Abstract
A critical step in the activation of integrin receptors is the binding of talin to the cytoplasmic domain of the β subunits. This interaction leads to separation of the integrin α and β trans membrane domain and significant conformational changes in the extracellular domains, resulting in a dramatic increase in the affinity of integrin affinity for ligands. It has long been shown that the membrane bilayer also plays a critical role in the talin - integrin interaction, where anionic lipids are required for proper interaction, yet the specificity for specific anionic headgroups is not clear. In this report we document talin-membrane interactions in solution to membranes of controlled composition using Nanodiscs and a new FRET assay. We show that recruitment of the talin head domain to the membrane surface is governed by charge in the absence of other adapter proteins. In addition, measurement of the donor acceptor distances reveals that anionic lipids promote a conformational change in the talin head domain allowing interaction of the F3 domain with the phospholipid bilayer. The magnitude of this conformational change is regulated by the identity of the phospholipid headgroup with phosphatidylinositides promoting the largest change. This emphasizes the importance of phoshpaidylinositol-4,5-bisphosphate (PIP2) in converting talin to a conformation optimized for interactions with integrin cytoplasmic tails.
Keywords: talin, lipid bilayer, conformational change, phosphatidylinositol, phosphatidylserine
INTRODUCTION
Talin is a large adapter protein that plays a key role in connecting the actin cytoskeleton to the extracellular matrix (ECM)1 via integrin receptors. Talin was discovered nearly 30 years ago and since that time many studies have shown that it is involved in the final step of integrin inside-out activation (1-3). Talin is composed of a globular head domain (THD) that is homologous to the domain found in band 4.1/ezrin/radixin/moesin family of proteins (FERM domain). THD differs from a canonical FERM domain by the presence of an additional 85 amino acid at the C-terminus termed F0. In comparison to canonical FERM domains, THD adopts a linear configuration of F0-F3 as opposed to the standard cloverleaf structure seen in other FERM domains (4). Talin also contains a large rod domain which consists of 13 helical bundles and a small dimerization helix (4,5). Functionally, the head domain is critical for the activation of integrin through interactions with the cytoplasmic tails of β integrins and the cytoplasmic membrane surface, while the rod domain provides a link to the actin cytoskeleton via interactions with actin and other adapter proteins such as vinculin (6-9). The rod domain also plays a role in the regulation of talin through a self association with the head domain thus creating an autoinhibited form of talin (10,11). A recent study has shown that talin exists as a dimer and adopts a complex donut-like structure in what would presumably be the auto inhibited form (12). Several mechanisms of talin activation have been proposed; interaction with rap1 interacting adapter molecule (RIAM), calpain, cleavage or through interaction with PIP2 (13-15). Talin activation will be the focus of future studies.
Integrins are heterodimeric membrane receptors that link the ECM to the actin cytoskeleton. Integrins consist of one α- subunit and one β-subunit which associate via an interaction between the transmembrane (TM) helices within the phospholipid bilayer (16,17). In platelet integrin αIIbβ3, key interactions that stabilize the heterodimer consist of the outer membrane clasp (OMC) which involves interaction of closely packed glycine residues, and the inner membrane clasp (IMC) that is defined by the packing of two Phe residues on the α-TM domain with the β-TM domain and the formation of a salt bridge between α-D723 and β-R995 (18). It is the association of the TM domains that stabilize the inactive form of integrins, where the extracellular domain is in a conformation that has low affinity for extracellular ligands (19). Integrins are activated through “inside out signaling” converting the ECM domains to a high affinity state, allowing the interaction with extracellular ligands and the subsequent transmission of signals across the cell membrane (“outside in signaling”). There is an abundance of evidence that shows talin to be a key activator of integrin during inside-out signaling (3,20-23).
The mechanism of talin mediated activation is known to involve the interaction of the THD with the cytoplasmic tail of β-integrins (20,23,24). This interaction is believed to separate the α and β tails by disrupting key interactions in the IMC. Interaction of the F3 domain with the Phospho-Tyrosine Binding (PTB) NPxY motif on the β-cytoplasmic tail and disruption of the α - β salt bridge promote the separation of the integrin TM helices (18,25-28). It is becoming clearer that precise anchoring of the head domain to the membrane surface is critical for proper activation of integrin. Kim et al., using a novel fluorescence assay, have shown that interaction of THD with anionic lipid promotes the tilt of membrane embedded β3 tails (29). Charge reversal mutations on the F2 and F3 domains which inhibit membrane association decreased the tilting of the β-TM domain emphasizing the importance of a tight association of talin with the bilayer surface.
In recent years several laboratories have directed their attention toward the role of the lipid environment, particularly anionic lipids, in the structural mechanism of inside out signaling (14,30,31). The crystal structure of the THD shows a large dipole moment with a positively charged face primarily located on the F2 and F3 domains. The charge asymmetry is required for recruiting talin to the membrane surface. A positively charged patch of residues residing on the F2 domain have been named the membrane orientation patch (MOP) which steer the head domain toward negatively charged membrane surfaces as well as orient the F3 domain in close proximity for efficient interaction with integrin tails (32,33).
Although the overall membrane charge has been shown to control the affinity of talin head domain for the bilayer surface, it has been suggested that phoshpaidylinositol-4,5-bisphosphate (PIP2) plays a specific role in the talin mediated integrin activation by promoting conformational changes and activating the autoinhibited form (4,14,34). A recent study has suggested a push pull mechanism that allows PIP2 to promote the dissociation of the positively charged regulatory segment of the rod domain (34). Adding more evidence to the regulatory role is the interaction of phosphatidylinositol phosphate kinase type Iγ (PIPKIγ) with talin that promote the recruitment of talin and PIPKIγ to the membrane surface (35,36). PIPKIγ is responsible for the generation of PIP2 resulting in elevated PIP2 levels at the sites of adhesion (37).
