Abstract
Today COVID-19 pandemic articulates high stress on clinical resources around the world. At present, physical and viral tests are slowly emerging, and there is a need for robust pandemic detection that biomedical sensors can aid. The utility of biomedical sensors is correlated with the medical instruments with physiological metrics. These Biomedical sensors are integrated with the systematic device to track the target analytes with a biomedical component. The COVID-19 patients' samples are collected, and biomarkers are detected using four sensors: blood pressure sensor, G-FET based biosensor, electrochemical sensor, and potentiometric sensor with different quantifiable measures. The imputed data is then profiled with chest X-ray images from the Covid-19 patients.Multi-Layer Perceptron (MLP), an AI model, is deployed to identify the hidden signatures with biomarkers. The performance of the biosensor is measured with three parameters such as sensitivity, specificity and detection limit by generating the calibration plots that accurately fits the model.
Keywords: Biomedical sensors, COVID-19, Medical instruments, Quantifiable Measures, Biomarkers, Hidden signatures, Artificial intelligence
1. Introduction
The COVID-19 outbreak caused a severe acute respiratory syndrome coronavirus that was proclaimed a pandemic in 2020. It increased the mortality rate and led to a fortuitous burden in the worldwide socioeconomic crisis [1]. The transmission of this coronavirus occurred through the air, breathing, touching certain abiotic superficies. The transfer of the coronavirus has also occurred through asymptotic victims that have been contemplated in many patients [2], [3]. In this scenario, robust and efficient diagnosis has become an essential tool to overcome the outbreak and take instantaneous decisions for better diagnosis.
The precise way of detecting target virus can be compassed using biosensor-based techniques [4]. The technique behind the biosensors is the method of implementing bio perception components and target molecules on the biosensing platforms that detect the biological analytes [5], [6]. This integration of bio components with the biosensors acts as transducers and generates the signals directly or indirectly through perception components like enzymes and relevant compounds [7], [8]. Nowadays, biomedical sensors are applied in various medical diagnoses, environmental monitoring and processing food and agricultural products [9]. These biosensors act as analytical devices that detect the presence of the analyte and explore the structure of the cell and the drug molecules present on the cell [10], [11]. Accordingly, the biosensors are designed in conjunction with the target to achieve the measurable signal with suitable resolution and differentiate even the slightest change in the target analyte concentration [12], [13]. This type of biosensor involves the biosensing platforms that identify viral infection and analyze analytes' presence in viral nucleic acids like DNA and RNA, proteins and antibodies/antigens generated in the patient's immune response against this virus [14].
In the proposed model, the four types of biosensors are deployed to collect the covid-19 samples from the patients and fed accordingly in the biosensing platforms to analyze the bio perception components. Biosensors contribute a lot to the detection of viral disease. The role of immune sensors is very efficient in isolating the antibodies with expanded analytes [15]. Potentiometric sensors draw less current, and it serves as circuit opposition to the current flow and acts as an essential bio-recognition process in electrochemical biosensors [16], [17]. These biosensors help exhibit the spread of covid disease even with a low quantity of analytes. The FET-based biosensors are used in clinical diagnosis that senses the surroundings' changes and surface, susceptible to noise detection.[18]. By implementing AI techniques, hidden Signatures of the coronavirus are identified. The paper is organized into five sections: Section 1 is the introduction. This section presents the need and importance of biomedical sensors in detecting the COVID-19 disease. Section 2 provides relevant works in this scenario and the biosensors' implementation. Section 3 describes the materials and methods implemented in the proposed work. Section 3.1 portrays biosensors' deployment and the biological process's layout. The integration of the bio perception components with biomedical sensors is introduced in Section 3.2. Section 3.3 describes the AI-based detection of hidden features, while Section 4 provides statistical analysis and measured samples and methods. Section 5 ends with a conclusion of the paper.
2. Related works
Kumar R et al. [19] proposed Adaptive Neuro-fuzzy Inference System (ANFIS), a machine learning technique to predict the outbreak and track the COVID-19 disease. The system tracks the epidemic growth based on the prior datasets harnessed from cloud computing showing an accuracy of 86%.
Wang, Q. et al. [20] devised a model that dealt with edge and structure information for medical image inpainting (ESMII). Features are extracted from the image based on three scales using multi-scale residual blocks (MRB). This model generated more structures with good textures when compared to other models. The system also solved the distortion of medical images.
Sitharthan, R. and Rajesh, M [21] presented an IOT based machine learning system to monitor the infected persons from the preliminary data to isolate themselves from the uninfected persons. This combined technology deploys parallel computing to track the pandemic disease using Artificial intelligence, thus preventing the spread. This system provides 93% accuracy in monitoring and tracking the victims.
Ganesh Babu R. and Chellaswamy [22] deployed a machine learning technique with a hyperspectral image to identify the three stages: early, middle and critical stage diseases in squash plants. A new innovative uncrewed aerial vehicle (UAV) was implemented to collect information from the field. The model also implemented Locality Preserving Discriminative Broad Learning (LPDBL) to distinguish healthy and diseased plants.
Kumar R et al. [23] proposed a new methodology to identify the hidden feature of lung sickness and monitor the growth in the early stage itself to prevent further growth and save the patient from lung sickness. The system used P-SVM calculation for sequencing high dimensional lung datasets. The model produced 83% of accurately predicting the characterization.
Karthickraja, R et al. [24] implemented sensor devices with wearable sensors to predict the COVID-19 cases. The model also incorporates clinical therapy with wearable devices that monitor the symptoms and test the suspicious cases that integrate the IoT elements. The paper also includes the risk factor analysis that acknowledges the oxygen saturation (Spo2) with chi-square distribution.
