Summary
Brain-derived neurotrophic factor (BDNF) plays a pivotal role in neuronal growth and differentiation, neuronal plasticity, learning, and memory. Using CRISPR/Cas9 technology, we generated a vital Bdnf null mutant line in zebrafish and carried out its molecular and behavioral characterization. Although no defects are evident on a morphological inspection, 66% of coding genes and 37% of microRNAs turned out to be differentially expressed in bdnf−/− compared with wild type sibling embryos. We deeply investigated the circadian clock pathway and confirmed changes in the rhythmic expression of clock (arntl1a, clock1a and clock2) and clock-controlled (aanat2) genes. The modulatory role of Bdnf on the zebrafish circadian clock was then validated by behavioral tests highlighting the absence of circadian activity rhythms in bdnf−/− larvae. The circadian behavior was partially rescued by pharmacological treatment. The bdnf−/− zebrafish line presented here is the first valuable and stable vertebrate model for the study of BDNF-related neurodevelopmental diseases
Subject areas: Behavioral neuroscience, Molecular neuroscience, Developmental neuroscience, Cellular neuroscience
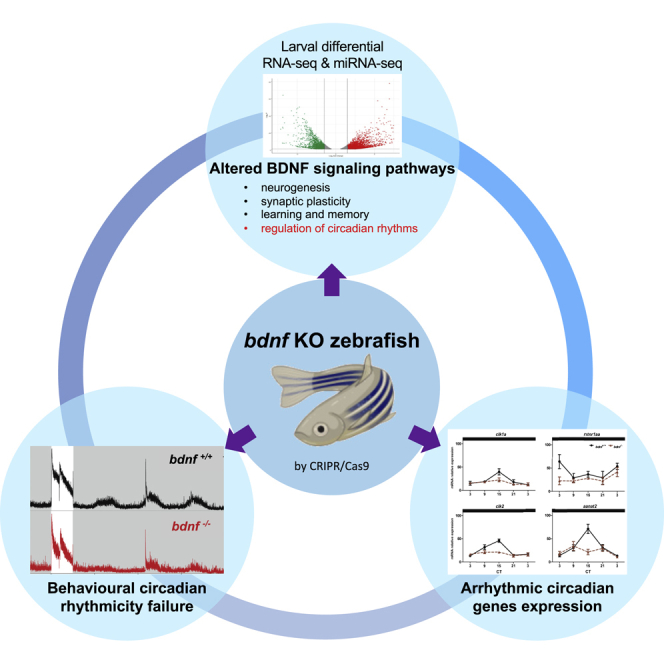
Graphical abstract
Highlights
-
•
Generation of a viable bdnf KO line in zebrafish
-
•
Bdnf deficiency affects locomotor activity and thigmotaxis in larvae
-
•
Differential RNA-seq analysis shows changes in expression of circadian clock genes
-
•
Bdnf mutant fails in the generation of the behavioral circadian rhythmicity
Behavioral neuroscience; Molecular neuroscience; Developmental neuroscience; Cellular neuroscience
Introduction
Brain-derived neurotrophic factor (BDNF) is a small, secreted protein belonging to the neurotrophin growth factor family (Leibrock et al., 1989). Since its discovery (Barde et al., 1982), BDNF has been considered critical to regulating the differentiation and survival of discrete neuronal populations during the development of the CNS (Acheson and Lindsay, 1996; Pencea et al., 2001; Binder and Scharfman, 2004). Its action is exerted through the binding of two classes of transmembrane receptors: the pan-neurotrophin p75 receptor (Ibáñez and Simi, 2012) and the tropomyosin-related kinase (Trk) family receptors (Barbacid, 1994; Segal, 2003; Reichardt, 2006), with higher affinity for the TrkB subtype (Soppet et al., 1991). The interaction with these molecules leads to the activation of major signaling pathways that modulate the expression of downstream genes involved in the regulation of specific neuronal targets’ growth, maintenance, and survival (Kaplan and Miller, 2000; Huang and Reichardt, 2003).
Besides its well-established role in nervous system development, the ability to enhance synaptic plasticity in the processes of learning and memory is an additional function ascribed to BDNF (Binder and Scharfman, 2004). A large body of evidence indicates that BDNF exerts a critical role in long-term potentiation (Cunha et al., 2010), a form of synaptic plasticity widely considered a cellular model for studying long-term memory formation (Bliss and Collingridge, 1993). BDNF is also involved in the regulation of circadian rhythms. It is expressed rhythmically in the suprachiasmatic nucleus (SCN) of the hypothalamus of rodents (Liang et al., 1998) and plays an important role in photic entrainment (Liang et al., 2000).
To date, most of the available information about the roles of BDNF is derived from studies performed in mice, but the whole scenario has not yet been fully delineated. The major obstacle is the early postnatal lethality of bdnf homozygous deletion (Ernfors et al., 1994a, 1994b). Therefore, all results are limited to heterozygous animals that do not show strong evidence of impaired phenotypes (Jones et al., 1994; Erickson et al., 1996).
BDNF is well-conserved in vertebrates, with a high sequence homology (>90%) between mammals and fish. Although Bdnf and other neurotrophic factors have been studied in zebrafish (De Felice et al., 2014; Cacialli et al., 2016; Nittoli et al., 2018), they are poorly investigated at mechanistic level in fish lineage. Recently, a role in brain regeneration, appetite, and metabolism in zebrafish was attributed to this neurotrophin (Wurzelmann et al., 2017; Blanco et al., 2020). In contrast with mammals, in zebrafish, it is possible to generate a viable knockout mutant. In particular, in this study, we generated a bdnf−/− zebrafish line with a stable and heritable homozygous 40-base pair (bp) deletion in the bdnf locus using CRISPR/Cas9 technology and assayed transcriptional and behavioral phenotypes in mutants versus controls. We observed that Bdnf mutants display evident defects in swimming activity, consistent with the well-established role of BDNF in complex behaviors such as locomotion and learning (Bekinschtein et al., 2014). Differential expression analysis (RNA-seq and qPCR) revealed alteration in the expression of key genes implicated in the circadian timekeeping system. Moreover, in the present study, we aimed to investigate daily and circadian activity in vivo in bdnf mutant larvae. Although photic entrainment is not impaired in mutant larvae, rhythmicity was strongly dampened in constant darkness, demonstrating the role of Bdnf in the generation of circadian activity. Interestingly, we also found that some miRNAs involved in the regulation of clock gene expression are expressed differentially in mutants.
Based on our findings, we propose the bdnf−/− zebrafish as an ideal vertebrate model for shedding light on the role of Bdnf in complex behaviors such as locomotion, feeding, and learning, as well as depressive and anxiety-related diseases.
Results
Zebrafish bdnf CRISPR/Cas9 knockout
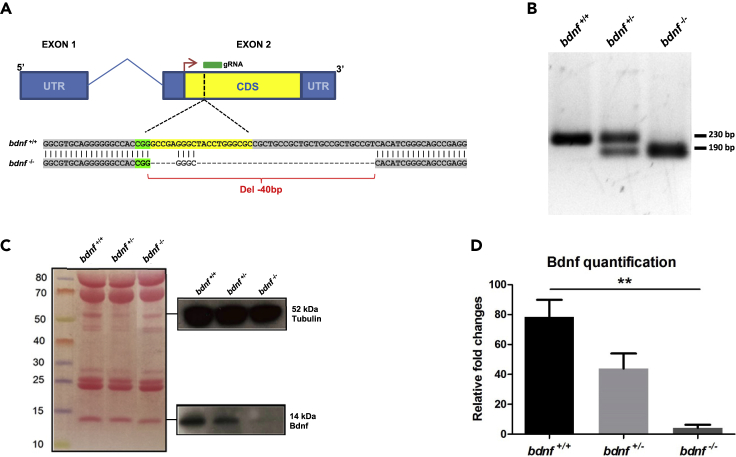
To investigate the role of Bdnf during the development of the zebrafish nervous system, we generated a CRISPR/Cas9-mediated knockout line with a 40 bp deletion in bdnf exon 2, immediately downstream of the start codon affecting all five ORF gene isoforms (Figure 1A; Table S1). The efficiency of the gRNA mutagenesis targeting the selected region was assessed using the T7 endonuclease I (T7EI) assay performed on genomic DNA extracted from a pool of microinjected embryos (Figures S1A and S1B). F0 mutation carrier fish were outcrossed with wild type (wt) fish. The heterozygous population (bdnf+/-) was bred for one generation (F2) to reduce mosaicism and unintended off-target mutations. Using T7EI assay and Sanger sequencing, we verified the absence of mutations in the nova2 gene, the unique off-target gene predicted by the CRISPR DESIGN tool (Figures S1C–S1E; Tables S2 and S3). The final F3 generation reflected the Mendelian ratio of 25% bdnf+/+, 50% bdnf+/−, and 25% bdnf−/− fish. An example of PCR-based screening of the bdnf target region from bdnf+/+, bdnf+/−, and bdnf−/− fish is shown in Figure 1B. Finally, Western blotting analysis performed on total protein lysates from embryo pools for each condition (n = 50; at 48 h postfertilization - hpf) confirmed that Bdnf protein levels are approximately half in bdnf+/- fish compared to bdnf+/+ controls and almost depleted in the homozygous bdnf−/− (Kruskall-Wallis ANOVA K3 = 6.49, p < 0.04; Figures 1C and 1D). Although we did not expect any positive signal in the full mutant’s protein lysate, we observed a faint band in the Western blotting (Figures 1C and 1D), which could be because of maternal Bdnf or to a cross-reaction of the heterologous anti-BDNF antibody with other neurotrophic factors.
Figure 1.
Generation of bdnf−/− zebrafish line
(A) The bdnf gene structure in Danio rerio is composed of two exons (blue box) and one intron (blue line), with the mRNA coding sequence (CDS) (yellow box) fully present in the exon 2. gRNA designed to target the CDS binds a genomic region immediately after the start codon (red arrow) and determines a 40 bp deletion.