Anionic lipids not only enhance the affinity of THD/bilayer interactions, they have been shown to promote functionally important conformational changes in the talin structure. Evidence for lipid induced conformational changes were first described by Martel et al. that showed a differential sensitivity to proteolysis upon binding to phosphatidylinositides (14). More recently advanced Molecular Dynamics (MD) simulations have shown that interaction of talin with the lipid bilayer converts the linear arrangement of the F0-F3 domains to a “V” conformation having an angle 60 degrees between F0F1 and F2F3 (38). This conformation is shown to optimize membrane contacts and increase the number of H-bonds upon binding to β-tails. In yet another MD simulation, Arcario and Tajkhorshid used a Highly Mobile Membrane Mimetic (HMMM) to capture snapshots of the interaction of the F2F3 domain to an a ionic membrane surface (33). They confirmed the role of the MOP in steering the head domain toward the bilayer in an orientation conducive to interaction with integrin tails. Their simulations also identified a buried phenylalanine rich area that opens up and allows the embedding of the phenylalanine residues forming a membrane anchor. This conformational change is triggered via the snorkeling of the lysine residues in the MOP upon interaction with anionic lipids. Interestingly the formation of the membrane anchor also promotes a conformational change in the F3 domain, bringing K325 and K327 (K322 and K324 in talin 1) to the membrane surface and in an optimal conformation for interaction with integrin tails.
In this study we have developed a FRET based solution assay using Nanodiscs as a controlled membrane surface to investigate the influences of the lipid headgroup on THD /bilayers interactions. We also compare how anionic lipids can control the conformational equilibrium of the THD-Nanodisc complex by calculating the FRET donor acceptor distances. We report for the first time an experimentally observable conformational change consistent with previous MD simulations that is sensitive to the identity of the lipid headgroup present in the THD binding site.
EXPERIMENTAL PROCEDURES
Materials:
Phospholipids DMPC (PC), DMPS (PS), DMPG (PG), DMPA (PA), brain PI(4,5)P2 (PIP2) and brain phoshpaidylinositol-4-phosphate (PIP) were purchased from Avanti Polar Lipids. Membrane scaffold proteins (MSP) were expressed in E. coli BL21 DE3 and purified as previously described (39). Tetra-methyl rhodamine (5 and 6) maleimide (TAMRA) was purchased from Anaspec. Uniblue A (UA) was obtained form Sigma-Aldrich. iProof polymerase was obtained from BioRad. pET30A-THD for expression of talin 1 head domain was a kind gift from Dr. Mark Ginsberg.
Talin Head Domain (THD) Mutagenesis:
THD I398C and D201C mutants were generated by using pET30a-THD as the template plasmid. THD K322E, K324E, mutants were made using pET30a-THD-I398C as template for PCR mutagenesis. iProof polymerase was used for all PCR reactions. Primers were designed using the SerialCloner software package and purchased from Integrated DNA Technologies. Sequencing was performed by ACGT inc.
Preparation of TAMRA Labeled MSP:
MSP1 D73C was labeled with TAMRA by first incubating with Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 20 mM Tris pH 7.4, 100 mM NaCl to reduce any disulfides bonds that may have formed. After 10 minutes a 10 fold molar excess of TAMRA dissolved in dry DMSO was added dropwise to the protein solution with stirring under an argon atmosphere. Reactions were allowed to proceed for 4 hours at room temperature, followed by overnight incubation at 4 C. Excess TAMRA was removed by incubation with Amberlite XAD hyrdophiobic beads followed by passage over a Sephadex G25 column equilibrated in 20 mM Tris pH 7.4 100 mM NaCl. Labeling efficiency was calculated using ε280nm = 21 mM−1 for MSP, and ε557nm = 60 mM−1 for TAMRA. An A280 correction factor of 0.34 was used to subtract the absorbance of the dye at 280 nm, allowing determination of the protein concentration. Final Dye to protein ratios were >80%.
Nanodisc Preparation:
Noanodisc were prepared as previously decribed with slight modifications (39). Briefly phospholipids mixtures dissolved in cholorform were dried under vacuum overnight. Lipids were solubilized in 200 mM sodium cholate and TAMRA labeled MSP was added to achieve a 100:1 lipid to MSP ratio. After mixing, the sample was incubated with Amberlite XAD hydrophobic beads to remove the detergent and initiate disc assembly. The discs were purified by passing over an S200 Increase size exclusion column (GE Healthcare) equilibrated in 20 mM HEPES pH 7.2 4 mM KH2PO4, 125 mM KCl, 14 mM NaCl, 1 mM MgCl2, 0.02 mM EGTA.
Preparation of (UA) Labeled THD:
Four equivalents of TCEP (pH 7.4) were added to THD I398C or D201C (~100 μM) in 20 mM Tris pH 7.0, 100 mM NaCl. After 10 minutes, 2 equivalents of UniBlueA dissolved in DMSO were added drop wise to the sample. 100 μL aliquots were taken every 15 minutes and the excess dye was removed dye by gel filtration on a 2 ml Sephadex G25 column. The absorbances at 280 nm and 595 nm were measured and the dye to protein ratio was calculated using ε280 nm = 42 mM−1 for THD, and ε595 nm = 11 mM−1 for UA, and a 280 nm correction factor of 1.45. The reaction was allowed to proceed until the labeling efficiency was >95% after which the reaction was stopped by addition of dithiothreitol to a final concentration of 10 mM. Free dye was removed by passing the sample over a G25 column equilibrated in 20 mM HEPES pH 7.2, 4 mM KH2PO4, 125 mM KCl, 14 mM NaCl, 1 mM MgCl2, 0.02 mM EGTA. Final dye to protein ratios were > 95%.
FRET binding assay:
The FRET based assay was adopted from that of Bayburt et al (40). Fluorescence quenching experiments were performed in a Hitachi 3010 fluorometer equipped with a circulating water bath for temperature control using an excitation wavelength of 557 nm and an emission wavelength of 578 nm. Typically 50 μM UA labeled THD was titrated into 100 nM TAMRA labeled Nanodisc solutions in buffer containing 20 mM HEPES pH 7.2 4 mM KH2PO4, 125 mM KCl, 14 mM NaCl, 1 mM MgCl2, 0.02 mM EGTA at 20 °C. After mixing, samples were allowed to equilibrate and the florescence intensities were recorded after the signal stabilized, typically after 3 – 5 minutes. FRET Efficiency is calculated using the following equation:
| (1) |
where F0 is the fluorescence intensity of TAMRA labeled Nanodiscs in the absence of UB labeled talin and F is the fluorescence at each titration point.