Nandagopal, V et al. [25] proposed a computation method, namely fuzzy-based logistic regression, for predicting the gene expression data. The feature selection is carried out using LASSO Logistic Regression (LLR). Maximum likelihood estimation (MLE) is integrated with the regression model for continuous monitoring of the gene data. The system explores mining technology to classify the cancer data by evaluating the training and testing data.
Koushik, SS and Srinivasa, KG [26] proposed Nesterov accelerated Adaptive moment estimation to predict pneumonia in two phases: bacterial phase and viral phase from chest X-rays deploying convolution neural networks. The model achieved 94% and 93% validation accuracy with training data and 87% test accuracy.
Casaccia, S et al. [27] devised a real-time location system (RTLS) integrated with inertial measurement unit (IMU)sensors implemented for social distancing and to measure physical activities. This system monitors and tracks the social distancing using this instrumentation and continuously assess their interpersonal distance does not exceed the maximum of 1.54 min. The collected data through the accelerometers are filtered by deploying discrete wavelet transform and measures the ageing people by fixing the threshold value.
3. Materials and methods
3.1. Biosensor
Today biosensors play a vital role in analyzing the molecular interactions in the human body. Biosensors generally comprise biological receptors and transducers. The former detects the target analyte and converts the recognized biomolecules into electrical signals and then is processed and displayed in an electronic system [28]. The transducer and electronics are combined, known as complementary metal-oxide semiconductors [29], [30]. This technology comes with a solution that designs silicon integrated with various potential at a low cost. This sensor provides varied ion sensitivity with different outputs based on the charge transfer process. These biosensors provide digital output by determining the recognized biological molecules, including enzymes, antibodies/antigens, proteins, viral RNA, and DNA interacting with a transducer. This interaction is computed by the transducer that produces a measurable signal equivalent to a target analyte.
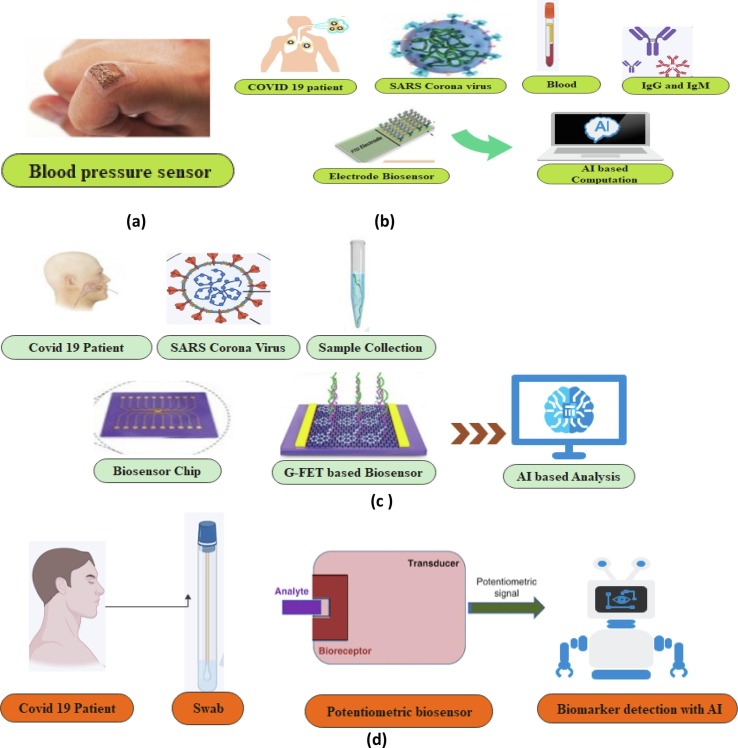
The biosensor's relevance concentrates on the place of interest [31], [32] when detecting biological sources, and specific biosensors are used in clinical diagnostics to determine the virus proportion in the air. A few molecular-based biosensors are used to detect the virus in the environment and detect the air pollutant [33]. The Covid-19 samples are collected through various biosensors such as blood pressure sensor, G-FET based biosensor, Electrode Biosensor and Potentiometric sensor by identifying various biomarkers [ Fig. 1 ]. The proposed AI model improves the correlation performance and discriminates between COVID-19 + ve, COVID-19 –ve and viral pneumonia. Finally, AI-based evaluation measures provide better results in detecting the hidden signatures of COVID-19 viruses.
Fig. 1.
Illustrative Representation of Four Bio Sensors in COVID-19 Detection. (a) Softsonics- Blood Pressure Sensor Patch. (b) Schematic Diagram of Electrode biosensor detecting blood samples and IgG, IgM antibody. (c) Structure of G-FET-based diagnostic technique to detect the COVID-19 samples. (d) A biosensing platform with Potentiometric Biosensor detecting the swab sample.
The proposed framework is focused on quick diagnosis by detecting the hidden signatures. The dataset is patterned into severe, non-severe and hidden signatures following the demographic features in Table 1 . The decision-making diagnosis is mainly based on these hidden signatures using the MLP classifier.
Table 1.
