Abstract
Doxorubicin is a chemotherapeutic medication commonly used to treat many types of cancers, but it has side effects including vomiting, rash, hair loss, and bone marrow suppression. The most dangerous side effects are cardiomyopathy, cardiofibrosis, and heart failure, as doxorubicin generates cytotoxicity and stops DNA replication. There is no treatment to block these side effects. We have developed a transgenic mouse line overexpressing the circular RNA circNlgn and shown that circNlgn is a mediator of doxorubicin-induced cardiofibrosis. Increased expression of circNlgn decreased cardiac function and induced cardiofibrosis by upregulating Gadd45b, Sema4C, and RAD50 and activating p38 and pJNK in circNlgn transgenic heart. Silencing circNlgn decreased the effects of doxorubicin on cardiac cell activities and prevented doxorubicin-induced expression of fibrosis-associated molecules. The protein (Nlgn173) translated by circNlgn could bind and activate H2AX, producing γH2AX, resulting in upregulation of IL-1b, IL-2Rb, IL-6, EGR1, and EGR3. We showed that silencing these molecules in the signaling pathway prevented doxorubicin-induced cardiomyocyte apoptosis, increased cardiomyocyte viability, decreased cardiac fibroblast proliferation, and inhibited collagen production. This mechanism may hold therapeutic implications for mitigating the side effects of doxorubicin therapy in cancer patients.
Keywords: circular RNA, circRNA, RNA-seq, circNlgn, doxorubicin, cardiac fibrosis, cardiac remodeling, heart function
Graphical abstract
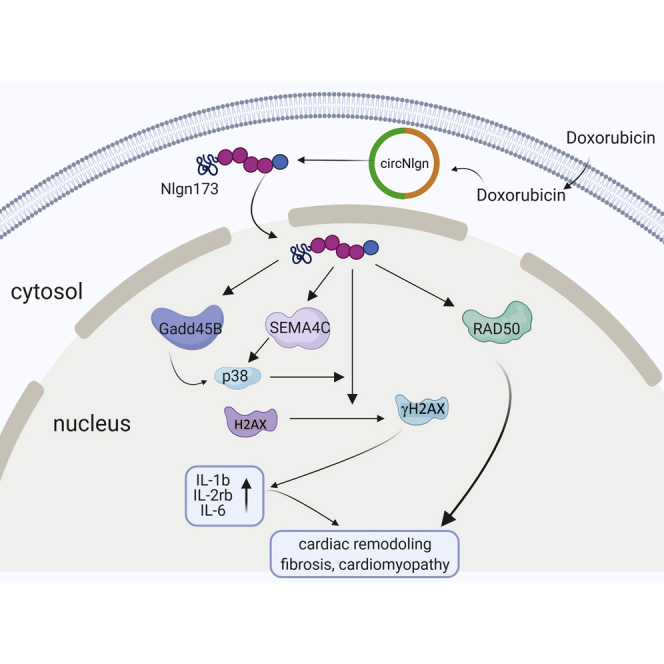
We found that transgenic expression of circNlgn mediates doxorubicin-induced cardiofibrosis. The protein Nlgn173, translated by circNlgn, interacts and activates H2AX, resulting in upregulation of IL-1b, IL-2Rb, IL-6, EGR1, and EGR3. This mechanism may hold therapeutic implications for mitigating the side effects of doxorubicin in cancer treatment.
Introduction
Despite extensive use as a chemotherapeutic drug in treating patients with various types of cancers, doxorubicin generates severe side effects because of its cardiotoxicity.1, 2, 3 The cardiotoxicity of doxorubicin may cause reactive oxidative stress, alter DNA functions and gene expression, stop DNA replication, and induce cell apoptosis.2 Extensive cellular dysfunction and death induce cardiac remodeling, accompanied by aberrant accumulation of type I and type III collagens, and dysregulation of transforming growth factor β (TGF-β) signaling.4,5 Over time, the remodeling induces cardiofibrosis, hypertrophy, dilation, cardiomyopathy, and decreased contractile strength that is unable to meet the volume and pressure demands of the heart.6,7 This remodeling is irreversible and leads to myocardial dysfunction and heart failure.8,9 The mechanism is not well understood. We recently found that a circular RNA, circNlgn, appeared to be implicated in mediating the effects of doxorubicin, as transgenic mice expressing circNlgn displayed cardiac remodeling, cardiofibrosis, and heart failure.
Circular RNAs (circRNAs) are non-coding RNAs generated from genomic transcripts with covalently closed circles at the 5′ and 3′ ends that occur through a splice acceptor site upstream linked to a splice donor site downstream.10 Some circRNAs are expressed specifically in certain cell types and developmental stages, suggesting specific functions of these circRNAs.11, 12, 13 Most circRNAs are expressed at very low levels, while some have high abundancy. The copy number of some circRNAs can be much greater than the associated linear RNAs, suggesting potential biological functions.11,14 circNlgn is an example that is highly expressed in the heart, where its parental mRNA, Nlgn, is hardly detected. Studies have reported that circRNAs play roles in the cardiovascular system,15, 16, 17, 18, 19 neural cells and tissues,20, 21, 22 and cancer progression.23, 24, 25, 26 circRNAs may function by sponging microRNAs (miRNAs), regulating transcription, splicing, interacting with proteins in the associated signal pathways, and translating to protein peptides.27 circRNAs were initially reported to function as miRNA sponges, thereby reducing miRNA-mediated functions.28 Some circRNAs were found to bind to the promoters of their parental genes and their transcription elements to enhance transcription of their parental genes.29 Since circRNAs share the same canonical splicing sites with their linear counterparts, circRNA production may compete with linear splicing.30 Due to their three-dimensional structure, circRNAs may bind to proteins independent of their parental mRNAs.31, 32, 33 Since many circRNAs contain the second exon and translation initiation site, they have the potential for protein peptide translation.34 Several circRNAs have been reported to encode functional proteins with oncogenic or tumor-suppressing activities.25,35, 36, 37 These circRNA-encoded small protein peptides are functionally associated with their parental genes. In this study, we found that in transgenic mice overexpressing circNlgn, circNlgn and its translated product Nlgn173 mediated the side effects of doxorubicin, and we thus used this mouse model to study the downstream effects of doxorubicin with the aim of developing approaches to reduce its side effects.
Results
circNlgn mediates doxorubicin-induced cardiac fibrosis
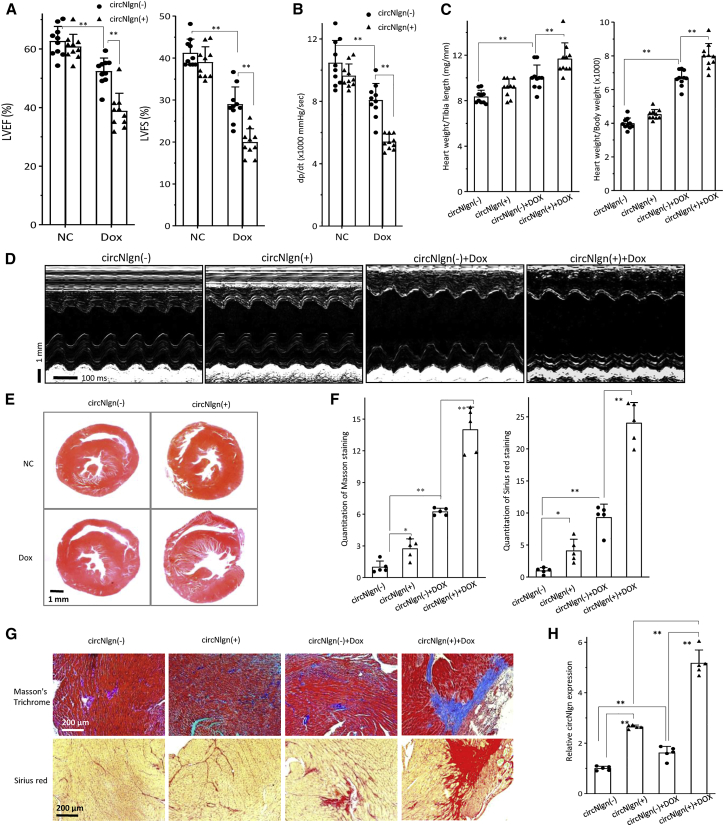
Treatment with doxorubicin is known to induce cardiomyopathy and cardiac fibrosis. Our recent study showed that expression of circNlgn impaired heart function and induced cardiac fibrosis via its translated protein Nlgn173.38 We wondered whether Nlgn173 could mediate doxorubicin-induced cardiomyopathy and cardiac fibrosis. Eight-week-old transgenic mice expressing circNlgn were treated with doxorubicin and subjected to echocardiographic assessment (treatment scheme shown in Figure S1A). While there was no significant difference in heart rates (Figure S1B), we detected a significant decrease in the left-ventricular ejection fraction (LVEF) and left-ventricular fractional shortening (LVFS) in the circNlgn-transgenic mice relative to the untreated controls (Figure 1A). We also detected a significant decrease in left-ventricular pressure (dp/dt) (Figure 1B) and in left-ventricular end diastolic diameter (LVEDD) and left-ventricular end systolic diameter (LVESD) (Figure S1C) in the circNlgn-transgenic mice, but a significant increase in heart weight (Figures 1C and S1D). We also detected decreased ventricular contractile ability in the circNlgn-transgenic mice (Figure 1D). Hematoxylin and eosin staining of cross views of the heart tissues displayed hypertrophy of the heart sections in the doxorubicin-treated mice (Figure 1E). Consistent with this, we detected decreased left-ventricular posterior wall thickness (LVPW) in the circNlgn transgenic mice relative to the litter-matched negative mice after doxorubicin treatment (Figure S1E).
