Abstract
Most therapeutic proteins are glycosylated with N-glycans and/or O-glycans. N-glycans on therapeutic proteins have been extensively studied for their control strategy and impact on drug product quality. However, knowledge of O-glycosylation in therapeutic protein production and its impact on product quality remains elusive. To address this gap, we generated an O-glycoengineered Chinese Hamster Ovary (CHO) cell line platform to modulate O-glycosylation of therapeutic proteins and investigated the impact of O-glycans on the physicochemical and biological properties of etanercept. Our results demonstrate that this CHO cell line platform produces controlled O-glycosylation profiles containing either truncated O-glycans (sialylTn and/or Tn), or sialylCore 3 alone, or sialylCore 1 with sialylTn or sialylCore 3 O-glycans on endogenous and recombinant proteins. Moreover, the platform demonstrated exclusive modulation of O-glycosylation without affecting N-glycosylation. Importantly, certain O-glycans on etanercept enhanced tumor necrosis factor-α binding affinity and consequent potency. This is the first report that describes the systematic establishment of an O-glycoengineered CHO cell line platform with direct evidence that supports the applicability of the platform in the production of engineered proteins with desired O-glycans. This platform is valuable for identifying O-glycosylation as a critical quality attribute of biotherapeutics using the quality by design principle.
Keywords: glycosylation, etanercept, cell line engineering, CHO cells, biopharmaceuticals, O-glycans, therapeutic fusion proteins
Graphical abstract

The impact of serine or threonine linked glycans (O-glycans) on the safety and efficacy of therapeutic proteins remains unclear. Here, an O-glycoengineered CHO cell platform was developed and confirmed by producing six different etanercept O-glycovariants. One etanercept O-glycovariant carrying SialylTn and sialylCore 1 possess increased binding affinity and enhanced potency.
Introduction
Protein glycosylation is a post-translational enzymatic process that modifies certain amino acid side chains through covalent linkage of saccharides. The glycosylation process occurs in cellular compartments of the endoplasmic reticulum and Golgi apparatus during protein translocation to the plasma membrane or extracellular space.1 There are two major glycosylation types: N-linked glycans (N-glycans) on asparagine (N) residues, and O-linked glycans (O-glycans) on serine, threonine, and tyrosine residues. N-glycosylation is well known to impact the safety and efficacy of therapeutic proteins.2 For example, the core fucosylation status of the Asn297-linked glycans within the CH2 domain of IgG1 monoclonal antibodies modulates antibody effector functions, such as antibody-dependent cellular cytotoxicity.2 To improve the control of N-glycosylation heterogeneity during therapeutic protein production, Chinese Hamster Ovary (CHO) cell lines have been genetically engineered to produce specific N-glycans,3,4 which is part of pharmaceutical quality by design practice.5,6 In contrast with N-glycans, the impact of O-glycans on therapeutic protein safety and efficacy requires more evaluation with only a few studies implicating O-glycans to modulation of immunogenicity.7, 8, 9, 10
CHO cells are the most common cell substrate for manufacturing recombinant therapeutic proteins.11 Although CHO cells are unique for their capability to produce therapeutic proteins with “human-like” glycans, the capability to produce different glycan structures is limited owing to silenced transcription of numerous genes encoding enzymes within the glycosylation pathways.12 There are at least eight human O-glycan core structures (cores 1–8), and the majority of O-glycans are built from Core 1 (Galβ1-3GalNAcα-), Core 2 (GlcNAc β1-6[Galβ1-3]GalNAcα-), Core 3 (GlcNAcβ1-3GalNAcα-), and Core 4 (GlcNAc β1-6[GlcNAcβ1-3]GalNAcα-) structures (Figure S1A).13 Particularly, CHO cells are unable to produce the Core 2, extended Core 1 (GlcNAcβ1-3Galβ1-3GalNAcα-), or Core 3 complex O-glycans, but exclusively produce mono-sialylated Core 1 (SAα2-3Galβ1-3GalNAcα-, SAα2-6 [Galβ1-3]GalNAcα-) and di-sialylated Core 1 (SAα2-3Galβ1-3[SAα2-6]GalNAcα-) structures (Figure S1A).14 Studies have demonstrated that O-glycans on endogenous glycoproteins play many critical roles in biological systems, such as signal transduction, cell adhesion, development, and immunity.15,16 Additionally, altered O-linked glycan structures, such as the Tn antigen (GalNAcα-Ser/Thr) and its sialylated version, sialylTn (STn) antigen (SAα2-6GalNAcα-Ser/Thr), have been identified in human cancers.15 Here we have established clonally derived O-glycoengineered CHO cell lines that have distinct O-glycosylation machinery to produce glycoproteins with the Tn antigen with or without STn antigen, sialylCore 3 structures alone, or sialylCore 1 structures with STn antigen or sialylCore 3 structures.
Etanercept, a tumor necrosis factor alpha (TNF-α) inhibitor, is an U.S. Food and Drug Administration-approved biotherapeutic to treat autoimmune diseases that are caused by an overactive immune responses, including rheumatoid arthritis, juvenile idiopathic arthritis, and psoriatic arthritis, plaque psoriasis, and ankylosing spondylitis by inhibiting TNF-α. Etanercept is a dimeric fusion protein that contains the extracellular ligand binding portion of human p75 TNF receptor 2 linked to the Fc portion of human IgG1 with 3 N-linked and 13 O-linked glycosylation sites.17 Previous structural, physicochemical, and biological characterizations of etanercept and its biosimilar etanercept-ykto in combination with data from clinical studies suggest that increasing the sialylated O-glycans content on etanercept may improve safety.7, 8, 9, 10 Therefore, etanercept was chosen as a proof-of-principle therapeutic protein to evaluate the O-glycoengineered CHO cell line platform. Here, using an O-glycoengineered CHO cell line platform, etanercept was produced with different O-glycan profiles (O-glycovariants) and TNF-α binding, potency, and stability were evaluated for each variant as compared with the reference product, etanercept. The data show that changes in O-glycans on etanercept altered its isoelectric point, enhanced TNF-α binding and increased the potency without affecting the protein stability under oxidative stress conditions.
Results
Establishment of an O-glycoengineered CHO cell line platform
Cosmc, Core 1 β3-galactosyltransferase-specific molecular chaperone, assists the correct folding of Core 1 glycoprotein-α-N-acetylgalactosamine β1,3-galactosyltransferase (core 1 β3-galactosyltransferase, T synthase), which transfers galactose from UDP-Gal to the Tn antigen to form the Core 1 structure (Figure S1A).18 To generate Cosmc knockout (KO) CHO cells that produce the Tn antigen (Figure S1B), CRISPR/Cas9 technology was used to mutate Cosmc, also known as C1GalT1C1. In comparison with the wild-type (WT) cell line, Cosmc-KO cells had undetectable levels of the Cosmc protein (Figure 1A) and a loss in T synthase activity (Figure 1B), which suggests the Cosmc protein was dysfunctional. Consistent with a previous report,19 KO of Cosmc in CHO cells correlated with increased levels of glycoproteins with the Tn antigen, but not the STn antigen (Figure 1C). These data confirm the generation of a stable Cosmc-KO CHO cell line that produces the Tn antigen on endogenous proteins.
Figure 1.