(B) Genomic screening by PCR shows a single band of 230 bp in bdnf+/+ fish, one band of 230 bp and one of 190 bp in the bdnf+/-, and a single 190 bp band in the bdnf−/−.
(C and D) Western Blot analysis and (D) Bdnf quantification show that Bdnf protein is reduced by approximately 50% in bdnf+/- and almost completely in bdnf−/− in respect to bdnf+/+. Tubulin antibody was used as loading control. Densitometric analysis of Bdnf protein from three biological replicates is normalized with respect to Tubulin.
Mean values ±SEM; Dunn’s Multiple Comparison Test: ∗∗p < 0.01.
Therefore, we concluded that we successfully established a stable and heritable bdnf−/− zebrafish line. Importantly, morphological abnormalities and lethality during development were not observed in mutant embryos. Male and female mutant fish are able to reach adulthood and exhibit normal reproductive success.
Behavior in bdnf knockout larvae
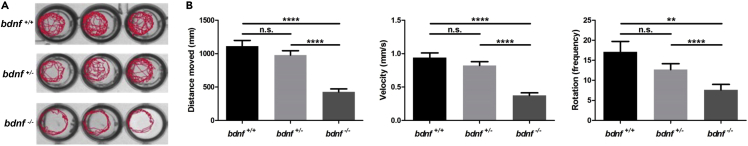
In addition to its known action in neuronal, survival and differentiation, BDNF also plays a role in the regulation of synaptic plasticity (Binder and Scharfman, 2004). Therefore, we speculated that its absence could affect neural circuits that regulate the locomotor activity in bdnf mutants. To test this hypothesis, we measured the swimming behavior of 4 days-postfertilization (dpf) larvae in response to light stimulation. As shown in Figure 2, mutant larvae showed locomotor defects ranging from an insignificant reduction in activity in the heterozygous bdnf+/- (n = 32) larvae to a strong locomotor inhibition in the homozygous bdnf−/− (n = 32) larvae when compared to the bdnf+/+ (n = 32) control larvae. Movement deficiency was measured by three parameters: total distance traveled, velocity, and frequency of rotation during a 10-min period after light stimulation. As shown in Figure 2B, for all three measured parameters, bdnf+/- larvae presented an insignificant reduction when compared to the bdnf+/+ larvae, whereas bdnf−/− larvae were strongly affected (Distance moved: F2,95 = 32.11, p < 0.0001; Velocity: F2,95 = 30.88, p < 0.0001; Rotation: F2,191 = 6.86, p < 0.001). Furthermore, analysis of tracks (Figure 2A), representing the path of the zebrafish inside the well during the test indicated that bdnf−/− larvae displayed marked thigmotaxis, i.e., the tendency of an animal to avoid the center of an arena and move toward its edge or periphery (Kalueff et al., 2013), whereas bdnf+/+ and bdnf+/- larvae covered almost the whole arena. Because thigmotaxis is one of the well-validated indices of anxiety in animals, this result also confirms the role of Bdnf in stress-related behavior in zebrafish (Miao et al., 2020).
Figure 2.
Behavior of bdnf+/- and bdnf−/− zebrafish larvae
(A) Reduced swimming activity of bdnf−/− mutant larvae as shown by locomotory tracking with DanioVision.
(B) bdnf−/− larvae (n = 96 per strain) assayed for distance, velocity, and rotations at four dpf showed significantly lower scores.
Mean values ±SEM; Dunn’s multiple comparison test: n. s. = not significant; ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
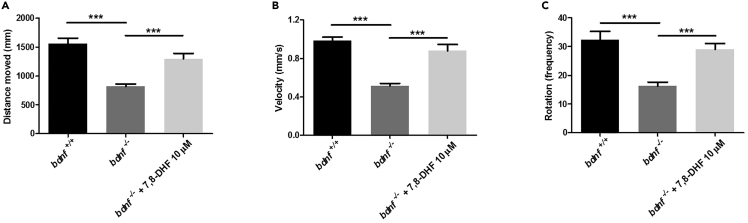
To demonstrate that the observed phenotype was related to the lack of Bdnf, we performed a pharmacological rescue experiment by treating 22 hpf bdnf−/− embryos with 10 μM of 7,8-dihydroxyflavone hydrate (7,8-DHF), a synthetic molecule that mimics the action of Bdnf by activating its TrkB receptor. The behavioral test on 7,8-DHF-treated bdnf−/− mutant larvae (n = 32) demonstrated a statistically significant recovery of locomotor activity compared to the bdnf−/− larvae (n = 89) (Figures 3A–3C). These results confirm that Bdnf deficiency affects locomotor activity and thigmotaxis in zebrafish larvae.
Figure 3.
Behavioral tests for pharmacological rescue of bdnf−/− zebrafish larvae
(A–C). Normal swimming behavior was rescued by pharmacologic treatments. Distance (A), velocity (B), and rotation (C) was measured for bdnf+/+ larvae, bdnf−/− larvae, bdnf−/− + 7,8-DHF (10 μM).
Mean ± SEM; Kruskal-Wallis ANOVA, Distance moved: K3 = 48.26, p < 0.0001; Velocity: K3 = 77.44, p < 0.0001; Rotation: K3 = 33.05, p < 0.0001; Dunn’s Multiple Comparison test: ∗∗∗p < 0.0001.
Differential gene expression analysis of the bdnf−/− line using RNA-seq
We conducted deep RNA sequencing (RNA-seq) of two biological replicates of equivalent pools of 48 hpf bdnf−/− mutants and bdnf+/+ embryos (see STAR Methods). An average of 28 million short paired-end fragments per sample was generated, of which approximately 89% were mapped unambiguously to the Danio rerio genome.
Using the DESeq2 method, we identified 2614 upregulated and 1653 downregulated genes between 48 hpf bdnf−/− mutants and bdnf+/+ embryos (BH Corrected p-value ≤ 0.10 and log2 FoldChange ≥ 0.6), suggesting remarkable transcript differences related to bdnf null mutation (Figure S2; Tables S4 and S5). It is noteworthy that the different expression levels of key genes involved in the neurotrophic pathway, such as the upregulation of Trk2a (the main Bdnf and Nt4/5 receptor), Trk3a (the Nt3 receptor), and p75 (the universal neurotrophin receptor for Ngf, Bdnf, Nt3, Nt4/5, and Nt6/7) (Table S4). This aspect is particularly appealing because it may indicate that an essential rebalancing of the neuronal survival pathway (i.e., upregulation of Trk2a and Trk3a) versus the death pathway (i.e., upregulation of p75) occurs during development in the bdnf-missing genetic background.
To gain insights into the functions of differentially expressed genes, we performed a Gene Ontology (GO) enrichment analysis for upregulated and downregulated genes. As expected, among the biological processes (BP) enriched terms, we identified several categories related to bdnf pathways such as nervous system development and functioning, photoreception, swimming behavior, and interestingly, circadian clock (Tables S4 and S5).
These results confirm the biological changes at transcriptomic level induced in the bdnf null mutated lines.
Bdnf regulates the circadian rhythm
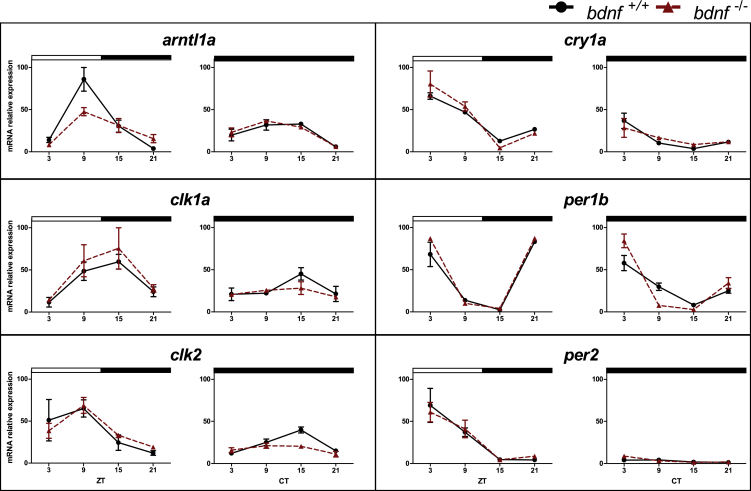
To validate the results of the transcriptomic analysis, we studied the expression pattern of a set of clock and clock-related genes using qPCR in bdnf+/+ and bdnf−/− larvae at eight dpf under 12:12 light-dark (LD) cycles or constant darkness (DD). First, we tested clock-regulated (arntl1a, clock1a, clock2, per1b) and light-regulated (per2, cry1a) gene expressions (Idda et al., 2012). Larvae from both genotypes showed daily changes in the expression levels of all genes investigated (Figure 4 and Table S6; p < 0.05), with differences in amplitude only for arntl1a. Indeed, the increase of arntl1a expression level at ZT9 was dampened significantly in mutant larvae (Figure 4; Pair-wise comparison after Kruskal-Wallis ANOVA test, p < 0.0005). In DD condition, as expected, bdnf+/+ larvae maintained a rhythmic pattern of clock controlled-gene expression (Figure 4; p < 0.005) and showed markedly damped oscillations of light-inducible genes per2 and cry1a mRNAs. Conversely, in bdnf−/− larvae, the expression profiles of the cardinal positive clock regulators clock1a and clock2 became arrhythmic during the 24 h (Table S6; p > 0.05).
