Abstract
Dengue is one of the most dominant arthropod-borne viral diseases, infecting at least 390 million people every year throughout the world. Despite this, there is no effective treatment against dengue, and the only available vaccine has already been withdrawn owing to the significant adverse effects. Therefore, passive immunotherapy using monoclonal antibodies is now being sought as a therapeutic option. To date, many dengue monoclonal antibodies have been identified, most of which are serotype-specific, and only a few of which are cross-reactive. Furthermore, antibodies that cross-react within serotypes are weakly neutralizing and frequently induce antibody-dependent enhancement, which promotes viral entry and replication. Therefore, broadly neutralizing antibodies with no risk of antibody-dependent enhancement are required for the treatment of dengue. Here, we developed a single-chain variable fragment (scFv) antibody from an anti-fusion loop E53 antibody (PDB: 2IGF). We introduced previously predicted favorable complementarity-determining region (CDR) mutations into the gene encoding the scFv antibody for affinity maturation, and the resultant variants were tested in vitro against the highly conserved fusion and bc epitope of the dengue virus envelope protein. We show some of these scFv variants with two to three substitution mutations in three different CDRs possess affinity constants (KD) ranging from 20 to 200 nM. The scFv-mutant15, containing D31L, Y105W, and S227W substitutions, showed the lowest affinity constant, (KD = 24 ± 7 nM), approximately 100-fold lower than its parental construct. We propose that the scFv-derivative antibody may be a good candidate for the development of an effective and safe immunotherapy.
Keywords: scFv antibody, Fu-bc epitope, DENV, antibody fragment, affinity maturation, therapeutic antibody
Abbreviations: ADE, antibody-dependent enhancement; BLI, bio-layer interferometry; DENV, dengue virus; Fu-bc, fusion and bc loop; RBS, receptor-binding site; scFv, single chain variable fragment antibody; SPR, surface plasmon resonance
Dengue virus is one of the most prevalent mosquito-borne human pathogens, affecting about half of the world's population (1). Dengue virus-1 (DENV-1) through 4, are four distinct dengue virus serotypes that vary by 25 to 40% in the amino acid sequences (2). Infection can be caused by any of the four serotypes, although primary infection is self-limiting and recovers after a systematic illness (3). Secondary infection with the homologous serotype also confers life-long immunity; however, secondary infection with the heterologous serotype confers only partial or transitory immunity, which frequently results in dengue with warning signs and severe dengue, according to the revised WHO dengue case classification (4, 5). Nonetheless, there are no effective vaccines or drugs that can completely protect the dengue virus infection. The most promising vaccine candidate (Dengvaxia) recently demonstrated an increased risk of severe dengue in the people who had never been infected with DENV, and there is no significant protection against DENV-2 (6, 7).
Monoclonal antibodies are now being sought as an alternative therapy for dengue fever patients. Passive immunotherapy using monoclonal antibodies has already been proven to reduce influenza, severe acute respiratory syndrome, middle east respiratory syndrome, Ebola, and HIV viral titers significantly (8, 9, 10, 11). Even so, the food and drug administration has recently approved Emergency Use Authorization to some monoclonal antibody therapies for the treatment of mild-to-moderate COVID-19 in nonhospitalized patients (12, 13). Antibody-based therapy presents a unique set of challenges in the treatment of dengue due to the ability to both mediate protection and exacerbate the disease via the process of antibody-dependent enhancement (ADE) (14, 15). As a result, finding therapeutically safe antibodies is challenging, and producing them in adequate quantities using B cells from infected persons is costly. For cost-effective manufacture and to address ADE's biosafety issues, many expression systems and protein engineering techniques are now being researched (16, 17).
The dengue virus is a member of the Flaviviridae family that forms both mature and immature states during its life cycle. The outer surface of a mature virus is made up of 180 copies of the E glycoprotein (18, 19), of which 90 homodimers are organized in icosahedral symmetry to generate a smooth surface (18), whereas the glycoprotein E and prM are arranged into heterodimers and form 60 trimeric spikes on the surface of the immature virion (20). Each of the E glycoproteins has three unique beta-barrel ecto-domains: DI, DII, and DIII (21). At the apex of DII, two conserved hydrophobic loops (fusion and bc) are essential for pH-mediated membrane fusion (21, 22). The majority of antibodies are produced against the E glycoprotein during the normal course of infection, and the most of these antibodies are specific to the fusion loop (23, 24).
Antibodies that recognize the Flavivirus fusion loop are often strongly cross-reactive but poorly neutralizing (25, 26). E53 is an anti-fusion loop antibody that primarily binds to the Flavivirus E protein's fusion loop and, to a lesser extent, the bc loop (27). The E53 antibody protects mice from deadly WNV infection by inhibiting viral attachment, according to preliminary functional tests (26, 28). Further structural studies have revealed that E53 has the capacity to sterically prevent the conformational transition from an immature to mature virus (27). However, the anti-fusion loop antibody might induce ADE by promoting FcγR-mediated entry of the partially immature virions into the monocytes or phagocytes (29). By considering these facts, exploring the engineered version of such Fc-free Fab or single-chain variable (scFv) antibody fragments can provide a critical understanding of the means to limit viral progression by locking them into immature conformation and avoiding the ADE.
Recently, we reported the development of a scFv antibody fragment from the E53 antibody. A virtual scFv mutant library was constructed and tested against the fusion and bc (Fu-bc) region of the dengue envelope protein using a structure-guided approach (30). A recombinant Fu-bc subunit protein was also developed and evaluated for immunogenicity in BALB/c mice for in vitro binding of the scFv mutant library (31). In this study, we have created a series of scFv mutants by substitution mutations in the complementarity-determining regions (CDRs) and tested them in vitro against the Fu-bc subunit protein. The favorable CDR mutations screened from the in vitro binding assay were further recombined to create synergistically powerful scFv variants. Detailed structural analysis also enabled us to explore significant insights about the inter-residue atomic interaction between the altered CDR residues of the scFv antibody and the targeted epitope (Fu-bc) residues.
