Abstract
Duchenne muscular dystrophy (DMD) is a lethal muscle disease caused by mutations in the dystrophin gene. CRISPR/Cas9 genome editing has been used to correct DMD mutations in animal models at young ages. However, the longevity and durability of CRISPR/Cas9 editing remained to be determined. To address these issues, we subjected ΔEx44 DMD mice to systemic delivery of AAV9-expressing CRISPR/Cas9 gene editing components to reframe exon 45 of the dystrophin gene, allowing robust dystrophin expression and maintenance of muscle structure and function. We found that genome correction by CRISPR/Cas9 confers lifelong expression of dystrophin in mice and that corrected skeletal muscle is highly durable and resistant to myofiber necrosis and fibrosis, even in response to chronic injury. In contrast, when muscle fibers were ablated by barium chloride injection, we observed a loss of gene edited dystrophin expression. Analysis of on- and off-target editing in aged mice confirmed the stability of gene correction and the lack of significant off-target editing at 18 months of age. These findings demonstrate the long-term durability of CRISPR/Cas9 genome editing as a therapy for maintaining the integrity and function of DMD muscle, even under conditions of stress.
Keywords: CRISPR/Cas9, gene editing, exon reframing, Duchenne muscular dystrophy, AAV
Graphical abstract

CRISPR/Cas9 genome editing is used to correct Duchenne muscular dystrophy (DMD)-causing mutations. Here, Karri et al. demonstrate the durability and longevity of CRISPR/Cas9 genome editing in a mouse model of DMD, highlighting its potential as a long-term therapeutic for DMD patients.
Introduction
Duchenne muscular dystrophy (DMD) is an X-linked disorder that affects 1 in 5,000 boys worldwide. The disease is caused by the loss of dystrophin, a large sarcolemma protein that connects the actin cytoskeleton network to the extracellular matrix.1,2 In addition, dystrophin interacts with sarcospan, the sarcoglycan complex, dystroglycan, syntrophin, alpha-dystrobrevin, and nitric oxide synthase to form the dystrophin-glycoprotein complex (DGC), which participates in mechanotransduction, muscle growth, and calcium handling.3 The dystrophin protein consists of a N-terminal actin binding domain, 24 spectrin-like repeats, a cysteine-rich region, and a C-terminal scaffold. During muscle contraction, dystrophin functions to transduce force across the sarcolemma and maintains membrane integrity. In the absence of dystrophin, muscle fibers degenerate and are especially susceptible to contraction-induced injury.2 DGC components are also lost or mislocalized in muscles affected by DMD, and signaling downstream of the DGC is disrupted.4, 5, 6, 7 Consequently, DMD muscle is unable to adapt to increased use and undergoes progressive damage and degeneration. As a result of these abnormalities, most DMD patients lose ambulation by the early teenage years and die in the third decade of life due to either dilated cardiomyopathy or respiratory failure.
Over 4,000 mutations in the dystrophin gene (DMD) have been identified in DMD patients.8,9 Approximately 70% of these mutations are exon deletions or duplications that disrupt the DMD open reading frame and introduce a premature stop codon, which prevents the production of functional dystrophin protein.8,9 Many of these mutations are found in a hotspot region containing exons 43–55, which encode the spectrin repeats within the dystrophin protein.10,11 In contrast to DMD, Becker muscular dystrophy (BMD) patients contain in-frame deletions in exons that encode the spectrin repeats. As a result, these patients express functional, truncated forms of dystrophin and can present with mild or no muscle symptoms.12 Thus, exon skipping or reframing strategies that bypass or correct out-of-frame exons potentially allow for restoration of the dystrophin open reading frame and conversion of DMD to the milder BMD.
We and others have worked to correct DMD mutations based on CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) gene editing (reviewed in Chemello et al13; Coi and Koo14; and Olson15). In “single-cut” CRISPR editing, a Cas9 nuclease induces a double-strand DNA break (DSB) near the mutation causing DMD. Subsequently, repair of the DSB by non-homologous end-joining (NHEJ) results in the introduction of insertions or deletions (INDELs) that either reframe an out-of-frame exon or cause exon skipping, allowing the preceding exon to splice into downstream in-frame exons.4,16 These permanent alterations to the mutant DMD allele result in production of a functional dystrophin protein with truncations in the spectrin-like repeats. Using this approach, we have corrected a variety of DMD mutations in animals and cardiomyocytes from patient-derived induced pluripotent stem cells (iPSCs).5,17, 18, 19, 20
Skeletal muscle injury from DMD, as well as chronic use, results in activation of satellite cells residing below the muscle basal lamina, culminating in myofiber regeneration.21 The extent to which satellite cells contribute to the maintenance of muscle structure and function following CRISPR/Cas9 gene editing is currently unresolved. Efficient gene editing in satellite cells following systemic delivery of gene editing components with adeno-associated virus (AAV) would require efficient AAV infectivity as well as the use of gene regulatory elements that allow for Cas9 expression in this specialized stem cell population.
Recently, it has been shown that dystrophin expression is sustained for 18 months in CRISPR-corrected mdx mice, which harbor a nonsense mutation in exon 23 of the dystrophin gene.22,23 However, the effects of chronic injury on corrected DMD muscle are unknown. This is a particularly important issue because of the cycles of degeneration and regeneration and known turnover of skeletal muscle fibers in response to injury. To address these issues, we monitored the longevity of dystrophin expression in gene edited mice with a deletion of Dmd exon 44 (ΔEx44), which represents a prominent deletion mutation in humans. Here, we demonstrate that gene edited dystrophin expression is sustained throughout the lifespan of corrected ΔEx44 mice. In addition, corrected ΔEx44 muscle was resistant to damage induced by chronic injury resulting from high intensity downhill running. In contrast, severe injury induced by BaCl2 injection, which causes loss of muscle fibers, resulted in the loss of gene edited dystrophin expression. These results highlight the potential of single-cut CRISPR/Cas9 genome editing for long-term maintenance of skeletal muscle structure and function in DMD.
Results
CRISPR/Cas9-mediated "single-cut" gene editing in ΔEx44 mice prevents skeletal muscle damage induced by downhill running
Forced downhill treadmill running causes severe muscle injury over time to mdx mice.24,25 To determine whether CRISPR/Cas9 gene edited muscle is resistant to this form of chronic injury, we used a DMD mouse model harboring a deletion of exon 44 in the Dmd gene (ΔEx44).17 We corrected ΔEx44 mice by intraperitoneal administration of AAV9-SpCas9 and AAV9-single-guide RNA (sgRNA) at a 1:1 ratio of 8 × 1013 vector genomes (vg)/kg each on postnatal day 4 (P4), as previously described.19 SpCas9 expression was controlled by the creatine kinase 8 promoter (CK8e), a muscle-specific regulatory element from the MCK gene,26,27 and sgRNA expression was controlled by three different RNA polymerase III promoters, as previously described.4 This strategy induces a single DSB in exon 45 of the Dmd gene, and the INDELs generated by NHEJ-mediated DNA repair reframe exon 45, restoring the open reading frame of dystrophin. Exons 44 and 45 encode for spectrin-like repeat 17 of the central rod domain of dystrophin, which is contained within the neuronal nitric oxide synthase (nNOS) binding site. Therefore, gene editing predominantly produces a near-full-length dystrophin protein that is missing 48 amino acids of the nNOS binding domain.
