Abstract
The rodent visual thalamus has served as a powerful model to elucidate the cellular and molecular mechanisms that underlie sensory circuit formation and function. Despite significant advances in our understanding of the role of axon-target interactions and neural activity in orchestrating circuit formation in visual thalamus, the role of non-neuronal cells, such as astrocytes, is less clear. In fact, we know little about the transcriptional identity and development of astrocytes in mouse visual thalamus. To address this gap in knowledge, we studied the expression of canonical astrocyte molecules in visual thalamus using immunostaining, in situ hybridization, and reporter lines. While our data suggests some level of heterogeneity of astrocytes in different nuclei of the visual thalamus, the majority of thalamic astrocytes appeared to be labelled in Aldh1l1-EGFP mice. This led us to use this transgenic line to characterize the neonatal and postnatal development of these cells in visual thalamus. Our data show that not only have the entire cohort of astrocytes migrated into visual thalamus by eye-opening but they also have acquired their adult-like morphology, even while retinogeniculate synapses are still maturing. Furthermore, ultrastructural, immunohistochemical, and functional approaches revealed that by eye-opening, thalamic astrocytes ensheath retinogeniculate synapses and are capable of efficient uptake of glutamate. Taken together, our results reveal that the morphological, anatomical, and functional development of astrocytes in visual thalamus occurs prior to eye-opening and the emergence of experience-dependent visual activity.
Keywords: Astrocytes, thalamus, retinogeniculate synapse, dorsal lateral geniculate nucleus, ventral lateral geniculate nucleus
Graphical Abstract
Graphical Abstract Text. Morphological and functional development of astrocytes in the visual thalamus In the present study, we show that by eye-opening, astrocytes not only possess adult-like morphology but also completely enwrap retinogeniculate synapses and efficiently uptake extracellular glutamate
1. INTRODUCTION
Retinal ganglion cells (RGCs) innervate over forty regions in the rodent brain and more than ten of those are within the thalamus (Lawrence & Studholme, 2014; Martersteck et al., 2017; Monavarfeshani et al., 2017). Of those ten, three adjacent nuclei at the border of dorsal and ventral thalamus receive the densest retinal projections: the dorsal lateral geniculate nucleus (dLGN), ventral lateral geniculate nucleus (vLGN), and intergeniculate leaflet (IGL) (Monavarfeshani et al., 2017). The dLGN, which receives, processes, and transmits image-forming visual information, has received the most attention and has served as a powerful model for dissecting mechanisms of sensory neural circuit assembly and refinement (Guido, 2018). The assembly of synapses between presynaptic retinal axons and thalamocortical (TC) relay cells (termed retinogeniculate [RG] synapses) begin to form during pre- and neonatal periods in nocturnal rodents (Godement et al., 1984; Hammer et al., 2014; R. Singh et al., 2012). Retinal axons also form synapses onto local GABAergic interneurons, however, these cells are not present in dLGN at birth and migrate in from thalamic and tectal sources during the first week of postnatal development (Golding et al., 2014; Jager et al., 2016; Su et al., 2020). Coinciding with the arrival of migrating interneurons during postnatal dLGN development, retinal projections are refined based on both retinotopy and eye-of-origin (Chen & Regehr, 2000; Hong & Chen, 2011; Huberman et al., 2008; Jaubert-Miazza et al., 2005). Beginning shortly before eye-opening (~P12-P14), synaptic elimination, synaptic strengthening, and bouton clustering each contribute to the eventual emergence of adult-like RG synapses (>P25), in terms of both anatomy and function (Bickford et al., 2010; Chen & Regehr, 2000; Hong & Chen, 2011; Hooks & Chen, 2006; Monavarfeshani et al., 2018).
The processes of synaptic targeting, assembly, maturation, and refinement at RG synapses is orchestrated by interactions between cell surface molecules on developing retinal axons and TC relay cells. For example, members of Eph family of receptor tyrosine kinase and cell surface-bound ephrins expressed by retinal axons and dLGN TC relay cells contribute to the mapping and refinement of retinal axons into topographic maps and eye-specific domains in dLGN (Cang et al., 2008; Huberman et al., 2008; Pfeiffenberger et al., 2005, 2006). In addition to such interactions between retinal axons and thalamic target neurons, developmentally relevant signals from adjacent astrocytes likely contribute to synapse assembly, maturation, and function (Chung et al., 2015; Eroglu & Barres, 2010). While such roles for astrocytes are not entirely clear for the initial formation of synapses in visual thalamus, they have been identified in other retinorecipient nucleus such as the superior colliculus (Christopherson et al., 2005; Kucukdereli et al., 2011; Ullian et al., 2001). However, astrocytes have been reported to contribute to developmental refinement of retinal axons and RG synapses in dLGN (Chung et al., 2013; Stevens et al., 2007). In addition, neonatal astrocytes induce the long distance migration of interneurons into the developing visual thalamus by generating and releasing fibroblast growth factor 15 (FGF15) (Su et al., 2020). Together with the well-described encapsulation of RG synapses by astrocytic processes, these studies suggest that astrocytes play significant role in the development and function of circuits in visual thalamus (Hammer et al., 2014; Litvina & Chen, 2017; Szentágothai, 1963). However, at present the transcriptional, morphological, and functional development of astrocytes in this brain region has not been well addressed.
To address these gaps, we used immunohistochemistry (IHC), in situ hybridization (ISH), and genetic reporter lines to study astrocyte development in dLGN, IGL, and vLGN. Results from these studies and our previous studies (Su et al. 2020) suggest that based on gene and protein expression, astrocytes represent a heterogenous group of cells in mouse visual thalamus. However, here we show that tools that label Fgfr3, Gja1, and Aldh1l1 expression all appear to label most, if not all, astrocytes in dLGN and vLGN despite this heterogeneity. To characterize the development of astrocyte distribution and morphology in visual thalamus, we took advantage of two transgenic reporter lines (Aldh1l1-EGFP and mGfap-Cre::ROSA-Stop-tdT). Surprisingly, even as RG synapses are still maturing, thalamic astrocytes appear adult-like in their morphology. Moreover, immunohistochemical, ultrastructural, and functional studies revealed that by eye-opening, astrocytes ensheath RG synapses and are adult-like in their ability to uptake glutamate from the synaptic cleft. Overall, these studies reveal that astrocytes achieve adult-like morphology and function at RG synapses early in thalamic development, by eye-opening and prior to visual experience.
2. MATERIALS AND METHODS
2.1. Animals
C57BL/6J mice, ROSA-Stop-tdT mice (JAX#:007909, RRID:IMSR_JAX:007909, genetic background: C57BL/6J mice) and Cx3cr1-GFP mice (JAX#:005582, RRID:IMSR_JAX:005582, genetic background: C57BL/6J mice) were obtained from The Jackson Laboratory. Aldh1l1-EGFP (RRID:MMRRC_011015-UCD, genetic background: FVB/N mice) and mGFAP-Cre (JAX#:024098,RRID:IMSR_JAX:024098, genetic background: (BALB/c x C57BL/6NHsd)F1) were obtained from S. Robel (Virginia Tech). The following primers were used for genotyping: tdT_forward: 5′-ACC TGG TGG AGT TCA AGA CCA TCT-3’; tdT_reverse: 5′-TTG ATG ACG GCC ATG TTG TTG TCC-3′; GFP_forward: 5′-AAG TTC ATC TGC ACC ACC G-3’; GFP_reverse: 5′-TCC TTG AAG AAG ATG GTG CG-3’; Cre_forward: 5′-CGT ACT GAC GGT GGG AGA AT-3′; and Cre_reverse: 5′-TGC ATG ATC TCC GGT ATT GA-3′. Both females and males were used in all the experiments. Mice had ad libitum access to food and water and were housed in a 12 hr dark/light cycle and in a temperature-controlled environment. All experiments were carried out in compliance with National Institutes of Health protocols and guidelines and were approved by Virginia Tech Institutional Animal Care and Use Committee.
2.2. Reagents
The chemicals and reagents used in these studies were obtained from Sigma, Fisher, or Tocris Bioscience unless otherwise noted. All DNA primers were purchased from Integrated DNA Technologies.
2.3. Antibodies
All monoclonal and polyclonal antibodies used in the present study are listed in Table 1. Fluorescently conjugated secondary antibodies were purchased from Invitrogen Life Technologies.
Table 1.
Antibody information
| Antigen | Manufacturer & catalog no. | Dilution | Species raised in, Isotype, Clonality | Immunogen | RRID |
|---|---|---|---|---|---|
| GFP | Thermo Fisher Scientific, A-11122 | 1:250 | Rabbit polyclonal IgG | GFP isolated directly from the jellyfish Aequorea victoria | AB_221569 |
| WFA | Vector Laboratories, B-1355–2 | 1:1000 | Biotinylated | AB_2336874 | |
| VGLUT 2 | Synaptic Systems, 135402 | 1:300 | Rabbit polyclonal | Synthetic peptide corresponding to AA 566 to 582 from rat VGLUT2 | AB_2187539 |
| GLT1 | Millipore, AB1783 | 1:1000 | Guinea pig polyclonal | Synthetic peptide from the carboxy-terminus of rat GLT1 | AB_90949 |
| GFAP | Dako, Z0334 | 1:1000 | Rabbit polyclonal | GFAP isolated from cow spinal cord | AB_10013382 |
| SOX9 | Millipore, AB5535 | 1:1000 | Rabbit polyclonal | KLH-conjugated linear peptide corresponding to the C-terminal sequence of human Sox9 | AB_2239761 |
| S100β | Dako, Z0311 | 1:200 | Rabbit polyclonal | S100 isolated from cow brain | AB_10013383 |
| HRP-DIG antibody | Roche, 11207733910 | 1:1000 | Sheep polyclonal | Digoxigenin (DIG) | AB_514500 |
| HRP-FL antibody | Roche, 11426346910 | 1:1000 | Sheep polyclonal | Fluorescein (FL) | AB_840257 |
2.4. Antibody characterizations
GFP
The GFP antibody (Thermo Fisher Scientific, A-11122) made against GFP isolated from the Aequorea victoria has been widely used to detect native GFP and GFP variants such as YFP (Briner et al., 2010). It has been used in previous studies (from Allen mouse brain atlas: www.alleninstitute.org; Fu et al., 2013) to specifically detect GFP-positive cells in the brains of transgenic mice expressing GFP reporter.
VGLUT2
The VGLUT2 antibody (Synaptic Systems, 135 402) was generated against a peptide corresponding to amino acids 566 to 582 of rat VGLUT2. It detects a single 60-kDa band from synaptic vesicle fraction of rat brains using Western blot (manufacturer’s technical sheet), and previous studies have shown that IHC staining using this antibody is specific to retinal terminals in dLGN (Hammer et al., 2014; Monavarfeshani et al., 2018).
GLT1
The GLT1 antibody (Millipore, AB1783) was generated against carboxy-terminus of rat GLT1. It detects an expected band at 62 kDa on Western blot of mouse brain membrane lysates (Pasquettaz et al., 2020). Previous IHC studies in mice have shown that this antibody reliably detects astrocytic GLT1 (Campbell et al., 2020; Zoltowska et al., 2018).
GFAP
The GFAP antibody (Dako, Z0334) was generated against GFAP isolated from cow spinal cord. It recognizes several bands of ~50 kDA in brain extracts, which correspond to different isoforms of GFAP (Kamphuis et al., 2012). In our hands, while it detects ~50 kDA proteins in brain extracts, it fails to detect protein in cerebellar synaptosome fractions (Su et al., 2012, 2016). Previous IHC studies have shown that immunostaining with this antibody specifically labels astrocytes in vitro or in vivo (Kamphuis et al., 2012; Su et al., 2010).
SOX9
The SOX9 antibody (Millipore, AB5535) was generated against C-terminal sequence of human SOX9. It recognizes a single band of ~65 kDA on Western blot of fetal mouse brain lysates (manufacturer’s technical sheet). Previous studies have used this antibody to stain nucleus of astrocytes using IHC (Barnabé-Heider et al., 2010; Yoshioka et al., 2012).