Results
Effects of scFv-CDR mutations on the binding with the Fu-bc epitope
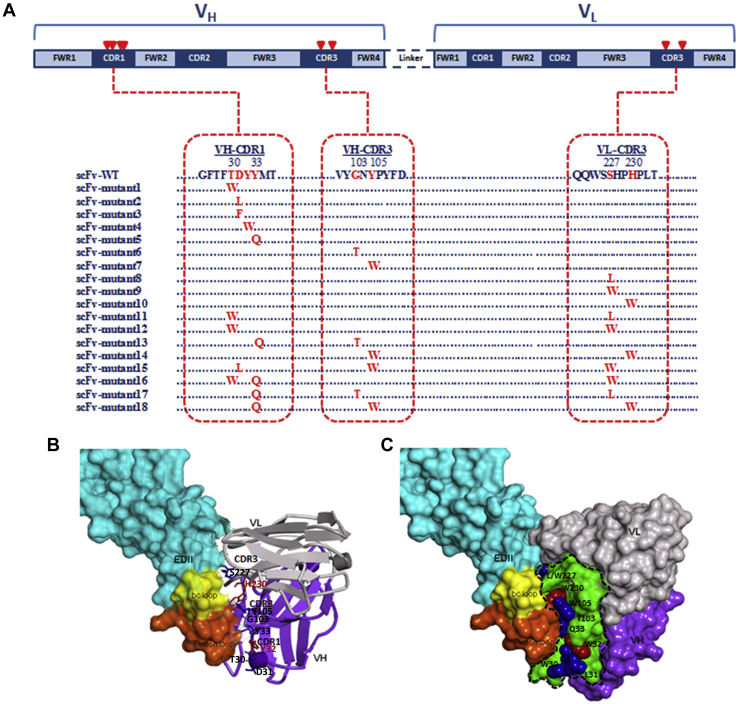
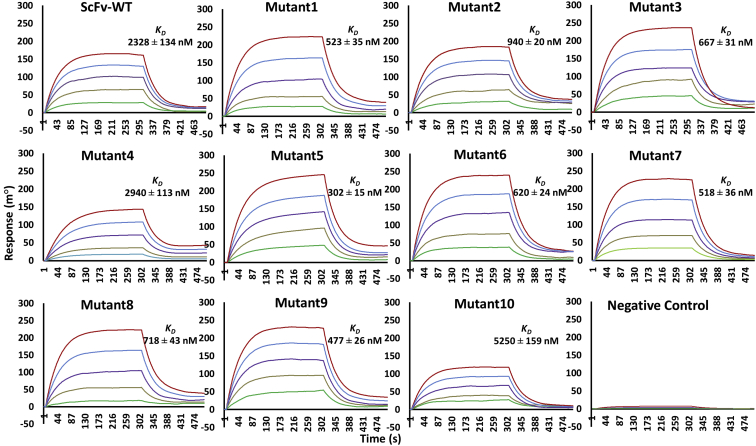
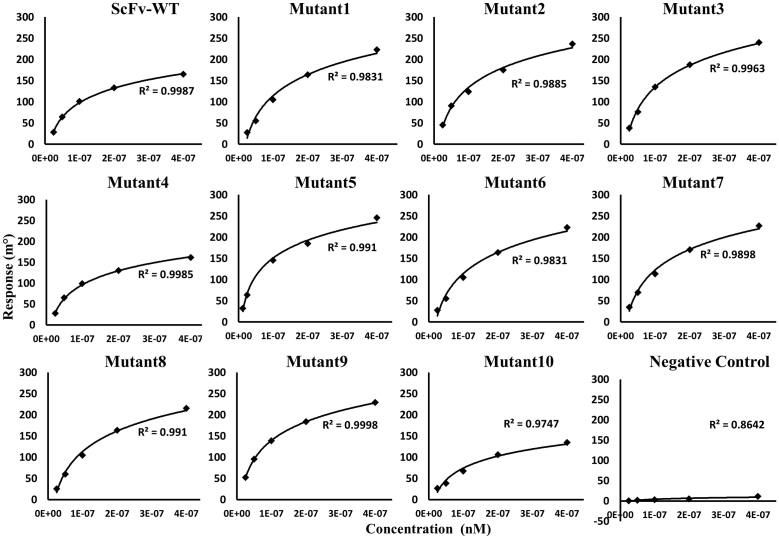
All of the scFv mutants and Fu-bc epitopic proteins were expressed in Escherichia coli (BL21) and purified via size-exclusion chromatography to a high purity level. Both of the proteins behaved as monomers by size exclusion chromatography (Figs. S1 and S2) and, previously, we confirmed their activity by an in vitro binding assay (30, 31). As determined by surface plasmon resonance (SPR), the WT scFv binds to the immobilized Fu-bc protein with a KD of 2.3 μM, which is consistent with our current estimation by the same technique (30). To improve the binding affinity, further saturation mutagenesis was performed in the epitope–paratope interface by using Discovery Studio 4.0, and the top 10 substitutions (T30W, D31L, D31F, Y32W, Y33Q, G103T, Y105W, S227L, S227W, and H230W) at seven hotspots in the three CDRs (VH-CDR1, VH-CDR3, and VL-CDR3) of scFv were selected based on higher negative mutation energy in single, double, and triple mutation combinations (30). The CDR alterations and sites (Fig. 1A) that lead to more favorable epitope interactions as determined by Discovery Studio 4.0 were created in vitro using site-directed mutagenesis, and their affinity was determined using SPR (Figs. 2 and S3). Except for Y32W (Mutant4) in VH-CDR1 and H230W (Mutant10) in VL-CDR3 of the scFv protein, the affinity constant, KD, was observed to decrease (up to 300 nM) with all of the chosen substitution mutations (Table 1). In the 3D structure of the DENV–scFv complex, the positions of both the affinity enhancing and decreasing substitution mutations are shown in Figure 1, B and C.
Figure 1.
Sites of substitution mutations modeled on the dengue virus E protein.A, depicts the putative affinity-enhancing mutation sites in the three complementarity determining regions (CDRs) of scFv's primary structure (selected from our previous in silico study (30)). B, in the 3D complex model structure, the mutation sites in VH-CDR1, CDR3, and VL-CDR3 are located in close proximity to the Fu-bc epitope (orange and yellow surface, respectively). The residues that were substituted by site-directed mutagenesis are shown as blue and red sticks on the dengue virus EDII cartoon-surface structure, where the blue stick shows affinity-enhancing substitution sites (VH-CDR1: T30, D31, and Y33; VH-CDR3: G103 and Y105; VL-CDR3: S227), and the red stick shows affinity-reducing substitution sites (VH-CDR1: Y33 and VL-CDR3: H230). C, both the beneficial and detrimental substitution sites are located on the periphery of the epitope-binding site. The green surface area enclosed by the black dotted line includes residues of structural paratope of the scFv antibody. The blue and red colors include the beneficial and detrimental substitution residues, respectively. Else, the variable heavy and light chains are shown in gray and violet surface areas, respectively. EDII, envelope Domain II; Fu-bc, fusion and bc loop; scFv, single chain variable fragment antibody.
Figure 2.
Kinetics of scFv WT and single mutant proteins’ binding to the Fu-bc subunit protein. The Fu-bc subunit protein was coupled to the SPR biosensor chip, and scFv-WT, 10 scFv single mutants (Mutant1-10), and one negative control (other than scFv protein) were applied at graded concentrations (25–500 nM) over the coupled surface (from t = 0 to t = 300 s), followed by buffer washout (dissociation) for another 180 s and measurement of net binding (in RU). The association (ka) and dissociation rate constants (kd) were derived by fitting the recorded sensograms to (1:1) Langumir-binding rate equations, and the affinity constant KD was derived by dividing the dissociation constant (kd) with the association constant (ka). The overlay of curves for different concentrations of scFv protein was fitted into a numerical model by global analysis using Autolab kinetic evaluation software 5.1. The color of each fitted curve shown is a representative response of different concentrations of analytes (i.e., green, 25 nM; golden, 50 nM; violet, 100 nM; blue, 200 nM; and red, 400 nM). Fu-bc, fusion and bc loop; scFv, single chain variable fragment antibody; SPR, surface plasmon resonance.
Table 1.
Binding affinity of scFv-CDR mutants for Fu-bc subunit protein
 |
The affinity constant (KD) of the scFv variants was determined by surface plasmon resonance (Autolab ESPRIT). The fold change in the KD values is the ratio KDWT/KDmutant. The errors of ka and kd measurements were between 0.001 and 0.009 E5 M−1S−1 and between 0.001 and 0.0001 S−1, respectively.