Immunohistochemistry (IHC) of the tibialis anterior (TA) muscle at 1 month of age showed that >90% of corrected ΔEx44 muscle fibers expressed dystrophin (Figure S1A). In addition, corrected ΔEx44 mice exhibited a 90% reduction in serum creatine kinase (CK) activity, an indicator of muscle damage, when compared with uncorrected ΔEx44 mice (Figure S1B). These results validate the efficient rescue of dystrophin expression in corrected ΔEx44 mice.
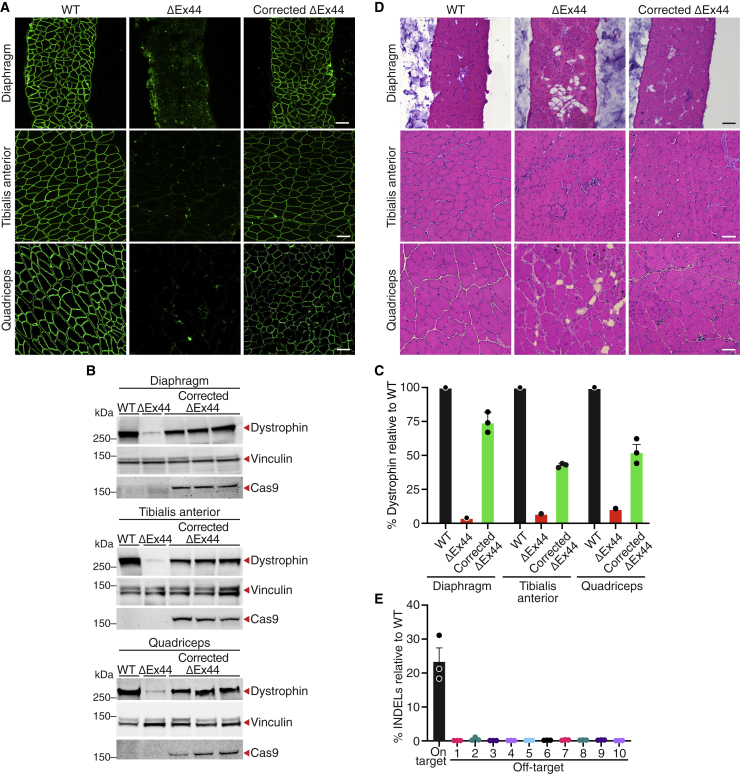
To induce chronic injury, 4-week-old wild-type (WT), ΔEx44, and corrected ΔEx44 mice were subjected to a protocol of forced downhill running modified from TREAT-Neuromuscular Disease guidelines (Figure 1A).24 After downhill running, WT mice did not show centralized nuclei, a marker of skeletal muscle regeneration, or necrosis (Figures 1B–1D and S2). In contrast, ΔEx44 mice exhibited prominent centralized nuclei and necrosis in the quadriceps, TA, and diaphragm muscles following downhill running (Figures 1B–1D and S2). The ΔEx44 quadriceps muscle was the most affected, with a 20% increase in centralized nuclei and a 15% increase in necrotic area (Figures 1C, 1D, and S2). Therefore, high-speed downhill running selectively induced skeletal muscle damage and subsequent necrosis in ΔEx44 mice.
Figure 1.
CRISPR/Cas9 genome editing prevents skeletal muscle injury induced by downhill running in ΔEx44 mice
(A) ΔEx44 mice were injected intraperitoneally (IP) with ssAAV-SpCas9 and scAAV-sgRNA, each at 8 × 1013 vg/kg on postnatal day 4 (P4) (corrected ΔEx44). At 4 weeks of age, mice were subjected to sedentary or downhill running conditions for 4 weeks. (B) H&E staining of the quadriceps muscle from WT, ΔEx44, and corrected ΔEx44 mice that were either sedentary or run downhill. Scale bar, 100 μm. (C) Quantification of centralized nuclei in sedentary or downhill run WT, ΔEx44, and corrected ΔEx44 quadriceps muscle. Data are shown as mean ± SEM. Unpaired Student’s t test was performed. ∗∗∗p < 0.001 (n = 6). (D) Quantification of necrotic area in sedentary or downhill run WT, ΔEx44, and corrected ΔEx44 quadriceps muscle. Data are shown as mean ± SEM. Unpaired Student’s t test was performed. ∗∗∗∗p < 0.0001 (n = 6). (E) Hindlimb grip strengths of WT, ΔEx44, and corrected ΔEx44 mice, measured following 4 weeks of downhill running. Hindlimb grip strength was normalized to body weight. Data are shown as mean ± SEM. A one-way ANOVA and a post hoc Tukey’s multiple comparison test were performed. ∗∗∗p < 0.001 (n ≥ 5). (F) Specific force generated by the extensor digitorum longus (EDL) and soleus muscles of WT, ΔEx44, and corrected ΔEx44 mice following 4 weeks of downhill running. Data are shown as mean ± SEM. A one-way ANOVA and a post hoc Tukey’s multiple comparison test were performed. ∗p < 0.05, ∗∗p < 0.005 (n ≥ 5).
After CRISPR/Cas9-mediated "single-cut" genome editing, skeletal muscles of ΔEx44 mice were protected from damage. Under sedentary conditions, corrected ΔEx44 muscle contained <5% centralized nuclei and <1% necrosis (Figures 1B–1D and S2). After downhill running, there was no significant increase in centralized nuclei or necrosis in corrected ΔEx44 skeletal muscle (Figures 1B–1D and S2). In addition, following downhill running, the serum CK activity of corrected ΔEx44 mice was 10% of uncorrected ΔEx44 mice (Figure S3). Because serum CK activity is a marker for muscle fiber integrity, these results demonstrate that corrected ΔEx44 muscle fibers are resistant to damage induced by downhill running.
To assess corrected ΔEx44 skeletal muscle function following chronic injury, we performed hindlimb grip strength tests and ex vivo electrophysiological analyses of the extensor digitorum longus (EDL) and soleus muscles. Following downhill running, we observed a 40% reduction in the grip strength of ΔEx44 mice relative to WT mice (Figure 1E). However, the grip strengths of corrected ΔEx44 and WT mice were indistinguishable after downhill running (Figure 1E). Consistent with these results, following downhill running, the maximum specific forces of ΔEx44 EDL and soleus muscles were 65% of the forces generated by WT muscles (Figure 1F). In contrast, the maximum specific forces of corrected ΔEx44 EDL and soleus muscles were equivalent to the WT controls (Figure 1F). Together, these results demonstrate that CRISPR/Cas9 gene editing provides functional benefit to ΔEx44 muscle subjected to chronic injury.
IHC for dystrophin revealed the lack of dystrophin-positive fibers in sedentary and downhill run ΔEx44 skeletal muscle (Figure 2A). In contrast, >75% of muscle fibers expressed dystrophin in the diaphragm, TA, quadriceps, EDL, and soleus muscles of sedentary and downhill-run corrected ΔEx44 mice (Figures 2A, S4, S5A, and S5B). Western blot analysis indicated the absence of dystrophin expression in sedentary and downhill run ΔEx44 mice (Figures 2B, 2C, S5C, and S5D). In sedentary corrected ΔEx44 mice, there was >30% rescue of dystrophin protein across various muscle groups relative to WT (Figures 2B, 2C, S5C, and S5D). After downhill running, gene edited dystrophin protein levels remained constant in corrected ΔEx44 mice (Figures 2B, 2C, S5C, and S5D). Consistent with these findings, we observed no decrease in on-target INDELs or AAV9-SpCas9 viral genomes in the quadriceps muscles of corrected ΔEx44 mice following downhill running (Figures S6 and S7). Thus, CRISPR/Cas9-mediated gene editing in ΔEx44 mice restores dystrophin expression, prevents muscle damage, and provides functional benefit following chronic injury.