S100β
The S100β antibody (Dako, Z0311) was generated by immunizing rabbits with S100 protein isolated from cow brain. According to manufacturer’s technical sheet, the antibody detects S100β strongly and other members of S100 family weakly using Western blotting of purified human recombinant S100 proteins. Previous studies from us and others have reliably used this antibody for IHC staining of astrocytes in mouse visual thalamus (Su et al., 2020; Xiao et al., 2002).
2.5. Tissue Preparation and IHC
Mice were intraperitoneally injected with 12.5 μg/mL tribromoethanol (Avertin) and transcardially perfused with PBS and 4% paraformaldehyde (PFA, pH 7.4) as previously described (Su et al., 2010, 2020). Brains were extracted, kept in 4% PFA overnight at 4°C, and then transferred to 30% sucrose in PBS for at least 48 hr. Fixed brains were embedded in Tissue Freezing Medium (Electron Microscopy Sciences) and cryosectioned at 16 μm sections on a Leica CM1850 cryostat. Slides were air-dried for 30 min and then incubated in IHC blocking buffer (5% normal goat serum serum, 2.5% bovine serum albumin, 0.1% Triton-X in PBS) for 30 min. Primary antibodies diluted in blocking buffer were incubated on the sections for >18 hr at 4°C. After washing with PBS, anti-rabbit or anti-guinea pig fluorophore-conjugated secondary antibodies (1:1000 dilution in blocking buffer) were incubated on tissue sections for 1 hr at room temperature. After several washes with PBS, sections were stained with 4′,6-diamidino-2-phenylindole (DAPI, 1:5000 in PBS) and mounted with Vectashield (Vector Laboratories). Slides were visualized on a Zeiss LSM 700 confocal microscope.
2.6. Riboprobe production and ISH
Riboprobes were generated as previously described (Levy et al., 2015; Monavarfeshani et al., 2018; Su et al., 2010). Plasmids carrying Fgfr3 (cat #MMM1013–202798065), Fgf15 (cat #5066286), Syt1 (cat # MM1013–9199901), and Syt2 (cat #MMM1013– 7512379) were purchased from GE Dharmacon. Gja1 1.1Kb cDNA (Gja1-F: CGTGAAGGGAAGAAGCGA and Gja1-R: GCCTGCAAACTGCCAAGT, corresponding to nucleotides 714–1854) and Hevin 1.093Kb cDNA (Hevin-F: TCGGAGTGCTTTCATTCCCG and Hevin-R: TCGGAGTGCTTTCATTCCCG; corresponding to nucleotides 777–1850) were generated using SuperScript II Reverse Transcriptase First Strand cDNA Synthesis kit (Invitrogen, cat #18064014) according to the manufacturer’s protocol, amplified by PCR using primers mentioned above, gel purified, and then cloned into the pGEM-T Easy vector (Promega, cat #A1360) according to the manufacturer’s protocol. Sense and antisense riboprobes against Hevin, Gja1, Syt1, Syt2, Fgfr3, and Fgf15 were synthesized from linearized plasmids (5 μg) using DIG- or FL-labeled uridylyltransferase (Roche, cat #11685619910 and cat #11277073910) and MAXIscript in vitro Transcription Kit (Ambion, cat #AM1312) according to the manufacturer’s protocol. Riboprobes were then hydrolyzed into ~0.5 Kb fragments by adding 4 μl of NaHCO3 (1M), 6 μl of Na2CO3 (1M), and 80 μl of water, and incubating the reaction mixture at 60°C. Riboprobes were precipitated in ethanol and dissolved in RNAase-free water. ISH using the generated riboprobes was performed on 16 μm PFA-fixed cryosectioned tissue as previously described (Monavarfeshani et al., 2018; Su et al., 2020). Briefly, sections were fixed in 4% PFA for 10 min, washed with PBS, and incubated in the proteinase K solution (i.e., 1 μg/mL in 50mM Tris pH 7.5, 5mM EDTA) for 10 min. Sections were then washed with PBS, incubated for 5 min in 4% PFA, washed with PBS, and incubated in acetylation solution (1.33% triethanolamine, 20 mM hydrochloric acid, and 0.25% acetic anhydride) for 10 min. Later, they were washed with PBS, incubated in 1% triton in PBS for 30 min for permeabilization, and washed with PBS. To block endogenous peroxidase, they were incubated in 0.3% hydrogen peroxide in PBS for 30 min and washed with PBS. Sections were then equilibrated in hybridization solution (50 mL of prehyb solution, 25 mg Roche yeast RNA, and 1.6 ml of 5 mg/ml heparin) for 1 hr and incubated with ~50 μl of heat-denatured (10 min at 70°C) diluted riboprobes overnight at 65°C. On the subsequent day, slides were washed with 0.2x saline-sodium citrate solution and then with tris-buffered saline (TBS). Slides were incubated for 1 hr in blocking buffer (10% lamb serum, 0.2% Roche blocking reagent in TBS) and were then incubated overnight at 4°C with HRP-conjugated anti-DIG or anti-FL antibodies. On day 3, riboprobes were detected using Tyramide Signal Amplification system (PerkinElmer, cat #NEL75300 1KT) and the signal was visualized on a Zeiss LSM 700 confocal microscope.
2.7. Serial block-face scanning electron microscopy (SBFSEM)
C57BL/6J mice were perfused with 0.1M sodium cacodylate buffer containing 2.5% glutaraldhehyde and 4% PFA. The dLGN was microdissected and was processed, embedded, and imaged by Renovo Neural Inc as previously described (Carrillo et al., 2020; Hammer et al., 2014, 2015; Mukherjee et al., 2016). Datasets represented dLGN regions that were 35–50 μm × 40–50 μm and consisted of 250–500 serial sections (with each section being 50–80 nm thick). The images were imaged at a 5 nm/pixel resolution. TrakEM2 in Fiji was used to analyze serial image stacks (Cardona et al., 2012).
2.8. Whole-cell patch-clamp recording
Whole-cell patch-clamp recordings were performed in acute brain slices as previously described (Campbell et al., 2014; Robel et al., 2015). Aldh1l1-EGFP mice of P14-P15 and >P60 ages were decapitated and the brains were immediately immersed in the ice-cold cutting solution (containing, all in mM; 1.5 KCl, 135 N-methyl-D-gluconate, 23 choline bicarbonate, 1.5 KH2PO4, 25 D-glucose, 3.5 MgSO4, and 0.5 CaCl2; pH 7.4, 310±5mosm) saturated with carbogen (95% O2 + 5% CO2). 300 μm coronal brain slices were generated with a vibrating blade microtome (Leica VT1200) and were incubated in artificial cerebrospinal fluid (ACSF containing, all in mM, 3 KCl, 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 25 D-glucose, and 1.3 MgSO4, pH 7.4, 310±5mosm) at 32°C for 1 hr. Afterwards, slices were kept at room temperature until used for recordings. All the recordings were performed in a recording chamber continuously superfused with ACSF at a flow rate of ~2 ml/min while bath temperature was maintained at 32–33°C throughout recordings.
dLGN astrocytes expressing EGFP were identified under an upright microscope (Leica DMLFSA) with 5X and 40X water immersion lens and appropriate fluorescent filters combinations and infrared illumination. An axopatch 200B amplifier (Molecular Devices) in combination with Clampex 10.4 software and Axon Digidata 1550A interface (Molecular Devices) was used to acquire whole cell recordings. Data were filtered at 5 kHz, digitized at 10–20 kHz and analyzed using Clampfit 10.6 software (Molecular Devices). Patch pipettes of 7–10 MΩ open-tip resistance were created from standard borosilicate capillaries (WPI, 4 in Thin Wall Gl 1.5OD/1.12ID) using HEKA PIP 6 vertical pipette puller. Patch pipettes were filled with an intracellular solution of 134 mM potassium gluconate, 1 mM KCl, 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2 mM adenosine 5′-triphosphate magnesium salt (Mg-ATP), 0.2 mM guanosine 5′-triphosphate sodium salt (Na-GTP) and 0.5 mM ethylene glycol tetraacetic acid (EGTA) (pH 7.4, 290–295 mOsm).
Tight seals (~2–5GΩ) were made on astrocytes followed by breaking the seal with gentle suction to achieve the whole cell configuration and clamping astrocytes at −80mV. We measured resting membrane potential (I = 0 mode), whole-cell capacitance, and series resistance directly from the amplifier outputs within ~1 min of achieving the whole cell configuration. To isolate astrocytic glutamate transporter responses, the following drugs were added to ACSF recording solution: 500 nM TTX, 20 μM bicuculline, 100 μM CdCl2, 50 μM AP5, and 50 μM CNQX. A 500 msec puff of 200 μM glutamate (2PSI pressure using a Pico-liter Injector PLI-10 from Warner Instruments) was applied onto the whole-cell voltage clamped EGFP+ astrocytes from a distance of about 70–100μm. We calculated the peak uptake current, decay time of current, and total charge transfer of glutamate current from an average trace of minimum 3 recordings. Glutamate uptake current was blocked in presence of 100μM mM DL-threo-β-benzyloxyaspartic acid (TBOA), and 300μM mM dihydrokainic acid (DHK) suggesting that it is mediated by astrocytic glutamate transporters.
2.9. Imaging, quantification and statistics
All the representative images in the figures are maximum intensity projections from z-stacks, however, colocalization was confirmed by visualizing single-planes. All quantification was performed on experiments with at least three biological replicates per age and genotype. Student’s T-test or ANOVA analysis were used to determine significant differences between groups. Unless otherwise stated, data are plotted as mean ± SEM. To quantify the astrocytes as percentage of total cells in dLGN and vLGN (magnification used to obtain images was 10x and at least three thalamic sections per animal), we counted Aldh1l1+ (by Aldh1l1-EGFP transgenic reporter), SOX9+ (by IHC), Fgfr3+ (by ISH), and Gja1+ (by ISH) cells and divided by all DAPI+ cells counted in that section. To calculate the density of Aldh1l1+ astrocytes (magnification used to obtain images was 10x and at least three thalamic sections per animal), we quantified genetically labelled Aldh1l1+ cells (using Aldh1l1-EGFP transgenic reporter mice) in either dLGN or vLGN before normalizing to the area of the respective brain area. Areas were measured by manually outlining the border of dLGN or vLGN using ImageJ (version 1.52n, NIH). To quantify percentage of dLGN area covered by Aldh1l1-EGFP (magnification used to obtain images was 40x and three fields of view per animal), all images from different ages were taken at same confocal settings and these grayscale images were then binarized to measure percentage of area fraction using ImageJ. To quantify the spatial distribution of GFAP immunoreactivity within the visual thalamus, we performed line scan analysis on the confocal images of GFAP-stained coronal sections. Background was subtracted from these images using the “Rolling ball radius” (10 pixels) function on the ImageJ. To obtain the fluorescent intensity along the dorsomedial-ventrolateral axis of the visual thalamus, lines parallel to the optic tract were drawn on ImageJ. The “Multi Plot” profile values were then averaged to plot the signal intensity across each coordinate. Based on observed variability and previous experience, at three animals were used as biological replicates and at least three dLGN and vLGN sections per mouse brain were averaged to obtain the mean for that biological replicate. DAPI counter-staining was used to determine the boundaries of dLGN and vLGN.
