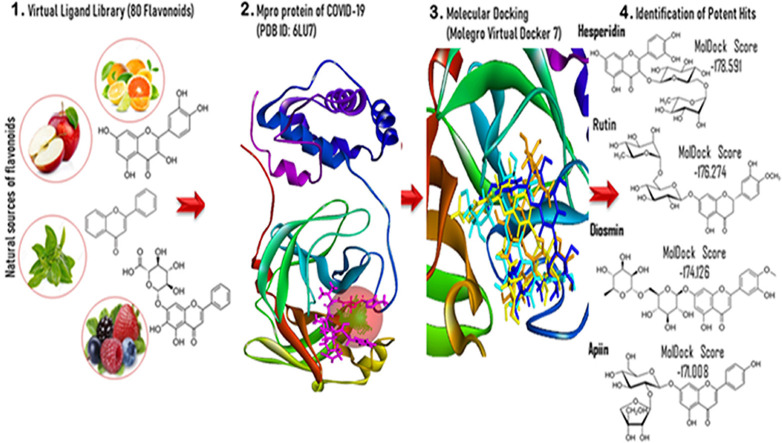
Abstract
SARS-CoV-2, a rapidly spreading new strain of human coronavirus, has affected almost all the countries around the world. The lack of specific drugs against SARS-CoV-2 is a significant hurdle towards the successful treatment of COVID-19. Thus, there is an urgent need to boost up research for the development of effective therapeutics against COVID-19. In the current study, we investigated the efficacy of 81 medicinal plant-based bioactive compounds against SARS-CoV-2 Mpro by using various in silico techniques. The interaction affinities of polyphenolic compounds towards SARS-CoV-2 Mpro was assessed via intramolecular (by Quantum Mechanic), intermolecular (by Molecular Docking), and spatial (by Molecular Dynamic) simulations. Our obtained result demonstrate that Hesperidin, rutin, diosmin, and apiin are most effective compounds agents against SARS-CoV-2 Mpro as compared to Nelfinavir (positive control). This study will hopefully pave a way for advanced experimental research to evaluate the in vitro and in vivo efficacy of these compounds for the treatment of COVID-19.
Keywords: COVID-19, SARS CoV-2 Mpro, Molecular docking, Molecular dynamics simulation, Quantum mechanic, Flavonoids
Graphical abstract
1. Introduction
An acute respiratory disorder caused by 2019-novel coronavirus [2019-nCoV, now known as SARS-CoV-2 (severe acute respiratory syndrome coronavirus-2)] has emerged as a serious public health issue at the end of 2019 [1]. During the 21st century, COVID-19 marked the history with the third large-scale coronavirus epidemic into the human population after SARS-CoV in 2002 in china [2] and Middle East respiratory syndrome coronavirus (MERS-CoV) in the Middle East region in 2012 [3]. The potentially fatal virus has affected millions of humans around the world and this pandemic is still steadily spreading [4]. Researchers have developed vaccines for COVID-19, however, research suggests that developed vaccines will not completely enough to fight against COVID-19 due to high mutation rate of SARS CoV2 [5]. The recommended HIV protease inhibitors (Nelfinavir, Lopinavir, and Ritonavir), antibody cocktail, and other virus-fighting drugs have very limited potential to cure COVID-19 [5,6]. Thus, there is an urgent need to develop therapeutics for COVID-19.
The structure of SARS CoV-2 has been identified as β-coronavirus, a non-segmented enveloped positive-sense RNA virus, with ∼30 kb genome [7,8]. SARS CoV-2 causes severe respiratory tract infection and utilizes angiotensin-converting enzyme 2 (ACE2) receptors to infect human cells [9]. The crystallized form of SARS CoV-2 virus main protease (Mpro) was demonstrated by a Chinese researcher Liu et al. [10], which is a potential drug target for the inhibition of SARS CoV-2 replication [11]. The Mpro is an essential protein required for the proteolytic cleavage of viral polypeptide [12] to release functional proteins such as endoribonuclease, exoribonuclease, and RNA polymerase [13]. The functional significance of Mpro in the SARS CoV-2 life cycle [15], as well as the absence of its close homologs in humans, identify Mpro as an attractive target for antiviral drug discovery [14].
The growing evidence has established the worth of polyphenols as cheaper and safer drug candidates for drug discovery against various human diseases [16,17]. In silico based screening has proven to be an excellent tool to meet the challenges of drug discovery [18,19]. Additionally, Molecular Dynamics Simulation (MDS) is extremely useful in drug design against different viruses, where protein dynamics are assessed in the nano to picosecond time interval [[20], [21], [22], [23], [24], [25], [26]]. Molecular dynamic theory is based on the spatial conformation of molecular interactions from their active sides by intermolecular interactions like weak van der Waals interactions, London forces or hydrogen bonding, etc [27,28]. So, MD simulations are performed to obtain information about docked agents to the real (in vitro) experimental results. Many computational approaches have been developed to predict the absorption, distribution, metabolism, and excretion (ADME) properties of drug candidates from their physicochemical properties by comparing them with compounds. These methods, which reduce costs and save time, have now become an integral part of drug research and development studies [29].
In the current study, we have investigated the potential of 81 polyphenolic compounds against SARS CoV-2 Mpro by molecular docking via frontier molecular orbital theory by Material Studio 7.0 (MS), ADMET, and molecular dynamics by NAMD assessments. All small molecules used for docking studies were obtained from https://pubchem.ncbi.nlm.nih.gov/as SDF form and in the 3D Conformer. QM assessment of molecules was performed to elaborate the molecular docking studies. The best-fitted geometry of the molecules, highest occupied molecular orbital (HOMO), lowest unoccupied molecular orbital (LUMO), and other intermolecular parameters were calculated by MS using density functional theory (DFT). The investigated molecules were evaluated for their metabolic activities and pharmacokinetic parameter in the cell membrane, blood-brain barrier, etc. by ADMET online platform. Our obtained results indicate that hesperidin, diosmin, rutin, and apiin are best four potent inhibitors against SARS CoV-2 Mpro. These compounds were further tested against the SARS CoV-2 Mpro at different dynamics states by NAMD using CHARMM 36 forcefield in 100 ns MDS run to prove their biological activities in computer simulation conditions.
2. Materials and methods
2.1. Molecular docking studies
2.1.1. Target protein model preparation
The docking of the compounds inside SARS-CoV-2 Mpro was performed using Molegro Virtual Docker (MVD) and AutoDock Vina software [30,31]. The crystal structure of SARS-CoV-2 Mpro was retrieved from the protein data bank website (http://www.rcsb.org/pdb) (PDB ID: 6LU7 and 6Y84: Resolution 2.16 Å and 1.39 Å, respectively) [32,33]. Proteins were imported to Molegro Virtual Docker and prepared for docking. Water molecules at crystal structure were removed, protein structure errors were checked. The structure errors of amino acid residues were checked, repaired, optimized with the neighborhood residues.
2.1.2. Small molecules preparation
Small molecules or ligands used in docking studies were obtained from https://pubchem.ncbi.nlm.nih.gov/as SDF form and in the 3D Conformer [34]. They were prepared with Molegro Virtual Docker (MVD) for molecular docking.
2.1.3. Molecular docking
The selected cavity of SARS-CoV-2 Mpro was the binding site of natural inhibitor N3. The cavity is centered at (−10.85, 15.32, 68.39) with 15 Å radius. AutoDock Vina box size (20 × 20 × 20 Å) was centered at the active site residues around (−10.85, 15.32, 68.39) Å. Ten docking trials were performed in both software, and the compounds with best binding affinity were selected for further evaluation. Nelfinavir was utilized as a positive control. Nelfinavir is a viral protease inhibitor used for the treatment of the human immunodeficiency virus (HIV), which has been previously reported as a SARS CoV-2 Mpro inhibitor [35]. Possible docking modes between compounds and the COVID-19 virus Mpro were analyzed using the Discovery Studio 2020 Client and PyMOL software [36].
2.2. Molecular dynamic simulation
The best resolution of SARS-CoV-2 Mpro (PDB ID: 6Y84) was subjected to 100 ns MDS after the minimization and equilibration steps. Firstly, the protein is solvated using the TIP3P water model then ionized with NaCl [37]. NAMD was used for the minimization via using the CHARMM 36 force field [20,38,39]. After minimizing the whole system, temperature was slowly adjusted to 310 K to resemble the physiological temperature of the human body (37 °C). A small NPT MDS run was performed to adjust the system volume, followed by the production run at the NVT ensemble for 100 ns. VMD software was utilized in the analysis of the data [40].
2.3. Frontier molecular orbital modeling
The modeling of HOMO, LUMO orbitals of the most effective small molecules and target protein are calculated by density functional theory (DFT) on Material Studio 7.0 Code embedded in the Material Studio package (Accelrys, SanDiego, CA) [41] by using DMol3 module [[42], [43], [44]].
2.4. ADMET assessment
The ADMET properties of selected compounds were analyzed by using the http://biosig.unimelb.edu.au/pkcsm/prediction website. The molecular structures of the natural compounds were uploaded using isomeric SMILES into webtool. Important results were retrieved from website for analysis [45].