Figure 1.
Doxorubicin-induced cardiac fibrosis and impaired heart function were exacerbated by circNlgn expression
(A) Left, circNlgn transgenic mice, circNlgn (+), showed decreased left-ventricular ejection fraction (LVEF) relative to the litter-matched negative mice, circNlgn (−), after doxorubicin treatment. ∗∗p < 0.01 versus circNlgn (−) (n = 10). Right, circNlgn (+) mice showed decreased left-ventricular fractional shortening (LVFS) compared with circNlgn (−) after doxorubicin treatment. ∗∗p < 0.01 versus circNlgn (−) (n = 10). (B) circNlgn (+) mice displayed lower left-ventricular pressure (dp/dt) than circNlgn (−) mice after doxorubicin treatment. ∗∗p < 0.01 versus circNlgn (−) (n = 10). (C) circNlgn (+) mice displayed higher heart weight/tibia length (left) and heart weight/body weight (right) than circNlgn (−) mice after doxorubicin treatment. ∗∗p < 0.01 versus circNlgn (−) (n = 10). (D) Representative echocardiography photographs showing heart function of circNlgn (+) and circNlgn (−) mice with or without doxorubicin treatment.
(E) Representative H&E staining of cross sections of heart tissues. circNlgn (+) mice displayed enlarged heart chamber. (F) circNlgn (+) mouse hearts displayed increased Masson’s trichrome (left) and Sirius red (right) staining compared with circNlgn (−) controls. Doxorubicin treatment significantly increased Masson’s trichrome and Sirius red staining in mouse hearts. ∗p < 0.05, ∗∗p < 0.01 (n = 10). (G) Representative Masson’s trichrome and Sirius red staining of cross sections of heart tissues. circNlgn (+) mouse hearts displayed increased Masson’s trichrome and Sirius red staining after doxorubicin treatment. (H) circNlgn (+) mouse hearts expressed higher levels of circNlgn than circNlgn (−) controls measured by real-time PCR. Doxorubicin treatment promoted circNlgn expression in mouse hearts. ∗∗p < 0.01 (n = 10). It is not necessary to start a new paragraph between (E) and (D). It may be good to prinit this figure in one page. I have provided a new figure (condensed a bit), so that the legend and the figure can be prinited in one page.
Our previous study showed that expression of circNlgn induced cardiac fibrosis.38 We examined the effect of doxorubicin on cardiac fibrosis by Masson’s trichrome staining and Sirius red staining. While we confirmed promotion of Masson’s trichrome and Sirius red staining in the circNlgn-transgenic mouse heart relative to the wild-type mouse heart, we detected an additive effect of doxorubicin treatment on both types of staining (Figure 1F), suggesting the promotion of cardiac fibrosis by doxorubicin treatment in the circNlgn-transgenic mice. Examination of the tissue sections revealed large areas of Masson’s trichrome and Sirius red staining in the circNlgn-transgenic mouse hearts treated with doxorubicin (Figure 1G). Interestingly, doxorubicin treatment increased circNlgn levels in both wild-type and circNlgn-transgenic mouse hearts (Figure 1H).
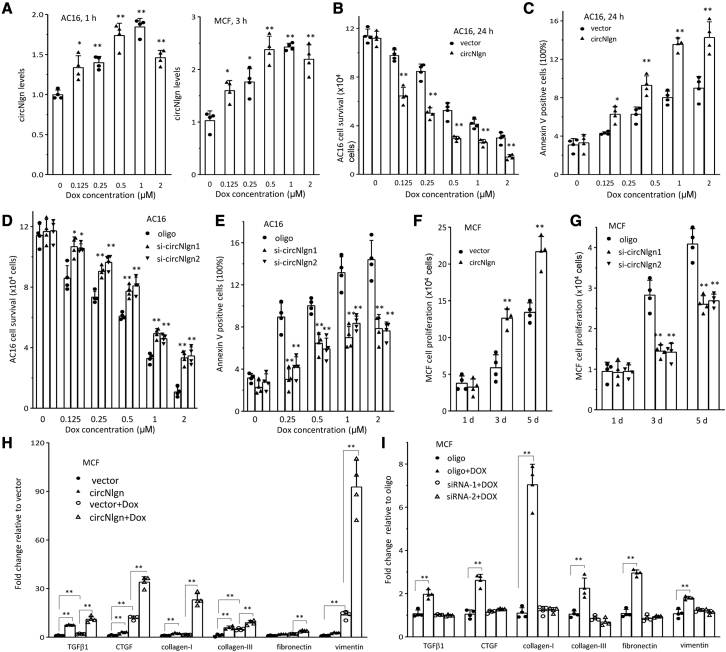
To test whether circNlgn was a downstream molecule of doxorubicin, we performed a number of experiments in cardiomyocyte cell line AC16 and cardiac fibroblast cell line MCF. In both types of cells, treatment with doxorubicin increased circNlgn expression, which was dose related (Figure 2A). AC16 cells transfected with the circNlgn expression construct showed decreased cell survival (Figure 2B) and increased cell apoptosis (Figure 2C) when treated with different concentrations of doxorubicin. Similar results were obtained in the cells treated for different periods of time (Figure S2A and S2B) and measured by MTT assay (Figures S2C and S2D). To corroborate these results, we knocked down circNlgn by transfecting the cells with two small interfering RNAs (siRNAs) targeting circNlgn (Figure S3). Silencing circNlgn enhanced AC16 survival (Figures 2D and S4A) and decreased apoptosis (Figures 2E and S4B). In MCF cells, transfection with circNlgn enhanced cell proliferation (Figures 2F and S5A), while silencing circNlgn decreased proliferation (Figures 2G and S5B).
Figure 2.
Silencing circNlgn decreased the effects of doxorubicin on cardiac cell activity and prevented doxorubicin-induced expression of fibrosis-associated molecules
(A) Left, real-time PCR showed that AC16 cells expressed increased circNlgn when cultured in basal medium and treated with doxorubicin for 1 h. ∗p < 0.05, ∗∗p < 0.01 versus doxorubicin 0 μM (n = 4). Right, real-time PCR showed that MCFs expressed increased levels of circNlgn when treated with doxorubicin. ∗p < 0.05, ∗∗p < 0.01 versus doxorubicin 0 μM (n = 4). (B) AC16 cells transfected with circNlgn or the vector were cultured in basal medium treated with doxorubicin for survival assay. circNlgn expression repressed cell survival. ∗∗p < 0.01 versus vector (n = 4). (C) Expression of circNlgn promoted cell apoptosis when treated with doxorubicin, which was dose dependent. ∗p < 0.05, ∗∗p < 0.01 versus vector (n = 4). (D) Silencing circNlgn enhanced AC16 cell survival. ∗p < 0.05, ∗∗p < 0.01 versus oligo (n = 4). (E) Silencing circNlgn repressed AC16 apoptosis. ∗∗p < 0.01 versus oligo (n = 4). (F) MCF cells transfected with circNlgn or the vector were cultured in basal medium for the indicated time points. Expression of circNlgn enhanced MCF cell proliferation. ∗∗p < 0.01 versus vector (n = 4). (G) Silencing circNlgn suppressed MCF cell proliferation. ∗∗p < 0.01 versus oligo (n = 4). (H) RNAs were isolated from circNlgn- or vector-transfected MCF cells treated with or without 0.25 μM doxorubicin for 24 h and subjected to RT-PCR. Real-time PCR showed that transfection with circNlgn increased expression of fibrosis markers TGF-β1, CTGF, collagen I, collagen III, fibronectin, and vimentin, which were further enhanced by doxorubicin treatment. ∗∗p < 0.01 (n = 4). (I) Control oligo- and circNlgn siRNA-transfected MCF cells were cultured in basal medium treated with 0.1 μM doxorubicin for 24 h, harvested, and subjected to RT-PCR. MCF cells showed increased expression of fibrosis markers TGF-β1, CTGF, collagen I, collagen III, fibronectin, and vimentin, which could be prevented by silencing circNlgn. ∗∗p < 0.01 versus oligo (n = 4).
We then analyzed the effects of doxorubicin on cardiac fibrosis. MCF cells transfected with circNlgn or the control vector were treated with or without doxorubicin, followed by measurement of circNlgn levels. While we confirmed increased expression of molecules associated with cardiac fibrosis, including TGF-β1, CTGF, collagen I, collagen III, fibronectin, and vimentin, we observed significant additive effect of doxorubicin treatment in promoting expression of these molecules. As expected, treatment with doxorubicin increased expression of these fibrotic molecules (Figure 2H). Importantly, silencing circNlgn by transfecting MCFs with two siRNAs targeting circNlgn prevented the increase in expression of these fibrotic molecules induced by doxorubicin treatment (Figure 2I). This suggests that targeting circNlgn may serve as an approach to alleviate doxorubicin-induced cardiomyopathy and cardiac fibrosis in clinical applications.
