Abstract
Background & Aims
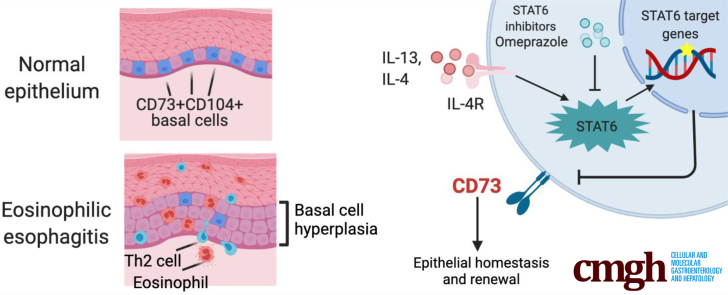
Although basal cell hyperplasia is a histologic hallmark of eosinophilic esophagitis (EoE), little is known about the capabilities of epithelial renewal and differentiation in the EoE inflammatory milieu. In murine esophageal epithelium, there are self-renewing and slowly proliferating basal stem-like cells characterized by concurrent expression of CD73 (5'-nucleotidase ecto) and CD104 (integrin β4). Here, we investigated CD73+CD104+ cells within the basal population of human esophageal epithelium and clarified the biological significance of these cells in the EoE epithelium.
Methods
We performed flow cytometry on esophageal biopsy samples from EoE and non-EoE patients to determine the quantity of CD73+CD104+ cells in the epithelium. Simulating the EoE milieu we stimulated primary patient-derived and immortalized cell line–derived esophageal organoids with interleukin (IL)4 and IL13 and analyzed by flow cytometry, immunohistochemistry, and quantitative reverse-transcription polymerase chain reaction. We performed single-cell RNA sequencing on primary organoids in the setting of IL13 stimulation and evaluated the CD73+CD104+ population. We performed fluorescent-activated cell sorting to purify CD73+CD104+ and CD73- CD104+ populations and seeded these groups in organoid culture to evaluate the organoid formation rate and organoid size. We used RNA interference to knock down CD73 in esophageal organoids to evaluate organoid formation rates and size. We evaluated the effects of signal transducer and activator of transcription 6 (STAT6) signaling inhibition by RNA interference, a STAT6 inhibitor, AS1517499, as well as the proton pump inhibitor omeprazole.
Results
EoE patients showed decreased epithelial CD73+CD104+ cell content. IL4 and IL13 stimulation depleted this population in 3-dimensional organoids with a recapitulation of basal cell hyperplasia as corroborated by single-cell RNA sequencing of the organoids, which suggests depletion of CD73+CD104+ cells. The CD73+CD104+ population had enhanced organoid formation compared with the CD73-CD104+ population. Similarly, knock-down of CD73 resulted in decreased organoid formation rate. Genetic and pharmacologic inhibition of STAT6 prevented T helper 2 cytokine-induced depletion of CD73+CD104+ cells. Lastly, omeprazole treatment prevented the effects of IL4 and IL13 on the CD73+CD104+ population.
Conclusions
This study addressed the role of CD73+CD104+ cells in epithelial renewal and homeostasis in the context of EoE. The depletion of the CD73+CD104+ self-renewal population by helper T cell 2 cytokines in EoE milieu may be perpetuating epithelial injury. Future therapies targeting epithelial restitution in EoE could decrease the need for immune modulation and steroid therapy.
Keywords: Eosinophilic Esophagitis, Epithelium, Organoids, CD73, CD104
Abbreviations used in this paper: BCH, basal cell hyperplasia; DAPI, 4’,6-diamidino-2-phenylindole; DEG, differentially expressed gene; EdU, ethynyldeoxyuridine; EGD, esophagogastroduodenoscopy; EoE, eosinophilic esophagitis; eos/hpf, esophageal mucosal eosinophils per high-power field; FACS, fluorescence-activated cell sorting; GO, Gene Ontology; IHC, immunohistochemistry; IL, interleukin; mRNA, messenger RNA; OFR, organoid formation rate; OVA, ova-albumin; PCA, principal component analysis; PDO, patient-derived organoid; PPI, proton pump inhibitor; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; siRNA, small interfering RNA; STAT6, signal transducer and activator of transcription 6; 3D, 3-dimensional; Th, helper T cell
Graphical abstract
Summary.
We show that the CD73+CD104+ progenitor population regulates epithelial renewal and homeostasis in the basal zone of esophageal epithelium. T helper 2 cytokines, interleukins 4 and 13 deplete this CD73+CD104+ self-renewing population via the signal transducer and activator of transcription 6 pathway, which may be perpetuating epithelial injury in the context of eosinophilic esophagitis.
The stratified squamous epithelium of the esophagus comprises basal, parabasal, and suprabasal cell layers that display an exquisite proliferation–differentiation gradient. The basal/parabasal cell layers contain proliferative basal cells (keratinocytes) that undergo postmitotic terminal differentiation within the suprabasal cell layer. Differentiated keratinocytes form intercellular bridges (desmosomes, tight junctions, and so forth) to provide the first line of defense against the chemical and biological milieu of luminal contents. Although exposure to acid, carcinogens, and allergens perturbs squamous cell differentiation,1 it is unknown how these insults affect epithelial renewal and proliferation. Disruption of this homeostatic differentiation gradient or barrier function is linked to multiple human pathologies including gastroesophageal reflux disease and eosinophilic esophagitis (EoE).
EoE is an allergen-induced chronic inflammatory disease of the esophagus, characterized by dysphagia, food impactions, and fibrotic strictures.2 Histologically, the most conspicuous finding in EoE, in addition to the eosinophilic infiltrate, is basal cell hyperplasia (BCH).3,4 BCH is induced by injury or inflammation and involves an expansion of basal cells (>20% of epithelial height) with limited formation of intercellular bridges, a hallmark of squamous cell differentiation. The EoE transcriptome suggests stalled differentiation of the esophageal epithelium, leading to epithelial barrier defects and ongoing antigen exposure.5, 6, 7 Multiple EoE-relevant cytokines such as interleukin (IL)5 and IL13 induce BCH in murine models of EoE.4,8 These cytokines mediate the functional interplay between basal cells and fibroblasts to facilitate lamina propria fibrosis.3,9, 10, 11, 12 Detailing the populations that exist within the basal epithelium and how these are perturbed in the EoE epithelium would allow for broader understanding of epithelial responses to inflammation. Similarly, therapies aiming to re-establish epithelial homeostasis represent an unexplored area that potentially could spare immune suppression or steroid exposure in EoE.
Basal cells are heterogeneous with variable expression of markers such as CD104 (integrin β4) and CD73 (5'-nucleotidase ecto), as well as variable proliferation and differentiation capabilities.13, 14, 15, 16 DeWard et al14 described heterogeneity within the murine esophageal basal epithelium, identifying CD73+CD104+ basal cells as a distinct stem cell–like population with self-renewal and epithelial formation capabilities. However, CD73+CD104+ cells remain uncharacterized in the human esophageal epithelium, and the effect of inflammation on homeostatic epithelial renewal remains unknown.
Herein, we observed alterations in the heterogeneity within proliferative and undifferentiated basal or basaloid (defined as proliferative and undifferentiated basal-like cells present in the suprabasal cell layers population17) of the human esophageal epithelium and assessed perturbations that occur with allergic inflammation. Using patient biopsy specimens and single-cell–derived esophageal 3-dimensional (3D) organoids, a novel modeling platform in esophageal epithelial homeostasis, pathobiology, and personalized medicine,7,18 we show that the stem-like CD73+CD104+ population is diminished markedly in EoE, implicating helper T cell (Th)2 cytokines and signal transducer and activator of transcription 6 (STAT6) signaling as the effector and a promising target for therapy.
Results
CD73+ Cells Are Under-Represented in Active EoE Epithelium
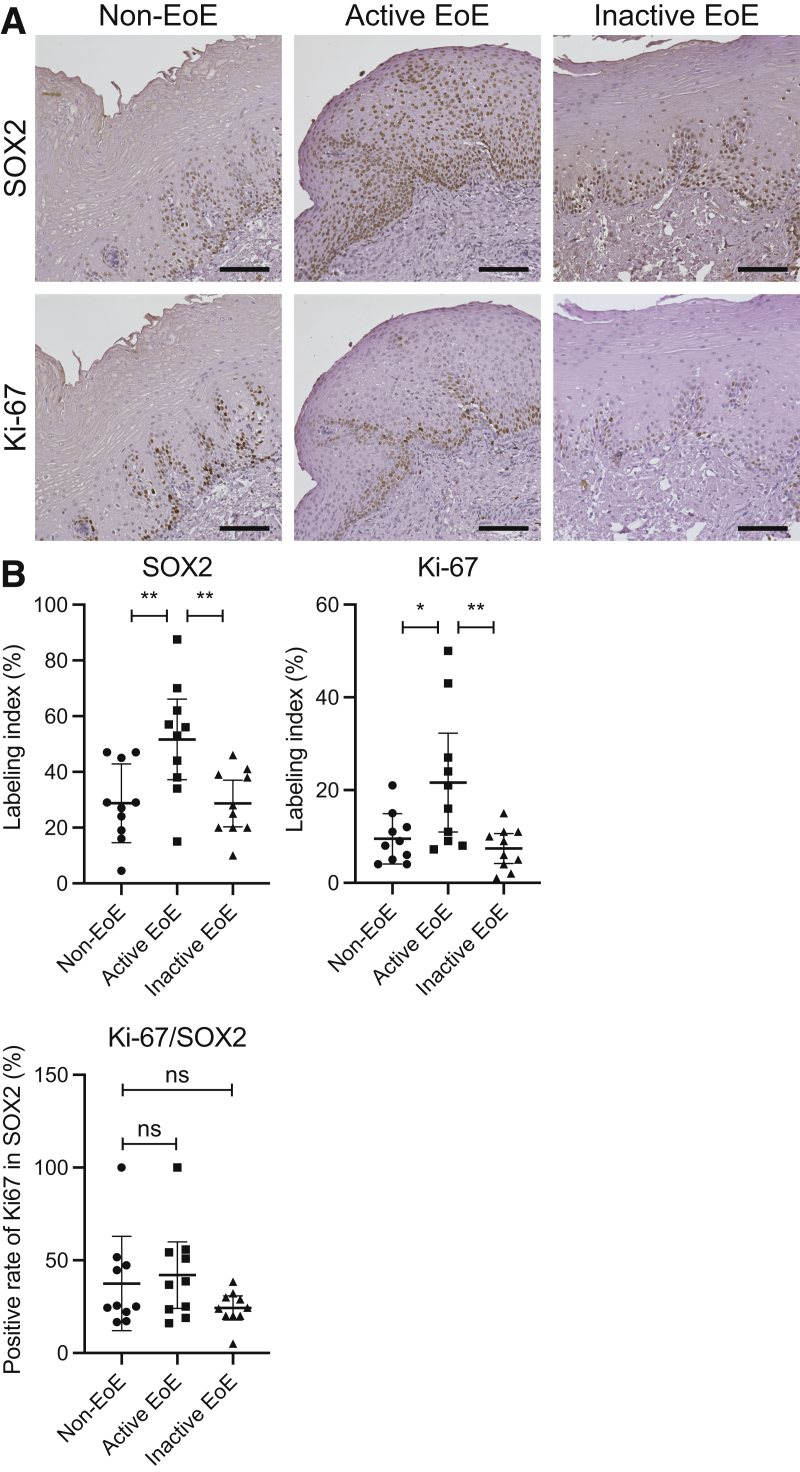
To assess the proliferation of the basal cells in the esophageal epithelium biopsy specimens, we performed immunohistochemistry (IHC) for Ki-67 and SOX2, which are recognized as markers of cell proliferation and basal cells, respectively.4,19,20 As previously shown,3 active EoE biopsy specimens are characterized by marked expansion of the basal cells, as shown by increased SOX2 staining in the active EoE epithelium (Figure 1A). We also found that Ki-67+ cells were increased in the esophageal epithelium in active EoE specimens, as has been described previously.4 Interestingly, we noted that the Ki-67/SOX2 positivity ratio remains unchanged in active EoE (Figure 1B). This indicates that not all SOX2-positive cells in the basal zone are proliferative. These findings suggest that within the basal zone of the epithelium there are distinct subpopulations, and despite an increase in the total number of SOX2-positive cells in EoE, only a distinct subset remains proliferative.