3. Results and discussion
3.1. Molecular docking studies
Coronaviruses have a long history of infecting humans and animals and causing respiratory, digestive, liver, and central nervous system diseases [46]. A novel newly emerged virus, SARS-CoV-2, is presenting significant threats to human health nowadays. Currently, no specific clinical therapeutics are available for the treatment of SARS-CoV-2-mediated respiratory infections [47]. Thus, the need of the hour is to identify and characterize novel drug candidates to overcome the health losses caused by SARS-CoV-2. In this context, natural products have gained importance as potent antiviral agents during recent years [48,49]. Several computational studies suggest the significance of phytochemicals and nutraceuticals in drug development against SARS-CoV-2 [22,[50], [51], [52], [53], [54]]. Considering the immediate need for therapeutics against COVID-19 and services of natural products in drug discovery [55], we have screened flavonoids against a novel drug target, SARS-CoV-2 Mpro, for the identification of natural scaffolds for drug development against COVID-19.
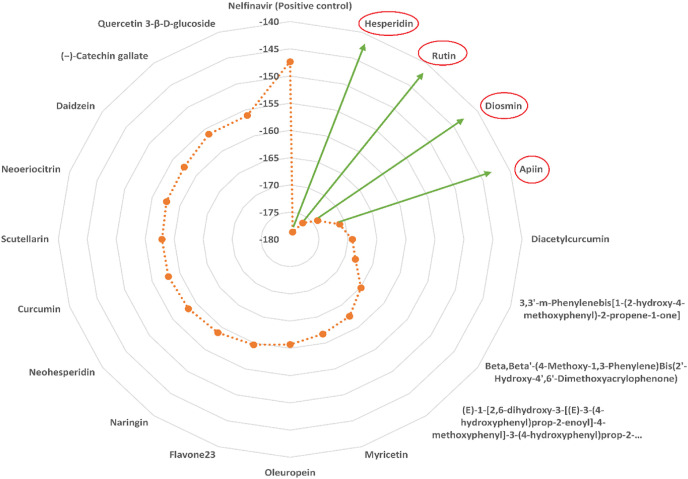
The binding energies obtained after docking of the compounds into SARS-CoV-2 Mpro (PDB ID: 6LU7) are presented in Table 1 . According to the in silico results, 24 of the compounds have a better affinity against COVID-19 virus Mpro than Nelfinavir (Fig. 1 ). Hesperidin exhibited the highest affinity to the active site of SARS-CoV-2 Mpro followed by rutin, diosmin, and apiin respectively (Fig. 1).
Table 1.
Results of the docking of some phenolic compounds on the crystal structure of COVID-19 main protease.
| Ligands |
MolDock Score | HBond | Ligands |
MolDock Score | HBond | ||
|---|---|---|---|---|---|---|---|
| PubChem CID | Name | PubChem CID | Name | ||||
| N3 (Reference ligand) | −162.17 | −8.19 | 5280343 | Quercetin | −122.99 | −13.97 | |
| 64143 | Nelfinavir (Positive control) | −147.38 | −6.87 | 5280551 | Xenognosin B | −119.14 | −7.85 |
| 10621 | Hesperidin | −178.59 | −20.26 | 5280373 | Biochanin a | −118.80 | −9.65 |
| 5280805 | Rutin | −176.27 | −21.24 | 5280863 | Kaempferol | −118.63 | −14.28 |
| 5281613 | Diosmin | −174.13 | −27.26 | 3764 | Isoformononetin | −117.02 | −7.10 |
| 5280746 | Apiin | −171.01 | −10.19 | 31425 | Vat Yellow 2 | −116.61 | −1.86 |
| 6441419 | Diacetylcurcumin | −169.26 | −9.57 | 5280378 | Formononetin | −115.58 | −7.39 |
| 101502236 | 3,3′-m-Phenylenebis[1-(2-hydroxy-4-methoxyphenyl)-2-propene-1-one] | −168.14 | −7.74 | 439533 | (±)-Taxifolin | −115.47 | −12.37 |
| 101526067 | Beta, Beta'-(4-Methoxy-1,3-Phenylene)Bis(2′-Hydroxy-4′,6′-Dimethoxyacrylophenone) | −164.87 | −6.68 | 5280445 | Luteolin | −114.90 | −10.76 |
| 46702071 | (E)-1-[2,6-dihydroxy-3-[(E)-3-(4-hydroxyphenyl)prop-2-enoyl]-4-methoxyphenyl]-3-(4-hydroxyphenyl)prop-2-en-1-one | −162.53 | −13.65 | 5281670 | Morin | −114.74 | −14.28 |
| 44259442 | Myricetin | −161.72 | −16.62 | 97214 | Eupatorin | −113.99 | −12.52 |
| 5281544 | Oleuropein | −160.69 | −26.89 | 188323 | Cirsimaritin | −113.98 | −5.63 |
| 3035266 | Flavone23 | −159.64 | −2.21 | 5281612 | Diosmetin | −113.51 | −8.07 |
| 442428 | Naringin | −158.82 | −8.59 | 445154 | Resveratrol | −113.09 | −5.76 |
| 442439 | Neohesperidin | −158.33 | −16.12 | 5281628 | Hispidulin | −112.01 | −7.60 |
| 969516 | Curcumin | −157.92 | −10.50 | 54685921 | Hispidin | −110.03 | −11.94 |
| 185617 | Scutellarin | −157.90 | −14.20 | 5281708 | Daidzein | −109.72 | −7.58 |
| 114627 | Neoeriocitrin | −157.58 | −18.62 | 5281616 | Galangin | −109.69 | −10.31 |
| Daidzein | −157.41 | −7.08 | 5281697 | Scutellarein | −108.52 | −14.50 | |
| 6419835 | (−)-Catechin gallate | −156.13 | −16.87 | 5280443 | Apigenin | −108.11 | −11.77 |
| 44259136 | Quercetin 3-β-D-glucoside | −156.08 | −17.24 | 68077 | Tangeritin | −107.75 | −2.09 |
| 64982 | Baicalin | −153.60 | −12.10 | 5281894 | 7-Hydroxyflavone | −106.62 | −5.58 |
| 168849 | Pectolinarin | −152.42 | −9.01 | 932 | Naringenin | −106.38 | −12.13 |
| 5281718 | Polydatin | −151.85 | −13.86 | 5281607 | Chrysin | −105.48 | −5.74 |
| 5282151 | Vitexin 2-o-rhamnoside | −150.02 | −17.63 | 72276 | (−)-epicatechin | −103.49 | −11.21 |
| 71602340 | Balsacone A | −149.70 | −14.08 | 72281 | Hesperitin | −102.36 | −8.96 |
| 5469424 | Scutellaric Acid | −102.01 | −2.17 | 5281703 | Wogonin | −98.56 | −8.71 |
| 46781931 | Quercetin 3-d-galactoside | −148.42 | −11.77 | 637775 | Sinapinic acid | −98.54 | −2.49 |
| 5315472 | Bisdemethoxycurcumin | −144.47 | −8.52 | 11349 | 3-Hydroxyflavone | −97.65 | −3.33 |
| 73447 | Uvaretin | −140.06 | −5.08 | 5281605 | Baicalein | −95.74 | −11.12 |
| 23640558 | Curcumin Pyrazole | −139.95 | −8.77 | 68112 | 5-Hydroxyflavone | −95.71 | −2.45 |
| 56842347 | Oleuropein aglycone | −138.79 | −14.31 | 72279 | 6-Hydroxyflavone | −94.09 | −3.46 |
| 151670 | Isouvaretin | −133.17 | −9.08 | 5281855 | Ellagic acid | −93.64 | −13.37 |
| 11652416 | Oleocanthal | −132.88 | −15.19 | 10680 | Flavone | −91.48 | 0.00 |
| 5320945 | Rhamnazin | −132.22 | −11.87 | 445858 | ferulic | −87.97 | −4.89 |
| 18684078 | Oleacein | −130.73 | −11.41 | 10742 | syringic acid | −86.68 | −5.44 |
| 4303567 | Acacetin Diacetate | −130.16 | −12.80 | 689043 | Caffeic acid | −85.71 | −6.71 |
| 638278 | Isoliquiritigenin | −128.09 | −10.42 | 114850 | Oxymatrine | −84.27 | −0.56 |
| 31553 | Silibinin | −127.04 | −18.58 | 637542 | p-Coumaric acid | −80.83 | −4.11 |
| 65084 | (+)-gallocatechin | −126.55 | −17.54 | 8468 | Vanillic | −78.61 | −4.56 |
| 162464 | Cirsilineol | −126.48 | −11.81 | 54670067 | l-Ascorbic acid | −77.10 | −15.98 |
| 5281377 | Genistin | −126.28 | −12.71 | 8742 | Shikimic acid | −76.85 | −6.74 |
| 5281614 | Fisetin | −123.53 | −13.67 | 370 | Gallic acid | −75.87 | −10.65 |
| 124052 | Glabridin | −123.00 | −5.16 | 1491 | 3,4-Dihydroxybenzoic acid | −72.59 | −4.00 |
Fig. 1.