Synergistic effects of scFv CDR mutations
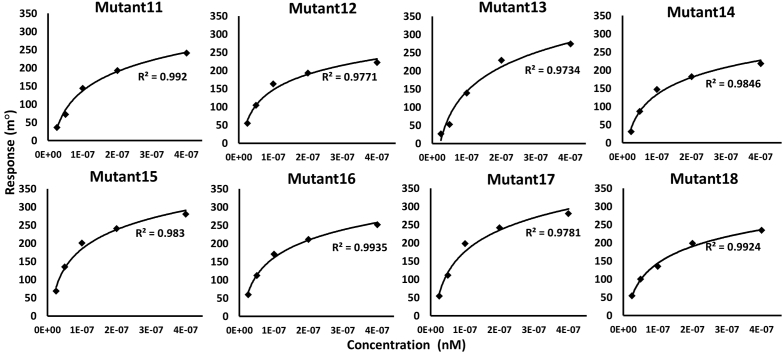
Four sets of double substitution mutants were created prior to the generation of scFv triple combinatorial mutants to assess their synergistic effects on each other. These were created through the combination of two primary affinity-enhancing single mutations in the two alternative CDRs. The SPR data indicated that all of the four sets of double mutants increased the affinity with KD values ranging from 50 to 200 nM (Figs. 3 and S4). Notably, when combined with primary affinity-enhancing substitutions G103T in VH-CDR3 (Mutant6), the scFv VH-CDR1_Y33Q (Mutant5) showed 50-fold higher affinity than the VH-CDR1_T30W (Mutant1) when combined with VL-CDR3_S227L (Mutant8) and S227W (Mutant9). On the other hand, the VH-CDR3_Y105W (Mutant7), when combined with the other affinity-enhancing mutation VL-CDR3_S227W (Mutant9), showed only 13-fold higher affinity (Table 1). Since each of the CDR substitution mutations in a combination of alternative CDR mutations shows only a positive epistatic effect, these combinations were further utilized for the creation of triple combinatorial mutants.
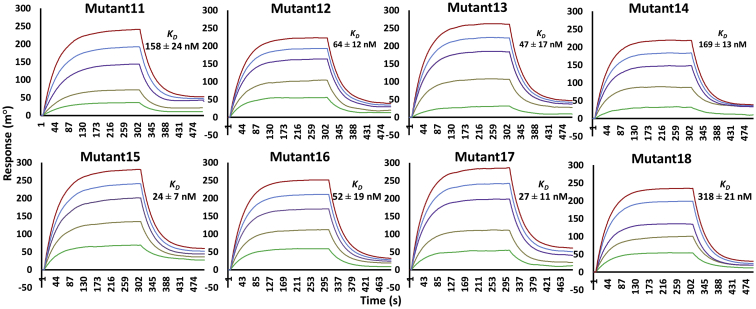
Figure 3.
Kinetics of scFv double and triple mutant proteins’ binding to the Fu-bc subunit protein. The Fu-bc subunit protein was coupled to the SPR biosensor chip, and scFv double (Mutant11–14) and triple (Mutant14–18) mutants were applied at graded concentrations (25–500 nM) over the coupled surface (from t = 0 to t = 300 s), followed by buffer washout (dissociation) for another 180 s and measurement of net binding (in RU). The association (ka) and dissociation rate constants (kd) were derived by fitting the recorded sensograms to (1:1) Langumir-binding rate equations, and the affinity constant KD was derived by dividing the dissociation constant (kd) with the association constant (ka). The overlay of curves for different concentrations of scFv protein was fitted into a numerical model by global analysis using Autolab kinetic evaluation software 5.1. The color of each fitted curve shown is a representative response of different concentrations of analytes (i.e., green, 25 nM; golden, 50 nM; violet, 100 nM; blue, 200 nM; and red, 400 nM). Fu-bc, fusion and bc loop; scFv, single chain variable fragment antibody; SPR, surface plasmon resonance.
Combinatorial effects of scFv triple mutations
Since the substitution mutation Y33Q (Mutant5) enhances affinity the most among the VH-CDR1 mutations while also having a good synergistic impact with the VH-CDR3 mutations, it was further considered to recombine with the other double mutants viz. Mutant12 and Mutant14 to acquire more affinity. Moreover, the affinity-enhancing VL-CDR3 substitution mutation S227L (Mutant8) was incorporated into the double mutant13 (Y33Q, G103T) to investigate a mutation combination within the three alternative CDRs. Another triple mutation combination (Mutant15) was created by introducing the highest affinity enhancing mutation (Y105W) in the VH-CDR3 on the backbone of double mutant12 (T30W, S227W). Surface plasmon resonance experiments revealed that the resulting three sets of triple substitution combination (Mutant15–17) increased the affinity with KD values 20 to 52 nM (Fig. 3). The triple combination mutant15 (D31L, Y105W, and S2227l) increases the highest affinity in terms of KD value change, approximately 100-fold higher than its WT variant, whereas the fourth set of triple combination variant (Mutant18) showed a moderate affinity enhancement (only ∼7 fold) among all the double and triple combinations, because it consisted the most detrimental substitution mutation, H230W, along with the two affinity-enhancing substitution mutations Y33Q and Y105W. On the other hand, scFv-mutant16 (T30W, Y33Q, and S227W) and mutant17 (Y33Q, G103T, and S227L) showed higher affinity, though at a moderate level of fold change in the KD values, that are ∼45 fold and ∼86 fold higher than the WT, respectively (Table 1).
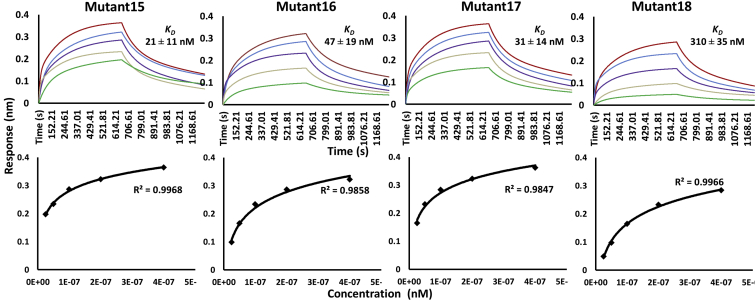
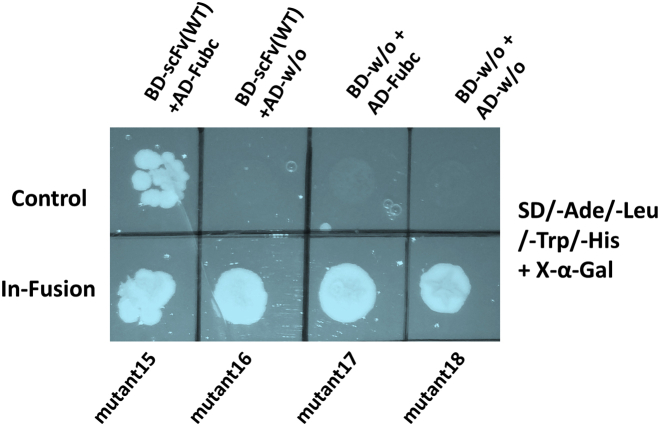
To corroborate the binding affinity of these scFv triple mutants, the KD values were further analyzed by using bio-layer interferometry (BLI). According to BLI, all the four sets of triple combination mutants (Mutant15, 16, 17, and 18) bind to the immobilized Fu-bc protein with KD values of 21 ± 11 nM, 47 ± 19 nM, 31 ± 14 nM, and 310 ± 35 nM, respectively (Fig. 4 and Table S1), which are almost consistent with the KD values determined by SPR. Further, to confirm the binding affinity, in vivo binding analysis was performed by the yeast two-hybrid assay. For this, the Fu-bc subunit gene was cloned in the pGADT7 (bait) vector, and the four sets of scFv triple combination variants were cloned in the pGBKT7 (prey) vector. The qualitative results of the yeast two-hybrid assay have shown that all of the scFv triple combination variants are also capable of interacting with the Fu-bc in the cellular conditions (Fig. S5).
Figure 4.
