh S., Parsy M.L., Simpson P.J., O’Dwyer K., Matthews S.J., Paget M.S. The actinobacterial transcription factor RbpA binds to the principal sigma subunit of RNA polymerase. Nucleic Acids Res. 2013;41:5679–5691. doi: 10.1093/nar/gkt277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galburt E.A. The calculation of transcript flux ratios reveals single regulatory mechanisms capable of activation and repression. Proc. Natl. Acad. Sci. U. S. A. 2018;115:11604–11613. doi: 10.1073/pnas.1809454115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen D., Galburt E.A. The context-dependent influence of promoter sequence motifs on transcription initiation kinetics and regulation. J. Bacteriol. 2021;203 doi: 10.1128/JB.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gnatt A.L., Cramer P., Fu J., Bushnell D.A., Kornberg R.D. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 19.Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: Structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuznedelov K., Minakhin L., Niedziela-Majka A., Dove S.L., Rogulja D., Nickels B.E., Hochschild A., Heyduk T., Severinov K. A role for interaction of the RNA polymerase flap domain with the σ subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 21.Vishwakarma R.K., Brodolin K. The σ subunit-remodeling factors: An emerging paradigms of transcription regulation. Front. Microbiol. 2020;11:1798. doi: 10.3389/fmicb.2020.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dove S.L., Darst S.A., Hochschild A. Region 4 of σ as a target for transcription regulation. Mol. Microbiol. 2003;48:863–874. doi: 10.1046/j.1365-2958.2003.03467.x. [DOI] [PubMed] [Google Scholar]
- 23.Toulokhonov I., Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J. Mol. Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 24.Naryshkina T., Kuznedelov K., Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 25.Cashel M., Hsu L.M., Hernandez V.J. Changes in conserved region 3 of Escherichia coli σ70 reduce abortive transcription and enhance promoter escape. J. Biol. Chem. 2003;278:5539–5547. doi: 10.1074/jbc.M211430200. [DOI] [PubMed] [Google Scholar]
- 26.Kulbachinskiy A., Mustaev A. Region 3.2 of the σ subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J. Biol. Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- 27.Pupov D., Kuzin I., Bass I., Kulbachinskiy A. Distinct functions of the RNA polymerase σ subunit region 3.2 in RNA priming and promoter escape. Nucleic Acids Res. 2014;42:4494–4504. doi: 10.1093/nar/gkt1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pupov D., Petushkov I., Esyunina D., Murakami K.S., Kulbachinskiy A. Region 3.2 of the factor controls the stability of rRNA promoter complexes and potentiates their repression by DksA. Nucleic Acids Res. 2018;46:11477–11487. doi: 10.1093/nar/gky919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi W., Zhou W., Zhang B., Huang S., Jiang Y., Schammel A., Hu Y., Liu B. Structural basis of bacterial σ 28 -mediated transcription reveals roles of the RNA polymerase zinc-binding domain. EMBO J. 2020;39:1–15. doi: 10.15252/embj.2020104389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuzenkova Y., Tadigotla V.R., Severinov K., Zenkin N. A new basal promoter element recognized by RNA polymerase core enzyme. EMBO J. 2011;30:3766–3775. doi: 10.1038/emboj.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin W., Das K., Degen D., Mazumder A., Duchi D., Wang D., Ebright Y.W., Ebright R.Y., Sineva E., Gigliotti M., Srivastava A., Mandal S., Jiang Y., Liu Y., Yin R., et al. Structural basis of transcription inhibition by fidaxomicin (Lipiarmycin A3) Mol. Cell. 2018;70:60–71. doi: 10.1016/j.molcel.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morichaud Z., Chaloin L., Brodolin K. Regions 1.2 and 3.2 of the RNA polymerase σ subunit promote DNA melting and attenuate action of the antibiotic Lipiarmycin. J. Mol. Biol. 2016;428:463–476. doi: 10.1016/j.jmb.2015.12.017. [DOI] [PubMed] [Google Scholar]