TAMRA labeled MSP quantum Yield and Forster distance determination:
The quantum yield of the TAMRA labeled Nanodiscs was determined by comparing the fluorescence intensity to that of a quantum yield standard using the following equation:
| (2) |
where QR is the quantum yield of rhodamine B in water (0.31) I, IR n, nR are the intensities and indices of refraction of the sample and reference respectively. In our case n = nR. For a more accurate measure of the quantum yield we used 5 samples of TAMRA labeled Nanodiscs and rhodamine B prepared having optical densities ranging from 0.005 to 0.01 at 540nm. Emission spectra were collected from 560 nm to 650 nm using 540 nm excitation. The integrated fluorescence intensity vs. optical density was fit to a gradient function. The quantum yield is then calculated as follows:
| (3) |
The Forster radius (R0) for the TAMRA UA donor acceptor pair was determined using the following equation:
| (4) |
where κ2 is an constant related to the orientation of the transition dipoles of the donor and acceptor and is assumed to be 2/3, ΦD is the quantum yield of the donor, n is the index of refraction of the solution, and J(λ) is the overlap integral of the donor’s emission spectrum and the acceptor’s absorbance spectrum and is equal to:
| (5) |
where εA (λ)is the absorption spectrum of the acceptor in units of M−1cm−1 and FD(λ) is the emission spectrum of the donor normalized to an area of 1 (41). Fluorescence emission spectra of TAMRA labeled MSP1 were measured using 540 nm excitation and normalized to an area of 1. The absorbance spectra of Uniblue A labeled Talin was measured and normalized to an extinction of 11,000 M−1 cm−1 at 595 nm. The a∣e - UV-Vis-IR Spectral Software (www.fluortools.com) was used to calculate the overlap integral from the normalized spectra. Using equations 3, 4 and 5 we calculated an R0 of 41 Å for the TAMRA – UA dye pair.
RESULTS
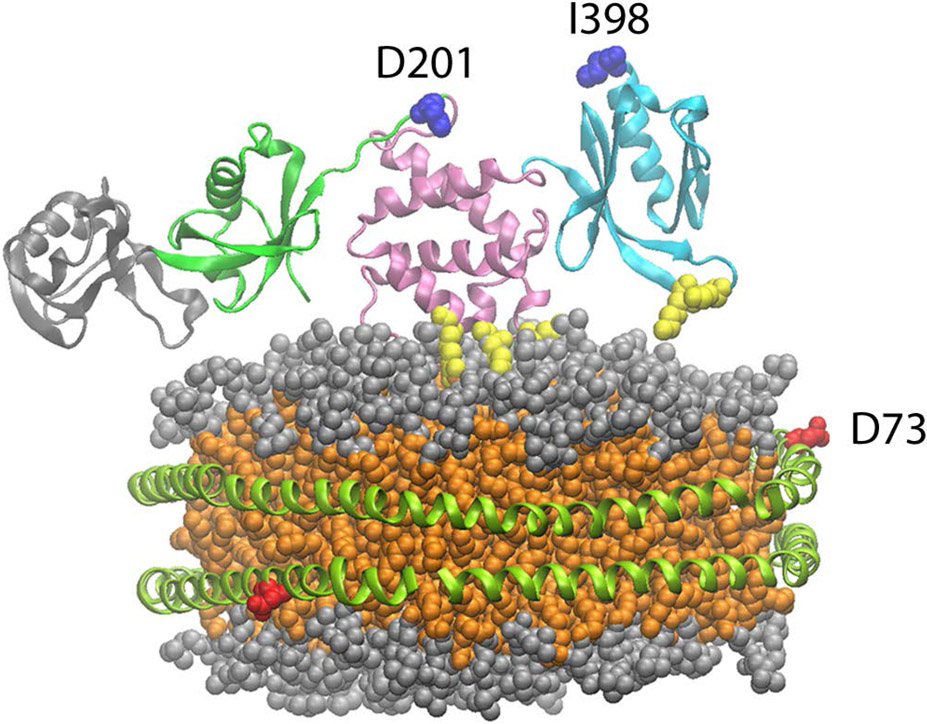
