Abstract
Humans differ in their susceptibility to infectious disease, partly due to variation in the immune response following infection. We used single-cell RNA-sequencing to quantify variation in the response to influenza infection in peripheral blood mononuclear cells from European- and African-ancestry males. Genetic ancestry effects are common but highly cell type-specific. Higher levels of European ancestry are associated with increased type I interferon pathway activity in early infection, which predicts reduced viral titers at later time points. Substantial population-associated variation is explained by cis-expression quantitative trait loci that are differentiated by genetic ancestry. Furthermore, genetic ancestry-associated genes are enriched among genes correlated with COVID-19 disease severity, suggesting that the early immune response contributes to ancestry-associated differences for multiple viral infection outcomes.
Keywords: Genetic ancestry, single-cell RNA-sequencing, immune responses, influenza infection, SARS-CoV-2 infection, COVID-19, expression quantitative trait loci (eQTL)
One sentence summary:
Genetic ancestry and genetic variation explain population differences in the immune response to influenza infection in a cell type-dependent manner.
Pathogenic viruses are among the strongest sources of selection pressure in human evolution (1, 2). Prior to the modern era, however, global pandemics on the scale of the 1918 Spanish influenza or the SARS-CoV-2 pandemic were probably rare due to the restricted potential for long-distance exchange (3). If past viral epidemics were geographically stratified, they may have driven population divergence in the frequencies of polymorphisms that mediate the immune response to viral infection. Testing this hypothesis is therefore valuable both for understanding human evolutionary history and for explaining differential susceptibility to viral epidemics in the present-day.
Indeed, genetic effects on the response to viruses are well-known in human populations (4). For example, over 120 genetic variants have been identified in humans that predict the gene regulatory response to influenza A virus (IAV) in dendritic cells (5). Variation in the transcriptional response to IAV in vitro is also correlated with genetic ancestry in monocytes derived from individuals of African and European descent (6). These results suggest that genetic divergence between human populations, especially at loci that are moderately differentiated by genetic ancestry, plays an important role in shaping the immune response to viral infection. However, because studies to date focus on isolated cell types (5, 6), they fail to capture interactions between immune cell types necessary to mount an efficient antiviral response. They also leave unclear whether genetic ancestry effects are unique to, or generalize across, distinct immune cell types.
To address these limitations, here we combined single-cell RNA-sequencing with in vitro IAV infection assays in peripheral blood mononuclear cells from study subjects with varying degrees of European versus African genetic ancestry.
Single-cell profiling of the transcriptional response to influenza infection
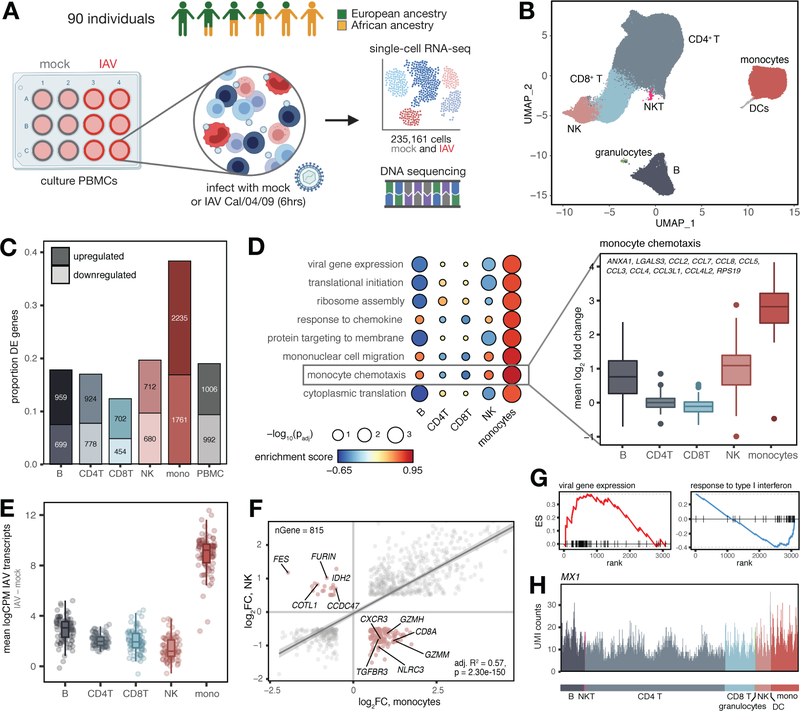
We exposed peripheral blood mononuclear cells (PBMCs) from a diverse panel of humans (Table S1) to either a mock treatment or the pandemic H1N1 Cal/04/09 influenza A virus (IAV) strain (multiplicity of infection 0.5) (n = 180 samples, paired mock-exposed and IAV-infected samples from each of 90 males). We focused on males to avoid potential effects of sex-specific differences in expression (7), which would reduce the power of our study. Following 6 hours of viral exposure, we performed single-cell RNA-sequencing on all samples (Fig. 1A). In total, we captured 255,731 single-cell transcriptomes across all individuals and conditions (n = 235,161 high-quality cells, Table S1). We also performed whole-genome sequencing to estimate the proportion of African and European ancestry for each individual (n = 89 individuals who were successfully genotyped; Fig. S1A, Table S1). Clustering revealed eight distinct immune cell types (Fig. 1B), with five major cell clusters corresponding to the main PBMC cell types (CD4+T cells, CD8+T cells, B cells, natural killer (NK) cells, and monocytes).