Nlgn173 binds and activates H2AX
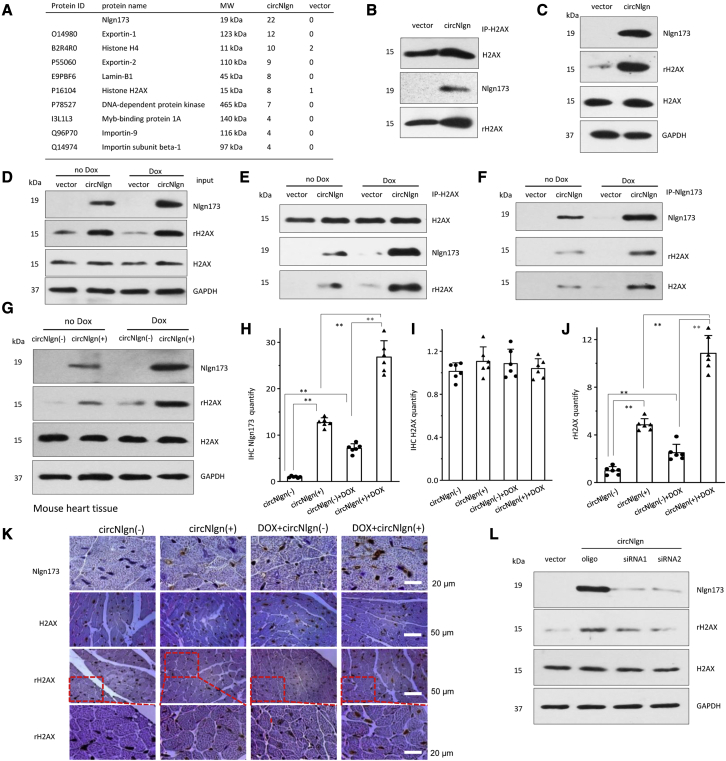
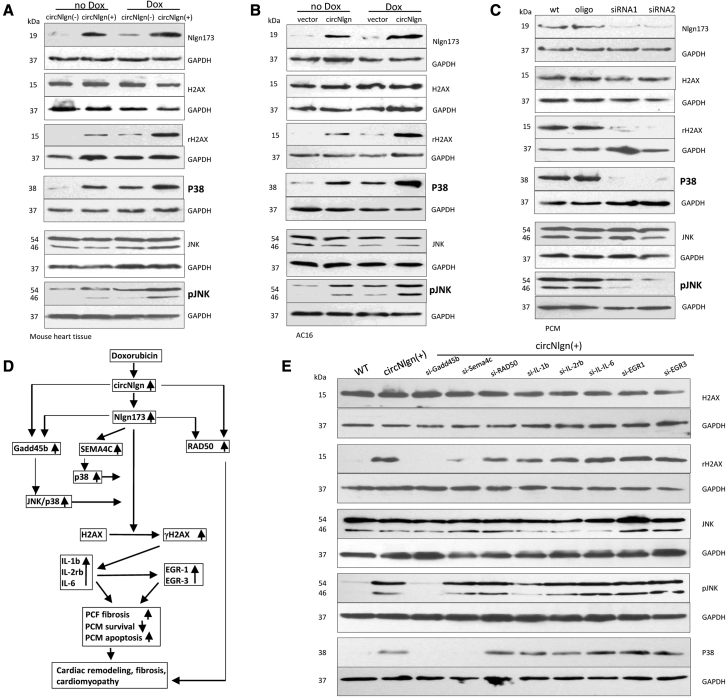
To dissect out the downstream protein molecules that mediated circNlgn’s effect, we precipitated the circNlgn-translated protein Nlgn173 with the antibody against the N terminus of Nlgn that recognizes Nlgn173. The precipitated proteins were subjected to mass spectrophotometry. Since Nlgn173 was translocated to nuclei by LaminB1,38 we focused on nuclear proteins that displayed increased levels in the circNlgn-transfected cells. While there were a number of proteins co-precipitated by the anti-Nlgn antibody, the one that caught our attention was H2AX (Figure 3A). H2AX is a histone protein that plays roles in DNA repair. When there is a double-strand break in DNA, the chromatin structure changes and induces phosphorylation of H2AX to become γH2AX.39 We confirmed the binding of Nlgn173 with H2AX using the anti-Nlgn antibody. Anti-H2AX antibody detected H2AX and γH2AX precipitated by the anti-Nlgn antibody (Figure 3B). On the other hand, the anti-Nlgn antibody detected Nlgn173 precipitated by the anti-H2AX antibody.
Figure 3.
Nlgn173 binds and activates H2AX, producing γH2AX
(A) Mass spectrophotometry showed proteins precipitated by antibody against Nlgn173. (B) AC16 cells transfected with circNlgn or the vector were lysed and subjected to immunoprecipitation with anti-Nlgn antibody followed by western blotting. Precipitation of Nlgn173 pulled down H2AX, and vice versa. (C) The interaction of Nlgn173 and H2AX promoted phosphorylation of H2AX (γH2AX). (D) AC16 cells transfected with circNlgn expressed higher levels of Nlgn173 and γH2AX, which were further enhanced by doxorubicin treatment. H2AX expression was not affected. (E) Precipitation of H2AX pulled down Nlgn173. H2AX phosphorylation (γH2AX) was higher in the sample where Nlgn173 interacted with H2AX. (F) Precipitation of Nlgn173 pulled down H2AX. The interaction of Nlgn173 and H2AX promoted phosphorylation of H2AX (γH2AX). (G) Western blot showed that circNlgn (+) mouse tissues expressed high levels of Nlgn173 and γH2AX, which were further enhanced by doxorubicin treatment. (H) Quantitation of IHC staining of heart tissues showed increased expression of Nlgn173 in the circNlgn transgenic heart, which could be further promoted by doxorubicin treatment. ∗∗p < 0.01 (n = 6). (I) Quantitation of IHC staining of heart tissues showed that H2AX expression was not affected by circNlgn expression nor by doxorubicin treatment. (J) Quantitation of IHC staining of heart tissues showed increased expression of γH2AX in the circNlgn transgenic heart, which could be further promoted by doxorubicin treatment. ∗∗p < 0.01 (n = 6). (K) Representative IHC photographs of heart tissues. circNlgn (+) mouse hearts displayed increased Nlg173 and γH2AX (Ser139) expression levels, which were further enhanced by doxorubicin treatment. H2AX expression was not affected. (L) Western blot showed that silencing circNlgn suppressed Nlgn173 and γH2AX expression.
Lysates from AC16 cells transfected with circNlgn and a vector were analyzed on western blot. Transfection with circNlgn did not affect H2AX expression but promoted phosphorylation of H2AX to become γH2AX (Figure 3C), suggesting that interaction of Nlgn173 with H2AX induces H2AX activation, and a similar phenomenon is observed when H2AX interacts with double-strand breaks in DNA. The cells were then treated with or without doxorubicin to induce DNA breaks. As expected, treatment with doxorubicin increased Nlgn173 levels, which also increased the difference in γH2AX levels between transfection with circNlgn or the vector (Figure 3D). The lysates were subjected to immunoprecipitation with an antibody against H2AX. Western blotting confirmed increased precipitation of γH2AX and Nlgn173 (Figure 3E). The interaction of Nlgn173 and H2AX promoted phosphorylation of H2AX (γH2AX). An antibody against Nlgn173 precipitated increased levels of H2Ax and γH2AX, which was further enhanced by doxorubicin treatment (Figure 3F).
Similar experiments were then performed in the circNlgn-transgenic and wild-type heart tissues of mice treated with or without doxorubicin. Western blotting detected similar patterns of γH2AX levels relative to Nlgn173 levels: expression of circNlgn increased Nlgn173 and γH2AX levels, which were further promoted by doxorubicin treatment (Figure 3G). Heart tissue sections were subjected to immunohistochemistry (IHC) staining followed by quantitation. While expression of Nlgn173 increased in the circNlgn-transgenic mice and was further promoted by doxorubicin treatment (Figure 3H), H2AX levels were unaffected (Figure 3I). The increased expression pattern of γH2AX appeared similar to that of Nlgn173 (Figure 3J), both of which were detected in the nuclei of heart sections (Figure 3K). To confirm the effect of circNlgn on H2AX activation, we silenced circNlgn using two siRNAs targeting circNlgn. While silencing circNlgn showed no effect on H2AX expression, we detected decreased levels of γH2AX (Figure 3L).