Hidden Signatures of COVID-19 Cases.
| Demographic Features | Severe Cases | Non-Severe Cases | Hidden Signatures |
|---|---|---|---|
| Transmission | Throat infection | Cough or sneezes Touching infected objects |
Skin problems, Dizziness |
| Systematic | High Fever | Fever | Brain Fog |
| Fatigue | Fatigue | ||
| Eye problems | |||
| Circulatory | Cardiovascular damage | Decreased white blood cells | No Signature |
| Respiratory | Pneumonia | Sneezing and runny nose | Dry Coughing |
| Acute Respiratory Syndrome | Shortness of breath | ||
| Lungs inflammation | Mild breathing difficulties | ||
| Dry Coughing | Sore throat | ||
| Digestive | Diarrhea | Gastrointestinal issues | |
| Excretory | Kidney failure | Decreased kidney function | No Signature |
3.1.1. Blood pressure sensor
Hypertension is a common cause of COVID-19 infection. Softsonics (Figure) is a flexible patch-like device that measures blood pressure using ultrasound pulses. This device can easily be worn on the skin over the jugular vein that produces 24 h continuous measurement. It delivers continuous reading essential in ICU wards and daily.
3.1.2. G-FET based biosensor
Graphene-based field-effect transistor biosensors are utilized for sensitive and effective measurements with few analytes [34], [35]. These FET-based biosensors are substantially used in clinical diagnosis on-spot detection specifically for testing in point of care aspects. Graphene-based F.E.T. biosensor comprises carbon atoms arranged hexagonally with two-dimensional sheets exposing towards the surface [36]. It senses environmental changes and detects low noise with its ultrasensitive nature. So, it is mainly considered for pathological diagnosis [37], [38]. In the proposed work, Graphene sheets act as a detection platform against COVID-19 spike antibody, and the sensor detects the target antigen protein and the detection limit is measured as 1 fg/ml. This sensor illustrated the better synthesize of COVID-19 spike antibody with that of the graphene sheet producing sensitive detection with the samples. The graphene sheet produces ultrasensitive with low noise detection and provides instant measurement periodically.
3.1.3. Electrochemical biosensor
An electrochemical biosensor is an electrochemical integrated biosensor that provides quantitative information based on analytes and mainly based on the current, field-effect that interacts with the target molecule with the bio perception components placed on the sensing platform. The enzymes on the biosensors initiate the process and increase the electron transfer towards the surface of the electrode. Then these electrical signals are then transferred to the reference electrode, and the current output is measured. The applied potential induces the electrochemical reaction in the blood sample solution [39], [40]. This biosensor provides a limited current with varied potential and a quantifiable current that helps the biorecognition process.
3.1.4. Potentiometric biosensor
This EC is categorized into different metrics based on the transducer deployed in the electrochemical biosensor. The potentiometric biosensor is one of the potentiometry of the electrochemical sensor. This sensor is an alternate of EC where it measures the electrode's potential in the case of failure of current flow in the EC cell [41]. The potentiometry produces an output of redox (reduction and oxidation). The Bio perception components are conjugated into potentiometric sensors where the electrodes detect the catalysts [42]. The swab of the COVID-19 patients is fed into the sensor plates that act as an analyte.
3.2. Bio perception components
The bio perception components (BPC) are strapped on the surface of the transducer to interact with the analytes. Bio perception elements are classified into biocatalytic in the proposed work, including enzymes, antibodies, antigen, and nucleic acid sequence [43]. To sense the analyte, a nanomaterial-based surface is utilized. Perhaps, very sensitive and selectivity perception components are not used. However, the equilibrium between physical parameters is maintained by Morales and Halpern's systematic design of biosensors [44], [45]. The BPCs used in this proposed work is as follows:
3.2.1. Antibody/Antigen
It is a protein-based BPC possessing 3D recognized pattern to identify the analyte combining antigen–antibody immune compound. This antibody-antigen is observed by the electrochemical and potentiometric sensor [46], [47], [48]. Different types of antibodies the deployed in the biosensor in appropriate proportion to increase the sensitivity of the biosensor [49].
3.2.2. Enzymes and RNA
It is one of the biocatalysts used for its specificity, and it induces to fit the model. Enzyme-based biosensors act as biocatalytic, and analytes are converted into the product measured when the color changes [50]. The enzymes are located very close to the surface of the transducer to enhance signal generation. These enzymes are integrated into the antibody to generate the signal [51]. The glucose detection is mainly used enzyme in the biosensor [52]. RNA is an influential biomarker used to detect the COVID-19 virus. RNA expression genes are considered the intended targets for biosensors and PCR-based tests [5], [53]. The diagnosis is mainly made based on the target samples integrated with the biomarkers immobilized on the biosensing elements [ Fig. 2 ].
Fig. 2.
COVID-19 Target Samples and Biomarkers.
