Abstract
SARS-CoV-2 is one of the greatest threats to global human health. Point-of-care diagnostic tools for SARS-CoV-2 could facilitate rapid therapeutic intervention and mitigate transmission. In this work, we report CRISPR-Cas13a cascade-based viral RNA (Cas13C) assay for label-free and isothermal determination of SARS-CoV-2 and its mutations in clinical samples. Cas13a/crRNA was utilized to directly recognize the target of SARS-CoV-2 RNA, and the recognition events sequentially initiate the transcription amplification to produce light-up RNA aptamers for output fluorescence signal. The recognition of viral RNA via Cas13a-guide RNA ensures a high specificity to distinguish SARS-CoV-2 from MERS-CoV and SARS-CoV, as well as viral mutations. A post transcription amplification strategy was triggered after CRISPR-Cas13a recognition contributes to an amplification cascade that achieves high sensitivity for detecting SARS-CoV-2 RNA, with a limit of detection of 0.216 fM. In addition, the Cas13C assay could be able to discriminate single-nucleotide mutation, which was proven with N501Y in SARS-Cov-2 variant. This method was validated by a 100% agreement with RT-qPCR results from 12 clinical throat swab specimens. The Cas13C assay has the potential to be used as a routine nucleic acid test of SARS-CoV-2 virus in resource-limited regions.
Nonstandard Abbreviations: COVID-19, coronavirus disease 2019; CRISPR, clustered regularly interspaced short palindromic repeats; Cas, CRISPR associated proteins; crRNA, CRISPR RNA; LOD, limit of detection; NASBA, nucleic acid sequence-based amplification; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; T4 PNK, T4 Polynucleotide Kinase
Keywords: COVID-19, SARS-CoV-2, CRISPR-Cas13, RNA aptamer, Nucleic acid tests
Graphical Abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) through coronavirus disease 2019 (COVID-19) has caused a high mortality and morbidity, significantly affecting both human health and the global economy. By July 13th 2021, more than ~184 million confirmed cases of COVID-19 infection have been reported, together with over 4 million of death[1]. Recently, confirmed daily cases are still high, such the reported 412,262 cases in a single day in India, which causes a huge burden on healthcare systems. Factors such as the long incubation period and the high percentage of asymptomatic carriers make contact tracing difficult, thus affecting outbreak prevention and control[2]. In addition, SARS-CoV-2 variants have emerged and caused higher transmission rates and stronger immune-escape responses[3], [4], [5]. Currently, RT-qPCR performed on nasopharyngeal and/or oropharyngeal swabs, remains the mainstream approach for SARS-CoV-2 diagnostic[6], [7], [8], [9]. While it is sensitive and accurate, it requires well-trained personnel, highly purified samples, as well as expensive equipment such as a thermocycler, which limits its application in the under-equipped laboratories. Therefore, it is necessary to establish sensitive and reliable point-of-care analytical tools to distinguish infected from healthy individuals.
CRISPR (clustered regularly interspaced short palindromic repeats) and Cas (CRISPR associated proteins) can recognize and degrade exogenous nucleic acid under the specific guidance of single stranded RNA, which are self-adaptive immune monitoring systems widespread in bacteria and archaea[10], [11], [12], [13]. The newly discovered collateral cleavage activity of Cas enzymes (encompassing Cas12a[13], [14], [15], Cas12b, Cas13a[16], [17], [18], Cas13b, and Cas14[19], [20], [21] after recognizing their specific targets turned CRISPR/Cas system into a perfect candidate for next generation diagnostic platforms[22], [23], [24], [25], [26]. Among them, CRISPR/Cas13a is the only RNA-targeted CRISPR effector, and possesses the capacity of transcendent signal amplification[12], [27], [28]. A Cas13a protein (RNA guided RNase) and a crRNA (“CRISPR RNA”: the target RNA recognizer) are the component of CRISPR-Cas13a system. Target RNA is specifically recognized by the crRNA and cut by Cas13a[29], [30]. The activated Cas13a collaterally cleaves the RNA probes provide, which fluorophore-quencher labeled RNA reporter for trans cleavage activity with a turnover efficiency of ~4854[31], thereby enabling the amplification of the target RNA recognition events. Zhang et al. developed a diagnostic tools based on Cas13, named SHERLOCK and SHERLOCKv2 to detect viruses such as dengue and Zika virus, with the LOD (limit of detection) as low as 50 fM and 2 aM, respectively, after the introduction of RPA (recombinase polymerase amplification)[16], [22]. The CRISPR-Cas13a system is a promising tool for SARS-CoV-2 virus diagnostic[32], [33], [34], [35]. The elimination of expensive probes (e.g., Quencher- and fluorophore- modified RNA probes) and costly instruments (such as thermocyclers), is expected to promote its use in the routine test of SARS-CoV-2 virus in resources-constrained regions.
Here, we report a CRISPR-Cas13a cascade-based viral RNA (Cas13C) assay, allowing label-free and isothermal detection of SARS-CoV-2 and its mutations in clinical samples. Cas13a/crRNA was utilized to directly recognize SARS-CoV-2 RNA and trigger the transcription amplification to produce light-up RNA aptamers for output fluorescence signal[36], [37], [38]. The Cas13C assay makes full use of the RNA-specific recognition ability of Cas13a/crRNA and integrates its trans cleavage activity in a high-efficient cascade amplification. The specific amplification of NASBA and the single base recognition ability of Cas13a guarantee the high specificity of the method, enabling the discrimination of highly homologous coronaviruses and allowing to discriminate single-nucleotide mutation in the SARS-Cov-2 variant, N501Y. Also, the Cas13C assay showed a low probability to get a false positive than CRISPR-Cas12a-Based methods[39], [40], [41], [42] because DNA cannot be used as template in NASBA. Lastly, the detection protocol does not involve labeling probes and expensive technical equipment (e.g., thermocyclers or real-time PCR platforms), thereby dramatically reducing the complexity of detection and cost of equipment. The capacity of Cas13C assay for detecting SARS-CoV-2 in throat swab samples was also evaluated, indicating its promise in clinical use.