Fig. 1.
Shared and cell type-specific responses to IAV infection. (A) Study design. (B) UMAP of 235,161 mock and IAV-infected cells across individuals. (C) Numbers and proportions of differentially expressed genes upon infection. (D) Upregulated (FDR < 0.10) monocyte-specific GO pathways following infection (Table S3). “Monocyte chemotaxis” genes display greater upregulation in monocytes (plotted means for each individual across genes in IAV minus mock condition, t-tests, all p-values < 1×10−10 compared to each other cell type). (E) Distribution of IAV transcripts across cell types. (F) Correlation between global infection effect sizes in monocytes and NK cells among DE genes in both cell types (n = 815). P-value and best-fit slope was obtained from a linear regression model. Highlighted genes (pink) display discordant responses. (G) Example pathways enriched among genes with high (viral gene expression) and low (response to type I interferon) specificity scores. Genes are rank-ordered by specificity score (x-axis, highest to lowest). (H) UMI counts per cell in the IAV-infected condition for an example IFN-inducible gene (MX1) with a ubiquitous expression pattern.
We first investigated the overall signature of IAV infection by collapsing the single-cell gene expression values for each of the five main clusters and all cells together (i.e., “PBMCs”) to generate pseudobulk estimates for each sample. Principal component analysis (PCA) of the PBMC pseudobulk data revealed a marked separation of mock and IAV-infected samples on PC1, which explains 43% of the variance in the dataset (Fig. S1B, paired t-test, p < 1×10−10). Monocytes were the most responsive to IAV infection (n = 3,996 differentially expressed (DE) genes identified using limma (8) [38.3% of those tested compared to 12.4 – 19.6% in other cell types], |log2 fold-change| > 0.5, false discovery rate [FDR] < 0.05) (Figs. 1C and 1D, Tables S2 and S3). Monocytes also exhibited the highest levels of intracellular IAV transcripts (i.e., influenza-derived transcripts generated and processed by infected host cells; 3–6-fold increase in IAV transcript levels in monocytes relative to all other cell types, all t-test p-values < 1×10−10) (Fig. 1E). This observation is consistent with previous work showing that, among blood mononuclear cells, monocytes are particularly susceptible to viral infections (9).
We then explored the extent to which the infection response was shared across cell types. Overall, responses were strongly correlated (Pearson’s r range 0.65 – 0.95 for pairwise effect size correlations across cell types among DE genes following IAV infection, Fig. S1C). However, discordant responses were also observed. For example, among differentially expressed genes shared by monocytes and NK cells (n = 815), 135 genes (16.6%, Fig. S1D) responded to IAV infection in opposite directions (Fig. 1F). These findings underscore the importance of considering immune responses in a cell type-specific manner. Not only does this approach better capture the biological origins of variation in the response to viral infection, but it also avoids false negative or potentially misleading results that can emerge from bulk analysis.
To further dissect cell type-specific versus shared responses, we generated a specificity score based on variation in the strength of responses across cell types for all genes significantly differentially expressed in at least one cell type (Table S4, see (10) for details). Genes with highly cell type-specific response patterns were enriched for roles in translational initiation and viral gene expression (FDR < 1×10−10 for both terms, Fig. 1G, left, Table S4). In contrast, genes with low specificity scores were enriched for pathways related to type I interferon (IFN) signaling (FDR < 1×10−10) and response to type I IFN (FDR = 2.9×10−3) (Fig. 1G, right, Table S4). Thus, concordant with previous work in mice (11), our data show that the induction of IFN-related genes is a fundamental component of the antiviral response shared across immune cell types (Fig. 1H).
Increased European genetic ancestry predicts a stronger type I/II IFN response following IAV infection
We next identified genes for which expression levels are correlated with quantitative genetic ancestry estimates (i.e., proportion of estimated African ancestry) at baseline, following infection, or both (controlling for age, batch, and other technical covariates). To increase power and improve our effect size estimates for these “population differentially expressed” (popDE) genes, we applied a multivariate adaptive shrinkage method (mash) (12), which leverages the correlation structure of genetic ancestry effect sizes across cell types (see (10) for details of statistical models). Across conditions and cell types, we identified 1,949 unique popDE genes (local false sign rate [lfsr] < 0.10), ranging from 830 in NK cells to 1,235 genes in CD4+T cells (Figs. 2A and S2A for distribution of effect sizes, Table S5). Within each cell type, most popDE genes were shared between conditions (52.9% in monocytes – 77.4% in CD8+T cells, Fig. 2A). In contrast, across cell types, genetic ancestry effects on gene expression were highly cell type-specific, such that the majority of popDE genes were identified in only one or two cell types (52.2% in mock, 51.4% in IAV-infected, Figs. 2B. and S2B, left). Only 17.8% (mock) and 24.7% (IAV-infected) of popDE genes exhibited shared genetic ancestry effects across all five cell types (Figs. 2B. and S2B, right). Notably, despite differences in study subject country of origin, IAV strain, and experimental design, our popDE effect size estimates for monocytes were largely concordant with those derived from an independent bulk RNA-seq dataset of IAV-infected monocytes from European- and African-ancestry individuals (6) (Pearson’s r = 0.662 [mock], Pearson’s r = 0.499 [IAV], p < 1.0×10−10 in both conditions, Figure S2C).