Parallel sensor kinetics and equilibrium analyses of scFv triple mutant proteins’ binding to the Fu-bc subunit protein. The Fu-bc subunit protein was coupled to the BLI biosensor chip, and scFv triple mutants (Mutant14–18) were applied at graded concentrations (25–400 nM) over the coupled surface (from t = 0 to t = 660 s), followed by buffer washout (dissociation) for another 490 s and measurement of net binding (in RU). The association (ka) and dissociation rate constants (kd) were derived by fitting the recorded sensograms to a (1:1) kinetic titration series model, and the affinity constant KD was derived by dividing the dissociation constant (kd) with the association constant (ka). The overlay of curves for different concentrations of scFv protein was fitted into a numerical model by global analysis using BioEvaluation software 4.1. The color of each fitted curve shown is a representative response of different concentrations of analytes (i.e., green, 25 nM; golden, 50 nM; violet, 100 nM; blue, 200 nM; and red, 400 nM). Furthermore, the BLI responses at equilibrium were plotted against each of the injected protein concentrations (lower panel). The curves were fitted by nonlinear least squares regression. All the R2 values indicate that the regression model fits the data much better than the null hypothesis. Fu-bc, fusion and bc loop; scFv, single chain variable fragment antibody.
Superimposition of scFv antibody on WNV–Fab crystal complex
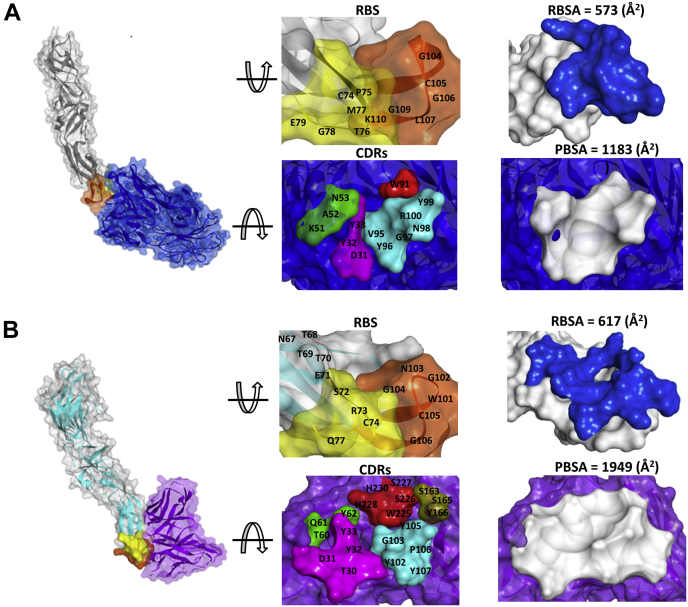
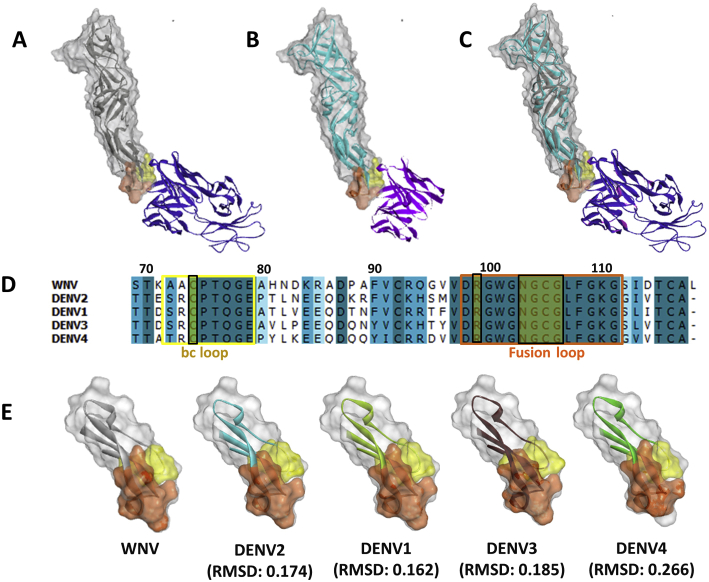
The WNV/DENV–scFv chimeric complex was iteratively built and superposed on the WNV-Fab crystal complex structure (PDB: 3I50) to analyze the structural basis for the capacity of Fab-derived (PDB: 2IGF) scFv to interact with the receptor-binding site (Fu-bc loop) of DENV E protein (Fig. S6C). The epitope and paratope regions of the WNV/DENV–scFv chimeric complex were compared to those of the WNV–Fab complex to acquire a better understanding of the structural ramifications of the interactions between the scFv protein and the DENV E protein. The WNV–Fab complex has 573 Å2 receptor-binding surface area and 1183 Å2 paratope-binding surface area, but the WNV/DENV–scFv chimeric interface has 617 Å2 receptor-binding surface area and 1949 Å2 paratope-binding surface area, which are significantly larger. The overall binding surface area of the WNV/DENV–scFv chimeric complex (2199 Å2) is likewise bigger than that of WNV-Fab (1365 Å2), implying a broader surface range interaction (Fig. 5). The detail interactions of the WNV-Fab and WNV/DENV–scFv complexes are summarized in Tables S2 and S3, respectively.
Figure 5.
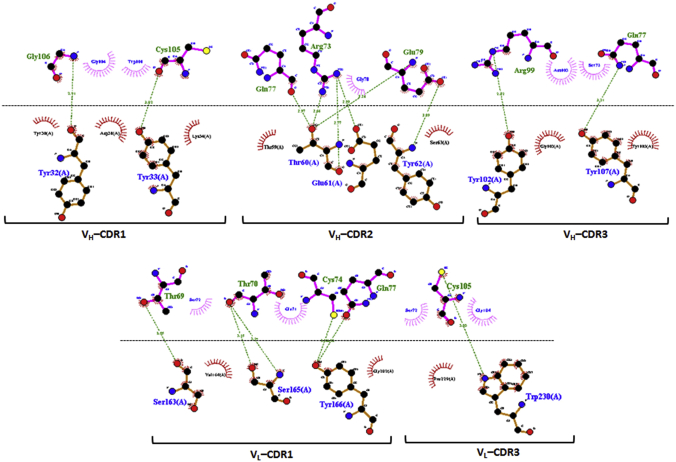
Comparison of interaction interphase between WNV-Fab and WNV/DENV-scFv chimeric complex.A, WNV_RBS-Fab; B, DENV_RBS-scFv. (Individual contacting residues are listed in Tables S2 and S3). In the RBS, the epitopic residues that contact the complementarity determining regions (CDRs) are located in fusion and bc loop. Fusion and bc loop residues are colored in orange and yellow, respectively. VH-CDR1, VH-CDR2, VH-CDR3, VL-CDR2, and VL-CDR3 are painted pink, green, cyan, brawn, and red, respectively. Receptor-binding surface area and PBSA are painted blue and lime color, respectively. PBS, sparatope-binding surface area; RBS, receptor-binding site; scFv, single chain variable fragment antibody.
The interphase of the WNV–Fab and DENV–scFv complexes were two dimensionally plotted using Discovery Studio 4.0 to get further insight into the interaction. In the case of the WNV–Fab complex, four CDRs (VH-CDR1, VH-CDR2, VH-CDR3, and VL-CDR3) contact the receptor-binding site (RBS) (Fu-bc epitope), whereas five CDRs (VH-CDR1, VH-CDR2, VH-CDR3, VL-CDR2, and VL-CDR3) interact in the WNV/DENV–scFv chimeric complex, which also suggests a broader range of interaction. Furthermore, the interface of the WNV–Fab complex forms at least 11 potential H-bonds or other nonbonds (i.e., other than covalent bonds), whereas the interface of the WNV/DENV–scFv chimeric complex forms 16 potential H-bonds or other nonbonds without any steric interference, indicating stronger interaction (Tables S2 and S3). The fusion and bc loop residues (in RBS) that make contact with Fab or scFv CDR residues in both the WNV–Fab and DENV–scFv complexes are completely conserved not just between DENV2 and WNV but also throughout all DENV serotypes (Fig. S6D). Despite this, the three-dimensional structures of the RBS (Fu-bc loop) of all four dengue virus serotypes (DENV2, DENV1, DENV3, and DENV4) are superimposable on the WNV RBS with reduced RMSD (Fig. S6E). All of these findings indicate that the Fu-bc loop specific antibody must be cross-reactive, and its scFv format provides an advantage for improved epitope binding.