2. Materials and methods
2.1. Materials and reagents
All of the DNA oligonucleotides listed in Table S1 (in the Supporting Information), 50 × TAE buffer, 6 × Loading buffer, and agarose were compounded from Sangon Biotech (Shanghai, China), and purified by HPLC or PAGE. LwaCas13a protein and its corresponding buffer (#32117) were procured from Tolo Biotech (Shanghai, China). Klenow exo- (10 U/μL, #EP0422), Phi29 DNA Polymerase (10 U/μL, #EP0094), RNAase H (5 U/μL, #EN0202), T4 Polynucleotide Kinase (T4 PNK, 10 U/μL, #EK0032), T7 RNA Polymerase (20 U/μL, #EP0111), RT buffer (#K1691) and qPCR SuperMix-UDG with ROX (Platinum® SYBR® Green) (#11744) were procured from Thermo Fisher Scientific (Waltham, USA). ProtoScriptⅡ Reverse Transcriptase (200 U/μL, #M0368S), dNTPs (#N0447S) and rNTPs mixes (#N0466S) were procured from NEB (Ipswich, USA). Hifair® Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR and 10000 × YeaRed Nucleic Acid Gel Stain were procured from YeaSen (Shanghai, China). DFHBI-1 T (5-difluoro-4-hydroxybenzylidene imidazolidinone) (#410–1 mg) was from Lucerna (Brooklyn, USA). E.Z.N.A. ®Viral RNA Kit (#R6874–01) was from OMEGA Biotek (Georgia, USA). Total RNA purified in H9N2, H7N9 and H1N1 from influenza viruses were kindly provided by Prof. Yi Shi group (Chinese Academy of Sciences, Beijing, China). SARS-CoV-2, MERS-CoV, SARS-CoV and SARS-CoV-2(N501Y) pseudovirus were procured from Jiman Biotechnology Co., Ltd. (Shanghai, China). All solutions were configured into RNase-free water (Corning, New York, USA).
2.2. In vitro transcription (IVT reaction) of crRNA, target RNA and Broccoli
IVT reaction was conducted in the mixture of 4 μL 10 × phi29 DNA polymerase buffer (pH 7.9 at 37 ℃), 4 μL L-crRNA or L-Broccoli (10 µM) as DNA templates (Table S1), 4 μL T7 promoter (10 µM) and 19.5 μL H2O, incubating at 90 ℃, 3 min firstly, followed closely by an incubation at 25 ℃, 15 min. Then, 1 μL dNTPs mixes and 0.5 μL phi29 DNA polymerase (10 U/μL) were added and incubated at 30 ℃, 30 min. Phi29 was subsequently inactivated by heating at 75 ℃, 10 min. Finally, 2 μL rNTP, 1 μL T7 RNA polymerase (20 U/μL) and 8 μL its corresponding buffer were put into the mixture and they were incubated at 37 ℃ for the transcription reaction.
2.3. Viral RNA detection
RNAs were either extracted from pseudovirus using commercialized test kit (E.Z.N.A.® Viral RNA Kit, OMEGA) according to manufacturer’s protocol or obtained in IVT reaction. The extracted viral RNAs were detected after the isothermal NASBA (nucleic acid sequence-based amplification) as described in the Supplementary Material. 4 μL RNA samples, 4 μL crRNA (2 μM), 8 μL RT buffer, 4 μL Pre-primer (2.4 μM), 4 μL templates (3 μM), 0.5 μL dNTPs (10 mM), 2 μL rNTPs (25 mM), 1 μL Klenow exo- (10 U/μL), 1 μL T4 PNK (10 U/μL), 0.4 μL Lwacas13a (10 μM), 4 μL DFHBI-1 T (100 μM) and 5.1 μL RNase-free H2O were blended- then added 2 μL T7 RNA Polymerase (20 U/μL) onto the lid of the reaction tube and incubated at 37 ℃ in metal bath for 30 min to obtained a double-stranded DNA template containing the T7 promoter. Then terminated the reaction through heating at 65 ℃ in a metal bath for 3 min. After that, the T7 RNA polymerase was centrifuged to the reaction solution with an instantaneous centrifuge and then mixed at 37 ℃, 2 h. Finally, termination of the detection reaction through heating at 65 ℃,10 min. Fluorescent signals were detected using the excitation and emission wavelength of 468 nm and 498–650 nm respectively with a Synergy H1 microplate reader (BioTek, USA).
2.4. Gel electrophoresis analysis
1 μL 6 × DNA loading buffer was added into each 5 μL reaction, analyzed by agar gel electrophoresis on a 3% concentration of 1 × TAE buffer using 150 V for 45 min, and then visualized with a Gel Doc XR+ system (Bio-Rad, USA).
2.5. RT-qPCR analysis
Isolated RNA was converted into cDNA via reverse transcription according to the manufactures instructions of commercialized test kit (Hifair® Ⅲ 1st Strand cDNA Synthesis SuperMix for qPCR kit). Gene-specific primers, which are listed in Table S1, were designed using Primer 5.0. Each qPCR reaction contained 1 μL of cDNA, 10 μL qPCR SuperMix-UDG with ROX (Platinum® SYBR® Green), 0.4 μL of each forward and reverse primers (10 µM) in 8.2 μL RNase-free H2O (final volume, 20 μL). All RT-qPCR analyses were done in 4 times. The RT-qPCR was carried with 40 cycles using the following conditions: 95 ℃, 10 s, 60 ℃, 10 s, and 72 ℃, 15 s. The qPCR reaction was performed by using QuantStudio® 3 Real-Time PCR System (Thermo Fisher Scientific).