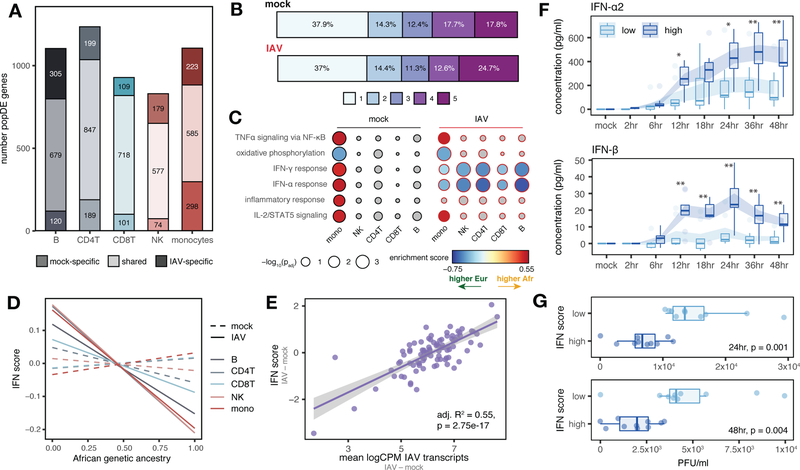
Fig. 2.
Genetic ancestry influences the immune response to IAV infection. (A) Number of shared and condition-specific popDE genes. (B) Cell type sharing of popDE effects (1 = detected in a single cell type, 5 = detected across all cell types). (C) GO enrichments for popDE effects in the mock- and IAV-infected conditions. Colored circles represent pathways with FDR < 0.10. IFN pathways are among the most divergent between European and African-ancestry individuals in monocytes, with 26% (42 out of 163) of all IFN genes tested classified as popDE after infection. (D) Correlation between African genetic ancestry proportion and IFN score in mock (dotted lines) and IAV-infected conditions (solid lines). (E) IAV transcript levels are associated with IFN response in PBMCs. (F) Secreted IFN-α2 and IFN-β levels in low versus high IFN responders over a 48 hour time course. Shaded area represents the mean ± SE. *p < 0.02, **p < 0.009 (Mann-Whitney U tests). (G) Viral titers (plaque-forming units, PFU/ml) detected in supernatant 24 and 48 hpi. In (D) and (E), p-values and best-fit slopes were obtained from linear regression models.
To identify the functional pathways most closely associated with genetic ancestry, we performed gene set enrichment analysis for the MSigDB Hallmark gene sets (13) (Fig. 2C, Table S6). In monocytes, we identified significant enrichments for multiple immune pathways prior to infection, including IFN-α response (FDR = 1.9×10−3), IFN-γ response (FDR = 5.4×10−4), TNFα signaling via NF-κB (FDR = 6.1×10−4), IL-2/STAT5 signaling (FDR = 2.1×10−3), and inflammatory response (FDR = 0.012) (Fig. 2C). In these cases, the enrichments were identified for genes more highly expressed at baseline in individuals with a greater proportion of African ancestry. Intriguingly, in IAV-infected monocytes, this pattern reversed: post-infection, we observed an enrichment of type I and II IFN pathways (IFN-α response FDR = 0.014, IFN-γ response FDR = 0.040 in monocytes) in genes more highly expressed with increasing European ancestry (Fig. 2C). Notably, this enrichment of type I/II IFN pathways among genes more highly expressed with greater European ancestry after infection was even more clear in the other four cell types (FDR range: 0.03 – 4.1×10−4, Table S6). To further characterize genetic ancestry-associated differences in the IFN response, we constructed a per-sample score of interferon signaling activity, the “IFN score,” which provides an estimate of the average expression of genes belonging to the hallmark IFN-α and IFN-γ gene sets for each individual (10). Again, increased European ancestry was strongly correlated with increased IFN score, but only following infection (mean Pearson’s r across cell types = −0.26, Fisher’s meta-p = 2.9×10−6 for IAV-infected; mean Pearson’s r = −0.0045, Fisher’s meta-p = 0.746 for mock) (Figs. 2D and S2D for cell type-specific associations).
These findings suggest that genetic ancestry may also predict the magnitude of the response to IAV infection. In support of this idea, we identified 445 genes for which genetic ancestry was associated with the magnitude of the response to infection (i.e., “population differentially-responsive” [popDR] genes, lfsr < 0.10). PopDR genes were found for all five cell types but were most common in monocytes (popDR genes: nmonocytes = 272 versus range = 53 – 181 in other cell types). A core set of 21 popDR genes was shared across all cell types (Fig. S3A, Table S7). Increased European genetic ancestry predicted a stronger type I/II IFN response (measured as the difference in IFN score between the IAV-infected and mock conditions per individual) across cell types (mean Pearson’s r = −0.23, Fisher’s meta-p = 6.0×10−5, Fig. S3B). This observation was not explained by baseline levels of Cal/04/09-specific serum IgG antibodies (a proxy for prior exposure to IAV), which were uncorrelated with genetic ancestry, the transcriptional response to IAV (Figs. S3C, D), and HLA genotype (10). However, stronger type I/II IFN responses predicted increased intracellular IAV transcript levels in PBMCs (adj. R2 = 0.55, p = 2.8×10−17, Figs. 2E and S3E for cell type-specific effects). IAV transcript levels were also significantly higher in individuals with increased European ancestry (Pearson’s r = −0.32, p = 0.002, Fig. S3F).