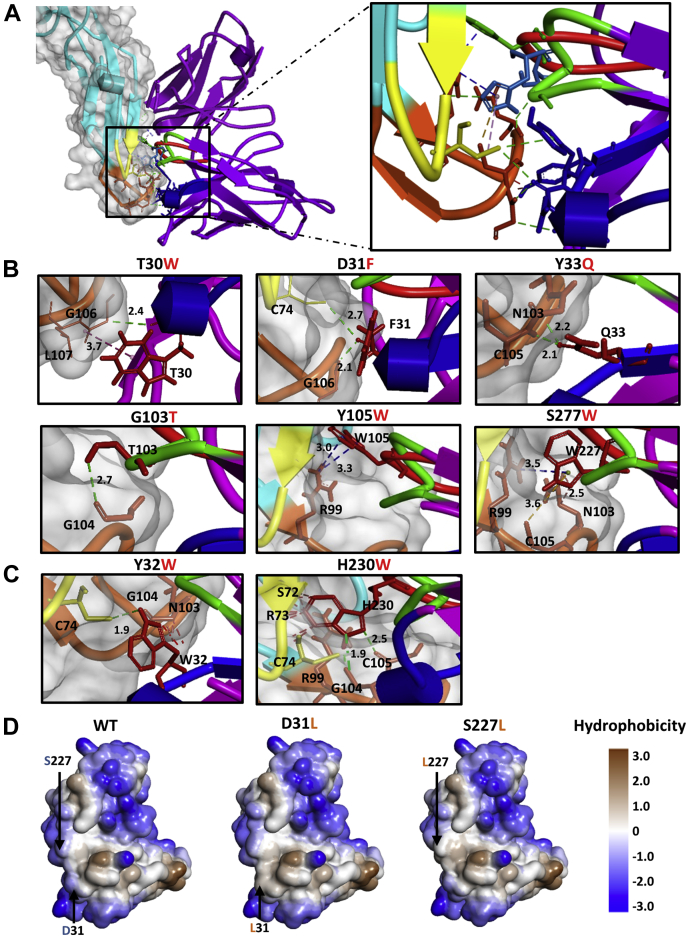
Complementarity determining region mutations alter the binding interface of WNV/DENV–scFv chimeric complex
The WNV/DENV–scFv chimeric complex was visualized using DimPlot to analyze the effect of substitution at each of the SPR-validated CDR positions. Except for VH-CDR1 residues Tyr32, Tyr33, and VL-CDR3 residue His230, all of the targeted CDR residues reside apart from interaction with the RBS (Fu-bc loop) (Fig. S7). Interestingly, the SPR data revealed that, with the exception of Tyr32Trp and His-230Trp, all the targeted CDR substitutions resulted in the reduced affinity constant KD relative to the WT version (Fig. 2). As a result, Tyr32 and His230 are likely to be critical for receptor binding, as their replacement disturbs the interaction with the RBS, and raises the affinity constant KD compared to the WT variant. Although Tyr33 of VH-CDR1 interacts with RBS (Fu-bc loop), and its replacement with Gln reduced affinity constant KD when compared to its parental form, this suggests that Gln plays another role in the interaction interface.
The interaction interface of the WNV/DENV–scFv chimeric complex was examined using the Analyze Protein Interface function of Discovery Studio 4.0 to find further impacts of each of the CDR substitution mutations. Table S4 summarizes the alterations in the epitope–paratope interaction caused by each substitution mutation, which are illustrated in Figure 6. The substitution of T30W, D31F, Y33Q, G103T, Y105W, and S227W at the epitope-binding interface enhanced one or more nonbond interactions (Fig. 6B), which might be playing a key role in strong epitope binding. The substitutions of Y32W and H230W, on the other hand, increased certain nonbond interactions at the interface, but it also caused steric clashes with neighboring epitope residues, which might prevent epitope binding (Fig. 6C). In addition, D31L and S227L substitutions have no direct effect on nonbond interactions, but they do enhance the hydrophobicity of the receptor-binding site, which is thought to be important for binding initiation (Fig. 6D) (32).
Figure 6.
Effects of scFv-CDR mutations on binding with Fu-bc loop of dengue virus E.A, close-up view of the binding interface of DENV-Fubc and scFv-complementarity determining regions (CDRs) complex. Binding sites of Fu-bc and putative affinity-enhancing mutational sites in scFv-CDRs are rendered in ball and stick format. DENV EDII is colored in cyan, while Fusion and bc loops are colored in orange and yellow, respectively. Otherwise, scFv VH-CDR1, VH-CDR3, and VL-CDR3 are painted blue, green, and red, respectively. B, bonding effects of affinity-enhancing CDR mutations. C, bonding and steric effects of affinity-reducing CDR mutations. D, hydrophobic effects of affinity-enhancing CDR mutations. Using the Discovery Studio 3.0 program, the interaction interface was analyzed after substitution with each of the putative affinity-enhancing amino acids in Coot, with local energy minimization (within 10–20 Å). In silico mutagenesis was performed using the WNV/DENV-scFv complex model. Discontinuous lines with different colors between residues represent different types of nonbonds (i.e., green, H-bond; violet, electrostatic; pink, hydrophobic, orange, other non-bonds, and the red lines denoting the steric hindrances). EDII, envelope Domain II; Fu-bc, fusion and bc loop; scFv, single chain variable fragment antibody.
Discussion
The E53 antibody is a highly cross-reactive fusion loop–specific Fab antibody that binds to a conserved region of the WNV and dengue virus E proteins. These antibodies produced during primary DENV infection may cross-react during secondary infection and may promote ADE. An earlier study suggests that E53 poorly binds with mature viruses but has the ability to sterically hinder the transition from immature to mature virus (27). As a result, this E53 (Fab) has been reconstructed into a small version of scFv with the goal of achieving more epitope binding, affinity maturation, and therapeutic testing trial. Others have pointed out that its smaller size, flexibility of VH-VL chains, and ability to be produced in E. coli expression systems offer advantages over Fab (33, 34). However, in spite of the numerous advantages, the engineered WT scFv antibody which was derived from the anti-fusion loop E53 (2IGF) antibody, retains low affinity (KD = 2.3 μM) for the recombinant Fu-bc protein. The interacting residues in the receptor-binding site (Fu and bc loop) are entirely conserved across all the DENV strains and WNV, according to conservation studies. By superimposition, scFv completely overlaps the VH and VL chains of Fab (E53) without creating any steric clash with the RBS of the WNV–Fab complex structure. Furthermore, structural analyses have revealed that scFv captured a greater epitope region than Fab (Fig. 5), indicating that scFv is an excellent candidate for affinity maturation and the production of cross-reactive and highly potent antibody fragments. We recently identified certain hotspots in the scFv CDR and demonstrated higher negative mutation energy by altering them with particular residues (30). After in silico verification of inter-residue atomic interaction between scFv CDR residues and their adjacent Fu-bc residues, the CDR mutations that contributed to more favorable antigen-antibody binding were investigated here experimentally.