Figure 1.
The heterogeneity of proliferation in the SOX2-positive basal zone. IHC staining for SOX2 or Ki-67 on biopsy specimens from non-EoE controls (n = 10), and patients with active (n = 10) or inactive EoE (n = 10). (A) Representative images are shown. Scale bars: 100 μm. (B) The positive rate of SOX2 or Ki-67 in total cells, or the positive rate of Ki-67 in SOX2-positive cells are compared. The data are shown as means ± SD. ∗P < .05, ∗∗P < .01, analysis of variance.
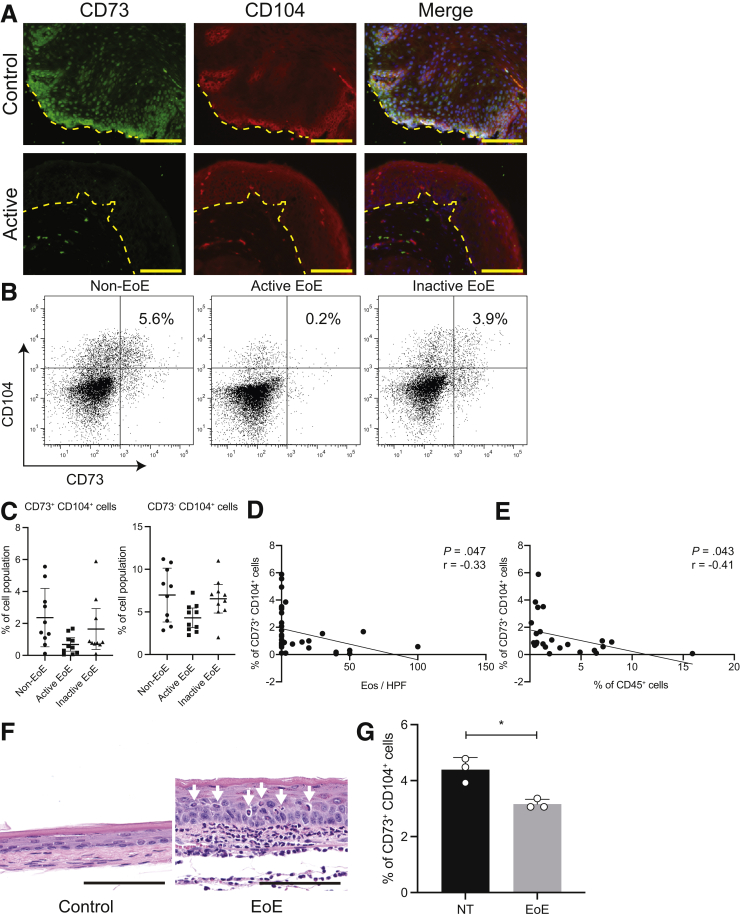
Even though murine and human epithelial populations are fundamentally different,21,22 we hypothesized that the observation of basal zone heterogeneity may be related to the phenomenon described by DeWard et al14 in the mouse esophagus, and therefore we sought to characterize these populations using the cell surface markers CD73 and CD104Click or tap here to enter text. We performed immunofluorescence for CD73 and CD104 with esophageal biopsy samples collected from active EoE, as well as non-EoE controls, and found CD73+CD104+ cells are decreased in EoE (Figure 2A). We also performed flow cytometry on esophageal biopsy samples from active or inactive EoE patients, as well as non-EoE controls. After negatively selecting leukocytes (CD45+), we classified all cells into the following 3 groups: CD73+CD104+, CD73-CD104+, and CD73-CD104-. The representative flow cytometry plots of non-EoE controls and patients with active or inactive EoE are shown (Figure 2B). CD73+CD104+ cells are decreased significantly in active EoE (Figure 2C) (2.4% in non-EoE, 0.7% in active EoE, and 1.7% in inactive EoE; P = .0062). We found a weak negative correlation between CD73+CD104+ and CD45+ cells and esophageal mucosal eosinophils per high-power field (eos/hpf) (r = 0.41 and 0.33, respectively), signifying that the degree of inflammation and disease activity may be promoting these changes in the EoE esophagus (Figure 2D and E).
Figure 2.
The CD73+CD104+population is depleted in the esophagus of active EoE patients and a murine model of EoE. (A) Immunofluorescence staining for CD73 and CD104 on biopsy specimens from non-EoE controls and patients with active EoE. Representative images are shown. Dotted line represents the basement membrane. Scale bars: 100 μm. (B–E) Biopsy specimens from non-EoE controls (n = 10), and patients with active (n = 10) or inactive EoE (n = 10) were dissociated for flow cytometry. The representative flow cytometry plots are shown in panel B. (C) The percentage of CD73+CD104+ or CD73-CD104+ cells in non-EoE controls (n = 10), and patients with active (n = 10) or inactive EoE (n = 10). (D and E) Pearson correlation coefficient analyses comparing the percentage of CD73+CD104+ cells detected by flow cytometry and the number of eosinophils (eos/hpf) in (D) H&E-stained biopsy specimens or (E) CD45+ cells detected by flow cytometry. (F and G) After BALB/c mice were treated with the EoE protocol, their esophagi were resected for histologic sections or flow cytometry. (F) Histologic sections (H&E staining) of the esophagus from control (nontreated [NT]) or EoE protocol–treated mice. Arrowheads identify tissue-infiltrating eosinophils. (G) The percentage of CD73+ cells in esophageal epithelium of EoE mice (n = 3), compared with NT mice (n = 3). The data are shown as means ± SD. A 2-tailed Student t test or analysis of variance was performed for comparing 2 or multiple comparisons, respectively. ∗P < .05.
These findings were validated in our established murine model of EoE.23,24 In mice with EoE, the esophagus is characterized by basal cell hyperplasia and infiltrating eosinophils in the mucosa (Figure 2F), and we found a similarly decreased proportion of CD73+CD104+ cells in the esophageal epithelium in EoE mice (Figure 2G).
Th2 Cytokines Shift Basaloid Population From CD73+ to CD73- in Human Esophageal 3D Organoids
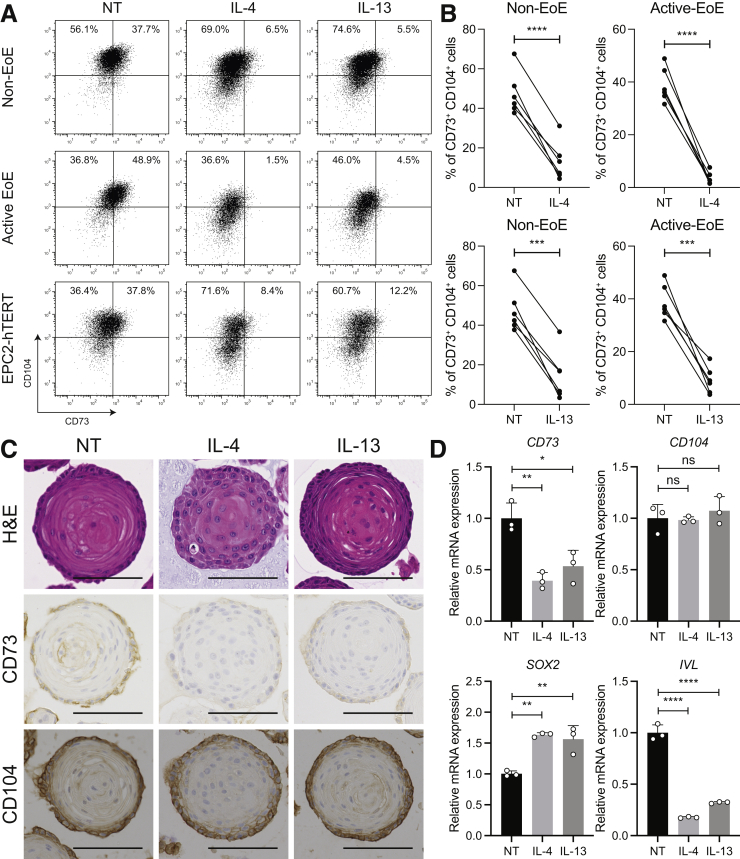
Because inflammation was weakly correlated with a diminished presence of CD73+ cells in EoE epithelium, we investigated the impact on the basaloid epithelium18 of the major effector Th2-type cytokines involved in EoE, IL4, and IL13. To that end, we treated patient-derived organoids (PDOs), as well as immortalized non-transformed cell line EPC2-hTERT organoids, with IL4 or IL13, followed by flow cytometry analysis (Figure 3A). We found the percentage of CD73+CD104+ cells decreased in organoids treated with IL4 or IL13, compared with nontreated organoids, accompanied by a significant increase of CD73-CD104+ cells by flow cytometry and IHC staining (Figure 3A–C, respectively).
Figure 3.
Th2 cytokines shift the basaloid population from CD73+to CD73-in human esophageal 3D organoids. PDOs from non-EoE controls (n = 6) or patients with active EoE (n = 6), or immortalized esophageal epithelial (EPC2-hTERT) organoids were treated with IL4 (10 ng/mL) or IL13 (10 ng/mL) from day 7 to day 11. Nontreated (NT) organoids were used as control. (A) Representative flow cytometry plots showing the percentage of CD73+CD104+ cells in PDOs from non-EoE controls or patients with active EoE, or EPC2-hTERT organoids, treated with IL4 or IL13. (B) The percentage of CD73+CD104+ cells in each PDO treated with IL4 or IL13, paired with NT control. The data are shown as means ± SD. ∗∗∗P < .001, ∗∗∗∗P < .0001, 2-tailed Student t test. (C) The representative images of H&E staining, and IHC for CD73 or CD104 in EPC2-hTERT organoids treated with IL4 or IL13. Scale bars: 100 μm. (D) CD73, CD104, SOX2, and IVL gene expression in EPC2-hTERT organoids treated with IL4 or IL13 were validated by qRT-PCR. The relative expression over NT is represented. The data are shown as means ± SD (n = 3). ∗P < .05, ∗∗P < .01, and ∗∗∗∗P < .0001, analysis of variance.