3. RESULTS
3.1. Expression of astrocytic markers in visual thalamus
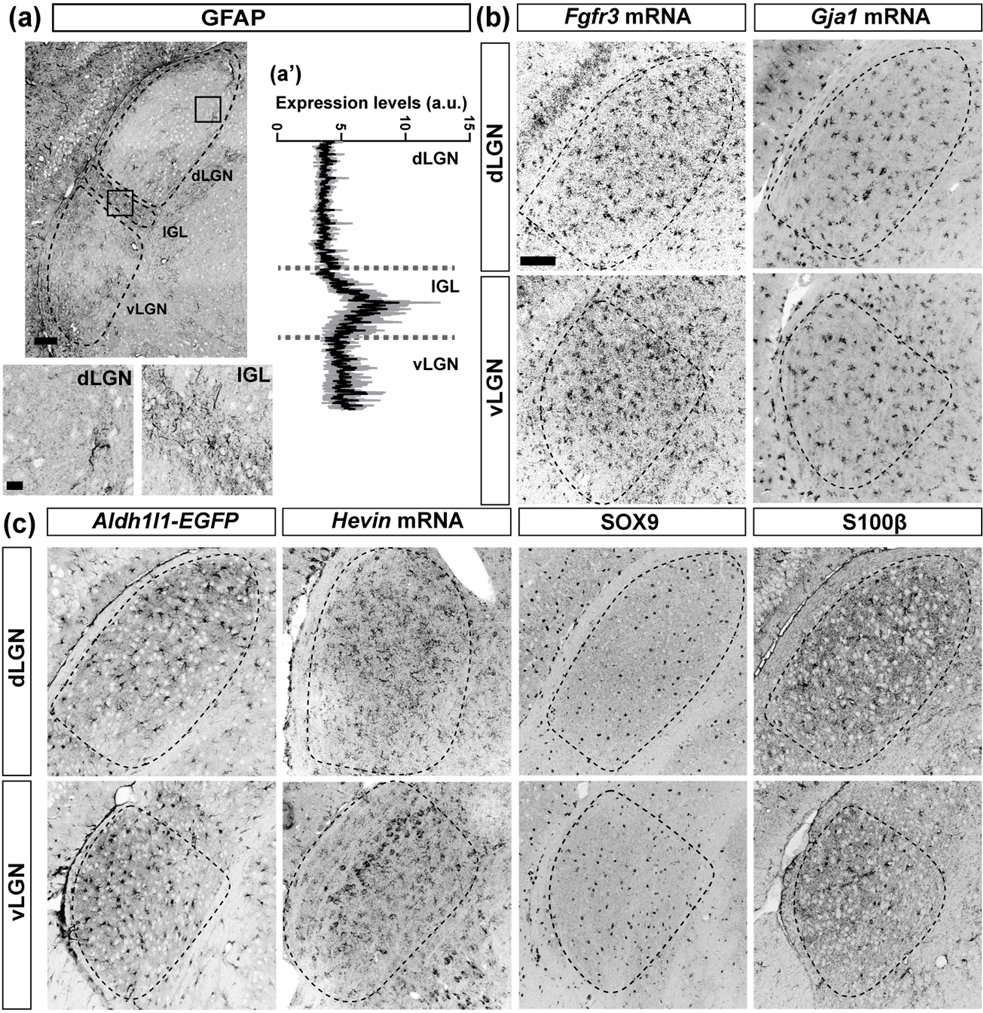
Our previous studies identified transcriptional differences in a cohort of neonatal astrocytes in visual thalamus (Su et al., 2020), therefore, we started by identifying molecules appropriate to broadly study astrocytes in visual thalamus. Previously published transcriptomic data from bulk-sequencing of whole P25 dLGN and vLGN revealed the most commonly used molecular markers for astrocytes in other brain regions (Slc1a3, Aldoc, Gfap, Aldh1l1, S100ß, Sox9, Glul, Hevin, Fgfr3, and Gja1) are expressed in visual thalamus (Monavarfeshani et al., 2018). The expression levels of most of these canonical astrocyte genes appeared similar in these regions, except for Gfap, the gene that encodes Glial Fibrillary Acidic Protein (GFAP). Gfap mRNA was significantly higher in vLGN/IGL (RNAseq raw reads: 4967) compared to dLGN, with RNAseq raw reads: 1537 (Monavarfeshani et al., 2018). Immunostaining for GFAP supported this difference by revealing its enrichment in IGL, compared to its very low protein expression level in the adjacent dLGN (Fig. 1(a–a’)) (Su et al., 2011). Thus, while GFAP-immunoreactivity may be useful for assessing astrocyte development, distribution, and morphology in many brain regions (including the hippocampus which can be seen in Fig. 1(a)), it is not useful for studying astrocytes in dLGN. Therefore, we next assessed the distribution of several other astrocyte genes/markers in visual thalamus with immunohistochemistry (IHC), genetic reporters, and in situ hybridization (ISH) (Fig. 1(b–c)). The distribution of cells labelled with antibodies (against proteins SOX9 or S100ß), riboprobes (Fgfr3, Gja1, or Hevin), or genetic reporter proteins (Aldh1l1-EGFP) appeared different, suggesting either heterogeneous expression by astrocytes or expression by other cell types in these regions. To distinguish between these possibilities, we assessed the co-expression of these six markers with each other and with other cell-type specific markers in visual thalamus. For example, to label microglia we used Cx3cr1-GFP (Jung et al., 2000) and to label neurons we used riboprobes against Syt1 and Syt2 mRNAs, which encode the synaptic vesicle proteins Synaptotagmin 1 and 2, respectively.
Fig. 1. Expression of different astrocyte associated molecules in visual thalamus.
(a) Immunostaining for GFAP in adult WT dLGN, IGL, and vLGN. Boxes provide higher magnification examples of GFAP immunoreactivity in dLGN and IGL. (a’) Line scan analysis for the expression of GFAP immunoreactivity in dLGN, IGL, and vLGN. Arbitrary fluorescence units (a.u.) are presented against distance from the dorsal part to the ventral region of the visual thalamus. Solid black line represents mean and shaded gray area represents SEM. (b-c) Immunostaining, ISH, and a genetic reporter line depict the expression of astrocyte genes and proteins in adult dLGN and vLGN. Scale bar in (a)=100μm, in high-mag (a)=20μm, and in (b-c)=100μm
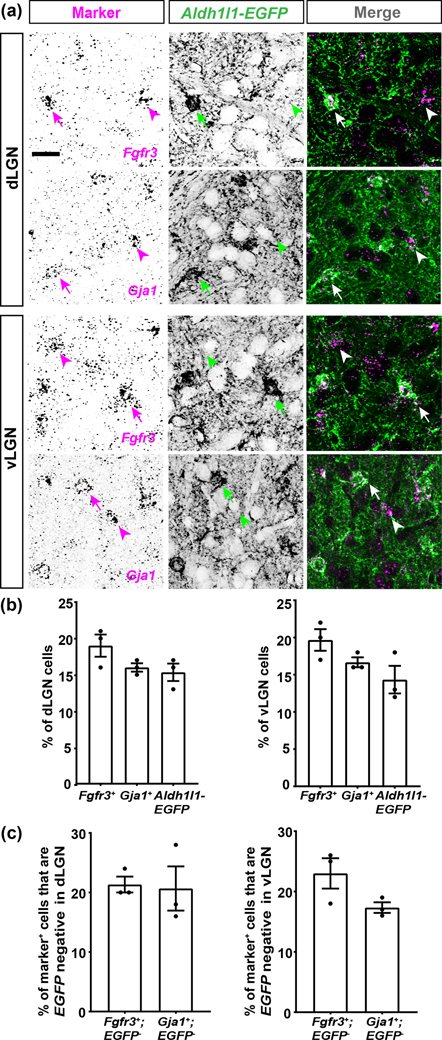
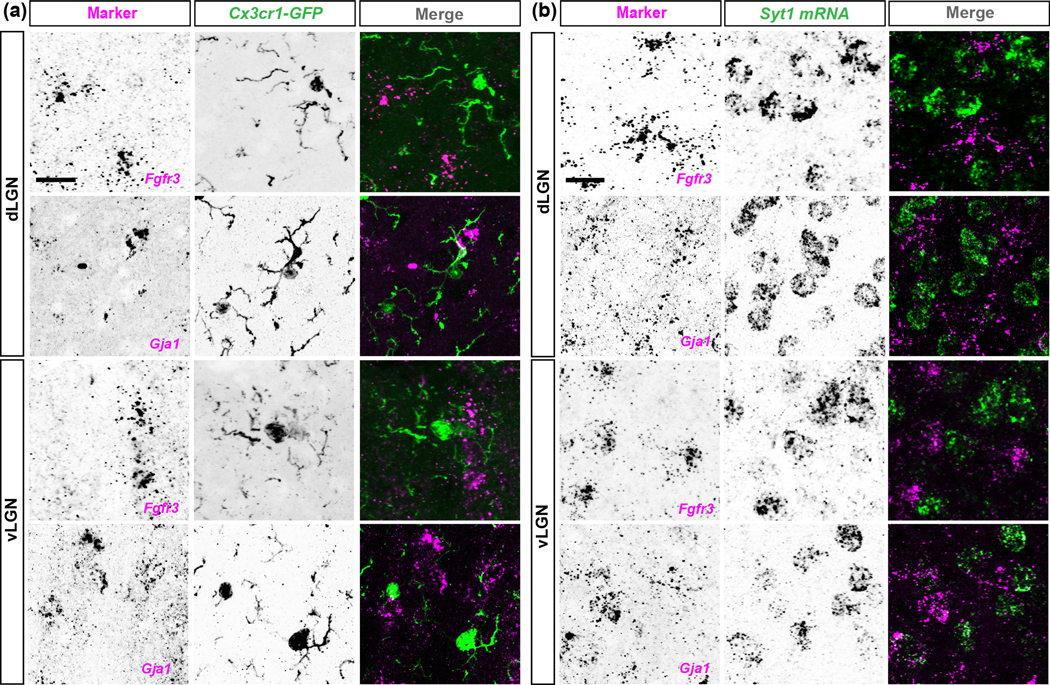
We first focused our attention on Aldh1l1, Fgfr3, and Gja1 (which encodes Connexin 43). Each of these genes has been reported to widely label astrocytes in other brain regions (Agius et al., 2004; Giaume et al., 1991; Kang et al., 2014; Pringle et al., 2003; Wallraff et al., 2006; Yamamoto et al., 1990; Young et al., 2010; Zhang et al., 2016). To test whether these molecules labelled the same cells in visual thalamus, we performed ISH in thalamic slices from Aldh1l1-EGFP mice. We observed that all EGFP+ (i.e. Aldh1l1+) astrocytes expressed Fgfr3 and Gja1 mRNAs in both dLGN and vLGN (Fig. 2(a–c)). There were cases, however, in which cells positive for Fgfr3 or Gja1 mRNAs did not express EGFP (Fig. 2(a–c)). One possibility is that these cells represent astrocytes not labelled in this BAC transgenic line. An alternative is that Fgfr3 and Gja1 may be generated by other cell types in these brain regions. To test this latter possibility, we assessed their expression in other cell types, such as microglia and neurons. ISH for Fgfr3 and Gja1 mRNAs in Cx3cr1-GFP mice revealed that microglia do not express these genes in visual thalamus (Fig. 3(a)). Similarly, double ISH for Fgfr3 or Gja1 and the neuronal mRNA Syt1 revealed no co-expression of Syt1 and Fgfr3 or Gja1, suggesting that neurons also do not express these genes (Fig. 3(b)). We interpret these results to show that Fgfr3 and Gja1 are specifically expressed by astrocytes and are expressed by most, if not all, astrocytes in the adult visual thalamus (including in the developing visual thalamus, fig. 6). Although not all astrocytes may be labelled in Aldh1l1-EGFP mice, all EGFP+ astrocytes are Fgfr3+ and Gja1+, making this transgenic reporter line a reliable tool to broadly study astrocytes in visual thalamus.
Fig. 2. Fgfr3, Gja1, and Aldh1l1 are expressed by large subsets of astrocytes in visual thalamus.