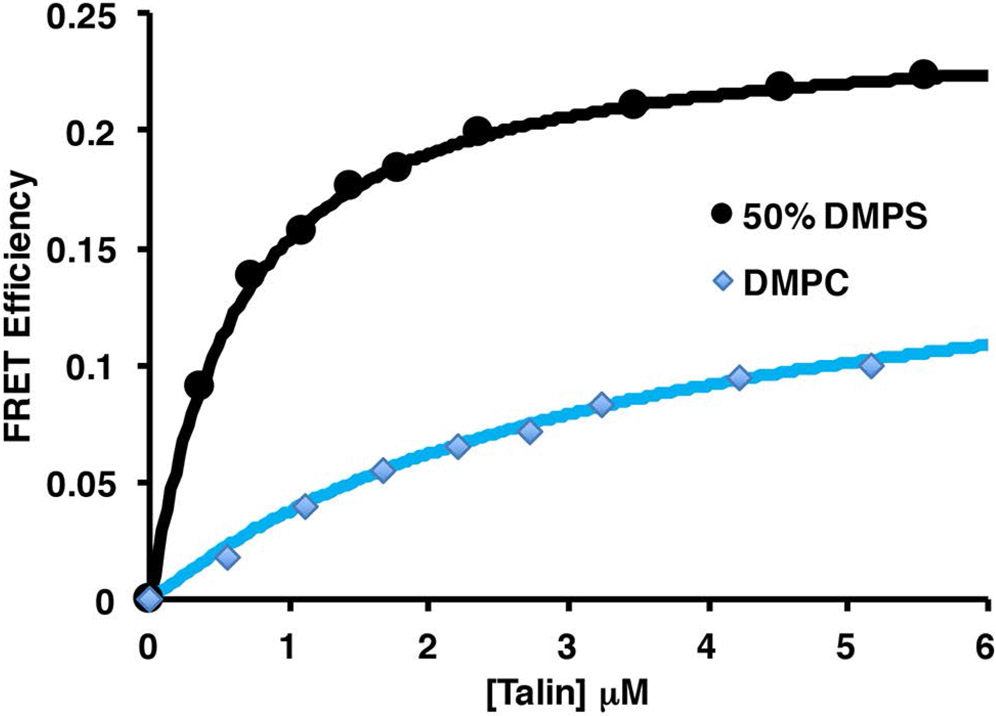
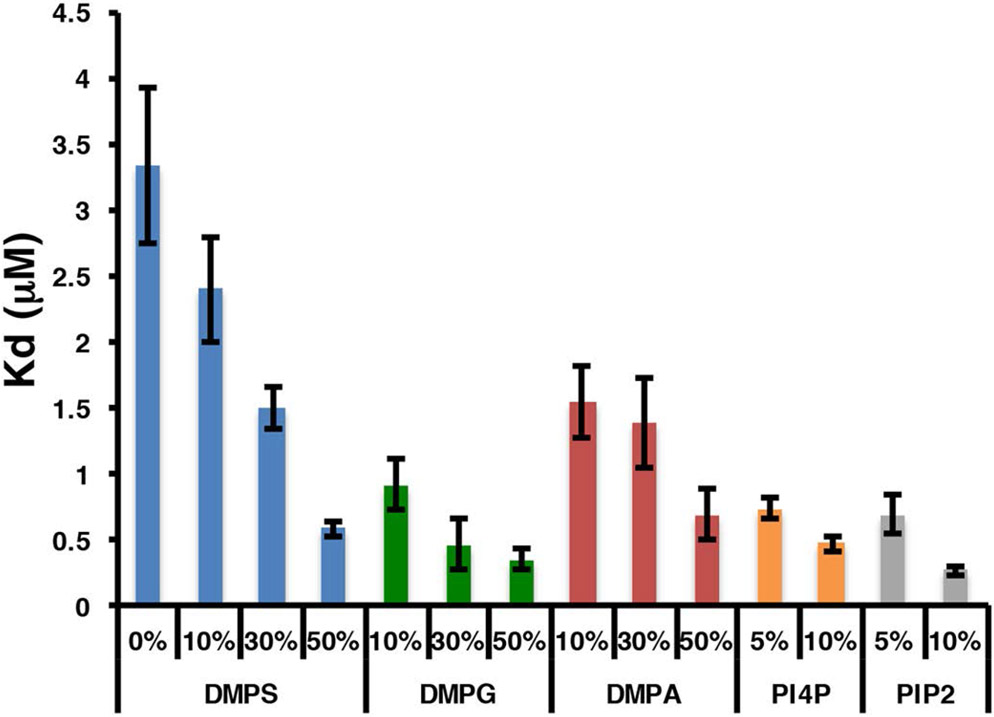
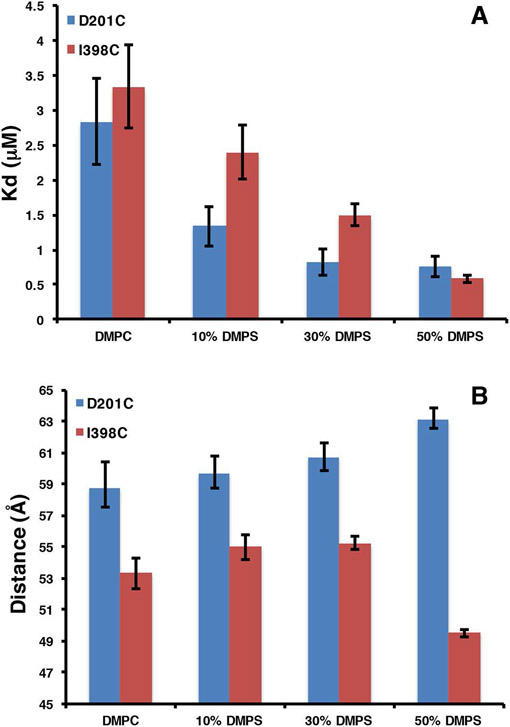
Talin has long been known to interact with negatively charged bilayers, yet it is not clear if there exists a preference for a specific negatively charged headgroup. What is also not clear is the functional role of the headgroup identity. PIP2 has been reported to be an activator of the auto inhibited form of talin suggesting specificity over other negatively charged headgroups (10,34). To investigate further the role of the phospholipid headgroup in talin – membrane interactions we have used a FRET based assay to measure the interaction of talin with negatively charged phospholipids. Figure 1 shows a molecular model of talin docked the surface of a 10 nm Nanodisc (4,42). In our system the MSP belt of the Nanodsic is labeled with the Fluorophore TAMRA at position 73 and Talin is labeled with the dark quencher Uniblue A at either position 398 in the F3 domain or 201 in the F2 domain. We prepared TAMRA labeled Nanodiscs containing 0, 10, 30, and 50% PS, PA and PG as well as 5% and 10% PIP and PIP2. Figure 2 shows a representative binding isotherm for the interaction of THD labeled at position 398 with Uniblue A (THD-398-UA) with TAMRA labeled Nanodiscs containing 50% DMPS / 50% DMPC and 100% DMPC. Binding isotherms fit well to a single binding site model assuming one talin binds per Nanodisc leaflet (solid line). Figure 3 summarizes the dissociation constants for all lipids tested in our study. As expected there exists a clear trend of increased affinity with increasing anionic lipid content. The dissociation constant decreases 5 – 10 fold when in the presence of 50% DMPS, DMPA or DMPG. Interestingly in the binding of talin to Nanodiscs containing PIP2 or PIP only 5% is needed to reach a 5 to 10 fold increase in affinity. This clearly points to specificity towards phosphatidylinositides.
Figure 1.