Figure 2.
Expression of dystrophin is retained in corrected ΔEx44 mice following chronic injury
(A) Immunohistochemistry shows retention of dystrophin-positive fibers in the diaphragm, tibialis anterior, and quadriceps muscles of corrected ΔEx44 mice following 4 weeks of downhill running. Dystrophin is shown in green. Scale bar, 100 μm. (B) Western blot analysis shows retention of dystrophin protein in the diaphragm, tibialis anterior, and quadriceps muscles of corrected ΔEx44 mice following 4 weeks of downhill running. Vinculin was loading control. (C) Quantification of dystrophin protein in diaphragm, tibialis anterior, and quadriceps muscles. Dystrophin protein levels were first normalized to vinculin loading control and then to WT sedentary controls. Data are shown as mean ± SEM. Unpaired Student’s t test was performed (n = 3).
CRISPR/Cas9 gene editing normalizes the transcriptome of corrected ΔEx44 skeletal muscle
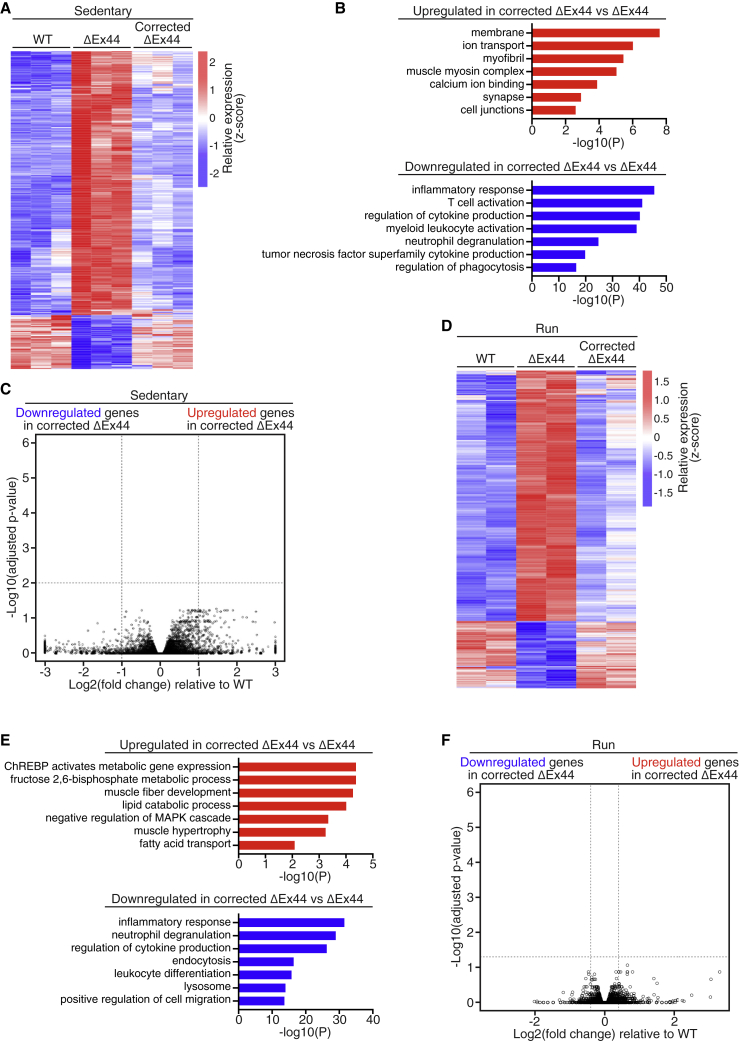
The DGC preserves muscle fiber integrity and participates in mechanical signal transduction.3,6 Previous work demonstrated that CRISPR/Cas9 correction of dystrophin mutations restores the DGC to the sarcolemma.4,5,28,29 However, in corrected DMD skeletal muscle, it is unknown whether gene expression pathways downstream of the DGC are properly restored. Therefore, we compared gene expression profiles of quadriceps muscles from WT, ΔEx44, and corrected ΔEx44 mice by RNA sequencing (RNA-seq) analysis.
In quadriceps muscles from 4-week-old ΔEx44 mice, we identified 1,222 upregulated and 216 downregulated genes (>2-fold difference, adjusted p < 0.01) compared with WT muscle (Figure 3A). Gene ontology (GO) analysis revealed that the upregulated genes were enriched in terms such as innate immune response, chemotaxis, cellular response to interferon gamma, and the adaptive immune response (Figure S8A). In DMD, membrane damage elicits uptake of cellular debris by the innate immune system, and subsequent cytokine release activates the adaptive immune response.30 Meanwhile, GO analysis of the downregulated genes revealed terms related to cell junctions, Z disc, I band, and skeletal muscle thin filament assembly, consistent with the degeneration of DMD skeletal muscle (Figure S8A).
Figure 3.
Transcriptional homeostasis is maintained in quadriceps muscle of corrected ΔEx44 mice following downhill running
(A) Heatmap of differentially expressed genes in the quadriceps muscles of 4-week-old WT, ΔEx44, and corrected ΔEx44 mice. Gene expression is represented as a transformed Z score (n = 3). (B) Selected GO terms for up- and downregulated genes in corrected ΔEx44 quadriceps muscle relative to ΔEx44 quadriceps muscle. (C) Volcano plot of differentially expressed genes between corrected ΔEx44 and WT quadriceps muscles. (D) Heatmap of differentially expressed genes in the quadriceps of 8-week-old WT, ΔEx44, and corrected ΔEx44 mice following 4 weeks of downhill running. Gene expression is represented as a transformed Z score (n = 2). (E) Selected GO terms for up- and downregulated genes in downhill-run-corrected ΔEx44 quadriceps muscle relative to ΔEx44 quadriceps muscle. (F) Volcano plot of differentially expressed genes between downhill-run-corrected ΔEx44 and WT quadriceps muscles.
To determine the effects of gene editing, we compared the transcriptomes of WT, ΔEx44, and corrected ΔEx44 quadriceps muscles (Figure 3A). Compared with ΔEx44 muscle, there were 187 upregulated and 1,142 downregulated genes (>2-fold difference, adjusted p < 0.01) in corrected ΔEx44 quadriceps muscle. Upregulated genes were related to myofibril, muscle myosin complex, and integral components of the membrane (Figure 3B). Downregulated genes were related to T cell activation, regulation of cytokine production, tumor necrosis factor superfamily cytokine production, and regulation of phagocytosis (Figure 3B). Therefore, CRISPR/Cas9 genome editing of the ΔEx44 mutation normalizes the DMD phenotype at the transcriptomic level.
Differential gene expression analysis of the transcriptomes of WT and corrected ΔEx44 quadriceps muscles revealed no significant differentially expressed genes between the two groups (Figure 3C). Consistent with these findings, principal-component analysis (PCA) revealed high divergence between the transcriptomes of WT and ΔEx44 quadriceps muscles (Figure S8B), whereas the transcriptomes of WT and corrected ΔEx44 quadriceps muscles were highly similar (Figure S8B).
Next, we sought to determine whether the normalization of gene expression of DMD muscle by gene editing was maintained following chronic stress from forced downhill running. Therefore, we conducted RNA-seq analysis of quadriceps muscles from 8-week-old WT, ΔEx44, and corrected ΔEx44 mice that had undergone 4 weeks of downhill running (Figure 3D). Differential gene expression analysis revealed 679 upregulated and 149 downregulated genes (>2-fold difference, adjusted p < 0.05) in the downhill run ΔEx44 quadriceps muscles relative to WT (Figure 3D). GO analysis of upregulated genes revealed terms related to immune system processes, phagocytosis, and the external side of the plasma membrane (Figure S8C). Downregulated genes were related to cell junctions, membrane, and neuron projections (Figure S8C). These transcriptional changes are consistent with the increased muscle damage and necrosis observed in the downhill run ΔEx44 quadriceps muscles.