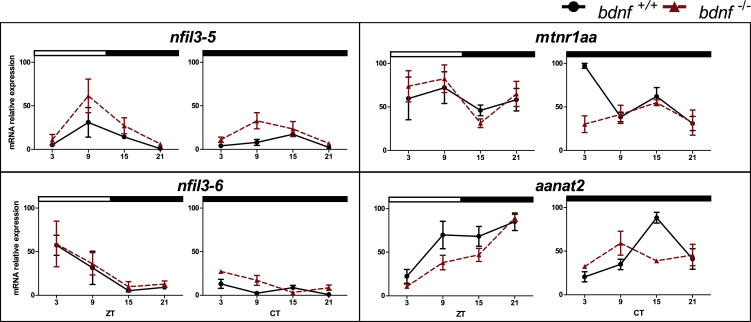
Figure 4.
Daily and circadian expression levels of clock genes in zebrafish larvae
qPCR analysis of clock and light-regulated clock gene expression at eight dpf in zebrafish larvae exposed to LD cycles and DD. For all panels, each point represents the mean ± SEM (n = 5). Relative expression levels (100% is the maximum level detected for each gene in LD and DD condition) are plotted on the y axis, whereas on the x axes, time is expressed as zeitgeber time (ZT, where ZT0 represents lights on; CT, where CT0 represents beginning of the subjective day). White and black bars represent light and dark periods, respectively. Solid lines indicate bdnf+/+; dotted lines indicate bdnf−/−.
Afterward, we investigated clock-controlled genes that mediate downstream circadian clock processes and feedback onto the core oscillator in zebrafish, such as those encoding Nfil3, a bZIP transcription factor. During the LD cycle, nfil3-5 and nfil3-6 showed a time-of-day dependent expression (Figure 5 and Table S6; p < 0.001), oscillating with a different phase (nfil3-5: peaks at ZT9; nfil3-6: peaks at ZT3) in bdnf+/+ and bdnf−/− larvae. In DD condition, both genotypes’ profiles remained rhythmic (Figure 5 and Table S6; p < 0.001).
Figure 5.
Daily and circadian expression levels of clock-controlled genes in zebrafish larvae
qPCR analysis of clock-controlled gene expression in larvae of zebrafish exposed to LD cycles or DD. For all panels, each point represents the mean ± SEM (n = 5). bdnf+/+ = solid line; bdnf−/− = dotted line. For more details, see Figure 4.
Clock-controlled genes involved in the synthesis and regulation of melatonin also play a cardinal role in the fish circadian timekeeping system (Ben-Moshe et al., 2014). The transcript level of aanat2, the enzyme that catalyzes melatonin synthesis in the pineal gland, was low at the beginning of the light phase and then increased, reaching its peak at the end of the night (ZT21) in both genotypes (Figure 5 and Table S6; p < 0.0001). In DD, aanat2 expression levels in bdnf+/+ larvae remained rhythmic (Figure 5 and Table S6; p < 0.0001), with a shift of the peak at CT15, whereas aanat2 mRNA in bdnf−/− was arrhythmic (p > 0.05). Similarly, melatonin receptor mtnr1aa mRNA was expressed rhythmically in control larvae in LD and DD (Figure 5 and Table S6; p < 0.05), whereas its oscillation was abolished in mutant larvae in DD (p > 0.8).
We also investigated whether Bdnf signaling plays a role in regulating circadian rhythms by analyzing the rhythmic locomotor activity of mutant and wt larvae maintained for 3 days under the LD cycle, followed by 3 days of DD. During the LD cycle, bdnf+/+, and bdnf−/− larvae displayed daily rhythms in locomotor activity (Cosinor, p < 0.001), as well as the typical diurnal pattern of zebrafish, with higher activity during the light phase (Figure 6). Interestingly, larvae showed significant differences in the overall amount of activity during the light, but not during the dark period (Figure 6; U-test, L: p < 0.03; D: p = 0.4). Furthermore, wt and mutant genotypes showed an increase in activity after feeding (Figure 6).
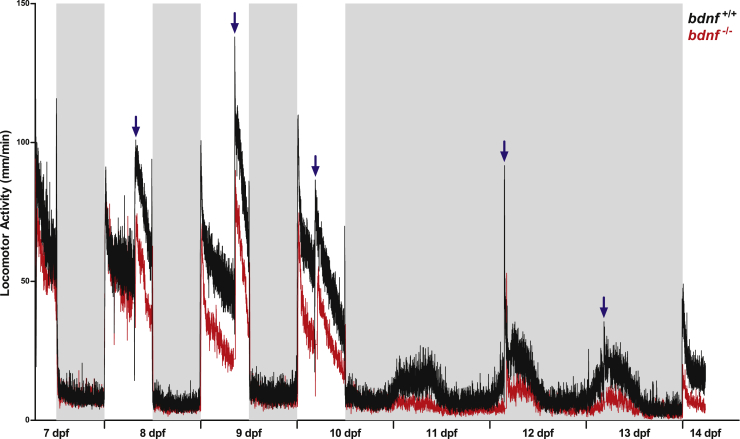
Figure 6.
Daily and circadian activity rhythms of bdnf+/+ and bdnf−/− zebrafish larvae
Mean waveform of locomotor activity under 12:12 LD cycles from 7 to 10 dpf and DD from 10 to 13 dpf (n = 32/genotype). Vertical axis shows the average distance moved (mm/1 min), whereas X axis indicates time in recording. White and gray bars show light and dark phases, respectively. Blue arrows indicate feeding time. Data are expressed as mean ± SEM.
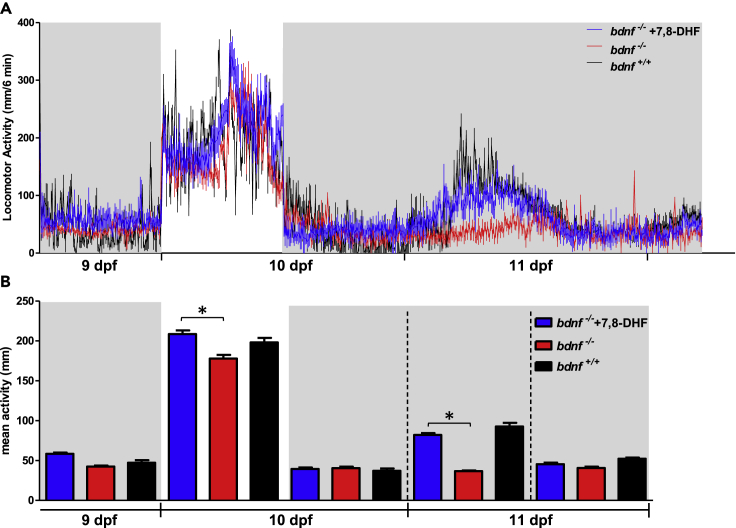
To test the presence of a circadian timekeeping system in bdnf−/− mutants, we recorded circadian locomotor activity in larvae kept in DD for 3 days. In the constant condition, bdnf+/+ larvae displayed circadian locomotor activity (Figure 6; Cosinor, p = 0.0003) with a free-running period of 24 h and peaks during the subjective day. Conversely, bdnf−/− became immediately arrhythmic during the first day in DD (Figure 6; Cosinor, p > 0.2), indicating the absence of behavioral circadian rhythmicity. Furthermore, the overall activity during the subjective day and night of the first day in DD was not statistically different from the dark phases of the LD cycles (Figure 6; Kruskal-Wallis ANOVA test, p = 0.4). The increase inactivity during the second day and third day in DD after feeding, which also occurred in mutant larvae, indicates the ability to respond to environmental signals. Finally, after three days in DD, larvae from both genotypes responded to light with increased activity. To confirm the role of Bdnf in the generation of behavioral circadian rhythm in larvae and to exclude any bias because of the feeding procedure, we repeated the test in mutants, measuring locomotor activity in DD and starvation for more than 80 h and confirmed the absence of rhythmicity in mutant larvae (Figure S3). To confirm that the arrhythmic behavioral phenotype was actually related to the lack of Bdnf, we performed the pharmacological rescue in nine dpf bdnf−/− larvae using 10 μM of 7,8-DHF and recorded daily and circadian locomotor activity. The behavioral test on 7,8-DHF-treated bdnf−/− larvae (n = 32) demonstrated a statistically significant recovery of locomotor activity and circadian pattern, compared to untreated mutant larvae (n = 32) (Figures 7A and 7B). In fact, in the light phase of LD cycle and in the subjective day in DD, 7,8-DHF-treated bdnf−/− larvae showed higher level of activity respected to untreated mutant larvae (Figure 7B; Kruskal-Wallis test, light phase: K3 = 23.89, p < 0.0001; subjective day K3 = 141.5, p < 0.0001), at levels comparable to bdnf+/+ larvae (Dunn’s Multiple Comparison Test, p > 0.05). Furthermore, in DD, the pharmacological treatment was able to rescue the circadian rhythmicity in bdnf−/− larvae (Figure 7A; Cosinor, p = 0.0001).
Figure 7.
Daily and circadian activity rhythms of pharmacological rescued bdnf−/− zebrafish larvae
(A) Mean waveform of locomotor activity under 12:12 LD cycles from 9 to 10 dpf and DD from 10 to 11 dpf (n = 32/group). See Figure 6 for details.
(B) Mean activity in the light and dark phases and in subjective day and subjective night in DD from 9 to 11 dpf. Kruskal-Wallis test; ∗p < 0.0001. Data are expressed as mean ± SEM.
Differentially expressed miRNAs upon loss of bdnf signaling
To shed light on the regulatory mechanisms that might have caused a shift in the expression levels of circadian genes in the bdnf−/− zebrafish, we looked for differentially expressed miRNAs. For this purpose, we prepared small RNA libraries at two stages of development, 24 and 48 hpf, for NGS sequencing. Several miRNAs were expressed differently when comparing the bdnf mutant embryos to wt embryos, indicating that they could be active regulatory intermediates in the Bdnf signaling cascade, and therefore could be responsible for the observed circadian rhythm alterations. Importantly, among the many miRNAs that surely deserve further investigation in the future, we highlighted a short list that has been described in the literature for playing a key role in the regulation of the circadian rhythm (Figure 8; Table S7. Differentially expressed miRNA analysis at embryonic stage 24 hpf, related to Figure 8, Table S8. Gene Ontology (GO) enrichment analysis results for identified differentially expressed miRNAs at embryonic stage 24 hpf, related to Figure 8, Table S9. Differentially expressed miRNA analysis at embryonic stage 48 hpf, related to Figure 8, Table S10. Gene Ontology (GO) enrichment analysis results for identified differentially expressed miRNAs at embryonic stage 48 hpf, related to Figure 8), thus supporting the miRNA-seq data shown here.