Model of talin binding to a Nanodisc. MSP is site specifically labeled at position 73 with TAMRA. Talin head domain is labeled at position 398 on the F3 domain or at position 201 on the F2 domain. Colors used: Talin; F0-gray, F1-green, F2-pink, F3-cyan, MOP and FAP residues – yellow, Nanodisc; MSP-green, lipid headgroups -gray, Lipid acyl chains-orange. Talin coordinates from PDB3IVF (4).
Figure 2.
FRET efficiency of TAMRA 50% DMPS and 100% DMPC Nanodiscs as a function of UA labeled THD. Data is fit to a Lamgmuir binding isotherm using a single binding site model.
Figure 3.
Dissociation constants of THD binding to anionic bilayers.
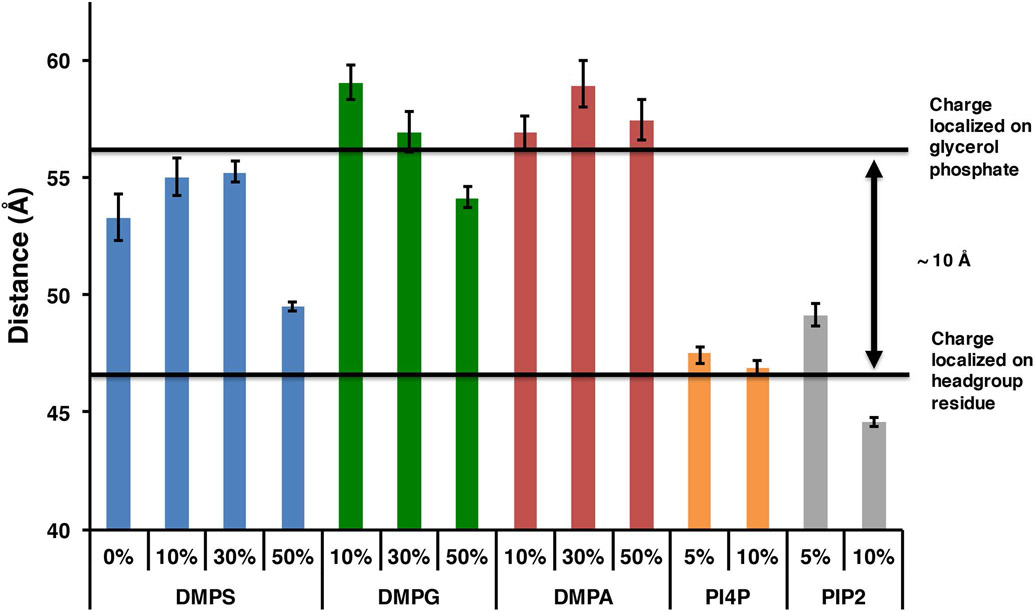
Since our assay is based on FRET and our acceptor dye is a dark quencher, we can easily measure the average donor acceptor distances from the FRET efficiencies. We determined the Forster distance of the TAMRA – Uniblue A pair to be 41 Å making this dye pair ideal for measuring donor acceptor distances between 30 and 60 Å where the FRETefficiency is expected to be greater than 10%. In our Nanodisc binding assay the measured dye separation distances represent the spacing between the labeled residue on talin and position 73 on the MSP belt. (figure 1). We assume THD can bind to both sides of the Nanodisc in any azimuthal orientation thus the measured distance represents an average of all orientations in the complex. Figure 4 summarizes the donor acceptor distances for all the phospholipids tested when THD is labeled in the F3 domain at position 398. On bilayers with high anionic lipid content, the donor acceptor distances become shorter. Since our label resides on the F3 domain, the shorter distances suggest that increased anionic lipid content favors a conformation of the complex in which the F3 domain is closer to the bilayer. Interestingly PIP2 and PIP promote this conformational change at just 5% mole fraction. In order to confirm that we are indeed probing a large conformational change in the F3 domain, the labeling site was moved to position 201 (THD-201-UA ) located on the back side of the F2 domain. Figure 5A shows that the affinity of THD398-UA and THD-201-UA for DMPS bilayers are similar. In contrast, inspection of the dye separation distance shows that it does not decrease but rather increases from 59 to 63 Å (figure 5B).
Figure 4.
Dye separation distances of THD-398-UA.
Figure 5.
Compairison of THD-398-UA and THD-201UA binding to DMPS bilayers. A, dissociation constants. B, dye separation distances.
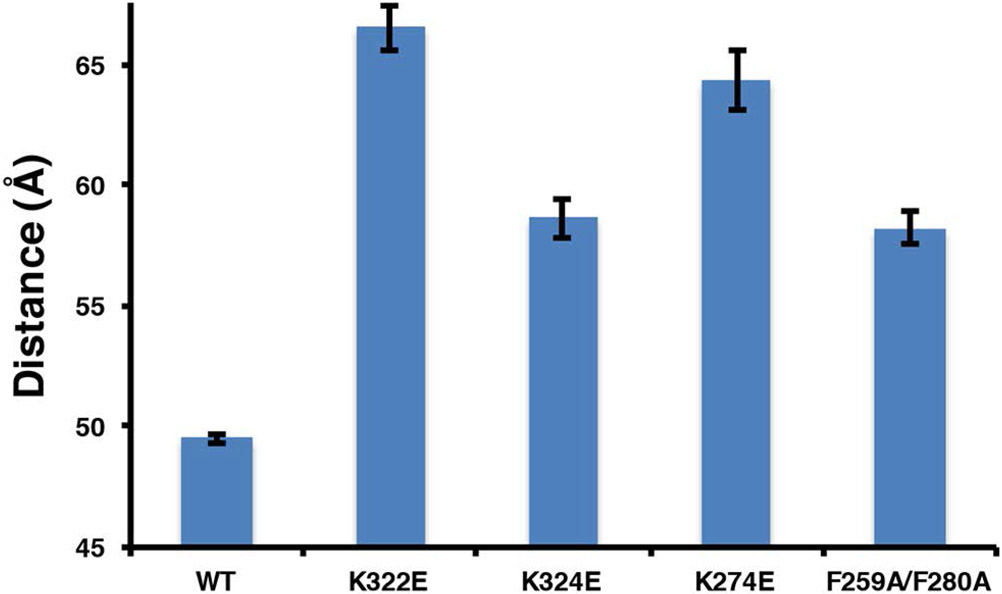
Recent MD simulations have suggested key residues in the F2 and F3 domains that promote and stabilize this proposed conformational change (33). To further test this hypotheses, we generated the charge reversal mutants K322E and K324E in the F3 domain, and K274E in the F2 domain. In addition we also mutated the proposed membrane anchor F259 and F280 to Alanine (F259A, F280A). K322 is believed to interact with the anionic membrane surface while K324 has been shown to form a key salt bridge with an acidic residue in the integrin β-tail during talin mediated integrin activation (30). The mutations have only a modest effect on the dissociation constants (data not shown), consistent with membrane binding being driven by the overall electrostatics of the THD – membrane interactions (30). Figure 6 shows the dye separation distances of the mutants and despite the fact that the dissociation constants are similar, we see that the label in the F3 domain of the mutants are 10 – 15 Å further away from the TAMRA label suggesting a major change in the conformation of the complex compared to wild type.