Comparison of the transcriptomes of the downhill run ΔEx44 and corrected ΔEx44 quadriceps muscles revealed 510 dysregulated genes between the two groups (>2-fold difference, adjusted p < 0.05) (Figure 3D). GO analysis of the downregulated genes in corrected ΔEx44 quadriceps muscles revealed terms related to inflammatory response, lysosome, and extracellular matrix deposition (Figure 3E). These downregulated genes indicated that CRISPR/Cas9 genome editing preserved muscle fiber integrity following downhill running. These results are consistent with histology and muscle function findings (Figures 1 and S2). Interestingly, GO analysis of the upregulated genes in corrected ΔEx44 quadriceps muscles revealed terms related to mitogen-activated protein kinase (MAPK) activity, glucose and lipid metabolism, and muscle fiber development (Figure 3E). In normal muscle, running has been shown to activate MAPK and downstream signaling to promote fat and glucose utilization to meet increased metabolic demands.31,32 In DMD muscle, repeated damage to the sarcolemma does not allow muscle fibers to properly adapt to running.33 Therefore, our results suggest that gene edited dystrophin expression is sufficient to preserve muscle fiber integrity and to permit proper adaptations to downhill running.
Finally, to assess the degree of transcriptional rescue in downhill-run corrected ΔEx44 mice, we compared differentially expressed genes between WT and corrected ΔEx44 quadriceps muscles following downhill running. Differential gene expression analysis revealed no significant differentially expressed genes between the two groups (Figure 3F). To validate these findings, we conducted PCA of downhill run WT, ΔEx44, and corrected ΔEx44 quadriceps muscles (Figure S8D). While ΔEx44 and corrected ΔEx44 quadriceps muscles were highly divergent, WT and corrected ΔEx44 quadriceps muscles clustered closely (Figure S8D). Therefore, our RNA-seq results indicate that CRISPR/Cas9 genome editing of the ΔEx44 mutation rescues the transcriptome of DMD muscle, and that transcriptional homeostasis is preserved following chronic injury caused by downhill running.
Destruction of myofibers results in the loss of gene edited dystrophin expression
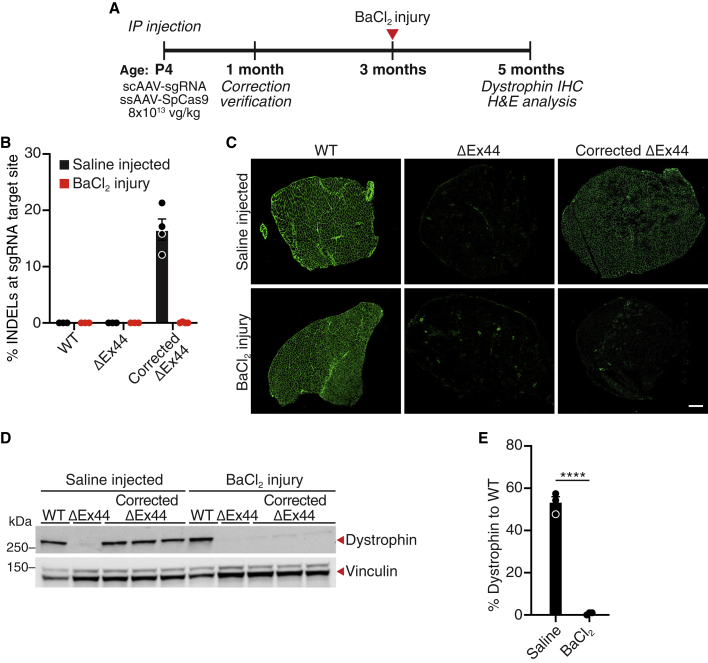
Recently, it was reported that AAV9 can transduce satellite cells.34, 35, 36 The muscle-specific CK8e promoter has also been suggested to be active in satellite cells, although there is debate as to the level of activity of this promoter in these cells.36 Injection of muscle with BaCl2 results in rapid ablation of muscle fibers and is followed by regeneration via satellite cell activation.37 To determine whether activated satellite cells were capable of restoring dystrophin expression following myofiber ablation, we injected TA muscles of 3-month-old WT, ΔEx44, and corrected ΔEx44 mice with BaCl2 to induce acute injury (Figure 4A). Two months following BaCl2 injection, H&E analysis of WT TA muscle showed muscle fibers with centralized nuclei and no necrosis, indicative of robust regeneration (Figure S9). In contrast, corrected ΔEx44 TA muscle exhibited small necrotic foci similar to those of uncorrected ΔEx44 TA muscle, suggesting impaired muscle regeneration (Figure S9). To determine whether this impaired muscle regeneration was due to a loss in gene edited muscle nuclei, we performed genomic analysis of BaCl2 injected, corrected ΔEx44 TA muscle and saline injected contralateral controls (Figure 4B). In the saline injected, corrected ΔEx44 TA muscle, we observed ∼20% INDELs within exon 45 (Figure 4B). However, we did not detect INDELs in corrected ΔEx44 TA muscle injected with BaCl2, suggesting a complete loss of CRISPR/Cas9 genome editing (Figure 4B). Consistent with these results, IHC showed a loss of almost all dystrophin-positive fibers in BaCl2 treated, corrected ΔEx44 TA muscle when compared with saline injected controls (Figure 4C). Western blot analysis showed <5% of WT dystrophin protein levels in corrected ΔEx44 TA muscle following acute injury (Figure 4D). In addition, we were unable to detect ssAAV-SpCas9 viral genomes in BaCl2 treated, corrected ΔEx44 TA muscle (Figure S10). Together, these results suggest that satellite cells are not efficiently edited by the AAV delivery strategy deployed in these experiments. As a result, following BaCl2 induced injury, CRISPR/Cas9 gene edited myofiber nuclei were replaced by unedited satellite cells, resulting in the loss of gene edited dystrophin expression.
Figure 4.
BaCl2-induced acute injury results in the loss of gene edited dystrophin
(A) ΔEx44 mice were injected intraperitoneally (IP) with ssAAV-SpCas9 and scAAV-sgRNA, each at 8 × 1013 vg/kg on postnatal day 4 (P4) (corrected ΔEx44). At 3 months of age, the corrected ΔEx44 tibialis anterior muscle was injected with BaCl2. Two months after BaCl2 injury, tissues were harvested for analysis. (B) TIDE analysis of Dmd exon 45 in saline-injected or BaCl2 treated WT, ΔEx44, and corrected ΔEx44 tibialis anterior muscle. (C) Immunohistochemistry shows the loss of dystrophin-positive fibers in corrected ΔEx44 tibialis anterior muscle following BaCl2 injury. Dystrophin is shown in green. Scale bar, 250 μm (n = 4). (D) Western blot analysis shows a loss of dystrophin protein in the tibialis anterior muscle of corrected ΔEx44 mice following BaCl2 injury. Vinculin was loading control. (E) Quantification of dystrophin protein in saline or BaCl2 injected, corrected ΔEx44 TA muscle. Dystrophin protein levels were first normalized to vinculin loading control and then to corresponding WT controls. Data are shown as mean ± SEM. Unpaired Student’s t test was performed. ∗∗∗∗p < 0.0001 (n = 3).