Generation of an O-glycoengineered CHO K1 cell line platform
(A) Representative immunoblot of Cosmc, ST6 N-acetylgalactosaminide α2,6-sialyltransferase 1 (ST6GalNAc1), and Core 3 β1,3 -N-acetylglucosaminyltransferase (β3GnT-6, C3GnT) using proteins harvested from WT CHO cells with ST6GalNAc1 (WS) or C3GnT (WC), CosmcKO CHO cells, and Cosmc-KO cells with ST6GalNAc1 (KS) or C3GnT (KC). (B) Relative fluorescence unit measurements of T synthase activity in CHO cell lysates from WT, KO, WS, KS, WC, and KC CHO cell lines. Dots represent biological replicates. (One-way ANOVA, n = 6, brackets indicate significant differences between groups according to a Tukey's multiple comparison test, ∗p < 0.05). (C) Representative T neoantigen (Tn antigen) and STn antigen immunoblots using cell lysates. (D) Representative histograms of Tn and STn antigens, and C3GnT using the different O-glycoengineered CHO K1 cells.
α-N-acetylgalactosaminyl-O-glycoprotein α2,6-sialyltransferase 1 (ST6GalNAc1) and α-N-acetylgalactosaminyl-O-glycoprotein β1,3-N-acetylglucosaminyltransferase (core 3 β1,3-N-Acetylglucosaminyltransferase, Core 3 GnT, C3GnT, or β1,3-N-Acetylglucosaminyltransferase 6, β3GnT6) are two enzymes in the O-glycosylation pathway that mediate the synthesis of STn antigen and Core 3 O-glycan structures, respectively (Figure S1A).15 The STn antigen on endogenous proteins has been reported to have an essential role in the ability of cancer cells to evade immune cells, and Core 3 glycans on endogenous proteins play critical tumor suppression role.20 A thorough understanding of the effects of O-glycosylation on product quality requires further study. Therefore, we set out to generate CHO cells that produce O-glycans on experimental therapeutic proteins to identify the structure/function relationships between O-glycan types and product quality attributes. To generate CHO cells that produce the STn antigen and Core 3 O-glycans, WT and Cosmc-KO cells were engineered to stably express ST6GalNAc1 or C3GnT (Figure S1B). Using immunoblotting, the overexpression of ST6GalNAc1 or C3GnT in WT and Cosmc-KO cells was confirmed (Figure 1A) and the loss of T synthase activity was identified to be exclusive in the Cosmc-KO lines expressing ST6GalNAc1 (KS) or C3GnT (KC) to confirm the sustained depletion of Cosmc in these cell lines (Figure 1B). T synthase activity was lower in WT cells expressing ST6GALNAC1 (WS) or C3GnT (WC), which could be due to two possible mechanisms: (1) exogenous expression of ST6GalNAC1 or C3GnT downregulated the expression level endogenous T synthase protein and activity; or (2) expression of the endogenous Tsynthase in those ST6GalNAC1 and C3GnT-expressing clones was lower than the WT CHO cells, which may be a heterogeneous population of CHO cells with variable T synthase expression and activity.
To demonstrate that ST6GalNAc1 was functional, Tn and STn antigen immunoblotting was performed and revealed that levels of endogenous proteins containing STn antigen increased in both WT and Cosmc CHO cells expressing ST6GalNAc1, and only Cosmc-KO CHO cells expressing ST6GalNAc1 (KS) produced both Tn and STn antigen on endogenous proteins (Figures 1C and S2A). In contrast with ST6GalNAc1, monitoring the activity of C3GnT directly through immunoblotting is technically limited and more advanced methods were required to confirm the presence of Core 3 structures. However, the loss of the Tn antigen in Cosmc-KO CHO cells expressing C3GnT (KC) provided indirect evidence that Core 3 O-glycan structures were synthesized by the cells (Figure 1C). To ensure that all the clonally derived cell lines maintained a single normally distributed population of cells, fluorescence-activated cell sorting (FACS) analysis was performed using the antibodies for the Tn antigen, STn antigen, and C3GnT (Figure 1D) and confirmed the anticipated staining profiles. Interestingly, there was no significant difference in the mean fluorescence intensity of anti-Tn staining between KO and KS cells (Figures 1D and S2A), which could be most likely due to the saturated staining of those cells with an excess amount of anti-Tn antibody, and/or possible recognition of the “Tn” moiety within STn by the anti-Tn antibody (mouse IgM). The modulation of Cosmc, ST6GalNAc 1, and C3GnT did not significantly impact cell growth or proliferation (Figure S2B). Collectively, these data demonstrate the establishment of an O-glycoengineered CHO cell line platform that was constituted by the following six cell lines: WT CHO cells, Cosmc-KO CHO cells, CHO cells overexpressing ST6GalNAc1 (WS) or C3GnT (WC), and Cosmc-KO CHO cells overexpressing ST6GalNAc1 (KS) or C3GnT (KC).
N- and O-glycan characterization of the CHO endogenous proteins using mass spectrometry
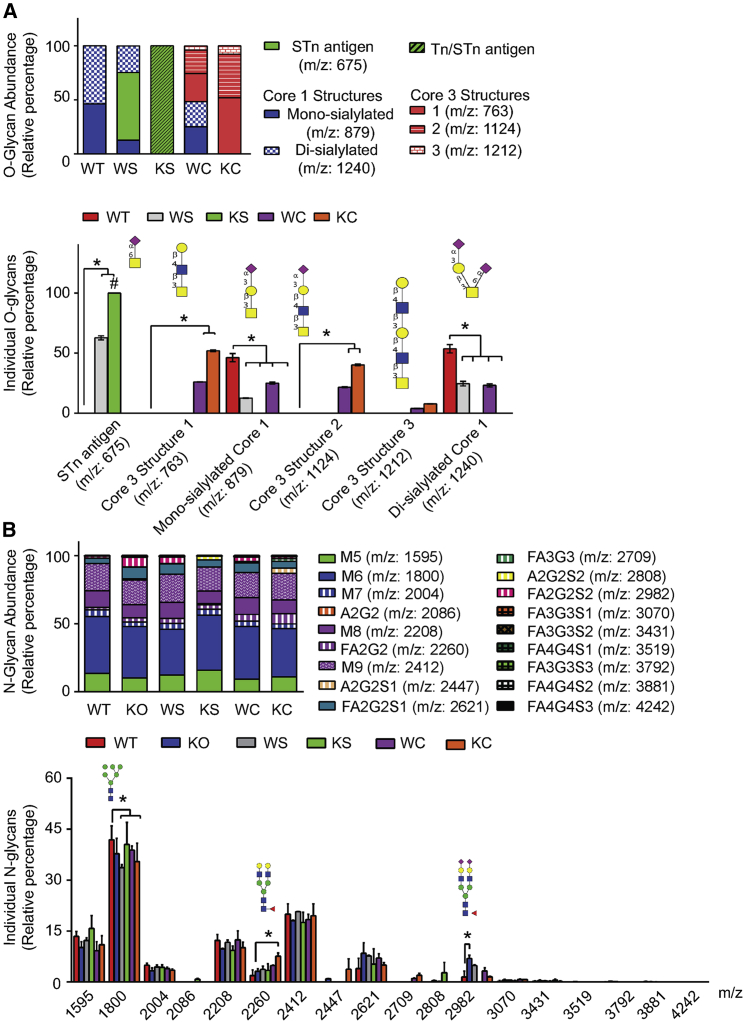
To characterize the O- and N-glycan profiles of the CHO cell line platform, MALDI-TOF/TOF- mass spectrometry (MS) was performed using lysates from the CHO cell line platform to structurally and quantitatively analyze permethylated O- and N-glycans released from endogenous proteins (Figure 2). Each of these O-glycoengineered CHO cell lines had structurally and quantitatively different O-glycan profiles when compared with the WT cell line or each other (Figures 2A, S3, and Table S2). WT cells exclusively produced mono- and di-sialylated Core 1 structures as the major O-glycans, and Cosmc-KO cells did not produce any STn antigen, Core 1, or Core 3 structures (Table S2). While the immunoblotting data (Figure 1C) confirmed that Cosmc-KO cells expressed Tn antigens, they were not detected in the MS analysis possibly owing to the sample loss during the chloroform extraction of permethylated glycans (Figure S3). Unlike other larger glycans, the permethylated Tn may not have been hydrophobic enough to be extracted into chloroform. The WT CHO cells expressing ST6GalNAc1 (WS) produced STn in addition to di-sialylCore 1 structures. Interestingly, the WS cells did not produce mono-sialylCore 1 structures as seen in the WT cells, indicating that sialyltransferases in the WS cells sufficiently sialylated Core 1 structures. The STn structure was observed only in Cosmc-KO cells expressing ST6GalNAc1 (Figure S3) and, as expected, MS did not detect the presence of the immunoblot-confirmed Tn antigen (Figure 1C). Therefore, the relative abundance of STn and Tn antigens in WS cells could not be accurately determined. WT cells expressing C3GnT (WC) produced Core 3 glycans in addition to sialylCore 1 O-glycans at a decreased level as compared to the WT cell line (Figure 2A). In contrast, Cosmc-KO cells expressing C3GnT (KC) exclusively produced Core 3-based structures with and without sialylation (Figure 2A). Collectively, these data confirm the establishment of an O-glycoengineered CHO cell line production platform that can modulate Tn antigen, STn antigen, sialylCore 1 structures, and sialylCore 3-based structures.