Figure 6.
Dye separation distances for MOP and FAP mutants binding to 50% DMPS bilayers.
DISCUSSION
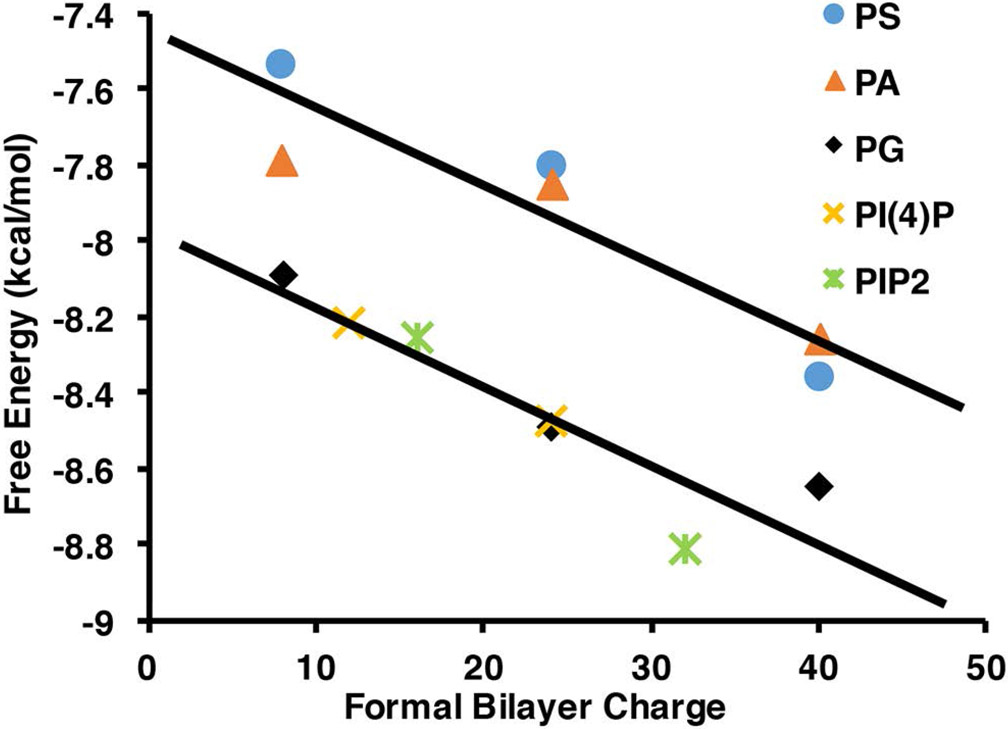
We have utilized Nanodiscs and their ability to precisely control the phospholipid content to elucidate the mechanism of talin interactions with the membrane surface. Previous studies have shown that anionic lipids are required for efficient talin mediated integrin activation (15). Although the importance of PIP2 in the activation of auto inhibited talin has been suggested, it is still unclear if this preference is based solely on the differences in the charge of the membrane bilayer or if the structure of the headgroup is providing some specificity. Figure 3 shows the dissociation constants of THD binding to bilayers containing various anionic lipids. The dissociation constant (Kd) decreases as anionic lipid content increases. The weakest interaction is between pure DMPC bilayers with a Kd of 3.3 μM and the tightest being interactions with membranes containing 10% PIP2 displaying a Kd of 0.25 μM. Comparing identical mole % anionic lipids we see that the Kd differs by no more than a factor of 3 for PS, PA and PG while 5% and 10% PIP and PIP2 affinities are comparable to 50% PS, PG, and PA. This difference is presumably due to the multiple charges present on the PIP and PIP2 headgroups and a specific headgroup interaction. For a more direct comparison we have plotted the free energies of association vs. formal bilayer charge of the Nanodisc samples (figure 7). The formal bilayer charge is calculated by multiplying the headgroup charge by the mole % anionic lipids. Since the experiments are performed at pH 7.2 we use a charge of −1 for PS, PA, and PG, −3 for PIP, and −4 for PIP2 (15). There is an obvious linear grouping of the free energies of PG, PIP, and PIP2 lying on a line that is 0.75 kcal /mol lower in free energy of association than the line formed by PS and PA headgroups. The differences here could be explained by the identity of the charged moiety on the headgroup. Those that have the lower free energy of association possess a negatively charged phosphate group and a sugar based headgroup. This points to a subtle free energy preference to headgroups providing some polar interactions in addition to the main electrostatic driving force.
Figure 7.
Comparison of free energy differences of talin binding to anionic lipid headgroups.
In addition to the determinations of the binding free energy the FRET efficiency provides us a measure of the donor – acceptor dye separation distances. With the label located on the back of the F3 domain at residue 398, the shorter FRET distances are representative of the closer approach of the F3 domain toward the bilayer in the presence of anionic lipids. These results are in complete agreement with MD simulations that predict a significant conformational change in the THD upon engaging a negatively charged bilayer (33). In this model the F3 domain moves down 5 – 10 Å after initial docking of the MOP and promotes the interaction of K325 (K322 in talin 1) and K327 (K324 in talin 1) with the surface of the membrane. To confirm our interpretation, we moved the labeling site to the back of the F2 domain at residue 201. In this case the dye separation as a function of PS content slightly increases, which in itself may be a result of the F3 domain engaging the membrane at high anionic lipid content (figure 5B). Despite the differences in the dye separation, the measured dissociation constant is the same regardless of the THD labeling site (figure 5A).