An early-induced type I IFN response is associated with decreased viral titers at later time points
To functionally validate our findings, we infected PBMCs from the 20 individuals with the strongest (n = 10, “high responders”) and weakest (n = 10, “low responders”) transcriptional type I/II IFN responses at 6 hours post infection (hpi) with IAV. We collected secreted cytokine measurements across 8 time points over 48 hours and viral titer measurements at 24 and 48 hpi. High responders produced significantly more secreted IFN-α2 (Fig. 2F, top) and IFN-β (Fig. 2F, bottom) than low responders beginning at 12 hpi, an effect that was exacerbated over time to 4-fold (IFN-α2) and 11.6 fold (IFN-β) more by 48 hpi (p < 0.007 for both cytokines, Mann-Whitney U tests). Viral titers quantified from supernatant at 24 and 48 hpi were also reduced in high responders compared to low responders (Mann-Whiney U tests, p = 0.001 for 24 hpi, p = 0.004 for 48 hpi, Fig. 2G). None of the 20 study subjects in this experiment harbored predicted loss-of-function mutations among genes associated with defects in type I IFN signaling (14, 15), suggesting that these results are not driven by rare genetic variants (Table S1). Taken together, these results indicate that individuals better able to mount type I IFN responses shortly after infection also displayed a greater capacity to limit productive viral replication later in infection/at later time points. These observations are consistent with the finding that individuals with rare immunodeficiencies leading to defects in type I IFN signaling restrict viral replication poorly and, subsequently, are at increased risk for severe influenza (16, 17).
Cis-regulatory genetic variation explains ancestry-associated differences in gene regulation
To assess the contribution of genetic variation to genetic ancestry-associated differences in the transcriptional response to IAV infection, we mapped expression quantitative trait loci (eQTL) in the mock and IAV-infected samples. We focused on cis-eQTL, which we defined as SNPs located either within or flanking (±100 kilobases) each gene tested. We identified at least one cis-eQTL for 2,234 genes (lfsr < 0.10, hereafter referred to as eGenes) across all cell types and conditions (Fig. 3A, Table S8). Independent bulk RNA-sequencing generated from the same samples validated our eGene discovery in the scRNA-seq data (Figs. S4A, B; average adj. R2 = 0.71 for eGene effect sizes in the pseudobulk scRNA-seq and bulk RNA-seq datasets).
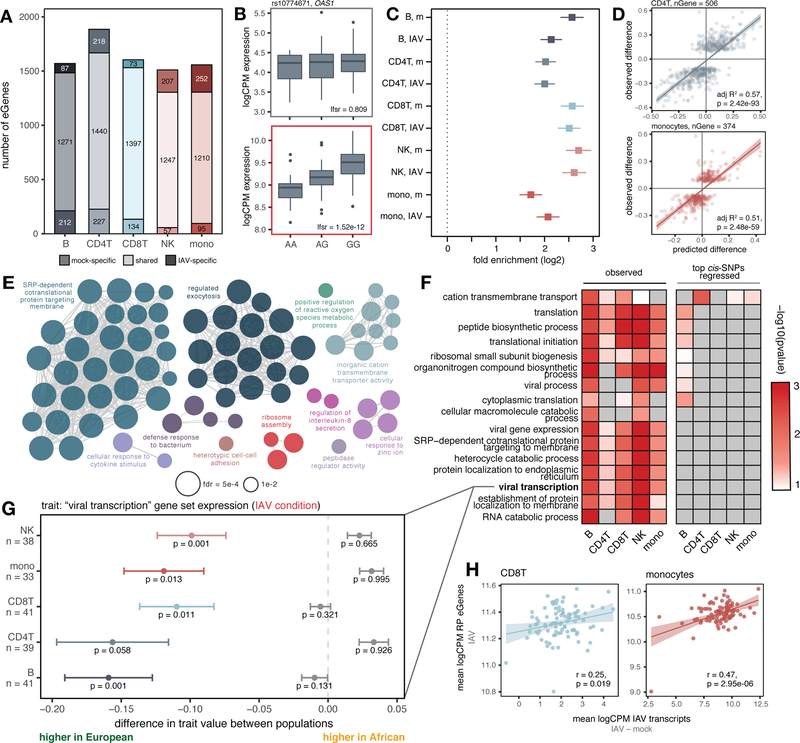
Fig. 3.