3. Results
3.1. Working principle
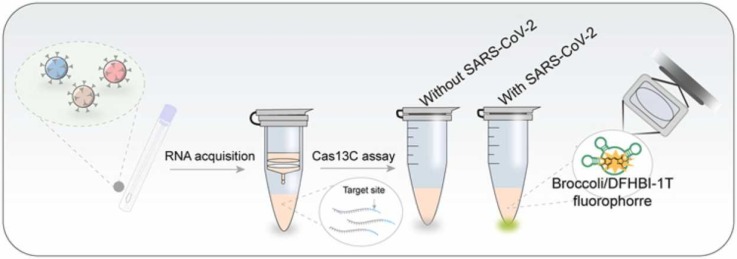
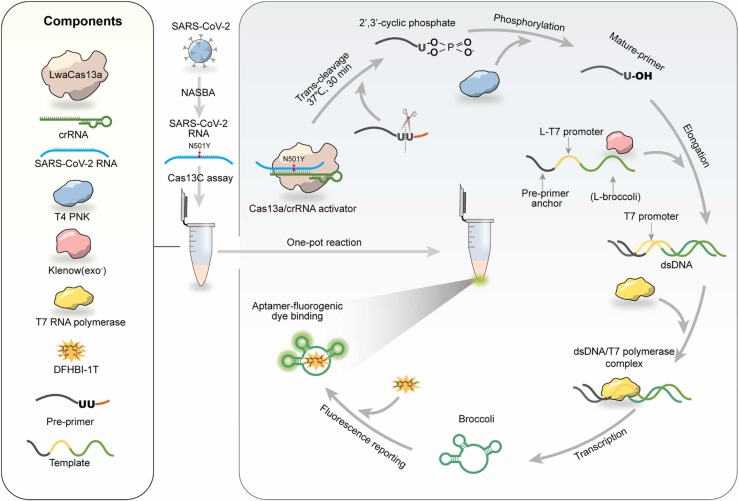
The SARS-CoV-2 RNA detection reaction scheme is illustrated in Fig. 1. The Cas13C assay is composed of Cas13a/crRNA system, a Pre-primer, a transcription template and the required enzymes (klenowexo-, T4 PNK and T7 RNA polymerase). We chose LwaCas13a because it is prone to cleave uracil ribonucleotide (rU) flanked phosphodiester linkage. Thus, we designed a pre-primer bore two “rU” as the substrate of Cas13a/crRNA. The pre-primer is consisted of transcription template anchor, two “rU” and 3' overhang. The 3' overhang can prevent the chain elongation when the target is absent. The transcription template is consisted of pre-primer anchor, a 5-nt space, a T7 promoter complementary sequence (L-T7 promoter) and an aptamer complementary sequence (L-broccoli).
Fig. 1.
Schematic diagram of the Cas13C assay for detecting SARS-CoV-2 RNA in collected samples.
Upon recognition of the SARS-CoV-2 RNA target, Cas13a/crRNA collaterally cleaves pre-primer from the “rU” cut site releasing the mature primer. Then, the remanent 5’-end of the pre-primer fragment which anchored in the transcription template can further initiate klenow (exo-)-mediated chain elongation after its 3’-end dephosphorylated by T4 PNK. Next, transcription amplification is initiated by T7 RNA polymerase, which generates abundant light-up RNA aptamers, broccoli. The fluorescence emitted from DFHBI-1 T-broccoli aptamer complex would indicate the presence of SARS-CoV-2 RNA. In this design, instead of using a labeled RNA probe, the transcription template, containing an aptamer sequence, is introduced into the system to generate a versatile reporting aptamer probe able to monitor the activation of CRISPR-Cas13a. The proposed Cas13C assay enables a high sensitivity (sub-femtomolar concentrations) and specificity for single-nucleotide resolution as a consequence of the dual amplification steps including NASBA (nucleic acid sequence-based amplification), transcription and direction mismatch recognition of Cas13a-crRNA.
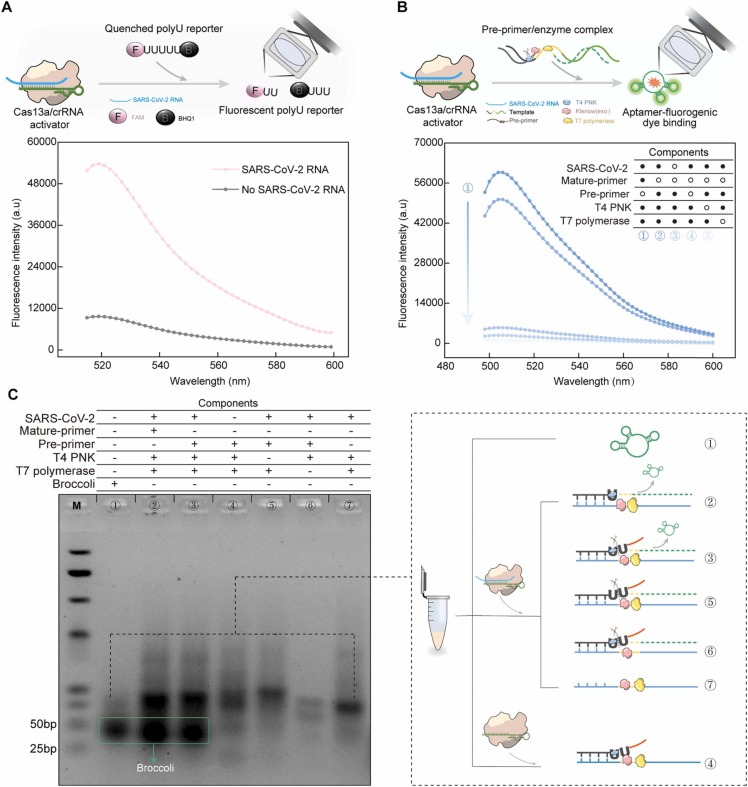
Firstly, we demonstrated the feasibility of using broccoli aptamers to quantificationally monitor SARS-CoV-2 RNA by measuring fluorescence intensity. The measured fluorescence intensities showed a linear relationship with the concentrations of broccoli aptamers in the range of 0 – 30.00 µM (Supplemental Fig. 1). We used a synthesized RNA sequence in the N gene of SRAS-CoV-2 serving as the target RNA sequence to test the Cas13C assay. The target RNA-activated trans-cleavage activity of Cas13a/crRNA was verified using FAM-BHQ1-modified RNA reporter, polyU reporter ( Fig. 2, A). Fluorescence analysis was further used to investigate each step of Cas13C assay. Lack of transcription amplification either in the absence of T7 polymerase, pre-primer or SRAS-CoV-2 gene showed a fluorescence close to 0 (Fig. 2, B). The presence of the mature-primer increased the fluorescence to be ~60,000 a.u, indicating that the mature-primer allowed to initiate the transcription amplification. In Cas13C assay, the transcription amplification was initiated using the mature primer which comes from the pre-primer, cleaved via target-activated Cas13a/crRNA. However, the 5’-end of the pre-primer fragment retains a 2’,3’-cyclic phosphate at its 3’-end, which might impede its extension to form a transcription template using DNA polymerase[22]. We found that the addition of T4 PNK to convert the 5’-end of the pre-primer fragment into a mature primer via a dephosphorylation allowed to initiate the DNA extension reaction and transcription amplification (Fig. 2, B).