Gene edited dystrophin expression is sustained in 18-month-old corrected ΔEx44 mice
To determine whether gene edited dystrophin was expressed throughout the lifespan of corrected ΔEx44 mice, we analyzed 18-month-old corrected ΔEx44 mice for gene edited dystrophin expression. IHC showed no dystrophin-positive fibers in the ΔEx44 diaphragm, TA, and quadriceps muscles (Figure 5A). In contrast, >90% of muscle fibers in the corrected ΔEx44 diaphragm, TA, and quadriceps muscles were dystrophin positive (Figures 5A and S11). Consistent with these findings, western blot analysis did not detect dystrophin in the ΔEx44 skeletal muscle, whereas there was >40% rescue of dystrophin protein expression in the corrected ΔEx44 diaphragm, TA, and quadriceps muscles, relative to WT (Figures 5B and 5C).
Figure 5.
Gene edited dystrophin is retained in 18-month-old corrected ΔEx44 mice
(A) Immunohistochemistry shows dystrophin-positive muscle fibers in the diaphragm and tibialis anterior of 18-month-old corrected ΔEx44 mice. Dystrophin is shown in green. Scale bar, 100 μm. (B) Western blot analysis shows sustained dystrophin and SpCas9 expression in the diaphragm, tibialis anterior, and quadriceps muscles of 18-month-old corrected ΔEx44 mice. Vinculin was loading control. (C) Quantification of dystrophin protein in the diaphragm, tibialis anterior, and quadriceps muscles. Dystrophin protein levels were first normalized to vinculin loading control and then to WT controls. Data are shown as mean ± SEM. (D) H&E staining of the diaphragm, tibialis anterior, and quadriceps muscles from 18-month-old WT, ΔEx44, and corrected ΔEx44 mice. Scale bar, 100 μm. (E) INDEL analysis of the on-target and top 10 off-target sites in 18-month-old corrected ΔEx44 tibialis anterior muscle. INDELs have been normalized to a WT control (n = 3).
To determine whether CRISPR/Cas9 genome editing could rescue muscle degeneration seen in 18-month-old ΔEx44 mice, we performed H&E analysis (Figure 5D). In corrected ΔEx44 mice, we observed <1% necrosis and <10% centrally nucleated muscle fibers in the diaphragm, TA, and quadriceps muscles (Figures 5D and S12). Together, these findings demonstrate that sustained gene edited dystrophin expression prevents skeletal muscle degeneration in 18-month-old corrected ΔEx44 mice.
Analysis of SpCas9 expression and gene editing in aged mice
In addition to sustained dystrophin protein expression, we observed significant SpCas9 protein expression in corrected ΔEx44 mice at 18 months of age (Figure 4B). To determine the long-term effects of SpCas9 in skeletal muscle, we performed amplicon-based deep sequencing on genomic DNA isolated from 18-month-old corrected ΔEx44 TA. We observed, on average, 23.5% INDEL formation in exon 45 of the Dmd gene (Figure 5E). Tracking of Indels by Decomposition (TIDE) analysis indicated that there was a 15% increase in the total on-target INDEL formation compared with 4-week-old corrected ΔEx44 TA (Figure S13). Approximately 80% of these INDELs were a single-nucleotide insertion or a two-nucleotide deletion at the predicted double-stranded DNA cleavage site, which results in productive reframing of the dystrophin transcript (Figure S14 and Table S1).17
Also, using CRISPR design tools (Benchling), we determined the top 10 potential off-target sites, as previously described.17 We did not observe significant INDELs at these predicted off-target sites (Figure 5E). In addition, we observed <1% AAV genome integration at the target site and none at the potential off-target sites (Table S1). The rates of off-target editing and AAV integration were similar to those previously reported in 4-week-old corrected ΔEx44 TA, suggesting that sustained expression of SpCas9 does not result in significant off-target editing.17
Transcriptional homeostasis is retained in 18-month-old corrected ΔEx44 skeletal muscle
To determine the longevity of CRISPR/Cas9-mediated transcriptional homeostasis, we performed RNA-seq of quadriceps muscles from 18-month-old mice (Figure 6A). In ΔEx44 quadriceps muscle, there were 1,571 upregulated and 524 downregulated genes when compared with WT (>2-fold change, p < 0.01). GO analysis of upregulated genes revealed terms related to immune system processes, positive regulation of tumor necrosis factor, and signal transduction (Figure S15A). Downregulated genes were related to membrane, ion channel activity, and muscle contraction (Figure S15A). These findings are consistent with the severe muscle degeneration observed in 18-month-old ΔEx44 skeletal muscle.
Figure 6.
Transcriptional homeostasis is maintained in 18-month-old corrected ΔEx44 quadriceps muscle
(A) Heatmap of differentially expressed genes in 18-month-old WT, ΔEx44, and corrected ΔEx44 quadriceps muscles. Gene expression is represented as a transformed Z score (n = 3). (B) Selected GO terms for up- and downregulated genes in 18-month-old corrected ΔEx44 quadriceps muscle relative to ΔEx44 quadriceps muscle. (C) Volcano plot of differentially expressed genes between 18-month-old corrected ΔEx44 and WT quadriceps muscles.
Next, we compared the transcriptomes of 18-month-old corrected ΔEx44 and ΔEx44 quadriceps muscles. Compared with ΔEx44 quadriceps muscles, there were 193 upregulated and 526 downregulated genes in corrected ΔEx44 quadriceps muscles (>2-fold change, p < 0.01). GO analysis of upregulated genes revealed terms related to glycogen metabolic processes, cell junctions, ion channel activity, sarcoplasmic reticulum, and synapses (Figure 6B). Downregulated genes were related to the extracellular region, cell adhesion, and fat cell differentiation (Figure 6B). These findings suggest that corrected ΔEx44 muscle fibers are largely intact and are not replaced by adipose tissue and fibrosis, which is also observed histologically (Figure 5D).
To evaluate the extent of this transcriptional rescue, we compared the transcriptomes of 18-month-old corrected ΔEx44 and WT quadriceps muscles. Only 18 genes were dysregulated in corrected ΔEx44 quadriceps muscles compared with WT (>2-fold change, p < 0.05). Upregulated genes were related to immune system processes (Timp1, Lcn2, Mt2, Ccl6, and Nox4) and to regenerating muscle fibers (Myh3, Myl4, and Actc1) (Figure 6C). Downregulated genes were involved in energy utilization (Plin1 and Tiam1) (Figure 6C). Additionally, we observed a downregulation in Cd28, which is also observed in chronic inflammation (Figure 6C).38 These results indicate activation of the immune system in response to muscle fiber damage. However, considering the small number of dysregulated genes, the inflammation in 18-month-old corrected ΔEx44 quadriceps muscle is very mild. Consistent with this conclusion, there was only a small distance between corrected ΔEx44 and WT samples in a PCA plot (Figure S15B), suggesting that the transcriptional rescue observed in corrected ΔEx44 skeletal muscle persists with aging.
Discussion
Multiple studies have utilized CRISPR/Cas9 gene editing to rescue dystrophin expression in mouse models of DMD (reviewed in Chemello et al13; Coi and Koo14; and Olson15). However, the durability of gene edited DMD skeletal muscle following chronic injury has not been fully explored. Because mice lacking dystrophin maintain mobility and display a relatively modest phenotype,39 we used high-speed downhill treadmill running to exacerbate the DMD skeletal muscle phenotype. Here, we demonstrate that CRISPR/Cas9 gene edited ΔEx44 skeletal muscle is resistant to damage induced by downhill running. Expression of gene edited dystrophin protein was also maintained following chronic injury.