(a) ISH for Fgfr3 or Gja1 in the dLGN and vLGN of >P25 Aldh1l1-EGFP transgenic mice revealed most Fgfr3+ or Gja1+ cells overlap with Aldh1l1-EGFP+ cells (arrows). Only a few Fgfr3+ or Gja1+ cells did not coexpress EGFP (arrowheads). (b) Quantification of the percentage of DAPI+ cells that are Fgfr3+ or Gja1+ or Aldh1l1-EGFP+ in >P25 dLGN and vLGN. (c) Quantification of the percentage of Fgfr3+ or Gja1+ cells that do not express EGFP in >P25 dLGN and vLGN. Each data point in (b-c) represents one biological replicate and data is shown as mean ± SEM. Scale bar in (a)=20μm
Fig. 3. Fgfr3 and Gja1 are not expressed by microglia or neurons in visual thalamus.
(a) ISH for Fgfr3 or Gja1 in the dLGN and vLGN of >P25 Cx3cr1-GFP transgenic mice revealed no expression of Fgfr3 or Gja1 by GFP+ microglia. (b) ISH for Fgfr3 or Gja1 in the dLGN and vLGN of >P25 C57/BL6 revealed no expression of Fgfr3 or Gja1 by Syt1+ neurons. Scale bar in (a-b)=20μm
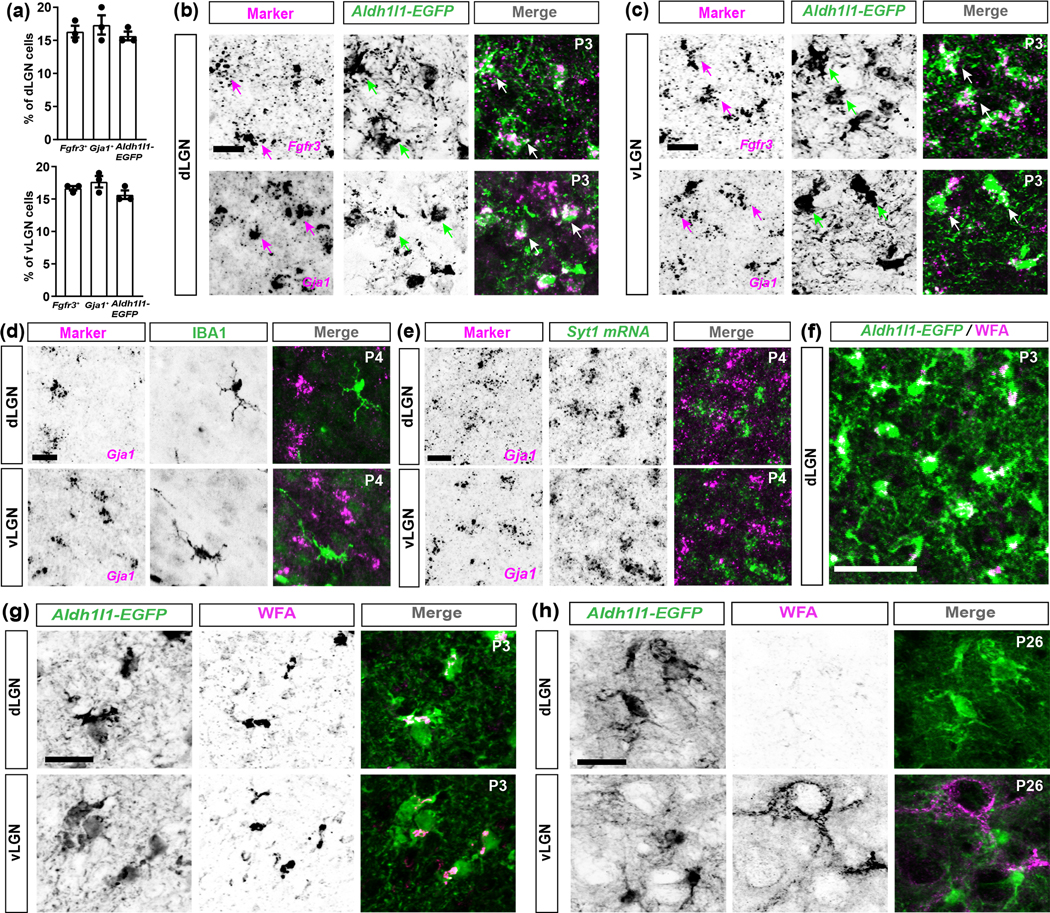
Fig. 6. Fgfr3, Gja1, and WFA are reliable markers for astrocytes in the developing visual thalamus.
(a) Quantification of the percentage of DAPI+ cells that are Fgfr3+ or Gja1+ or Aldh1l1-EGFP+ in P3 dLGN and vLGN. Each data point represents one biological replicate and data is shown as mean ± SEM. (b-c) ISH of Fgfr3 or Gja1 in the dLGN (b) and vLGN (c) of P3 Aldh1l1-EGFP transgenic mice revealed Fgfr3+ or Gja1+ cells overlap with Aldh1l1-EGFP+ cells (arrows). (d) ISH of Gja1 with IHC of IBA1 in the P4 C57/BL6 dLGN and vLGN revealed no expression of Gja1 by microglia. (e) ISH of Gja1 in the dLGN and vLGN of P4 C57/BL6 revealed no expression of Gja1 by Syt1+ neurons. (f) Low magnification example of WFA staining in the dLGN of P3 Aldh1l1-EGFP mice. (g-h) Immunostaining for WFA in the dLGN and vLGN of P3 (g) and P26 (h) Aldh1l1-EGFP mice. Scale bar in (b-e)=20μm, (f)=50μm, and (g-h)=20μm
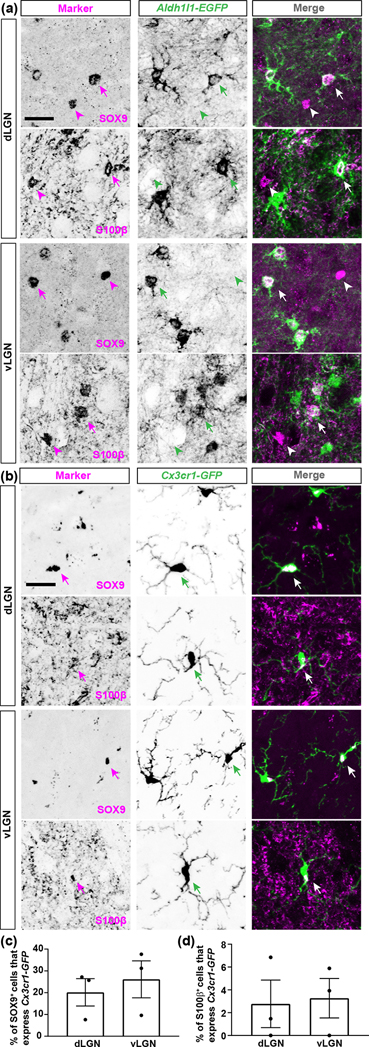
We next studied the expression of SOX9 and S100β, two proteins commonly used to label and identify astrocytes. Using IHC, we observed a widespread expression of these two proteins in dLGN and vLGN (Fig. 1(c)). In fact, we found that immunostaining for SOX9 labelled a greater percentage of cells in thalamus than those labelled in Aldh1l1-EGFP mice (e.g., in dLGN ~21% of DAPI+ cells express SOX9 versus ~15% that express EGFP in Aldh1l1-EGFP mice). Immunostaining for SOX9 and S100β in Aldh1l1-EGFP thalamic sections revealed that although all EGFP+ astrocytes were SOX9+ in visual thalamic nuclei, many SOX9+ or S100β+ cells lacked EGFP (Fig. 4(a)). Although widely used to label astrocytes, a few studies have reported that SOX9 and S100β are also expressed by other cell types in the brain (Adami et al., 2001). This led us to ask whether microglia or neurons in visual thalamus express SOX9 and S100β. We failed to observe SOX9 or S100β expression in Syt1+ thalamic neurons (data not shown), however, we found a subset of Cx3cr1-GFP+ microglia contain SOX9 or S100β in both dLGN and vLGN (Fig. 4(b–d)). This suggests that a subset of microglia generate these proteins. Alternatively, as microglia are capable of phagocytosing astrocytes during development, it is possible that SOX9 or S100β immunoreactivity within Cx3cr1-GFP+ microglia could represent such phagocytosis. However, it is important to highlight that we observed co-localization of SOX9 and S100β in microglia in healthy adult animals. Thus, while antibodies against SOX9 and S100β do label a significant number of astrocytes, their presence in other glial cell types such as microglia (and also potentially in oligodendrocytes that we did not stain for in this study) in visual thalamus suggests they are not a reliable marker for identifying astrocytes (Deloulme et al., 2004).
Fig. 4. Expression of SOX9 and S100ß by astrocytes and microglia in visual thalamus.
(a) IHC for SOX9 or S100ß in the dLGN and vLGN of >P25 Aldh1l1-EGFP transgenic mice revealed two observations: SOX9 or S100ß expression in Aldh1l1-EGFP+ astrocytes (arrows), and SOX9 or S100ß protein in cells not labelled in Aldh1l1-EGFP mice (arrowheads). (b) IHC for SOX9 or S100ß in the dLGN and vLGN of >P25 Cx3cr1-GFP transgenic mice revealed expression of SOX9 or S100ß by GFP+ microglial cells (arrows). (c) Quantification of the percentage of SOX9+/GFP+ or S100ß+/GFP+ cells in >P25 Cx3cr1-GFP dLGN and vLGN. Each data point in (c) represents one biological replicate and data is shown as mean ± SEM. Scale bar for (a-b)=20μm.
The final molecule we explored was Hevin, a synaptogenic extracellular matrix protein (Cahoy et al., 2008; Eroglu, 2009). Although neurons also generate this molecule, in other regions of the rodent visual system such as superior colliculus, Hevin is largely generated by astrocytes (Kucukdereli et al., 2011; Mongrédien et al., 2019). The distribution of Hevin+ cells in visual thalamus was strikingly different than the distribution of Fgfr3+, Gja1+, Aldh1l1+, SOX9+, and S100β+ cells, especially in vLGN. In fact, Hevin+ cells appear to be stratified in vLGN, where we recently reported the discovery of neuronal subtype-specific laminae (Sabbagh et al., 2020) (Fig. 1(c)). This suggested to us that Hevin labels neurons in visual thalamus. Colocalization studies in Aldh1l1-EGFP confirmed this possibility, revealing sparse expression of Hevin by EGFP+ astrocytes in adult dLGN and vLGN (Fig. 5(a)). Hevin+ cells that were not astrocytes were also not microglia, as they did not colocalize with GFP+ cells in Cx3cr1-GFP+ mice (data not shown). In contrast, we observed significant co-expression of Hevin mRNA with Syt1 or Syt2 in both dLGN and vLGN (Fig. 5(b–c)), confirming that this gene was generated mainly by neurons in visual thalamus (see also Kalish et al, 2018).
Fig. 5. Expression of Hevin by neurons in both dLGN and vLGN.
(a) ISH for Hevin in the dLGN and vLGN of >P25 Aldh1l1-EGFP transgenic mice revealed sparse expression of Hevin by EGFP+ astrocytes (arrows). Most Hevin+ cells did not coexpress EGFP in these mice (arrowheads). (b-c) ISH for Hevin in the dLGN and vLGN of >P25 WT mice revealed significant expression of Hevin in Syt1+ (b) or Syt2+ (c) neurons (arrows). Scale bar for (a-c)=20μm.
Taken together, these expression studies indicate that riboprobes against Fgfr3 and Gja1 and the transgenic mouse line Aldh1l1-EGFP are the most appropriate tools to broadly study astrocyte development in visual thalamus.