3.3. AI-Based techniques in the detection of biomarkers
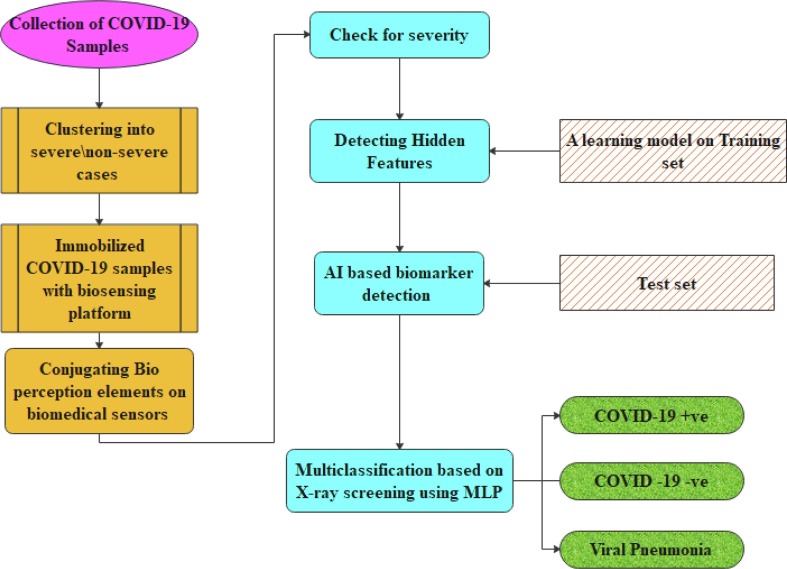
Artificial intelligence-based diagnosis is a great weapon to use large data models. Few data restrict the exploitation of AI for disease diagnosis [54]. Though the usage of CT and X-rays explores some pathological solutions, the visibility of the images does not lead to the proper diagnosis of disease for further treatment, and the spread of infection are not reported due to abnormal pattern [55]. The COVID samples are collected and clustered into severe and non-severe categories concerning their demographic features in the suggested work. Then the COVID−19 samples are immobilized with biosensing platforms. To identify the biomarkers, the target analytes are conjugated with the bio perception components (BPC), thus analyzing the progression of the infection. With this BPC, the severity of the virus is determined. The proposed model also deploys AI techniques to identify and recognize the CT X-ray images of COVID-19 using an MLP classifier with a neural network. AI-based X-ray screening is implemented to identify hidden signatures during COVID-19 testing and categorize the multi-class probes from the Kaggle Chest X-rays into COVID + Ve, COVID –Ve and Viral Pneumonia. The architecture of the proposed model is shown in Fig. 3 . The two phases of the MLP are used for error propagation and classification. The forward process is implemented for classification and backward procedure for error identification with nonlinear activation with hidden layers solving nonlinear paradigms.
Fig. 3.
Proposed Architecture of detecting and diagnosing the COVID-19.
4. Results and discussions
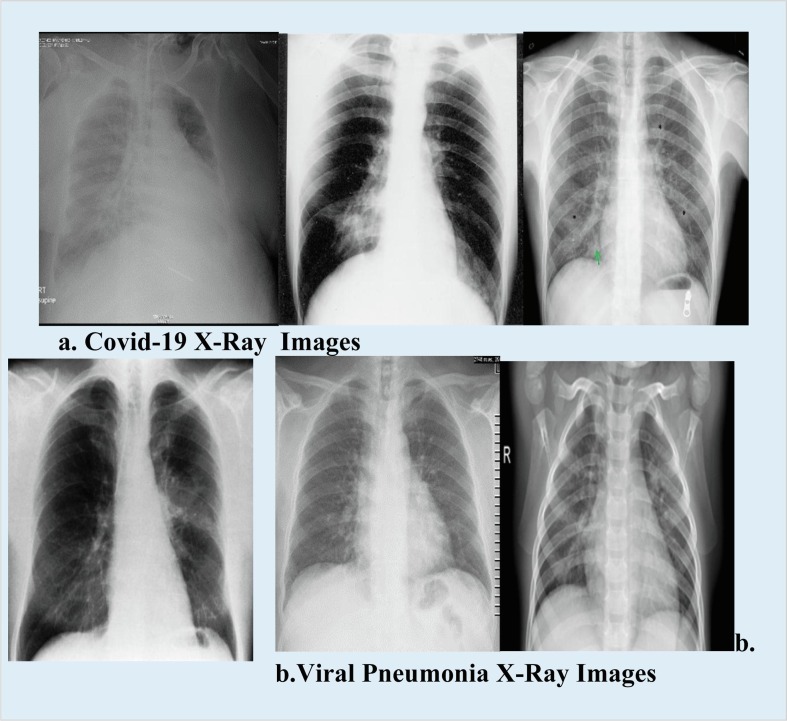
The propounded work is deployed with three biomedical sensors associated with bio perception components. The extracted samples are synthesized with the bio perception components. Then target samples are identified for further diagnosis. The significant issue of diagnosis lies in the sample collection and transportation from COVID 19 patients. The detection limit is computed for the various biosensors and bio perception components (BPC) associated with COVID 19 virus testing and validating, shown in Table 2 . The detection limit of the biomedical sensors is measured for each sample. The Kaggle dataset was harnessed, and these COVID-19 datasets were split into training and test datasets. MLP classifier model is implemented to distinguish multiple classes. The dataset is divided into three classes with COVID-19 positive, COVID-19 negative and Viral Pneumonia influenced by the COVID−19 viruses. The dataset is comprised of chest images of pneumonia patients. Fig. 4 shows some samples of COVID-19 cases obtained from the kaggle X-ray dataset. After synthesizing the samples, the biosensors are evaluated for the analytical measures. At the initial stage, the sensors are assessed with sensitivity and selectivity based on the target analyte [56].
Table 2.
Diagnosing Target Samples with Bio Perception Components(BPC).
| Biomedical Sensors | Bio Perception Components | Extracting Sample Type | Target Sample | Diagnosis | Detection Limit |
|---|---|---|---|---|---|
| Blood Pressure Sensor | Blood Pressure | Blood | Blood | yes | 1 Pa. |
| G-FET-Based biosensor | Antibody/Antigen | (IgG, IgM) | Antibody | yes | 1 fg/ml |
| Electrochemical Bio Sensor | Viral RNA separated from molecules | (IgG, IgM), Saliva Sputum | Spike Protein | yes | 10 µg mL−1 |
| Potentiometric Biosensor | Viral RNA, Protein | Swab, Saliva Sputum | Anti-Spike Protein | yes | 101 cfu mL−1 |
Fig. 4.
X-Ray Samples of COVID-19 and Viral Pneumonia images.
A regression plot is generated under the signal output with different concentrations of enzymes and molecules, which is further considered for the sensing process. Based on this calibration plot, the performance of the biosensors are measured with specific parameters:
-
i.