- 33.Weiss L.A., Harrison P.G., Nickels B.E., Glickman M.S., Campbell E.A., Darst S.A., Stallings C.L. Interaction of CarD with RNA polymerase mediates Mycobacterium tuberculosis viability, rifampin resistance, and pathogenesis. J. Bacteriol. 2012;194:5621–5631. doi: 10.1128/JB.00879-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pashley C.A., Parish T. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 2003;229:211–215. doi: 10.1016/S0378-1097(03)00823-1. [DOI] [PubMed] [Google Scholar]
- 35.Garner A.L., Rammohan J., Huynh J.P., Onder L.M., Chen J., Bae B., Jensen D., Weiss L.A., Manzano A.R., Darst S.A., Campbell E.A., Nickels B.E., Galburt E.A., Stallings C.L. Effects of increasing the affinity of CarD for RNA polymerase on Mycobacterium tuberculosis growth, rRNA transcription, and virulence. J. Bacteriol. 2017;199 doi: 10.1128/JB.00698-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis E., Chen J., Leon K., Darst S.A., Campbell E.A. Mycobacterial RNA polymerase forms unstable open promoter complexes that are stabilized by CarD. Nucleic Acids Res. 2015;43:433–445. doi: 10.1093/nar/gku1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hubin E.A., Lilic M., Darst S.A., Campbell E.A. Structural insights into the mycobacteria transcription initiation complex from analysis of X-ray crystal structures. Nat. Commun. 2017;8:16072. doi: 10.1038/ncomms16072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyaci H., Chen J., Jansen R., Darst S.A., Campbell E.A. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature. 2019;565:382–385. doi: 10.1038/s41586-018-0840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stefan M.A., Velazquez G.M., Garcia G.A. High-throughput screening to discover inhibitors of the CarD·RNA polymerase protein–protein interaction in Mycobacterium tuberculosis. Sci. Rep. 2020;10:21309. doi: 10.1038/s41598-020-78269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee R., Rudra P., Prajapati R.K., Sengupta S., Mukhopadhyay J. Optimization of recombinant Mycobacterium tuberculosis RNA polymerase expression and purification. Tuberculosis. 2014;94:397–404. doi: 10.1016/j.tube.2014.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, alignment of RbpA proteins from actinobacterial species. Conserved RbpA NTT residues targeted in this analysis are indicated by an asterisk, and the four RbpA structural domains are indicated. B, structural modeling of RbpA NTT interactions with the RNAP from PDB structure 6BZO. RbpA NTT residues targeted in this analysis and the M. tuberculosis RNAP-σA residues positioned to interact with RbpA are shown with PyMol stick representation while the rest of the structure is shown with PyMol cartoon representation. Polar interactions are indicated by red dashed lines. C, zones of inhibition (ZOI) by Fdx on bacterial lawns of M. smegmatis expressing RbpAMtbWT, RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E, or RbpAMtbS15A as the only copy of rbpA. Mean radii of ZOI ± SD from at least two experiments with at least two replicates at 100 μM, 250 μM, and 500 μM Fdx is plotted. D, doubling times of M. smegmatis strains expressing RbpAMtbWT, RbpAMtbR4A, RbpAMtbR4E, RbpAMtbL6A, RbpAMtbR7A, RbpAMtbR7E, or RbpAMtbS15A normalized to the average doubling time of M. smegmatis expressing RbpAMtbWT. The mean ± SD from at least two independent experiments with at least two replicates per experiment. For both (C) and (D), statistical significance of differences was analyzed by ANOVA and Tukey’s multiple comparison test. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. All comparisons to RbpAMtbWT were included in the analysis, but only statistically significant comparisons are indicated in the figure. RbpAMtbWT growth rate and ZOI data included in this figure are the same data included in Figure 2, A and C. Fdx, fidaxomicin; NTT, N-terminal tail; RbpA, RNA polymerase binding protein A; RNAP, RNA polymerase.
Data Availability Statement
All data are contained in the manuscript and the supporting information file.