Previous work showed that restoration of ∼15% of dystrophin protein is sufficient to maintain muscle fiber integrity in mdx mice.40,41 In corrected ΔEx44 skeletal muscle, we observed rescue of dystrophin expression to >30% of normal levels in almost all muscle groups. Thus, there was sufficient dystrophin to stabilize the sarcolemma of corrected ΔEx44 muscle fibers. We and others have reported that CRISPR/Cas9 gene editing can restore the DGC at the sarcolemma.4,5,17,28,29 In the present study, corrected ΔEx44 and WT skeletal muscle transcriptomes were indistinguishable after chronic injury. Therefore, we hypothesize that mechanical signal transduction downstream of the DGC was restored in corrected ΔEx44 skeletal muscle, allowing adaptation to the eccentric contractions induced by chronic injury. Together, these results suggest that CRISPR/Cas9 genome editing can preserve muscle fiber integrity. Thus, the durability of gene edited DMD muscle demonstrates the promise of CRISPR/Cas9 genome editing as a potential therapeutic for DMD.
Remarkably, we observed >40% restoration of dystrophin protein after 18 months, demonstrating the stability of gene edited myofibers. Our in vivo CRISPR/Cas9 strategy utilizes the muscle-specific CK8e promoter to drive SpCas9 expression. The CK8e promoter is not highly active in the resident muscle stem cells.36 Therefore, we would expect a decrease in dystrophin expression with aging due to the fusion of unedited resident stem cells with corrected DMD muscle fibers. However, we observed an increase in on-target INDEL formation with age. Also, we observed significant SpCas9 expression at 18 months. It is possible that sustained SpCas9 expression resulted in editing of newly incorporated muscle nuclei.
The sustained expression of SpCas9 at 18 months raises a potential concern of increased off-target genome editing over time. Importantly, however, we observed <1% off-target genome editing in 18-month-old corrected ΔEx44 TA muscle. In addition, there was <1% of AAV integration at the target locus and no observable AAV integration at the top 10 predicted off-target sites. These low off-target editing rates, combined with the postmitotic nature of skeletal muscle, diminish the likelihood of malignant transformations resulting from long-term systemic gene editing.
Our results add to a growing body of work suggesting that CRISPR/Cas9 gene editing may confer long-term benefits in DMD.22,23 The clinical efficacy and safety of AAV9 were demonstrated in successful SMA gene-therapy trials.42 However, AAV can only be effectively delivered once due to the development of a humoral immune response.43 Importantly, our findings suggest that a single CRISPR AAV dose is sufficient to maintain long-term gene edited dystrophin expression. It will be of interest to monitor gene edited dystrophin expression over longer times in larger mammalian DMD models in which the progression of muscle fiber degeneration is much more severe than in mice.
In this study, we showed that CRISPR/Cas9 genome editing protected ΔEx44 skeletal muscle from chronic injury caused by downhill running. However, acute injury induced by BaCl2 resulted in the loss of gene edited dystrophin. Recent studies have shown that AAV9 can transduce satellite cells, albeit relatively inefficiently.35,36 However, the CK8e promoter shows low activity in satellite cells.36 We hypothesize that low promoter activity, combined with relatively low tropism of AAV9 for satellite cells, resulted in inefficient satellite cell genome editing in corrected ΔEx44 mice. Therefore, following BaCl2 injury, muscle was likely reconstituted by unedited satellite cells; hence, we observed little or no dystrophin expression in this setting. In the future, it will be of interest to design a promoter that is highly active in skeletal muscle and satellite cells. Ubiquitously expressed promoters such as CMV are not desirable due to accumulation of Cas9 and editing of non-target tissues.
While acute injury resulted in the loss of gene edited dystrophin, these findings do not diminish the potential of CRISPR/Cas9 genome editing as a long-term therapeutic for DMD. Injury induced by BaCl2 is more acute and severe than in DMD. Under physiologically relevant stress, as in our chronic injury model and in aging, corrected muscle nuclei are retained and gene edited dystrophin is sustained.
Finally, we recently developed a base editing strategy to induce exon skipping in DMD by disrupting exon splice sites.44 In addition, new genome editing technologies, such as prime editing, have been developed.44,45 In the future, it will be of interest to determine the durability of gene edited dystrophin resulting from these new strategies.
Materials and methods
Study design
This study aimed to determine the durability and longevity of "single-cut" CRISPR/Cas9 gene editing in a mouse model of DMD. Exclusion, randomization, or blinding was not used to assign mice to experiments. Animal work described in this manuscript has been approved and conducted under the oversight of the UT Southwestern Institutional Animal Care and Use Committee. Sample sizes for each experiment are stated in the figure legends and represent independent biological replicates.
AAV9 production
Single-stranded (ss) and self-complementary (sc) AAV9 were produced and purified by Boston Children’s Hospital Viral Core, as previously described.19 These AAV9 vectors expressed CRISPR/Cas9 genome editing components that target the ΔEx44 mutation in the Dmd gene. AAV9 titers were determined with qPCR, as previously described.17
In vivo delivery of CRISPR AAV9 to ΔEx44 mice
On P4, ΔEx44 male pups were injected intraperitoneally with 80 uL of a viral mixture containing AAV-SpCas9 and AAV-sgRNA, as previously described.17,19 For long-term studies, viruses were delivered at doses of 5 × 1013 vg/kg for ssAAV-SpCas9 and 2.5 × 1014 vg/kg for ssAAV-sgRNA. While these longevity studies were in progress, our group discovered that packaging the sgRNA expression cassette in scAAV increases sgRNA expression and subsequently enhances gene editing efficiency.19 Therefore, for injury studies and transcriptome analyses in younger mice, viruses were delivered at doses of 8 × 1013 vg/kg for ssAAV-SpCas9 and 8 × 1013 vg/kg for scAAV-sgRNA. At various time points following systemic AAV9 delivery, mice were sacrificed for physiology, histology, and molecular analysis.
Downhill running
Prior to the forced running regime, WT, ΔEx44, and corrected ΔEx44 mice were acclimatized to the treadmill (C Exer-3/6M, Columbus Instruments), as previously described.24 Then, 4-week-old mice were run (12 m/min at a 25° decline) on alternating days for 1 month. The duration of each running session was 30 min.
Histological analysis of skeletal muscle
Skeletal muscle was cryosectioned into 8 μm sections, and IHC or H&E staining was conducted, as previously described.17 For IHC, mouse anti-dystrophin antibody (1:1000, MANDYS8, Sigma-Aldrich, D8168) and Mouse on Mouse biotinylated anti-mouse IgG (BMK-2202, Vector Laboratories) were used.
Western blot for dystrophin and SpCas9
Skeletal muscle was crushed and lysed in 10% SDS, 62.5 mM Tris (pH 6.8), 1 mM EDTA, and protease inhibitor. Protein, 50 μm, was loaded onto a 4%–20% Criterion TGX Stain-Free Protein Gel. SDS-PAGE electrophoresis, membrane transfer, and membrane blotting were conducted as previously described.17 Antibodies used for western blot analysis were mouse anti-dystrophin antibody (1:1000, MANDYS8, Sigma-Aldrich, D8168), mouse anti-SpCas9 Antibody (1:1000, clone 7A9, Millipore, MAC133), and mouse anti-vinculin antibody (1:1000, Sigma-Aldrich, V9131). Secondary antibody was goat anti-mouse horseradish peroxidase (HRP) antibody (1:10,000, BioRad).