Fig. 2.
Validation of the work principle of the Cas13C assay. (A) Fluorescence response of Cas13a/crRNA induced-cleavage of FAM-BHQ1-modified RNA reporter with or without SARS-CoV-2 RNA. (B) Fluorescence analysis for each reaction in the Cas13C assay. The concentrations of mature-primer, pre-primer, Cas13a protein, T4 PNK, T7 polymerase and SARS-CoV-2 RNA were 240 nM, 240 nM, 100 nM, 0.5 U/μL, 0.56 U/μL, and 10 nM, respectively. (C) Electrophoresis analysis of each reaction product in the Cas13C assay, M is the nucleic acid marker.
Gel electrophoresis was used to further validate each step of Cas13C assay (Fig. 2, C). The analysis of the gel showed a new band close to the band of the transcription product, which corresponds to broccoli (line 1) in addition to the SARS-CoV-2 RNA (lane 3). No broccoli product was found in the group with no target RNA (lane 4). No addition of T4 PNK, T7 polymerase and pre-primer (lane 5, 6, 7) also did not release any broccoli.
3.2. Optimization of the Cas13C assay
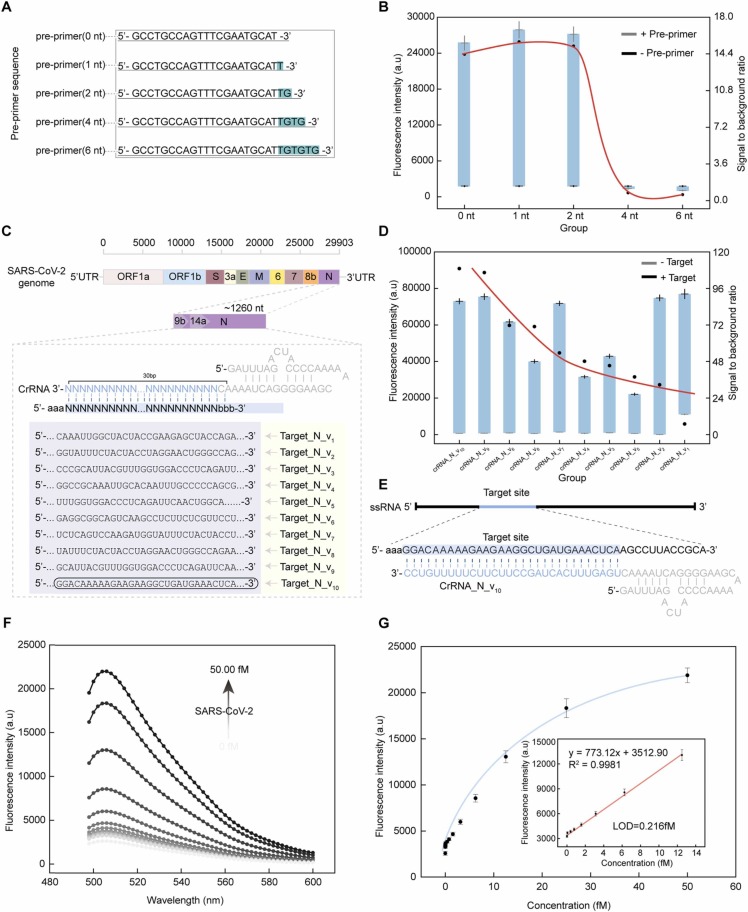
The pre-primer was designed with two rU to serve as the substrate of the activated crRNA/ Cas13a. It is essential to test whether the 3’ overhang of the pre-primer can effectively prevent self-elongation in the absence of targets. A series of pre-primers with an overhang of 0, 1, 2, 4 and 6 nucleotides (nt) were designed ( Fig. 3, A). When the extension of the overhang nucleotides was 4 nt, the fluorescence after transcription amplification in the presence of Pre-primer was very close to the fluorescence in the absence of pre-primer (Fig. 3, B), indicating that the addition these bases can hinder the elongation reaction and the transcription amplification, and only the cleavage pre-primer can initiate the transcription amplification process.
Fig. 3.
Quantification performance of the Cas13C assay for SARS-CoV-2 determination. (A) The design of Pre-primer sequences with an overhang of 0, 1, 2, 4 and 6 nucleotides (nt). (B) Fluorescence intensity analysis of the Cas13C assay using Pre-primer designed in (A). (C) Genomes of SARS-CoV-2 and sequences targeted by Cas13a/crRNA. (D) Fluorescence response of the polyU cleavage reaction using crRNAs designed in (C). (E) The optimized Cas13a/crRNA for SARS-CoV-2 RNA detection. The target site is highlighted in blue. (F) Typical fluorescence spectrum of the Cas13C assay upon the addition of different concentrations of SARS-CoV-2 RNA (0.00 fM, 0.006 fM, 0.01 fM,0.02 fM, 0.05 fM, 0.39 fM, 0.78 fM, 1.56 fM, 3.12 fM, 6.25 fM, 12.50 fM, 25.00 fM, and 50.00 fM). (G) The relationship between the concentration of SARS-CoV-2 pseudovirus and the fluorescence intensity of the Cas13C assay; Inset: Linear relationship between the concentration of SARS-CoV-2 pseudovirus and fluorescence response of the Cas13C assay. Quantitative data were shown as means ± SD (n = 3).