Our current SPR experiment has revealed that except for the two substitution mutations Y32W and H230W (scFv-mutant4 and 10), the rest of the predicted scFv CDR mutations exhibited improve binding affinity as they displayed lower affinity constant KD with respect to the WT variant. That is why mutant4 (Y32W) was not pursued further, and mutant10 (H230W) was tested as a negative control. In terms of minimal affinity constant (KD = 302 ± 15 nM), the single mutant Y33Q (Mutant5) had the greatest overall binding profile. The higher affinity-enhancing substitution mutations in each of the CDRs were recombined with each other to analyze their epistatic effects. As determined by SPR, each of the four double combination mutants (Mutant11–14) has good synergistic effects and displayed binding affinity about 10- to 45-fold greater than the WT. Mutant13, which comprises substitution mutations Y33Q (Mutant5) in VH-CDR1 and G103T (Mutant6) in VH-CDR, showed the greatest synergistic effects, with a 45-fold greater affinity than the WT. Due to the positive synergistic effects of all four double combination mutations, these mutants were recombined with other affinity-enhancing mutations to identify the optimum triple combination. The final round of SPR data revealed that three of the four triple combination mutants (Mutant15–17) exhibited higher affinity by 30- to 100-fold over the WT. The best triple combination mutant is mutant15, which has D31L, Y105W, and S227W substitution mutations, and has the lowest affinity constant (KD = 24 ± 7 nM). Mutant16 and 17, the other two triple combination mutants, bind Fu-bc subunit protein with KD values 52 ± 17 nM and 27 ± 11 nM, respectively. The fourth set of triple substitution mutant (Mutant18, Y33Q, Y105W, and H230W), however, showed the least binding affinity among all double and triple combinations because it combines two affinity-enhancing substitution mutations (Y33Q and Y105W) with the most detrimental substitution mutation H230W (Mutant10), implying that His230 is located at the mutation disallowed site and may play a role in epitope-paratope surface complementarity (Table 1).
According to the structural study, the VH and VL chain CDRs of scFv interact with the Fu and the bc loop, respectively. The Fu-bc epitope was located near to the residues that were substituted by the site-directed mutagenesis. Both the affinity-enhancing and affinity-reducing substitution sites were found on the periphery of the epitope-binding site (Fig. 6). The structure of the WNV/DENV–scFv chimeric complex is similar to that of the WNV–Fab complex (PDB: 3I50), and the two complexes are virtually superimposable with RMSD of 0.321. According to a two-dimensional reduction plot (DimPlot) (Fig. S7), most of the targeted residues do not come into direct contact with the Fu-bc residues. Because His230 and Tyr32 interact directly with the fusion loop residues Cys105 and Gly106, respectively, changes to them resulted in the overall reduced affinity, as demonstrated by the SPR experiment (Table 1). Although the alteration of the directly interacting Tyr33 residue enhances affinity, probably because changing Tyr33 to Gln creates an additional H-bond with a pre-existing one, both of which aid epitope binding (Fig. 6B).
Further examination of inter-residue atomic interactions of the epitope–paratope interface revealed that 10 H-bonds and six other nonbonds are involved in the surface complementarity of the DENV–scFv complex. However, 12 H-bonds and nine other nonbonds are involved in the surface complementarity of the Fu-bc and scFv-mutant15, which might be the primary reason for its improved binding. In addition, mutants 16 and 17 showed at least three to five more H-bonds in the binding interface, thereby having higher affinity than the other mutants. The surface complementarity of Fu-bc and scFv-mutant18 contact surfaces, on the other hand, involved 12 H-bonds, and a comparable number of favorable nonbonds as the WT; nevertheless, it induced 10 additional bumps, which might be the major cause of its weaker binding (Tables S3 and S4). The detailed changes in binding interface and nonbond interactions generated by each of the substitution mutations are summarized in Table S4 and shown in Figure 6, respectively. Overall, structural and binding investigations suggest that each of the substitution mutations in the scFv CDRs have significant role in epitope binding.
In summary, the structure-guided redesign of the scFv antibody provides a useful platform to engineer a cross-reactive and potential antibody candidate. In silico antigen–antibody interface analysis has enabled the successful prediction of mutational hotspots in the CDRs of scFv antibodies. Further, site-directed mutagenesis of scFv antibody validates the structure-guided mutational approach with regards to enhancement of in vitro binding affinity, which provides an alternative to conventional genetic approaches to affinity maturation. Ultimately, some of the resulting scFv variants exhibit nM range of the binding affinity, which could be cross-reactive because their parental E53 antibody is highly cross-reactive and deviates from ADE effect due to the lack of the Fc part. Taken together, these data suggest that the newly generated scFv variants might be valuable in the development of therapeutic antibodies and diagnostic tools.
Experimental procedures
Chemicals and instruments
The principal components of bacterial culture: LB broth, tryptone, and yeast extract; antibiotics: kanamycin; protein expression and extraction reagents: IPTG, Tris–HCL and base, glycine, lysozyme, PMSF, SDS, acrylamide/bis-acrylamide, ammonium persulfate, tetramethylethylenediamine (TEMED), DTT, and EDTA, and Coomassie G-250 were purchased from Himedia and VWR life science. Inclusion body solubilizing and protein refolding reagents: guanidine hydrochloride, urea, arginine hydrochloride, and isopropanol were from Sigma-Aldrich. PCR cloning kit: pJET1.2; Q5 high fidelity DNA polymerase and restriction enzymes, T4-DNA ligase, and rapid protein assay BCA kit were from New England Biolabs (NEB) and Thermo Scientific. Protein purification was performed by AKTA FPLC systems using superose 12 10/300 size-exclusion column which were purchased from GE Healthcare. Surface Plasmon Resonance instrument (Autolab ESPRIT) and BLI systems (Octet RED96) were used to measure the binding affinity.
Site-directed mutagenesis
From our previous in silico studies, top ten (T30W, D31L, D31F, Y32W, Y33Q, G103T, Y105W, S227L, S227W and H230W) substitution mutations at seven hotspots in three different CDRs of scFv were selected based on higher negative mutation energy in single as well as in different double and triple mutation combination formats (30). In order to in vitro creation of these substitutions by site-directed mutagenesis, 10 sets of partially overlapping primers were designed by altering the target mutational site (Table S5). These primers are synthesized by Integrated DNA Technology. Our previously cloned scFv gene in the pET28a vector (scFv-pET28a) (33) was used as a template for full-length PCR amplification of the scFv-pET28a clone using Q5 high-fidelity DNA polymerase. The PCR product was treated with DpnI for the digestion of WT template DNA, and the remaining sample was used for transformation in E. coli (XL10). After plasmid isolation, mutations were confirmed by DNA sequencing. Furthermore, double and triple mutants were created by a similar procedure and confirmed by DNA sequencing.
Expression and purification of scFv-WT, scFv-mutants, and Fu-bc subunit protein
Positive mutants and WT scFv were further transformed into E. coli (BL21). A single colony for each was chosen for inoculation with 10 ml of LB growth media and allowed to grow overnight at 37 °C in the presence of 50 mg/ml of kanamycin (Sigma). The next day, 500 ml of LB growth media was inoculated with the previously grown 5 ml primary culture, which was allowed to grow at 37 °C until the A600 was around 0.5. The secondary culture was further induced by 0.5 mM IPTG and allowed for overnight growth at a temperature of 30 °C. The resulting growth culture was centrifuged at 4000 rpm for 30 min to harvest the bacterial cell pellet. Further, scFv-WT and scFv mutant proteins were solubilized and refolded by following the procedure as described earlier (33). Soluble scFv proteins were then purified by AKTA FPLC systems using a superose 12 (10/300) size-exclusion column. On the other hand, Fu-bc subunit protein (GenBank Accession no: MN781186) was generated and purified using a similar procedure as described previously (31).