Upregulation of Gadd45b, Sema4C, and RAD50 by circNlgn expression
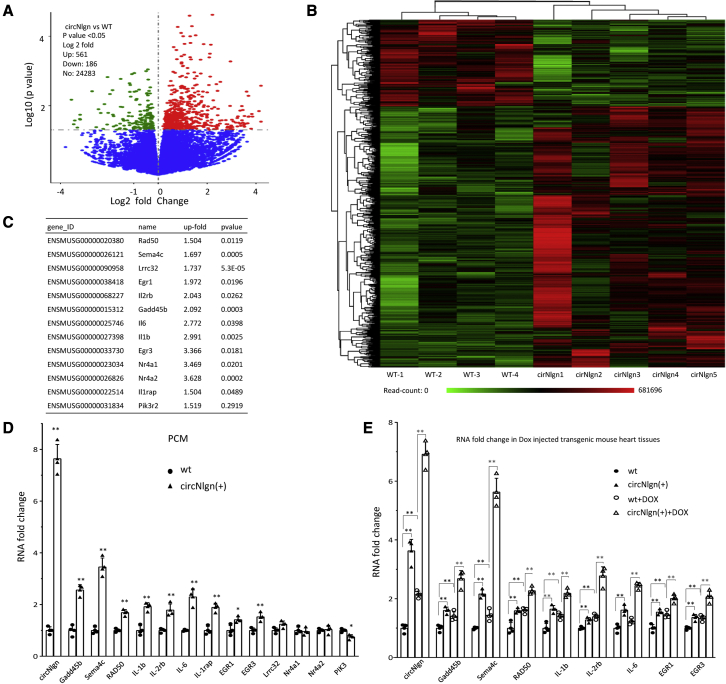
To examine the molecules mediating circNlgn functions, we harvested the heart tissues from circNlgn-transgenic and wild-type mice and had the mRNA sequenced by Novogene. In a total of 24,283 mRNAs detected, 3% were differentially expressed, and the distribution as shown by volcano analysis followed a bell shape (Figure 4A). Heatmap analysis was performed to group the differentially expressed RNAs (Figure 4B). Among the 3% differentially expressed RNAs, 474 reached 1.5-fold and 307 were from protein-coding genes (Table S1). We analyzed these 307 mRNAs and selected several interesting mRNAs that might play roles in regulating cell proliferation, survival, differentiation, apoptosis, and fibrosis (Figure 4C), since these genes might be involved in cardiac remodeling and fibrosis.
Figure 4.
Upregulation of Gadd45b, Sema4C, and RAD50 in circNlgn transgenic heart
(A) Volcano analysis of 24,283 mRNAs detected by RNA sequencing. Approximately 3% of the RNAs were differentially expressed. (B) Heatmap analysis showing the differentially expressed RNAs. (C) From the 307 differentially expressed mRNAs, the most up- and downregulated were selected. (D) PCR showed that PCMs from the hearts of the circNlgn-transgenic mice expressed significantly higher levels of circNlgn, Gadd45b, Sema4C, RAD50, IL-1b, IL-1rap, IL-2Rb, IL-6, EGR1, and EGR3. ∗p < 0.05, ∗∗p < 0.01 versus wild type (n = 4). (E) PCR showed that circNlgn (+) mouse heart tissues expressed high levels of circNlgn, Gadd45b, Sema4C, RAD50, IL-1b, IL-2Rb, IL-6, EGR1, and EGR3, which were further enhanced by delivery of doxorubicin. ∗∗p < 0.01 (n = 4).
To test the effects of the selected candidates, we isolated primary cardiomyocytes (PCMs) from the hearts of circNlgn-transgenic and litter-matched circNlgn-negative mice. Expression of these genes was determined by real-time PCR. circNlgn, Gadd45b, Semaphorin-4C (Sema4C), RAD50, IL-1b, IL-1rap, IL-2Rb, IL-6, early growth response (EGR) 1, and EGR3 were significantly upregulated (Figure 4D), which confirmed the results obtained by RNA sequencing. We did not detect significant upregulation of Lrrc32, Nr4a1, Nr4a2, and PIK3, and thus, these molecules were excluded from the following experiments. Since we did not set the cutoff at higher levels of differential expression, it is reasonable that we detected some false positives.
We further validated this result by measuring the expression of these molecules in circNlgn-transgenic and litter-matched circNlgn-negative mice treated with or without doxorubicin. circNlgn-transgenic mouse heart tissues expressed significantly higher levels of circNlgn, Gadd45b, Sema4C, RAD50, IL-1b, IL-2Rb, IL-6, IL-1rap, EGR1, and EGR3 relative to the litter-matched negative mouse hearts, which were further enhanced by doxorubicin treatment (Figure 4E). Ectopic expression of circNlgn in AC16 cells showed similar results (Figure S6).
Dissecting out downstream signal molecules of H2AX
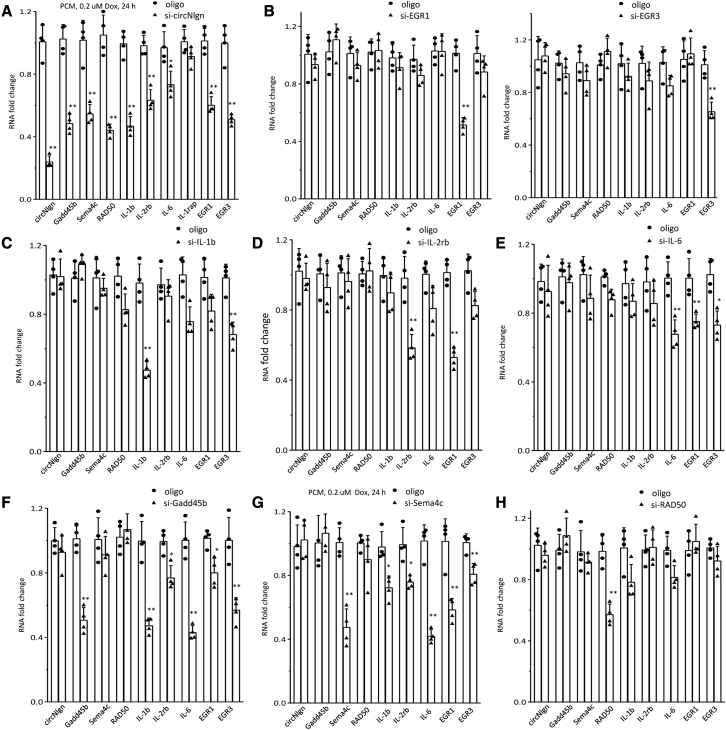
We then silenced circNlgn in the PCMs to confirm the effects of circNlgn in regulating the expression of these molecules. Real-time PCR revealed decreased levels of Gadd45b, Sema4C, RAD50, IL-1b, IL-2Rb, IL-6, EGR1, and EGR3 (Figure 5A). IL-1rap was not downregulated upon circNlgn silencing, and it was excluded from the following experiments.
Figure 5.
Dissecting out the downstream signal molecules of γH2AX
(A) PCR showed that PCMs transfected with circNlgn siRNA repressed expression of circNlgn, Gadd45b, Sema4C, RAD50, IL-1b, IL-2Rb, IL-6, EGR1, and EGR3. IL-1rap did not show downregulation upon circNlgn silencing. ∗p < 0.05, ∗∗p < 0.01 versus oligo (n = 4). (B) Silencing EGR1 (left) and EGR3 (right) did not affect expression of any other molecules. ∗∗p < 0.01 versus oligo (n = 4). (C and D) Silencing IL-1b decreased expression of EGR3 (C), while silencing IL-2Rb decreased expression of EGR1 (D). ∗∗p < 0.01 versus oligo (n = 4). (E) Silencing IL-6 decreased expression of EGR1 and EGR3. ∗p < 0.05, ∗∗p < 0.01 versus oligo (n = 4). (F and G) Silencing Gadd45b (F) or Sema4C (G) repressed expression of all these molecules (IL-1b, IL-2Rb, IL-6, EGR1, and EGR3). ∗p < 0.05, ∗∗p < 0.01 versus oligo (n = 4). (H) Silencing RAD50 did not affect expression of any molecules tested. ∗∗p < 0.01 versus oligo (n = 4).
To validate the effects of these molecules, we designed siRNAs targeting each of them. The isolated PCMs were transfected with these siRNAs and a control oligo. PCM transfection with siRNAs targeting EGR1 and EGR3 did not affect the expression of any other molecules (Figure 5B), suggesting that EGR1 and EGR3 are at the bottom of the signaling pathway associated with these molecules. Silencing IL-1b decreased expression of EGR3 (Figure 5C). Silencing IL-2Rb decreased expression of EGR1 (Figure 5D). Silencing IL-6 decreased expression of EGR1 and EGR3 (Figure 5E). These results suggest that IL-1b, IL-2Rb, and IL-6 are upstream of EGR1 and EGR3.
Silencing Gadd45b or Sema4C repressed the expression of all these molecules (IL-1b, IL-2Rb, IL-6, EGR1, and EGR3), but Gadd45b and Sema4C did not affect each other (Figures 5F and 5G). This suggests that Gadd45b and Sema4C are upstream of IL-1b, IL-2Rb, IL-6, EGR1, and EGR3. Silencing RAD50 did not affect the expression of any molecules tested (Figure 5H), suggesting that RAD50 mediates the effects of circNlgn in a separate pathway.