Although there is a general trend of decreasing dye separation with increasing anionic lipids, not all anionic lipid headgroups promote the same conformational change. Comparing DMPS, DMPA and DMPG bilayers which all have identical formal charges, we see that on 50% PS membranes the F3 domain is 5 – 8 Å closer than on 50% PA and 50% PG membranes. Similarly on 5 and 10% PIP and PIP2 bilayers the F3 distances are 10 Å closer than on 50% PA and 50% PG membranes. The key differences between the 50% PS, PIP, PIP2 bilayers and the PA, PG bilayers are two-fold. First there is the charge localization; PS, PIP, and PIP2 membranes carry the charge on the headgroup residue, while PA and PG carry the negative charge on the glycerophosphate. Second, the PIP lipids have a charge of −3 and −4 creating a much higher local charge density in the binding site. Taken as a whole there is a requirement for a high charge density and that the charge is localized out away from the glycerol backbone to trigger this conformation change. 50% PS , 5% PIP, 10% PIP, 5% PIP2 and 10% PIP2 all fit this requirement. We see that when a bilayer has a high charge density, either local or in bulk, and that the charge is localized on the headgroup residue, the F3 domain binds in a geometry nearly 10 Å closer than when the charge is localized on the glycerol phosphate (4). We suggest that in the case of DMPS, only 50% PS bilayers present a local charge density comparable to that of PIP and PIP2 thus triggering this large conformational change.
MD simulations have identified key residues in the F2 MOP and the F3 FAP that are important in triggering the conformational change (33). These are the lysine residues on the F2 domain which are proposed to snorkel into the anionic bilayer, exposing a phenaylalanine rich pocket that forms a stable membrane anchor. The formation of the membrane anchor promotes a large conformational change in the F3 domain, bringing it to the bilayer surface. We have made the mutations K322E and K324E in the F3 association patch (FAP), and K276 in the MOP, F259A/F280A in the membrane anchor to see the affects on the measured conformational change. All four mutants display a only modest change in Kd, increasing by no more than a factor of 2 (WT = 0.6 μM, K324E = 1.2 μM, K322E = 0.4 μM, K274E = 0.9 μM, and F259A/F280A = 0.7 μM) indicating that the remaining positive charges provide a significant portion of the overall free energy of association. Although the affinities are minimally affected, the lysine to glutamate mutations at positions 322, 324, and 276, alter the conformation of the THD bilayer complex. The dye separation distances are nearly 10 Å longer than in the wild type complex. This can be explained in two ways. First, the clash of the negatively charged glutamates at position 322 and 324 with the negatively charged bilayer does not allow the close association of the F3 domain. Second, mutation of K276E prevents the snorkeling required to open up the membrane anchor pocket, inhibiting the trigger of the large conformational change. Similarly, when we mutate the phenylalanine residues implicated in the formation of the membrane anchor, we once again see that the F3 domain remains 10 Å further from the bilayer surface. It is important to note that the F259A/F280A mutant presumably has the same electrostatic interactions as the wild-type yet, the conformational change is inhibited.
In summary, our studies utilized a novel FRET and Nanodiscs based assay to detect protein-membrane interactions. Particularly, our results reveal a rich interplay between electrostatics, and conformational equilibrium that governs the formation of the complex between phospholipid bilayers and talin FERM domain. We show that altering membrane compositions modulates affinities of talin and phospholipid bilayers as well as induces different talin binding geometry. We propose that PIP2 plays a critical role in integrin activation by stimulating a conformation in the THD that is optimized for interaction with integrin tails similar to that proposed by Arcario et al. on PS membranes (33). These results emphasize the importance of the interaction of talin with PIPKIγ and production of PIP2 at the site of adhesion.
Acknowledgments:
This work was funded by NIH grant GM101048 and GM33775. We would like to thank Dr. Mark H. Ginsberg and Dr. Feng Ye for the gift of the THD expression vector.
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest with the contents of this article
Abbreviations used: ECM, extra cellular matrix; PIP2, phoshpaidylinositol-4,5-bisphosphate; PIP, phoshpaidylinositol-4-phosphate; FERM, band 4.1/ezrin/radixin/moesin; THD, talin head domain; TM, trans-membrane; OMC, outer membrane clasp; IMC, inner membrane clasp; MD, Molecular Dynamics; TAMRA, Tetra-methyl rhodamine (5 and 6) maleimide; MSP, Membrane Scaffold Protein; Uniblue A , UA;
REFERENCES
- 1.Burridge K, and Connell L (1983) A new protein of adhesion plaques and ruffling membranes. The Journal of cell biology 97, 359–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthis NJ, and Campbell ID (2011) The tail of integrin activation. Trends in biochemical sciences 36, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye F, Hu G, Taylor D, Ratnikov B, Bobkov AA, McLean MA, Sligar SG, Taylor KA, and Ginsberg MH (2010) Recreation of the terminal events in physiological integrin activation. The Journal of cell biology 188, 157–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elliott PR, Goult BT, Kopp PM, Bate N, Grossmann JG, Roberts GC, Critchley DR, and Barsukov IL (2010) The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure 18, 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GC, Ballestrem C, Critchley DR, and Barsukov IL (2013) RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem 288, 8238–8249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, and Critchley DR (2010) Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem 285, 29577–29587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gingras AR, Vogel KP, Steinhoff HJ, Ziegler WH, Patel B, Emsley J, Critchley DR, Roberts GC, and Barsukov IL (2006) Structural and dynamic characterization of a vinculin binding site in the talin rod. Biochemistry 45, 1805–1817 [DOI] [PubMed] [Google Scholar]
- 8.Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, and Emsley J (2005) Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem 280, 37217–37224 [DOI] [PubMed] [Google Scholar]
- 9.Goldmann WH, Senger R, Kaufmann S, and Isenberg G (1995) Determination of the affinity of talin and vinculin to charged lipid vesicles: a light scatter study. FEBS letters 368, 516–518 [DOI] [PubMed] [Google Scholar]