In addition, an optimal crRNA candidate that can stably bind both the Cas13a and the viral RNA is critical for the Cas13C assay. All crRNAs were designed targeting the conserved and specific domain (~30 nt) of the SARS-CoV-2 (N gene) by searching the NCBI gene database (Fig. 3, C). The sequences of the crRNAs are listed in Fig. 3, C and Table S1. To intuitively reflect the trans cleavage activity of the LwaCas13a-equiped different crRNAs, PolyU was administered as substrates with the crRNA-Cas13a complex. We defined a signal-to-background ratio as the ratio of the fluorescence of the polyU cleavage reaction in the presence of target to that in the absence of target. These crRNA candidates showed different response to SARS-CoV-2 RNA. For examples, crRNA-4, 5 and 8 generated a relatively low fluorescence in the presence of the target (Fig. 3, D). crRNA-1 and 2 showed a higher fluorescence response intensity to the reaction, but they also had a high signal-to-noise ratio due to their relatively high background fluorescence (Fig. 3, D). The failure of these crRNAs for efficient recognition of SARS-CoV-2 could be attributed to the complex secondary structure of SARS-CoV-2 genes as well as the sequence bias of Cas13a activation. Among the 10 crRNA candidates, the crRNA-10 bound to Cas13a achieved the highest signal-to-background ratio, therefore, we chose it for the subsequent studies. In addition, we also examined whether crRNA/Cas13a ratio influenced the cleavage capacity of the crRNA/Cas13a activator. The result showed that the use of the crRNA with a ratio of crRNA to Cas13a of 1:1 could fully cleave the polyU reporter (Supplemental Fig. 2, A), and the signal-to-background-ratio was the highest when the ratio was 2:1. An optimal ratio of Pre-primer to template as 0.8:1 was also determined based on the measurement of the signal-to-background-ratio (Supplemental Fig. 2, B).
3.3. Quantification performance
To test the quantification performance of the Cas13C assay, we constructed a lentivirus containing the S, E and N genes from SARS-CoV-2 to serve as SARS-CoV-2 pseudovirus (S-E-N, Supplemental Fig. 3). We first amplified the RNA sequences extracted from SARS-CoV-2 pseudoviruses by NASBA (Supplemental Fig. 4 and Fig. 5). At the optimized conditions, we validated the analytical performance of the Cas13C assay by utilizing a series of concentrations (0.0 fM ~ 50.00 fM) of SARS-CoV-2 pseudoviruses. The fluorescence intensity of the Cas13C assay was gradually enhanced with the increasing copy number of SARS-CoV-2 pseudovirus as showed in Fig. 3, F. Moreover, the measured fluorescence intensities showed a linear relationship with SARS-CoV-2 pseudoviruses in the range of 0.006 – 12.50 fM (y = 773.12x + 3512.90; R 2=0.9981) (Fig. 3, G), where y is the fluorescence intensity of the Cas13C assay and x is the concentration of SARS-CoV-2 RNA (fM). The limit of detection (LOD) for the Cas13C assay was 0.216 fM per reaction for detecting SARS-CoV-2 pseudoviruses (calculated at 3 times the standard deviation of the background), and the sensitivity is comparable to SHERLOCK (with a LOD of 50 fM).
3.4. Specificity tests
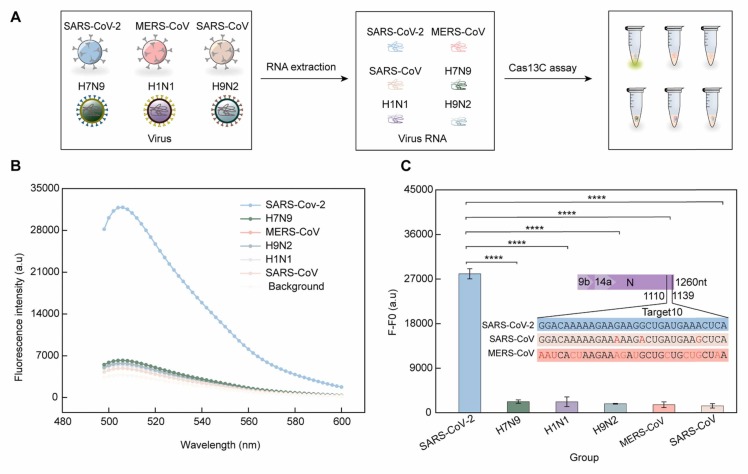
We next chose two coronavirus species (SARS-CoV and MERS-CoV) and three influenza viruses (H7N9, H1N1 and H9N2), to evaluate the specificity of the proposed methods for SARS-CoV-2 RNA detection. Fig. 4, A shows the experimental workflow of the specificity tests. As showed in Fig. 4, B and Fig. 4, C, despite highly similarity of the sequences of the N gene in SARS-CoV-2, SARS-CoV and MERS-CoV (Fig. 4, C), only SARS-CoV-2 RNA produced high fluorescence. In contrast, the response to SARS-CoV, MERS-CoV, H7N9, H1N1 and H9N2 was close to the background (with no addition of viruses). This result indicates that the Cas13C assay allowed to specifically recognize SARS-CoV-2.
Fig. 4.
Specificity tests of the Cas13C assay for detecting SARS-CoV-2 RNA. (A) A schematic of the Cas13C assay workflow from detecting pseudovirus samples. (B-C) Fluorescence response of the Cas13a assay towards different viral RNAs (SARS-CoV-2, H7N9, H1N1, MERS-CoV, H9N2, and SARS-CoV); (Inset) The sequences of the N gene of SARS-CoV-2, SARS-CoV and MERS-CoV. The different sections in SARS-CoV and MERS-CoV compared to SARS-CoV-2 are indicated in red. F is the fluorescence signal of the Cas13C assay upon the addition of each viral RNA; F0 is the fluorescence signal without any viral RNAs. The concentrations of each pseudoviruses were 100 nM. Quantitative data were shown as means ± SD, ****p ≤ 0.0001 was deemed to have statistically significant.