3.2. Development of astrocytes in visual thalamus
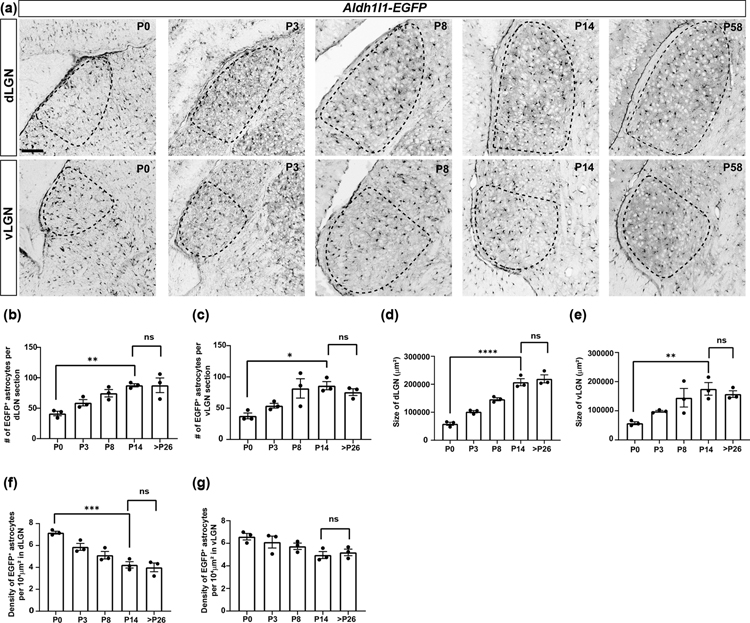
In addition to labelling astrocytes in the adult visual thalamus, ISH for Fgfr3 and Gja1 and the Aldh1l1-EGFP mouse line all labelled astrocytes in the neonatal and developing dLGN and vLGN (Fig. 6(a–e) and 7(a)). We also discovered that fluorescently conjugated Wisteria floribunda agglutinin (WFA), a lectin typically used to label perineuronal nets in the adult brain, labels neonatal astrocytes in the developing visual thalamus (Fig. 6(f–h)). However, while ISH for Fgfr3 and Gja1 and staining with WFA labels astrocyte somas, they fail to reliably label astrocyte processes or provide accurate morphological data on this cell type. Therefore, we focused our attention on using Aldh1l1-EGFP mice to characterize astrocyte development. During early development, we observed a significant increase in the number of EGFP+ astrocytes in dLGN and vLGN of Aldh1l1-EGFP mice. Subsequent to eye-opening (P14), there was no further change in EGFP+ astrocyte number in visual thalamus (Fig. 7(b–c)). Developmental increases in EGFP+ astrocytes coincided with a dramatic increase in the area of developing dLGN and vLGN, therefore we normalized the number of EGFP+ astrocytes to the cross-sectional area of these retinorecipient regions (Fig. 7(d–e)) (El-Danaf et al., 2015).
Fig. 7. Distribution of Aldh1l1-EGFP+ astrocytes in the developing visual thalamus.
(a) Distribution of EGFP+ cells in the dLGN and vLGN of neonatal and postnatal Aldh1l1-EGFP mice (b-c) Age-related changes in EGFP+ astrocyte number per section of the dLGN (b) and vLGN (c) of Aldh1l1-EGFP mice. (d-e) Age-related changes in size of the dLGN (b) and vLGN (c) of Aldh1l1-EGFP mice. (f-g) Age-related changes in the density of EGFP+ astrocytes in the dLGN (b) and vLGN (c) of Aldh1l1-EGFP mice. Each data point in (b-g) represents one biological replicate and data is shown as mean ± SEM. Asterisks (*) represent significance (****p<0.0001,***p<0.001, **p<0.01,*p<0.05, ns=not significant) by one-way ANOVA. Scale bar for (a)=100μm.
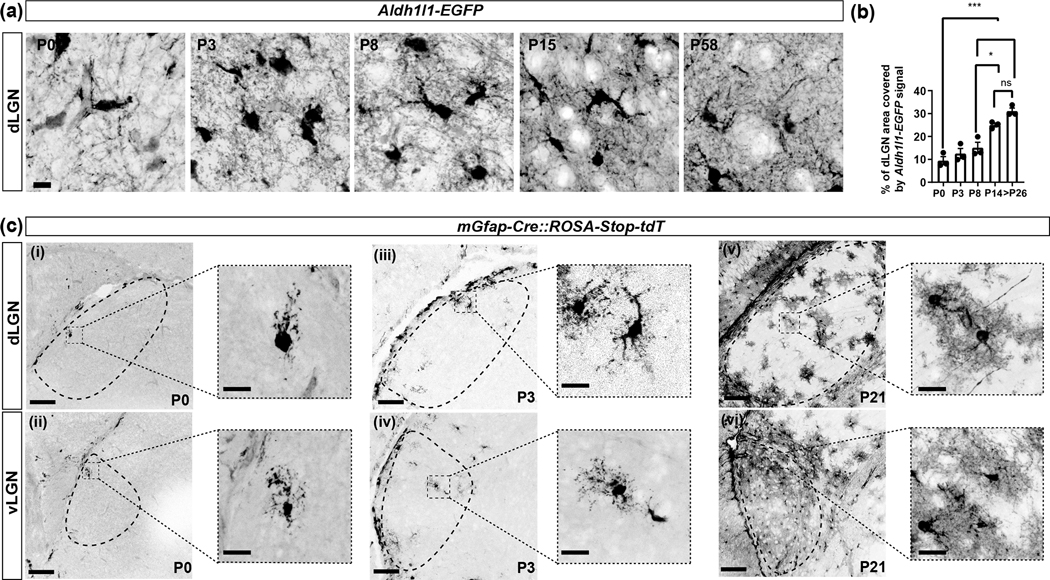
Imaging astrocyte morphology in Aldh1l1-EGFP mice revealed that at early postnatal ages (P0, P3, and P8), EGFP+ astrocytes generated few processes and significant regions of the thalamic neuropil appeared to lack EGFP+ astrocytic processes (Fig. 8(a)). By eye-opening, astrocytes appeared to have developed a highly branched morphology and their processes appeared to densely fill the thalamic neuropil, similar to what is observed in adults (P58) (Fig. 8(a)). To confirm this finding, we quantified the percentage of dLGN area occupied by EGFP fluorescence. This demonstrated significant increase in the area of dLGN containing EGFP+ signal at each age, leading up to eye-opening. No significant differences were observed between the percent occupancy of EGFP+ signal between P14 and adult dLGN (Fig. 8(b)).
Fig. 8. Morphological development of astrocytes in visual thalamus.
(a) High magnification images showing the morphology of EGFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice at different ages. (b) Quantification of the percentage of dLGN area covered by EGFP fluorescence. Each data point represents one biological replicate and data is shown as mean ± SEM. Asterisks (*) represent significance (****p<0.0001,***p<0.001, **p<0.01,*p<0.05, ns=not significant) by one-way ANOVA (c) Morphology of tdT+ astrocytes in the dLGN and vLGN of mGfap-Cre::ROSA-Stop-tdT mice. Scale bar in (a)=10μm, in (c) (i-vi)=100μm, and in high magnification images in (c)=20μm
The approach described above labels almost all astrocytes with EGFP, making single cell morphology difficult to discern at later ages. To circumvent this, we generated mGfap-Cre::ROSA-Stop-tdT mice. Although little GFAP protein is made in visual thalamus (Fig. 1(a)), Gfap mRNA is expressed (see RNA-seq datasets in Monavarfeshani et al., 2018), making this a viable tool to study astrocytic morphology in visual thalamus. At early ages, this transgenic tool labelled sparse astrocytes in dLGN and vLGN (Fig. 8(c)). Analysis in mGfap-Cre::ROSA-Stop-tdT confirmed the immature appearance of tdT+ astrocytes in neonatal thalamus and the emergence of their adult-like morphology by eye-opening (Fig. 8(c)).
3.3. dLGN astrocytes can uptake extracellular glutamate by eye-opening
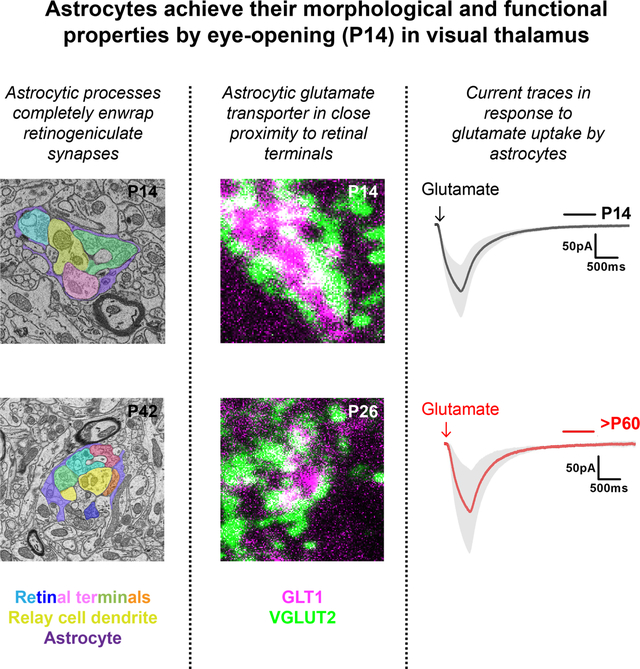
Since astrocytes in visual thalamus appear to have acquired their mature morphology by eye-opening, we hypothesized that they may have acquired their mature function, such as ensheathing retinogeniculate (RG) synapses and removing glutamate from the synaptic cleft. We addressed both of these possibilities with immunohistochemical, ultrastructural, and functional studies. We used two approaches to investigate the proximity of astrocytic processes to retinal terminals in the developing and adult dLGN: IHC and serial block-face scanning electron microscopy (SBFSEM). Immunostaining for vesicular glutamate transporter 2 (VGLUT2), a presynaptic protein specifically enriched in retinal terminals in dLGN revealed that processes of EGFP+ astrocytes are in close contact to VGLUT2+ terminals by eye-opening in dLGN of Aldh1l1-EGFP mice (Fig. 9(a–b)) (Hammer et al., 2014; Monavarfeshani et al., 2018). In fact, the association of EGFP+ processes at RG synapses appeared similar at eye-opening to that at mature P26 dLGN synapses.
Fig. 9. Astrocytic processes enwrap RG synapses in dLGN by eye-opening.
(a-b) Immunostaining for VGLUT2 in the dLGN of P14 (eye-opening) (a) and P26 (b) Aldh1l1-EGFP mice. Arrows show examples of close proximity of VGLUT2+ retinal terminals to EGFP+ astrocytic processes. Insets depict the high magnification of these locations. (c-f) SBFSEM revealed complete encapsulation of simple (c-d) and complex (e-f) RG synapses by astrocytic processes in the dLGN of P14 (c, e) and P42 (d, f) mice. At the bottom of each are color legends to identify different cellular components of each synapse. Scale bar in (a-b)=10μm and (c-f)=0.5μm.
IHC lacks the resolution to truly assess synaptic ensheathment by astrocytic processes, therefore, we next turned to SBFSEM. We previously reported that two morphologically distinct types of RG synapses (simple and complex) are present in rodent dLGN as early as eye-opening (Hammer et al., 2015; Monavarfeshani et al., 2018). Ultrastructural features (such as synaptic vesicles and pale mitochondria) were used to identify retinal terminals in both types of RG synapses (Hammer et al., 2014, 2015). Likewise, morphological features such as glycogen granules were used to identify astrocytes and their processes in SBFSEM datasets. Analysis of SBFSEM datasets from P14 and P42 dLGN revealed that at both ages, perisynaptic astrocytic processes (PAPs) enwrap nerve terminals in both simple and complex RG synapses (Fig. 9(c–f)). Taken together, these data provide evidence that astrocytic processes ensheath retinal terminals as early as eye-opening.