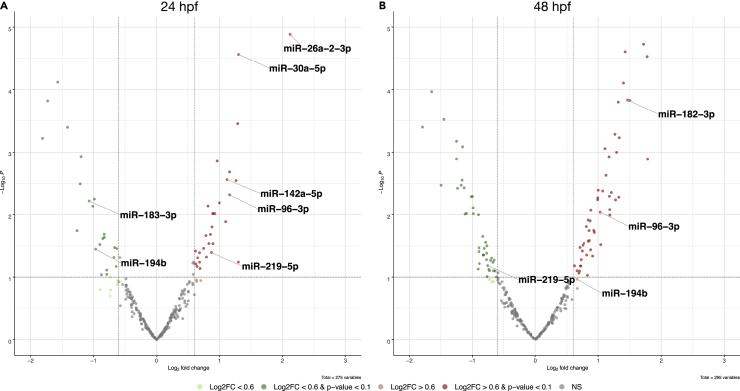
Figure 8.
Volcano plots showing differentially expressed miRNAs
(A and B) Analysis of the differentially expressed miRNAs at two embryonic stages, 24 hpf (A) and 48 hpf (B), displays changes in the expression level of several miRNAs known to have a role in the regulation of circadian genes.
We found that miR-183, a regulator of the clock-controlled anaat2 in the zebrafish pineal gland (Ben-Moshe et al., 2014), and miR-194, an inhibitor of the period gene family in cell cultures (Nagel et al., 2009), are downregulated at 24 hpf in bdnf mutants. Moreover, miR-219, known as a target of the Clock/Bmal complex (Cheng et al., 2007), resulted in reduced expression at 48 hpf in mutant fish embryos compared to controls (Figure 8). Similarly, miR-26, which has a critical function in circadian input entrainment and output pathways in the SCN and retina (Shi et al., 2009), is less expressed than in controls. However, our miRNA-seq results highlighted the significantly higher levels of expression of miR-96 and miR-182, which are known to control clock and the clock-controlled gene Adenylyl cyclase type 6 (Adcy6) in mice (Zhou et al., 2021). The brain-specific miR-219, known to modulate the circadian period length, is upregulated at 24 hpf and downregulated at 48 hpf (Saus et al., 2010) (Figure 8).
Gene Ontology (GO) enrichment analysis for upregulated and downregulated miRNAs at 24 and 48 hpf confirmed the implications of such differentially expressed miRNAs in several biological processes related to locomotory behavior, development regulation, and circadian rhythm (Table S7. Differentially expressed miRNA analysis at embryonic stage 24 hpf, related to Figure 8, Table S8. Gene Ontology (GO) enrichment analysis results for identified differentially expressed miRNAs at embryonic stage 24 hpf, related to Figure 8, Table S9. Differentially expressed miRNA analysis at embryonic stage 48 hpf, related to Figure 8, Table S10. Gene Ontology (GO) enrichment analysis results for identified differentially expressed miRNAs at embryonic stage 48 hpf, related to Figure 8).
Discussion
In this study, we investigated Bdnf’s role during zebrafish development through the generation of a 40 bp germline deletion using the CRISPR/Cas9 system, the most innovative technology for specifically editing a target gene (D'Agostino and D'Aniello, 2017). After confirming that the selected mutation in bdnf exon-2 resulted in the loss of Bdnf, we assayed transcriptional and behavioral phenotypes associated with bdnf−/− mutants, shedding light on the impairment of key genes involved in the regulation of the circadian rhythm. Currently, few data are available on Bdnf’s role in the regulation of the circadian rhythm. The experiments described in the present work made it possible to validate what emerged from the transcriptomic analyses, as behavioral tests under different light stimulation conditions (i.e., LD or DD cycle) confirmed an alteration of the circadian rhythm. Furthermore, drug treatments of bdnf−/− embryos at 22 hpf with synthetic BDNF molecules (7,8-DHF) resulted in the partial rescue of the mutant phenotype when subjected to locomotor analysis tests, thus confirming that the mutant behavior was actually because of the lack of the Bdnf protein.
In vivo experiments in animal models that recapitulate specific features of human disease offer an opportunity to understand the developmental origin of the disorder. In the neurodevelopmental context, zebrafish represents a powerful biological tool for investigating specific mechanisms connected to genetic aberrations (Kalueff et al., 2014). In particular, modeling how a mutation can affect nervous system development in animal models is critical to highlighting the mechanisms underlying neurological disorders such as depression and anxiety, which are strongly associated with Bdnf deficiency or circadian rhythm alteration (Yi et al., 2015). Moreover, because bdnf is highly conserved across vertebrates (Tettamanti et al., 2010) with the deduced amino acid sequence as zebrafish, which is 91% identical to that of mammalian orthologs (Heinrich and Pagtakhan, 2004), we assume that molecular and behavioral phenotypes observed in our mutant fish might be translated to human research.
Previous studies reported that mouse bdnf mutants exhibit reduction in neurons of the vestibular ganglion (Ernfors et al., 1994a, 1994b) responsible for locomotion, balance, and postural control (Jones et al., 1994; Kondo et al., 2008); reduction of tyrosine hydroxylase (TH)-positive neuron in the substantia nigra (Baquet et al., 2005); lack of a subset of afferents involved in ventilator control that causes severely irregular breathing that might contribute to premature death (Erickson et al., 1996); aggressiveness; increased anxiety and hyperphagia accompanied by weight gain (Rios, 2013).
In the mouse model, a major obstacle in elucidating the role of BDNF is the early postnatal lethality of the homozygous mutant, which generally occurs within 2 days after birth; only a very small fraction of animals live for 2–3 weeks at most (Jones et al., 1994). Therefore, all the results available until now derive from studies on heterozygous animals that reach the adult state and appear fertile. Because low doses of BDNF might be able to exert their own function, a complete knockout of bdnf is pivotal to fully understand the many biological functions played by this neurotrophin. The vital bdnf−/− zebrafish line that we generated could help to compensate for this gap and offer new insights. Although we still know little about its physiology, the zebrafish mutant line here presented is able to survive to adulthood, and a possible explanation could come from the activation of “compensatory network effect” exerted by paralogues genes that have been demonstrated to buffer against deleterious mutations induced by genome editing (Rossi et al., 2015).
Neurotrophins, including BDNF, have beenshown to be involved in the regulation of the SCN circadian pacemaker to light pulse. In bdnf+/- mice, a marked dampened expression of BDNF in the SCN and in the amplitude of light-induced phase shifts of locomotor activity in DD indicated that this neurotrophin plays a role in the regulation of photic entrainment (Liang et al., 2000). Similar results have been obtained in rats in which infusions of exogenous BDNF into the SCN altered the behavioral response to light pulses (Liang et al., 2000).
Because this critical issue has not been studied previously, we also investigated the role of Bdnf in the zebrafish circadian clock system. Using zebrafish null mutants, we showed that Bdnf plays an important role in the generation of the circadian rhythmicity. In fact, in mutants, the locomotor activity was arrhythmic in DD condition, and rhythmic behavior was partially recovered by pharmacological treatment, demonstrating the validity of the model. In addition, differences in the pattern of mRNA expression in mutant larvae in DD indicate a modulatory role of Bdnf on clock1a and clock2, two positive elements of the circadian feedback loops (Di Rosa et al., 2015; Morbiato et al., 2019), as well as on aanat2, which encodes the enzyme catalyzing melatonin synthesis (Saha et al., 2019).
One major output of the fish circadian clock is the rhythmic synthesis and secretion of the hormone melatonin by the pineal gland (Ben-Moshe et al., 2014). Melatonin is produced at night and its production rate is determined by the enzymatic activity of AANAT. High melatonin levels at night reflect increased AANAT synthesis and activity, whereas the termination of melatonin production during the day reflects proteasomal degradation of this enzyme (Falcón et al., 2001).
Our data indicate the elimination of the circadian rhythm expression of the clock and aanat2 genes and locomotor activity in bdnf−/− larvae in DD, corroborating previous findings (Ben-Moshe Livne et al., 2016). Using the dominant-negative strategy to selectively block clock rhythmic expression in the zebrafish pineal gland, Ben-Moshe et al. showed disruption of the rhythm of melatonin production in the pineal gland and marked attenuation of the behavioral rhythms in zebrafish larvae under DD conditions. These findings support the cardinal role of melatonin in the generation of behavioral circadian rhythms (Ben-Moshe Livne et al., 2016; Gandhi et al., 2015; Oikonomou and Prober, 2017).
Our results regarding the circadian rhythm phenotype are also supported by the deregulation in bdnf−/− of some miRNAs involved in the modulation of circadian clock target mRNAs (Cheng et al., 2007; Pegoraro and Tauber, 2008; Nagel et al., 2009; Shi et al., 2009; Saus et al., 2010; Hansen et al., 2011; Ben-Moshe et al., 2014). In particular, the downregulation of miR-183 and alteration of aanat2 and nfil3 gene expression in bdnf−/− larvae indicate the involvement of Bdnf in the regulation of this miRNA, which in turn modulates the rhythmic mRNA levels of aanat2 and nfil3, as previously demonstrated (Ben-Moshe et al., 2014). It is noteworthy that a number of miRNAs, which are known to regulate circadian core genes, did not change their expression levels in our miRNA-seq analysis. This category of miRNAs comprises miR-24, a known repressor of Per2 protein accumulation (Yoo et al., 2017); miR-340, which targets Clock, Per1 and Cry2; miR-669, which targets Per2; miR-374, which targets Per3; miR-338, which targets Nr1d1 (Wang et al., 2019), and miR-132, which is induced by photic entrainment cues, modulates clock gene expression, and attenuates the entraining effects of light (Cheng et al., 2007). Recently, the circadian regulatory function of the cluster miR-183/96/182 has been validated in vitro and in vivo (Zhou et al., 2021).