Muscle physiology
Following 4 weeks of downhill running, the EDL and soleus muscles were isolated from WT, ΔEx44, or corrected ΔEx44 mice, as previously described.17 Contraction-induced specific force (mN/mm2) was measured as previously described.17
Serum creatine measurement
Blood was collected via the submandibular vein and then centrifuged at 15,000 × g at 4°C to collect the serum. CK activity was measured using VITROS Chemistry 7 slides, as per manufacturer instructions.
Grip strength measurements
In a blinded study, grip strengths of WT, ΔEx44, and corrected ΔEx44 mice were measured using a grip strength meter (Columbus Instruments), as previously described.18
BaCl2 induced injury
Mice were anesthetized with 5% Avertin. The right TA muscle was injected with 50 μL of 1.2% (w/v) BaCl2 in saline. As an uninjured control, in the same mouse, the left TA muscle was injected with 50 uL of saline. Two months following injury, the TA muscles were harvested and analyzed for genome editing, dystrophin, and histology.
Viral genome quantification in skeletal muscle
AAV-SpCas9 viral copy numbers were quantified with qPCR, as previously described.19 Primers used for qPCR annealed to the SpCas9 gene and are listed in Table S2.
RNA sequencing
Cryopreserved quadriceps muscle was homogenized in TRIzol (Thermo Fisher Scientific). RNA was isolated using QIAGEN RNeasy columns, as per manufacturer instructions. mRNA library preparation was conducted using a KAPA mRNA HyperPrep Kit (Roche). Deep sequencing and analysis were performed as previously described.46
TIDE analysis
PCR was conducted to amplify a 500 bp region around the target site in exon 45 of the Dmd gene. PCR primers are listed in Table S2. Then, Sanger sequencing was conducted. INDELs were analyzed using the TIDE package (https://www.tide.deskgen.com), as previously described.17,19
Amplicon deep sequencing analysis of genomic DNA
Genomic DNA was isolated from skeletal muscle using a DNeasy and Blood & Tissue Kit (QIAGEN). PCR was conducted to amplify genomic regions containing the CRISPR/Cas9 target site and predicted off-target sites, as previously described.17 A second round of PCR was performed to add Illumina flow cell binding sequences and barcodes. Primers are listed in Table S3. Deep sequencing and analysis were conducted as previously described.17
Data availability
Data to evaluate the conclusions in this paper are presented in the paper figures and supplemental data. RNA-seq data have been uploaded to GEO. GEO profile:GSE180771. Additional data pertaining to this paper can be requested from the authors.
Statistics
Data are represented as mean ± SEM. To compare two groups, an unpaired Student’s t test was performed. To compare multiple groups, one-way ANOVA and Tukey’s multiple comparison tests were performed; p < 0.05 was considered statistically significant.
Acknowledgments
We thank A. Mireault for tissue harvesting and performing the grip strength assay; E. Sanchez-Ortiz for performing IHC; H. Li for TIDE analysis and protocols for amplicon-based deep sequencing; X. Menédez Caravia for help with the BaCl2 study; J. Cabrera for graphics; the Boston Children’s Hospital Viral Core for AAV production; the Metabolic Phenotyping Core for serum CK analysis; the Sanger Sequencing Core for sequencing services; and the Histology Core for H&E staining. We are grateful to S. Hauschka (University of Washington) for providing the muscle-specific CK8e promoter, to D. Grimm (Heidelberg University Hospital, Germany) for providing TRISPR-sgRNA expression plasmid, and S. Gray (University of Texas Southwestern Medical Center) for providing the self-complementary AAV plasmid. This work was supported by the National Institutes of Health (NIH) (grants HL130253), Senator Paul D. Wellstone Muscular Dystrophy Specialized Research Center (grant P50 HD 087351), and Robert A. Welch Foundation (grant 1-0025 to E.N.O.).
Author contributions
D.R.K., N.L., R.B.-D., and E.N.O. wrote and edited the manuscript. D.R.K., Y.Z, R.B.-D., and E.N.O. designed the experiments. D.R.K. performed downhill running studies and BaCl2 injury and analyzed data. D.R.K. and F.C. performed the older mouse study and analyzed data. Y.-L.M. generated the mouse model. Y.-L.M. and Y.Z. designed the genome editing strategy. J.H. performed the electrophysiology studies. P.P.A.M. provided oversight of the electrophysiology analysis. J.K. performed bioinformatic analysis. L.X. provided oversight of the bioinformatic analysis.
Declaration of interests
E.N.O. is a consultant for Vertex Therapeutics. Y.-L.M. is an employee at Vertex Pharmaceuticals. The other authors declare that they have no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.03.004.
Supplemental information
References
- 1.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Fairclough R.J., Wood M.J., Davies K.E. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat. Rev. Genet. 2013;14:373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 3.Allen D.G., Whitehead N.P., Froehner S.C. Absence of Dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol. Rev. 2016;96:253–305. doi: 10.1152/physrev.00007.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amoasii L., Long C., Li H., Mireault A.A., Shelton J.M., Sanchez-Ortiz E., McAnally J.R., Bhattacharyya S., Schmidt F., Grimm D., et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl Med. 2017;9:eaan8081. doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amoasii L., Hildyard J.C.W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., Harron R., Stathopoulou T.R., Massey C., Shelton J.M., et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell K.P., Kahl S.D. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 7.Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 8.Aartsma-Rus A., Van Deutekom J.C., Fokkema I.F., Van Ommen G.J., Den Dunnen J.T. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 9.Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T., et al. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Ma H., Zhang F., Chen L., Xing X., Wang S., Zhang X., Luo Y. Screening of Duchenne muscular dystrophy (DMD) mutations and investigating its mutational mechanism in Chinese patients. PLoS One. 2014;9:e108038. doi: 10.1371/journal.pone.0108038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuffery-Giraud S., Beroud C., Leturcq F., Yaou R.B., Hamroun D., Michel-Calemard L., Moizard M.P., Bernard R., Cossee M., Boisseau P., et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum. Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 12.Ginjaar I.B., Kneppers A.L., v d Meulen J.D., Anderson L.V., Bremmer-Bout M., van Deutekom J.C., Weegenaar J., den Dunnen J.T., Bakker E. Dystrophin nonsense mutation induces different levels of exon 29 skipping and leads to variable phenotypes within one BMD family. Eur. J. Hum. Genet. 2000;8:793–796. doi: 10.1038/sj.ejhg.5200535. [DOI] [PubMed] [Google Scholar]