We further validated these findings by assessing gene expression in esophageal organoids treated with IL4 or IL13: the CD73 gene was down-regulated by cytokine treatment whereas CD104 gene expression was not affected by IL4 or IL13. Of note, similar to in vivo EoE, these organoids show basal zone expansion and diminished differentiation, assessed both morphologically (delayed keratinization in IL4/IL13 organoids) and transcriptionally by changes in expression of IVL and SOX2 genes (Figure 3D).
CD73+CD104+ Cells Represent a Less-Differentiated Basaloid Cell Population
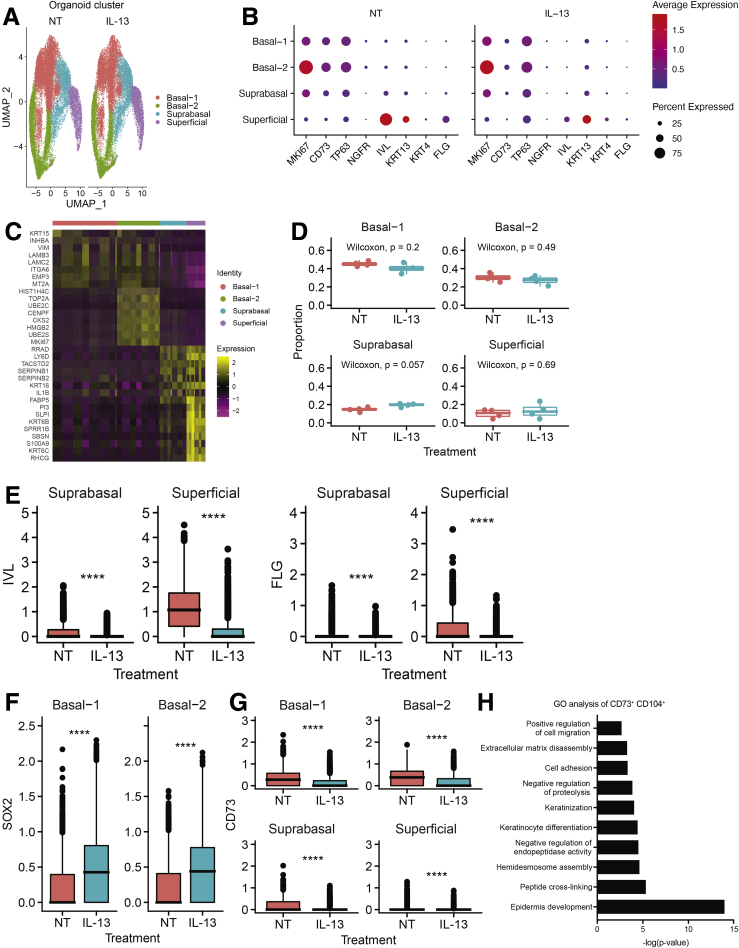
To investigate the molecular heterogeneity of human esophageal organoids, we performed single-cell RNA sequencing on esophageal PDOs from 4 non-EoE control patients. Seurat’s (Version 4.0) unsupervised dimensionality reduction and clustering along with uniform manifold approximation and projection visualization showed 4 distinct cell clusters in esophageal PDOs cultures with or without IL13 (Figure 4A), and the expression of population marker genes and CD73 in each cell cluster are shown (Figure 4B). The cell clusters then were categorized as basal-1, basal-2, suprabasal, and superficial based on the expression of epithelial marker genes MKI67, TP63, NGFR, IVL, KRT13, KRT4, and FLG. For example, basal-1 and basal-2 clusters were defined by moderate expression of basal cell marker gene TP63 in more than 50% of cells and lack of expression of differentiation marker genes IVL, KRT13, KRT4, and FLG, with the distinguishing feature being high expression of proliferation marker gene MKI67 in more than 75% of cells in basal-2 cluster, with only moderate expression of MKI67 in more than 50% of cells in the basal-1 cluster (Figure 4B). Expression patterns of the top 8 differentially expressed genes (DEGs) in each cluster in untreated PDOs are presented in Figure 4C.
Figure 4.
Single-cell RNA sequencing analysis of PDOs from 4 non-EoE patients. PDOs from 4 non-EoE patients were treated with IL13 (10 ng/mL) or vehicle (phosphate-buffered saline) from day 7 to day 11, and harvested and dissociated for single-cell RNA sequencing analysis on day 11. (A) Seurat’s Uniform Manifold Approximation and Projection (UMAP) visualization of 4 classified cell groups, basal-1, basal-2, suprabasal, and superficial, within PDOs from non-EoE controls grown with or without IL13 (10 ng/mL). (B) Average normalized gene expression values of MKI67, CD73, basal markers (TP63, NGFR), and differentiated markers (IVL, KRT13, FLG) in each cell cluster. Circle size reflects the percentage of cells with a nonzero expression level for the indicated genes. Color intensity reflects the average expression level across all cells within each cluster. (C) Expression z-scores for the top 8 up-regulated genes in each cluster. (D) The proportions of each cell cluster were compared in PDOs (n = 4) with IL13 or vehicle. (E–G) The normalized expression of (E) IVL and FLG, (F) SOX2, and (G) CD73 in nontreated (NT) PDOs was compared with that in PDOs treated with IL13 for each cluster. ∗∗∗∗P < .0001, Wilcoxon signed-rank test. (H) GO analysis of differentially expressed gene profiles of CD73+CD104+ cells, compared with CD73-CD104+ cells in NT PDOs.
Interestingly, although we did not observe significant differences in the abundance of each cluster in PDOs in response to IL13 treatment (Figure 4D), we found significantly decreased expression of differentiation marker genes IVL and FLG in suprabasal and superficial clusters (Figure 4E). The SOX2 gene was up-regulated in basal-1 and basal-2 clusters in response to IL13 (Figure 4F).
We found CD73 expression, as well as other basal cell markers, up-regulated in basal-1 and basal-2 clusters (Figure 4B). The expression of CD73 in nontreated PDOs was compared with that in PDOs treated with IL13 for each cluster (Figure 4G). We found CD73 expression was down-regulated in PDOs treated with IL13. We then compared gene expression profiles in the population of cells expressing both CD73 and CD104 messenger RNA (mRNA) (CD73+CD104+) with the population expressing only CD104 (CD73-CD104+). We subjected the list of differentially expressed genes from each population to Gene Ontology (GO) analysis, which showed that pathways related to squamous cell differentiation were differentially regulated (Figure 4H).
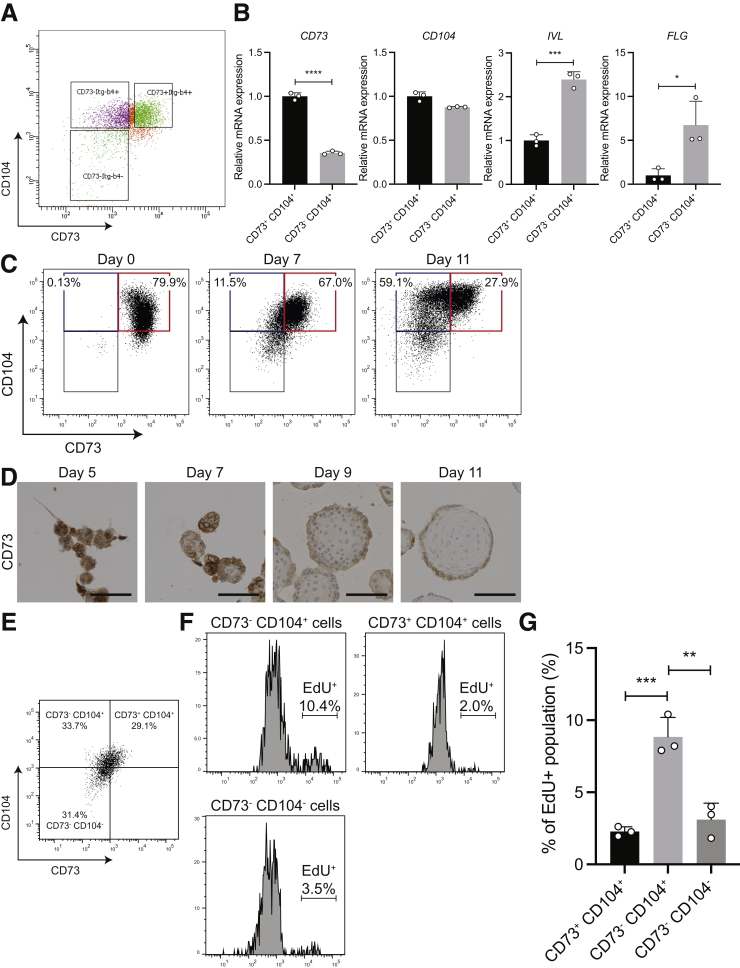
To validate the findings from GO analysis, we sorted 3 populations (CD73+CD104+, CD73-CD104+, and CD73-CD104-) from the EPC2-hTERT organoids with fluorescence-activated cell sorting (FACS), followed by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis for markers of squamous cell differentiation (Figure 5A and B). CD73+CD104+ cells were overall less differentiated, with decreased expression of IVL and FLG. We confirmed this by evaluating CD73 protein expression over 11 days in organoid growth. We found that as cells differentiated into mature organoids, CD73 expression decreased (Figure 5C and D). Thus, although both CD73+CD104+ and CD73-CD104+ cells are basal in nature, CD73-CD104+ represent a more differentiated population.
Figure 5.
CD73+CD104+cells represent a less-differentiated and slow-cycling cell population within the basal zone. (A and B) EPC2-hTERT organoids were harvested on day 11 and dissociated for FACS. (A) The cells in EPC2-hTERT organoids were sorted based on CD73 and CD104 expression. (B) CD73, CD104, IVL, and FLG relative gene expression in CD73+CD104+ and CD73+CD104- populations was validated by qRT-PCR. (C) EPC2-hTERT cells in 2-dimensional culture (organoid day 0) and EPC2-hTERT organoid (day 7 and day 11) were harvested for flow cytometry and the representative flow cytometry plots for CD73 and CD104 expression are shown. (D) Representative IHC images for CD73 staining in EPC2-hTERT organoids harvested on day 5, day 7, day 9, and day 11. Scale bars: 100 μm. (E–G) EPC2-hTERT organoids were harvested on day 11 after incubation in culture medium with EdU for 2 hours, and dissociated for flow cytometry. (E) A representative FACS dot plot of cells from EPC2-hTERT organoids subjected to EdU, with the frequencies of each population according to CD73 and CD104 expression, is shown. (F) Representative histograms of EdU staining in each cell population. The percentage of EdU+ cells in each population is shown. (G) Percentages of EdU+ cells in each population. All data are shown as means ± SD (n = 3). A 2-tailed Student t test or analysis of variance were performed for comparing 2 or multiple comparisons, respectively. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
To characterize the proliferative capacity of the CD73+CD104+ basal zone cells in human esophageal epithelium, we investigated the cell-cycle profile in each of the 3 populations defined earlier by using the Click-it ethynyldeoxyuridine (EdU) assay Thermo Fisher Scientific (Watham, MA). EPC2-hTERT organoids were harvested after incubation in culture medium with EdU and dissociated into single cells, and evaluated for EdU incorporation by flow cytometry. We found a greater percentage of cells with EdU incorporation (10.4%) in CD73-CD104+ cells, compared with CD73+CD104+ cells (2%; P < .01) (Figure 5E–G). This indicates a higher proliferative capacity of CD73-CD104+ cells in human esophageal epithelium.