Figure 2.
N- and O-linked glycans of CHO cells
(A and B) WT and glycoengineered (KO, WS, KS, WC, and KC) CHO cells were analyzed for changes in the relative quantity of (A) O-glycan and (B) N- glycan structures summarized from the data of MS analysis. Bars represent the mean percentage ±standard deviation. (Two-way ANOVAs, n = 3, Tukey multiply comparison to WT cells, ∗p < 0.05). # indicates sample with Tn and STn antigens.
To determine if O-glycoengineering might impact N-glycosylation, the N-glycans on endogenous proteins of the CHO cell lysates were also analyzed by MS (Figures 2B, S4, and Table S3). Among the 18 N-glycans identified in this CHO cell line platform, high mannose type N-glycans were the most abundant (Man6 [41.8%–33.7%], Man9 [20.7%–17.5%], Man8 [12.4%–9.3%], and Man5 ([15.7%–9.2%]) and the major complex N-glycans were identified to be FA2G2S1 (8.4%–4.0%) and FA2G2 (7.6%–1.9%) for all cell lines (Figures 2B, S4, and Table S3). Overall, the N-glycan profiles among these cell lines were similar and only two minor complex N-glycans had cell-specific changes in the relative abundance. As compared with the WT cell line, FA2G2 increased in the Cosmc-KO cells expressing C3GnT (KC) cell line, and FA2G2S2 increased in the Cosmc-KO CHO cell line (KO) (Figures 2B, S4, and Table S3). Importantly, the non-human αGal or Neu5Gc glycans were not detected in the glycoproteins from either WT CHO cells or the O-glycoengineered CHO cells using MS analysis. Collectively, these data suggested that O-glycoengineering in CHO cells had a minor impact on N-glycosylation and the changes in N-glycan levels were cell-type specific.
Generation of O-glycoengineered CHO cell line platform expressing etanercept
To identify potential clones that stably express etanercept, the etanercept gene sequence was cloned into a reporter vector that used an internal ribosomal entry site to drive the expression of green fluorescent protein (GFP). Stable cell lines were generated by enriching the GFP-positive population using FACS, followed by one round of limiting dilution. Flow cytometry was performed to verify a single population of cells expressing GFP and to monitor the distribution of GFP expression in O-glycoengineered CHO cells (Figure 3A). To confirm that the insertion of the etanercept gene did not disrupt the glycosylation pathway modulation in the cells, the Tn and STn antigen levels were measured by flow cytometry. Neither Tn nor STn antigen on endogenous proteins increased in WT expressing etanercept (EW) cells; however, the Tn antigen, but not the STn antigen, levels increased in KO expressing etanercept (EK) cells (Figure 3A). Similar to WS cells, the STn antigen levels in WS-expressing etanercept cells (EWS) increased without an increase in Tn antigen levels. Both the Tn and STn antigen levels increased in KS-expressing etanercept (EKS) cells. As previously discussed, identifying the presence of Core 3 O-glycans was limited and technically challenging; thus, additional studies are required to confirm the WT and Cosmc-KO cells expressing etanercept and C3GnT retained Core 3-based structures.
Figure 3.
Generation of etanercept-expressing O-glycoengineered CHO cell platform
(A) Representative histograms of etanercept using GFP as a surrogate marker, Tn antigen, and STn antigen using flow cytometry and stably expressing etanercept cells from the O-glycoengineered CHO cell platform (WT, KO, WS, KS, WC, and KC). (B) Representative immunoblots of an IgG1-Fc protein, potentially etanercept, in 10 μL media on cultivation day 7 with and without fed media.
To confirm that etanercept was expressed and secreted in each O-glycoengineered CHO cell line, culture vessels were sampled at days 5 and 7 during production and subjected to anti-human Fc immunoblotting. Media from each O-glycoengineered cell line demonstrated the presence of an IgG1 Fc-containing protein with a molecular weight comparable with etanercept reference material (Etan-Ref) (Figure 3B). As expected, a modest molecular weight shift owing to the formation of only the Tn antigen occurred in etanercept secreted from the EK cell line. These data demonstrated that six stable etanercept-expressing CHO cell lines with modified O-glycosylation pathways were successfully developed.
Production of O-glycovariants of etanercept using the CHO cell line platform
After the production of each etanercept glycovariant from the corresponding cell line, the harvested media with etanercept was subject to sequential chromatography steps: protein A, anion exchange, and size exclusion chromatography. The purified etanercept O-glycovariants were then analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analytical size exchange chromatography (ultra-high performance liquid chromatography-size exclusion chromatography) to confirm size and relative purity. Coomassie-stained SDS-PAGE gels under reducing and non-reducing conditions demonstrated the presence of single bands at approximately 75 kDa and 150 kDa, respectively, that were comparable with the Etan-Ref (Figure 4A). These data support the etanercept O-glycovariants were forming disulfide bond-dependent dimers. Size exclusion high-performance liquid chromatography revealed the purity of the main peak ranged between 90% and 99% for all etanercept O-glycovariants with overall fewer aggregates than reference product, but higher fragment levels (Table 1).
Figure 4.
Purity and identity of etanercept purified from CHO cell harvest media
(A) Coomassie-stained SDS-PAGE gels using etanercept reference material (Etan-Ref) and purified proteins from each O-glycoengineered cell line. WT cells were expected to produce etanercept with sialylCore 1 structures (EW-Etan-C1), while Cosmc-KO cells would only produce etanercept with Tn antigen (EK-Etan-Tn). Overexpressing ST6GalNAc1 in WT and Cosmc-KO cells should produce both STn antigen and sialylCore 1 structures (EWS-Etan-STn-C1) and STn antigens (EKS-Etan-S/Tn), respectively. C3GnT over expression in WT cells was anticipated to produce sialylCore 1- and Core 3-based structures (EWC-Etan-C1-C3), while Cosmc-KO cells expressing C3GnT was expected to produce etanercept with Core 3-based structures alone (EKC-Etan-C3). (B) Representative IgG-Fc, Tn antigen, and STn antigen immunoblots using 100 ng purified Etan-Ref, EW-Etan-C1, EK-Etan-Tn, EWS-Etan-STn-C1, EKS-Etan-S/Tn, EWC-Etan-C1-C3, and EKC-Etan-C3.
Table 1.