3.5. Mutant strain-discrimination
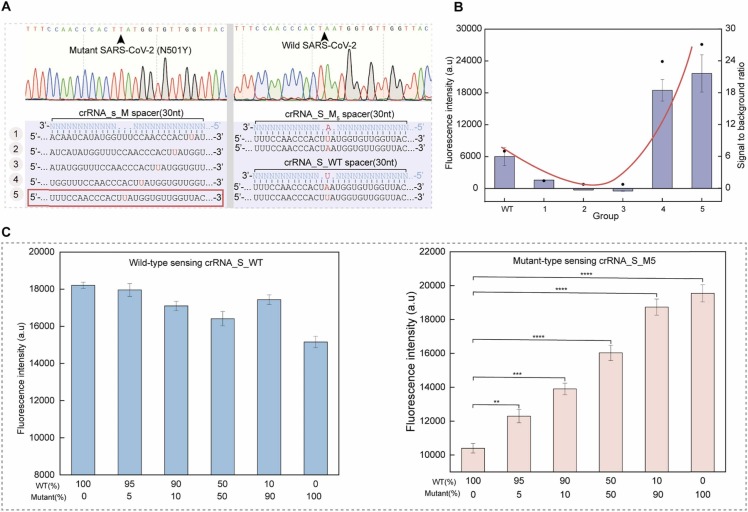
Spike glycoprotein mutations, such as the N501Y mutation, are present in many SARS-CoV-2 variants. These mutations can lead to the change of the RBD domain in spike protein, which is associated with strengthening the interaction with the ACE2 receptor, possibly contributing to increased transmissibility[43], [44]. It has been reported that using the only ancestral crRNA can be directly discriminate the single-nucleotide variations[16]. Inspired by that, we engineered a series of crRNAs with single mutations at various positions ( Fig. 5, A), to test their capability of distinguishing the N501Y mutant from the wild type. As we can see in Fig. 5B, the high fluorescent response of the polyU cleavage reaction was observed when using crRNA_S4/5. The best discrimination rate was found with crRNA_S5, so we chose it for the subsequent experiments. Then, we sought to determine if the Cas13C assay had the ability to discriminate N501Y mutations in the presence of varying proportions of wild type RNAs. We found that the Cas13C assay was able to discriminate the N501Y-mutated variant when it was 5% of the variants (Fig. 5, C). These results demonstrate that the Cas13C assay can be used to detect SARS-CoV-2 variants owes with single-base discrimination capacity.
Fig. 5.
Identify N501Y mutations of SARS-CoV-2 variants. (A) Design of crRNA_S (1−5) for identifying N501Y mutations. (B) Fluorescence response of the crRNA_S_Mn-assembled Cas13a assay to polyU. (C) The Cas13C assay can detect the mutant N501Y in wild SARA-CoV-2 samples. Quantitative data were shown as means ± SD, * * p ≤ 0.01; * ** p ≤ 0.001; * ** * p ≤ 0.0001 was deemed to have statistically significant.
3.6. SARS-CoV-2 detection in clinical throat swab samples
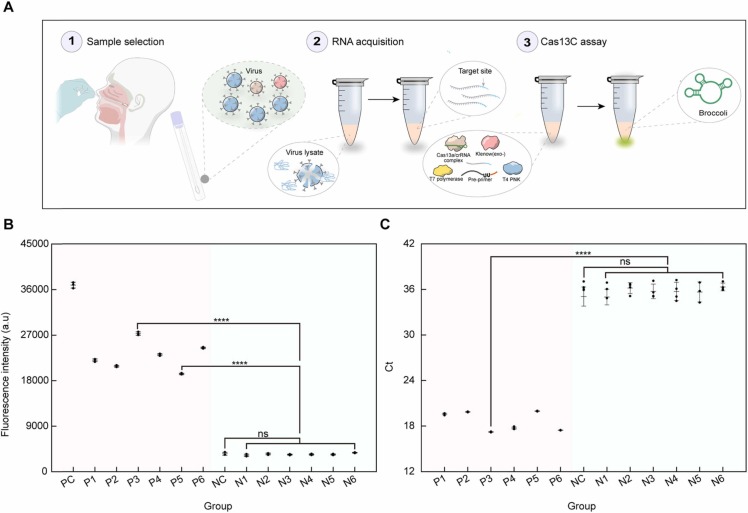
Oropharyngeal swab is the most commonly used sampling method for the diagnose of COVID-19 in the clinic. In order to test the feasibility of the Cas13C assay to detect SARS-CoV-2 from clinical samples, throat swab samples of COVID-19 patients were collected from the West China Hospital (Ethical Approval, 2020(100)). The extracted RNA was divided into two parts, one for analysis by the Cas13a assay and the other for RT-qPCR. The extraction of SARS-CoV-2 and RT-qPCR method were performed following the guidelines from WHO[6]. The overall workflow implemented for the detection of throat swab samples by the Cas13C assay is depicted in Fig. 6, A. Fig. 6, B illustrates that the samples from the six confirmed cases resulted in a clear increase in fluorescence compared with the background, while no significant statistical difference with the background were found (P > 0.05) between the signals of the non-infected cases. The RT-qPCR results show an increased abundance of the N gene in patients in comparison to the non-infected cases (Fig. 6, C). Therefore, the results from the Cas13C assay were in good accordance with the RT-qPCR results, indicating the potential of the Cas13C assay to be used as a complementary tool for SARS-CoV-2 diagnostic.
Fig. 6.
SARS-CoV-2 detection in clinical throat swab samples. (A) Extraction and detection protocol of SARS-CoV-2 RNA from patients or non-infected persons. (B) Fluorescence response of the Cas13C assay towards SARS-CoV-2 RNA extracted from patients or non-infected persons. PC, P, NC and N indicates positive control, patient, negative control and non-infected persons, respectively. (C) Signal response of the RT-qPCR towards SARS-CoV-2 RNA extracted from patients or non-infected persons. Quantitative data were showed as means ± SD, * ** * p ≤ 0.0001 was deemed to have statistically significant; ns considered no significant (p > 0.05).
4. Conclusions
In summary, we constructed a label-free and isothermal method based on CRISPR-Cas13a for SARS-CoV-2 diagnosis with the capacity to discriminate between variant strains. The assay, termed Cas13C, uses Cas13a/crRNA to directly recognize viral RNAs. Instead of using labeled RNA probes, the target RNA can be quantified using RNA aptamers generated after transcription amplification. The Cas13C assay can provide a linear response between 0.006 fM to 12.50 fM and achieves a LOD of 0.216 fM, which is comparable to that of SHERLOCK (50 fM)[16]. Importantly, highly homologous coronaviruses can be discriminated using the Cas13C assay. The strategy was demonstrated to detect SARS-CoV-2 variants with single-base mutations. We showed that SARS-CoV-2 can be detected from throat swabs, which can indicate the potential use of this technique in clinical application. Moreover, the Cas13C assay is a homogeneous and isothermal method which does not need a labeled nucleic acid probe and expensive temperature-control equipment, facilitating its use in resource-limited regions.