CD73+ Cells Mediate Epithelial Renewal in Human Esophageal Epithelium
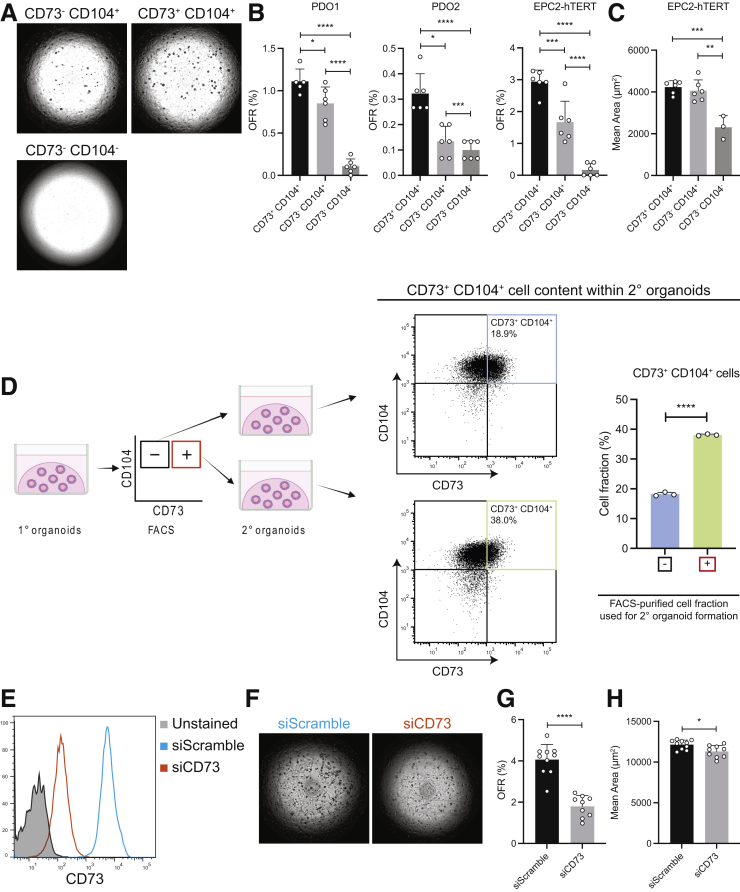
We then sought to determine the role of CD73 in epithelial renewal in the esophageal epithelium. First, we fluorescence-activated cell–sorted single-cell suspensions from patient-derived and immortalized esophageal organoids into 3 populations (CD73+CD104+, CD73-CD104+, and CD73-CD104-) and compared organoid formation rate (OFR) among the 3 groups (Figure 6A). CD73+CD104+ cells had the highest capacity to generate organoids, followed by CD73-CD104+ cells and CD73-CD104- cells, respectively (Figure 6B). Despite the differences in OFR between the CD73+CD104+ and CD73-CD104+ cells, organoid size did not differ between these populations. However, CD73-CD104- cells formed significantly smaller organoids compared with organoids from CD73-CD104+ and CD73+CD104+ cell populations (Figure 6C). To determine if there are any long-lasting differences in cell-type frequencies within organoids established from CD73+ or CD73- cells, we have analyzed the cells from these organoids by flow cytometry. The organoids established from CD73-CD104+ cells had significantly smaller fractions of CD73+CD104+ cells compared with the organoids established from the CD73+CD104+ cells (Figure 6D).
Figure 6.
CD73+cells mediate epithelial renewal in human esophageal epithelium. (A–D) PDOs or EPC2-hTERT organoids were harvested on day 11 (1° organoids) and dissociated for FACS. The cells were sorted based on CD73 and CD104 expression and resuspended in Matrigel for organoid establishment and growth. OFR and organoid size (mean area) of 2° organoids were (B and C) quantified on a Celigo image cytometer and (D) harvested on day 11 and again dissociated for flow cytometry. (A) Representative phase-contrast images of organoids formed by corresponding cell populations. (B) OFR and (C) mean size were compared among organoids from the 3 populations. (D) The scheme of this experiment and the representative flow cytometry plots of 2° organoids are shown. The percentage of CD73+ cells in EPC2-hTERT were compared. The data are shown as means ± SD (n = 3). (E–H) EPC2-hTERT cells were treated for 48 hours with CD73 siRNA or nonsilencing (scramble) siRNA in 2D culture, followed by seeding into organoid cultures. (E) CD73 silencing efficiency was evaluated by flow cytometry. (F) Representative phase-contrast images of organoids formed by corresponding cells. (G) OFR and (H) mean size of organoids from EPC2-hTERT cells transfected with CD73 or scramble siRNA. The data are shown as means ± SD (n = 10). Two-tailed Student t test or analysis of variance were performed for single or multiple comparisons, respectively. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, and ∗∗∗∗P < .0001.
We further assessed the functional consequences of CD73 knockdown in human esophageal epithelial cells by RNA interference. After validation of silencing efficiency by flow cytometry (Figure 6E), small interfering RNA (siRNA)-transfected cells were seeded in Matrigel (Corning, Tewksbury, MA) to form organoids. CD73 knockdown significantly decreased the OFR and organoid size compared with control cells transfected with scrambled nonsilencing siRNA (Figure 6F and G).
STAT6 Inhibition Prevents Th2-Mediated Depletion of CD73+CD104+ Population
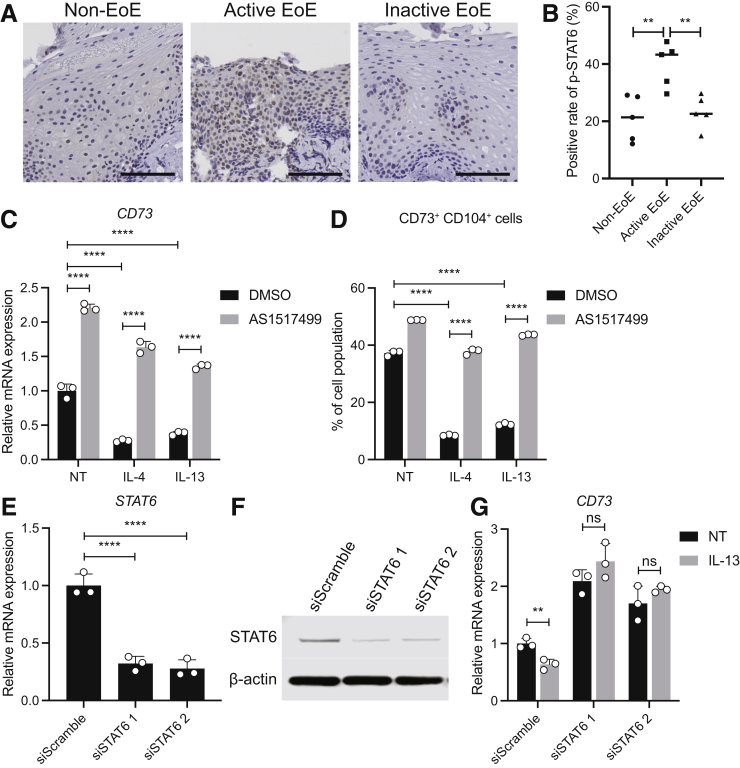
Upon binding of IL4 and IL13 to the IL4 receptor, STAT6 is phosphorylated and translocates to the nucleus to induce downstream inflammation characteristic of EoE.25 We hypothesized that STAT6 inhibition may prevent depletion of CD73+ cells in esophageal epithelium. To assess STAT6 activity in the esophageal epithelium, we performed IHC staining for phosphorylated STAT6 with esophageal biopsy specimens from active or inactive EoE patients, as well as non-EoE controls. We found that STAT6 became phosphorylated and translocated to the nucleus in the esophageal epithelium in the active EoE specimens (Figure 7A and B).
Figure 7.
The therapeutic potential of STAT6 inhibition in EoE. (A) IHC staining for phosphorylated STAT6 was performed, with representative images shown. Scale bars: 100 μm. (B) The positive rate of phosphorylated STAT6 in esophageal epithelium are compared. The data are shown as means ± SD (n = 5). (C) Relative expression of CD73 gene in EPC2-hTERT organoids treated with STAT6 inhibitor (AS151749, 400 nmol/L), IL4 (10 ng/mL), or IL13 (10 ng/mL). (D) The frequencies of CD73+CD104+ cells in organoids treated with AS151749 or dimethyl sulfoxide (DMSO) under the presence of Th2 cytokines. STAT6 silencing efficiency was evaluated by (E) qRT-PCR and (F) Western blot. (G) CD73 gene expression in EPC2-hTERT organoids treated with STAT6 siRNA or nonsilencing (scramble) siRNA with or without IL13. The data are shown as means ± SD (n = 3). Analysis of variance was performed with multiple comparisons. ∗∗P < .01, ∗∗∗∗P < .0001.
To evaluate the impact of STAT6 inhibition on the CD73+CD104+ population, we treated esophageal organoids with AS1517499, a small-molecule STAT6 inhibitor,25 in addition to stimulation with IL4 or IL13. Indeed, we found that AS1517499 prevented cytokine-induced inhibition of CD73 expression by qRT-PCR (Figure 7C). Similarly, AS1517499 treatment rescued the Th2 cytokine-mediated decrease in the percentage of CD73+CD104+ cells by flow cytometry (Figure 7D). Inhibition of STAT6 lead to a 2-fold increase in CD73 in the untreated state.
To confirm that the effect of AS1517499 treatment is dependent on inhibition of STAT6, we transduced EPC2-hTERT cells grown in high-calcium medium (differentiation conditions) with siRNA targeting STAT6, followed by IL4 or IL13 treatment. Down-regulation of STAT6 by siRNA was confirmed at the mRNA and protein levels (Figure 7E and F). Importantly, STAT6 knockdown rescued the cytokine-mediated decrease in CD73 mRNA (Figure 7G). These data confirm that STAT6 mediates the Th2 cytokine-induced decrease in the CD73+CD104+ cell population in esophageal epithelium. Furthermore, the fact that STAT6 inhibition in the untreated state increased CD73 expression may suggest that STAT6 regulates the expression of CD73 not only in the setting of inflammation but also in homeostasis.