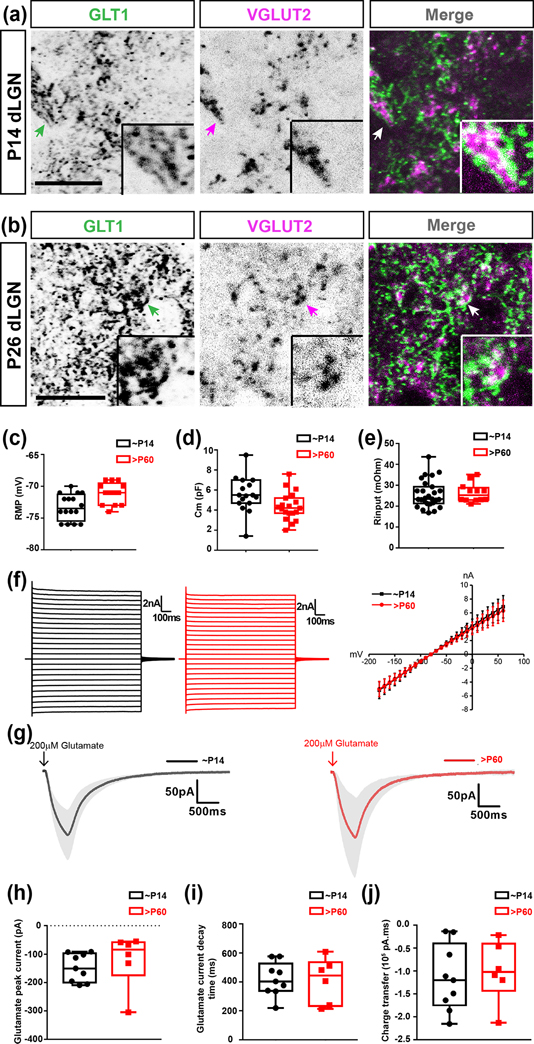
The proximity of astrocytic processes to RG synapses during eye-opening does not indicate they are functionally able to clear glutamate from the synaptic cleft of these synapses. To test whether astrocytes can uptake extracellular glutamate at eye-opening, we used two approaches. First, we assessed the proximity of glutamate transporter-1 (GLT1), a glutamate transporter enriched in astrocyte processes to retinal terminals in dLGN. At both ages, GLT1+ astrocyte processes were observed in close proximity to, and even completely ensheating, VGLUT2+ retinal terminals (Fig. 10(a–b)). This suggested that by eye opening, astrocyte processes contain the machinery to efficiently clear glutamate from the synaptic cleft of RG synapses. Second, we performed whole-cell patch clamp recordings from dLGN astrocytes in acute slices from Aldh1l1-EGFP mice to examine their biophysical properties and glutamate currents as a readout of GLT1’s functional activity. Randomly patched EGFP+ astrocytes in dLGN at eye-opening (P14) and in adult dLGN (>P60) showed similar resting membrane potential (RMP) membrane capacitance, and input resistance (Fig. 10(c–e)). The hyperpolarizing RMP, low input resistance, and linear IV curve suggested that astrocytes in both age groups belong to same functional category and exhibit typical passive glial phenotype (Fig. 10(c–f)). dLGN astrocytes at eye-opening (P14) showed typical glutamate uptake current upon puffing 200μM glutamate from a distance of ~70–100μm from the recorded astrocytes. This glutamate uptake current was nearly eliminated upon application of a cocktail of TBOA (100μM) and DHK (300μM), the inhibitors of astrocyte glutamate transporters GLT1 and GLAST, respectively. The magnitude and kinetic properties of glutamate currents, including uptake current decay time and total charge transfer, for astrocytes during eye-opening were not significantly different from that of adult astrocytes (>P60), suggesting equally efficient glutamate transport in both ages (Fig. 10(g–i)). Taken together, these data suggest that at eye-opening, astrocytes possess similar biophysical properties to those in adult dLGN and are equally capable of clearing excess extracellular glutamate around RG synapses.
Fig. 10. Astrocytic GLT1 is functionally capable of clearing glutamate spillover in dLGN by eye-opening.
(a-b) Immunostaining for GLT1 in the dLGN of P14 (a) and P26 (b) mice. Arrows show examples of close proximity of VGLUT2+ retinal terminals to GLT1. Insets depict the high magnification of these locations. (c) Resting membrane potential of GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice (n=16 cells for P14; n=13 cells for >P60). (d) Membrane capacitance of GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice (n=17 cells for P14; n=19 cells for >P60). (e) Input resistance of GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice (n=27 cells for P14; n=12 cells for >P60). (f) Representative voltage clamp traces (left and center), and I-V plot (right) of GFP+ astrocytes on applying step currents of different polarity and magnitude showing passive changes in current. (g) Glutamate uptake currents of GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice. Dark traces represent the mean current and associated gray areas present the standard deviation of the mean currents (n=8 cells from P14; n=5 cells for >P60). (h) Glutamate uptake peak current in GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice after applying glutamate puffs. (n=9 cells for P14; n=6 cells for >P60). (i) Glutamate uptake current’s decay time in GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice (n=9 cells for P14; n=6 cells for >P60). (j) Total charge transfer during puffed glutamate uptake in GFP+ astrocytes in the dLGN of Aldh1l1-EGFP mice (n=9 cells for P14; n=6 cells for >P60). Scale bar in (a-b)=20μm. Data in (c-e), and (h-j) are represented as box and whisker plots. The central lines in the box represent medians; the two ends of the rectangles represent first and third quartiles. The upper and lower whiskers extend to the highest and lowest values in the data set, respectively. Individual data points are represented by dots.
4. DISCUSSION
In this study, we took advantage of genetic and molecular tools to study astrocyte development in the developing visual thalamus. From our data, we report that the tools that label Fgfr3 and Gja1 transcripts or the transgenic mouse line Aldh1l1-EGFP are the most reliable approaches to broadly label astrocytes in the developing and adult dLGN and vLGN. With this knowledge, we assessed the astrocyte morphogenesis in the developing visual thalamus and found as early as eye-opening (when RG synapses are still maturing), astrocytes achieve their adult-like morphology and elaborate PAPs to form tripartite synapses. In addition to anatomically ensheathing these synapses, by the end of the second week of postnatal development, thalamic astrocytes express high levels of GLT1 and are functionally able to buffer glutamate. We interpret these results to suggest that by eye-opening, and prior to RG synapse maturation, thalamic astrocytes are capable of preventing excessive glutamate spillover at RG synapses.
4.1. Development of astrocytes in visual thalamus
The development of astrocytes has been well-described in visual cortex where they are generated prenatally but continue to grow until cortical synapses are stable in the fourth postnatal week of development (Farhy-Tselnicker et al., 2020; Farhy-Tselnicker & Allen, 2018). Our observations in visual thalamus reveal that the development of thalamic astrocytes is slightly different from what has been reported in visual cortex (Farhy-Tselnicker et al., 2020). The rapid expansion of astrocytes into visual thalamus occurred prior to eye-opening, after which the number of astrocytes in this region plateaued (Fig 7(b–c)). We also observed that these astrocytes achieve their adult-like morphology (with highly branched processes) by eye-opening, earlier than in many other brain regions (Bushong et al., 2004; Clavreul et al., 2019; Holt et al., 2019). Not only do astrocytes have their process-bearing adult morphology by eye-opening, they also contact and ensheathe RG synapses at these early ages. As RG synapses are still maturing at these ages, this puts these astrocyte processes in prime locations to contribute to the strengthening or refining of synaptic connections between retinal axons and relay cells. Because of the close proximity of astrocyte processes to RG synapses, it could be either secreted factors or membrane-bound adhesion molecules on these astrocyte processes that could signal to the developing RG synapses. Such mechanisms have been demonstrated for astrocytes in other parts of the developing brain (Eroglu, 2009; Kucukdereli et al., 2011; Singh et al., 2016).
Our results suggest that the development of astrocytes in visual thalamus occurs prior to and independent of the experience-dependent visual activity. However, experience-dependent activity strongly influences the expression and function of glutamate transporters in astrocytes in other parts of the developing visual system (Sipe et al., 2020). This difference could reflect differences in the development of cortical and subcortical brain structures. For example, the critical period of visual thalamus ends prior to that of the visual cortex and the advanced development of thalamic astrocytes might be crucial to help serve the early needs of visual thalamus in processing and transmitting visual information (Espinosa & Stryker, 2012; Liang & Chen, 2020). Of course, it is also possible that experience-dependent visual activity might play a yet-to-be-identified role in the fine-tuning of astrocytic functions in visual thalamus that we were not able to capture in these studies.
Assuming an adult-like morphology or function prior to eye-opening does not preclude the importance of contributions by retinal inputs and/or activity in contributing to astrocyte development. In fact, we previously reported that retinal inputs are important for inducing the expression of FGF15 in astrocytes in the neonatal visual thalamus (Su et al., 2020). These studies were done in mice lacking retinal inputs to visual thalamus, therefore, it remains unclear if FGF15 expression is dependent on retinal activity or molecular cues by RGC axons. Both possibilities have been suggested in other brain regions. In visual cortex, the developmental regulation of astrocyte genes (such as chordin-like 1 and glypican 4) depends on visually-evoked neuronal activity (Farhy-Tselnicker et al., 2020). Whereas, in retina, RGC-derived morphogens serve to promote the proliferation and migration of astrocytes (D’Souza & Lang, 2020). Finally, other key possibilities to consider are the spontaneous cholinergic waves and glutamatergic waves that occur before eye-opening. Exploring their importance for astrocyte development is crucial, especially since these waves have been reported to influence other aspects of dLGN development such as eye-specific segregation (Tiriac et al., 2018).
4.2. Astrocytic heterogeneity in the visual thalamus
A decade ago Zhang and Barres stated that the diversity of astrocytes was an underappreciated topic in the field of neurobiology (Zhang & Barres, 2010). We now recognize astrocytes in many brain regions as being heterogeneous based on morphology, gene expression, and function (Bayraktar et al., 2015; Matias et al., 2019; Meldolesi, 2020; Miller, 2018; Pestana et al., 2020). For example, in neocortex, Bayraktar and colleagues recently reported that distinct subtypes of astrocytes are organized into different cortical layers (Bayraktar et al., 2020). Although, distinct cytoarchitectural layers and regions have been described in vLGN and dLGN (Krahe et al., 2011; Sabbagh et al., 2020), we did not see stratification of astrocyte types in visual thalamus. We did however see nucleus-specific differences, with the IGL having GFAP+ astrocytes that were absent from the adjacent dLGN (Fig. 1(a)). This matches what has previously been reported for GFAP+ cells in the hamster visual thalamus (Botchkina & Morin, 1995). Interestingly, our previous work revealed a subset of astrocytes in visual thalamus generate Fgf15 mRNA, however we failed to identify many Fgf15+ astrocytes in IGL (Su et al., 2020), further highlighting the differences between astrocytes in IGL and those in dLGN and vLGN. More astrocyte heterogeneity may exist than we show here (or in our previous studies) since we selected canonical astrocyte-associated molecules that were highly expressed in our datasets. This potentially may have limited our ability to identify transcriptionally distinct subtypes of astrocytes in visual thalamus. Other studies have used antibody cocktails against cell-surface markers in Aldh1l1-EGFP mice and have reported the existence of at least five subpopulations of astrocytes in mouse thalamus (John Lin et al., 2017). Therefore, future studies involving more unbiased approaches, such as single-cell transcriptional profiling, will be needed to delineate the full cohort of astrocytic diversity in visual thalamus.
How does astrocytic heterogeneity translate into functionality in visual thalamus? Subtypes of astrocytes have been suggested to contribute to their involvement in different developmental processes (Farhy-Tselnicker & Allen, 2018; John Lin et al., 2017; Matias et al., 2019; Pestana et al., 2020). For example, GFAP+ astrocytes in IGL create a pathway using their long processes, along which neuropeptide Y-positive cells migrate into this region (Botchkina & Morin, 1995). Likewise, Fgf15, generated by a different subset of astrocytes in dLGN and vLGN, is also involved in the recruitment of GABAergic interneurons into the developing visual thalamus (Su et al., 2020). Interneurons in this study were labeled in Gad67-GFP mice and, perhaps unsurprisingly, these GFP+ interneurons are largely absent from the IGL where FGF15 is not generated by astrocytes. Thus, at least two types of molecularly distinct astrocytes in visual thalamus are required for orchestrating the recruitment of different types of neurons.