- 10.Goksoy E, Ma YQ, Wang X, Kong X, Perera D, Plow EF, and Qin J (2008) Structural basis for the autoinhibition of talin in regulating integrin activation. Molecular cell 31, 124–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GC, and Critchley DR (2009) The structure of an interdomain complex that regulates talin activity. J Biol Chem 284, 15097–15106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, and Hanein D (2013) Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. Journal of structural biology 184, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan B, Calderwood DA, Yaspan B, and Ginsberg MH (2001) Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J Biol Chem 276, 28164–28170 [DOI] [PubMed] [Google Scholar]
- 14.Martel V, Racaud-Sultan C, Dupe S, Marie C, Paulhe F, Galmiche A, Block MR, and Albiges-Rizo C (2001) Conformation, localization, and integrin binding of talin depend on its interaction with phosphoinositides. J Biol Chem 276, 21217–21227 [DOI] [PubMed] [Google Scholar]
- 15.Moore DT, Nygren P, Jo H, Boesze-Battaglia K, Bennett JS, and DeGrado WF (2012) Affinity of talin-1 for the beta3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc Natl Acad Sci U S A 109, 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C, Lau TL, Ulmer TS, and Ginsberg MH (2009) Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood 113, 4747–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Ma YQ, Page RC, Misra S, Plow EF, and Qin J (2009) Structure of an integrin alphaIIb beta3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc Natl Acad Sci U S A 106, 17729–17734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau TL, Kim C, Ginsberg MH, and Ulmer TS (2009) The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. The EMBO journal 28, 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo BH, Springer TA, and Takagi J (2004) A specific interface between integrin transmembrane helices and affinity for ligand. PLoS biology 2, e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calderwood DA, Zent R, Grant R, Rees DJ, Hynes RO, and Ginsberg MH (1999) The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J Biol Chem 274, 28071–28074 [DOI] [PubMed] [Google Scholar]
- 21.Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RA, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, and Ginsberg MH (2007) Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. The Journal of experimental medicine 204, 3103–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, and Campbell ID (2007) Structural basis of integrin activation by talin. Cell 128, 171–182 [DOI] [PubMed] [Google Scholar]
- 23.Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, and Calderwood DA (2003) Talin binding to integrin beta tails: a final common step in integrin activation. Science 302, 103–106 [DOI] [PubMed] [Google Scholar]
- 24.Campbell ID, and Ginsberg MH (2004) The talin-tail interaction places integrin activation on FERM ground. Trends in biochemical sciences 29, 429–435 [DOI] [PubMed] [Google Scholar]
- 25.Luo BH, Carman CV, Takagi J, and Springer TA (2005) Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci U S A 102, 3679–3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo BH, Carman CV, and Springer TA (2007) Structural basis of integrin regulation and signaling. Annual review of immunology 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalli AC, Campbell ID, and Sansom MS (2011) Multiscale simulations suggest a mechanism for integrin inside-out activation. Proc Natl Acad Sci U S A 108, 11890–11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalli AC, Wegener KL, Goult BT, Anthis NJ, Campbell ID, and Sansom MS (2010) The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure 18, 1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim C, Ye F, Hu X, and Ginsberg MH (2012) Talin activates integrins by altering the topology of the beta transmembrane domain. The Journal of cell biology 197, 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthis NJ, Wegener KL, Ye F, Kim C, Goult BT, Lowe ED, Vakonakis I, Bate N, Critchley DR, Ginsberg MH, and Campbell ID (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. The EMBO journal 28, 3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orlowski A, Kukkurainen S, Poyry A, Rissanen S, Vattulainen I, Hytonen VP, and Rog T (2015) PIP2 and Talin Join Forces to Activate Integrin. The journal of physical chemistry. B 119, 12381–12389 [DOI] [PubMed] [Google Scholar]
- 32.Saltel F, Mortier E, Hytonen VP, Jacquier MC, Zimmermann P, Vogel V, Liu W, and Wehrle-Haller B (2009) New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. The Journal of cell biology 187, 715–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arcario MJ, and Tajkhorshid E (2014) Membrane-Induced Structural Rearrangement and Identification of a Novel Membrane Anchor in Talin F2F3. Biophysical journal 107, 2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, and Qin J (2012) A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell research 22, 1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Pereda JM, Wegener KL, Santelli E, Bate N, Ginsberg MH, Critchley DR, Campbell ID, and Liddington RC (2005) Structural basis for phosphatidylinositol phosphate kinase type Igamma binding to talin at focal adhesions. J Biol Chem 280, 8381–8386 [DOI] [PubMed] [Google Scholar]
- 36.Di Paolo G, Pellegrini L, Letinic K, Cestra G, Zoncu R, Voronov S, Chang S, Guo J, Wenk MR, and De Camilli P (2002) Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420, 85–89 [DOI] [PubMed] [Google Scholar]
- 37.Legate KR, Takahashi S, Bonakdar N, Fabry B, Boettiger D, Zent R, and Fassler R (2011) Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. The EMBO journal 30, 4539–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalli AC, Campbell ID, and Sansom MS (2013) Conformational changes in talin on binding to anionic phospholipid membranes facilitate signaling by integrin transmembrane helices. PLoS computational biology 9, e1003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denisov IG, Grinkova YV, Lazarides AA, and Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. Journal of the American Chemical Society 126, 3477–3487 [DOI] [PubMed] [Google Scholar]
- 40.Bayburt TH, Vishnivetskiy SA, McLean MA, Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG, and Gurevich VV (2011) Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J Biol Chem 286, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lakowicz JR (2006). in Principles of Fluorescence Spectroscopy, Third Ed., Springer US, New York. pp 954 [Google Scholar]
- 42.Shih AY, Denisov IG, Phillips JC, Sligar SG, and Schulten K (2005) Molecular dynamics simulations of discoidal bilayers assembled from truncated human lipoproteins. Biophysical journal 88, 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]