Relative peak percentages and main peak retention time by ultra-high performance liquid chromatography-size exclusion chromatography of etanercept reference material (Etan-Ref) and O-glycovariants
| Relative peak percentage |
Main peak |
|||
|---|---|---|---|---|
| HMWS | Main | LMWS | Retention time | |
| Etan-Ref | 2.17 | 97.1 | 0.73 | 3.851 |
| EW-Etan-C1 | 0.32 | 98.61 | 1.07 | 3.833 |
| EK-Etan-Tn | 1.36 | 95.04 | 3.58 | 3.96 |
| EWS-Etan-S/Tn-C1 | 0 | 99 | 1 | 3.895 |
| EKS-Etan-S/Tn | 0.35 | 98.26 | 1.39 | 3.877 |
| EWC-Etan-C1-C3 | 0 | 90.83 | 9.17 | 3.835 |
| EKC-Etan-C3 | 0.71 | 97.9 | 1.38 | 3.827 |
HMWS, high-molecular-weight species; LMWS, low-molecular-weight species.
To confirm the identity of the etanercept glycovariants, purified proteins were subjected to immunoblotting with antibodies against the IgG1 Fc region, Tn antigen, and STn antigen under reducing and non-reducing conditions (Figure 4B). While all of the purified proteins contained an IgG1 Fc region, indicative of etanercept, the levels of Tn and STn antigen varied by the O-glycoengineered cell type used for production. Etanercept produced in WT CHO cells did not contain Tn or STn antigen (EW-Etan-C1), similar to Etan-Ref. Cosmc-KO CHO cells produced etanercept with Tn Antigen (EK-Etan-Tn). WT CHO cells overexpressing ST6GalNAc1 produced etanercept with mainly sialylCore 1 and STn (EWS-Etan-STn-C1), but trace amounts of Tn were only detectable under reducing conditions, which may suggest the Tn antigen was inaccessible to the antibody under non-denature conditions. Cosmc-KO cells overexpressing ST6GalNAc1 produced etanercept containing both STn and Tn antigens (EKS-Etan-S/Tn). WT and Cosmc-KO CHO cells expressing C3GnT produced etanercept (EWC-Etan-C1-C3 and EKC-Etan-C3) that did not contain Tn or STn antigens. These data supported the hypothesis that this O-glycoengineered CHO cell line platform could produce the anticipated O-glycovariants of experimental therapeutic proteins, such as etanercept.
Glycan structures of etanercept O-glycovariants analyzed by MS
A one-pot MS method was used to analyze the glycan structures on purified etanercept O-glycovariants and Etan-Ref (Figure S5). Characterization of the released O-glycan profiles demonstrated that the O-glycoengineered platform successfully produced the O-glycans on etanercept (Table S4 and Figure S6). Etan-Ref, EW-Etan-C1, EKS-Etan-STn-C1, and EWC-Etan-C1-C3 contained mono- and di-sialylCore 1 structures, while EK-Etan-Tn, EKS-Etan-S/Tn, and EKC-Etan-C3 did not contain any Core 1-based structure (Figure 5A). In contrast with Etan-Ref and EW-Etan-C1, the STn antigen structure was only found on EWS-Etan-STn-C1 and EKS-Etan-S/Tn, and the Core 3-based O-glycans were only on EWC-Etan-C1-C3 and EKC-Etan-C3 (Figure 5A). The proposed structures of all major O-glycans were confirmed by the MALDI-TOF-MS/MS (Figure S7). As anticipated, the N-glycan compositions/structures were mostly conserved among the O-glycovariants of etanercept and Etan-Ref (Table S5 and Figure S8). A subtle difference in FA2 and Man5 was identified to be less than 5% on EW-Etan-C1 as compared with Etan-Ref (Figure 5B), indicating that the reference product etanercept-producing CHO cell line generates more immature N-glycans than the CHO cell line platform. To compare the N-glycan and O-glycan compositions/structures between Etan-Ref and the etanercept O-glycovariants, the relative abundances of all the glycans were calculated and compared (Table S6). The results demonstrated that while etanercept proteins among the O-glycoengineered CHO platform have very similar N-glycan profiles, each of them carries distinct O-glycans as designed. Collectively, these data confirmed that the O-glycoengineering CHO cell line platform could modulate the O-glycan structures on therapeutic proteins, such as etanercept.
Figure 5.
N- and O-linked glycans of etanercept
(A and B) Etanercept reference material (Etan-Ref) and O-glycovariants (EW-Etan-C1, EK-Etan-Tn, EWS-Etan-STn-C1, EKS-Etan-S/Tn, EWC-Etan-C1-C3, and EKC-Etan-C3) were analyzed for changes in the relative quantity of (A) O-linked and (B) N-linked glycan structures. Bars represent the mean percentage ± standard deviation. (Two-way ANOVAs, n = 3, Tukey multiply comparison with Etan-C1, ∗p < 0.05). # indicates sample with Tn and STn antigen.
Consistent with the endogenous glycoproteins, non-human glycans of neither αGal nor Neu5Gc were identified in any of the etanercept O-glycovariants from WT or the O-glycoengineered CHO cell platforms.
Physicochemical and biological characterization of etanercept O-glycovariants
To investigate the impact of O-glycans on etanercept's physicochemical and biological properties, the Etan-Ref and O-glycovariants were characterized for changes in isoelectric point, protein stability, TNF-α binding, and potency. Using a gel-based method for isoelectric focusing, a qualitative difference in the isoelectric point was observed for EK-Etan-Tn as compared with EW-Etan-C1 and Etan-Ref, and smaller shifts can be observed in the other O-glycovariants (EWS-Etan-STn-C1, EKS-Etan-S/Tn, EWC-Etan-C1-C3, and EKC-Etan-C3) (Figure 6A). These differences could potentially be caused by sialic acid content. A decrease in the levels of negatively charged sialic acid is known to correlate with an increase in the isoelectric point.21 These data confirm that different O-glycans can alter etanercept's net charge, which is attributed to differences in the sialic acid content.
Figure 6.
Partial characterization of the physicochemical, structural, and biological properties of etanercept
(A) IEF: Representative Coomassie-stained IEF gel using etanercept reference material (Etan-Ref) and O-glycovariants (EW-Etan-C1, EK-Etan-Tn, EWS-Etan-STN-C1, EKS-Etan-S/Tn, EWC-Etan-C1-C3, and EKC-Etan-C3). (B) Stability: Representative Coomassie-stained SDS-PAGE gel of etanercept Etan-Ref and O-glycovariants with and without a 0.3% hydrogen peroxide (H2O2) treatment for 4 h at 4°C. (C) Representative electropherograms of Etanercept and TNF- α Binding using biolayer interferometry (BLI) and a TNF- α concentration gradient. (D) Potency: EC50 values for TNF- α neutralization were determined for etanercept reference and the series of O-glycovariants. Open circles represent the average of duplicate samples for each biological replicate, while the line with bars represent the mean ± standard deviation. (One-way ANOVA, n = 6, Sidak's multiple comparison test, ∗p < 0.05).
To determine if the change in glycans altered oxidative stress induced protein instability, the purified proteins were incubated at 4°C in the presence and absence of hydrogen peroxide for 4 h. While fragmentation was observed for all etanercept variants, there were no significant differences in the banding pattern identified by SDS-PAGE (Figure 6B).
The binding affinity of etanercept Etan-Ref and O-glycovariants for TNF-α was determined using biolayer interferometry (Figure 6C and Table 2). Consistent with a previous report,22 the TNF-α binding affinity for Etan-Ref was 250 ± 56.9 pM, which was a much lower ligand binding affinity than that of EWS-Etan-STn-C1 and EKS-Etan-S/Tn with dissociation constants (KD) of 24.6 ± 6.1 pM and 18.9 ± 4.8 pM, respectively. Moreover, EKC-Etan-C3 demonstrated a lower binding affinity than the Etan-Ref with a KD of 585 ± 212 pM. The TNF- α binding affinity of other variants, EW-Etan-C1, EK-Etan-Tn, and EWC-Etan-C1-C3, were comparable with Etan-Ref. These data support that differences in O-glycan structures on etanercept can impact TNF- α binding affinity.