MolDock docking experiment (A) MolDock scores calculated for the best 24 natural polyphenols and Nilfinavir against the SARS-CoV-2 Mpro. (B) Summary of the in silico based screening of natural product library and identification of potential natural inhibitors of SARS-CoV-2 Mpro.
The amino acid residues that contribute to interactions between the reference ligand and hit compounds are demonstrated in Table 2 . Glu 189, Glu 166, and Met165 are the main residues contributing for interactions between target proteins and ligands.
Table 2.
The ten amino acid residues at the active cavity interacting with the reference inhibitor and ligands.
| Hesperidin |
Rutin |
Diosmin |
Apiin |
N3 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Residue | ID | Total | Residue | ID | Total | Residue | ID | Total | Residue | ID | Total | Residue | ID | Total |
| Glu | 166 | −37.06 | Gln | 189 | −31.49 | Glu | 166 | −28.64 | Gln | 189 | −24.03 | Glu | 166 | −28.623 |
| Gln | 189 | −20.72 | Met | 165 | −21.92 | Met | 165 | −27.68 | Met | 165 | −18.34 | Gln | 189 | −26.6855 |
| Met | 165 | −19.9 | Glu | 166 | −17.37 | Gln | 189 | −20.89 | His | 41 | −15.55 | Met | 165 | −21.0185 |
| Asn | 142 | −16.47 | Asn | 142 | −17.08 | His | 164 | −14.33 | Ser | 144 | −13.43 | Asn | 142 | −14.016 |
| Cys | 145 | −14.9 | His | 41 | −14.81 | Arg | 188 | −13.65 | Glu | 166 | −11.42 | His | 41 | −13.3386 |
| His | 41 | −12.35 | Arg | 188 | −14.45 | Asp | 187 | −13.42 | Cys | 145 | −10.96 | Thr | 190 | −12.4393 |
| Ser | 144 | −12.24 | Thr | 26 | −11.25 | Gln | 192 | −10.91 | Met | 49 | −10.96 | Pro | 168 | −11.4904 |
| Gln | 192 | −11.93 | Asp | 187 | −10.21 | Asn | 142 | −10.91 | Asn | 142 | −10.76 | His | 164 | −11.258 |
| Arg | 188 | −11.37 | Gly | 143 | −9.154 | Leu | 167 | −10.31 | Gln | 192 | −10.59 | Gly | 143 | −10.6192 |
| Thr | 190 | −9.416 | Cys | 145 | −9.015 | His | 41 | −9.586 | Gly | 143 | −9.827 | Leu | 141 | −9.58516 |
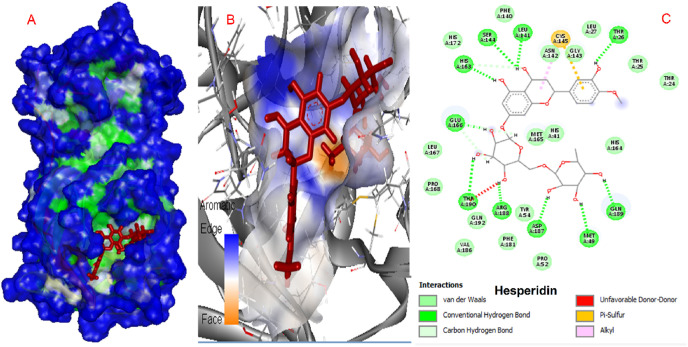
Hesperidin exhibited the highest binding energy at the active site of SARS-CoV-2. 2D and 3D interaction diagrams are provided in Fig. 2 . Hesperidin formed hydrogen bond interactions with Thr 26, Glu 166, Arg 188, Gln 189, Met 49, Asp 187, His 163, Leu 141, and Ser 144 residues. B ring of flavonoid structure formed Pi-Sulfur interacts with Cys 145. Further about 10 amino acids contributed to the stabilization of the molecule in the active site via van der Waals interactions.
Fig. 2.
Interactions of hesperidin and COVID-19 virus Mpro.
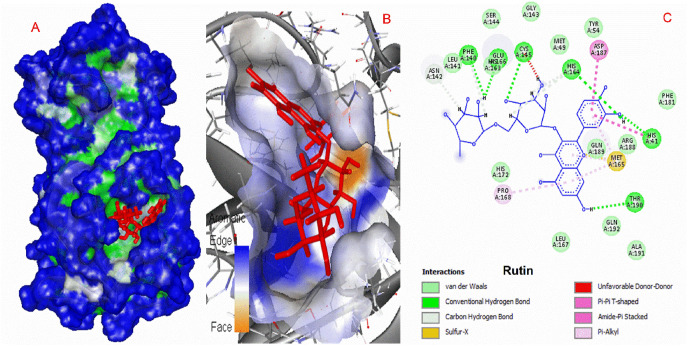
The obtained results showed that His 41, Cys 145, Phe 140, Glu 166, Gln 192, Thr 190, and His 164 are critical residues for the hydrogen bonding interactions at the binding site of rutin to protease protein (Fig. 3 ). Hydrophobic interactions such as Pi-Pi T-shaped, Amide-Pi Stacked, and Pi-Alkyl were responsible for interactions between target protein and ligands. Cys 145 interacted with both phenyl rings and a heterocyclic ring to form pi-sulfur interaction.
Fig. 3.
Interactions of rutin and COVID-19 virus Mpro.
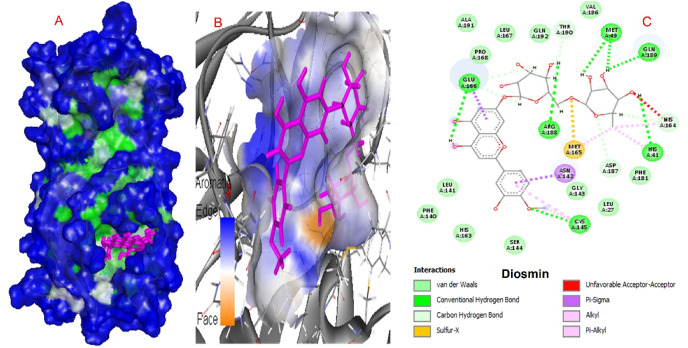
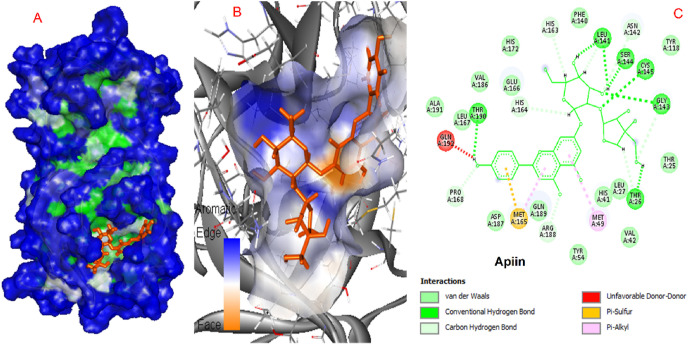
The interacting amino acids for diosmin and apiin are presented in Fig. 4, Fig. 5 .
Fig. 4.
Interactions of apiin and COVID-19 virus Mpro.
Fig. 5.
Interactions of diosmin and COVID-19 virus Mpro.
SARS main protease enzymes have a protease Cys-His dyad created by Cys145 and His41 residues. The reported protease inhibitors interact with Cys145 [56]. Diosmin, Apiin, and Rutin form a hydrogen bond with this catalytic amino acid, while Hesperidin interacts via pi-sulfur interaction. Table 1 demonstrates that these natural compounds form strong interactions with His41. 2D interaction models showed that Hesperidin and Apiin forms van der Waals interaction with His41 and Diosmin and rutin form hydrogen bond interactions.