Overall, these data, which were collected in bdnf−/− zebrafish larvae for the first time, offer a new framework for contextualizing the pleiotropic effect of Bdnf in the circadian rhythm. However, further experiments are needed to identify intermediate factors thatcould directly or indirectly connect Bdnf to the molecular circadian clock network. Targeting specific Bdnf signaling components downstream to the Bdnf-TrkB pathway will be necessary to unravel the complex roles of this intriguing molecule.
In conclusion, we describe the first viable bdnf−/− vertebrate model that provides unique insights into the molecular and cellular mechanisms of neurotrophic factors. We demonstrated that Bdnf in zebrafish larvae is crucial for the generation of behavioral circadian rhythmicity and could pave the way to a better understanding of neurodevelopmental and circadian regulation diseases in humans. This study confirmed the numerous pathways in which Bdnf is involved, and the generated zebrafish null mutant line might serve as a unique tool for future investigations aimed at understanding the role of Bdnf in different biological processes including neuron development and synaptic transmission, visual light perception, regeneration, and metabolism.
Limitations of the study
In the present work, we studied the role of Bdnf in the circadian gene expression and behavior in zebrafish. Future research will provide important insight into the direct or indirect regulatory effect of Bdnf on the circadian clock mechanisms occurring during development. One limitation of this study is the partial characterization of the impact of Bdnf loss on CNS development and function, which is particularly important for a better interpretation of the KO phenotype.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Primary human BDNF | Santa Cruz Biotechnology | Cat#sc-546, RRID: AB_630940 |
| α-tubulin | Sigma-Aldrich | Cat#T5168, AB_477579 |
| Chemicals, peptides, and recombinant proteins | ||
| Phenol Red tracer | Sigma-Aldrich | P3532 |
| T7 endonuclease I | New England BioLabs | M0302 |
| Ponceau S Solution | Sigma-Aldrich | P7170 |
| TRIzol™ Reagent | Thermo Fisher Scientific | 15596026 |
| 7,8-dihydroxyflavone hydrate | Sigma-Aldrich | D5446 |
| SsoAdvanced Universal SYBR Green Supermix | BIO-RAD | 1725270 |
| Critical commercial assays | ||
| MEGAshortscript T7 Kit | Thermo Fisher Scientific | AM1354 |
| mirVana™ miRNA Isolation Kit | Thermo Fisher Scientific | AM1560 |
| mMESSAGE mMACHINE™ T3 Transcription Kit | Thermo Fisher Scientific | AM1348 |
| RNeasy Mini Kit | QUIAGEN | 74104 |
| QIAquick PCR purification Kit | QUIAGEN | 28104 |
| Quick Start™ Bradford Protein Assay Kit | BIO-RAD | 5000201 |
| NuPAGE™ Large Protein Blotting Kit | Thermo Fisher Scientific | LP0001 |
| TruSeq SmallRNA Sample Prep Kit | ILLUMINA | RS-200-0012 |
| TruSeq Stranded Total RNA Library Prep Kit | ILLUMINA | 20020596 |
| iScript™ gDNA Clear cDNA Synthesis Kit | BIO-RAD | 1725034 |
| Deposited data | ||
| RNA-seq and miRNA-seq of bdnf+/+ and bdnf−/− zebrafish larvae at 24 and 48 hpf | NCBI Sequence Read Archive (SRA) | SUB8904035 |
| Experimental models: Organisms/strains | ||
| Zebrafish AB: CRISPR/Cas9 bdnf | Bertolucci/D’Aniello | N/A |
| Oligonucleotides | ||
| Dr_bdnf_gRNA Forward TAGGCGGCGCCCAGGTAGCCCT |
This paper | N/A |
| Dr_bdnf_gRNA Reverse AAACAGGGCTACCTGGGCGCCG |
This paper | N/A |
| Dr_bdnf Forward x PCR screening GAAGAGTGATGACCATCCTG |
This paper | N/A |
| Dr_bdnf Reverse x PCR screening ATGACCTGCTCGAAAGTGTCCG |
This paper | N/A |
| Dr_bdnf Forward x T7 endonuclease I assay GAAGAGTGATGACCATCCTG |
This paper | N/A |
| Dr_bdnf Reverse x T7 endonuclease I assay GTGTACACTATCTGCCCC |
This paper | N/A |
| Dr_nova2 Forward x T7 endonuclease I assay ACAGCCTCCAGTCTCCTGGG |
This paper | N/A |
| Dr_nova2 Reverse x T7 endonuclease I assay TAGCCCACCTTCTGCGGATTGG |
This paper | N/A |
| Primers for qPCR, see Table S5 | This paper | N/A |
| Recombinant DNA | ||
| pT7-gRNA vector (Chen lab) | Jao et al., 2013 | Addgene # 46759 |
| pCS2-nCas9n (Chen lab) | Jao et al., 2013 | Addgene # 47929 |
| Software and algorithms | ||
| Guide Design Resources (Zhang lab) | https://zlab.bio/guide-design-resources | N/A |
| ImageJ Software | Wayne Rasband (NIH) | N/A |
| ViiA™ 7 Software | Thermo Fisher Scientific | N/A |
| FASTX Toolkit (Hannon lab) | Affero GPL (AGPL) | N/A |
| HTseq tool | Anders et al., 2015a, 2015b | http://htseq.readthedocs.io |
| miRdeep2 | Friedländer et al., 2011 | http://www.mdc-berlin.de/content/mirdeep2-documentation |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| edgeR | Robinson et al., 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| clusterProfiler | Yu et al., 2012 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| EthoVision XT Software | NOLDUS | N/A |
| Prism 5 | GraphPad Inc. | N/A |
| R 3.6.1 | www.R-project.org/ | N/A |
| Cosinor | Refinetti et al., 2007 | N/A |
| Other | ||
| RIPA Lysis and Extraction Buffer | Thermo Fisher Scientific | 89900 |
| Complete Protease Inhibitor Cocktail | MERK | 11697498001 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Prof. Cristiano Bertolucci (bru@unife.it).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Next-generation sequencing data (RNA-seq and miRNA-seq) of bdnf+/+ and bdnf−/− zebrafish larvae at 24 and 48 hpf have been deposited in the NCBI Sequence Read Archive (SRA): SUB8904035.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
Experimental model and subject details
Adult wild-type zebrafish (AB strain), bdnf+/- and bdnf−/− lines were kept in a constant re-circulating water system at 28°C, on a 14 h light/10 h dark cycle, according to standard procedures (Westerfield, 2000). Embryos were obtained by natural spawning, and staged according to timing-post fertilization and morphological criteria reported by Kimmel et al. (1995). All handling and breading protocols were performed in accordance to the methods published in the Zebrafish Book (Nüsslein-Volhard and Dahm, 2012). We have bred many generations of mutant zebrafish, therefore we believe that the mutation is stable in the fish line and the lifespan and reproductive fitness are not compromised.
Heterozygous bdnf fish were generated by clustered regularly interspaced short palindromic repeats (CRISPR)-mediated knockout, as described (Jao et al., 2013). The plasmids coding for the Cas9 protein (pT3TS-nls-zCas-nls) and the gRNA backbone were provided from Addgene (Catalog #46759). The genomic target site was identified within the coding sequence of zebrafish bdnf gene, using the Guide Design Resources (Zhang Lab, https://zlab.bio/guide-design-resources). The bdnf-specific gRNA was designed in order to insert mutation(s) in all 5 splicing isoforms and to avoid residual peptide production. To synthesize the gRNA standard complement and reverse insert-oligonucleotides (Sigma-Aldrich) (Table S1) were re-suspended in TE buffer (10 mM Tris-HCl, 0.1 mM EDTA), annealed in 1X NEB buffer (New England BioLab) and ligated to the pT7-gRNA vector (Addgene #46759). Then the linearized pT7-bdnf-gRNA vector was directionally transcribed in vitro using the MEGAshortscript T7 kit (Thermo Fisher Scientific) and purified with the mirVana miRNA isolation kit (Thermo Fisher Scientific), following the manufacturer’s instructions.
To synthesize the capped nls-zCas9-nls mRNA, the linearized pT3TS-nls-zCas-nls vector (Addgene #47929) was directionally transcribed in vitro using the mMESSAGE mMachine T3 Transcription kit (Thermo Fisher Scientific) and purified with the RNeasy Mini kit (Qiagen), following the manufacturer’s instructions. One nl of a mix containing gRNA (80 ng/μl) and Cas9 mRNA (150 ng/μl) was microinjected into 1-cell stage zygotes. The volume injected was estimated to be about 5% of the egg volume by controlling the amount of co-injected Phenol Red tracer (Sigma-Aldrich). The needles used for the microinjection were made from capillary tubes (Microcaps, Drummond Scientific Company) appropriately pulled with a PN-3 Micro-Electrode Puller (Narishige).
Ethical approval
All husbandry and experimental procedures were performed in accordance with European Legislation for the Protection of Animals used for Scientific Purposes (Directive 2010/63/EU) and the Italian animal protection standards (D.lgs. 26/2014). The research project was approved by the Institutional Animal Care and Use Committees of the Universities of Bologna and Ferrara, and by the Italian Ministry of Health (Authorization numbers 563/2018-PR and 340/2019-PR). License for fish maintenance and breeding at the University of Bologna is n. 6089/15 and at the University of Ferrara is n. 18/2017-UT.