CRediT authorship contribution statement
Yuxi Wang: Conceptualization, Funding acquisition, Writing – review & editing. Ting Xue: Conceptualization, Investigation, Formal analysis, Methodology, Writing – original draft. Minjin Wang: Investigation, Methodology, Resources, Writing – review & editing. Rodrigo Ledesma-Amaro: Supervision, Writing – review & editing. Ying Lu: Methodology, Formal analysis. Xinyue Hu: Methodology, Formal analysis. Ting Zhang: Investigation, Methodology, Validation, Formal analysis. Ming Yang: Methodology, Formal analysis. Yalun Li: Resources, Supervision, Methodology. Jin Xiang: Resources, Formal analysis. Ruijie Deng: Conceptualization, Investigation, Supervision, Funding acquisition, Writing – review & editing. Binwu Ying: Conceptualization, Investigation, Supervision, Writing – review & editing. Weimin Li: Conceptualization, Methodology, Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Sichuan Science and Technology Program [grant numbers 2021YFS0403, 2019YFS0003, 2020YFS0572, and 2020YFS0573], Major Science and Technology Innovation Project of Chengdu City [grant numbers 2020-YF08-00080-GX], National Major Scientific and Technological Special Project for ‘Significant New Drugs Development’ [grant numbers 2018ZX09201018–021, 2018ZX09201018–020], Special Funds for COVID-19 Prevention of West China Hospital [grant numbers HX-2019-nCoV-003].
Biography
Ruijie Deng Professor. B.Sc. from Central South University in 2012, and Ph.D. from Tsinghua University in 2017. Professor at Sichuan University from July 2017. Current research interests include single cell and single molecule analysis, DNA nanotechnology, fluorescence imaging, isothermal amplification development and biosensors.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.131765.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Coronavirus disease 2019 (COVID-19) situation report −73, (2021). https://covid19.who.int/.
- 2.Li Q., Guan X.H., Wu P., Wang X.Y., Zhou L., Tong Y.Q., Ren R.Q., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X.S., Xiang N.J., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W.X., Chen C.D., Jin L.M., Yang R., Wang Q., Zhou S.H., Wang R., Liu H., Luo Y.B., Liu Y., Shao G., Li H., Tao Z.F., Yang Y., Deng Z.Q., Liu B.X., Ma Z.T., Zhang Y.P., Shi G.Q., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z.J. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Zhang Y., Kang J.-Y., Chen S., He Y., Han B., Liu M.-F., Lu L., Li L., Yi Z., Chen L. Potential transmission chains of variant B.1.1.7 and co-mutations of SARS-CoV-2. Cell Disco. 2021;7 doi: 10.1038/s41421-021-00282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L.J. Abu-Raddad, H. Chemaitelly, H.H. Ayoub, H.M. Yassine, F.M. Benslimane, H.A. al Khatib, P. Tang, M.R. Hasan, P. Coyle, S. AlMukdad, Z. al Kanaani, E. al Kuwari, A. Jeremijenko, A.H. Kaleeckal, A.N. Latif, R.M. Shaik, H.F. Abdul Rahim, G.K. Nasrallah, M.G. al Kuwari, A.A. Butt, H.E. al Romaihi, M.H. Al-Thani, A. al Khal, R. Bertollini, Severity, criticality, and fatality of the SARS-CoV-2 Beta variant, MedRxiv. (2021) 2021.08.02.21261465. 10.1101/2021.08.02.21261465. [DOI]
- 5.Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., Kruger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M.S., Schulz S., Jack H.M., Jahrsdorfer B., Schrezenmeier H., Muller M., Kleger A., Munch J., Pohlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, (2020). https://apps.who.int/iris/handle/10665/331329.
- 7.Feng W., Newbigging A.M., Le C., Pang B., Peng H.Y., Cao Y.R., Wu J.J., Abbas G., Song J., Wang D.B., Cui M.M., Tao J., Tyrrell D.L., Zhang X.E., Zhang H.Q., Le X.C. Molecular diagnosis of COVID-19: challenges and research needs. Anal. Chem. 2020;92:10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 8.Yan S., Ahmad K.Z., Warden A.R., Ke Y., Maboyi N., Zhi X., Ding X. One-pot pre-coated interface proximity extension assay for ultrasensitive co-detection of anti-SARS-CoV-2 antibodies and viral RNA. Biosens. Bioelectron. 2021;193 doi: 10.1016/j.bios.2021.113535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S., Huang S., Ke Y., Chen H., Dang J., Huang C., Liu W., Cui D., Wang J., Zhi X., Ding X., HiPAD A. Integrated with rGO/MWCNTs Nano-Circuit Heater for Visual Point-of-Care Testing of SARS-CoV-2. Adv. Funct. Mater. 2021;31 doi: 10.1002/adfm.202100801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 11.Marraffini L.A., Sontheimer E.J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., Severinov K., Regev A., Lander E.S., v Koonin E., Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:557–565. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J.S., Ma E.B., Harrington L.B., da Costa M., Tian X.R., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding X., Yin K., Li Z.Y., v Lalla R., Ballesteros E., Sfeir M.M., Liu C.C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020;11:4711. doi: 10.1038/s41467-020-18575-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang B., Xu J.Y., Liu Y.M., Peng H.Y., Feng W., Cao Y.R., Wu J.J., Xiao H.Y., Pabbaraju K., Tipples G., Joyce M.A., Saffran H.A., Tyrrell D.L., Zhang H.Q., Le X.C. Isothermal amplification and ambient visualization in a single tube for the detection of SARS-CoV-2 using loop-mediated amplification and CRISPR technology. Anal. Chem. 2020;92:16204–16212. doi: 10.1021/acs.analchem.0c04047. [DOI] [PubMed] [Google Scholar]
- 16.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., v Koonin E., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myhrvold C., Freije C.A., Gootenberg J.S., Abudayyeh O.O., Metsky H.C., Durbin A.F., Kellner M.J., Tan A.L., Paul L.M., Parham L.A., Garcia K.F., Barnes K.G., Chak B., Mondini A., Nogueira M.L., Isern S., Michael S.F., Lorenzana I., Yozwiak N.L., MacInnis B.L., Bosch I., Gehrke L., Zhang F., Sabeti P.C. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruch R., Baaske J., Chatelle C., Meirich M., Madlener S., Weber W., Dincer C., Urban G.A. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv. Mater. 2019;31 doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 19.Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song F.G., Wei Y.D., Wang P., Ge X.L., Li C.Y., Wang A.M., Yang Z.Q., Wan Y., Li J.H. Combining tag-specific primer extension and magneto-DNA system for Cas14a-based universal bacterial diagnostic platform. Biosens. Bioelectron. 2021;185 doi: 10.1016/j.bios.2021.113262. [DOI] [PubMed] [Google Scholar]