3.2. Quantum mechanic studies
3.2.1. General chemical reactivities by HOMO-LUMO estimation
Global Reactivity Descriptors based on the HOMO and LUMO energy differences are very important indicators to determine the reactive affinity of molecules against nucleophilic electrophilic and radicalic attacks. All descriptors or indexes were calculated by well-known equations (Eqs. (Eq.1), (Eq.2), (Eq.3), (Eq.4), (Eq.5)) according to the literature [57,58].
| η = 1/2 (ELUMO − EHOMO) | (Eq.1) |
| S = 1/2 η | (Eq.2) |
| ω = μ2/2 | (Eq.3) |
| μ = − χ = 1/2 (ELUMO + EHOMO) | (Eq.4) |
| χ = − 1/2 (ELUMO + EHOMO) | (Eq.5) |
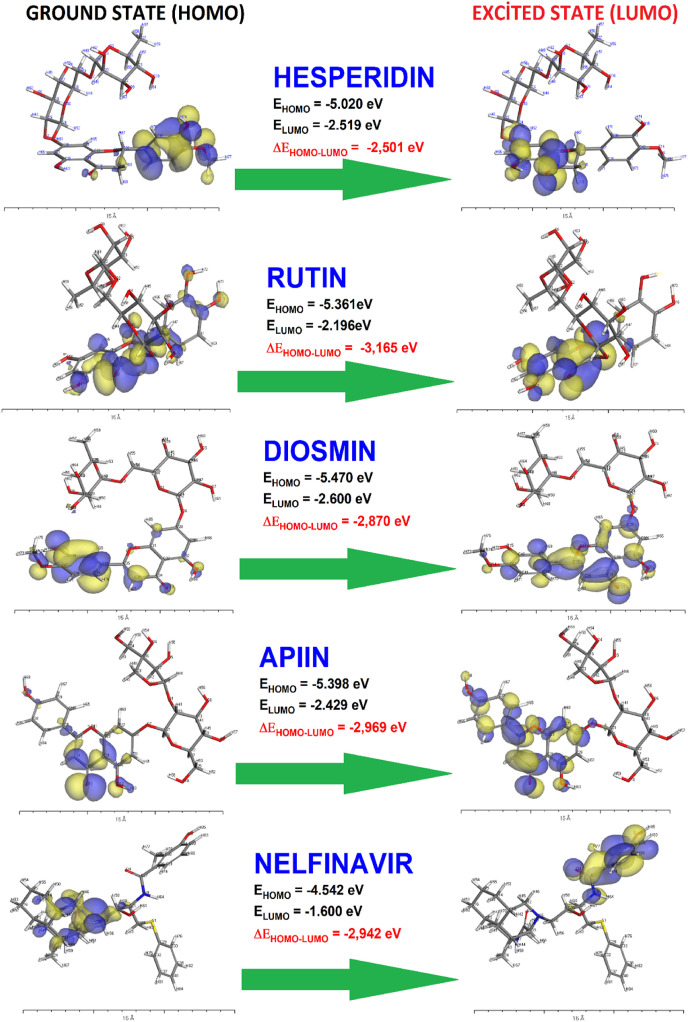
The global electrophilicity (ω) chemical potential (μ), global hardness (η), global softness (S), electronegativity (χ) were calculated from the simulation results are presented in Table 3 . The frontier orbital representations are shown in Fig. 6 for hesperidin, rutin, diosmin, apiin, and positive control nelfinavir. According to docking results, the following fitting rank among the polyphenols is determined: hesperidin > rutin > diosmin > apiin. The comparison of HOMO and LUMO energy values of polyphenols with nelfinavir, showed that the energy level of nelfinavir is lower than the polyphenols significantly for each of the frontier orbitals. Whereas, the bandgap value of nelfinavir is nearly equal to the polyphenolic compounds. Best-docked polyphenolic compound's eV values are approximately equal to each other and are in the range of −2.196 to −2.600 for LUMO and −5.020 to −5.470 for HOMO. While the eV values are 1.600 for LUMO and 4.542 for HOMO for nelfinavir.
Table 3.
Physical characteristics of computed for polyphenols (eV).
| NELFINAVIR | HESPERIDIN | RUTIN | DIOSMIN | APIIN | |
|---|---|---|---|---|---|
| Total DFT-D energy (Ha) | −2107.65 | −2213.32 | −2248.04 | −2212.14 | −2058.34 |
| Df binding energy (eV) | −390.767 | −364.522 | −350.602 | −359.267 | −325.608 |
| EHOMO(eV) | −4.542 | −5.020 | −5.361 | −5.470 | −5.398 |
| ELUMO(eV) | −1.600 | −2.519 | −2.196 | −2.600 | −2.429 |
| Band gap (EHOMO − LUMO) (eV) | −2.942 | −2.501 | −3.165 | −2.870 | −2.969 |
| ΔE(ELUMO-HOMO) (eV) | 2.942 | 2.501 | 3.165 | 2.870 | 2.969 |
| Electronegativity (χ) (eV) | 3.071 | 3.770 | 3.779 | 4.035 | 3.914 |
| Chemical potential (μ) (eV) | −3.071 | −3.770 | −3.779 | −4.035 | −3.914 |
| Global hardness (η) (eV) | 1.471 | 1.251 | 1.583 | 1.435 | 1.485 |
| Global softness (S) (eV-1) | 0.736 | 0.625 | 0.791 | 0.718 | 0.742 |
| Global electrophilicity index (ω) (eV) | 4.716 | 7.105 | 7.139 | 8.141 | 7.658 |
Fig. 6.
3D plots frontier orbital energies using DFT method for hesperidin, rutin, diosmin, apiin, and nelfinavir compounds.
HOMO propagation on the molecule is attributed to its ability for nucleophilic attack. LUMO is another parallel consideration can be done for the reactive ability [59]. The difference between energy values of LUMO and HOMO in interaction reveals the stimulation of the residues of the protein, which facilitates electron transitions between protein and ligand. This difference governs the magnitude of reactivity (reaction kinetic) and the stability of the molecule [60,61]. Under the light of the phenomenons, ω, μ, and χ values are significantly higher for the polyphenol compounds depending on the HOMO and LUMO energy values than nelfinavir. These values determine the global reactivity of all molecules against the target protein as seen from Table 3. The higher global electrophilicity and softness values in ligand-protein interactions can be attributed good reactivities of ligands that are expected particularly in biological system-based studies.
The propagation of HOMO and LUMO through the compounds is provided in Fig. 6. According to the figure, nucleophilic and electrophilic reactivity sides of molecules are commonly on the aromatic or phenolic sites of the compounds. As it can be seen that the HOMO&LUMO-based reactivity of compounds against protein is compatible with the docking results especially in the conventional hydrogen bond, π- π, amide-π and π-alkyl interaction formations.
3.2.2. Local chemical reactivities by Fukui function
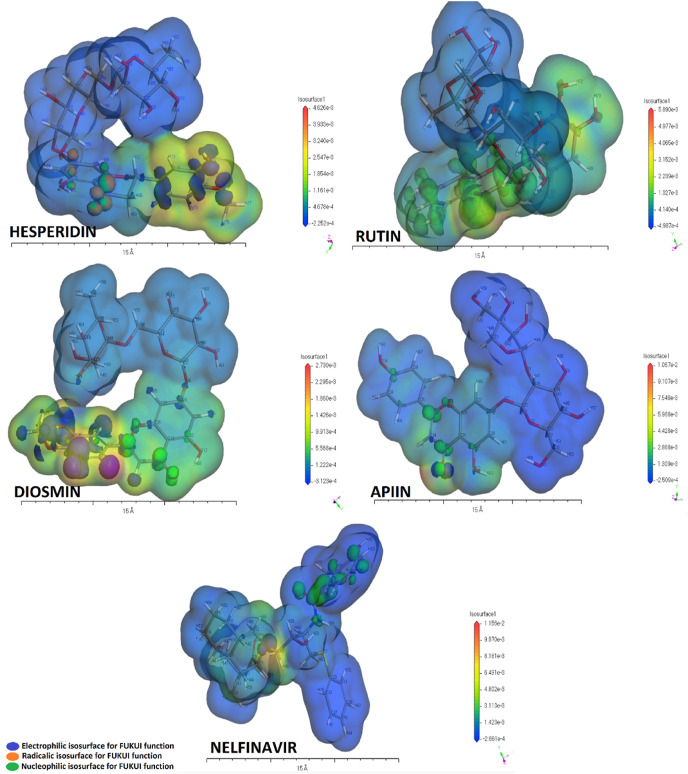
The Fukui function describes the electron density in the frontier orbitals depending on electrophilic, nucleophilic, and radicalic propagation. It is a useful tool to discuss the reactivity of the compounds in researches [43,44]. In the current study, the Fukui indices-based spaces were given inside the electrostatic potential map together in Fig. 7 . The effective electrophilic, nucleophilic, and radicalic Fukui indices; ƒ - , ƒ + , and ƒ 0 are tabulated in Table 4 identifying local reactivities of compounds. All Fukui indices containing local electrophilicity (W − , W + , and W 0) and local softness (S − , S + , and S 0) for each atom of the best effective compounds against Covid 19 are given in the supplementary documents in the range of Tables 1S–5S Also, Muliiken's atomic charges for each of the compounds are tabulated in Table 6S and the atomic charge propagation is shown Figs. 6S–10S. 2D molecular structures for easy comparison of the most effective compounds are given in supplementary files in Figs. 6S–10S. Along with the simulation results from Table 4, it can be seen that the atom number of electrophilic, nucleophilic, and radicalic character and their propagation over the hesperidin molecule are widely distributed as compared with the other molecules. Also, these characteristic atoms of the molecules are commonly located on the aromatic and phenolic regions of the molecules and they are dominant to form strong interactions against target regions of the protein. Whereas, aliphatic regions of molecules are mostly recessive and they are associated with the formation of weak interactions. So, the interaction possibility of hesperidin molecules with SARS-CoV-2 main protease is higher than the other effective molecules.
Fig. 7.
Electrophilic surface-based electrostatic potential map containing the Fukui surfaces.
Table 4.