Table 2.
Steady State etanercept O-glycovariant binding affinity to TNF-α with association and dissociation rates
| KD (M) |
Ka (1/Ms) |
Kd (1/s) |
||||
|---|---|---|---|---|---|---|
| Avg. | S.D. | Avg. | S.D. | Avg. | S.D. | |
| Ref | 2.50E-10 | 5.69 × 10−11 | 1.58 × 106 | 4.94 × 105 | 1.74 × 10−4 | 6.36 × 10−5 |
| Etan-C1 | 2.28E-10 | 9.26 × 10−11 | 1.30 × 106 | 3.11 × 104 | 1.87 × 10−4 | 5.80 × 10−5 |
| Etan-Tn | 1.33E-10 | 4.60 × 10−11 | 1.82 × 106 | 4.45 × 105 | 1.14 × 10−4 | 7.42 × 10−5 |
| Etan-C1-STn | 2.46 × 10−11 | 6.08 × 10−12 | 1.84 × 106 | 5.04 × 105 | 1.84 × 10−4 | 9.79 × 10−5 |
| Etan-S/Tn | 1.89 × 10−11 | 4.81 × 10−12 | 2.07 × 106 | 1.77 × 105 | 7.38 × 10−5 | 9.72 × 10−5 |
| Etan-C1-C3 | 3.97E-10 | 6.22 × 10−11 | 1.38 × 106 | 4.88 × 104 | 1.93 × 10−4 | 1.35 × 10−4 |
| Etan-C3 | 5.85E-10 | 2.12–10 | 8.75 × 105 | 5.89 × 104 | 2.64 × 10−4 | 8.26 × 10−5 |
Avg, Average; SD, standard deviation.
To identify differences in potency between the Etan-Ref and O-glycovariants, a TNF- α neutralization assay was performed and revealed that the median effective concentration (EC50) value for EWS-Etan-STn-C1 significantly decreased as compared with EW-Etan-C1 (Figure 6D). The EC50 value for TNF- α neutralization using EW-Etan-C1 was 65 ± 10.17 ng/mL, while the EC50 value of EWS-Etan-STn-C1 decreased by approximately 38%, to 40.7 ± 9.7 ng/mL. The calculated EC50 values for Etan-Ref, EK-Etan-Tn, EKS-Etan-S/Tn, EWC-Etan-C1-C3, and EKC-Etan-C3 were comparable with the EW-Etan-C1 EC50 value. Collectively, these data suggest that changes in the O-glycan structures of etanercept can affect TNF- α neutralization.
Discussion
Using etanercept as an example, our study reveals a novel and practical manufacturing strategy for the production of O-glycoengineered therapeutic proteins from an O-glycoengineered CHO cell line platform. While the impact of O-glycans on therapeutic safety, efficacy, and quality remain to be defined, this O-glycoengineered CHO cell line platform provides a novel approach for evaluating the influence of O-glycans on engineered therapeutic proteins with different O-glycan structures. Notably, changes in the structures of O-glycans, particularly STn, and Core 3 O-glycans, on etanercept were associated with altered TNF- α binding affinity. Collectively, this is the first systematic study that describes a CHO cell manufacturing strategy to produce O-glycoengineered therapeutic proteins to define quality attributes and evaluate the role of O-glycans in therapeutic efficacy, quality, and perhaps safety (Graphical Abstract).
Studies have demonstrated that different types of O-glycans play different biological roles.15,16 SialylCore 1 O-glycans are broadly expressed on all cell types and tissues and play variety of biological functions. Core 3 O-glycans on mucins from gastrointestinal epithelial cells play an important protection role and suppress colorectal carcinogenesis and progression.23, 24, 25 The STn antigen, which is normally expressed in certain tissues or cell types only, is thought to modulate immune responses and often is highly expressed in carcinomas.26,27 The Tn antigen is not usually found in normal tissues or cells, but is highly expressed on tumor cells.15,20 The expression of Tn antigens and STn antigens are thought to benefit tumor survival and escape from immune surveillance.26, 27, 28 As many therapeutic proteins, such as fusion proteins etanercept, abatacept, atacicept, hormone erythropoietin (EPO), recombinant coagulation factor VIII, granulocyte macrophage colony stimulating factor, and others are both O-glycosylated and N-glycosylated.19,29, 30, 31, 32, 33, 34 The role of N-glycosylation in quality and safety of those protein drugs are well known. However, the effects of O-glycosylation on critical quality attributes of these therapeutic proteins are currently largely uninvestigated.
The enzymes responsible for synthesizing STn and Core 3 O-glycan structures on proteins are suppressed at the messenger RNA level in CHO cells.12 The Cosmc-KO CHO cells can only produce the Tn antigen owing to an inactive T synthase. Stably expressing C3GnT in the Cosmc-KO cells caused the loss of the Tn antigen on endogenous proteins, but stably expressing ST6GalNAc1 in the Cosmc-KO cells lead to the produce of STn antigen with sustained Tn antigen levels on proteins. These data suggest that C3GnT have converted all the Tn antigen to Core 3-based O-glycan structures, but ST6GalNAc 1 was incapable of completely transforming the Tn antigen to the STn antigen, most likely owing to its expression/activity level. Other possibilities include that C3GnT and ST6GalNAc1 may have different acceptor substrate (glycoprotein) preferences, and limitation of availability of CMP-sialic acid, the donor for ST6GalNAc1 and other sialyltransferases. The incomplete conversion of Tn to STn antigen O-glycans in the Cosmc KO CHO cells expressing ST6GalNAc 1 warrants for additional investigations.
CHO cell bioengineering has delivered quality by design products and improved product quality.35 Genetic modulation of the glycosylation pathway in CHO cells has primarily focused on the production of N-linked glycans owing to their well characterized role in monoclonal antibody effector function and clearance.36 In contrast with N-linked glycans, the impact of O-linked glycans on therapeutic proteins is less clear and studies of the O-glycosylation pathway in CHO cells are limited.37,38 Previous studies have modulated the O-glycosylation pathway in CHO cells by knocking out Cosmc, which resulted in proteins, including endogenous proteins and EPO, with only the Tn antigen.19 However, the N-linked glycan profile was not analyzed. It is unknown if these changes were specific for O-glycans, and if they impact the quality and safety of EPO. In the current study, we demonstrated that our five key glycan engineered plus the WT CHO cells produced major O-glycoforms of etanercept as expected, while the N-linked glycosylation profiles of these six different etanercept O-glycovariants were similar. These results demonstrate that engineering O-glycosylation in CHO cells did not lead to significant changes in the N-glycosylation of etanercept. Using this platform, novel O-glycovariants of therapeutic proteins can be manufactured that may potentially improve the safety, efficacy, and quality of drug products. Additional applications for this platform include the generation of O-glycovariants of ligands to improve the specificity of bioassays targeted against specific glycoproteins.