It has been reported that Gadd45b enhances pJNK and p38 expression,40,41 and this pathway is associated with H2AX phosphorylation in DNA repair. We examined the effect of Gadd45b promotion by circNlgn on pJNK/p38 expression and found increased levels of pJNK/p38 in the heart tissues of the circNlgn-transgenic mice relative to the litter-matched negative mice (Figure 6A). Treatment with doxorubicin, while increasing Nlgn173 levels, further increased the expression of pJNK/p38. Similar results were obtained in AC16 cells transfected with circNlgn or the vector (Figure 6B). PCMs isolated from circNlgn-transgenic mouse heart were transfected with two siRNAs targeting circNlgn. Silencing circNlgn decreased the expression of pJNK/p38 and Nlgn173, leading to decreased phosphorylation of H2AX (Figure 6C).
Figure 6.
Effects of the signaling molecules in mediating cellular functions of circNlgn
(A) Western blot showed that circNlgn (+) heart tissues expressed high levels of Nlgn173, γH2AX, pJNK, and p38, which were further promoted by delivery of doxorubicin. (B) PCMs were transfected with circNlgn siRNAs and cultured with basal medium with 0.2 μM doxorubicin for 36 h. Silencing circNlgn suppressed Nlgn173, γH2AX, pJNK, and p38 expression. (C) Control vector- and circNlgn-transfected cells were treated with or without 0.25 μM doxorubicin for 24 h, lysed, and subjected to western blotting. circNlgn transfection increased the levels of Nlgn173, γH2AX, pJNK, and p38, which were further promoted by doxorubicin treatment. (D) Signaling pathway of circNlgn for mediating doxorubicin effect on cardiac remodeling, fibrosis, and cardiomyopathy. (E) Western blot showed that circNlgn (+) PCMs expressed high levels of γH2AX, pJNK, and p38. Silencing Gadd45b prevented circNlgn-enhanced γH2AX, pJNK, and p38. Silencing Sema4C prevented circNlgn-enhanced γH2AX and p38, but not pJNK.
Activation of p38 can also enhance H2AX phosphorylation, and Sema4C is known to play roles in tissue differentiation.42,43 We tested the effects of Sema4C promotion by circNlgn on p38 phosphorylation and found increased levels of p38 in the heart tissues of the circNlgn-transgenic mice relative to the litter-matched negative mice (Figure 6A). Treatment with doxorubicin further activated JNK and p38. Similar results were seen in circNlgn- or vector-transfected AC16 cells (Figure 6B). Silencing circNlgn in PCMs decreased levels of pJNK/p38 (Figure 6C).
The above results are outlined in a diagram, in which all molecules involved in cardiac fibrosis are shown (Figure 6D). Most importantly, we found that circNlgn and its translated protein Nlgn173 mediated the cardiac fibrotic effect of doxorubicin treatment. Our results showed that the immediate downstream molecules of Nlgn173 were Gadd45b, Seman4C, and RAD50, which were initially detected by RNA sequencing, as well as activation of H2AX. The activated H2AX, γH2AX, induced expression of the cytokines IL-1b, IL-2Rb, and IL-6. These cytokines promoted expression of EGR1 and EGR3, which altogether with the cytokines could induce primary cardiac fibroblast (PCF) fibrosis, decrease PCM survival, and increase PCM apoptosis. Increased Gadd45b could enhance p38 and pJNK expression, leading to activation of H2AX, while Sema4C was able to activate p38 and JNK, leading to H2AX phosphorylation. RAD50 appeared to be in a separate sub-pathway that could directly increase PCF proliferation, leading to cardiac fibrosis.
We then performed western blot to validate the effects of Gadd45b and Sema4C on the expression of the other molecules by silencing each of the molecules involved in the pathway in PCMs isolated from the circNlgn-transgenic mouse heart. While we confirmed increased levels of Nlgn173, γH2AX, pJNK, and p38, silencing Gadd45b prevented circNlgn-enhanced γH2AX, pJNK, and p38 expression (Figure 6E). Silencing Sema4C could also prevent circNlgn-enhanced γH2AX and p38, but not pJNK expression.
Involvement of the signaling pathway in regulating cellular activities
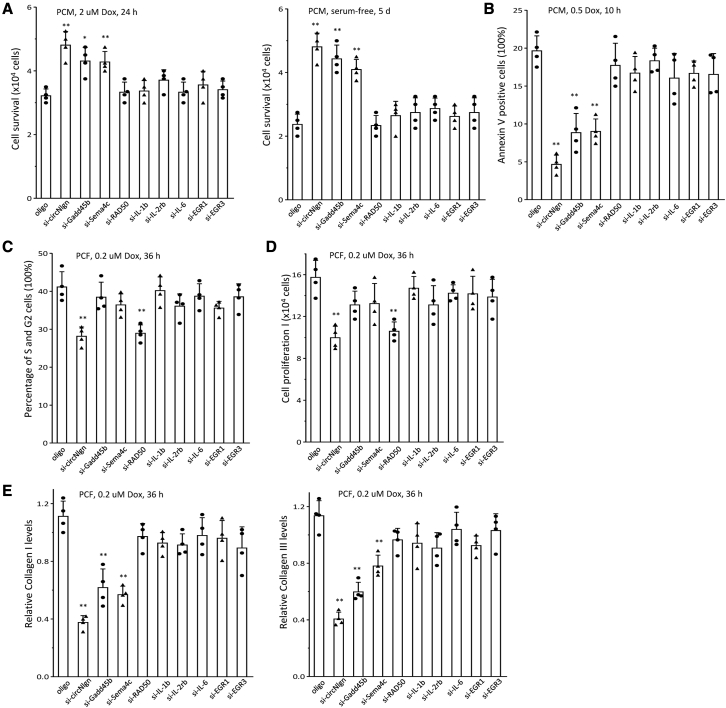
Finally, we examined how the signaling pathway played roles in cardiac fibrosis by isolating PCMs from the hearts of circNlgn-transgenic and litter-matched circNlgn-negative mice. Isolated PCMs were transfected with siRNAs targeting molecules in the signaling pathway and a control oligo, followed by incubation with doxorubicin or culture in serum-free medium. Only silencing with siRNAs targeting Gadd45b and Sema4C, similar to siRNA targeting circNlgn, increased cell survival under both culture conditions (Figure 7A). Consistently, only silencing Gadd45b and Sema4C decreased cell apoptosis (Figure 7B).
Figure 7.
Activation of H2AX by p38 and pJNK
(A) PCMs were transfected with siRNAs and cultured in basal medium with 2 μM doxorubicin for 24 h (left) or serum free for 5 days (right). Silencing circNlgn, Gadd45b, or Sema4C enhanced PCM survival. ∗p < 0.05, ∗∗p < 0.01 versus oligo (n = 4). (B) The transfected PCMs were cultured in basal medium with 0.5 μM doxorubicin for 24 h and subjected to Annexin staining followed by flow cytometry. Silencing circNlgn, Gadd45b, or Sema4C repressed PCM apoptosis. ∗∗p < 0.01 versus oligo (n = 4). (C) PCFs were cultured in basal medium treated with 0.2 μM doxorubicin for 36 h and then transfected with siRNAs. After 48 h, the cells were harvested and subjected to flow cytometry. Silencing circNlgn or RAD50 repressed PCF cycle entry. ∗∗p < 0.01 versus oligo (n = 4). (D) Cell proliferation assays showed that silencing circNlgn or RAD50 repressed PCF proliferation. ∗∗p < 0.01 versus oligo (n = 4). (E) PCR showed that circNlgn, Gadd45b, or Sema4C repressed collagen I (left) and collagen III (right) in the above doxorubicin-treated PCFs. ∗∗p < 0.01 versus oligo (n = 4). We found that transgenic expression of circNlgn mediates doxorubicin-induced cardiofibrosis. The protein Nlgn173 translated by circNlgn interacts and activates H2AX, resulting in upregulation of IL-1b, IL-2Rb, IL-6, EGR1, and EGR3. This mechanism may hold therapeutic implications for mitigating the side effects of doxorubicin in cancer treatment.
We also isolated PCFs from the hearts of circNlgn-transgenic and litter-matched circNlgn-negative mice. PCFs were transfected with the same siRNAs and treated with doxorubicin. We detected decreased cell-cycle progression (Figure 7C) and cell proliferation (Figure 7D) in PCFs transfected with siRNA targeting CRD50, similar to the results obtained by silencing circNlgn. Since cardiac fibroblasts play crucial roles in cardiac fibrosis, we measured the expression of the fibrotic markers collagen I and collagen III and found that expression of both collagen molecules decreased when PCFs were transfected with siRNAs targeting Gadd45b and Sema4C, similar to the results obtained by siRNA targeting circNlgn (Figure 7E).