Fukui (ƒ−, ƒ+, ƒ0) indices for electrophilic, nucleophilic and radical attack of best effective compounds against Covid 19.
| THE EFFECTIVE FUKUI INDICES OF HESPERIDIN |
THE EFFECTIVE FUKUI INDICES OF RUTIN |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ƒ- | ƒ+ | ƒ+ | ƒ- | ƒ+ | ƒ+ | ||||||
| O14 | 0.126 | O11 | 0.042 | O13 | 0.089 | O6 | 0.056 | O11 | 0.065 | O6 | 0.039 |
| O15 | 0.127 | O12 | 0.06 | O14 | 0.065 | O12 | 0.177 | O12 | 0.160 | O11 | 0.065 |
| C38 | 0.043 | O13 | 0.164 | O15 | 0.066 | O15 | 0.043 | O14 | 0.043 | O12 | 0.160 |
| C39 | 0.078 | C28 | 0.057 | C36 | 0.073 | O16 | 0.047 | C30 | 0.072 | C30 | 0.072 |
| C40 | 0.059 | C30 | 0.053 | C39 | 0.042 | C31 | 0.105 | C31 | 0.105 | ||
| C41 | 0.051 | C35 | 0.057 | C35 | 0.057 | ||||||
| C42 | 0.059 | C36 | 0.141 | H65 |
0.042 |
||||||
| H70 | 0.052 | H65 | 0.041 |
THE EFFECTIVE FUKUI INDICES OF APIIN |
|||||||
| H71 | 0.064 | H66 | 0.044 |
ƒ- |
ƒ+ |
ƒ+ |
|||||
| H73 | 0.055 | H68 | 0.053 | O13 | 0.299 | O12 | 0.041 | O11 | 0.048 | ||
| H75 | 0.047 | H69 | 0.061 | C34 | 0.053 | O13 | 0.286 | O13 | 0.133 | ||
|
H76 |
0.047 |
H62 | 0.058 | C32 | 0.063 | C30 | 0.042 | ||||
|
THE EFFECTIVE FUKUI INDICES OF DIOSMIN |
C33 | 0.042 | C32 | 0.078 | |||||||
|
ƒ- |
ƒ+ |
ƒ+ |
C34 | 0.054 | C33 | 0.074 | |||||
| O14 | 0.042 | O11 | 0.047 | O13 | 0.078 | C34 | 0.043 | ||||
| O15 | 0.136 | O12 | 0.040 | O15 | 0.077 | H62 |
0.053 |
||||
| C36 | 0.043 | O13 | 0.123 | C34 | 0.048 |
THE EFFECTIVE FUKUI INDICES OF NELFINAVIR |
|||||
| C38 | 0.051 | C34 | 0.087 | C35 | 0.048 |
ƒ- |
ƒ+ |
ƒ+ |
|||
| C39 | 0.078 | C35 | 0.077 | C36 | 0.042 | N6 | 0.154 | O4 | 0.130 | O4 | 0.071 |
| C41 | 0.041 | C36 | 0.041 | C38 | 0.042 | H57 | 0.118 | C27 | 0.082 | N6 | 0.075 |
| C42 | 0.059 | H67 | 0.051 | C39 | 0.054 | H56 | 0.068 | C30 | 0.058 | C27 | 0.042 |
| H69 | 0.048 | C42 | 0.045 | H51 | 0.068 | C39 | 0.099 | C39 | 0.051 | ||
| H70 | 0.056 | H67 | 0.040 | H46 | 0.071 | H74 | 0.051 | H57 | 0.063 | ||
| H71 | 0.047 | H70 | 0.042 | H45 | 0.075 | H80 | 0.050 | ||||
| H83 | 0.054 | ||||||||||
3.2.3. Electron density analysis by Muliken's population analysis
Electron distribution over atoms inside molecules, can not be ignored because of its significant effects on the bond angles and lengths [58]. Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5S describes the atomic charges depending on Muliken's algorithm. The evaluation of the relationship between the molecular structures and atomic charges shows that electron density in all molecules is higher in the aromatic side of the molecules especially on (C O, C C, C–O–H) bond species. The higher charges of the H atoms can be attributed to (C = = C, H–O–C = = C) structures. Oxygen atoms and double-bonded oxygen atoms neighboring carbon atoms in all molecules commonly have negative charges. While the other carbon atoms have strong or slight positive charges depending on the electronegativity of the neighboring atom. Electrophilic surface-based electrostatic potential maps in Fig. 7 is coherent with electrostatic assessment of each atom in molecules. This evaluation also can be verified for nucleophilic and radicalic attacks.
3.3. Molecular dynamic studies
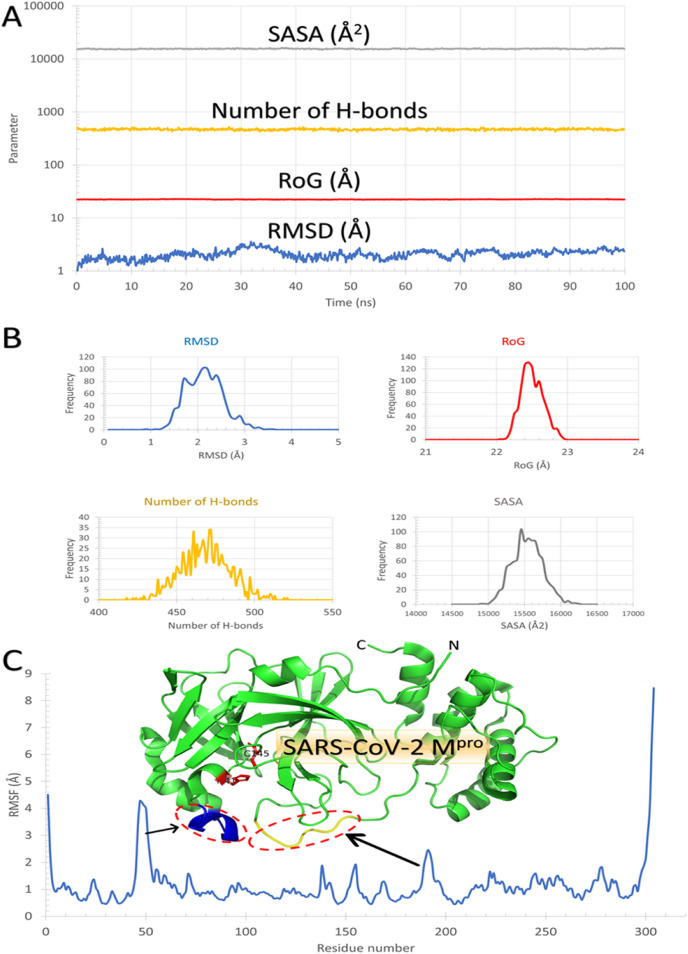
To further analyze the binding behavior of the natural polyphenols, molecular dynamics was utilized with the aid of NAMD software. The SARS-CoV-2 Mpro (PDB ID: 6Y84) with the best resolution structure was subjected to 100 ns MDS. Fig. 8 A, B, and 8C summarize the simulated dynamics of the protein during the 100 ns time domain. Root Mean Square Deviation (RMSD) in Ǻ (blue), Radius of Gyration (RoG) in Å (red), the total number of H-bonds (orange), and Surface Accessible Surface Area (SASA) in Å2 (gray) are represented in Fig. 8A versus time of the simulation in nanoseconds (ns). The histogram of each parameter is depicted in Fig. 8B with the same color code. As shown from RMSD versus time and the RMSD histogram, the system is equilibrated from the first ten ns of the simulation, reaching a value of 2.2 ± 0.8 Å. Both RoG and SASA are stable during the simulation period with values 22.5 ± 0.4 Å and 15600 ± 600 Å2, respectively. The total number of the H-bonds is also stable ranging from 418 to 522 during the 100 ns simulation. These values reflect the stability of the protein system during the simulation.
Fig. 8.
Molecular Dynamics Simulation of SARS-CoV-2 Mpro. (A) RMSD (in Å) in blue, RoG (Å) in red, Number of H-bonds in yellow, and SASA (in Å2) in gray versus time in ns. (B) Histogram of the four parameters mentioned in A with the same color code. (C) The per-residue RMSF of SARS-CoV-2 Mpro with the protein structure represented in the green cartoon. The most movable parts are marked in the RMSF chart and the protein structure (the encircled blue helix and yellow coil).
On the other hand, the per-residue Root Mean Square Fluctuations (RMSF) in Å are depicted in Fig. 8C. The structure of the SARS-CoV-2 Mpro is shown in the colored representation in Fig. 8C. As expected, the N and C terminals of the protein are highly flexible, where RMSF values are reaching 4.5 and 8.5 Å, respectively. The entire protein (green cartoon) has RMSF values ranging between 0.5 and 2 Å except for the marked two regions (residues 46–53 (blue helix) and residues 189–194 (yellow coil)). These two regions have RMSF values reaching 4.2 Å and 2.4 Å, respectively. These highly movable regions are located apart from the two active site residues (H41 and C145), represented by the red stick in Fig. 8C. In order to check the effect of protein dyanmics on binding, AutoDock Vina was utilized to dock the best four compounds from Fig. 1 into H41 and C145 of SARS-CoV-2 Mpro.