- 13.Chemello F., Bassel-Duby R., Olson E.N. Correction of muscular dystrophies by CRISPR gene editing. J. Clin. Invest. 2020;130:2766–2776. doi: 10.1172/JCI136873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi E., Koo T. CRISPR technologies for the treatment of Duchenne muscular dystrophy. Mol. Ther. 2021;29:3179–3191. doi: 10.1016/j.ymthe.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson E.N. Toward the correction of muscular dystrophy by gene editing. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2004840117. e2004840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long C., Li H., Tiburcy M., Rodriguez-Caycedo C., Kyrychenko V., Zhou H., Zhang Y., Min Y.-L., Shelton J.M., Mammen P.P.A., et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018;4:eaap9004. doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Min Y.-L., Li H., Rodriguez-Caycedo C., Mireault A.A., Huang J., Shelton J.M., McAnally J.R., Amoasii L., Mammen P.P.A., Bassel-Duby R., et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci. Adv. 2019;5:eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Li H., Min Y.-L., Sanchez-Ortiz E., Huang J., Mireault A.A., Shelton J.M., Kim J., Mammen P.P.A., Bassel-Duby R., et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci. Adv. 2020;6:eaay6812. doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Min Y.-L., Chemello F., Li H., Rodriguez-Caycedo C., Sanchez-Ortiz E., Mireault A.A., McAnally J.R., Shelton J.M., Zhang Y., Bassel-Duby R., et al. Correction of three prominent mutations in mouse and human models of duchenne muscular dystrophy by single-cut genome editing. Mol. Ther. 2020;28:2044–2055. doi: 10.1016/j.ymthe.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumont N.A., Bentzinger C.F., Sincennes M.C., Rudnicki M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015;5:1027–1059. doi: 10.1002/cphy.c140068. [DOI] [PubMed] [Google Scholar]
- 22.Nelson C.E., Wu Y., Gemberling M.P., Oliver M.L., Waller M.A., Bohning J.D., Robinson-Hamm J.N., Bulaklak K., Castellanos Rivera R.M., Collier J.H., et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hakim C.H., Wasala N.B., Nelson C.E., Wasala L.P., Yue Y., Louderman J.A., Lessa T.B., Dai A., Zhang K., Jenkins G.J., et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight. 2018;3:e124297. doi: 10.1172/jci.insight.124297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Luca A., Nico B., Liantonio A., Didonna M.P., Fraysse B., Pierno S., Burdi R., Mangieri D., Rolland J.F., Camerino C., et al. A multidisciplinary evaluation of the effectiveness of cyclosporine a in dystrophic mdx mice. Am. J. Pathol. 2005;166:477–489. doi: 10.1016/S0002-9440(10)62270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brussee V., Tardif F., Tremblay J.P. Muscle fibers of mdx mice are more vulnerable to exercise than those of normal mice. Neuromuscul. Disord. 1997;7:487–492. doi: 10.1016/s0960-8966(97)00115-6. [DOI] [PubMed] [Google Scholar]
- 26.Himeda C.L., Chen X., Hauschka S.D. Design and testing of regulatory cassettes for optimal activity in skeletal and cardiac muscles. Methods Mol. Biol. 2011;709:3–19. doi: 10.1007/978-1-61737-982-6_1. [DOI] [PubMed] [Google Scholar]
- 27.Ramos J.N., Hollinger K., Bengtsson N.E., Allen J.M., Hauschka S.D., Chamberlain J.S. Development of novel micro-dystrophins with enhanced functionality. Mol. Ther. 2019;27:623–635. doi: 10.1016/j.ymthe.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X., et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L., Park K.H., Zhao L., Xu J., El Refaey M., Gao Y., Zhu H., Ma J., Han R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosenberg A.S., Puig M., Nagaraju K., Hoffman E.P., Villalta S.A., Rao V.A., Wakefield L.M., Woodcock J. Immune-mediated pathology in Duchenne muscular dystrophy. Sci. Transl Med. 2015;7:299rv4. doi: 10.1126/scitranslmed.aaa7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bengal E., Aviram S., Hayek T. p38 MAPK in glucose metabolism of skeletal muscle: beneficial or harmful? Int. J. Mol. Sci. 2020;21:6480. doi: 10.3390/ijms21186480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer H.F., Goodyear L.J. Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. J. Appl. Physiol. 2007;103:388–395. doi: 10.1152/japplphysiol.00085.2007. [DOI] [PubMed] [Google Scholar]
- 33.Camerino G.M., Cannone M., Giustino A., Massari A.M., Capogrosso R.F., Cozzoli A., De Luca A. Gene expression in mdx mouse muscle in relation to age and exercise: aberrant mechanical-metabolic coupling and implications for pre-clinical studies in Duchenne muscular dystrophy. Hum. Mol. Genet. 2014;23:5720–5732. doi: 10.1093/hmg/ddu287. [DOI] [PubMed] [Google Scholar]
- 34.Nance M.E., Shi R., Hakim C.H., Wasala N.B., Yue Y., Pan X., Zhang T., Robinson C.A., Duan S.X., Yao G., et al. AAV9 edits muscle stem cells in normal and dystrophic adult mice. Mol. Ther. 2019;27:1568–1585. doi: 10.1016/j.ymthe.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldstein J.M., Tabebordbar M., Zhu K., Wang L.D., Messemer K.A., Peacker B., Ashrafi Kakhki S., Gonzalez-Celeiro M., Shwartz Y., Cheng J.K.W., et al. In situ modification of tissue stem and progenitor cell genomes. Cell Rep. 2019;27:1254–1264.e7. doi: 10.1016/j.celrep.2019.03.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon J.B., Ettyreddy A.R., Vankara A., Bohning J.D., Devlin G., Hauschka S.D., Asokan A., Gersbach C.A. Gene editing of muscle stem cells with adeno-associated viral vectors in a mouse model of duchenne muscular dystrophy. Mol. Ther. Methods Clin. Dev. 2020;19:320–329. doi: 10.1016/j.omtm.2020.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morton A.B., Norton C.E., Jacobsen N.L., Fernando C.A., Cornelison D.D.W., Segal S.S. Barium chloride injures myofibers through calcium-induced proteolysis with fragmentation of motor nerves and microvessels. Skelet Muscle. 2019;9:27. doi: 10.1186/s13395-019-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bryl E., Vallejo A.N., Weyand C.M., Goronzy J.J. Down-regulation of CD28 expression by TNF-alpha. J. Immunol. 2001;167:3231–3238. doi: 10.4049/jimmunol.167.6.3231. [DOI] [PubMed] [Google Scholar]
- 39.Yucel N., Chang A.C., Day J.W., Rosenthal N., Blau H.M. Humanizing the mdx mouse model of DMD: the long and the short of it. NPJ Regen. Med. 2018;3:4. doi: 10.1038/s41536-018-0045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godfrey C., Muses S., McClorey G., Wells K.E., Coursindel T., Terry R.L., Betts C., Hammond S., O'Donovan L., Hildyard J., et al. How much dystrophin is enough: the physiological consequences of different levels of dystrophin in the mdx mouse. Hum. Mol. Genet. 2015;24:4225–4237. doi: 10.1093/hmg/ddv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Putten M., Hulsker M., Nadarajah V.D., van Heiningen S.H., van Huizen E., van Iterson M., Admiraal P., Messemaker T., den Dunnen J.T., 't Hoen P.A., et al. The effects of low levels of dystrophin on mouse muscle function and pathology. PLoS One. 2012;7:e31937. doi: 10.1371/journal.pone.0031937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 43.Calcedo R., Wilson J.M. Humoral immune response to AAV. Front Immunol. 2013;4:341. doi: 10.3389/fimmu.2013.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chemello F., Chai A.C., Li H., Rodriguez-Caycedo C., Sanchez-Ortiz E., Atmanli A., Mireault A.A., Liu N., Bassel-Duby R., Olson E.N. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci. Adv. 2021;7:eabg4910. doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anzalone A.V., Randolph P.B., Davis J.R., Sousa A.A., Koblan L.W., Levy J.M., Chen P.J., Wilson C., Newby G.A., Raguram A., et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramirez-Martinez A., Zhang Y., Chen K., Kim J., Cenik B.K., McAnally J.R., Cai C., Shelton J.M., Huang J., Brennan A., et al. The nuclear envelope protein Net39 is essential for muscle nuclear integrity and chromatin organization. Nat. Commun. 2021;12:690. doi: 10.1038/s41467-021-20987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data to evaluate the conclusions in this paper are presented in the paper figures and supplemental data. RNA-seq data have been uploaded to GEO. GEO profile:GSE180771. Additional data pertaining to this paper can be requested from the authors.