Cis-regulatory variation drives differences in the antiviral response. (A) Number of shared and condition-specific eGenes. (B) Condition-specific eQTL example (rs10774671) in CD4+T cells (top: mock, bottom: IAV-infected). (C) Enrichment of eGenes among popDE genes in each cell type/condition determined using logistic regression (log2-fold enrichment with 95% confidence interval; “m” = mock). (D) Correlation of cis-predicted (x-axis) versus observed (y-axis) population differences in expression among popDE genes with an eQTL in CD4+T cells and monocytes. (E) Significant ClueGO enrichments (hypergeometric test, FDR < 0.01) for popDE eGenes across cell types in the IAV-infected condition. (F) Heatmap of −log10 p-values in support of median ancestry-associated differences in gene expression among a subset of enriched GO terms (left) and a model estimating this effect after regressing out the effects of the top cis-SNPs for all genes contained in the term (right). (G) Example of a GO term for which patterns of population variation are compatible with polygenic selection. PopDE genes with an eQTL that belong to the GO term “viral transcription” (n range = 33 – 41 genes) show consistently higher expression levels in European-ancestry individuals (median observed ancestry-associated difference (x-axis) < 0, colored points +/− SE). Following cis-SNP regression (gray points +/− SE), the overall trend for higher expression of viral transcription genes in European- compared to African-ancestry individuals is no longer significant. Empirical p-values were calculated using a permutation-based approach for (F) and (G) ((10) for details). (H) Correlation between IAV transcripts and ribosomal protein eGene expression in CD8+T cells and monocytes. In (D) and (H), p-values and best-fit slopes were obtained from linear regression models.
Although many variants are shared across cell types and conditions (45%, Fig. S4C), 13 – 24% of the eGenes identified within each cell type were only detected in one condition even after probing shared effects with mash (12). A small set of 29 eGenes were also only detectable following infection, including the key IFN-inducible genes OAS1 (Fig. 3B), IFI44L, IFIT1, IRF1, and ISG15 (Fig. S4C).
We next tested whether eGenes were likely to be differentially expressed by genetic ancestry. Across cell types and conditions, eGenes (lfsr < 0.10) were 3.2 to 6.5-fold more likely to be classified as popDE (lfsr < 0.10) than expected by chance (Fig. 3C), and 1.3 to 5.0-fold more likely to specifically belong to the set of IFN-associated popDE genes (Fig. S4D). These enrichments suggest that ancestry-associated differences in gene expression are likely to have a substantial genetic component, perhaps due to divergence in allele frequencies at the causal eQTL. To test this hypothesis, we calculated the correlation between 1) the estimated genetic ancestry effect from our popDE analysis, and 2) the predicted genetic ancestry effect from the effect size of the top eQTL per eGene and the dosage genotype for this SNP across individuals (restricted to popDE genes that were also eGenes in at least one cell type, n = 835 genes; see (10) for details). The genotype and eQTL effect size for the top eQTL alone explained an average of 52.5% (mock) and 53.6% (IAV-infected) of the variance in genetic ancestry effect sizes across cell types (Figs. 3D and S4E). Thus, among popDE genes with an eQTL, over 50% of population differences are explained by differences in the frequency of cis-regulatory variants.
Polygenic selection on ribosomal protein gene expression
We next sought to evaluate if the intersection set of popDE genes and eGenes clustered into specific biological pathways. Among popDE genes where we also observed eQTL, we identified a strong enrichment for many Gene Ontology (GO) terms related to transcriptional and translational processes, including ribosomal small subunit biogenesis and viral transcription (FDR < 3×10−10 in mock and IAV-infected, Fig. 3E, Table S9). Consistent population differences in the expression of genes within the same pathway/gene set could be explained by two hypotheses. First, genes in a given gene set may have evolved under relaxed evolutionary constraint, allowing cis-regulatory variants for these genes to diverge in frequency across populations due to genetic drift. Alternatively, if variants within a given pathway have been a repeated target of selection, they may have experienced directionally concordant shifts in allele frequencies across populations – a pattern consistent with polygenic selection.
We tested for such a pattern in each of the popDE eGene-enriched pathways in all cell type-condition combinations (n = 10: five cell types in the mock and IAV-infected conditions). To do so, we calculated the median genetic ancestry-associated effect on gene expression (i.e., popDE effect size) across all popDE genes in the gene set that also had an eQTL. Under the hypothesis of neutrality, we expect the direction of ancestry-associated effects to be randomly distributed: some genes will be more highly expressed in European-ancestry individuals whereas others will be more highly expressed in African-ancestry individuals. In contrast, under polygenic selection, we expect to find a directional effect, such that most genes for a given pathway show higher expression in one ancestry group versus the other (18). Consistent with a history of polygenic selection, most of the GO terms for ribosomal protein (RP)-related pathways (e.g., ribosomal biogenesis, viral transcription, etc.) show gene expression levels that are consistently higher in individuals with increased European ancestry across cell types (Figs. 3F, “observed”; 3G, colored bars). This pattern holds in both mock-exposed (Fig. S4F) and IAV-infected cells (Figs. 3F, 3G).