Fig. 8A shows the binding energies of the docking of Nelfinavir, diosmin, rutin, hesperidin, and apiin into the active site of SARS-CoV-2 Mpro. Here 12 different conformations of the protein are selected after clustering analysis using Chimera software. The 12 representative conformations are taken at 8.4, 10.8, 14.2, 22.7, 40.1, 44.7, 54.7, 65.0, 68.3, 75.7, 79.2, and 90.6 ns. As shown in Fig. 8C, the RMSF is stable near the active site pocket suggesting no major effect on the ligand binding. This is reflected in the average binding energies calculated from the different conformations of SARS-CoV-2 Mpro depicted in Fig. 8A, with the standard deviation values as the error bars. At least four compounds show better average binding affinities to SARS-CoV-2 Mpro compared to the positive control compound, Nelfinavir (−7.4 ± 0.6 kcal/mol). These four compounds are diosmin (−8.7 ± 0.5 kcal/mol), rutin (−8.6 ± 0.6 kcal/mol), hesperidin (−8.5 ± 0.4 kcal/mol) and apiin (−8.2 ± 0.4 kcal/mol).
The most common interactions established between the polyphenols and the SARS-CoV-2 Mpro are the H-bonding and hydrophobic interaction. Fig. 8B shows the interactions formed between the polyphenols (Diosmin, Rutin, Hesperidin, and Apiin) and SARS-CoV-2 Mpro active site residues H41 and C145 after docking. Diosmin forms seven H-bonds (L141, S144, E166, R188, Q189, T190, and Q192) and two hydrophobic contacts (E166 and P168), while both Rutin and Apiin, form eight H-bonds (T26, N142, G143, S144(2), C145, E166 and Q189 for Rutin, and T24, T26, N119, N142, G143, S144(2), and C145 for Apiin). Rutin forms three hydrophobic contacts (M165, E166, and Q189), while Apiin doesn't form any hydrophobic interactions with SARS-CoV-2 Mpro. Hesperidin creates fewer hydrophobic contacts (one with T25), while it establishes six H-bonds (N142, G143, S144(2), Q189, and T190) with SARS-CoV-2 Mpro.
As shown in Fig. 8B, the most dominant interactions are the H-bonding (blue line) in all the studied polyphenols. The most conserved H-bonds that are established in all the four polyphenols are through S144, while Rutin, Hesperidin, and Apiin form H-bonds to N142 and G143. E166 and Q189 are engaged in both H-bonds (blue lines) and hydrophobic interactions (dashed-gray lines) with Diosmin and Rutin.
3.4. ADMET studies
ADMET analysis of hit compounds is provided in Table 5 . The volume of distribution at steady state (VDSS) was predicted to range from 1.663 to 0.996. They can be found at high concentrations in plasma rather than tissue. If the target is not in the brain, protecting the brain from the toxic effects of drugs is pharmacologically very important. In this model, values between 0.4- (−1) for logBB were defined as the molecule medium-level could cross the BBB, and lower than −1 poorly distributed to the brain. Nelfinavir (−0.522) indicates a low BBB permeability and these compounds (−1.715 - (−1.899) also display a poor BBB permeability. Similar results were obtained for CNS permeability studies. These results have demonstrated that the most active compounds against SARS-CoV-2 main protease had less toxicity than nelfinavir for the brain. In the evaluation of the maximum tolerated dose suggested for the human phase I study, 0.477 logs (mol/kg/day) equal or less concentration is considered low, and above it is considered high. While Rutin and Apiin have suitable values for the human phase I study suggested that, Hesperidin and Diosmin have values slightly above the recommended toxicity value. Also, their maximum tolerated dose (human) is lower Nelfinavir than. Four compounds were predicted as no potent inhibitors for CYP1A2, CYP2C19, CYP2C9, CYP2D6, CYP3A4 enzymes. Therefore, these flavonoids were predicted as nontoxic for the liver. But, Nelfinavir was found as a potential inhibitor for CYP2C19, CYP2D6, CYP3A4 enzymes, and predicted to have hepatotoxicity. In terms of other toxicity values such as hERG I inhibitor, oral rat acute toxicity (LD50), oral rat chronic toxicity (LOAEL), skin sensitization, these compounds falls within acceptable range.
Table 5.
Important ADMET properties for some flavonoids with high binding affinity.
| Property | Model Name | Predicted Value |
||||
|---|---|---|---|---|---|---|
| Hesperidin | Rutin | Diosmin | Apiin | Nelfinavir | ||
| Absorption | Water solubility (log mol/L) | −3.014 | −2.892 | −2.929 | −2.851 | −3.894 |
| Caco2 permeability (log Papp in 10−6 cm/s) | 0.505 | −0.949 | 0.305 | −0.966 | 0.693 | |
| Intestinal absorption (human) (% Absorbed) | 31.481 | 23.446 | 29.319 | 17.411 | 70.888 | |
| Skin Permeability(log Kp) | −2.735 | −2.735 | −2.735 | −2.735 | −2.737 | |
| P-glycoprotein substrate | Yes | Yes | Yes | Yes | Yes | |
| P-glycoprotein I inhibitor | No | No | No | No | Yes | |
| P-glycoprotein II inhibitor | No | No | No | No | Yes | |
| Distribution | VDss (human) (log L/kg) | 0.996 | 1.663 | 1.428 | 1.004 | 0.563 |
| Fraction unbound (human) (Fu) | 0.101 | 0.187 | 0.105 | 0.171 | 0.094 | |
| BBB permeability (log BB) | −1.715 | −1.899 | −1.795 | −1.793 | −0.522 | |
| CNS permeability (log PS) | −4.807 | −5.178 | −4.836 | −4.972 | −2.245 | |
| Metabolism | CYP2D6 substrate | No | No | No | No | No |
| CYP3A4 substrate | No | No | No | No | Yes | |
| CYP1A2 inhibitior | No | No | No | No | No | |
| CYP2C19 inhibitior | No | No | No | No | Yes | |
| CYP2C9 inhibitior | No | No | No | No | No | |
| CYP2D6 inhibitior | No | No | No | No | Yes | |
| CYP3A4 inhibitior | No | No | No | No | Yes | |
| Excretion | Total Clearance(log ml/min/kg) | 0.211 | −0.369 | −0.113 | −0.054 | 0.399 |
| Renal OCT2 substrate | No | No | No | No | No | |
| Toxicity | Max. tolerated dose (human) (log mg/kg/day) | 0.525 | 0.452 | 0.565 | 0.446 | −0.576 |
| hERG I inhibitor | No | No | No | No | No | |
| hERG II inhibitor | Yes | Yes | Yes | Yes | Yes | |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 2.506 | 2.491 | 2.512 | 2.49 | 2.54 | |
| Oral Rat Chronic Toxicity (LOAEL) (log mg/kg_bw/day) | 3.167 | 3.673 | 3.343 | 4.574 | 3.911 | |
| Hepatotoxicity | No | No | No | No | Yes | |
| Skin Sensitization | No | No | No | No | No | |
The antiviral effects of flavonoids have been the subject matter of several reports [[62], [63], [64]]. It has been previously reported that flavonoids exert their antiviral effects via blockage of cellular receptors, inhibiting viral antigenic determinants, loss of enzymatic functions, and/or inhibition of particle biosynthesis, which is consistent with our findings [[65], [66], [67]]. Furthermore, the antiviral activity of specific flavonoid subclass groups such as catechins, flavanones, flavonols has been reported previously against various viral strains [68,69]. Song et al. (2005) reported reduced viral infectivity by catechins [68]. Antiviral natural product-based medicines have also been used for two previous coronavirus outbreaks of SARS-CoV and MERS-CoV, which suggests that nature has tremendous potential to provide treatment for the ongoing epidemic of COVID-19 [[70], [71], [72]].
Previous reports also suggested the anti-influenza virus potential of Hesperidin and Apiin [73,74]. Additionally, the anti-Dengue virus (DENV) activity of Rutin [75], the anti-rotavirus potential of Diosmin, was reported [76]. Hesperidin and diosmin are flavanone glycoside, which is richly found in the citrus, including lemons, grapefruits, and sweet oranges [77,78]. Among all the flavonoids, Diosmin, Rutin, Hesperidin, and Apiin are found to possess very promising binding interactions with SARS-CoV-2 Mpro.