Sensitivity: The sensitivity in the biosensor is a regression line representing the slope. This slope highlights the changes in the output signal concerning the concentration of the analyte. A steeper slope in the biosensor is considered more sensitive and can easily detect a slight change in a solution. The equation of the sensitivity is given as:
| (1) |
Whereas and are signals of the analyte concentrations and and are the two analytes.
-
ii.
Limit of detection (LOD): The limit of detection (LOD) is measured in the minimum quantity of analyte that produces a discernable signal in the biosensor. LOD is computed as:
| (2) |
where is the detection limit, is the standard deviation of blank, and is the slope that shows the probe sensitivity for the analyte. During the clinical detection, some sample loss affects the LOD. This loss can be expressed as:
| (3) |
Where is a quantile function with k-1 degrees of freedom, defines the confidence interval k at various analyte concentrations. The limit detection can also be extracted by adjusting the inverse function and represented as:
| (4) |
Moreover, the detection limit can also be calculated in terms of sensor resolution and surface mass concentration concerning the analyte.
| 5) |
Where Rs is the resolution of the sensor; resultant between the change in sensor and surface mass concentration of the analyte.
-
iii.
Selectivity: This measure identifies and differentiates the target analyte from other molecules present in the sample. Furthermore, it produces the output signal corresponding to the target analyte. This parameter gives the ideal solution to recognize one target analyte in the complex solution. Specificity is achieved by deploying antibodies and antigens, whereas selectivity is the property of analyzing the nearest molecules group.
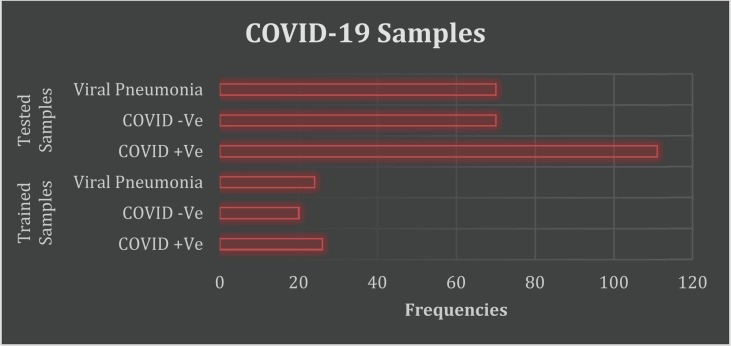
The hidden features of COVID-19 are detected using MLP classifiers. Totally 321 COVID-19 samples are taken for analysis. Among which 70 samples are used for training, and 251 samples are taken for testing. The proposed model MLP classifier is deployed to optimize the clinical trials for medications as the classifier supports multiple classifications. With AI-based techniques, the CT scans and X-rays are generated to diagnose the COVID-19 samples in three categories: positive, negative, and viral pneumonia, shown in Table 3 . The corresponding graphical representation (Fig. 5 ) is portrayed.
Table 3.
Trained and Tested COVID-19 Samples.
| COVID −19 Samples | Categories | Frequencies |
|---|---|---|
| Trained Samples | COVID + Ve | 26 |
| COVID -Ve | 20 | |
| Viral Pneumonia | 24 | |
| Tested Samples | COVID + Ve | 111 |
| COVID -Ve | 70 | |
| Viral Pneumonia | 70 |
Fig. 5.
Classification of COVID-19 Dataset using MLP.
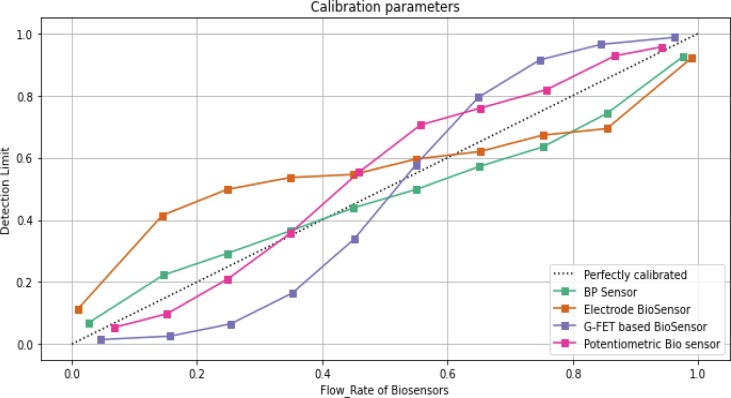
MLP regression deploys the backpropagation without activation function in the output layer. This classifier uses the square error as a loss function, and the continuous values are set as output. The correlation coefficient and the error rate are computed for the MLP classifier with its parameters Mean Absolute Error (MAE), Relative Absolute Error (RAE) and Root Mean Square Error (RMSE) for all three class labels ( Table 4 ). Calibration of the many biosensors is quantified in terms of limit detection and each sensor's flow rate in seconds, as seen in Fig. 6 .
Table 4.
MLP Classification with Error Rate.
| Classes | Covid-19 samples | Correlation Coefficient | MAE | RAE | RMSE |
|---|---|---|---|---|---|
| Class1 | Covid-19 - -ve | 1 | 0.0025 | 0.001 | 0.004 |
| Class 2 | Covid-19 - +ve | 0.98 | 0.0021 | 0.005 | 0.0056 |
| Class 3 | Viral Pneumonia | 0.85 | 0.1004 | 0.02 | 0.004 |
Fig. 6.
Calilibration Parameters vs Biomedical Sensors.