Acknowledgements:
We are grateful to the members of the M.A.F. lab for scientific discussion and comments on the manuscript. We thank Dr. Robel for generously providing Aldh1l1-EGFP and mGFAP-Cre mice.
Funding statement:
This work was supported by NIH grants EY021222 and EY030568 (M.A.F.) NS113459 (U.S.), and NS120596 (G.L.C.).
Footnotes
Conflicts of interest disclosure: The authors have no conflicts of interest to declare.
Ethics approval and patient consent: Not applicable.
Data availability statement:
RNAseq experiment on the developing visual thalamus has been made publicly available and previously described in Monavarfeshani et al., 2018. All other data presented here are included in the main text and supporting information and are available for sharing upon request to the authors.
REFERENCES
- Adami C, Sorci G, Blasi E, Agneletti AL, Bistoni F, & Donato R (2001). S100B expression in and effects on microglia. Glia, 33(2), 131–142. [DOI] [PubMed] [Google Scholar]
- Agius E, Soukkarieh C, Danesin C, Kan P, Takebayashi H, Soula C, & Cochard P (2004). Converse control of oligodendrocyte and astrocyte lineage development by Sonic hedgehog in the chick spinal cord. Developmental Biology, 270(2), 308–321. 10.1016/j.ydbio.2004.02.015 [DOI] [PubMed] [Google Scholar]
- Barnabé-Heider F, Göritz C, Sabelström H, Takebayashi H, Pfrieger FW, Meletis K, & Frisén J (2010). Origin of new glial cells in intact and injured adult spinal cord. Cell Stem Cell, 7(4), 470–482. 10.1016/j.stem.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Bartels T, Holmqvist S, Kleshchevnikov V, Martirosyan A, Polioudakis D, Ben Haim L, Young AMH, Batiuk MY, Prakash K, Brown A, Roberts K, Paredes MF, Kawaguchi R, Stockley JH, Sabeur K, Chang SM, Huang E, Hutchinson P, … Rowitch DH (2020). Astrocyte layers in the mammalian cerebral cortex revealed by a single-cell in situ transcriptomic map. Nature Neuroscience, 23(4), 500–509. 10.1038/s41593-020-0602-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar OA, Fuentealba LC, Alvarez-Buylla A, & Rowitch DH (2015). Astrocyte development and heterogeneity. Cold Spring Harbor Perspectives in Biology, 7(1), 1–16. 10.1101/cshperspect.a020362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickford ME, Slusarczyk A, Dilger EK, Krahe TE, Kucuk C, & Guido W (2010). Synaptic development of the mouse dorsal lateral geniculate nucleus. Journal of Comparative Neurology, 518(5), 622–635. 10.1002/cne.22223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchkina GI, & Morin LP (1995). Specialized neuronal and glial contributions to development of the hamster lateral geniculate complex and circadian visual system. Journal of Neuroscience, 15(1 I), 190–201. 10.1523/jneurosci.15-01-00190.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner A, De Roo M, Dayer A, Muller D, Kiss JZ, & Vutskits L (2010). Bilateral whisker trimming during early postnatal life impairs dendritic spine development in the mouse somatosensory barrel cortex. Journal of Comparative Neurology, 518(10), 1711–1723. 10.1002/cne.22297 [DOI] [PubMed] [Google Scholar]
- Bushong EA, Martone ME, & Ellisman MH (2004). Maturation of astrocyte morphology and the establishment of astrocyte domains during postnatal hippocampal development. International Journal of Developmental Neuroscience, 22(2), 73–86. 10.1016/j.ijdevneu.2003.12.008 [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, Thompson WJ, & Barres BA (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. Journal of Neuroscience, 28(1), 264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SC, Muñoz-Ballester C, Chaunsali L, Mills WA, Yang JH, Sontheimer H, & Robel S (2020). Potassium and glutamate transport is impaired in scar-forming tumor-associated astrocytes. Neurochemistry International, 133(July 2019), 104628. 10.1016/j.neuint.2019.104628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell SL, Hablitz JJ, & Olsen ML (2014). Functional changes in glutamate transporters and astrocyte biophysical properties in a rodent model of focal cortical dysplasia. Frontiers in Cellular Neuroscience, 8(DEC), 1–13. 10.3389/fncel.2014.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang J, Niell CM, Liu X, Pfeiffenberger C, Feldheim DA, & Stryker MP (2008). Selective Disruption of One Cartesian Axis of Cortical Maps and Receptive Fields by Deficiency in Ephrin-As and Structured Activity. Neuron, 57(4), 511–523. 10.1016/j.neuron.2007.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, & Douglas RJ (2012). TrakEM2 software for neural circuit reconstruction. PLoS ONE, 7(6). 10.1371/journal.pone.0038011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo GL, Ballard VA, Glausen T, Boone Z, Teamer J, Hinkson CL, Wohlfert EA, Blader IJ, & Fox MA (2020). Toxoplasma infection induces microglia-neuron contact and the loss of perisomatic inhibitory synapses. Glia, 68(10), 1968–1986. 10.1002/glia.23816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, & Regehr WG (2000). Developmental remodeling of the retinogeniculate synapse. Neuron, 28(3), 955–966. 10.1016/S0896-6273(00)00166-5 [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CCA, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, & Barres BA (2005). Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell, 120(3), 421–433. 10.1016/j.cell.2004.12.020 [DOI] [PubMed] [Google Scholar]
- Chung W, Allen NJ, & Eroglu C (2015). Astrocyte & synapse. Cold Spring Harbor Laboratory Press, 7(a020370), 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, Smith SJ, & Barres BA (2013). Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature, 504(7480), 394–400. 10.1038/nature12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavreul S, Abdeladim L, Hernández-Garzón E, Niculescu D, Durand J, Ieng SH, Barry R, Bonvento G, Beaurepaire E, Livet J, & Loulier K (2019). Cortical astrocytes develop in a plastic manner at both clonal and cellular levels. Nature Communications, 10(1), 1–14. 10.1038/s41467-019-12791-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza S, & Lang RA (2020). Retinal ganglion cell interactions shape the developing mammalian visual system. Development (Cambridge, England), 147(23), 1–13. 10.1242/dev.196535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deloulme JC, Raponi E, Gentil BJ, Bertacchi N, Marks A, Labourdette G, & Baudier J (2004). Nuclear expression of S100B in oligodendrocyte progenitor cells correlates with differentiation toward the oligodendroglial lineage and modulates oligodendrocytes maturation. Molecular and Cellular Neurosciences, 27(4), 453–465. 10.1016/j.mcn.2004.07.008 [DOI] [PubMed] [Google Scholar]
- El-Danaf RN, Krahe TE, Dilger EK, Bickford ME, Fox MA, & Guido W (2015). Developmental remodeling of relay cells in the dorsal lateral geniculate nucleus in the absence of retinal input. Neural Development, 10(1), 1–17. 10.1186/s13064-015-0046-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C (2009). The role of astrocyte-secreted matricellular proteins in central nervous system development and function. Journal of Cell Communication and Signaling, 3(3–4), 167–176. 10.1007/s12079-009-0078-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroglu C, & Barres BA (2010). Regulation of synaptic connectivity by glia. Nature, 468(7321), 223–231. 10.1038/nature09612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, & Stryker MP (2012). Review Development and Plasticity of the Primary Visual Cortex. 10.1016/j.neuron.2012.06.009 [DOI] [PMC free article] [PubMed]
- Farhy-Tselnicker I, Boisvert MM, Liu H, Dowling C, Erikson GA, Blanco-Suarez E, Farhy C, Shokhirev M, Ecker JR, & Allen NJ (2020). Activity-Dependent Modulation of Synapse-Regulating Genes in Astrocytes. BioRxiv, 2020.12.30.424365. 10.1101/2020.12.30.424365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhy-Tselnicker Isabella, & Allen NJ (2018). Astrocytes, neurons, synapses: A tripartite view on cortical circuit development. Neural Development, 13(1), 1–12. 10.1186/s13064-018-0104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Tvrdik P, Makki N, Machold R, Paxinos G, & Watson C (2013). The interfascicular trigeminal nucleus: A precerebellar nucleus in the mouse defined by retrograde neuronal tracing and genetic fate mapping. Journal of Comparative Neurology, 521(3), 697–708. 10.1002/cne.23200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaume C, Fromaget C, El Aoumari A, Cordier J, Glowinski J, & Grost D (1991). Gap junctions in cultured astrocytes: Single-channel currents and characterization of channel-forming protein. Neuron, 6(1), 133–143. 10.1016/0896-6273(91)90128-M [DOI] [PubMed] [Google Scholar]
- Godement P, Salaün J, & Imbert M (1984). Prenatal and postnatal development of retinogeniculate and retinocollicular projections in the mouse. Journal of Comparative Neurology, 230(4), 552–575. 10.1002/cne.902300406 [DOI] [PubMed] [Google Scholar]
- Golding B, Pouchelon G, Bellone C, Murthy S, Di Nardo AA, Govindan S, Ogawa M, Shimogori T, Lüscher C, Dayer A, & Jabaudon D (2014). Retinal input directs the recruitment of inhibitory interneurons into thalamic visual circuits. Neuron, 81(5), 1057–1069. 10.1016/j.neuron.2014.01.032 [DOI] [PubMed] [Google Scholar]
- Guido W (2018). Development, form, and function of the mouse visual thalamus. Journal of Neurophysiology, 120(1), 211–225. 10.1152/jn.00651.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer S, Carrillo GL, Govindaiah G, Monavarfeshani A, Bircher JS, Su J, Guido W, & Fox MA (2014). Nuclei-specific differences in nerve terminal distribution, morphology, and development in mouse visual thalamus. Neural Development, 9(1). 10.1186/1749-8104-9-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer S, Monavarfeshani A, Lemon T, Su J, & Fox MA (2015). Multiple Retinal Axons Converge onto Relay Cells in the Adult Mouse Thalamus. Cell Reports, 12(10), 1575–1583. 10.1016/j.celrep.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LM, Hernandez RD, Pacheco NL, Torres Ceja B, Hossain M, & Olsen ML (2019). Astrocyte morphogenesis is dependent on BDNF signaling via astrocytic TrkB.T1. ELife, 8, 1–27. 10.7554/eLife.44667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YK, & Chen C (2011). Wiring and rewiring of the retinogeniculate synapse. Current Opinion in Neurobiology, 21(2), 228–237. 10.1016/j.conb.2011.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, & Chen C (2006). Distinct Roles for Spontaneous and Visual Activity in Remodeling of the Retinogeniculate Synapse. Neuron, 52(2), 281–291. 10.1016/j.neuron.2006.07.007 [DOI] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, & Barres BA (2008). Architecture and Activity-Mediated Refinement of Axonal Projections from a Mosaic of Genetically Identified Retinal Ganglion Cells. Neuron, 59(3), 425–438. 10.1016/j.neuron.2008.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager P, Ye Z, Yu X, Zagoraiou L, Prekop HT, Partanen J, Jessell TM, Wisden W, Brickley SG, & Delogu A (2016). Tectal-derived interneurons contribute to phasic and tonic inhibition in the visual thalamus. Nature Communications, 7, 1–14. 10.1038/ncomms13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaubert-Miazza L, Green E, Lo FS, Bui K, Mills J, & Guido W (2005). Structural and functional composition of the developing retinogeniculate pathway in the mouse. Visual Neuroscience, 22(5), 661–676. 10.1017/S0952523805225154 [DOI] [PubMed] [Google Scholar]