An alternative explanation for this observation is that global ancestry is correlated with consistent, directionally biased environmental effects on the expression of genes in RP-related pathways. If so, controlling for local genetic effects on gene expression (e.g., cis-eQTL where allele frequencies are not strongly correlated with ancestry) should not affect the ancestry-gene expression relationship. However, we find the opposite pattern. Specifically, when the effect of the top cis-eQTL for each gene is regressed out, the directional bias towards higher expression with increased European ancestry disappears for all RP-related enriched pathways (Figs. 3F, “top cis-SNPs regressed”; 3G, gray bars). Thus, our results suggest that the higher expression of transcription and translation-related pathways in European-ancestry individuals is driven by the cumulative effect of cis-regulatory variants that affect the regulation of genes within these pathways. This shift may in turn be explained by viral infection-induced selection pressures. In support of this possibility, we observed a strong correlation between the average expression of RP eGenes and IAV transcript expression in both CD8+T cells (Pearson’s r = 0.32, p = 0.002) and monocytes (Pearson’s r = 0.58, p < 1×10−10, Fig. 3H).
Genes differentially expressed between African- and European-ancestry individuals are enriched among genes associated with COVID-19 severity
The immune pathways activated in response to IAV largely overlap those triggered by other single-stranded RNA viruses (19). Thus, our dataset provides an opportunity to evaluate whether differences in COVID-19 susceptibility (caused by SARS-CoV-2, another single-stranded RNA virus) in African Americans and non-Hispanic white Americans (20) could be partially explained by differences in population genetic history. We reasoned that if the genetic ancestry-associated differences in gene expression identified in our in vitro infection model also affect susceptibility to COVID-19, those genes should be enriched among genes associated with COVID-19 disease severity in vivo. To test this hypothesis, we re-analyzed a publicly-available single-cell RNA-sequencing dataset consisting of 505,616 PBMCs across 129 COVID-19 patients with varying degrees of disease severity (21) based on the World Health Organization Ordinal Scale (WOS) for Clinical Improvement (see (10) for details). Using a model adjusting for age, sex, and self-identified race and ethnicity, we identified genes where expression levels correlated with severity (“COVID severity-associated genes”) within each of the five PBMC cell types included in the IAV data set. Monocytes, by far, displayed the largest number of genes associated with severity (n = 839, lfsr < 0.01) (Fig. 4A, Table S10).
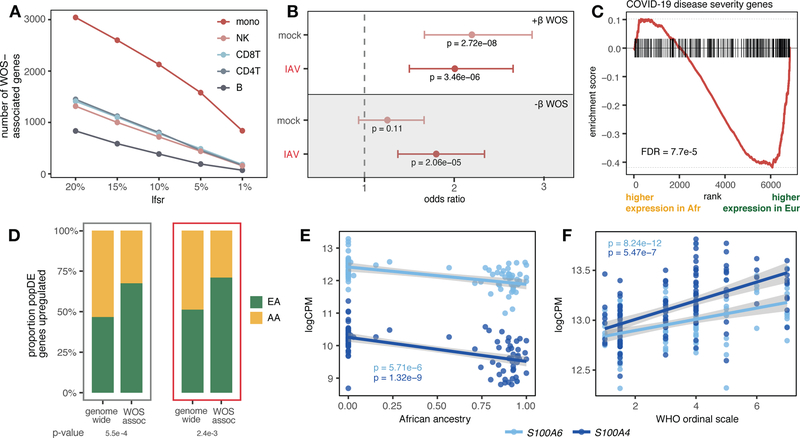
Fig. 4.
Genes associated with COVID-19 severity display population-associated variation in expression. (A) Number of COVID severity-associated genes by cell type for different significance thresholds (x-axis). (B) Enrichment of popDE genes identified in mock and IAV-infected conditions among genes positively (white) and negatively (gray) associated with severity in monocytes (odds ratio with 95% confidence interval). (C) Enrichment plot for genes positively associated with COVID severity in monocytes among the IAV-infection popDE effect sizes in monocytes (x-axis). (D) Proportion of genome-wide popDE and severity-associated popDE genes upregulated by individuals with a higher level of European (green) or African (yellow) genetic ancestry in mock (gray) and IAV-infected conditions (red). (E) Correlation between African genetic ancestry proportion and S100A4/S100A6 expression in monocytes after IAV infection. (F) Correlation between WOS and S100A4/S100A6 expression in COVID-19 patients. In (E) and (F), p-values and best-fit slopes were obtained from linear regression models.
Genes where higher expression was associated with COVID-19 severity in monocytes (lfsr < 0.01) were 2.0 to 2.2 times more likely to be identified as popDE genes in our single-cell IAV dataset (lfsr < 0.10) compared to genome-wide expectations (Fisher’s exact test and permutations, p-values = 2.7×10−8 [mock] and 3.5×10−6 [IAV], Figs. 4B and S5A). These genes also tended to be more highly expressed in monocytes from individuals with more European ancestry (FDRs = 9.8×10−5 [mock], 7.7×10−5 [IAV], Figs. 4C and S5B). Consequently, an average of 69% of COVID severity-associated genes across conditions in monocytes showed increased expression with greater European ancestry, a significantly higher proportion than the 49% observed among all popDE genes (Chi-square test, p-values = 5.5×10−4 [mock] and 2.4×10−3 [IAV], Fig. 4D). Finally, we identified several S100 family genes among those most strongly associated with both genetic ancestry (Fig. 4E) and COVID-19 disease severity (Fig. 4F). Members of this gene family encode proteins that regulate inflammation and can endogenously activate and amplify inflammatory responses in phagocytes (22). S100A4/A6/A8 expression has been associated with patient improvement when upregulated early in the course of COVID-19 infection (21), and S100A8/A9 are systemically upregulated in immune cells, particularly monocytes, in severe, late-stage COVID-19 patients (23). In our data, S100A4, S100A6, and S100A8 are all significantly more highly expressed with greater European ancestry early after IAV infection (Fig. 4E, Table S5), consistent with a potential contribution of genetic ancestry to observed differences in COVID-19 susceptibility between African Americans and European Americans.