Omeprazole Treatment Prevents Th2 Cytokine-Mediated Depletion of the CD73+ Population
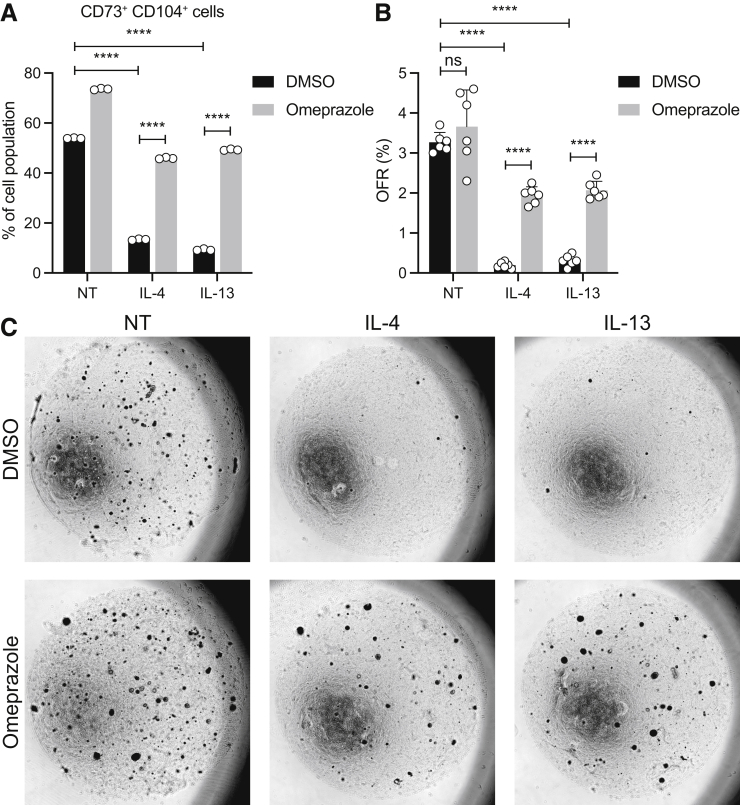
Proton pump inhibitors (PPIs) are used frequently as a first-line therapy in EoE, and, therefore, we sought to determine if PPIs can prevent Th2 cytokine-induced depletion of the self-renewing CD73+CD104+ population in EoE. Indeed, we found that omeprazole treatment reversed the effects of IL4 and IL13 on the CD73+CD104+ population (Figure 8A). To investigate the impact of omeprazole on self-renewal capacity of esophageal keratinocytes in the EoE milieu, we treated EPC2-hTERT organoids with omeprazole, followed by dissociation to single cells and seeding in Matrigel for organoid formation. These organoids then were treated with IL4 or IL13, and the OFR was assessed. Strikingly, pretreatment with omeprazole prevented a cytokine-induced decrease in OFR (Figure 8B and C). These data indicate that PPI therapy may act in EoE in part through restoration of epithelial cell renewal, in addition to its described role in modulation of secretion of gastric acid and eotaxin-3 from the epithelium.
Figure 8.
Omeprazole treatment prevents Th2-mediated depletion of CD73+CD104+population. (A) EPC2-hTERT organoids were treated with omeprazole (50 μmol/L), IL4 (10 ng/mL), or IL13 (10 ng/mL). The frequencies of CD73+CD104+ cells in organoids are shown as means ± SD (n = 3). (B and C) The treated EPC2-hTERT organoids were harvested on day 11 and dissociated into single cells, followed by seeding in Matrigel to assess organoid formation capacity. (B) OFR was evaluated manually on day 11. (C) Representative phase-contrast images of organoids from corresponding groups. The data are shown as means ± SD (n = 6). Analysis of variance was performed with multiple comparisons. ∗∗∗∗P < .0001. DMSO, dimethyl sulfoxide.
Discussion
Damage to the epithelium from the inflammation in EoE drives symptomatology, as well as progression of esophageal fibrostenosis,2,11,26 leading to food impactions and stricture. Moreover, PPI, steroids, or diet modification are the only treatment options currently available to EoE patients. Understanding how the basal zone cell subpopulations are affected and working to restore homeostasis in EoE would provide an alternative pharmacologic approach.
Herein, we describe unique subpopulations in the basal zone epithelium of the esophagus and the shift that occurs within that basal zone in the setting of allergic inflammation. We have found that in EoE patients there is decreased CD73 expression, and we were able to recapitulate these changes in our MC903/ova-albumin (OVA)-induced mouse model of EoE, as well as primary 3D organoid system with EoE-relevant cytokines. Functionally, the CD73+ population shows renewal capabilities in our system with enhanced organoid formation rate from a single-cell suspension and represents a less-differentiated basaloid population that is slow-cycling. We found that STAT6 mediates decreased CD73 expression in response to Th2 cytokines. Finally, treatment with omeprazole, a PPI used ubiquitously in EoE, prevents decreased CD73 expression and restores the organoid formation rate.
There is an ongoing debate regarding the composition of the basal layer and whether a true slow-cycling stem cell population exists. Pan et al27 showed a 5-iodo-2’-deoxyuridine retainment in the human esophageal epithelium in both histologically normal patients and those with dysplasia. Multiple murine studies have proposed the existence of specialized subpopulations within the basal layer.14 Label-retaining studies have offered a myriad of murine candidates for a stem cell subpopulation among the basal cells in murine esophagus including expression of cytokeratin 15,15 co-expression of CD73 and CD104, and co-expression of a6 integrin and CD71.13,14,27,28 Although the existence of a single esophageal stem progenitor may be controversial, we show that the CD73+CD104+ fraction in human basal epithelium has renewal capabilities, slower proliferation, and are phenotypically less differentiated. The shift of the EoE epithelium from CD73+CD104+ to CD73- CD104+ marks the shift away from a stem-like progenitor toward a more proliferative population.29,30
CD73, or ecto-5’-nucleotidase, is a membrane-bound ectoenzyme responsible for catalyzing the conversion of extracellular adenosine monophosphate into free adenosine. Adenosine acts in a largely anti-inflammatory capacity by inhibiting IL2 production, which in turn reduces CD4+ T-cell activation and proliferation.31,32 It may be that in EoE, decreased levels of CD73 and adenosine lead to unchecked activation of lymphocytes and trigger sustained inflammation. Interestingly, in the lungs, adenosine seems to have proinflammatory/profibrotic effects, especially in activating myofibroblasts and extracellular matrix remodeling.33 In addition, CD73 is known to be increased in various forms of cancer including breast, pancreas, and lung, and treatment with anti-CD73 antibodies slows tumor growth in vitro and in vivo.33, 34, 35, 36 It has been postulated that increased adenosine within the tumor microenvironment assists the tumor in evading the local immune cells and allows for malignant growth. Thus, there is likely an organ, context, and cell type–specific role for CD73 and adenosine. In the context of EoE, despite the fact that there is chronic inflammation and increased cell turn over, there are paradoxically low rates of malignant transformation.37, 38, 39 Depletion of CD73/adenosine may represent an unexplored mechanism by which allergic inflammation is protective against malignancy in the esophagus. Future work will delve into the functional significance of CD73 in the esophageal epithelium and determine if its role in renewal is based on enzymatic activity.
Reports have shown a protective role of CD73 in the lung by pharmacologically restoring CD73 function in knock-out mice in acute lung injury models.40 Paradoxically, although there is an expansion of the basal epithelium in EoE, there are fewer CD73+ cells with renewal capabilities. Other studies have shown that Th2 cytokines increase proliferation in the esophageal epithelium in EoE,8,29 with the total number of Ki-67+ cells increasing. Although the total number of Ki-67+ cells is increased overall, the fraction of basal cells that are Ki-67+ remains unchanged in EoE esophagus (Figure 1A and B). This suggests a cycle of ongoing insult in which the esophagus is not able to re-establish its homeostatic gradient of renewal, proliferation, and differentiation, resulting in barrier disruption and continued bombardment with antigens. Restoration of the epithelial homeostasis offers a new direction in the development of EoE therapeutics. We found that STAT6 activation mediates decreased CD73 expression in response to Th2 cytokines IL4 and IL13. We have not yet defined a molecular mechanism for this regulation, but experiments are ongoing to show the responsible components and may provide insight into the epithelial healing that has been shown to occur when inhibiting this pathway with biologics currently undergoing clinical trials.
A phenomenon exists whereby a subset of EoE patients respond to PPIs alone without the need for topical steroids. We have shown that PPIs can reverse the effects of IL4 and IL13 on the CD73+ population. Previous work has shown that PPIs inhibit eotaxin-3 expression, thus reducing the inflammatory infiltrate.41,42 We now show another mechanism, intrinsic to the epithelium, through which PPIs may act to induce healing by re-establishing homeostasis. It may also be inhibition of CD73 transcription, epigenetic regulation, or post-transcriptional protein modification. The mechanism by which PPIs maintain the CD73+CD104+ population in the esophageal epithelium in Th2 inflammation also needs to be evaluated further. Rochman et al30 recently compared the EoE transcriptome with that of esophageal epithelial cells treated with PPI. They found that with PPI treatment, 70 genes were differentially expressed compared with the EoE transcriptome, with GO analysis enriched for biological processes involved in cell-cycle and microtubule organization. These data support our focus on the progenitor and proliferative populations in EoE and suggest a mechanism still at large.
In this work we rely heavily on patient-derived organoids as a model of human esophageal epithelium, and although it is a powerful scientific tool, it is important to recognize its limitations. By design, esophageal organoids are composed of only epithelial cells, which prevents us from evaluating cell–cell interactions with other cell types such as immune cells, fibroblasts, or endothelium. We have mimicked the EoE milieu by treating the organoids with IL13 and IL14, the 2 cytokines known to be involved in EoE pathogenesis.5,43,44 However, in follow-up studies to evaluate the effect of CD73+ cells on other cell types, we would use co-culture approaches, adding on cell types such as eosinophils or fibroblasts, to dissect these interactions.
In our clinical studies looking at CD73/CD104 expression in the EoE esophagus, the inactive patients show expression of CD73/CD104 that is between the controls and the active patients. This may be owing to the fact that the current cut-off value for active EoE is 15 eos/hpf, with inactive patients having anywhere from 0 to 14 eos/hpf. This likely represents a wide margin of disease activity within the inactive population. Alternatively, it may show an inherent difference in epithelial renewal in the EoE compared with non-EoE. In our recent report,3 25% of inactive patients have endoscopic findings, 29.1% had basal zone hyperplasia, and 73.6% had spongiosis, suggesting that there are ongoing and significant epithelial changes while under the current definition of fewer than 15 eos/hpf. There has been some movement in the field to consider pushing down the eosinophil count in this population, and recent clinical trials45,46 are using fewer than 6 eos/hpf in their analysis to account for this.
The biological significance of basal cell heterogeneity is unknown in EoE. This study sheds some light on heterogeneity within the basal zone population of the esophagus and the perturbations that occur within that population during allergic inflammation. We provide a mechanism for how the EoE milieu mediates the changes in basal cell compartment, as well as show strategies for therapeutic targeting of this mechanism in EoE.
Methods
Human Subjects and Endoscopic Esophageal Biopsy Specimens
In accordance with Institutional Review Board standards and guidelines at The Children's Hospital of Philadelphia, esophageal biopsy specimens were collected from patients undergoing esophagogastroduodenoscopy (EGD) as part of routine care, and informed consent was obtained from all patients before EGD. Subjects were classified according to international consensus diagnostic criteria for EoE.47 Subjects who met the clinical criteria of EoE with the histologic presence of 15 or more eos/hpf were classified as active EoE. EoE subjects who showed resolution of histologic inflammation and eosinophilia documented on previous endoscopy were designated as inactive EoE (<15 eos/hpf). Non-EoE subjects had no previous diagnosis of EoE and reported symptoms warranting EGD but showed no histopathologic abnormalities. Subjects with a history of inflammatory bowel disease, celiac disease, gastrointestinal bleeding, or any other acute or chronic intestinal disorders were excluded as described previously.