While EK-Etan-Tn and EWC-Etan-C1-C3 had comparable affinity to TNF-α and potency, our findings demonstrated that EWS-Etan-STn-C1 and EKS-Etan-S/Tn etanercept glycovariants had an enhanced TNF- α binding, but the EKS-Etan-S/Tn enhanced binding affinity did not translate to an increase in potency relative to etanercept from WT cells (Figures 6C and 6D). Interestingly, EKC-Etan-C3 had a lower affinity for TNF-α, but similar bioactivity. This finding warrants further investigation. We speculate that differences in O-glycans, such as STn and Core 3 structures at specific glycosites, could potentially modulate interactions of etanercept with TNF-α. A possible explanation of the higher affinity of EKS-Etan-S/Tn to TNF-α without higher bioactivity could be due to (1) the cleavage of etanercept with truncated O-glycans, (2) the Tn antigen on either the linker between TNFR domain and Fc resulting in monomer of etanercept, or (3) the Tn carrying sites within the function domain of TNFR cleaved during the cell potency assay in which bovine serum was present. It is plausible that proteases in serum may have caused the proteolytic cleavage of etanercept. Nevertheless, our results suggested that the etanercept O-glycovariant, EWS-Etan-STn-C1, may be a biobetter of etanercept with greater potency and perhaps better safety; the STn antigens on the product could suppress its immunogenicity and host immunity that have been seen in the tumor biology studies.26, 27, 28 Future studies will characterize the O-glycosylation sites and O-glycan structures on the TNFR domain and the linker to identify the O-glycans and O-glycosylation site(s) contributing to the increase in potency to serve as a critical quality attribute. Furthermore, the immunogenicity profiles of those etanercept O-glycovariants will also be assessed using in vitro cell models and appropriate animal models. Moreover, some evidence also suggested that the Fc region within fusion proteins could also be O-glycosylated, and O-glycosylation of Fc may affect binding to FcRs. These future studies will further clarify the relationship between glycan structures and protein function. This comprehensive study demonstrated a manufacturing strategy to control O-glycans on therapeutic proteins and that modulation of O-glycans on etanercept can affect physicochemical properties and biological activity.
There are two more types of O-glycans, Core 2 and extended Core 1 O-glycans often found in hematopoietic derived cells, which were not included in our current O-glycoengineered CHO cell line platform. These O-glycan structures may impact the biological activity and safety of certain therapeutic glycoproteins. Future development will include generating those additional CHO lines to make a full array of O-glycovariants of therapeutic proteins.
In summary, we established a manufacturing relevant O-glycoengineered CHO cell line platform that can be used for the production of recombinant proteins, but leveraging this platform technology for commercial drug production requires additional developmental and characterization studies. As a proof of principle study, etanercept produced from this platform was used to demonstrate that differences in O-glycans could alter the therapeutic protein's critical quality attributes as certain O-glycovariants of etanercept enhanced binding activity to TNF- α and potency in the cell-based assay. Notably, this work may shed the new light on quality by design of biobetters for etanercept which itself is an effective biotech drug on the market with billions of dollars of revenue each year. We propose that this CHO cell line platform can be used for a variety of different applications, such as the production of O-glycan-specific ligands for assay development, the production of O-glycovariants of growth factors for cell culture, and the manufacturing of therapeutic proteins with designed O-glycan structures. This platform offers a tool to define the quality and safety, such as immunogenicity attributes of O-glycosylation, of O-glycosylated therapeutic proteins.
Materials and methods
Generation of clonally derived O-glycoengineered CHO cell line platform
CHO cells were obtained from ATCC (Manassas, VA, Cat # CCL-61) and cultivated according to their instructions. Transfections were performed using HilyMAX (Dojindo Mol Tech. Inc, Rockville, MD, Cat# H357-15) as described by the manufacturer. The plasmid constructs used for the transfection are described in the Supplemental Information. To generate stable clones, transfected cells were exposed to antibiotic selection for 14 days before two sequential FACS cycles enriched for Tn antigen- or STn antigen-positive populations. To support clonal derivation, the enriched positive population of cells were subjected to one round of limiting dilution cloning at 0.5 cells per well.
Production and purification of experimental and O-glycovariants of etanercept
Etanercept expressing cells were seeded in T150 vented flasks at 1 × 106 cells on day 0 with 50 mL growth media. After 5 days at 37°C and 5% CO2, the media were collected and replaced with 75 mL fresh media for an additional 2 days. All the harvested media were pooled and centrifuged at 500 × g for 10 min before passage through a 0.22-micron filter. To purify the O-glycovariants of etanercept, several chromatographic methods were performed as described in the Supplemental Information.
T synthase activity assay
The T synthase activity assay was performed as previously described.39
MS analysis of O-linked and N-linked glycans
An one-pot approach to simultaneously analyze N- and O-glycans was performed and will be described in a second report. In brief, a dry aliquot consisting of 50 μg of protein from whole cell lysates or purified etanercept was reconstituted in 25 μL of buffer (10 mM sodium phosphate, 0.1% SDS, and 0.1% β-mercaptoethanol at pH 7.5) and incubated for 1 h at 60°C. After cooling, 2.5 μL of a 10% nonidet-P40 solution was added, followed by incubation at room temperature for 10 min. The N-glycans were released using 1 μL of a 1:10 solution of recombinant PNGase F (Prozyme, Hayward CA, Cat# GKE-5006B) diluted in 10 mM sodium phosphate buffer, pH 7.5 (0.25 mU/μL) and incubated overnight at 37°C. The PNGase F-treated samples were digested with 1 μL of a 20 mg/mL proteinase K solution (Viagen, Los Angeles CA, Cat# 502-PK) and incubated for 18 h at 55°C. Released N-glycans and small O-glycopeptides were purified in tandem using active charcoal microspin columns. The eluent was then dried under vacuum and treated with 10 μL of ammonia borane complex in H2O (10 mg/mL) for 1 h at 60°C. Excess reactants were removed by a series of methanolic evaporations. Finally, the samples were processed by solid phase permethylation releasing O-glycans from the peptide backbone, and simultaneously methylating free glycans. The mixture of reduced N-glycans and unreduced O-glycans were resuspended in 10 μL of 50% methanol:water, mixed 1:1 with a 2,5 dihydroxybenzoic acid matrix (10 mg/mL with 1 mM sodium acetate in 50% methanol) and spotted on a target plate for analysis using a Bruker UltrafleXtreme MALDI-TOF/TOF mass spectrometer as previously described.40 Mass measurements were obtained in positive ion reflector mode, and peaks corresponding to sodiated glycans were fragmented in TOF/TOF mode. Reduced N-glycans containing an increased mass of 16 Da were easily differentiated from unreduced O-glycans. Glycan intensity was normalized to percent abundance based on total glycan identifications, and the average and standard deviation were calculated for all structures.
SDS-PAGE, isoelectric focusing, and immunoblotting
Purified proteins (1 μg) were used to qualitatively determine the isoelectric point using Novex IEF gels (Thermofisher, Waltham, MA, Cat# EC6655BOX) as described by the manufacturer. Purity and protein stability were determined using SDS-PAGE (Any Kda Protein Gel, Bio Rad, Hercules, CA, Cat# 4569036), 1 μg protein, and EZBlue Coomassie stain (Sigma Aldrich, St. Louis, MO, Cat# G1041-500 ML) as described by the manufacturer. Immunoblotting was performed using up to 60 μg of cell lysate, 10 μL of harvest media, or 200 ng of purified protein. Using Azure 600 (Azure Biosystems, Dublin, CA), the proteins were imaged using the following primary and secondary antibodies: TAG-72 (Santa Cruz biotech, Santa Cruz, CA, Cat# sc-20042), beta actin (Cell Signaling Technology, Danvers, MA, Cat# 4970 and 3700), ST6GalNAc 1 and C3GNT (Proteintech, Rosemont, IL, Cat# 15363-1-AP and 21291-1-AP), anti-human Fc (Novus Biologicals, Centennial, CO, Cat# NBP1-40876), and several near infrared secondary antibodies (Licor, Bad Homburg, Germany, Cat# 926-68072, 926-68-021, and 926-32232).
Flow cytometry
Two million cells were incubated for one hour in growth media (no phenol red) containing 2% FBS with a primary antibody at a 1:200 ratio at 4°C. After two washes with PBS, cells were incubated with appropriate Alexa Fluor antibodies at a 1:1,000 ratio for 1 h at 4°C. Using a BD LSRII, 30k-50k cellular events were counted; single cells were identified, and quantitative analyses were performed using FloJo version 10.1 (BD Biosciences, San Jose, CA).