- John Lin CC, Yu K, Hatcher A, Huang TW, Lee HK, Carlson J, Weston MC, Chen F, Zhang Y, Zhu W, Mohila CA, Ahmed N, Patel AJ, Arenkiel BR, Noebels JL, Creighton CJ, & Deneen B (2017). Identification of diverse astrocyte populations and their malignant analogs. Nature Neuroscience, 20(3), 396–405. 10.1038/nn.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, & Littman DR (2000). Analysis of Fractalkine Receptor CX3CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Molecular and Cellular Biology, 20(11), 4106–4114. 10.1128/mcb.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AHP, Verveer M, de Groot LR, Smith VD, Rangarajan S, Rodríguez JJ, Orre M, & Hol EM (2012). GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS ONE, 7(8). 10.1371/journal.pone.0042823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Lee SW, Han JE, Choi JW, & Song MR (2014). The complex morphology of reactive astrocytes controlled by fibroblast growth factor signaling. Glia, 62(8), 1328–1344. 10.1002/glia.22684 [DOI] [PubMed] [Google Scholar]
- Krahe TE, El-Danaf RN, Dilger EK, Henderson SC, & Guido W (2011). Morphologically distinct classes of relay cells exhibit regional preferences in the dorsal lateral geniculate nucleus of the mouse. Journal of Neuroscience, 31(48), 17437–17448. 10.1523/JNEUROSCI.4370-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, & Eroglu C (2011). Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins hevin and SPARC. Proceedings of the National Academy of Sciences of the United States of America, 108(32). 10.1073/pnas.1104977108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence PM, & Studholme KM (2014). Retinofugal projections in the mouse. Journal of Comparative Neurology, 522(16), 3733–3753. 10.1002/cne.23635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Brooks JM, Chen J, Su J, & Fox MA (2015). Cell-specific and developmental expression of lectican-cleaving proteases in mouse hippocampus and neocortex. Journal of Comparative Neurology, 523(4), 629–648. 10.1002/cne.23701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, & Chen C (2020). Organization, Function, and Development of the Mouse Retinogeniculate Synapse. Annual Review of Vision Science, 6, 261–285. 10.1146/annurev-vision-121219-081753 [DOI] [PubMed] [Google Scholar]
- Litvina EY, & Chen C (2017). An evolving view of retinogeniculate transmission. Visual Neuroscience, 34(2017). 10.1017/S0952523817000104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martersteck EM, Hirokawa KE, Evarts M, Bernard A, Duan X, Li Y, Ng L, Oh SW, Ouellette B, Royall JJ, Stoecklin M, Wang Q, Zeng H, Sanes JR, & Harris JA (2017). Diverse Central Projection Patterns of Retinal Ganglion Cells. Cell Reports, 18(8), 2058–2072. 10.1016/j.celrep.2017.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias I, Morgado J, & Gomes FCA (2019). Astrocyte Heterogeneity: Impact to Brain Aging and Disease. Frontiers in Aging Neuroscience, 11(March), 1–18. 10.3389/fnagi.2019.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J (2020). Astrocytes: News about brain health and diseases. Biomedicines, 8(10), 1–14. 10.3390/biomedicines8100394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SJ (2018). Astrocyte heterogeneity in the adult central nervous system. Frontiers in Cellular Neuroscience, 12(November), 1–6. 10.3389/fncel.2018.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monavarfeshani A, Sabbagh U, & Fox MA (2017). Not a one-trick pony: Diverse connectivity and functions of the rodent lateral geniculate complex. Visual Neuroscience, 34, E012. 10.1017/S0952523817000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monavarfeshani A, Stanton G, Van Name J, Su K, Mills WA, Swilling K, Kerr A, Huebschman NA, Su J, & Fox MA (2018). LRRTM1 underlies synaptic convergence in visual thalamus. ELife, 7. 10.7554/eLife.33498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongrédien R, Erdozain AM, Dumas S, Cutando L, del Moral AN, Puighermanal E, Rezai Amin S, Giros B, Valjent E, Meana JJ, Gautron S, Callado LF, Fabre V, & Vialou V (2019). Cartography of hevin-expressing cells in the adult brain reveals prominent expression in astrocytes and parvalbumin neurons. Brain Structure and Function, 224(3), 1219–1244. 10.1007/s00429-019-01831-x [DOI] [PubMed] [Google Scholar]
- Mukherjee K, Clark HR, Chavan V, Benson EK, Kidd GJ, & Srivastava S (2016). Analysis of brain mitochondria using serial block-face scanning electron microscopy. Journal of Visualized Experiments, 2016(113), 1–10. 10.3791/54214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri EM, Menga A, Lebrun A, Hooper DC, Butterfield DA, Mazzone M, & Castegna A (2017). Blockade of Glutamine Synthetase Enhances Inflammatory Response in Microglial Cells. Antioxidants and Redox Signaling, 26(8), 351–363. 10.1089/ars.2016.6715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquettaz R, Kolotuev I, Rohrbach A, Gouelle C, Pellerin L, & Langlet F (2020). Peculiar protrusions along tanycyte processes face diverse neural and nonneural cell types in the hypothalamic parenchyma. Journal of Comparative Neurology, January 2020, 553–575. 10.1002/cne.24965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestana F, Edwards-Faret G, Belgard TG, Martirosyan A, & Holt MG (2020). No longer underappreciated: The emerging concept of astrocyte heterogeneity in neuroscience. Brain Sciences, 10(3), 1–21. 10.3390/brainsci10030168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Cutforth T, Woods G, Yamada J, Rentería RC, Copenhagen DR, Flanagan JG, & Feldheim DA (2005). Ephrin-As and neural activity are required for eye-specific patterning during retinogeniculate mapping. Nature Neuroscience, 8(8), 1022–1027. 10.1038/nn1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffenberger C, Yamada J, & Feldheim DA (2006). Ephrin-As and patterned retinal activity act together in the development of topographic maps in the primary visual system. Journal of Neuroscience, 26(50), 12873–12884. 10.1523/JNEUROSCI.3595-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, & Richardson WD (2003). Fgfr3 expression by astrocytes and their precursors: Evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development, 130(1), 93–102. 10.1242/dev.00184 [DOI] [PubMed] [Google Scholar]
- Robel S, Buckingham SC, Boni JL, Campbell SL, Danbolt NC, Riedemann T, Sutor B, & Sontheimer H (2015). Reactive astrogliosis causes the development of spontaneous seizures. Journal of Neuroscience, 35(8), 3330–3345. 10.1523/JNEUROSCI.1574-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbagh U, Govindaiah G, Somaiya RD, Ha RV, Wei JC, Guido W, & Fox MA (2020). Diverse GABAergic neurons organize into subtype-specific sublaminae in the ventral lateral geniculate nucleus. Journal of Neurochemistry, April, 1–19. 10.1111/jnc.15101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Su J, Brooks J, Terauchi A, Umemori H, & Fox MA (2012). Fibroblast growth factor 22 contributes to the development of retinal nerve terminals in the dorsal lateral geniculate nucleus. Frontiers in Molecular Neuroscience, 4(JANUARY 2012), 1–13. 10.3389/fnmol.2011.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz LJ, Kim IH, Manhaes AC, Rodrigues WS, Pamukcu A, Enustun E, Ertuz Z, Scheiffele P, Soderling SH, Silver DL, Ji RR, Medina AE, & Eroglu C (2016). Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1α and NL1 via Hevin. Cell, 164(1–2), 183–196. 10.1016/j.cell.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe G, Petravicz J, Rikhye R, Garcia R, Mellios N, & Sur M (2020). Astrocytic glutamate uptake coordinates experience-dependent, eye-specific refinement in developing visual cortex. BioRxiv. 10.1101/2020.05.25.113613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, Sher A, Litke AMM, Lambris JD, Smith SJ, John SWM, & Barres BA (2007). The Classical Complement Cascade Mediates CNS Synapse Elimination. Cell, 131(6), 1164–1178. 10.1016/j.cell.2007.10.036 [DOI] [PubMed] [Google Scholar]
- Su J, Charalambakis NE, Sabbagh U, Somaiya RD, Monavarfeshani A, Guido W, & Fox MA (2020). Retinal inputs signal astrocytes to recruit interneurons into visual thalamus. Proceedings of the National Academy of Sciences of the United States of America, 117(5), 2671–2682. 10.1073/pnas.1913053117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Chen J, Lippold K, Monavarfeshani A, Carrillo GL, Jenkins R, & Fox MA (2016). Collagen-derived matricryptins promote inhibitory nerve terminal formation in the developing neocortex. Journal of Cell Biology, 212(6), 721–736. 10.1083/jcb.201509085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Gorse K, Ramirez F, & Fox MA (2010). Collagen XIX is expressed by interneurons and contributes to the formation of hippocampal synapses. Journal of Comparative Neurology, 518(2), 229–253. 10.1002/cne.22228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Haner CV, Imbery TE, Brooks JM, Morhardt DR, Gorse K, Guido W, & Fox MA (2011). Reelin is required for class-specific retinogeniculate targeting. Journal of Neuroscience, 31(2), 575–586. 10.1523/JNEUROSCI.4227-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J, Stenbjorn RS, Gorse K, Su K, Hauser KF, Ricard-Blum S, Pihlajaniemi T, & Fox MA (2012). Target-Derived Matricryptins Organize Cerebellar Synapse Formation through α3β1 Integrins. Cell Reports, 2(2), 223–230. 10.1016/j.celrep.2012.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szentágothai J (1963). The Structure of the Synapse in the Lateral Geniculate Body. Cells Tissues Organs, 55(1–2), 166–185. 10.1159/000142468 [DOI] [PubMed] [Google Scholar]
- Tiriac A, Smith BE, & Feller MB (2018). Light Prior to Eye Opening Promotes Retinal Waves and Eye-Specific Segregation. Neuron, 100(5), 1059–1065.e4. 10.1016/j.neuron.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, & Barres BA (2001). Control of synapse number by glia. Science, 291(5504), 657–661. 10.1126/science.291.5504.657 [DOI] [PubMed] [Google Scholar]
- Wallraff A, Köhling R, Heinemann U, Theis M, Willecke K, & Steinhäuser C (2006). The impact of astrocytic gap junctional coupling on potassium buffering in the hippocampus. Journal of Neuroscience, 26(20), 5438–5447. 10.1523/JNEUROSCI.0037-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Wu H, Pandolfi PP, Louis DN, & Van Dyke T (2002). Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell, 1(2), 157–168. 10.1016/S1535-6108(02)00029-6 [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Ochalski A, Hertzberg EL, & Nagy JI (1990). On the organization of astrocytic gap junctions in rat brain as suggested by LM and EM immunohistochemistry of connexin43 expression. Journal of Comparative Neurology, 302(4), 853–883. 10.1002/cne.903020414 [DOI] [PubMed] [Google Scholar]
- Yoshioka N, Asou H, Hisanaga SI, & Kawano H (2012). The astrocytic lineage marker calmodulin-regulated spectrin-associated protein 1 (Camsap1): Phenotypic heterogeneity of newly born Camsap1-expressing cells in injured mouse brain. Journal of Comparative Neurology, 520(6), 1301–1317. 10.1002/cne.22788 [DOI] [PubMed] [Google Scholar]
- Young KM, Mitsumori T, Pringle N, Grist M, Kessaris N, & Richardson WD (2010). An Fgfr3-iCreERT2 transgenic mouse line for studies of neural stem cells and astrocytes. Glia, 58(8), 943–953. 10.1002/glia.20976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, & Barres BA (2010). Astrocyte heterogeneity: An underappreciated topic in neurobiology. Current Opinion in Neurobiology, 20(5), 588–594. 10.1016/j.conb.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MSB, Li G, Duncan JA, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MGH, & Barres BA (2016). Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron, 89(1), 37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoltowska KM, Maesako M, Meier J, & Berezovska O (2018). Novel interaction between Alzheimer’s disease-related protein presenilin 1 and glutamate transporter 1. Scientific Reports, 8(1), 1–9. 10.1038/s41598-018-26888-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNAseq experiment on the developing visual thalamus has been made publicly available and previously described in Monavarfeshani et al., 2018. All other data presented here are included in the main text and supporting information and are available for sharing upon request to the authors.