- 21.Ge X., Meng T., Tan X., Wei Y., Tao Z., Yang Z., Song F., Wang P., Wan Y. Cas14a1-mediated nucleic acid detectifon platform for pathogens. Biosens. Bioelectron. 2021;189 doi: 10.1016/j.bios.2021.113350. [DOI] [PubMed] [Google Scholar]
- 22.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding X., Yin K., Li Z.Y., Sfeir M.M., Liu C.C. Sensitive quantitative detection of SARS-CoV-2 in clinical samples using digital warm-start CRISPR assay. Biosens. Bioelectron. 2021;184 doi: 10.1016/j.bios.2021.113218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yue H., Shu B., Tian T., Xiong E., Huang M., Zhu D., Sun J., Liu Q., Wang S., Li Y., Zhou X. Droplet Cas12a assay enables DNA quantification from unamplified samples at the single-molecule level. Nano Lett. 2021;21:4643–4653. doi: 10.1021/acs.nanolett.1c00715. [DOI] [PubMed] [Google Scholar]
- 25.Hatoum-Aslan A. CRISPR methods for nucleic acid detection herald the future of molecular diagnostics. Clin. Chem. 2018;64:1681–1683. doi: 10.1373/clinchem.2018.295485. [DOI] [PubMed] [Google Scholar]
- 26.Nouri R., Tang Z., Dong M., Liu T., Kshirsagar A., Guan W. CRISPR-based detection of SARS-CoV-2: a review from sample to result. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tambe A., East-Seletsky A., Knott G.J., Doudna J.A., O’Connell M.R. RNA binding and HEPN-nuclease activation are decoupled in CRISPR-Cas13a. Cell Rep. 2018;24:1025–1036. doi: 10.1016/j.celrep.2018.06.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian T., Shu B.W., Jiang Y.Z., Ye M.M., Liu L., Guo Z.H., Han Z.P., Wang Z., Zhou X.M. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. Acs Nano. 2021;15:1167–1178. doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- 29.Liu L., Li X.Y., Ma J., Li Z.Q., You L.L., Wang J.Y., Wang M., Zhang X.Z., Wang Y.L. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 30.East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shan Y.Y., Zhou X.M., Huang R., Xing D. High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity. Anal. Chem. 2019;91:5278–5285. doi: 10.1021/acs.analchem.9b00073. [DOI] [PubMed] [Google Scholar]
- 32.Fozouni P., Son S., Díaz de León Derby M., Knott G.J., Gray C.N., D’Ambrosio M. v, Zhao C., Switz N.A., Kumar G.R., Stephens S.I., Boehm D., Tsou C.L., Shu J., Bhuiya A., Armstrong M., Harris A.R., Chen P.Y., Osterloh J.M., Meyer-Franke A., Joehnk B., Walcott K., Sil A., Langelier C., Pollard K.S., Crawford E.D., Puschnik A.S., Phelps M., Kistler A., DeRisi J.L., Doudna J.A., Fletcher D.A., Ott M. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184 doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arizti-Sanz J., Freije C.A., Stanton A.C., Petros B.A., Boehm C.K., Siddiqui S., Shaw B.M., Adams G., Kosoko-Thoroddsen T.S.F., Kemball M.E., Uwanibe J.N., Ajogbasile F. v, Eromon P.E., Gross R., Wronka L., Caviness K., Hensley L.E., Bergman N.H., MacInnis B.L., Happi C.T., Lemieux J.E., Sabeti P.C., Myhrvold C. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020;4 doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- 35.Ke Y., Huang S., Ghalandari B., Li S., Warden A.R., Dang J., Kang L., Zhang Y., Wang Y., Sun Y., Wang J., Cui D., Zhi X., Ding X. Hairpin-Spacer crRNA-enhanced CRISPR/Cas13a system promotes the specificity of single nucleotide polymorphism (SNP) identification. Adv. Sci. 2021;8 doi: 10.1002/advs.202003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paige J.S., Nguyen-Duc T., Song W.J., Jaffrey S.R. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335:1194. doi: 10.1126/science.1218298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paige J.S., Wu K.Y., Jaffrey S.R. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ying Z.M., Wu Z., Tu B., Tan W.H., Jiang J.H. Genetically encoded fluorescent RNA sensor for ratiometric imaging of microRNA in living tumor cells. J. Am. Chem. Soc. 2017;139:9779–9782. doi: 10.1021/jacs.7b04527. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y., Chen M., Liu C., Chen J., Luo X., Xue Y., Liang Q., Zhou L., Tao Y., Li M., Wang D., Zhou J., Wang J. Sensitive and rapid on-site detection of SARS-CoV-2 using a gold nanoparticle-based high-throughput platform coupled with CRISPR/Cas12-assisted RT-LAMP. Sens. Actuators, B: Chem. 2021;345 doi: 10.1016/j.snb.2021.130411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo X., Xue Y., Ju E., Tao Y., Li M., Zhou L., Yang C., Zhou J., Wang J. Digital CRISPR/Cas12b-based platform enabled absolute quantification of viral RNA. Anal. Chim. Acta. 2022;1192 doi: 10.1016/j.aca.2021.339336. [DOI] [PubMed] [Google Scholar]
- 41.Marqués M.C., Ruiz R., Montagud-Martínez R., Márquez-Costa R., Albert S., Domingo-Calap P., Rodrigo G. CRISPR-Cas12a-based detection of SARS-CoV-2 harboring the E484K mutation. ACS Synth. Biol. 2021;10 doi: 10.1021/acssynbio.1c00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu X., Chan C., Springs S.L., Lee Y.H., Lu T.K., Yu H. A warm-start digital CRISPR/Cas-based method for the quantitative detection of nucleic acids. Anal. Chim. Acta. 2022;1196 doi: 10.1016/j.aca.2022.339494. [DOI] [PubMed] [Google Scholar]
- 43.F. Tian, B. Tong, L. Sun, S. Shi, B. Zheng, Z. Wang, X. Dong, P. Zheng, Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2, BioRxiv. (2021) 2021.02.14.431117. 10.1101/2021.02.14.431117. [DOI]
- 44.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao N.Y., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenco J., Alcantara L.C.J., Pond S.K.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material