Primary Esophageal Epithelial Cultures and 3-Dimensional Organoids
The immortalized normal human esophageal epithelial cell line EPC2-hTERT48 was grown in keratinocyte-serum free media (Life Technologies, Carlsbad, CA) containing 0.09 mmol/L Ca2+.49,50 As previously described,18,50 we cultured 3D organoids from human esophageal biopsy specimens (PDOs) or EPC2-hTERT for 11 days. In brief, single cells were suspended in Matrigel (Corning, Tewksbery, MA) and modified keratinocyte-serum free media containing 0.6 mmol/L Ca2+ was added to support organoid formation. Three-dimensional organoids were grown for 7 days before treatment with IL4 (10 ng/mL) or IL13 (10 ng/mL) for 96 hours. To evaluate the therapeutic effect of STAT6 inhibitor (AS1517499; Axon Medchem) or the PPI omeprazole (Sigma-Aldrich), we pretreated with AS1517499 (400 nmol/L) 24 hours before the treatment of cytokines, or with omeprazole (50 μmol/L) for 2 hours before the treatment of cytokines. Of note, STAT6 inhibition with AS1517499 has been shown to cause successful down-regulation of the STAT6 signaling pathway, inhibiting canonical downstream targets such C-C Motif Chemokine Ligand 26 in response to IL13.25
Passaged organoids after treatment were assessed OFR, defined as the number of organoids/total cells seeded and organoid size (mean area) by the Celigo Imaging Cytometer (Nexcelom Bioscience, Lawrence, MA) to validate the self-renewal activities of putative stem cells in the treated cells.
Histology, Immunofluorescence, and Immunohistochemistry
Paraffin-embedded biopsy specimens and 3D organoid products were serially sectioned and subjected to H&E staining or IHC as described previously.12,18 In brief, sections were incubated overnight at 4°C with anti-CD73 antibody (rabbit monoclonal, ab133582, 1:100; Abcam, Cambridge, UK) and anti–Itg-β4 (mouse monoclonal, MAB4060, 1:100; R&D Systems, Minneapolis, MN) for immunofluorescence, and anti–Ki-67 (rabbit monoclonal, ab16667, 1:200; Abcam), anti-SOX2 (rabbit monoclonal, 14962S, 1:300; Cell Signaling Technology, Danvers, MA), or anti-STAT6 (phospho Y641, rabbit polyclonal, ab28829, 1:50; Abcam) for IHC. IHC of 3D organoids for CD73 and CD104 was performed by BOND RXm (Leica Biosystems, Nusslock, Germany) with anti-CD73 antibody (rabbit polyclonal, ab175396, 1:200; Abcam) or anti-CD104 monoclonal antibody (NBP2-37392, 1:1000, Novus Biologicals, Englewood, CA). Imaging was performed with the BZ-X710 Fluorescence Microscope (Keyence, Osaka, Japan).
Immunoblotting
Cells were lysed and subjected to immunoblotting, as previously described.12 The anti-STAT6 antibody (rabbit polyclonal, 9362S, 1:1000; Cell Signaling Technology) and the anti–β-actin antibody (mouse monoclonal, A5316, 1:5000; Sigma-Aldrich) were used as primary antibodies. β-actin served as a loading control.
Murine Model of EoE
Female BALB/c mice (Jackson Laboratories, Bar Harbor, ME) aged 8–10 weeks, were procured in accordance with the Institutional Animal Care and Use Committee of the University of Pennsylvania. Epicutaneous sensitization of mice was performed as previously described with modification.23 Briefly, mice were treated daily with 2 nmol MC903 (Tocris Bioscience, Bristol, UK) in 20 uL of 100% EtOH on ears in the presence of 1000 ug OVA (A5503-50G; Sigma-Aldrich) for 12 days. Mice were challenged intragastrically with 50 mg OVA on days 15 and 17. Upon first intragastric OVA challenge, mice were continuously provided with water containing 15 g/L OVA. All mice were killed on day 18, and the esophagi were resected and dissociated enzymatically for flow cytometry.
Single-cell RNA Sequencing
Organoids from 4 non-EoE control patients were grown for 7 days, followed by treatment with IL13 (10 ng/mL) or vehicle (phosphate-buffered saline) for 4 days. Organoids were harvested and dissociated enzymatically and mechanically into single cells on day 11. We used the Dead Cell Removal Kit (Miltenyi Biotec, Auburn, CA) to ensure viability. Single-cell RNA matrices from all samples were processed using Seurat (v4.0.0) on R. To remove dead or dying cells from the matrices, cells with more than 15% of their transcripts mapping to mitochondrial genes were excluded from further analyses. Each of the matrices then was log1p normalized and the variable features were calculated. To analyze shared cell states between all samples, the Seurat integration protocol was performed for dimensionality reduction and clustering: Briefly, the variable features (genes) in each matrix were used to find anchors of shared transcriptional states between each and all of the matrices using canonical correlation analysis. Principal component analysis (PCA) then was used on the integrated matrices as initial dimensionality reduction, and uniform manifold approximation and projection was used on the first 20 PCs from the PCA for visualization. Finally, clusters of cells were discovered using the shared nearest-neighbor graph constructed from the first 20 PCs of the PCA. Cell clusters were classified as basal-1 (low proliferation), basal-2 (high proliferation), suprabasal, or superficial based on the expression of markers MKI67, TP63, NGFR, IVL, KRT13, KRT4, and FLG. DEGs were discovered using Seurat's FindMarkers function while implementing the use of model-based analysis of single-cell transcriptomics, a Generalized Linear Models-based framework for DEG testing that uses transcript detection rate as a covariate.
Flow Cytometry and FACS
Flow cytometry and FACS were performed as described previously.18,50 In brief, cells were trypsinized and resuspended in FACS buffer (phosphate-buffered saline supplemented with 1% bovine serum albumin). 4’,6-Diamidino-2-phenylindole (DAPI) was used to eliminate cellular debris and nonviable cells present in the sample. Allophycocyanin-H7–anti-CD45 (560178, 1:20; BD Biosciences, Franklin Lakes, NJ) was used to detect and remove the population of leukocytes in biopsy specimens. Human epithelial cells were stained with Allophycocyanin–anti-CD73 (560847, 1:20; BD Biosciences) and Phycoerythrin–anti-CD104 (327808, 1:20; BioLegend, San Diego, CA). Murine cells were stained with BUV395–anti-CD45 (564279, 1:500; BD Biosciences), Phycoerythrin–Cy7–anti-CD73 (127224, 1:160; BioLegend), and Alexa Fluor 647–anti-CD104 (123608, 1:100; BioLegend). The top value of the unstained control was defined as the cut-off value of CD73 and CD104. LSRII (BD Biosciences) and FlowJo (TreeStar, Ashland, OR) were used for flow cytometry. Both the FACSAria Fusion Sorter (BD Biosciences) and the MoFlo Astrios Cell Sorter (Beckman Coulter, Brea, CA) were used for cell sorting according to the expression of CD73 and CD104.
Cell-Cycle (DNA Content) Analysis
EPC2-hTERT organoids were grown for 11 days and exposed to EdU for 2 hours before they were harvested. After incubation for 2 hours, organoids then were dissociated, and single cells were suspended in FACS buffer. The LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (L34975; Invitrogen, Waltham, MA) was used to determine cell viability. The Click-it Plus Alexa Fluor 488 Flow Cytometry Assay Kit (C10632; Invitrogen) was used to assess cell-cycle kinetics, following the manufacturer’s instructions. The intracellular DNA content was assessed by DAPI staining. Additional staining of cell-surface antigens for some experiments was performed before the fixing of cells. The population of cells in each cell-cycle phase was identified according to cell-cycle kinetics (EdU) and DNA content (DAPI). Cell-cycle assay was performed by LSRII and analyzed by FlowJo.
qRT-PCR Assays
RNA was isolated from esophageal 3D organoids and complementary DNA synthesis was performed as described previously.18 qRT-PCR was performed using TaqMan Gene Expression Assays (Thermo Fisher Scientific, Waltham, MA) for NT5E (CD73) (Hs00159686), ITGB4 (CD104) (Hs00173995), SOX2 (Hs01053049), IVL (Hs00846307), and GAPDH (Hs02786624), using the StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA). The relative level of each mRNA was normalized to GAPDH as an internal control gene.
RNA Interference
We performed RNA interference with siRNA directed against CD73 (sc-42862; Santa Cruz Biotechnology, Dallas, TX) or STAT6 (Invitrogen), each consisting of 3 target-specific sequences. We used siRNA-A (sc-37007; Santa Cruz Biotechnology; or Silencer Select Negative Control No. 1; Invitrogen) as nonsilencing control siRNA. EPC2-hTERT cells grown in a monolayer were incubated with siRNA (10 nmol/L) for 48 hours using the Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer’s instructions. EPC2-hTERT cells were harvested 48 hours after transfection, and the silencing efficiency was assessed by flow cytometry or qRT-PCR assay. In addition, RNA interference of STAT6 has been shown to cause successful down-regulation of C-C Motif Chemokine Ligand 26 in response to IL13.51,52
Statistical Analysis
Data are shown as means ± SD, and continuous variables were analyzed by a 2-tailed Student t test for comparing 2 groups or 1-way analysis of variance and multiple comparisons for comparing more than 2 groups. Correlation analyses were estimated by the Pearson rank correlation coefficient. P values less than .05 were considered to indicate significance. Statistical analyses were performed using GraphPad Prism version 8 (GraphPad Software, Inc, San Diego, CA). All authors had access to the study data and reviewed and approved the final manuscript.
Acknowledgments
The authors thank Cameron Sabet and Gloria Soto for help in making the schematics, and the Molecular Pathology and Imaging Core (K. Bennett, L. Son, R. Ly, J. Katz) for technical support. Schematics were made with BioRender (Toronto, ON).
CRediT Authorship Contributions
Takeo Hara, MD, PhD (Conceptualization: Lead; Formal analysis: Lead; Investigation: Lead; Writing –original draft: Lead)
Yuta Kasagi, MD, PhD (Conceptualization: Supporting; Investigation: Supporting)
Joshua Wang, MS (Data curation: Supporting; Formal analysis: Supporting;Investigation: Supporting)
Masaru Sasaki, MD PhD (Data curation: Equal)
Bailey Aaron, BS (Data curation: Supporting; Formal analysis: Supporting;Investigation: Supporting)
Adam Karami, MS (Formal analysis: Lead)
Masataka Shimonosono, MD, PhD (Formal analysis: Supporting; Investigation:Supporting)
Rieko Shimonosono, MD (Data curation: Supporting)
Hisatsugu Maekawa, MD, PhD (Investigation: Supporting)
Lauren Dolinsky, BA (Data curation: Lead)
Benjamin Wilkins, MD, PhD (Formal analysis: Lead)
Jeremy Klein, BA (Data curation: Supporting)
Jane Wei, BA (Visualization: Equal)
Kathryn Nunes, BS (Data curation: Supporting)
Kristle Lynch, MD (Data curation: Supporting; Formal analysis: Supporting)
Jonathan M. Spergel, MD, PhD (Conceptualization: Supporting; Formal analysis: Supporting; Funding acquisition: Equal)
Kathryn E. Hamilton, PhD (Funding acquisition: Supporting)
Melanie A. Ruffner, MD, PhD (Data curation: Supporting; Formal analysis: Supporting)
Tatiana A. Karakasheva, PhD (Writing – original draft: Supporting; Writing – review & editing: Supporting)
Kelly A. Whelan, PhD (Funding acquisition: Supporting; Writing – original draft: Supporting)
Hiroshi Nakagawa, MD, PhD (Conceptualization: Lead; Funding acquisition: Equal;Writing – original draft: Equal; Writing – review & editing: Lead)
Amanda Muir, MD (Conceptualization: Lead; Funding acquisition: Lead; Investigation:Lead; Methodology: Lead; Supervision: Lead; Writing – original draft: Lead; Writing –review & editing: Lead)
Footnotes
Conflicts of interest Amanda Muir has recieved research funding from Allakos and Morphic. The remaining authors disclose no conflicts.