Hesperetin, a primary metabolite of hesperidin, has been shown to inhibit viral Mpro cleavage activity with IC50 of 8.3 μM [71]. Hesperidin is a readily bioavailable compound reaching a peak plasma concentration of 1.28 μmol/L in humans after intake of 1L orange juice [79]. Some studies also suggest its efficacy at the SARS-CoV-2 replication sites such as lungs. Hesperidin and Hesperetin were found to be effective in reducing the discharge of pro-inflammatory cytokines in the lungs. Hesperidin has also been reported as an effective antagonist of Th2 cytokines in the alveolar space where localized inflammatory cytokine storms arise during early phases of respiratory distress syndrome, eventually leading to organ damage [80,81]. A daily dose of 292 mg Hesperidin (500 mL of orange) resulted in significant anti-atherogenic and anti-inflammatory activities in a large-scale clinical trial [81]. As COVID-19 - patients suffer from severe inflammatory cytokine response in the later stages of infection; thus, dual antiviral/anti-inflammatory properties of this compound recommend its efficacy for the prevention and treatment of COVID-19. Interestingly, its toxicity is also low, with LD50 more than 4.5 g/kg [82]. Thus, hesperidin's efficacy, bioavailability, and low toxicity suggest its immense potential against COVID-19. COVID-19 patients also suffer from hypoxic pulmonary vasoconstriction and reduced lung ventilation [83]. Daflon (500 mg) is a flavonoid vasoprotective containing Diosmin (90%) and Hesperidin as active ingredients with no side effects and having LD50 > 3 g/kg [84]. Thus, combinatorial therapies of Daflon or Hesperidin supplements with other drugs might work effectively for the treatment of COVID-19 patients.
4. Conclusion
In this study, computer-aided molecular docking was performed using 81 flavonoid-based compounds against SARS-CoV-2 Mpro. The most effective compounds (Hesperidin, Rutin, Diosmin, and Apiin) were studied in multiple dimensions using computer-aided simulation and computational techniques to determine their affinity, binding stability, and the closest real condition responses against SARS-CoV-2 Mpro. We found that these compounds have a better binding affinity than Nelfinavir. All the compounds bearing good binding potency are components of dietary foods, further supporting the potential of these compounds as cheaper and safer candidates to develop therapeutics against COVID-19. The study provides a scientific basis for the utilization of dietary molecules as SARS-CoV-2 Mpro inhibitors, however, these in silico results need to be validated by in vitro protease activity assay and in vivo studies. Besides, the optimal dosing regimen for adults and the elderly based on reducing viral load and shortening the infectious period needs to be optimized for therapeutic implications.
Declaration of competing interest
The authors have no conflict of interest.
Acknowledgments
MDS calculations are conducted on the supercomputing facility of the Bibliotheca Alexandrina, Alexandria, Egypt. This work is done during the Junior associate award granted to the author for the period 2016–2021 from the Abdus Salam International Center for Theoretical Physics (ICTP), Trieste, Italy. The preprinted form of this paper was published in www.preprints.org with the DOI number; 10.20944/preprints202003.0333.v1 entitled “Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: an in silico strategy unveils a hope against CORONA”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105452.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Aanouz I., Belhassan A., El-Khatabi K., Lakhlifi T., El-Ldrissi M., Bouachrine M. Moroccan Medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1758790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta M.K., Vemula S., Donde R., Gouda G., Behera L., Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan R.J., Jha R.K., Amera G.M., Jain M., Singh E., Pathak A., Singh R.P., Muthukumaran J., Singh A.K. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose methyltransferase. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1753577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alméciga-Díaz C.J., Pimentel-Vera L.N., Caro A., Mosquera A., Moreno C.A.C., Rojas J.P.M., DíazTribaldos D.C. 2020. Virtual Screening of Potential Inhibitors for SARS-CoV-2 Main Protease. [Google Scholar]
- 6.Yamamoto N., Matsuyama S., Hoshino T., Yamamoto N. 2020. Nelfinavir Inhibits Replication of Severe Acute Respiratory Syndrome Coronavirus 2 in Vitro. [Google Scholar]
- 7.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 9.Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A.A., Falahati M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 12.Muralidharan N., Sakthivel R., Velmurugan D., Gromiha M.M. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J. Biomol. Struct. Dyn. 2020:1–6. doi: 10.1080/07391102.2020.1752802. [DOI] [PubMed] [Google Scholar]
- 13.Elmezayen A.D., Al-Obaidi A., Sahin A.T., Yelekci K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., et al. 2020. Structure-based Drug Design, Virtual Screening and High-Throughput Screening Rapidly Identify Antiviral Leads Targeting COVID-19. [Google Scholar]
- 15.Grosso G. Effects of polyphenol-rich foods on human health. Nutrients. 2018;10 doi: 10.3390/nu10081089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomford N.E., Senthebane D.A., Rowe A., Munro D., Seele P., Maroyi A., Dzobo K. Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19061578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elfiky A.A., Azzam E.B. Novel guanosine derivatives against MERS CoV polymerase: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murgueitio M.S., Bermudez M., Mortier J., Wolber G. In silico virtual screening approaches for anti-viral drug discovery, Drug discovery today. Technologies. 2012;9:e219–225. doi: 10.1016/j.ddtec.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leach A. second ed. Prentice Hall; 2001. Molecular Modelling: Principles and Applications. [Google Scholar]
- 21.Elfiky A.A., Elshemey W.M. Molecular dynamics simulation revealed binding of nucleotide inhibitors to ZIKV polymerase over 444 nanoseconds. J. Med. Virol. 2018;90:13–18. doi: 10.1002/jmv.24934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elfiky A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elfiky A.A., Azzam E.B. Novel guanosine derivatives against MERS CoV polymerase: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1758789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elfiky A.A., Ismail A.M., Elshemey W.M. Recognition of gluconeogenic enzymes; Icl1, Fbp1, and Mdh2 by Gid4 ligase: a molecular docking study. J. Mol. Recogn. 2020;33 doi: 10.1002/jmr.2831. [DOI] [PubMed] [Google Scholar]
- 26.Elfiky A.A., Ismail A. Molecular dynamics and docking reveal the potency of novel GTP derivatives against RNA dependent RNA polymerase of genotype 4a HCV. Life Sci. 2019;238:116958. doi: 10.1016/j.lfs.2019.116958. [DOI] [PubMed] [Google Scholar]
- 27.Nabati M., Bodaghi-Namileh V. In silico study of the active components (17 alpha-ethinyl estradiol and segesterone acetate) of annovera as a novel vaginal contraceptive system by docking of their binding to estrogen and progesterone receptors. Eurasian Chem. Commun. 2020;2:234–246. [Google Scholar]
- 28.Formagio A.S.N., Volobuff C.R.F., Kassuya C.A.L., Cardoso C.A.L., do Carmo Vieira M., Pereira Z.V., Bagatin M.C., de Freitas Gauze G. Psychotria leiocarpa extract and vincosamide reduce chemically-induced inflammation in mice and inhibit the acetylcholinesterase activity. Inflammation. 2019;42:1561–1574. doi: 10.1007/s10753-019-01018-w. [DOI] [PubMed] [Google Scholar]
- 29.Ferreira L.L., Andricopulo A.D. ADMET modeling approaches in drug discovery. Drug Discov. Today. 2019;24:1157–1165. doi: 10.1016/j.drudis.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 30.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molegro A. DK-8000 Aarhus C; Denmark: 2019. MVD 7.0 Molegro Virtual Docker. [Google Scholar]
- 32.Liu X., Zhang B., Jin Z., Yang H., Rao Z. 2020. The Crystal Structure of COVID-19 Main Protease in Complex with an Inhibitor N3. [Google Scholar]
- 33.COVID-19 main protease with unliganded active site. http://www.rcsb.org/structure/6Y84
- 34.Kim S., Thiessen P.A., Bolton E.E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B.A. PubChem substance and compound databases. Nucleic Acids Res. 2015;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. 2020. Potential Inhibitor of COVID-19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Molecular Docking Study. [Google Scholar]
- 36.V. 1.7.6, The PyMOL Molecular Graphics System, Version 1.7.6 Schrödinger, LLC.