Etanercept potency assay
U937 cells were obtained from and cultivated as described by ATCC (Cat # CRL-1593.2). Etanercept (200 ng/mL to 1 ng/mL) was incubated with 10 ng/mL TNF-α (Biolegend, San Diego, CA, Cat# 570104) for 1 h at 37°C before addition to U937 cells (15,000 cells per well). Cells were then incubated for 2.5 h at 37°C. Induction of TNF-α-mediated caspase 3 signaling was measured by luciferase-based assay using Caspase-Glo 3/7 (Promega, Madison, WI) as described by the manufacturer. The EC50 values were then calculated.
Etanercept binding assay
Biolayer interferometry was performed using 1 μg of etanercept, purified TNF-α (200 ng–1 ng), anti-human Fc-capture biosensors, and an Octet Red96e from Sartorius (Goettingen, Germany) as described by the manufacturer. Briefly, etanercept-loaded Fc-capture biosensors were allowed to associate with TNF- α for 5 min before dissociation for 5 min.
Statistics and reagents
All other reagents were purchased from Sigma Aldrich. The etanercept reference material (Etan-Ref) was commercially manufactured by Amgen Inc. and purchased through a pharmacy. Statistical analyses were performed using GraphPad Prism 6 from at least two independently performed experiments. When 1 factor was analyzed, a one-way ANOVA was performed with a Sidak's multiple comparison test to identify statistical differences. When more than one factor was analyzed, a two-way ANOVA was performed with a Tukey's multiple comparison post-hoc test to identify significant differences. A p value of less than 0.05 indicated statistical differences. The degree of freedom, p values, and test methods are provided in Table S1.
Acknowledgments
We would like to thank Tao Wang (FDA), Francesca Mascia (FDA), and Vineet Kewal Ramani (NIH) for the scientific guidance and/or the critical review of the manuscript. This project was supported by the CDER Domestic Manufacturing Initiatives, OPQ Centers of Excellence, and CDER Regulatory Science and Research Committee at the United States Food and Drug Administration (US FDA). The project was also partially supported by Office of Women's Health/FDA (H.X. and T.J).
Author contributions
The manuscript's contributor roles using credit taxonomy is as follows: T.G.B. - conceptualization, methodology, software, validation, formal analysis, investigation, writing—original draft and editing, visualization, project administration, funding acquisition; T. F. – methodology, software, investigation, writing – review and editing; A. M. – investigation, methodology, writing – review and editing; G. Z. – investigation, methodology, writing – review and editing; U. O. – investigation, methodology, writing – review and editing; M. P. – investigation, validation, writing – review and editing; N. A. – investigation, methodology; F. G. – conceptualization, writing - review and editing; S. R. – writing – review & editing; S. J. – writing – review and editing; K. C. – writing – review and editing; H. X. – funding acquisition, resources, supervision, writing– review and editing; C. A.i. – resources, supervision, writing – review and editing; A. R. – resources, supervision, writing– review and editing; T. J. – conceptualization, methodology, resources, writing – review and editing, visualization, supervision, project administration, funding acquisition.
Declaration of interests
The authors have no interests to declare. The funding source had no part in the study design, data collection/analysis, decision to publish, or preparation of the manuscript. The authors have no conflicts of interests. The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the United States Food and Drug Administration and the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the United States Government.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtm.2022.03.002.
Supplemental information
References
- 1.Neelamegham S., Mahal L.K. Multi-level regulation of cellular glycosylation: from genes to transcript to enzyme to structure. Curr. Opin. Struct. Biol. 2016;40:145–152. doi: 10.1016/j.sbi.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mimura Y., Katoh T., Saldova R., O'Flaherty R., Izumi T., Mimura-Kimura Y., Utsunomiya T., Mizukami Y., Yamamoto K., Matsumoto T., et al. Glycosylation engineering of therapeutic IgG antibodies: challenges for the safety, functionality and efficacy. Protein Cell. 2018;9:47–62. doi: 10.1007/s13238-017-0433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malphettes L., Freyvert Y., Chang J., Liu P.Q., Chan E., Miller J.C., Zhou Z., Nguyen T., Tsai C., Snowden A.W., et al. Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol. Bioeng. 2010;106:774–783. doi: 10.1002/bit.22751. [DOI] [PubMed] [Google Scholar]
- 4.Yuan Y., Zong H., Bai J., Han L., Wang L., Zhang X., Zhang X., Zhang J., Xu C., Zhu J., et al. Bioprocess development of a stable FUT8(-/-)-CHO cell line to produce defucosylated anti-HER2 antibody. Bioproc. Biosyst Eng. 2019;42:1263–1271. doi: 10.1007/s00449-019-02124-7. [DOI] [PubMed] [Google Scholar]
- 5.Yu L.X., Amidon G., Khan M.A., Hoag S.W., Polli J., Raju G.K., Woodcock J. Understanding pharmaceutical quality by design. AAPS J. 2014;16:771–783. doi: 10.1208/s12248-014-9598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alt N., Zhang T.Y., Motchnik P., Taticek R., Quarmby V., Schlothauer T., Beck H., Emrich T., Harris R.J. Determination of critical quality attributes for monoclonal antibodies using quality by design principles. Biologicals. 2016;44:291–305. doi: 10.1016/j.biologicals.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Emery P., Vencovsky J., Sylwestrzak A., Leszczynski P., Porawska W., Baranauskaite A., Tseluyko V., Zhdan V.M., Stasiuk B., Milasiene R., et al. A phase III randomised, double-blind, parallel-group study comparing SB4 with etanercept reference product in patients with active rheumatoid arthritis despite methotrexate therapy. Ann. Rheum. Dis. 2017;76:51–57. doi: 10.1136/annrheumdis-2015-207588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Girolomoni G., Feldman S.R., Emery P., Ghil J., Keum J.W., Cheong S.Y., Hong E. Comparison of injection-site reactions between the etanercept biosimilar SB4 and the reference etanercept in patients with rheumatoid arthritis from a phase III study. Br. J. Dermatol. 2018;178:e215–e216. doi: 10.1111/bjd.16032. [DOI] [PubMed] [Google Scholar]
- 9.Cho I.H., Lee N., Song D., Jung S.Y., Bou-Assaf G., Sosic Z., Zhang W., Lyubarskaya Y. Evaluation of the structural, physicochemical, and biological characteristics of SB4, a biosimilar of etanercept. mAbs. 2016;8:1136–1155. doi: 10.1080/19420862.2016.1193659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelechas E., Drosos A.A. Etanercept biosimilar SB-4. Expert Opin. Biol. Ther. 2019;19:173–179. doi: 10.1080/14712598.2019.1566456. [DOI] [PubMed] [Google Scholar]
- 11.Kunert R., Reinhart D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 2016;100:3451–3461. doi: 10.1007/s00253-016-7388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Nagarajan H., Lewis N.E., Pan S., Cai Z., Liu X., Chen W., Xie M., Wang W., Hammond S., et al. The genomic sequence of the Chinese hamster ovary (CHO)-K1 cell line. Nat. Biotechnol. 2011;29:735–741. doi: 10.1038/nbt.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells L. In: Handbook of Glycomics. Cumming R.D., Pierce J.M., editors. Acadenuc Oress; 2010. Chapter 2 - O-glycan complexity and analysis; pp. 45–57. [Google Scholar]