Interaction study by SPR
To measure the changes in binding affinity of scFv proteins, purified Fu-bc subunit protein was covalently immobilized on the biosensor chip in a dual-channel kinetic evaluation instrument, Autolab ESPRIT Twingle Biosensor platform, using amine coupling strategy. Initially, the sensor surface was activated for both of the channels by passing a mixture (1:1 v/v) of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and N-hydroxysuccinimide for a period of 300 s. The Fu-bc protein dissolved in coupling buffer (10 mM sodium acetate buffers pH 5.0) around 150 μg/ml concentration was immobilized in channel-1, whereas, only coupling buffer was passed in channel-2 for 600 s. The remaining activated -COOH groups in both the channels were blocked with 1 M ethanolamine (pH 8.5) for a period of 600 s, and chip surface was washed twice with running buffer (PBS pH 7.4). After preparation of SPR sensor surface, each of the analytic scFv protein samples were injected through channel-1, and the running buffer (PBS pH 7.4) was injected through channel-2 over the immobilized surface and allowed 300 s for association followed by buffer washout (dissociation) for another 180 s. Interaction between surface-bound Fu-bc and scFv analytic proteins were recorded as a differential of response in channel-1 and channel-2, thereby deducting the contribution of running buffer (PBS, pH 7.4) in each run. The overall binding response was acquired using data acquisition software and analyzed using kinetic evaluation software version 5.1 provided with system. The association (ka) and dissociation rate constants (kd) were derived by fitting the recorded sensograms to (1:1) Langumir binding rate equations, and the affinity constant KD was derived by dividing the dissociation constant (kd) with the association constant (ka) (Table 1). The overlay of curves for different concentrations of scFv protein was fitted into a numerical model using a global analysis technique. Furthermore, the SPR responses at equilibrium were plotted against each of the injected protein concentrations. The curves were fitted by nonlinear least squares regression and are shown in Figs. S3 and S4.
Parallel sensor kinetics of scFv triple mutants with BLI
To measure the affinity between Fu-bc subunit protein and scFv triple mutants, Fu-bc protein was also immobilized via amine coupling reaction on AR2G biosensors. In an Octet RED96 BLI instrument, 12 AR2G biosensors were used for this parallel sensor kinetics. A set of six sensors were allotted for the samples which were preincubated in ddH20 for 10 min, activated in a 1:1 mixture of 0.1 M N-hydroxysuccinimide and 0.4 M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide for 800 s and incubated in the binding buffer PBS for 900 s. Another set of six sensors were also activated but not incubated in binding buffer, which was allotted for reference sensors. All of the sensors were blocked with 1 M ethanolamine for 180 s and stored in ddH20 before further usage. To measure the binding titration, the association and dissociation phases were recorded for 480 s and 600 s, respectively for each of the scFv triple mutants. Five dilutions of each of the analytes were applied for parallel sensor kinetics in the Octet RED96 BLI instrument. Five sample sensors were recorded in the kinetic titration series, and one sensor was recorded as the reference signal for buffer PBS (pH 7.4). Additional six sensors were used as a reference. All procedures were carried out at 25 °C with an agitation speed 1000 rpm. All of the recorded sensorgrams were double referenced against the buffer reference signal and empty sensors by the data acquisition ForteBio software 7.01.0.92. The double referenced signals of each association and dissociation phase were combined and exported into a BioEvaluation 4.1 compatible “csv”-format using a python script (SI: Scripts). The association (ka) and dissociation rate constants (kd) were derived by fitting the recorded sensograms to (1:1) kinetic titration series model, and the affinity constant KD was derived by dividing the dissociation constant (kd) with the association constant (ka) and are tabulated in Table S1. Furthermore, the BLI responses at equilibrium were plotted against each of the injected protein concentrations. The curves were fitted by nonlinear least squares regression.
Yeast two-hybrid assay
Our previously created scFv triple mutant DNA sequences in the pET28a vector were digested with NcoI and EcoRI, and the Fu-bc antigen sequence in the pET28a vector were digested with EcoRI and BamHI. The resulting scFv triple mutant sequences and Fu-bc sequence were cloned in pGBKT7 vector with the fusion of a GAL4 DNA-binding domain and in the pGADT7 vector with the fusion of a GAL4 activation domain, respectively. The bait and prey recombinant vectors were transformed into the yeast Gold and AH109 strains, respectively. Initially, the bait and prey transformants were grown on SD/-Trp and SD/-Leu medium, respectively, at 30 °C for 5 days. Positive clones from each of the bait and prey transformants were grown on SD/-Trp and SD/-Leu liquid medium, respectively, at 30 °C for 2 days, and allowed for overnight mating and then grown on SD/-Leu/-Trp medium at 30 °C for 5 days. Positive clones from each of the transformants were mixed with 20 μl of double distilled water, then 2 μl of the resulting mixture was dropped onto SD/-Trp/-Leu and SD/-Leu/-Trp/-His/-Ade selection medium containing 40 mg/l x-α-Gal and 15 mM 3-Amino-1,2,4-triazole and grown at 30 °C for 2 to 3 days.
Structural study of Fubc–scFv interaction interface
Initially, the DENV–Fab (3IXY) complex was created via molecular replacement using the crystal structure coordinates of the WNV–Fab complex (3I50) (27). However, due to a resolution issue, the DENV E (1OAN) protein's RBS (Fu-bc loop) was colocated in the WNV–Fab complex (3I50), and the VH (1–120) and VL (221–324) chains of Fab (PDB: 2IGF) were joined with a flexible linker of (G4S)3, resulting in the formation of the WNV/DENV–scFv chimeric complex. We overlaid the WNV/DENV–scFv complex on the crystal structure of the WNV–Fab complex to determine the structural basis for the capacity of Fab-derived (PDB: 2IGF) scFv to engage with the RBS (Fu-bc loop) of DENV E protein. Using the Discovery Studio 4.0 tool, the RBS, paratope binding sites, and interaction of these two complexes were comprehensively examined and compared to each other. To demonstrate their identity, the amino acid sequence and 3D structures of RBS (Fu-bc) from the four DENV serotypes were aligned and superposed on the WNV-RBS sequence and structure, respectively. The interaction interface of the WNV/DENV–scFv complex was examined using the Analyze Protein Interface programme of Discovery Studio 4.0 to gain deeper insight into the interactions of each of the scFv-CDR substitution mutations.
Data availability
All the relevant data and plasmids used to support the findings of this study are available for sharing upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
R. D. G. conceptualization; A. S., A. S. R., and R. D. G. data curation; A. S. and R. D. G. formal analysis; R. D. G. funding acquisition; A. S., A. S. R., M. F. K., and R. D. G. investigation; A. S., A. S. R., M. F. K., and R. D. G. resources; R. D. G. supervision; A. S., A. S. R., M. F. K., and R. D. G. validation; A. S. and R. D. G. writing–original draft; A. S. and R. D. G. writing–reviewing and editing.
Funding and additional information
R. D. G. thanks SERB for funding (Project Number: EMR/2016/007246). A. S., A. S. R., and M. F. K. are thankful to South Asian University for PhD fellowships.
Edited by Craig Cameron
Supporting information
Supplemental Figure S1.
Supplemental Figure S2.
Supplemental Figure S3.
Supplemental Figure S4.
Supplemental Figure S5.
Supplemental Figure S6.
Supplemental Figure S7.