Discussion
Together, our results provide a detailed characterization of the genetic determinants that shape inter-individual and genetic ancestry-associated differences during the early response to viral infection in immune cells. Our findings expand on previous work measuring genetic ancestry effects in isolated cell types (5, 6) by showing that the majority of ancestry effects on the immune response to IAV are cell type-specific. One clear exception to this overall pattern was genetic ancestry-associated differences in the IFN response. Our analysis reveals that, across all cell types, increased European ancestry is associated with a stronger type I IFN response shortly after influenza infection, which in turn predicts reduced viral titers at later time points. Given the central role played by interferons in conferring antiviral activity to host cells (16), our findings have potential clinical implications not only for influenza infection but also for other viruses, including SARS-CoV-2, for which the timing and magnitude of IFN-mediated antiviral responses are associated with disease progression and severity (24).
Many of the genetic ancestry-associated differences in immune regulation we observe are driven by allele frequency differences at cis-regulatory variants. Among popDE genes in which we identify at least one cis-eQTL across cell types and conditions, we estimate that, on average, cis-eQTLs explain approximately 53% of the variance in the observed ancestry-associated differences. Our results stress the key role played by genetics in shaping population differences in immune responses, including that these differences are overwhelmingly due to variants found across populations, but segregating at different frequencies (6, 25). We note, however, that for about half of popDE genes, we were not able to identify an eQTL, pointing to additional, co-acting drivers of genetic ancestry-correlated gene expression. These may include other genetic effects (either cis-acting effects or trans-acting effects we are underpowered to map (26, 27)) or unmeasured environmental factors that are stratified by genetic ancestry.
Viruses have been shown to be among the strongest sources of selection pressure in human evolution (1, 2). Among the different forms of natural selection in humans, polygenic selection is thought to be the most pervasive (18), but specific examples of polygenic selection in humans remain rare. Our results provide novel evidence for ancestry-associated directional shifts in molecular traits (i.e., gene expression phenotypes related to specific biological pathways) that are under cis-regulatory genetic control, highlighting the potential role of polygenic selection in the history of these phenotypes. The best candidate for polygenic selection was observed for RP genes, in which we consistently found that alleles associated with higher expression are also more prevalent in individuals with more European ancestry. This observation represents one of the few instances of polygenic selection in humans that is supported by functional genomic data. The signature of selection at ribosomal protein genes is particularly interesting in the context of viral infections, as ribosomal proteins facilitate translation initiation of viral transcripts (28) and directly interact with viral mRNA and proteins to enable viral protein synthesis (29). Further, a subset of ribosomes, known as immunoribosomes, has been hypothesized to preferentially synthesize antigenically-relevant cellular and viral peptides for immunosurveillance by the MHC class I system, which may allow immune cells to more quickly recognize and eliminate infected cells (30). Together, these observations raise the possibility that polygenic selection on ribosomal pathways, acting heterogeneously on different human populations, has contributed to present-day variation in viral control.
Finally, our results show that genes differentially expressed by genetic ancestry are enriched among genes associated with COVID-19 disease severity. Our findings suggest that immune response variation may therefore interact with or exacerbate environmentally-driven health disparities in viral susceptibility and morbidity, which occur for both influenza and COVID-19 (20, 31). An important goal for future work is to evaluate whether the variation we observe early in the viral response translates to differences in COVID-19 patient outcomes. Indeed, time course studies (32, 33) highlight the importance of temporal dynamics in the immune response to infection, which can include time-dependent reversals of effects. For example, the early upregulation of antiviral and proinflammatory genes shortly after initial infection has been associated with protection but their delayed induction is a hallmark of severe illness (34). Our results motivate further studies that investigate whether genetic ancestry-linked effects on innate immunity extend to influence the adaptive immune response as well, and, ultimately, viral clearance and disease severity over the course of viral infections in vivo.
Supplementary Material
Acknowledgements:
We thank J. Tung, B. Mittleman, G. Harrison, and members of the Barreiro lab for their constructive comments and feedback. We thank P. Carbonetto and M. Stephens for advice regarding the mash analyses. We thank J. Sanz for guidance with statistics and modeling. We thank J. Ayroles for providing us with the Tn5 transposase used to generate the TM3’seq libraries. Computational resources were provided by the University of Chicago Research Computing Center. We thank the University of Chicago Cytometry Antibody Technologies Facility (RRID: SCR_017760), particularly D. Leclerc and L. Johnston, for their assistance with the Luminex cytokine assays, and the University of Chicago Genomics Facility (RRID: SCR_019196), especially P. Faber, for their assistance with RNA-sequencing. Figure 1A was created with BioRender.com.