5. Conclusion
Biomedical sensors are mainly devised for the sensitivity of the target analytes with bioanalytical instruments. The proposed model exploits three types of biosensors: blood pressure biosensor, Electrochemical biosensor, G-FET-based biosensor, and potentiometric biosensor. These biosensors are developed to detect the sensitivity of the microbes or viruses with a meagre amount of analyte in very complex bio perception compounds. The proposed model segregates the severe and non-severe cases under the demographic features. These targets are extracted and conjugated with bio perception compounds (BPC) and then immobilized on the biosensing platforms. Each biosensor comprises its detection limit based upon the target analyte. The suggestion model identifies the target analyte and is synthesized with biomarker elements. The Bio perception compounds such as blood samples, antibody/antigen, viral RNA are integrated with the target analyte to detect the infection range of the COVID-19 cases. The hidden signatures of the COVID-19 are detected using AI technique with the Chest X-rays, and an MLP classifier is deployed to classify the virus in three categories comprising COVID + ve, COVID –ve and viral pneumonia. The flow rate of biosensors is measured in terms of bio perception compounds integrated with the target analyte, and calibration parameters of biomedical sensors are compared in terms of detection of limit. Besides these biosensors, technologically developed innovative devices-based biosensors need to be developed to target the antibodies or antigens that give quick remedy when compared to these devices. Usage of biosensors provides high throughput sensing background. The large scale of testing includes a variety of characterizations to explicit the biosensors' specificity and sensitivity, which becomes a significant challenge in deploying the biosensors in this pandemic crisis.
CRediT authorship contribution statement
V. Hemamalini: Writing – original draft, Supervision, Writing – review & editing. L. Anand: Methodology, Resources. S. Nachiyappan: Validation. S. Geeitha: Project administration, Writing – review & editing, Methodology, Conceptualization. Venkata Ramana Motupalli: Validation. R. Kumar: Investigation, Validation. A. Ahilan: Visualization. M. Rajesh: Formal analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Santiago I. Trends and innovations in biosensors for COVID-19 mass testing. Chembiochem: Eur. J. ChemBiol. 2020;21(20):2880–2889. doi: 10.1002/cbic.202000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoehl S., Rabenau H., Berger A., Kortenbusch M., Cinatl J., Bojkova D., Behrens P., Böddinghaus B., Götsch U., Naujoks F., Neumann P., Schork J., Tiarks-Jungk P., Walczok A., Eickmann M., Vehreschild M.J.G.T., Kann G., Wolf T., Gottschalk R., Ciesek S. Evidence of SARS-CoV-2 infection in returning travelers from Wuhan, China. New Engl. J. Med. 2020;382(13):1278–1280. doi: 10.1056/NEJMc2001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu P., Zhu J., Zhang Z., Han Y. Han: A familial cluster of infection associated with the 2019 novel coronavirus indicating possible person-to-person transmission during the incubation period. J. Infect. Dis. 2020;221(11):1757–1761. doi: 10.1093/infdis/jiaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asif M., Ajmal M., Ashraf G., Muhammad N., Aziz A., Iftikhar T., Wang J., Liu H. The role of biosensors in coronavirus disease-2019 outbreak. Curr. Opin. Electrochem. 2020;23:174–184. doi: 10.1016/j.coelec.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., et al. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. ClinChem. 2020;66:549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asif M., Haitao W., Shuang D., Aziz A., Zhang G., Xiao F., et al. Metal oxide intercalated layered double hydroxide nanosphere: with enhanced electrocatalytic activity towards H2O2 for biological applications. Sensor Actuator B Chem. 2017;239:243–252. [Google Scholar]
- 7.Asif M., Liu H., Aziz A., Wang H., Wang Z., Ajmal M., Xiao F., Liu H. Core shell iron oxide-layered double hydroxide: high electrochemical sensing performance of H2O2 biomarker in live cancer cells with plasma therapeutics. Biosensbioelectron. 2017;97:352–359. doi: 10.1016/j.bios.2017.05.057. [DOI] [PubMed] [Google Scholar]
- 8.Dincer C., Bruch R., Costa‐Rama E., Fernández‐Abedul M.T., Merkoçi A., Manz A., Urban G.A., Güder F. Disposable sensors in diagnostics, food, and environmental monitoring. Adv. Mater. 2019:1806739. doi: 10.1002/adma.201806739. [DOI] [PubMed] [Google Scholar]
- 9.Rodovalho V., Alves L., Castro A., et al. Biosensors applied to diagnose infectious diseases—an update. Austin J. BiosensBioelectron. 2015;1:1–12. [Google Scholar]