- 37.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001;105:9954–9960. [Google Scholar]
- 38.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noorbatcha Ibrahim A., Khan A.M., Salleh H.M. Molecular dynamics studies of human ?-Glucuronidase. Am. J. Appl. Sci. 2010;7:823–828. [Google Scholar]
- 40.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(33–38):27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 41.A.s. Inc . 2009. Modeling and Simulation Solutions for Chemicals and Materials Research, Materials Studio (Version 5.0) San Diego, USA. [Google Scholar]
- 42.Wu X., Ray A.K. Density-functional study of water adsorption on the (formula presented) surface. Phys. Rev. B Condens. Matter. 2002;65:1–7. [Google Scholar]
- 43.Delley B. An all-electron numerical method for solving the local density functional for polyatomic molecules. J. Chem. Phys. 1990;92:508–517. [Google Scholar]
- 44.Delley B. From molecules to solids with the DMol3 approach. J. Chem. Phys. 2000;113:7756–7764. [Google Scholar]
- 45.Pires D.E., Blundell T.L., Ascher D.B. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58:4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.To K.K., Hung I.F., Chan J.F., Yuen K.Y. From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis. 2013;5(Suppl 2):S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pant S., Singh M., Ravichandiran V., Murty U.S.N., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martinez J.P., Sasse F., Bronstrup M., Diez J., Meyerhans A. Antiviral drug discovery: broad-spectrum drugs from nature. Nat. Prod. Rep. 2015;32:29–48. doi: 10.1039/c4np00085d. [DOI] [PubMed] [Google Scholar]
- 49.Lin L.T., Hsu W.C., Lin C.C. Antiviral natural products and herbal medicines. J. Tradit., Complementary Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T., Zhang Y.J., Chinnasamy S., Wei D.Q. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2. A molecular dynamic study. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundu Umesh D., Selvaraj C., Singh S.K., Dubey V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1763202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinha S.K., Shakya A., Prasad S.K., Singh S., Gurav N.S., Prasad R.S., Gurav S.S. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sk M.F., Roy R., Jonniya N.A., Poddar S., Kar P. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J. Biomol. Struct. Dyn. 2020:1–21. doi: 10.1080/07391102.2020.1768149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abdelli I., Hassani F., Bekkel Brikci S., Ghalem S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J. Biomol. Struct. Dyn. 2020:1–7. doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Refaey R.H., El-Ashrey M.K., Nissan Y.M. Repurposing of renin inhibitors as SARS-COV-2 main protease inhibitors: a computational study. Virology. 2021;554:48–54. doi: 10.1016/j.virol.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah R., Alharbi A., Hameed A.M., Saad F., Zaky R., Khedr A.M., El-Metwaly N. Synthesis and structural elucidation for new schiff Base complexes; conductance, conformational, MOE-docking and biological studies. J. Inorg. Organomet. Polym. 2020;30:3595–3607. [Google Scholar]
- 58.Al-Hazmi G.A.A., Abou-Melha K.S., El-Metwaly N.M., Althagafi I., Zaki R., Shaaban F. Green synthesis for 3-(2-Benzoylhydrazono)-N-(pyridin-2-yl)butanamide complexes: spectral, analytical, modelling, MOE docking and biological studies. J. Inorg. Organomet. Polym. 2019;30:1519–1536. [Google Scholar]
- 59.Mumit M.A., Pal T.K., Alam M.A., Islam M.A., Paul S., Sheikh M.C. DFT studies on vibrational and electronic spectra, HOMO-LUMO, MEP, HOMA, NBO and molecular docking analysis of benzyl-3-N-(2,4,5-trimethoxyphenylmethylene)hydrazinecarbodithioate. J. Mol. Struct. 2020;1220:128715. doi: 10.1016/j.molstruc.2020.128715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Z., Xie Y., Hao F., Liu P., Luo H.a. Catalytic oxidation of cyclohexane by substituted metalloporphyrins: experimental and molecular simulation. RSC Adv. 2015;5:101593–101598. [Google Scholar]
- 61.Li K., Li N., Yan N., Wang T., Zhang Y., Song Q., Li H. Adsorption of small hydrocarbons on pristine, N-doped and vacancy graphene by DFT study. Appl. Surf. Sci. 2020;515 [Google Scholar]
- 62.Chen C., Qiu H., Gong J., Liu Q., Xiao H., Chen X.W., Sun B.L., Yang R.G. (-)-Epigallocatechin-3-gallate inhibits the replication cycle of hepatitis C virus. Arch. Virol. 2012;157:1301–1312. doi: 10.1007/s00705-012-1304-0. [DOI] [PubMed] [Google Scholar]
- 63.Gescher K., Hensel A., Hafezi W., Derksen A., Kuhn J. Oligomeric proanthocyanidins from Rumex acetosa L. inhibit the attachment of herpes simplex virus type-1. Antivir. Res. 2011;89:9–18. doi: 10.1016/j.antiviral.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 64.Roh C., Jo S.K. (-)-Epigallocatechin gallate inhibits hepatitis C virus (HCV) viral protein NS5B. Talanta. 2011;85:2639–2642. doi: 10.1016/j.talanta.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 65.Bae E.A., Han M.J., Lee M., Kim D.H. In vitro inhibitory effect of some flavonoids on rotavirus infectivity. Biological & pharmaceutical bulletin. 2000;23:1122–1124. doi: 10.1248/bpb.23.1122. [DOI] [PubMed] [Google Scholar]
- 66.Chang L.K., Wei T.T., Chiu Y.F., Tung C.P., Chuang J.Y., Hung S.K., Li C., Liu S.T. Inhibition of Epstein-Barr virus lytic cycle by (-)-epigallocatechin gallate. Biochem. Biophys. Res. Commun. 2003;301:1062–1068. doi: 10.1016/s0006-291x(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 67.Calzada F., Cedillo-Rivera R., Bye R., Mata R. Geranins C and D, additional new antiprotozoal A-type proanthocyanidins from Geranium niveum. Planta Med. 2001;67:677–680. doi: 10.1055/s-2001-17358. [DOI] [PubMed] [Google Scholar]
- 68.Song J.M., Lee K.H., Seong B.L. Antiviral effect of catechins in green tea on influenza virus. Antivir. Res. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 69.Savi L.A., Caon T., de Oliveira A.P., Sobottka A.M., Werner W., Reginatto F.H., Schenkel E.P., Barardi C.R., Simoes C.M. Evaluation of antirotavirus activity of flavonoids. Fitoterapia. 2010;81:1142–1146. doi: 10.1016/j.fitote.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li S.Y., Chen C., Zhang H.Q., Guo H.Y., Wang H., Wang L., Zhang X., Hua S.N., Yu J., Xiao P.G., Li R.S., Tan X. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir. Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin C.W., Tsai F.J., Tsai C.H., Lai C.C., Wan L., Ho T.Y., Hsieh C.C., Chao P.D. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antivir. Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lau K.M., Lee K.M., Koon C.M., Cheung C.S., Lau C.P., Ho H.M., Lee M.Y., Au S.W., Cheng C.H., Lau C.B., Tsui S.K., Wan D.C., Waye M.M., Wong K.B., Wong C.K., Lam C.W., Leung P.C., Fung K.P. Immunomodulatory and anti-SARS activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118:79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong W., Wei X., Zhang F., Hao J., Huang F., Zhang C., Liang W. A dual character of flavonoids in influenza A virus replication and spread through modulating cell-autonomous immunity by MAPK signaling pathways. Sci. Rep. 2014;4:7237. doi: 10.1038/srep07237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu A.L., Liu B., Qin H.L., Lee S.M., Wang Y.T., Du G.H. Anti-influenza virus activities of flavonoids from the medicinal plant Elsholtzia rugulosa. Planta Med. 2008;74:847–851. doi: 10.1055/s-2008-1074558. [DOI] [PubMed] [Google Scholar]
- 75.Zandi k., Teoh B.T., Sam S.S., F W.P., Mustafa M.R., Bakar S.A. In vitro antiviral activity of Fisetin, Rutin and Naringenin against Dengue virus type-2. J. Med. Plants Res. 2011;5:5534–5539. [Google Scholar]
- 76.Lipson S.M. Flavonoid-associated direct loss of rotavirus antigen/antigen activity in cell-free suspension. Journal of Medicinally Active Plants. 2013;2 [Google Scholar]
- 77.Hsu C.C., Lin M.H., Cheng J.T., Wu M.C. Diosmin, a citrus nutrient, activates imidazoline receptors to alleviate blood glucose and lipids in type 1-like diabetic rats. Nutrients. 2017;9 doi: 10.3390/nu9070684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hajialyani M., Hosein Farzaei M., Echeverria J., Nabavi S.M., Uriarte E., Sobarzo-Sanchez E. Hesperidin as a neuroprotective agent: a review of animal and clinical evidence. Molecules. 2019:24. doi: 10.3390/molecules24030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Manach C., Morand C., Gil-Izquierdo A., Bouteloup-Demange C., Remesy C. Bioavailability in humans of the flavanones hesperidin and narirutin after the ingestion of two doses of orange juice. Eur. J. Clin. Nutr. 2003;57:235–242. doi: 10.1038/sj.ejcn.1601547. [DOI] [PubMed] [Google Scholar]
- 80.Pugin J., Verghese G., Widmer M.C., Matthay M.A. The alveolar space is the site of intense inflammatory and profibrotic reactions in the early phase of acute respiratory distress syndrome. Crit. Care Med. 1999;27 doi: 10.1097/00003246-199902000-00036. [DOI] [PubMed] [Google Scholar]
- 81.Meneguzzo F., Ciriminna R., Zabini F., Pagliaro M. 2020. Hydrodynamic Cavitation-Based Rapid Expansion of Hesperidin-Rich Products from Waste Citrus Peel as a Potential Tool against COVID-19. [Google Scholar]
- 82.Li Y., Kandhare A.D., Mukherjee A.A., Bodhankar S.L. Acute and sub-chronic oral toxicity studies of hesperidin isolated from orange peel extract in Sprague Dawley rats. Regul. Toxicol. Pharmacol. : RTP (Regul. Toxicol. Pharmacol.) 2019;105:77–85. doi: 10.1016/j.yrtph.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 83.Solaimanzadeh I. Acetazolamide, nifedipine and phosphodiesterase inhibitors: rationale for their utilization as adjunctive countermeasures in the treatment of coronavirus disease 2019 (COVID-19) Cureus. 2020;12 doi: 10.7759/cureus.7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyer O.C. Safety and security of Daflon 500 mg in venous insufficiency and in hemorrhoidal disease. Angiology. 1994;45:579–584. doi: 10.1177/000331979404500614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.