T7 endonuclease I assay
The efficiency of mutagenesis of the gRNA was assessed using the T7 endonuclease I assay (New England BioLabs) (Figure S1). Genomic DNA was extracted from a pool of injected and control embryos at 7 dpf. gRNA target site was PCR amplified using primers designed to anneal 200 bp upstream and downstream from the expected cut site (Table S2). The PCR amplicons were purified using the QIAquick PCR purification kit (Qiagen). A total of 200 ng of purified PCR was denatured in a thermo-block at 95 °C for 10 min and slowly renatured to allow hetero-duplex formation. Re-annealed amplicons were digested with the T7 endonuclease I enzyme following the manufacturer’s instructions and resolved by electrophoresis in a 1.5% agarose gel.
Zebrafish genotyping
F0 generation fish (deriving from 50 microinjected embryos) were mated with wt fish, producing F1 generation fish with different types of mutations that were genotyped by genomic DNA extraction from the caudal fin and directional sequencing of the target region in order to select the desirable insertion/deletion (InDel). The selected F1 fish carrying a 40 bp deletion in bdnf locus was mated with a wt fish in order to obtain heterozygous fish with the same mutation in the F2 generation. Finally, two heterozygous fishes were inter-crossed to have homozygous fish (F3) in a percentage of 25%. Once the −40 bp deletion was fixed in the genome of the knock-out fish line, two further generations were screened by standard PCR using primers overlapping the mutated site or by qPCR on genomic DNA (Table S2) and derivative melting curves analysis using the ViiA™ 7 Software (Thermo Fisher Scientific), as described in D’Agostino et al., 2016.
Off-target validation
To test the specificity of the injected gRNA, the off-target region (nova2 gene) predicted by the “CRISPR DESIGN” tool was analysed by T7 endonuclease I assay on three different embryos from F1 generation and on a pool of injected embryos. To further confirm the absence of InDels, the putative off-target site was also directionally sequenced by Sanger chromatography (Table S3).
Method details
Protein extraction and western blotting
Total proteins were extracted from a pool of 50 embryos in 250 μL of RIPA buffer [1 mM EDTA, 150 mM NaCl, 10 mM Tris-HCl, pH 7.4, 1% Triton-100X, 0.1% SDS, protease inhibitor cocktail tablet (Roche)]. After incubation for 20 min at 4°C, homogenization was obtained using the TissueLyser II (Qiagen). A centrifugation step was performed at 4°C for 20 min at 14.000 g to remove cell debris. The supernatant containing the proteins was transferred in a new tube and stored at −80°C until use. The Quick Start™ Bradford Protein Assay Kit (BioRad) was used for determining protein concentration (595 nm). Comparison using a standard curve with known amount of bovine serum albumin (BSA) provided a relative measurement of protein concentration.
Protein samples from three biological replicates were heated at 70°C for 10 min in presence of NuPage LDS Sample buffer 4X containing 1 μl NuPage reducing agent 10X (Thermo Fisher Scientific). Total proteins were fractionated by SDS-PAGE on NuPage 4–12% Bis-Tris Gel (Thermo Fisher Scientific) in MES buffer (2-[N-morpholino] ethanesulfonic acid) at a constant 200 V and transferred for 2 h at 200 mA to a 0.20 μm nitrocellulose membrane paper sandwich included with the NuPAGE Large Protein Blotting Kit (Thermo Fisher Scientific).
The proteins were stained with Ponceau-Red solution (Sigma-Aldrich) and after blocking with 5% non-fat milk in TBST (10 mM Tris pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 60 min, the membrane was incubated overnight at 4°C with primary antibody against human BDNF 1:800 (sc-546, Santa Cruz Biotechnology) and α-tubulin 1:5000 (T5168, Sigma-Aldrich). Membranes were washed three times with TBST for 10 min and incubated with a dilution of horseradish peroxidase-conjugated anti-mouse or anti-rabbit antibodies for 1 h at room temperature. Blots were washed with TBST three times and revealed with the ECL Western Blotting System (Amersham), according to the manufacturer’s protocols.
RNA library preparation and sequencing
Total RNA was extracted in RNase free environment from 50 pooled whole zebrafish embryos at 24 and 48 hpf (three biological replicates for each condition) using TRIzol™ Reagent solution (Thermo Fisher Scientific). The RNA integrity number (RIN) was measured with an Agilent 2100 Bioanalyzer (Agilent Technologies). Next generation sequencing (NGS) experiments, comprising samples quality control, were performed by Genomix4Life (www.genomix4life.com). Indexed libraries were prepared from 1 μg/ea purified RNA with TruSeq SmallRNA Sample Prep Kit and TruSeq Stranded Total RNA Library Prep Kit (Illumina), according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer and pooled such that each index-tagged sample was present in equimolar amounts, with final concentration of the pooled samples of 2 nM. The pooled samples were subject to cluster generation and sequencing using an Illumina HiSeq 2500 System (Illumina) in a 1 × 50 single read (SmallRNA) and 2 × 100 paired-end (RNA-Seq) format at a final concentration of 10 pmol. The raw sequence files generated (.fastq files) underwent quality control analysis using FastQC (Babraham Bioinformatics).
Differential gene expression analysis
Sequenced libraries raw reads were trimmed from the Illumina adapters using fastx_trimmer tool of FASTX Toolkit (Hannon Lab, https://zlab.bio/guide-design-resources). For RNA-seq library preparation, adapter-trimmed reads were aligned to the zebrafish reference genome (GRCz10/danRer10) using STAR version 2.5.4 (Dobin et al., 2013). Read counts per gene were computed using HTseq tool v0.7.0 (Anders et al. 2015a, 2015b). For smallRNA-seq library, adapter-trimmed reads were mapped to zebrafish miRNA sequences from miRBase version 21 and miRNA expression profiles were computed using miRdeep2 (Friedländer et al., 2011). Downstream analysis was performed in the R statistical environment.
Differential expression analysis between the two considered conditions was performed using the robust approaches DESeq2 v1.16.2 (Love et al., 2014) for RNA-seq and edgeR v3.18.0 (Robinson et al., 2010) for smallRNA-seq, respectively.
Normalized read counts were computed by dividing the raw read counts by size factors and fitting to a negative binomial (Gamma-Poisson) distribution. Genes with an adjusted p value (Benjamini & Hochberg correction) ≤0.10 and log2 FoldChange ≥ 0.6 were considered significantly differentially expressed.
Rescue experiment
Starting from 22 hpf, a pool of bdnf−/− embryos (n = 32) were dechorionated and treated with 10 μM of 7,8-dihydroxyflavone hydrate (7,8-DHF; Sigma) as reported in Daly et al. (2017). In parallel, a pool of bdnf−/− (n = 65) and of bdnf+/+ embryos (n = 89) were grown in 0.1% dimethyl sulfoxide (DMSO) in E3 solution (v/v). For each treatment larvae were transferred in Petri dishes containing 10 ml of drug solution and incubated under standard conditions until 5 dpf. Then control and null mutant treated larvae were removed from the drug solution, washed in fresh E3 solution and transferred in 96 multi-well plates to perform locomotor analysis.
qPCR of clock and clock-controlled genes
Larval zebrafish (bdnf−/− n = 750, bdnf +/+ n = 750) were raised on a 12:12 LD cycle (light on: 6 a.m., light off: 6 p.m.). At 6 dpf, half larvae (bdnf−/− n = 180 and bdnf+/+ n = 180) were turned into DD and the second half were maintained in LD. It is conventional to divide the 24-h LD cycle in 24 Zeitgeber hours (ZT) and indicate time of lights on as ZT0 and time of lights off as ZT12. In DD the 24-h circadian cycle is divided in 24 circadian hours (CT) and it is conventional to use as reference point the phase that would normally (i.e., in LD 12:12) coincide with the onset of light and to call this CT0. After 2 days (8 dpf) larvae were sampled at five different time points (ZT/CT3, 9, 15, 21 and ZT/CT3 of the following day). For each time point, 15 larvae were sampled and pooled (n = 5 pooled samples per ZT/CT). Total RNA was then isolated using Trizol reagent (Thermo Fisher Scientific). Following the manufacturer’s instructions. The amount, quality and composition of isolated RNA were analysed by BioSpec-nano (Shimadzu). cDNA was synthesized from 1 μg of Dnase-treated RNA using iScript™ cDNA Synthesis Kit (Biorad) and quantitative PCR was carried out using SsoAdvanced Universal SYBR Green Supermix (Biorad) in triplicate on a CFX Connect Real-Time PCR Detection System (Biorad) instrument.
Gene-specific primers for per1b, per2, clock1a, clock2, cry1a, arntl1a, nfil3-5, nfil3-6, aanat2 and mtnr1aa are reported in Table S11. We verified the efficiency of the primers by constructing standard curves for all genes investigated. Moreover, the dissociation curve was used to confirm the specificity of the amplicon. The relative levels of each sample were calculated by the 2–ΔΔCT method (where CT is the cycle number at which the signal reaches the threshold of detection) (Livak and Schmittgen, 2001). As housekeeping genes we used ef1α and 18S, validated reference genes for zebrafish (Di Rosa et al., 2015). Primers for housekeeping genes are reported in Table S5. Nearly identical results were observed with both housekeeping genes. Each CT value used for these calculations is the mean of three replicates of the same reaction.
Data base analysis
RNA-seq data availability and Gene Ontology analysis
Next-generation sequencing data (RNA-seq and miRNA-seq) of bdnf+/+ and bdnf−/− zebrafish larvae at 24 and 48 hpf have been deposited in the NCBI Sequence Read Archive (SRA) under the project accession number: SUB8904035. Gene Ontology (GO) enrichment analysis of differentially expressed genes (DE-Gs) and miRNAs (DE-Ms) was computed using the cluster Profiler v3.3.6 package (Yu et al., 2012) with a p value cut-off of 0.05 and GO enrichment results were visualized using an R custom script. In order to simplify the enriched results, the similarity of GO terms has been computed using the simplify function of cluster Profiler package and highly similar GO terms (similarity cutoff of 0.6) were removed and kept only one representative and most informative term.