Discussion
Since the discovery of doxorubicin in the 1950s and its approval for medical application in the United States in 1974, it has been used in the treatment of a number of cancers, including breast carcinoma, lymphoma, acute lymphocytic leukemia, bladder cancer, and Kaposi’s sarcoma by injecting into the patient’s vein.1 Doxorubicin inhibits tumor growth by interfering with the DNA function of tumor cells. Doxorubicin binds to DNA by intercalating between two base pairs, which blocks DNA transcription and replication.44 By interacting with DNA, doxorubicin induces histone eviction from the chromatin and inhibits transcription activity.45 It also promotes free radical production and induces tumor cell death. While these effects of doxorubicin are beneficial to cancer patients in reducing cancer progression, the side effects can be deadly. The most dangerous side effects include cardiomyopathy and congestive heart failure, as doxorubicin induces cardiomyocyte apoptosis and decreases cardiomyocyte viability, leading to cardiomyopathy and heart failure. Thus, it is essential to dissect the signaling pathway that mediates doxorubicin effects. It is ideal to retain its function in inhibiting cancer progression, while preventing or reducing its side effects. Our study showed that the circRNA circNlgn and its translated product Nlgn173 are downstream of doxorubicin. Silencing endogenous circNlgn decreased doxorubicin-induced cardiomyocyte apoptosis and increased cardiomyocyte viability. While cardiomyocyte death is promoted by doxorubicin, empty space becomes available to allow cardiac fibroblasts to move in and grow. The hallmark of this process is cardiac remodeling, characterized by increased deposition of collagens and expression of fibrosis markers. Our study showed that silencing circNlgn prevented collagen deposition and increased expression of fibrosis markers. This suggests that targeting circNlgn may relieve the side effects of doxorubicin. It has not been reported that circNlgn plays roles in cancer progression, but this awaits further investigation. circNlgn is generated by the parental neuroligin pre-mRNA. Neuroligin is a type I membrane protein that forms synapses between neurons.46, 47, 48 A novel protein isoform translated by circNlgn was detected in the nuclei because the translated protein acquired a motif for nuclear translocation. This motif contained nine amino acids due to the extension of translation of the circRNA beyond the back-splicing junction point. The nuclear protein functioned as a transcription factor to activate SGK3 expression, resulting in enhanced cardiac fibroblast proliferation and collagen deposition, and ING4 expression, resulting in decreased cardiomyocyte viability. As a consequence, circNlgn and the translated protein promoted cardiac remodeling and fibrosis.
It has been reported that doxorubicin induces H2AX eviction and DNA double-strand breaks, leading to DNA damage and cell apoptosis.45 When DNA double-strand breaks occur, proteins necessary for DNA repair are recruited, and H2AX is phosphorylated to form γH2AX.39 Phosphorylation of H2AX further de-condenses chromatin structure, promoting DNA double-strand breaks. Our study showed that increased expression of circNlgn facilitated H2AX phosphorylation. Silencing circNlgn decreased H2AX phosphorylation, while the total levels of H2AX were unaffected. circNlgn acted by directly binding to H2AX and activated this substrate. However, silencing circNlgn only partially reversed H2AX phosphorylation. This suggests that there might be another pathway mediating the effects of circNlgn on reducing doxorubicin-induced cardiomyocyte apoptosis and fibrosis.
To have a global picture of circNlgn functions, we examined gene expression in the heart tissues of circNlgn-transgenic mice by mRNA sequencing and found upregulation of a number of genes involved in DNA repair and cell survival. RAD50, a gene involved in DNA repair by binding to double-stranded DNA, was upregulated in circNlgn-transgenic mice. Upregulation of DNA sensor protein RAD50 was recently shown to be responsible for lipopolysaccharide (LPS)-induced inflammation.49 In the cytosol of LPS-stimulated macrophages, the upregulated RAD50 interacted with double-stranded DNA released from mitochondria and mediated LPS-induced acute lung injury. RAD50 also plays roles in enhancing fibroblast proliferation.50 Thus, in circNlgn-transgenic mice, the upregulated RAD50 appears to be responsible for the enhanced cardiac fibrosis.
We also detected upregulation of two other genes, Sema4C and Gadd45b. Sema4C is expressed in a number of tissues, including the vascular system.51 While the functions of Sema4C have been mainly reported in cancer progression, Sema4C appears to be involved in tissue differentiation by regulating the p38 MAPK pathway.42 Our results showed that Sema4C also plays roles in cardiomyocyte viability in mediating circNlgn functions. Treatment with doxorubicin promoted Sema4C expression, which was further promoted by circNlgn expression. Doxorubicin-induced cardiomyocyte apoptosis could be reduced by silencing circNlgn or Sema4C. Similarly, Gadd45b regulates cell apoptosis by binding to p38.40 It can also be activated by oxidative stress.52 Gadd45b enhances podocyte injury and apoptosis by activating the p38 MAPK pathway.41 Our results showed that Gadd45b played a role similar to that of Sema4C in mediating circNlgn’s effects on cardiomyocyte survival and apoptosis, which appeared to be mediated by the p38 MAPK pathway.
In dissecting the downstream pathway of activation of H2AX, Sema4C, Gadd45b, pJNK, and p38 MAPK, we found that the cytokines IL-1b, IL-2Rb, and IL-6 were activated. Activation of these cytokines appeared to be essential in mediating the induction of cardiomyopathy by doxorubicin. Consistent with these observation, we detected upregulation of the EGR members. EGR1 is a transcription factor and master regulator of tissue remodeling and fibrosis.53,54 It plays crucial roles in the progression of cardiovascular disease such as cardiac hypertrophy, myocardial ischemia/reperfusion, and atherosclerosis.55 Targeting EGR1 treatment after acute myocardial ischemia reduces inflammation, oxidative damage, and myocardial apoptosis and improves heart function.56 Similarly, EGR3 also plays roles in tissue remodeling.54 EGR1 and EGR3 regulate the expression of genes involved in cell proliferation and differentiation by activating the transcription cascade.57 EGR3 can also induce expression of TGF-β and other fibrotic genes, resulting in tissue remodeling and progression of wound repair.58 Our results showed that EGR1 and EGR3 were induced by doxorubicin and upregulated by circNlgn expression. Since EGRs are master regulators of tissue remodeling and fibrosis and they are downstream signaling molecules in mediating the effects of circNlgn, these molecules may be potential targets for reducing the side effects of doxorubicin in cancer patients.
Material and methods
Materials
Rabbit polyclonal antibody against the Nlg173 junction peptide (GYRPAANWI) was generated by GenScript (Nanjing, China). The antibody was purified with histidine-tagged Nlg173 protein that bound Ni-NTA resins. The monoclonal antibodies against Nlgn (N terminus), JNK, pJNK, and p38 were obtained from Santa Cruz Biotechnology. The monoclonal antibodies against H2AX, γH2AX, and GAPDH were purchased from Cell Signaling. Horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG were purchased from Sigma. RNA and DNA extraction kits and RNA RT and PCR kits were obtained from Qiagen. The Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit was from Biovision.
Constructs, siRNAs, transfection, and analysis
The plasmid expressing human circNlgn was generated and reported in our previous study.38 In the study, we also designed two siRNAs complementary to the junction sequence of circNlgn. Both siRNAs could silence circNlgn in vivo and in vitro. Expression and circularization of circNlgn were validated by primers flanking the junction sequence of circNlgn.38
Animal model and cardiac functional assessment
All animal experiments were performed in accordance with relevant guidelines and regulations. All procedures conformed to the guidelines of the NIH Guide for the Care and Use of Laboratory Animals. The animal use protocol was approved by the Animal Care Committee of Sunnybrook Research Institute. A priori power calculation was used in the determination of experimental mouse group sizes with the software G∗Power.59 Both male and female animals were randomly grouped in the experiments.
The circNlgn-transgenic mouse model was generated by pronuclear microinjection of a circNlgn-containing DNA fragment into C57BL/6J, performed by the Toronto Centre for Phenogenomics (TCP). All transgenic mice were ear tagged and genotyped after weaning. The genotyping primer sequences are provided in the supplementary information.
The animal model of doxorubicin-induced cardiomyopathy and fibrosis was induced in 8-week-old mice by intraperitoneal injection of doxorubicin (3 mg/kg, three times per week, with a total cumulative dose of doxorubicin of 21 mg/kg). Each group had 10 mice. In the negative control group, the mice were injected with an equal volume of saline.
Three weeks after doxorubicin injection, all the mice were anesthetized with 2% isoflurane inhalation to perform transthoracic echocardiography. Transthoracic echocardiography was performed and analyzed in a blinded manner, using a Vevo 2000 high-resolution imaging system to assay LVEDD, LVESD, LVEF, LVFS, and dp/dt.
All mice were sacrificed following cardiac functional assessment. Hearts were harvested and cut into two halves. The upper half of the heart was kept frozen for PCR or processed to obtain frozen sections, followed by immunofluorescence, while the lower half was fixed with 10% buffered formalin and embedded in paraffin and then sectioned into 5 μm slides. H&E, Masson’s trichrome, and Sirius red staining were performed.
Heart fibrosis staining
Masson’s trichrome and Sirius red staining were performed with a Masson’s trichrome kit (StatLab, American MasterTech, Lodi, CA, USA) and a Sirius red staining kit (Millipore Sigma, Burlington, MA, USA). Masson’s trichrome staining was performed according to the manufacturer’s instructions. The formalin-fixed paraffin-embedded tissue sections were de-paraffinized and washed with 100% ethanol and water. The slides were incubated with Bouin’s fluid at 60°C for 1–2 h. After being washed with water, the slides were incubated with Weigert’s working hematoxylin (5 min), Biebrich scarlet-acid fuchsin (15 min), phosphomolybdic/phosphotungstic acid (15 min), aniline blue stain (10 min), and 1% acetic acid (5 min). The slides were dehydrated with 100% ethanol and clean xylene and mounted with mounting coverslips and permanent mounting medium.