- 14.Sasaki H., Bothner B., Dell A., Fukuda M. Carbohydrate structure of erythropoietin expressed in Chinese hamster ovary cells by a human erythropoietin cDNA. J. Biol. Chem. 1987;262:12059–12076. [PubMed] [Google Scholar]
- 15.Ju T., Otto V.I., Cummings R.D. The Tn antigen-structural simplicity and biological complexity. Angew. Chem. Int. Ed. Engl. 2011;50:1770–1791. doi: 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schjoldager K.T., Narimatsu Y., Joshi H.J., Clausen H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020;21:729–749. doi: 10.1038/s41580-020-00294-x. [DOI] [PubMed] [Google Scholar]
- 17.Montacir O., Montacir H., Springer A., Hinderlich S., Mahboudi F., Saadati A., Parr M.K. Physicochemical characterization, glycosylation pattern and biosimilarity assessment of the fusion protein etanercept. Protein J. 2018;37:164–179. doi: 10.1007/s10930-018-9757-y. [DOI] [PubMed] [Google Scholar]
- 18.Ju T., Cummings R.D. Protein glycosylation: chaperone mutation in Tn syndrome. Nature. 2005;437:1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z., Halim A., Narimatsu Y., Jitendra Joshi H., Steentoft C., Schjoldager K.T., Alder Schulz M., Sealover N.R., Kayser K.J., Paul Bennett E., et al. The GalNAc-type O-Glycoproteome of CHO cells characterized by the SimpleCell strategy. Mol. Cell Proteomics. 2014;13:3224–3235. doi: 10.1074/mcp.M114.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stowell S.R., Ju T., Cummings R.D. Protein glycosylation in cancer. Annu. Rev. Pathol. 2015;10:473–510. doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrabes S., Sarrats A., Fort E., De Llorens R., Rudd P.M., Peracaula R. Effect of sialic acid content on glycoprotein pI analyzed by two-dimensional electrophoresis. Electrophoresis. 2010;31:2903–2912. doi: 10.1002/elps.200900764. [DOI] [PubMed] [Google Scholar]
- 22.Cui X., Chang L., Li Y., Lv Q., Wang F., Lin Y., Li W., Meade J.D., Walden J.C., Liang P. Trivalent soluble TNF Receptor, a potent TNF-alpha antagonist for the treatment collagen-induced arthritis. Sci. Rep. 2018;8:7327. doi: 10.1038/s41598-018-25652-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai T., Kudo T., Kawamoto R., Kubota T., Togayachi A., Hiruma T., Okada T., Kawamoto T., Morozumi K., Narimatsu H. Core 3 synthase is down-regulated in colon carcinoma and profoundly suppresses the metastatic potential of carcinoma cells. Proc. Natl. Acad. Sci. U S A. 2005;102:4572–4577. doi: 10.1073/pnas.0407983102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S.H., Hatakeyama S., Yu S.Y., Bao X., Ohyama C., Khoo K.H., Fukuda M.N., Fukuda M. Core3 O-glycan synthase suppresses tumor formation and metastasis of prostate carcinoma PC3 and LNCaP cells through down-regulation of alpha2beta1 integrin complex. J. Biol. Chem. 2009;284:17157–17169. doi: 10.1074/jbc.M109.010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An G., Wei B., Xia B., McDaniel J.M., Ju T., Cummings R.D., Braun J., Xia L. Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 2007;204:1417–1429. doi: 10.1084/jem.20061929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearce O.M.T. Cancer glycan epitopes: biosynthesis, structure and function. Glycobiology. 2018;28:670–696. doi: 10.1093/glycob/cwy023. [DOI] [PubMed] [Google Scholar]
- 27.RodrIguez E., Schetters S.T.T., van Kooyk Y. The tumour glyco-code as a novel immune checkpoint for immunotherapy. Nat. Rev. Immunol. 2018;18:204–211. doi: 10.1038/nri.2018.3. [DOI] [PubMed] [Google Scholar]
- 28.Jiang Y., Wen T., Yan R., Kim S.R., Stowell S.R., Wang W., Wang Y., An G., Cummings R.D., Ju T. O-glycans on death receptors in cells modulate their sensitivity to TRAIL-induced apoptosis through affecting on their stability and oligomerization. FASEB J. 2020;34:11786–11801. doi: 10.1096/fj.201900053RR. [DOI] [PubMed] [Google Scholar]
- 29.Yang Q., An Y., Zhu S., Zhang R., Loke C.M., Cipollo J.F., Wang L.X. Glycan remodeling of human erythropoietin (EPO) through combined mammalian cell engineering and chemoenzymatic transglycosylation. ACS Chem. Biol. 2017;12:1665–1673. doi: 10.1021/acschembio.7b00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caval T., Tian W., Yang Z., Clausen H., Heck A.J.R. Direct quality control of glycoengineered erythropoietin variants. Nat. Commun. 2018;9:3342. doi: 10.1038/s41467-018-05536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L., Guo Q., Guo H., Liu T., Zheng Y., Gu P., Chen X., Wang H., Hou S., Guo Y. Versatile characterization of glycosylation modification in CTLA4-Ig fusion proteins by liquid chromatography-mass spectrometry. mAbs. 2014;6:1474–1485. doi: 10.4161/mabs.36313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stavenhagen K., Gahoual R., Dominguez Vega E., Palmese A., Ederveen A.L.H., Cutillo F., Palinsky W., Bierau H., Wuhrer M. Site-specific N- and O-glycosylation analysis of atacicept. mAbs. 2019;11:1053–1063. doi: 10.1080/19420862.2019.1630218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai J.D., Swystun L.L., Cartier D., Nesbitt K., Zhang C., Hough C., Dennis J.W., Lillicrap D. N-linked glycosylation modulates the immunogenicity of recombinant human factor VIII in hemophilia A mice. Haematologica. 2018;103:1925–1936. doi: 10.3324/haematol.2018.188219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim J., Park H., Park B.T., Hwang H.S., Kim J.I., Kim D.K., Kim H.H. O-glycans and O-glycosylation sites of recombinant human GM-CSF derived from suspension-cultured rice cells, and their structural role. Biochem. Biophys. Res. Commun. 2016;479:266–271. doi: 10.1016/j.bbrc.2016.09.057. [DOI] [PubMed] [Google Scholar]
- 35.Fischer S., Otte K. In: Cell Culture Engineering:Recombinant Protein Production. Lee G.M., Kildegaard H.F., editors. Wiley-VCH Verlag GmbH & Co.; 2020. CHO cell engineering for improved process performance and product quality; pp. 207–250. [Google Scholar]
- 36.Boune S., Hu P., Epstein A.L., Khawli L.A. Principles of N-linked glycosylation variations of IgG-based therapeutics: pharmacokinetic and functional considerations. Antibodies (Basel) 2020;9:22. doi: 10.3390/antib9020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narimatsu Y., Joshi H.J., Nason R., Van Coillie J., Karlsson R., Sun L., Ye Z., Chen Y.H., Schjoldager K.T., Steentoft C., et al. An atlas of human glycosylation pathways enables display of the human glycome by gene engineered cells. Mol. Cell. 2019;75:394–407.e5. doi: 10.1016/j.molcel.2019.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian W., Ye Z., Wang S., Schulz M.A., Van Coillie J., Sun L., Chen Y.H., Narimatsu Y., Hansen L., Kristensen C., et al. The glycosylation design space for recombinant lysosomal replacement enzymes produced in CHO cells. Nat. Commun. 2019;10:1785. doi: 10.1038/s41467-019-09809-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ju T., Xia B., Aryal R.P., Wang W., Wang Y., Ding X., Mi R., He M., Cummings R.D. A novel fluorescent assay for T-synthase activity. Glycobiology. 2011;21:352–362. doi: 10.1093/glycob/cwq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou G., Kosikova M., Kim S.R., Kotian S., Wu W.W., Shen R., Powers D.N., Agarabi C., Xie H., Ju T. Comprehensive analysis of N-glycans in IgG purified from ferrets with or without influenza A virus infection. J. Biol. Chem. 2018;293:19277–19289. doi: 10.1074/jbc.RA118.005294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.