Locomotor analysis
Locomotor analysis of zebrafish larvae (n = 96 per strain) was performed using the DanioVision (Noldus Information Technology). DanioVision is equipped with an IR-sensitive camera, a temperature controller unit and a power supply to control light intensity. All the behavioural tests were performed between 10.00 and -12.00 am. The analysis was performed in a 96-well plate at 96 hpf and repeated three times. The wells were filled with 150 μl of E3 medium and introduced into the DanioVision observation chamber for video tracking. Locomotor activity was registered for 10 min in response to light stimulation, after 10 min of acclimatization in dark to reduce anxiety-like behaviour. The EthoVision XT Software (Noldus Information Technology) was used to analyse video tracks. The thresholds for movement detection were 0.2 mm/s (start velocity) and 0.1 mm/s (stop velocity).
Recording of Circadian Locomotor Activity
bdnf−/− and bdnf+/+ embryos were collected immediately after spawning and raised in E3 medium in a light-and temperature-controlled incubator. At 7 dpf larvae were placed in 96-well plate (n = 32 per genotype, 1 larva per well, 0.5 mL of E3 medium) in the observation chamber of the DanioVision tracking system (Noldus Information Technology). Larvae locomotor activity was tracked for 5-7 consecutive days and then analysed by Ethovision v.11 software (Noldus Information Technology). Three biological repeats were conducted. The IR-sensitive camera was set to 25 frames per second. Locomotor activity of each larva was calculated as the total distance moved during a 1 min time window. A minimal distance moved of 0.2 mm was used. Larvae were kept under 12:12 LD cycles (lights on at 06:00, lights of 18:00) or under DD conditions. For light sources, an array of LED strips was used and irradiances were set at 0.17 W/m2.
Rescue of the circadian locomotor activity
bdnf−/− embryos were collected immediately after spawning, raised in E3 medium and kept under 12:12 LD cycles. At 9 dpf a group of bdnf−/− larvae (n = 32) were treated with 10 μM of 7,8-DHF and placed in a 96-well plate in the observation chamber of the DanioVision tracking system. In parallel control groups of bdnf−/− and bdnf+/+ larvae (n = 32 each genotype) were bred in 0.1% dimethyl sulfoxide (DMSO) in E3 solution (v/v) under the same light conditions. Larval locomotor activity was tracked for 3 consecutive days under 12:12 LD cycles (lights on at 06:00, lights off 18:00) or under DD conditions, and analysed as described above.
Quantification and statistical analysis
All the results were expressed as means ± SEM. Data were analysed by parametric and non-parametric tests to determine significant differences using the software Prism 5 (GraphPad Inc.) or R 3.6.1 (https://www.R-project.org/). p values <0.05 were considered statistically significant. Daily and circadian expression gene expression profiles (periodicity and phase of oscillation) were evaluated by the RAIN algorithm (Rhythmicity Analysis Incorporating Non-parametric) (Thaben and Westermark, 2014). The presence of daily and circadian periodicity in the activity rhythms was determined by means of Cosinor (Refinetti et al., 2007).
Acknowledgments
Salvatore D’Aniello is grateful to Josè Luis Gomez-Skarmeta for his contagious enthusiasm for science and the fruitful discussions on an embryonic version of the project. This work was funded by a FIRB grant (RBFR12QW4I) to Salvatore D’Aniello and a PON grant (A3_00239) to Paolo Sordino from the Italian Ministry of Education, University and Research (MIUR). Ylenia D’Agostino was supported by a PhD fellowship from the RBFR12QW4I grant. Cristiano Bertolucci and Elena Frigato were supported by University of Ferrara Research Grants (FAR2019, FAR2020, FAR2021, and FIR2020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceptualization, C.B., S.D.A.; Data Curation, Y.D.A., E.F., T.M.R.N., M.C., and L.C.; Formal Analysis, Y.D.A., E.F., T.M.R.N., M.C., L.C., C.B., and S.D.A.; Funding Acquisition, P.S., C.B., S.D.A.; Investigation, Y.D.A., E.F., T.M.R.N., M.T., F.F., Lu.C, M.C., P.S., L.C., C.B., S.D.A.; Project Administration, S.D.A.; Resources, P.S., C.B., S.D.A.; Supervision, C.B. and S.D.A.; Writing – Original Draft, Y.D.A., C.B., S.D.A.; Writing – Review & Editing, Y.D.A., E.F., T.M.R.N., M.T., F.F., Lu.C, M.C., P.S., L.C., C.B., and S.D.A.
Declaration of interests
The authors declare no competing interests.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104054.
Contributor Information
Cristiano Bertolucci, Email: bru@unife.it.
Salvatore D’Aniello, Email: salvatore.daniello@szn.it.
Supplemental information
References
- Acheson A., Lindsay R.M. Non target-derived roles of the neurotrophins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1996;35:417–422. doi: 10.1098/rstb.1996.0037. [DOI] [PubMed] [Google Scholar]
- Anders C., Niewoehner O., Jinek M. In vitro reconstitution and crystallization of Cas9 endonuclease bound to a Guide RNA and a DNA target. Methods Enzymol. 2015;558:515–537. doi: 10.1016/bs.mie.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq - a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baquet Z.C., Bickford P.C., Jones K.R. Brain-derived neurotrophic factor is required for the establishment of the proper number of dopaminergic neurons in the substantia nigra pars compacta. J. Neurosci. 2005;25:6251–6259. doi: 10.1523/JNEUROSCI.4601-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M. The Trk family of neurotrophin receptors. J. Neurobiol. 1994;25:1386–1403. doi: 10.1002/neu.480251107. [DOI] [PubMed] [Google Scholar]
- Barde Y.A., Edgar D., Thoenen H. Purification of a new neurotrophic factor from mammalian brain. EMBO J. 1982;1:549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekinschtein P., Cammarota M., Medina J.H. BDNF and memory processing. Neuropharmacology. 2014;76 Pt:677–683. doi: 10.1016/j.neuropharm.2013.04.024. [DOI] [PubMed] [Google Scholar]
- Ben-Moshe Z., Foulkes N.S., Gothilf Y. Functional development of the circadian clock in the zebrafish pineal gland. Biomed. Res. Int. 2014;2014:235781. doi: 10.1155/2014/235781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Moshe Livne Z., Alon S., Vallone D., Bayleyen Y., Tovin A., Shainer I., Nisembaum L.G., Aviram I., Smadja-Storz S., Fuentes M., et al. Genetically blocking the zebrafish pineal clock affects circadian behavior. PLoS Genet. 2016;12:e1006445. doi: 10.1371/journal.pgen.1006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder D.K., Scharfman H.E. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco A.M., Bertucci J.I., Hatef A., Unniappan S. Feeding and food availability modulate brain-derived neurotrophic factor, an orexigen with metabolic roles in zebrafish. Sci. Rep. 2020;10:10727. doi: 10.1038/s41598-020-67535-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss T.V., Collingridge G.L. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Cacialli P., Gueguen M.M., Coumailleau P., D'Angelo L., Kah O., Lucini C., Pellegrini E. BDNF expression in larval and adult zebrafish brain: distribution and cell identification. PLoS One. 2016;11:e0158057. doi: 10.1371/journal.pone.0158057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.Y., Papp J.W., Varlamova O., Dziema H., Russell B., Curfman J.P., Nakazawa T., Shimizu K., Okamura H., Impey S., Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C., Brambilla R., Thomas K.L. A simple role for BDNF in learning and memory? Front Mol. Neurosci. 2010;3:1. doi: 10.3389/neuro.02.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino Y., D’Aniello S. Molecular basis, applications and challenges of CRISPR/Cas9: a continuously evolving tool for genome editing. Brief. Funct. Genomics. 2017;6:211–216. doi: 10.1093/bfgp/elw038. [DOI] [PubMed] [Google Scholar]
- D'Agostino Y., Locascio A., Ristoratore F., Sordino P., Spagnuolo A., Borra M., D'Aniello S. A rapid and cheap methodology for CRISPR/Cas9 zebrafish mutant screening. Mol. Biotechnol. 2016;58:73–78. doi: 10.1007/s12033-015-9905-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly C., Shine L., Heffernan T., Deeti S., Reynolds A.L., O'Connor J.J., Dillon E.T., Duffy D.J., Kolch W., Cagney G., Kennedy B.N. A brain-derived neurotrophic factor mimetic is sufficient to restore cone photoreceptor visual function in an inherited blindness model. Sci. Rep. 2017;7:11320. doi: 10.1038/s41598-017-11513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice E., Porreca I., Alleva E., De Girolamo P., Ambrosino C., Ciriaco E., Germanà A., Sordino P. Localization of BDNF expression in the developing brain of zebrafish. J. Anat. 2014;224:564–574. doi: 10.1111/joa.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rosa V., Frigato E., Lopez-Olmeda J.F., Sanchez-Vazquez F.J., Bertolucci C. The light wavelength Affects the ontogeny of clock gene expression and activity rhythms in zebrafish larvae. PLoS One. 2015;10:e0132235. doi: 10.1371/journal.pone.0132235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Mark C., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson J.T., Conover J.C., Borday V., Champagnat J., Barbacid M., Yancopoulos G., Katz D.M. Mice lacking brain-derived neurotrophic factor exhibit visceral sensory neuron losses distinct from mice lacking NT4 and display a severe developmental deficit in control of breathing. J. Neurosci. 1996;16:5361–5371. doi: 10.1523/JNEUROSCI.16-17-05361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernfors P., Lee K.F., Jaenisch R. Mice lacking brain-derived neurotrophic factor develop with sensory deficits. Nature. 1994;368:147–150. doi: 10.1038/368147a0. [DOI] [PubMed] [Google Scholar]