Sirius red staining was performed according to the manufacturer’s instructions. Briefly, the formalin-fixed paraffin-embedded tissue sections were de-paraffinized and washed with 100% ethanol and then water. The slides were stained with Weigert’s working hematoxylin for 5 min and washed 10 min in running tap water. The slides were stained with picro-Sirius red for 1–4 h to obtain sufficient staining and washed with 0.5% acetic acid for 10–60 min to de-stain the background. The slides were then dehydrated with 100% ethanol and clean xylene, followed by mounting with mounting coverslips and permanent mounting medium.
Immunohistochemistry staining
The formalin-fixed paraffin-embedded tissue sections were de-paraffinized, heated in a pressure cooker, and treated for 30 min with peroxidase-blocking reagent to quench the endogenous peroxidase activity. The sections were incubated with 10% goat serum and incubated with primary antibody in TBS containing 10% goat serum overnight. The sections were then washed and incubated with biotinylated secondary antibody, followed by avidin-conjugated horseradish peroxidase provided by the Vectastain ABC kit, and with diaminobenzidine (DAB) followed by Mayer’s hematoxylin for counterstaining. The slides were then immersed in DAB (DAKO, Glostrup, Denmark) substrate for 5 min, followed by washing in distilled water for 1 h. After being counterstained with Harris hematoxylin, the slides were dehydrated and mounted.
Isolation of primary mouse cardiomyocytes and cardiac fibroblasts
The enzymatic dispersion technique was used to isolate PCMs and PCFs from neonatal mouse heart. Briefly, animals were heparinized, anesthetized with isoflurane, and sacrificed by cervical dislocation. The hearts were rapidly removed; washed in PBS with 20 mM BDM; transferred into a drop of HEPES-buffered Tyrode solution containing 130 mM NaCl, 5.4 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 0.33 mM Na2HPO4, 10 mM HEPES, and 5.5 mM glucose (pH 7.4); and minced into small pieces. Cardiac tissues were fragmented and incubated in 25 mL Tyrode solution (0.012 g collagenase D, 0.009 g collagenase B, and 0.001 g protease XIV from Streptomyces griseus) at 37°C for 20–30 min. Digested products were filtered and centrifuged at 600 rpm for 5 min. The cell pellet was resuspended in DMEM/F12 medium containing 10% FBS and 20 mM BDM and plated onto a 10 cm cell culture dish and incubated for 1–3 h in a cell culture incubator. This pre-plating step isolated the PCFs, which adhered to the uncoated cell culture dish. Non-adherent cells were transferred to a cell culture dish coated with 1% gelatin solution to allow PCM adhesion and collection.
Cell-cycle analysis
The cells were detached by trypsin/EDTA, washed and resuspended in cold PBS, incubated in 0.5 mL ice-cold 70% ethanol, and kept at −20°C for 2 h. The cells were then centrifuged at 1,500 rpm for 10 min and washed with cold PBS twice. The pellets were resuspended in 0.5 mL PBS containing 40 μg/mL propidium iodide and 100 μg/mL RNase, and incubated at room temperature for 30 min. The propidium iodide-stained cells were kept on ice before analysis. The percentages of S and G2 population cells were analyzed with flow cytometry.
Cell-proliferation assay
Cells were cultured in 10% FBS basal medium in 12 well culture dishes (1 × 104 to 2 × 104 cells/well) at 37°C overnight in an incubator containing 5% CO2. After cell attachment, the cells were maintained 37°C for 1–6 days. Cells were detached with trypsin/EDTA, washed with PBS, and suspended in 0.5 mL culture medium daily or at specific time points (1–6 days). The cells were harvested and stained with trypan blue. Proliferating cells were counted by a Coulter counter under an inverted microscope.
Cells were also cultured in 10% FBS basal medium in 96 well plates (1 × 103 to 5 × 103 cells/well) for indicated time points. Cell viability was measured with an MTT kit. Briefly, the cells were plated in 96-well culture dishes in 200 μL of culture medium and maintained at 37°C in a humidified chamber for the indicated time points. Fifty microliters of serum-free medium and 50 μL of MTT solution were added to each well. After incubation for 3 h, 100 μL of solubilization solution was added to each well and incubated overnight in a humidified atmosphere. The absorbance was read at 570 nm using a microplate reader (Bio-Rad, CA, USA).
Cell-survival assay
Cells were cultured in 10% FBS basal medium in 12 well culture dishes (5 × 104 to 8 × 104 cells/well) at 37°C overnight in an incubator containing 5% CO2. After cell attachment, the medium was replaced with serum-free basal medium or 10% FBS basal medium containing 0.2–2 μM doxorubicin. Cells were detached with trypsin/EDTA, washed with PBS, and suspended in 0.5 mL culture medium daily or at specific time points (12 h–6 days). The harvested cells were stained with trypan blue and the number of surviving cells was counted by Coulter counter under an inverted microscope.
The cell viability was also determined using an MTT assay kit (Sigma). Briefly, cells were plated in 96 well culture dishes (1 × 104 cells) in 200 μL of culture medium and maintained at 37°C for 12 h. After cell attachment, the medium was replaced with DMEM containing 10% FBS and doxorubicin and cultured at 37°C for the indicated time points in a humidified chamber. Fifty microliters of serum-free medium and 50 μL of MTT solution were added to each well. After incubation for 3 h, 100 μL of solubilization solution was added to each well and the cultures were incubated overnight in a humidified atmosphere. The absorbance was read at 570 nm using a microplate reader (Bio-Rad, CA, USA).
Annexin V assay
An Annexin V-APC apoptosis detection kit (BioVision) was used to detect cell apoptosis. Cells (1 × 106) were cultured in serum-free basal medium or 10% FBS basal medium with 0.2–2 μM doxorubicin for 12–48 h. The cells were detached with trypsin/EDTA, washed with PBS, and suspended in 100 μL binding buffer. The cultures were incubated with Annexin V-APC at room temperature for 30 min. The cells were washed and resuspended in 200 μL binding buffer with 5 μL propidium iodide and kept on ice before analysis. Staining of Annexin V-APC activity was determined by flow cytometry using an APC signal detector (FL1) and propidium staining by the phycoerythrin emission signal detector (FL2).
Immunoprecipitation assay
Cells (107) were harvested and washed in ice-cold PBS followed by lysis in 1 mL co-immunoprecipitation (coIP) buffer. Two micrograms of primary antibodies was incubated with 200 mL of magnetic beads (Bio-Rad) at room temperature for 1 h. Antibody-conjugated magnetic beads were washed with PBS with 0.1% Tween three times. Equal amounts of protein were incubated with the antibody-conjugated magnetic beads at room temperature for 2 h. The beads were washed three times with PBS and resuspended in 2× Laemmli buffer (0.125 M Tris-Cl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromophenol blue [pH 6.8]) and incubated at 70°C for 10 min followed by western blotting.
RT-PCR and real-time PCR
Cells (2 × 105) were harvested, and total RNA was extracted with the Qiagen RNeasy mini kit. One microgram of total RNA was used to synthesize cDNA, a portion of which (1 μL, equal to 0.2 μg cDNA) was used in a PCR with two appropriate primers. Real-time PCR was performed with the miScript SYBR Green PCR kit (Qiagen) using 1 μL cDNA as the template. Thermocycler conditions were 40 cycles of denaturation at 95°C for 15 s, annealing at 56°C for 10 s, and extension at 72°C for 5 s. The ΔΔCT method was used to quantify all relative mRNA levels using small nuclear U6 RNA as the reference and an internal standard for quantitation.
Western blot analysis
Cells were lysed and subjected to SDS-PAGE containing 5%–12% acrylamide depending on the size of the protein of interest. Separated proteins were transferred onto a nitrocellulose membrane in 1× Tris/glycine buffer containing 20% methanol at 80 V at 4°C for 2 h. The membrane was blocked in TBST (10 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 5% non-fat dry milk powder (TBSTM) for 0.5 h, and then incubated with primary antibodies at 4°C overnight. The membranes were washed with TBST (3 × 30 min) and incubated with secondary antibodies for 1 h. After washing, the bound antibodies were visualized with an ECL detection kit.
Statistical analysis
Data are presented as the mean (bar) with standard deviation (SD). For multiple group analyses, we used a one-way ANOVA followed by Tukey analysis. The presented p values are corrected. Student’s t test was performed to assess the difference between two groups with a single independent factor. These statistical analyses were calculated using Prism 8 (GraphPad Software, La Jolla, CA, USA). Significance was defined as ∗p < 0.05 or ∗∗p < 0.01 or as specified in the figures.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (PJT-153105, PJT-155962, and PJT-166107) to B.B.Y. Our study complies with the Declaration of Helsinki in that the Animal Care Committee of Sunnybrook Research Institute approved the research protocol and informed consent was obtained from the subjects. Analysis of human heart tissues was approved by the Ethics Committee of Guangdong General Hospital.
Author contributions
J.X., W.W.D., N.W., F.L., and X.L. performed the experiments. J.X., W.W.D., and B.B.Y. analyzed the results. S.W. and B.B.Y. designed and supervised the project. W.W.D. and B.B.Y. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.03.007
Contributor Information
Yizhen Xie, Email: xieyizhen@126.com.