Funding This study was supported by National Institutes of Health grants P01CA098101, U54CA163004, R01DK114436, and R01AA026297 (H.N.), P30CA013696 (Columbia University Herbert Irving Comprehensive Cancer Center Shared Resources), and K01DK103953 and R01DK121159 (K.A.W.); University of Pennsylvania Transdisciplinary Awards Program in Translational Medicine and Therapeutics (A.B.M.); National Institute of Health R01 DK124369-01 (K.E.H.), K08AI148456 (M.A.R.), K08DK106444, R03DK118310, and R01DK124266-01 (A.B.M.); and the Children’s Hospital of Philadelphia Gastrointestinal Epithelial Modeling Program (A.B.M., T.A.K., and K.E.H.). CEGIR (U54 AI117804) (A.B.M., J.M.S., and M.A.R.) is part of the Rare Disease Clinical Research Network, an initiative of the Office of Rare Diseases Research, National Center for Advancing Translational Sciences, and is funded through collaboration between the National Institute of Allergy and Infectious Diseases, National Institute of Diabetes and Digestive and Kidney Diseases, and National Center for Advancing Translational Sciences. Consortium of Eosinophilic Gastrointestinal Disease Researchers also is supported by patient advocacy groups including the American Partnership for Eosinophilic Disorders, Campaign Urging Research for Eosinophilic Diseases, and Eosinophilic Family Coalition.
References
- 1.Katzka D.A., Ravi K., Geno D.M., Smyrk T.C., Iyer P.G., Alexander J.A., Mabary J.E., Camilleri M., Vaezi M.F. Endoscopic mucosal impedance measurements correlate with eosinophilia and dilation of intercellular spaces in patients with eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:1242–1248.e1. doi: 10.1016/j.cgh.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 2.Muir A., Falk G.W. Eosinophilic esophagitis: a review. JAMA. 2021;326:1310–1318. doi: 10.1001/jama.2021.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whelan K.A., Godwin B.C., Wilkins B., Elci O.U., Benitez A., DeMarshall M., Sharma M., Gross J., Klein-Szanto A.J., Liacouras C.A., Dellon E.S., Spergel J.M., Falk G.W., Muir A.B., Nakagawa H. Persistent Basal Cell Hyperplasia Is Associated With Clinical and Endoscopic Findings in Patients With Histologically Inactive Eosinophilic Esophagitis. Clinical Gastroenterology and Hepatology. 2020;18:1475–1482.e1. doi: 10.1016/j.cgh.2019.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang M., Ku W.Y., Zhou Z., Dellon E.S., Falk G.W., Nakagawa H., Wang M.L., Liu K., Wang J., Katzka D.A., Peters J.H., Lan X., Que J. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 2015;125:1557–1568. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Omori-Miyake M., Yamashita M., Tsunemi Y., Kawashima M., Yagi J. In vitro assessment of IL-4- or IL-13-mediated changes in the structural components of keratinocytes in mice and humans. J Invest Dermatol. 2014;134:1342–1350. doi: 10.1038/jid.2013.503. [DOI] [PubMed] [Google Scholar]
- 6.Sherrill J.D., Kc K., Wu D., Djukic Z., Caldwell J.M., Stucke E.M., Kemme K.A., Costello M.S., Mingler M.K., Blanchard C., Collins M.H., Abonia J.P., Putnam P.E., Dellon E.S., Orlando R.C., Hogan S.P., Rothenberg M.E. Desmoglein-1 regulates esophageal epithelial barrier function and immune responses in eosinophilic esophagitis. Mucosal Immunol. 2014;7:718–729. doi: 10.1038/mi.2013.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masterson J.C., Biette K.A., Hammer J.A., Nguyen N., Capocelli K.E., Saeedi B.J., Harris R.F., Fernando S.D., Hosford L.B., Kelly C.J., Campbell E.L., Ehrentraut S.F., Ahmed F.N., Nakagawa H., Lee J.J., McNamee E.N., Glover L.E., Colgan S.P., Furuta G.T. Epithelial HIF-1α/claudin-1 axis regulates barrier dysfunction in eosinophilic esophagitis. J Clin Invest. 2019;129:3224–3235. doi: 10.1172/JCI126744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masterson J.C., McNamee E.N., Hosford L., Capocelli K.E., Ruybal J., Fillon S.A., Doyle A.D., Eltzschig H.K., Rustgi A.K., Protheroe C.A., Lee N.A., Lee J.J., Furuta G.T. Local hypersensitivity reaction in transgenic mice with squamous epithelial IL-5 overexpression provides a novel model of eosinophilic oesophagitis. Gut. 2014;63:43–53. doi: 10.1136/gutjnl-2012-303631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollis M., Hindley J. Satellite II DNA of human lymphocytes: tandem repeats of a simple sequence element. Nucleic Acids Res. 1988;16:363. doi: 10.1093/nar/16.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muir A.B., Lim D.M., Benitez A.J., Modayur Chandramouleeswaran P., Lee A.J., Ruchelli E.D., Spergel J.M., Wang M.L. Esophageal epithelial and mesenchymal cross-talk leads to features of epithelial to mesenchymal transition in vitro. Exp Cell Res. 2013;319:850–859. doi: 10.1016/j.yexcr.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muir A.B., Wang J.X., Nakagawa H. Epithelial-stromal crosstalk and fibrosis in eosinophilic esophagitis. J Gastroenterol. 2019;54:10–18. doi: 10.1007/s00535-018-1498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasagi Y., Dods K., Wang J.X., Chandramouleeswaran P.M., Benitez A.J., Gambanga F., Kluger J., Ashorobi T., Gross J., Tobias J.W., Klein-Szanto A.J., Spergel J.M., Cianferoni A., Falk G.W., Whelan K.A., Nakagawa H., Muir A.B. Fibrostenotic eosinophilic esophagitis might reflect epithelial lysyl oxidase induction by fibroblast-derived TNF-α. J Allergy Clin Immunol. 2019;144:171–182. doi: 10.1016/j.jaci.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbera M., Pietro M.D., Walker E., Brierley C., MacRae S., Simons B.D., Jones P.H., Stingl J., Fitzgerald R.C. The human squamous oesophagus has widespread capacity for clonal expansion from cells at diverse stages of differentiation. Gut. 2015;64:11–19. doi: 10.1136/gutjnl-2013-306171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeWard A.D., Cramer J., Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giroux V., Lento A.A., Islam M., Pitarresi J.R., Kharbanda A., Hamilton K.E., Whelan K.A., Long A., Rhoades B., Tang Q., Nakagawa H., Lengner C.J., Bass A.J., Wileyo E.P., Klein-Szanto A.J., Wang T.C., Rustgi A.K. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J Clin Invest. 2017;127:2378–2391. doi: 10.1172/JCI88941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalabis J., Oyama K., Okawa T., Nakagawa H., Michaylira C.Z., Stairs D.B., Figueiredo J.L., Mahmood U., Diehl J.A., Herlyn M., Rustgi A.K. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J Clin Invest. 2008;118:3860–3869. doi: 10.1172/JCI35012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa H., Kasagi Y., Karakasheva T.A., Hara T., Aaron B., Shimonosono M., Kijima T., Giroux V., Bailey D., Wilkins B., Abrams J.A., Falk G.W., Aceves S.S., Spergel J.M., Hamilton K.E., Whelan K.A., Muir A.B. Modeling epithelial homeostasis and reactive epithelial changes in human and murine three-dimensional esophageal organoids. Curr Protoc Stem Cell Biol. 2020;52 doi: 10.1002/cpsc.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasagi Y., Chandramouleeswaran P.M., Whelan K.A., Tanaka K., Giroux V., Sharma M., Wang J., Benitez A.J., DeMarshall M., Tobias J.W., Hamilton K.E., Falk G.W., Spergel J.M., Klein-Szanto A.J., Rustgi A.K., Muir A.B., Nakagawa H. The esophageal organoid system reveals functional interplay between notch and cytokines in reactive epithelial changes. Cell Mol Gastroenterol Hepatol. 2018;5:333–352. doi: 10.1016/j.jcmgh.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Travers J., Rochman M., Caldwell J.M., Besse J.A., Miracle C.E., Rothenberg M.E. IL-33 is induced in undifferentiated, non-dividing esophageal epithelial cells in eosinophilic esophagitis. Sci Rep. 2017;7 doi: 10.1038/s41598-017-17541-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denning K.L., Al-Subu A., Elitsur Y. Immunoreactivity of p53 and Ki-67 for dysplastic changes in children with eosinophilic esophagitis. Pediatr Dev Pathol. 2013;16:331–336. doi: 10.2350/13-03-1306-OA.1. [DOI] [PubMed] [Google Scholar]
- 21.Dunaway S., Rothaus A., Zhang Y., Luisa Kadekaro A., Andl T., Andl C.D. Divide and conquer: two stem cell populations in squamous epithelia, reserves and the active duty forces. Int J Oral Sci. 2019;11:26. doi: 10.1038/s41368-019-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayakawa Y., Nakagawa H., Rustgi A.K., Que J., Wang T.C. Stem cells and origins of cancer in the upper gastrointestinal tract. Cell Stem Cell. 2021;28:1343–1361. doi: 10.1016/j.stem.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noti M., Wojno E.D.T., Kim B.S., Siracusa M.C., Giacomin P.R., Nair M.G., Benitez A.J., Ruymann K.R., Muir A.B., Hill D.A., Chikwava K.R., Moghaddam A.E., Sattentau Q.J., Alex A., Zhou C., Yearley J.H., Menard-Katcher P., Kubo M., Obata-Ninomiya K., Karasuyama H., Comeau M.R., Brown-Whitehorn T., de Waal Malefyt R., Sleiman P.M., Hakonarson H., Cianferoni A., Falk G.W., Wang M.L., Spergel J.M., Artis D. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013;19:1005–1013. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whelan K.A., Merves J.F., Giroux V., Tanaka K., Guo A., Chandramouleeswaran P.M., Benitez A.J., Dods K., Que J., Masterson J.C., Fernando S.D., Godwin B.C., Klein-Szanto A.J., Chikwava K., Ruchelli E.D., Hamilton K.E., Muir A.B., Wang M.L., Furuta G.T., Falk G.W., Spergel J.M., Nakagawa H. Autophagy mediates epithelial cytoprotection in eosinophilic oesophagitis. Gut. 2017;66:1197–1207. doi: 10.1136/gutjnl-2015-310341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng E., Zhang X., Wilson K.S., Wang D.H., Park J.Y., Huo X., Yu C., Zhang Q., Spechler S.J., Souza R.F. JAK-STAT6 pathway inhibitors block eotaxin-3 secretion by epithelial cells and fibroblasts from esophageal eosinophilia patients: promising agents to improve inflammation and prevent fibrosis in EOE. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muir A.B., Dods K., Noah Y., Toltzis S., Chandramouleeswaran P.M., Lee A., Benitez A., Bedenbaugh A., Falk G.W., Wells R.G., Nakagawa H., Wang M.L. Esophageal epithelial cells acquire functional characteristics of activated myofibroblasts after undergoing an epithelial to mesenchymal transition. Exp Cell Res. 2015;330:102–110. doi: 10.1016/j.yexcr.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Q., Nicholson A.M., Barr H., Harrison L.A., Wilson G.D., Burkert J., Jeffery R., Alison M.R., Looijenga L., Lin W.R., McDonald S.A.C., Wright N.A., Harrison R., Peppelenbosch M.P., Jankowski J.A. Identification of lineage-uncommitted, long-lived, label-retaining cells in healthy human esophagus and stomach, and in metaplastic esophagus. Gastroenterology. 2013;144:761–770. doi: 10.1053/j.gastro.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Croagh D., Phillips W.A., Redvers R., Thomas R.J.S., Kaur P. Identification of candidate murine esophageal stem cells using a combination of cell kinetic studies and cell surface markers. Stem Cells. 2007;25:313–318. doi: 10.1634/stemcells.2006-0421. [DOI] [PubMed] [Google Scholar]