Funding:
This work was supported by grant R01-GM134376 to L.B.B. H.E.R was supported by a National Science Foundation Graduate Research Fellowship (DGE-1746045).
Footnotes
Competing interests: Authors have no competing interests to declare.
Data and materials availability:
Fastq and RNA-sequencing count files are available at GEO under accession GSE162632. Genome sequencing data are available at SRA under accession PRJNA736483. Processed data files, scripts, and associated documentation are available at (35).
References and Notes:
- 1.Fumagalli M et al. , PLOS Genetics. 7, e1002355 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Enard D, Petrov DA, Cell. 175, 360–371.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enard D, Petrov DA, Philosophical Transactions of the Royal Society B: Biological Sciences. 375, 20190575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenney AD et al. , Annu Rev Genet. 51, 241–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MN et al. , Science. 343 (2014), doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quach H et al. , Cell. 167, 643–656.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliva M et al. , Science. 369 (2020), doi: 10.1126/science.aba3066. [DOI] [Google Scholar]
- 8.Ritchie ME et al. , Nucleic Acids Res. 43, e47–e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hou W et al. , Blood. 119, 3128–3131 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Methods are available as supplementary materials.
- 11.Steuerman Y et al. , Cell Systems. 6, 679–691.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urbut SM, Wang G, Carbonetto P, Stephens M, Nat Genet. 51, 187–195 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A et al. , cels. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Hum Genet. 139, 941–948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Q et al. , Science. 370 (2020), doi: 10.1126/science.abd4570. [DOI] [Google Scholar]
- 16.Ciancanelli MJ, Abel L, Zhang S-Y, Casanova J-L, Current Opinion in Immunology. 38, 109–120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomsen MM et al. , European Journal of Immunology. 49, 2111–2114 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Pritchard JK, Pickrell JK, Coop G, Curr Biol. 20, R208–215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen S, Thomsen AR, Journal of Virology. 86, 2900–2910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko JY et al. , Clinical Infectious Diseases. 72, e695–e703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su Y et al. , Cell. 183, 1479–1495.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia C, Braunstein Z, Toomey AC, Zhong J, Rao X, Front. Immunol. 8 (2018), doi: 10.3389/fimmu.2017.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X et al. , Cell. 184, 1895–1913.e19 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee JS, Shin E-C, Nat Rev Immunol. 20, 585–586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nédélec Y et al. , Cell. 167, 657–669.e21 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Lappalainen T et al. , Nature. 501, 506–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westra H-J et al. , Nat Genet. 45, 1238–1243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J-Y, Su W-C, Jeng K-S, Chang T-H, Lai MMC, PLoS Pathog. 8 (2012), doi: 10.1371/journal.ppat.1002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Cells. 8 (2019), doi: 10.3390/cells8050508. [DOI] [Google Scholar]
- 30.Wei J, Yewdell JW, Molecular Immunology. 113, 38–42 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekhar R et al. , Influenza and Other Respiratory Viruses. 11, 479–488 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C et al. , Cell. 184, 1836–1857.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardes JP et al. , Immunity. 53, 1296–1314.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S et al. , Nature Medicine, 1–9 (2021). [Google Scholar]
- 35.Randolph HE, Influenza A response variation scripts (Zenodo, 2021; doi: 10.5281/zenodo.4273999). [DOI] [Google Scholar]
- 36.Aguet F et al. , Nature. 550, 204–213 (2017).29022597 [Google Scholar]
- 37.Fodor E et al. , J Virol. 73, 9679–9682 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG, PNAS. 97, 6108–6113 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hai R et al. , Journal of Virology. 84, 4442–4450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pallares LF, Picard S, Ayroles JF, G3: Genes, Genomes, Genetics. 10, 143–150 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H et al. , Bioinformatics. 30, 1006–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DePristo MA et al. , Nat Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Durbin R, Bioinformatics. 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cingolani P et al. , Fly (Austin). 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sancho-Shimizu V et al. , J Clin Invest. 121, 4889–4902 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Privé F, Aschard H, Ziyatdinov A, Blum MGB, Bioinformatics. 34, 2781–2787 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auton A et al. , Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander DH, Lange K, BMC Bioinformatics. 12, 246 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin M, EMBnet.journal. 17, 10–12 (2011). [Google Scholar]
- 50.Bray NL, Pimentel H, Melsted P, Pachter L, Nature Biotechnology. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Soneson C, Love MI, Robinson MD, F1000Res. 4, 1521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK, Bioinformatics. 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng GXY et al. , Nat Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heaton H et al. , Nature Methods. 17, 615–620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuart T et al. , Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hafemeister C, Satija R, Genome Biology. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storey JD, Tibshirani R, PNAS. 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shabalin AA, Bioinformatics. 28, 1353–1358 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang CC et al. , Gigascience. 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng X et al. , Bioinformatics. 28, 3326–3328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z, BMC Bioinformatics. 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bindea G et al. , Bioinformatics. 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A et al. , Proc Natl Acad Sci U S A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P et al. , Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fastq and RNA-sequencing count files are available at GEO under accession GSE162632. Genome sequencing data are available at SRA under accession PRJNA736483. Processed data files, scripts, and associated documentation are available at (35).