Sheng Wang, Email: shengwang_gz@163.com.
Burton B. Yang, Email: byang@sri.utoronto.ca.
Supplemental information
References
- 1.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: an update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 2.Zhang S., Liu X., Bawa-Khalfe T., Lu L.S., Lyu Y.L., Liu L.F., Yeh E.T. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nat. Med. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 3.Burridge P.W., Li Y.F., Matsa E., Wu H., Ong S.G., Sharma A., Holmstrom A., Chang A.C., Coronado M.J., Ebert A.D., et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat. Med. 2016;22:547–556. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X., Javan H., Li L., Szucsik A., Zhang R., Deng Y., Selzman C.H. A modified murine model for the study of reverse cardiac remodelling. Exp. Clin. Cardiol. 2013;18 e115–117. [PMC free article] [PubMed] [Google Scholar]
- 5.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keating M.T., Sanguinetti M.C. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 8.Minotti G., Ronchi R., Salvatorelli E., Menna P., Cairo G. Doxorubicin irreversibly inactivates iron regulatory proteins 1 and 2 in cardiomyocytes: evidence for distinct metabolic pathways and implications for iron-mediated cardiotoxicity of antitumor therapy. Cancer Res. 2001;61:8422–8428. [PubMed] [Google Scholar]
- 9.Bostrom P., Mann N., Wu J., Quintero P.A., Plovie E.R., Panakova D., Gupta R.K., Xiao C., MacRae C.A., Rosenzweig A., et al. C/EBPbeta controls exercise-induced cardiac growth and protects against pathological cardiac remodeling. Cell. 2010;143:1072–1083. doi: 10.1016/j.cell.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 12.Hentze M.W., Preiss T. Circular RNAs: splicing's enigma variations. EMBO J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misir S., Wu N., Yang B.B. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481–491. doi: 10.1038/s41418-022-00948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilusz J.E., Sharp P.A. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., et al. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lavenniah A., Luu T.D.A., Li Y.P., Lim T.B., Jiang J., Ackers-Johnson M., Foo R.S. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. J. Am. Soc. Gene Ther. 2020;28:1506–1517. doi: 10.1016/j.ymthe.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S.K., Garg A., Bar C., Chatterjee S., Foinquinos A., Milting H., Streckfuss-Bomeke K., Fiedler J., Thum T. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aufiero S., Reckman Y.J., Pinto Y.M., Creemers E.E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 2019;16:503–514. doi: 10.1038/s41569-019-0185-2. [DOI] [PubMed] [Google Scholar]
- 19.Lu D., Thum T. RNA-based diagnostic and therapeutic strategies for cardiovascular disease. Nat. Rev. Cardiol. 2019;16:661–674. doi: 10.1038/s41569-019-0218-x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y., Du L., Bai Y., Han B., He C., Gong L., Huang R., Shen L., Chao J., Liu P., et al. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol. Psychiatry. 2020;25:1175–1190. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang R., Zhang Y., Bai Y., Han B., Ju M., Chen B., Yang L., Wang Y., Zhang H., Zhang H., et al. N(6)-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry. 2020;88:392–404. doi: 10.1016/j.biopsych.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 22.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., Du L., Xu L., Wu F., et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sang Y., Chen B., Song X., Li Y., Liang Y., Han D., Zhang N., Zhang H., Liu Y., Chen T., et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2019;27:1638–1652. doi: 10.1016/j.ymthe.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X., Zhang N., Han P., Moon B.S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu N., Qadir J., Yang B.B. CircRNA perspective: new strategies for RNA therapy. Trends Mol. Med. 2022;S1471-4914:00041–00047. doi: 10.1016/j.molmed.2022.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 30.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cel. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 31.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 33.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z., et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cel. 2017;66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang W.C., Wong C.W., Liang P.P., Shi M., Cao Y., Rao S.T., Tsui S.K., Waye M.M., Zhang Q., Fu W.M., et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84. doi: 10.1186/s13059-019-1685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 37.Zheng X., Chen L., Zhou Y., Wang Q., Zheng Z., Xu B., Wu C., Zhou Q., Hu W., Wu C., et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du W.W., Xu J., Yang W., Wu N., Li F., Zhou L., Wang S., Li X., He A.T., Du K.Y., et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ. Res. 2021;129:568–582. doi: 10.1161/CIRCRESAHA.120.318364. [DOI] [PubMed] [Google Scholar]
- 39.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 40.Cho H.J., Park S.M., Hwang E.M., Baek K.E., Kim I.K., Nam I.K., Im M.J., Park S.H., Bae S., Park J.Y., et al. Gadd45b mediates Fas-induced apoptosis by enhancing the interaction between p38 and retinoblastoma tumor suppressor. J. Biol. Chem. 2010;285:25500–25505. doi: 10.1074/jbc.M109.091413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z., Wan X., Hou Q., Shi S., Wang L., Chen P., Zhu X., Zeng C., Qin W., Zhou W., et al. GADD45B mediates podocyte injury in zebrafish by activating the ROS-GADD45B-p38 pathway. Cell Death Dis. 2016;7:e2068. doi: 10.1038/cddis.2015.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu H., Wang X., Liu S., Wu Y., Zhao T., Chen X., Zhu L., Wu Y., Ding X., Peng X., et al. Sema4C participates in myogenic differentiation in vivo and in vitro through the p38 MAPK pathway. Eur. J. Cel. Biol. 2007;86:331–344. doi: 10.1016/j.ejcb.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Sluss H.K., Davis R.J. H2AX is a target of the JNK signaling pathway that is required for apoptotic DNA fragmentation. Mol. Cel. 2006;23:152–153. doi: 10.1016/j.molcel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Momparler R.L., Karon M., Siegel S.E., Avila F. Effect of adriamycin on DNA, RNA, and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36:2891–2895. [PubMed] [Google Scholar]
- 45.Pang B., Qiao X., Janssen L., Velds A., Groothuis T., Kerkhoven R., Nieuwland M., Ovaa H., Rottenberg S., van Tellingen O., et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheiffele P., Fan J., Choih J., Fetter R., Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 47.Sudhof T.C. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhan X., Cui R., Geng X., Li J., Zhou Y., He L., Cao C., Zhang C., Chen Z., Ying S. LPS-induced mitochondrial DNA synthesis and release facilitate RAD50-dependent acute lung injury. Signal Transduction Targeted Therapy. 2021;6:103. doi: 10.1038/s41392-021-00494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkening L., Vatselia A., Asgedom G., Bastians H., Lavin M., Schindler D., Schambach A., Bousset K., Dork T. RAD50 regulates mitotic progression independent of DNA repair functions. FASEB J. Official Publ. Fed. Am. Societies Exp. Biol. 2020;34:2812–2820. doi: 10.1096/fj.201902318R. [DOI] [PubMed] [Google Scholar]
- 51.Zielonka M., Xia J., Friedel R.H., Offermanns S., Worzfeld T. A systematic expression analysis implicates Plexin-B2 and its ligand Sema4C in the regulation of the vascular and endocrine system. Exp. Cel. Res. 2010;316:2477–2486. doi: 10.1016/j.yexcr.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 52.Kim J.H., Qu A., Reddy J.K., Gao B., Gonzalez F.J. Hepatic oxidative stress activates the Gadd45b gene by way of degradation of the transcriptional repressor STAT3. Hepatology. 2014;59:695–704. doi: 10.1002/hep.26683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bhattacharyya S., Wu M., Fang F., Tourtellotte W., Feghali-Bostwick C., Varga J. Early growth response transcription factors: key mediators of fibrosis and novel targets for anti-fibrotic therapy. Matrix Biol. J. Int. Soc. Matrix Biol. 2011;30:235–242. doi: 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L., Carter J., Gao X., Whitehead J., Tourtellotte W.G. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol. Cell. Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ramadas N., Rajaraman B., Kuppuswamy A.A., Vedantham S. Early growth response-1 (EGR-1) - a key player in myocardial cell injury. Cardiovasc. Hematological Agents Med. Chem. 2014;12:66–71. doi: 10.2174/1871525713666150123152131. [DOI] [PubMed] [Google Scholar]
- 56.Rayner B.S., Figtree G.A., Sabaretnam T., Shang P., Mazhar J., Weaver J.C., Lay W.N., Witting P.K., Hunyor S.N., Grieve S.M., et al. Selective inhibition of the master regulator transcription factor Egr-1 with catalytic oligonucleotides reduces myocardial injury and improves left ventricular systolic function in a preclinical model of myocardial infarction. J. Am. Heart Assoc. 2013;2:e000023. doi: 10.1161/JAHA.113.000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumbrink J., Kirsch K.H., Johnson J.P. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J. Cell. Biochem. 2010;111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang F., Shangguan A.J., Kelly K., Wei J., Gruner K., Ye B., Wang W., Bhattacharyya S., Hinchcliff M.E., Tourtellotte W.G., et al. Early growth response 3 (Egr-3) is induced by transforming growth factor-beta and regulates fibrogenic responses. Am. J. Pathol. 2013;183:1197–1208. doi: 10.1016/j.ajpath.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faul F., Erdfelder E., Lang A.G., Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.