References
- 1.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R.W., Simmons C.P., et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasilakis N., Weaver S.C. The history and evolution of human dengue emergence. Adv. Virus Res. 2008;72:1–76. doi: 10.1016/S0065-3527(08)00401-6. [DOI] [PubMed] [Google Scholar]
- 3.Martina B.E.E., Koraka P., Osterhaus A.D.M.E. Dengue virus pathogenesis: An integrated view. Clin. Microbiol. Rev. 2009;22:564–581. doi: 10.1128/CMR.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaño-Osorio J.C., Giraldo A.M.G.-G., I M. Current status of vaccines against dengue virus. Dengue Fever - a Resilient Threat in the Face of Innovation. 2018 doi: 10.5772/intechopen.80820. [DOI] [Google Scholar]
- 5.Guzman M.G., Alvarez M., Halstead S.B. Secondary infection as a risk factor for dengue hemorrhagic fever/dengue shock syndrome: An historical perspective and role of antibody-dependent enhancement of infection. Arch. Virol. 2013;158:1445–1459. doi: 10.1007/s00705-013-1645-3. [DOI] [PubMed] [Google Scholar]
- 6.Capeding M.R., Tran N.H., Hadinegoro S.R.S., Ismail H.I.H.J.M., Chotpitayasunondh T., Chua M.N., Luong C.Q., Rusmil K., Wirawan D.N., Nallusamy R., Pitisuttithum P., Thisyakorn U., Yoon I.-K., van der Vliet D., Langevin E., et al. CYD14 Study Group Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in asia: A phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384:1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 7.Villar L., Dayan G.H., Arredondo-García J.L., Rivera D.M., Cunha R., Deseda C., Reynales H., Costa M.S., Morales-Ramírez J.O., Carrasquilla G., Rey L.C., Dietze R., Luz K., Rivas E., Miranda Montoya M.C., et al. Efficacy of a tetravalent dengue vaccine in children in Latin America. New Engl. J. Med. 2015;372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- 8.Walker L.M., Burton D.R. Passive immunotherapy of viral infections: “super-antibodies” enter the fray. Nat. Rev. Immunol. 2018;18:297–308. doi: 10.1038/nri.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos E.L., Mitcham J.L., Koller T.D., Bonavia A., Usner D.W., Balaratnam G., Fredlund P., Swiderek K.M. Efficacy and safety of treatment with an anti-m2e monoclonal antibody in experimental human influenza. J. Infect. Dis. 2015;211:1038–1044. doi: 10.1093/infdis/jiu539. [DOI] [PubMed] [Google Scholar]
- 10.Caskey M., Klein F., Lorenzi J.C.C., Seaman M.S., West A.P., Buckley N., Kremer G., Nogueira L., Braunschweig M., Scheid J.F., Horwitz J.A., Shimeliovich I., Ben-Avraham S., Witmer-Pack M., Platten M., et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler A.M., Koepsell S.A. The use of convalescent plasma to treat emerging infectious diseases: Focus on ebola virus disease. Curr. Opin. Hematol. 2015;22:521–526. doi: 10.1097/MOH.0000000000000191. [DOI] [PubMed] [Google Scholar]
- 12.Deb P., Molla Md.M.A., Saif-Ur-Rahman K.M. An update to monoclonal antibody as therapeutic option against COVID-19. Biosafety and Health. 2021;3:87–91. doi: 10.1016/j.bsheal.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandt W.E., McCown J.M., Gentry M.K., Russell P.K. Infection enhancement of dengue type 2 virus in the U-937 human monocyte cell line by antibodies to flavivirus cross-reactive determinants. Infect. Immun. 1982;36:1036. doi: 10.1128/iai.36.3.1036-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M., Yang F., Huang D., Huang Y., Zhang X., Wang C., Zhang S., Zhang R. Anti-idiotypic antibodies specific to prM monoantibody prevent antibody dependent enhancement of dengue virus infection. Front. Cell Infect. Microbiol. 2017;7:157. doi: 10.3389/fcimb.2017.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nandi S., Kwong A.T., Holtz B.R., Erwin R.L., Marcel S., McDonald K.A. Techno-economic analysis of a transient plant-based platform for monoclonal antibody production. MAbs. 2016;8:1456–1466. doi: 10.1080/19420862.2016.1227901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramadhany R., Hirai I., Sasaki T., Ono K., Ramasoota P., Ikuta K., Kurosu T. Antibody with an engineered Fc region as a therapeutic agent against dengue virus infection. Antiviral Res. 2015;124:61–68. doi: 10.1016/j.antiviral.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G., Baker T.S., Strauss J.H. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X., Ge P., Yu X., Brannan J.M., Bi G., Zhang Q., Schein S., Zhou Z.H. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Corver J., Chipman P.R., Zhang W., Pletnev S.V., Sedlak D., Baker T.S., Strauss J.H., Kuhn R.J., Rossmann M.G. Structures of immature flavivirus particles. EMBO J. 2003;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modis Y., Ogata S., Clements D., Harrison S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 22.Huang C.Y.-H., Butrapet S., Moss K.J., Childers T., Erb S.M., Calvert A.E., Silengo S.J., Kinney R.M., Blair C.D., Roehrig J.T. The dengue virus type 2 envelope protein fusion peptide is essential for membrane fusion. Virology. 2010;396:305–315. doi: 10.1016/j.virol.2009.10.027. [DOI] [PubMed] [Google Scholar]
- 23.Crill W.D., Hughes H.R., Delorey M.J., Chang G.-J.J. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS One. 2009;4 doi: 10.1371/journal.pone.0004991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai C.-Y., Tsai W.-Y., Lin S.-R., Kao C.-L., Hu H.-P., King C.-C., Wu H.-C., Chang G.-J., Wang W.-K. Antibodies to envelope glycoprotein of dengue virus during the natural course of infection are predominantly cross-reactive and recognize epitopes containing highly conserved residues at the fusion loop of domain II. J. Virol. 2008;82:6631–6643. doi: 10.1128/JVI.00316-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crill W.D., Chang G.-J.J. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 2004;78:13975–13986. doi: 10.1128/JVI.78.24.13975-13986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliphant T., Nybakken G.E., Engle M., Xu Q., Nelson C.A., Sukupolvi-Petty S., Marri A., Lachmi B.-E., Olshevsky U., Fremont D.H., Pierson T.C., Diamond M.S. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherrier M.V., Kaufmann B., Nybakken G.E., Lok S.-M., Warren J.T., Chen B.R., Nelson C.A., Kostyuchenko V.A., Holdaway H.A., Chipman P.R., Kuhn R.J., Diamond M.S., Rossmann M.G., Fremont D.H. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009;28:3269–3276. doi: 10.1038/emboj.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nybakken G.E., Oliphant T., Johnson S., Burke S., Diamond M.S., Fremont D.H. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goncalvez A.P., Engle R.E., St. Claire M., Purcell R.H., Lai C.-J. Monoclonal antibody-mediated enhancement of dengue virus infection in vitro and in vivo and strategies for prevention. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9422–9427. doi: 10.1073/pnas.0703498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathore A.S., Sarker A., Gupta R.D. Designing antibody against highly conserved region of dengue envelope protein by in silico screening of scFv mutant library. PLoS One. 2019;14 doi: 10.1371/journal.pone.0209576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rathore A.S., Sarker A., Gupta R.D. Production and immunogenicity of Fubc subunit protein redesigned from DENV envelope protein. Appl. Microbiol. Biotechnol. 2020;104:4333–4344. doi: 10.1007/s00253-020-10541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma B., Elkayam T., Wolfson H., Nussinov R. Protein–protein interactions: Structurally conserved residues distinguish between binding sites and exposed protein surfaces. PNAS. 2003;100:5772–5777. doi: 10.1073/pnas.1030237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarker A., Rathore A.S., Gupta R.D. Evaluation of scFv protein recovery from E. coli by in vitro refolding and mild solubilization process. Microb. Cell Factories. 2019;18:5. doi: 10.1186/s12934-019-1053-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bird R.E., Hardman K.D., Jacobson J.W. Single-chain antigen-binding proteins. Science. 1988;241:423–426. doi: 10.1126/science.3140379. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the relevant data and plasmids used to support the findings of this study are available for sharing upon request.