- Ernfors P., Lee K.F., Jaenisch R. Target derived and putative local actions of neurotrophins in the peripheral nervous system. Prog. Brain Res. 1994;103:43–54. doi: 10.1016/s0079-6123(08)61125-5. [DOI] [PubMed] [Google Scholar]
- Falcón J., Galarneau K.M., Weller J.L., Ron B., Chen G., Coon S.L., Klein D.C. Regulation of arylalkylamine N-acetyltransferase-2 (AANAT2, EC 2.3.1.87) in the fish pineal organ: evidence for a role of proteasomal proteolysis. Endocrinology. 2001;142:1804–1813. doi: 10.1210/endo.142.5.8129. [DOI] [PubMed] [Google Scholar]
- Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi A.V., Mosser E.A., Oikonomou G., Prober D.A. Melatonin is required for the circadian regulation of sleep. Neuron. 2015;85:1193–1199. doi: 10.1016/j.neuron.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.F., Sakamoto K., Obrietan K. MicroRNAs: a potential interface between the circadian clock and human health. Genome Med. 2011;3:10. doi: 10.1186/gm224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich G., Pagtakhan C.J. Both 5' and 3' flanks regulate Zebrafish brain-derived neurotrophic factor gene expression. BMC Neurosci. 2004;5:19. doi: 10.1186/1471-2202-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E.J., Reichardt L.F. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Ibáñez C.F., Simi A. p75 neurotrophin receptor signaling in nervous system injury and degeneration: paradox and opportunity. Trends Neurosci. 2012;35:431–440. doi: 10.1016/j.tins.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Idda M.L., Bertolucci C., Vallone D., Gothilf Y., Sanchez-Vazquez F.J., Foulkes N.S. Circadian clocks: lessons from fish. Prog. Brain Res. 2012;199:41–57. doi: 10.1016/B978-0-444-59427-3.00003-4. [DOI] [PubMed] [Google Scholar]
- Jao L.E., Wente S.R., Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.R., Fariñas I., Backus C., Reichardt L.F. Targeted disruption of the BDNF gene perturbs brain and sensory neuron development but not motor neuron development. Cell. 1994;76:989–999. doi: 10.1016/0092-8674(94)90377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff A.V., Gebhardt M., Stewart A.M., Cachat J.M., Brimmer M., Chawla J.S., Craddock C., Kyzar E.J., Roth A., Landsman S., et al. Zebrafish Neuroscience Research Consortium Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish. 2013;10:70–86. doi: 10.1089/zeb.2012.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff A.V., Stewart A.M., Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014;35:63–75. doi: 10.1016/j.tips.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.R., Miller F.D. Neurotrophin signal transduction in the nervous system. Curr. Opin. Neurobiol. 2000;10:381–391. doi: 10.1016/s0959-4388(00)00092-1. [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Kondo M., Gray L.J., Pelka G.J., Christodoulou J., Tam P.P.L., Hannan A.J. Environmental enrichment ameliorates a motor coordination deficit in a mouse model of Rett syndrome--Mecp2 gene dosage effects and BDNF expression. Eur. J. Neurosci. 2008;27:3342–3350. doi: 10.1111/j.1460-9568.2008.06305.x. [DOI] [PubMed] [Google Scholar]
- Leibrock J., Lottspeich F., Hohn A., Hofer M., Hengerer B., Masiakowski P.O., Thoenen H., Barde Y.A. Molecular cloning and expression of brain-derived neurotrophic factor. Nature. 1989;341:149–152. doi: 10.1038/341149a0. [DOI] [PubMed] [Google Scholar]
- Liang F.Q., Walline R., Earnest D.J. Circadian rhythm of brain-derived neurotrophic factor in the rat suprachiasmatic nucleus. Neurosci. Lett. 1998;242:89–92. doi: 10.1016/s0304-3940(98)00062-7. [DOI] [PubMed] [Google Scholar]
- Liang F.Q., Allen G., Earnest D.J. Role of brain-derived neurotrophic factor in the circadian regulation of the suprachiasmatic pacemaker by light. J. Neurosci. 2000;20:2978–2987. doi: 10.1523/JNEUROSCI.20-08-02978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., Wang Y., Sun Z. The relationships between stress, mental disorders, and epigenetic regulation of BDNF. Int. J. Mol. Sci. 2020;21:1375. doi: 10.3390/ijms21041375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbiato E., Frigato E., Dinarello A., Maradonna F., Facchinello N., Argenton F., Carnevali O., Dalla Valle L., Bertolucci C. Feeding entrainment of the zebrafish circadian clock is regulated by the glucocorticoid receptor. Cells. 2019;8:1342. doi: 10.3390/cells8111342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel R., Clijsters L., Agami R. The miRNA-192/194 cluster regulates the Period gene family and the circadian clock. FEBS J. 2009;276:5447–5455. doi: 10.1111/j.1742-4658.2009.07229.x. [DOI] [PubMed] [Google Scholar]
- Nittoli V., Sepe R.M., Coppola U., D'Agostino Y., De Felice E., Palladino A., Vassalli Q.A., Locascio A., Ristoratore F., Spagnuolo A., et al. A comprehensive analysis of neurotrophins and neurotrophin tyrosine kinase receptors expression during development of zebrafish. J. Comp. Neurol. 2018;526:1057–1072. doi: 10.1002/cne.24391. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Dahm R. First Edition. Vol. 82. Oxford University Press; 2012. Zebrafish: a practical approach; p. 79. (Genetical Research). [Google Scholar]
- Oikonomou G., Prober D.A. Attacking sleep from a new angle: contributions from zebrafish. Curr. Opin. Neurobiol. 2017;44:80–88. doi: 10.1016/j.conb.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro M., Tauber E. The role of microRNAs (miRNA) in circadian rhythmicity. J. Genetics. 2008;87:505–511. doi: 10.1007/s12041-008-0073-8. [DOI] [PubMed] [Google Scholar]
- Pencea V., Bingaman K.D., Wiegand S.J., Luskin M.B. Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J. Neurosci. 2001;21:6706–6717. doi: 10.1523/JNEUROSCI.21-17-06706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R., Lissen G.C., Halberg F. Procedures for numerical analysis of circadian rhythms. Biol. Rhythm Res. 2007;38:275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt L.F. Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios M. BDNF and the central control of feeding: accidental bystander or essential player? Trends Neurosci. 2013;36:83–90. doi: 10.1016/j.tins.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A., Kontarakis Z., Gerri C., Nolte H., Hölper S., Krüger M., Stainier D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Saha S., Singh K.M., Gupta B.B.P. Melatonin synthesis and clock gene regulation in the pineal organ of teleost fish compared to mammals: similarities and differences. Gen. Comp. Endocrinol. 2019;279:27–34. doi: 10.1016/j.ygcen.2018.07.010. [DOI] [PubMed] [Google Scholar]
- Saus E., Soria V., Escaramís G., Vivarelli F., Crespo J.M., Kagerbauer B., Menchón J.M., Urretavizcaya M., Gratacòs M., Estivill X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum. Mol. Genet. 2010;19:4017–4025. doi: 10.1093/hmg/ddq316. [DOI] [PubMed] [Google Scholar]
- Segal R.A. Selectivity in neurotrophin signaling: theme and variations. Annu. Rev. Neurosci. 2003;26:299–330. doi: 10.1146/annurev.neuro.26.041002.131421. [DOI] [PubMed] [Google Scholar]
- Shi L., Ko M.L., Ko G.Y.P. Rhythmic expression of MicroRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J. Biol. Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppet D., Escandon E., Maragos J., Middlemas D.S., Reid S.W., Blair J., Burton L.E., Stanton B.R., Kaplan D.R., Hunter T., et al. The neurotrophic factors brain-derived neurotrophic factor and neurotrophin-3 are ligands for the trkB tyrosine kinase receptor. Cell. 1991;65:895–903. doi: 10.1016/0092-8674(91)90396-g. [DOI] [PubMed] [Google Scholar]
- Tettamanti G., Cattaneo A.G., Gornati R., de Eguileor M., Bernardini G., Binelli G. Phylogenesis of brain-derived neurotrophic factor (BDNF) in vertebrates. Gene. 2010;450:85–93. doi: 10.1016/j.gene.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Thaben P.F., Westermark P.O. Detecting rhythms in time series with RAIN. J. Biol. Rhythms. 2014;29:391–400. doi: 10.1177/0748730414553029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lv K., Zhao M., Chen H., Ji G., Zhang Y., Wang T., Cao H., Li Y., Qu L. Analysis of miRNA expression profiles in the liver of ClockΔ19 mutant mice. PeerJ. 2019;7:e8119. doi: 10.7717/peerj.8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. University of Oregon Press; 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- Wurzelmann M., Romeika J., Sun D. Therapeutic potential of brain-derived neurotrophic factor (BDNF) and a small molecular mimics of BDNF for traumatic brain injury. Neural Regen. Res. 2017;12:7–12. doi: 10.4103/1673-5374.198964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L.T., Luo L., Wu Y.J., Liu B.B., Liu X.L., Geng D., Liu Q. Circadian variations in behaviors, BDNF and cell proliferation in depressive mice. Metab. Brain Dis. 2015;30:1495–1503. doi: 10.1007/s11011-015-9710-0. [DOI] [PubMed] [Google Scholar]
- Yoo S.H., Kojima S., Shimomura K., Koike N., Buhr E.D., Furukawa T., Ko C.H., Gloston G., Ayoub C., Nohara K., et al. Period2 3’-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. U S A. 2017;114:E8855–E8864. doi: 10.1073/pnas.1706611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L., Han Y., He Q. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Miller C., Miraglia L.J., Romero A., Mure L.S., Panda S., Kay S.A. A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2020454118. e2020454118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Next-generation sequencing data (RNA-seq and miRNA-seq) of bdnf+/+ and bdnf−/− zebrafish larvae at 24 and 48 hpf have been deposited in the NCBI Sequence Read Archive (SRA): SUB8904035.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.