- 10.Chandra P., Son N.X., Noh H.-B., Goyal R.N., Shim Y.-B. Investigation on the downregulation of dopamine by acetaminophen administration based on their simultaneous determination in urine. Biosens. Bioelectron. 2013;39(1):139–144. doi: 10.1016/j.bios.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Kumar A., Purohit B., Mahato K., Mandal R., Srivastava A., Chandra P. Gold-iron bimetallic nanoparticles impregnated reduced graphene oxide-based nanosensor for label-free detection of biomarker related to non-alcoholic fatty liver disease. Electroanalysis. 2019;31(12):2417–2428. doi: 10.1002/elan.201900337. [DOI] [Google Scholar]
- 12.Carpenter A., Paulsen I., Williams T. Blueprints for biosensors: design, limitations, and applications. Genes. 2018;9:375. doi: 10.3390/genes9080375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.K. Mahato, P.K. Maurya, P. Chandra, Fundamentals and commercial aspects of nanosensors in point-of-care clinical diagnostics 3 (2018) 1–14, https:// doi.org/10.1007/s13205-018-1148-8. Biotech. 8. [DOI] [PMC free article] [PubMed]
- 14.Ozer T., Geiss B.J., Henry C.S. Review—Chemical and Biological Sensors for Viral Detection. J. Electrochem. Soc. 2020;167(3):037523. doi: 10.1149/2.0232003JES. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobed A., Sepehri Shafigh E. Biosensors promising bio-device for pandemic screening “COVID-19“. Microchem. J. 2021;164:106094. doi: 10.1016/j.microc.2021.106094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bäcker M., Koch C., Eiben S., Geiger F., Eber F., Gliemann H., Poghossian A., Wege C., Schöning M.j. Tobacco mosaic virus as enzyme nanocarrier for electrochemical biosensors. Sens. Actuators, B. 2017;238:716–722. [Google Scholar]
- 17.Kaushik A., Tiwari S., Jayant R.D., Vashist A., Nikkhah-Moshaie R., El-Hage N., Nair M. Electrochemical biosensors for early stage Zika diagnostics. Trends Biotechnol. 2017;35(4):308–317. doi: 10.1016/j.tibtech.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang K.-H., Chang T.-J., Wang M.-L., Tsai P.-H., Lin T.-H., Wang C.-T., Yang D.-M. Novel biosensor platforms for the detection of coronavirus infection and SARS-CoV-2 J. Chin. Med. Assoc. 2020 doi: 10.1097/JCMA.0000000000000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R., Al-Turjman F., Srinivas L.N.B., Braveen M., Ramakrishnan J. ANFIS for prediction of epidemic peak and infected cases for COVID-19 in India. Neural Comput. Appl. 2021:1–14. doi: 10.1007/s00521-021-06412-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q., Chen Y.i., Zhang N., Gu Y. Medical image inpainting with edge and structure priors. Measurement. 2021;185:110027. doi: 10.1016/j.measurement.2021.110027. [DOI] [Google Scholar]
- 21.Sitharthan R., Rajesh M. Application of machine learning (ML) and internet of things (IoT) in healthcare to predict and tackle pandemic situations. Distributed and Parallel Databases. 2021:1–19. doi: 10.1007/s10619-021-07358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesh Babu R., Chellaswamy C. Different stages of disease detection in squash plants based on machine learning. J. Biosci. 2022;47(1):1–14. [PubMed] [Google Scholar]
- 23.Kumar R., Al-Turjman F., Anand L., Kumar A., Magesh S., Vengatesan K., Sitharthan R., Rajesh M. Genomic sequence analysis of lung infections using artificial intelligence technique. Interdiscipl. Sci.: Computational Life Sci. 2021;13(2):192–200. doi: 10.1007/s12539-020-00414-3. [DOI] [PubMed] [Google Scholar]
- 24.Karthickraja R., Kumar R., Kirubakaran S., Manikandan R. COVID-19 prediction and symptom analysis using wearable sensors and IoT. Int. J. Pervasive Computing Commun. 2020 [Google Scholar]
- 25.Nandagopal V., Geeitha S., Kumar K.V., Anbarasi J. Feasible analysis of gene expression–a computational based classification for breast cancer. Measurement. 2019;140:120–125. [Google Scholar]
- 26.Koushik S.S., Srinivasa K.G. Detection of respiratory diseases from chest X rays using Nesterov accelerated adaptive moment estimation. Measurement. 2021;176 [Google Scholar]
- 27.Casaccia S., Naccarelli R., Moccia S., Migliorelli L., Frontoni E., Revel G.M. Development of a measurement setup to detect the level of physical activity and social distancing of ageing people in a social garden during COVID-19 pandemic. Measurement. 2021;184:109946. doi: 10.1016/j.measurement.2021.109946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui F., Zhou H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020:112349. doi: 10.1016/j.bios.2020.112349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arora R.K., Saini R.P. Biosensors: way of diagnosis. Int. J. Pharmaceut. Sci. Res. 2013;4(7):2517–2527. [Google Scholar]
- 30.Dincer C., Bruch R., Kling A., Dittrich P.S., Urban G.A. Multiplexed point-of- Care testing–xPOCT. Trends Biotechnol. 2017;35(8):728–742. doi: 10.1016/j.tibtech.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tech Desk, Meet the world's first smart wrist band that can detect body temperature, 14, 2020, p. 5. https://indianexpress.com/article/technology/tech-ne ws-technology/meet-the-worlds-first-smart-wrist-band-that-can-detect-body- temperature/.