- 29.Vanoni S., Zeng C., Marella S., Uddin J., Wu D., Arora K., Ptaschinski C., Que J., Noah T., Waggoner L., Barski A., Kartashov A., Rochman M., Wen T., Martin L., Spence J., Collins M., Mukkada V., Putnam P., Naren A., Chehade M., Rothenberg M.E., Hogan S.P. Identification of anoctamin 1 (ANO1) as a key driver of esophageal epithelial proliferation in eosinophilic esophagitis. J Allergy Clin Immunol. 2020;145:239–254.e2. doi: 10.1016/j.jaci.2019.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rochman M., Xie Y.M., Mack L., Caldwell J.M., Klingler A.M., Osswald G.A., Azouz N.P., Rothenberg M.E. Broad transcriptional response of the human esophageal epithelium to proton pump inhibitors. J Allergy Clin Immunol. 2021;147:1924–1935. doi: 10.1016/j.jaci.2020.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haskó G., Linden J., Cronstein B., Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov. 2008;7:759–770. doi: 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sevigny C.P., Li L., Awad A.S., Huang L., McDuffie M., Linden J., Lobo P.I., Okusa M.D. Activation of adenosine 2A receptors attenuates allograft rejection and alloantigen recognition. J Immunol. 2007;178:4240–4249. doi: 10.4049/jimmunol.178.7.4240. [DOI] [PubMed] [Google Scholar]
- 33.Chunn J.L., Molina J.G., Mi T., Xia Y., Kellems R.E., Blackburn M.R. Adenosine-dependent pulmonary fibrosis in adenosine deaminase-deficient mice. J Immunol. 2005;175:1937–1946. doi: 10.4049/jimmunol.175.3.1937. [DOI] [PubMed] [Google Scholar]
- 34.Novak I., Yu H., Magni L., Deshar G. Purinergic signaling in pancreas—from physiology to therapeutic strategies in pancreatic cancer. Int J Mol Sci. 2020;21:1–21. doi: 10.3390/ijms21228781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian Z., Dixon J., Guo X., Deal B., Liao Q., Zhou Y., Cheng F., Allen-Gipson D.S. Co-inhibition of CD73 and ADORA2B improves long-term cigarette smoke induced lung injury. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.614330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin R., Liu L., Xing Y., Meng T., Ma L., Pei J., Cong Y., Zhang X., Ren Z., Wang X., Shen J., Yu K. Dual mechanisms of novel CD73-targeted antibody and antibody–drug conjugate in inhibiting lung tumor growth and promoting antitumor immune-effector function. Mol Cancer Ther. 2020;19:2340–2352. doi: 10.1158/1535-7163.MCT-20-0076. [DOI] [PubMed] [Google Scholar]
- 37.Muir A.B., Whelan K.A., Dougherty M.K., Aaron B., Navarre B., Aceves S.S., Dellon E.S., Jensen E.T. The potential for malignancy from atopic disorders and allergic inflammation: a systematic review and meta-analysis. Clin Exp Allergy. 2020;50:147–159. doi: 10.1111/cea.13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Syed A., Maradey-Romero C., Fass R. The relationship between eosinophilic esophagitis and esophageal cancer. Dis Esophagus. 2017;30:1–5. doi: 10.1093/dote/dox050. [DOI] [PubMed] [Google Scholar]
- 39.Takashima S., Tanaka F., Otani K., Hosomi S., Nagami Y., Kamata N., Taira K., Yamagami H., Tanigawa T., Fukumoto S., Watanabe T., Fujiwara Y. Barrett’s esophagus is negatively associated with eosinophilic esophagitis in Japanese subjects. Esophagus. 2019;16:168–173. doi: 10.1007/s10388-018-0648-2. [DOI] [PubMed] [Google Scholar]
- 40.Eckle T., Füllbier L., Wehrmann M., Khoury J., Mittelbronn M., Ibla J., Rosenberger P., Eltzschig H.K. Identification of ectonucleotidases CD39 and CD73 in innate protection during acute lung injury. J Immunol. 2007;178:8127–8137. doi: 10.4049/jimmunol.178.12.8127. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X., Cheng E., Huo X., Yu C., Zhang Q., Pham T.H., Wang D.H., Spechler S.J., Souza R.F. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. 2012;7 doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng E., Zhang X., Huo X., Yu C., Zhang Q., Wang D.H., Spechler S.J., Souza R.F. Omeprazole blocks eotaxin-3 expression by oesophageal squamous cells from patients with eosinophilic oesophagitis and GORD. Gut. 2013;62:824–832. doi: 10.1136/gutjnl-2012-302250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanchard C., Stucke E.M., Rodriguez-Jimenez B., Burwinkel K., Collins M.H., Ahrens A., Alexander E.S., Buckmeier Butz B.K., Jameson S.C., Kaul A., Franciosi J.P., Kushner J.P., Putnam P.E., Abonia J.P., Rothenberg M.E. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–217. doi: 10.1016/j.jaci.2010.10.039. 217.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo L., Fulkerson P.C., Finkelman F.D., Mingler M., Fischetti C.A., Blanchard C., Rothenberg M.E. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13Rα2–inhibited pathway. J Immunol. 2010;185:660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirano I., Dellon E.S., Hamilton J.D., Collins M.H., Peterson K., Chehade M., Schoepfer A.M., Safroneeva E., Rothenberg M.E., Falk G.W., Assouline-Dayan Y., Zhao Q., Chen Z., Swanson B.N., Pirozzi G., Mannent L., Graham N.M.H., Akinlade B., Stahl N., Yancopoulos G.D., Radin A. Efficacy of dupilumab in a phase 2 randomized trial of adults with active eosinophilic esophagitis. Gastroenterology. 2020;158:111–122.e10. doi: 10.1053/j.gastro.2019.09.042. [DOI] [PubMed] [Google Scholar]
- 46.Hirano I., Collins M.H., Katzka D.A., Mukkada V.A., Falk G.W., Morey R., Desai N.K., Lan L., Williams J., Dellon E.S. Budesonide oral suspension improves outcomes in patients with eosinophilic esophagitis: results from a phase 3 trial. Clin Gastroenterol Hepatol. 2022;20:475–476. doi: 10.1016/j.cgh.2021.04.022. [DOI] [PubMed] [Google Scholar]
- 47.Dellon E.S., Liacouras C.A., Molina-Infante J., Furuta G.T., Spergel J.M., Zevit N., Spechler S.J., Attwood S.E., Straumann A., Aceves S.S., Alexander J.A., Atkins D., Arva N.C., Blanchard C., Bonis P.A., Book W.M., Capocelli K.E., Chehade M., Cheng E., Collins M.H., Davis C.M., Dias J.A., di Lorenzo C., Dohil R., Dupont C., Falk G.W., Ferreira C.T., Fox A., Gonsalves N.P., Gupta S.K., Katzka D.A., Kinoshita Y., Menard-Katcher C., Kodroff E., Metz D.C., Miehlke S., Muir A.B., Mukkada V.A., Murch S., Nurko S., Ohtsuka Y., Orel R., Papadopoulou A., Peterson K.A., Philpott H., Putnam P.E., Richter J.E., Rosen R., Rothenberg M.E., Schoepfer A., Scott M.M., Shah N., Sheikh J., Souza R.F., Strobel M.J., Talley N.J., Vaezi M.F., Vandenplas Y., Vieira M.C., Walker M.M., Wechsler J.B., Wershil B.K., Wen T., Yang G.Y., Hirano I., Bredenoord A.J. Updated international consensus diagnostic criteria for eosinophilic esophagitis: proceedings of the AGREE conference. Gastroenterology. 2018;155:1022–1033.e10. doi: 10.1053/j.gastro.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harada H., Nakagawa H., Oyama K., Takaoka M., Andl C.D., Jacobmeier B., von Werder A., Enders G.H., Opitz O.G., Rustgi A.K. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol Cancer Res. 2003;1:729–738. [PubMed] [Google Scholar]
- 49.Ohashi S., Natsuizaka M., Naganuma S., Kagawa S., Kimura S., Itoh H., Kalman R.A., Nakagawa M., Darling D.S., Basu D., Gimotty P.A., Klein-Szanto A.J., Diehl J.A., Herlyn M., Nakagawa H. A NOTCH3-mediated squamous cell differentiation program limits expansion of EMT-competent cells that express the ZEB transcription factors. Cancer Res. 2011;71:6836–6847. doi: 10.1158/0008-5472.CAN-11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kijima T., Nakagawa H., Shimonosono M., Chandramouleeswaran P.M., Hara T., Sahu V., Kasagi Y., Kikuchi O., Tanaka K., Giroux V., Muir A.B., Whelan K.A., Ohashi S., Naganuma S., Klein-Szanto A.J., Shinden Y., Sasaki K., Omoto I., Kita Y., Muto M., Bass A.J., Diehl J.A., Ginsberg G.G., Doki Y., Mori M., Uchikado Y., Arigami T., Avadhani N.G., Basu D., Rustgi A.K., Natsugoe S. Three-dimensional organoids reveal therapy resistance of esophageal and oropharyngeal squamous cell carcinoma cells. Cell Mol Gastroenterol Hepatol. 2019;7:73–91. doi: 10.1016/j.jcmgh.2018.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fujimoto T., Imaeda H., Takahashi K., Nishida A., Shioya M., Inatomi O., Bamba S., Shiomi H., Tani M., Andoh A. Eotaxin-3 (CCL26) expression in human pancreatic myofibroblasts. Pancreas. 2016;45:420–424. doi: 10.1097/MPA.0000000000000480. [DOI] [PubMed] [Google Scholar]
- 52.Walker W., Healey G.D., Hopkin J.M. RNA interference of STAT6 rapidly attenuates ongoing interleukin-13-mediated events in lung epithelial cells. Immunology. 2009;127:256–266. doi: 10.1111/j.1365-2567.2008.02951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]