- 32.Gjoreski M., Luštrek M., Gams M., Gjoreski H. Monitoring stress with a wrist Device using context. J. Biomed. Inf. 2017;73:159–170. doi: 10.1016/j.jbi.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Qiu G., Gai Z., Tao Y., Schmitt J., Kullak-Ublick G.A., Wang J. Dual-functional plasmonicphotothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14(5):5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 34.Janissen R., Sahoo P.K., Santos C.A., da Silva A.M., von Zuben A.A.G., Souto D.E.P., Costa A.D.T., Celedon P., Zanchin N.I.T., Almeida D.B., Oliveira D.S., Kubota L.T., Cesar C.L., Souza A.P., Cotta M.A. InP Nanowire Biosensor with Tailored Biofunctionalisation: Ultrasensitive and Highly Selective Disease Biomarker Detection. NanoLett. 2017;17:5938–5949. doi: 10.1021/acs.nanolett.7b01803. [DOI] [PubMed] [Google Scholar]
- 35.Liu J., Chen X., Wang Q., Xiao M., Zhong D., Sun W., Zhang G., Zhang Z. Ultrasensitive Monolayer MoS2 Field-Effect Transistor Based DNA Sensors for Screening of Down Syndrome. NanoLett. 2019;19:1437–1444. doi: 10.1021/acs.nanolett.8b03818. [DOI] [PubMed] [Google Scholar]
- 36.Seo G., Lee G., Kim M.J., Baek S.-H., Choi M., Ku K.B., Lee C.-S., Jun S., Park D., Kim H.G., Kim S.-J., Lee J.-O., Kim B.T., Park E.C., Kim S.I. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14(4):5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 37.Lei Y.M., Xiao M.M., Li Y.T., Xu L., Zhang H., Zhang Z.Y., Zhang G.J. Detection of Heart Failure-Related Biomarker in Whole Blood with Graphene Field Effect Transistor Biosensor. Biosens. Bioelectron. 2017;91:1–7. doi: 10.1016/j.bios.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L., Mao H., Wu C., Tang L., Wu Z., Sun H., Zhang H., Zhou H., Jia C., Jin Q., Chen X., Zhao J. Label-Free Graphene Biosensor Targeting Cancer Molecules Based on Non-Covalent Modification. Biosens. Bioelectron. 2017;87:701–707. doi: 10.1016/j.bios.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 39.Meirinho S.G., Dias L.G., Peres A.M., Rodrigues L.R. Voltammetricaptasensors for protein disease biomarkers detection: a review. Biotechnol. Adv. 2016;34(5):941–953. doi: 10.1016/j.biotechadv.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Gupta V.K., Jain R., Radhapyari K., Jadon N., Agarwal S. Voltammetric techniques for the assay of pharmaceuticals-A review. Anal. Biochem. 2011;408(2):179–196. doi: 10.1016/j.ab.2010.09.027. [DOI] [PubMed] [Google Scholar]
- 41.Ding J., Qin W. Recent advances in potentiometric biosensors. TrAC Trends Anal. Chem. (Reference Ed.) 2020;124:115803. doi: 10.1016/j.trac.2019.115803. [DOI] [Google Scholar]
- 42.Juang D.S., Lin C.H., Huo Y.R., Tang C.Y., Cheng C.R., Wu H.S., Huang S.F., Kalnitsky A., Lin C.C. Proton-ELISA: electrochemical immunoassay on a dual Gated ISFET array. Biosens. Bioelectron. 2018;117:175–182. doi: 10.1016/j.bios.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Purohit B., Kumar A., Mahato K., Roy S., Chandra P. Elsevier; Mod. Anim. Biotechnol.: 2019. Cancer cytosensing approaches in miniaturized settings based on advanced nanomaterials and biosensors, in Nanotechnol; pp. 133–147. [Google Scholar]
- 44.Morales M.A., Halpern J.M. Guide to selecting a biorecognition element for biosensors. Bioconjug. Chem. 2018;29(10):3231–3239. doi: 10.1021/acs.bioconjchem.8b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.JAB Tech, A. Prasad, K. Mahato, PK Maurya, P. Chandra, Analytical &Bioanalytical Techniques Biomaterials for Biosensing Applications, 2016, 7(2) pp. 1-2.
- 46.Mahato K., Purohit B., Kumar A., Chandra P. Clinically comparable impedimetricimmunosensor for serum alkaline phosphatase detection based on electrochemically engineered Au-nano-Dendroids and graphene oxide nanocomposite. Biosens. Bioelectron. 2020;148:111815. doi: 10.1016/j.bios.2019.111815. [DOI] [PubMed] [Google Scholar]
- 47.Chandra P., Noh H.-B., Pallela R., Shim Y.-B. Ultrasensitive detection of drug-resistant cancer cells in biological matrixes using an amperometricnanobiosensor. Biosens. Bioelectron. 2015;70:418–425. doi: 10.1016/j.bios.2015.03.069. [DOI] [PubMed] [Google Scholar]
- 48.Pallela R., Chandra P., Noh H.-B., Shim Y.-B. An amperometric nano biosensor using a biocompatible conjugate for early detection of metastatic cancer cells in biological fluid. Biosens. Bioelectron. 2016;85:883–890. doi: 10.1016/j.bios.2016.05.092. [DOI] [PubMed] [Google Scholar]
- 49.Estrela P., Sharma S., Byrne H., O'Kennedy R. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016;60(1):9–18. doi: 10.1042/EBC20150002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahato K., Chandra P. based miniaturized immunosensor for naked eye ALP detection based on digital image colourimetry integrated with smartphone. Biosens. Bioelectron. 2019;128:9–16. doi: 10.1016/j.bios.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Kondzior M., Grabowska I. Antibody-Electroactive Probe Conjugates Based Electrochemical Immunosensors. Sensors. 2020;20(7):2014. doi: 10.3390/s20072014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y., Wang M., Zhao F., Xu Z., Dong S. The direct electron transfer of glucose oxidase and glucose biosensor based on carbon nanotubes/chitosan matrix. Biosens. Bioelectron. 2005;21(6):984–988. doi: 10.1016/j.bios.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Asif M., Haitao W., Shuang D., Aziz A., Zhang G., Xiao F., Liu H. Metal oxide intercalated layered double hydroxide nanosphere: with enhanced electrocatalytic activity towards H2O2 for biological applications. Sens. Actuators, B. 2017;239:243–252. [Google Scholar]
- 54.Naudé W. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI & Soc. 2020;35(3):761–765. doi: 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest CT for typical coronavirus disease 2019 (COVID-19) pneumonia: relationship to negative RT-PCR testing. Radiology. 2020;296(2):E41–E45. doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purohit B., Vernekar P.R., Shetti N.P., Chandra P. Biosensor nanoengineering: Design, operation, and implementation for biomolecular analysis. Sensors Int. 2020;1:100040. doi: 10.1016/j.sintl.2020.100040. [DOI] [Google Scholar]